- 1The Key Laboratory of Carcinogenesis of the Chinese Ministry of Health, Xiangya Hospital, Central South University, Changsha, Hunan, China
- 2The Key Laboratory of Carcinogenesis and Cancer Invasion of the Chinese Ministry of Education, Cancer Research Institute, Central South University, Changsha, Hunan, China
- 3Hunan Key Laboratory of Non-resolving Inflammation and Cancer, Disease Genome Research Center, The Third Xiangya Hospital, Central South University, Changsha, Hunan, China
Regulatory T (Treg) cells are a group of cells that are heterogeneous in origin and in functional activity. Treg cells comprise a necessary balance to adaptive immune responses. As key regulators of self-tolerance, Treg cells have been involved in a series of pathologic processes and considered as therapeutic targets. Here, we summarize recent research regarding Treg cell origins and their functional classification, highlight the role of exosomes and non-coding RNA in modulating Treg cell homeostasis, and discuss the current understanding of resident Treg cells.
Introduction
Immune tolerance regulation is a critical aspect of immunology. Distinct populations of T cells with suppressor functions make a major contribution to such regulation. Regulatory T (Treg) cells are important for preventing inappropriate responses by the immune system (1). Treg cells exert their suppressive role from triggering of innate immune cells to adaptive cell-mediated responses. Treg cells have been implicated in a number of pathologic processes involving severe systemic autoimmunity and many malignancies (2, 3). As critical regulator of immune tolerance and homeostasis, Treg cells have been regarded as immunotherapeutic targets. Manipulation of the number and/or suppressive activity of Treg cells has been shown to be impactful in the treatment of autoimmune disorders, allograft rejection, and cancer (4–6). Thus, understanding the local immune regulation and regulatory mechanisms of Treg cells is essential. In this review, the characteristics of Treg cells and tissue-resident Treg cells are summarized. The regulatory mechanisms of Treg cells are also discussed, focusing on exosomes and non-coding RNA.
Characterization of Treg Cells
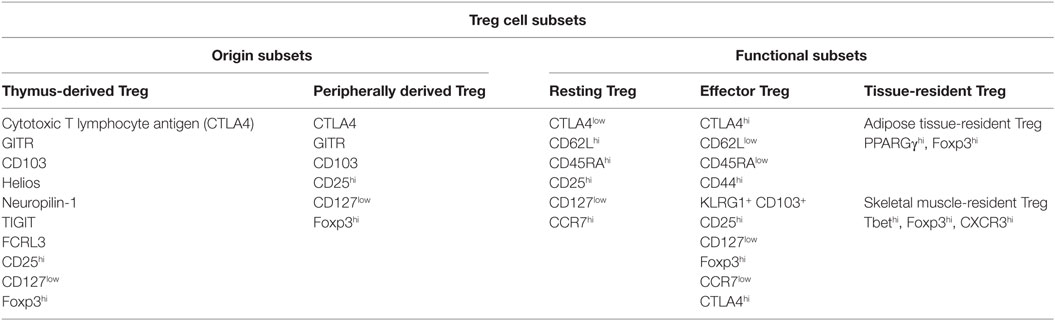
Regulatory T cells are distinguished from other lymphocytes by several characteristics, including surface marker, transcription factor, origin, and function (Table 1). Treg cells express CD25 (IL-2 receptor α chain) and are dependent on stimulation by IL-2 for their function (7). Foxp3 was discovered to be a “master regulator” of Treg cell development and function (8–11). Mutations of the Foxp3 gene in humans result in Treg deficiencies and are responsible for immunodysregulation polyendocrinopathy enteropathy X-linked syndrome (12). Foxp3 and CD25 are reliable and constitutive markers that have been used to isolate and characterize Treg cells. In addition to CD25 and Foxp3, Treg cells express co-stimulatory and co-inhibitory molecules that are involved in their suppressive function, such as CD28 and cytotoxic T lymphocyte antigen 4 (CTLA4), tumor necrosis factor (TNF), and TNF receptor family members, including RANKL and GITR, and Toll-like receptors (4). It has been shown that CD127 expression inversely correlates with Foxp3 expression and CD4+ Treg cells suppressive function (13) and that the combined use of Foxp3+, CD25+, and CD127−, might better define the Treg cell population with suppressive functions (14).
Treg cells can be separated according to their two possible origins: tTreg (thymus-derived Treg) cells and pTreg (peripherally derived Treg) cells, also called natural Treg cells and induced Treg cells, respectively (15). Most Treg cells arise in the thymus, where the expression of Foxp3 is initiated via a combination of self-antigen recognition with moderate- to high-avidity and microenvironmental influences, and these tTreg cells migrate to the periphery to maintain self-tolerance (16). Moreover, tTreg cells can also be induced in the periphery from Foxp3– recent thymic emigrants (17). Another way of Treg generation is in the periphery, where CD4+ T cells develop into pTreg cells upon encountering antigens under certain conditions (18, 19). Two populations of peripherally induced CD4+ Treg cells have been described: Tr1 cells and Th3 cells, they are induced in peripheral, secrete interleukin 10 (IL-10) and/or transforming growth factor beta (TGF-beta), and exert suppress function via a cytokine-dependent mechanism (20–22). Both thymic-derived and peripherally induced Treg cells are antigen specific, possess T-cell receptors, and are selected with a suppressive function. A variety of molecular markers can be used to distinguish different Treg populations. Transcription factor Helios and cell surface glycoprotein neuropilin-1 are usually highly expressed by tTreg cells but poorly expressed by pTreg cells, as thus, both these molecular markers can be applied to distinguish tTreg from pTreg cells; nevertheless, pTreg cells may upregulate these factors expression depending on local inflammatory conditions or the type of antigen-presenting cells and activation signals that are present (15, 23, 24). Furthermore, a study of human Treg subsets described an important role for T cell immunoreceptor with Ig and ITIM domains (TIGIT) and FcR-like 3 (FCRL3) in distinguishing tTreg cells from pTreg cells (25).
Regulatory T cells can also be divided into functional subpopulations as well as into origin subsets (26–28). (1) Resting Treg cells (CD62LhiCCR7+ or CD45RAhiCD25low Treg cells), also known as central or naive Treg cells, conprise the great number of Treg cells in secondary lymphoid organs and in circulation. Resting Treg cells have a history of antigen exposure and baseline suppressive function, and they share circulation patterns and activation markers with naive and memory conventional T cells. (2) Effector Treg cells (CD45RAlowCD25hi or CD62LlowCCR7lowCD44hiKLRG1+CD103+ Treg cells), also known as activated Treg cells, constitute a small part of Treg cells in circulation and in secondary lymphoid organs (29). This subset has enhanced function and signs of recent antigen encounter and shares phenotypic features with activated conventional T cells. It remains unclear whether effector Treg cells are capable of reverting to resting Treg cells or are terminally differentiated. (3) Recently, a greater emphasis has been placed on a specific subset of tissue-resident Treg cells that take part in immune processes as well as in the maintenance of tissue homeostasis (27, 28, 30, 31). The phenotype and function of tissue-resident Treg cells are different from those of the classical lymphoid Treg cells. Each tissue might have its own unique tissue-resident Treg cells, which have good sensitivity and a high turnover rate in response to a number of environment signals (30). These characteristics of tissue-resident Treg cells enable rapid adjustments in Treg cell location and number that are required to effectively react to immune dynamics (27, 30). Moreover, to be able to optimally control the immune response in dynamic tissue microenvironments, Treg cells can afford a certain degree of functional plasticity. Treg cells preserve their core immunosuppressive characteristics and alter their transcriptional program to achieve functional plasticity. Recent work has demonstrated that tissue-resident Treg cells often have distinct transcription programs from lymphoid organ Treg cells. For instance, visceral adipose tissue Treg cells show high expression of the transcription factor peroxisome proliferator-activated receptor γ, which acts as a crucial regulator of adipocyte differentiation. Similarly, skeletal muscle-resident Treg cells display a transcriptional program that sustains their repair function following acute injury (32). Furthermore, to control the Teff cell response, Treg cells can express distinct transcription factors and immunosuppressive molecules associated with that type of Teff cell. For example, Tbet+ Treg cells, induced by type 1 inflammatory conditions, express chemokine (C–X–C motif) receptor 3 and accumulate at T helper 1 (Th1) cell-mediated inflammation sites. CXCR3 is a key molecule on Th1 cells that mediates the accumulation of Th1 cells at sites of local inflammation. Thus, the function of Treg cells partially depends on the degree of plasticity that they exhibit in response to the microenvironment (32–34).
Treg Cells and Exosomes
Exosomes are small membrane vesicles derived from multivesicular bodies or from the plasma membrane (35). Exosomes play critical roles in intercellular communication, as they transfer RNAs, proteins, and other type of molecules between donor and receptor cells (36). Exosome protein contents mostly reflect that of the parent cells, and exosomes are enriched in cytoskeleton molecules, cytoplasmic enzymes, signal transduction proteins, and so on (37). Furthermore, exosomes contain a variety of non-coding RNAs (ncRNAs), involving microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circRNAs (36–39).
Exosomes participate in important biological functions and are involved in numerous physiological processes. Immune and non-immune cell-derived exosomes play critical roles in immunity regulation; these exosomes can mediate immune homeostasis and can drive inflammation, autoimmunity, and infectious disease pathology (40–44). Exosomes derived from Treg cells appear to be greater in quantity than those from other type of T cells and are regulated by changes in intracellular calcium, synthesis of the sphingolipid ceramide, hypoxia, and the presence of IL-2 (45–48). Exosomes make great contribution to Treg cells function (Figure 1), as inhibiting the release of exosomes can reverse the suppressive capabilities of Treg cells, such as effector T cells (Teffs) suppression and disease prevention. Rab27-DKO Treg cells that failed to release exosomes also failed to prevent disease, resulting in colon shortening, weight loss, and increased IFNγ expression. The failure of these cells to control Teffs also led to significant colonic and systemic inflammation and IFNγ expression (49). Recently, the transfer of miRNAs, including Let-7d, Let-7b, and miR-155, via Treg cell exosomes to conventional T cells has been shown. Let-7d-containing Treg cell-derived exosomes contributed to the suppression of Th1 cell proliferation and IFNγ secretion via Cox-2 (40). However, upregulation of Let-7b in Th1 cells had little impact on proliferation or IFNγ production. Treg cells transfer miR-155 to conventional T cells with a concomitant upregulation of several Treg-cell-associated genes in recipient cells (40). Furthermore, several immunomodulatory molecules, including CD73, CD25, and CTLA4, were also found in Treg exosomes (43, 49). Treg cell-derived CD73-expressing exosomes contribute to their suppressive activity through the production of the anti-inflammatory mediator adenosine (43). Adenosine can bind to adenosine receptors, triggering intracellular cAMP, leading to the inhibition of cytokine production by activated Teffs. CD25 and CTLA4 are expressed in Treg cell-derived exosomes, but these molecules may not contribute to the suppressive function of Treg cells. Several molecules, including FasL, CD39, PDL1, and Galectin1, are present on exosomes derived from other type of cells. FasL- and CD39-containing exosomes have been shown to have immunomodulatory properties. Lymphoblastoid cell-derived MHCII+FasL+exosomes induced apoptosis in CD4+ T cells. Tumor exosomes express CD39 and CD73, which are capable to suppress T cells (50–53). Whether they are also present on Treg cell-derived exosomes and play a role in Treg cells suppressive function is yet to be validated.
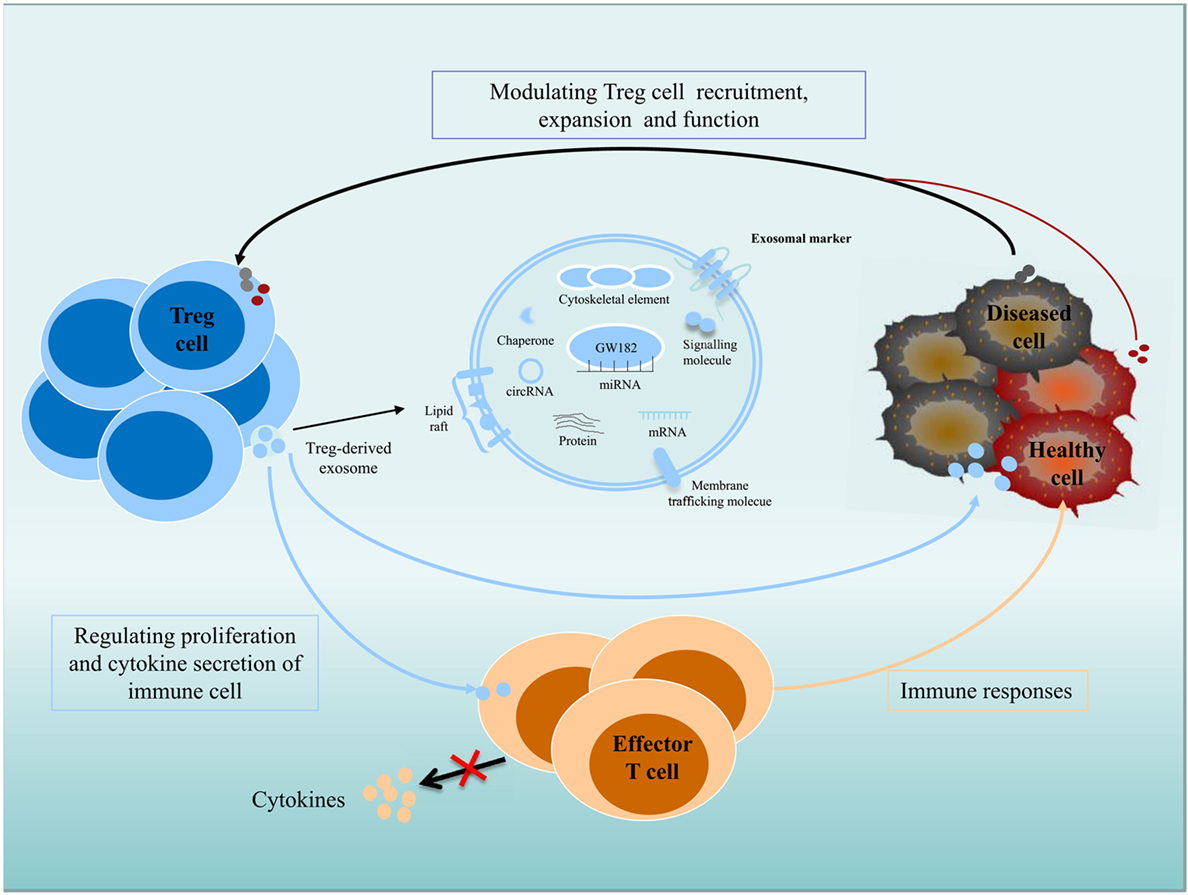
Figure 1. Role of exosomes in intercellular communication between Treg cells and recipient or donor cells. Exosomes transfer their contents, including proteins, lipids, and RNAs, between cells. Immune and non-immune cell-derived exosomes have important roles in the regulation of immunity. Exosomes contribute significantly to the function of Treg cells, and Treg cell-derived exosomes can be delivered to immune cells and diseased or healthy tissue cells; regulate the proliferation and cytokine secretion of effector T cells; and modulate the immune response. Diseased cells, including tumor cells, can modulate Treg cell recruitment, expansion, and function via an exosome-based pathway.
In addition, recent studies have investigated that exosomes derived from tumor cells exert widespread detrimental effects on the immune system (54). Secreted exosomes can serve as signaling tools in mediating tumor cell–Treg cell communication (55). In nasopharyngeal carcinoma, tumor cell-derived exosomes facilitated the expansion of Treg cells and upregulated their suppressive functions. The tumor cell-derived exosomes also promoted the conversion of conventional CD4+CD25− T cells into Treg cells and enhanced the chemoattraction of Treg cells through CCL20 (54). Extracellular vesicles derived from colorectal cancer cell induced a phenotypic change of T cells to Treg-like cells, which had remarkable tumor-growth promoting activity by activating TGF-β/Smad signaling and inactivating SAPK signaling (56). In lung carcinoma, tumor-derived miR-214 reduced PTEN expression (phosphatase and tensin homolog) and promoted the expansion of Treg cells, and miR-214-induced higher secretion of IL-10 in Treg cells and promoted tumor growth (55). It is possible that cancer cells can actively control the immune cells antitumor activities by transfer tumor-specific molecules to recipient immune cells, including Treg cells, via an exosome-based pathway (55). Clearly, exosomes are vital mediators of immunity, for which there will be extensive therapeutic applications (36, 49).
Treg Cells and Non-Coding RNAs
Non-coding RNAs comprise multiple classes of RNA transcripts that are not transcribed into proteins but have been shown to regulate the transcription, stability, or translation of protein-coding genes. To date, there have been many studies of miRNAs and lncRNAs, and other classes of experimentally identified ncRNAs with various lengths and characteristics have also been reported (57). ncRNAs are key regulators of the immune system and regulate important aspects of Treg cells, including Treg cells development, homeostasis, and function. Dynamic homeostatic processes maintain the diverse pool of Treg cells and preserve their number in a normal range. Treg cells can tailor their functions and homeostatic properties to a wide range of conditions (27). Treg homeostasis and function is governed by a number of factors; here, we discuss miRNA- and lncRNA-mediated regulation of Treg cell homeostasis and function (Figure 2).
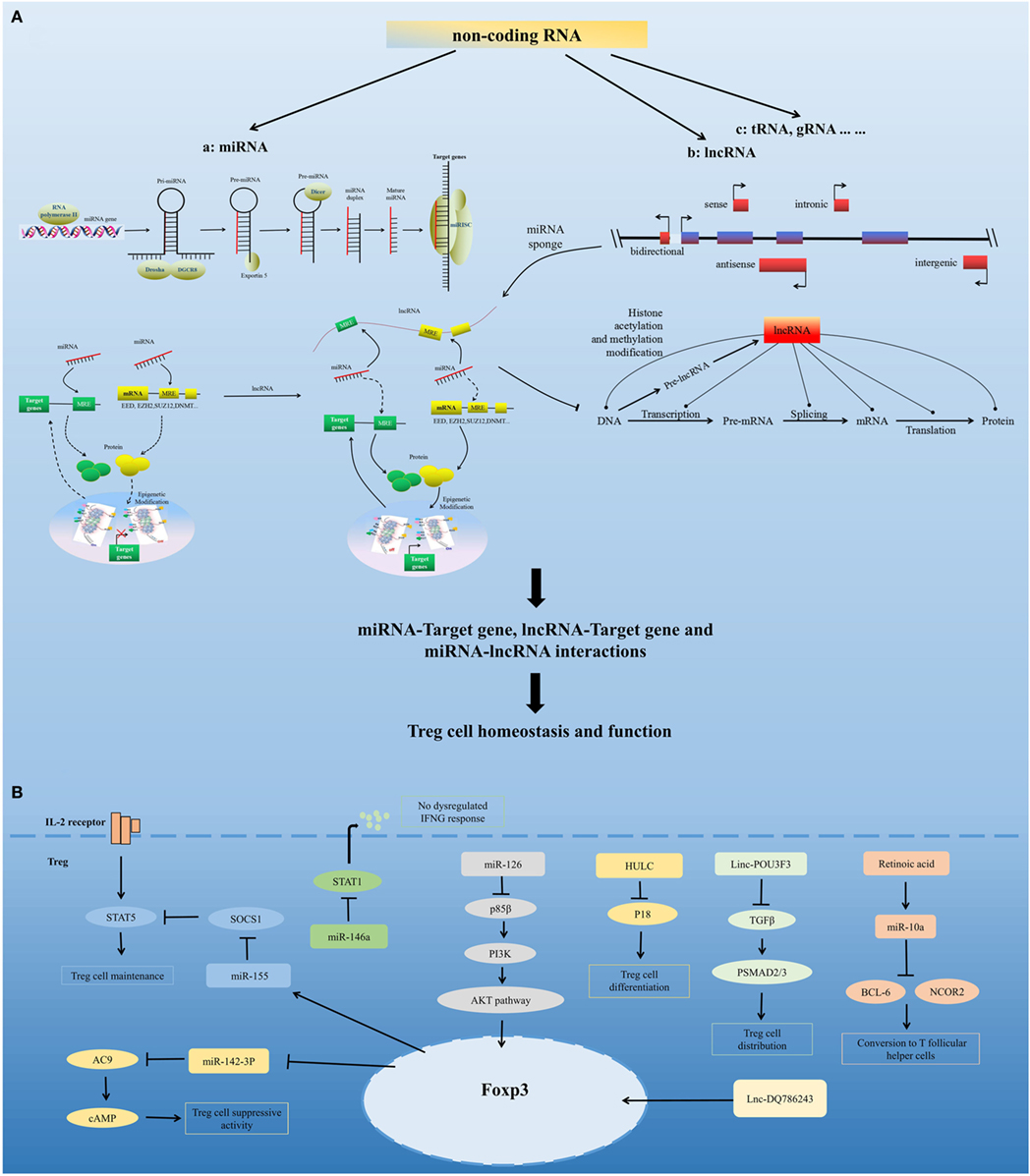
Figure 2. Non-coding RNA-mediated regulation of Treg cell homeostasis and function. (A) Model for non-coding RNAs (ncRNA)-mediated regulation of Treg cell homeostasis and function. ncRNAs include highly abundant and functionally important RNAs, such as microRNAs (miRNA), long non-coding RNAs (lncRNA), and tRNA. miRNAs are sequentially processed from longer transcripts by the RNase III enzymes Drosha and Dicer. Pri-miRNAs are processed by Drosha into hairpin structures (pre-miRNAs). Exportin 5 shuttles pre-miRNAs from the nucleus into the cytoplasm, where the RNase III Dicer cleaves off the hairpin loop of the pre-miRNA. The duplex segregates, and the mature single-stranded miRNA associates with argonaute proteins and other accessory proteins to form the miRNA-induced silencing complex, which directly mediates the translational repression and the increased degradation of its mRNA targets. miRNA can also indirectly regulate gene expression by repressing the expression of several key enzymes involved in epigenetic modification processes, such as DNA methylation and histone modification. Based on the position of lncRNA relative to the neighboring protein-coding genes in the genome, lncRNAs can be divided into five categories, namely, sense, antisense, bidirectional, intronic, and intergenic. lncRNAs can modulate chromatin modification, mRNA stability, miRNA activity, and the function of proteins by interacting with chromatin, RNA, and protein. lncRNA functions as a miRNA sponge, sequestering miRNAs to regulate the expression level of other transcripts sharing common miRNA response elements. This process leads to fewer miRNA molecules available to bind to target mRNA and, thus, an increase in its protein expression level. miRNA, lncRNA, and mRNA form a well-regulated interacting network and play critical regulatory roles in Treg cell homeostasis and function. (B) Regulation of Treg cells by several representative miRNAs and lncRNAs, including miR-155, miR-146a, miR-126, miR-10a, miR-142-3p, HULC, Linc-POU3F3, and Lnc-DQ786243.
miRNA-Mediated Regulation of Treg Cell Homeostasis and Function
MicroRNAs are a group of evolutionarily conserved small non-coding RNAs. They carry out their function by guiding the miRNA-induced silencing complex to target mRNAs (58, 59). miRNAs can directly regulate the expression of target genes by sequence-specific binding to the 3′ untranslated region (3′ UTR) or other regions, and they can also indirectly regulate gene expression by repressing the expression of several key enzymes involved in epigenetic processes, such as DNA methylation and histone modification (60, 61). Research has confirmed the requirements for miRNA expression in Treg cells and shown that miRNAs are important for the maintenance of Treg cell homeostasis and their immunosuppressive function (62).
Studies have shown that depletion of thymus-miRNAs downregulated the number of Treg cells in the thymus, lymph nodes, and spleen, with normal development of conventional T cells in the thymus (63). miRNA-deficient CD4+ T cells fail to develop into tTreg cells and have reduced potential to differentiate into iTreg cells (62). Dicer-deficient Treg cells showed inferior proliferative potential, impaired suppressor function, and impaired peripheral homeostasis. Foxp3 downregulation interrupts Treg cell lineage stability in Dicer-deletion mice (64, 65). Similar to Dicer, deletion of Drosha in Treg cells leads to defective suppressive activity (66). In addition, Treg cell-specific miRNA-deficient mice show fatal early-onset lymphoproliferative syndrome, and the conditional deficiency of Dicer or Drosha in Foxo3+ Treg cells gives rise to the early onset of severe spontaneous autoimmunity (64–66). These findings confirmed the critical role of miRNA in Treg cell development and function and in preventing immune disease.
Several miRNAs have been found that affect Treg cell homeostasis and function. miR-155 is highly expressed in Treg cells. Foxp3 binds to the B cell integration cluster (encodes the primary miR-155 transcript), and controls this high expression of miR-155 in Treg cells (67–70). Recent studies have shown that miR-155 contribute to the development and homeostasis of Treg cell, but not the function of Treg cell. miR-155 deletion mice show a decreased number of tTreg cells and pTreg cells, due to defective development (63). miR-155 facilitates Treg cell homeostasis by targeting suppressor of cytokine signaling 1 (69), a negative regulator of signal transduction and activation of transcription (STAT) 5 that has a crucial role in the IL-2 signaling pathway and in Treg cell development. miR-155 deletion also downregulates the IL-2 production of CD4+ T cells (71), which demonstrates that miR-155 might control IL-2-directed Treg cell homeostasis via both cell-intrinsic and cell-extrinsic pathway (62). miR-10a contributes to Treg cell stability by maintaining high levels of Foxp3 expression. TGF-beta and retinoic acid, which boost the Treg cell phenotype, are need for maximal induction of miR-10a in pTreg cells (72, 73). In addition, miR-10a inhibits pTreg cells conversion into T follicular helper cells by directly targeting BCL-6 and its co-repressor NCOR2. The expression of miR-10a in Treg cells is inversely correlated with susceptibility to autoimmune disease (74, 75). miR-146a has a marked effect on Treg cell function and plays a critical role in Treg cell-mediated immunological tolerance. miR-146a-deficient mice developed severe lympho- and myeloproliferative syndrome (76) and had an elevated number of Treg cells in the periphery that had a modest increase in activation markers and heightened proliferative activity. The restriction of miR-146a deficiency mainly to Treg cells resulted in IFNγ-dependent immune-mediated lesions and a Th1 cell-mediated pathology (77, 78), which was similar to the disease observed in Treg cell-specific Dicer- or Drosha-deficient mice. In addition, miR-146a controls Treg-mediated suppression of IFNγ-dependent Th1 responses and inflammation by targeting STAT1 (78). miR-126 is highly expressed in Treg cells. Silencing of miR-126 could attenuate the suppressive activity of Treg cells. miR-126 regulates the induction and function of Treg cells through the p85β/PI3K/Akt pathway. miR-142-3p may be a unique molecule in Treg cell function. It has been found that Treg cells exert their suppressor function by transferring cAMP to responder T cells (79). miR-142-3p restricts cAMP production in Treg cells by targeting adenylate cyclase 9 (AC9), whereas Foxp3 could maintain the activity of the AC9/cAMP pathway by downregulating miR-142-3p in Treg cells (80). In addition to the miRNAs mentioned above, many other miRNAs play important roles in the regulation of Treg cells, including miR-21 (81), miR210 (82), miR15a/16 (83), among others.
Regulation of Treg Cells by lncRNAs
Long non-coding RNAs are transcripts of more than 200 bp that are often expressed with higher cell specificity than protein-coding genes despite having lower expression levels. Recent studies have found that several lncRNAs can affect Treg cells. The lncRNA HULC, which is upregulated in hepatocellular carcinoma, affects the differentiation of Treg cells by downregulating the level of p18 directly in HBV-related liver cirrhosis (84). Linc-POU3F3 was able to facilitate the distribution of Treg cells among peripheral T cells, which caused increased cell proliferation of gastric cancer cells through recruiting TGF-beta and activating TGF-beta pathway (85). The lncRNA DQ786243 affects the expression of cAMP response element binding protein and Foxp3 by Treg cells in Crohn’s disease (86). Further studies are needed to identify the mechanisms of lncRNAs in the regulation of Treg cells.
Tissue-Resident Treg Cells
Regulatory T cells are present in various non-lymphoid tissues in health and disease. Each tissue might have its own unique tissue-resident Treg cells, and the phenotype and function of tissue Treg cells are different from those of classical lymphoid Treg cells (30). These cells not only display some activated and/or effector cell features but also show some unique properties, such as specific chemokine receptors, transcription factors and adhesion molecules or distinct T cell antigen receptor repertoires, mechanisms of action, targets, and migration patterns (23). Treg cells have been found in several non-lymphoid tissues, including adipose tissue, skeletal muscle, intestinal mucosa, skin, and tumor tissue (30, 87–89). Understanding the development and the maintenance of tissue-resident Treg cells provides important insights into local immune regulation and tissue-specific biological therapies. Here, we review the current state of knowledge of intestine-resident Treg cells and tumor tissue-resident Treg cells.
Intestine-Resident Treg Cells
There are a large number of Treg cells in the intestines because of the exposure to food-derived antigens and commensal microflora. Intestine-resident Treg cells are different from other organ Treg cells and have intestine-specific phenotypes, TCR repertoires, and functions (90, 91). High levels of microbe-derived TLRs and metabolites of the commensal flora can dramatically influence the development, function, and maintenance of intestine-resident Treg cells (19). In addition, specialized CD103+ DCs, together with TGF-beta and retinoic acid, can enhance pTreg cell development in the intestine and also induce Treg cells to express intestinal homing receptors (92, 93). Intestine-resident Treg cells are a self-renewing and stable population, with a few proportions derived from the periphery after their initial development and seeding early in life. Further understanding of the molecular mechanisms responsible for the tissue-specific and condition-adapted development of stable Treg cell populations in the intestines could supply new treatment approaches for many diseases (91).
Tumor-Resident Treg Cells
Depressed cellular immunity has been demonstrated in patients with a variety of lymphoreticular and non-lymphoreticular neoplasms. Given the recent successes of immunomodulatory antitumor strategies, there is growing interest in the more heterogeneous group of tumor-infiltrating Treg cells (30, 94). Elevated percentages of Treg cells are found in the total T cell population isolated from tumor tissue, and these cells can account for 30–50% of CD4+ T cells, depending on the tumor type (95). Tumor-resident Treg cells have been identified and characterized, and, similar to tissue-resident Treg cells, the phenotypes of tumor-resident Treg cells are different from those found in lymphoid organs or in the circulation. Foxp3+CD4+ T cells from the tumor environment show upregulation of cell-surface markers, including CTLA4, TIM3, and PD1, as well as a variety of chemokine receptors and suppressive cytokines. Tumor-resident Treg cells represent an important cellular mechanism by which tumors evade immunosurveillance, as these cells are capable of restricting the proliferation and cytokine production of a wide range of immune cells and suppressing the antitumor activity of CD4+ T cells, CD8+ T cells, and NK cells (5, 95–99). In addition to these functions, tumor-resident Treg cells can promote tumor growth, angiogenesis, and metastasis. Treg cells also exert antitumor effects by inhibiting the immune response in the tumor microenvironment. In many cases, rich Treg cell infiltration into tumor microenvironment is correlated with poor prognosis (95). However, Treg cells play controversial roles in some cancers, in which abundance Foxp3+ Treg cells promised relatively good prognosis. A recent study found that in colorectal cancer, functionally distinct subsets of tumor-infiltrating Foxp3+ T cells contribute in opposing ways to measuring outcomes (100). Dissection of the pathways regulated by tumor-resident Treg cells is critical for immunotherapies that aim to modulate Treg cells in cancer.
Conclusion
Recent studies have provided new insights into Treg cell regulation and homeostasis. Dynamic homeostatic processes maintain the diverse pool of Treg cells and preserve the number of Treg cells within steady-state conditions. Treg cells have two origins and can be divided into functional subsets. Tissue-resident Treg cells are a relatively new subtype; thus, it is not surprising that there are some questions that remain unanswered. Here, we highlight three questions that need to be explored. First, which type of Treg cells play a predominant role in the regulation of the immune response compared with non-resident Treg cells? Second, are tumor-specific Treg cells mainly derived from tissue-resident Treg cells or from tTreg cells and pTreg cells? Third, can Treg cells be classified into new subsets, and can the degree of infiltration of these subpopulations contribute to disease prognosis?
Regulatory T cell homeostasis is governed by a number of factors. In this review, we focused on the exosome- and ncRNA-mediated regulation of Treg cell homeostasis. The study of Treg cell-derived exosomes is a relatively new area of Treg cell biology. The exosome output by Treg cells changes with cell status and reflect intracellular events. Exosomes, therefore, provide an enriched pool of information and could be considered to be potential biomarkers. In addition to the effects of Treg cell-derived exosomes on immune responses, exosomes could be used as therapeutic agents in various conditions. ncRNAs are crucial to the homeostasis of Treg cells. There are many studies of miRNA and lncRNA, but the roles and mechanisms of other classes of ncRNAs, including circRNA, in the regulation of Treg cells remain unclear and will require further study.
Author Contributions
All the authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
This study was supported by the National Key Technology Research and Development program of the Ministry of Science and Technology of China (Grant 2014BAI04B02), Graduate Research and Innovation Projects in Hunan Province (Grant CX2015B056), and the Mittal Innovative Entrepreneurial Project of Central South University (Grant 15MX43).
References
1. Sakaguchi S. Naturally arising Foxp3-expressing CD25+CD4+ regulatory T cells in immunological tolerance to self and non-self. Nat Immunol (2005) 6(4):345–52. doi: 10.1038/ni1178
2. Linehan DC, Goedegebuure PS. CD25+ CD4+ regulatory T-cells in cancer. Immunol Res (2005) 32(1–3):155–68. doi:10.1385/IR:32:1-3:155
3. Belkaid Y. Regulatory T cells and infection: a dangerous necessity. Nat Rev Immunol (2007) 7(11):875–88. doi:10.1038/nri2189
4. Loser K, Beissert S. Regulatory T cells: banned cells for decades. J Invest Dermatol (2012) 132(3 Pt 2):864–71. doi:10.1038/jid.2011.375
5. Penaloza-MacMaster P, Kamphorst AO, Wieland A, Araki K, Iyer SS, West EE. Interplay between regulatory T cells and PD-1 in modulating T cell exhaustion and viral control during chronic LCMV infection. J Exp Med (2014) 211(9):1905–18. doi:10.1084/jem.20132577
6. Tan T, Xiang Y, Chang C, Zhou Z. Alteration of regulatory T cells in type 1 diabetes mellitus: a comprehensive review. Clin Rev Allergy Immunol (2014) 47(2):234–43. doi:10.1007/s12016-014-8440-0
7. Furtado GC, Curotto DLM, Kutchukhidze N, Lafaille JJ. Interleukin 2 signaling is required for CD4(+) regulatory T cell function. J Exp Med (2002) 196(6):851–7. doi:10.1084/jem.20020190
8. Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol (2003) 4(4):330–6. doi:10.1038/ni904
9. Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science (2003) 299(5609):1057–61. doi:10.1126/science.1079490
10. Sun L, Wu J, Yi S. Foxp3 is critical for human natural CD4+CD25+ regulatory T cells to suppress alloimmune response. Transpl Immunol (2012) 26(2–3):71–80. doi:10.1016/j.trim.2011.10.005
11. Zhao M, Liang GP, Tang MN, Luo SY, Zhang J, Cheng WJ. Total glucosides of paeony induces regulatory CD4(+)CD25(+) T cells by increasing Foxp3 demethylation in lupus CD4(+) T cells. Clin Immunol (2012) 143(2):180–7. doi:10.1016/j.clim.2012.02.002
12. Gavin MA, Torgerson TR, Houston E, DeRoos P, Ho WY, Stray-Pedersen A. Single-cell analysis of normal and FOXP3-mutant human T cells: FOXP3 expression without regulatory T cell development. Proc Natl Acad Sci U S A (2006) 103(17):6659–64. doi:10.1073/pnas.0509484103
13. Liu W, Putnam AL, Xu-Yu Z, Szot GL, Lee MR, Zhu S. CD127 expression inversely correlates with FoxP3 and suppressive function of human CD4+ T reg cells. J Exp Med (2006) 203(7):1701–11. doi:10.1084/jem.20060772
14. Schwab M, Barth S, Boffetta P, Colditz GA, Duhé RJ, Hunter K, et al. Encyclopedia of Cancer. Berlin, Heidelberg: Springer (2009).
15. Li Z, Li D, Tsun A, Li B. FOXP3+ regulatory T cells and their functional regulation. Cell Mol Immunol (2015) 12(5):558–65. doi:10.1038/cmi.2015.10
16. Liston A, Rudensky AY. Thymic development and peripheral homeostasis of regulatory T cells. Curr Opin Immunol (2007) 19(2):176–85. doi:10.1016/j.coi.2007.02.005
17. Paiva RS, Lino AC, Bergman ML, Caramalho I, Sousa AE, Zelenay S. Recent thymic emigrants are the preferential precursors of regulatory T cells differentiated in the periphery. Proc Natl Acad Sci U S A (2013) 110(16):6494–9. doi:10.1073/pnas.1221955110
18. Chen W, Jin W, Hardegen N, Lei KJ, Li L, Marinos N. Conversion of peripheral CD4+CD25- naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J Exp Med (2003) 198(12):1875–86. doi:10.1084/jem.20030152
19. Gratz IK, Campbell DJ. Organ-specific and memory treg cells: specificity, development, function, and maintenance. Front Immunol (2014) 5:333. doi:10.3389/fimmu.2014.00333
20. Roncarolo MG, Gregori S, Battaglia M, Bacchetta R, Fleischhauer K, Levings MK. Interleukin-10-secreting type 1 regulatory T cells in rodents and humans. Immunol Rev (2006) 212:28–50. doi:10.1111/j.0105-2896.2006.00420.x
21. Weiner HL. Induction and mechanism of action of transforming growth factor-beta-secreting Th3 regulatory cells. Immunol Rev (2001) 182:207–14. doi:10.1034/j.1600-065X.2001.1820117.x
22. Wang HY, Wang RF. Regulatory T cells and cancer. Curr Opin Immunol (2007) 19(2):217–23. doi:10.1016/j.coi.2007.02.004
23. Thornton AM, Korty PE, Tran DQ, Wohlfert EA, Murray PE, Belkaid Y. Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+ T regulatory cells. J Immunol (2010) 184(7):3433–41. doi:10.4049/jimmunol.0904028
24. Yadav M, Louvet C, Davini D, Gardner JM, Martinez-Llordella M, Bailey-Bucktrout S. Neuropilin-1 distinguishes natural and inducible regulatory T cells among regulatory T cell subsets in vivo. J Exp Med (2012) 209(10):S1–19. doi:10.1084/jem.20120822
25. Bin DK, D’Hennezel E, Nashi E, Bar-Or A, Rieder S, Shevach EM. Coexpression of TIGIT and FCRL3 identifies Helios+ human memory regulatory T cells. J Immunol (2015) 194(8):3687–96. doi:10.4049/jimmunol.1401803
26. Rosenblum MD, Way SS, Abbas AK. Regulatory T cell memory. Nat Rev Immunol (2016) 16(2):90–101. doi:10.1038/nri.2015.1
27. Liston A, Gray DH. Homeostatic control of regulatory T cell diversity. Nat Rev Immunol (2014) 14(3):154–65. doi:10.1038/nri3605
28. Campbell DJ, Koch MA. Phenotypical and functional specialization of FOXP3+ regulatory T cells. Nat Rev Immunol (2011) 11(2):119–30. doi:10.1038/nri2916
29. Mendes F, Domingues C, Rodrigues-Santos P, Abrantes AM, Goncalves AC, Estrela J. The role of immune system exhaustion on cancer cell escape and anti-tumor immune induction after irradiation. Biochim Biophys Acta (2016) 1865(2):168–75. doi:10.1016/j.bbcan.2016.02.002
30. Burzyn D, Benoist C, Mathis D. Regulatory T cells in nonlymphoid tissues. Nat Immunol (2013) 14(10):1007–13. doi:10.1038/ni.2683
31. Lu J, Meng H, Zhang A, Yang J, Zhang X. Phenotype and function of tissue-resident unconventional Foxp3-expressing CD4(+) regulatory T cells. Cell Immunol (2015) 297(1):53–9. doi:10.1016/j.cellimm.2015.06.005
32. Li X, Zheng Y. Regulatory T cell identity: formation and maintenance. Trends Immunol (2015) 36(6):344–53. doi:10.1016/j.it.2015.04.006
33. Barnaba V, Schinzari V. Induction, control, and plasticity of Treg cells: the immune regulatory network revised? Eur J Immunol (2013) 43(2):318–22. doi:10.1002/eji.201243265
34. Kleinewietfeld M, Hafler DA. The plasticity of human Treg and Th17 cells and its role in autoimmunity. Semin Immunol (2013) 25(4):305–12. doi:10.1016/j.smim.2013.10.009
35. Booth AM, Fang Y, Fallon JK, Yang JM, Hildreth JE, Gould SJ. Exosomes and HIV Gag bud from endosome-like domains of the T cell plasma membrane. J Cell Biol (2006) 172(6):923–35. doi:10.1083/jcb.200508014
36. Robbins PD, Morelli AE. Regulation of immune responses by extracellular vesicles. Nat Rev Immunol (2014) 14(3):195–208. doi:10.1038/nri3622
37. Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol (2002) 2(8):569–79. doi:10.1038/nri855
38. Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res (2015) 25(8):981–4. doi:10.1038/cr.2015.82
39. Balaj L, Lessard R, Dai L, Cho YJ, Pomeroy SL, Breakefield XO. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat Commun (2011) 2:180. doi:10.1038/ncomms1180
40. Okoye IS, Coomes SM, Pelly VS, Czieso S, Papayannopoulos V, Tolmachova T. MicroRNA-containing T-regulatory-cell-derived exosomes suppress pathogenic T helper 1 cells. Immunity (2014) 41(1):89–103. doi:10.1016/j.immuni.2014.05.019
41. Wang GJ, Liu Y, Qin A, Shah SV, Deng ZB, Xiang X. Thymus exosomes-like particles induce regulatory T cells. J Immunol (2008) 181(8):5242–8. doi:10.4049/jimmunol.181.8.5242
42. Mittelbrunn M, Gutierrez-Vazquez C, Villarroya-Beltri C, Gonzalez S, Sanchez-Cabo F, Gonzalez MA. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Commun (2011) 2:282. doi:10.1038/ncomms1285
43. Smyth LA, Ratnasothy K, Tsang JY, Boardman D, Warley A, Lechler R. CD73 expression on extracellular vesicles derived from CD4+ CD25+ Foxp3+ T cells contributes to their regulatory function. Eur J Immunol (2013) 43(9):2430–40. doi:10.1002/eji.201242909
44. Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol (2007) 9(6):654–9. doi:10.1038/ncb1596
45. Savina A, Furlan M, Vidal M, Colombo MI. Exosome release is regulated by a calcium-dependent mechanism in K562 cells. J Biol Chem (2003) 278(22):20083–90. doi:10.1074/jbc.M301642200
46. King HW, Michael MZ, Gleadle JM. Hypoxic enhancement of exosome release by breast cancer cells. BMC Cancer (2012) 12:421. doi:10.1186/1471-2407-12-421
47. Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science (2008) 319(5867):1244–7. doi:10.1126/science.1153124
48. Fontenot JD, Rasmussen JP, Gavin MA, Rudensky AY. A function for interleukin 2 in Foxp3-expressing regulatory T cells. Nat Immunol (2005) 6(11):1142–51. doi:10.1038/ni1263
49. Agarwal A, Fanelli G, Letizia M, Tung SL, Boardman D, Lechler R. Regulatory T cell-derived exosomes: possible therapeutic and diagnostic tools in transplantation. Front Immunol (2014) 5:555. doi:10.3389/fimmu.2014.00555
50. Clayton A, Al-Taei S, Webber J, Mason MD, Tabi Z. Cancer exosomes express CD39 and CD73, which suppress T cells through adenosine production. J Immunol (2011) 187(2):676–83. doi:10.4049/jimmunol.1003884
51. Kim SH, Bianco N, Menon R, Lechman ER, Shufesky WJ, Morelli AE. Exosomes derived from genetically modified DC expressing FasL are anti-inflammatory and immunosuppressive. Mol Ther (2006) 13(2):289–300. doi:10.1016/j.ymthe.2005.09.015
52. Mokarizadeh A, Delirezh N, Morshedi A, Mosayebi G, Farshid AA, Mardani K. Microvesicles derived from mesenchymal stem cells: potent organelles for induction of tolerogenic signaling. Immunol Lett (2012) 147(1–2):47–54. doi:10.1016/j.imlet.2012.06.001
53. Buzas EI, Gyorgy B, Nagy G, Falus A, Gay S. Emerging role of extracellular vesicles in inflammatory diseases. Nat Rev Rheumatol (2014) 10(6):356–64. doi:10.1038/nrrheum.2014.19
54. Mrizak D, Martin N, Barjon C, Jimenez-Pailhes AS, Mustapha R, Niki T. Effect of nasopharyngeal carcinoma-derived exosomes on human regulatory T cells. J Natl Cancer Inst (2015) 107(1):363. doi:10.1093/jnci/dju363
55. Yin Y, Cai X, Chen X, Liang H, Zhang Y, Li J. Tumor-secreted miR-214 induces regulatory T cells: a major link between immune evasion and tumor growth. Cell Res (2014) 24(10):1164–80. doi:10.1038/cr.2014.121
56. Yamada N, Kuranaga Y, Kumazaki M, Shinohara H, Taniguchi K, Akao Y. Colorectal cancer cell-derived extracellular vesicles induce phenotypic alteration of T cells into tumor-growth supporting cells with transforming growth factor-beta1-mediated suppression. Oncotarget (2016) 7(19):27033–43. doi:10.18632/oncotarget.7041
57. Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov (2013) 12(11):847–65. doi:10.1038/nrd4140
58. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell (2009) 136(2):215–33. doi:10.1016/j.cell.2009.01.002
59. Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem (2010) 79:351–79. doi:10.1146/annurev-biochem-060308-103103
60. Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature (2008) 455(7216):1124–8. doi:10.1038/nature07299
61. Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5’ UTR as in the 3’ UTR. Proc Natl Acad Sci U S A (2007) 104(23):9667–72. doi:10.1073/pnas.0703820104
62. Baumjohann D, Ansel KM. MicroRNA-mediated regulation of T helper cell differentiation and plasticity. Nat Rev Immunol (2013) 13(9):666–78. doi:10.1038/nri3494
63. Zhou L, Park JJ, Zheng Q, Dong Z, Mi Q. MicroRNAs are key regulators controlling iNKT and regulatory T-cell development and function. Cell Mol Immunol (2011) 8(5):380–7. doi:10.1038/cmi.2011.27
64. Liston A, Lu LF, O’Carroll D, Tarakhovsky A, Rudensky AY. Dicer-dependent microRNA pathway safeguards regulatory T cell function. J Exp Med (2008) 205(9):1993–2004. doi:10.1084/jem.20081062
65. Zhou X, Jeker LT, Fife BT, Zhu S, Anderson MS, McManus MT. Selective miRNA disruption in T reg cells leads to uncontrolled autoimmunity. J Exp Med (2008) 205(9):1983–91. doi:10.1084/jem.20080707
66. Chong MM, Rasmussen JP, Rudensky AY, Littman DR. The RNAseIII enzyme Drosha is critical in T cells for preventing lethal inflammatory disease. J Exp Med (2008) 205(9):2005–17. doi:10.1084/jem.20081219
67. Marson A, Kretschmer K, Frampton GM, Jacobsen ES, Polansky JK, MacIsaac KD. Foxp3 occupancy and regulation of key target genes during T-cell stimulation. Nature (2007) 445(7130):931–5. doi:10.1038/nature05478
68. Zheng Y, Josefowicz SZ, Kas A, Chu TT, Gavin MA, Rudensky AY. Genome-wide analysis of Foxp3 target genes in developing and mature regulatory T cells. Nature (2007) 445(7130):936–40. doi:10.1038/nature05563
69. Lu LF, Thai TH, Calado DP, Chaudhry A, Kubo M, Tanaka K. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity (2009) 30(1):80–91. doi:10.1016/j.immuni.2008.11.010
70. Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR. Requirement of bic/microRNA-155 for normal immune function. Science (2007) 316(5824):608–11. doi:10.1126/science.1139253
71. Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y. Regulation of the germinal center response by microRNA-155. Science (2007) 316(5824):604–8. doi:10.1126/science.1141229
72. Jeker LT, Zhou X, Gershberg K, de Kouchkovsky D, Morar MM, Stadthagen G. MicroRNA 10a marks regulatory T cells. PLoS One (2012) 7(5):e36684. doi:10.1371/journal.pone.0036684
73. Tang X, Tang R, Xu Y, Wang Q, Hou Y, Shen S. MicroRNA networks in regulatory T cells. J Physiol Biochem (2014) 70(3):869–75. doi:10.1007/s13105-014-0348-x
74. Tsuji M, Komatsu N, Kawamoto S, Suzuki K, Kanagawa O, Honjo T. Preferential generation of follicular B helper T cells from Foxp3+ T cells in gut Peyer’s patches. Science (2009) 323(5920):1488–92. doi:10.1126/science.1169152
75. Takahashi H, Kanno T, Nakayamada S, Hirahara K, Sciume G, Muljo SA. TGF-beta and retinoic acid induce the microRNA miR-10a, which targets Bcl-6 and constrains the plasticity of helper T cells. Nat Immunol (2012) 13(6):587–95. doi:10.1038/ni.2286
76. Boldin MP, Taganov KD, Rao DS, Yang L, Zhao JL, Kalwani M. miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. J Exp Med (2011) 208(6):1189–201. doi:10.1084/jem.20101823
77. Lu LF, Boldin MP, Chaudhry A, Lin LL, Taganov KD, Hanada T. Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell (2010) 142(6):914–29. doi:10.1016/j.cell.2010.08.012
78. Tang Y, Luo X, Cui H, Ni X, Yuan M, Guo Y. MicroRNA-146A contributes to abnormal activation of the type I interferon pathway in human lupus by targeting the key signaling proteins. Arthritis Rheum (2009) 60(4):1065–75. doi:10.1002/art.24436
79. Qin A, Wen Z, Zhou Y, Li Y, Li Y, Luo J. MicroRNA-126 regulates the induction and function of CD4(+) Foxp3(+) regulatory T cells through PI3K/AKT pathway. J Cell Mol Med (2013) 17(2):252–64. doi:10.1111/jcmm.12003
80. Huang B, Zhao J, Lei Z, Shen S, Li D, Shen GX. miR-142-3p restricts cAMP production in CD4+CD25- T cells and CD4+CD25+ TREG cells by targeting AC9 mRNA. EMBO Rep (2009) 10(2):180–5. doi:10.1038/embor.2008.224
81. Hu Y, Wang C, Li Y, Zhao J, Chen C, Zhou Y. MiR-21 controls in situ expansion of CCR6(+) regulatory T cells through PTEN/AKT pathway in breast cancer. Immunol Cell Biol (2015) 93(8):753–64. doi:10.1038/icb.2015.37
82. Zhao M, Wang LT, Liang GP, Zhang P, Deng XJ, Tang Q. Up-regulation of microRNA-210 induces immune dysfunction via targeting FOXP3 in CD4(+) T cells of psoriasis vulgaris. Clin Immunol (2014) 150(1):22–30. doi:10.1016/j.clim.2013.10.009
83. Liu X, Robinson SN, Setoyama T, Tung SS, D’Abundo L, Shah MY. FOXP3 is a direct target of miR15a/16 in umbilical cord blood regulatory T cells. Bone Marrow Transplant (2014) 49(6):793–9. doi:10.1038/bmt.2014.57
84. Zhao J, Fan Y, Wang K, Ni X, Gu J, Lu H. LncRNA HULC affects the differentiation of Treg in HBV-related liver cirrhosis. Int Immunopharmacol (2015) 28(2):901–5. doi:10.1016/j.intimp.2015.04.028
85. Xiong G, Yang L, Chen Y, Fan Z. Linc-POU3F3 promotes cell proliferation in gastric cancer via increasing T-reg distribution. Am J Transl Res (2015) 7(11):2262–9.
86. Qiao YQ, Huang ML, Xu AT, Zhao D, Ran ZH, Shen J. LncRNA DQ786243 affects Treg related CREB and Foxp3 expression in Crohn’s disease. J Biomed Sci (2013) 20:87. doi:10.1186/1423-0127-20-87
87. Dudda JC, Perdue N, Bachtanian E, Campbell DJ. Foxp3+ regulatory T cells maintain immune homeostasis in the skin. J Exp Med (2008) 205(7):1559–65. doi:10.1084/jem.20072594
88. Denning TL, Kim G, Kronenberg M. Cutting edge: CD4+CD25+ regulatory T cells impaired for intestinal homing can prevent colitis. J Immunol (2005) 174(12):7487–91. doi:10.4049/jimmunol.174.12.7487
89. Cipolletta D. Adipose tissue-resident regulatory T cells: phenotypic specialization, functions and therapeutic potential. Immunology (2014) 142(4):517–25. doi:10.1111/imm.12262
90. Lathrop SK, Bloom SM, Rao SM, Nutsch K, Lio CW, Santacruz N. Peripheral education of the immune system by colonic commensal microbiota. Nature (2011) 478(7368):250–4. doi:10.1038/nature10434
91. Tanoue T, Atarashi K, Honda K. Development and maintenance of intestinal regulatory T cells. Nat Rev Immunol (2016) 16(5):295–309. doi:10.1038/nri.2016.36
92. Coombes JL, Siddiqui KR, Arancibia-Carcamo CV, Hall J, Sun CM, Belkaid Y. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-beta and retinoic acid-dependent mechanism. J Exp Med (2007) 204(8):1757–64. doi:10.1084/jem.20070590
93. Jaensson E, Uronen-Hansson H, Pabst O, Eksteen B, Tian J, Coombes JL. Small intestinal CD103+ dendritic cells display unique functional properties that are conserved between mice and humans. J Exp Med (2008) 205(9):2139–49. doi:10.1084/jem.20080414
94. Gajewski TF, Schreiber H, Fu YX. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol (2013) 14(10):1014–22. doi:10.1038/ni.2703
95. Tanchot C, Terme M, Pere H, Tran T, Benhamouda N, Strioga M. Tumor-infiltrating regulatory T cells: phenotype, role, mechanism of expansion in situ and clinical significance. Cancer Microenviron (2013) 6(2):147–57. doi:10.1007/s12307-012-0122-y
96. Park HJ, Kusnadi A, Lee EJ, Kim WW, Cho BC, Lee IJ. Tumor-infiltrating regulatory T cells delineated by upregulation of PD-1 and inhibitory receptors. Cell Immunol (2012) 278(1–2):76–83. doi:10.1016/j.cellimm.2012.07.001
97. Gao X, Zhu Y, Li G, Huang H, Zhang G, Wang F. TIM-3 expression characterizes regulatory T cells in tumor tissues and is associated with lung cancer progression. PLoS One (2012) 7(2):e30676. doi:10.1371/journal.pone.0030676
98. Gobert M, Treilleux I, Bendriss-Vermare N, Bachelot T, Goddard-Leon S, Arfi V. Regulatory T cells recruited through CCL22/CCR4 are selectively activated in lymphoid infiltrates surrounding primary breast tumors and lead to an adverse clinical outcome. Cancer Res (2009) 69(5):2000–9. doi:10.1158/0008-5472.CAN-08-2360
99. Strauss L, Bergmann C, Szczepanski M, Gooding W, Johnson JT, Whiteside TL. A unique subset of CD4+CD25highFoxp3+ T cells secreting interleukin-10 and transforming growth factor-beta1 mediates suppression in the tumor microenvironment. Clin Cancer Res (2007) 13(15 Pt 1):4345–54. doi:10.1158/1078-0432.CCR-07-0472
Keywords: Treg cell, exosome, non-coding RNA, resident Treg cell, Treg homeostasis
Citation: Li P, Liu C, Yu Z and Wu M (2016) New Insights into Regulatory T Cells: Exosome- and Non-Coding RNA-Mediated Regulation of Homeostasis and Resident Treg Cells. Front. Immunol. 7:574. doi: 10.3389/fimmu.2016.00574
Received: 18 July 2016; Accepted: 23 November 2016;
Published: 06 December 2016
Edited by:
Bruno Laugel, Cardiff University, UKReviewed by:
António Gil Castro, University of Minho, PortugalCosima T. Baldari, University of Siena, Italy
Copyright: © 2016 Li, Liu, Yu and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Minghua Wu, d3VtaW5naHVhNTU0JiN4MDAwNDA7YWxpeXVuLmNvbQ==
 Peiyao Li
Peiyao Li Changhong Liu
Changhong Liu Zhibin Yu
Zhibin Yu Minghua Wu
Minghua Wu