- 1Pathogen Biology Laboratory, Department of Biotechnology and Bioinformatics, University of Hyderabad, Hyderabad, India
- 2Laboratory Sciences and Services Division, International Centre for Diarrhoeal Disease Research Bangladesh (icddr,b), Dhaka, Bangladesh
Tuberculosis (TB) caused by the intracellular pathogen, Mycobacterium tuberculosis (Mtb), claims more than 1.5 million lives worldwide annually. Despite promulgation of multipronged strategies to prevent and control TB, there is no significant downfall occurring in the number of new cases, and adding to this is the relapse of the disease due to the emergence of antibiotic resistance and the ability of Mtb to remain dormant after primary infection. The pathology of Mtb is complex and largely attributed to immune-evading strategies that this pathogen adopts to establish primary infection, its persistence in the host, and reactivation of pathogenicity under favorable conditions. In this review, we present various biochemical, immunological, and genetic strategies unleashed by Mtb inside the host for its survival. The bacterium enables itself to establish a niche by evading immune recognition via resorting to masking, establishment of dormancy by manipulating immune receptor responses, altering innate immune cell fate, enhancing granuloma formation, and developing antibiotic tolerance. Besides these, the regulatory entities, such as DosR and its regulon, encompassing various putative effector proteins play a vital role in maintaining the dormant nature of this pathogen. Further, reactivation of Mtb allows relapse of the disease and is favored by the genes of the Rtf family and the conditions that suppress the immune system of the host. Identification of target genes and characterizing the function of their respective antigens involved in primary infection, dormancy, and reactivation would likely provide vital clues to design novel drugs and/or vaccines for the control of dormant TB.
Introduction
Tuberculosis (TB), a chronic infectious disease caused by Mycobacterium tuberculosis (Mtb), is one of the major drivers of human mortality worldwide since many decades with an estimated global burden of 10.4 million new TB cases and 1.4 million TB deaths in the year 2015 (1). Due to the growing efficiency of case finding and in the aftermath of DOTS regimen, the mortality rate decreased worldwide by 22% during 2000–2015 (1). There has been a decrease in the prevalence of TB cases dramatically from 4 million to 2.8 million cases in the last decade with a decrease in TB mortality form 330,000 cases to 220,000 cases, annually, in India (2). However, the burden of disease in the form of active TB still persists at an alarming rate in low and middle income countries with an estimated 580,000 new cases due to multiple drug resistant TB (MDR-TB), globally (1). TB infection is caused by the inhalation of aerosolized particles harboring Mtb. Various factors such as host’s immune status, inhaled bacillary load, the closeness of contact, and infectiousness of the source case play a primary role in TB transmission (3). The ability of inhaled Mtb aerosolized particles to infect the phagocytic immune cells [dendritic cells (DCs) and macrophages] and the non-phagocytic alveolar endothelial cells such as M cells and type 1 and type 2 epithelial cells (pneumocytes) (4) allows Mtb to replicate within the macrophages and spread to pulmonary lymph nodes and to several extra pulmonary sites before the adaptive immunity sets in (5). Hence, multiple possibilities exist where there could be (a) bacterial clearance by host immune activation, (b) multiplication of bacteria leading to primary infection, (c) dormant survival of bacteria rendering the host non-contagious and asymptomatic, and (d) reactivation of bacteria by infringement of dormancy causing re-emergence of the infection (6). Dormancy of the Mtb in the host is largely attributed to its sophisticated immune-evading capability that allows it to persist indefinitely. The key strategies adopted by Mtb to maintain its dormant phase include manifestation of immune [manipulation of toll-like receptor (TLR), cytokine, and immune cell function], biochemical (development of resistance to reactive intermediates and antibiotics), and genetic (activation of dormancy-associated genes) mechanisms. Despite vaccination with BCG (which is effective only in children) and the availability of powerful drugs to treat Mtb, there has been no decrease in the global burden. Epidemiological studies indicate that 90–95% of new Mtb infections could become dormant, and this dictates the immunological poise between the pathogen and the host (7). Besides persistent infections due to evolution of multidrug resistant and extensive drug resistant Mtb, a large reservoir of population hosting Mtb in the dormant stage represent the prime cause of new TB cases throughout the world (8). Hence, diagnosis and treatment of individuals hosting Mtb in a dormant stage is one of the crucial strategies to be adopted for the prevention of TB. Diagnostic methods such as tuberculin skin test (TST) and cell-mediated immune response-dependent approaches were developed based on the current understanding of the mechanisms that contribute to the establishment of persistent infection (9). The latest developments in understanding the cellular, biochemical, and molecular mechanisms that are employed for the establishment of dormant stage by Mtb are discussed in this review.
Immunology of Dormant Mtb
Evading Immune Detection
Masking: An Immunological Disguise
Mycobacteria adopt multiple strategies to avoid the attack from macrophages. They express surface lipids such as phthiocerol dimycoceroserate, which can mask the pathogen-associated molecular patterns (PAMPs), thereby going “unnoticed” by the innate immune system (10). The phenolic glycolipid produced by these bacilli induces the production of the chemokine CCL2 to recruit macrophages for further infection (10). In the upper airway where a constant and heavy recruitment of macrophages occurs due to the presence of TLR stimulating bacteria, thus posing a very hostile environment, Mtb adopts a different immune evasion strategy by forming small infection droplets that allow them to be delivered directly into the alveolar spaces of the lower lung, which anchorages a few microbicidal macrophages (11).
Manipulating the TLR Responses
In the macrophages, which are the crucial niche for replication, Mtb interacts with various receptors to initiate phagocytosis. Despite the bactericidal properties of the macrophages, Mtb employs phagocytosis as a primary mode of gaining entry to establish the niche. The opsonization of the bacillus by the complement or antibodies determines the nature of receptors engaged and also the nature of events that are involved in the outcome of the infection. Recognition of Mtb through its cell wall glycolipids involves the formation of TLR heterodimers (12). The importance of TLR-mediated signaling during Mtb infection is well proven in various TLR knockout animal models (13). Mycobacterial components such as lipomannan, lipoarabinomannan (LAM), 38- and 19-kDa mycobacterial glycoproteins, and phosphatidylinositol mannoside (PIM) induce the formation of TLR1/6 heterodimer (12). The 38- and 19-kDa mycobacterial glycoproteins, PIM, and triacylated lipoproteins favor the formation of TLR2/TLR1, whereas the diacylated lipoprotein induces TLR2/TLR6 dimerization (13). The susceptibility to Mtb infection is also due to genetic polymorphisms in the host genes (14). It is well established that Mtb has the ability to modulate the immune responses to its advantage. Exposure of THP-1 cells to Mtb cell wall components results in the de novo synthesis of TLR4, thereby decreasing the production of Th1 cytokines (15). Induction of apoptosis in bystander cells during Mtb infection of macrophages is a classic example of how this pathogen causes immunosuppression in infected individuals, thereby gaining the survival advantage (16). Interaction of Mtb cell wall components with TLRs modulates a number of events that include antigen presentation (17), phagolysosomal fusion (13), apoptosis of macrophages (12), and production of reactive oxygen and nitrogen intermediates (18).
Although Myd88-dependent signaling of TLRs is well established in mycobacterial pathogenesis, recent studies indicate independent roles for Mal (the TLR adaptor) and Myd88. Individuals with the single-nucleotide polymorphisms, D96N and S180L, in the TIRAP gene (that codes for Mal) display differential susceptibility to Mtb. Heterozygous genotype was associated with a protection toward TB, whereas the homozygous genotype was related to susceptibility (19). Using a murine model that carried the human equivalent mutation in TIRAP gene, it was demonstrated that in the homozygous genotype for the mutation, the mycobacterial load was higher and this was independent of the macrophage cytokine production (20). Further, in vitro studies indicated that mutation in TIRAP gene affected phagosome maturation and intracellular killing of Mtb (20).
Antigen Presentation by MHC
The TLR2-dependent surface expression of MHC class II receptor and their antigen-presenting ability was found to be inhibited by either Mtb infected or 19-kDa lipoprotein (LpqH) exposed macrophages (12, 21). Class II transactivator (CIITA), a TLR-2-dependent regulator of MHC class II a, b, invariant chains contributes to antigen processing and its expression was found to be decreased during Mtb infection (22–24). Its importance is further strengthened with the observation that CIITA knockout mice could not survive Mtb infection (25). Mtb also inhibits the expression of genes involved in MHC class II processing and presentation (26, 27) and the posttranslational function of these molecules. Another interesting feature by which Mtb evades TLR-mediated immune response is by the differential antigen-presenting ability of MHC class II. It is demonstrated in Mtb-infected lung DCs of mice that despite normal levels of MHC class II molecules, antigen presentation capability was decreased, whereas in macrophages, both the MHC class II molecules expression and antigen presentation capacity were found to be decreased (28–31).
Phagolysosomal Fusion/Fission
During chronic infections, repeated stimulation of TLRs by the Mtb components such as mannosylated-LAM (ManLAM) and PIM causes phagosomal maturation and arrest allowing persistence of mycobacteria inside the phagosome (32) (Figure 1). Among the successful strategies adopted by Mtb to establish a niche in the host, inhibition of macrophage maturation is best characterized. The mycobacterial products (ManLAM, trehalose dimycolate, and sulfolipids), phosphatase SapM, kinase PknG, and early secretory antigenic target-6 (ESAT-6) have been implicated in the inhibition of macrophage maturation (33, 34). ManLAM inhibits Ca+ surge that modulates the calmodulin- and Ca2+/calmodulin-dependent kinase II-dependent delivery of early endosomal autoantigen1 (35–37), which in turn is necessary for the delivery of lysosomal hydrolases and vacuolar H+-ATPases into phagosomes. ManLAM blocks ESAT-6 recruitment by inhibiting PI3K hVPS34 to block PIP3 production as well as SapM-mediated dephosphorylation of phosphatidylinositol 3-phosphate (37–39). Further, phosphorylation of unknown substrates by the kinase PknG and activation of p38MAPK by LAM to reduce the levels of Rab5 are some other mechanisms by which Mtb inhibits macrophage maturation, though the exact role of ManLAM in these two mechanisms is not clear (40–42). Further, Mtb also disrupts the scaffolding of endosomes required for phagosome–endosome interactions leading to delay in phagosomal maturation (43, 44). Because of the abovementioned mechanisms, the phagosomal compartment formed in the macrophage is devoid of acidification and lysosomal enzymes, thereby allowing Mtb to successfully establish a niche for its growth and replication.
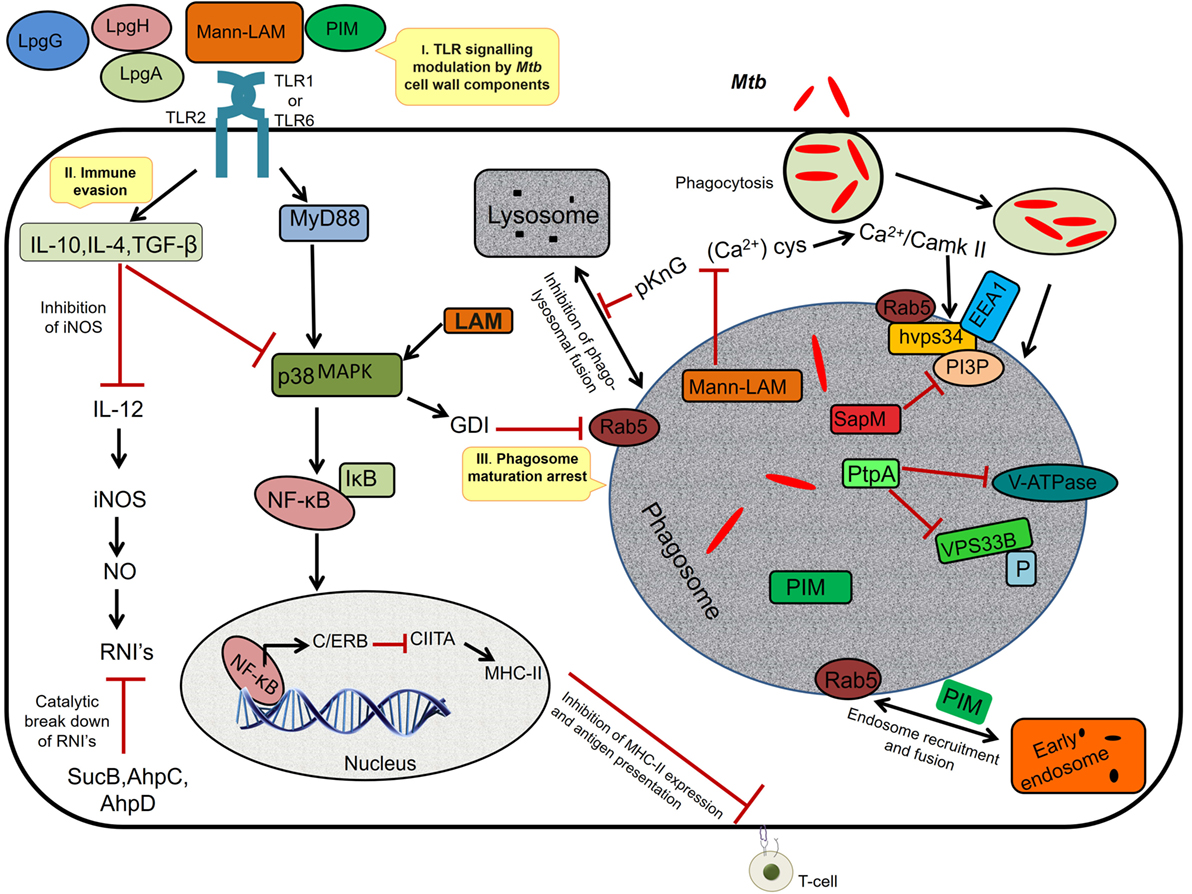
Figure 1. I. TLR signaling modulation. The Mycobacterium tuberculosis (Mtb) cell wall components interact with TLR-2 and modulate the host cell signaling via p38 MAPK, resulting in activation of NF-κB and synthesis of C/ERB that binds to class II transactivator (CIITA) promoter and inhibits CIITA production leading to decreased expression of MHC-II, thus inhibiting antigen presentation. II. Immune evasion: prolonged signaling by cell wall components induces the anti-inflammatory cytokines, TGF-β, interleukin (IL)-10, and IL-4 (Th2-dependent manner), which inhibits IL-12. IL-12 is required for the production of interferon-γ, iNOS, and NO, a major defense of the host against Mtb. The intracellular pathogen secretes SucB, AhpC, and AhpD, which catalyzes the breakdown of reactive nitric intermediates (RNIs). III. Phagosome maturation inhibition: mannosylated-LAM (ManLAM) activates GDI via p38MAPK leading to the inhibition of Rab5 activity that is required for the recruitment of early endosomal autoantigen1 (EEA1); it also inhibits the increase in cytosolic Ca2+ flux required for the hvps34 activity. pknG prevents phagolysosomal fusion and sapM, ptpA inactivates the phosphatidylinositol 3-phosphate (PI3P) and VPS33B through dephosphorylation. Phosphatidylinositol mannoside (PIM) mediates the early endosomal fusion through which bacilli gains access to nutrients such as iron required for its survival.
Apoptosis of Immune Cells and Redefining the Immune Cell Fate: A Process of Exploitation
Mycobacterial infection leads to various cellular fates such as apoptosis, necroptosis (type of programed necrosis), and autophagy (45–47). In the macrophages, apoptosis and autophagy are the natural defense mechanisms that operate to eliminate microbial infection and invasion. The spread of these bacilli is lowered (46) by tumor necrosis factor (TNF)-α activated caspase 8-mediated extrinsic cell death pathway that involves kinases p38, ASK1, and c-Abl (48). Additionally, autophagy also promotes the clearance of Mtb (47, 49), which is supported by the observation that the survival of Mtb depends on the expression pattern of factors predominantly involved in autophagy (50). However, mycobacterial species have evolved mechanisms that prevent apoptosis and the autophagy of immune cells, so that they can survive in these cells and remain dormant for longer durations (51). Interestingly, the extent of alveolar macrophage apoptosis and Mtb virulence are inversely correlated. Mtb H37Rv, a virulent strain, inhibits apoptosis by enhancing the release of membrane-bound TNFR2 receptors (52, 53) and also by upregulating the expression of Mcl-1 protein, a member of antiapoptotic B-cell lymphoma/leukemia 2 family (54). The products encoded by certain Mtb genes also influence the apoptosis of infected macrophages. Overexpression of the Mtb type I NADH dehydrogenase (nuoG) neutralizes NOX2-derived reactive oxygen species (ROS) (55, 56) and thereby inhibits apoptosis in macrophages. Such a role was also observed in PknE and SecA2 genes using knockout strategies (57, 58).
Apoptosis is induced in host cells in response to pathogen infection, which determines the initiation of infection, survival, and escape from the host. An interesting feature during this process is the induction of apoptosis in uninfected cells due to the non-specific activation of cytokines (59). Thus, the induction of apoptosis of T cells (bystander cells) by Mtb is beneficial for its survival (16). In T cells stimulated with non-specific phytohemagglutinin (PHA) or specific culture filtrate protein of Mtb, apoptosis was evident in non-specifically stimulated T-cells that were dependent on Fas–Fas ligand interactions. Further, a significant release of TNF-α indicated its association with specific T-cell apoptosis during Mtb infection of macrophages (16).
Mtb exerts altogether a different kind of action on T-cells (CD4+ and CD8+) to delay T-cell responses by inhibiting apoptosis. Evidence for the ability of Mtb to inhibit apoptosis is indicated by (a) the promotion of development of CD8+ T cell responses by the Sec2A-deficient mutant strain of Mtb; (b) manipulation of eicosanoid metabolism of T cells (60); (c) increased frequency of macrophage apoptosis, accelerated CD4+ and CD8+ T-cell responses, and enhanced control of bacterial burden in Alox5 (5-lipoxygenase, required for generation of LXA4) deficient mice infected with virulent Mtb (61); and (d) enhanced susceptibility to Mtb infection due to polymorphisms in Alox5 and lta4h (62, 63). Another important mechanism by which Mtb enhances its survival in the host is to delay the expansion of Foxp3+ regulatory T (Treg) cells and thereby delay adaptive immunity (64). Although inflammation and cytokines produced during Mtb infection play a role in Treg cell proliferation, Treg cells that recognize Mtb antigens expand preferentially. It is reported that mice infected with wild-type Mtb displayed higher Treg cell proliferation than those infected with a virulent strain of Mtb deficient in expression of the specific antigen, Ag85B (64). Further, increased bacterial load associated with delay in priming of effector T cells was observed in mice receiving Mtb-specific Treg cells and such an effect was not observed in mice that received Mtb-specific Foxp3− CD4+ T cells (64). These observations clearly indicate that Mtb expresses certain antigens and allows Treg cells specific to these antigens to proliferate rapidly and limit the rate of effector T-cell priming and expansion at this site.
Migration of DCs from the lungs to the lymph node precedes CD4+ T-cell responses during Mtb infection, and this was demonstrated in interleukin (IL)-12p40-deficient mice (65). Further, the restoration of the ability of DCs and activation of CD4+ T cells upon treatment with IL-12p40 (65) and presence of the same subset of DCs in the lymph nodes that were primed in the lungs (30) provides further evidence for the acquisition of bacilli and trafficking by DCs as a rate limiting step in the initiation of adaptive immunity. An inflammatory stimulus that resulted in migration of DCs from the lung to the lymph node failed to accelerate the delivery of Mtb from lung to lymph nodes, suggesting that DCs infected with Mtb are intrinsically impaired to migrate (65).
Manipulation of Host Cytokine Responses
Mtb infection manipulates host cytokine responses in different directions to create a balance and take advantage for its survival. Prolonged signaling of TLR2 by Mtb cell wall components results in increased production of IL-10, IL-4, and TGF-β, which then inhibits the IFN-γ-mediated activation of macrophages (27, 66–69), thereby the immune surveillance of T cells is evaded. Recognition of mycobacterial PAMPs by macrophages stimulates the production of cytokines like type I interferons (IFNs) and TNF (70, 71) through the TLR2 pathway, and these two cytokines promote apoptosis and necroptosis. Host cytokine responses are manipulated by various molecules during Mtb infection (72). It is demonstrated that IFN-γ and ESAT-6 inhibits TNF-α, IL-17 production, and early expression markers on T cells (73). Tim-3 inhibits the expansion of Th1 cells to prevent production of excess pro-inflammatory cytokines (74). Further, programed death-1 (PD-1) inhibits CTL function during Mtb infection (75). Mtb evolved mechanisms to control the expression of ESTAT-6, Tim-3, and PD-1 to control the manipulation of cytokine responses.
Increased production of TNF-α by macrophages induces generation of mitochondrial ROS, which confers the antimicrobial properties and necroptosis of these cells during Mtb infection (76–78). TNF-mediated ROS generation is brought through receptor-interacting protein 1 (RIP1), receptor-interacting protein 3 (RIP3), phosphoglycerate mutase family member 5, mixed lineage kinase domain-like protein (MLKL), and dynamin-related protein-1-dependent pathways (77). In brief, binding of TNF-α to its receptor results in the formation a membrane-proximal super-molecular structure complex 1 [containing TNF receptor-associated death domain (TRADD) that binds to RIP1, TNF receptor-associated factor 2/5 (TRAF2/5), and cellular inhibitor of apoptosis 1/2 (cIAP1/2)] followed by polyubiquitination of RIP1 or TRAF2 by cIAPs, which then allows NF-κB translocation into nucleus to initiate transcription of A20 and cylindromatosis (CYLD) (79, 80). Deubiquitination of RIP1 by A20 and CYLD (79) results in complex I getting converted to complex II [containing RIP1, Fas-associated protein with death domain (FADD), caspase-8, and TRADD] (80). Apoptosis is initiated by the activated caspase-8 of this complex and in situations wherein this activity is abrogated, RIP1 and RIP3 come together in complex III also with FADD, caspase-8, and TRADD to form a necrosome, in which RIP1 phosphorylates RIP3 and further engages MLKL, leading to necroptosis (80). Mtb targets the caspase-8 activity and leading thereby the macrophages to undergo necroptosis instead of apoptosis, since the former process is favorable to its survival (81). Inducing a very high level of TNF-α and promoting the secretion of a biological factor that can block caspase-8 activity have been proposed to be reasons for the ability of virulent Mtb to favor occurrence of necroptosis (82).
Other cytokines that are implicated in Mtb pathophysiology are IL-10 and IL-4/IL-13. IL-10, also referred to as “cytokine synthesis inhibitory factor,” produced by Th2 cells regulates macrophage and DC function in response to Mtb infection. The production of IL-10 during Mtb infection is more of an advantage to the pathogen than the host. IL-10 facilitates Mtb survival by inhibiting phagosome maturation thorough a STAT3-dependent and p38-independent mechanism (83), IFN-γ-mediated production of reactive oxygen and nitrogen intermediates (84), blocking antigen presentation by downregulating the expression of major histocompatibility complex molecules (85), DC migration (86), and recruitment of Th1 cells to the lungs by modulating CXCL10 production (87). In humans, IL-10 is responsible for limiting immune responses during Mtb infection (88). Further, an association between IL-10 gene polymorphism and susceptibility to TB was demonstrated (89). On the other hand, the Mtb strains HN878 and CH subvert the immune response via induction of IL-10 (90).
Besides the classical activation of macrophages modulated by many cytokines during Mtb infection, IL-4/IL-13 facilitates alternative activation of these cells. Mtb exploits the alternatively activated macrophages to divert the microbial actions of classically activated macrophages. Alternative activation results in induction of Arg1 gene whose protein product competes with iNOS for the substrate l-arginine (91). This results in lower production of NO reactive intermediates. Upregulation of IL-4/IL-13 was observed in patients with progressive pulmonary TB and in PBMCs infected with HN878 Mtb strain (92–94). IL-4/IL-13 induces Arg1 in alternatively activated macrophages to subvert the host NO-based mycobactericidal activity and could be a tactic by Mtb to thrive inside classically activated macrophages. On the other hand, the enhanced production of IL-4/IL-13 due to alternate activation of macrophages inhibits autophagy to facilitate the survival of Mtb (95).
Resistance to Reactive Nitrogen Intermediates
Generation of reactive nitrogen intermediates (RNIs) by the macrophages through nitric oxide synthase 2-dependent pathway mediated by IFN-γ is an antimicrobial strategy displayed by these cells and this process has been shown to be vital for the control of TB (96). In the macrophages, the inducible form of NOS is activated by the cytokines produced by Th1 lymphocytes that stimulate the production of nitric oxide (97), which reacts with superoxide radicals to form RNIs. The RNIs thus produced, attack bacterial macromolecules to aid in killing. Although the role of RNIs in the control of TB in humans is not yet clear, some studies provide substantial evidence that RNIs play a role in innate immunity mounted during mycobacterial infection. The susceptibility to TB was found to be associated with genetic alterations in the NOS2A gene (98). Further, the negative correlation of mycobacterial growth and NO production in human alveolar macrophages, elevated expression of NOS2 in the lungs of TB patients, and reactivation of dormant TB due NOS2 inhibition support the role of the RNIs mycobacterial pathogenesis (99). However, Mtb has developed mechanisms that can subvert the antimicrobial actions of the macrophages, which allows these bacilli to establish a niche and remain in the host for a long time. ManLAM was also described to trigger Th2 cytokines such as IL-4 and IL-10 that inhibit the action of inducible NO synthase, an enzyme critical for the production of NO (9).
The alkyl hydroperoxide reductase subunit C encoded by the mycobacterial gene AhpC, in association with peroxidase, peroxinitrite reductase, dihydrolipoamide dehydrogenase (Lpd), dihydrolipoamide succinyltransferase (SucB), and thioredoxin-like AhpD catalyzes the breakdown of RNIs to protect Mtb from the antimicrobial actions of the macrophages (100, 101). Further, the mycobaterial gene MsrA encodes methionine sulfoxide reductase, an enzyme that converts methionine sulfoxide [produced out of a reaction between peroxynitrite (ONOO−) and methionine residues of proteins] to methionine, protects bacteria against RNIs (102). Rv1205, a pupylated proteasome substrate, catalyzes the production of cytokinins and helps Mtb to defend against NO (103). To summarize, TLR agonists present in the thick cell wall of Mtb causes prolonged TLR signaling resulting in various immune evasion mechanisms. Among these mechanisms, inhibition of MHC class II appears to be predominant that allows preventing detection by CD4+ T cells.
Antibiotic Tolerance: An Acquired Fitness Advantage
Besides the fixed genetic mutations that the Mtb undergo to develop antibiotic tolerance, another interesting feature adopted by Mtb to remain dormant in the host is the development of “phenotypic drug resistance” or simply called “drug resistance,” during which a transient resistance to antibiotics is developed (104, 105). This is achieved by induction of specific macrophage-induced efflux pumps (106). Adding to this, the same pumps promote intracellular bacterial growth, thereby providing a double advantage to the Mtb.
The role of toxin–antitoxin (TA) systems in conferring the “non-classical” antibiotic resistance to allow Mtb to remain in a non-replicating phase for longer durations is very interesting. This system encoded by two genes is composed of two proteins, namely, the long-lived protein “toxin” and the short-lived protein “antitoxin.” In Mtb, under normal physiological conditions, the toxin is neutralized by the antitoxin. To remain in dormant stage, Mtb represses the expression of the antitoxin protein resulting in the accumulation of toxin. Under such conditions, the toxin protein acts as a ribonuclease to cleave free and ribosomal bound single-stranded mRNA resulting in inhibition of protein synthesis and bacterial growth. This allows the Mtb to persist in the host without any signs of infection state for a long time. Multiple TA systems, approximately 88, exist in the bacilli and their numbers are ever increasing (107–109), with H37Rv strain found to harbor 38 modules of 5 TA systems (3 relBE, 24 vapBC, 8 mazEF, 1 higBA, and 2 parDE) (108). Over expression of these genes belonging to different modules in Mycobacterium smegmatis stopped the growth of the recipient cells to remain in latent phase. In general, the mycobacterial TA proteins are homologous to the proteins of other bacterial species and some of them exhibit certain special features. The MazF protein besides cleaving mRNA can also interact with DNA topoisomerase (110), thus indicating that differential mechanisms may exist in the way the TA proteins act to contribute to persistent infection in the host.
Granuloma Formation: A Finely Negotiated Refuge?
Granulomas are formed in response to infectious and non-infectious stimuli and are associated with various diseases (111). They are aggregates of macrophages whose membranes are interlocked and their occurrence is more prevalent in TB worldwide. Although it was initially reported that granulomas are complex protective structures that contain host cells to wall off bacteria (104) and can also sterilize infection (112), many studies have indicated that they are conducive to heavy bacterial burdens in TB (113). In case of active disease, some of the lesions are cleansed by the host even though there is progression of other lesions indicating that lesional heterogeneity persists after the initiation of adaptive immunity where differential killing of the bacteria take place, deciding the outcome of the clinical infection (112). Mtb in fact enhances the formation of granulomas (114) for their expansion and dissemination. This is accomplished by spreading of Mtb from dying macrophages to newly recruited ones. Macrophages undergo apoptosis when the bacterial load reaches a threshold and the nascent granulomas are presented to the uninfected macrophages. Thus, the Mtb numbers are phenomenally expanded by way of macrophage death and re-phagocytosis (114).
In the granuloma lesions, either the infection is cleared or viability is promoted by creating a favorable niche to the bacterium (Figure 2) (115). The immune status of the human host and transcriptional signature of the bacteria directs early granuloma establishment and consequence of the disease (116). However, it is shown that the immunological responses to Mtb by infected individuals vary to a great extent and is dependent on the granulomatous lesion formed. Two kinds of granulomatous lesions were demonstrated in cynomolgus macaques that were infected with low doses of Mtb (117). The classical ones are the caseous granulomas with low oxygen availability that is characterized by fibroblasts surrounding the epithelial macrophages and neutrophils in the periphery and dead macrophages in the center (118), whereas fibrotic lesions are associated with latent TB and exclusively filled with fibroblasts with sparsely distributed macrophages (119). In the event of an immunocompromised condition, granulomas liquefy and the bacteria are released from degraded granuloma to re-infect lung tissue and spread to new hosts (120).
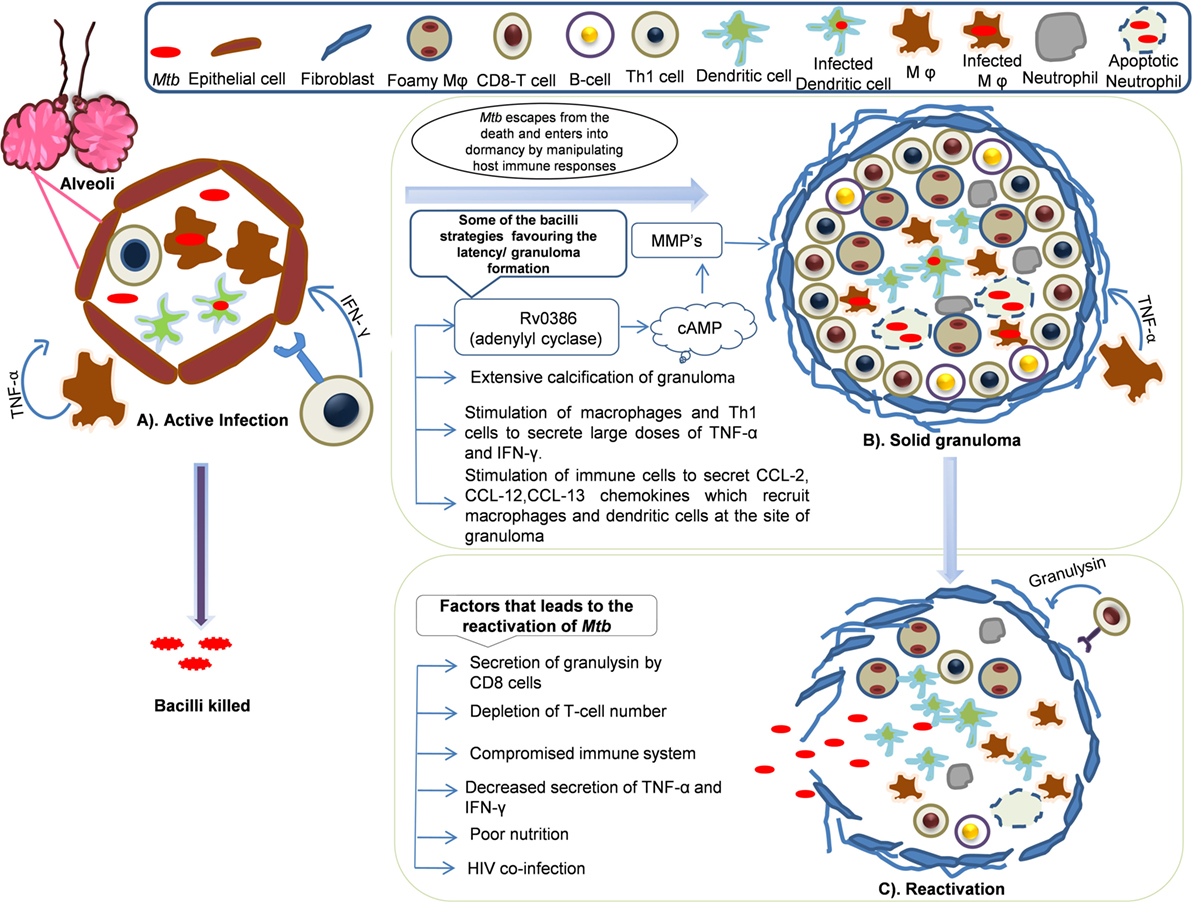
Figure 2. Dynamics of granuloma formation, maintenance, and reactivation: Mycobacterium tuberculosis primarily harbors lungs and infects alveolar macrophages and establishes its niche. The host defense sets in to counteract the actions Mtb. In this process, multiple possibilities exist where there could be (A) active infection/clearance: macrophages (Mφ) and Th1 cells secrete tumor necrosis factor (TNF)-α, IFN-γ that recruits other immune cells like neutrophils, dendritic cells (DCs), and B-cells that might clear the infection or bacilli may multiply leading to primary infection. However, some of the bacilli might escape the host’s immune actions and enter into dormancy. (B) Solid granuloma: solid granuloma is composed of macrophages, lymphocytes (B-cells and T-cells), DCs, and neutrophils. The solid granuloma is usually encircled by fibroblasts. During latent infection, Mtb encourages the immune system to form granuloma by manipulating host immune responses for its survival. Some of the Mtb survival strategies include stimulation of macrophages and T-cells to secrete large doses of TNF-α, chemokines such as CCL-2, CCL-12, and CCL-13, which are crutial for the recruitment of other immune cells and maintenance of granuloma. Extensive calcification of granuloma by Mtb leads to prevention of apoptosis. Secretion of Mtb antigens such as Rv0386 (adenylyl cyclase) produce cAMP which signal the synthesis of matrix metalloproteinases (MMPs) that are involved in the maintenance of granuloma by unknown mechanism. (C) Reactivation: Mtb reactivates and exits from the granuloma when immune system compromises and bacilli spread to new sites of infection. Poor nutrition, decrease in the number of T-cells, HIV coinfection are some of the contributing factor for reactivation.
Although granuloma formation is a basic immune response elicited by the host against an infection, it is also promoted by Mtb as part of its virulence program (121, 122). TNF-α plays an important role in mounting a response that is cytotoxic to the pathogen as well as maintaining the structural integrity of the granuloma and this is evident by the fact that neutralization of TNF-α leads to fatal reactivation of the bacterium and increased burden of the bacilli in the lung tissues (123, 124). It is demonstrated that the lipid-rich mycobacterial cell wall composed of trehalose dimycolate induces TNF-α induction and thus the granulomatous inflammatory response (120). Eliciting an excessive TNF-α response is another mechanism of virulence of Mtb to favor its existence in the host without being detected. This is accomplished by increasing the levels of TNF-α in the macrophages by directly interfering with cAMP mediated responses. In the mutant strain of Mtb lacking Rv0386 (encodes an adenylate cyclase), reduced protein kinase A and cAMP response element-binding protein activation was observed, which results in a significant reduction of macrophage TNF-α secretion (125). Although excess production of TNF-α is required for the containment of Mtb, this pathogen, however, uses adenylate cyclase to deliver excess cAMP to macrophage cytoplasm and acts to subvert host cell signal transduction so as to result in a pro-granulomatous response with excess TNF-α secretion (125).
The mechanisms of granuloma formation are not well studied. However, evidence points to the role of matrix metalloproteinases (MMPs) and tissue inhibitors of metalloproteinases (TIMPs) in Mtb for induction of granuloma formation (126). The expression of MMPs is largely dependent on cAMP-mediated signaling in the host cell (127) and thus the levels of these enzymes can be modulated by bacterial mechanisms (for example, the Mtb Rv0386 pathway) that regulate cAMP levels. Secretion of MMP-1, -2, -7, and -9 and decreased expression of TIMP-1, -2, and -3 (126, 128) have been shown in the peripheral blood mononuclear cells and human airway epithelial cells. Governing granuloma formation is also exploited by this bacterium to increase its numbers and dissemination systemically. In the Mycobacterium marinum-zebrafish model, it was demonstrated that the infected macrophages attract new cells and they in turn ingest the bacterium and allow them to sustain, thus allowing the granuloma to expand (114). Thus, it is clear that Mtb pathogenesis involves subversion of host signaling in a pro-inflammatory manner to create an environment that is favorable for its dormancy.
Regulation of Granuloma Maintenance
Calcification
Neutrophils on the other hand also display antimycobacterial activity and the mechanism involves generation of ROS (129, 130). Further, infected neutrophils undergo apoptosis (131, 132), thus contributing to an effective adaptive immunity during Mtb infection (133). However, Mtb adopts strategies to inhibit apoptosis and promotes necrosis of these cells by various mechanisms by expressing the region of difference 1 (RD1) and a type VII secretion system (ESX) (134). Apoptotic neutrophils are also subjected to necrosis because of the activation of Calpain, a Ca2+-activated protease (135) due to Ca2+ influx, and this process is generally termed as Ca2+-induced necrosis. The Ca2+ influx and the subsequent necrosis of neutrophils are promoted by ESAT, a mycobacterial leukocidin (136). Further, the dormancy regulator of DosR activates the Ca2+ ATPase of the plasma membrane of the microvesicles of mycobacteria to create hypoxic conditions (137). Thus, the calcification of neutrophils is an important survival strategy adopted by Mtb at the cellular level. On the other hand, it was reported that feeding Mycobacterium paratuberculosis-infected rats with low calcium enhanced their ability to clear the infection (138). Besides utilizing Ca2+ for its survival at the cellular level, Mtb infection causes extensive calcification in various host tissues. Calcification is observed in myocardial (139), pulmonary (140), musculoskeletal, central nervous, abdominal (141) and genitourinary systems (142), and in lymphatic tissue and more importantly the granulomas. During the granuloma formation, the parenchymal tissues calcify to form caseous structures, especially during chronic or latent infection (143). Calcification allows the formation of various granuloma structures within a single host, thus creating different microenvironments that are unique among them to allow the survival of Mtb (143). The molecular mechanisms of granuloma calcification remain unclear. However, calcification of granulomas allows a safe environment for Mtb to remain dormant for extended periods of time.
Fibroblasts
Besides macrophages, granulomas also contain other cells such as eosinophils, neutrophils, and fibroblasts. In the mature granulomas, fibroblasts are found in the periphery and embedded within collagen fibers. TB inflammation is associated with increased number of fibroblasts and the fibroblastic activity (144). These cells produce collagen bundles such that the fibrotic capsule separates the bacteria from the surrounding tissue. This process is a consequence of intra-granulomatous cytokine secretion (145). On the other hand, fibroblasts also participate in the burning out of granuloma (in the event of the pathogen completely eliminated) by secreting tissue inhibitors of metalloproteases (145). The dynamics of granuloma formation depends on the number and degree of differentiation of fibroblasts (144). Interferon-γ primed fibroblasts present bacterial antigens to Th1 cells for further processing. However, it was demonstrated that Mtb-infected fibroblasts fail to present the antigens (146), thus enabling these bacilli to evade immune response of Th1 cells. Mtb also directly affects the collagen turnover and expression of matrix metalloproteases and tissue inhibitors of matrix metalloproteases in fibroblasts (147). Thus, the manipulation of fibroblast function is one of the key strategies of Mtb for the establishment and sustenance of persistent infection.
Genetics of Host and Pathogen in Mycobacterial Dormancy: A Perfect Storm?
The pathogenesis of Mtb, especially its ability to remain in dormant state and reactivation, depends on both the genetics of the host and the pathogen (148). This is evidenced by the fact that (a) dormant mycobacteria do not replicate at all and their cell divisions occur at a very low rate (149, 150) and (b) a high level of genomic stability was observed in these bacterium isolated from human populations. Hence, studies on the genetic variations of the Mtb and the host have invited a lot of interest in the past few years and the same are discussed in this section.
Host gene–environment interactions play a crucial role in determining the outcome of TB and these are very important to evolve strategies to prevent mycobacterial infection at the genomic level. Interestingly, the ability of the host genetic control depends on the mycobacterial strain encountered and the exposure intensity (151). Two major loci on chromosomal regions, 18q11.2 and 11p13, were found to be associated with incidence of TB (152, 153). TST reactivity and interferon-γ release assay responses during Mtb exposure were found to be hereditarily controlled (154, 155). Since heritability is related to latent tuberculosis infection (LTBI), the phenotype of certain genes seems to govern mycobacterial infection and the associated dormancy. For example, in a Ghanaian population, IL-10 promoter haplotype (−2849G/−1082G/−819C/−592C) in TST non-responders was significantly more compared to TST responders (15.3 vs. 9.7%, OR = 2.09, p = 0.01) (156). In individuals carrying GG genotype at 1082A>G, the prevalence of TST non-response was 1.5 times than those carrying the AA and AG genotypes (156). Further, SNPs in IL4 (−590T>C, p = 0.007) and IFN-γ (+874A>T, p = 0.02) genes are associated with TST response (156). A linkage of persistent TST negativity with chromosomal regions 2q21-2q24, 5p13-5q22 (157), 11p14 (also called TST1 locus), and 5p15 (also called TST2 locus) was identified (158). Thus, the genetics of the host determines the susceptibility to mycobacterial infection and the development of LTBI.
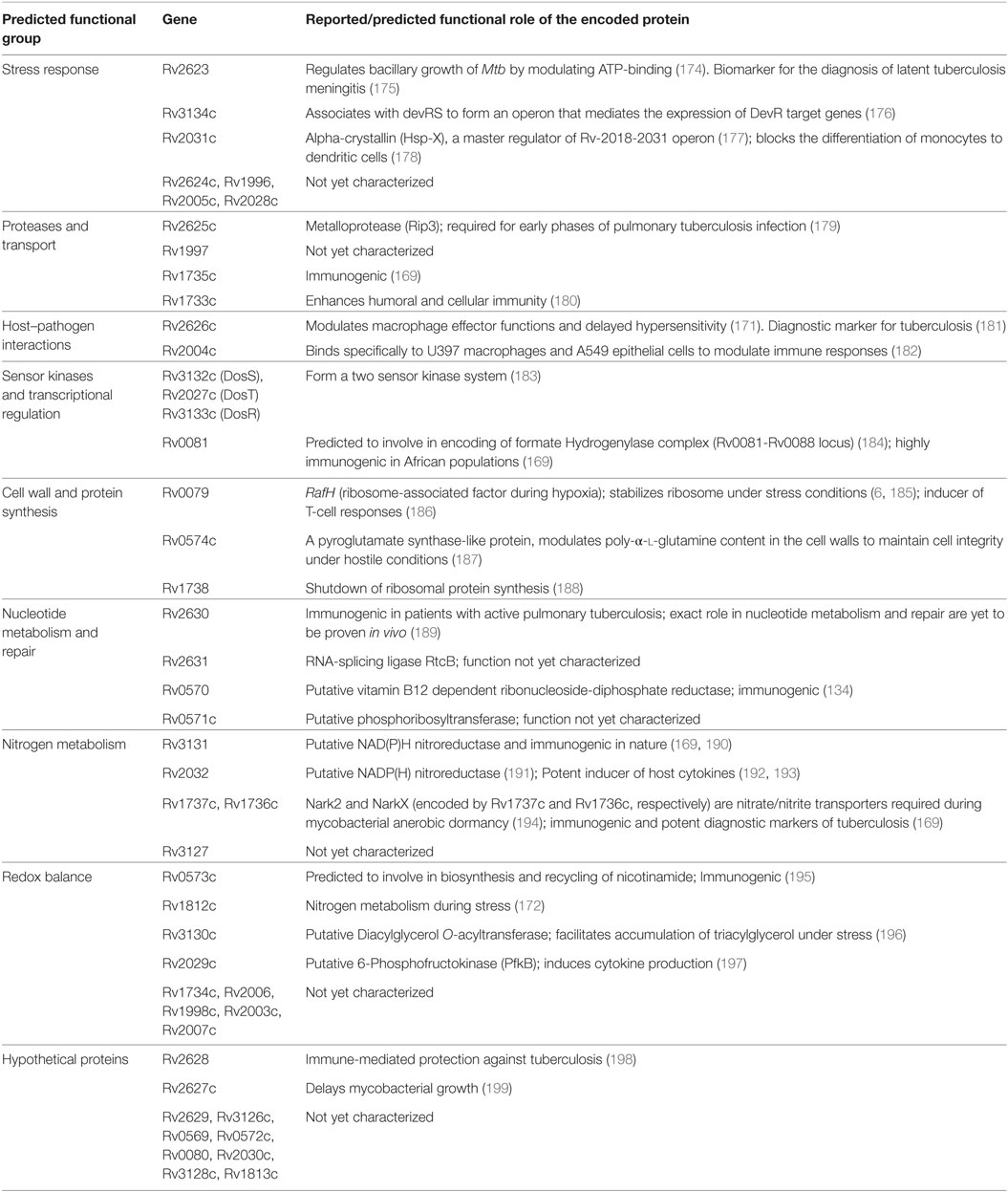
Dormancy in Mtb is regulated by a set of approximately 50 genes, the DosR regulon, under the tight control of the dormancy survival regulator transcription factor (159). The genes of the DosR regulon are distributed in nine blocks in the genome: (block 1) Rv0079–Rv0081, (block 2) Rv0569–Rv0574c, (block 3) Rv1733c–Rv1738, (block 4) Rv1812c–Rv1813c, (block 5) Rv1996–Rv1998c, (block 6) Rv2003c–Rv2007c, (block 7) Rv2028c–Rv2032, (block 8) Rv2623–Rv2631, and (block 9) Rv3126c–Rv3134c (160). Evolutionarily, these genes are conserved across various pathogenic, non-pathogenic, and environmental bacteria of diverse habitats (160), and this could have occurred due to horizontal gene transfer mechanism during adaptation to challenging environmental conditions (161). Functionally, they emerged primarily to assist Mtb to adapt for anaerobic environment, thus enabling its survival in the host granuloma (162). Their expression is induced under hypoxia (163, 164) and under conditions where mycobacterial growth is inhibited by external growth factors both in vitro (macrophages) (165) and in vivo (mice and guinea pigs) (166, 167), suggesting their role in maintaining a low profile of bacterial growth under unfavorable conditions. Interestingly, the protein products of many of these genes seem to be good T-cell antigens and involved in many physiological processes of both the bacilli and the host.
The protein products encoded by DosR genes are predicted/reported to be involved in various functions, and on this basis, they were classified into eight groups (160). The functional roles of these genes are outlined in Table 1. Although they were predicted to have diverse functions, immune regulation and modulation of host responses have gained importance in the recent years. Out of the DosR genes expressed during dormancy, 18 are T cell responders that trigger strong IFN-γ response in TB patients. Further, it was demonstrated that Rv1733c, Rv2029c, Rv2627c, and Rv2628 are strong IFN-γ responders in latently infected individuals (168). Further, Rv2032, Rv1998c, Rv2031c, Rv2623, and Rv3132c exhibit strong T-cell response, whereas Rv0079, Rv0080, Rv3127, Rv2626c, and Rv2029c exhibit strong humoral immune response (169, 170). It was identified that Rv2626c is a secretory protein, which binds to macrophage affecting its function and also elicit TNF-α and strong B-cell responses (171). In aerosol-mediated murine TB model, decreased induction of pro-inflammatory cytokines (IL-1 and IL-12) and decreased bacterial load and delayed death was observed when infected with Rv1813c deletion mutant of Mtb (172). It is to be noted that evidence for the role of DosR genes in Mtb dormancy stems out from murine models and no concrete evidence exists in higher animal models. In a recent study, using macaque as the model system, it was demonstrated that during hypoxic conditions DosR regulon modulates the timing and magnitude of adaptive immune responses to favor persistence of infection by Mtb (173).
It is very interesting to note that mutations in dosR does not induce Mtb death under hypoxic conditions, indicating that other factors beyond dosR are important for the dormancy and survival of Mtb in the host. Further, the massive expression of dosR-independent genes during hypoxic conditions and the variation in the expression profiles of dosR genes in different strains of mycobacteria of different virulence indicates that further studies are required to warrant the exclusive role of these genes in mycobacterial dormancy (200–202).
Environmental Signals of Sleep
The complex life cycle of Mtb involves adaptation to various stresses and to accomplish this it encodes about 190 regulatory proteins among which 11 form the two-component signal transduction systems (TCSSs) (134, 203). TCSSs found in Mtb are conserved in other closely related mycobacterial species in terms of genetic arrangement and location (134, 204). However, the number of functional TCSSs seems to vary between the species, wherein the Mycobacterium leprae had only four TCSSs (204). The TCSSs identified in Mtb are phoP-phoR, regX3-senX3, dosR-dosS (dosT), Rv0600c-Rv0601-ctcrA, narL-Rv0845, tcrX-tcrY, mprA-mprB, prrA-prrB, trcR-trcS, pdtaR-pdtaS, mtrA-mtrB, and kdpD-kdpE (205).
A typical TCSS comprises a histidine sensor kinase and a response regulator that are localized in the plasma membrane and cytoplasm, respectively. Both of them have specific domains through which they sense environmental cues. The sensor kinase comprises a sensor domain, one or more transmembrane domains, and a cytoplasmic transmitter containing a dimerization motif and a kinase domain, and the latter can be again divided into two subdomains possessing a histidine phosphorylation box and an ATP-binding pocket (made of N, D, F, and G boxes that have highly conserved amino acids); signal recognition results in dimerization and auto-phosphorylation followed by the transfer of this phosphate to the response regulator, thus enabling it to promote transcriptional, translational, and functional aspects (206–209). The environmental cues that activate the TCSSs are not yet defined for all of them. Inorganic phosphate (regX3-senX3); SDS, triton X-100 alkaline pH, and nutrient limitation (mprA-mprB); low oxygen, nitric oxide, carbon monoxide, and ascorbate (DosS-DosR) are some of the signals that are identified to activate the TCSSs in model organisms such as M. smegmatis. However, whether the same signals also activate these TCSSs in Mtb is not yet clear. The signaling mechanisms of some of the characterized TCSSs remain more or less similar as described above for a typical TCSS, with some variations. One of the downstream events of TCSS activation is gene regulation. SenX3-RegX3 activation leads to the upregulation of several genetic determinants such as phoA (alkaline phosphatase) (210), pstSCAB and phnDCE (encodes phosphate-specific transporter systems) (210, 211), and phnF (encodes a negative regulator of phnDCE) (211) and SenX3-RegX3 itself (210, 212). PhoP-PhoR regulates about 150 genes that are involved in general and lipid metabolism, and respiration (213, 214). Further, a number of genes that code for membrane proteins, genes of dosRS regulon, genes of the PE/PPE/PE-PGRS protein families, and genes of the virulence-associated RD1 such as espB and espR (214, 215). The narL-Rv0845 TCSS regulates genes involved in nitrate metabolism during anaerobic respiration (216). mprA-mprB differentially regulates about 200 genes (217, 218). Examples include expression of its own gene, pepD and moaB2 and Acr2 (alpha-crystallin-like protein) (219, 220). The trcR-trcS TCSS regulates about 50 genes (221). dosR-dosS (dosT), one of the well-characterized TCSS, regulates about 48 genes (collectively called the DosR regulon), predominantly having a role in hypoxia. MtrA-MtrB seems to be an important TCSS since it regulates genes involved in DNA replication and cell wall integrity (222, 223). The gene expression regulated by pdtaR binding to RNA involves prevention of stem-loop structure formation by acting as an anti-terminator (224). The huge variety of genes regulated by TCSSs gives an edge for Mtb to create favorable conditions for its survival in the host in a dormant condition for longer durations.
Evidence to strengthen the crucial role of TCSS is demonstrated in animal models of TB infected with mycobacterial strains that harbor mutation in the TCSS genes. Attenuation of bacterial growth in lung, liver, and spleen and delayed time to death of Mtb-infected animals are some of the phenotypes described in studies that used TCSS mutants [reviewed in Ref. (205)].
Reactivation
Reactivation and Liquefaction of the Granuloma
The reactivation of dormant Mtb is governed by a group of proteins belonging to the resuscitation-promoting factor (RPF) family and their genes were found to be upregulated during this process (149, 225, 226) (Figure 3). In general, these genes are upregulated when the stress is removed. The importance of these genes was demonstrated by the fact that Mtb in which Rpf genes were knocked down, were unable to undergo reactivation even after immune suppression of the host (227, 228). The five Rpf genes (A to E), though, not required for general viability, are crucial for the induction of reactivation of Mtb and the bacilli can survive multiple mutations across the underlying genes. The Rpf proteins conserve a domain that is structurally close to lytic transglycosylases and are thought to participate in cell wall hydrolysis, an essential early phase step in the reactivation or resuscitation process (229). The action of these proteins breaks the peptidoglycan strands of the highly impermeable cell wall of the granulomatous cells. During the reactivation process, increase in cAMP levels due to activation of adenylate cyclase by free fatty acids is evident and the RPF proteins do not seem to be involved in this process (230). However, gene expression analyses indicated that the RPF biosynthesis is active in the later phase of reactivation, suggesting that they may mediate the early and late events (230, 231). Thus, each of these genes is considered as a potential drug target that would allow Mtb to exit the dormant stage for further treatment by conventional drugs.
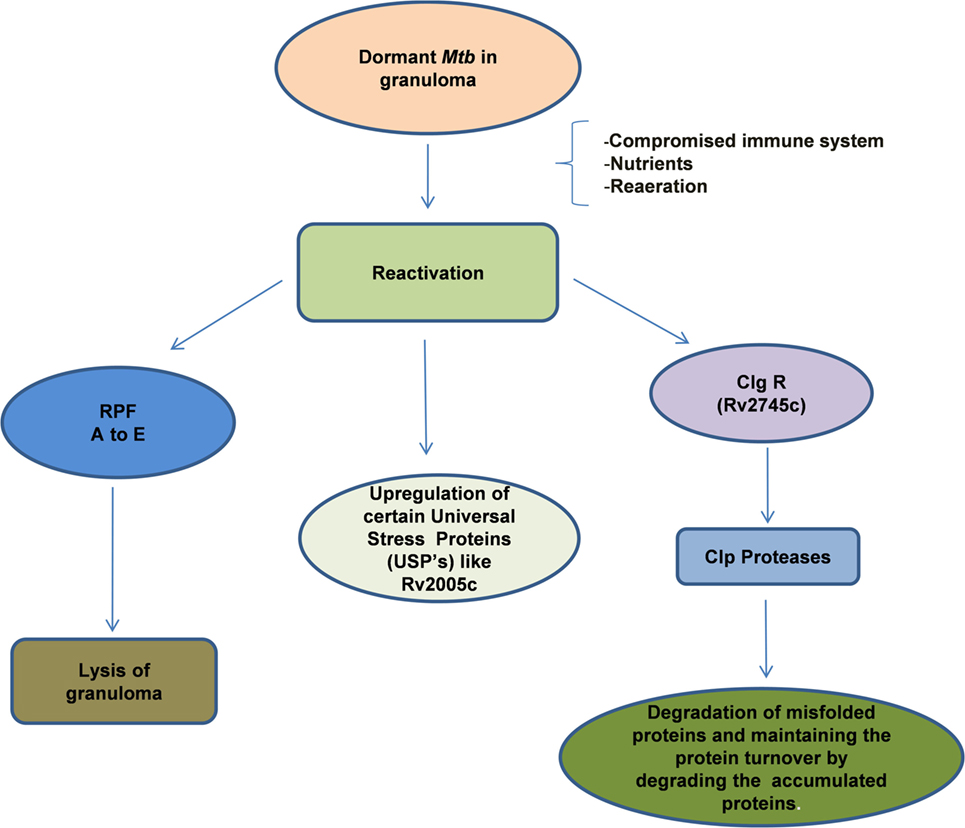
Figure 3. Reactivation of latent bacilli: Mycobacterium tuberculosis (Mtb) persisting under hostile conditions in granuloma reactivates when favorable conditions prevail such as availability of nutrients, re-aeration, and immune compromization of the host leading to activation of different set of genes that are required for the re-growth of the bacilli. During reactivation, Mtb mainly expresses Clp protease gene Regulator (ClpgR) and its inducible genes Clp Proteases. Clp proteases play a vital role in the maintenance of protein turnover, degradation of misfolded or accumulated proteins, helping the bacteria to shift to metabolic phase. On the other hand, Mtb expresses a family of five resuscitation-promoting factors (RPF A to E) that are essential for the lysis of granulomatous cell wall. Certain Universal stress proteins (like Rv2005c) are also upregulated during resuscitation which might play an essential role for the re-growth.
Besides the Rpf genes, the DosR regulon gene (Rv3133c) also contributes to the rapid resumption of Mtb growth especially when they transit from the non-respiring conditions to the respiring conditions (200). The dosR regulon also regulates the expression of non-coding short RNAs that are involved in both dormancy and reactivation (232). A recent study indicates that Clp protease gene regulator, Rv2745c (clgR) is required for in vitro reactivation from hypoxia-induced dormancy (233). In the isogenic mutant Mtb:ΔRv2745c, genes in the σ(H)/σ(E) regulon as well as the dosR regulon were dysregulated suggesting that the DosR genes are regulated at a different level to contribute to the reactivation process (233). It thus appears that Mtb reactivation involves biochemical, immunological, and genetic aspects all of which are potential drug targets for the development of treatment strategies to prevent dormancy and reactivation of the bacilli.
Exiting the Granuloma: In Search of a New Niche
Besides the biochemical aspects that favor apoptosis or necrosis, genetic reprograming also plays a vital role in the reactivation of Mtb from its inactive state (234). The prime necessity of any pathogen to enter a dormant phase is to establish its niche and re-infect when the conditions are favorable. Evolutionarily, the pathogens exit the host in which they have resided to infect a new host. In the case of Mtb, the bacilli exit from the granulomas that have undergone necrosis in to the bronchial tree and this seems to be most efficient method of transmission to the new host (235, 236). Reactivation generally occurs when the host immune machinery is weakened or suppressed due to physiological or pathological factors. For example, in the case of HIV+ individuals, increased risk of Mtb reactivation is possible due to the low levels of CD4+ T cells (237). Further, the immune responses during reactivation differ from that of primary infection with CD8+ T cells taking the lead than the CD4+ T cells (238). In a primate model exhibiting latent TB and HIV coinfection, it was observed that animals with low CD4+ T cells show higher incidence of TB suggesting that T-cell depletion is one of the major triggering factor for Mtb reactivation (239). Though these observations were confirmed in smaller animal models, controversy still exists in the primates and humans (240).
Further the immunological factors, cellular events also determine the efficiency of reactivation. The nature of cell death experienced by the granuloma cells defines the extent of infection (111). Apoptosis of the macrophages allows the bacilli to remain encased in the macrophages, which then are phagocytosed by new macrophages and thus allowing bacterial expansion or maintenance in the new granuloma cells. On the other hand, necrosis of macrophages in the granuloma releases the bacilli into the extracellular milieu allowing multiplication in high numbers and these new bacilli structurally appear as serpentine cords. It is very interesting to note that the serpentine cords are not readily recognized by macrophages, thus allowing them to successfully get transmitted to a new individual (241). What factors and conditions that allows a granuloma to undergo apoptosis or necrosis is still a matter of debate and is being actively investigated. Once in the airway mileu, the new bacterial cells are aerosolized in cough droplets.
Anti-Cytokine Inhibitors: Contribution to Reactivation
As discussed earlier, TNF is a potent inflammatory cytokine that controls the dynamics of pathogen survival and host immune response. Among the cytokines implicated in Mtb pathogenesis, TNF is a potent inflammatory cytokine that controls the dynamics of pathogen survival and host immune response. TNF confers anti-TB immunity to the host by manipulating the levels of other cytokines, adhesion molecules, and apoptosis of macrophages. In the clinical settings, TNF blockers such as infliximab, adalimumab, certolizumab pegol, and etanercept are routinely used for the treatment of various autoimmune disorders. An emerging concern is the association between the use of TNF inhibitors and increased risk of Mtb reactivation. It is observed that treatment with TNF inhibitors resulted in progression of Mtb from latent to reactivation (242, 243). Recent studies project a higher risk of TB in rheumatoid arthritis patients receiving TNF inhibitor treatment (244). Further, in various animal models, it is demonstrated that neutralization of TNF resulted in increased susceptibility to primary infection of Mtb (245, 246). TNF inhibitors interfere with innate and adaptive immune responses such as increased T-cell activity, complement-mediated lysis, apoptosis of immune cells, and phagosomal maturation (247). The changes that occur in the immune responses due to TNF inhibitors give an opportunity for Mtb to reactivate. A thorough multistep screening for Mtb is proposed for individuals who are subjected to anti-TNF therapy to treat autoimmune diseases (248).
Clinical Implications of Dormancy/Therapeutic Manipulations of Dormancy
The unique feature of Mtb is its ability to maintain a persistent infection without being detected under different conditions has a lot of impact on the clinical implications. Mtb, because of its ability to create a secure environment for itself is not susceptible to certain antibiotics and also resistant to strong antibiotics such as isoniazid (249). Development of drug resistance during dormancy is mainly due to chromosomal mutations in genes required for antibiotic action. Isoniazid (INH) and rifampicin (RIF) are the front line drugs for the treatment of TB. However, over a period of time, Mtb developed multidrug resistance and currently a combination of 8–10 drugs are being used for treating MDR-TB. Development of multidrug resistance by Mtb complicates the clinical interventions. Serious side effects such as nephrotoxicity, ototoxicity, and dysglycaemia due to the use of powerful anti-TB drugs such as aminoglycosides, ethionamide, and gatifloxacin are some of the indirect clinical implications caused by the ability of Mtb to acquire drug resistance (250). Another clinical implication that is very serious is the re-emergence of TB when host immune responses fail in conditions such as HIV infection (237) and the increased risk of developing TB in patients treated with anti-TNF (251). Further, corticosteroid therapy, deficiency of vitamin D, and other possible conditions that affect T cell function increases the risk of TB (252). The main therapeutic implication of mycobacterial dormancy is development of drug resistance and this is mainly due to chromosomal mutations in genes required for antibiotic action.
Conclusion
Despite global efforts to tackle incidence and transmission of TB, about 10 million people are diagnosed with this disease each year, leading to approximately 2 million deaths. A minority of the infected patients enjoy total elimination of the pathogen but, a majority of the cases appear to contain Mtb in dormant phase. Identification and diagnosis of individuals with latent TB has been an active area of investigation even in the era of advanced molecular and cell biology. Although the role of many genes and their protein products that contribute to dormancy and reactivation of Mtb were studied and proposed as potential targets/antigens for the development of drugs and vaccines, the application of the same at the ground level in containing TB still remains a major challenge. Whether or not the functional applicability identified for these genes is sufficient to develop control strategies, such as development of vaccines against TB will need to be tested, evaluated, and addressed. If so, would the vaccines be designed with multiple antigens encoded by the genes involved in dormancy and reactivation of Mtb? On the same lines, should multiple antigen strategy be adopted in designing highly sensitive diagnostics that can detect stage-specific antigens of Mtb to allow the option of a strategic treatment protocol and prevent the “under representation” of dormant Mtb cases as healthy individuals? On the other hand, can the immune system of the host be manipulated to efficiently prevent dormancy of Mtb? What is the relevance of the functional aspects of genes involved in latency and reactivation for the development of extreme drug resistance in Mtb? Answering all these questions, although difficult form a practical point of view, would enable us tackle Mtb by a multipronged approach that involves prevention, timely detection of infection, and also the identification of the stage of infection through the development of novel drugs that can target the pathogen at all the three stages, namely, infection, dormancy, and reactivation.
Author Contributions
VP: collected the data and developed the entire manuscript. SD: sketched the figures and participated in discussions for developing the ideas centered on dormancy regulon gene functions. NA: provided overall concept and thought leadership and edited the final version of the manuscript. All the authors have read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
Work in NA’s lab is funded by grants from the Department of Biotechnology (DBT; BT/PR12817/COE/34/23/2015). VP was supported by the DST Women Scientist—A Project [SR/WOS-A/LS-126/2013 (G)]. SD was supported by a CSIR fellowship (CSIR-SRF). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
1. WHO. Global Tuberculosis Report 2015. World Health Organization (2016). Available from: http://www.who.int/tb/publications/factsheet_global.pdf
2. CTBD. TB India 2015, Annual Status Report. (2016). Available from: www.tbcindia.nic.in
3. Mathema B, Kurepina N, Fallows D, Kreiswirth BN. Lessons from molecular epidemiology and comparative genomics. Semin Respir Crit Care Med (2008) 29:467–80. doi: 10.1055/s-0028-1085699
4. Ryndak MB, Singh KK, Peng Z, Laal S. Transcriptional profile of Mycobacterium tuberculosis replicating in type II alveolar epithelial cells. PLoS One (2015) 10:e0123745. doi:10.1371/journal.pone.0123745
5. Teitelbaum R, Schubert W, Gunther L, Kress Y, Macaluso F, Pollard JW, et al. The M cell as a portal of entry to the lung for the bacterial pathogen Mycobacterium tuberculosis. Immunity (1999) 10:641–50. doi:10.1016/S1074-7613(00)80063-1
6. Trauner A, Lougheed KE, Bennett MH, Hingley-Wilson SM, Williams HD. The dormancy regulator DosR controls ribosome stability in hypoxic mycobacteria. J Biol Chem (2012) 287:24053–63. doi:10.1074/jbc.M112.364851
7. Gideon HP, Flynn JL. Latent tuberculosis: what the host “sees”? Immunol Res (2011) 50:202–12. doi:10.1007/s12026-011-8229-7
8. Lillebaek T, Dirksen A, Baess I, Strunge B, Thomsen VO, Andersen AB. Molecular evidence of endogenous reactivation of Mycobacterium tuberculosis after 33 years of latent infection. J Infect Dis (2002) 185:401–4. doi:10.1086/338342
9. Ahmad S. Pathogenesis, immunology, and diagnosis of latent Mycobacterium tuberculosis infection. Clin Dev Immunol (2011) 2011:814943. doi:10.1155/2011/814943
10. Cambier CJ, Takaki KK, Larson RP, Hernandez RE, Tobin DM, Urdahl KB, et al. Mycobacteria manipulate macrophage recruitment through coordinated use of membrane lipids. Nature (2014) 505:218–22. doi:10.1038/nature12799
11. El Kebir D, Jozsef L, Pan W, Wang L, Filep JG. Bacterial DNA activates endothelial cells and promotes neutrophil adherence through TLR9 signaling. J Immunol (2009) 182:4386–94. doi:10.4049/jimmunol.0803044
12. Harding CV, Boom WH. Regulation of antigen presentation by Mycobacterium tuberculosis: a role for toll-like receptors. Nat Rev Microbiol (2010) 8:296–307. doi:10.1038/nrmicro2321
13. Kleinnijenhuis J, Oosting M, Joosten LA, Netea MG, Van Crevel R. Innate immune recognition of Mycobacterium tuberculosis. Clin Dev Immunol (2011) 2011:405310. doi:10.1155/2011/405310
14. Thada S, Valluri VL, Gaddam SL. Influence of toll-like receptor gene polymorphisms to tuberculosis susceptibility in humans. Scand J Immunol (2013) 78:221–9. doi:10.1111/sji.12066
15. Saraav I, Singh S, Sharma S. Outcome of Mycobacterium tuberculosis and toll-like receptor interaction: immune response or immune evasion? Immunol Cell Biol (2014) 92:741–6. doi:10.1038/icb.2014.52
16. Sharma S, Sharma M, Bose M. Mycobacterium tuberculosis infection of human monocyte-derived macrophages leads to apoptosis of T cells. Immunol Cell Biol (2009) 87:226–34. doi:10.1038/icb.2008.97
17. Mahajan S, Dkhar HK, Chandra V, Dave S, Nanduri R, Janmeja AK, et al. Mycobacterium tuberculosis modulates macrophage lipid-sensing nuclear receptors PPARgamma and TR4 for survival. J Immunol (2012) 188:5593–603. doi:10.4049/jimmunol.1103038
18. Stamm CE, Collins AC, Shiloh MU. Sensing of Mycobacterium tuberculosis and consequences to both host and bacillus. Immunol Rev (2015) 264:204–19. doi:10.1111/imr.12263
19. Liu Q, Li W, Li D, Feng Y, Tao C. TIRAP C539T polymorphism contributes to tuberculosis susceptibility: evidence from a meta-analysis. Infect Genet Evol (2014) 27:32–9. doi:10.1016/j.meegid.2014.06.025
20. Ni Cheallaigh C, Sheedy FJ, Harris J, Munoz-Wolf N, Lee J, West K, et al. A common variant in the adaptor Mal regulates interferon gamma signaling. Immunity (2016) 44:368–79. doi:10.1016/j.immuni.2016.01.019
21. Noss EH, Pai RK, Sellati TJ, Radolf JD, Belisle J, Golenbock DT, et al. Toll-like receptor 2-dependent inhibition of macrophage class II MHC expression and antigen processing by 19-kDa lipoprotein of Mycobacterium tuberculosis. J Immunol (2001) 167:910–8. doi:10.4049/jimmunol.167.2.910
22. Chang CH, Guerder S, Hong SC, Van Ewijk W, Flavell RA. Mice lacking the MHC class II transactivator (CIITA) show tissue-specific impairment of MHC class II expression. Immunity (1996) 4:167–78. doi:10.1016/S1074-7613(00)80681-0
23. Pennini ME, Liu Y, Yang J, Croniger CM, Boom WH, Harding CV. CCAAT/enhancer-binding protein beta and delta binding to CIITA promoters is associated with the inhibition of CIITA expression in response to Mycobacterium tuberculosis 19-kDa lipoprotein. J Immunol (2007) 179:6910–8. doi:10.4049/jimmunol.179.10.6910
24. Ghorpade DS, Holla S, Sinha AY, Alagesan SK, Balaji KN. Nitric oxide and KLF4 protein epigenetically modify class II transactivator to repress major histocompatibility complex II expression during Mycobacterium bovis bacillus Calmette-Guerin infection. J Biol Chem (2013) 288:20592–606. doi:10.1074/jbc.M113.472183
25. Repique CJ, Li A, Brickey WJ, Ting JP, Collins FM, Morris SL. Susceptibility of mice deficient in the MHC class II transactivator to infection with Mycobacterium tuberculosis. Scand J Immunol (2003) 58:15–22. doi:10.1046/j.1365-3083.2003.01266.x
26. Chang CH, Flavell RA. Class II transactivator regulates the expression of multiple genes involved in antigen presentation. J Exp Med (1995) 181:765–7. doi:10.1084/jem.181.2.765
27. Pai RK, Pennini ME, Tobian AA, Canaday DH, Boom WH, Harding CV. Prolonged toll-like receptor signaling by Mycobacterium tuberculosis and its 19-kilodalton lipoprotein inhibits gamma interferon-induced regulation of selected genes in macrophages. Infect Immun (2004) 72:6603–14. doi:10.1128/IAI.72.11.6603-6614.2004
28. Geijtenbeek TB, Van Vliet SJ, Koppel EA, Sanchez-Hernandez M, Vandenbroucke-Grauls CM, Appelmelk B, et al. Mycobacteria target DC-SIGN to suppress dendritic cell function. J Exp Med (2003) 197:7–17. doi:10.1084/jem.20021229
29. Gehring AJ, Dobos KM, Belisle JT, Harding CV, Boom WH. Mycobacterium tuberculosis LprG (Rv1411c): a novel TLR-2 ligand that inhibits human macrophage class II MHC antigen processing. J Immunol (2004) 173:2660–8. doi:10.4049/jimmunol.173.4.2660
30. Wolf AJ, Linas B, Trevejo-Nunez GJ, Kincaid E, Tamura T, Takatsu K, et al. Mycobacterium tuberculosis infects dendritic cells with high frequency and impairs their function in vivo. J Immunol (2007) 179:2509–19. doi:10.4049/jimmunol.179.4.2509
31. Mihret A. The role of dendritic cells in Mycobacterium tuberculosis infection. Virulence (2012) 3:654–9. doi:10.4161/viru.22586
32. Vergne I, Gilleron M, Nigou J. Manipulation of the endocytic pathway and phagocyte functions by Mycobacterium tuberculosis lipoarabinomannan. Front Cell Infect Microbiol (2014) 4:187. doi:10.3389/fcimb.2014.00187
33. Tan T, Lee WL, Alexander DC, Grinstein S, Liu J. The ESAT-6/CFP-10 secretion system of Mycobacterium marinum modulates phagosome maturation. Cell Microbiol (2006) 8:1417–29. doi:10.1111/j.1462-5822.2006.00721.x
34. Rohde K, Yates RM, Purdy GE, Russell DG. Mycobacterium tuberculosis and the environment within the phagosome. Immunol Rev (2007) 219:37–54. doi:10.1111/j.1600-065X.2007.00547.x
35. Malik ZA, Denning GM, Kusner DJ. Inhibition of Ca(2+) signaling by Mycobacterium tuberculosis is associated with reduced phagosome-lysosome fusion and increased survival within human macrophages. J Exp Med (2000) 191:287–302. doi:10.1084/jem.191.2.287
36. Malik ZA, Thompson CR, Hashimi S, Porter B, Iyer SS, Kusner DJ. Cutting edge: Mycobacterium tuberculosis blocks Ca2+ signaling and phagosome maturation in human macrophages via specific inhibition of sphingosine kinase. J Immunol (2003) 170:2811–5. doi:10.4049/jimmunol.170.6.2811
37. Vergne I, Chua J, Deretic V. Tuberculosis toxin blocking phagosome maturation inhibits a novel Ca2+/calmodulin-PI3K hVPS34 cascade. J Exp Med (2003) 198:653–9. doi:10.1084/jem.20030527
38. Saleh MT, Belisle JT. Secretion of an acid phosphatase (SapM) by Mycobacterium tuberculosis that is similar to eukaryotic acid phosphatases. J Bacteriol (2000) 182:6850–3. doi:10.1128/JB.182.23.6850-6853.2000
39. Pieters J, McKinney J. Preface.pathogenesis of Mycobacterium tuberculosis and its interaction with the host organism. Curr Top Microbiol Immunol (2013) 374:v–vi.
40. Tse HM, Josephy SI, Chan ED, Fouts D, Cooper AM. Activation of the mitogen-activated protein kinase signaling pathway is instrumental in determining the ability of Mycobacterium avium to grow in murine macrophages. J Immunol (2002) 168:825–33. doi:10.4049/jimmunol.168.2.825
41. Fratti RA, Chua J, Deretic V. Induction of p38 mitogen-activated protein kinase reduces early endosome autoantigen 1 (EEA1) recruitment to phagosomal membranes. J Biol Chem (2003) 278:46961–7. doi:10.1074/jbc.M305225200
42. Mishra AK, Driessen NN, Appelmelk BJ, Besra GS. Lipoarabinomannan and related glycoconjugates: structure, biogenesis and role in Mycobacterium tuberculosis physiology and host-pathogen interaction. FEMS Microbiol Rev (2011) 35:1126–57. doi:10.1111/j.1574-6976.2011.00276.x
43. Guerin I, de Chastellier C. Pathogenic mycobacteria disrupt the macrophage actin filament network. Infect Immun (2000) 68:2655–62. doi:10.1128/IAI.68.5.2655-2662.2000
44. Podinovskaia M, Lee W, Caldwell S, Russell DG. Infection of macrophages with Mycobacterium tuberculosis induces global modifications to phagosomal function. Cell Microbiol (2013) 15:843–59. doi:10.1111/cmi.12092
45. Behar SM, Divangahi M, Remold HG. Evasion of innate immunity by Mycobacterium tuberculosis: is death an exit strategy? Nat Rev Microbiol (2010) 8:668–74. doi:10.1038/nrmicro2387
46. Behar SM, Martin CJ, Booty MG, Nishimura T, Zhao X, Gan HX, et al. Apoptosis is an innate defense function of macrophages against Mycobacterium tuberculosis. Mucosal Immunol (2011) 4:279–87. doi:10.1038/mi.2011.3
47. Bradfute SB, Castillo EF, Arko-Mensah J, Chauhan S, Jiang S, Mandell M, et al. Autophagy as an immune effector against tuberculosis. Curr Opin Microbiol (2013) 16:355–65. doi:10.1016/j.mib.2013.05.003
48. Kundu M, Pathak SK, Kumawat K, Basu S, Chatterjee G, Pathak S, et al. A TNF- and c-Cbl-dependent FLIP(S)-degradation pathway and its function in Mycobacterium tuberculosis-induced macrophage apoptosis. Nat Immunol (2009) 10:918–26. doi:10.1038/ni.1754
49. Yu X, Li C, Hong W, Pan W, Xie J. Autophagy during Mycobacterium tuberculosis infection and implications for future tuberculosis medications. Cell Signal (2013) 25:1272–8. doi:10.1016/j.cellsig.2013.02.011
50. Kumar D, Nath L, Kamal MA, Varshney A, Jain A, Singh S, et al. Genome-wide analysis of the host intracellular network that regulates survival of Mycobacterium tuberculosis. Cell (2010) 140:731–43. doi:10.1016/j.cell.2010.02.012
51. Forrellad MA, Klepp LI, Gioffre A, Sabio y Garcia J, Morbidoni HR, de la Paz Santangelo M, et al. Virulence factors of the Mycobacterium tuberculosis complex. Virulence (2013) 4:3–66. doi:10.4161/viru.22329
52. Balcewicz-Sablinska MK, Keane J, Kornfeld H, Remold HG. Pathogenic Mycobacterium tuberculosis evades apoptosis of host macrophages by release of TNF-R2, resulting in inactivation of TNF-alpha. J Immunol (1998) 161:2636–41.
53. Rakotosamimanana N, Doherty TM, Andriamihantasoa LH, Richard V, Gicquel B, Soares JL, et al. Expression of TNF-alpha-dependent apoptosis-related genes in the peripheral blood of Malagasy subjects with tuberculosis. PLoS One (2013) 8:e61154. doi:10.1371/journal.pone.0061154
54. Sly LM, Hingley-Wilson SM, Reiner NE, Mcmaster WR. Survival of Mycobacterium tuberculosis in host macrophages involves resistance to apoptosis dependent upon induction of antiapoptotic Bcl-2 family member Mcl-1. J Immunol (2003) 170:430–7. doi:10.4049/jimmunol.170.1.430
55. Velmurugan K, Chen B, Miller JL, Azogue S, Gurses S, Hsu T, et al. Mycobacterium tuberculosis nuoG is a virulence gene that inhibits apoptosis of infected host cells. PLoS Pathog (2007) 3:e110. doi:10.1371/journal.ppat.0030110
56. Miller JL, Velmurugan K, Cowan MJ, Briken V. The type I NADH dehydrogenase of Mycobacterium tuberculosis counters phagosomal NOX2 activity to inhibit TNF-alpha-mediated host cell apoptosis. PLoS Pathog (2010) 6:e1000864. doi:10.1371/journal.ppat.1000864
57. Jayakumar D, Jacobs WR Jr, Narayanan S. Protein kinase E of Mycobacterium tuberculosis has a role in the nitric oxide stress response and apoptosis in a human macrophage model of infection. Cell Microbiol (2008) 10:365–74. doi:10.1111/j.1462-5822.2007.01049.x
58. Parandhaman DK, Hanna LE, Narayanan S. PknE, a serine/threonine protein kinase of Mycobacterium tuberculosis initiates survival crosstalk that also impacts HIV coinfection. PLoS One (2014) 9:e83541. doi:10.1371/journal.pone.0083541
59. Kelly DM, Ten Bokum AM, O’Leary SM, O’Sullivan MP, Keane J. Bystander macrophage apoptosis after Mycobacterium tuberculosis H37Ra infection. Infect Immun (2008) 76:351–60. doi:10.1128/IAI.00614-07
60. Chen M, Divangahi M, Gan H, Shin DS, Hong S, Lee DM, et al. Lipid mediators in innate immunity against tuberculosis: opposing roles of PGE2 and LXA4 in the induction of macrophage death. J Exp Med (2008) 205:2791–801. doi:10.1084/jem.20080767
61. Divangahi M, Desjardins D, Nunes-Alves C, Remold HG, Behar SM. Eicosanoid pathways regulate adaptive immunity to Mycobacterium tuberculosis. Nat Immunol (2010) 11:751–8. doi:10.1038/ni.1904
62. Herb F, Thye T, Niemann S, Browne EN, Chinbuah MA, Gyapong J, et al. ALOX5 variants associated with susceptibility to human pulmonary tuberculosis. Hum Mol Genet (2008) 17:1052–60. doi:10.1093/hmg/ddm378
63. Tobin DM, Vary JC Jr, Ray JP, Walsh GS, Dunstan SJ, Bang ND, et al. The lta4h locus modulates susceptibility to mycobacterial infection in zebrafish and humans. Cell (2010) 140:717–30. doi:10.1016/j.cell.2010.02.013
64. Shafiani S, Tucker-Heard G, Kariyone A, Takatsu K, Urdahl KB. Pathogen-specific regulatory T cells delay the arrival of effector T cells in the lung during early tuberculosis. J Exp Med (2010) 207:1409–20. doi:10.1084/jem.20091885
65. Khader SA, Partida-Sanchez S, Bell G, Jelley-Gibbs DM, Swain S, Pearl JE, et al. Interleukin 12p40 is required for dendritic cell migration and T cell priming after Mycobacterium tuberculosis infection. J Exp Med (2006) 203:1805–15. doi:10.1084/jem.20052545
66. Shi S, Nathan C, Schnappinger D, Drenkow J, Fuortes M, Block E, et al. MyD88 primes macrophages for full-scale activation by interferon-gamma yet mediates few responses to Mycobacterium tuberculosis. J Exp Med (2003) 198:987–97. doi:10.1084/jem.20030603
67. Pecora ND, Gehring AJ, Canaday DH, Boom WH, Harding CV. Mycobacterium tuberculosis LprA is a lipoprotein agonist of TLR2 that regulates innate immunity and APC function. J Immunol (2006) 177:422–9. doi:10.4049/jimmunol.177.1.422
68. Nigou J, Vasselon T, Ray A, Constant P, Gilleron M, Besra GS, et al. Mannan chain length controls lipoglycans signaling via and binding to TLR2. J Immunol (2008) 180:6696–702. doi:10.4049/jimmunol.180.10.6696
69. Cyktor JC, Turner J. Interleukin-10 and immunity against prokaryotic and eukaryotic intracellular pathogens. Infect Immun (2011) 79:2964–73. doi:10.1128/IAI.00047-11
70. Bhatt K, Salgame P. Host innate immune response to Mycobacterium tuberculosis. J Clin Immunol (2007) 27:347–62. doi:10.1007/s10875-007-9084-0
71. Killick KE, Ni Cheallaigh C, O’Farrelly C, Hokamp K, Machugh DE, Harris J. Receptor-mediated recognition of mycobacterial pathogens. Cell Microbiol (2013) 15:1484–95. doi:10.1111/cmi.12161
72. Cooper AM, Mayer-Barber KD, Sher A. Role of innate cytokines in mycobacterial infection. Mucosal Immunol (2011) 4:252–60. doi:10.1038/mi.2011.13
73. Wang X, Barnes PF, Dobos-Elder KM, Townsend JC, Chung YT, Shams H, et al. ESAT-6 inhibits production of IFN-gamma by Mycobacterium tuberculosis-responsive human T cells. J Immunol (2009) 182:3668–77. doi:10.4049/jimmunol.0803579
74. Jayaraman P, Sada-Ovalle I, Beladi S, Anderson AC, Dardalhon V, Hotta C, et al. Tim3 binding to galectin-9 stimulates antimicrobial immunity. J Exp Med (2010) 207:2343–54. doi:10.1084/jem.20100687
75. Jurado JO, Alvarez IB, Pasquinelli V, Martinez GJ, Quiroga MF, Abbate E, et al. Programmed death (PD)-1:PD-ligand 1/PD-ligand 2 pathway inhibits T cell effector functions during human tuberculosis. J Immunol (2008) 181:116–25. doi:10.4049/jimmunol.181.1.116
76. Galluzzi L, Kroemer G. Necroptosis: a specialized pathway of programmed necrosis. Cell (2008) 135:1161–3. doi:10.1016/j.cell.2008.12.004
77. Wang Z, Jiang H, Chen S, Du F, Wang X. The mitochondrial phosphatase PGAM5 functions at the convergence point of multiple necrotic death pathways. Cell (2012) 148:228–43. doi:10.1016/j.cell.2011.11.030
78. Roca FJ, Ramakrishnan L. TNF dually mediates resistance and susceptibility to mycobacteria via mitochondrial reactive oxygen species. Cell (2013) 153:521–34. doi:10.1016/j.cell.2013.03.022
79. Vandenabeele P, Declercq W, Van Herreweghe F, Vanden Berghe T. The role of the kinases RIP1 and RIP3 in TNF-induced necrosis. Sci Signal (2010) 3:re4. doi:10.1126/scisignal.3115re4
80. Han J, Zhong CQ, Zhang DW. Programmed necrosis: backup to and competitor with apoptosis in the immune system. Nat Immunol (2011) 12:1143–9. doi:10.1038/ni.2159
81. Jayaraman P, Sada-Ovalle I, Nishimura T, Anderson AC, Kuchroo VK, Remold HG, et al. IL-1beta promotes antimicrobial immunity in macrophages by regulating TNFR signaling and caspase-3 activation. J Immunol (2013) 190:4196–204. doi:10.4049/jimmunol.1202688
82. Xu G, Wang J, Gao GF, Liu CH. Insights into battles between Mycobacterium tuberculosis and macrophages. Protein Cell (2014) 5:728–36. doi:10.1007/s13238-014-0077-5
83. O’Leary S, O’Sullivan MP, Keane J. IL-10 blocks phagosome maturation in Mycobacterium tuberculosis-infected human macrophages. Am J Respir Cell Mol Biol (2011) 45:172–80. doi:10.1165/rcmb.2010-0319OC
84. Redpath S, Ghazal P, Gascoigne NR. Hijacking and exploitation of IL-10 by intracellular pathogens. Trends Microbiol (2001) 9:86–92. doi:10.1016/S0966-842X(00)01919-3
85. Moore KW, De Waal Malefyt R, Coffman RL, O’Garra A. Interleukin-10 and the interleukin-10 receptor. Annu Rev Immunol (2001) 19:683–765. doi:10.1146/annurev.immunol.19.1.683
86. Demangel C, Bertolino P, Britton WJ. Autocrine IL-10 impairs dendritic cell (DC)-derived immune responses to mycobacterial infection by suppressing DC trafficking to draining lymph nodes and local IL-12 production. Eur J Immunol (2002) 32:994–1002. doi:10.1002/1521-4141(200204)32:4<994::AID-IMMU994>3.3.CO;2-Y
87. Redford PS, Boonstra A, Read S, Pitt J, Graham C, Stavropoulos E, et al. Enhanced protection to Mycobacterium tuberculosis infection in IL-10-deficient mice is accompanied by early and enhanced Th1 responses in the lung. Eur J Immunol (2010) 40:2200–10. doi:10.1002/eji.201040433
88. Boussiotis VA, Tsai EY, Yunis EJ, Thim S, Delgado JC, Dascher CC, et al. IL-10-producing T cells suppress immune responses in anergic tuberculosis patients. J Clin Invest (2000) 105:1317–25. doi:10.1172/JCI9918
89. Pacheco AG, Cardoso CC, Moraes MO. IFNG +874T/A, IL10 -1082G/A and TNF -308G/A polymorphisms in association with tuberculosis susceptibility: a meta-analysis study. Hum Genet (2008) 123:477–84. doi:10.1007/s00439-008-0497-5
90. Qualls JE, Neale G, Smith AM, Koo MS, Defreitas AA, Zhang H, et al. Arginine usage in mycobacteria-infected macrophages depends on autocrine-paracrine cytokine signaling. Sci Signal (2010) 3:ra62. doi:10.1126/scisignal.2000955
91. Martinez FO, Helming L, Gordon S. Alternative activation of macrophages: an immunologic functional perspective. Annu Rev Immunol (2009) 27:451–83. doi:10.1146/annurev.immunol.021908.132532
92. Ashenafi S, Aderaye G, Bekele A, Zewdie M, Aseffa G, Hoang AT, et al. Progression of clinical tuberculosis is associated with a Th2 immune response signature in combination with elevated levels of SOCS3. Clin Immunol (2014) 151:84–99. doi:10.1016/j.clim.2014.01.010
93. Nolan A, Fajardo E, Huie ML, Condos R, Pooran A, Dawson R, et al. Increased production of IL-4 and IL-12p40 from bronchoalveolar lavage cells are biomarkers of Mycobacterium tuberculosis in the sputum. PLoS One (2013) 8:e59461. doi:10.1371/journal.pone.0059461
94. Manca C, Reed MB, Freeman S, Mathema B, Kreiswirth B, Barry CE III, et al. Differential monocyte activation underlies strain-specific Mycobacterium tuberculosis pathogenesis. Infect Immun (2004) 72:5511–4. doi:10.1128/IAI.72.9.5511-5514.2004
95. Harris J, De Haro SA, Master SS, Keane J, Roberts EA, Delgado M, et al. T helper 2 cytokines inhibit autophagic control of intracellular Mycobacterium tuberculosis. Immunity (2007) 27:505–17. doi:10.1016/j.immuni.2007.09.003
96. Mishra BB, Rathinam VA, Martens GW, Martinot AJ, Kornfeld H, Fitzgerald KA, et al. Nitric oxide controls the immunopathology of tuberculosis by inhibiting NLRP3 inflammasome-dependent processing of IL-1beta. Nat Immunol (2013) 14:52–60. doi:10.1038/ni.2474
97. Yang CS, Yuk JM, Jo EK. The role of nitric oxide in mycobacterial infections. Immune Netw (2009) 9:46–52. doi:10.4110/in.2009.9.2.46
98. Lin PL, Flynn JL. Understanding latent tuberculosis: a moving target. J Immunol (2010) 185:15–22. doi:10.4049/jimmunol.0903856
99. Jung JY, Madan-Lala R, Georgieva M, Rengarajan J, Sohaskey CD, Bange FC, et al. The intracellular environment of human macrophages that produce nitric oxide promotes growth of mycobacteria. Infect Immun (2013) 81:3198–209. doi:10.1128/IAI.00611-13
100. Bryk R, Lima CD, Erdjument-Bromage H, Tempst P, Nathan C. Metabolic enzymes of mycobacteria linked to antioxidant defense by a thioredoxin-like protein. Science (2002) 295:1073–7. doi:10.1126/science.1067798
101. Lee HN, Lee NO, Han SJ, Ko IJ, Oh JI. Regulation of the ahpC gene encoding alkyl hydroperoxide reductase in Mycobacterium smegmatis. PLoS One (2014) 9:e111680. doi:10.1371/journal.pone.0111680
102. Lee WL, Gold B, Darby C, Brot N, Jiang X, De Carvalho LP, et al. Mycobacterium tuberculosis expresses methionine sulphoxide reductases A and B that protect from killing by nitrite and hypochlorite. Mol Microbiol (2009) 71:583–93. doi:10.1111/j.1365-2958.2008.06548.x
103. Samanovic MI, Tu S, Novak O, Iyer LM, Mcallister FE, Aravind L, et al. Proteasomal control of cytokinin synthesis protects Mycobacterium tuberculosis against nitric oxide. Mol Cell (2015) 57:984–94. doi:10.1016/j.molcel.2015.01.024
104. Rittershaus ES, Baek SH, Sassetti CM. The normalcy of dormancy: common themes in microbial quiescence. Cell Host Microbe (2013) 13:643–51. doi:10.1016/j.chom.2013.05.012
105. Sala A, Bordes P, Genevaux P. Multiple toxin-antitoxin systems in Mycobacterium tuberculosis. Toxins (Basel) (2014) 6:1002–20. doi:10.3390/toxins6031002
106. Pompei L, Jang S, Zamlynny B, Ravikumar S, Mcbride A, Hickman SP, et al. Disparity in IL-12 release in dendritic cells and macrophages in response to Mycobacterium tuberculosis is due to use of distinct TLRs. J Immunol (2007) 178:5192–9. doi:10.4049/jimmunol.178.8.5192
107. Buts L, Lah J, Dao-Thi MH, Wyns L, Loris R. Toxin-antitoxin modules as bacterial metabolic stress managers. Trends Biochem Sci (2005) 30:672–9. doi:10.1016/j.tibs.2005.10.004
108. Pandey DP, Gerdes K. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res (2005) 33:966–76. doi:10.1093/nar/gki201
109. Van Melderen L, Saavedra De Bast M. Bacterial toxin-antitoxin systems: more than selfish entities? PLoS Genet (2009) 5:e1000437. doi:10.1371/journal.pgen.1000437
110. Huang F, He ZG. Characterization of an interplay between a Mycobacterium tuberculosis MazF homolog, Rv1495 and its sole DNA topoisomerase I. Nucleic Acids Res (2010) 38:8219–30. doi:10.1093/nar/gkq737
111. Ramakrishnan L. Revisiting the role of the granuloma in tuberculosis. Nat Rev Immunol (2012) 12:352–66. doi:10.1038/nri3211
112. Lin PL, Ford CB, Coleman MT, Myers AJ, Gawande R, Ioerger T, et al. Sterilization of granulomas is common in active and latent tuberculosis despite within-host variability in bacterial killing. Nat Med (2014) 20:75–9. doi:10.1038/nm.3412
113. Silva Miranda M, Breiman A, Allain S, Deknuydt F, Altare F. The tuberculous granuloma: an unsuccessful host defence mechanism providing a safety shelter for the bacteria? Clin Dev Immunol (2012) 2012:139127. doi:10.1155/2012/139127
114. Davis JM, Ramakrishnan L. The role of the granuloma in expansion and dissemination of early tuberculous infection. Cell (2009) 136:37–49. doi:10.1016/j.cell.2008.11.014
115. Young DB, Gideon HP, Wilkinson RJ. Eliminating latent tuberculosis. Trends Microbiol (2009) 17:183–8. doi:10.1016/j.tim.2009.02.005
116. Guirado E, Mbawuike U, Keiser TL, Arcos J, Azad AK, Wang SH, et al. Characterization of host and microbial determinants in individuals with latent tuberculosis infection using a human granuloma model. MBio (2015) 6:e2537–2514. doi:10.1128/mBio.02537-14
117. Lin PL, Rodgers M, Smith L, Bigbee M, Myers A, Bigbee C, et al. Quantitative comparison of active and latent tuberculosis in the cynomolgus macaque model. Infect Immun (2009) 77:4631–42. doi:10.1128/IAI.00592-09
118. Via LE, Lin PL, Ray SM, Carrillo J, Allen SS, Eum SY, et al. Tuberculous granulomas are hypoxic in guinea pigs, rabbits, and nonhuman primates. Infect Immun (2008) 76:2333–40. doi:10.1128/IAI.01515-07
119. Barry CE III, Boshoff HI, Dartois V, Dick T, Ehrt S, Flynn J, et al. The spectrum of latent tuberculosis: rethinking the biology and intervention strategies. Nat Rev Microbiol (2009) 7:845–55. doi:10.1038/nrmicro2236
120. Kim MJ, Wainwright HC, Locketz M, Bekker LG, Walther GB, Dittrich C, et al. Caseation of human tuberculosis granulomas correlates with elevated host lipid metabolism. EMBO Mol Med (2010) 2:258–74. doi:10.1002/emmm.201000079
121. Bold TD, Ernst JD. Who benefits from granulomas, mycobacteria or host? Cell (2009) 136:17–9. doi:10.1016/j.cell.2008.12.032
122. Rubin EJ. The granuloma in tuberculosis – friend or foe? N Engl J Med (2009) 360:2471–3. doi:10.1056/NEJMcibr0902539
123. Mohan VP, Scanga CA, Yu K, Scott HM, Tanaka KE, Tsang E, et al. Effects of tumor necrosis factor alpha on host immune response in chronic persistent tuberculosis: possible role for limiting pathology. Infect Immun (2001) 69:1847–55. doi:10.1128/IAI.69.3.1847-1855.2001
124. Allie N, Grivennikov SI, Keeton R, Hsu NJ, Bourigault ML, Court N, et al. Prominent role for T cell-derived tumour necrosis factor for sustained control of Mycobacterium tuberculosis infection. Sci Rep (2013) 3:1809. doi:10.1038/srep01809
125. Agarwal N, Lamichhane G, Gupta R, Nolan S, Bishai WR. Cyclic AMP intoxication of macrophages by a Mycobacterium tuberculosis adenylate cyclase. Nature (2009) 460:98–102. doi:10.1038/nature08123
126. Elkington PT, Ugarte-Gil CA, Friedland JS. Matrix metalloproteinases in tuberculosis. Eur Respir J (2011) 38:456–64. doi:10.1183/09031936.00015411
127. Lai WC, Zhou M, Shankavaram U, Peng G, Wahl LM. Differential regulation of lipopolysaccharide-induced monocyte matrix metalloproteinase (MMP)-1 and MMP-9 by p38 and extracellular signal-regulated kinase 1/2 mitogen-activated protein kinases. J Immunol (2003) 170:6244–9. doi:10.4049/jimmunol.170.12.6244
128. Rand L, Green JA, Saraiva L, Friedland JS, Elkington PT. Matrix metalloproteinase-1 is regulated in tuberculosis by a p38 MAPK-dependent, p-aminosalicylic acid-sensitive signaling cascade. J Immunol (2009) 182:5865–72. doi:10.4049/jimmunol.0801935
129. Jones GS, Amirault HJ, Andersen BR. Killing of Mycobacterium tuberculosis by neutrophils: a nonoxidative process. J Infect Dis (1990) 162:700–4. doi:10.1093/infdis/162.3.700
130. Lowe DM, Redford PS, Wilkinson RJ, O’Garra A, Martineau AR. Neutrophils in tuberculosis: friend or foe? Trends Immunol (2012) 33:14–25. doi:10.1016/j.it.2011.10.003
131. Corleis B, Korbel D, Wilson R, Bylund J, Chee R, Schaible UE. Escape of Mycobacterium tuberculosis from oxidative killing by neutrophils. Cell Microbiol (2012) 14:1109–21. doi:10.1111/j.1462-5822.2012.01783.x
132. Yang CT, Cambier CJ, Davis JM, Hall CJ, Crosier PS, Ramakrishnan L. Neutrophils exert protection in the early tuberculous granuloma by oxidative killing of mycobacteria phagocytosed from infected macrophages. Cell Host Microbe (2012) 12:301–12. doi:10.1016/j.chom.2012.07.009
133. Briken V. “With a little help from my friends”: efferocytosis as an antimicrobial mechanism. Cell Host Microbe (2012) 12:261–3. doi:10.1016/j.chom.2012.08.008
134. Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature (1998) 393:537–44. doi:10.1038/31159
135. Francis RJ, Kotecha S, Hallett MB. Ca2+ activation of cytosolic calpain induces the transition from apoptosis to necrosis in neutrophils with externalized phosphatidylserine. J Leukoc Biol (2013) 93:95–100. doi:10.1189/jlb.0412212
136. Francis RJ, Butler RE, Stewart GR. Mycobacterium tuberculosis ESAT-6 is a leukocidin causing Ca2+ influx, necrosis and neutrophil extracellular trap formation. Cell Death Dis (2014) 5:e1474. doi:10.1038/cddis.2014.394
137. Pulido PA, Novoa-Aponte L, Villamil N, Soto CY. The DosR dormancy regulator of Mycobacterium tuberculosis stimulates the Na(+)/K (+) and Ca (2+) ATPase activities in plasma membrane vesicles of mycobacteria. Curr Microbiol (2014) 69:604–10. doi:10.1007/s00284-014-0632-6
138. Stabel JR, Goff JP, Whipple DL, Ackermann MR, Reinhardt TA. Low calcium diet and 1,25-dihydroxyvitamin D(3) infusion modulate immune responses during Mycobacterium paratuberculosis infection in beige mice. Vet Immunol Immunopathol (1996) 50:127–43. doi:10.1016/0165-2427(95)05484-7
139. Hitsumoto T, Ikeda S, Matsukage S, Hamada M. Extensive myocardial calcinosis due to Mycobacterium tuberculosis. Eur Heart J (2015) 37:1195. doi:10.1093/eurheartj/ehv604
140. Bendayan D, Barziv Y, Kramer MR. Pulmonary calcifications: a review. Respir Med (2000) 94:190–3. doi:10.1053/rmed.1999.0716
141. Skoura E, Zumla A, Bomanji J. Imaging in tuberculosis. Int J Infect Dis (2015) 32:87–93. doi:10.1016/j.ijid.2014.12.007
142. Merchant S, Bharati A, Merchant N. Tuberculosis of the genitourinary system-urinary tract tuberculosis: renal tuberculosis-part I. Indian J Radiol Imaging (2013) 23:46–63. doi:10.4103/0971-3026.113617
143. Guirado E, Schlesinger LS. Modeling the Mycobacterium tuberculosis granuloma – the critical battlefield in host immunity and disease. Front Immunol (2013) 4:98. doi:10.3389/fimmu.2013.00098
144. Shkurupiy VA, Kim LB, Potapova OV, Cherdantseva LA, Putyatina AN, Nikonova IK. Fibrogenesis in granulomas and lung interstitium in tuberculous inflammation in mice. Bull Exp Biol Med (2014) 156:731–5. doi:10.1007/s10517-014-2435-y
145. Azouz A, Razzaque MS, El-Hallak M, Taguchi T. Immunoinflammatory responses and fibrogenesis. Med Electron Microsc (2004) 37:141–8. doi:10.1007/s00795-004-0255-2
146. Mariotti S, Sargentini V, Pardini M, Giannoni F, De Spirito M, Gagliardi MC, et al. Mycobacterium tuberculosis may escape helper T cell recognition by infecting human fibroblasts. Hum Immunol (2013) 74:722–9. doi:10.1016/j.humimm.2013.02.005
147. Gonzalez-Avila G, Sandoval C, Herrera MT, Ruiz V, Sommer B, Sada E, et al. Mycobacterium tuberculosis effects on fibroblast collagen metabolism. Respiration (2009) 77:195–202. doi:10.1159/000163064
148. Russell DG. Mycobacterium tuberculosis and the intimate discourse of a chronic infection. Immunol Rev (2011) 240:252–68. doi:10.1111/j.1600-065X.2010.00984.x
149. Gengenbacher M, Kaufmann SH. Mycobacterium tuberculosis: success through dormancy. FEMS Microbiol Rev (2012) 36:514–32. doi:10.1111/j.1574-6976.2012.00331.x
150. Raffetseder J, Pienaar E, Blomgran R, Eklund D, Patcha Brodin V, Andersson H, et al. Replication rates of Mycobacterium tuberculosis in human macrophages do not correlate with mycobacterial antibiotic susceptibility. PLoS One (2014) 9:e112426. doi:10.1371/journal.pone.0112426
151. Di Pietrantonio T, Hernandez C, Girard M, Verville A, Orlova M, Belley A, et al. Strain-specific differences in the genetic control of two closely related mycobacteria. PLoS Pathog (2010) 6:e1001169. doi:10.1371/journal.ppat.1001169
152. Thye T, Vannberg FO, Wong SH, Owusu-Dabo E, Osei I, Gyapong J, et al. Genome-wide association analyses identifies a susceptibility locus for tuberculosis on chromosome 18q11.2. Nat Genet (2010) 42:739–41. doi:10.1038/ng.639
153. Thye T, Owusu-Dabo E, Vannberg FO, Van Crevel R, Curtis J, Sahiratmadja E, et al. Common variants at 11p13 are associated with susceptibility to tuberculosis. Nat Genet (2012) 44:257–9. doi:10.1038/ng.1080
154. Sepulveda RL, Heiba IM, King A, Gonzalez B, Elston RC, Sorensen RU. Evaluation of tuberculin reactivity in BCG-immunized siblings. Am J Respir Crit Care Med (1994) 149:620–4. doi:10.1164/ajrccm.149.3.8118628
155. Cobat A, Gallant CJ, Simkin L, Black GF, Stanley K, Hughes J, et al. High heritability of antimycobacterial immunity in an area of hyperendemicity for tuberculosis disease. J Infect Dis (2010) 201:15–9. doi:10.1086/648611
156. Thye T, Browne EN, Chinbuah MA, Gyapong J, Osei I, Owusu-Dabo E, et al. IL10 haplotype associated with tuberculin skin test response but not with pulmonary TB. PLoS One (2009) 4:e5420. doi:10.1371/journal.pone.0005420
157. Stein CM, Zalwango S, Malone LL, Won S, Mayanja-Kizza H, Mugerwa RD, et al. Genome scan of M. tuberculosis infection and disease in Ugandans. PLoS One (2008) 3:e4094. doi:10.1371/journal.pone.0004094
158. Cobat A, Gallant CJ, Simkin L, Black GF, Stanley K, Hughes J, et al. Two loci control tuberculin skin test reactivity in an area hyperendemic for tuberculosis. J Exp Med (2009) 206:2583–91. doi:10.1084/jem.20090892
159. Park HD, Guinn KM, Harrell MI, Liao R, Voskuil MI, Tompa M, et al. Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol Microbiol (2003) 48:833–43. doi:10.1046/j.1365-2958.2003.03474.x
160. Selvaraj S, Sambandam V, Sardar D, Anishetty S. In silico analysis of DosR regulon proteins of Mycobacterium tuberculosis. Gene (2012) 506:233–41. doi:10.1016/j.gene.2012.06.033
161. Becq J, Gutierrez MC, Rosas-Magallanes V, Rauzier J, Gicquel B, Neyrolles O, et al. Contribution of horizontally acquired genomic islands to the evolution of the tubercle bacilli. Mol Biol Evol (2007) 24:1861–71. doi:10.1093/molbev/msm111
162. Bartek IL, Rutherford R, Gruppo V, Morton RA, Morris RP, Klein MR, et al. The DosR regulon of M. tuberculosis and antibacterial tolerance. Tuberculosis (Edinb) (2009) 89:310–6. doi:10.1016/j.tube.2009.06.001
163. Voskuil MI, Schnappinger D, Visconti KC, Harrell MI, Dolganov GM, Sherman DR, et al. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J Exp Med (2003) 198:705–13. doi:10.1084/jem.20030205
164. Galagan JE, Minch K, Peterson M, Lyubetskaya A, Azizi E, Sweet L, et al. The Mycobacterium tuberculosis regulatory network and hypoxia. Nature (2013) 499:178–83. doi:10.1038/nature12337
165. Schnappinger D, Ehrt S, Voskuil MI, Liu Y, Mangan JA, Monahan IM, et al. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J Exp Med (2003) 198:693–704. doi:10.1084/jem.20030846
166. Karakousis PC, Yoshimatsu T, Lamichhane G, Woolwine SC, Nuermberger EL, Grosset J, et al. Dormancy phenotype displayed by extracellular Mycobacterium tuberculosis within artificial granulomas in mice. J Exp Med (2004) 200:647–57. doi:10.1084/jem.20040646
167. Sharma D, Bose A, Shakila H, Das TK, Tyagi JS, Ramanathan VD. Expression of mycobacterial cell division protein, FtsZ, and dormancy proteins, DevR and Acr, within lung granulomas throughout guinea pig infection. FEMS Immunol Med Microbiol (2006) 48:329–36. doi:10.1111/j.1574-695X.2006.00160.x
168. Bivas-Benita M, Lin MY, Bal SM, Van Meijgaarden KE, Franken KL, Friggen AH, et al. Pulmonary delivery of DNA encoding Mycobacterium tuberculosis latency antigen Rv1733c associated to PLGA-PEI nanoparticles enhances T cell responses in a DNA prime/protein boost vaccination regimen in mice. Vaccine (2009) 27:4010–7. doi:10.1016/j.vaccine.2009.04.033
169. Black GF, Thiel BA, Ota MO, Parida SK, Adegbola R, Boom WH, et al. Immunogenicity of novel DosR regulon-encoded candidate antigens of Mycobacterium tuberculosis in three high-burden populations in Africa. Clin Vaccine Immunol (2009) 16:1203–12. doi:10.1128/CVI.00111-09
170. Yasuhiro Y, Kunio T, Shintaro S, Masato U, Hironao H, Toshi N, et al. Immunogenicity of latency-associated antigens of Mycobacterium tuberculosis in DNA-vaccinated mice. Vaccinology (2010) 3:19–26. doi:10.1016/j.provac.2010.11.004
171. Bashir N, Kounsar F, Mukhopadhyay S, Hasnain SE. Mycobacterium tuberculosis conserved hypothetical protein rRv2626c modulates macrophage effector functions. Immunology (2010) 130:34–45. doi:10.1111/j.1365-2567.2009.03196.x
172. Bretl DJ, He H, Demetriadou C, White MJ, Penoske RM, Salzman NH, et al. MprA and DosR coregulate a Mycobacterium tuberculosis virulence operon encoding Rv1813c and Rv1812c. Infect Immun (2012) 80:3018–33. doi:10.1128/IAI.00520-12
173. Mehra S, Foreman TW, Didier PJ, Ahsan MH, Hudock TA, Kissee R, et al. The DosR regulon modulates adaptive immunity and is essential for Mycobacterium tuberculosis persistence. Am J Respir Crit Care Med (2015) 191:1185–96. doi:10.1164/rccm.201408-1502OC
174. Drumm JE, Mi K, Bilder P, Sun M, Lim J, Bielefeldt-Ohmann H, et al. Mycobacterium tuberculosis universal stress protein Rv2623 regulates bacillary growth by ATP-binding: requirement for establishing chronic persistent infection. PLoS Pathog (2009) 5:e1000460. doi:10.1371/journal.ppat.1000460
175. Jain RK, Nayak AR, Husain AA, Panchbhai MS, Chandak N, Purohit HJ, et al. Mycobacterial dormancy regulon protein Rv2623 as a novel biomarker for the diagnosis of latent and active tuberculous meningitis. Dis Markers (2013) 35:311–6. doi:10.1155/2013/309816
176. Chauhan S, Tyagi JS. Cooperative binding of phosphorylated DevR to upstream sites is necessary and sufficient for activation of the Rv3134c-devRS operon in Mycobacterium tuberculosis: implication in the induction of DevR target genes. J Bacteriol (2008) 190:4301–12. doi:10.1128/JB.01308-07
177. Siddiqui KF, Amir M, Gurram RK, Khan N, Arora A, Rajagopal K, et al. Latency-associated protein Acr1 impairs dendritic cell maturation and functionality: a possible mechanism of immune evasion by Mycobacterium tuberculosis. J Infect Dis (2014) 209:1436–45. doi:10.1093/infdis/jit595
178. Mushtaq K, Sheikh JA, Amir M, Khan N, Singh B, Agrewala JN. Rv2031c of Mycobacterium tuberculosis: a master regulator of Rv2028-Rv2031 (HspX) operon. Front Microbiol (2015) 6:351. doi:10.3389/fmicb.2015.00351
179. Izzo AA, Izzo LS, Kasimos J, Majka S. A matrix metalloproteinase inhibitor promotes granuloma formation during the early phase of Mycobacterium tuberculosis pulmonary infection. Tuberculosis (Edinb) (2004) 84:387–96. doi:10.1016/j.tube.2004.07.001
180. Zhang W, Jiang H, Bai YL, Kang J, Xu ZK, Wang LM. Construction and immunogenicity of the DNA vaccine of Mycobacterium tuberculosis dormancy antigen rv1733c. Scand J Immunol (2014) 79:292–8. doi:10.1111/sji.12160
181. Prabhavathi M, Pathakumari B, Raja A. IFN-gamma/TNF-alpha ratio in response to immuno proteomically identified human T-cell antigens of Mycobacterium tuberculosis – the most suitable surrogate biomarker for latent TB infection. J Infect (2015) 71:238–49. doi:10.1016/j.jinf.2015.04.032
182. Forero M, Puentes A, Cortes J, Castillo F, Vera R, Rodriguez LE, et al. Identifying putative Mycobacterium tuberculosis Rv2004c protein sequences that bind specifically to U937 macrophages and A549 epithelial cells. Protein Sci (2005) 14:2767–80. doi:10.1110/ps.051592505
183. Chen T, He L, Deng W, Xie J. The Mycobacterium DosR regulon structure and diversity revealed by comparative genomic analysis. J Cell Biochem (2013) 114:1–6. doi:10.1002/jcb.24302
184. He H, Bretl DJ, Penoske RM, Anderson DM, Zahrt TC. Components of the Rv0081-Rv0088 locus, which encodes a predicted formate hydrogenlyase complex, are coregulated by Rv0081, MprA, and DosR in Mycobacterium tuberculosis. J Bacteriol (2011) 193:5105–18. doi:10.1128/JB.05562-11
185. Kumar A, Lewin A, Rani PS, Qureshi IA, Devi S, Majid M, et al. Dormancy associated translation inhibitor (DATIN/Rv0079) of Mycobacterium tuberculosis interacts with TLR2 and induces proinflammatory cytokine expression. Cytokine (2013) 64:258–64. doi:10.1016/j.cyto.2013.06.310
186. Leyten EM, Lin MY, Franken KL, Friggen AH, Prins C, Van Meijgaarden KE, et al. Human T-cell responses to 25 novel antigens encoded by genes of the dormancy regulon of Mycobacterium tuberculosis. Microbes Infect (2006) 8:2052–60. doi:10.1016/j.micinf.2006.03.018
187. Garg R, Tripathi D, Kant S, Chandra H, Bhatnagar R, Banerjee N. The conserved hypothetical protein Rv0574c is required for cell wall integrity, stress tolerance, and virulence of Mycobacterium tuberculosis. Infect Immun (2015) 83:120–9. doi:10.1128/IAI.02274-14
188. Bunker RD, Mandal K, Bashiri G, Chaston JJ, Pentelute BL, Lott LS, et al. A functional role of Rv1738 in Mycobacterium tuberculosis persistence suggested by racemic protein crystallography. Proc Natl Acad Sci U S A (2015) 112:4310–15.
189. Kassa D, Ran L, Geberemeskel W, Tebeje M, Alemu A, Selase A, et al. Analysis of immune responses against a wide range of Mycobacterium tuberculosis antigens in patients with active pulmonary tuberculosis. Clin Vaccine Immunol (2012) 19:1907–15. doi:10.1128/CVI.00482-12
190. Peddireddy V, Doddam SN, Qureshi IA, Yerra P, Ahmed N. A putative nitroreductase from the DosR regulon of Mycobacterium tuberculosis induces pro-inflammatory cytokine expression via TLR2 signaling pathway. Sci Rep (2016) 6:24535. doi:10.1038/srep24535
191. Purkayastha A, Mccue LA, Mcdonough KA. Identification of a Mycobacterium tuberculosis putative classical nitroreductase gene whose expression is coregulated with that of the acr aene within macrophages, in standing versus shaking cultures, and under low oxygen conditions. Infect Immun (2002) 70:1518–29. doi:10.1128/IAI.70.3.1518-1529.2002
192. Essone PN, Chegou NN, Loxton AG, Stanley K, Kriel M, Van Der Spuy G, et al. Host cytokine responses induced after overnight stimulation with novel M. tuberculosis infection phase-dependent antigens show promise as diagnostic candidates for TB disease. PLoS One (2014) 9:e102584. doi:10.1371/journal.pone.0102584
193. Derrick SC, Yabe IM, Yang A, Kolibab K, Hollingsworth B, Kurtz SL, et al. Immunogenicity and protective efficacy of novel Mycobacterium tuberculosis antigens. Vaccine (2013) 31:4641–6. doi:10.1016/j.vaccine.2013.07.032
194. Honaker RW, Stewart A, Schittone S, Izzo A, Klein MR, Voskuil MI. Mycobacterium bovis BCG vaccine strains lack narK2 and narX induction and exhibit altered phenotypes during dormancy. Infect Immun (2008) 76:2587–93. doi:10.1128/IAI.01235-07
195. Hozumi H, Tsujimura K, Yamamura Y, Seto S, Uchijima M, Nagata T, et al. Immunogenicity of dormancy-related antigens in individuals infected with Mycobacterium tuberculosis in Japan. Int J Tuberc Lung Dis (2013) 17:818–24. doi:10.5588/ijtld.12.0695
196. Sirakova TD, Dubey VS, Deb C, Daniel J, Korotkova TA, Abomoelak B, et al. Identification of a diacylglycerol acyltransferase gene involved in accumulation of triacylglycerol in Mycobacterium tuberculosis under stress. Microbiology (2006) 152:2717–25. doi:10.1099/mic.0.28993-0
197. Mensah GI, Addo KK, Tetteh JA, Sowah S, Loescher T, Geldmacher C, et al. Cytokine response to selected MTB antigens in Ghanaian TB patients, before and at 2 weeks of anti-TB therapy is characterized by high expression of IFN-gamma and granzyme B and inter-individual variation. BMC Infect Dis (2014) 14:495. doi:10.1186/1471-2334-14-495
198. Goletti D, Butera O, Vanini V, Lauria FN, Lange C, Franken KL, et al. Response to Rv2628 latency antigen associates with cured tuberculosis and remote infection. Eur Respir J (2010) 36:135–42. doi:10.1183/09031936.00140009
199. Flores Valdez MA, Schoolnik GK. DosR-regulon genes induction in Mycobacterium bovis BCG under aerobic conditions. Tuberculosis (Edinb) (2010) 90:197–200. doi:10.1016/j.tube.2010.04.001
200. Leistikow RL, Morton RA, Bartek IL, Frimpong I, Wagner K, Voskuil MI. The Mycobacterium tuberculosis DosR regulon assists in metabolic homeostasis and enables rapid recovery from nonrespiring dormancy. J Bacteriol (2010) 192:1662–70. doi:10.1128/JB.00926-09
201. Malhotra V, Tyagi JS, Clark-Curtiss JE. DevR-mediated adaptive response in Mycobacterium tuberculosis H37Ra: links to asparagine metabolism. Tuberculosis (Edinb) (2009) 89:169–74. doi:10.1016/j.tube.2008.12.003
202. Minch K, Rustad T, Sherman DR. Mycobacterium tuberculosis growth following aerobic expression of the DosR regulon. PLoS One (2012) 7:e35935. doi:10.1371/journal.pone.0035935
203. Tekaia F, Gordon SV, Garnier T, Brosch R, Barrell BG, Cole ST. Analysis of the proteome of Mycobacterium tuberculosis in silico. Tuber Lung Dis (1999) 79:329–42. doi:10.1054/tuld.1999.0220
204. Zahrt TC, Deretic V. Mycobacterium tuberculosis signal transduction system required for persistent infections. Proc Natl Acad Sci U S A (2001) 98:12706–11. doi:10.1073/pnas.221272198
205. Bretl DJ, Demetriadou C, Zahrt TC. Adaptation to environmental stimuli within the host: two-component signal transduction systems of Mycobacterium tuberculosis. Microbiol Mol Biol Rev (2011) 75:566–82. doi:10.1128/MMBR.05004-11
206. Hess JF, Bourret RB, Simon MI. Histidine phosphorylation and phosphoryl group transfer in bacterial chemotaxis. Nature (1988) 336:139–43. doi:10.1038/336139a0
207. Weiss V, Magasanik B. Phosphorylation of nitrogen regulator I (NRI) of Escherichia coli. Proc Natl Acad Sci U S A (1988) 85:8919–23. doi:10.1073/pnas.85.23.8919
208. Stock JB, Ninfa AJ, Stock AM. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol Rev (1989) 53:450–90.
209. Gao R, Mack TR, Stock AM. Bacterial response regulators: versatile regulatory strategies from common domains. Trends Biochem Sci (2007) 32:225–34. doi:10.1016/j.tibs.2007.03.002
210. Glover RT, Kriakov J, Garforth SJ, Baughn AD, Jacobs WR Jr. The two-component regulatory system senX3-regX3 regulates phosphate-dependent gene expression in Mycobacterium smegmatis. J Bacteriol (2007) 189:5495–503. doi:10.1128/JB.00190-07
211. Gebhard S, Cook GM. Differential regulation of high-affinity phosphate transport systems of Mycobacterium smegmatis: identification of PhnF, a repressor of the phnDCE operon. J Bacteriol (2008) 190:1335–43. doi:10.1128/JB.01764-07
212. Himpens S, Locht C, Supply P. Molecular characterization of the mycobacterial SenX3-RegX3 two-component system: evidence for autoregulation. Microbiology (2000) 146(Pt 12):3091–8. doi:10.1099/00221287-146-12-3091
213. Walters SB, Dubnau E, Kolesnikova I, Laval F, Daffe M, Smith I. The Mycobacterium tuberculosis PhoPR two-component system regulates genes essential for virulence and complex lipid biosynthesis. Mol Microbiol (2006) 60:312–30. doi:10.1111/j.1365-2958.2006.05102.x
214. Gonzalo-Asensio J, Mostowy S, Harders-Westerveen J, Huygen K, Hernandez-Pando R, Thole J, et al. PhoP: a missing piece in the intricate puzzle of Mycobacterium tuberculosis virulence. PLoS One (2008) 3:e3496. doi:10.1371/journal.pone.0003496
215. Wang S, Engohang-Ndong J, Smith I. Structure of the DNA-binding domain of the response regulator PhoP from Mycobacterium tuberculosis. Biochemistry (2007) 46:14751–61. doi:10.1021/bi700970a
216. Stewart V. Nitrate regulation of anaerobic respiratory gene expression in Escherichia coli. Mol Microbiol (1993) 9:425–34. doi:10.1111/j.1365-2958.1993.tb01704.x
217. He H, Hovey R, Kane J, Singh V, Zahrt TC. MprAB is a stress-responsive two-component system that directly regulates expression of sigma factors SigB and SigE in Mycobacterium tuberculosis. J Bacteriol (2006) 188:2134–43. doi:10.1128/JB.188.6.2134-2143.2006
218. Pang X, Vu P, Byrd TF, Ghanny S, Soteropoulos P, Mukamolova GV, et al. Evidence for complex interactions of stress-associated regulons in an mprAB deletion mutant of Mycobacterium tuberculosis. Microbiology (2007) 153:1229–42. doi:10.1099/mic.0.29281-0
219. He H, Zahrt TC. Identification and characterization of a regulatory sequence recognized by Mycobacterium tuberculosis persistence regulator MprA. J Bacteriol (2005) 187:202–12. doi:10.1128/JB.187.1.202-212.2005
220. White MJ, He H, Penoske RM, Twining SS, Zahrt TC. PepD participates in the mycobacterial stress response mediated through MprAB and SigE. J Bacteriol (2010) 192:1498–510. doi:10.1128/JB.01167-09
221. Wernisch L, Kendall SL, Soneji S, Wietzorrek A, Parish T, Hinds J, et al. Analysis of whole-genome microarray replicates using mixed models. Bioinformatics (2003) 19:53–61. doi:10.1093/bioinformatics/19.1.53
222. Li Y, Zeng J, Zhang H, He ZG. The characterization of conserved binding motifs and potential target genes for M. tuberculosis MtrAB reveals a link between the two-component system and the drug resistance of M. smegmatis. BMC Microbiol (2010) 10:242. doi:10.1186/1471-2180-10-242
223. Rajagopalan M, Dziedzic R, Al Zayer M, Stankowska D, Ouimet MC, Bastedo DP, et al. Mycobacterium tuberculosis origin of replication and the promoter for immunodominant secreted antigen 85B are the targets of MtrA, the essential response regulator. J Biol Chem (2010) 285:15816–27. doi:10.1074/jbc.M109.040097
224. Morth JP, Feng V, Perry LJ, Svergun DI, Tucker PA. The crystal and solution structure of a putative transcriptional antiterminator from Mycobacterium tuberculosis. Structure (2004) 12:1595–605. doi:10.1016/j.str.2004.06.018
225. Kesavan AK, Brooks M, Tufariello J, Chan J, Manabe YC. Tuberculosis genes expressed during persistence and reactivation in the resistant rabbit model. Tuberculosis (Edinb) (2009) 89:17–21. doi:10.1016/j.tube.2008.08.004
226. Kana BD, Mizrahi V. Resuscitation-promoting factors as lytic enzymes for bacterial growth and signaling. FEMS Immunol Med Microbiol (2010) 58:39–50. doi:10.1111/j.1574-695X.2009.00606.x
227. Biketov S, Potapov V, Ganina E, Downing K, Kana BD, Kaprelyants A. The role of resuscitation promoting factors in pathogenesis and reactivation of Mycobacterium tuberculosis during intra-peritoneal infection in mice. BMC Infect Dis (2007) 7:146. doi:10.1186/1471-2334-7-146
228. Russell-Goldman E, Xu J, Wang X, Chan J, Tufariello JM. A Mycobacterium tuberculosis Rpf double-knockout strain exhibits profound defects in reactivation from chronic tuberculosis and innate immunity phenotypes. Infect Immun (2008) 76:4269–81. doi:10.1128/IAI.01735-07
229. Hett EC, Chao MC, Rubin EJ. Interaction and modulation of two antagonistic cell wall enzymes of mycobacteria. PLoS Pathog (2010) 6:e1001020. doi:10.1371/journal.ppat.1001020
230. Shleeva M, Goncharenko A, Kudykina Y, Young D, Young M, Kaprelyants A. Cyclic AMP-dependent resuscitation of dormant mycobacteria by exogenous free fatty acids. PLoS One (2013) 8:e82914. doi:10.1371/journal.pone.0082914
231. Kondratieva T, Azhikina T, Nikonenko B, Kaprelyants A, Apt A. Latent tuberculosis infection: what we know about its genetic control? Tuberculosis (Edinb) (2014) 94:462–8. doi:10.1016/j.tube.2014.06.009
232. Arnvig KB, Comas I, Thomson NR, Houghton J, Boshoff HI, Croucher NJ, et al. Sequence-based analysis uncovers an abundance of non-coding RNA in the total transcriptome of Mycobacterium tuberculosis. PLoS Pathog (2011) 7:e1002342. doi:10.1371/journal.ppat.1002342
233. McGillivray A, Golden NA, Kaushal D. The Mycobacterium tuberculosis Clp gene regulator is required for in vitro reactivation from hypoxia-induced dormancy. J Biol Chem (2015) 290:2351–67. doi:10.1074/jbc.M114.615534
234. Cambier CJ, Falkow S, Ramakrishnan L. Host evasion and exploitation schemes of Mycobacterium tuberculosis. Cell (2014) 159:1497–509. doi:10.1016/j.cell.2014.11.024
235. Bekker LG, Wood R. The changing natural history of tuberculosis and HIV coinfection in an urban area of hyperendemicity. Clin Infect Dis (2010) 50(Suppl 3):S208–14. doi:10.1086/651493
236. Huang CC, Tchetgen ET, Becerra MC, Cohen T, Hughes KC, Zhang Z, et al. The effect of HIV-related immunosuppression on the risk of tuberculosis transmission to household contacts. Clin Infect Dis (2014) 58:765–74. doi:10.1093/cid/cit948
237. Pawlowski A, Jansson M, Skold M, Rottenberg ME, Kallenius G. Tuberculosis and HIV co-infection. PLoS Pathog (2012) 8:e1002464. doi:10.1371/journal.ppat.1002464
238. Flynn JL, Chan J. Tuberculosis: latency and reactivation. Infect Immun (2001) 69:4195–201. doi:10.1128/IAI.69.7.4195-4201.2001
239. Diedrich CR, Mattila JT, Klein E, Janssen C, Phuah J, Sturgeon TJ, et al. Reactivation of latent tuberculosis in cynomolgus macaques infected with SIV is associated with early peripheral T cell depletion and not virus load. PLoS One (2010) 5:e9611. doi:10.1371/journal.pone.0009611
240. Shen Y, Shen L, Sehgal P, Huang D, Qiu L, Du G, et al. Clinical latency and reactivation of AIDS-related mycobacterial infections. J Virol (2004) 78:14023–32. doi:10.1128/JVI.78.24.14023-14032.2004
241. Bernut A, Herrmann JL, Kissa K, Dubremetz JF, Gaillard JL, Lutfalla G, et al. Mycobacterium abscessus cording prevents phagocytosis and promotes abscess formation. Proc Natl Acad Sci U S A (2014) 111:E943–52. doi:10.1073/pnas.1321390111
242. Keane J, Gershon S, Wise RP, Mirabile-Levens E, Kasznica J, Schwieterman WD, et al. Tuberculosis associated with infliximab, a tumor necrosis factor alpha-neutralizing agent. N Engl J Med (2001) 345:1098–104. doi:10.1056/NEJMoa011110
243. Nunez Martinez O, Ripoll Noiseux C, Carneros Martin JA, Gonzalez Lara V, Gregorio Maranon HG. Reactivation tuberculosis in a patient with anti-TNF-alpha treatment. Am J Gastroenterol (2001) 96:1665–6. doi:10.1016/S0002-9270(01)02399-1
244. Arkema EV, Jonsson J, Baecklund E, Bruchfeld J, Feltelius N, Askling J, et al. Are patients with rheumatoid arthritis still at an increased risk of tuberculosis and what is the role of biological treatments? Ann Rheum Dis (2015) 74:1212–7. doi:10.1136/annrheumdis-2013-204960
245. Champsi J, Young LS, Bermudez LE. Production of TNF-alpha, IL-6 and TGF-beta, and expression of receptors for TNF-alpha and IL-6, during murine Mycobacterium avium infection. Immunology (1995) 84:549–54.
246. Bean AG, Roach DR, Briscoe H, France MP, Korner H, Sedgwick JD, et al. Structural deficiencies in granuloma formation in TNF gene-targeted mice underlie the heightened susceptibility to aerosol Mycobacterium tuberculosis infection, which is not compensated for by lymphotoxin. J Immunol (1999) 162:3504–11.
247. Harris J, Keane J. How tumour necrosis factor blockers interfere with tuberculosis immunity. Clin Exp Immunol (2010) 161:1–9. doi:10.1111/j.1365-2249.2010.04146.x
248. Munoz L, Casas S, Juanola X, Bordas X, Martinez C, Santin M, et al. Prevention of anti-tumor necrosis factor-associated tuberculosis: a 10-year longitudinal cohort study. Clin Infect Dis (2015) 60:349–56. doi:10.1093/cid/ciu796
249. Fox W, Ellard GA, Mitchison DA. Studies on the treatment of tuberculosis undertaken by the British Medical Research Council tuberculosis units, 1946-1986, with relevant subsequent publications. Int J Tuberc Lung Dis (1999) 3:S231–79.
250. Shehzad A, Rehman G, Ul-Islam M, Khattak WA, Lee YS. Challenges in the development of drugs for the treatment of tuberculosis. Braz J Infect Dis (2013) 17:74–81. doi:10.1016/j.bjid.2012.10.009
251. Beham AW, Puellmann K, Laird R, Fuchs T, Streich R, Breysach C, et al. A TNF-regulated recombinatorial macrophage immune receptor implicated in granuloma formation in tuberculosis. PLoS Pathog (2011) 7:e1002375. doi:10.1371/journal.ppat.1002375
Keywords: Mycobacterium tuberculosis, dormancy, DosR regulon, granuloma, antibiotic resistance, alveolar macrophages, latency
Citation: Peddireddy V, Doddam SN and Ahmed N (2017) Mycobacterial Dormancy Systems and Host Responses in Tuberculosis. Front. Immunol. 8:84. doi: 10.3389/fimmu.2017.00084
Received: 28 October 2016; Accepted: 18 January 2017;
Published: 15 February 2017
Edited by:
Raj Wadgaonkar, SUNY Downstate Medical Center, USAReviewed by:
James Harris, Monash University, AustraliaGiovanni Delogu, Università Cattolica del Sacro Cuore, Italy
Javed Naim Agrewala, Institute of Microbial Technology (CSIR), India
Copyright: © 2017 Peddireddy, Doddam and Ahmed. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Niyaz Ahmed, YWhtZWQubml6aUBnbWFpbC5jb20=, bml5YXouYWhtZWRAaWNkZHJiLm9yZw==
†These authors have contributed equally to this work.
 Vidyullatha Peddireddy
Vidyullatha Peddireddy Sankara Narayana Doddam
Sankara Narayana Doddam Niyaz Ahmed
Niyaz Ahmed