- 1Viral Pseudotype Unit, Medway School of Pharmacy, The Universities of Greenwich and Kent at Medway, Chatham Maritime, Kent, UK
- 2Department of Zoology, University of Oxford, Oxford, UK
- 3The Jenner Institute Laboratories, University of Oxford, Oxford, UK
The use of vaccination against the influenza virus remains the most effective method of mitigating the significant morbidity and mortality caused by this virus. Antibodies elicited by currently licensed influenza vaccines are predominantly hemagglutination-inhibition (HI)-competent antibodies that target the globular head of hemagglutinin (HA) thus inhibiting influenza virus entry into target cells. These antibodies predominantly confer homosubtypic/strain specific protection and only rarely confer heterosubtypic protection. However, recent academia or pharma-led R&D toward the production of a “universal vaccine” has centered on the elicitation of antibodies directed against the stalk of the influenza HA that has been shown to confer broad protection across a range of different subtypes (H1–H16). The accurate and sensitive measurement of antibody responses elicited by these “next-generation” influenza vaccines is, however, hampered by the lack of sensitivity of the traditional influenza serological assays HI, single radial hemolysis, and microneutralization. Assays utilizing pseudotypes, chimeric viruses bearing influenza glycoproteins, have been shown to be highly efficient for the measurement of homosubtypic and heterosubtypic broadly neutralizing antibodies, making them ideal serological tools for the study of cross-protective responses against multiple influenza subtypes with pandemic potential. In this review, we will analyze and compare literature involving the production of influenza pseudotypes with particular emphasis on their use in serum antibody neutralization assays. This will enable us to establish the parameters required for optimization and propose a consensus protocol to be employed for the further deployment of these assays in influenza vaccine immunogenicity studies.
Influenza Pseudotypes
Influenza is a respiratory syndrome caused by three of six genera in the orthomyxoviridae family, influenza A, B, and C. A putative fourth genus (influenza D) has recently been characterized and proposed (1). Influenza A is the most widespread, its various subtypes are classified according to their antigenically variable surface glycoproteins: hemagglutinin (HA, H1–H18) and neuraminidase (NA, N1–N11). The virion consists of a segmented negative sense genome encapsidated in ribonucleoprotein complexes, which are surrounded by a matrix shell and lipid envelope containing the two surface glycoproteins and the M2 ion channel. Influenza A is the primary source of the human seasonal form of the disease, responsible for up to 500,000 deaths per annum as well as deaths caused by pandemics such as those occurring in 1918, 1957, 1968, and 2009 (2). Consequently, vaccines against influenza need to be regularly updated to match predicted circulating strains that are constantly escaping from vaccine protection through a mechanism known as antigenic drift. Influenza A is primarily associated with wild fowl/birds in the case of the majority of subtypes and can reassort with human strains through antigenic shift to yield human compatible viruses with previously un-encountered surface epitopes. Pigs are usually considered to be the mixing vessel for reassortment as they express a mixture of α-2,3 and α-2,6 sialic acid linkages. Influenza virus research is often hindered by the requirement for expensive biosafety precautions, especially in the case of the highly pathogenic avian influenza (HPAI, e.g. H5N1, H7N1) or pandemic strains.
Pseudotypes or pseudotype particles are chimeric “viruses” consisting of a surrogate virus core surrounded by a lipid envelope with the surface glycoproteins of another virus, such as HA. By removing the genetic element of the virus being studied and replacing it with a suitable reporter, viruses, especially HPAI, can be studied in this safer, single cycle system. The comparative safety of pseudotype viruses circumvents the need for restrictive, expensive, and widely unavailable high-category biosafety facilities, increasing access to research groups interested in highly pathogenic viruses.
This review is a systematic analysis encompassing a wide range of peer-reviewed literature in English concerning the production and use of pseudotypes bearing influenza glycoproteins to date. For the purpose of this review, pseudotypes will be defined as replication-deficient viruses containing a viral core from one species and bearing glycoproteins from another that are not represented in the genome. Literature was gathered by searching for “influenza pseudotypes” using Google Scholar and NCBI PubMed. The resulting list of publications was expanded by following up cited references and finally, those falling outside of our pseudotype definition or not specifically using influenza pseudotypes were excluded from the sections on production, transduction, and neutralization.
This review will be useful to those interested in the production of pseudotypes for use in immunogenicity testing of pre-clinical influenza vaccines, whether in human or animal settings, and including “universal vaccine” candidates. Influenza serological studies such as the measurement of seroprevalence will benefit from this manuscript, which will also help to inform the process of validation of pseudotype-based assays to clinical end-point. Furthermore, studies utilizing chimeric HA proteins in order to differentiate between stalk and head directed antibodies will be discussed.
Pseudotype Components
Cores and Reporters
The core and its associated genome containing a reporter are the backbone of the pseudotype system, which can be used to study the properties of selected entry proteins. The use of cores from lentiviral human immunodeficiency virus (HIV) and gammaretroviruses such as murine leukemia virus (MLV) predominate in the influenza pseudotype literature. Recent development of systems involving rhabdoviruses, in particular the vesicular stomatitis virus (VSV), has also been used to produce pseudotype cores with promising results (3, 4).
Retroviral and Lentiviral Cores and Vectors
Retroviral and lentiviral vectors are complex systems, which will be explained in simple terms specific to the production and use of pseudotypes. Pseudotype core and vector systems have been reviewed in detail (5, 6).
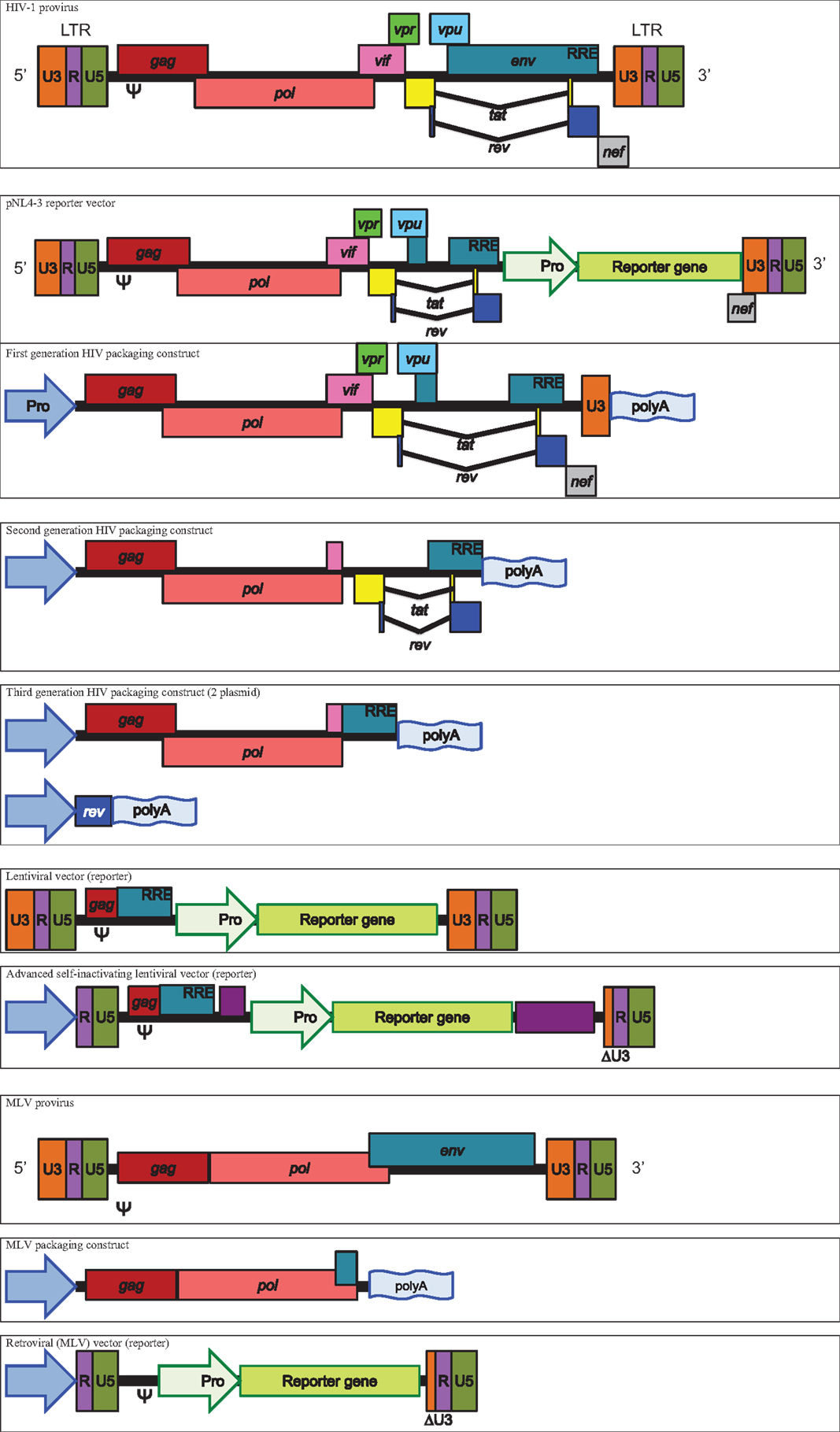
The primary genes provided by retroviral and lentiviral systems are gag and pol. In the case of HIV, gag provides the structural proteins p18, p24, and p15, whereas pol provides the integrase and reverse transcriptase in conjunction with the p10 protease required for cleavage and maturation of each distinct protein from their respective polypeptide chain (7, 8). Reporter constructs are associated with their respective cores based on the Psi (Ψ) packaging element incorporated in the vector design process, making them specific to the surrogate species used.
Human immunodeficiency virus cores are derived from several different origins between laboratory groups. First generation pNL4-3 vectors are well represented and the pNL4-3-Luc.E-R-variant is the most commonly used (9–14). The pNL4-3.Luc.E-R-replication deficient proviral HIV-1 clone is derived from the pNL precursor but has inhibitory frame shifts in the env and vpr genes as well as a luciferase reporter gene cloned into nef and the entire construct is incorporated into progeny pseudotypes. The vector’s life cycle mimics that of HIV, using the Ψ element to allow encapsidation into nascent pseudotypes and long terminal repeat (LTR) regions bearing the U3 promoter, which with the aid of tat, permit the expression of the viral proteins after integration into the host genome. The rev responsive element (RRE) allows nuclear export of viral messenger RNA (mRNA), including the reporter gene transcript, which is the measure of output for this system. Due to the incorporation of the HIV core genes into the same integrated construct as the reporter, transduced cells may possibly produce luciferase containing cores alongside its transcribed enzyme, which could potentially interfere with luciferase activity.
Another commonly used HIV core vector is pCMV ΔR8.2, a relation of pCMV ΔR8.9, which still contains intact vif, vpr, vpu, and nef genes (15–20).
A further approach uses the second generation HIV vector p8.91 that also originates from pCMV ΔR8.9 and ΔR9 (15, 21). The p8.91 vector is a modified HIV-1 clone, lacking the Ψ sequence as well as the env, vif, nef, vpu, and vpr genes and is widely used in the articles studied (22–25). The cytomegalovirus promoter is used in lieu of LTR-based promotion, meaning that p8.91 provides the necessary genes for the production of the core but the proviral and packaging elements (LTRs, RRE, and Ψ) are transferred to a separate plasmid bearing the reporter gene. Thus, the reporter construct will be incorporated into nascent virions and integrated into the transduced cell’s genome, whereupon the LTRs and RRE will act to enhance expression. In the case of the commonly used firefly luciferase or green fluorescent protein (GFP) plasmids pCSFLW or pCSGW, a safety component is incorporated through a deletion in the 3′ LTR (U3 promoter region), creating so called self-inactivating (SIN) vectors (26, 27).
Third generation vectors have also been used. In this instance, HIV structural and accessory genes are separated from rev, which is provided in cis on an additional plasmid. The third generation Invitrogen ViraPower Lentiviral Expression System was used in several cases using the plasmids pLP1 and pLP2 (28–31).
Murine leukemia virus cores are less widely used but provide similar gag and pol elements to HIV vectors (32–38). One MLV core used consists of gag and pol under the effect of a CMV promoter, a vector which has been shared across various laboratories (39–41). In this instance, the vector originates from pCI G3 N, B, or NB, which are differentially restricted in certain murine cells based on the mouse resistant gene alleles Fv1N and Fv1B (42). The reporters used in this system are derived from CLONTECH vectors LNCX and pIRES2-EGFP (39, 41). Another described MLV plasmid, pkatgagpolATG originates from the ecotropic Moloney MLV and strain 4070A (17).
Minor differences have been observed when pseudotyping HIV or MLV cores with influenza glycoproteins (43). Therefore, the question of which core to use to produce pseudotypes is often down to choice, preference, and availability (44).
See Figure 1 for schematic representations of packaging constructs and vectors.
Rhabdoviruses
Recombinant VSV viruses are produced expressing GFP in place of the resident VSV envelope glycoprotein (VSV-G). In certain cases, HA and NA or simply HA are also added to the VSV genome. These additions produce a replication-competent virus, which will promote GFP production in infected cells (4). As these recombinant viruses are not limited to a single cycle of replication, they lack the safety element found within other systems.
A safer VSV-based alternative involves transfection of surface protein encoding plasmids (HA/NA) into cells and subsequent infection with a recombinant VSV. In this way, one can produce VSV pseudotyped with influenza surface proteins, which lack entry-glycoproteins in its resident genome, rendering the second generation of virus infection-incompetent (3).
Reporter Systems
The output of the pseudotype system is based on the incorporated reporter, which mimics the genome of the surrogate virus. In the case of HIV or MLV surrogates, the reporter will often be incorporated into the pseudotype in RNA form, which upon transduction will be reverse transcribed, translocated to the nucleus, and integrated into the host cell genome. The reporter will then be produced by the host cell and can be used to measure transduction efficiency.
The primary reporter used in influenza pseudotypes is firefly derived luciferase (45–53). Relative luminescence units (RLU) or relative luciferase activity (RLA) are used as output, measured by lysing transduced cells and adding substrate for the luciferase enzyme, the signal from which is then read using a luminometer.
Green fluorescent protein is also commonly used, in which case transduction efficiency is determined by counting the number of fluorescing cells via epifluorescence microscopy or fluorescence-activated cell sorter (FACS) (54, 55).
Other reporters such as lacZ (29, 54, 56, 57) as well as Gaussia (58) and Renilla (59, 60) luciferase are also used to a lesser extent.
Influenza Envelope Proteins: Hemagglutinin
The trimeric attachment and fusion protein HA is the principal constituent of the influenza virus envelope, alongside NA and M2. Attachment to sialic acid residues on target cell membranes triggers endocytosis and pH-dependent exposure and engagement of the fusion peptide, mediating entry of the virus (61). This process is the basis on which influenza neutralization assays are founded – the exploitation of attachment and entry for the study of HA-directed antibodies and their neutralizing ability. Analysis has permitted classification of influenza A subtypes into two distinct groups: group 1 containing subtypes 1, 2, 5, 6, 8, 9, 11, 12, 13, 16, 17, and 18 and group 2 containing 3, 4, 7, 10, 14, and 15 (62–64). Subtypes within each group are often subdivided into clades with further sequence dissimilarity.
See Figure 2 for a phylogeny of influenza groups and Figure 3 for influenza strains pseudotyped with HA compared to HA sequence entries currently in NCBI GenBank. A wide variety of influenza A strains exist and have been pseudotyped, influenza B is grouped into two distinct lineages (Yamagata and Victoria) and has yet to be pseudotyped. Influenza C pseudotypes have been produced using a VSV core (65).
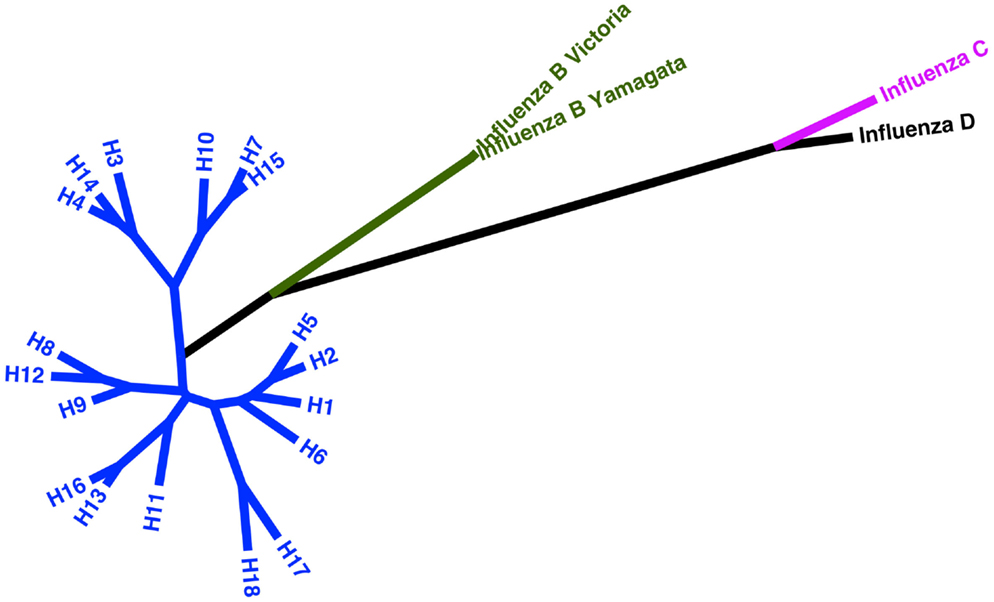
Figure 2. Phylogeny of current influenza subtypes using the HA glycoprotein. Maximum likelihood tree representing amino acid sequences of the HA glycoprotein for influenza A, B, and C virus as well as putative influenza D. The tree inferred is based on MUSCLE alignment of downloaded sequences conducted using MEGA 5.2 under the WAG + G model (four categories). The phylogenetic tree with the highest log likelihood (−16773.4044) is shown. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 22 amino acid sequences. All positions containing gaps and missing data were eliminated. There were a total of 538 positions in the final dataset (66, 67).
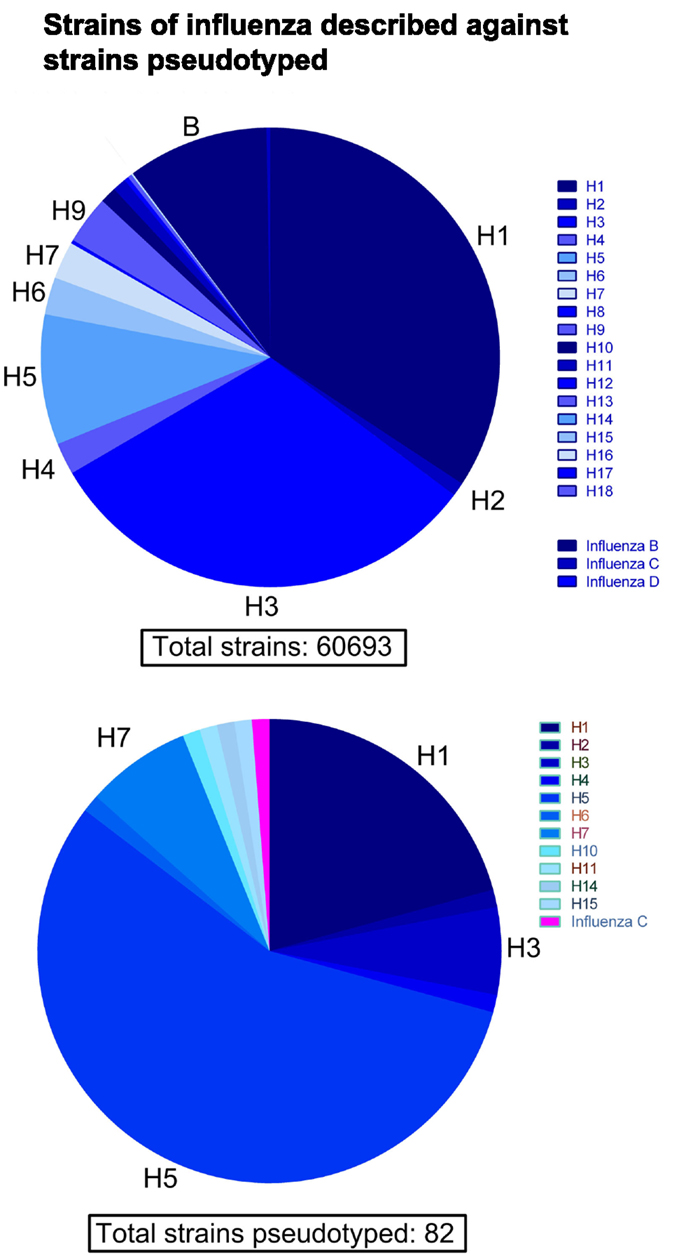
Figure 3. Comparison of influenza HA sequences described against strains pseudotyped. Out of a total of 60,693 HA amino acid sequences extracted from NCBI GenBank, the vast majority come from subtypes H1, H3, H5, and influenza B. Conversely, the current number of different subtypes and strains of HA used to produce pseudotypes is 82. The majority of pseudotyped strains come from subtypes H1, H3, and especially H5.
Codon optimization, synthesized genes
Codon optimization has been employed for several commercially synthesized genes, which are sometimes used concurrently with extracted wild type viral sequences depending on availability [Genscript, Gene Art, Integrated DNA technologies (54, 68–73)]. Recursive PCR has been used in some cases to produce the same end product (16, 68, 74, 75). In the context of pseudotype production, codon optimization is performed based on the assumption that conforming to codon-bias within producer cells will increase production of proteins and pseudotype yields.
Influenza Envelope Proteins: Neuraminidase
As with wild type influenza virus, NA is required for the exit of influenza pseudotypes via its cleavage of surface sialic acid molecules on producer cells. However, it is common to circumvent the requirement of NA expression for pseudotype production by the treatment of cultured cell lines with commercial exogenous bacterial NA 24 h after transfection (59, 76–78). This 24 h time period requires optimization to allow maximal budding of pseudotypes and minimal loss through transduction of producing cells. Exogenous NA treatment is often used in neutralization studies in order to prevent NA directed antibodies from providing a neutralization signal. However, several studies opt to incorporate an NA plasmid such as that from influenza B/Yamagata/16/88, A/Shanghai/37T/2009, A/Thailand/1(KAN)-1/04, or A/Puerto Rico/8/1934 (16, 58, 79–85).
Several recent articles have characterized sialic acid binding attributes of neuraminidases sharing particular genetic characteristics. New mutations have been characterized such as G147R in the A/WSN/33 strain that has been shown to rescue HA-binding deficient viruses. The G147R mutation is present in a range of strains including representatives of pandemic H1N1 and chicken H5N1 (86, 87).
Influenza Envelope Proteins: M2
It is also possible to incorporate the M2 ion channel into influenza pseudotypes in order to study its effect on the production process. However, the M2 role in acidification of the wild type influenza virus core is not required for the dissociation of pseudotype cores as they are derived from non-influenza viruses, which achieve release of their genetic material (i.e., a luciferase reporter gene transcript) through different mechanisms. Therefore, M2 is not required for the production of influenza pseudotypes despite being shown to have an effect on yields and infectivity (88, 89). There are reports of M2 incorporation increasing pseudotype particle yields such as H7 A/FPV/Rostock, and for H1N1 pseudotypes (29, 88). M2 has been shown to influence the budding of wild type influenza and consequently, this may be the mechanism through which M2 expression increases the reported pseudotype yields (90).
Proteases
As HA is produced and trafficked through the secretory pathway it requires proteolytic cleavage in order to become fusion competent. Proteolytic cleavage is mediated by certain host cell proteases and restricts certain subtypes to epithelial cells where these required proteases are expressed. While this is achieved naturally in wild type infection, a cleavage component must be incorporated into pseudotype production workflows in order to achieve optimal yields. This is because in producer cell lines the required proteases are either not expressed or are expressed, but not at sufficient levels to make the pseudotypes fusion competent.
In order to mimic the proteolytic properties of the natural host cells of influenza, protease encoding plasmids can be transfected alongside the other requisite plasmids in order to induce transient expression within the same timeframe as the production of pseudotypes. The serine transmembrane protease (TMPRSS2) and the human airway trypsin (HAT), which cleave wild type influenza (91) have been used successfully in several studies for pseudotype production (17, 28, 70, 78, 92–96). TMPRSS4, another serine protease has also been used to successfully cleave wild type and influenza lentiviral pseudotypes (97).
However, the addition of a protease encoding plasmid can be side-stepped through cleavage post-production using tosyl phenylalanyl chloromethyl ketone (TPCK) treated trypsin (17, 28, 92, 98). TPCK inhibits the less specific proteolytic elements of chymotrypsin, restricting the treatment process to the cleavage of peptide bonds required for HA maturation (99, 100).
TPCK-trypsin concentrations used for the production of pseudotypes generally ranged from 1 to 50 μg/ml. However, one study reported increased transduction when used at concentrations above 40 μg/ml for H1N1 pseudotypes (101). Incubation ranged from 10 min at room temperature to the more usual 1 h at 37°C. TPCK-trypsin treatment is typically carried out an hour before transduction. The enzyme is then neutralized before transduction using commercial trypsin inhibitors, in some cases originating from soybean (28, 92).
HA derived from HPAI strains that contain a polybasic cleavage sequence in the HA0 protein are cleaved by a wider range of proteases that are ubiquitous in cells. This allows the omission of protease plasmids or TPCK-trypsin treatment in HPAI pseudotype production (102). In some cases, the polybasic cleavage site of HPAI strains have been integrated into other HAs in an attempt to produce pseudotypes without the protease plasmid requirement, or to give strains similar entry characteristics (60, 73).
See Figures 4 and 5 for representative drawings of the pseudotype production process and different cores used.
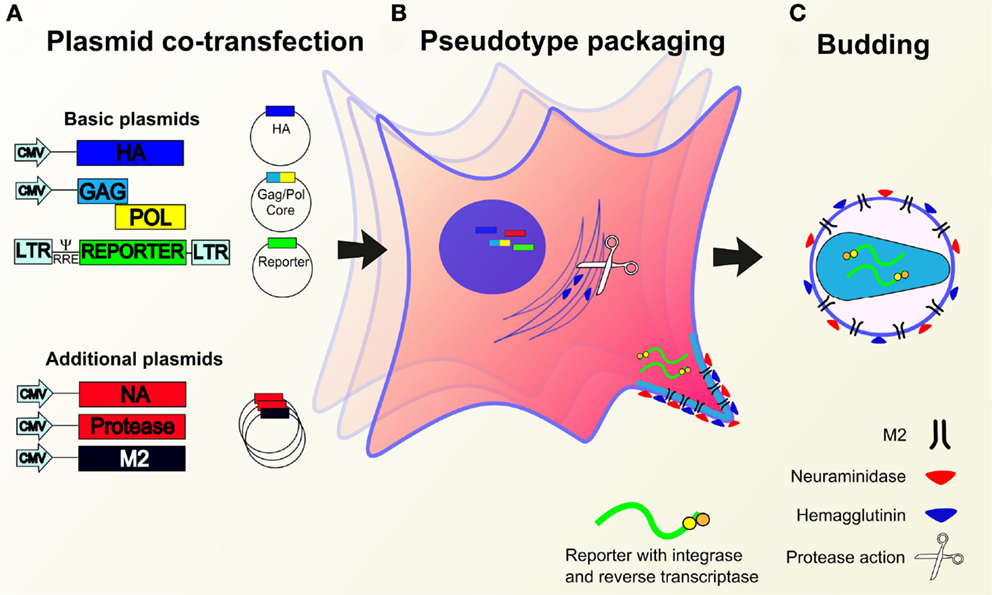
Figure 4. Production of lentiviral or retroviral pseudotypes. (A) Essential (containing HA, packaging construct gag pol, reporter construct) and/or additional (NA, protease, M2) expression plasmids are co-transfected into HEK293T producer cells. (B) Plasmids migrate to the nucleus whereupon genes are expressed leading to the production of pseudotype proteins and the reporter RNA construct. Cleavage of HA is mediated by transfected or cellular proteases. (C) Pseudotype proteins are packaged by the cell and budding occurs at the cell membrane to yield pseudotypes bearing desired glycoproteins and incorporated reporter.
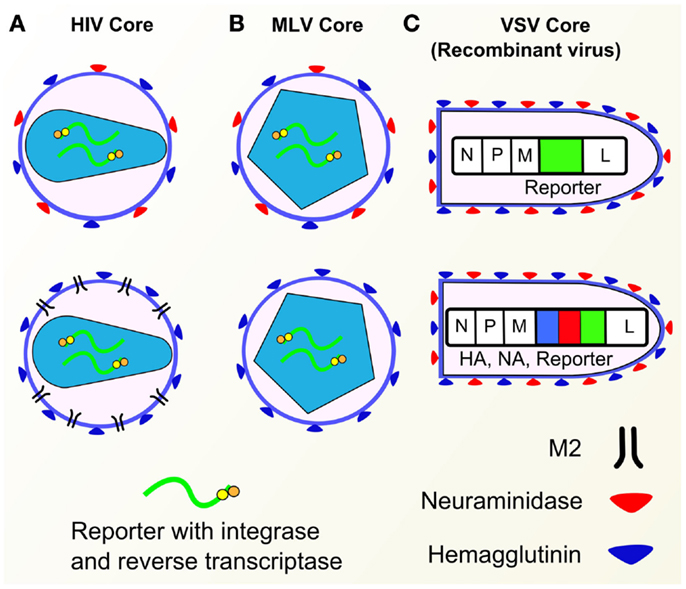
Figure 5. Pseudotype cores. (A) HIV cores with various envelope glycoproteins (HA, NA, M2). (B) MLV cores with HA or HA and NA. (C) Recombinant VSV containing GFP gene (top) and HA/NA/GFP genes (bottom). Components of influenza pseudotypes can be varied according to need. Pseudotypes have been produced with HA, NA, and M2 influenza envelope proteins, with a range of core packaging constructs (HIV, MLV, VSV shown) as well as different reporters.
Production Methods
Plasmids Ratios and Amounts
There is considerable variation between studies regarding choice of expression plasmids as particular systems are established within research groups and networks, inherited from previous studies and are often dependent on collaborations or gifts. The most popular system employed involves a multiple plasmid co-transfection approach using separate plasmids for the HA, reporter and retroviral gag and pol core genes. These genes are cloned into a range of expression plasmids such as pI.18, pcDNA3.1, phCMV, and pCAGGS (43, 74, 103, 104). Kozak consensus sequences are very rarely mentioned and only defined in one study, in which a kozak consensus sequence derived from the pHW2000-N1 (Kan) plasmid was used (3).
Additional plasmids encoding NA and M2 are sometimes used when studying the relevant aspects of influenza infection or pseudotype production, but more rarely in the case of neutralization (93, 105). In one study, a 10- to 30-fold increase in pseudotype production was achieved through expression of M2 using lentiviral cores and a 5-fold increase was achieved when an MLV core was used (29).
Plasmid ratios are crucial to pseudotype production but specific to plasmids used as well as transfection methods. In order to attain the highest quality and yields, optimization is required. Typically, the “core:HA:reporter” plasmid ratio is 1:1:1.5. However, the “HA:NA” ratio ranges from 3:1 to 8:1 and protease gene bearing plasmids (HAT, TMPRSS2) are often present at 50% (or below) the concentration of HA (e.g., 1 μg HA plasmid to 0.5 or 0.25 μg protease plasmid). Calcium phosphate precipitation requires the highest plasmid input, with as much as 25 μg of each plasmid per 100 mm dish used, whereas other methods [Fugene, polyethylenimine (PEI), Lipofectamine] require quantities of between 1 and 5 μg for each plasmid per 100 mm dish (20, 29, 69). Plasmid ratios are differentially affected by the composition of plasmids and therefore the quantities used to produce pseudotypes in the literature are justified based on optimization carried out by particular laboratories (17, 72, 88).
Producer Cells
The producer cell lines used for pseudotype production are predominantly Human Embryonic Kidney 293 cells transformed with the SV40 large T antigen (HEK293T, 293T). These cells are highly susceptible to transfection and make good retroviral packaging cells (106). The clone 17 (HEK293T/17) of this cell line is also extensively used to produce high-titer influenza pseudotypes. Other cell lines used include 293FT cells (Invitrogen) used in the production of VSV–HA–NA pseudotypes (3).
Where mentioned, cell confluency at transfection varies between 60 and 90% with cells subcultured 24 h before transfection (74). Cell monolayers are grown on dishes ranging from 60 to 150 mm with the occasional study using T75 Flasks or multi-well plates (74, 107). Transfections are usually carried out using medium with serum such as fetal bovine serum (FBS) at concentrations of up to 10% (83, 108, 109).
Transfection Reagent/Method
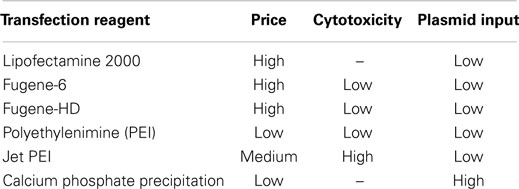
The methods studied use the following chemical transfection reagents: Lipofectamine, Lipofectamine 2000 (Thermo Fisher Scientific), Fugene-6, Fugene-HD (Promega), PEI, jetPEI (Polyplus Transfection), or calcium phosphate precipitation. The choice of reagent is based on optimized lab protocol, cost, as well as the cytotoxicity of each reagent depending on requirements of pseudotype production. Of the above reagents, calcium phosphate precipitation and Fugene-6 are the most popular.
Calcium phosphate precipitation is a well-established transfection method of mammalian cells, developed in 1973 by Graham and van der Eb. This method involves mixing a comparatively high amount (5–25 μg) of plasmid DNA with calcium chloride and then adding this mixture slowly to a buffered saline solution. The mixture is incubated at room temperature whereupon a positively charged DNA and calcium phosphate precipitate is formed. The charge allows the precipitate to associate with the negatively charged cell membrane, entering by endocytosis or phagocytosis. The calcium phosphate precipitation process is sensitive to small differences in pH (20, 110, 111).
Polyethylenimine is a polymeric cation, which was first evaluated for its transfection capabilities in 1995. PEI acts at a range of pH values and associates with DNA to produce a complex with an overall positive charge that can then allow interaction with the cell membrane. Entry is by endocytosis and PEI has been shown to aid the delivery of nucleic acids to the cell nucleus of transfected cells. The original report states that PEI is non-cytotoxic at optimal concentration for transfection (112). However, when using PEI for transfection, it is commonplace to change cell culture medium within 24 h of transfection. JetPEI is a manufactured linear form of PEI, which is suited to high-throughput assays (84, 113).
Lipofectamine (or Lipofectamine 2000) are cationic lipids sold by Invitrogen that allow delivery of nucleic acids such as vectors into host cells through the formation of positively charged liposomes. The liposomes containing the pseudotype vectors are then able to fuse with cell membranes due to their positive charge and lipid constitution (114). Lipofectamine is among the most expensive transfection reagents used in influenza pseudotype production. The benefits of using this method are not readily apparent when the cost of the reagent is considered (107, 115).
Fugene-6 and Fugene-HD are cationic lipid complexes, which have low cell cytotoxicity. This allows laboratories to avoid replacing the transfection medium that may allow an increase in final titers of pseudotype when harvested. Fugene-HD has been shown to be more efficient than other transfection reagents (17, 96, 116, 117).
Table 1 shows a list of transfection reagents, their cytotoxicity, cost, and plasmid input required.
Cell Washes and Medium Replenishment
In transfections where cytotoxic reagents are used, medium is replenished 6–24 h post-transfection, with most studies stating that media is typically replenished after overnight incubation (73). Media replacement can also be accompanied by a PBS wash. Where rhabdoviruses are used for pseudotyping, cell lines are washed using PBS 12 h after transfection with influenza surface glycoprotein plasmids. Helper virus is then added and 4 h later the helper virus containing medium is replaced after a further PBS wash step (3).
Sodium Butyrate
Sodium butyrate, a compound that can increase cell proliferation and pseudotype production is used in several studies with the concentrations ranging from 10 μM to 10 mM (18, 28, 57, 68).
Harvest
Pseudotypes are harvested at various time intervals, typically 48 h post-transfection but sometimes also at 24 or 72 h. The supernatant is taken from the transfected cell monolayer and passed through a 0.45 μM filter to remove cell debris before being stored at −80°C. In many cases, centrifugation at low or high speed is used to concentrate harvested virus (20, 29, 57, 68, 77, 89, 105, 115, 118, 119).
One study has demonstrated that influenza pseudotypes are stable after five freeze–thaw cycles, retaining over 80% infectivity. Keeping pseudotype supernatant at −20°C for 6 months had a similar effect. However, storage at −4 or 20°C led to a reduction in infectivity of 50% in both cases (120). In environments lacking reliable refrigeration facilities, pseudotypes can be lyophilized and stored at a range of increased temperatures and humidity, maintaining viability and concentrations adequate for use in neutralization assays (121).
See Figure 6 for a detailed depiction of methods used for production based on all pseudotype employing articles cited in this review.
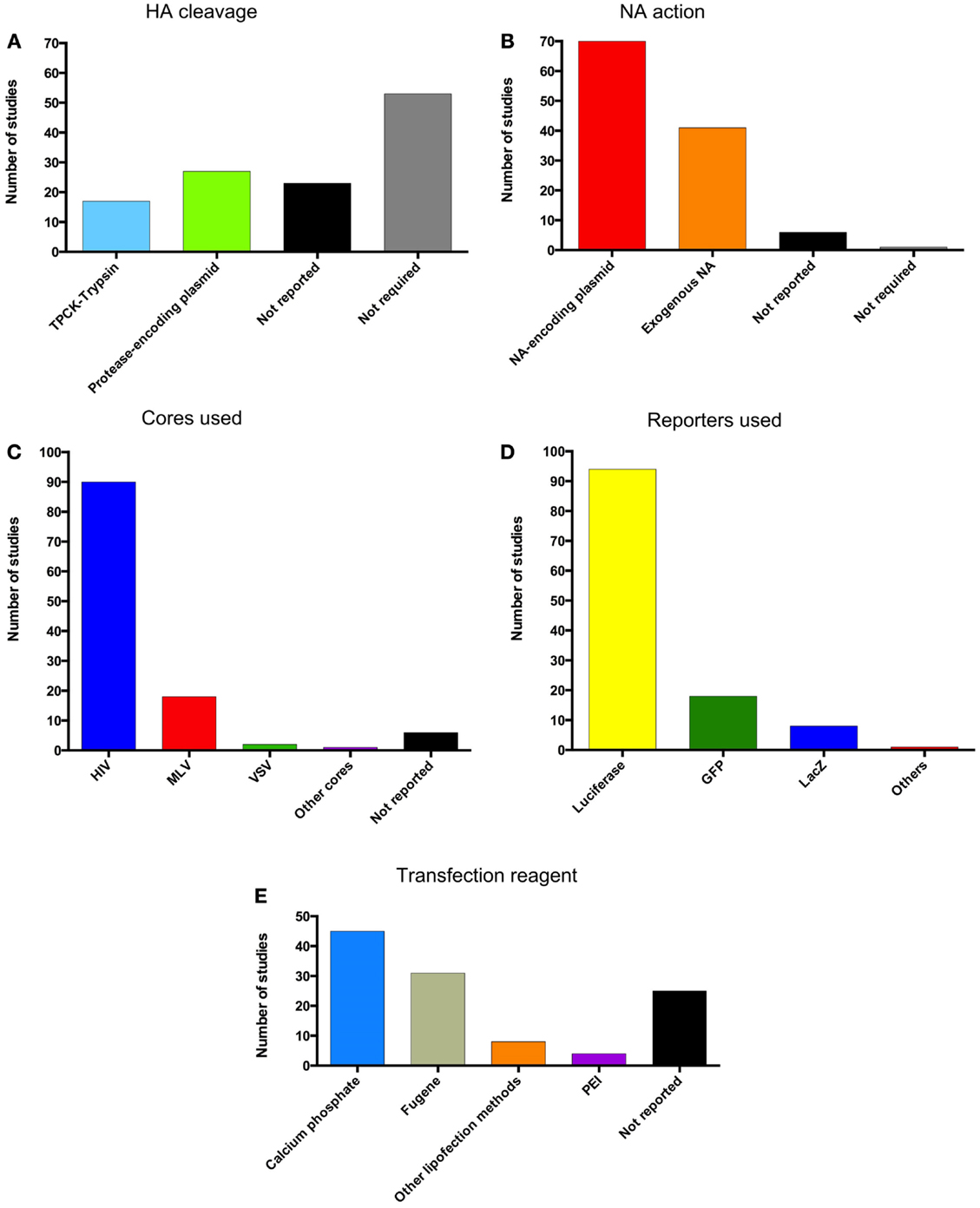
Figure 6. Pseudotype production methods. Graphical representation of the methods used for pseudotype production in the literature cited in this review. (A) Method of HA cleavage used. (B) Method of NA action used. (C) Pseudotype cores used. (D) Reporters incorporated into pseudotypes. (E) Transfection reagents for the production of pseudotypes.
Transduction
Titration
As previously mentioned, with luciferase reporter pseudotypes RLU readings derived from titrations can be used as a secondary measure of pseudotype concentration within a sample. However, RLU readings are dependent on many variables surrounding the cells and the particular luminometer used.
Pseudotypes are titrated by 2-fold serially diluting 100 μl of harvested supernatant in a 96-well plate. After an incubation of 48 or 72 h, RLU can be measured by lysing the transduced cells and adding luciferin (luciferase substrate). This can then be used to calculate the RLU per well and the RLU/ml of the original sample.
Reverse transcriptase quantitative PCR (qRT-PCR) has also been employed in order to estimate transfected gene copies as well as mRNA copies in cells. This method is often used in conjunction with others described in this section in order to have comparative measurements of pseudotype quantity (55, 109, 122).
In many studies, pseudotype input is normalized via enzyme-linked immunosorbent assay (ELISA) detection of the principal component of the HIV core, p24 (16, 17, 28, 54, 59, 88, 89, 92, 95, 103, 109, 113, 115, 123, 124). However, as core budding is independent of surface HA, this method will detect cores lacking envelope glycoproteins as well as cores belonging to transduction competent pseudotypes. Pseudotype HA has also been detected using ELISA and used to normalize pseudotype input (82, 98, 125).
Quantification through hemagglutination assay has also been used frequently (28, 55, 58, 82, 84, 95, 98, 101, 122, 126–128).
Western blotting is used in some cases to determine the amount of glycoprotein or HIV p24 in a pseudotype sample (59, 72, 109). It is also used in a wider range of studies to ascertain glycoprotein or HIV p24 expression (17, 28, 55, 109, 122).
Cell Input
The vast majority of studies involving neutralization assays titrate and transduce in 96-well plates with 1 × 104 cells (HEK293, HEK293T/17, or MDCK) per well. However, the amount of cells can range from 5 × 103 to 1 × 105. In some instances, 293A and MDCK-London cells are also used, whereas BHK-21 cells are frequently used for VSV-based pseudotype infection due to their comparative susceptibility (65, 73, 93, 95, 129–132). Specialized cells overexpressing α2,6-linked sialic acid (MDCK-SIAT) have also been used and compared to parental cells in the presence of soluble HA (77).
In one case, transduction was carried out in 96-well transparent culture plates, before lysates were then transferred to 96-well luminometer plates for analysis (69). The importance of pseudotype input in batch to batch variation is highlighted in Garcia et al. (133), the study suggests that an RLU of at least 1 × 105 per well should be used to ensure that antibody titer is independent of pseudotype input.
Substrates
Steady-Glo or Bright-Glo (Promega) are the most common sources of luciferin. While expensive, these two substrates also serve a secondary purpose of lysing cells and releasing any expressed luciferase enzyme.
Equipment: 96-Well Plates and Luminometers
There is some disparity in the recording of equipment used in the articles studied for this review. Without this required information, reproduction of each study is hampered by these further variables relating to plate reading. Information relating to the color and manufacturer of 96-well plates is very important in the quantification of viable pseudotypes in order to prevent introduction of further variables between laboratories. While logistically difficult, the standardization of neutralization assay equipment across laboratories studying influenza would bring benefits to the interpretation of research data. Standardization of plate reading equipment is also required in order to ensure comparable data are obtained from different machines when reading the same experiment.
High-Throughput Approaches
A high-throughput approach has been used to evaluate antiviral compound effects on pseudotype transduction, testing a wide range of unique compounds in a single assay performed with 96- or 384-well plates (115, 134).
Increased Transduction Efficiency
Polybrene (hexamethrine bromide) and polyfect (Qiagen) are used in several studies in order to increase transduction efficiency (17, 29, 58, 68, 76, 79, 80, 98, 135). 1 μg/ml, 8 μg/ml, or 16 mg/ml of polybrene is added to virus or virus/antibody mixes before the addition of cells in titration and neutralization assays or during incubation.
In two studies, spinoculation was used to increase transduction rate. To achieve the increased transduction rates, the pseudotypes and cells were centrifuged at 1250 rpm for 2 h or 3000 rpm for 1 h (3, 92).
Pseudotype Neutralization Assays
Protocol
Pseudotype neutralization assays (pMN) are usually carried out in 96-well white plates. A measured amount of antibody in medium is serially diluted across the plate and incubated with a set amount of quantified virus in medium, usually at a 1:1 virus:antibody ratio. Incubation is carried out at between 20 and 37°C for between 30 min to 2 h (43, 82, 96, 123, 133, 136). About 1 × 104 target cells are then added to each well, subsequently the plate is left to incubate at 37°C in 5% CO2 for 48 or 72 h. A cell-only control as well as known positive and negative sera standards should be used as benchmarks for the neutralization assay (95, 113). See Figure 7 for a depiction of the pMN assay.
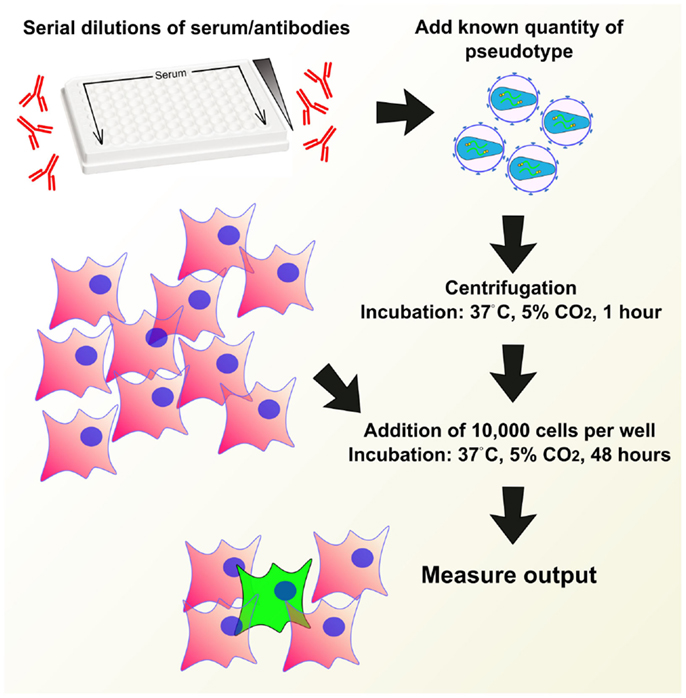
Figure 7. Example of a pseudotype neutralization assay (pMN). Serum or antibodies are serially diluted across a 96-well plate, a known quantity of pseudotype is added and the plate is centrifuged and incubated to allow antibody binding. A set quantity of cells are added and plates are incubated for 48 h. Output is measured in a manner depending on reporter used.
Pseudotype Input
The quantities of pseudotype used in neutralization assays, which were normalized based on p24 ELISA ranged from 6.25 to 50 ng/ml (17, 95). RLU or RLA values of between 1 × 104 and 1 × 106 per well were used (in a 96-well plate), sometimes in conjunction or normalized with p24 or qPCR methods (89, 121, 127). Estimates of copy number per set volume of original viral supernatant can also be used. It is important to note that RLU based values are affected by the make-up of the plasmid bearing the HA gene, as well as a multitude of factors such as the luminometer, which is used to measure transduction.
Serum/Antibody Dilutions and Start Points
Antibody input varies depending on availability, especially when taking into account the possibility of repeats and replicates. Antibodies are primarily diluted 2-fold in Dulbecco’s modified eagle medium (DMEM), with or without FBS, across a 96-well plate, with the occasional three, four, or 5-fold dilution experiment (3, 18, 58, 137, 138). Where mentioned, starting antibody concentration ranged between 1:4 and 1:40.
Incubation Times and Time Periods
When stated, serum complement inactivation varies from 30 min to 1 h at 56°C (68, 133). Pseudotype-antibody incubation times are generally consistent between studies, at 37°C for 1 h. Transduction times vary in 24 h increments at 24, 48, and 72 h, after which output is measured.
Controls
Positive sera or specific commercial antibodies are required as positive controls, which can be compared to tested sera and used to normalize between assays (see Approaches Toward Validation and Standardization). Reference sera from the National Institute for Biological Standards and Control (NIBSC), Office International des Epizooties (OIE), Animal and Plant Health Agency (APHA, previously AHVLA), and US Food and Drug Administration (FDA) are regularly used (88, 89, 127, 139, 140).
Neutralizing Antibody Titer Determination
Antibody effect is displayed using one of many inhibitory concentrations (IC50, IC80, IC90, and IC95). The numerical value relates to the percentage point each particular study is calculating. For example, the IC50 value can represent the concentration of an antibody that reduces RLU reading by 50%, when compared to 100 and 0% transduction controls (48, 68, 141, 142). These controls are essential to the calculation. About 100% inhibition can be benchmarked by a cell-only control and 0% by incubation of cells and virus in the absence of sera.
Hemagglutination-Inhibition Assay
Hemagglutination-inhibition assay (HI) assays using pseudotypes utilize the same procedures as with wild type virus. A quantified amount of viral sample (as determined by hemagglutination assay) in phosphate buffered saline is added to serially diluted sera in a 96-well plate, to which 50 μl of a 0.5–1% chicken/turkey red blood cell suspension is added. After 30 min to 1 h, the HI plates are scored for agglutination. Pseudotype input is adjusted according to WHO guidelines at four hemagglutination units and sera is treated with receptor destroying enzyme to inactivate non-specific inhibition of agglutination (37, 98).
Post-Attachment Assay
The post-attachment neutralization assay is used to identify antibodies that neutralize HA after it has bound to sialic acid. Oh et al. (143) modified the post-attachment assays, originally developed by Edwards and Dimmock (144), to allow wild type influenza virus to be replaced by influenza pseudotype particles.
In this assay, pseudotype particles are incubated at 4°C with cells to enable the synchronization of the attachment of virus to sialic acid on the cell surface and to block viral endocytosis. A diluted serum is then added, and following another 4°C incubation, plates are transferred to 37°C to permit transduction (143). Transduction is then measured using the same approach as that taken in a neutralization assay.
Antibodies detected by this assay have neutralizing activity via their ability to impede the endocytosis step and subsequent HA conformational changes necessary for virus–endosome fusion (143, 144). Antibodies that have neutralizing activity through impeding viral attachment will produce negative results in this assay. The post-attachment assay is useful for evaluating the neutralizing capacity of stalk-directed antibodies that do not inhibit viral attachment (143, 145).
Cross Reactivity
The issue of cross-reactive sera has been raised previously in traditional serological assays, serum samples produced by injection of wild-type virus into mice have been shown to lead to the presence of interfering antibodies directed toward NA or M2 epitopes (146, 147). It is expected that pMN will suffer from the same problems of cross-reactivity, an important issue, which must be addressed in the future in order to strengthen the usefulness of this assay as a competitor to the current gold standards.
Reproducibility
Reproducibility is a major issue in the field of serology. Serum samples are often finite, leading to an inability to reproduce experiments or results in the same context as they were originally published. However, by standardizing methods for production, titration, and neutralization and the use of common reference standards it is possible to minimize variation between experiments and research groups.
Correlation with Other Serological Assays
Comparisons have been made between pMN assays and traditional serological assays with mixed results. Several articles report increases of between 31.9 and 200% in human antibody titers in comparison to microneutralization (MN) based results (148, 149). Buchy et al. (148) show a correlation between H5 pseudotypes and MN (spearman 0.79, p < 0.001), which is also seen in Du et al. (69) and Wang et al. (89), the latter presenting r2 values of 0.9802 for A/Vietnam/1203/2004, 0.8193 for A/Anhui/1/2005, and 0.5244 for A/turkey/Turkey/1/2005 strains.
Alberini et al. (137) compared pMN assays to hemagglutination-inhibition (HI), single radial hemolysis (SRH), and MN assays using 226 different human serum samples. The Pearson correlation test produced significant correlation (p < 0.001) between the antibody titers calculated from each assay. The correlation coefficients between pMN and HI, SRH, and MN assays were 0.73, 0.70, and 0.78, respectively. Furthermore, the correlation between H5 MN and H5 pMN allowed the establishment of a threshold from which pMN titers could be based. pMN data were then analyzed based on the threshold, which showed protective titers in patients of 38–43, 54, and 79% after adjuvanted vaccination, second dose and booster, respectively (137).
Qiu et al. (81) show a range of correlations between HI and pMN using different HA subtypes. A/Moscow/10/1999 (H3N2) correlates well (r = 0.8454, p < 0.0001), A/Brisbane/59/2007 (H1N1), and A/Japan/305/57 (H2N2) poorly (r = 0.1171, p = 0.7472 and r = 0.1171, p = 7472) whereas A/Vietnam/1203/2004 (H5N1) correlates (r = 0.7921, p = 0.0029). In an additional study, HI and pMN (IC50) correlate well in Qiu et al. (107) in the case of A/Shanghai/4664T/2013 (H7N9) (spearman r = 0.88, p < 0.0001) as well as in Whittle et al. (126) (r2 = 0.6491, p < 0.0001).
A significant correlation of 65% (p = 0.002, r = 0.65) has also been reported between SRH and pMN using equine influenza pseudotypes and sera and another study showed the relationship between RLU and HA content (78, 119).
Approaches toward Validation and Standardization
Approaches toward the standardization of pMN should follow the procedure that was required for MN standardization. Standardization of MN in general has focused on the use of pooled serum samples as reference standards. A/California/7/2009 (pandemic H1N1, pdm) standard was established by the WHO in 2010 with an assignment of potency of 13,000 IU/ml. A second pooled sera reference standard for H5N1 exists and has successfully been used in a number of studies (89, 137, 150). A cut off value for positive and negative H5N1 neutralizing sera exists for this set of H5N1 reference standards (137).
Chimeric Hemagglutinin and Stalk-Directed Antibodies
There has been considerable research into the stalk region of HA in relation to vaccine design and immunity to influenza. Various stalk-directed monoclonal antibodies (mAB) such as CR6261 have been characterized, opening up the potential use of chimeric HA to test for the presence of similar antibodies in serum samples (151, 152).
Stalk-directed antibodies were first identified in 1994 when the cross-reactive C179 mouse monoclonal antibody was identified and found to inhibit fusion of several HA subtypes (153). Since then many studies have focused on stalk-directed antibodies and their neutralization of multiple diverse subtypes of influenza (145, 152, 154–156). However, this range of heterosubtypic immunity is dependent on the characteristics of the epitope of each antibody tested, which will influence which subtypes, clades and whether they neutralize group 1 or 2 influenza.
The stalk region of HA is more conserved than the variable globular head to which the vast majority of neutralizing antibodies are directed. While residues in the head mediate attachment of the virus to target cells by binding to sialic acid, the fusion peptide in the stalk of HA is just as crucial to the HA function (157, 158). In order to test for neutralizing stalk antibodies, studies have employed a variety of chimeric HA constructs bearing stalks and heads from different subtypes. The concept behind this revolves around the use of HA heads that are largely unreactive to the antibodies used in the assay. Utilizing this approach, a neutralizing response can be detected in the absence of head-directed neutralization.
Several hybrids have been constructed and pseudotyped using HIV cores, these are generally constructed through PCR amplification and incorporation of complementary restriction sites, allowing ligation of different segments of HA genes. A wider variety has been used in reverse genetics approaches toward development of wild type virus bearing chimeric HAs (159–161). These chimeric HA are promising candidates for the testing of “universal” vaccines.
Table 2 displays the regions and subtypes used in the construction of chimeric hemagglutinins. Figure 8 is a visualization of chimeric HA construction in the form of a computer model.
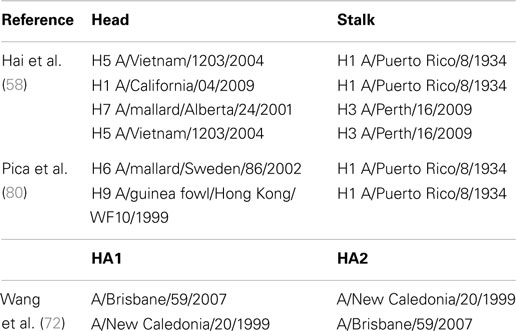
Table 2. Examples of chimeric hemagglutinins originating from divergent subtypes and used for pseudotype production.
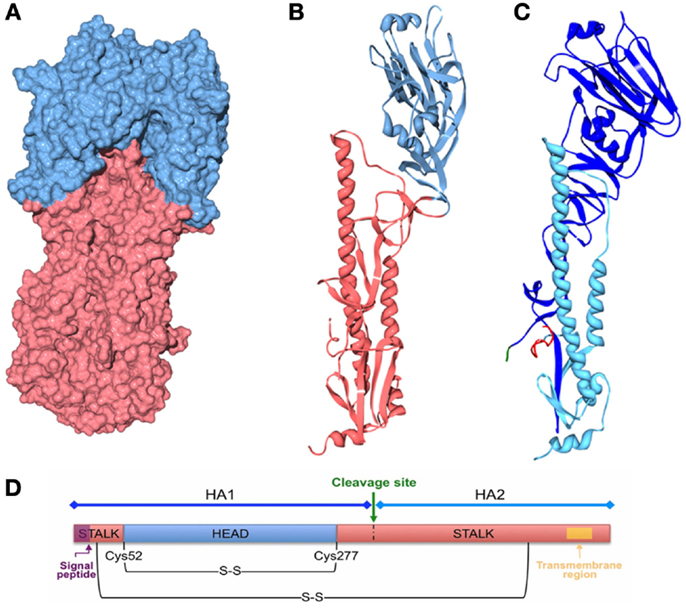
Figure 8. Computer models of chimeric HA. Three-dimensional structures were generated with Swiss PDB viewer and POV-Ray 3.7 using the structure of the recombinant virus A/Hong Kong/1/1968 X-31 H3 [PDB ID: 2VIU (162)]. The signal peptide is not present in the HA. The transmembrane region is not resolved by X-ray crystallography. (A) Three-dimensional structure of the influenza HA trimer, showing the HA surface of the head (blue) and stalk (red) regions. (B) Three-dimensional ribbon structure of the influenza HA monomer showing the head (blue) and stalk (red) regions. (C) Three-dimensional ribbon structure of the influenza HA monomer showing HA1 (blue) and HA2 (light blue) subunits, the cleavage site and the fusion peptide are also shown in green and red, respectively. (D) Schematic of the HA polypeptide.
Future of Influenza Pseudotypes
Pseudotype neutralization assay offers the safety of using pseudotypes and the sensitivity of the MN assay. Further validation and standardization of the assay are required but once established, the assay should offer a robust and sensitive means of interrogating influenza vaccine trials for head and stalk-targeting antibodies. The production of vaccines that elicit stalk-targeting antibodies may in time lead to a universal vaccine, preventing 250,000–500,000 deaths from seasonal influenza and the emergence of pandemic strains, most recently the H1N1 2009 pdm, which caused an estimated 284,500 deaths (163). pMN currently offers the opportunity to batch test vaccines or commercialized antibodies in the absence of standardization.
Furthermore, the ability of the pMN assay to include chimeric HA, and also NA and M2 allows the pMN to be used to explain the pathogenicity of seasonal and pandemic influenza strains and perhaps elucidate the antigenic evolution of influenza further.
Other Uses of Pseudotyping Influenza
Gene Therapy and Vaccines
As the field of gene therapy progresses, influenza pseudotyping will benefit from the design of even safer and more effective vectors. As more sophisticated systems are developed they may become more easily standardized and comparable to wild type virus.
One aspect of gene therapy that may benefit the field of influenza is the use of viral entry proteins to target delivery of nucleic acids into specific cells, as vaccines or delivery systems. One delivery system study used influenza pseudotypes to transduce the respiratory epithelial cells of mice after nasal administration with promising results indicating that the method could be used in the treatment of cystic fibrosis (118). A similar study presented the rescue of ciliary function using influenza pseudotypes containing therapeutic cDNA (164).
Pseudotype-based influenza gene delivery vaccines are also becoming more widespread, with several candidates already cited in this review. Baculovirus pseudotyped with VSV-G has been used successfully to express HA in mammalian cells and provided an efficacious vaccine when tested in chickens and mice (165). Originally a popular vector for transgene expression in insect cells, baculovirus has been shown to be a useful tool for vaccine production in mammalian cells (166). In Wu et al. (165), delivery was achieved through VSV-G incorporation into baculovirus under the effect of the polyhedron promoter and HA under the effect of the CMV promoter in order to achieve expression and subsequent infection of mammalian cells. This is an interesting gene delivery system, which could be used as a method for the introduction of pseudotype genes into cells through a VSV-G bearing baculovirus in lieu of cytotoxic transfection reagents.
A further pseudotype vaccine has been developed which contains a modified HA gene, allowing expression in transduced cells but lacking the viral RNA sequences required for replication. This approach yields a particle bearing the desired glycoproteins, in this case A/Puerto Rico/8/1934 (H1) that consequently induces a robust T-cell response when given to mice via inhalation. Reduction in the severity of symptoms was also seen in mice infected with a different subtype: H3N2, A-X31 (71). While these approaches demonstrate the flexibility of the pseudotype platform, other more established methods including adenovirus or modified vaccinia viruses (e.g., modified vaccinia Ankara) may present a more attractive option for the delivery of influenza genes, and have been reviewed in great depth (167).
Pseudotypes used as immunogens, such as those bearing H5 have been tested in mice as a candidate vaccine, eliciting high levels of anti-HA antibodies as determined by HI. Mice that were vaccinated survived despite weight loss of approximately 12.8–21.1% whereas the non-vaccinated group lost approximately 25.5–26.2% of their bodyweight and perished 6 days after H5N1 virus challenge (20). A similar approach is taken by Szécsi et al. (168) in the production of H5 and H7 pseudotyped virus-like particles as immunogens tested in mice.
Influenza pseudotypes could also be used in vaccine design through the use of integrase defective lentiviral vector technology. Defective lentiviral vector technology allows transduction of target cells through maintenance of an episomal reporter construct without integration into the genome. This approach may bring benefits by reducing the chance of interrupting host genes and the eventual dilution of the delivered gene over time (169, 170).
See Figure 9 for a depiction of the various pseudotype-based vaccines and immunogens.
Figure 9. Pseudotypes used for gene delivery or as immunogens. Pseudotypes can be employed as immunogens bearing the antigen of choice or as delivery systems for genes of choice. (A) HA-based pseudotype/virus-like particle immunogen. (B) VSV-G pseudotype delivery system for HA gene. (C) HA pseudotype delivery system for HA gene.
Recommended Consensus Protocol from Synthesis of Published Articles
Production Protocol
A HEK293T cell monolayer of 60–90% confluence should be transfected using Fugene-6 or calcium phosphate precipitation in medium containing 10% FBS. Plasmid ratios should be optimized based on the plasmids used. The use of second generation HIV packaging constructs is recommended. An NA encoding plasmid can be used or exogenous NA can be added at 24 h post-transfection to induce release of pseudotypes. The supernatant should be harvested at 48 h post-transfection and filtered through a 0.45 μm filter. Filtered supernatant should be kept at −80°C in single use aliquots if long-term storage is required.
Titration Protocol
Titration should be carried out using luciferase-based transduction in 96-well white plates, by p24 ELISA or other methods of quantification. Quantification of pseudotype particles using luciferase-based transduction involves the 2-fold serial dilution of 100 μl of pseudotype in 10% FBS medium. About 1 × 104 cells are then added in a 50 μl volume and the resulting solution is incubated for 48 h. After the 48-h incubation period luciferase substrate is added to each well and RLU values are read. Cell only, ΔEnv and VSV-G bearing pseudotypes can be used as negative and positive controls.
Pseudotype-Based Neutralization Protocol
Serum samples are serially diluted across a 96-well plate in 50 μl of media. Pseudotype virus should be added in a 50 μl volume at a concentration of 1 × 106 RLU. After 1 h incubation at 37°C, 1 × 104 HEK293T or MDCK cells should be added in a 50 μl volume. The plate is then incubated at 37°C for 48 h before luciferase substrate is added to each well, after which RLU values are read. Standards should ideally be used in the form of neutralizing antibodies or pooled serum samples.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was partially funded by the European Research Council under the European Union’s Seventh Framework Programme (FP7/2007-2013)/ERC grant agreement no. 268904 – DIVERSITY.
References
1. Hause BM, Collin EA, Liu R, Huang B, Sheng Z, Lu W, et al. Characterization of a novel influenza virus in cattle and swine: proposal for a new genus in the orthomyxoviridae family. MBio (2014) 5:1–10. doi:10.1128/mBio.00031-14
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
2. World Health Organisation. WHO Fact Sheet (2014). p. 5–8. Available from: http://www.who.int/mediacentre/factsheets/fs211/en/
3. Cheresiz SV, Kononova AA, Razumova YV, Dubich TS, Chepurnov AA, Kushch AA, et al. A vesicular stomatitis pseudovirus expressing the surface glycoproteins of influenza A virus. Arch Virol (2014) 159:2651–8. doi:10.1007/s00705-014-2127-y
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
4. Zimmer G, Locher S, Berger Rentsch M, Halbherr SJ. Pseudotyping of vesicular stomatitis virus with the envelope glycoproteins of highly pathogenic avian influenza viruses. J Gen Virol (2014) 95:1634–9. doi:10.1099/vir.0.065201-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
5. Blesch A. Lentiviral and MLV based retroviral vectors for ex vivo and in vivo gene transfer. Methods (2004) 33:164–72. doi:10.1016/j.ymeth.2003.11.005
6. Sakuma T, Barry MA, Ikeda Y. Lentiviral vectors: basic to translational. Biochem J (2012) 443:603–18. doi:10.1042/BJ20120146
7. Jacks T, Power MD, Masiarz FR, Luciw PA, Barr PJ, Varmus HE. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature (1988) 331:280–3. doi:10.1038/331280a0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
8. Engelman A, Cherepanov P. The structural biology of HIV-1: mechanistic and therapeutic insights. Nat Rev Microbiol (2012) 10:279–90. doi:10.1038/nrmicro2747
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
9. Pappas L, Foglierini M, Piccoli L, Kallewaard NL, Turrini F, Silacci C, et al. Rapid development of broadly influenza neutralizing antibodies through redundant mutations. Nature (2014) 516:418–22. doi:10.1038/nature13764
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
10. Bertram S, Heurich A, Lavender H, Gierer S, Danisch S, Perin P, et al. Influenza and SARS-coronavirus activating proteases TMPRSS2 and HAT are expressed at multiple sites in human respiratory and gastrointestinal tracts. PLoS One (2012) 7:e35876. doi:10.1371/journal.pone.0035876
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
11. Chen M-W, Liao H-Y, Huang Y, Jan J-T, Huang C-C, Ren C-T, et al. Broadly neutralizing DNA vaccine with specific mutation alters the antigenicity and sugar-binding activities of influenza hemagglutinin. Proc Natl Acad Sci U S A (2011) 108:3510–5. doi:10.1073/pnas.1019744108
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
12. Chen M-W, Cheng T-JR, Huang Y, Jan J-T, Ma S-H, Yu AL, et al. A consensus-hemagglutinin-based DNA vaccine that protects mice against divergent H5N1 influenza viruses. Proc Natl Acad Sci U S A (2008) 105:13538–43. doi:10.1073/pnas.0806901105
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
13. Zmora P, Blazejewska P, Moldenhauer A-S, Welsch K, Nehlmeier I, Wu Q, et al. DESC1 and MSPL activate influenza A viruses and emerging coronaviruses for host cell entry. J Virol (2014) 88:12087–97. doi:10.1128/JVI.01427-14
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
14. Labrosse B, Tourdjman M, Porcher RL, Legoff J, de Lamballerie X, Simon F, et al. Detection of extensive cross-neutralization between pandemic and seasonal A/H1N1 influenza viruses using a pseudotype neutralization assay. PLoS One (2010) 5:e11036. doi:10.1371/journal.pone.0011036
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
15. Zufferey R, Nagy D, Mandel RJ, Naldini L, Trono D. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat Biotechnol (1997) 15:871–5. doi:10.1038/nbt0997-871
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
16. Zhou F, Wang G, Buchy P, Cai Z, Chen H, Chen Z, et al. A triclade DNA vaccine designed on the basis of a comprehensive serologic study elicits neutralizing antibody responses against all clades and subclades of highly pathogenic avian influenza H5N1 viruses. J Virol (2012) 86:6970–8. doi:10.1128/JVI.06930-11
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
17. Wang W, Butler EN, Veguilla V, Vassell R, Terrig Thomas J, Moos M, et al. Establishment of retroviral pseudotypes with influenza hemagglutinins from H1, H3, and H5 subtypes for sensitive and specific detection of neutralizing antibodies. J Virol Methods (2008) 153:111–9. doi:10.1016/j.jviromet.2008.07.015
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
18. Ding H, Tsai C, Zhou F, Buchy P, Deubel V, Zhou P. Heterosubtypic antibody response elicited with seasonal influenza vaccine correlates partial protection against highly pathogenic H5N1 virus. PLoS One (2011) 6:e17821. doi:10.1371/journal.pone.0017821
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
19. Hashem AM, Van Domselaar G, Li C, Wang J, She YM, Cyr TD, et al. Universal antibodies against the highly conserved influenza fusion peptide cross-neutralize several subtypes of influenza A virus. Biochem Biophys Res Commun (2010) 403:247–51. doi:10.1016/j.bbrc.2010.11.030
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
20. Zhang S, Xiao L, Zhou H, Yu Z, Chen H, Guo A, et al. Generation and characterization of an H5N1 avian influenza virus hemagglutinin glycoprotein pseudotyped lentivirus. J Virol Methods (2008) 154:99–103. doi:10.1016/j.jviromet.2008.08.016
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
21. Naldini L, Blömer U, Gage FH, Trono D, Verma IM. Efficient transfer, integration, and sustained long-term expression of the transgene in adult rat brains injected with a lentiviral vector. Proc Natl Acad Sci U S A (1996) 93:11382–8. doi:10.1073/pnas.93.21.11382
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
22. Ahmed MS, Jacques LC, Mahallawi W, Ferrara F, Temperton NJ, Upile N, et al. Cross-reactive immunity against influenza viruses in children and adults following 2009 pandemic H1N1 infection. Antiviral Res (2015) 114:106–12. doi:10.1016/j.antiviral.2014.12.008
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
23. Smith SE, Gibson MS, Wash RS, Ferrara F, Wright E, Temperton NJ, et al. Chicken interferon-inducible transmembrane protein 3 restricts influenza viruses and lyssaviruses in vitro. J Virol (2013) 87:12957–66. doi:10.1128/JVI.01443-13
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
24. Molesti E, Ferrara F, Lapini G, Montomoli E, Temperton NJ. Discordant correlation between serological assays observed when measuring heterosubtypic responses against avian influenza H5 and H7 viruses in unexposed individuals. Biomed Res Int (2014) 2014:231365. doi:10.1155/2014/231365
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
25. Benfield C, Smith SE, Wright E, Wash RS, Ferrara F, Temperton NJ, et al. Bat and pig interferon-induced transmembrane protein 3 restrict cell entry by influenza virus and lyssaviruses. J Gen Virol (2015) pii:vir.0.000058. doi:10.1099/vir.0.000058
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
26. Wright E, Hayman DTS, Vaughan A, Temperton NJ, Wood JLN, Cunningham AA, et al. Virus neutralising activity of African fruit bat (Eidolon helvum) sera against emerging lyssaviruses. Virology (2010) 408:183–9. doi:10.1016/j.virol.2010.09.014
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
27. Zufferey R, Dull T, Mandel RJ, Bukovsky A, Quiroz D, Naldini L, et al. Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J Virol (1998) 72:9873–80.
28. Sawoo O, Dublineau A, Batéjat C, Zhou P, Manuguerra J-C, Leclercq I. Cleavage of hemagglutinin-bearing lentiviral pseudotypes and their use in the study of influenza virus persistence. PLoS One (2014) 9:e106192. doi:10.1371/journal.pone.0106192
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
29. McKay T, Patel M, Pickles RJ, Johnson LG, Olsen JC. Influenza M2 envelope protein augments avian influenza hemagglutinin pseudotyping of lentiviral vectors. Gene Ther (2006) 13:715–24. doi:10.1038/sj.gt.3302715
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
30. Finer MH, Dull TJ, Qin L, Farson D, Roberts MR. kat: a high-efficiency retroviral transduction system for primary human T lymphocytes. Blood (1994) 83:43–50.
31. Farrell KB, Ting Y-T, Eiden MV. Fusion-defective gibbon ape leukemia virus vectors can be rescued by homologous but not heterologous soluble envelope proteins. J Virol (2002) 76:4267–74. doi:10.1128/JVI.76.9.4267-4274.2002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
32. Wallerström S, Lagerqvist N, Temperton NJ, Cassmer M, Moreno A, Karlsson M, et al. Detection of antibodies against H5 and H7 strains in birds: evaluation of influenza pseudovirus particle neutralization tests. Infect Ecol Epidemiol (2014) 4:1–8. doi:10.3402/iee.v4.23011
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
33. Oh S, Selleck P, Temperton NJ, Chan PKS, Capecchi B, Manavis J, et al. Neutralizing monoclonal antibodies to different clades of influenza A H5N1 viruses. J Virol Methods (2009) 157:161–7. doi:10.1016/j.jviromet.2008.12.016
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
34. Wang S-Y, Su C-Y, Lin M, Huang S-Y, Huang W-I, Wang C-C, et al. HA-pseudotyped retroviral vectors for influenza antagonist screening. J Biomol Screen (2009) 14:294–302. doi:10.1177/1087057108330786
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
35. Huang IC, Bailey CC, Weyer JL, Radoshitzky SR, Becker MM, Chiang JJ, et al. Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog (2011) 7:e1001258. doi:10.1371/journal.ppat.1001258
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
36. Tao L, Chen J, Meng J, Chen Y, Li H, Liu Y, et al. Enhanced protective efficacy of H5 subtype influenza vaccine with modification of the multibasic cleavage site of hemagglutinin in retroviral pseudotypes. Virol Sin (2013) 28:136–45. doi:10.1007/s12250-013-3326-5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
37. Haynes JR, Dokken L, Wiley JA, Cawthon AG, Bigger J, Harmsen AG, et al. Influenza-pseudotyped Gag virus-like particle vaccines provide broad protection against highly pathogenic avian influenza challenge. Vaccine (2009) 27:530–41. doi:10.1016/j.vaccine.2008.11.011
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
38. Hatziioannou T, Delahaye E, Martin F, Russell SJ, Cosset FL. Retroviral display of functional binding domains fused to the amino terminus of influenza hemagglutinin. Hum Gene Ther (1999) 10:1533–44. doi:10.1089/10430349950017860
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
39. Bock M, Bishop KN, Towers G, Stoye JP. Use of a transient assay for studying the genetic determinants of Fv1 restriction. J Virol (2000) 74:7422–30. doi:10.1128/JVI.74.16.7422-7430.2000
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
40. Soneoka Y, Cannon PM, Ramsdale EE, Griffiths JC, Romano G, Kingsman SM, et al. A transient three-plasmid expression system for the production of high titer retroviral vectors. Nucleic Acids Res (1995) 23:628–33. doi:10.1093/nar/23.4.628
41. Towers G, Bock M, Martin S, Takeuchi Y, Stoye JP, Danos O. A conserved mechanism of retrovirus restriction in mammals. Proc Natl Acad Sci U S A (2000) 97:12295–9. doi:10.1073/pnas.200286297
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
42. Hartley JW, Rowe WP, Huebner RJ. Host-range restrictions of murine leukemia viruses in mouse embryo cell cultures. J Virol (1970) 5:221–5.
43. Temperton NJ, Hoschler K, Major D, Nicolson C, Manvell R, Hien VM, et al. A sensitive retroviral pseudotype assay for influenza H5N1-neutralizing antibodies. Influenza Other Respi Viruses (2007) 1:105–12. doi:10.1111/j.1750-2659.2007.00016.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
44. Garcia J-M, Lai JCC. Production of influenza pseudotyped lentiviral particles and their use in influenza research and diagnosis: an update. Expert Rev Anti Infect Ther (2011) 9:443–55. doi:10.1586/eri.11.25
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
45. Mullarkey CE, Boyd A, Van Laarhoven A, Lefevre EA, Veronica Carr B, Baratelli M, et al. Improved adjuvanting of seasonal influenza vaccines: preclinical studies of MVA-NP+M1 coadministration with inactivated influenza vaccine. Eur J Immunol (2013) 43:1940–52. doi:10.1002/eji.201242922
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
46. Lingwood D, McTamney PM, Yassine HM, Whittle JRR, Guo X, Boyington JC, et al. Structural and genetic basis for development of broadly neutralizing influenza antibodies. Nature (2012) 489:566–70. doi:10.1038/nature11371
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
47. Su Y, Zhu X, Wang Y, Wu M, Tien P. Evaluation of Glu11 and Gly8 of the H5N1 influenza hemagglutinin fusion peptide in membrane fusion using pseudotype virus and reverse genetics. Arch Virol (2008) 153:247–57. doi:10.1007/s00705-007-1088-9
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
48. Rao SS, Kong WP, Wei CJ, Van Hoeven N, Patrick Gorres J, Nason M, et al. Comparative efficacy of hemagglutinin, nucleoprotein, and matrix 2 protein gene-based vaccination against H5N1 influenza in mouse and ferret. PLoS One (2010) 5:e9812. doi:10.1371/journal.pone.0009812
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
49. Loureiro S, Ren J, Phapugrangkul P, Colaco CA, Bailey CR, Shelton H, et al. Adjuvant-free immunization with hemagglutinin-Fc fusion proteins as an approach to influenza vaccines. J Virol (2011) 85:3010–4. doi:10.1128/JVI.01241-10
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
50. Wei C-J, Yassine HM, McTamney PM, Gall JGD, Whittle JRR, Boyington JC, et al. Elicitation of broadly neutralizing influenza antibodies in animals with previous influenza exposure. Sci Transl Med (2012) 4:ra114–47. doi:10.1126/scitranslmed.3004273
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
51. Ledgerwood JE, Zephir K, Hu Z, Wei CJ, Chang L, Enama ME, et al. Prime-boost interval matters: a randomized phase 1 study to identify the minimum interval necessary to observe the H5 DNA influenza vaccine priming effect. J Infect Dis (2013) 208:418–22. doi:10.1093/infdis/jit180
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
52. Wei C-J, Xu L, Kong W-P, Shi W, Canis K, Stevens J, et al. Comparative efficacy of neutralizing antibodies elicited by recombinant hemagglutinin proteins from avian H5N1 influenza virus. J Virol (2008) 82:6200–8. doi:10.1128/JVI.00187-08
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
53. Wang W, Peng H, Tao Q, Zhao X, Tang H, Tang Z, et al. Serologic assay for avian-origin influenza A (H7N9) virus in adults of Shanghai, Guangzhou and Yunnan, China. J Clin Virol (2014) 60:305–8. doi:10.1016/j.jcv.2014.04.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
54. Ao Z, Patel A, Tran K, He X, Fowke K, Coombs K, et al. Characterization of a trypsin-dependent avian influenza H5N1-pseudotyped HIV vector system for high throughput screening of inhibitory molecules. Antiviral Res (2008) 79:12–8. doi:10.1016/j.antiviral.2008.02.001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
55. Wu J, Zhang F, Wang M, Xu C, Song J, Zhou J, et al. Characterization of neuraminidases from the highly pathogenic avian H5N1 and 2009 pandemic H1N1 influenza a viruses. PLoS One (2010) 5:e15825. doi:10.1371/journal.pone.0015825
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
56. Hatziioannou T, Valsesia-Wittmann S, Russell SJ, Cosset FL. Incorporation of fowl plague virus hemagglutinin into murine leukemia virus particles and analysis of the infectivity of the pseudotyped retroviruses. J Virol (1998) 72:5313–7.
57. Konishi M, Kawamoto K, Izumikawa M, Kuriyama H, Yamashita T. Gene transfer into guinea pig cochlea using adeno-associated virus vectors. J Gene Med (2008) 10:610–8. doi:10.1002/jgm.1189
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
58. Hai R, Krammer F, Tan GS, Pica N, Eggink D, Maamary J, et al. Influenza viruses expressing chimeric hemagglutinins: globular head and stalk domains derived from different subtypes. J Virol (2012) 86:5774–81. doi:10.1128/JVI.00137-12
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
59. Su B, Wurtzer S, Rameix-Welti MA, Dwyer D, van der Werf S, Naffakh N, et al. Enhancement of the influenza a hemagglutinin (HA)-mediated cell-cell fusion and virus entry by the viral neuraminidase (NA). PLoS One (2009) 4:e8495. doi:10.1371/journal.pone.0008495
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
60. Ascione A, Capecchi B, Campitelli L, Imperiale V, Flego M, Zamboni S, et al. Human monoclonal antibodies in single chain fragment variable format with potent neutralization activity against influenza virus H5N1. Antiviral Res (2009) 83:238–44. doi:10.1016/j.antiviral.2009.05.005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
61. Wiley DC, Skehel JJ. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Annu Rev Biochem (1987) 56:365–94. doi:10.1146/annurev.bi.56.070187.002053
62. Fouchier RAM, Munster V, Wallensten A, Bestebroer TM, Herfst S, Smith D, et al. Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls. J Virol (2005) 79:2814–22. doi:10.1128/JVI.79.5.2814-2822.2005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
63. Tong S, Li Y, Rivailler P, Conrardy C, Castillo DAA, Chen L-M, et al. A distinct lineage of influenza A virus from bats. Proc Natl Acad Sci U S A (2012) 109:4269–74. doi:10.1073/pnas.1116200109
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
64. Tong S, Zhu X, Li Y, Shi M, Zhang J, Bourgeois M, et al. New world bats harbor diverse influenza A viruses. PLoS Pathog (2013) 9:e1003657. doi:10.1371/journal.ppat.1003657
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
65. Hanika A, Larisch B, Steinmann E, Schwegmann-Weßels C, Herrler G, Zimmer G. Use of influenza C virus glycoprotein HEF for generation of vesicular stomatitis virus pseudotypes. J Gen Virol (2005) 86:1455–65. doi:10.1099/vir.0.80788-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
66. Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics (2004) 5:113. doi:10.1186/1471-2105-5-113
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
67. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol (2011) 28:2731–9. doi:10.1093/molbev/msr121
68. Tsai C, Caillet C, Hu H, Zhou F, Ding H, Zhang G, et al. Measurement of neutralizing antibody responses against H5N1 clades in immunized mice and ferrets using pseudotypes expressing influenza hemagglutinin and neuraminidase. Vaccine (2009) 27:6777–90. doi:10.1016/j.vaccine.2009.08.056
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
69. Du L, Zhao G, Zhang X, Liu Z, Yu H, Zheng BJ, et al. Development of a safe and convenient neutralization assay for rapid screening of influenza HA-specific neutralizing monoclonal antibodies. Biochem Biophys Res Commun (2010) 397:580–5. doi:10.1016/j.bbrc.2010.05.161
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
70. Ferrara F, Molesti E, Böttcher-Friebertshäuser E, Cattoli G, Corti D, Scott SD, et al. The human transmembrane protease serine 2 is necessary for the production of group 2 influenza A virus pseudotypes. J Mol Genet Med (2013) 7:309–14.
71. Powell TJ, Silk JD, Sharps J, Fodor E, Townsend ARM. Pseudotyped influenza A virus as a vaccine for the induction of heterotypic immunity. J Virol (2012) 86:13397–406. doi:10.1128/JVI.01820-12
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
72. Wang W, Anderson CM, de Feo CJ, Zhuang M, Yang H, Vassell R, et al. Cross-neutralizing antibodies to pandemic 2009 H1N1 and recent seasonal H1N1 influenza a strains influenced by a mutation in hemagglutinin subunit 2. PLoS Pathog (2011) 7:e1002081. doi:10.1371/journal.ppat.1002081
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
73. Kong W-P, Hood C, Yang Z-Y, Wei C-J, Xu L, García-Sastre A, et al. Protective immunity to lethal challenge of the 1918 pandemic influenza virus by vaccination. Proc Natl Acad Sci U S A (2006) 103:15987–91. doi:10.1073/pnas.0607564103
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
74. Huang I-C, Li W, Sui J, Marasco W, Choe H, Farzan M. Influenza A virus neuraminidase limits viral superinfection. J Virol (2008) 82:4834–43. doi:10.1128/JVI.00079-08
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
75. Prodromou C, Pearl LH. Recursive PCR: a novel technique for total gene synthesis. Protein Eng (1992) 5:827–9. doi:10.1093/protein/5.8.827
76. Lin AH, Cannon PM. Use of pseudotyped retroviral vectors to analyze the receptor-binding pocket of hemagglutinin from a pathogenic avian influenza A virus (H7 subtype). Virus Res (2002) 83:43–56. doi:10.1016/S0168-1702(01)00407-5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
77. Tang DJ, Lam YM, Siu YL, Lam CH, Chu SL, Peiris JSM, et al. A single residue substitution in the receptor-binding domain of H5N1 hemagglutinin is critical for packaging into pseudotyped lentiviral particles. PLoS One (2012) 7:e43596. doi:10.1371/journal.pone.0043596
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
78. Scott S, Molesti E, Temperton NJ, Ferrara F, Böttcher-Friebertshäuser E, Daly J. The use of equine influenza pseudotypes for serological screening. J Mol Genet Med (2012) 6:304–8.
79. Krammer F, Pica N, Hai R, Tan GS, Palese P. Hemagglutinin stalk-reactive antibodies are boosted following sequential infection with seasonal and pandemic H1N1 influenza virus in mice. J Virol (2012) 86:10302–7. doi:10.1128/JVI.01336-12
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
80. Pica N, Hai R, Krammer F, Wang TT, Maamary J, Eggink D, et al. Hemagglutinin stalk antibodies elicited by the 2009 pandemic influenza virus as a mechanism for the extinction of seasonal H1N1 viruses. Proc Natl Acad Sci U S A (2012) 109:2573–8. doi:10.1073/pnas.1200039109
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
81. Qiu C, Huang Y, Wang Q, Tian D, Zhang W, Hu Y, et al. Boosting heterosubtypic neutralization antibodies in recipients of 2009 pandemic H1N1 influenza vaccine. Clin Infect Dis (2012) 54:17–24. doi:10.1093/cid/cir753
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
82. Li Z, Ma C, Liu Z, He W. Serologic cross-reactivity among humans and birds infected with highly pathogenic avian influenza A subtype H5N1 viruses in China. Immunol Lett (2011) 135:59–63. doi:10.1016/j.imlet.2010.09.012
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
83. Hu H, Voss J, Zhang G, Buchy P, Zuo T, Wang L, et al. A human antibody recognizing a conserved epitope of H5 hemagglutinin broadly neutralizes highly pathogenic avian influenza H5N1 viruses. J Virol (2012) 86:2978–89. doi:10.1128/JVI.06665-11
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
84. Tisoncik JR, Guo Y, Cordero KS, Yu J, Wang J, Cao Y, et al. Identification of critical residues of influenza neuraminidase in viral particle release. Virol J (2011) 8:14. doi:10.1186/1743-422X-8-14
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
85. Margine I, Krammer F, Hai R, Heaton NS, Tan GS, Andrews SA, et al. Hemagglutinin stalk-based universal vaccine constructs protect against group 2 influenza A viruses. J Virol (2013) 87:10435–46. doi:10.1128/JVI.01715-13
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
86. Hooper KA, Crowe JE, Bloom JD. Influenza viruses with receptor-binding N1 neuraminidases occur sporadically in several lineages and show no attenuation in cell culture or mice. J Virol (2015) 89(7):3737–45. doi:10.1128/JVI.00012-15
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
87. Hooper KA, Bloom JD. A mutant influenza virus that uses an N1 neuraminidase as the receptor-binding protein. J Virol (2013) 87:12531–40. doi:10.1128/JVI.01889-13
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
88. Wang W, Castelán-Vega JA, Jiménez-Alberto A, Vassell R, Ye Z, Weiss CD. A mutation in the receptor binding site enhances infectivity of 2009 H1N1 influenza hemagglutinin pseudotypes without changing antigenicity. Virology (2010) 407:374–80. doi:10.1016/j.virol.2010.08.027
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
89. Wang W, Xie H, Ye Z, Vassell R, Weiss CD. Characterization of lentiviral pseudotypes with influenza H5N1 hemagglutinin and their performance in neutralization assays. J Virol Methods (2010) 165:305–10. doi:10.1016/j.jviromet.2010.02.009
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
90. Rossman JS, Xianghong J, Leser GP, Lamb RA. Influenza virus M2 protein mediates ESCRT-independent membrane scission. Cell (2011) 142:902–13. doi:10.1016/j.cell.2010.08.029
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
91. Böttcher E, Matrosovich T, Beyerle M, Klenk H-D, Garten W, Matrosovich M. Proteolytic activation of influenza viruses by serine proteases TMPRSS2 and HAT from human airway epithelium. J Virol (2006) 80:9896–8. doi:10.1128/JVI.01118-06
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
92. Chaipan C, Kobasa D, Bertram S, Glowacka I, Steffen I, Tsegaye TS, et al. Proteolytic activation of the 1918 influenza virus hemagglutinin. J Virol (2009) 83:3200–11. doi:10.1128/JVI.02205-08
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
93. O’Donnell CD, Wright A, Vogel LN, Wei C-J, Nabel GJ, Subbarao K. Effect of priming with H1N1 influenza viruses of variable antigenic distances on challenge with 2009 pandemic H1N1 virus. J Virol (2012) 86:8625–33. doi:10.1128/JVI.00147-12
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
94. Molesti E, Cattoli G, Ferrara F, Böttcher-Friebertshäuser E, Terregino C, Temperton NJ. The production and development of H7 Influenza virus pseudotypes for the study of humoral responses against avian viruses. J Mol Genet Med (2013) 7:315–20.
95. Wei C, Boyington JC, Dai K, Houser KV, Melissa B, Kong W, et al. Cross-neutralization of 1918 and 2009 influenza viruses: role of glycans in viral evolution and Va. Sci Transl Med (2011) 2:1–16. doi:10.1126/scitranslmed.3000799
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
96. Mallajosyula VVA, Citron M, Ferrara F, Lu X, Callahan C, Heidecker GJ, et al. Influenza hemagglutinin stem-fragment immunogen elicits broadly neutralizing antibodies and confers heterologous protection. Proc Natl Acad Sci U S A (2014) 111:1–10. doi:10.1073/pnas.1402766111
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
97. Bertram S, Glowacka I, Blazejewska P, Soilleux E, Allen P, Danisch S, et al. TMPRSS2 and TMPRSS4 facilitate trypsin-independent spread of influenza virus in Caco-2 cells. J Virol (2010) 84:10016–25. doi:10.1128/JVI.00239-10
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
98. Yang J, Li W, Long Y, Song S, Liu J, Zhang X, et al. Reliability of pseudotyped influenza viral particles in neutralizing antibody detection. PLoS One (2014) 9:e113629. doi:10.1371/journal.pone.0113629
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
99. Kostka V, Carpenter FH. Inhibition of chymotrypsin activity in crystalline trypsin preparations. J Biol Chem (1964) 239:1799–803.
100. Walsh KA, Neurath H. Trypsinogen and chymotrypsinogen as homologous proteins. Proc Natl Acad Sci U S A (1964) 52:884–9. doi:10.1073/pnas.52.4.884
101. Zhang Y, Lin X, Zhang F, Wu J, Tan W, Bi S, et al. Hemagglutinin and neuraminidase matching patterns of two influenza A virus strains related to the 1918 and 2009 global pandemics. Biochem Biophys Res Commun (2009) 387:405–8. doi:10.1016/j.bbrc.2009.07.040
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
102. Garten W, Klenk HD. Cleavage activation of the influenza virus hemagglutinin and its role in Pathogenesis. In: Klenk H-D, Matrosovich MN, Stech J, editors. Avian Influenza (Monographs in Virology). Basel: Karger (2008). p. 156–67. doi:10.1159/000151618
103. Nefkens I, Garcia J-M, Ling CS, Lagarde N, Nicholls J, Tang DJ, et al. Hemagglutinin pseudotyped lentiviral particles: characterization of a new method for avian H5N1 influenza sero-diagnosis. J Clin Virol (2007) 39:27–33. doi:10.1016/j.jcv.2007.02.005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
104. Li Z, Liu Z, Ma C, Zhang L, Su Y, Gao GF, et al. Identification of amino acids in highly pathogenic avian influenza H5N1 virus hemagglutinin that determine avian influenza species specificity. Arch Virol (2011) 156:1803–12. doi:10.1007/s00705-011-1056-2
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
105. Alvarado-Facundo E, Gao Y, Ribas-Aparicio RM, Jiménez-Alberto A, Weiss CD, Wang W. Influenza virus M2 protein ion channel activity helps to maintain pandemic 2009 H1N1 virus hemagglutinin fusion competence during transport to the cell surface. J Virol (2014) 89:1975–85. doi:10.1128/JVI.03253-14
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
106. Pear WS, Nolan GP, Scott ML, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci U S A (1993) 90:8392–6. doi:10.1073/pnas.90.18.8392
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
107. Qiu C, Huang Y, Zhang A, Tian D, Wan Y, Zhang X, et al. Safe pseudovirus-based assay for neutralization antibodies against influenza A(H7N9) virus. Emerg Infect Dis (2013) 19:1685–7. doi:10.3201/eid1910.130728
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
108. Szécsi J, Drury R, Josserand V, Grange MP, Boson B, Hartl I, et al. Targeted retroviral vectors displaying a cleavage site-engineered hemagglutinin (HA) through HA-protease interactions. Mol Ther (2006) 14:735–44. doi:10.1016/j.ymthe.2006.04.007
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
109. Du N, Zhou J, Lin X, Zhang Y, Yang X, Wang Y, et al. Differential activation of NK cells by influenza A pseudotype H5N1 and 1918 and 2009 pandemic H1N1 viruses. J Virol (2010) 84:7822–31. doi:10.1128/JVI.00069-10
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
110. Loyter A, Scangos GA, Ruddle FH. Mechanisms of DNA uptake by mammalian cells: fate of exogenously added DNA monitored by the use of fluorescent dyes. Proc Natl Acad Sci U S A (1982) 79:422–6. doi:10.1073/pnas.79.2.422
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
111. Graham FL, van der Eb AJ. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology (1973) 52:456–67. doi:10.1016/0042-6822(73)90341-3
112. Boussif O, Lezoualc’h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, et al. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc Natl Acad Sci U S A (1995) 92:7297–301. doi:10.1073/pnas.92.16.7297
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
113. Zhao R, Cui S, Guo L, Wu C, Gonzalez R, Paranhos-Baccalà G, et al. Identification of a highly conserved H1 subtype-specific epitope with diagnostic potential in the hemagglutinin protein of influenza A virus. PLoS One (2011) 6:e23374. doi:10.1371/journal.pone.0023374
114. Felgner PL, Gadek TR, Holm M, Roman R, Chan HW, Wenz M, et al. Lipofection: a highly efficient, lipid-mediated DNA-transfection procedure. Proc Natl Acad Sci U S A (1987) 84:7413–7. doi:10.1073/pnas.84.21.7413
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
115. Basu A, Antanasijevic A, Wang M, Li B, Mills DM, Ames JA, et al. New small molecule entry inhibitors targeting hemagglutinin-mediated influenza A virus fusion. J Virol (2013) 88:1447–60. doi:10.1128/JVI.01225-13
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
116. Yamano S, Dai J, Moursi AM. Comparison of transfection efficiency of nonviral gene transfer reagents. Mol Biotechnol (2010) 46:287–300. doi:10.1007/s12033-010-9302-5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
117. Arnold AS, Laporte V, Dumont S, Appert-Collin A, Erbacher P, Coupin G, et al. Comparing reagents for efficient transfection of human primary myoblasts: FuGENE 6, effectene and ExGen 500. Fundam Clin Pharmacol (2006) 20:81–9. doi:10.1111/j.1472-8206.2005.00344.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
118. Patel M, Giddings AM, Sechelski J, Olsen JC. High efficiency gene transfer to airways of mice using influenza hemagglutinin pseudotyped lentiviral vectors. J Gene Med (2013) 15:51–62. doi:10.1002/jgm.2695
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
119. Su CY, Wang SY, Shie JJ, Jeng KS, Temperton NJ, Fang JM, et al. In vitro evaluation of neuraminidase inhibitors using the neuraminidase-dependent release assay of hemagglutinin-pseudotyped viruses. Antiviral Res (2008) 79:199–205. doi:10.1016/j.antiviral.2008.03.002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
120. Molesti E, Wright E, Terregino C, Rahman R, Cattoli G, Temperton NJ. Multiplex evaluation of influenza neutralizing antibodies with potential applicability to in-field serological studies. J Immunol Res (2014) 2014:457932. doi:10.1155/2014/457932
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
121. Mather ST, Wright E, Scott SD, Temperton NJ. Lyophilisation of influenza, rabies and Marburg lentiviral pseudotype viruses for the development and distribution of a neutralisation -assay-based diagnostic kit. J Virol Methods (2014) 210:51–8. doi:10.1016/j.jviromet.2014.09.021
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
122. Zhang Y, Lin X, Wang G, Zhou J, Lu J, Zhao H, et al. Neuraminidase and hemagglutinin matching patterns of a highly pathogenic avian and two pandemic H1N1 influenza A viruses. PLoS One (2010) 5:e9167. doi:10.1371/journal.pone.0009167
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
123. Schmeisser F, Friedman R, Besho J, Lugovtsev V, Soto J, Wang W, et al. Neutralizing and protective epitopes of the 2009 pandemic influenza H1N1 hemagglutinin. Influenza Other Respi Viruses (2013) 7:480–90. doi:10.1111/irv.12029
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
124. Bosch V, Kramer B, Pfeiffer T, Stärck L, Steinhauer DA. Inhibition of release of lentivirus particles with incorporated human influenza virus haemagglutinin by binding to sialic acid-containing cellular receptors. J Gen Virol (2001) 82:2485–94.
125. Wang H, Ma C, Lu Y, Ji X, Pang Y, Hua F, et al. Generation of human neutralizing monoclonal antibodies against the 2009 pandemic H1N1 virus from peripheral blood memory B lymphocytes. Cell Mol Immunol (2013) 10:1–10. doi:10.1038/cmi.2013.25
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
126. Whittle JRR, Wheatley AK, Wu L, Lingwood D, Kanekiyo M, Ma SS, et al. Flow cytometry reveals that H5N1 vaccination elicits cross-reactive stem-directed antibodies from multiple Ig heavy-chain lineages. J Virol (2014) 88:4047–57. doi:10.1128/JVI.03422-13
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
127. Hultberg A, Temperton NJ, Rosseels V, Koenders M, Gonzalez-Pajuelo M, Schepens B, et al. Llama-derived single domain antibodies to build multivalent, superpotent and broadened neutralizing anti-viral molecules. PLoS One (2011) 6:e17665. doi:10.1371/journal.pone.0017665
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
128. Yang D-G, Chung Y-C, Lai Y-K, Lai C-W, Liu H-J, Hu Y-C. Avian influenza virus hemagglutinin display on baculovirus envelope: cytoplasmic domain affects virus properties and vaccine potential. Mol Ther (2007) 15:989–96. doi:10.1038/sj.mt.6300236
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
129. Ledgerwood JE, Hu Z, Gordon IJ, Yamshchikov G, Enama ME, Plummer S, et al. Influenza virus H5 DNA vaccination is immunogenic by intramuscular and intradermal routes in humans. Clin Vaccine Immunol (2012) 19:1792–7. doi:10.1128/CVI.05663-11
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
130. Ledgerwood JE, Wei CJ, Hu Z, Gordon IJ, Enama ME, Hendel CS, et al. DNA priming and influenza vaccine immunogenicity: two phase 1 open label randomised clinical trials. Lancet Infect Dis (2011) 11:916–24. doi:10.1016/S1473-3099(11)70240-7
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
131. Lin X, Zhou J, Zhang Y, Wu J, Zhang F, Li Z, et al. Oseltamivir boosts 2009 H1N1 virus infectivity in vitro. Biochem Biophys Res Commun (2009) 390:1305–8. doi:10.1016/j.bbrc.2009.10.142
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
132. Rao S, Kong WP, Wei CJ, Yang ZY, Nason M, Styles D, et al. Multivalent HA DNA vaccination protects against highly pathogenic H5N1 avian influenza infection in chickens and mice. PLoS One (2008) 3:e2432. doi:10.1371/journal.pone.0002432
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
133. Garcia J-M, Lagarde N, Ma ESK, de Jong MD, Peiris JSM. Optimization and evaluation of an influenza A (H5) pseudotyped lentiviral particle-based serological assay. J Clin Virol (2010) 47:29–33. doi:10.1016/j.jcv.2009.10.009
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
134. Wang J, Cheng H, Ratia K, Varhegyi E, Hendrickson WG, Li J, et al. A comparative high-throughput screening protocol to identify entry inhibitors of enveloped viruses. J Biomol Screen (2013) 19:100–7. doi:10.1177/1087057113494405
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
135. Sandrin V, Cosset FL. Intracellular versus cell surface assembly of retroviral pseudotypes is determined by the cellular localization of the viral glycoprotein, its capacity to interact with Gag, and the expression of the Nef protein. J Biol Chem (2006) 281:528–42. doi:10.1074/jbc.M506070200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
136. Shelton H, Roberts KL, Molesti E, Temperton NJ, Barclay WS. Mutations in haemagglutinin that affect receptor binding and pH stability increase replication of a PR8 influenza virus with H5 HA in the upper respiratory tract of ferrets and may contribute to transmissibility. J Gen Virol (2013) 94:1220–9. doi:10.1099/vir.0.050526-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
137. Alberini I, Del Tordello E, Fasolo A, Temperton NJ, Galli G, Gentile C, et al. Pseudoparticle neutralization is a reliable assay to measure immunity and cross-reactivity to H5N1 influenza viruses. Vaccine (2009) 27:5998–6003. doi:10.1016/j.vaccine.2009.07.079
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
138. Mahallawi WH, Kasbekar AV, McCormick MS, Hoschler K, Temperton NJ, Leong SC, et al. Infection with 2009 H1N1 influenza virus primes for immunological memory in human nose-associated lymphoid tissue, offering cross-reactive immunity to H1N1 and avian H5N1 viruses. J Virol (2013) 87:5331–9. doi:10.1128/JVI.03547-12
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
139. Molesti E, Milani A, Terregino C, Cattoli G, Temperton NJ. Comparative serological assays for the study of H5 and H7 avian influenza viruses. Influenza Res Treat (2013) 2013:286158. doi:10.1155/2013/286158
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
140. Sui J, Hwang W, Perez S, Wei G, Aird D, Chen L, et al. Strutural and Functional bases for broad spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol (2009) 16:265–73. doi:10.1038/nsmb.1566
141. Kanekiyo M, Wei C-J, Yassine HM, McTamney PM, Boyington JC, Whittle JRR, et al. Self-assembling influenza nanoparticle vaccines elicit broadly neutralizing H1N1 antibodies. Nature (2013) 499:102–6. doi:10.1038/nature12202
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
142. Gorres JP, Lager KM, Kong WP, Royals M, Todd JP, Vincent AL, et al. DNA vaccination elicits protective immune responses against pandemic and classic swine influenza viruses in pigs. Clin Vaccine Immunol (2011) 18:1987–95. doi:10.1128/CVI.05171-11
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
143. Oh H-LJ, Akerström S, Shen S, Bereczky S, Karlberg H, Klingström J, et al. An antibody against a novel and conserved epitope in the hemagglutinin 1 subunit neutralizes numerous H5N1 influenza viruses. J Virol (2010) 84:8275–86. doi:10.1128/JVI.02593-09
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
144. Edwards MJ, Dimmock NJ. Hemagglutinin 1-specific immunoglobulin G and Fab molecules mediate postattachment neutralization of influenza A virus by inhibition of an early fusion event. J Virol (2001) 75:10208–18. doi:10.1128/JVI.75.21.10208-10218.2001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
145. Corti D, Voss J, Gamblin SJ, Codoni G, Macagno A, Jarrossay D, et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science (2011) 333:850–6. doi:10.1126/science.1205669
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
146. Shahsavandi S, Salmanian AH, Ghorashi SA, Masoudi S, Fotouhi F, Ebrahimi MM. Specific subtyping of influenza A virus using a recombinant hemagglutinin protein expressed in baculovirus. Mol Biol Rep (2011) 38:3293–8. doi:10.1007/s11033-010-0434-2
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
147. Lee C, Senne DA, Suarez DL. Development and application of reference antisera against 15 hemagglutinin subtypes of influenza virus by DNA vaccination of chickens. Clin Vaccine Immunol (2006) 13:395–402. doi:10.1128/CVI.13.3.395-402.2006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
148. Buchy P, Vong S, Chu S, Garcia J-M, Hien TT, Hien VM, et al. Kinetics of neutralizing antibodies in patients naturally infected by H5N1 virus. PLoS One (2010) 5:e10864. doi:10.1371/journal.pone.0010864
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
149. Garcia J-M, Pepin S, Lagarde N, Ma ESK, Vogel FR, Chan KH, et al. Heterosubtype neutralizing responses to influenza A (H5N1) viruses are mediated by antibodies to virus haemagglutinin. PLoS One (2009) 4:e7918. doi:10.1371/journal.pone.0007918
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
150. Stephenson I, Heath A, Major D, Newman RW, Hoschler K, Junzi W, et al. Reproducibility of serologic assays for influenza virus A (H5N1). Emerg Infect Dis (2009) 15:1252–9. doi:10.3201/eid1508.081754
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
151. Ekiert DC, Bhabha G, Elsliger M, Friesen RHE, Throsby M, Goudsmit J, et al. Antibody recognition of a highly conserved influenza virus epitope : implications for universal prevention and therapy. Science (2010) 324:246–51. doi:10.1126/science.1171491
152. Throsby M, van den Brink E, Jongeneelen M, Poon LLM, Alard P, Cornelissen L, et al. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS One (2008) 3:e3942. doi:10.1371/journal.pone.0003942
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
153. Okuno Y, Matsumoto K, Isegawa Y, Ueda S. Protection against the mouse-adapted A/FM/1/47 strain of influenza A virus in mice by a monoclonal antibody with cross-neutralizing activity among H1 and H2 strains. J Virol (1994) 68:517–20.
154. Ekiert DC, Friesen RH, Bhabha G, Kwaks T, Jongeneelen M, Yu W, et al. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science (2012) 333:843–50. doi:10.1126/science.1204839
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
155. Wrammert J, Koutsonanos D, Li G-M, Edupuganti S, Sui J, Morrissey M, et al. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J Exp Med (2011) 208:181–93. doi:10.1084/jem.20101352
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
156. Corti D, Suguitan AL, Pinna D, Silacci C, Fernandez-Rodriguez BM, Vanzetta F, et al. Heterosubtypic neutralizing antibodies are produced by individuals immunized with a seasonal influenza vaccine. J Clin Invest (2010) 120:1663–73. doi:10.1172/JCI41902
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
157. Wei C-J, Boyington JC, McTamney PM, Kong W-P, Pearce MB, Xu L, et al. Induction of broadly neutralizing H1N1 influenza antibodies by vaccination. Science (2010) 329:1060–4. doi:10.1126/science.1192517
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
158. Sui J, Sheehan J, Hwang WC, Bankston LA, Burchett SK, Huang CY, et al. Wide prevalence of heterosubtypic broadly neutralizing human anti-influenza a antibodies. Clin Infect Dis (2011) 52:1003–9. doi:10.1093/cid/cir121
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
159. Krammer F, Hai R, Yondola M, Tan GS, Leyva-Grado VH, Ryder AB, et al. Assessment of influenza virus hemagglutinin stalk-based immunity in ferrets. J Virol (2014) 88:3432–42. doi:10.1128/JVI.03004-13
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
160. Krammer F, Margine I, Hai R, Flood A, Hirsh A, Tsvetnitsky V, et al. H3 stalk-based chimeric hemagglutinin influenza virus constructs protect mice from H7N9 challenge. J Virol (2013) 88:2340–3. doi:10.1128/JVI.03183-13
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
161. Krammer F, Pica N, Hai R, Margine I, Palese P. Chimeric hemagglutinin influenza virus vaccine constructs elicit broadly protective stalk-specific antibodies. J Virol (2013) 87:6542–50. doi:10.1128/JVI.00641-13
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
162. Fleury D, Wharton SA, Skehel JJ, Knossow M, Bizebard T. Antigen distortion allows influenza virus to escape neutralization. Nat Struct Biol (1998) 5:119–23. doi:10.1038/nsb0298-119
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
163. Ferrara F, Molesti E, Temperton NJ. The application of pseudotypes to influenza pandemic preparedness. Future Virol (2015).
164. Ostrowski LE, Yin W, Patel M, Sechelski J, Rogers T, Burns K, et al. Restoring ciliary function to differentiated primary ciliary dyskinesia cells with a lentiviral vector. Gene Ther (2014) 21:253–61. doi:10.1038/gt.2013.79
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
165. Wu Q, Fang L, Wu X, Li B, Luo R, Yu Z, et al. A pseudotype baculovirus-mediated vaccine confers protective immunity against lethal challenge with H5N1 avian influenza virus in mice and chickens. Mol Immunol (2009) 46:2210–7. doi:10.1016/j.molimm.2009.04.017
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
166. Hofmann C, Sandig V, Jennings G, Rudolph M, Schlag P, Strauss M. Efficient gene transfer into human hepatocytes by baculovirus vectors. Proc Natl Acad Sci U S A (1995) 92:10099–103. doi:10.1073/pnas.92.22.10099
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
167. Coughlan L, Mullarkey C, Gilbert S. Adenoviral vectors as novel vaccines for influenza. J Pharm Pharmacol (2015) 67(3):382–99. doi:10.1111/jphp.12350
168. Szécsi J, Boson B, Johnsson P, Dupeyrot-Lacas P, Matrosovich M, Klenk H-D, et al. Induction of neutralising antibodies by virus-like particles harbouring surface proteins from highly pathogenic H5N1 and H7N1 influenza viruses. Virol J (2006) 3:70. doi:10.1186/1743-422X-3-70
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
169. Banasik MB, McCray PB. Integrase-defective lentiviral vectors: progress and applications. Gene Ther (2010) 17:150–7. doi:10.1038/gt.2009.135
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: influenza, hemagglutinin, pseudotype, neutralization assay, universal vaccine, lentiviral vector, retroviral vector
Citation: Carnell GW, Ferrara F, Grehan K, Thompson CP and Temperton NJ (2015) Pseudotype-based neutralization assays for influenza: a systematic analysis. Front. Immunol. 6:161. doi: 10.3389/fimmu.2015.00161
Received: 28 February 2015; Paper pending published: 20 March 2015;
Accepted: 25 March 2015; Published online: 29 April 2015.
Edited by:
Arun Kumar, GlaxoSmithKline Vaccines, ItalyReviewed by:
Yasu Takeuchi, University College London, UKGhazi Kayali, St. Jude Children’s Research Hospital, USA
Copyright: © 2015 Carnell, Ferrara, Grehan, Thompson and Temperton. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nigel James Temperton, Viral Pseudotype Unit, Medway School of Pharmacy, Chatham Maritime, Kent ME4 4TB, UK e-mail:bi50ZW1wZXJ0b25Aa2VudC5hYy51aw==
 George William Carnell
George William Carnell Francesca Ferrara
Francesca Ferrara Keith Grehan
Keith Grehan Craig Peter Thompson
Craig Peter Thompson Nigel James Temperton
Nigel James Temperton