- 1Jewish General Hospital, Lady Davis Institute, McGill University, Montreal, QC, Canada
- 2Division of Experimental Medicine, McGill University, Montreal, QC, Canada
- 3Department of Microbiology and Immunology, McGill University, Montreal, QC, Canada
Innate immunity is the first line of defense against invading pathogens. Rapid and efficient detection of pathogen-associated molecular patterns via pattern-recognition receptors is essential for the host to mount defensive and protective responses. Retinoic acid-inducible gene-I (RIG-I) is critical in triggering antiviral and inflammatory responses for the control of viral replication in response to cytoplasmic virus-specific RNA structures. Upon viral RNA recognition, RIG-I recruits the mitochondrial adaptor protein mitochondrial antiviral signaling protein, which leads to a signaling cascade that coordinates the induction of type I interferons (IFNs), as well as a large variety of antiviral interferon-stimulated genes. The RIG-I activation is tightly regulated via various posttranslational modifications for the prevention of aberrant innate immune signaling. By contrast, viruses have evolved mechanisms of evasion, such as sequestrating viral structures from RIG-I detections and targeting receptor or signaling molecules for degradation. These virus–host interactions have broadened our understanding of viral pathogenesis and provided insights into the function of the RIG-I pathway. In this review, we summarize the recent advances regarding RIG-I pathogen recognition and signaling transduction, cell-intrinsic control of RIG-I activation, and the viral antagonism of RIG-I signaling.
Introduction
Eukaryotic organisms rely on the host innate immune system to defend against viruses or other pathogenic microbes in early phases of infection. The innate antiviral immune response starts with the detection of evolutionarily conserved structures, termed pathogen-associated molecular patterns (PAMPs), by a set of germline-encoded pattern-recognition receptors (PRRs). With respect to their cellular localization, ligand specificity, and functions, PRRs are categorized into distinct families including the toll-like receptors, nucleotide-binding oligomerization domain-like receptors, C-type lectin receptors, retinoic acid-inducible gene-I (RIG-I)-like receptors (RLRs) (1–5), as well as cytosolic viral DNA sensors such as cyclic GMP-AMP synthase (6, 7). Following the detection of specific viral PAMPs, PRRs trigger the activation of intracellular signaling cascades, ultimately leading to the production of type I interferons (IFNs), as well as pro-inflammatory cytokines. Secreted IFNs are crucial for the induction of numerous interferon-stimulated genes (ISGs); the products of which are major forces in controlling and restricting viral infections, thereby establishing a cellular antiviral state as well as helping to shape the adaptive immune response (8). Recent studies showed that viruses have evolved complex strategies to affect multiple stages of the host antiviral defense, from inhibiting the viral detection to manipulating components of the signaling pathways (9, 10). To ensure successful antiviral defenses and to avoid aberrant or dysregulation of host immune signaling, antiviral pathways need to be tightly regulated at each level. In this review, we will summarize the cell-intrinsic regulation of RIG-I receptor activity, as well as the viral strategies to subvert the RIG-I signaling machinery.
RIG-I Structure and Ligand Interactions
The three members of the RLR family: RIG-I, MDA5 (melanoma differentiation factor 5), and LGP2 (laboratory of genetics and physiology 2) are expressed in most cell and tissue types. They function as cytoplasmic sensors for the recognition of a variety of RNA viruses and subsequent activation of downstream signaling to drive type I IFN production and antiviral gene expressions. These three RLR proteins are RNA-dependent ATPases belonging to the DExD/H-box family of helicases (11). Structurally, RLRs have a similar central helicase core that is comprised of two helicase domains, Hel1 and Hel2 with an insertion termed Hel2i. In addition, they all have a C-terminal domain (CTD). However, only RIG-I and MDA5 contain two N-terminal caspase activation and recruitment domains (CARDs) (3) (Figure 1A). Among these three, RIG-I is the founding member and hence the most intensively studied member of this family. Each domain of RIG-I plays unique roles during RIG-I autorepression and activation. In brief, the CTD and helicase domain are involved in RNA ligand binding and ATP hydrolysis-involved conformational changes (12–14), whereas the RIG-I CARDs facilitate interaction with other downstream CARD containing molecules (15).
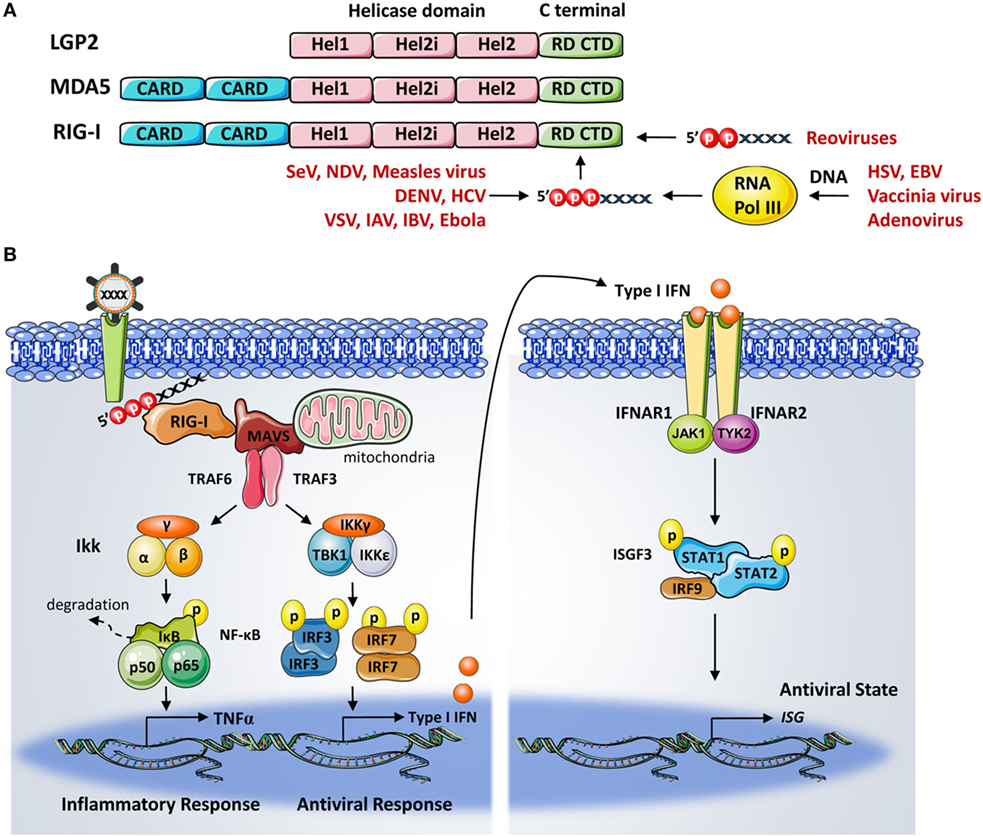
Figure 1. (A) Domain structure of retinoic acid-inducible gene-I (RIG-I). RIG-I belongs to the DExD/H-box family of helicases and is comprised of helicase domains 1 (Hel1) and 2 (Hel2) with a Hel2i insertion, N-terminal caspase activation and recruitment domains, and a C-terminal domain (CTD) or repressor domain. RIG-I CTD is responsible for recognizing a plethora of RNA viruses with short 5′ triphosphate (5′ppp) RNA and 5′-diphosphate-bearing RNA structures. RIG-I also detects 5′ppp RNA species synthesized through the transcription of viral DNA by RNA polymerase III. (B) The cytoplasmic pattern-recognition receptor RIG-I is essential for recognizing RNA viruses with a 5′ppp signature. Upon viral RNA recognition, RIG-I recruits the adaptor protein mitochondrial antiviral signaling protein to activate the TBK1–IKKϵ complex and IKKα–IKKβ complex, which are responsible for the activation of transcription factors interferon regulator factor (IRF) 3, IRF7, and nuclear factor-κB. These transcription factors then translocate to the nucleus and coordinate the induction of type I interferons (IFNs). This is followed by the binding of the IFNs α and β to their cognate receptor, which will lead to the transcriptional activation of interferon-stimulated genes (ISGs) by the JAK/STAT signaling pathway. The products of ISGs are key factors in limiting pathogen spreading.
Retinoic acid-inducible gene-I has been shown to be involved in the recognition of a variety of RNA viruses in the cytoplasm, such as the Sendai virus, influenza A and B viruses (IAV, IBV), vesicular stomatitis virus, measles virus (MV), Newcastle disease virus, Ebola virus (EBOV), dengue virus (DENV), and hepatitis C virus (HCV) (16–19). The short double-stranded (ds) RNA with a triphosphate (ppp) motif at the 5′-end, as found in these viral genomes, were shown to be a key signature recognized by RIG-I (20, 21). The 5′ppp dsRNA of viral nucleocapsids has also been characterized as stimulating RIG-I (22). 5′-Diphosphate-bearing RNA (5′ppRNA), either naturally contained in viruses, produced by in vitro transcription, or via chemical synthesis, were all shown to bind to RIG-I and were sufficient to activate RIG-I (20, 23). Physiologically, the control of in vitro and in vivo infections of reoviruses, which bear the 5′ppRNA genome, relies on RIG-I functionality (24). It is worth noting that the in vitro-synthesized 5′pppRNA sequences also trigger RIG-I activation (25). These agonists have demonstrated their therapeutic potential as broad-spectrum antiviral agents and could be optimized as vaccine adjuvant candidates (26–30). Furthermore, the recognition of several DNA viruses, including herpes simplex virus type 1 (HSV-1), Epstein–Barr virus (EBV), vaccinia virus (VACV), and adenovirus, via the RNA polymerase III were found to be RIG-I-dependent (31, 32). Interestingly, the RIG-I-mediated upregulation of STING is required for protection against the HSV-1 by the RIG-I agonist, offering new evidence of the overlapping between RIG-I signaling and the host response to DNA viral infection (33). Notably, viral RNA triggered RIG-I signaling also mediates the inflammatory response via distinct pathways. The first involves the formation of the RIG-I inflammasome through interactions between RIG-I, ASC, and caspase-1 and the stimulation of IL-1β release. The second involves the adaptor proteins CARD9, Bcl-10, mitochondrial antiviral signaling protein (MAVS), and the activation of nuclear factor-κB (NF-κB) (34, 35). Upon RNA ligand binding, RIG-I undergoes a series of conformational changes and posttranslational modifications (PTMs) to achieve full activation (further detail below).
RIG-I Signaling Transduction
Activated RIG-I recruits its downstream adaptor molecule MAVS (also known as IPS-1, CARDIF, and VISA) through CARD–CARD-mediated interactions (36, 37). The oligomeric RIG-I CARD assembly and the polymeric formation of MAVS, together serve as a signaling platform for protein complexes that mediate the bifurcation of signaling into two branches. One branch recruits tumor necrosis factor receptor-associated factors (TRAF)-2/6 and the receptor-interacting protein 1 to subsequently activate the IKK complex, resulting in NF-κB activation (38). The other branch signals through TRAF3 and activates the TANK/IKKγ/IKKϵ/TBK1 complex, leading to the phosphorylation and dimerization of interferon regulator factors (IRF)-3 and -7 (39, 40). Activated IRF3/7 and NF-κB then translocate to the nucleus, together with ATF2, c-Jun, and the transcription coactivator CREB-binding protein/p300, to coordinate the IFN and pro-inflammatory gene expressions (41). Once secreted, IFNs bind to specific cell surface receptors and activate the JAK–STAT pathway. The activated transcription factors STAT1, STAT2, and IRF9 form the interferon-stimulated gene factors (ISGF3) complex. ISGF3 then translocates to the nucleus and coordinates the transcription of hundreds of ISGs including RIG-I, thus generating an amplifying loop leading to the accumulation of RIG-I during several types of infections (8) (Figure 1B).
Mechanisms of RIG-I Activation
RIG-I Autorepression
Structural and biochemical studies have demonstrated that the activation of RIG-I is a multi-step process and is primarily regulated by conformational changes and PTMs. When initially identified as a dsRNA sensor, it was hypothesized that RIG-I was under negative regulation in physiological conditions. The over expression of the CARD domain of RIG-I alone demonstrated superior signaling activity than full length RIG-I in absence of viral PAMPs (2). Studies by Saito et al. showed that the deletion of CARD was dominant-negative for RIG-I signaling. By contrast, the deletion of repressor domain (RD) resulted in constitutive signaling, whereas RD expression alone ablated RIG-I signaling actions. Together, these findings provided the model of RIG-I autoregulation in which the RD is predicted to mask CARDs for signaling transduction in uninfected cells (42). The crystal structural analysis further delineated the models of autorepressed and ligand activated states of RIG-I, respectively. In a ligand-free state, CARDs and Hel2i interactions hinder dsRNA binding and inactivate RIG-I (14). The binding of 5′ppp dsRNA to RD leads to a conformational switch of RIG-I, which releases the autorepressed CARDs and exposes the helicase domain for ATP binding (14, 43). ATP hydrolysis is essential for RIG-I signaling. It enables RIG-I to translocate along the dsRNA, and further promotes the oligomerization of RIG-I CARDs. These processes assemble RIG-I into a filamentous architecture which facilitates the CARD–CARD interactions with the mitochondrial MAVS, leading to the subsequent signaling transduction for IFN production (44, 45). Importantly, RIG-I ATPase activity also plays a role in distinguishing self-RNA from non-self-RNA (46). It was reported that RIG-I ATP hydrolysis increases the binding affinity of RIG-I and dsRNA ligands; whereas the RIG-I mutants deficient in ATP hydrolysis promotes the interaction of RIG-I and self-dsRNA and results in unintentional immune signaling (47).
Posttranslational Control of RIG-I
Ubiquitination
One of the first PTMs of RIG-I following the initial ligand recognition is performed by the robust ubiquitination machinery (Figure 2). Mass spectrometry analysis revealed that TRIM25, a member of the tripartite motif (TRIM) protein family possessing E3 ligase activity, induces the covalent Lys63-linked ubiquitination of RIG-I. Mechanistically, the C-terminal SPRY domain of TRIM25 interacts with CARD1 and facilitates the ubiquitination of CARD2 at K172 (48). The RIG-I–TRIM25 ubiquitination complex, associates with the adaptor protein 14-3-3ϵ and translocates to mitochondria for MAVS binding (49). Mutation of K172 disrupts the interaction between RIG-I and MAVS thus abrogating downstream signaling and IFNs production (50). Furthermore, a RIG-I splice variant which lacks the TRIM25 interaction domain acts as a feedback inhibitor of RIG-I signaling transduction upon viral infections (48). In addition, Riplet (RING-finger protein leading to RIG-I activation, also named RNF135 or REUL), another E3 ubiquitin ligase, also promotes RIG-I ubiquitination. Multiple sites within the CARDs, as well as within the CTD of RIG-I, were identified as the crucial ubiquitin anchoring residues (51–53). Among which, K63-linked polyubiquitination (pUb) at Lys788, is demonstrated as being critical for RIG-I activation. However, unlike TRIM25-induced ubiquitination, Riplet induced RIG-I pUb is dispensable for RIG-I-RNA binding but is essential for releasing CARD from its autorepressed state. This enhances TRIM25 functionality as well as promoting the oligomerization of RIG-I and the activation of MAVS (54). MEX3C (Mex-3 RNA binding family member C), another recently identified E3 ligase, also mediates Lys63-Ub at K99 and K169 of CARD, playing a critical role in RIG-I activation (55). In addition, the oligoadenylate synthetases-like (OASL) protein, although not an E3 ubiquitin ligase itself, contains a dsRNA-binding groove and enhances RIG-I activation by mimicking the K63-linked pUb through its ubiquitin-like (UBL) domain (56, 57). Non-covalent binding of K63-ubiquitin chains to CARDs also potently activates RIG-I (58). Recent structural analysis suggests that covalent and non-covalent binding of ubiquitin synergistically stabilize RIG-I tetramerization and enhance polymerization of MAVS CARDs (59).
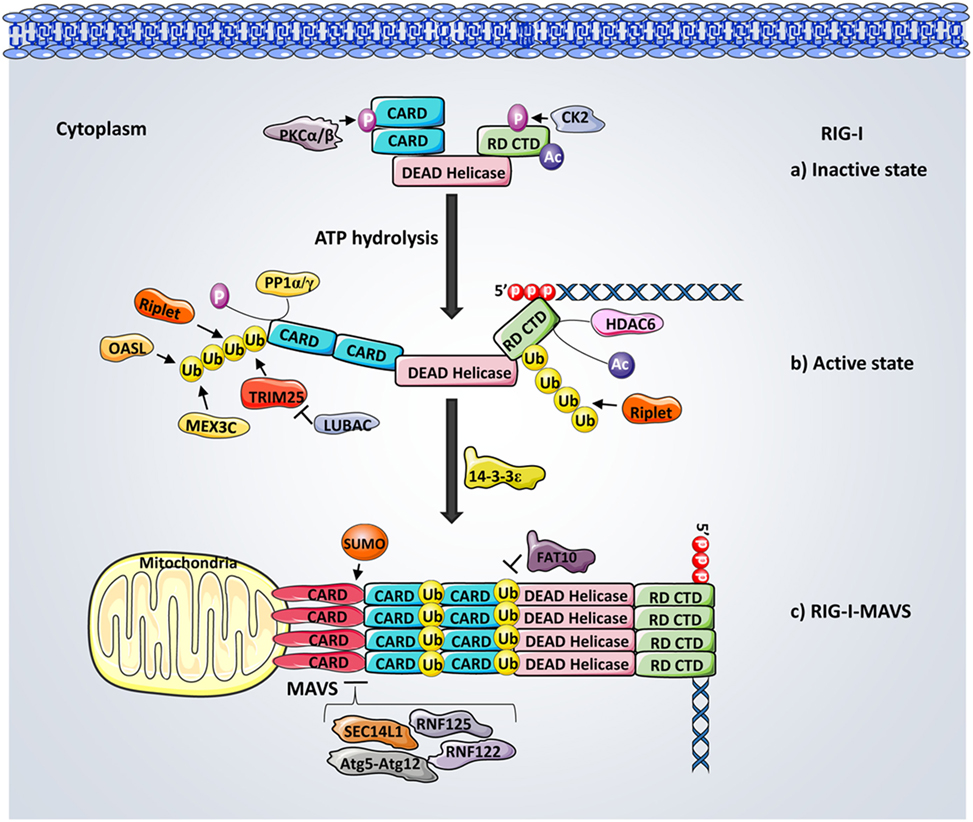
Figure 2. Regulation of retinoic acid-inducible gene-I (RIG-I) activation. (a) In resting cells, RIG-I is kept inactivated through the phosphorylation of caspase activation and recruitment domains (CARDs) and C-terminal domain (CTD) mediated by casein kinase II and protein kinase C-α/β, respectively. (b) Following the binding of 5′ triphosphate (5′ppp) RNA and ATP hydrolysis, RIG-I is dephosphorylated by phosphoprotein phosphatase 1-α/γ and results in a conformational change that opens CARDs. HDAC6-mediated deacetylation of RIG-I CTD is critical for RIG-I and 5′pppRNA binding. The Lys63-linked ubiquitination of RIG-I mediated by TRIM25, Riplet, oligoadenylate synthetases-like protein, and MEX3C at both CARDs and CTD further activate RIG-I and facilitate its tetramerization. (c) Interactions between RIG-I–TRIM25 complex and 14-3-3ϵ promote RIG-I translocation to mitochondrial mitochondrial antiviral signaling protein (MAVS) for downstream signaling, leading to interferon production. Interactions between TRIM25, RIG-I, and MAVS are further negatively regulated by the Lys48-linked ubiquitination, which is meditated by LUBAC, RNF125, and RNF122. SEC14L1 and Atg5–Atg12 both inhibit the signaling by interrupting RIG-I–MAVS interactions, whereas SUMOylation promotes RIG-I–MAVS binding.
On the other hand, several deubiquitinating enzymes (DUBs) were identified to remove K63-linked pUb chains from RIG-I, thus dampening RIG-I signaling. The tumor suppressor protein cylindromatosis (CYLD) removes K63-linked pUb chains from RIG-I as well as TBK1 and IKKϵ to inhibit the IRF3 response, serving as a pathway negative regulator (60). Syndecan-4, a newly identified negative regulator of RIG-I, functions through attracting CYLD to RIG-I complex, thus potentiating the K63-mediated deubiquitination of RIG-I (61). In addition, the ubiquitin-specific protease (USP) family members, such as USP3 and USP21, were also identified as inhibitors of RIG-I activation by deubiqutinating RIG-I (62, 63).
In contrast to K63-linked ubiquitination, which promotes protein activation, K48-linked ubiquitination triggers proteasomal degradation of its target. For instance, the RING-finger protein 125 (RNF125), together with the ubiquitin E2 ligase UbcH5, conjugate K48-linked ubiquitin to RIG-I and MAVS, targeting them for proteasomal degradation and thereby inhibiting downstream signaling (64). Similarly, RNF122 was recently demonstrated to mediate the proteasomal degradation of RIG-I by delivering the K48-linked ubiquitin to RIG-I CARDs (65). The linear ubiquitin assembly complex (LUBAC) has been shown to promote K48 pUb of TRIM25, leading to its degradation (66). Conversely, the deubiquitinase USP15 antagonizes LUBAC by removing K48-linked ubiquitin from TRIM25, leading to its stabilization and thereby promoting RIG-I-mediated antiviral signaling (67).
Phosphorylation
In parallel with ubiquitination, phosphorylation has emerged in the past several years as a critical regulator of the RIG-I signaling transduction (Figure 2). Protein purification and mass spectrometry analysis identified that phosphorylation of Thr170 in the CARDs antagonizes RIG-I signaling by inhibiting TRIM25-mediated Lys172 ubiquitination and MAVS binding (68). Ser8 phosphorylation of CARDs also serves as a negative regulator of RIG-I (69). In addition, the CTD of RIG-I is constitutively phosphorylated at Thr770 and Ser854/855 by casein kinase II to promote intermolecular interactions between CTD and CARDs, thereby maintaining RIG-I at an autorepressive state to prevent premature downstream signaling (70). A recent mass spectrometry analysis revealed that IKK phosphorylates RIG-I at Ser855, thereby providing a negative feedback regulation of RIG-I (71). Furthermore, conventional protein kinase C-α (PKC-α) and PKC-β have also been shown to phosphorylate CARDs, thus suppressing RIG-I–TRIM interaction and subsequent antiviral responses (72). In fact, RIG-I signaling activity is controlled by a dynamic balance between phosphorylation and dephosphorylation. Dephosphorylation of RIG-I occurs rapidly with the presence of viral RNA. A functional siRNA screen identified phosphoprotein phosphatase 1-α (PP1α) and PP1γ as essential phosphatases responsible for CARDs dephosphorylation at Ser8 and Thr170, leading to RIG-I signal activation and viral inhibition (73).
Acetylation
In addition to the ubiquitination and phosphorylation described above, acetylation modulation has recently started to gain more acknowledgment for controlling RIG-I activity (Figure 2). Mass spectrometry has identified the acetylation of two lysine residues (K858 and K909) in the CTD of RIG-I at its inactivate state and are deacetylated during viral infection (74). The mutation of these two sites restricts RIG-I from undergoing the virus-induced interaction with MAVS. K858 and K909 acetylation of RIG-I has also been shown to control the PAMP RNA-induced RIG-I oligomerization (75). The cytoplasmic deacetylase HDAC6-mediated removal of K909 acetylation has been shown as critical for RIG-I binding to dsRNA during viral infections (76). Furthermore, HDAC6-dependent RIG-I deacetylation also regulates RIG-I oligomerization upon ligand binding, thus facilitating RIG-I activation (75).
Other Regulatory Mechanisms
RIG-I signal transduction is further regulated by additional PTMs, regulatory proteins, and other cellular processes (Figure 2). It is worth noting that a number of UBL proteins including SUMO, ISG15, FAT10, and Atg8–Atg12 are involved in these positive or negative regulatory mechanisms (77). SUMOylation serves as a positive regulator of RIG-I by enhancing the RIG-I and MAVS binding (78). On the contrary, the HLA-F adjacent transcription 10 (FAT10), an UBL modifier protein, was shown to negatively regulate RIG-I by modulating RIG-I solubility through a non-covalent association with CARDs (79). In addition, IFN-induced ISG15 negatively regulates the RIG-I mediated signaling in a feedback-loop control manner (80). SEC14L1 has been observed competing with MAVS for RIG-I CARD binding (81). Furthermore, autophagy has been reported to be involved in RIG-I modulation through its key regulator, the Atg5–Atg12 conjugate. Atg5–Atg12 has been found to suppress RIG-I–MAVS interaction, thereby inhibiting downstream signaling (82). Recently, deamidation of CTD has been described as a distinct means to induce RIG-I activation. For examples, vGAT (glutamine amidotransferase), from KSHV (kaposi’s sarcoma-associated herpesvirus) and γHV68 (murine gamma herpesvirus 68), recruits cellular phosphoribosylformyglycinamide synthase to deamindate and activate RIG-I (83, 84).
Viral Antagonism of RIG-I Signaling
In order to establish infections, viruses have developed sophisticated mechanisms to counteract host immune responses. With regard to RIG-I signaling, these include mechanisms such as altering viral genomes and their intermediate transcripts to avoid detection, manipulating the activation and degradation of RIG-I and MAVS, as well as modulating downstream signaling cascades. Studying these antagonistic viral strategies has greatly broadened our understanding of RIG-I activation and regulation.
Sequestration of Viral RNAs
Since 5′ triphosphate (5′ppp) is an important feature recognized by RIG-I, modification of this motif has long been described as one of the major mechanisms for viruses to antagonize RIG-I signaling. Crimean–Congo hemorrhagic fever virus, Borna disease virus (BDV), and hantavirus (HTNV) remove the 5′ppp group on their genome posttranscriptionally, make RIG-I unable to bind to viral RNA, and therefore incapable of triggering RIG-I activation (85). Mechanistically, HTNV uses the “prime and realign” strategy to generate a 5′-terminal monophosphorylate (86, 87). BDV on the other hand, employs genome trimming to form a 3′-terminal overhang as well as convert 5′ppp to 5′p to avoid detection by RIG-I (88). The arenavirus presents an unpaired 5′ppp-nucleotide overhang to evade recognition by RIG-I (89). The 5′-end of viral RNA can also be modified through RNA-capping pathways. For example, the genomic RNA of polioviruses linked to Vpg (viral protein genome-linked) to cap the 5′-end from exposure to RIG-I (90). The 5′-end capping with 7-methyl guanosine and methylation of 5′ppp dsRNA at the 2′-O position makes viral RNA non-distinguishable from the host mRNAs, and therefore does not stimulate RIG-I (91, 92).
By contrast, some viruses encode viral proteins to prevent RNA recognition. The EBOV utilizes its VP35 protein to sequester viral RNA (18). The crystal structural analysis indicates that the VP35 interferon inhibitory domain competes with RIG-I for dsRNA binding by forming an “end-cap” complex with dsRNA, resulting in substantially diminished activation of RIG-I (93). Similarly, the marburg virus VP35 spirals around the dsRNA backbone and end-caps the dsRNA to escape from RIG-I detection (94, 95). The IAV non-structural protein 1 (NS1) possesses dsRNA-binding properties to shield viral RNA from RIG-I (96). IAV has also been shown to antagonize RIG-I activation via its viral polymerase subunit PB2. PB2 position 627K in the mammalian strain increases PB2-nucleocapids binding affinity, thus inhibiting RIG-I interaction with the nucleoprotein-encapsidated 5′ppp RNA (22, 97).
In addition to altering and concealing their genome to prevent RNA binding, viruses also re-localize viral RNA to specific cellular compartments, such as mitochondria, endoplasmic reticulum (ER), and Golgi, to avoid cytosolic surveillance by RIG-I. For instance, the DENV conceals dsRNA in the intracellular membrane as an escape strategy (98). ER is an important organelle for viral entry, replication, and assembly. The severe acute respiratory syndrome (SARS) coronavirus (SARS-CoV) has been shown to induce a modified ER to hide its replicating RNA from detection (99). These viral antagonism strategies highlight the importance of cellular organelle localization in viral–host interactions during innate antiviral responses.
Manipulation of RIG-I–MAVS Signaling
Modulation of the PTMs
As reviewed above, ubiquitination represents one critical PTM mechanism of RIG-I activation and, not surprisingly, is an attractive target for viral manipulation (Figure 3A). Viruses have evolved ways to inhibit K63-linked ubiquitination of RIG-I by interacting with the E3 ligases TRIM25 and Riplet. For instance, IAV NS1 from various strains has been shown to suppress TRIM25-mediated RIG-I CARDs ubiquitination. Among all the TRIM25 binding amino acids identified in NS1, R38/K41 and E96/E97 were described as critical in interfering with the coil-coiled domain of TRIM25. These interactions resulted in an inhibition of TRIM25 multimerization and therefore blocked the RIG-I CARDs ubiquitination (100). Intriguingly, NS1-TRIM25 binding is found to be preserved in human and avian, but lost in mouse, indicating a species-specific manner of inhibition. This study further demonstrates that the NS1 suppression of RIG-I ubiquitination in mouse is Riplet-dependent (101). Conversely, phosphorylation of NS1 at Thr49 was recently identified as impairing the NS1–TRIM25 interaction, thereby suppressing its antagonistic activity of RIG-I signaling (102). Phosphorylation of another site on NS1, Thr80, has also been reported to disrupt NS1 binding affinity with RIG-I (103). Similar to IAV, the IBV non-structural NS protein (NS1-B) has recently been described as inhibiting RIG-I ubiquitination, which involves TRIM25-NS1 C-terminal effector domain interaction and the RIG-I/TRIM25/NS1-B complex formation (104). By contrast, the protease NS3-4A of HCV functions differently, rather than inhibiting TRIM25, it is thought to target the E3 ligase Riplet. NS3-4A directly disrupts Riplet, abolishes Riplet-mediated RIG-I ubiquitination, and further reduces the interaction between TRIM25 and RIG-I (54).
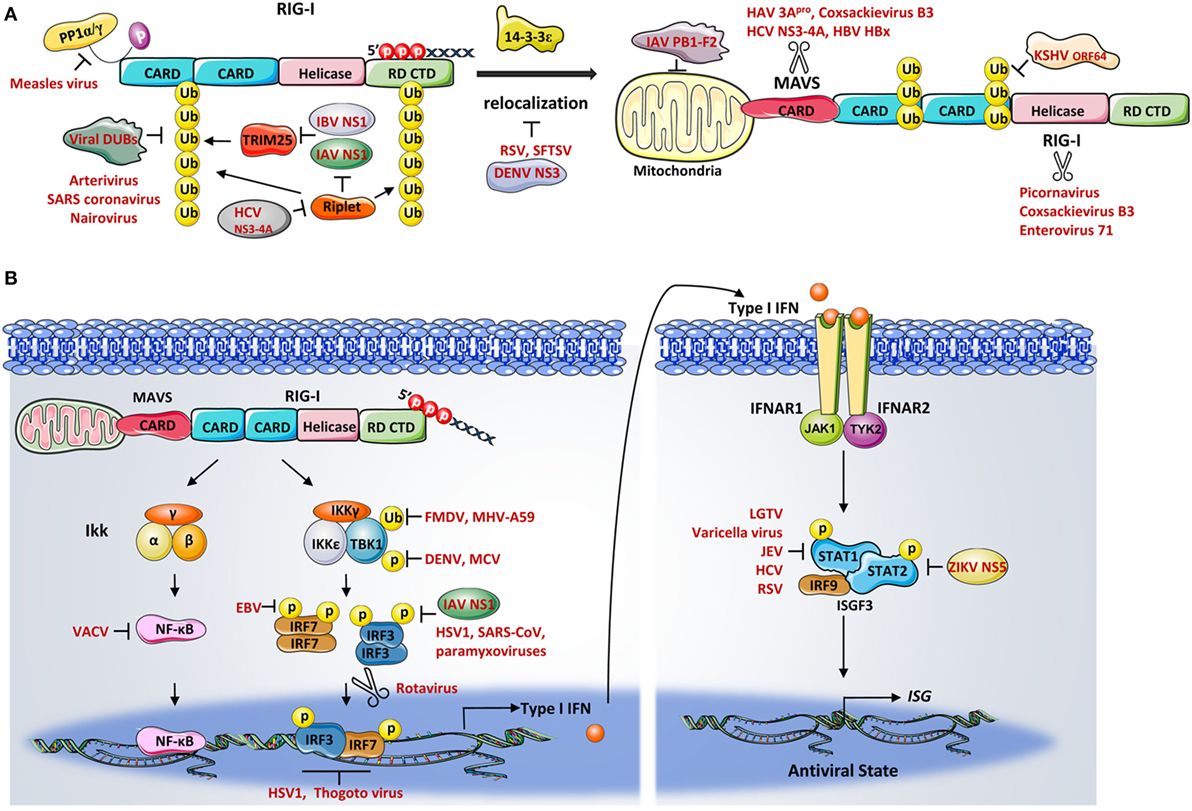
Figure 3. (A) Viral inhibition of retinoic acid-inducible gene-I (RIG-I)– mitochondrial antiviral signaling protein (MAVS) signaling. Numerous viral proteins have been described as RIG-I signaling antagonists which inhibit the activation of RIG-I and its adaptor protein MAVS. Influenza A virus (IAV) NS1 and influenza B virus NS1 were both shown to suppress the TRIM25-mediated ubiquitination of RIG-I caspase activation and recruitment domains. The protease NS3-4A of hepatitis C virus functions by targeting Riplet. A number of virus-encoded deubiquitination enzymes from various viruses remove Lys63-linked ubiquitin from RIG-I, resulting in signaling suppression. The measles virus efficiently blocks RIG-I dephosphorylation to prevent RIG-I activation. Dengue virus (DENV), respiratory syncytial virus, and thrombocytopenia syndrome virus (SFTSV) were involved in disrupting RIG-I translocation to the mitochondria. Numerous virus-encoded proteases were shown to directly cleave or degrade RIG-I and MAVS. Additionally, the PB1-F2 of IAV decreases the mitochondrial membrane potential thereby inhibiting RIG-I–MAVS interaction. (B) Viral modulation of downstream signaling components. Ubiquitination- and phosphorylation-mediated TBK1 activation were shown be antagonized by a number of viruses, including foot-and-mouth disease virus, mouse hepatitis virus A59, molluscum contagiosum virus, and DENV. IAV NS1, herpes simplex virus type 1, severe acute respiratory syndrome coronavirus (SARS-CoV), and several paramyxoviruses have been shown to interfere with interferon regulator factor (IRF) 3 phosphorylation. Epstein–Barr virus suppresses IRF7 transcriptional activity. The rotavirus targets both IRF3 and IRF7 for degradation. The binding of transcription factors to the IFNB promoter is also challenged by herpes simplex virus and the thogoto virus. Other viruses subvert the JAK–STAT signaling to inhibit the production of interferon-stimulated genes. STAT1 and STAT2 are therefore often targets by a number of viruses.
On the other hand, some viruses encode enzymes that directly deubiquitinate RIG-I. For instance, KSHV encoded deubiquitinase ORF64 cleaves Lys63-ubiquination chains on CARDs, blocks CARDs interaction between RIG-I and MAVS, thereby downregulating RIG-I signaling (105). Other viruses including arterivirus, nairovirus, SARS-CoV, and foot-and-mouth disease virus (FMDV) have also been reported to downregulate RIG-I ubiquitination through their viral encoded DUBs (106, 107).
Few viruses have been shown to manipulate RIG-I regulation with regards to targeting the phosphorylation or dephosphorylation process of RIG-I. Nevertheless, it was reported that MV efficiently escapes antiviral response via suppressing RIG-I dephosphorylation in dendritic cells (DCs). In this study, the growth arrest and DNA damage protein (GADD34) was shown to form complexes with PP1 to facilitate RIG-I activation. The MV infection induced DC-SIGN signaling results in an inhibition of GADD34-PP1 phosphatases activity and thereby impairs RIG-I activation (108).
Degradation of RIG-I and MAVS
Another distinct strategy used by viruses to antagonize RIG-I signaling is the direct cleavage or degradation of the receptor and multiple members of the signaling cascade (Figure 3A). RIG-I has been reported in some studies to be cleaved by the proteinase 3Cpro during infections with picornavirus, coxsackievirus B3 (CVB3), and enterovirus 71 (EV71) (109, 110). The encephalomyocarditis virus directs both caspase- and proteasome-dependent degradation of RIG-I (111). Intriguingly, the NS1–NS2 degradasome of the respiratory syncytial virus (RSV) has been shown to mediate the proteasomal degradation of RIG-I (112).
Mitochondrial antiviral signaling protein is also a well-studied molecule which is often targeted by many types of viral-induced cleavage. For example, the hepatitis A virus (HAV) cleaves MAVS for proteolysis by its protease 3Cpro (113). Both CVB3 proteinase 2Apro and 3Cpro trigger MAVS cleavage at different sites during infection, and the cleavage of MAVS by EV71 is accomplished via its 2Apro activity (110, 114). In addition, serine protease NS3-4A of HCV cleaves MAVS, removing it from the mitochondria, thereby inhibiting downstream signaling (36, 115). In a parallel fashion, many viruses mediate cellular proteolytic degradation of MAVS to attenuate RIG-I antiviral responses. Hepatitis B virus viral protein HBx triggers the proteasome-mediated degradation of MAVS through Lys136 ubiquitination (116). Another study reported that the HAV cysteine protease ABC targets MAVS for proteolysis at mitochondrial membrane (113). Additionally, viral modulation of cellular organelles such as mitochondria also affects RIG-I–MAVS signaling. The PB1-F2 of IAV, for instance, has been described as decreasing the mitochondrial membrane potential, resulting in the acceleration of mitochondrial fragmentation, thereby inhibiting RIG-I–MAVS signaling (117–119).
It is important to note that the proper localization of RIG-I and MAVS is a prerequisite for effective signaling transduction. MAVS resides on the mitochondrial membrane, peroxisomes, and mitochondria-associated membranes for antiviral signaling. In fact, a RIG-I translocon has been identified to direct RIG-I redistribution from cytosol to membranes during viral infection (49). Studies have shown that several viruses encode proteins to disrupt the proper localization of RIG-I or MAVS as a novel mechanism of regulation, such as NS3 of DENV (113), nucleoprotein of RSV (120), and non-structural proteins of thrombocytopenia syndrome virus (SFTSV) (121).
Modulation of Downstream Signaling Components
To ensure successful RIG-I signaling transduction, the kinase activities of TBK1 and IKKϵ are tightly controlled via various regulatory mechanisms and are common targets of viruses (Figure 3B). For example, both the leader proteinase (Lpro) of FMDV (122) and the non-structural protein 3 (ns3) of the mouse hepatitis virus A59 (123) inhibit ubiquitination of TBK1. Dengue virus serotype4 non-structural proteins, NS2A and NS4B, as well as the FLIPs proteins encoded by the molluscum contagiosum virus (MCV), all reduce TBK1 phosphorylation, thereby preventing its activation (124, 125). Several viruses have been shown to prevent the formation of functional TBK1-containing complexes. The K7 protein of the VACV prevents TBK1/IKKϵ complex-induced IRF activation by targeting host DEAD box protein 3 (DDX3) (126). Two other viruses, the NY-1 HTNV and SARS-CoV, disrupt the TBK1–TRAF3 and TANK–TBK1/IKKϵ complex, respectively (127, 128). Moreover, SFTSV has been shown to irreversibly re-localize TBK1 and IKK from mitochondria and sequester the TBK1/IKKϵ/IRF3 complex via the formation of inclusion bodies, causing signaling cascade termination (129).
Viral regulation of the transcription factors, IRFs and NF-κB, further serve as points of control in RIG-I signaling (Figure 3B). One of the best studied examples is the inhibition of IRF3 activity by the IAV NS1 protein (130). Besides this, the HSV-1, rabies virus, SARS-CoV, as well as several paramyxoviruses have been demonstrated to interfere with the phosphorylation state of IRF3, thereby blocking IFN induction (131–134). The EBV conjugates SUMO to IRF7 at lysine 452 to decrease IRF7 transcriptional activity (135). The rotavirus NS1, targets both IRF3 and IRF7 for degradation to prevent IRFs from undergoing dimerization (136). Viruses have also developed various means to suppress the IRF3 DNA binding ability. Herpes simplex virus, thogoto virus, and KSHV, all developed strategies to downregulate IRF3 transcriptional activity by either disrupting IRF3 binding complex formations or competing binding regions on the IFNB promoter (137–139). Viral strategies in inhibiting cytoplasmic or transcriptional activities of NF-κB have been extensively studied during the VACV infection. Studies reported that multiple proteins encoded by VACV and HSV-1 suppress NF-κB activation (140–143).
Viruses have also developed multiple inhibitory mechanisms to counteract the IFN stimulation of ISGs by targeting STAT1 and/or STAT2 (Figure 3B). For example, the langat virus was shown to inhibit the phosphorylation of both STAT1 and STAT2 (144). Varicella viruses and the Japanese encephalitis virus, both block the JAK/STAT1 pathway through multiple mechanisms including inhibiting STAT proteins phosphorylation and nucleotranslocation (145, 146). The non-structural protein NS5 of several flaviviruses, have been shown to target STAT proteins via distinct mechanisms. For example, MNV NS5 inhibits STAT1 phosphorylation, whereas DENV NS5 interacts with UBR4 to promote STAT2 degradation (147, 148). By contrast, the Zika virus NS5 induced proteasomal degradation of STAT2 was recently identified as UBR4 independent (149). Furthermore, other viruses, such as HCV (150), RSV (151), and paramyxovirus (152), also demonstrate negative regulation of the JAK–STAT pathway.
Concluding Remarks
Studies from the past decade have well established RIG-I as one of the principal PRRs for the recognition of cytoplasmic viral RNA, as well as defining its critical role in the induction of IFNs during viral infections. Our understanding of the RIG-I-mediated antiviral response has been greatly expanded with the key discoveries made regarding the molecular mechanism of RIG-I regulation, such as ubiquitination, phosphorylation, and acetylation. Meanwhile, investigating viral strategies to manipulate RIG-I responses not only allow us to understand the viral pathogenesis, but also significantly contributed to our knowledge of how RIG-I is activated and regulated. These new insights into the viral-mediated RIG-I regulations are important for vaccine and drug development aiming to suppress infectious diseases and enhance immune responses.
Author Contributions
YL wrote the manuscript. RL and DO revised and approved the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Alexandre Sze for his critical reading and editing of the manuscript. This research was supported by grant from Canadian Institutes of Health Research (MOP130401) to RL; DO was supported by a Peter Quinlan McGill postdoctoral fellowship. The authors would like to acknowledge all the colleagues in the field and apologies to those whose important contributions could not be included in the review due to space constraints. The figures of the review were illustrated using the Servier Medical Art library, http://www.servier.com/Powerpoint-image-bank.
References
1. Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on toll-like receptors. Nat Immunol (2010) 11:373–84. doi:10.1038/ni.1863
2. Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol (2004) 5:730–7. doi:10.1038/ni1087
3. Loo YM, Gale M Jr. Immune signaling by RIG-I-like receptors. Immunity (2011) 34:680–92. doi:10.1016/j.immuni.2011.05.003
4. Takeda K, Akira S. TLR signaling pathways. Semin Immunol (2004) 16:3–9. doi:10.1016/j.smim.2003.10.003
5. Elinav E, Strowig T, Henao-Mejia J, Flavell RA. Regulation of the antimicrobial response by NLR proteins. Immunity (2011) 34:665–79. doi:10.1016/j.immuni.2011.05.007
6. Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science (2013) 339:786–91. doi:10.1126/science.1232458
7. Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, et al. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol (2010) 11:997–1004. doi:10.1038/ni.1932
8. Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol (2005) 5:375–86. doi:10.1038/nri1604
9. Bowie AG, Unterholzner L. Viral evasion and subversion of pattern-recognition receptor signalling. Nat Rev Immunol (2008) 8:911–22. doi:10.1038/nri2436
10. Chan YK, Gack MU. Viral evasion of intracellular DNA and RNA sensing. Nat Rev Microbiol (2016) 14:360–73. doi:10.1038/nrmicro.2016.45
11. Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell (2010) 140:805–20. doi:10.1016/j.cell.2010.01.022
12. Luo D, Ding SC, Vela A, Kohlway A, Lindenbach BD, Pyle AM. Structural insights into RNA recognition by RIG-I. Cell (2011) 147:409–22. doi:10.1016/j.cell.2011.09.023
13. Jiang F, Ramanathan A, Miller MT, Tang GQ, Gale M Jr, Patel SS, et al. Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature (2011) 479:423–7. doi:10.1038/nature10537
14. Kowalinski E, Lunardi T, McCarthy AA, Louber J, Brunel J, Grigorov B, et al. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell (2011) 147:423–35. doi:10.1016/j.cell.2011.09.039
15. Belgnaoui SM, Paz S, Hiscott J. Orchestrating the interferon antiviral response through the mitochondrial antiviral signaling (MAVS) adapter. Curr Opin Immunol (2011) 23:564–72. doi:10.1016/j.coi.2011.08.001
16. Yoneyama M, Fujita T. Recognition of viral nucleic acids in innate immunity. Rev Med Virol (2010) 20:4–22. doi:10.1002/rmv.633
17. Yoneyama M, Fujita T. RNA recognition and signal transduction by RIG-I-like receptors. Immunol Rev (2009) 227:54–65. doi:10.1111/j.1600-065X.2008.00727.x
18. Cardenas WB, Loo YM, Gale M Jr, Hartman AL, Kimberlin CR, Martinez-Sobrido L, et al. Ebola virus VP35 protein binds double-stranded RNA and inhibits alpha/beta interferon production induced by RIG-I signaling. J Virol (2006) 80:5168–78. doi:10.1128/JVI.02199-05
19. Nasirudeen AM, Wong HH, Thien P, Xu S, Lam KP, Liu DX. RIG-I, MDA5 and TLR3 synergistically play an important role in restriction of dengue virus infection. PLoS Negl Trop Dis (2011) 5:e926. doi:10.1371/journal.pntd.0000926
20. Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science (2006) 314:994–7. doi:10.1126/science.1132505
21. Schlee M, Roth A, Hornung V, Hagmann CA, Wimmenauer V, Barchet W, et al. Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity (2009) 31:25–34. doi:10.1016/j.immuni.2009.05.008
22. Weber M, Gawanbacht A, Habjan M, Rang A, Borner C, Schmidt AM, et al. Incoming RNA virus nucleocapsids containing a 5′-triphosphorylated genome activate RIG-I and antiviral signaling. Cell Host Microbe (2013) 13:336–46. doi:10.1016/j.chom.2013.01.012
23. Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science (2006) 314:997–1001. doi:10.1126/science.1132998
24. Goubau D, Schlee M, Deddouche S, Pruijssers AJ, Zillinger T, Goldeck M, et al. Antiviral immunity via RIG-I-mediated recognition of RNA bearing 5′-diphosphates. Nature (2014) 514:372–5. doi:10.1038/nature13590
25. Goulet ML, Olagnier D, Xu Z, Paz S, Belgnaoui SM, Lafferty EI, et al. Systems analysis of a RIG-I agonist inducing broad spectrum inhibition of virus infectivity. PLoS Pathog (2013) 9:e1003298. doi:10.1371/journal.ppat.1003298
26. Olagnier D, Scholte FE, Chiang C, Albulescu IC, Nichols C, He Z, et al. Inhibition of dengue and chikungunya virus infections by RIG-I-mediated type I interferon-independent stimulation of the innate antiviral response. J Virol (2014) 88:4180–94. doi:10.1128/JVI.03114-13
27. Beljanski V, Chiang C, Kirchenbaum GA, Olagnier D, Bloom CE, Wong T, et al. Enhanced influenza virus-like particle vaccination with a structurally optimized RIG-I agonist as adjuvant. J Virol (2015) 89:10612–24. doi:10.1128/JVI.01526-15
28. Pinto AK, Brien JD, Lam CY, Johnson S, Chiang C, Hiscott J, et al. Defining new therapeutics using a more immunocompetent mouse model of antibody-enhanced dengue virus infection. MBio (2015) 6:e1316–1315. doi:10.1128/mBio.01316-15
29. Chiang C, Beljanski V, Yin K, Olagnier D, Ben Yebdri F, Steel C, et al. Sequence-specific modifications enhance the broad-spectrum antiviral response activated by RIG-I agonists. J Virol (2015) 89:8011–25. doi:10.1128/JVI.00845-15
30. Hochheiser K, Klein M, Gottschalk C, Hoss F, Scheu S, Coch C, et al. Cutting edge: the RIG-I ligand 3pRNA potently improves CTL cross-priming and facilitates antiviral vaccination. J Immunol (2016) 196:2439–43. doi:10.4049/jimmunol.1501958
31. Melchjorsen J, Rintahaka J, Soby S, Horan KA, Poltajainen A, Ostergaard L, et al. Early innate recognition of herpes simplex virus in human primary macrophages is mediated via the MDA5/MAVS-dependent and MDA5/MAVS/RNA polymerase III-independent pathways. J Virol (2010) 84:11350–8. doi:10.1128/JVI.01106-10
32. Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell (2009) 138:576–91. doi:10.1016/j.cell.2009.06.015
33. Liu Y, Goulet ML, Sze A, Bel Hadj S, Belgnaoui SM, Lababidi RR, et al. RIG-I mediated STING up-regulation restricts HSV-1 infection. J Virol (2016) 90:9406–19. doi:10.1128/JVI.00748-16
34. Poeck H, Bscheider M, Gross O, Finger K, Roth S, Rebsamen M, et al. Recognition of RNA virus by RIG-I results in activation of CARD9 and inflammasome signaling for interleukin 1 beta production. Nat Immunol (2010) 11:63–9. doi:10.1038/ni.1824
35. Pothlichet J, Meunier I, Davis BK, Ting JP, Skamene E, von Messling V, et al. Type I IFN triggers RIG-I/TLR3/NLRP3-dependent inflammasome activation in influenza A virus infected cells. PLoS Pathog (2013) 9:e1003256. doi:10.1371/journal.ppat.1003256
36. Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, et al. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature (2005) 437:1167–72. doi:10.1038/nature04193
37. Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell (2005) 122:669–82. doi:10.1016/j.cell.2005.08.012
38. Vallabhapurapu S, Karin M. Regulation and function of NF-kappaB transcription factors in the immune system. Annu Rev Immunol (2009) 27:693–733. doi:10.1146/annurev.immunol.021908.132641
39. Paz S, Vilasco M, Werden SJ, Arguello M, Joseph-Pillai D, Zhao T, et al. A functional C-terminal TRAF3-binding site in MAVS participates in positive and negative regulation of the IFN antiviral response. Cell Res (2011) 21:895–910. doi:10.1038/cr.2011.2
40. Zhao T, Yang L, Sun Q, Arguello M, Ballard DW, Hiscott J, et al. The NEMO adaptor bridges the nuclear factor-kappaB and interferon regulatory factor signaling pathways. Nat Immunol (2007) 8:592–600. doi:10.1038/ni1465
41. Yoneyama M, Suhara W, Fukuhara Y, Fukuda M, Nishida E, Fujita T. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J (1998) 17:1087–95. doi:10.1093/emboj/17.4.1087
42. Saito T, Hirai R, Loo YM, Owen D, Johnson CL, Sinha SC, et al. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc Natl Acad Sci U S A (2007) 104:582–7. doi:10.1073/pnas.0606699104
43. Cui S, Eisenacher K, Kirchhofer A, Brzozka K, Lammens A, Lammens K, et al. The C-terminal regulatory domain is the RNA 5′-triphosphate sensor of RIG-I. Mol Cell (2008) 29:169–79. doi:10.1016/j.molcel.2007.10.032
44. Patel JR, Jain A, Chou YY, Baum A, Ha T, Garcia-Sastre A. ATPase-driven oligomerization of RIG-I on RNA allows optimal activation of type-I interferon. EMBO Rep (2013) 14:780–7. doi:10.1038/embor.2013.102
45. Peisley A, Wu B, Yao H, Walz T, Hur S. RIG-I forms signaling-competent filaments in an ATP-dependent, ubiquitin-independent manner. Mol Cell (2013) 51:573–83. doi:10.1016/j.molcel.2013.07.024
46. Anchisi S, Guerra J, Garcin D. RIG-I ATPase activity and discrimination of self-RNA versus non-self-RNA. MBio (2015) 6:e02349. doi:10.1128/mBio.02349-14
47. Lässig C, Matheisl S, Sparrer KMJ, de Oliveira Mann CC, Moldt M, Patel JR, et al. Correction: ATP hydrolysis by the viral RNA sensor RIG-I prevents unintentional recognition of self-RNA. Elife (2016) 5:e14954. doi:10.7554/eLife.14954
48. Gack MU, Kirchhofer A, Shin YC, Inn KS, Liang C, Cui S, et al. Roles of RIG-I N-terminal tandem CARD and splice variant in TRIM25-mediated antiviral signal transduction. Proc Natl Acad Sci U S A (2008) 105:16743–8. doi:10.1073/pnas.0804947105
49. Liu HM, Loo YM, Horner SM, Zornetzer GA, Katze MG, Gale M Jr. The mitochondrial targeting chaperone 14-3-3epsilon regulates a RIG-I translocon that mediates membrane association and innate antiviral immunity. Cell Host Microbe (2012) 11:528–37. doi:10.1016/j.chom.2012.04.006
50. Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature (2007) 446:916–20. doi:10.1038/nature05732
51. Oshiumi H, Matsumoto M, Hatakeyama S, Seya T. Riplet/RNF135, a RING finger protein, ubiquitinates RIG-I to promote interferon-beta induction during the early phase of viral infection. J Biol Chem (2009) 284:807–17. doi:10.1074/jbc.M804259200
52. Oshiumi H, Miyashita M, Inoue N, Okabe M, Matsumoto M, Seya T. The ubiquitin ligase Riplet is essential for RIG-I-dependent innate immune responses to RNA virus infection. Cell Host Microbe (2010) 8:496–509. doi:10.1016/j.chom.2010.11.008
53. Gao D, Yang YK, Wang RP, Zhou X, Diao FC, Li MD, et al. REUL is a novel E3 ubiquitin ligase and stimulator of retinoic-acid-inducible gene-I. PLoS One (2009) 4:e5760. doi:10.1371/journal.pone.0005760
54. Oshiumi H, Miyashita M, Matsumoto M, Seya T. A distinct role of Riplet-mediated K63-linked polyubiquitination of the RIG-I repressor domain in human antiviral innate immune responses. PLoS Pathog (2013) 9:e1003533. doi:10.1371/journal.ppat.1003533
55. Kuniyoshi K, Takeuchi O, Pandey S, Satoh T, Iwasaki H, Akira S, et al. Pivotal role of RNA-binding E3 ubiquitin ligase MEX3C in RIG-I-mediated antiviral innate immunity. Proc Natl Acad Sci U S A (2014) 111:5646–51. doi:10.1073/pnas.1401674111
56. Zhu J, Zhang Y, Ghosh A, Cuevas RA, Forero A, Dhar J, et al. Antiviral activity of human OASL protein is mediated by enhancing signaling of the RIG-I RNA sensor. Immunity (2014) 40:936–48. doi:10.1016/j.immuni.2014.05.007
57. Ibsen MS, Gad HH, Andersen LL, Hornung V, Julkunen I, Sarkar SN, et al. Structural and functional analysis reveals that human OASL binds dsRNA to enhance RIG-I signaling. Nucleic Acids Res (2015) 43:5236–48. doi:10.1093/nar/gkv389
58. Zeng W, Sun L, Jiang X, Chen X, Hou F, Adhikari A, et al. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell (2010) 141:315–30. doi:10.1016/j.cell.2010.03.029
59. Peisley A, Wu B, Xu H, Chen ZJ, Hur S. Structural basis for ubiquitin-mediated antiviral signal activation by RIG-I. Nature (2014) 509:110–4. doi:10.1038/nature13140
60. Friedman CS, O’Donnell MA, Legarda-Addison D, Ng A, Cardenas WB, Yount JS, et al. The tumour suppressor CYLD is a negative regulator of RIG-I-mediated antiviral response. EMBO Rep (2008) 9:930–6. doi:10.1038/embor.2008.136
61. Lin W, Zhang J, Lin H, Li Z, Sun X, Xin D, et al. Syndecan-4 negatively regulates antiviral signalling by mediating RIG-I deubiquitination via CYLD. Nat Commun (2016) 7:11848. doi:10.1038/ncomms11848
62. Cui J, Song Y, Li Y, Zhu Q, Tan P, Qin Y, et al. USP3 inhibits type I interferon signaling by deubiquitinating RIG-I-like receptors. Cell Res (2014) 24:400–16. doi:10.1038/cr.2013.170
63. Fan Y, Mao R, Yu Y, Liu S, Shi Z, Cheng J, et al. USP21 negatively regulates antiviral response by acting as a RIG-I deubiquitinase. J Exp Med (2014) 211:313–28. doi:10.1084/jem.20122844
64. Arimoto K, Takahashi H, Hishiki T, Konishi H, Fujita T, Shimotohno K. Negative regulation of the RIG-I signaling by the ubiquitin ligase RNF125. Proc Natl Acad Sci U S A (2007) 104:7500–5. doi:10.1073/pnas.0611551104
65. Wang W, Jiang M, Liu S, Zhang S, Liu W, Ma Y, et al. RNF122 suppresses antiviral type I interferon production by targeting RIG-I CARDs to mediate RIG-I degradation. Proc Natl Acad Sci U S A (2016) 113:9581–6. doi:10.1073/pnas.1604277113
66. Inn KS, Gack MU, Tokunaga F, Shi M, Wong LY, Iwai K, et al. Linear ubiquitin assembly complex negatively regulates RIG-I- and TRIM25-mediated type I interferon induction. Mol Cell (2011) 41:354–65. doi:10.1016/j.molcel.2010.12.029
67. Pauli EK, Chan YK, Davis ME, Gableske S, Wang MK, Feister KF, et al. The ubiquitin-specific protease USP15 promotes RIG-I-mediated antiviral signaling by deubiquitylating TRIM25. Sci Signal (2014) 7:ra3. doi:10.1126/scisignal.2004577
68. Gack MU, Nistal-Villan E, Inn KS, Garcia-Sastre A, Jung JU. Phosphorylation-mediated negative regulation of RIG-I antiviral activity. J Virol (2010) 84:3220–9. doi:10.1128/JVI.02241-09
69. Nistal-Villan E, Gack MU, Martinez-Delgado G, Maharaj NP, Inn KS, Yang H, et al. Negative role of RIG-I serine 8 phosphorylation in the regulation of interferon-beta production. J Biol Chem (2010) 285:20252–61. doi:10.1074/jbc.M109.089912
70. Sun Z, Ren H, Liu Y, Teeling JL, Gu J. Phosphorylation of RIG-I by casein kinase II inhibits its antiviral response. J Virol (2011) 85:1036–47. doi:10.1128/JVI.01734-10
71. Zhang X, Yu H, Zhao J, Li X, Li J, He J, et al. IKK negatively regulates RIG-I via direct phosphorylation. J Med Virol (2016) 88:712–8. doi:10.1002/jmv.24376
72. Maharaj NP, Wies E, Stoll A, Gack MU. Conventional protein kinase C-alpha (PKC-alpha) and PKC-beta negatively regulate RIG-I antiviral signal transduction. J Virol (2012) 86:1358–71. doi:10.1128/JVI.06543-11
73. Wies E, Wang MK, Maharaj NP, Chen K, Zhou S, Finberg RW, et al. Dephosphorylation of the RNA sensors RIG-I and MDA5 by the phosphatase PP1 is essential for innate immune signaling. Immunity (2013) 38:437–49. doi:10.1016/j.immuni.2012.11.018
74. Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science (2009) 325:834–40. doi:10.1126/science.1175371
75. Liu HM, Jiang F, Loo YM, Hsu S, Hsiang TY, Marcotrigiano J, et al. Regulation of retinoic acid inducible gene-I (RIG-I) activation by the histone deacetylase 6. EBioMedicine (2016) 9:195–206. doi:10.1016/j.ebiom.2016.06.015
76. Choi SJ, Lee HC, Kim JH, Park SY, Kim TH, Lee WK, et al. HDAC6 regulates cellular viral RNA sensing by deacetylation of RIG-I. EMBO J (2016) 35:429–42. doi:10.15252/embj.201592586
77. van der Veen AG, Ploegh HL. Ubiquitin-like proteins. Annu Rev Biochem (2012) 81:323–57. doi:10.1146/annurev-biochem-093010-153308
78. Mi Z, Fu J, Xiong Y, Tang H. SUMOylation of RIG-I positively regulates the type I interferon signaling. Protein Cell (2010) 1:275–83. doi:10.1007/s13238-010-0030-1
79. Nguyen NT, Now H, Kim WJ, Kim N, Yoo JY. Ubiquitin-like modifier FAT10 attenuates RIG-I mediated antiviral signaling by segregating activated RIG-I from its signaling platform. Sci Rep (2016) 6:23377. doi:10.1038/srep23377
80. Kim MJ, Hwang SY, Imaizumi T, Yoo JY. Negative feedback regulation of RIG-I-mediated antiviral signaling by interferon-induced ISG15 conjugation. J Virol (2008) 82:1474–83. doi:10.1128/JVI.01650-07
81. Li MT, Di W, Xu H, Yang YK, Chen HW, Zhang FX, et al. Negative regulation of RIG-I-mediated innate antiviral signaling by SEC14L1. J Virol (2013) 87:10037–46. doi:10.1128/JVI.01073-13
82. Jounai N, Takeshita F, Kobiyama K, Sawano A, Miyawaki A, Xin KQ, et al. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc Natl Acad Sci U S A (2007) 104:14050–5. doi:10.1073/pnas.0704014104
83. He S, Zhao J, Song S, He X, Minassian A, Zhou Y, et al. Viral pseudo-enzymes activate RIG-I via deamidation to evade cytokine production. Mol Cell (2015) 58:134–46. doi:10.1016/j.molcel.2015.01.036
84. Zhao J, Li J, Xu S, Feng P. Emerging roles of protein deamidation in innate immune signaling. J Virol (2016) 90:4262–8. doi:10.1128/JVI.01980-15
85. Habjan M, Andersson I, Klingstrom J, Schumann M, Martin A, Zimmermann P, et al. Processing of genome 5′ termini as a strategy of negative-strand RNA viruses to avoid RIG-I-dependent interferon induction. PLoS One (2008) 3:e2032. doi:10.1371/journal.pone.0002032
86. Garcin D, Lezzi M, Dobbs M, Elliott RM, Schmaljohn C, Kang CY, et al. The 5′ ends of Hantaan virus (Bunyaviridae) RNAs suggest a prime-and-realign mechanism for the initiation of RNA synthesis. J Virol (1995) 69:5754–62.
87. Wang H, Vaheri A, Weber F, Plyusnin A. Old World hantaviruses do not produce detectable amounts of dsRNA in infected cells and the 5′ termini of their genomic RNAs are monophosphorylated. J Gen Virol (2011) 92:1199–204. doi:10.1099/vir.0.029405-0
88. Schneider U, Schwemmle M, Staeheli P. Genome trimming: a unique strategy for replication control employed by Borna disease virus. Proc Natl Acad Sci U S A (2005) 102:3441–6. doi:10.1073/pnas.0405965102
89. Marq JB, Kolakofsky D, Garcin D. Unpaired 5′ ppp-nucleotides, as found in arenavirus double-stranded RNA panhandles, are not recognized by RIG-I. J Biol Chem (2010) 285:18208–16. doi:10.1074/jbc.M109.089425
90. Lee YF, Nomoto A, Detjen BM, Wimmer E. A protein covalently linked to poliovirus genome RNA. Proc Natl Acad Sci U S A (1977) 74:59–63. doi:10.1073/pnas.74.1.59
91. Decroly E, Ferron F, Lescar J, Canard B. Conventional and unconventional mechanisms for capping viral mRNA. Nat Rev Microbiol (2012) 10:51–65. doi:10.1038/nrmicro2675
92. Devarkar SC, Wang C, Miller MT, Ramanathan A, Jiang F, Khan AG, et al. Structural basis for m7G recognition and 2′-O-methyl discrimination in capped RNAs by the innate immune receptor RIG-I. Proc Natl Acad Sci U S A (2016) 113:596–601. doi:10.1073/pnas.1515152113
93. Leung DW, Prins KC, Borek DM, Farahbakhsh M, Tufariello JM, Ramanan P, et al. Structural basis for dsRNA recognition and interferon antagonism by Ebola VP35. Nat Struct Mol Biol (2010) 17:165–72. doi:10.1038/nsmb.1765
94. Bale S, Julien JP, Bornholdt ZA, Kimberlin CR, Halfmann P, Zandonatti MA, et al. Marburg virus VP35 can both fully coat the backbone and cap the ends of dsRNA for interferon antagonism. PLoS Pathog (2012) 8:e1002916. doi:10.1371/journal.ppat.1002916
95. Ramanan P, Edwards MR, Shabman RS, Leung DW, Endlich-Frazier AC, Borek DM, et al. Structural basis for Marburg virus VP35-mediated immune evasion mechanisms. Proc Natl Acad Sci U S A (2012) 109:20661–6. doi:10.1073/pnas.1213559109
96. Donelan NR, Basler CF, Garcia-Sastre A. A recombinant influenza A virus expressing an RNA-binding-defective NS1 protein induces high levels of beta interferon and is attenuated in mice. J Virol (2003) 77:13257–66. doi:10.1128/JVI.77.24.13257-13266.2003
97. Weber M, Sediri H, Felgenhauer U, Binzen I, Banfer S, Jacob R, et al. Influenza virus adaptation PB2-627K modulates nucleocapsid inhibition by the pathogen sensor RIG-I. Cell Host Microbe (2015) 17:309–19. doi:10.1016/j.chom.2015.01.005
98. Uchida L, Espada-Murao LA, Takamatsu Y, Okamoto K, Hayasaka D, Yu F, et al. The dengue virus conceals double-stranded RNA in the intracellular membrane to escape from an interferon response. Sci Rep (2014) 4:7395. doi:10.1038/srep07395
99. Knoops K, Kikkert M, Worm SH, Zevenhoven-Dobbe JC, van der Meer Y, Koster AJ, et al. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol (2008) 6:e226. doi:10.1371/journal.pbio.0060226
100. Gack MU, Albrecht RA, Urano T, Inn KS, Huang IC, Carnero E, et al. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe (2009) 5:439–49. doi:10.1016/j.chom.2009.04.006
101. Rajsbaum R, Albrecht RA, Wang MK, Maharaj NP, Versteeg GA, Nistal-Villan E, et al. Species-specific inhibition of RIG-I ubiquitination and IFN induction by the influenza A virus NS1 protein. PLoS Pathog (2012) 8:e1003059. doi:10.1371/journal.ppat.1003059
102. Kathum OA, Schrader T, Anhlan D, Nordhoff C, Liedmann S, Pande A, et al. Phosphorylation of influenza A virus NS1 protein at threonine 49 suppresses its interferon antagonistic activity. Cell Microbiol (2016) 18:784–91. doi:10.1111/cmi.12559
103. Zheng W, Cao S, Chen C, Li J, Zhang S, Jiang J, et al. Threonine 80 phosphorylation of non-structural protein 1 regulates the replication of influenza A virus by reducing the binding affinity with RIG-I. Cell Microbiol (2016). doi:10.1111/cmi.12643
104. Jiang J, Li J, Fan W, Zheng W, Yu M, Chen C, et al. Robust Lys63-linked ubiquitination of RIG-I promotes cytokine eruption in early influenza B virus infection. J Virol (2016) 90:6263–75. doi:10.1128/JVI.00549-16
105. Inn KS, Lee SH, Rathbun JY, Wong LY, Toth Z, Machida K, et al. Inhibition of RIG-I-mediated signaling by Kaposi’s sarcoma-associated herpesvirus-encoded deubiquitinase ORF64. J Virol (2011) 85:10899–904. doi:10.1128/JVI.00690-11
106. Chiang JJ, Davis ME, Gack MU. Regulation of RIG-I-like receptor signaling by host and viral proteins. Cytokine Growth Factor Rev (2014) 25:491–505. doi:10.1016/j.cytogfr.2014.06.005
107. van Kasteren PB, Beugeling C, Ninaber DK, Frias-Staheli N, van Boheemen S, Garcia-Sastre A, et al. Arterivirus and nairovirus ovarian tumor domain-containing deubiquitinases target activated RIG-I to control innate immune signaling. J Virol (2012) 86:773–85. doi:10.1128/JVI.06277-11
108. Mesman AW, Zijlstra-Willems EM, Kaptein TM, de Swart RL, Davis ME, Ludlow M, et al. Measles virus suppresses RIG-I-like receptor activation in dendritic cells via DC-SIGN-mediated inhibition of PP1 phosphatases. Cell Host Microbe (2014) 16:31–42. doi:10.1016/j.chom.2014.06.008
109. Barral PM, Sarkar D, Fisher PB, Racaniello VR. RIG-I is cleaved during picornavirus infection. Virology (2009) 391:171–6. doi:10.1016/j.virol.2009.06.045
110. Feng Q, Langereis MA, Lork M, Nguyen M, Hato SV, Lanke K, et al. Enterovirus 2Apro targets MDA5 and MAVS in infected cells. J Virol (2014) 88:3369–78. doi:10.1128/JVI.02712-13
111. Papon L, Oteiza A, Imaizumi T, Kato H, Brocchi E, Lawson TG, et al. The viral RNA recognition sensor RIG-I is degraded during encephalomyocarditis virus (EMCV) infection. Virology (2009) 393:311–8. doi:10.1016/j.virol.2009.08.009
112. Goswami R, Majumdar T, Dhar J, Chattopadhyay S, Bandyopadhyay SK, Verbovetskaya V, et al. Viral degradasome hijacks mitochondria to suppress innate immunity. Cell Res (2013) 23:1025–42. doi:10.1038/cr.2013.98
113. Yang Y, Liang Y, Qu L, Chen Z, Yi M, Li K, et al. Disruption of innate immunity due to mitochondrial targeting of a picornaviral protease precursor. Proc Natl Acad Sci U S A (2007) 104:7253–8. doi:10.1073/pnas.0611506104
114. Mukherjee A, Morosky SA, Delorme-Axford E, Dybdahl-Sissoko N, Oberste MS, Wang T, et al. The coxsackievirus B 3C protease cleaves MAVS and TRIF to attenuate host type I interferon and apoptotic signaling. PLoS Pathog (2011) 7:e1001311. doi:10.1371/journal.ppat.1001311
115. Li XD, Sun L, Seth RB, Pineda G, Chen ZJ. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc Natl Acad Sci U S A (2005) 102:17717–22. doi:10.1073/pnas.0508531102
116. Wei C, Ni C, Song T, Liu Y, Yang X, Zheng Z, et al. The hepatitis B virus X protein disrupts innate immunity by downregulating mitochondrial antiviral signaling protein. J Immunol (2010) 185:1158–68. doi:10.4049/jimmunol.0903874
117. Varga ZT, Grant A, Manicassamy B, Palese P. Influenza virus protein PB1-F2 inhibits the induction of type I interferon by binding to MAVS and decreasing mitochondrial membrane potential. J Virol (2012) 86:8359–66. doi:10.1128/JVI.01122-12
118. Varga ZT, Ramos I, Hai R, Schmolke M, Garcia-Sastre A, Fernandez-Sesma A, et al. The influenza virus protein PB1-F2 inhibits the induction of type I interferon at the level of the MAVS adaptor protein. PLoS Pathog (2011) 7:e1002067. doi:10.1371/journal.ppat.1002067
119. Yoshizumi T, Ichinohe T, Sasaki O, Otera H, Kawabata S, Mihara K, et al. Influenza A virus protein PB1-F2 translocates into mitochondria via Tom40 channels and impairs innate immunity. Nat Commun (2014) 5:4713. doi:10.1038/ncomms5713
120. Lifland AW, Jung J, Alonas E, Zurla C, Crowe JE Jr, Santangelo PJ. Human respiratory syncytial virus nucleoprotein and inclusion bodies antagonize the innate immune response mediated by MDA5 and MAVS. J Virol (2012) 86:8245–58. doi:10.1128/JVI.00215-12
121. Santiago FW, Covaleda LM, Sanchez-Aparicio MT, Silvas JA, Diaz-Vizarreta AC, Patel JR, et al. Hijacking of RIG-I signaling proteins into virus-induced cytoplasmic structures correlates with the inhibition of type I interferon responses. J Virol (2014) 88:4572–85. doi:10.1128/JVI.03021-13
122. Wang D, Fang L, Li P, Sun L, Fan J, Zhang Q, et al. The leader proteinase of foot-and-mouth disease virus negatively regulates the type I interferon pathway by acting as a viral deubiquitinase. J Virol (2011) 85:3758–66. doi:10.1128/JVI.02589-10
123. Zheng D, Chen G, Guo B, Cheng G, Tang H. PLP2, a potent deubiquitinase from murine hepatitis virus, strongly inhibits cellular type I interferon production. Cell Res (2008) 18:1105–13. doi:10.1038/cr.2008.294
124. Dalrymple NA, Cimica V, Mackow ER. Dengue virus NS proteins inhibit RIG-I/MAVS signaling by blocking TBK1/IRF3 phosphorylation: dengue virus serotype 1 NS4A is a unique interferon-regulating virulence determinant. MBio (2015) 6:e553–515. doi:10.1128/mBio.00553-15
125. Randall CM, Biswas S, Selen CV, Shisler JL. Inhibition of interferon gene activation by death-effector domain-containing proteins from the molluscum contagiosum virus. Proc Natl Acad Sci U S A (2014) 111:E265–72. doi:10.1073/pnas.1314569111
126. Schroder M, Baran M, Bowie AG. Viral targeting of DEAD box protein 3 reveals its role in TBK1/IKKepsilon-mediated IRF activation. EMBO J (2008) 27:2147–57. doi:10.1038/emboj.2008.143
127. Alff PJ, Sen N, Gorbunova E, Gavrilovskaya IN, Mackow ER. The NY-1 hantavirus Gn cytoplasmic tail coprecipitates TRAF3 and inhibits cellular interferon responses by disrupting TBK1-TRAF3 complex formation. J Virol (2008) 82:9115–22. doi:10.1128/JVI.00290-08
128. Chen X, Yang X, Zheng Y, Yang Y, Xing Y, Chen Z. SARS coronavirus papain-like protease inhibits the type I interferon signaling pathway through interaction with the STING-TRAF3-TBK1 complex. Protein Cell (2014) 5:369–81. doi:10.1007/s13238-014-0026-3
129. Wu X, Qi X, Qu B, Zhang Z, Liang M, Li C, et al. Evasion of antiviral immunity through sequestering of TBK1/IKKepsilon/IRF3 into viral inclusion bodies. J Virol (2014) 88:3067–76. doi:10.1128/JVI.03510-13
130. Talon J, Horvath CM, Polley R, Basler CF, Muster T, Palese P, et al. Activation of interferon regulatory factor 3 is inhibited by the influenza A virus NS1 protein. J Virol (2000) 74:7989–96. doi:10.1128/JVI.74.17.7989-7996.2000
131. Irie T, Kiyotani K, Igarashi T, Yoshida A, Sakaguchi T. Inhibition of interferon regulatory factor 3 activation by paramyxovirus V protein. J Virol (2012) 86:7136–45. doi:10.1128/JVI.06705-11
132. Matthews K, Schafer A, Pham A, Frieman M. The SARS coronavirus papain like protease can inhibit IRF3 at a post activation step that requires deubiquitination activity. Virol J (2014) 11:209. doi:10.1186/s12985-014-0209-9
133. Rieder M, Brzozka K, Pfaller CK, Cox JH, Stitz L, Conzelmann KK. Genetic dissection of interferon-antagonistic functions of rabies virus phosphoprotein: inhibition of interferon regulatory factor 3 activation is important for pathogenicity. J Virol (2011) 85:842–52. doi:10.1128/JVI.01427-10
134. Wang S, Wang K, Lin R, Zheng C. Herpes simplex virus 1 serine/threonine kinase US3 hyperphosphorylates IRF3 and inhibits beta interferon production. J Virol (2013) 87:12814–27. doi:10.1128/JVI.02355-13
135. Bentz GL, Shackelford J, Pagano JS. Epstein-Barr virus latent membrane protein 1 regulates the function of interferon regulatory factor 7 by inducing its sumoylation. J Virol (2012) 86:12251–61. doi:10.1128/JVI.01407-12
136. Arnold MM, Barro M, Patton JT. Rotavirus NSP1 mediates degradation of interferon regulatory factors through targeting of the dimerization domain. J Virol (2013) 87:9813–21. doi:10.1128/JVI.01146-13
137. Jennings S, Martinez-Sobrido L, Garcia-Sastre A, Weber F, Kochs G. Thogoto virus ML protein suppresses IRF3 function. Virology (2005) 331:63–72. doi:10.1016/j.virol.2004.10.015
138. Lefort S, Soucy-Faulkner A, Grandvaux N, Flamand L. Binding of Kaposi’s sarcoma-associated herpesvirus K-bZIP to interferon-responsive factor 3 elements modulates antiviral gene expression. J Virol (2007) 81:10950–60. doi:10.1128/JVI.00183-07
139. Melroe GT, Silva L, Schaffer PA, Knipe DM. Recruitment of activated IRF-3 and CBP/p300 to herpes simplex virus ICP0 nuclear foci: potential role in blocking IFN-beta induction. Virology (2007) 360:305–21. doi:10.1016/j.virol.2006.10.028
140. Bravo Cruz AG, Shisler JL. The vaccinia virus K1 ankyrin repeat protein inhibits NF-kB activation by preventing RelA acetylation. J Gen Virol (2016) 97:2691–702. doi:10.1099/jgv.0.000576
141. Ember SW, Ren H, Ferguson BJ, Smith GL. Vaccinia virus protein C4 inhibits NF-kappaB activation and promotes virus virulence. J Gen Virol (2012) 93:2098–108. doi:10.1099/vir.0.045070-0
142. Xing J, Ni L, Wang S, Wang K, Lin R, Zheng C. Herpes simplex virus 1-encoded tegument protein VP16 abrogates the production of beta interferon (IFN) by inhibiting NF-kappaB activation and blocking IFN regulatory factor 3 to recruit its coactivator CBP. J Virol (2013) 87:9788–801. doi:10.1128/JVI.01440-13
143. Wang K, Ni L, Wang S, Zheng C. Herpes simplex virus 1 protein kinase US3 hyperphosphorylates p65/RelA and dampens NF-kappaB activation. J Virol (2014) 88:7941–51. doi:10.1128/JVI.03394-13
144. Best SM, Morris KL, Shannon JG, Robertson SJ, Mitzel DN, Park GS, et al. Inhibition of interferon-stimulated JAK-STAT signaling by a tick-borne flavivirus and identification of NS5 as an interferon antagonist. J Virol (2005) 79:12828–39. doi:10.1128/JVI.79.20.12828-12839.2005
145. Lin RJ, Chang BL, Yu HP, Liao CL, Lin YL. Blocking of interferon-induced Jak-Stat signaling by Japanese encephalitis virus NS5 through a protein tyrosine phosphatase-mediated mechanism. J Virol (2006) 80:5908–18. doi:10.1128/JVI.02714-05
146. Verweij MC, Wellish M, Whitmer T, Malouli D, Lapel M, Jonjic S, et al. Varicella viruses inhibit interferon-stimulated JAK-STAT signaling through multiple mechanisms. PLoS Pathog (2015) 11:e1004901. doi:10.1371/journal.ppat.1004901
147. Laurent-Rolle M, Boer EF, Lubick KJ, Wolfinbarger JB, Carmody AB, Rockx B, et al. The NS5 protein of the virulent West Nile virus NY99 strain is a potent antagonist of type I interferon-mediated JAK-STAT signaling. J Virol (2010) 84:3503–15. doi:10.1128/JVI.01161-09
148. Morrison J, Laurent-Rolle M, Maestre AM, Rajsbaum R, Pisanelli G, Simon V, et al. Dengue virus co-opts UBR4 to degrade STAT2 and antagonize type I interferon signaling. PLoS Pathog (2013) 9:e1003265. doi:10.1371/journal.ppat.1003265
149. Grant A, Ponia SS, Tripathi S, Balasubramaniam V, Miorin L, Sourisseau M, et al. Zika virus targets human STAT2 to inhibit type I interferon signaling. Cell Host Microbe (2016) 19:882–90. doi:10.1016/j.chom.2016.05.009
150. Stevenson NJ, Bourke NM, Ryan EJ, Binder M, Fanning L, Johnston JA, et al. Hepatitis C virus targets the interferon-alpha JAK/STAT pathway by promoting proteasomal degradation in immune cells and hepatocytes. FEBS Lett (2013) 587:1571–8. doi:10.1016/j.febslet.2013.03.041
151. Elliott J, Lynch OT, Suessmuth Y, Qian P, Boyd CR, Burrows JF, et al. Respiratory syncytial virus NS1 protein degrades STAT2 by using the Elongin-Cullin E3 ligase. J Virol (2007) 81:3428–36. doi:10.1128/JVI.02303-06
Keywords: innate immunity, antiviral, infection, RIG-I, type I IFNs, virus–host interaction
Citation: Liu Y, Olagnier D and Lin R (2017) Host and Viral Modulation of RIG-I-Mediated Antiviral Immunity. Front. Immunol. 7:662. doi: 10.3389/fimmu.2016.00662
Received: 07 October 2016; Accepted: 16 December 2016;
Published: 03 January 2017
Edited by:
Uday Kishore, Brunel University London, United KingdomReviewed by:
Winfried Barchet, University of Bonn, GermanyNicola Tamassia, University of Verona, Italy
Copyright: © 2017 Liu, Olagnier and Lin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rongtuan Lin, cm9uZ3R1YW4ubGluQG1jZ2lsbC5jYQ==
†Present address: David Olagnier, Department of Biomedicine, Aarhus Research Center for Innate Immunology, Aarhus University, Aarhus, Denmark
 Yiliu Liu
Yiliu Liu David Olagnier1,2†
David Olagnier1,2† Rongtuan Lin
Rongtuan Lin