- 1Research Center, Novartis Vaccines and Diagnostics S.r.l. (a GSK Company), Siena, Italy
- 2Dipartimento di Biologia e Biotecnologie “Charles Darwin”, Università degli Studi di Roma “La Sapienza”, Rome, Italy
Vaccination against influenza is the most effective way to protect the population. Current vaccines provide protection by stimulating functional B- and T-cell responses; however, they are poorly immunogenic in particular segments of the population and need to be reformulated almost every year due to the genetic instability of the virus. Next-generation influenza vaccines should be designed to induce cross-reactivity, confer protection against pandemic outbreaks, and promote long-lasting immune responses among individuals at higher risk of infection. Multiple strategies are being developed for the induction of broad functional humoral immunity, including the use of adjuvants, heterologous prime-boost strategies, and epitope-based antigen design. The basic approach is to mimic natural responses to influenza virus infection by promoting cross-reactive neutralizing antibodies that directly prevent the infection. This review provides an overview of the mechanisms underlying humoral responses to influenza vaccination or natural infection, and discusses promising strategies to control influenza virus.
Introduction
Influenza virus alone causes over 40,000 deaths every year in the United States, and even more during pandemics, like in 2009 with pandemic A/California/07/09 H1N1 virus strain (1, 2). Influenza viruses contain eight single stranded RNA segments and are classified in three different types (A, B, and C), on the basis of major antigenic differences; only influenza A and B are responsible for annual human epidemics. All influenza virus subtypes circulating in non-human species have the potential to infect humans, and transmissions from animals to humans may occur, albeit rarely, with dramatic scenarios for the public health; this was the case of the avian H5N1 strain that appeared for the first time in human in 1998 and re-appeared in 2004–2005 with a mortality rate of 50% among infected patients and thousands of deaths are reported until today (3, 4). Treatment of influenza infections is a major challenge for clinics and public health institutions because available antiviral drugs are often ineffective due to antigenic mutations or are given too late after infection (5). The most effective intervention that we have today to combat influenza is the vaccination that reduces virus infection and spreading, even if some levels of morbidity and mortality remain due to the suboptimal efficacy of the current vaccines and mismatch between the vaccine and the circulating virus strain.
Most of the current seasonal influenza vaccines are produced with live attenuated or inactivated (split or subunit) virus and both types of vaccines reduce virus infectivity and restrict viral replication by inducing functional antibodies against the virus. The antibodies generated against the virus represent the primary correlate of immunity, whereas cell-mediated immunity can contribute to reduce the clinical symptoms (6). Although existing vaccines confer acceptable levels of protection in the general population, they are suboptimal, and are associated with some important limitations: (i) antigen composition needs to be updated every year in order to match the new seasonal circulating viruses, (ii) mismatch between the vaccine and the circulating virus can always happen, and (iii) people with a reduced ability to mount an immune response, infants, the elderly, and pregnant women respond suboptimally to these vaccines, requiring a tailored vaccine formulations (7–11).
Current influenza vaccines consist of three different virus strains: two influenza A strains (usually H1N1 and H3N2) and one influenza B strain. More recently, quadrivalent influenza vaccines have been developed, which are composed of influenza B strains of both lineages (12). Unfortunately, influenza strains acquire mutations every 1–3 years in their genome segments expressing the antibody-binding regions, a process named antigenic drift, and give raise to new circulating strains. Antigenic drift represents the principal immune evasion mechanism of influenza virus and has two major consequences: first, the humoral immunity developed in response to previous infections/vaccinations is usually non-fully effective against the new emerging strains, and second, manufacturers need to update the vaccine every year with increasing costs and risks of delays in the release of the lots. The virus can also undergo major antigenic changes in his hemagglutinin (HA) and neuraminidase (NA), referred to as antigenic shift, which consists of an ample reassortment of viral gene segments between different viruses of human or zoonotic origin, leading to the emergence of totally new and potentially dangerous virus strains, as happened during the pandemics of the last century and more recently in 2009 with the H1N1 virus of swine origin (13).
In this review, we summarize the mechanisms eliciting humoral responses against influenza infection or vaccination, and discuss the approaches that are today under evaluation to develop broadly protective and, hopefully, universal vaccines against influenza.
Learning from Antibody Responses Against Influenza
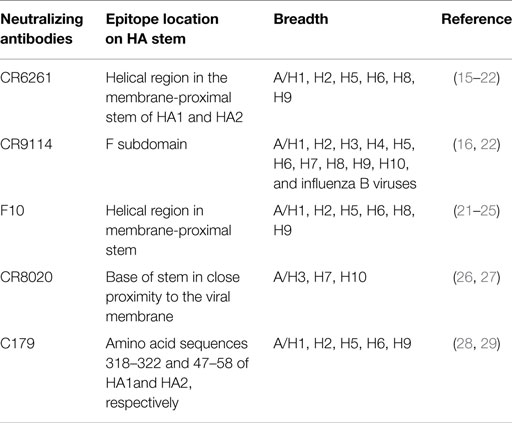
Immune responses, generated against influenza by vaccination and by natural infection, consist of neutralizing and non-neutralizing antibodies. Non-neutralizing antibodies make the most part of the antibody pool generated during the immune response, but only a small fraction is functional and participates in the clearance of infected cells through interaction with other immune cells. On the other hand, neutralizing antibodies specifically bind epitopes crucial for viral function and are extremely important to confer immunity. Most of the neutralizing antibodies recognize surface proteins of the virus, in particular, the trimeric HA, which is critical during the process of cell invasion. The overall structure of HA can be segmented in a globular head and a stem region (Figure 1). The globular head is responsible for the sialic acid-dependent binding on the extracellular surface of target cells, and allows for a conformational change of the protein for membrane fusion. Neutralizing antibodies against HA interfere during both steps of the process, in particular, they bind to the sialic acid-binding site (or in close proximity) of the globular head, thus preventing attachment of the virus to the cells. Antibodies against the stem region may restrict the conformational changes required for the membrane cell fusion. Although both kinds of antibodies are functional, only those against the stem region can have the intrinsic ability to confer broad protection against different influenza strains because this region is much less susceptible to antigenic changes as compared to the globular head (Table 1). Unfortunately, stem-specific neutralizing antibodies are rare and difficult to induce because vaccination with the seasonal vaccine formulations typically skews the specificity of B cell responses toward non-neutralizing epitopes of the stem region or, depending on the formulation of the vaccines, toward immunodominant epitopes of the HA globular head (14).
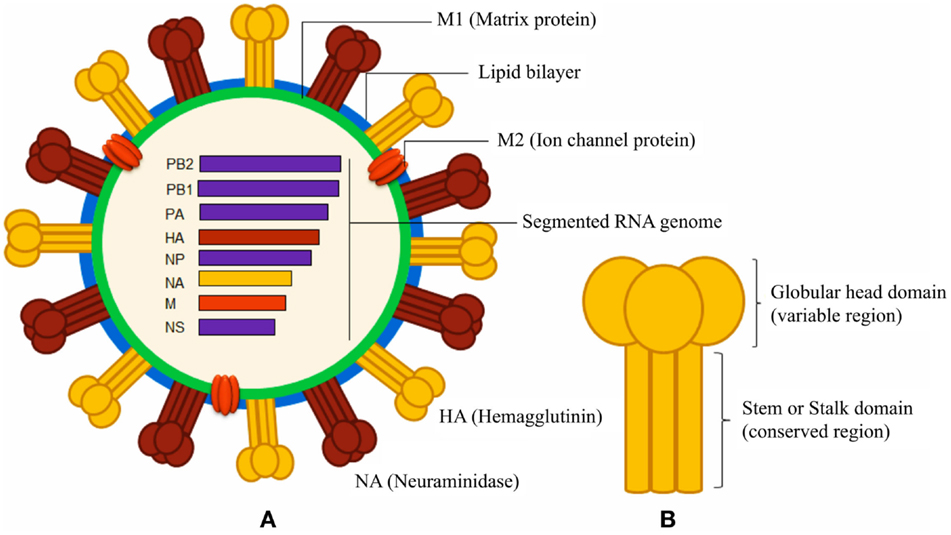
Figure 1. Schematic diagrams of influenza A virus and surface hemagglutinin protein. (A) The segmented negative-sense RNA genome of influenza A virus encodes three envelope proteins (hemagglutinin, neuraminidase, and ion channel M2 protein), and internal nucleoprotein (NP), polymerases (PA, PB1, and PB2), matrix protein 1 (M1), and non-structural proteins (NS). The lipid bilayer is derived from host cell membrane. (B) The cylindrical HA is a homo-trimeric protein consisting of a variable globular head and a conserved stem domain.
Natural responses against influenza elicit also non-neutralizing antibodies, which are specific not only to HA and NA (30) but also against M1, M2, and NP proteins (Figure 1). Such non-neutralizing antibodies, typically, can promote the clearance of the virus, relying on their Fc portion after the interaction of the variable-region of the antibody with its epitope. Several cell populations, including phagocytic cells and natural killer cells, express Fc receptors (FcRs) and may mediate the clearance of virions or virus-infected cells (31, 32). Natural killer cells express FcRs and may participate in the killing of the virus-infected cells by a mechanism called antibody-dependent cell-mediated cytotoxicity (ADCC) (33); ADCC has been observed to participate in the control of the H1N1 influenza virus infection in macaques, and a mix of intravenous antibodies that may mediate ADCC has been suggested as therapeutic for humans (34, 35). Also, the complement system may participate in clearing the virus by a mechanism called complement-dependent cytotoxicity (CDC) involving influenza-specific antibodies; in particular, IgG antibodies that bind to target surface proteins may activate complement factors in the host serum that ultimately puncture the lipid membrane of pathogen or infected host cells. Two studies have clearly shown that C3, a critical component of the complement system, may participate in reducing viral titers and in clearing the virus with a mechanism that involves M2-specific antibodies of the IgG1 or IgG2a subclasses (36, 37).
B Cell Responses to Influenza
Most of the responding B cells after influenza vaccination or infection are specific to HA, and they are difficult to isolate and characterize, especially by flow cytometry (38), because of the binding of HA to any sialic acid residue on the host cell. Taking advantage of ex vivo ELISpot assay, several studies have shown that adults or older children possess low but consistent base line levels of influenza-specific IgG memory B cells, in the range of 0.1–0.6% of the total IgG memory B cells (39). Those cells respond to further antigen encounter by quickly differentiating in antibody-secreting cells, they mostly produce isotype switched antibodies and show high frequencies of mutation in their Ig genes (40, 41). Pre-existing immunity in adults makes the characterization of the responses after seasonal vaccination challenging, so the 2009 H1N1 pandemic (pH1N1) influenza virus was a great opportunity to better understand the immune responses to influenza. Indeed, the pH1N1 HA was remarkably divergent from the HAs of the seasonal vaccines (even with a stem region quite conserved). Surprisingly, the highest numbers of deaths during the 2009 H1N1 pandemic were registered among the younger population, while the older population showed pre-existing protective immunity. How to explain the unexpected level of deaths among adults that is typically the most resistant group to influenza infections? It was suggested that adults had too low frequencies of cross-specific B cells to generate protective levels of cross-neutralizing antibodies against HA (42). On the contrary, the older population (over 65 years old) showed a very low incidence of infection and hospitalization (42–45), probably due to their life-long accumulation of an expanded reservoir of stem-specific cross-reactive memory B cells that efficiently responded to the 2009 pH1N1 virus (42). In addition, a close antigenic relation was found between the HA of the 2009 pH1N1 virus and the HA of influenza viruses that had circulated before 1950; hence, neutralizing antibodies against the HA globular head may also have contributed to protect the elderly population (46, 47).
In 2010, Lanzavecchia et al. reported that some individuals who received the seasonal influenza vaccine developed cross-reactive antibodies able to neutralize viruses belonging to different HA subtypes (H1, H2, H5, H6, and H9), including the pH1N1 isolate. By immortalizing IgG-expressing B cells, Lanzavecchia et al. showed that heterosubtypic monoclonal antibodies bound to acid-sensitive epitopes in the HA stem region, used different VH genes and carried high frequency of somatic mutations (24, 48, 49). More recently, the same group showed that most of the HA stem-specific antibodies are characterized by the use of the heavy-chain variable-region VH1-69 gene, only few polymorphisms are functional, and that few single somatic mutations are sufficient to promote high-affinity HA-specific antibodies (50).
The above studies have enhanced our understanding of influenza-specific B cell responses, and helped to set the primary goals in the development of next-generation anti-influenza therapies and vaccines. A major objective is to promote the generation of HA-specific broadly neutralizing antibodies in order to target cross-protective epitopes that are present among multiple strains. A second objective is to promote long-lasting memory B cells and plasma cells, hopefully for the entire life. Several strategies are today evaluated to achieve such goals including the use of adjuvants in vaccine formulation, heterologous prime-boost strategies, and antigen design with a “minimalistic-approach.”
Cutting-Edge Strategies for Inducing Protective Anti-Influenza Immune Responses
How to translate our knowledge of the influenza-specific humoral responses into novel strategies that specifically elicit the ideal protective immunity? As primary goals, successful vaccination strategies should confer cross-protection against multiple strains of influenza virus, and should boost long-lasting protective immunity in subjects with weakened immunity, as well as in younger and elderly populations.
A very promising strategy to meet those purposes is based on the use of particular adjuvant formulations. Adjuvants have been used in influenza vaccines for decades, usually in combination with split or subunit vaccines with the major goal to enhance their intrinsic immunogenicity (51). Although aluminum salts are potent adjuvants for most of the subunit antigens present in licensed vaccines, they seem not to be good adjuvants for influenza antigens. Instead, oil-in-water emulsions, like MF59, have been successfully used in influenza vaccines for the past 20 years with outstanding results (52, 53). MF59 not only induces high titers of influenza-specific antibodies but also cross-reactive responses against different clades of influenza viruses (54–56). Khurana et al. showed that MF59 adjuvant promotes high titers of HA-specific antibodies and expands the overall diversity of the influenza-specific antibody repertoire (14, 57). MF59 also promotes persistence of long-lasting memory B cells and increases the affinity of the antibody responses, not only in adults but also in younger and elderly (14, 52, 56–58); such evidences have shed light on the use of oil-in-water emulsion as adjuvants for influenza vaccines. Furthermore, oil-in-water adjuvants may prevent the effect of the “original antigenic sin” that is the propensity of the immune system to preferentially utilize immunological memory instead of inducing novel responses, hence limiting the development of an expanded B cell repertoire (59–61). Although their mechanisms of action are still not fully understood, MF59 and AS03 (62), the other oil-in-water adjuvant used for pandemic vaccines, represent an important tool on the way to develop broadly protective influenza vaccines. An increased risk of narcolepsy was found few years ago following vaccination with AS03-adjuvanted split influenza vaccine used in several European countries during the A/California/07/09 H1N1 influenza pandemic, but multiple subsequent studies have not confirmed any possible association between vaccination and narcolepsy (63–66).
An alternative strategy consists of heterologous prime/boost vaccinations. When the immune system encounters for the first time an influenza antigen, it generates specific antibodies and long-lasting memory B cells. Many influenza epitopes shift every year, so a second encounter with the antigen will recruit naïve B cells, which are specific for the new shifted epitopes and will also expand the pre-existing pool of memory B cells that is specific for the most conserved epitopes (30, 67, 68). Subsequent immunizations with divergent influenza antigens, the “prime/boost strategy” might expand the memory B cells specific for the most conserved epitopes that usually are under-represented in the B cell repertoire, hence inducing cross-protective immunity. This approach has been shown to be successful by Wang et al. who used a gene-based heterologous prime/boost strategy to induce cross-protection. Mice were sequentially immunized with DNA coding for the HA of different influenza A H3 virus strains (A/Hong Kong/1/1968, A/Alabama/1/1981 or A/Beijing/47/1992) and boosted with another H3 virus, A/Wyoming/3/2003; mice developed cross-neutralizing antibodies and protective capacity against multiple subtypes of H3 viruses (69). In a similar study, Wei et al. immunized mice twice with the same HA strains, but using a different delivery system for priming and for boosting. Mice primed with a DNA plasmid encoding H1N1 HA or H3N2 HA from the 2006/2007 vaccine strains and boosted with the trivalent 2006/2007 seasonal vaccine, developed enhanced neutralizing antibodies against diverse H1N1 strains compared to mice receiving only DNA or seasonal vaccine and showed higher levels of protection after infection (18). The above studies provided the proof-of-concept that a prime/boost strategy can increase the production of broadly neutralizing antibodies, and suggested that a combined strategy involving nucleic acids/proteins may have the benefit of expanding the antibody repertoire as well as inducing a different type of cellular immunity (18, 70). We further speculate that sequential immunizations with different HA proteins properly formulated with oil-in-water emulsion adjuvants (MF59 or AS03), may truly maximize the broadly neutralizing repertoire against influenza compared to non-adjuvanted vaccine formulations.
The minimalist approach is an innovative strategy that is evaluated today to promote cross-protective humoral responses. It is based on the design of antigens composed only of cross-protective epitopes, in order to focus the immune system on the desired response and generate cross-protective immunity. This approach is strongly supported by the fact that most of the broadly neutralizing antibodies identified until today are directed against the stem region of HA, and very few against its globular head (17, 23, 24, 26, 28). The minimalistic approach for antigen design has demonstrated to be successful in mice immunized with a “headless” HA, an antigen composed by the complete HA2 polypeptide and some regions of HA1 that both form the stem part of HA. Such antigen maintained the structural integrity of the conserved stem domain, but lacked the globular head with its immunodominant strain-specific epitopes (71). Sera form mice receiving the “headless HA” showed broader reactivity against heterologous strains than sera from mice vaccinated with the full-length HA and were protected against lethal virus challenge. Similar findings were obtained by using a stabilized HA2 peptide (72). Furthermore, Wang et al. designed a 60-amino-acid peptide to reproduce a long α-helix (LAH) of HA2 recognized by a broadly neutralizing monoclonal antibody, the clone 12D1 (73). The LAH peptide was not much immunogenic by itself, but when coupled to a carrier protein (KLH)-induced protection in mice challenged with divergent subtypes of influenza viruses, including H3N2, H5N1, and H1N1 strains; this work represents the most important proof that a carefully designed immunogen can be used in influenza vaccines to skew the B cell responses toward the epitope of interests. However, some concerns have been raised regarding the development of a vaccine to elicit HA2 stem-targeting antibodies, not only because the stem region is poorly immunogenic by itself (requiring further optimization of the formulation with adjuvants or protein carriers) but also, in some circumstances, anti-stem antibodies have been observed to be detrimental for the host. Indeed, in a swine experimental model, Khurana et al. showed that a vaccine inducing anti-stem antibodies may have the risk to worsen the outcome of the influenza infection (74, 75).
Other groups have characterized cross-protective epitopes included in the HA globular head; in particular, Whittle et al. have identified a broadly neutralizing antibody that recognizes the receptor-binding pocket of HA and have suggested that a modified HA globular head could be used for epitope-based antigen design to promote broadly neutralizing antibodies (76, 77).
Although not strictly related to the “minimalistic approach” for antigen design, some work recently published by Giles et al. described a new computationally optimized broadly reactive antigen (COBRA) based on the structure of the HA from H5N1 subtype; mice and non-human primates immunized with this antigen develop broadly reactive antibodies and are protected from H5N1 challenge (78–80).
Conclusion
Current influenza vaccines confer limited cross-protection against different strains of influenza and often fail to promote protective immunity in high-risk populations. Scientists are today evaluating multiple strategies to develop a universal influenza vaccine able to confer cross-protection, long-lasting immunity, and to be effective in subjects with weakened immunity. Such strategies include the use of oil-in-water emulsion adjuvants, heterologous prime/boost strategies, and antigen design. All these new strategies aim at inducing influenza-specific neutralizing antibodies that would confer sterilizing immunity in vaccinated hosts, and HA is the ideal antigen candidate to meet this purpose. Some groups are also evaluating alternative antigen candidates, such as NA, NP and M2, which are well conserved in multiple influenza strains and generate protective immunity through non-neutralizing antibodies helping to control the infection; hence, a multi-component vaccine not limited to HA antigen can be also considered. Each of the above strategies is promising to be successful, and most likely a combination of them will provide a universal influenza treatment in the future.
Conflict of Interest Statement
Oretta Finco, Giuseppe Del Giudice and Sylvie Bertholet are employees of Novartis Vaccines and Diagnostics S.r.l. (a GSK company). Giuseppe Lofano and Arun Kumar have no conflict of interest to declare.
References
1. Beigel JH. Concise definitive review: influenza. Crit Care Med (2008) 36:2660–6. doi:10.1097/CCM.0b013e318180b039
2. Available from: http://www.who.int/en/
3. Nicholson KG, Colegate AE, Podda A, Stephenson I, Wood J, Ypma E, et al. Safety and antigenicity of non-adjuvanted and MF59-adjuvanted influenza A/Duck/Singapore/97 (H5N3) vaccine: a randomised trial of two potential vaccines against H5N1 influenza. Lancet (2001) 357:1937–43. doi:10.1016/S0140-6736(00)05066-2
4. Subbarao K, Joseph T. Scientific barriers to developing vaccines against avian influenza viruses. Nat Rev Immunol (2007) 7:267–78. doi:10.1038/nri2054
5. Poland GA, Jacobson RM, Ovsyannikova IG. Influenza virus resistance to antiviral agents: a plea for rational use. Clin Infect Dis (2009) 48:1254–6. doi:10.1086/598989
6. Wilkinson TMA, Li CK, Chui CSC, Huang AKY, Perkins M, Liebner JC, et al. Preexisting influenza-specific CD4+ T cells correlate with disease protection against influenza challenge in humans. Nat Med (2012) 18:274–80. doi:10.1038/nm.2612
7. Carrat F, Flahault A. Influenza vaccine: the challenge of antigenic drift. Vaccine (2007) 25:6852–62. doi:10.1016/j.vaccine.2007.07.027
8. Chen WH, Kozlovsky BF, Effros RB, Grubeck-Loebenstein B, Edelman R, Sztein MB. Vaccination in the elderly: an immunological perspective. Trends Immunol (2009) 30:351–9. doi:10.1016/j.it.2009.05.002
9. Dormitzer PR, Galli G, Castellino F, Golding H, Khurana S, Del Giudice G, et al. Influenza vaccine immunology. Immunol Rev (2011) 239:167–77. doi:10.1111/j.1600-065X.2010.00974.x
10. Del Giudice G, Weinberger B, Grubeck-Loebenstein B. Vaccines for the elderly. Clin Microbiol Infect (2012) 18:100–8. doi:10.1111/j.1469-0691.2012.03944.x
11. Christian LM. Optimizing benefits of influenza virus vaccination during pregnancy: potential behavioral risk factors and interventions. Vaccine (2014) 32:2958–64. doi:10.1016/j.vaccine.2014.03.075
12. Hannoun C. The evolving history of influenza viruses and influenza vaccines. Expert Rev Vaccines (2013) 12:1085–94. doi:10.1586/14760584.2013.824709
13. Gatherer D. The 2009 H1N1 influenza outbreak in its historical context. J Clin Virol (2009) 45:174–8. doi:10.1016/j.jcv.2009.06.004
14. Khurana S, Chearwae W, Castellino F, Manischewitz J, King LR, Honorkiewicz A, et al. Vaccines with MF59 adjuvant expand the antibody repertoire to target protective sites of pandemic avian H5N1 influenza virus. Sci Transl Med (2010) 2:15ra5. doi:10.1126/scitranslmed.3000624
15. Ekiert DC, Bhabha G, Elsliger M, Friesen RHE, Jongeneelen M, Throsby M, et al. Antibody recognition of a highly conserved influenza virus epitope. Science (2009) 324:246–51. doi:10.1126/science.1171491
16. Dreyfus C, Laursen NS, Kwaks T, Zuijdgeest D, Khayat R, Ekiert DC, et al. Highly conserved protective epitopes on influenza B viruses. Science (2012) 337:1343–8. doi:10.1126/science.1222908
17. Throsby M, van den Brink E, Jongeneelen M, Poon LLM, Alard P, Cornelissen L, et al. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS One (2008) 3:e3942. doi:10.1371/journal.pone.0003942
18. Wei C-J, Boyington JC, McTamney PM, Kong W-P, Pearce MB, Xu L, et al. Induction of broadly neutralizing H1N1 influenza antibodies by vaccination. Science (2010) 329:1060–4. doi:10.1126/science.1192517
19. Lingwood D, McTamney PM, Yassine HM, Whittle JRR, Guo X, Boyington JC, et al. Structural and genetic basis for development of broadly neutralizing influenza antibodies. Nature (2012) 489:566–70. doi:10.1038/nature11371
20. Kanekiyo M, Wei C-J, Yassine HM, McTamney PM, Boyington JC, Whittle JRR, et al. Self-assembling influenza nanoparticle vaccines elicit broadly neutralizing H1N1 antibodies. Nature (2013) 499:102–6. doi:10.1038/nature12202
21. Whittle JRR, Wheatley AK, Wu L, Lingwood D, Kanekiyo M, Ma SS, et al. Flow cytometry reveals that H5N1 vaccination elicits cross-reactive stem-directed antibodies from multiple Ig heavy-chain lineages. J Virol (2014) 88:4047–57. doi:10.1128/JVI.03422-13
22. Pappas L, Foglierini M, Piccoli L, Kallewaard NL, Turrini F, Silacci C, et al. Rapid development of broadly influenza neutralizing antibodies through redundant mutations. Nature (2014) 516:418–22. doi:10.1038/nature13764
23. Sui J, Hwang WC, Perez S, Wei G, Aird D, Chen L, et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol (2009) 16:265–73. doi:10.1038/nsmb.1566
24. Corti D, Voss J, Gamblin SJ, Codoni G, Macagno A, Jarrossay D, et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science (2011) 333:850–6. doi:10.1126/science.1205669
25. Wrammert J, Koutsonanos D, Li GM, Edupuganti S, Sui J, Morrissey M, et al. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J Exp Med (2011) 208:181–93. doi:10.1084/jem.20101352
26. Ekiert DC, Friesen RHE, Bhabha G, Kwaks T, Jongeneelen M, Yu W, et al. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science (2011) 333:843–50. doi:10.1126/science.1204839
27. Tharakaraman K, Subramanian V, Cain D, Sasisekharan V, Sasisekharan R. Broadly neutralizing influenza hemagglutinin stem-specific antibody CR8020 targets residues that are prone to escape due to host selection pressure. Cell Host Microbe (2014) 15:644–51. doi:10.1016/j.chom.2014.04.009
28. Okuno Y, Isegawa Y, Sasao F, Ueda S. A common neutralizing epitope conserved between the hemagglutinins of influenza A virus H1 and H2 strains. J Virol (1993) 67:2552–8.
29. Dreyfus C, Ekiert DC, Wilson IA. Structure of a classical broadly neutralizing stem antibody in complex with a pandemic H2 influenza virus hemagglutinin. J Virol (2013) 87:7149–54. doi:10.1128/JVI.02975-12
30. Webster RG, Laver WG. Preparation and properties of antibody directed specifically against the neuraminidase of influenza virus. J Immunol (1967) 99:49–55.
31. Nimmerjahn F, Ravetch JV. Antibody-mediated modulation of immune responses. Immunol Rev (2010) 236:265–75. doi:10.1111/j.1600-065X.2010.00910.x
32. DiLillo DJ, Tan GS, Palese P, Ravetch JV. Broadly neutralizing hemagglutinin stalk-specific antibodies require FcγR interactions for protection against influenza virus in vivo. Nat Med (2014) 20:143–51. doi:10.1038/nm.3443
33. Jegerlehner A, Schmitz N, Storni T, Bachmann MF. Influenza A vaccine based on the extracellular domain of M2: weak protection mediated via antibody-dependent NK cell activity. J Immunol (2004) 172:5598–605. doi:10.4049/jimmunol.172.9.5598
34. Jegaskanda S, Vandenberg K, Laurie KL, Loh L, Kramski M, Winnall WR, et al. Cross-reactive influenza-specific antibody-dependent cellular cytotoxicity in intravenous immunoglobulin as a potential therapeutic against emerging influenza viruses. J Infect Dis (2014) 210:1811–22. doi:10.1093/infdis/jiu334
35. Jegaskanda S, Weinfurter JT, Friedrich TC, Kent SJ. Antibody-dependent cellular cytotoxicity is associated with control of pandemic H1N1 influenza virus infection of macaques. J Virol (2013) 87:5512–22. doi:10.1128/JVI.03030-12
36. Wang R, Song A, Levin J, Dennis D, Zhang NJ, Yoshida H, et al. Therapeutic potential of a fully human monoclonal antibody against influenza A virus M2 protein. Antiviral Res (2008) 80:168–77. doi:10.1016/j.antiviral.2008.06.002
37. Mozdzanowska K, Maiese K, Furchner M, Gerhard W. Treatment of influenza virus-infected SCID mice with nonneutralizing antibodies specific for the transmembrane proteins matrix 2 and neuraminidase reduces the pulmonary virus titer but fails to clear the infection. Virology (1999) 254:138–46. doi:10.1006/viro.1998.9534
38. Bardelli M, Alleri L, Angiolini F, Buricchi F, Tavarini S, Sammicheli C, et al. Ex vivo analysis of human memory B lymphocytes specific for a and B influenza hemagglutinin by polychromatic flow-cytometry. PLoS One (2013) 8:e70620. doi:10.1371/journal.pone.0070620
39. Sasaki S, Jaimes MC, Holmes TH, Dekker CL, Mahmood K, Kemble GW, et al. Comparison of the influenza virus-specific effector and memory B-cell responses to immunization of children and adults with live attenuated or inactivated influenza virus vaccines. J Virol (2007) 81:215–28. doi:10.1128/JVI.01957-06
40. Pinna D, Corti D, Jarrossay D, Sallusto F, Lanzavecchia A. Clonal dissection of the human memory B-cell repertoire following infection and vaccination. Eur J Immunol (2009) 39:1260–70. doi:10.1002/eji.200839129
41. Wrammert J, Smith K, Miller J, Langley WA, Kokko K, Larsen C, et al. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature (2008) 453:667–71. doi:10.1038/nature06890
42. Ellebedy AH, Ahmed R. Re-engaging cross-reactive memory B cells: the influenza puzzle. Front Immunol (2012) 3:53. doi:10.3389/fimmu.2012.00053
43. Jain S, Kamimoto L, Bramley AM, Schmitz AM, Benoit SR, Louie J, et al. Hospitalized patients with 2009 H1N1 influenza in the United States, April-June 2009. N Engl J Med (2009) 361:1935–44. doi:10.1056/NEJMoa0906695
44. Mehta SR, Granger CB, Boden WE, Steg PG, Bassand J-P, Faxon DP, et al. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N Engl J Med (2009) 360:2165–75. doi:10.1056/NEJMoa0903810
45. Chowell G, Bertozzi SM, Colchero MA, Lopez-Gatell H, Alpuche-Aranda C, Hernandez M, et al. Severe respiratory disease concurrent with the circulation of H1N1 influenza. N Engl J Med (2009) 361:674–9. doi:10.1056/NEJMoa0904023
46. Hancock K, Veguilla V, Lu X, Zhong W, Butler EN, Sun H, et al. Cross-reactive antibody responses to the 2009 pandemic H1N1 influenza virus. N Engl J Med (2009) 361:1945–52. doi:10.1056/NEJMoa0906453
47. Skowronski DM, Hottes TS, McElhaney JE, Janjua NZ, Sabaiduc S, Chan T, et al. Immuno-epidemiologic correlates of pandemic H1N1 surveillance observations: higher antibody and lower cell-mediated immune responses with advanced age. J Infect Dis (2011) 203:158–67. doi:10.1093/infdis/jiq039
48. Corti D, Suguitan AL, Pinna D, Silacci C, Fernandez-Rodriguez BM, Vanzetta F, et al. Heterosubtypic neutralizing antibodies are produced by individuals immunized with a seasonal influenza vaccine. J Clin Invest (2010) 120:1663–73. doi:10.1172/JCI41902
49. Corti D, Lanzavecchia A. Broadly neutralizing antiviral antibodies. Annu Rev Immunol (2013) 31:705–42. doi:10.1146/annurev-immunol-032712-095916
50. Pappas L, Foglierini M, Piccoli L, Kallewaard NL, Turrini F, Silacci C, et al. Rapid development of broadly influenza neutralizing antibodies through redundant mutations. Nature (2014) 516:418–22. doi:10.1038/nature13764
52. O’Hagan DT, Ott GS, Van Nest G, Rappuoli R, Del Giudice G. The history of MF59(®) adjuvant: a phoenix that arose from the ashes. Expert Rev Vaccines (2013) 12:13–30. doi:10.1586/erv.12.140
53. O’Hagan DT, Rappuoli R, De Gregorio E, Tsai T, Del Giudice G. MF59 adjuvant: the best insurance against influenza strain diversity. Expert Rev Vaccines (2011) 10:447–62. doi:10.1586/erv.11.23
54. Stephenson I, Bugarini R, Nicholson KG, Podda A, Wood JM, Zambon MC, et al. Cross-reactivity to highly pathogenic avian influenza H5N1 viruses after vaccination with nonadjuvanted and MF59-adjuvanted influenza A/duck/Singapore/97 (H5N3) vaccine: a potential priming strategy. J Infect Dis (2005) 191:1210–5. doi:10.1086/428948
55. Banzhoff A, Pellegrini M, Del Giudice G, Fragapane E, Groth N, Podda A. MF59-adjuvanted vaccines for seasonal and pandemic influenza prophylaxis. Influenza Other Respi Viruses (2008) 2:243–9. doi:10.1111/j.1750-2659.2008.00059.x
56. Galli G, Hancock K, Hoschler K, DeVos J, Praus M, Bardelli M, et al. Fast rise of broadly cross-reactive antibodies after boosting long-lived human memory B cells primed by an MF59 adjuvanted prepandemic vaccine. Proc Natl Acad Sci U S A (2009) 106:7962–7. doi:10.1073/pnas.0903181106
57. Khurana S, Verma N, Yewdell JW, Hilbert AK, Castellino F, Lattanzi M, et al. MF59 adjuvant enhances diversity and affinity of antibody-mediated immune response to pandemic influenza vaccines. Sci Transl Med (2011) 3:85ra48. doi:10.1126/scitranslmed.3002336
58. Khurana S, Coyle EM, Dimitrova M, Castellino F, Nicholson K, Del Giudice G, et al. Heterologous prime-boost vaccination with MF59-adjuvanted H5 vaccines promotes antibody affinity maturation towards the hemagglutinin HA1 domain and broad H5N1 cross-clade neutralization. PLoS One (2014) 9:e95496. doi:10.1371/journal.pone.0095496
59. Kim JH, Skountzou I, Compans R, Jacob J. Original antigenic sin responses to influenza viruses. J Immunol (2009) 183:3294–301. doi:10.1073/pnas.0912458109
60. Francis T, Salk JE, Quilligan JJ. Experience with vaccination against influenza in the spring of 1947; a preliminary report. Am J Public Health Nations Health (1947) 37:1013–6. doi:10.2105/AJPH.37.8.1013
61. Wu J, Zhong X, Li CK, Zhou J, Lu M, Huang K-Y, et al. Optimal vaccination strategies for 2009 pandemic H1N1 and seasonal influenza vaccines in humans. Vaccine (2011) 29:1009–16. doi:10.1016/j.vaccine.2010.11.058
62. Garçon N, Vaughn DW, Didierlaurent AM. Development and evaluation of AS03, an adjuvant system containing α-tocopherol and squalene in an oil-in-water emulsion. Expert Rev Vaccines (2012) 11:349–66. doi:10.1586/erv.11.192
63. Del Giudice G, Rappuoli R. Inactivated and adjuvanted influenza vaccines. In: Oldstone MBA, Compans RW, editor. Influenza Pathogenesis and Control, Current Topics in Microbiology and Immunology (2014). Vol. 386, p. 151–80.
64. Fox CB, Haensler J. An update on safety and immunogenicity of vaccines containing emulsion-based adjuvants. Expert Rev Vaccines (2013) 12:747–58. doi:10.1586/14760584.2013.811188
65. Miller E, Andrews N, Stellitano L, Stowe J, Winstone AM, Shneerson J, et al. Risk of narcolepsy in children and young people receiving AS03 adjuvanted pandemic A/H1N1 2009 influenza vaccine: retrospective analysis. Br Med J (2013) 346:f794–794. doi:10.1136/bmj.f794
66. Available from: http://www.cdc.gov
67. Manicassamy B, Medina RA, Hai R, Tsibane T, Stertz S, Nistal-Villán E, et al. Protection of mice against lethal challenge with 2009 H1N1 influenza A virus by 1918-like and classical swine H1N1 based vaccines. PLoS Pathog (2010) 6:e1000745. doi:10.1371/journal.ppat.1000745
68. Medina RA, Manicassamy B, Stertz S, Seibert CW, Hai R, Belshe RB, et al. Pandemic 2009 H1N1 vaccine protects against 1918 Spanish influenza virus. Nat Commun (2010) 1:28. doi:10.1038/ncomms1026
69. Wang TT, Tan GS, Hai R, Pica N, Petersen E, Moran TM, et al. Broadly protective monoclonal antibodies against H3 influenza viruses following sequential immunization with different hemagglutinins. PLoS Pathog (2010) 6:e1000796. doi:10.1371/journal.ppat.1000796
70. Wu L, Kong W-P, Nabel GJ. Enhanced breadth of CD4 T-cell immunity by DNA prime and adenovirus boost immunization to human immunodeficiency virus Env and Gag immunogens. J Virol (2005) 79:8024–31. doi:10.1128/JVI.79.13.8024-8031.2005
71. Steel J, Lowen AC, Wang TT, Yondola M, Gao Q, Haye K, et al. Influenza virus vaccine based on the conserved hemagglutinin stalk domain. MBio (2010) 1:1–9. doi:10.1128/mBio.00018-10
72. Bommakanti G, Citron MP, Hepler RW, Callahan C, Heidecker GJ, Najar TA, et al. Design of an HA2-based Escherichia coli expressed influenza immunogen that protects mice from pathogenic challenge. Proc Natl Acad Sci U S A (2010) 107:13701–6. doi:10.1073/pnas.1007465107
73. Wang TT, Tan GS, Hai R, Pica N, Ngai L, Ekiert DC, et al. Vaccination with a synthetic peptide from the influenza virus hemagglutinin provides protection against distinct viral subtypes. Proc Natl Acad Sci U S A (2010) 107:18979–84. doi:10.1073/pnas.1013387107
74. Khurana S, Loving CL, Manischewitz J, King LR, Gauger PC, Henningson J, et al. Vaccine-induced anti-HA2 antibodies promote virus fusion and enhance influenza virus respiratory disease. Sci Transl Med (2013) 5:200ra114. doi:10.1126/scitranslmed.3006366
75. Crowe JE. Universal flu vaccines: primum non nocere. Sci Transl Med (2013) 5:200fs34. doi:10.1126/scitranslmed.3007118
76. Schmidt AG, Xu H, Khan AR, O’Donnell T, Khurana S, King LR, et al. Preconfiguration of the antigen-binding site during affinity maturation of a broadly neutralizing influenza virus antibody. Proc Natl Acad Sci U S A (2013) 110:264–9. doi:10.1073/pnas.1218256109
77. Whittle JRR, Zhang R, Khurana S, King LR, Manischewitz J, Golding H, et al. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc Natl Acad Sci U S A (2011) 108:14216–21. doi:10.1073/pnas.1111497108
78. Giles BM, Bissel SJ, DeAlmeida DR, Wiley CA, Ross TM. Antibody breadth and protective efficacy are increased by vaccination with computationally optimized hemagglutinin but not with polyvalent hemagglutinin-based H5N1 virus-like particle vaccines. Clin Vaccine Immunol (2012) 19:128–39. doi:10.1128/CVI.05533-11
79. Giles BM, Ross TM. A computationally optimized broadly reactive antigen (COBRA) based H5N1 VLP vaccine elicits broadly reactive antibodies in mice and ferrets. Vaccine (2011) 29:3043–54. doi:10.1016/j.vaccine.2011.01.100
Keywords: influenza, hemagglutinin, functional antibody responses, universal influenza vaccine, neutralizing antibodies, vaccination strategies
Citation: Lofano G, Kumar A, Finco O, Del Giudice G and Bertholet S (2015) B cells and functional antibody responses to combat influenza. Front. Immunol. 6:336. doi: 10.3389/fimmu.2015.00336
Received: 11 April 2015; Accepted: 15 June 2015;
Published: 30 June 2015
Edited by:
Marc H. V. Van Regenmortel, University of Strasbourg, FranceReviewed by:
Susanne Modrow, University of Regensburg, GermanyJose Esparza, University of Maryland School of Medicine, USA
Copyright: © 2015 Lofano, Kumar, Finco, Del Giudice and Bertholet. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Giuseppe Lofano, Ragon Institute of MGH, MIT, and Harvard, 400 Technology Square, Cambridge, MA 02139, USA,Z2xvZmFub0BtZ2guaGFydmFyZC5lZHU=;
Sylvie Bertholet, Novartis Vaccines and Diagnostics S.r.l. (A GSK Company), Research Center, via Fiorentina 1, Siena 53100, Italy,c3lsdmllLmMuYmVydGhvbGV0LWdpcmFyZGluQGdzay5jb20=
†Present address: Giuseppe Lofano, Ragon Institute of MGH, MIT and Harvard, Cambridge, MA, USA
 Giuseppe Lofano
Giuseppe Lofano Arun Kumar
Arun Kumar Oretta Finco
Oretta Finco Giuseppe Del Giudice
Giuseppe Del Giudice Sylvie Bertholet
Sylvie Bertholet