- Department of Microbiology and Immunology, Graduate School of Medicine, Osaka University, Suita, Japan
Hyaluronan is a major component of the extracellular matrix and plays pivotal roles in inflammation and cancer. Hyaluronan oligomers are frequently found in these pathological conditions, in which they exert their effects via association with the transmembrane receptor CD44. Lipid rafts are cholesterol- and glycosphingolipid-enriched membrane microdomains that may regulate membrane receptors while serving as platforms for transmembrane signaling at the cell surface. This article focuses on the recent discovery that lipid rafts regulate the interaction between CD44 and hyaluronan, which depends largely on hyaluronan’s size. Lipid rafts regulate CD44’s ability to bind hyaluronan in T cells, control the rolling adhesion of lymphocytes on vascular endothelial cells, and regulate hyaluronan- and CD44-mediated cancer cell migration. The implications of these findings for preventing inflammatory disorders and cancer are also discussed.
Introduction
Hyaluronan is a linear glycosaminoglycan consisting of repeating disaccharide units of d-glucuronic acid (GlcUA) and N-acetyl-d-glucosamine (GlcNAc) with the structure [β1,4-GlcUA-β-1,3-GlcNAc-]n, and a physiological molecular weight (relative molecular mass) ranging from 1 × 105 to 1 × 107 with polydispersity (1) (Figure 1). Hyaluronan was first purified from bovine vitreous humor in 1934 (2). It is now known to be ubiquitous in vertebrate tissues, with particular abundance in connective tissues, such as synovial fluid, Wharton’s jelly in the umbilical cord, and the vitreous humor of the eye, where it plays a mechanical role determined by its viscous features. While hyaluronan is traditionally regarded as a space filling, structural molecule involved in lubricating joints or holding connective tissues in place (3), it also functions as a microenvironmental cue in inflammation and cancer (4).
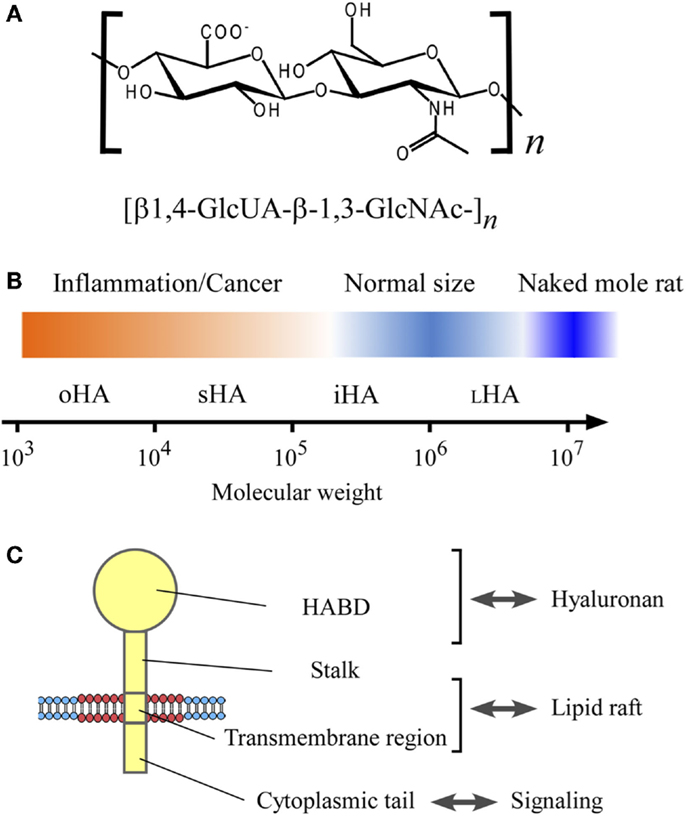
Figure 1. The structure and size of hyaluronan, and its interaction with CD44 in the lipid raft. (A) Chemical structure of hyaluronan. Hyaluronan is a linear glycosaminoglycan consisting of repeating disaccharide units of d-glucuronic acid (GlcUA) and N-acetyl-glucosamine (GlcNAc) with the structure [β1,4-GlcUA-β-1,3-GlcNAc-]n. (B) Hyaluronan size and its relevance to inflammation and cancer. The sizes of hyaluronan are categorized according to Weigel’s nomenclature (5), and shown in the log scale of the molecular weight: oligomeric hyaluronan (oHA; 1 × 103–1 × 104), small hyaluronan (sHA; 1 × 104–1 × 105), intermediate hyaluronan (iHA; 1 × 105–1 × 106), and large hyaluronan (LHA; 1 × 106–1 × 107). Hyaluronan’s molecular weight in normal physiological conditions ranges from 1 × 105 to 1 × 107, whereas the low-molecular weight hyaluronan has relevance to inflammation and cancer. The very high-molecular weight hyaluronan from naked mole rat (molecular weight, 6 × 106–1.2 × 107) has an anti-malignant activity (6). (C) CD44 structure. CD44 consists of four regions; hyaluronan-binding domain (HABD), stalk domain, transmembrane region, and cytoplasmic tail.
Hyaluronan: A Size-Dependent Bioactive Molecule
Structure and Physicochemical Properties
Hyaluronan has a simple structure that lacks a core protein linkage or sulfation. It is synthesized as a large, negatively charged linear polymer with multiple carboxyl groups on GlcUA residues. Both the GlcUA and GlcNAc residues are in the β configuration, which allows their bulky groups, including the hydroxyl and carboxyl groups, to reside in sterically unhindered equatorial positions, and thus hyaluronan forms particularly stable tertiary structures in aqueous solution that exhibit remarkable hydrodynamic properties, including non-Newtonian viscosity and water retention. In dilute solution, hyaluronan forms an expanded random coil due to the mutual repulsion of its carboxyl groups, and at higher concentrations it forms an entangled meshwork, the size of which depends on its concentration and molecular weight, and on the ionic strength and pH of the solution (7). At physiological ionic strengths, hyaluronan’s polyanionic structure causes the partition and diffusion of monovalent ions, such as Na+ and Cl− as well as the divalent cation Ca2+ at a nearly ideal Donnan equilibrium (8).
Biosynthesis
While most glycosaminoglycans are synthesized in the Golgi apparatus, hyaluronan is synthesized at the cell surface, from uridine 5′-diphosphate (UDP)-GlcUA and UDP-GlcNAc by hyaluronan synthases (HASs), a class of membrane-integrated gylcosyltransferases (EC 2.4.1.212). There are three HAS isoforms in mammals, such as HAS1, HAS2, and HAS3 (9), which have different tissue- and cell-specific expression patterns and Km values for their substrates; they also synthesize hyaluronan of different sizes in vitro (10). Various growth factors, including epidermal growth factor (EGF), platelet-derived growth factor (PDGF), and transforming growth factor-β (TGF-β), induce the transcription of HAS genes and enhance hyaluronan synthesis (11, 12). Dysregulated HAS expression or activity is sometimes associated with tissue injury and immune diseases (13).
Degradation
Hyaluronan is enzymatically degraded mainly by hyaluronidases. The mammalian hyaluronidases (EC 3.2.1.35) are endo-β-acetyl-hexosaminidases, which hydrolyze the hexosaminidic β1,4-linkages between GlcNAc and GlcUA residues (14). Six hyaluronidase-like sequences are present in the human genome; the five genes, i.e., HYAL1, HYAL2, HYAL3, HYAL4, and SPAM1 genes, which encode Hyal-1, Hyal-2, Hyal-3, Hyal-4, and PH-20, respectively, and a pseudogene PHYAL1 that is transcribed but not translated (15). Among these isoforms, Hyal-1 and Hyal-2 are predominantly active in somatic tissues (16). Hyal-1 is an acid-active lysosomal enzyme, and catalyzes the hydrolytic degradation of hyaluronan with any molecular weight, generating predominantly tetrasaccharides (17). Hyal-2 is an acid-active glycosylphosphatidylinositol (GPI)-anchored enzyme, and digests hyaluronan polymers to products with a molecular weight of approximately 2 × 104, i.e., 100 saccharides (18). In addition to enzymatic degradation, hyaluronan can be depolymerized by reactive oxygen species generated by oxidative stress (and/or reactive nitrogen species), which cause random cleavage of the endoglycosidic linkages (19).
The degradation of large hyaluronan to low-molecular weight hyaluronan occurs at sites of inflammation including atherosclerosis, rheumatoid arthritis, and tumorigenesis, and low-molecular weight hyaluronan promotes inflammation (20). Low-molecular weight hyaluronan arises by the action of hyaluronidases, and the upregulation of expression and activity of hyaluronidases have been noticed in such inflammation conditions (13). Reactive oxygen species accumulate at sites of inflammation, where low-molecular weight hyaluronan can arise also by oxidative degradation.
Size
Hyaluronan is a major component of extracellular matrix and plays important roles in development and tissue remodeling. Under normal physiological conditions, hyaluronan has a high average molecular weight (>1 × 106) and exhibits immunosuppressive effects: high-molecular weight hyaluronan suppresses septic responses to lipopolysaccharides (21), inhibits lymphocyte-mediated cytolysis (22), and has anti-angiogenic effects (23). Under pathological conditions, such as inflammation and cancer, extracellular matrix remodeling is upregulated. In this situation, the hyaluronan is more polydispersed, showing a preponderance of low-molecular weight forms (13). In general, low-molecular weight hyaluronan is highly immunostimulatory, inflammatory, and angiogenic.
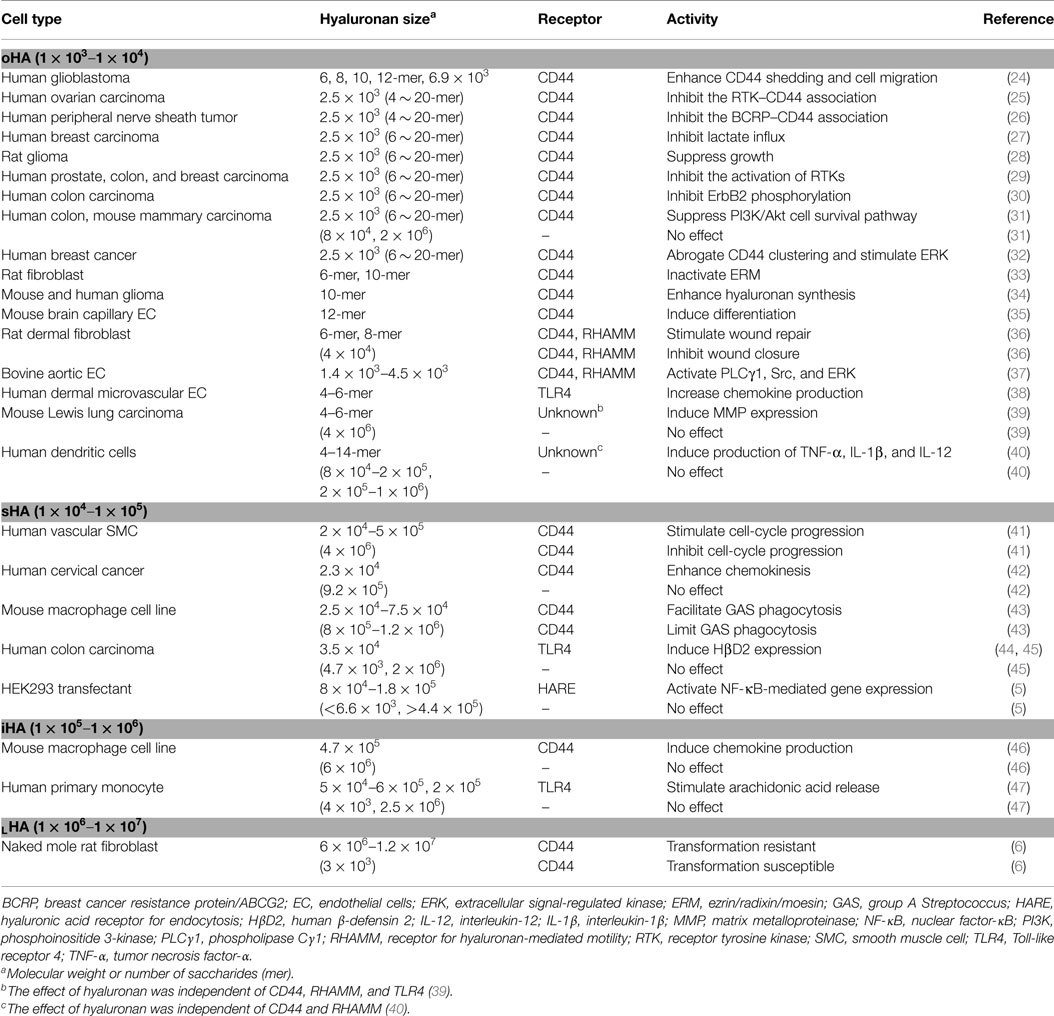
Table 1 summarizes the biological activities associated with different sizes of hyaluronan. The terms used in the literature to describe hyaluronan’s sizes are confusing and inconsistent. Therefore, this article uses the system proposed by Weigel (5), in which hyaluronan’s sizes are categorized according to the log of the molecular weight: oligomeric hyaluronan (oHA; 1 × 103–1 × 104), small hyaluronan (sHA; 1 × 104–1 × 105), intermediate hyaluronan (iHA; 1 × 105–1 × 106), and large hyaluronan (LHA; 1 × 106–1 × 107) (Figure 1B). As shown in Table 1, low-molecular weight hyaluronans (oHA, sHA, and iHA) generally exhibit inflammation- and cancer-promoting activities (5, 6, 24–47). The other effects of low-molecular weight hyaluronan on gene expression are well summarized elsewhere (13). Notably, studies in the naked mole rat (Heterocephalus glaber), an extraordinarily long-lived rodent with low cancer incidence, show that while low-molecular weight hyaluronan has pro-malignant or pro-inflammatory effects, very high-molecular weight hyaluronan (6 × 106–1.2 × 107) has an anti-malignant activity (6) (Figure 1B). Another study shows that oligomeric hyaluronan of 6–40 saccharides, which is frequently found in tumor-bearing patients, enhances cleavage of the hyaluronan receptor CD44 in malignant tumor cells, and concomitantly upregulates CD44-dependent tumor cell migration, whereas larger polymers of hyaluronan fail to enhance CD44 cleavage and migration (24). Collectively, low-molecular weight hyaluronan tends to function as a “danger signal” (48).
Receptors for Hyaluronan
The major cell-surface receptor for hyaluronan is CD44, a widely distributed type-I transmembrane glycoprotein that is implicated in a wide variety of biological processes, including cell adhesion and migration, as well as in inflammation and cancer (49). CD44 mediates the adhesion and dissemination of immune and cancer cells through its association with hyaluronan (50, 51) (Figure 1C). In addition to hyaluronan, CD44’s interaction with certain growth factors also plays important roles in cancer progression (52). Receptor for hyaluronan-mediated motility (RHAMM)/CD168 is another major hyaluronan receptor expressed in a variety of cell types, and it plays important roles in tissue injury and repair and in tumor cell motility (53). Other hyaluronan receptors include lymphatic vessel endothelial hyaluronan receptor 1 (LYVE-1) (54) and hyaluronan receptor for endocytosis (HARE)/stabilin-2 (55). LYVE-1 is mainly restricted to the endothelium of lymph nodes and lymphatic vessels, while HARE is expressed in sinusoidal endothelial cells of the liver, spleen, and lymph nodes, and it mediates the systemic clearance of hyaluronan from the vascular and lymphatic circulatory systems.
Lipid Rafts
Lipid Raft Structure
The plasma membrane is a dynamic mixture of proteins and lipids that forms the boundary and interface between the intracellular space and the cellular environment. The traditional model of cellular membrane structure was the fluid mosaic model proposed by Singer and Nicolson, in which globular proteins float in a lipid bilayer with an amphipathic structure (56). Later, non-homogenously distributed assemblies of lipids were found in the plasma membrane of many cell types, and the model was improved by Simons and van Meer, who suggested the existence of small domains called lipid rafts (57). In the understanding of the lipid raft model, cholesterol- and sphingolipid-enriched microdomains of the plasma membrane assumed a biophysical state resembling a liquid-ordered (Lo) phase floating within a liquid-disordered (Ld) membrane phase (58). In that model, the representative proteins with raft affinity were GPI-anchored proteins. The finding that GPI-anchored proteins were isolated in a low-density detergent resistant membrane (DRM) fractions contributed to the expectation of their residence in lipid rafts (59).
Since then, accumulating evidence has improved the understanding of lipid rafts, also called membrane rafts, and rafts are currently viewed as fluctuating nanoscale assemblies enriched in sphingolipid, cholesterol, and proteins that can be stabilized to coalesce, forming platforms that function in membrane signaling and trafficking (60).
Lipid Raft Function
The most important properties of lipid rafts are that they are small, dynamic, and heterogeneous, and can selectively recruit certain classes of proteins (61, 62). However, the underlying mechanism for the formation and functionality of lipid rafts has been largely unclear. Using single-molecule fluorescence tracking, Kusumi and colleagues recently found that GPI-anchored proteins formed dynamic, transient homodimer rafts in the plasma membrane, in a manner dependent on the interactions between their ectodomain protein portions (63). The homodimer formation seems to be the basic units for the organization and functions of membrane raft domains containing GPI-anchored proteins. Schütz and colleagues observed the relation between the physical state of the membrane domains and the partition of GPI-anchored proteins, and showed that GPI-anchored proteins do not reside in ordered domains (64). This report suggests that the phase partitioning is not a fundamental element of GPI-anchored protein organization in the plasma membrane, and also suggests the heterogeneity in the structure and function of membrane rafts.
Proteins with raft affinity include doubly acylated proteins such as Src family kinases, palmitoylated type-I transmembrane proteins, such as CD44 (65, 66), and receptor tyrosine kinases with two transmembrane subunits, such as inslulin receptor (67) and EGF receptor (68). Lipid rafts are implicated in many physiological cellular processes, such as protein membrane trafficking and signal transduction (62, 69). Cholesterol depletion is often used as a method for investigating the role of lipid rafts in vitro, although these studies are limited by non-specific effects. Nevertheless, these studies indicate that cholesterol is a crucial component of cell membranes that contributes to the organization of lipid rafts, and particularly to lipid rafts that contain large numbers of cancer-related signaling and adhesion molecules.
Hyaluronan–CD44 Interaction and Lipid Rafts in Cancer
The dynamics of extracellular matrix production, degradation, and remodeling are carefully regulated during organ development; the dysregulation of extracellular matrix turnover and maintenance leads to abnormal cell behaviors and to failure of organ homeostasis and function, one of the most severe clinical outcomes in cancer (70). Altered cell adhesion and enhanced cell migration are the most prominent features of malignant tumor cells (71). The migratory properties of invasive tumor cells are affected by the interaction of their adhesion molecules with the surrounding extracellular matrix, and by growth factor signaling (72). The proteolytic cleavage and release (shedding) of membrane proteins’ ectodomain is a critical regulatory step in both physiological and pathological processes (73, 74). It was recently reported that oligomeric hyaluronan induce CD44 shedding from tumor cells (24).
Ectopic hyaluronan production is a frequent feature of colorectal, gastric, and breast cancers (75–77). Under normal physiological conditions, hyaluronan exists as a long polymer with a molecular weight of around 1 × 105–1 × 107 (1), whereas low-molecular weight hyaluronan is frequently detected in certain pathological conditions, such as inflammation (78) and cancer (79, 80), possibly due to the dysregulated expression of HASs and hyaluronidases. Hyaluronidase expression in prostate cancer tissues increased with tumor grade and metastasis, suggesting that prostate tumor cell-derived hyaluronidase might help the accumulation of low-molecular weight hyaluronan (80).
A prominent abnormality of certain malignant tumor cells, e.g., gliomas, is overexpression of the EGF receptor, and EGF induces CD44 shedding, that concomitantly enhance hyaluronan-mediated cell migration (81). PDGF and bradykinin also induce CD44 shedding, indicating that the Rho family of small GTPases plays crucial roles in the regulation of CD44 cleavage (81). TGF-β induces CD44 shedding in breast cancer cells (82), and this cleavage is MT1-MMP-dependent as previously described (83, 84). Granulocyte-colony stimulating factor (G-CSF) stimulates the MT1-MMP-mediated CD44 proteolysis in hematopoietic progenitor cells (85). Although the molecular mechanisms of the intracellular signaling in the tumor microenvironment that lead to CD44 shedding have been partially clarified (81, 86), the mechanism that triggers CD44’s shedding at the membrane is not understood.
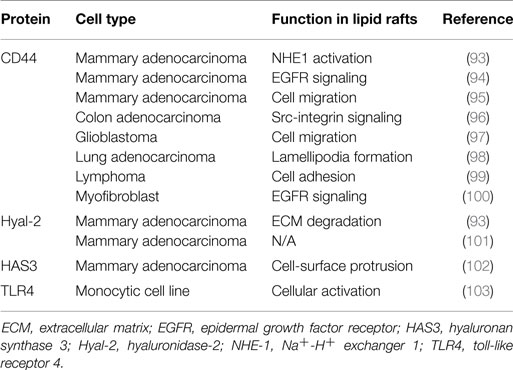
There is growing interest in targeting lipid rafts for cancer prevention and treatment, because of their role in regulating various steps of cancer progression, including cancer cell migration and invasion (87), and because cancer-related proteins were listed in an unbiased proteomics analysis of these structures (88). Cell adhesion is a key factor in the metastatic spread of cancer cells, and regulating this process holds promise as an important therapeutic intervention for cancer. CD44 is the principal cell adhesion receptor expressed in cancer cells and implicated in cancer cell migration, invasion, and metastasis (89). Several reports recently demonstrated that CD44 is present in lipid rafts (90–100) (Table 2), and the role of lipid rafts in cancer cell adhesion and migration is being elucidated.
Lipid rafts play a pivotal role in CD44’s localization and function (97). Cholesterol depletion form human glioma cells using methyl-β-cyclodextrin (MβCD), an agent frequently used to disrupt lipid rafts, results in increased CD44 shedding, which was mediated by a transmembrane protease a disintegrin and metalloproteinase 10 (ADAM10). The CD44 shedding induced by cholesterol depletion is also seen in other tumor cells, such as pancreatic cancer cells. CD44 shedding can also be induced by a polyene macrolide antibiotic filipin that binds cholesterol and disperses it in the membrane, thereby disrupting lipid rafts by a different mechanism from MβCD. The cholesterol-lowering medication simvastatin also enhances CD44 shedding; it also blocks the stimulation of glioma cell migration by oligomeric hyaluronan or EGF. Taken together, these results suggest that cholesterol-lowering causes disordered CD44 localization, raft-dependent CD44 shedding, and the suppression of tumor cell migration that is dependent on hyaluronan’s size. CD44’s affiliation with lipid rafts is likely to occur through its palmitoylation, which may play a role in breast cancer malignancy (95).
In addition to CD44, several hyaluronan-related proteins, Hyal-2, HAS3, and toll-like receptor 4 (TLR4), have been reported to be associated with lipid rafts in cell membranes (93, 101–103) (Table 2). These membrane proteins are also likely to be involved in the regulation of lipid raft-associated interactions between hyaluronan and CD44. In addition, CD147 was found to regulate the lipid raft-associated CD44 function in cancer cell invasion (94, 104).
Potential Roles of Hyaluronan–CD44 Interactions in Inflammation
The recruitment of lymphocytes from circulating blood to inflammatory sites or secondary lymphoid organs involves complementary receptor–ligand interactions between the lymphocytes and vascular endothelial cells. A multistep series of sequential receptor engagements enables the lymphocytes’ recognition of the endothelial surface and their subsequent extravasation (105). This process begins with the establishment of transient adhesive interactions that result in the rolling of lymphocytes along the endothelium under blood flow, and rolling is mediated by interactions between CD44 and hyaluronan (106, 107). The CD44–hyaluronan interaction is required for the extravasation of activated T cells from circulating blood to inflammatory sites (108). There is also evidence that the hyaluronan-binding ability of CD44 is correlated with the suppressor activity of CD4+CD25+ regulatory T cells (109).
CD44 does not bind hyaluronan constitutively in most immune cells, although CD44 is the principal receptor for hyaluronan in immune cells (89). Considering the ubiquitous distribution of CD44 and hyaluronan, tight regulation of the hyaluronan-binding ability of CD44 is likely to play a critical role in immunological responses: CD44 on resting T cells does not bind hyaluronan, but can be induced to bind it when the T cell is activated by antigen via the T-cell receptor (108–112). Various post-translational modifications on CD44, including glycosylation (113–115), chondroitin sulfate addition (116, 117), and sulfation (118, 119), are reported to affect its hyaluronan-binding ability. However, the membrane-based regulation of CD44’s hyaluronan-binding ability has not been clarified. A recent study demonstrated that the hyaluronan-binding ability of CD44 in T cells is upregulated by membrane cholesterol depletion, which causes CD44 to be dispersed from lipid rafts, although the effect is small (120). Cholesterol depletion also enhances the frequency of rolling adhesion under physiological flow conditions, suggesting that the CD44’s ligand-binding ability is governed by its cholesterol-dependent localization to lipid rafts.
Perspectives
Epidermal growth factor receptor is one of the first reported growth factor receptors that exhibit raft affinity, and EGF induced the coalescence of EGF receptor-containing rafts with different type of lipid rafts that contain GPI-anchored proteins (68). This coalescence of different types of nanoscale assemblies possibly leads to the formation of functional platforms for transmembrane signaling and the initiation of the internalization of EGF receptors. In the case of hyaluronan receptor CD44, the function of hyaluronan in the regulation of lipid rafts may be in a similar way as proposed for EGF. As oligomeric hyaluronan can displace large hyaluronan from cells (4), it may modulate the raft coalescence that leads to form signaling platforms toward inflammation and cancer progression. Competitive binding assay showed that the minimum length of hyaluronan that can compete large hyaluronan binding to CD44 is 6-mer, and the nuclear magnetic resonance spectroscopy confirmed that 6-mer is the shortest oligomeric hyaluronan to give essentially full perturbation of CD44’s spectra (121). The structure of CD44’s hyaluronan-binding domain (HABD) solved by X-ray crystallography revealed that CD44 forms two different conformations upon binding to hyaluronan (122). To understand the molecular mechanism and associated energetics underlying the hyaluronan–CD44 binding interaction, Guvench group performed extensive all-atom explicit-solvent molecular dynamics (MD) simulations employing the adaptive biasing force free-energy methodology (123). They determined a clear description for the conformation-dependent affinity switching of the hyaluronan–CD44 interactions by MD simulation. These results should help the development of novel small compounds to therapeutics in inflammation and cancer by modulating hyaluronan–CD44 interactions, which may regulate the functionality of lipid rafts.
There has been growing interest in lipid rafts, and the lipid raft is a potential novel target in inflammation and cancer therapy (66, 124). Targeting hyaluronan–CD44 axis is one of the principal ways, and the lipid raft-targeted delivery of hyaluronan-grafted liposomes could have important applications in cancer therapy (125, 126). The modulation of CD44’s partition to lipid rafts may also offer potential avenues in inflammation and cancer therapy. Thus, the regulation and manipulation of hyaluronan–CD44 interactions through lipid rafts have potential applications for the prevention of inflammatory disorders and cancer.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The author gratefully acknowledges the following financial support: Grants-in-Aid for Scientific Research (C), Grants-in-Aid for Young Scientists, and Grants-in-Aid for Scientific Research on Priority Areas, “System Cell Engineering by Multi-scale Manipulation” and “Functional Mechanism and Structural Organization of Biological Macromolecular Assemblies,” from the Ministry of Education, Culture, Sports, Science and Technology of Japan; the Nestlé Nutrition Council; the All Japan Coffee Association; and the Kieikai Research Foundation.
References
1. Fraser JRE, Laurent TC, Laurent UBG. Hyaluronan: its nature, distribution, functions and turnover. J Intern Med (1997) 242:27–33. doi: 10.1046/j.1365-2796.1997.00170.x
3. Balazs EA, Delinger JL. Sodium hyaluronate and joint function. J Equine Vet Sci (1985) 5:217–28. doi:10.1016/S0737-0806(85)80102-7
4. Toole BP. Hyaluronan: from extracellular glue to pericellular cue. Nat Rev Cancer (2004) 4:528–39. doi:10.1038/nrc1391
5. Pandey MS, Baggenstoss BA, Washburn J, Harris EN, Weigel PH. The hyaluronan receptor for endocytosis (HARE) activates NF-κB-mediated gene expression in response to 40-400-kDa, but not smaller or larger, hyaluronans. J Biol Chem (2013) 288:14068–79. doi:10.1074/jbc.M112.442889
6. Tian X, Azpurua J, Hine C, Vaidya A, Myakishev-Rempel M, Ablaeva J, et al. High-molecular-mass hyaluronan mediates the cancer resistance of the naked mole rat. Nature (2013) 499:346–9. doi:10.1038/nature12234
7. Scott JE, Cummings C, Brass A, Chen Y. Secondary and tertiary structures of hyaluronan in aqueous solution, investigated by rotary shadowing-electron microscopy and computer simulation. Hyaluronan is a very efficient network-forming polymer. Biochem J (1991) 274:699–705.
8. Maroudas A, Weinberg PD, Parker KH, Winlove CP. The distributions and diffusivities of small ions in chondroitin sulphate, hyaluronate and some proteoglycan solutions. Biophys Chem (1988) 32:257–70. doi:10.1016/0301-4622(88)87012-1
9. Weigel PH, Hascall VC, Tammi M. Hyaluronan synthases. J Biol Chem (1997) 272:13997–4000. doi:10.1074/jbc.272.22.13997
10. Itano N, Sawai T, Yoshida M, Lenas P, Yamada Y, Imagawa M, et al. Three isoforms of mammalian hyaluronan synthases have distinct enzymatic properties. J Biol Chem (1999) 274:25085–92. doi:10.1074/jbc.274.35.25085
11. Pienimäki JP, Rilla K, Fülöp C, Sironen RK, Karvinen S, Pasonen S, et al. Epidermal growth factor activates hyaluronan synthase 2 in epidermal keratinocytes and increases pericellular and intracellular hyaluronan. J Biol Chem (2001) 276:20428–35. doi:10.1074/jbc.M007601200
12. Li L, Asteriou T, Bernert B, Heldin CH, Heldin P. Growth factor regulation of hyaluronan synthesis and degradation in human dermal fibroblasts: importance of hyaluronan for the mitogenic response of PDGF-BB. Biochem J (2007) 404:327–36. doi:10.1042/BJ20061757
13. Jiang D, Liang J, Noble PW. Hyaluronan as an immune regulator in human diseases. Physiol Rev (2011) 91:221–64. doi:10.1152/physrev.00052.2009
14. Stern R. Hyaluronan catabolism: a new metabolic pathway. Eur J Cell Biol (2004) 83:317–25. doi:10.1078/0171-9335-00392
15. Stern R. Devising a pathway for hyaluronan catabolism: are we there yet? Glycobiology (2003) 13:105R–15R. doi:10.1093/glycob/cwg112
16. Stern R, Jedrzejas MJ. Hyaluronidases: their genomics, structures, and mechanisms of action. Chem Rev (2006) 106:818–39. doi:10.1021/cr050247k
17. Frost GI, Csóka TB, Wong T, Stern R. Purification, cloning, and expression of human plasma hyaluronidase. Biochem Biophys Res Commun (1997) 236:10–5. doi:10.1006/bbrc.1997.6773
18. Lepperdinger G, Strobl B, Kreil G. HYAL2, a human gene expressed in many cells, encodes a lysosomal hyaluronidase with a novel type of specificity. J Biol Chem (1998) 273:22466–70. doi:10.1074/jbc.273.35.22466
19. Gao F, Koenitzer JR, Tobolewski JM, Jiang D, Liang J, Noble PW, et al. Extracellular superoxide dismutase inhibits inflammation by preventing oxidative fragmentation of hyaluronan. J Biol Chem (2008) 283:6058–66. doi:10.1074/jbc.M709273200
20. Puré E, Assoian RK. Rheostatic signaling by CD44 and hyaluronan. Cell Signal (2009) 21:651–5. doi:10.1016/j.cellsig.2009.01.024
21. Muto J, Yamasaki K, Taylor KR, Gallo RL. Engagement of CD44 by hyaluronan suppresses TLR4 signaling and the septic response to LPS. Mol Immunol (2009) 47:449–56. doi:10.1016/j.molimm.2009.08.026
22. McBride WH, Bard JB. Hyaluronidase-sensitive halos around adherent cells. Their role in blocking lymphocyte-mediated cytolysis. J Exp Med (1979) 149:507–15. doi:10.1084/jem.149.2.507
23. Feinberg RN, Beebe DC. Hyaluronate in vasculogenesis. Science (1983) 220:1177–9. doi:10.1126/science.6857242
24. Sugahara KN, Murai T, Nishinakamura H, Kawashima H, Saya H, Miyasaka M. Hyaluronan oligosaccharides induce CD44 cleavage and promote cell migration in CD44-expressing tumor cells. J Biol Chem (2003) 278:32259–65. doi:10.1074/jbc.M300347200
25. Slomiany MG, Dai L, Tolliver LB, Grass GD, Zeng Y, Toole BP. Inhibition of functional hyaluronan-CD44 interactions in CD133-positive primary human ovarian carcinoma cells by small hyaluronan oligosaccharides. Clin Cancer Res (2009) 15:7593–601. doi:10.1158/1078-0432.CCR-09-2317
26. Slomiany MG, Dai L, Bomar PA, Knackstedt TJ, Kranc DA, Tolliver L, et al. Abrogating drug resistance in malignant peripheral nerve sheath tumors by disrupting hyaluronan-CD44 interactions with small hyaluronan oligosaccharides. Cancer Res (2009) 69:4992–8. doi:10.1158/0008-5472.CAN-09-0143
27. Slomiany MG, Grass GD, Robertson AD, Yang XY, Maria BL, Beeson C, et al. Hyaluronan, CD44, and emmprin regulate lactate efflux and membrane localization of monocarboxylate transporters in human breast carcinoma cells. Cancer Res (2009) 69:1293–301. doi:10.1158/0008-5472.CAN-08-2491
28. Gilg AG, Tye SL, Tolliver LB, Wheeler WG, Visconti RP, Duncan JD, et al. Targeting hyaluronan interactions in malignant gliomas and their drug-resistant multipotent progenitors. Clin Cancer Res (2008) 14:1804–13. doi:10.1158/1078-0432.CCR-07-1228
29. Misra S, Toole BP, Ghatak S. Hyaluronan constitutively regulates activation of multiple receptor tyrosine kinases in epithelial and carcinoma cells. J Biol Chem (2006) 281:34936–41. doi:10.1074/jbc.C600138200
30. Ghatak S, Misra S, Toole BP. Hyaluronan constitutively regulates ErbB2 phosphorylation and signaling complex formation in carcinoma cells. J Biol Chem (2005) 280:8875–83. doi:10.1074/jbc.M410882200
31. Ghatak S, Misra S, Toole BP. Hyaluronan oligosaccharides inhibit anchorage-independent growth of tumor cells by suppressing the phosphoinositide 3-kinase/Akt cell survival pathway. J Biol Chem (2002) 277:38013–20. doi:10.1074/jbc.M202404200
32. Yang C, Cao M, Liu H, He Y, Xu J, Du Y, et al. The high and low molecular weight forms of hyaluronan have distinct effects on CD44 clustering. J Biol Chem (2012) 287:43094–107. doi:10.1074/jbc.M112.349209
33. Duterme C, Mertens-Strijthagen J, Tammi M, Flamion B. Two novel functions of hyaluronidase-2 (Hyal2) are formation of the glycocalyx and control of CD44-ERM interactions. J Biol Chem (2009) 284:33495–508. doi:10.1074/jbc.M109.044362
34. Wiranowska M, Ladd S, Moscinski LC, Hill B, Haller E, Mikecz K, et al. Modulation of hyaluronan production by CD44 positive glioma cells. Int J Cancer (2010) 127:532–42. doi:10.1002/ijc.25085
35. Takahashi Y, Li L, Kamiryo M, Asteriou T, Moustakas A, Yamashita H, et al. Hyaluronan fragments induce endothelial cell differentiation in a CD44- and CXCL1/GRO1-dependent manner. J Biol Chem (2005) 280:24195–204. doi:10.1074/jbc.M411913200
36. Tolg C, Telmer P, Turley E. Specific sizes of hyaluronan oligosaccharides stimulate fibroblast migration and excisional wound repair. PLoS One (2014) 9:e88479. doi:10.1371/journal.pone.0088479
37. Slevin M, Kumar S, Gaffney J. Angiogenic oligosaccharides of hyaluronan induce multiple signaling pathways affecting vascular endothelial cell mitogenic and wound healing responses. J Biol Chem (2002) 277:41046–59. doi:10.1074/jbc.M109443200
38. Taylor KR, Trowbridge JM, Rudisill JA, Termeer CC, Simon JC, Gallo RL. Hyaluronan fragments stimulate endothelial recognition of injury through TLR4. J Biol Chem (2004) 279:17079–84. doi:10.1074/jbc.M310859200
39. Fieber C, Baumann P, Vallon R, Termeer C, Simon JC, Hofmann M, et al. Hyaluronan-oligosaccharide-induced transcription of metalloproteases. J Cell Sci (2004) 117:359–67. doi:10.1242/jcs.00831
40. Termeer CC, Hennies J, Voith U, Ahrens T, Weiss JM, Prehm P, et al. Oligosaccharides of hyaluronan are potent activators of dendritic cells. J Immunol (2000) 165:1863–70. doi:10.4049/jimmunol.165.4.1863
41. Kothapalli D, Flowers J, Xu T, Puré E, Assoian RK. Differential activation of ERK and Rac mediates the proliferative and anti-proliferative effects of hyaluronan and CD44. J Biol Chem (2008) 283:31823–9. doi:10.1074/jbc.M802934200
42. Saito T, Kawana H, Azuma K, Toyoda A, Fujita H, Kitagawa M, et al. Fragmented hyaluronan is an autocrine chemokinetic motility factor supported by the HAS2-HYAL2/CD44 system on the plasma membrane. Int J Oncol (2011) 39:1311–20. doi:10.3892/ijo.2011.1114
43. Schommer NN, Muto J, Nizet V, Gallo RL. Hyaluronan breakdown contributes to immune defense against group A Streptococcus. J Biol Chem (2014) 289:26914–21. doi:10.1074/jbc.M114.575621
44. Hill DR, Rho HK, Kessler SP, Amin R, Homer CR, McDonald C, et al. Human milk hyaluronan enhances innate defense of the intestinal epithelium. J Biol Chem (2013) 288:29090–104. doi:10.1074/jbc.M113.468629
45. Hill DR, Kessler SP, Rho HK, Cowman MK, de la Motte CA. Specific-sized hyaluronan fragments promote expression of human β-defensin 2 in intestinal epithelium. J Biol Chem (2012) 287:30610–24. doi:10.1074/jbc.M112.356238
46. McKee CM, Penno MB, Cowman M, Burdick MD, Strieter RM, Bao C, et al. Hyaluronan (HA) fragments induce chemokine gene expression in alveolar macrophages. The role of HA size and CD44. J Clin Invest (1996) 98:2403–13. doi:10.1172/JCI119054
47. Sokolowska M, Chen LY, Eberlein M, Martinez-Anton A, Liu Y, Alsaaty S, et al. Low molecular weight hyaluronan activates cytosolic phospholipase A2α and eicosanoid production in monocytes and macrophages. J Biol Chem (2014) 289:4470–88. doi:10.1074/jbc.M113.515106
48. Powell JD, Horton MR. Threat matrix: low-molecular-weight hyaluronan (HA) as a danger signal. Immunol Res (2005) 31:207–18. doi:10.1385/IR:31:3:207
49. Naor D, Sionov RV, Ish-Shalom D. CD44: structure, function, and association with the malignant process. Adv Cancer Res (1997) 71:241–319. doi:10.1016/S0065-230X(08)60101-3
50. Naor D, Nedvetzki S. CD44 in rheumatoid arthritis. Arthritis Res Ther (2003) 5:105–15. doi:10.1186/ar735
51. Naor D, Wallach-Dayan SB, Zahalka MA, Sionov RV. Involvement of CD44, a molecule with a thousand faces, in cancer dissemination. Semin Cancer Biol (2008) 18:260–7. doi:10.1016/j.semcancer.2008.03.015
52. Hertweck MK, Erdfelder F, Kreuzer KA. CD44 in hematological neoplasias. Ann Hematol (2011) 90:493–508. doi:10.1007/s00277-011-1161-z
53. Hardwick C, Hoare K, Owens R, Hohn HP, Hook M, Moore D, et al. Molecular cloning of a novel hyaluronan receptor that mediates tumor cell motility. J Cell Biol (1992) 117:1343–50. doi:10.1083/jcb.117.6.1343
54. Banerji S, Ni J, Wang SX, Clasper S, Su J, Tammi R, et al. LYVE-1, a new homologue of the CD44 glycoprotein, is a lymph-specific receptor for hyaluronan. J Cell Biol (1999) 144:789–801. doi:10.1083/jcb.144.4.789
55. Zhou B, Weigel JA, Saxena A, Weigel PH. Molecular cloning and functional expression of the rat 175-kDa hyaluronan receptor for endocytosis. Mol Biol Cell (2002) 13:2853–68. doi:10.1091/mbc.02-03-0048
56. Singer SJ, Nicolson GL. The fluid mosaic model of the structure of cell membranes. Science (1972) 175:720–31. doi:10.1126/science.175.4023.720
57. Simons K, van Meer G. Lipid sorting in epithelial cells. Biochemistry (1988) 27:6197–202. doi:10.1021/bi00417a001
58. Simons K, Ikonen E. Functional rafts in cell membranes. Nature (1997) 387:569–72. doi:10.1038/42408
59. Brown DA, Rose JK. Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell (1992) 68:533–44. doi:10.1016/0092-8674(92)90189-J
60. Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science (2010) 327:46–50. doi:10.1126/science.1174621
61. Resh MD. Fatty acylation of proteins: new insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim Biophys Acta (1999) 1451:1–16. doi:10.1016/S0167-4889(99)00075-0
62. Simons K, Toomre D. Lipid rafts and signal transduction. Nat Rev Mol Cell Biol (2000) 1:31–9. doi:10.1038/35036205
63. Suzuki KG, Kasai RS, Hirosawa KM, Nemoto YL, Ishibashi M, Miwa Y, et al. Transient GPI-anchored protein homodimers are units for raft organization and function. Nat Chem Biol (2012) 8:774–83. doi:10.1038/nchembio.1028
64. Sevcsik E, Brameshuber M, Fölser M, Weghuber J, Honigmann A, Schütz GJ. GPI-anchored proteins do not reside in ordered domains in the live cell plasma membrane. Nat Commun (2015) 6:6969. doi:10.1038/ncomms7969
65. Brown DA, London E. Structure and function of sphingolipid- and cholesterol-rich membrane rafts. J Biol Chem (2000) 275:17221–4. doi:10.1074/jbc.R000005200
66. Fessler MB, Parks JS. Intracellular lipid flux and membrane microdomains as organizing principles in inflammatory cell signaling. J Immunol (2011) 187:1529–35. doi:10.4049/jimmunol.1100253
67. Vainio S, Heino S, Månsson JE, Fredman P, Kuismanen E, Vaarala O, et al. Dynamic association of human insulin receptor with lipid rafts in cells lacking caveolae. EMBO Rep (2002) 3:95–100. doi:10.1093/embo-reports/kvf010
68. Hofman EG, Ruonala MO, Bader AN, van den Heuvel D, Voortman J, Roovers RC, et al. EGF induces coalescence of different lipid rafts. J Cell Sci (2008) 121:2519–28. doi:10.1242/jcs.028753
69. Hanzal-Bayer MF, Hancock JF. Lipid rafts and membrane traffic. FEBS Lett (2007) 581:2098–104. doi:10.1016/j.febslet.2007.03.019
70. Lu P, Weaver VM, Werb Z. The extracellular matrix: a dynamic niche in cancer progression. J Cell Biol (2012) 196:395–406. doi:10.1083/jcb.201102147
71. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell (2011) 144:646–74. doi:10.1016/j.cell.2011.02.013
72. Friedl P, Wolf K. Tumour-cell invasion and migration: diversity and escape mechanisms. Nat Rev Cancer (2003) 3:362–74. doi:10.1038/nrc1075
73. Huovila AP, Turner AJ, Pelto-Huikko M, Kärkkäinen I, Ortiz RM. Shedding light on ADAM metalloproteinases. Trends Biochem Sci (2005) 30:413–22. doi:10.1016/j.tibs.2005.05.006
74. Alfandari D, McCusker C, Cousin H. ADAM function in embryogenesis. Semin Cell Dev Biol (2009) 20:153–63. doi:10.1016/j.semcdb.2008.09.006
75. Ropponen K, Tammi M, Parkkinen J, Eskelinen M, Tammi R, Lipponen P, et al. Tumor cell-associated hyaluronan as an unfavorable prognostic factor in colorectal cancer. Cancer Res (1998) 58:342–7.
76. Setälä LP, Tammi MI, Tammi RH, Eskelinen MJ, Lipponen PK, Ågren UM, et al. Hyaluronan expression in gastric cancer cells is associated with local and nodal spread and reduced survival rate. Br J Cancer (1999) 79:1133–8. doi:10.1038/sj.bjc.6690180
77. Auvinen P, Tammi R, Parkkinen J, Tammi M, Ågren U, Johansson R, et al. Hyaluronan in peritumoral stroma and malignant cells associates with breast cancer spreading and predicts survival. Am J Pathol (2000) 156:529–36. doi:10.1016/S0002-9440(10)64757-8
78. Saari H, Konttinen YT. Determination of synovial fluid hyaluronate concentration and polymerisation by high performance liquid chromatography. Ann Rheum Dis (1989) 48:565–70. doi:10.1136/ard.48.7.565
79. Lokeshwar VB, Obek C, Soloway MS, Block NL. Tumor-associated hyaluronic acid: a new sensitive and specific urine marker for bladder cancer. Cancer Res (1997) 57:773–7.
80. Lokeshwar VB, Rubinowicz D, Schroeder GL, Forgacs E, Minna JD, Block NL, et al. Stromal and epithelial expression of tumor markers hyaluronic acid and HYAL1 hyaluronidase in prostate cancer. J Biol Chem (2001) 276:11922–32. doi:10.1074/jbc.M008432200
81. Murai T, Miyauchi T, Yanagida T, Sako Y. Epidermal growth factor-regulated activation of Rac GTPase enhances CD44 cleavage by metalloproteinase disintegrin ADAM10. Biochem J (2006) 395:65–71. doi:10.1042/BJ20050582
82. Kuo YC, Su CH, Liu CY, Chen TH, Chen CP, Wang HS. Transforming growth factor-β induces CD44 cleavage that promotes migration of MDA-MB-435s cells through the up-regulation of membrane type 1-matrix metalloproteinase. Int J Cancer (2009) 124:2568–76. doi:10.1002/ijc.24263
83. Kajita M, Itoh Y, Chiba T, Mori H, Okada A, Kinoh H, et al. Membrane-type 1 matrix metalloproteinase cleaves CD44 and promotes cell migration. J Cell Biol (2001) 153:893–904. doi:10.1083/jcb.153.5.893
84. Suenaga N, Mori H, Itoh Y, Seiki M. CD44 binding through the hemopexin-like domain is critical for its shedding by membrane-type 1 matrix metalloproteinase. Oncogene (2005) 24:859–68. doi:10.1038/sj.onc.1208258
85. Vagima Y, Avigdor A, Goichberg P, Shivtiel S, Tesio M, Kalinkovich A, et al. MT1-MMP and RECK are involved in human CD34+ progenitor cell retention, egress, and mobilization. J Clin Invest (2009) 119:492–503. doi:10.1172/JCI36541
86. Murai T, Miyazaki Y, Nishinakamura H, Sugahara KN, Miyauchi T, Sako Y, et al. Engagement of CD44 promotes Rac activation and CD44 cleavage during tumor cell migration. J Biol Chem (2004) 279:4541–50. doi:10.1074/jbc.M307356200
87. Murai T. The role of lipid rafts in cancer cell adhesion and migration. Int J Cell Biol (2012) 2012:763283. doi:10.1155/2012/763283
88. Murai T. Cholesterol lowering: role in cancer prevention and treatment. Biol Chem (2015) 396:1–11. doi:10.1515/hsz-2014-0194
89. Lesley J, Hyman R, Kincade PW. CD44 and its interaction with extracellular matrix. Adv Immunol (1993) 54:271–335. doi:10.1016/S0065-2776(08)60537-4
90. Ilangumaran S, Hoessli DC. Effects of cholesterol depletion by cyclodextrin on the sphingolipid microdomains of the plasma membrane. Biochem J (1998) 335:433–40.
91. Oliferenko S, Paiha K, Harder T, Gerke V, Schwärzler C, Schwarz H, et al. Analysis of CD44-containing lipid rafts: recruitment of annexin II and stabilization by the actin cytoskeleton. J Cell Biol (1999) 146:843–54. doi:10.1083/jcb.146.4.843
92. Gómez-Moutón C, Abad JL, Mira E, Lacalle RA, Gallardo E, Jiménez-Baranda S, et al. Segregation of leading-edge and uropod components into specific lipid rafts during T cell polarization. Proc Natl Acad Sci U S A (2001) 98:9642–7. doi:10.1073/pnas.171160298
93. Bourguignon LY, Singleton PA, Diedrich F, Stern R, Gilad E. CD44 interaction with Na+-H+ exchanger (NHE1) creates acidic microenvironments leading to hyaluronidase-2 and cathepsin B activation and breast tumor cell invasion. J Biol Chem (2004) 279:26991–7007. doi:10.1074/jbc.M311838200
94. Grass GD, Tolliver LB, Bratoeva M, Toole BP. CD147, CD44, and the epidermal growth factor receptor (EGFR) signaling pathway cooperate to regulate breast epithelial cell invasiveness. J Biol Chem (2013) 288:26089–104. doi:10.1074/jbc.M113.497685
95. Babina IS, McSherry EA, Donatello S, Hill AD, Hopkins AM. A novel mechanism of regulating breast cancer cell migration via palmitoylation-dependent alterations in the lipid raft affiliation of CD44. Breast Cancer Res (2014) 16:R19. doi:10.1186/bcr3614
96. Lee JL, Wang MJ, Sudhir PR, Chen JY. CD44 engagement promotes matrix-derived survival through the CD44-SRC-integrin axis in lipid rafts. Mol Cell Biol (2008) 28:5710–23. doi:10.1128/MCB.00186-08
97. Murai T, Maruyama Y, Mio K, Nishiyama H, Suga M, Sato C. Low cholesterol triggers membrane microdomain-dependent CD44 shedding and suppresses tumor cell migration. J Biol Chem (2011) 286:1999–2007. doi:10.1074/jbc.M110.184010
98. Kim KB, Yi JS, Nguyen N, Lee JH, Kwon YC, Ahn BY, et al. Cell-surface receptor for complement component C1q (gC1qR) is a key regulator for lamellipodia formation and cancer metastasis. J Biol Chem (2011) 286:23093–101. doi:10.1074/jbc.M111.233304
99. Singh V, Erb U, Zöller M. Cooperativity of CD44 and CD49d in leukemia cell homing, migration, and survival offers a means for therapeutic attack. J Immunol (2013) 191:5304–16. doi:10.4049/jimmunol.1301543
100. Midgley AC, Rogers M, Hallett MB, Clayton A, Bowen T, Phillips AO, et al. Transforming growth factor-β1 (TGF-β1)-stimulated fibroblast to myofibroblast differentiation is mediated by hyaluronan (HA)-facilitated epidermal growth factor receptor (EGFR) and CD44 co-localization in lipid rafts. J Biol Chem (2013) 288:14824–38. doi:10.1074/jbc.M113.451336
101. Andre B, Duterme C, Van Moer K, Mertens-Strijthagen J, Jadot M, Flamion B. Hyal2 is a glycosylphosphatidylinositol-anchored, lipid raft-associated hyaluronidase. Biochem Biophys Res Commun (2011) 411:175–9. doi:10.1016/j.bbrc.2011.06.125
102. Kultti A, Rilla K, Tiihonen R, Spicer AP, Tammi RH, Tammi MI. Hyaluronan synthesis induces microvillus-like cell surface protrusions. J Biol Chem (2006) 281:15821–8. doi:10.1074/jbc.M512840200
103. Triantafilou M, Miyake K, Golenbock DT, Triantafilou K. Mediators of innate immune recognition of bacteria concentrate in lipid rafts and facilitate lipopolysaccharide-induced cell activation. J Cell Sci (2002) 115:2603–11.
104. Grass GD, Dai L, Qin Z, Parsons C, Toole BP. CD147: regulator of hyaluronan signaling in invasiveness and chemoresistance. Adv Cancer Res (2014) 123:351–73. doi:10.1016/B978-0-12-800092-2.00013-7
105. Springer TA. Traffic signals for lymphocyte recirculation and leukocyte emigration: the multistep paradigm. Cell (1994) 76:301–14. doi:10.1016/0092-8674(94)90337-9
106. DeGrendele HC, Estess P, Picker LJ, Siegelman MH. CD44 and its ligand hyaluronate mediate rolling under physiologic flow: a novel lymphocyte-endothelial cell primary adhesion pathway. J Exp Med (1996) 183:1119–30. doi:10.1084/jem.183.3.1119
107. Clark RA, Alon R, Springer TA. CD44 and hyaluronan-dependent rolling interactions of lymphocytes on tonsillar stroma. J Cell Biol (1996) 134:1075–87. doi:10.1083/jcb.134.4.1075
108. DeGrendele HC, Estess P, Siegelman MH. Requirement for CD44 in activated T cell extravasation into an inflammatory site. Science (1997) 278:672–5. doi:10.1126/science.278.5338.672
109. Firan M, Dhillon S, Estess P, Siegelman MH. Suppressor activity and potency among regulatory T cells is discriminated by functionally active CD44. Blood (2006) 107:619–27. doi:10.1182/blood-2005-06-2277
110. Lesley J, Howes N, Perschl A, Hyman R. Hyaluronan binding function of CD44 is transiently activated on T cells during an in vivo immune response. J Exp Med (1994) 180:383–7. doi:10.1084/jem.180.1.383
111. Ariel A, Lider O, Brill A, Cahalon L, Savion N, Varon D, et al. Induction of interactions between CD44 and hyaluronic acid by a short exposure of human T cells to diverse pro-inflammatory mediators. Immunology (2000) 100:345–51. doi:10.1046/j.1365-2567.2000.00059.x
112. Maeshima N, Poon GF, Dosanjh M, Felberg J, Lee SSM, Cross JL, et al. Hyaluronan binding identifies the most proliferative activated and memory T cells. Eur J Immunol (2011) 41:1108–19. doi:10.1002/eji.201040870
113. Katoh S, Zheng Z, Oritani K, Shimozato T, Kincade PW. Glycosylation of CD44 negatively regulates its recognition of hyaluronan. J Exp Med (1995) 182:419–29. doi:10.1084/jem.182.2.419
114. English NM, Lesley JF, Hyman R. Site-specific de-N-glycosylation of CD44 can activate hyaluronan binding, and CD44 activation states show distinct threshold densities for hyaluronan binding. Cancer Res (1998) 58:3736–42.
115. Skelton TP, Zeng C, Nocks A, Stamenkovic I. Glycosylation provides both stimulatory and inhibitory effects on cell surface and soluble CD44 binding to hyaluronan. J Cell Biol (1998) 140:431–46. doi:10.1083/jcb.140.2.431
116. Levesque MC, Haynes BF. TNF-α and IL-4 regulation of hyaluronan binding to monocyte CD44 involves posttranslational modification of CD44. Cell Immunol (1999) 193:209–18. doi:10.1006/cimm.1999.1456
117. Ruffell B, Poon GF, Lee SS, Brown KL, Tjew SL, Cooper J, et al. Differential use of chondroitin sulfate to regulate hyaluronan binding by receptor CD44 in inflammatory and interleukin 4-activated macrophages. J Biol Chem (2011) 286:19179–90. doi:10.1074/jbc.M110.200790
118. Maiti A, Maki G, Johnson P. TNF-α induction of CD44-mediated leukocyte adhesion by sulfation. Science (1998) 282:941–3. doi:10.1126/science.282.5390.941
119. Brown KL, Maiti A, Johnson P. Role of sulfation in CD44-mediated hyaluronan binding induced by inflammatory mediators in human CD14+ peripheral blood monocytes. J Immunol (2001) 167:5367–74. doi:10.4049/jimmunol.167.9.5367
120. Murai T, Sato C, Sato M, Nishiyama H, Suga M, Mio K, et al. Membrane cholesterol modulates the hyaluronan-binding ability of CD44 in T lymphocytes and controls rolling under shear flow. J Cell Sci (2013) 126:3284–94. doi:10.1242/jcs.120014
121. Teriete P, Banerji S, Noble M, Blundell CD, Wright AJ, Pickford AR, et al. Structure of the regulatory hyaluronan binding domain in the inflammatory leukocyte homing receptor CD44. Mol Cell (2004) 13:483–96. doi:10.1016/S1097-2765(04)00080-2
122. Banerji S, Wright AJ, Noble M, Mahoney DJ, Campbell ID, Day AJ, et al. Structures of the Cd44-hyaluronan complex provide insight into a fundamental carbohydrate-protein interaction. Nat Struct Mol Biol (2007) 14:234–9. doi:10.1038/nsmb1201
123. Favreau AJ, Faller CE, Guvench O. CD44 receptor unfolding enhances binding by freeing basic amino acids to contact carbohydrate ligand. Biophys J (2013) 105:1217–26. doi:10.1016/j.bpj.2013.07.041
124. Hryniewicz-Jankowska A, Augoff K, Biernatowska A, Podkalicka J, Sikorski AF. Membrane rafts as a novel target in cancer therapy. Biochim Biophys Acta (2014) 1845:155–65. doi:10.1016/j.bbcan.2014.01.006
125. Dalla Pozza E, Lerda C, Costanzo C, Donadelli M, Dando I, Zoratti E, et al. Targeting gemcitabine containing liposomes to CD44 expressing pancreatic adenocarcinoma cells causes an increase in the antitumoral activity. Biochim Biophys Acta (2013) 1828:1396–404. doi:10.1016/j.bbamem.2013.01.020
Keywords: extracellular matrix remodeling, oligosaccharides, cholesterol, membrane raft, membrane dynamics, transmembrane signaling, a disintegrin and metalloproteinase, ectodomain shedding
Citation: Murai T (2015) Lipid raft-mediated regulation of hyaluronan–CD44 interactions in inflammation and cancer. Front. Immunol. 6:420. doi: 10.3389/fimmu.2015.00420
Received: 28 February 2015; Accepted: 01 August 2015;
Published: 20 August 2015
Edited by:
David Naor, Hebrew University of Jerusalem, IsraelReviewed by:
Ilpo Vattulainen, Tampere University of Technology, FinlandFlorence Burté, Newcastle University, UK
Bruno Flamion, University of Namur, Belgium
Copyright: © 2015 Murai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Toshiyuki Murai, Department of Microbiology and Immunology, Graduate School of Medicine, Osaka University, 2-2 Yamada-oka, Suita 565-0871, Japan, murai@orgctl.med.osaka-u.ac.jp
 Toshiyuki Murai
Toshiyuki Murai