
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 28 February 2017
Sec. Molecular Innate Immunity
Volume 8 - 2017 | https://doi.org/10.3389/fimmu.2017.00119
This article is part of the Research Topic Immunoregulatory mechanisms of Interferon View all 30 articles
Interferon lambdas (IFN-λs; IFNL1-4) modulate immunity in the context of infections and autoimmune diseases, through a network of induced genes. IFN-λs act by binding to the heterodimeric IFN-λ receptor (IFNLR), activating a STAT phosphorylation-dependent signaling cascade. Thereby hundreds of IFN-stimulated genes are induced, which modulate various immune functions via complex forward and feedback loops. When compared to the well-characterized IFN-α signaling cascade, three important differences have been discovered. First, the IFNLR is not ubiquitously expressed: in particular, immune cells show significant variation in the expression levels of and susceptibilities to IFN-λs. Second, the binding affinities of individual IFN-λs to the IFNLR varies greatly and are generally lower compared to the binding affinities of IFN-α to its receptor. Finally, genetic variation in the form of a series of single-nucleotide polymorphisms (SNPs) linked to genes involved in the IFN-λ signaling cascade has been described and associated with the clinical course and treatment outcomes of hepatitis B and C virus infection. The clinical impact of IFN-λ signaling and the SNP variations may, however, reach far beyond viral hepatitis. Recent publications show important roles for IFN-λs in a broad range of viral infections such as human T-cell leukemia type-1 virus, rotaviruses, and influenza virus. IFN-λ also potentially modulates the course of bacterial colonization and infections as shown for Staphylococcus aureus and Mycobacterium tuberculosis. Although the immunological processes involved in controlling viral and bacterial infections are distinct, IFN-λs may interfere at various levels: as an innate immune cytokine with direct antiviral effects; or as a modulator of IFN-α-induced signaling via the suppressor of cytokine signaling 1 and the ubiquitin-specific peptidase 18 inhibitory feedback loops. In addition, the modulation of adaptive immune functions via macrophage and dendritic cell polarization, and subsequent priming, activation, and proliferation of pathogen-specific T- and B-cells may also be important elements associated with infectious disease outcomes. This review summarizes the emerging details of the IFN-λ immunobiology in the context of the host immune response and viral and bacterial infections.
Patients with infectious diseases often show heterogeneous clinical courses with a range of associated morbidities and variable mortality. This is dependent on a series of factors covering the complex aspects of host–pathogen interactions (1–5). IFNs play a crucial role in these interactions—defining the outcome of many viral, bacterial, fungal, and parasitic infections (6–16) (see Figure 1). In addition, IFNs reduce tumor cell proliferation (17, 18) and show important immune regulatory functions in autoimmunity (19, 20). These broad effects are explained through the induction of hundreds of IFN-stimulated genes (ISGs) (21). Three types of IFNs have been described, which can induce ISG expression, and add further complexity: type I with mainly IFN-αs and -βs (22–26), type II with only IFN-γ (27), and type III with IFN-λs (28–31). Although most cells can induce and release various types of IFNs, specialized immune cells are the main producers during an inflammatory process. The effects induced by single or combined IFNs in exposed cells are very heterogeneous and range from differential patterns of ISG expression, regulation of cell proliferation (18), changes in cell surface molecules such as HLA DR (32), to the maturation of monocytes to dendritic cells (33). The effects depend on the plasticity of the various IFNs involved, including the peak concentrations, concentration changes over time, binding affinities of IFNs to the specific receptors, receptor expression, potentially induced feedback mechanisms, and the target cell type itself (34).
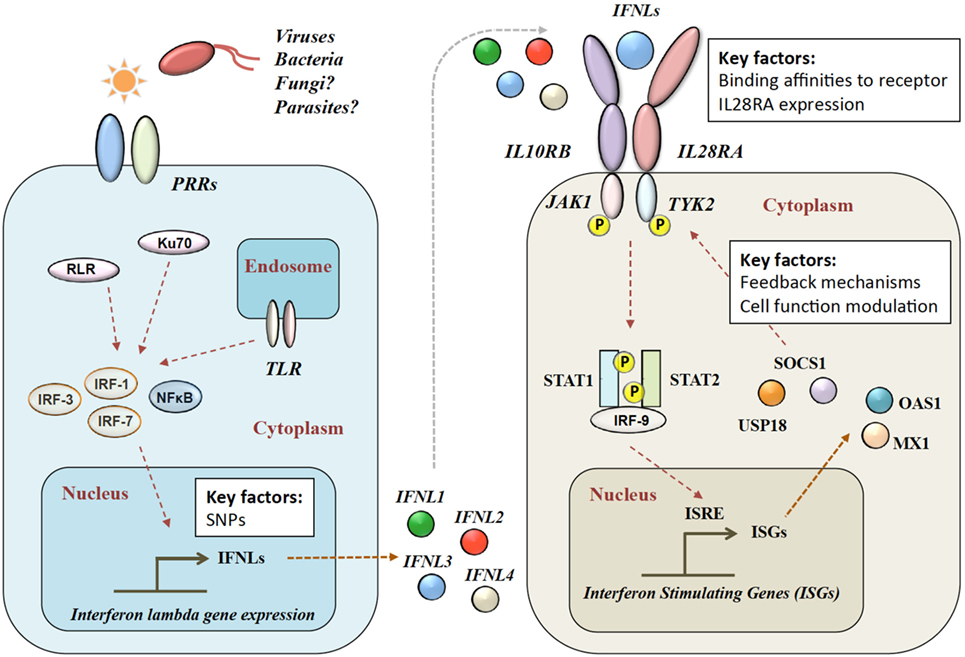
Figure 1. Type III IFN signaling pathway. Viral infection is sensed by pattern recognition receptors (PRRs), which induce IFN-λ production via various signaling pathways. IFN-λs bind to the heterodimeric IFN-λ receptor (IFNLR), which consists of IL28RA and IL10RB subunits. Upon binding, a JAK–STAT signaling cascade induces hundreds of IFN-stimulated genes (ISGs). RLR, RIG-1-like receptor; TLR, toll-like receptors; NF-κB, nuclear factor kappa-light-chain-enhancer of activated B cells; IL28RA, interleukin 28 receptor alpha; IL10RB, interleukin 10 receptor beta; JAK1, Janus Kinase 1; TYK2, tyrosine kinase 2; STAT, signal transducer and activator of transcription; IRF, interferon regulatory factor; ISRE, interferon-stimulated response element; MX1, interferon-induced GTP-binding protein Mx1; OAS1, 2′-5′-oligoadenylate synthetase.
Four IFN-λ ligands have been described: IFNL1–4, with each family member having antiviral effects on various viruses within different cell types (28). IFNL1–3 share high amino acid sequence homologies, whereas IFNL4 is more divergent with only 40.8% amino acid similarity to IFNL3 (35). The expression of IFN-λs is induced in a broad range of cell types by pattern recognition receptors including toll-like mediated (36–41), Ku70 (21398614) and RIG-1-like (24952503). Type 2 myeloid dendritic cells have been described as the main producers of IFN-λ (42–48). In mice, commonly used as a model organism for infectious disease and immune function, only IFNL2 and IFNL3 are functional, as IFNL1 and IFNL4 are present as inactive pseudogenes (49).
After release, IFN-λ binds to its heterodimeric IFN-λ receptor (IFNLR). The IFNLR consists of two subunits: α-subunit (IL28RA) and β-subunit (IL10RB) (35, 50–53). Despite high sequence homologies, binding affinities of the different IFN-λs to the IFNLR1 differ greatly. IFNL1 shows the highest binding affinity to IL28RA, and IFNL3 the lowest (54). The dimerization of the receptor subunits leads to activation of Janus Kinase 1 and tyrosine kinase 2 and phosphorylation of STAT-1 and -2, which induces the subsequent downstream signaling with the induction of hundreds of ISGs (31) (see Figure 1). IFN-α and IFN-λ both show a complex mechanism of positive and negative feedback loops, mainly modulated via the suppressor of cytokine signaling 1 and the ubiquitin-specific peptidase 18 (31, 55).
Two aspects are crucial to understanding the role of IFN-λs in the context of infectious diseases: (i) IFNLR distribution in infected cells and tissues and (ii) single-nucleotide polymorphisms (SNPs) in and around the genes encoding IFN-λs and IFNLR. Both aspects show important differences between humans and mice, which complicate studies and conclusions drawn from infectious disease models (56).
The IL10RB subunit is expressed in many cell types (57), whereas the IL28RA subunit expression is much more restricted. Expression of IL28RA mRNA has been detected in the lung, intestine, liver tissues, immune cells such as B cells, neutrophils, macrophages, and plasmacytoid dendritic cells (28, 29, 43, 58–62). Human NK cells seem not to express IFNLR (63), whereas mouse NK cells show deficient function in IL28R knockout animals (25901316). The effects of IFN-λ on cells and tissues are often measured in vitro via indirect markers, such as downstream expression of ISGs or changes in specific cellular phenotypes. Data on the induction of STAT phosphorylation, as the most direct measurement of signal induction, are still missing for some cell types and tissues. The IFNLR expression is regulated via transcription factors (31) and may show variability during an inflammatory process, which adds an additional level of complexity. Primary hepatocytes show relatively low baseline responsiveness to IFN-λs, yet upon IFN-α treatment a marked increase in IL28RA mRNA levels is observed (64, 65). Similarly, during cytomegalovirus (CMV) infection of fibroblasts, IL28RA mRNA levels increase by about twofold, but protein expression levels remain stable (66). A recent paper by Lazear et al. suggested that endothelial cells in the blood–brain barrier may be sensitive to IFN-λs, reducing permeability to West Nile virus in a mouse model (67).
Understanding which immune cells and subsets are responsive to IFN-λs in humans can be experimentally and technically challenging due to low target cell densities and less accessible cell types such as tissue resident cell types. In contrast, peripheral blood mononuclear cells (PBMCs) are relatively easy to access in order to explore responses to IFN-λs; therefore, most literature focuses on hepatocytes (from liver biopsies) and immune cells from the blood. The direct impact of IFN-λs on T-cells via surface expression of the specific IFNLR is subject to ongoing debate (58, 68–71). IFN-λs may also induce FOXP3-expressing regulatory T-cells (72), which may impact a series of immunoregulatory aspects during an infection as part of the inflammatory response. Several research groups confirm that IFN-λs influence the T-helper cell balance, which is shifted toward Th1 (70, 71, 73–76). The Th1/Th2 balance might be important for controlling specific infections such as helminths (6, 77, 78). In addition, the B-cell-driven humoral immune responses are also modulated by the presence of Th2 cytokines, e.g., during vaccination. We have recently shown that IFNL3 is a key regulator of the influenza virus-specific B-cell proliferation and antibody production (76). The exact mechanism of how Th1/Th2 balancing and B-cell activation is modulated by IFN-λs and how this impacts infectious disease outcome has to be explored in more detail in the future.
A series of SNPs in IFN-λ ligand and receptor genes have been described (see Figure 2). Most importantly, these SNPs have been associated with a series of important clinical phenotypes in the context of infectious diseases (see Table 1 for more details).
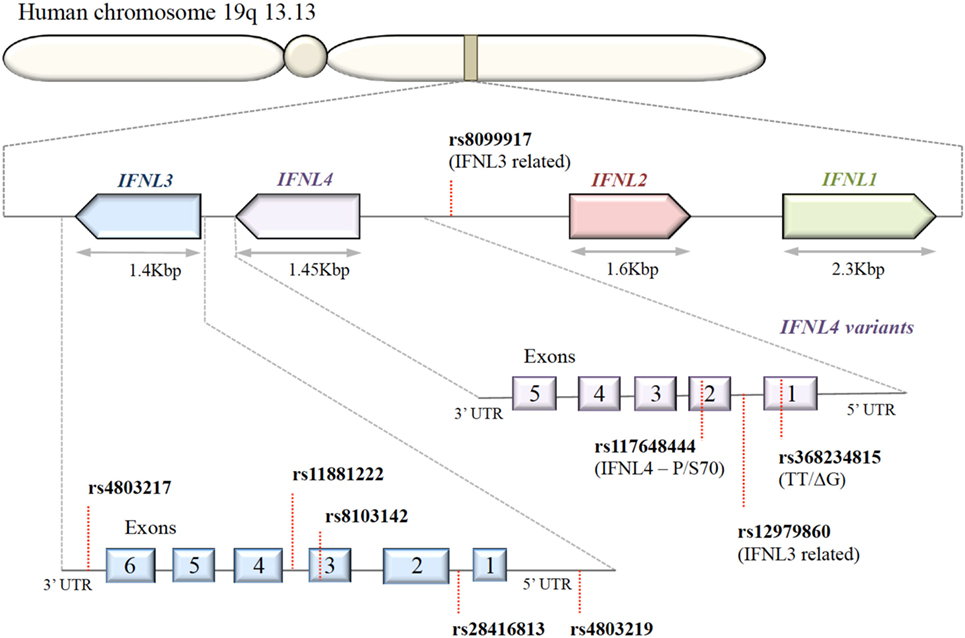
Figure 2. Organization of IFNL genes in the human genome. The IFN-λ genes are located in tandem on chromosome 19. Key single-nucleotide polymorphisms (SNPs) in coding and non-coding regions of IFN-λ genes are shown. IFNL1, IFNL2, and IFNL3 genes are functional; only a subset of the human population possess the SNP rs368234815 with ΔG frameshift mutation in exon 1, producing an in-frame IFNL4.
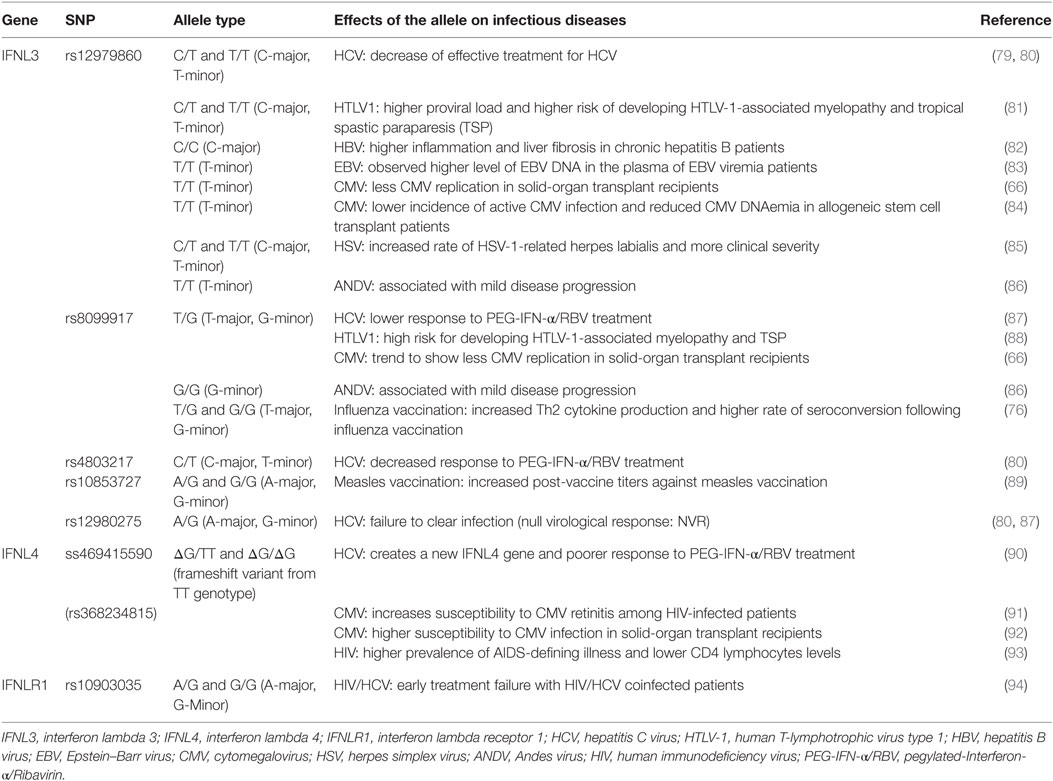
Table 1. Single-nucleotide polymorphisms (SNPs) within the IFNL3/IFNL4 gene locus and impact on infectious diseases.
Modulation of IFNLR expression may have a great impact on the effects of a particular IFN-λ ligand, and thereby influence the subsequent signaling pathway and the outcome of infectious diseases. Multiple SNPs in the gene encoding IL28RA have been described (94–97). The rs10903035 SNP is located within the 3′UTR of the IL28RA mRNA sequence, suggesting a potential microRNA binding site. This particular SNP was identified as an independent risk factor for IFN-α treatment failure against hepatitis C virus (HCV) (44, 98). In addition, this SNP has been associated with insulin resistance in HIV/HCV coinfected patients (94). Another SNP in this gene, rs4649203, has been linked to the risk of psoriasis in four independent populations (96), and to the development of systemic lupus erythematosus (97). These observations suggest an important influence of IL28RA on infectious and autoimmune diseases.
Expression of IFN-λ ligands is modulated by SNPs in both transcription factor binding sites and methylation sites of the promoter region, as well as frameshift mutations (99–102). The IFN-λ gene layout is shown in Figure 2. The clinical impact of SNPs in the IFNL3/4 locus was originally observed in the context of IFN-α treatment outcomes in patients with chronic HCV (79, 80, 87, 90, 103). SNPs within this locus are in high linkage disequilibrium, e.g., rs12979860 with ss469415590 (103, 104), which complicates the exploration of the effects of individual SNPs. Therefore, the impact of some SNPs on IFN-λ expression is still debated. Most studies have concluded that the minor alleles of SNPs rs12979860 (CT/TT) and rs8099917 (TG/GG) are associated with reduced IFNL3 expression during chronic HCV infection, observed in liver biopsies (80, 105–107), serum, and PBMCs stimulated with polyI:C-, CMV-, and influenza virus (66, 76, 108, 109). However, it has also been shown that the TT allele of rs12979860 in hepatocytes expresses higher levels of IFNL1 and IFNL3 (110). This minor allele genotype of rs12979860 (TT) has also been associated with a higher and prolonged ISG expression in HCV infection (79, 80, 87, 90, 103, 111). Interestingly, the same SNP of the IFNL3 gene is associated with a higher ISG expression in mothers after childbirth, suggesting that postpartum the normalization of physiological control of IFN signaling depends on the IFNL3 genotype (112). Although the rs12979860 SNPs have been specifically associated with IFNL3/L4 expression, these SNPs might also affect the expression of the other IFN-λ genes (80, 87, 113).
The impact of the ss469415590 SNP on the expression of IFNL4 is, in contrast, very well described: in the context of a delta-G polymorphism, a frameshift mutation generates a gene containing an alternative reading frame, which causes IFNL4 to be functionally expressed in about 40% of Caucasians (90). An amino acid substitution at residue 70 of IFNL4 (P70S) decreases the antiviral activity via a reduction in the ISG expression levels (111).
Beside the impact of SNPs on innate immune signaling via differences in ISG expression profiles, an important impact on adaptive immune functions has been noted. We have shown that IFN-λ decreases virus-induced B-cell proliferation and antibody secretion in a dose-dependent manner. In addition, IFN-λ increases influenza-induced Th1 cytokines (IFN-γ, IL6), whereas influenza-induced Th2 cytokines decrease (IL4, IL5, IL9, IL13). These effects can also be reproduced with specific allelic combinations. In particular, the TG/GG allele of rs8099917 shows significantly lower levels of IFN-α, IL2, and IL6 secretion in influenza-stimulated PBMCs. In an influenza vaccine cohort, vaccine recipients with the rs8099917 TG/GG (minor) allele showed significantly higher vaccine-induced humoral immune responses (76). Similarly, in a cohort of children vaccinated against measles, the post-vaccine antibody titers were significantly higher in the group with the rs10853727 SNP AG and GG (minor allele) (89). Both SNPs rs8099917 and rs10853727 lie within the IFNL3 promoter region and have been associated with lower IFNL3 expression (76, 89).
The dual role of IFN-λs, with direct antiviral effects (innate immunity) and more long-term immunomodulatory effects on T- and B-cell activation and modulation, can result in multiple possible interactions with different types of infectious disease. Table 2 summarizes the role of IFN-λs in several infectious diseases.
IFNs protect cells against viral infections. In response, every virus has evolved specific ways to counteract IFN signaling and its effects (139–143). Only a few studies have explored this in the context of IFN-λs. Parainfluenza virus 3 blocks antiviral mediators downstream of the IFNLR signaling by modulation of the STAT1 phosphorylation in BEAS 2B cells, a bronchial epithelial cell line (144). Dengue virus was recently shown to induce IFNL1 via its non-structural protein (NS1) in order to facilitate dendritic cell migration (114).
Using cell culture-based in vitro models, IFN-λs have been shown to play a role in controlling viral replication. In most studies, cultured cells were treated with IFN-λs and the impact of viral infection was assessed. These studies investigated human (66) and murine CMV (59), dengue virus (114, 145), encephalomyocarditis virus (28, 29, 146), herpes virus type 2 (120), hepatitis B virus (115), HCV (37, 60, 113, 115, 116, 147), HIV (40, 117, 118), human meta pneumovirus (121), influenza virus (122, 148–152), lymphocytic choriomeningitis virus (LCMV) (125), norovirus (124), respiratory syncytial virus (128, 153, 154), sendai virus (155–157), and vesicular stomatitis virus (131, 158, 159).
In vivo, the complexity of the role of IFN-λ within tissues and between various immune cells has been explored using an IL28RA−/− mouse model, leading to the discovery of multiple important aspects of IFN-λ signaling (122, 130, 150).
A recent study by Lin et al. demonstrated that the effects of type III IFNs change with increasing age. Rotavirus was controlled by both type I and III IFN in suckling mice, whereas epithelial cells in particular were responsive. In adult mice, epithelial cells were responsive only to type III and not type I IFNs, suggesting an orchestrated spatial and temporal organization of the IFN-α and IFN-λ responses in the aging murine intestinal tract (160). However, there is some controversy regarding the rotavirus data, as other researchers have shown that rotavirus is specifically controlled by type III and not type I IFN (21518880). Mahlakoiv et al. showed that leukocyte-derived IFN-α/β and epithelial IFN-λ constitute a compartmentalized mucosal defense system to restrict enteric viral infection in mice. The authors concluded that epithelial barriers to IFN-λ may have evolved to reduce frequent triggering of IFN-α/β and thus reduce exacerbated inflammation (161). A study by Baldridge et al. showed that antibiotics could prevent the persistence of enteric murine norovirus infection, but only in the presence of functional IFN-λ signaling. The IL28RA−/− mice showed a high rate of infection, despite the administration of antibiotics. This may suggest cross talk between the gut microbiota and IFN-λ signaling in modulating chronic viral infections (162). Important synergistic effects in the intestine have been described, with IL22-inducing IFN-λ expression in intestinal epithelial cells in a murine rotavirus infection model (163).
The role of IFN-λ during respiratory tract infections has also been explored using the IL28RA−/− mouse model. The studies so far have concentrated on the classical role of IFNs as antiviral cytokines. The IL28RA−/− mouse displayed a significantly higher burden of disease than wild-type mice during infections with influenza virus and SARS coronavirus (122, 130, 150). One study showed the immunoregulatory function of IFN-λ in an LCMV model. The authors noted that in an acute LCMV infection model, the IL28RA−/− mouse showed a greater than normal CD4+ and CD8+ T-cell response compare to the wild type, whereas in a chronic LCMV infection model, the IL28RA−/− mice showed a greater disease burden and a significantly reduced LCMV-specific T-cell response. The paper showed that germinal center B-cells were more frequent in peripheral blood in the IL28RA−/− mice than wild-type mice. However, the LCMV-induced memory B-cell response, in terms of frequencies and LCMV-specific antibodies, was comparable (164).
The immunoregulatory actions of IFN-λs have been explored in an ovalbumin (OVA)-induced asthma model. The IL28RA−/− mice showed a clear shift to increased Th2 cytokines and a more severe asthma phenotype. Importantly, IgE antibodies were also significantly increased (73). In this model, the IFNL2 (IL28A) immunoregulatory activity was dependent on lung CD11c+ dendritic cells to decrease OX40L, increase IL-12p70, and thereby promote Th1 differentiation (73). The potential role in infection-triggered asthma has also been explored in humans (72, 126).
Although these conclusions from mice studies are very important, a series of important differences to human effects have also been noted. In a human chimeric mouse model using human hepatocytes, the response rates of human and mice hepatocytes toward IFN-λs were very different, specifically in that mouse, hepatocytes did not respond to IFN-λ (56). In addition, the expression of IFNLR in immune cells seems to be strikingly different. Whereas B-cells in humans respond to IFN-λs, in B-cells from mice there seems to be no direct effect from IFN-λs (69, 164).
Studies on the impact of IFN-λs in clinical scenarios have been dominated by the strong association of IFNL3/L4 SNPs with spontaneous clearance of HCV and IFN-α treatment response (79, 80, 87, 90, 103, 111). Details on this important association have been reviewed in detail elsewhere (165–167). The association between IFN-λ SNPs and other infectious diseases is far less well explored. Not many studies have linked the genetic associations with mechanistic immunological assay.
Several studies have explored the association between SNPs in the IFNL3/L4 signaling and CMV replication. Transplant recipients with the rs8099917 GG allele demonstrate significantly less CMV primary replication. This SNP has been associated with reduced ISG expression upon infection (66). We postulate that this phenomenon has two reasons: (i) significant primary CMV replication is less likely due to a higher baseline ISG expression and (ii) naïve CMV-specific T cells from seronegative healthy blood donors show reduced proliferation capacity when pretreated with IFNL3 and stimulated with CMV lysate (66). In contrast, the rs368234815 ΔG SNP shows a higher risk for CMV retinitis in HIV-infected patients (91) and has been associated in a transplant cohort with an increased risk of CMV replication and disease, especially in patients receiving grafts from seropositive donors (92). Non-immunosuppressed patients with chronic periodontitis due to herpes virus infection show significant lower IFNL1 levels in gingival fluid compared to a healthy control group without viral replication (168), suggesting a protective effect of IFNL1 on virus replication, or CMV-induced antagonism of IFN-λ expression. These results highlight the different roles of IFN-λs in acute or chronic infection scenarios and viral reactivation.
The impact of IFN-λs on human T-cell leukemia type-1 virus has also been explored in several independent cohorts. The first evidence came from Kamihira et al. showing that the IFNL3 mRNA expression level was significantly higher in HTLV-1 mono-infection than HTLV-1/HCV coinfection. In addition, the high expression level was associated with the rs8099917 TT SNP (169). The impact of the rs8099917 GG SNP on the risk of HTLV-1 associated myelopathy/tropical spastic paraparesis (TSP) has since been confirmed (88). The impact of the rs12979860 SNP is more controversial. One study on the rs12979860 SNP showed that the CT/TT alleles were more frequent in patients with HTLV-1-associated myelopathy/TSP (81), although this finding was not replicated in two additional studies (170, 171). de Sa et al. reported that the major alleles of IFNL3 SNPs (rs12979860 CC and rs8099917 TT) are associated with a shift in the Th1/Th2 immune response toward a Th1 response (172). The Andes virus causes a hantavirus cardiopulmonary syndrome; in a cohort of Andes virus-infected patients, the minor alleles of rs12979860 and rs8099917 (TT and GG) were linked to milder disease compared to CT/CC and TG/TT (86).
The impact of the IFN-λ signaling on humoral immune function has been described in two vaccine cohorts: immunosuppressed patients vaccinated against influenza (76) and healthy children vaccinated against measles (89). These important observations hold promise for personalized vaccine strategies and adjuvant development (4).
The cytokine microenvironment of a tissue may have an impact on the rate at which a particular infectious bacterium can colonize and also influence the rate of infections. Planet et al. showed that IFN-λs might lead to important changes in the local microbiota during influenza infection. In a mouse model of influenza infection, the authors observed that mice with functional IL28 signaling showed more profound changes in their respiratory microbiota and subsequent higher colonization rates with Staphylococcus aureus compared to IL28RA−/− mice (173). These important findings should be confirmed in a human cohort, as S. aureus is an important source of bacterial superinfection after an influenza infection. In addition, microbiota changes upon common clinical scenarios such as antibiotic treatment may be modulated by IFN-λs and their genotypes.
Bacteria including M. tuberculosis induce IFN-α/β and IFN-γ; however, little is known about the effects of IFN-λs in epithelial immunity. Gram-positive bacteria such as S. aureus, Staphylococcus epidermidis, Enterococcus faecalis, and Listeria monocytogenes induce IFN-λs, whereas Salmonella enterica serovar Typhimurium, Shigella flexneri, and Chlamydia trachomatis do not substantially induce IFN-λs, in intestinal and placental cell lines (134). Others have reported that S. enterica serovar Typhumurium can induce IFN-λs in human DCs (137). IFN-λ gene expression can be increased within DCs upon stimulation with bacterial components such as lipopolysaccharide. In particular, during M. tuberculosis infection, IFN-α plays an important regulatory role in the pathogenesis (12, 174). M. tuberculosis in A549 lung epithelial cells stimulates expression of IFN-λs. In addition, the IFNL2 concentration in sputum of patients with pulmonary tuberculosis is significantly higher than that in the sputum of healthy controls (135). Although the impact of IFN-λs has not been explored in more detail, the cross talk between IFN-α and IFN-λs may play a crucial role in the pathogenesis of M. tuberculosis. The modulation of Th1/Th2 toward Th1 may be of additional importance.
Neutrophil functions are crucial in clearing bacterial infections and wound repair (175, 176). A major target of the effects of IFN-λs may be neutrophils (62, 177). A study by Blazek et al. showed that in a collagen-induced arthritis model, IFNL1 showed anti-inflammatory function by reducing the numbers of IL17-producing Th17 cells and the recruitment of IL-1b expressing neutrophils, which is important to amplify the inflammatory process (62). Similar effects on neutrophil recruitment to the lung have been observed in an OVA-based asthma mouse model (73). Although somewhat speculative, this may suggest an important modulatory function of IFN-λs via neutrophil recruitment toward sites of bacterial infection.
So far, only one study has linked SNPs in genes involved in the IFN-λ signaling pathway with an increased risk of bacterial infections. Xiao et al. showed that SNP rs10903035 with G allele in the IL28RA was associated with significantly less frequent urinary tract infection (178).
The role of IFN-λs in parasitic and fungal disease has not yet been explored. Although somewhat speculative, helminth infections in particular might be regulated by SNPs in the IFN-λ system, considering the profound evidence on the importance of Th1/Th2 balance (6, 77, 78). Furthermore, for parasite infections of the liver such as Plasmodium spp. there is important evidence on the importance of the IFN-α signaling (13, 179–182). Due the regulatory interactions of IFN-α and IFN-λ and the clinical importance of relevant SNPs (31), it is not unreasonable to postulate an impact.
IFN-λs, and their modulation via SNPs, are increasingly recognized as important players in a broad range of infectious diseases. Although the literature is still dominated by reports on HCV, work especially in mouse models has pointed out the important role in viral, respiratory, and gastrointestinal infections. Bacterial colonization and bacterial infections may also be modulated by IFN-λs. The important diversity in IFNs and the large number of SNPs adds a difficult-to-address layer of complexity. Therefore, further research on IFN-λs outside the HCV field is required to understand their roles and diagnostic and therapeutic potential. Most importantly, predictions of risks associated with infectious diseases have to be confirmed in independent cohorts to allow personalized medicine strategies.
Both the authors (AE and MS) have significantly contributed by writing the manuscript and designing the graphs.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors thank Dr. Helena Seth-Smith (Applied Microbiology Research, Department of Biomedicine, University of Basel) for critically reading the manuscript.
AE was supported by a research grant from the SNSF Ambizione Score PZ00P3_154709.
1. Bowness R. Systems medicine and infection. Methods Mol Biol (2016) 1386:107–18. doi: 10.1007/978-1-4939-3283-2_7
2. Subramanian N, Torabi-Parizi P, Gottschalk RA, Germain RN, Dutta B. Network representations of immune system complexity. Wiley Interdiscip Rev Syst Biol Med (2015) 7:13–38. doi:10.1002/wsbm.1288
3. Tsang JS. Utilizing population variation, vaccination, and systems biology to study human immunology. Trends Immunol (2015) 36:479–93. doi:10.1016/j.it.2015.06.005
4. Linnik JE, Egli A. Impact of host genetic polymorphisms on vaccine induced antibody response. Hum Vaccin Immunother (2016) 12:907–15. doi:10.1080/21645515.2015.1119345
5. Karlsson EK, Kwiatkowski DP, Sabeti PC. Natural selection and infectious disease in human populations. Nat Rev Genet (2014) 15:379–93. doi:10.1038/nrg3734
6. Coomes SM, Pelly VS, Kannan Y, Okoye IS, Czieso S, Entwistle LJ, et al. IFNgamma and IL-12 restrict Th2 responses during helminth/plasmodium co-infection and promote IFNgamma from Th2 cells. PLoS Pathog (2015) 11:e1004994. doi:10.1371/journal.ppat.1004994
7. Decker T, Muller M, Stockinger S. The yin and yang of type I interferon activity in bacterial infection. Nat Rev Immunol (2005) 5:675–87. doi:10.1038/nri1684
8. Gommerman JL, Browning JL, Ware CF. The Lymphotoxin Network: orchestrating a type I interferon response to optimize adaptive immunity. Cytokine Growth Factor Rev (2014) 25:139–45. doi:10.1016/j.cytogfr.2014.02.002
9. Moro K, Kabata H, Tanabe M, Koga S, Takeno N, Mochizuki M, et al. Interferon and IL-27 antagonize the function of group 2 innate lymphoid cells and type 2 innate immune responses. Nat Immunol (2016) 17:76–86. doi:10.1038/ni.3309
10. Reutterer B, Stockinger S, Pilz A, Soulat D, Kastner R, Westermayer S, et al. Type I IFN are host modulators of strain-specific Listeria monocytogenes virulence. Cell Microbiol (2008) 10:1116–29. doi:10.1111/j.1462-5822.2007.01114.x
11. Sandler NG, Bosinger SE, Estes JD, Zhu RT, Tharp GK, Boritz E, et al. Type I interferon responses in rhesus macaques prevent SIV infection and slow disease progression. Nature (2014) 511:601–5. doi:10.1038/nature13554
12. Teles RM, Graeber TG, Krutzik SR, Montoya D, Schenk M, Lee DJ, et al. Type I interferon suppresses type II interferon-triggered human anti-mycobacterial responses. Science (2013) 339:1448–53. doi:10.1126/science.1233665
13. Yu X, Cai B, Wang M, Tan P, Ding X, Wu J, et al. Cross-regulation of two type I interferon signaling pathways in plasmacytoid dendritic cells controls anti-malaria immunity and host mortality. Immunity (2016) 45(5):1093–107. doi:10.1016/j.immuni.2016.10.001
14. McNab F, Mayer-Barber K, Sher A, Wack A, O’Garra A. Type I interferons in infectious disease. Nat Rev Immunol (2015) 15:87–103. doi:10.1038/nri3787
15. Zuniga EI, Macal M, Lewis GM, Harker JA. Innate and adaptive immune regulation during chronic viral infections. Annu Rev Virol (2015) 2:573–97. doi:10.1146/annurev-virology-100114-055226
16. Snell LM, Brooks DG. New insights into type I interferon and the immunopathogenesis of persistent viral infections. Curr Opin Immunol (2015) 34:91–8. doi:10.1016/j.coi.2015.03.002
17. Galluzzi L, Buque A, Kepp O, Zitvogel L, Kroemer G. Immunogenic cell death in cancer and infectious disease. Nat Rev Immunol (2017) 17(2):97–111. doi:10.1038/nri.2016.107
18. Parker BS, Rautela J, Hertzog PJ. Antitumour actions of interferons: implications for cancer therapy. Nat Rev Cancer (2016) 16:131–44. doi:10.1038/nrc.2016.14
19. Teijaro JR. Type I interferons in viral control and immune regulation. Curr Opin Virol (2016) 16:31–40. doi:10.1016/j.coviro.2016.01.001
20. Crouse J, Kalinke U, Oxenius A. Regulation of antiviral T cell responses by type I interferons. Nat Rev Immunol (2015) 15:231–42. doi:10.1038/nri3806
21. Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol (2005) 5:375–86. doi:10.1038/nri1604
22. Allen G, Fantes KH. A family of structural genes for human lymphoblastoid (leukocyte-type) interferon. Nature (1980) 287:408–11. doi:10.1038/287408a0
23. Seto MH, Harkins RN, Adler M, Whitlow M, Church WB, Croze E. Homology model of human interferon-alpha 8 and its receptor complex. Protein Sci (1995) 4:655–70. doi:10.1002/pro.5560040406
24. Streuli M, Nagata S, Weissmann C. At least three human type alpha interferons: structure of alpha 2. Science (1980) 209:1343–7. doi:10.1126/science.6158094
25. Karpusas M, Nolte M, Benton CB, Meier W, Lipscomb WN, Goelz S. The crystal structure of human interferon beta at 2.2-A resolution. Proc Natl Acad Sci U S A (1997) 94:11813–8. doi:10.1073/pnas.94.22.11813
26. Senda T, Shimazu T, Matsuda S, Kawano G, Shimizu H, Nakamura KT, et al. Three-dimensional crystal structure of recombinant murine interferon-beta. EMBO J (1992) 11:3193–201.
27. Ealick SE, Cook WJ, Vijay-Kumar S, Carson M, Nagabhushan TL, Trotta PP, et al. Three-dimensional structure of recombinant human interferon-gamma. Science (1991) 252:698–702. doi:10.1126/science.1902591
28. Kotenko SV, Gallagher G, Baurin VV, Lewis-Antes A, Shen M, Shah NK, et al. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol (2003) 4:69–77. doi:10.1038/ni875
29. Sheppard P, Kindsvogel W, Xu W, Henderson K, Schlutsmeyer S, Whitmore TE, et al. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat Immunol (2003) 4:63–8. doi:10.1038/ni873
30. Gad HH, Hamming OJ, Hartmann R. The structure of human interferon lambda and what it has taught us. J Interferon Cytokine Res (2010) 30:565–71. doi:10.1089/jir.2010.0062
31. Egli A, Santer MD, O’Shea D, Tyrrell DL, Houghton M. The impact of the interferon-lambda family on the innate and adaptive immune response to viral infections. Emerg Microbes Infect (2014) 3:e51. doi:10.1038/emi.2014.51
32. Halloran PF, Urmson J, Van der Meide PH, Autenried P. Regulation of MHC expression in vivo. II. IFN-alpha/beta inducers and recombinant IFN-alpha modulate MHC antigen expression in mouse tissues. J Immunol (1989) 142:4241–7.
33. Pantel A, Teixeira A, Haddad E, Wood EG, Steinman RM, Longhi MP. Direct type I IFN but not MDA5/TLR3 activation of dendritic cells is required for maturation and metabolic shift to glycolysis after poly IC stimulation. PLoS Biol (2014) 12:e1001759. doi:10.1371/journal.pbio.1001759
34. Schreiber G, Piehler J. The molecular basis for functional plasticity in type I interferon signaling. Trends Immunol (2015) 36:139–49. doi:10.1016/j.it.2015.01.002
35. Miknis ZJ, Magracheva E, Li W, Zdanov A, Kotenko SV, Wlodawer A. Crystal structure of human interferon-lambda1 in complex with its high-affinity receptor interferon-lambdaR1. J Mol Biol (2010) 404:650–64. doi:10.1016/j.jmb.2010.09.068
36. Bowie AG, Unterholzner L. Viral evasion and subversion of pattern-recognition receptor signalling. Nat Rev Immunol (2008) 8:911–22. doi:10.1038/nri2436
37. Mihm S, Frese M, Meier V, Wietzke-Braun P, Scharf JG, Bartenschlager R, et al. Interferon type I gene expression in chronic hepatitis C. Lab Invest (2004) 84:1148–59. doi:10.1038/labinvest.3700135
38. Stoltz M, Klingstrom J. Alpha/beta interferon (IFN-alpha/beta)-independent induction of IFN-lambda1 (interleukin-29) in response to Hantaan virus infection. J Virol (2010) 84:9140–8. doi:10.1128/JVI.00717-10
39. Ank N, Iversen MB, Bartholdy C, Staeheli P, Hartmann R, Jensen UB, et al. An important role for type III interferon (IFN-lambda/IL-28) in TLR-induced antiviral activity. J Immunol (2008) 180:2474–85. doi:10.4049/jimmunol.180.4.2474
40. Hou W, Wang X, Ye L, Zhou L, Yang ZQ, Riedel E, et al. Lambda interferon inhibits human immunodeficiency virus type 1 infection of macrophages. J Virol (2009) 83:3834–42. doi:10.1128/JVI.01773-08
41. Chandra PK, Bao L, Song K, Aboulnasr FM, Baker DP, Shores N, et al. HCV infection selectively impairs type I but not type III IFN signaling. Am J Pathol (2014) 184:214–29. doi:10.1016/j.ajpath.2013.10.005
42. Zhang S, Kodys K, Li K, Szabo G. Human type 2 myeloid dendritic cells produce interferon-lambda and amplify interferon-alpha in response to hepatitis C virus infection. Gastroenterology (2013) 144:414.e–25.e. doi:10.1053/j.gastro.2012.10.034
43. Wolk K, Witte K, Witte E, Proesch S, Schulze-Tanzil G, Nasilowska K, et al. Maturing dendritic cells are an important source of IL-29 and IL-20 that may cooperatively increase the innate immunity of keratinocytes. J Leukoc Biol (2008) 83:1181–93. doi:10.1189/jlb.0807525
44. Yin Z, Dai J, Deng J, Sheikh F, Natalia M, Shih T, et al. Type III IFNs are produced by and stimulate human plasmacytoid dendritic cells. J Immunol (2012) 189:2735–45. doi:10.4049/jimmunol.1102038
45. Lauterbach H, Bathke B, Gilles S, Traidl-Hoffmann C, Luber CA, Fejer G, et al. Mouse CD8alpha+ DCs and human BDCA3+ DCs are major producers of IFN-lambda in response to poly IC. J Exp Med (2010) 207:2703–17. doi:10.1084/jem.20092720
46. Megjugorac NJ, Gallagher GE, Gallagher G. IL-4 enhances IFN-lambda1 (IL-29) production by plasmacytoid DCs via monocyte secretion of IL-1Ra. Blood (2010) 115:4185–90. doi:10.1182/blood-2009-09-246157
47. Hillyer P, Mane VP, Schramm LM, Puig M, Verthelyi D, Chen A, et al. Expression profiles of human interferon-alpha and interferon-lambda subtypes are ligand- and cell-dependent. Immunol Cell Biol (2012) 90(8):774–83. doi:10.1038/icb.2011.109
48. Nizzoli G, Krietsch J, Weick A, Steinfelder S, Facciotti F, Gruarin P, et al. Human CD1c+ dendritic cells secrete high levels of IL-12 and potently prime cytotoxic T-cell responses. Blood (2013) 122:932–42. doi:10.1182/blood-2013-04-495424
49. Donnelly RP, Kotenko SV. Interferon-lambda: a new addition to an old family. J Interferon Cytokine Res (2010) 30:555–64. doi:10.1089/jir.2010.0078
50. Gad HH, Dellgren C, Hamming OJ, Vends S, Paludan SR, Hartmann R. Interferon-lambda is functionally an interferon but structurally related to the interleukin-10 family. J Biol Chem (2009) 284:20869–75. doi:10.1074/jbc.M109.002923
51. Reboul J, Gardiner K, Monneron D, Uze G, Lutfalla G. Comparative genomic analysis of the interferon/interleukin-10 receptor gene cluster. Genome Res (1999) 9:242–50.
52. Josephson K, Logsdon NJ, Walter MR. Crystal structure of the IL-10/IL-10R1 complex reveals a shared receptor binding site. Immunity (2001) 15:35–46. doi:10.1016/S1074-7613(01)00169-8
53. Magracheva E, Pletnev S, Kotenko S, Li W, Wlodawer A, Zdanov A. Purification, crystallization and preliminary crystallographic studies of the complex of interferon-lambda1 with its receptor. Acta Crystallogr Sect F Struct Biol Cryst Commun (2010) 66:61–3. doi:10.1107/S1744309109048817
54. Syedbasha M, Linnik J, Santer D, O’Shea D, Barakat K, Joyce M, et al. An ELISA based binding and competition method to rapidly determine ligand-receptor interactions. J Vis Exp (2016) (109):e53575. doi:10.3791/53575
55. Francois-Newton V, Magno de Freitas Almeida G, Payelle-Brogard B, Monneron D, Pichard-Garcia L, Piehler J, et al. USP18-based negative feedback control is induced by type I and type III interferons and specifically inactivates interferon alpha response. PLoS One (2011) 6:e22200. doi:10.1371/journal.pone.0022200
56. Hermant P, Demarez C, Mahlakoiv T, Staeheli P, Meuleman P, Michiels T. Human but not mouse hepatocytes respond to interferon-lambda in vivo. PLoS One (2014) 9:e87906. doi:10.1371/journal.pone.0087906
57. Yoon SI, Jones BC, Logsdon NJ, Harris BD, Deshpande A, Radaeva S, et al. Structure and mechanism of receptor sharing by the IL-10R2 common chain. Structure (2010) 18:638–48. doi:10.1016/j.str.2010.02.009
58. Witte K, Gruetz G, Volk HD, Looman AC, Asadullah K, Sterry W, et al. Despite IFN-lambda receptor expression, blood immune cells, but not keratinocytes or melanocytes, have an impaired response to type III interferons: implications for therapeutic applications of these cytokines. Genes Immun (2009) 10:702–14. doi:10.1038/gene.2009.72
59. Brand S, Beigel F, Olszak T, Zitzmann K, Eichhorst ST, Otte JM, et al. IL-28A and IL-29 mediate antiproliferative and antiviral signals in intestinal epithelial cells and murine CMV infection increases colonic IL-28A expression. Am J Physiol Gastrointest Liver Physiol (2005) 289:G960–8. doi:10.1152/ajpgi.00126.2005
60. Doyle SE, Schreckhise H, Khuu-Duong K, Henderson K, Rosler R, Storey H, et al. Interleukin-29 uses a type 1 interferon-like program to promote antiviral responses in human hepatocytes. Hepatology (2006) 44:896–906. doi:10.1002/hep.21312
61. Maher SG, Sheikh F, Scarzello AJ, Romero-Weaver AL, Baker DP, Donnelly RP, et al. IFNalpha and IFNlambda differ in their antiproliferative effects and duration of JAK/STAT signaling activity. Cancer Biol Ther (2008) 7:1109–15. doi:10.4161/cbt.7.7.6192
62. Blazek K, Eames HL, Weiss M, Byrne AJ, Perocheau D, Pease JE, et al. IFN-lambda resolves inflammation via suppression of neutrophil infiltration and IL-1beta production. J Exp Med (2015) 212:845–53. doi:10.1084/jem.20140995
63. Morrison MH, Keane C, Quinn LM, Kelly A, O’Farrelly C, Bergin C, et al. IFNL cytokines do not modulate human or murine NK cell functions. Hum Immunol (2014) 75:996–1000. doi:10.1016/j.humimm.2014.06.016
64. Liu BS, Janssen HL, Boonstra A. IL-29 and IFNalpha differ in their ability to modulate IL-12 production by TLR-activated human macrophages and exhibit differential regulation of the IFNgamma receptor expression. Blood (2011) 117:2385–95. doi:10.1182/blood-2010-07-298976
65. Duong FH, Trincucci G, Boldanova T, Calabrese D, Campana B, Krol I, et al. IFN-lambda receptor 1 expression is induced in chronic hepatitis C and correlates with the IFN-lambda3 genotype and with nonresponsiveness to IFN-alpha therapies. J Exp Med (2014) 211(5):857–68. doi:10.1084/jem.20131557
66. Egli A, Levin A, Santer DM, Joyce M, O’Shea D, Thomas B, et al. Immunomodulatory function of interleukin-28B during primary infection with Cytomegalovirus. J Infect Dis (2014) 210(5):717–27. doi:10.1093/infdis/jiu144
67. Lazear HM, Daniels BP, Pinto AK, Huang AC, Vick SC, Doyle SE, et al. Interferon-lambda restricts West Nile virus neuroinvasion by tightening the blood-brain barrier. Sci Transl Med (2015) 7:284ra259. doi:10.1126/scitranslmed.aaa4304
68. Witte K, Witte E, Sabat R, Wolk K. IL-28A, IL-28B, and IL-29: promising cytokines with type I interferon-like properties. Cytokine Growth Factor Rev (2010) 21:237–51. doi:10.1016/j.cytogfr.2010.04.002
69. de Groen RA, Groothuismink ZM, Liu BS, Boonstra A. IFN-lambda is able to augment TLR-mediated activation and subsequent function of primary human B cells. J Leukoc Biol (2015) 98:623–30. doi:10.1189/jlb.3A0215-041RR
70. Dai J, Megjugorac NJ, Gallagher GE, Yu RY, Gallagher G. IFN-lambda1 (IL-29) inhibits GATA3 expression and suppresses Th2 responses in human naive and memory T cells. Blood (2009) 113:5829–38. doi:10.1182/blood-2008-09-179507
71. Jordan WJ, Eskdale J, Srinivas S, Pekarek V, Kelner D, Rodia M, et al. Human interferon lambda-1 (IFN-lambda1/IL-29) modulates the Th1/Th2 response. Genes Immun (2007) 8:254–61. doi:10.1038/sj.gene.6364382
72. Koch S, Finotto S. Role of interferon-lambda in allergic asthma. J Innate Immun (2015) 7:224–30. doi:10.1159/000369459
73. Koltsida O, Hausding M, Stavropoulos A, Koch S, Tzelepis G, Ubel C, et al. IL-28A (IFN-lambda2) modulates lung DC function to promote Th1 immune skewing and suppress allergic airway disease. EMBO Mol Med (2011) 3:348–61. doi:10.1002/emmm.201100142
74. Srinivas S, Dai J, Eskdale J, Gallagher GE, Megjugorac NJ, Gallagher G. Interferon-lambda1 (interleukin-29) preferentially down-regulates interleukin-13 over other T helper type 2 cytokine responses in vitro. Immunology (2008) 125:492–502. doi:10.1111/j.1365-2567.2008.02862.x
75. Gallagher G, Megjugorac NJ, Yu RY, Eskdale J, Gallagher GE, Siegel R, et al. The lambda interferons: guardians of the immune-epithelial interface and the T-helper 2 response. J Interferon Cytokine Res (2010) 30:603–15. doi:10.1089/jir.2010.0081
76. Egli A, Santer DM, O’Shea D, Barakat K, Syedbasha M, Vollmer M, et al. IL-28B is a key regulator of B- and T-cell vaccine responses against influenza. PLoS Pathog (2014) 10:e1004556. doi:10.1371/journal.ppat.1004556
77. Spellberg B, Edwards JE Jr. Type 1/Type 2 immunity in infectious diseases. Clin Infect Dis (2001) 32:76–102. doi:10.1086/317537
78. Harris N, Gause WC. To B or not to B: B cells and the Th2-type immune response to helminths. Trends Immunol (2011) 32:80–8. doi:10.1016/j.it.2010.11.005
79. Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature (2009) 461:399–401. doi:10.1038/nature08309
80. Tanaka Y, Nishida N, Sugiyama M, Kurosaki M, Matsuura K, Sakamoto N, et al. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet (2009) 41:1105–9. doi:10.1038/ng.449
81. Trevino A, Lopez M, Vispo E, Aguilera A, Ramos JM, Benito R, et al. Development of tropical spastic paraparesis in human T-lymphotropic virus type 1 carriers is influenced by interleukin 28B gene polymorphisms. Clin Infect Dis (2012) 55:e1–4. doi:10.1093/cid/cis343
82. Eslam M, Hashem AM, Leung R, Romero-Gomez M, Berg T, Dore GJ, et al. Interferon-lambda rs12979860 genotype and liver fibrosis in viral and non-viral chronic liver disease. Nat Commun (2015) 6:6422. doi:10.1038/ncomms7422
83. Akay E, Patel M, Conibear T, Chaggar T, Haque T. Interleukin 28B gene polymorphisms and Epstein-Barr virus-associated lymphoproliferative diseases. Intervirology (2014) 57:112–5. doi:10.1159/000357326
84. Bravo D, Solano C, Gimenez E, Remigia MJ, Corrales I, Amat P, et al. Effect of the IL28B Rs12979860 C/T polymorphism on the incidence and features of active cytomegalovirus infection in allogeneic stem cell transplant patients. J Med Virol (2013) 86(5):838–44. doi:10.1002/jmv.23865
85. Pica F, Volpi A, Gaziano R, Garaci E. Interferon-lambda in immunocompetent individuals with a history of recurrent herpes labialis. Antivir Ther (2010) 15:737–43. doi:10.3851/IMP1610
86. Angulo J, Pino K, Echeverria-Chagas N, Marco C, Martinez-Valdebenito C, Galeno H, et al. Association of single-nucleotide polymorphisms in IL28B, but not TNF-alpha, with severity of disease caused by Andes virus. Clin Infect Dis (2015) 61:e62–9. doi:10.1093/cid/civ830
87. Suppiah V, Moldovan M, Ahlenstiel G, Berg T, Weltman M, Abate ML, et al. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet (2009) 41:1100–4. doi:10.1038/ng.447
88. Assone T, de Souza FV, Gaester KO, Fonseca LA, Luiz Odo C, Malta F, et al. IL28B gene polymorphism SNP rs8099917 genotype GG is associated with HTLV-1-associated myelopathy/tropical spastic paraparesis (HAM/TSP) in HTLV-1 carriers. PLoS Negl Trop Dis (2014) 8:e3199. doi:10.1371/journal.pntd.0003199
89. Haralambieva IH, Ovsyannikova IG, Kennedy RB, Vierkant RA, Pankratz VS, Jacobson RM, et al. Associations between single nucleotide polymorphisms and haplotypes in cytokine and cytokine receptor genes and immunity to measles vaccination. Vaccine (2011) 29:7883–95. doi:10.1016/j.vaccine.2011.08.083
90. Prokunina-Olsson L, Muchmore B, Tang W, Pfeiffer RM, Park H, Dickensheets H, et al. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat Genet (2013) 45:164–71. doi:10.1038/ng.2521
91. Bibert S, Wojtowicz A, Taffe P, Manuel O, Bernasconi E, Furrer H, et al. The IFNL3/4 DeltaG variant increases susceptibility to cytomegalovirus retinitis among HIV-infected patients. AIDS (2014) 28:1885–9. doi:10.1097/QAD.0000000000000379
92. Manuel O, Wojtowicz A, Bibert S, Mueller NJ, van Delden C, Hirsch HH, et al. Influence of IFNL3/4 polymorphisms on the incidence of cytomegalovirus infection after solid-organ transplantation. J Infect Dis (2015) 211:906–14. doi:10.1093/infdis/jiu557
93. Machmach K, Abad-Molina C, Romero-Sanchez MC, Dominguez-Molina B, Moyano M, Rodriguez MM, et al. IFNL4 ss469415590 polymorphism is associated with unfavourable clinical and immunological status in HIV-infected individuals. Clin Microbiol Infect (2015) 21(289):e281–4. doi:10.1016/j.cmi.2014.10.012
94. Jimenez-Sousa MA, Berenguer J, Fernandez-Rodriguez A, Micheloud D, Guzman-Fulgencio M, Miralles P, et al. IL28RA polymorphism (rs10903035) is associated with insulin resistance in HIV/HCV-coinfected patients. J Viral Hepat (2014) 21:189–97. doi:10.1111/jvh.12130
95. Jin G, Kang H, Chen X, Dai D. Evaluation of the relationship between IL28B, IL10RB and IL28RA single-nucleotide polymorphisms and susceptibility to hepatitis C virus in Chinese Han population. Infect Genet Evol (2014) 21:8–14. doi:10.1016/j.meegid.2013.10.009
96. Genetic Analysis of Psoriasis Consortium & the Wellcome Trust Case Control Consortium 2, Strange A, Capon F, Spencer CC, Knight J, Weale ME, et al. A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nat Genet (2010) 42:985–90. doi:10.1038/ng.694
97. Li Y, Cheng H, Zuo XB, Sheng YJ, Zhou FS, Tang XF, et al. Association analyses identifying two common susceptibility loci shared by psoriasis and systemic lupus erythematosus in the Chinese Han population. J Med Genet (2013) 50:812–8. doi:10.1136/jmedgenet-2013-101787
98. Jimenez-Sousa MA, Berenguer J, Rallon N, Guzman-Fulgencio M, Lopez JC, Soriano V, et al. IL28RA polymorphism is associated with early hepatitis C virus (HCV) treatment failure in human immunodeficiency virus-/HCV-coinfected patients. J Viral Hepat (2013) 20:358–66. doi:10.1111/jvh.12041
99. Bibert S, Roger T, Calandra T, Bochud M, Cerny A, Semmo N, et al. IL28B expression depends on a novel TT/-G polymorphism which improves HCV clearance prediction. J Exp Med (2013) 210:1109–16. doi:10.1084/jem.20130012
100. Chinnaswamy S, Chatterjee S, Boopathi R, Mukherjee S, Bhattacharjee S, Kundu TK. A single nucleotide polymorphism associated with hepatitis C virus infections located in the distal region of the IL28B promoter influences NF-kappaB-mediated gene transcription. PLoS One (2013) 8:e75495. doi:10.1371/journal.pone.0075495
101. Fischer J, Bohm S, Scholz M, Muller T, Witt H, George J, et al. Combined effects of different interleukin-28B gene variants on the outcome of dual combination therapy in chronic hepatitis C virus type 1 infection. Hepatology (2012) 55:1700–10. doi:10.1002/hep.25582
102. Smith KR, Suppiah V, O’Connor K, Berg T, Weltman M, Abate ML, et al. Identification of improved IL28B SNPs and haplotypes for prediction of drug response in treatment of hepatitis C using massively parallel sequencing in a cross-sectional European cohort. Genome Med (2011) 3:57. doi:10.1186/gm273
103. Rauch A, Kutalik Z, Descombes P, Cai T, Di Iulio J, Mueller T, et al. Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology (2010) 138:e1331–7. doi:10.1053/j.gastro.2009.12.056
104. Duggal P, Thio CL, Wojcik GL, Goedert JJ, Mangia A, Latanich R, et al. Genome-wide association study of spontaneous resolution of hepatitis C virus infection: data from multiple cohorts. Ann Intern Med (2013) 158:235–45. doi:10.7326/0003-4819-158-4-201302190-00003
105. Honda M, Shirasaki T, Shimakami T, Sakai A, Horii R, Arai K, et al. Hepatic interferon-stimulated genes are differentially regulated in the liver of chronic hepatitis C patients with different interleukin 28B genotypes. Hepatology (2013) 59(3):828–38. doi:10.1002/hep.26788
106. Dill MT, Duong FH, Vogt JE, Bibert S, Bochud PY, Terracciano L, et al. Interferon-induced gene expression is a stronger predictor of treatment response than IL28B genotype in patients with hepatitis C. Gastroenterology (2011) 140:1021–31. doi:10.1053/j.gastro.2010.11.039
107. Rallon NI, Soriano V, Naggie S, Restrepo C, McHutchison J, Vispo E, et al. Impact of IL28B gene polymorphisms on interferon-lambda3 plasma levels during pegylated interferon-alpha/ribavirin therapy for chronic hepatitis C in patients coinfected with HIV. J Antimicrob Chemother (2012) 67:1246–9. doi:10.1093/jac/dkr598
108. Shi X, Pan Y, Wang M, Wang D, Li W, Jiang T, et al. IL28B genetic variation is associated with spontaneous clearance of hepatitis C virus, treatment response, serum IL-28B levels in Chinese population. PLoS One (2012) 7:e37054. doi:10.1371/journal.pone.0037054
109. Murakawa M, Asahina Y, Nakagawa M, Sakamoto N, Nitta S, Kusano-Kitazume A, et al. Impaired induction of interleukin 28B and expression of interferon lambda 4 associated with nonresponse to interferon-based therapy in chronic hepatitis C. J Gastroenterol Hepatol (2015) 30:1075–84. doi:10.1111/jgh.12902
110. Ferraris P, Chandra PK, Panigrahi R, Aboulnasr F, Chava S, Kurt R, et al. Cellular mechanism for impaired hepatitis C virus clearance by interferon associated with IFNL3 gene polymorphisms relates to intrahepatic interferon-lambda expression. Am J Pathol (2016) 186:938–51. doi:10.1016/j.ajpath.2015.11.027
111. Terczynska-Dyla E, Bibert S, Duong FH, Krol I, Jorgensen S, Collinet E, et al. Reduced IFNlambda4 activity is associated with improved HCV clearance and reduced expression of interferon-stimulated genes. Nat Commun (2014) 5:5699. doi:10.1038/ncomms6699
112. Price AA, Tedesco D, Prasad MR, Workowski KA, Walker CM, Suthar MS, et al. Prolonged activation of innate antiviral gene signature after childbirth is determined by IFNL3 genotype. Proc Natl Acad Sci U S A (2016) 113:10678–83. doi:10.1073/pnas.1602319113
113. Langhans B, Kupfer B, Braunschweiger I, Arndt S, Schulte W, Nischalke HD, et al. Interferon-lambda serum levels in hepatitis C. J Hepatol (2011) 54:859–65. doi:10.1016/j.jhep.2010.08.020
114. Hsu YL, Wang MY, Ho LJ, Lai JH. Dengue virus infection induces interferon-lambda1 to facilitate cell migration. Sci Rep (2016) 6:24530. doi:10.1038/srep24530
115. Robek MD, Boyd BS, Chisari FV. Lambda interferon inhibits hepatitis B and C virus replication. J Virol (2005) 79:3851–4. doi:10.1128/JVI.79.6.3851-3854.2005
116. Diegelmann J, Beigel F, Zitzmann K, Kaul A, Goke B, Auernhammer CJ, et al. Comparative analysis of the lambda-interferons IL-28A and IL-29 regarding their transcriptome and their antiviral properties against hepatitis C virus. PLoS One (2010) 5:e15200. doi:10.1371/journal.pone.0015200
117. Liu MQ, Zhou DJ, Wang X, Zhou W, Ye L, Li JL, et al. IFN-lambda3 inhibits HIV infection of macrophages through the JAK-STAT pathway. PLoS One (2012) 7:e35902. doi:10.1371/journal.pone.0035902
118. Tian RR, Guo HX, Wei JF, Yang CK, He SH, Wang JH. IFN-lambda inhibits HIV-1 integration and post-transcriptional events in vitro, but there is only limited in vivo repression of viral production. Antiviral Res (2012) 95:57–65. doi:10.1016/j.antiviral.2012.04.011
119. Griffiths SJ, Koegl M, Boutell C, Zenner HL, Crump CM, Pica F, et al. A systematic analysis of host factors reveals a Med23-interferon-lambda regulatory axis against herpes simplex virus type 1 replication. PLoS Pathog (2013) 9:e1003514. doi:10.1371/journal.ppat.1003514
120. Zhou L, Li JL, Zhou Y, Liu JB, Zhuang K, Gao JF, et al. Induction of interferon-lambda contributes to TLR3 and RIG-I activation-mediated inhibition of herpes simplex virus type 2 replication in human cervical epithelial cells. Mol Hum Reprod (2015) 21:917–29. doi:10.1093/molehr/gav058
121. Banos-Lara Mdel R, Harvey L, Mendoza A, Simms D, Chouljenko VN, Wakamatsu N, et al. Impact and regulation of lambda interferon response in human metapneumovirus infection. J Virol (2015) 89:730–42. doi:10.1128/JVI.02897-14
122. Mordstein M, Neugebauer E, Ditt V, Jessen B, Rieger T, Falcone V, et al. Lambda interferon renders epithelial cells of the respiratory and gastrointestinal tracts resistant to viral infections. J Virol (2010) 84:5670–7. doi:10.1128/JVI.00272-10
123. Davidson S, McCabe TM, Crotta S, Gad HH, Hessel EM, Beinke S, et al. IFNlambda is a potent anti-influenza therapeutic without the inflammatory side effects of IFNalpha treatment. EMBO Mol Med (2016) 8:1099–112. doi:10.15252/emmm.201606413
124. Nice TJ, Baldridge MT, McCune BT, Norman JM, Lazear HM, Artyomov M, et al. Interferon-lambda cures persistent murine norovirus infection in the absence of adaptive immunity. Science (2015) 347:269–73. doi:10.1126/science.1258100
125. Lukacikova L, Oveckova I, Betakova T, Laposova K, Polcicova K, Pastorekova S, et al. Antiviral effect of interferon lambda against lymphocytic choriomeningitis virus. J Interferon Cytokine Res (2015) 35:540–53. doi:10.1089/jir.2014.0083
126. Contoli M, Message SD, Laza-Stanca V, Edwards MR, Wark PA, Bartlett NW, et al. Role of deficient type III interferon-lambda production in asthma exacerbations. Nat Med (2006) 12:1023–6. doi:10.1038/nm1462
127. Ioannidis I, McNally B, Willette M, Peeples ME, Chaussabel D, Durbin JE, et al. Plasticity and virus specificity of the airway epithelial cell immune response during respiratory virus infection. J Virol (2012) 86:5422–36. doi:10.1128/JVI.06757-11
128. Villenave R, Broadbent L, Douglas I, Lyons JD, Coyle PV, Teng MN, et al. Induction and antagonism of antiviral responses in respiratory syncytial virus-infected pediatric airway epithelium. J Virol (2015) 89:12309–18. doi:10.1128/JVI.02119-15
129. Pott J, Mahlakoiv T, Mordstein M, Duerr CU, Michiels T, Stockinger S, et al. IFN-lambda determines the intestinal epithelial antiviral host defense. Proc Natl Acad Sci U S A (2011) 108:7944–9. doi:10.1073/pnas.1100552108
130. Mahlakoiv T, Ritz D, Mordstein M, Dediego ML, Enjuanes L, Muller MA, et al. Combined action of type I and type III interferon restricts initial replication of SARS-coronavirus in the lung but fails to inhibit systemic virus spread. J Gen Virol (2012) 93(Pt 12):2601–5. doi:10.1099/vir.0.046284-0
131. Guayasamin RC, Reynolds TD, Wei X, Fujiwara M, Robek MD. Type III interferon attenuates a vesicular stomatitis virus-based vaccine vector. J Virol (2014) 88:10909–17. doi:10.1128/JVI.01910-14
132. Ma D, Jiang D, Qing M, Weidner JM, Qu X, Guo H, et al. Antiviral effect of interferon lambda against West Nile virus. Antiviral Res (2009) 83:53–60. doi:10.1016/j.antiviral.2009.03.006
133. Cohen TS, Prince AS. Bacterial pathogens activate a common inflammatory pathway through IFNlambda regulation of PDCD4. PLoS Pathog (2013) 9:e1003682. doi:10.1371/journal.ppat.1003682
134. Bierne H, Travier L, Mahlakoiv T, Tailleux L, Subtil A, Lebreton A, et al. Activation of type III interferon genes by pathogenic bacteria in infected epithelial cells and mouse placenta. PLoS One (2012) 7:e39080. doi:10.1371/journal.pone.0039080
135. Travar M, Vucic M, Petkovic M. Interferon lambda-2 levels in sputum of patients with pulmonary Mycobacterium tuberculosis infection. Scand J Immunol (2014) 80:43–9. doi:10.1111/sji.12178
136. Lebreton A, Lakisic G, Job V, Fritsch L, Tham TN, Camejo A, et al. A bacterial protein targets the BAHD1 chromatin complex to stimulate type III interferon response. Science (2011) 331:1319–21. doi:10.1126/science.1200120
137. Pietila TE, Latvala S, Osterlund P, Julkunen I. Inhibition of dynamin-dependent endocytosis interferes with type III IFN expression in bacteria-infected human monocyte-derived DCs. J Leukoc Biol (2010) 88:665–74. doi:10.1189/jlb.1009651
138. Krupna-Gaylord MA, Liveris D, Love AC, Wormser GP, Schwartz I, Petzke MM. Induction of type I and type III interferons by Borrelia burgdorferi correlates with pathogenesis and requires linear plasmid 36. PLoS One (2014) 9:e100174. doi:10.1371/journal.pone.0100174
139. Fensterl V, Chattopadhyay S, Sen GC. No love lost between viruses and interferons. Annu Rev Virol (2015) 2:549–72. doi:10.1146/annurev-virology-100114-055249
140. Dankar SK, Miranda E, Forbes NE, Pelchat M, Tavassoli A, Selman M, et al. Influenza A/Hong Kong/156/1997(H5N1) virus NS1 gene mutations F103L and M106I both increase IFN antagonism, virulence and cytoplasmic localization but differ in binding to RIG-I and CPSF30. Virol J (2013) 10:243. doi:10.1186/1743-422X-10-243
141. Katze MG, He Y, Gale M Jr. Viruses and interferon: a fight for supremacy. Nat Rev Immunol (2002) 2:675–87. doi:10.1038/nri888
142. Tumpey TM, Szretter KJ, Van Hoeven N, Katz JM, Kochs G, Haller O, et al. The Mx1 gene protects mice against the pandemic 1918 and highly lethal human H5N1 influenza viruses. J Virol (2007) 81:10818–21. doi:10.1128/JVI.01116-07
143. Yang Y, Zhang L, Geng H, Deng Y, Huang B, Guo Y, et al. The structural and accessory proteins M, ORF 4a, ORF 4b, and ORF 5 of Middle East respiratory syndrome coronavirus (MERS-CoV) are potent interferon antagonists. Protein Cell (2013) 4:951–61. doi:10.1007/s13238-013-3096-8
144. Eberle KC, McGill JL, Reinhardt TA, Sacco RE. Parainfluenza virus 3 blocks antiviral mediators downstream of the interferon lambda receptor by modulating Stat1 phosphorylation. J Virol (2015) 90:2948–58. doi:10.1128/JVI.02502-15
145. Palma-Ocampo HK, Flores-Alonso JC, Vallejo-Ruiz V, Reyes-Leyva J, Flores-Mendoza L, Herrera-Camacho I, et al. Interferon lambda inhibits dengue virus replication in epithelial cells. Virol J (2015) 12:150. doi:10.1186/s12985-015-0383-4
146. Dellgren C, Gad HH, Hamming OJ, Melchjorsen J, Hartmann R. Human interferon-lambda3 is a potent member of the type III interferon family. Genes Immun (2009) 10:125–31. doi:10.1038/gene.2008.87
147. Marcello T, Grakoui A, Barba-Spaeth G, Machlin ES, Kotenko SV, MacDonald MR, et al. Interferons alpha and lambda inhibit hepatitis C virus replication with distinct signal transduction and gene regulation kinetics. Gastroenterology (2006) 131:1887–98. doi:10.1053/j.gastro.2006.09.052
148. Forbes RL, Wark PA, Murphy VE, Gibson PG. Pregnant women have attenuated innate interferon responses to 2009 pandemic influenza A virus subtype H1N1. J Infect Dis (2012) 206:646–53. doi:10.1093/infdis/jis377
149. Jewell NA, Cline T, Mertz SE, Smirnov SV, Flano E, Schindler C, et al. Lambda interferon is the predominant interferon induced by influenza A virus infection in vivo. J Virol (2010) 84:11515–22. doi:10.1128/JVI.01703-09
150. Mordstein M, Kochs G, Dumoutier L, Renauld JC, Paludan SR, Klucher K, et al. Interferon-lambda contributes to innate immunity of mice against influenza A virus but not against hepatotropic viruses. PLoS Pathog (2008) 4:e1000151. doi:10.1371/journal.ppat.1000151
151. Patel DA, You Y, Huang G, Byers DE, Kim HJ, Agapov E, et al. Interferon response and respiratory virus control are preserved in bronchial epithelial cells in asthma. J Allergy Clin Immunol (2014) 134(1402–1412):e1407. doi:10.1016/j.jaci.2014.07.013
152. Wei H, Wang S, Chen Q, Chen Y, Chi X, Zhang L, et al. Suppression of interferon lambda signaling by SOCS-1 results in their excessive production during influenza virus infection. PLoS Pathog (2014) 10:e1003845. doi:10.1371/journal.ppat.1003845
153. Spann KM, Tran KC, Chi B, Rabin RL, Collins PL. Suppression of the induction of alpha, beta, and lambda interferons by the NS1 and NS2 proteins of human respiratory syncytial virus in human epithelial cells and macrophages [corrected]. J Virol (2004) 78:4363–9. doi:10.1128/JVI.78.12.6705.2004
154. Ioannidis I, Ye F, McNally B, Willette M, Flano E. Toll-like receptor expression and induction of type I and type III interferons in primary airway epithelial cells. J Virol (2013) 87:3261–70. doi:10.1128/JVI.01956-12
155. Osterlund PI, Pietila TE, Veckman V, Kotenko SV, Julkunen I. IFN regulatory factor family members differentially regulate the expression of type III IFN (IFN-lambda) genes. J Immunol (2007) 179:3434–42. doi:10.4049/jimmunol.179.6.3434
156. Ank N, West H, Bartholdy C, Eriksson K, Thomsen AR, Paludan SR. Lambda interferon (IFN-lambda), a type III IFN, is induced by viruses and IFNs and displays potent antiviral activity against select virus infections in vivo. J Virol (2006) 80:4501–9. doi:10.1128/JVI.80.9.4501-4509.2006
157. Coccia EM, Severa M, Giacomini E, Monneron D, Remoli ME, Julkunen I, et al. Viral infection and Toll-like receptor agonists induce a differential expression of type I and lambda interferons in human plasmacytoid and monocyte-derived dendritic cells. Eur J Immunol (2004) 34:796–805. doi:10.1002/eji.200324610
158. Herman M, Ciancanelli M, Ou YH, Lorenzo L, Klaudel-Dreszler M, Pauwels E, et al. Heterozygous TBK1 mutations impair TLR3 immunity and underlie herpes simplex encephalitis of childhood. J Exp Med (2012) 209:1567–82. doi:10.1084/jem.20111316
159. Wongthida P, Diaz RM, Galivo F, Kottke T, Thompson J, Pulido J, et al. Type III IFN interleukin-28 mediates the antitumor efficacy of oncolytic virus VSV in immune-competent mouse models of cancer. Cancer Res (2010) 70:4539–49. doi:10.1158/0008-5472.CAN-09-4658
160. Lin JD, Feng N, Sen A, Balan M, Tseng HC, McElrath C, et al. Distinct roles of type I and type III interferons in intestinal immunity to homologous and heterologous rotavirus infections. PLoS Pathog (2016) 12:e1005600. doi:10.1371/journal.ppat.1005600
161. Mahlakoiv T, Hernandez P, Gronke K, Diefenbach A, Staeheli P. Leukocyte-derived IFN-alpha/beta and epithelial IFN-lambda constitute a compartmentalized mucosal defense system that restricts enteric virus infections. PLoS Pathog (2015) 11:e1004782. doi:10.1371/journal.ppat.1004782
162. Baldridge MT, Nice TJ, McCune BT, Yokoyama CC, Kambal A, Wheadon M, et al. Commensal microbes and interferon-lambda determine persistence of enteric murine norovirus infection. Science (2015) 347:266–9. doi:10.1126/science.1258025
163. Hernandez PP, Mahlakoiv T, Yang I, Schwierzeck V, Nguyen N, Guendel F, et al. Interferon-lambda and interleukin 22 act synergistically for the induction of interferon-stimulated genes and control of rotavirus infection. Nat Immunol (2015) 16:698–707. doi:10.1038/ni.3180
164. Misumi I, Whitmire JK. IFN-lambda exerts opposing effects on T cell responses depending on the chronicity of the virus infection. J Immunol (2014) 192:3596–606. doi:10.4049/jimmunol.1301705
165. Yoshio S, Kanto T. Host-virus interactions in hepatitis B and hepatitis C infection. J Gastroenterol (2016) 51:409–20. doi:10.1007/s00535-016-1183-3
166. Chinnaswamy S. Gene-disease association with human IFNL locus polymorphisms extends beyond hepatitis C virus infections. Genes Immun (2016) 17:265–75. doi:10.1038/gene.2016.24
167. Mihm S. Activation of type I and type III interferons in chronic hepatitis C. J Innate Immun (2015) 7:251–9. doi:10.1159/000369973
168. Muzammil, Jayanthi D, Faizuddin M, Noor Ahamadi HM. Association of interferon lambda-1 with herpes simplex viruses-1 and -2, Epstein-Barr virus, and human cytomegalovirus in chronic periodontitis. J Investig Clin Dent (2015). doi:10.1111/jicd.12200
169. Kamihira S, Usui T, Ichikawa T, Uno N, Morinaga Y, Mori S, et al. Paradoxical expression of IL-28B mRNA in peripheral blood in human T-cell leukemia virus type-1 mono-infection and co-infection with hepatitis C virus. Virol J (2012) 9:40. doi:10.1186/1743-422X-9-40
170. Sanabani SS, Nukui Y, Pereira J, da Costa AC, de Oliveira AC, Pessoa R, et al. Lack of evidence to support the association of a single IL28B genotype SNP rs12979860 with the HTLV-1 clinical outcomes and proviral load. BMC Infect Dis (2012) 12:374. doi:10.1186/1471-2334-12-374
171. Vallinoto AC, Santana BB, Sa KS, Ferreira TC, Sousa RC, Azevedo VN, et al. HTLV-1-associated myelopathy/tropical spastic paraparesis is not associated with SNP rs12979860 of the IL-28B gene. Mediators Inflamm (2015) 2015:804167. doi:10.1155/2015/804167
172. de Sa KS, Santana BB, de Souza Ferreira TC, Sousa RC, Caldas CA, Azevedo VN, et al. IL28B gene polymorphisms and Th1/Th2 cytokine levels might be associated with HTLV-associated arthropathy. Cytokine (2016) 77:79–87. doi:10.1016/j.cyto.2015.11.004
173. Planet PJ, Parker D, Cohen TS, Smith H, Leon JD, Ryan C, et al. Lambda interferon restructures the nasal microbiome and increases susceptibility to Staphylococcus aureus superinfection. MBio (2016) 7:e1939–1915. doi:10.1128/mBio.01939-15
174. Dorhoi A, Yeremeev V, Nouailles G, Weiner J III, Jorg S, Heinemann E, et al. Type I IFN signaling triggers immunopathology in tuberculosis-susceptible mice by modulating lung phagocyte dynamics. Eur J Immunol (2014) 44:2380–93. doi:10.1002/eji.201344219
175. de Oliveira S, Rosowski EE, Huttenlocher A. Neutrophil migration in infection and wound repair: going forward in reverse. Nat Rev Immunol (2016) 16:378–91. doi:10.1038/nri.2016.49
176. Soehnlein O, Lindbom L. Phagocyte partnership during the onset and resolution of inflammation. Nat Rev Immunol (2010) 10:427–39. doi:10.1038/nri2779
177. da Silva J, Hilzendeger C, Moermans C, Schleich F, Henket M, Kebadze T, et al. Raised interferon-beta, type 3 interferon and interferon-stimulated genes – evidence of innate immune activation in neutrophilic asthma. Clin Exp Allergy (2016):1–11. doi:10.1111/cea.12809
178. Xiao L, Gao LB, Wei Q. Association of polymorphism within the interleukin-28 receptor alpha gene, but not in interleukin-28B, with lower urinary tract symptoms (LUTS) in Chinese. Genet Mol Res (2015) 14:10682–91. doi:10.4238/2015.September.9.8
179. Montes de Oca M, Kumar R, Rivera FL, Amante FH, Sheel M, Faleiro RJ, et al. Type I interferons regulate immune responses in humans with blood-stage Plasmodium falciparum infection. Cell Rep (2016) 17:399–412. doi:10.1016/j.celrep.2016.09.015
180. Liehl P, Meireles P, Albuquerque IS, Pinkevych M, Baptista F, Mota MM, et al. Innate immunity induced by Plasmodium liver infection inhibits malaria reinfections. Infect Immun (2015) 83:1172–80. doi:10.1128/IAI.02796-14
181. Miller JL, Sack BK, Baldwin M, Vaughan AM, Kappe SH. Interferon-mediated innate immune responses against malaria parasite liver stages. Cell Rep (2014) 7:436–47. doi:10.1016/j.celrep.2014.03.018
Keywords: interferon lambda, immunity, immune cells, virus, infectious diseases, bacteria, fungi, parasites
Citation: Syedbasha M and Egli A (2017) Interferon Lambda: Modulating Immunity in Infectious Diseases. Front. Immunol. 8:119. doi: 10.3389/fimmu.2017.00119
Received: 12 November 2016; Accepted: 25 January 2017;
Published: 28 February 2017
Edited by:
Claudia U. Duerr, McGill University, CanadaReviewed by:
TImothy Nice, Oregon Health & Science University, USACopyright: © 2017 Syedbasha and Egli. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Adrian Egli, YWRyaWFuLmVnbGlAdXNiLmNo
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.