- Institute for Glycomics, Griffith University, Gold Coast, QLD, Australia
Infectious diseases are a leading cause of morbidity and mortality worldwide, and vaccines are one of the most successful and cost-effective tools for disease prevention. One of the key considerations for rational vaccine development is the selection of appropriate antigens. Antigens must induce a protective immune response, and this response should be directed to stably expressed antigens so the target microbe can always be recognized by the immune system. Antigens with variable expression, due to environmental signals or phase variation (i.e., high frequency, random switching of expression), are not ideal vaccine candidates because variable expression could lead to immune evasion. Phase variation is often mediated by the presence of highly mutagenic simple tandem DNA repeats, and genes containing such sequences can be easily identified, and their use as vaccine antigens reconsidered. Recent research has identified phase variably expressed DNA methyltransferases that act as global epigenetic regulators. These phase-variable regulons, known as phasevarions, are associated with altered virulence phenotypes and/or expression of vaccine candidates. As such, genes encoding candidate vaccine antigens that have no obvious mechanism of phase variation may be subject to indirect, epigenetic control as part of a phasevarion. Bioinformatic and experimental studies are required to elucidate the distribution and mechanism of action of these DNA methyltransferases, and most importantly, whether they mediate epigenetic regulation of potential and current vaccine candidates. This process is essential to define the stably expressed antigen target profile of bacterial pathogens and thereby facilitate efficient, rational selection of vaccine antigens.
Introduction
Infectious diseases are a leading cause of morbidity and mortality worldwide. An estimated 23% of all deaths and 52% of deaths in children under the age of 5 years are caused by pathogenic microorganisms (1, 2). Over the past two centuries, many vaccines have been developed that aim to prime the host immune system and protect against disease. Consequently, the morbidity and mortality of many diseases have been significantly reduced, such as polio (3), or even eradicated, such as small pox (4). Vaccination is often considered one of the greatest triumphs of medical science (5).
To date, vaccines are available against 26 pathogens; with at least a further 24 vaccines in the development pipeline (6). The manufacture and composition of these vaccines varies significantly (7): from killed-whole cell or virus vaccines [e.g., Salk’s original polio vaccine (8)] and live attenuated vaccines [e.g., the measles, mumps, and rubella vaccine (9)], to “rationally designed” vaccines, which are subunit formulations specifically developed against selected cellular targets [e.g., the polysaccharide capsule-based pneumococcal conjugate vaccines (10) and the multivalent recombinant protein-based serogroup B meningococcal vaccine (11)]. The majority of available vaccines induce antibody-mediated protective immunity and target microorganisms and antigens that have little or no antigenic diversity or variability. Unfortunately, development of vaccines has been more difficult for pathogens that are antigenically diverse, as well as those that cannot be cultured in the laboratory, lack suitable animal models of infection, and/or those that are controlled by mucosal or T cell-dependent immune responses. There is an increasing need for the development of rationally designed vaccines for these pathogens, which has been facilitated by improvements in molecular biology techniques (e.g., DNA sequencing and manipulation; protein and carbohydrate purification; and chemical conjugation methods for production of multivalent vaccines) and increased understanding of pathogen biology, host–pathogen interactions, and the requirements for immunogenicity (e.g., immune correlates of protection, and the adjuvants required to elicit this protection) (12–15).
The era of “omics” and “big data” projects has unleashed a wealth of information for bacterial vaccine development, facilitating the ability to rapidly select potential vaccine antigens from genome and proteome analyses (14–17). However, antigens with variable expression, due to environmental signals or phase variation (i.e., high frequency, random switching of expression), possess inbuilt immune evasion capacity and do not make ideal vaccine candidates. Phase variation is often mediated by the presence of highly mutagenic simple tandem DNA repeats [also known as simple sequence repeats (SSRs)], and genes with these sequence features need to be identified so that can be discounted as vaccine antigens. However, recent research has identified phase variably expressed DNA methyltransferases that act as epigenetic regulators in many bacterial pathogens (18). These global epigenetic regulators, called phasevarions, can switch expression of candidate vaccine antigens that heretofore have been assumed to be stably expressed.
In this review, we provide an overview of key aspects that are important during antigen selection for pathogenic bacteria and focus on the impact of phasevarions on vaccine development.
Key Considerations for Vaccine Antigen Selection
For rationally designed, subunit vaccines to succeed, the selection of appropriate vaccine antigens is critical. Key features of vaccine antigens include (1) immunogenicity (i.e., the ability to elicit an immune response), (2) the ability to induce protection (i.e., the ability of the elicited immune response to prevent proliferation and/or the induction of pathology by the pathogen), and (3) conservation (i.e., the presence and sequence similarity between many/all strains of the pathogen). However, the stable expression of antigens during infection is also a critical factor in antigen selection that is often overlooked.
Several “omics” approaches are now routinely used to perform systems-based screening of potential antigens, such as genome-based reverse vaccinology, proteomics, transcriptomics, glycomics, and metabolomics (14–16, 19–21). These approaches allow high throughput identification of the potential antigens of a pathogen. The subsequent analysis of antigen conservation is a relatively straightforward process and has been assisted by the increasing availability of genomes, driven by decreases in sequencing costs (22, 23). Sequence availability has also made it possible to assess antigenic drift (change by accumulation of mutations) and shift (complete replacement of antigens), both of which must be taken into account to select stable and effective vaccine antigens (24, 25).
Investigation of whether the target antigen is actually expressed by the pathogen during infection in vivo is a more complex task, due to regulation by environmental signals and the potential for expression to be influenced by stochastic mechanisms. The transcription and translation of cellular factors are often contingent on environmental signals (e.g., tissue tropism, pH, and temperature) and cellular conditions (e.g., cell cycle) (26–28). For example, for pathogens such as Escherichia coli and other enteric pathogens, entry to the site of infection induces the expression of a different antigen repertoire (29, 30) that is triggered by diverse environmental or host signals such as pH (31) and temperature (32). While methods exist that allow the identification of expressed RNA (transcriptome) or protein (proteome) content under selected conditions, data collected often only represent a single physiological state that does not always reflect conditions found in the host. Accordingly, it is important to understand when and how cellular factors are expressed, to ensure that the target antigen is expressed during infection and in the same location (i.e., during mucosal or systemic infection) as the immune response elicited by the vaccine.
Antigen Expression and the Complication of Phase Variation
Phase variation is defined as the high frequency, reversible ON/OFF, or graded switching of gene expression, which is mediated through either genetic [e.g., due to variations in the number of simple tandem DNA repeats, or genome rearrangements (33, 34)] or epigenetic [e.g., via deoxyadenosine methylase (Dam) (35)] mechanisms at individual promoters. Many antigens in bacterial pathogens are phase variably expressed. For most phase-variable genes, switching occurs randomly during genome replication, and thus antigen expression is impossible to predict. Consequently, phase-variable components are not ideal vaccine targets since cells that have low, or no, expression of the target antigen may be able to evade the immune system (Figure 1A).
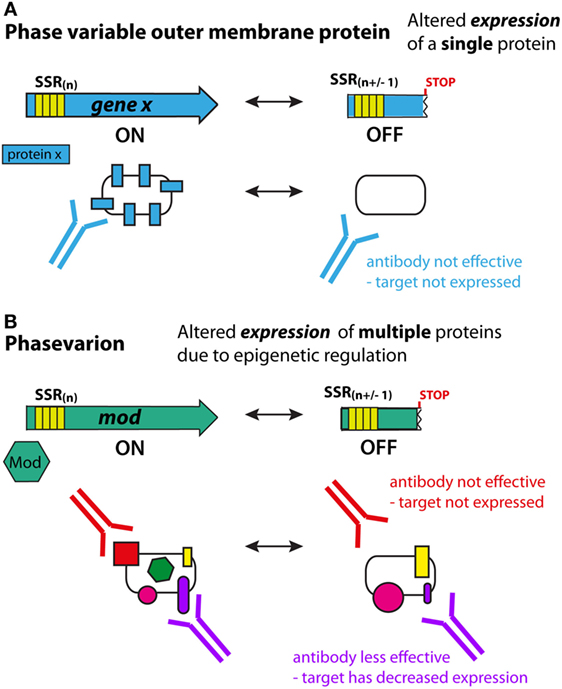
Figure 1. Phase variation and immune evasion. (A) For a phase-variable outer-membrane protein, slipped strand mispairing and changes in DNA sequence repeats in the gene during genome replication lead to ON/OFF expression of the encoded protein (blue). Antibodies to this antigen will not be effective if the protein has phased varied OFF. It is typically easy to predict phase-variable expression of these proteins due to the presence of DNA repeats (simple sequence repeat) in the coding region of the gene. (B) In phasevarions, phase-variable expression of a DNA methyltransferase causes genome-wide changes in DNA methylation, and expression differences in multiple genes due to epigenetic regulation. If these genes encode antigenic proteins/vaccine candidates, then methylation-dependent loss of expression (red protein) or reduced expression (purple protein) can lead to immune evasion as antibodies lose efficacy. However, due to the epigenetic nature of the phase-variable regulation, it is difficult to predict which proteins will have altered expression.
Many phase-variable genes can be identified bioinformatically, as the two main phase variation mechanisms, slipped strand mispairing and genome inversions, are well understood (36). Genes that are variable by slipped strand mispairing can be identified by the presence of multiple, tandem DNA repeats in the upstream or coding region of a gene. Slipped strand mispairing in DNA repeats causes loss or gain of repeats units, leading to frameshift mutations (ON/OFF switching) if located in the coding region, or altered expression levels if located within a promoter or operator region. In the case of genome inversions and recombination mediated mechanisms, phase-variable genes can be identified by the presence of various genetic markers such as recombinases, inverted sequence repeats, cryptic domains, and/or via genome comparisons for local reorganization (36, 37). Bioinformatic searches have been used successfully to identify numerous phase-variable genes in a variety of bacterial pathogens, such as Neisseria meningitidis (38–41), Neisseria gonorrhoeae (42), Campylobacter jejuni (43), Helicobacter pylori (44), and Haemophilus influenzae (45); and these genes are typically excluded from further screening of vaccine candidates. It is interesting to note that NadA, present in the meningococcal serogroup B vaccine (4CMenB, Bexsero), is phase variable. However, the variable expression of NadA is complex and was not easily identifiable in silico; the tandem repeats are distally located upstream of the nadA promoter and regulation involves both stochastic and classical mechanisms of gene regulation (46–48).
The DNA methyltransferase Dam is one of the best studied examples of epigenetic regulation in bacteria. While Dam itself is not phase variable or regulated, it is involved in phase variation of specific virulence genes in E. coli and Salmonella, such as pap (49, 50) and agn43 (51, 52). Dam is not believed to serve as a common transcriptional regulatory mechanism (35). Rather, competition between Dam and a particular DNA-binding regulatory protein provides opportunities for competitive stochastic switches that alter gene expression at specific target sites [reviewed in Ref. (35)].
Epigenetic Regulation of Antigens via Phase-Variable DNA Methyltransferases
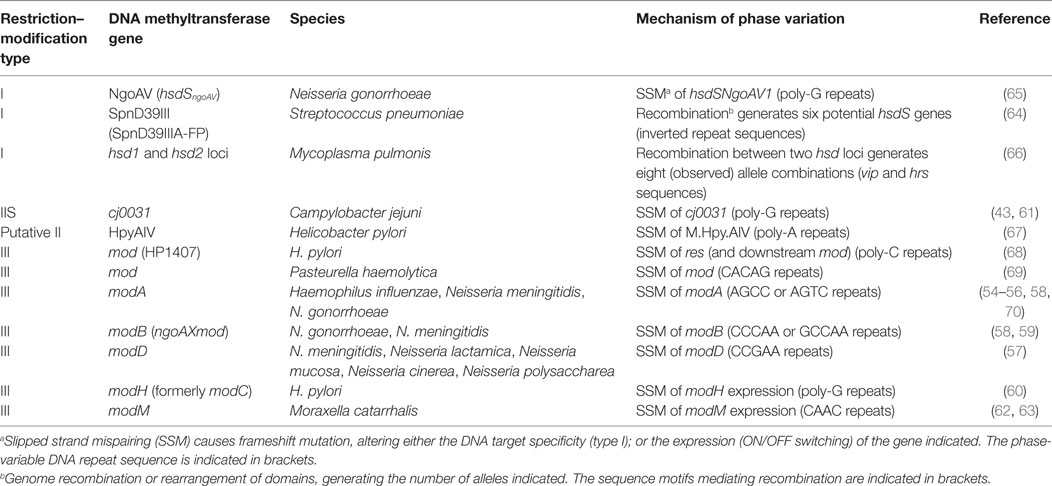
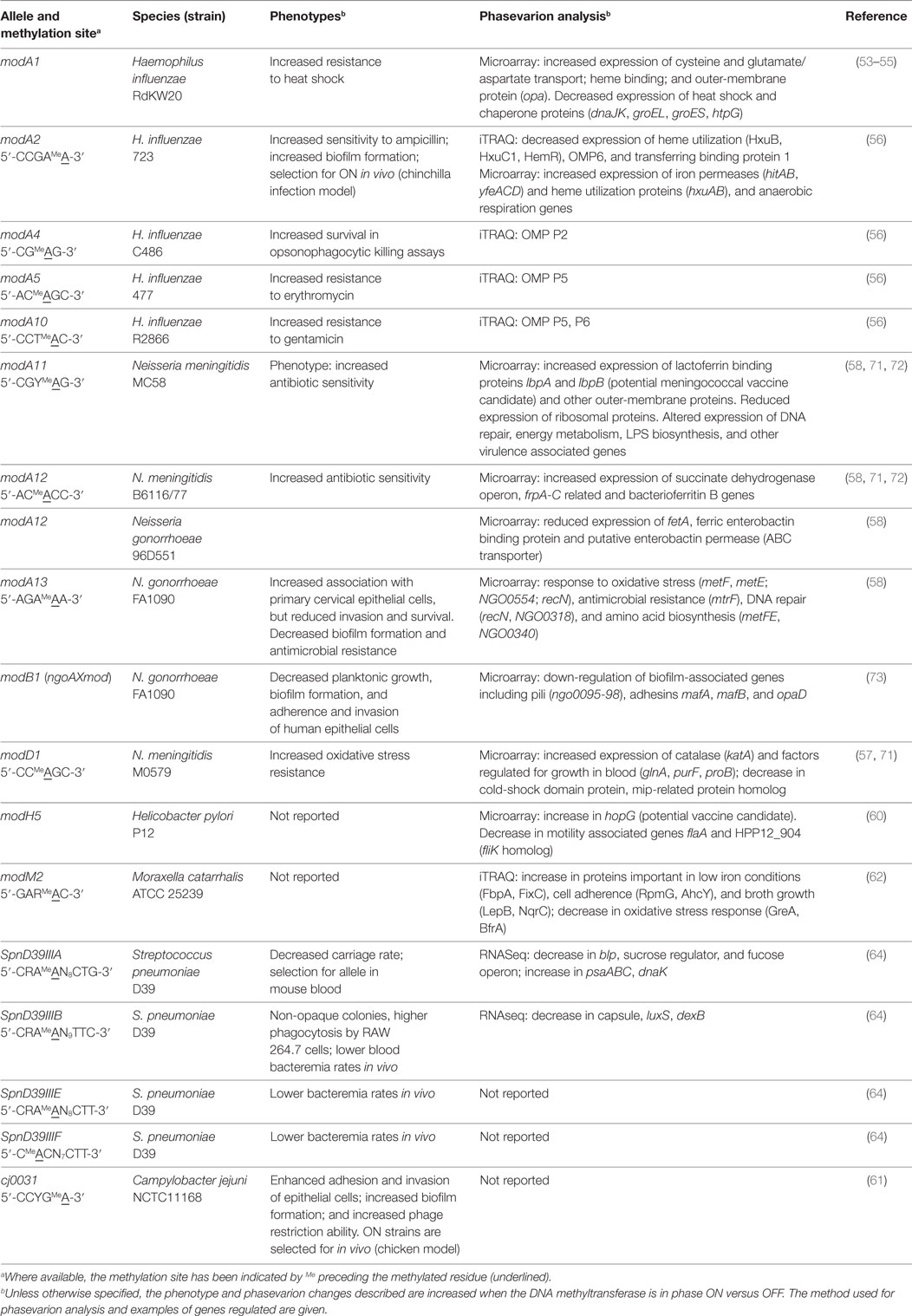
Phase-variable DNA methyltransferases, that act as global epigenetic regulators, have been identified in a number of pathogenic bacteria and add another layer of complexity to the process of antigen selection. Phase variation of these DNA methyltransferases results in coordinated, differential methylation of the entire genome in the DNA methyltransferases ON versus OFF variants. This leads to altered expression of a set of genes that is called a phasevarion, for phase-variable regulon (18, 53, 54) (Figure 1B). Phasevarions exert a pleiotropic effect and are associated with variable expression of proteins from diverse functional categories, such as metabolic processes, nutrient acquisition, stress responses, and virulence, as well controlling the variable expression of vaccine candidates. Phasevarions have been characterized in numerous pathogenic bacterial species, including H. influenzae (54–56); the pathogenic Neisseria (57–59); H. pylori (60), C. jejuni (43, 61), Moraxella catarrhalis (62, 63), and Streptococcus pneumoniae (64) (see Tables 1 and 2).
Phasevarions present a critical challenge for vaccine development, in that the genes controlled by phase-variable DNA methyltransferases do not have easily identifiable markers to indicate their phase-variable expression – these markers are only associated with the DNA methyltransferase and not the genes it regulates. Consequently, these components may be considered as potential vaccine candidates because their expression is erroneously assumed to be stable. This could potentially result in less effective, or completely ineffective, vaccines (Figure 1B).
Types of Phase-Variable DNA Methyltransferases
Phase-variable DNA methyltransferases have been described that are associated with all three major types of restriction–modification (R–M) systems (Figure 2A; Table 1). In type I R–M systems, the specificity of the DNA methyltransferase is dictated by a specificity subunit (HsdS). Phase variation is typically mediated by this locus, either by slipped strand mispairing [as with the NgoAV system of N. gonorrhoeae (65)] or by genome rearrangements of the hsdS subunit domains [as with the SpnD39III system of S. pneumoniae (64)]. In the SpnD39III system, genomic rearrangements result in recombination of one of two possible hsdS domain 1 sequences (TRD1.1 and TRD1.2) with one of three possible hsdS domain 2 sequences (TRD2.1, 2.2, and 2.3), which can result in the generation of six different hsdS alleles (SpnIIIA to SpnIIIF), producing six different HsdS specificity proteins (64) (Figure 2B). Accordingly, the SpnIII methyltransferase has six possible DNA specificities, each of which regulates expression of a distinct set of genes. While an individual cell expresses only one allele of each DNA methyltransferase at any particular time, populations of bacteria could express different mixtures of alleles.
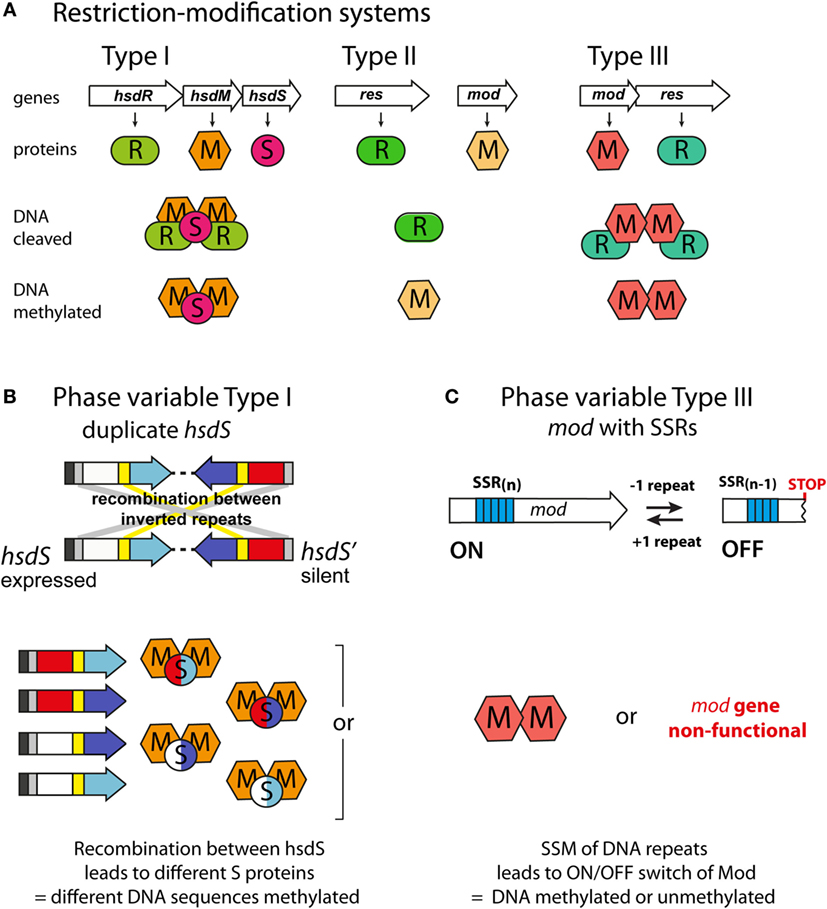
Figure 2. Phase-variable DNA methyltransferases. (A) The three main types of restriction–methylation (R–M) systems: type I consists of separate restriction (R), methyltransferase (M), and specificity (S) components, encoded by hsdR, hsdM, and hsdS genes, respectively. For restriction to occur, a pentameric R2M2S complex must form, but methylation can occur independently through a trimeric M2S complex. The HsdS subunits dictate the DNA sequences that are restricted and methylated. Type II systems are encoded by individual genes, often located separately on the chromosome. The resulting restriction (R) and methyltransferase (M) enzymes recognize and act independently upon the same DNA motif. Type III systems consist of colocalized mod [modification; encoding a methyltransferase, Mod (M)] and res [restriction; encoding a restriction enzyme, Res (R)] genes. Res proteins require Mod to restrict DNA (R2M2), but Mod enzymes are active as stand-alone methyltransferases (M2). (B) Phase variation of type I R–M systems via recombination between expressed (hsdS) and silent (hsdS') specificity genes. Each hsdS gene contains two target recognition domains (TRDs), each contributing half to the sequence recognized by the HsdS protein. Shuffling of each TRD via recombination between homologous inverted repeats (gray at 5′ end, yellow in center) leads to four possible combinations, and therefore, four different methyltransferase specificities in this example. (C) Phase variation of type III R–M systems via slipped strand mispairing (SSM) of simple sequence repeats in the open reading frame of the mod genes. Loss or gain of a repeat unit leads to variation in the open reading frame and either expression of a functional Mod protein (Mod ON), or transcriptional termination through the presence of a premature stop codon (Mod OFF).
In type II and III R–M systems, the DNA methyltransferases are independent proteins that dictate the specificity of the methylation site, and phase variation is typically mediated by slipped strand mispairing of SSRs in the coding sequence of the DNA methyltransferase (mod) gene (Table 1). Changes in repeat number cause frameshift mutations and switching of Mod protein expression between “ON” (expressed) or “OFF” (not expressed) states (Figure 2C). The type III Mod proteins are the most extensively studied (Table 1), and multiple allelic variants exist for each system, as determined by sequence differences in the DNA recognition domain responsible for methyltransferase specificity (18, 54, 56, 58, 60, 62, 71). For example, 21 modA alleles (56, 58, 70), 6 modB alleles (18, 58, 74), and 7 modD alleles (57, 74) have been identified to date. Unlike the type I systems described above, switching between alleles by genome rearrangement within a strain has not been reported and only one allele is present in a given strain. However, horizontal transfer of allele DNA recognition domains occurs and is postulated to generate novel DNA methyltransferase alleles over time (70, 75, 76).
Phasevarions and Vaccine Development
The challenge for vaccine development is to determine whether specific antigens are members of phasevarions prior to investing time in developing them as vaccine candidates. Previous studies have identified proposed vaccine candidates in phasevarions, including hopG (encoding a major outer-membrane protein) in the H. pylori ModH5 phasevarion (60), lbp (encoding lactoferrin binding protein) in the N. meningitidis ModA11 phasevarion (58), HMW adhesins in the H. influenzae ModA2, ModA4, and ModA5 phasevarions (56), and capsule in the S. pneumoniae SpnIIID39B phasevarion (64) (Table 2). Furthermore, many phasevarions are associated with virulence, which may be of concern as virulence determinants are often targets of vaccine development. For example, the ModA11 and ModA12 phasevarions in N. meningitidis (72), ModA13 and M.NgoAX in N. gonorrhoeae (58, 73), and ModA2, ModA5, and ModA10 in H. influenzae (56) all affect antimicrobial susceptibility. ModA2 (H. influenzae), ModA13 (N. gonorrhoeae) (58), and cj0031 (C. jejuni) (61) alter biofilm formation. N. meningitidis ModD1 can increase oxidative stress resistance and regulate factors important for growth and survival in blood (57). Different pneumococcal SpnIII alleles are associated with causing different phenotypes in S. pneumoniae, such as nasopharyngeal colonization (SpnIIIB) or bacteremia (SpnIIIA) (64) (Table 2).
Consequently, when considering the impact of phasevarions on vaccine development, it is important to know which allele(s) are present in the bacterial species, as well as the distribution of these alleles – that is, whether certain alleles predominate among the pathogenic strains that require targeting by the vaccine. Previous studies have used PCR and Sanger sequencing methods to identify and determine alleles (55–58, 60, 62); however, the increasing ease and lowered costs of full genome sequencing will enable the simple identification of phase-variable methyltransferases in broader, larger sample panels, as well as the identification of new or novel systems. For example, the availability of a large database of meningococcal genome sequences has recently been used to help survey the mod allele repertoire in over 1,600 isolates (74).
A bigger challenge lies in defining the proteins regulated within each phasevarion, as this must be determined experimentally. This has previously been accomplished by custom transcriptomic microarray analysis (57, 58, 60), but is being supplanted by next generation sequencing techniques [namely RNAseq, as in Ref. (64)] and proteomic analyses [e.g., iTRAQ, as in Ref. (56, 62)]. RNAseq allows the visualization of the full transcriptomic response to DNA methyltransferase phase variation, including differences in transcription of RNA genes (such as tRNAs) and non-coding RNAs (such as siRNAs and other regulatory RNAs). RNAseq will also provide valuable information about transcriptional start sites and upstream regulatory sequences for genes in the phasevarion, and possible transcription kinetics around methylation sites, enabling detailed mechanistic studies to be performed. In contrast, proteomic analyses will definitively identify the protein antigens differentially expressed by phasevarions under the conditions tested. This may differ from the transcriptomic data as RNA expression does not always correlate to protein translation, and so future studies should analyze expression data using multiple techniques in order to identify all members of each phasevarion. This will be invaluable for examining the actual changes in antigen levels and how this may affect vaccines.
The identification and analysis of genes controlled by phasevarions need to be carried out under conditions relevant to infection. This is because epigenetic regulation via DNA methylation is typically a multistep process, with DNA methylation affecting the action of regulatory proteins involved in transcription, rather than acting on transcriptional machinery itself [reviewed recently in Ref. (77)]. As such, conditions tested must be biologically relevant and allow these regulatory proteins to be active, in order to observe epigenetic regulation. This has been demonstrated by microarray analysis of the ModA11 phasevarion, where iron-limiting conditions were necessary to identify phasevarion members (mimicking iron limitation in the host, compared with standard laboratory culture conditions) (58). Unfortunately, the specific conditions that permit the full expression of the phasevarion can be difficult to determine, and bacteria should be grown under biologically relevant conditions, or if possible, collected directly from infection sites – such as from blood or mucosal surfaces. It is also critical that the whole, or representative, bacterial population is isolated and analyzed during phasevarion studies. This will allow the natural ON/OFF status and ratio of phase-variable DNA methyltransferases in the in vivo bacterial population to be understood.
Concluding Remarks
The development of bacterial vaccines depends on the selection of appropriate antigens. Ideal vaccine antigens are conserved, immunogenic, and protective. They should also be consistently expressed at high enough levels during infection to be targeted by the immune system. Transient and arbitrary expression makes antigen targeting by the immune system difficult and could lead to immune evasion via escape of a subpopulation that do not express the antigen. For this reason, phase-variable antigens do not make ideal vaccine candidates.
Phase-variable regulators complicate the prediction of stably expressed antigens, as the regulated genes within a phasevarion lack overt markers that indicate potential random switching of expression. While phasevarions have been studied in a range of pathogenic bacteria, important questions remain regarding allele variability, distribution, and regulatory mechanisms. More detailed understanding of these factors will help to elucidate the full complement of phase-variable genes in human pathogens for which vaccine development has been problematic, and help facilitate robust antigen selection for rational vaccine design in the future.
Author Contributions
All the authors contributed to drafting and revising the manuscript and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
This study was funded by Australian National Health and Medical Research Council (NHMRC) (Project Grants 2163530 and Career Development Fellowship to KS; Project Grant 1099279 to KS and JA; Program Grant 1071659 to MJ); Garnett Passe and Rodney Williams Memorial Foundation Research Training Fellowship to AT.
References
1. Liu L, Oza S, Hogan D, Perin J, Rudan I, Lawn JE, et al. Global, regional, and national causes of child mortality in 20-2013, with projections to inform post-2015 priorities: an updated systematic analysis. Lancet (2015) 385(9966):430–40. doi:10.1016/S0140-6736(14)61698-6
2. World Health Organisation. The Global Burden of Disease (2004 Update) (2004). Available from: http://www.who.int/healthinfo/global_burden_disease/2004_report_update/en/
3. Cochi SL, Freeman A, Guirguis S, Jafari H, Aylward B. Global polio eradication initiative: lessons learned and legacy. J Infect Dis (2014) 210(Suppl 1):S540–6. doi:10.1093/infdis/jiu345
4. Henderson DA. The eradication of smallpox – an overview of the past, present, and future. Vaccine (2011) 29(Suppl 4):D7–9. doi:10.1016/j.vaccine.2011.06.080
5. Greenwood B. The contribution of vaccination to global health: past, present and future. Philos Trans R Soc Lond B Biol Sci (2014) 369:1645.
6. World Health Organisation. World Health Organisation (2016). Available from: http://www.who.int/immunization/diseases/en/
7. Hajj Hussein I, Chams N, Chams S, El Sayegh S, Badran R, Raad M, et al. Vaccines through centuries: major cornerstones of global health. Front Public Health (2015) 3:269. doi:10.3389/fpubh.2015.00269
8. Salk JE, Krech U, Youngner JS, Bennett BL, Lewis LJ, Bazeley PL. Formaldehyde treatment and safety testing of experimental poliomyelitis vaccines. Am J Public Health Nations Health (1954) 44(5):563–70. doi:10.2105/AJPH.44.5.563
9. Lievano F, Galea SA, Thornton M, Wiedmann RT, Manoff SB, Tran TN, et al. Measles, mumps, and rubella virus vaccine (M-M-R™II): a review of 32 years of clinical and postmarketing experience. Vaccine (2012) 30(48):6918–26. doi:10.1016/j.vaccine.2012.08.057
10. Tromp KM, Campbell MW, Vazquez A. Recent developments and future directions of pneumococcal vaccine recommendations. Clin Ther (2015) 37(5):928–34. doi:10.1016/j.clinthera.2015.03.025
11. Esposito S, Principi N. Vaccine profile of 4CMenB: a four-component Neisseria meningitidis serogroup B vaccine. Expert Rev Vaccines (2014) 13(2):193–202. doi:10.1586/14760584.2014.874949
12. Nabel GJ. Designing tomorrow’s vaccines. New Engl J Med (2013) 368(6):551–60. doi:10.1056/NEJMra1204186
13. Rueckert C, Guzman CA. Vaccines: from empirical development to rational design. PLoS Pathog (2012) 8(11):e1003001. doi:10.1371/journal.ppat.1003001
14. Delany I, Rappuoli R, Seib KL. Vaccines, reverse vaccinology, and bacterial pathogenesis. Cold Spring Harb Perspect Med (2013) 3(5):a012476. doi:10.1101/cshperspect.a012476
15. Rinaudo CD, Telford JL, Rappuoli R, Seib KL. Vaccinology in the genome era. J Clin Invest (2009) 119(9):2515–25. doi:10.1172/JCI38330
16. Rappuoli R. Reverse vaccinology, a genome-based approach to vaccine development. Vaccine (2001) 19(17–19):2688–91. doi:10.1016/S0264-410X(00)00554-5
17. Adamczyk-Poplawska M, Markowicz S, Jagusztyn-Krynicka EK. Proteomics for development of vaccine. J Proteomics (2011) 74(12):2596–616. doi:10.1016/j.jprot.2011.01.019
18. Srikhanta YN, Fox KL, Jennings MP. The phasevarion: phase variation of type III DNA methyltransferases controls coordinated switching in multiple genes. Nat Rev Microbiol (2010) 8(3):196–206. doi:10.1038/nrmicro2283
19. Seib KL, Dougan G, Rappuoli R. The key role of genomics in modern vaccine and drug design for emerging infectious diseases. PLoS Genet (2009) 5(10):e1000612. doi:10.1371/journal.pgen.1000612
20. Geissner A, Anish C, Seeberger PH. Glycan arrays as tools for infectious disease research. Curr Opin Chem Biol (2014) 18:38–45. doi:10.1016/j.cbpa.2013.11.013
21. Schulz C, Hammerschmidt S. Exploitation of physiology and metabolomics to identify pneumococcal vaccine candidates. Expert Rev Vaccines (2013) 12(9):1061–75. doi:10.1586/14760584.2013.824708
22. Land M, Hauser L, Jun S-R, Nookaew I, Leuze MR, Ahn T-H, et al. Insights from 20 years of bacterial genome sequencing. Funct Integr Genomics (2015) 15(2):141–61. doi:10.1007/s10142-015-0433-4
23. Bentley SD, Parkhill J. Genomic perspectives on the evolution and spread of bacterial pathogens. Proc R Soc Lond B Biol Sci (2015) 282:1821.
24. Carrat F, Flahault A. Influenza vaccine: the challenge of antigenic drift. Vaccine (2007) 25(39–40):6852–62. doi:10.1016/j.vaccine.2007.07.027
25. White PA. Evolution of norovirus. Clin Microbiol Infect (2014) 20(8):741–5. doi:10.1111/1469-0691.12746
26. Browning DF, Busby SJ. The regulation of bacterial transcription initiation. Nat Rev Microbiol (2004) 2(1):57–65. doi:10.1038/nrmicro787
27. Buelow DR, Raivio TL. Three (and more) component regulatory systems – auxiliary regulators of bacterial histidine kinases. Mol Microbiol (2010) 75(3):547–66. doi:10.1111/j.1365-2958.2009.06982.x
28. Staron A, Sofia HJ, Dietrich S, Ulrich LE, Liesegang H, Mascher T. The third pillar of bacterial signal transduction: classification of the extracytoplasmic function (ECF) sigma factor protein family. Mol Microbiol (2009) 74(3):557–81. doi:10.1111/j.1365-2958.2009.06870.x
29. Winter SE, Winter MG, Godinez I, Yang H-J, Rüssmann H, Andrews-Polymenis HL, et al. A rapid change in virulence gene expression during the transition from the intestinal lumen into tissue promotes systemic dissemination of Salmonella. PLoS Pathog (2010) 6(8):e1001060. doi:10.1371/journal.ppat.1001060
30. Yang J, Tauschek M, Hart E, Hartland EL, Robins-Browne RM. Virulence regulation in Citrobacter rodentium: the art of timing. Microb Biotechnol (2009) 3(3):259–68. doi:10.1111/j.1751-7915.2009.00114.x
31. Schwan WR, Lee JL, Lenard FA, Matthews BT, Beck MT. Osmolarity and pH growth conditions regulate fim gene transcription and type 1 pilus expression in uropathogenic Escherichia coli. Infect Immun (2002) 70(3):1391–402. doi:10.1128/IAI.70.3.1391-1402.2002
32. Gophna U, Ron EZ. Virulence and the heat shock response. Int J Med Microbiol (2003) 292(7–8):453–61. doi:10.1078/1438-4221-00230
33. Henderson IR, Owen P, Nataro JP. Molecular switches – the on and off of bacterial phase variation. Mol Microbiol (1999) 33(5):919–32. doi:10.1046/j.1365-2958.1999.01555.x
34. Moxon R, Bayliss C, Hood D. Bacterial contingency loci: the role of simple sequence DNA repeats in bacterial adaptation. Annu Rev Genet (2006) 40(1):307–33. doi:10.1146/annurev.genet.40.110405.090442
35. Adhikari S, Curtis PD. DNA methyltransferases and epigenetic regulation in bacteria. FEMS Microbiol Rev (2016) 40(5):575–91. doi:10.1093/femsre/fuw023
36. van der Woude MW, Bäumler AJ. Phase and antigenic variation in bacteria. Clin Microbiol Rev (2004) 17(3):581–611. doi:10.1128/CMR.17.3.581-611.2004
37. Hallet B. Playing Dr Jekyll and Mr Hyde: combined mechanisms of phase variation in bacteria. Curr Opin Microbiol (2001) 4(5):570–81. doi:10.1016/S1369-5274(00)00253-8
38. Martin P, van de Ven T, Mouchel N, Jeffries AC, Hood DW, Moxon ER. Experimentally revised repertoire of putative contingency loci in Neisseria meningitidis strain MC58: evidence for a novel mechanism of phase variation. Mol Microbiol (2003) 50(1):245–57. doi:10.1046/j.1365-2958.2003.03678.x
39. Peng J, Yang L, Yang F, Yang J, Yan Y, Nie H, et al. Characterization of ST-4821 complex, a unique Neisseria meningitidis clone. Genomics (2008) 91(1):78–87. doi:10.1016/j.ygeno.2007.10.004
40. Saunders NJ, Jeffries AC, Peden JF, Hood DW, Tettelin H, Rappuoli R, et al. Repeat-associated phase variable genes in the complete genome sequence of Neisseria meningitidis strain MC58. Mol Microbiol (2000) 37(1):207–15. doi:10.1046/j.1365-2958.2000.02000.x
41. Snyder LAS, Butcher SA, Saunders NJ. Comparative whole-genome analyses reveal over 100 putative phase-variable genes in the pathogenic Neisseria spp. Microbiology (2001) 147(8):2321–32. doi:10.1099/00221287-147-8-2321
42. Jordan PW, Snyder LA, Saunders NJ. Strain-specific differences in Neisseria gonorrhoeae associated with the phase variable gene repertoire. BMC Microbiol (2005) 5:21. doi:10.1186/1471-2180-5-21
43. Bayliss CD, Bidmos FA, Anjum A, Manchev VT, Richards RL, Grossier J-P, et al. Phase variable genes of Campylobacter jejuni exhibit high mutation rates and specific mutational patterns but mutability is not the major determinant of population structure during host colonization. Nucleic Acids Res (2012) 40(13):5876–89. doi:10.1093/nar/gks246
44. Saunders NJ, Peden JF, Hood DW, Moxon ER. Simple sequence repeats in the Helicobacter pylori genome. Mol Microbiol (1998) 27(6):1091–8. doi:10.1046/j.1365-2958.1998.00768.x
45. Hood DW, Deadman ME, Jennings MP, Bisercic M, Fleischmann RD, Venter JC, et al. DNA repeats identify novel virulence genes in Haemophilus influenzae. Proc Natl Acad Sci U S A (1996) 93(20):11121–5. doi:10.1073/pnas.93.20.11121
46. Liguori A, Malito E, Lo Surdo P, Fagnocchi L, Cantini F, Haag AF, et al. Molecular basis of ligand-dependent regulation of NadR, the transcriptional repressor of meningococcal virulence factor NadA. PLoS Pathog (2016) 12(4):e1005557. doi:10.1371/journal.ppat.1005557
47. Martin P, Makepeace K, Hill SA, Hood DW, Moxon ER. Microsatellite instability regulates transcription factor binding and gene expression. Proc Natl Acad Sci U S A (2005) 102(10):3800–4. doi:10.1073/pnas.0406805102
48. Metruccio MME, Pigozzi E, Roncarati D, Berlanda Scorza F, Norais N, Hill SA, et al. A novel phase variation mechanism in the meningococcus driven by a ligand-responsive repressor and differential spacing of distal promoter elements. PLoS Pathog (2009) 5(12):e1000710. doi:10.1371/journal.ppat.1000710
49. Hernday AD, Braaten BA, Low DA. The mechanism by which DNA adenine methylase and PapI activate the pap epigenetic switch. Mol Cell (2003) 12(4):947–57. doi:10.1016/S1097-2765(03)00383-6
50. van der Woude M, Braaten B, Low D. Epigenetic phase variation of the pap operon in Escherichia coli. Trends Microbiol (1996) 4(1):5–9. doi:10.1016/0966-842X(96)81498-3
51. Haagmans W, Van Der Woude M. Phase variation of Ag43 in Escherichia coli: Dam-dependent methylation abrogates OxyR binding and OxyR-mediated repression of transcription. Mol Microbiol (2000) 35(4):877–87. doi:10.1046/j.1365-2958.2000.01762.x
52. Wallecha A, Munster V, Correnti J, Chan T, van der Woude M. Dam- and OxyR-dependent phase variation of agn43: essential elements and evidence for a new role of DNA methylation. J Bacteriol (2002) 184(12):3338–47. doi:10.1128/JB.184.12.3338-3347.2002
53. Fox KL, Srikhanta YN, Jennings MP. Phase variable type III restriction-modification systems of host-adapted bacterial pathogens. Mol Microbiol (2007) 65(6):1375–9. doi:10.1111/j.1365-2958.2007.05873.x
54. Srikhanta YN, Maguire TL, Stacey KJ, Grimmond SM, Jennings MP. The phasevarion: a genetic system controlling coordinated, random switching of expression of multiple genes. Proc Natl Acad Sci U S A (2005) 102(15):5547–51. doi:10.1073/pnas.0501169102
55. Fox KL, Dowideit SJ, Erwin AL, Srikhanta YN, Smith AL, Jennings MP. Haemophilus influenzae phasevarions have evolved from type III DNA restriction systems into epigenetic regulators of gene expression. Nucleic Acids Res (2007) 35(15):5242–52. doi:10.1093/nar/gkm571
56. Atack JM, Srikhanta YN, Fox KL, Jurcisek JA, Brockman KL, Clark TA, et al. A biphasic epigenetic switch controls immunoevasion, virulence and niche adaptation in non-typeable Haemophilus influenzae. Nat Commun (2015) 6:7828. doi:10.1038/ncomms8828
57. Seib KL, Pigozzi E, Muzzi A, Gawthorne JA, Delany I, Jennings MP, et al. A novel epigenetic regulator associated with the hypervirulent Neisseria meningitidis clonal complex 41/44. FASEB J (2011) 25(10):3622–33. doi:10.1096/fj.11-183590
58. Srikhanta YN, Dowideit SJ, Edwards JL, Falsetta ML, Wu HJ, Harrison OB, et al. Phasevarions mediate random switching of gene expression in pathogenic Neisseria. PLoS Pathog (2009) 5(4):e1000400. doi:10.1371/journal.ppat.1000400
59. Adamczyk-Poplawska M, Lower M, Piekarowicz A. Characterization of the NgoAXP: phase-variable type III restriction-modification system in Neisseria gonorrhoeae. FEMS Microbiol Lett (2009) 300(1):25–35. doi:10.1111/j.1574-6968.2009.01760.x
60. Srikhanta YN, Gorrell RJ, Steen JA, Gawthorne JA, Kwok T, Grimmond SM, et al. Phasevarion mediated epigenetic gene regulation in Helicobacter pylori. PLoS One (2011) 6(12):e27569. doi:10.1371/journal.pone.0027569
61. Anjum A, Brathwaite KJ, Aidley J, Connerton PL, Cummings NJ, Parkhill J, et al. Phase variation of a Type IIG restriction-modification enzyme alters site-specific methylation patterns and gene expression in Campylobacter jejuni strain NCTC11168. Nucleic Acids Res (2016) 44(10):4581–94. doi:10.1093/nar/gkw019
62. Blakeway LV, Power PM, Jen FE, Worboys SR, Boitano M, Clark TA, et al. ModM DNA methyltransferase methylome analysis reveals a potential role for Moraxella catarrhalis phasevarions in otitis media. FASEB J (2014) 28(12):5197–207. doi:10.1096/fj.14-256578
63. Seib KL, Peak IR, Jennings MP. Phase variable restriction-modification systems in Moraxella catarrhalis. FEMS Immunol Med Microbiol (2002) 32(2):159–65. doi:10.1016/S0928-8244(01)00294-2
64. Manso AS, Chai MH, Atack JM, Furi L, De Ste Croix M, Haigh R, et al. A random six-phase switch regulates pneumococcal virulence via global epigenetic changes. Nat Commun (2014) 5:5055. doi:10.1038/ncomms6055
65. Adamczyk-Poplawska M, Lower M, Piekarowicz A. Deletion of one nucleotide within the homonucleotide tract present in the hsdS gene alters the DNA sequence specificity of type I restriction-modification system NgoAV. J Bacteriol (2011) 193(23):6750–9. doi:10.1128/JB.05672-11
66. Dybvig K, Sitaraman R, French CT. A family of phase-variable restriction enzymes with differing specificities generated by high-frequency gene rearrangements. Proc Natl Acad Sci U S A (1998) 95(23):13923–8. doi:10.1073/pnas.95.23.13923
67. Skoglund A, Björkholm B, Nilsson C, Andersson AF, Jernberg C, Schirwitz K, et al. Functional analysis of the M.HpyAIV DNA methyltransferase of Helicobacter pylori. J Bacteriol (2007) 189(24):8914–21. doi:10.1128/JB.00108-07
68. de Vries N, Duinsbergen D, Kuipers EJ, Pot RGJ, Wiesenekker P, Penn CW, et al. Transcriptional phase variation of a Type III restriction-modification system in Helicobacter pylori. J Bacteriol (2002) 184(23):6615–23. doi:10.1128/JB.184.23.6615-6624.2002
69. Ryan KA, Lo RYC. Characterization of a CACAG pentanucleotide repeat in Pasteurella haemolytica and its possible role in modulation of a novel type III restriction-modification system. Nucleic Acids Res (1999) 27(6):1505–11. doi:10.1093/nar/27.6.1505
70. Gawthorne JA, Beatson SA, Srikhanta YN, Fox KL, Jennings MP. Origin of the diversity in DNA recognition domains in phasevarion associated modA genes of pathogenic Neisseria and Haemophilus influenzae. PLoS One (2012) 7(3):e32337. doi:10.1371/journal.pone.0032337
71. Seib KL, Jen FE, Tan A, Scott AL, Kumar R, Power PM, et al. Specificity of the ModA11, ModA12 and ModD1 epigenetic regulator N6-adenine DNA methyltransferases of Neisseria meningitidis. Nucleic Acids Res (2015) 43(8):4150–62. doi:10.1093/nar/gkv219
72. Jen FE, Seib KL, Jennings MP. Phasevarions mediate epigenetic regulation of antimicrobial susceptibility in Neisseria meningitidis. Antimicrob Agents Chemother (2014) 58(7):4219–21. doi:10.1128/AAC.00004-14
73. Kwiatek A, Mrozek A, Bacal P, Piekarowicz A, Adamczyk-Popławska M. Type III methyltransferase M.NgoAX from Neisseria gonorrhoeae FA1090 regulates biofilm formation and interactions with human cells. Front Microbiol (2015) 6:1426. doi:10.3389/fmicb.2015.01426
74. Tan A, Hill DMC, Harrison OB, Srikhanta YN, Jennings MP, Maiden MCJ, et al. Distribution of the type III DNA methyltransferases modA, modB and modD among Neisseria meningitidis genotypes: implications for gene regulation and virulence. Sci Rep (2016) 6:21015. doi:10.1038/srep21015
75. Bayliss CD, Callaghan MJ, Moxon ER. High allelic diversity in the methyltransferase gene of a phase variable type III restriction-modification system has implications for the fitness of Haemophilus influenzae. Nucleic Acids Res (2006) 34(14):4046–59. doi:10.1093/nar/gkl568
76. Furuta Y, Kobayashi I. Movement of DNA sequence recognition domains between non-orthologous proteins. Nucleic Acids Res (2012) 40(18):9218–32. doi:10.1093/nar/gks681
Keywords: phase variation, vaccine, DNA methyltransferase, DNA modification enzyme, gene expression, epigenetics
Citation: Tan A, Atack JM, Jennings MP and Seib KL (2016) The Capricious Nature of Bacterial Pathogens: Phasevarions and Vaccine Development. Front. Immunol. 7:586. doi: 10.3389/fimmu.2016.00586
Received: 09 October 2016; Accepted: 28 November 2016;
Published: 12 December 2016
Edited by:
Lee Mark Wetzler, Boston University School of Medicine, USAReviewed by:
Sanjay Ram, University of Massachusetts Medical Center, USAJosep Casadesús, Universidad de Sevilla, Spain
Copyright: © 2016 Tan, Atack, Jennings and Seib. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kate L. Seib, ay5zZWliQGdyaWZmaXRoLmVkdS5hdQ==
 Aimee Tan
Aimee Tan John M. Atack
John M. Atack Michael P. Jennings
Michael P. Jennings Kate L. Seib
Kate L. Seib