- Immunomodulation Research Group, School of Biotechnology, Dublin City University, Dublin, Ireland
Dendritic cells (DCs) provide an essential link between innate and adaptive immunity. At the site of infection, antigens recognized by DCs via pattern-recognition receptors, such as Toll-like receptors (TLRs), initiate a specific immune response. Depending on the nature of the antigen, DCs secrete distinct cytokines with which they orchestrate homeostasis and pathogen clearance. Dysregulation of this process can lead to unnecessary inflammation, which can result in a plethora of inflammatory diseases. Therefore, the secretion of cytokines from DCs is tightly regulated and this regulation is facilitated by highly conserved trafficking protein families. These proteins control the transport of vesicles from the Golgi complex to the cell surface and between organelles. In this review, we will discuss the role of soluble n-ethylmaleimide-sensitive factor attachment protein receptor proteins (SNAREs) in DCs, both as facilitators of secretion and as useful tools to determine the pathways of secretion through their definite locations within the cells and inherent specificity in opposing binding partners on vesicles and target membranes. The role of SNAREs in DC function may present an opportunity to explore these proteins as novel targets in inflammatory disease.
Introduction
Dendritic cells (DCs) were first described by Ralph Steinman and Zanvil Cohn as a rare murine splenocyte with dendrite-like protrusions that displayed phagocytic abilities (1). Following their discovery, work carried out by Steinman’s group demonstrated that DCs play a vital role in orchestrating immunity. In their immature state in the peripheral tissue, DCs are characterized by a high capacity for antigen capture and processing via phagocytosis. They display distinct chemokine receptor expression, chemokine responsiveness, and low T cell stimulatory capabilities (2). The ability of DCs to regulate immunity is dependent on their maturation, which is induced by the interaction of DCs with antigens that are subsequently presented to T cells. During this process, DCs secrete distinct profiles of cytokines and chemokines and express co-stimulatory molecules required to drive specific T cell responses (3). The control of these secretory pathways is an essential factor in regulating innate and adaptive immune homeostasis and controlling responses to infection (4). In this review, we will discuss the factors that induce DC maturation, the cytokines secreted by DCs that drive adaptive immunity, and the secretory pathways that have been identified and their potential as immunotherapeutic targets.
Dendritic Cell Activation
For maturation to occur, foreign antigens must be identified by DCs. DCs express pattern recognition receptors (PRRs) that allow them to recognize microbes that display pathogen-associated molecular patterns (PAMPs). PRRs can be divided into toll-like receptors (TLRs), C-type lectin receptors (CLRs), nucleotide-binding domain and leucine-rich repeat containing receptors (NLRs) and retinoic acid-inducible gene-I (RIG)-like receptors (RLRs) (5). Following engagement of these PRRs, signaling cascades are activated that induce the production of inflammatory cytokines (6). Inflammatory responses can also be triggered by endogenous damage-associated molecular patterns (DAMPs). These are intracellular markers that are also recognized by PRRs, and are released during tissue damage or when a cell is necrotic releasing intracellular material into its surroundings (7). Following PRR ligation and subsequent maturation, DCs become antigen presenting cells (APC) (8). The mature DC travels to the local lymph node (9), which facilitates the presentation of antigen to antigen-specific lymphocytes to prime naive T cells to become effector T cells. Three signals are required for this: antigen presentation, up-regulation of co-stimulatory markers, and secretion of cytokines; this review will focus on the third signal, cytokine secretion and their effects (10).
Cytokine Polarization of T Cell Subsets
Clonal immunity involves DC activation of T cells through presentation of processed antigen to naïve T cells in an immunogenic form and the secretion of immune mediators called cytokines. Cytokines are classified into interleukins (IL), interferons (IFNs), and colony-stimulating factors. Cytokine profiles secreted from DCs polarize naïve T cells into different phenotypes of CD4+ T cells. These differential cytokine secretion patterns are dependent on the PRR activation, and subsequently are the key signals required for the differentiation into different T helper (Th) cell phenotype subsets that are required to clear the antigen that trigger the immune response (11).
In order to clear bacterial and viral infections, naïve CD4+ T-cells require cytokine signaling from DCs to initiate differentiation into a Th1 phenotype. TLRs present on DCs recognize PAMPs, such as lipopolysaccharide and unmethylated dsDNA, through TLR4 and TLR9 respectively. Engagement of these TLRs results in secretion of the cytokines IL-1, IL-6, TNF-α, and IL-12p40 (10). Dimerization of IL-12p40 with IL-12p35 results in an active IL-12p70 heterodimer. IL-12p70 together with IFN-γ, which is produced in large amounts by activated T-cells, leads to differentiation of T-cells in to an IFN-γ producing Th1 cells subset (12). Th2 cells produce high levels of IL-4, IL-5, and IL-13. They are responsible for initiating the humoral immune response and are required to clear helminth infections. Th2 cells have been generated in vitro via stimulation of the TCR and the addition of IL-4 (13). IL-4 induces the induction of Th2 cells via STAT6, given the fact that DCs do not produce IL-4, the mechanisms through which Th2 cells initiate this subset remain unclear. However, cytokine signaling from DCs can indirectly affect the differentiation. DCs activated through TLR2, DC-SIGN, and OSCAR (an FcγR-associated receptor) lead to the activation of Erk1 and Erk2 pathways and phosphorylation of c-Fos (14). This leads to the inhibition of IL-12 and increased production of IL-10 in DCs, which is sufficient to induce a strong Th2 response in naïve T-cells. Cytokines produced by other cells can also influence DCs to drive a Th2 subset through chemokine production that recruits IL-4 producing cells and the activation of cytokine receptors (11).
Th17 cells were first described in 2006 and identified as a T helper subset distinct from Th1 and Th2 cells that produced the cytokine IL-17 (15). Th17 cells have been indicated to confer protection against extracellular bacteria and fungi by the secretion of effector molecules such as IL-17, IL-21, IL-22, GM-CSF, and CCL20 (16). However, Th17 cells have also been implicated in many autoimmune and inflammatory diseases, such as rheumatoid arthritis (RA), systemic lupus erythematosus, multiple sclerosis, psoriasis, and inflammatory bowel disease (IBD) (17). The conditions for differentiation of Th17 cells is more complex than Th1 and Th2 subsets, and there is evidence to support a role for a number of cytokines produced by DCs, including IL-1, IL-18, IL-6, and IL-23. These cytokines are produced by DCs in response to PRR recognition of bacteria and fungi.
In the case of Th17 differentiation, it is not just TLR-mediated activation that promotes the secretion of these cytokines. Dectin-1 and dectin-2 activate CLRs on DCs to promote section of IL-6 and IL-23, cytokines that play a major role in differentiating and stabilizing the Th17 response respectively (18). T-regulatory cells are essential for negative regulation of the immune response, as they possess immunosuppressive abilities that confer protection to the host from autoimmunity and prevention of excessive immunopathology. If this response becomes dysregulated, it can lead to autoimmune disorders due to loss of homeostasis. IL-27, IL-10, and TGF-β1 secretion by DCs have been heavily indicated in the induction of Treg cells (19).
Targeting Cytokines in Inflammatory Disease
Cytokines are essential in driving the adaptive immune response; thus, targeting cytokines associated with inflammatory disease has become a significant area for drug development (20). Indeed, secretory pathways are attractive to study as potential targets to block critical cytokines with the possibility of switching off inflammation without any detrimental side effects (21). A number of therapeutics currently exist that target key cytokines such as IL-23 and IL-6, which are associated with inflammatory disease.
IL-23, essential for stabilizing Th17 cells, has been implicated in the pathogenesis of multiple sclerosis, arthritis, and IBD. Using mouse models of diseases, the essential role of IL-23 has been established (16, 22, 23). A human monoclonal antibody, Ustekinumab, has been developed and licensed to target the IL-12/23 pathway. This antibody targets the p40 subunit of IL-12 and IL-23, and was originally developed to target IL-12 induction of Th1 cells, as IL-23 had still not yet been identified or recognized in its function of driving Th17 differentiation. These applications include psoriatic arthritis, multiple sclerosis, and Crohn’s disease, and the drug has displayed good efficacy except in the case of multiple sclerosis (24).
IL-6 has also been implicated as an important cytokine in driving the Th17 subset. Aberrant expression of IL-6 has been indicated in inflammatory diseases such as RA and juvenile idiopathic arthritis (JIA), leading to the development of a humanized IL-6 receptor (25). Tocilizumab blocks IL-6 receptor in both soluble and membrane bound forms with the result of blocking IL-6 function. In the clinic, tocilizumab has demonstrated amelioration of inflammatory diseases such as RA and JIA with improvement in symptoms such as joint destruction and a reduction in the number of flare incidents in JIA (26).
Secretion of Immune Mediators
It is clear that DC cytokine release is essential and tailored to clearing specific infections while also controlling immune homeostasis. Regulation of protein trafficking and release of immune mediators such as cytokines, chemokines, and lysosomal enzymes into the immediate environment from DCs is therefore highly regulated in order to prevent autoimmunity and inflammation. Indeed, a number of mutations or deficiencies resulting in dysregulated trafficking can have major implications in immune syndromes (27).
Depending on the required function of an immune mediator, different pathways exist to transport newly synthesized proteins out of the cell. These pathways are usually shared by a number of factors and their release can be constant (constitutive release) or triggered (regulated release) (4). Proteins produced in the endoplasmic reticulum (ER) can be soluble and contained within vesicles or membrane bound and associated within the plasma membrane of transported vesicles.
Thus, a number of proteins share transport to the plasma membrane to conserve energy (27). The release of these proteins can be basal or dependent on their transcription at gene level in response to cellular signaling (28).
Snare Proteins
The transport of vesicles between organelles, endosomes, and the cell surface plasma membrane is controlled by a complex network of cellular machinery of membrane associated proteins, lipids, and cytoplasmic proteins. Soluble-N-ethylmaleimide–sensitive factor accessory protein receptor proteins (SNAREs) are a large conserved family identified in the docking/fusion machinery that allow membranes to overcome opposing forces and fuse docked lipid bilayers (27).
SNARE proteins are all identified by a common conserved SNARE motif of 60–70 amino acids arranged in heptad repeats typical for coiled coils (29). Most SNAREs are type II integral membrane proteins that possess a single trans-membrane domain at their carboxyl (C-terminal) end that is connected to the SNARE motif by a flexible linker, with the majority of the protein extending into the cytoplasm (30). The conserved coiled-coil SNARE motif is necessary for complex formation and membrane fusion (31). When SNAREs form in a tight helical bundle, they form a conserved structure of an elongated coiled coil of four intertwined α-helices that correspond to the four interacting SNARE motifs occupying specific positions in the complex (32). There is a highly conserved layer of interacting amino acids in the central hydrophobic core of the helical bundle consisting of three glutamines and one arginine (31). Each of these contributing motifs contributes an amino acid and their structures are divided into sub-families used to further classify the SNARES as R-SNAREs or Q-SNARES (33).
R-SNARES are usually found on the membrane and have arginine (R) as the central functional residue in the SNARE motif. Q-SNARES are mainly found on the target membrane and are defined by a central glutamine (Q) residue. In order to form a complex and mediate membrane fusion, one R-SNARE and two or three Q-SNARE motifs are required to form a stable RQabc four helical bundle (30, 34). The SNARE proteins on opposing membranes interact to form the α-helix bundle that “zippers” together from the N-terminus to the C-terminus forming a trans-SNARE complex (or SNAREpin). The energy provided by the zippering complex formation pulls the membranes together displacing the water and hydrostatic pressure keeping them apart, thereby facilitating the mixing of hydrophobic lipids (35). Once the membranes have fused, the trans-SNARE conformation is converted to a highly stable inert cis-SNARE conformation on the resulting fused membrane. In order to dissociate this stable structure, metabolic energy is required and this is provided by two chaperone proteins; N-ethylmaleimide-sensitive factor (NSF), and Soluble NSF attachment protein (SNAP) (36, 37). This dissociation facilitates the recycling of SNARE proteins and prevents accumulation of SNAREs on the membrane. Many SNAREs operate predominantly in a specific subcellular compartment with their partners and some Qa SNAREs such as Syntaxin (STX)2 and STX4 are found on the plasma membrane and involved in fusion of vesicles and secretion of their contents from the cell. Other Qa SNAREs including STX5 and the R-SNARE vesicle-associated membrane protein (VAMP)4 are localized in the Golgi apparatus (38). These SNAREs are therefore found in the membranes of the donor compartment, acceptor compartment, and the intermediate trafficking vesicles (39).
The Role of SNAREs in Dendritic Cells
As the immune system consists of a plethora of cells that have specific functions, an understanding of the mechanism of trafficking is extremely important for when these functions become dysregulated and lead to disease. Cytokine signaling from DCs, in particular, is one of the most important types of signaling for the regulation of the adaptive immune response. However, while the role of SNAREs in secretion of key immune mediators has been addressed in many immune cells, the role of SNAREs in DCs function is poorly understood. Examples of SNAREs regulating critical function in other immune cells include STX11 which has been implicated in neutrophil degranulation and interferon-γ (IFN-γ) secretion from natural killer cells (40). Inhibition of SNAP23 or STX3 abolished chemokine release from mast cells and has been attributed to STX11. (41). While these individual SNAREs and their function have been identified in these cell types, a full elucidation of these trafficking pathways and/or SNARE partners have yet to be identified. However, in the last 10 years, Stow and colleagues have mapped the secretion of TNF-α from macrophages and fully elucidated an original secretion pathway through recycling endosomes that is implicitly linked to their phagocytic activity (42–44).
Furthermore, loss of STX11 in macrophages resulted in increased phagocytosis and TNF-α secretion, suggesting that this SNARE plays its part as a negative regulator (45). This hypothesis is supported in patients with the hyper-inflammatory disease, familial hemophagocytic lymphohistiocytosis type 4 (FHL-4), which results from a deletion or a mutation in STX11 (46). These patients possess high levels of pro-inflammatory cytokines (IFN-γ, IL-6, TNF-α, and IL-18) and have a higher number of macrophages that become over-activated and have increased phagocytic activity (47, 48). From their findings in macrophage, Offenhäuser et al. (45) accredited STX11’s regulatory ability due to it how it binds and inhibits other SNARE proteins via its conserved SNARE sequence and lack of a trans-membrane domain (45). A role for STX11 in regulation of DCs has also been suggested. STX11 was examined in Bone marrow-derived dendritic cells (BMDCs) by D’Orlando et al. (40), and it was reported that although mRNA levels were up-regulated in BMDCs, STX11 deficiency did not affect BMDC activities or TLR-induced secretion of IL-12, IL-6, or TNF-α (41). Work by our own group, in a paper published in this issue of Frontiers, shows a correlation of up-regulation of STX11 expression in BMDCs in response to TLR ligands. Further work is required to confirm the role of STX11 in DCs although it is possible that the role of STX11 in phagocytic APCs is conserved (49).
VAMP8 is an R-SNARE that has also been reported to significantly inhibit phagocytosis in DCs (50). In this study, the authors show that VAMP8 associates with lysosome-associated membrane protein-2 (LAMP2) at the phagosome in dendritic cells, following bacterial exposure. Use of VAMP8-/- DCs significantly increased phagocytic ability. Furthermore, over-expression of VAMP8 in a DC2.4 cell line inhibited phagocytosis. Ho et al. also suggest that VAMP7 also negatively regulates phagocytosis in the same manner, as both VAMP7 and VAMP8 can form SNARE complexes with STX7, STX8, and Vti1b (50). This complex has been show to mediate homotypic fusion of early and late endosomes (51).
SNARE expression in DC has been reported in some detail in a paper describing differential expression following treatment with acetylsalicylic acid (ASA), the active ingredient in aspirin. Cai et al. reported that SNARE expression of Vti1a, Vti1b, VAMP3, VAMP8, and STX8 were all up-regulated following treatment with ASA (52). The authors describe a decrease in phagocytosis in response to ASA, and attribute it to the up-regulation of Vti1a and Vti1b on phagosomes, and inhibition of SNARE complex formation. This study also describes the up-regulation of VAMP8 in cells that have decreased phagocytic ability, corroborating the findings by Ho et al. (50). Vti1b can also form a SNARE complex with VAMP 8 in the previously described STX7/STX8/Vti1b SNARE complex identified on endosomal membranes (51).
Cross-presentation is another essential function of DCs that is regulated by SNARE proteins. Sec22b was identified by Cebrian et al. (2011) as a specific regulator of cross-presentation. Sec22b is localized to the ER-Golgi intermediate compartment which interacts with STX4 on the phagosome. In the absence of Sec22b, DCs were unable to transfer antigen from phagosomes to the cytosol and degradation of the antigen in the phagosome increased (53). It was recently shown that while Sec22b-mediated phagosomal recruitment is required for cross presentation, it is independent of TLR signaling and cannot deliver MHC class I (MHC-I). Nair-Gupta et al. have shown that large quantities of MHC-I are stored in the endosomal recycling compartment (ERC) (which possess Rab11a, VAMP3, and VAMP8 on its surface). TLR signaling through the myeloid differentiation (MyD) marker, MyD88, results in phosphorylation of SNAP-23 present on the phagosome. This leads to stabilization of SNARE complexes, fusion of ERC and the phagosome, and ultimately cross-presentation (54). These findings regarding cross-presentation are summarized in Figure 1.
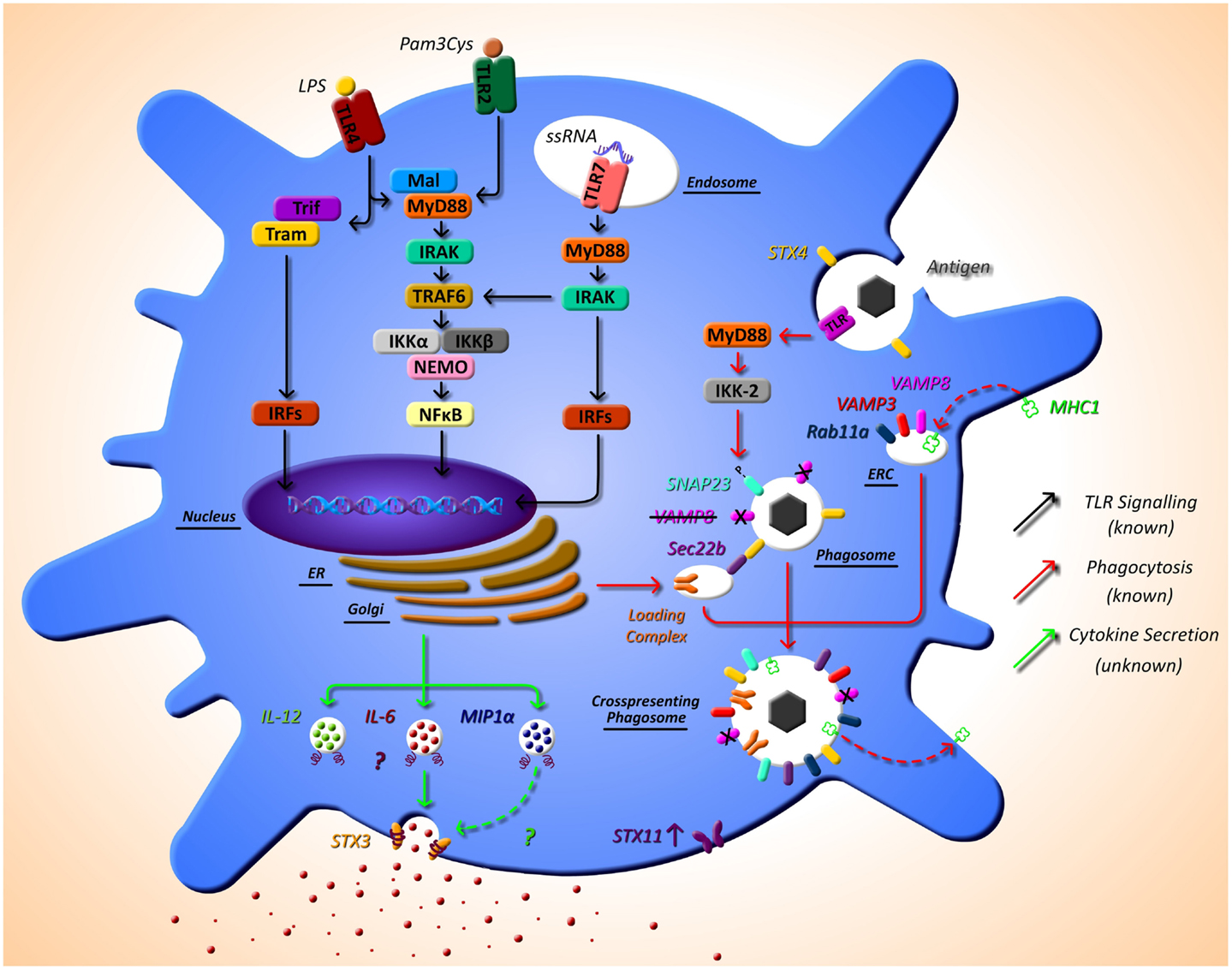
Figure 1. The as yet unknown side of DC secretion post-TLR activation, and trafficking molecules currently identified in DC function. Following TLR activation with TLR ligands, such as LPS, DCs activate intracellular pathways that lead to the transcription of inflammatory mediators, such as cytokines. While this pathway is extremely well defined (black arrows), very little is known of the post-transcriptional pathway and how proteins, in particular cytokines, are trafficked out of the DC. It is evident from other cell types that SNARE proteins play a role in secretion of cytokines; however, to date, their specific role has not yet been described. SNARE importance in DCs has been highlighted by studies that demonstrate that STX4 on the phagosome interacts with Sec22b on the ER-Golgi intermediate compartment, which is required for cross-presentation and MyD88-dependent TLR signaling, and results in phosphorylation of SNAP23 present on the phagosome. This leads to fusion with endosomal recycling compartment (ERC) and ultimately cross-presentation. Furthermore, VAMP8 has been shown to be a negative regulator of phagocytosis (red arrows). Our recent work has indicated a role for STX3 in IL-6 and possibly MIP1-α secretion (green arrows). This work and others also suggests a role for STX11 in DC function as it is highly up-regulated post TLR2, TLR4, and TLR7 activation.
Work in our own group is currently identifying the mechanisms of secretion of key immune mediators involved in inflammatory disease and we have recently identified a role for STX4 in IgE secretion from plasma cells (55). Due to the role of DCs in the direction of adaptive immune responses via secretion of cytokines, we aim to elucidate these pathways in detail in these cells. Another finding from the research paper published in this issue of Frontiers, we have described the effects of different TLR ligands on DCs and measured the cytokine profiles secreted by these cells and their concurrent SNARE expression (49). Specific TLR pathway activation allows DCs to respond as needed to specific challenges. For example, when TLR4 is activated by lipopolysaccharide (LPS), this in turn activates DCs to produce the cytokines necessary to orchestrate an adaptive immune response to clear a bacterial infection, as described above. Depending on the TLR agonist stimulation, different cytokine profiles are secreted by DC, and we demonstrate that these profiles correlate to differential expression of SNAREs. Assessing the secretion of cytokines from JAWSII DC’s following activation with TLR ligands revealed that IL-6 and MIP-1α secretion was significantly increased following stimulation with LPS (TLR4) and Loxoribrine (TLR7) but not PGN (TLR2). This correlated with a change in expression of STX3. Furthermore, knockdown of STX3 using siRNA against STX3 resulted in a significant decrease in IL-6 and MIP-1α secretion with no effect on IL-1β or TNF-α, indicating a role for STX3 in IL-6 secretion. To our knowledge, this is the first study to profile SNARE expression patterns following different TLR stimulations in DCs and demonstrates the correlation between cytokine secretion patterns and expression of SNAREs (49). This is overviewed in Figure 1, indicated by the green arrows.
Conclusion
Identifying specific SNARE complexes and their regulators will lead to a better understanding of DC function during inflammation and may present potential therapeutic targets in a wide range of inflammatory diseases. Current therapeutic monoclonal antibodies target aberrant and subsequent damaging cytokine production by blocking their activities. It is well known that prevention is better than a cure and the current therapeutic strategies do not meet this requirement, highlighting the need for more targeted therapies. Monoclonal antibodies treatments are also prone to off-target effects such as drug induced liver damage and the generation of anti-idiotypic antibodies, rendering them ineffective (56). While a number of combined antibody therapies and personalized medicine methodologies are being investigated, targeting trafficking proteins and pathways to inhibit cytokine release from the cell may prove to be a more effective therapeutic approach.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Steinman RM, Cohn ZA. Identification of a novel cell type in peripheral lymphoid organs of mice: i Morphology, quantitation, tissue distribution. J Exp Med (1973) 137:1142–62. doi: 10.1084/jem.137.5.1142
2. Banchereau J, Steinman RM. Dendritic cells and the control of immunity. Nature (1998) 392:245–52. doi:10.1038/32588
3. Scott K, Manunta M, Germain C, Smith P, Jones M, Mitchell P, et al. Qualitatively distinct patterns of cytokines are released by human dendritic cells in response to different pathogens. Immunology (2005) 116(2):245–54. doi:10.1111/j.1365-2567.2005.02218.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
4. Lacy P, Stow JL. Cytokine release from innate immune cells: association with diverse membrane trafficking pathways. Blood (2011) 118(1):9–18. doi:10.1182/blood-2010-08-265892
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
5. Creagh EM, O’Neill LAJ. TLRs, NLRs and RLRs: a trinity of pathogen sensors that co-operate in innate immunity. Trends Immunol (2006) 27:352–7. doi:10.1016/j.it.2006.06.003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
6. Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell (2004) 118:229–41. doi:10.1016/j.cell.2004.07.002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
7. Zhang Q, Kang R, Zeh HJ, Lotze MT, Tang D. DAMPs and autophagy: cellular adaptation to injury and unscheduled cell death. Autophagy (2013) 9:451–8. doi:10.4161/auto.23691
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
8. Sallusto F, Palermo B, Lenig D, Miettinen M, Matikainen S, Julkunen I, et al. Distinct patterns and kinetics of chemokine production regulate dendritic cell function. Eur J Immunol (1999) 29:1617–25. doi:10.1002/(SICI)1521-4141(199905)29:05<1617::AID-IMMU1617>3.0.CO;2-3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
9. Chan VW, Kothakota S, Rohan MC, Panganiban-Lustan L, Gardner JP, Wachowicz MS, et al. Secondary lymphoid-tissue chemokine (SLC) is chemotactic for mature dendritic cells. Blood (1999) 93:3610–6.
10. Walsh KP, Mills KHG. Dendritic cells and other innate determinants of T helper cell polarisation. Trends Immunol (2013) 34:521–30. doi:10.1016/j.it.2013.07.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
11. Pulendran B, Tang H, Manicassamy S. Programming dendritic cells to induce T(H)2 and tolerogenic responses. Nat Immunol (2010) 11(8):647–55. doi:10.1038/ni.1894
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
12. Schulz EG, Mariani L, Radbruch A, Höfer T. Sequential Polarization and Imprinting of Type 1 T Helper lymphocytes by Interferon-3 and Interleukin-12. Immunity (2009) 30:673–83. doi:10.1016/j.immuni.2009.03.013
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
13. Kaplan MH, Schindler U, Smiley ST, Grusby MJ. Stat6 is required for mediating responses to IL-4 and for the development of Th2 cells. Immunity (1996) 4:313–9. doi:10.1016/S1074-7613(00)80439-2
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
14. Dillon S, Agrawal A, Van Dyke T, Landreth G, McCauley L, Koh A, et al. A Toll-like receptor 2 ligand stimulates Th2 responses in vivo, via induction of extracellular signal-regulated kinase mitogen-activated protein kinase and c-Fos in dendritic cells. J Immunol (2004) 172:4733–43. doi:10.4049/jimmunol.172.8.4733
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
15. Park H, Li Z, Yang XO, Chang SH, Nurieva R, Wang YY-HH, et al. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat Immunol (2005) 6:1133–41. doi:10.1038/ni1261
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
16. Murphy CA, Langrish CL, Chen Y, Blumenschein W, McClanahan T, Kastelein RA, et al. Divergent pro- and antiinflammatory roles for IL-23 and IL-12 in joint autoimmune inflammation. J Exp Med (2003) 198:1951–7. doi:10.1084/jem.20030896
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
17. Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 Cells. Annu Rev Immunol (2009) 27:485–517. doi:10.1146/annurev.immunol.021908.132710
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
18. Ciofani M, Madar A, Galan C, Sellars M, Mace K, Pauli F, et al. A validated regulatory network for Th17 cell specification. Cell (2012) 151:289–303. doi:10.1016/j.cell.2012.09.016
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
19. Josefowicz SZ, Lu L-F, Rudensky AY. Regulatory T cells: mechanisms of differentiation and function. Annu Rev Immunol (2012) 30:531–64. doi:10.1146/annurev.immunol.25.022106.141623
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
20. Blanco P, Palucka AK, Pascual V, Banchereau J. Dendritic cells and cytokines in human inflammatory and autoimmune diseases. Cytokine Growth Factor Rev (2008) 19:41–52. doi:10.1016/j.cytogfr.2007.10.004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
21. Macdonald TT. New cytokine targets in inflammatory bowel disease. Gastroenterol Hepatol (N Y) (2011) 7:474–6.
22. Cua DJ, Sherlock J, Chen Y, Murphy CA, Joyce B, Seymour B, et al. Interleukin-23 rather than interleukin-12 is the critical cytokine for autoimmune inflammation of the brain. Nature (2003) 421:744–8. doi:10.1038/nature01355
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
23. Hue S, Ahern P, Buonocore S, Kullberg MC, Cua DJ, McKenzie BS, et al. Interleukin-23 drives innate and T cell-mediated intestinal inflammation. J Exp Med (2006) 203:2473–83. doi:10.1084/jem.20061099
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
24. Benson JM, Sachs CW, Treacy G, Zhou H, Pendley CE, Brodmerkel CM, et al. Therapeutic targeting of the IL-12/23 pathways: generation and characterization of ustekinumab. Nat Biotechnol (2011) 29:615–24. doi:10.1038/nbt.1903
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
25. Nishimoto N, Kishimoto T. Humanized antihuman IL-6 receptor antibody, tocilizumab. Handb Exp Pharmacol (2008) 181:151–60. doi:10.1007/978-3-540-73259-4_7
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
26. Frampton JE. Tocilizumab: a review of its use in the treatment of juvenile idiopathic arthritis. Paediatr Drugs (2013) 15:515–31. doi:10.1007/s40272-013-0053-1
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
27. Stow JL, Manderson AP, Murray RZ. SNAREing immunity: the role of SNAREs in the immune system. Nat Rev Immunol (2006) 6:919–29. doi:10.1038/nri1980
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
28. Kioumourtzoglou D, Gould GW, Bryant NJ. Insulin stimulates Syntaxin4 SNARE complex assembly via a novel regulatory mechanism. Mol Cell Biol (2014) 34(7):1271–9. doi:10.1128/MCB.01203-13
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
29. Benado A, Nasagi-Atiya Y, Sagi-Eisenberg R. Protein trafficking in immune cells. Immunobiology (2009) 214:403–21. doi:10.1016/j.imbio.2008.11.011
30. Jahn R, Scheller RH. SNAREs - engines for membrane fusion. Nat Rev Mol Cell Biol (2006) 7:631–43. doi:10.1038/nrm2002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
31. Jahn R, Südhof TC. Membrane fusion and exocytosis. Annu Rev Biochem (1999) 68:863–911. doi:10.1146/annurev.biochem.68.1.863
32. Fasshauer D. Structural insights into the SNARE mechanism. Biochim Biophys Acta (2003) 1641(2–3):87–97. doi:10.1016/S0167-4889(03)00090-9
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
33. Fasshauer D, Sutton RB, Brunger AT, Jahn R. Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc Natl Acad Sci USA (1998) 95(26):15781–6. doi:10.1073/pnas.95.26.15781
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
34. Stow JL, Murray RZ. Intracellular trafficking and secretion of inflammatory cytokines. Cytokine Growth Factor Rev (2013) 24:227–39. doi:10.1016/j.cytogfr.2013.04.001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
35. Sutton RB, Fasshauer D, Jahn R, Brunger AT. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 angstrom resolution. Nature (1998) 395:347–53. doi:10.1038/26412
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
36. Bock JB, Matern HT, Peden AA, Scheller RH. A genomic perspective on membrane compartment organization. Nature (2001) 409:839–41. doi:10.1038/35057024
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
37. Block MR, Rothman JE. Purification of N-ethylmaleimide-sensitive fusion protein. Methods Enzymol (1992) 219:300–9. doi:10.1016/0076-6879(92)19030-A
38. Clary DO, Rothman JE. Purification of soluble N-ethylmaleimide-sensitive fusion attachment proteins from bovine brain microsomes. Methods Enzymol (1992) 219:319–30. doi:10.1016/0076-6879(92)19032-2
39. Hong W. SNAREs and traffic. Biochim Biophys Acta (2005) 1744:120–44. doi:10.1016/j.bbamcr.2005.03.014
40. D’Orlando O, Zhao F, Kasper B, Orinska Z, Mueller J, Hermans-Borgmeyer I, et al. Syntaxin 11 is required for NK and CD8(+) T-cell cytotoxicity and neutrophil degranulation. Eur J Immunol (2013) 43(1):194–208. doi:10.1002/eji.201142343
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
41. Frank SPC, Thon K, Bischoff SC, Lorentz A. SNAP-23 and syntaxin-3 are required for chemokine release by mature human mast cells. Mol Immunol (2011) 49(1–2):353–8. doi:10.1016/j.molimm.2011.09.011
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
42. Pagan JK, Wylie FG, Joseph S, Widberg C, Bryant NJ, James DE, et al. The t-SNARE syntaxin 4 is regulated during macrophage activation to function in membrane traffic and cytokine secretion. Curr Biol (2003) 13(2):156–60. doi:10.1016/S0960-9822(03)00006-X
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
43. Murray RZ, Kay JG, Sangermani DG, Stow JL. A role for the phagosome in cytokine secretion. Science (2005) 310(5753):1492–5. doi:10.1126/science.1120225
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
44. Murray RZ, Wylie FG, Khromykh T, Hume DA, Stow JL. Syntaxin 6 and Vti1b form a novel SNARE complex, which is up-regulated in activated macrophages to facilitate exocytosis of tumor necrosis factor-alpha. J Biol Chem (2005) 280(11):10478–83. doi:10.1074/jbc.M414420200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
45. Offenhäuser C, Lei N, Roy S, Collins BM, Stow JL, Murray RZ. Syntaxin 11 binds Vti1b and regulates late endosome to lysosome fusion in macrophages. Traffic (2011) 12:762–73. doi:10.1111/j.1600-0854.2011.01189.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
46. zur Stadt U, Schmidt S, Kasper B, Beutel K, Diler AS, Henter JI, et al. Linkage of familial hemophagocytic lymphohistiocytosis (FHL) type-4 to chromosome 6q24 and identification of mutations in syntaxin 11. Hum Mol Genet (2005) 14:827–34. doi:10.1093/hmg/ddi076
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
47. Janka GE. Familial and acquired hemophagocytic lymphohistiocytosis. Eur J Pediatr (2007) 166:95–109. doi:10.1007/s00431-006-0258-1
48. Janka GE. Familial and acquired hemophagocytic lymphohistiocytosis. Annu Rev Med (2012) 63:233–46. doi:10.1146/annurev-med-041610-134208
49. Collins LE, DeCourcey J, Rochfort KD, Kristek M, Loscher CE. A role for syntaxin 3 in the secretion of IL-6 from dendritic cells following activation of toll-like receptors. Front Immunol (2015) 5:691. doi:10.3389/fimmu.2014.00691
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
50. Ho YHS, Cai DT, Wang C-C, Huang D, Wong SH. Vesicle-associated membrane protein-8/endobrevin negatively regulates phagocytosis of bacteria in dendritic cells. J Immunol (2008) 180:3148–57. doi:10.4049/jimmunol.180.5.3148
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
51. Wade N, Bryant NJ, Connolly LM, Simpson RJ, Luzio JP, Piper RC, et al. Syntaxin 7 complexes with mouse Vps10p tail interactor 1b, syntaxin 6, vesicle-associated membrane protein (VAMP)8, and VAMP7 in b16 melanoma cells. J Biol Chem (2001) 276(23):19820–7. doi:10.1074/jbc.M010838200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
52. Cai DT, Ho YHS, Chiow KH, Wee SH, Han Y, Peh MT, et al. Aspirin regulates SNARE protein expression and phagocytosis in dendritic cells. Mol Membr Biol (2011) 28:90–102. doi:10.3109/09687688.2010.525756
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
53. Cebrian I, Visentin G, Blanchard N, Jouve M, Bobard A, Moita C, et al. Sec22b regulates phagosomal maturation and antigen crosspresentation by dendritic cells. Cell (2011) 147:1355–68. doi:10.3109/09687688.2010.525756
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
54. Nair-Gupta P, Baccarini A, Tung N, Seyffer F, Florey O, Huang Y, et al. TLR signals induce phagosomal MHC-I delivery from the endosomal recycling compartment to allow cross-presentation. Cell (2014) 158(3):506–21. doi:10.1016/j.cell.2014.04.054
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
55. Rahman A, DeCourcey J, Ben Larbi N, Loughran ST, Walls D, Loscher CE. Syntaxin-4 is essential for IgE secretion by plasma cells. Biochem Biophys Res Commun (2013) 440:163–7. doi:10.1016/j.bbrc.2013.09.058
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: dendritic cells, SNAREs, IL-6, IL-23, IL-12
Citation: Collins LE, DeCourcey J, Soledad di Luca M, Rochfort KD and Loscher CE (2015) An emerging role for SNARE proteins in dendritic cell function. Front. Immunol. 6:133. doi: 10.3389/fimmu.2015.00133
Received: 24 September 2014; Accepted: 10 March 2015;
Published online: 31 March 2015.
Edited by:
Paige Lacy, University of Alberta, CanadaReviewed by:
Koichi S. Kobayashi, Texas A&M Health Science Center, USAShahram Salek-Ardakani, University of Florida, USA
Copyright: © 2015 Collins, DeCourcey, Soledad di Luca, Rochfort and Loscher. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christine E. Loscher, Immunomodulation Research Group, School of Biotechnology, Dublin City University, Collins Avenue, Dublin 9, Ireland e-mail:Y2hyaXN0aW5lLmxvc2NoZXJAZGN1Lmll
†Laura E. Collins and Joseph DeCourcey have contributed equally to this work.
 Laura E. Collins
Laura E. Collins Joseph DeCourcey
Joseph DeCourcey Mariana Soledad di Luca
Mariana Soledad di Luca Keith D. Rochfort
Keith D. Rochfort Christine E. Loscher
Christine E. Loscher