- 1Institute of Health and Wellbeing, School of Health, University of Northampton, Northampton, UK
- 2Department of Cancer Studies, University of Leicester, Leicester, UK
Defensins represent an evolutionary ancient family of antimicrobial peptides that play diverse roles in human health and disease. Defensins are cationic cysteine-containing multifunctional peptides predominantly expressed by epithelial cells or neutrophils. Defensins play a key role in host innate immune responses to infection and, in addition to their classically described role as antimicrobial peptides, have also been implicated in immune modulation, fertility, development, and wound healing. Aberrant expression of defensins is important in a number of inflammatory diseases as well as modulating host immune responses to bacteria, unicellular pathogens, and viruses. In parallel with their role in immunity, in other species, defensins have evolved alternative functions, including the control of coat color in dogs. Defensin genes reside in complex genomic regions that are prone to structural variations and some defensin family members exhibit copy number variation (CNV). Structural variations have mediated, and continue to influence, the diversification and expression of defensin family members. This review highlights the work currently being done to better understand the genomic architecture of the β-defensin locus. It evaluates current evidence linking defensin CNV to autoimmune disease (i.e., Crohn’s disease and psoriasis) as well as the contribution CNV has in influencing immune responses to HIV infection.
Introduction
The defensins represent a class of cationic antimicrobial peptides that play pivotal roles in innate and adaptive immunity as well as roles in non-immunological processes. They constitute an ancient and diverse gene family, present in most multicellular organisms ranging, from plants, fungi, insects, mollusks, and arachnids to mammals, including humans. During their evolutionary history, defensins have become highly diversified and have acquired novel functions in different species. Defensins have evolved to be highly efficient in their antimicrobial responses to a vast array of pathogens.
The term “Defensins” was coined in 1985 after granule rich sediments were purified from human and rabbit neutrophils. This resulted in the characterization of the primary structure of the first six neutrophils defensins (later known as α-defensins) (1–3). These early studies highlighted the structural hallmarks of defensins: that is, despite poor sequence identity across family members, all defensins possesses a highly conserved motif of six cysteine residues that is key to their antimicrobial function. Subsequently, peptides with similar structure were discovered in the early 1990s in bovine (4) and mouse airway first (5) and subsequently in the human intestinal epithelium (6), and became known as β-defensins. The recent ability to interrogate genomic and proteomic data from a diverse array of species allowed the discovery and characterization of further members of the defensin gene family, intensifying interest in unveiling the roles of defensins in physiological and pathological processes.
This review will primarily focus on the role of β-defensins in innate and adaptive immunity. We will highlight the methods currently employed to study the genomic architecture of this multifunctional gene family and how complex genetic variation has an impact on defensin host inflammatory responses.
Structure of β-Defensins
The β-defensin family members have poor sequence similarity, suggesting their antimicrobial activity is independent of their primary structure. Nuclear magnetic resonance (NMR) data have been used to evaluate the 3D structure of hBD1, hBD2, and hBD3 (7, 8). These data confirm a high degree of similarity in their tertiary structures, despite their diverged amino acid sequences. The major element of the mature peptides secondary structure is represented by three β-strands arranged in an antiparallel sheet. The strands are held together by the three intramolecular disulfide bonds, formed between the six cysteines. The order of the disulfide bridges can vary, characterizing each family member. The amino-terminal region contains a short α-helical loop (which is absent in α-defensins). α-helical structures are common for protein regions that are incorporated into cell membranes and it has been proposed that this region of the β-defensin protein may anchor to bacteria cell walls (9). This is supported by the presence of two sites under positive selection located in the N-terminal region that may contribute to β-defensin functional diversity (10).
Defensins do not appear to present a distinct hydrophobic core or a common pattern of charged or hydrophobic residues on the protein surface. This suggests peptide folding is driven and stabilized by disulfide bond formation alone. Moreover, the characteristic β-defensin 3D structure can be preserved and accommodates residues with different properties at most other positions. The first five amino acids of the mature peptide sequence are vital for correct protein folding under oxidative conditions. This favors the formation of the correct disulfide bonded pattern through the creation of a key intermediate (11).
The Evolution and Divergent Roles of β-Defensins
The evolutionary relationship between vertebrate and non-vertebrate defensins is still unclear; however, phylogeny indicates that a primordial β-defensin is the common ancestor of all vertebrate defensins and this gene family expanded throughout vertebrate evolution (12). This hypothesis is supported by the discovery of β-defensin-like genes in phylogenetically distant vertebrates, including reptiles (13), birds (14), and teleost fishes (15). α-defensins are mammalian specific genes, and in humans α-defensin genes and different β-defensin genes are present on adjacent loci on chromosome 8p22–p23. The organization of this cluster is consistent with a model of multiple rounds of duplication and divergence under positive selection from a common ancestral gene that produced a cluster of diversified paralogous (16, 17). This expansion occurred before the divergence of baboons and humans ~23–63 million years ago (18, 19). The present-day β-defensins probably evolved before mammals diverged from birds generating α-defensins in rodents, lagomorphs, and primates after their divergence from other mammals (20). Recent evidence suggests convergent evolution of β-defensin copy number (CN) in primates, where independent origins have been sponsored by non-allelic homologous recombination between repeat units. For rhesus macaques this resulted in only a 20 kb copy number variation (CNV) region containing the human ortholog of human β-defensin 2 gene. In humans, recent work suggests a repeat unit of 322 kb containing a number of β-defensin genes (21).
Defensin family members possess a plethora of non-immune activities and it is instructive to provide some examples of the diverged nature of defensins function. Some members of the β-defensin family have an important role in mammalian reproduction [reviewed in Ref. (22)]. For example, there are five human defensin genes (DEFB125–DEFB129) clustered on chromosome 20, which are highly expressed in the epithelial cell layer of the epididymal duct, which secretes factors responsible for sperm maturation (23). Moreover, human DEFB118 was shown to be a potent antimicrobial peptide able to bind to sperm, probably providing protection from microorganisms present in the sperm ducts (24). It is noticeable how in long tailed macaque (Macaca fascicularis) and in rhesus macaque (Macaca mulatta), there is a similar β-defensin, called DEFB126, which is the principal protein that coats sperm (25); this coating is lost in the oviduct allowing fertilization to occur. In support of this, the deletion of a cluster of nine beta defensin genes in a mouse model, resulted in male sterility (26). In human studies, a common mutation in DEFB126 has been shown to impair sperm function and fertility (27).
In a second example, recent studies have suggested that some β-defensin gene products including hBD1 and hBD3, can interact with a family of melanocortin receptors, modulating pigment expression in dogs and possibly in humans (28). Typically, there are two genes that control the switching of pigment types: the melanocortin receptor 1 (Mc1r) and Agouti, encoding a ligand for the Mc1r, which inhibits Mc1r signaling. Mc1r activation determines production of the dark pigment eumelanin exclusively, whereas Mc1r inhibition causes production of the lighter pigment pheomelanin. In dogs, it was discovered that a mutation in the canine DEFB103 is responsible for the dominant inheritance of black coat color, which does not signal directly through Mc1r; this insight revealed a previously uncharacterized role of β-defensins in controlling skin pigmentation. Further studies have been conducted on human melanocytes, discovering a novel role of hBD3 as an antagonist of the α-melanocyte-stimulating hormone (α-MSH, a known agonist of Mc1r, which stimulates cAMP signaling to induce eumelanin production). As hBD3 is produced by keratinocytes, it can act as a paracrine factor on melanocytes modulating α-MSH effects on human pigmentation and consequently responses to UV (29). Moreover, it is known that melanocortin receptors are also involved in inflammatory and immune response modulation (30).
Expression of β-Defensins
Different β-defensins are present in different epithelial and mucosal tissues and can be constitutively expressed or induced in response to various stimuli (31–52) (Table S1 in Supplementary Material). Their anatomical distribution clearly reflects their ability to neutralize different pathogens and they are more abundant at sites prone to the microbial infections they are specific for. For example, hBD2 is strongly expressed in lung (53); hBD4 is highly expressed in the stomach and testes (54), and hBD3 in the skin and tonsillar tissue (55). hBD1–hBD4 are expressed in the respiratory tract, with constitutive expression of hBD1 (56) and inducible expression of hBD2–hBD4 in response to inflammation or infection (57). In keratinocytes, there is constitutive mRNA expression of hBD1; conversely hBD2 expression is induced by lipopolysaccharides (LPS) or other bacterial epitopes in combination with interleukin-1β, released by resident monocyte-derived cells. hBD3 and hBD4 are inducible by stimulation with tumor necrosis factor (TNF), toll-like receptor ligands, interferon (IFN)-γ, or phorbolmyristate acetates (58). hBD3 is also induced in response to local release of surface-bound epidermal growth factor receptor (EGFR) ligands via activation of metalloproteinases (59, 60).
Antimicrobial Activity of β-Defensins
The most studied function for β-defensins is their direct antimicrobial activity, through permeabilization of the pathogen membrane. Their exact mechanism of action is incompletely understood and two different models have been proposed. The first is a carpet model, where several antimicrobial peptides opsonize the pathogen surface bringing about necrosis, possibly disrupting the electrostatic charge across the membrane (61). The latter is a pore model, with several peptides oligomerizing and forming pore-like membrane defects that allow efflux of essential ions and nutrients (55).
Defensins in vitro are active against gram negative and positive bacteria, unicellular parasites, viruses, and yeast. Cationic peptides including β-defensins are attracted to the overall net negative charge generated by the outer envelope of Gram negative bacteria by phospholipids and phosphate groups on LPS and to the teichoic acid present on the surface of Gram positive bacteria.
β-defensins also possess anti-viral activity, interacting directly with the virus and indirectly with its target cells. Noticeably, in mammals, β-defensins are also produced by the oral mucosa and they are active against HIV-1 virus: in particular, hBD1 is constitutively expressed whereas the presence of a low HIV-1 viral load can stimulate the expression of hBD2 and hBD3 gene products through direct interaction with the virus. More specifically, hBD2 has been shown to down-regulate the HIV transcription of early reverse-transcribed DNA products (62) and hBD2 and hBD3 can mediate CXCR4 down-regulation (but not CCR5) and internalization in immuno-stimulated peripheral blood mononuclear cells (63). This mechanism diminishes the chances of infection (64) and with other salivary gland components, could help to explain the oral mucosal natural resistance to HIV infection. hBD3 also possesses an inhibitory effect on the influenza virus blocking the fusion of the viral membrane with the endosome of the host cell, through cross linking of the viral glycoproteins (65).
Defensins have evolved to maximize their protective role, showing an extraordinary adaptation to different environmental challenges: for instance, plant defensins are particularly active against fungal infections [reviewed in Ref. (66)], slowing down hyphal elongation, and some of them also evolved to gain an α-amylase inhibitory activity that can confer protection against herbivores (67, 68).
Immune Modulatory Activity of β-Defensins
A role for defensins in pro-inflammatory responses and more recently immunosuppression [reviewed in Ref. (69)] has been delineated over the last two decades. An initial important observation was that β-defensins can recruit immature dendritic cells and memory T cells to sites of infection and/or inflammation providing a link between the innate and adaptive arms of the immune system. A mechanism for this was provided by Oppenheim’s group where they demonstrated that natural and recombinant hBD2 could chemoattract human immature dendritic cells and memory T cells in vitro in a dose-dependent manner. This response was inhibited with the Gαi inhibitor pertussis toxin and suggested the possible involvement of a chemokine receptor(s), which was confirmed using anti-CCR6 blocking antibodies.
TH17 cells express CCR6 and respond to β-defensins chemoattractant action. Furthermore, TH17 cytokines (i.e., IL-17 and IL-22) induce expression of defensins from relevant cell types including primary keratinocytes potentially resulting in an amplification of TH17 responses (70). Increased TH17 levels have been reported in different autoimmune diseases, such as multiple sclerosis (71), rheumatoid arthritis (72), and psoriasis (73), implicating β-defensin expression in autoimmunity. Given the role of defensins in chemoattracting monocytes and macrophages and the lack of CCR6 on these cell types other receptors were investigated that might mediate this chemoattractant activity. This resulted in the identification of CCR2 as a receptor for hBD2, hBD3, and their mouse orthologs (mBD4 and mBD14) (74).
In addition to signaling through chemokine receptors, defensins have been shown to function through toll-like receptors (75, 76). hBD2 has been shown to be a natural ligand for the toll-like-receptor-4 (TLR-4), present on immature DCs, up-regulating co-stimulatory molecules and leading to DC maturation, and on CD4+ T cells, possibly stimulating their proliferation and survival (77). On bone marrow-derived macrophages pre-treated with a recently identified mBD14 (78), TLR restimulation of these cells resulted in enhanced expression of pro-inflammatory mediators that was Gi protein dependent but independent of CCR2 or CCR6 signaling pathways (79).
β-Defensin Copy Number Variation and Disease Association Studies
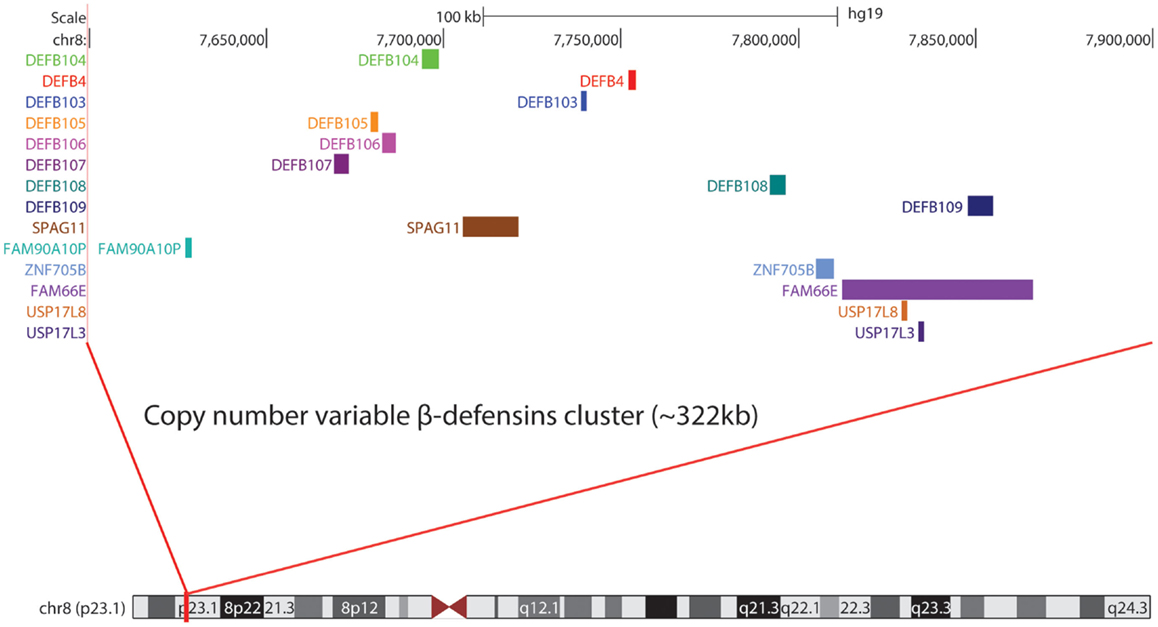
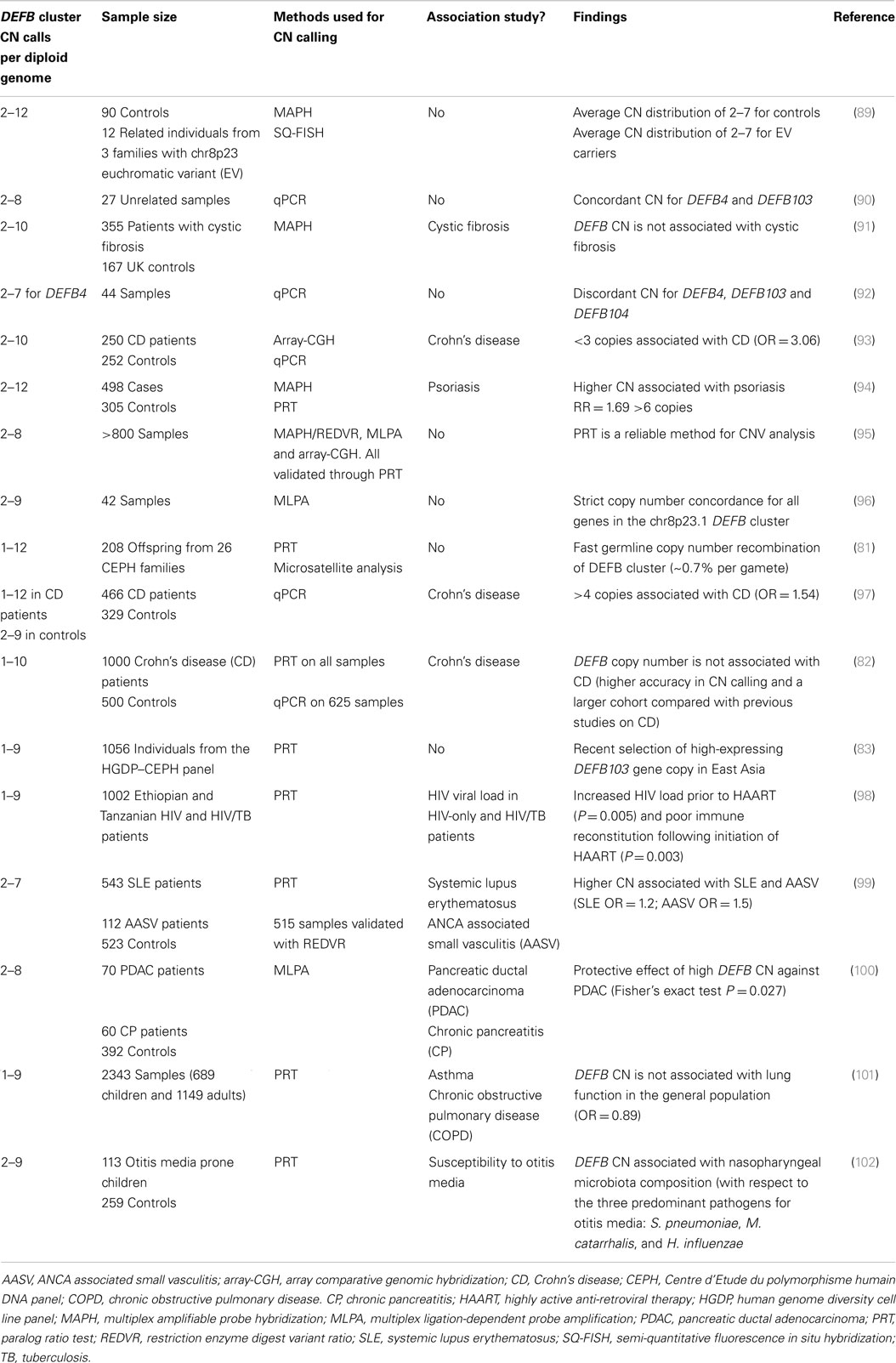
In humans, β-defensins genes are organized into three main clusters at 8p23.1, 20p13, and 20q11.1, with another likely small cluster on chromosome 6p12 (80). At 8p23.1, a number of β-defensins are found on a repeat unit that is typically present at 2–8 copies in the population, with a modal CN of 4. Each chromosome 8 copy can contain 1–8 copies of the repeat unit. The mutation rate at this locus is extremely fast (~0.7% per gamete) (81), indicative of the high level of plasticity in this genomic region. One-copy individuals are extremely rare (82, 83), and suggest that the presence of a null allele might be deleterious and selected against. At the other end of the DEFB, CN spectrum lies a proportion of high-copies individuals (9–12 copies) with a cytogenetically visible CN amplification at 8p23.1 that has no phenotypic effect (84). These first experimental observations ignited further interest into the chromosome 8 DEFB cluster. Within the repeat unit there is DEFB4, DEFB103, DEFB104, DEFB105, DEFB106, DEFB107, SPAG11, and PRR23D1 (21, 85) (Figure 1). The variation in the number of repeat units between individuals in the population and likely sequence variation between copies suggests that CNV of defensins may play a role in modulating defensin expression (86, 87) and function. The consequences of CNV have been explored for a number of years and may include increased gene product, the production of fusion genes, the formation of extra coding domains, or a position effect that alters expression of the gene product (88). This extensive structural genome variation in humans is particularly pertinent to diseases where defensins may be implicated in their pathology. This includes a number of autoimmune and infectious diseases (Table 1).
Mapping of the β-defensin CNV region has been challenging but recent data fixes the minimal length of the CNV at 157 kb (103) and a recent study using high density array comparative genomic hybridization combined with paralog ratio test (PRT) assays suggests it may be as large as 322 kb (21). Because of the extensive CNV of defensins, robust methods are required to accurately interrogate CN states in disease cohorts. Various locus specific techniques for CN determination have been utilized including multiplex amplifiable probe hybridization (MAPH) (104), multiple ligation probe amplification (MLPA) (105), and PRT (95). The advantage of such techniques is the ability to obtain data that clusters around integer CNs providing a high degree of concordance between the methods and confidence in the CN obtained. Association studies investigating some CNVs (i.e., CCL3L1/CCL4L2 in HIV) have provided conflicting results as the methods used did not generate data that clustered around integer CN values (106, 107). In some cases, initial findings have been replicated in subsequent studies that have utilized more robust methods (108).
In early association studies of multi-allelic CNV and disease, CNV of defensins was implicated in psoriasis. Individuals with more than five β-defensin copies presented a fivefold increased risk of developing psoriasis when compared to two copy individuals. In addition, there was a direct correlation between the number of copies and relative risk (odds ratio of 1.32) (94). This association was replicated (although with reduced odds ratio) in a subsequent study (109). In the case of an autoimmune condition, such as psoriasis, high CN may contribute to the strong induction of hBD2 and hBD3, conferring protection from bacterial infections of the psoriatic lesions (110).
Another disease strongly linked with defensin expression is Crohn’s disease (CD) where it has been demonstrated that reduced Paneth cell expression of defensins in the ileum results in ileal CD. Therefore, defensin expression at this site may be important in maintaining the mucosal microbiota. NOD2 has been strongly implicated in the pathogenesis of CD from GWAS (111) giving a 17.1-fold increased risk for CD in homozygous or compound heterozygous individuals. NOD2 is a nod like family receptor (NLR) member that controls expression of defensins in CD. Polymorphisms in NOD2 result in reduced α-defensin expression and exacerbated disease. Polymorphism of the DEFB1 (non-CNV gene) promoter has been associated with CD (112). So is there a role for CNV in CD? Previous studies indicated that α-defensin CN may be important (113). However, recent work that accurately measured CN using PRTs to determine CN of DEFA1A3 determined that a SNP (rs4300027) is associated with DEFA1A3 CN in Europeans (114). This SNP was then used to indirectly interrogate GWAS data and suggested that α-defensins CNV may not be important in CD. A similar outcome was obtained with β-defensin CN whereupon accurate measurement, there was no association with the CD (82) in contrast to previous reports (93, 97). These results, however, do not exclude the role of α and β-defensin expression in the pathogenesis of CD but suggest that the individuals CN state may not be important in this context.
Given the suspected anti-viral role of defensins, it was suggested that defensin CNV may be important in host responses to HIV infection. There are a number of conflicting reports of the association between defensin CN and HIV infection (114–116). A surprising finding from a cohort study that evaluated two sub-Saharan populations with HIV-1 or HIV-1/tuberculosis coinfection was that high CN of β-defensins did not result in the predicted low viral load and did not improve immune reconstitution in patients (98). The converse was found suggesting that the immune modulatory properties of defensins may be subverted during HIV-1 infection. A model suggested to explain this apparently paradoxical result was that high CN may promote increased recruitment of CCR6 expressing cell types that are highly permissive for HIV-1 infection thus amplifying the foci of HIV-1 infection.
Conclusion
Defensins play a key role in pathogen host interactions and are at the interface of innate and adaptive immunity. The complex genetic variation that underlies the evolutionary history of defensins and their biology is gradually being elucidated, suggesting defensin CNV is an important contributor to maximizing the host innate and adaptive response. The history of the defensin gene family is particularly paradigmatic given that many CNV loci in the human genome host immunity genes. Further studies should be conducted to better understand the genomic architecture of multi-allelic CNVs. This will aid the development of robust assays that evaluate the overall impact that CNV has on and both physiological and pathological mechanisms of immunity.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Dr. Edward Hollox (University of Leicester) for helpful discussions. This work was supported by a University of Leicester College of Medicine, Biological Sciences and Psychology Ph.D. studentship awarded to BO.
Supplementary Material
The Supplementary Material for this article can be found online at http://www.frontiersin.org/Journal/10.3389/fimmu.2015.00115/abstract
References
1. Ganz T, Selsted ME, Szklarek D, Harwig SS, Daher K, Bainton DF, et al. Defensins. Natural peptide antibiotics of human neutrophils. J Clin Invest (1985) 76:1427–35. doi: 10.1172/JCI112120
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
2. Selsted ME, Brown DM, DeLange RJ, Harwig SS, Lehrer RI. Primary structures of six antimicrobial peptides of rabbit peritoneal neutrophils. J Biol Chem (1985) 260:4579–84.
3. Selsted ME, Harwig SS, Ganz T, Schilling JW, Lehrer RI. Primary structures of three human neutrophil defensins. J Clin Invest (1985) 76:1436–9. doi:10.1172/JCI112121
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
4. Diamond G, Zasloff M, Eck H, Brasseur M, Maloy WL, Bevins CL. Tracheal antimicrobial peptide, a cysteine-rich peptide from mammalian tracheal mucosa: peptide isolation and cloning of a cDNA. Proc Natl Acad Sci U S A (1991) 88:3952–6. doi:10.1073/pnas.88.9.3952
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
5. Ouellette AJ, Lualdi JC. A novel mouse gene family coding for cationic, cysteine-rich peptides. Regulation in small intestine and cells of myeloid origin. J Biol Chem (1990) 265:9831–7.
6. Jones DE, Bevins CL. Paneth cells of the human small intestine express an antimicrobial peptide gene. J Biol Chem (1992) 267:23216–25.
7. Schibli DJ, Hunter HN, Aseyev V, Starner TD, Wiencek JM, McCray PB, et al. The solution structures of the human beta-defensins lead to a better understanding of the potent bactericidal activity of HBD3 against Staphylococcus aureus. J Biol Chem (2002) 277:8279–89. doi:10.1074/jbc.M108830200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
8. Bauer F, Schweimer K, Klüver E, Conejo-Garcia JR, Forssmann WG, Rösch P, et al. Structure determination of human and murine beta-defensins reveals structural conservation in the absence of significant sequence similarity. Protein Sci (2001) 10:2470–9. doi:10.1110/ps.ps.24401
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
9. Taylor K, Barran PE, Dorin JR. Structure-activity relationships in beta-defensin peptides. Biopolymers (2008) 90:1–7. doi:10.1002/bip.20900
10. Semple CAM, Maxwell A, Gautier P, Kilanowski FM, Eastwood H, Barran PE, et al. The complexity of selection at the major primate beta-defensin locus. BMC Evol Biol (2005) 5:32. doi:10.1186/1471-2148-5-32
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
11. Klüver E, Schulz-Maronde S, Scheid S, Meyer B, Forssmann W-G, Adermann K. Structure-activity relation of human beta-defensin 3: influence of disulfide bonds and cysteine substitution on antimicrobial activity and cytotoxicity. Biochemistry (2005) 44:9804–16. doi:10.1021/bi050272k
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
12. Xiao Y, Hughes AL, Ando J, Matsuda Y, Cheng J-F, Skinner-Noble D, et al. A genome-wide screen identifies a single beta-defensin gene cluster in the chicken: implications for the origin and evolution of mammalian defensins. BMC Genomics (2004) 5:56. doi:10.1186/1471-2164-5-56
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
13. Van Hoek ML. Antimicrobial peptides in reptiles. Pharmaceuticals (Basel) (2014) 7:723–53. doi:10.3390/ph7060723
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
14. Zhao C, Nguyen T, Liu L, Sacco RE, Brogden KA, Lehrer RI. Gallinacin-3, an inducible epithelial beta-defensin in the chicken. Infect Immun (2001) 69:2684–91. doi:10.1128/IAI.69.4.2684-2691.2001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
15. Zou J, Mercier C, Koussounadis A, Secombes C. Discovery of multiple beta-defensin like homologues in teleost fish. Mol Immunol (2007) 44:638–47. doi:10.1016/j.molimm.2006.01.012
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
16. Liu L, Zhao C, Heng HH, Ganz T. The human beta-defensin-1 and alpha-defensins are encoded by adjacent genes: two peptide families with differing disulfide topology share a common ancestry. Genomics (1997) 43:316–20. doi:10.1006/geno.1997.4801
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
17. Maxwell AI, Morrison GM, Dorin JR. Rapid sequence divergence in mammalian beta-defensins by adaptive evolution. Mol Immunol (2003) 40:413–21. doi:10.1016/S0161-5890(03)00160-3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
18. Semple CAM, Rolfe M, Dorin JR. Duplication and selection in the evolution of primate beta-defensin genes. Genome Biol (2003) 4:R31. doi:10.1186/gb-2003-4-5-r31
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
19. Glazko GV, Nei M. Estimation of divergence times for major lineages of primate species. Mol Biol Evol (2003) 20:424–34. doi:10.1093/molbev/msg050
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
20. Patil A, Hughes AL, Zhang G. Rapid evolution and diversification of mammalian alpha-defensins as revealed by comparative analysis of rodent and primate genes. Physiol Genomics (2004) 20:1–11. doi:10.1152/physiolgenomics.00150.2004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
21. Ottolini B, Hornsby MJ, Abujaber R, MacArthur JAL, Badge RM, Schwarzacher T, et al. Evidence of convergent evolution in humans and macaques supports an adaptive role for copy number variation of the β-defensin-2 gene. Genome Biol Evol (2014) 6:3025–38. doi:10.1093/gbe/evu236
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
22. Dorin JR, Barratt CLR. Importance of β-defensins in sperm function. Mol Hum Reprod (2014) 20:821–6. doi:10.1093/molehr/gau050
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
23. Rodríguez-Jiménez FJ, Krause A, Schulz S, Forssmann WG, Conejo-Garcia JR, Schreeb R, et al. Distribution of new human beta-defensin genes clustered on chromosome 20 in functionally different segments of epididymis. Genomics (2003) 81:175–83. doi:10.1016/S0888-7543(02)00034-4
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
24. Yenugu S, Hamil KG, Radhakrishnan Y, French FS, Hall SH. The androgen-regulated epididymal sperm-binding protein, human beta-defensin 118 (DEFB118) (formerly ESC42), is an antimicrobial beta-defensin. Endocrinology (2004) 145:3165–73. doi:10.1210/en.2003-1698
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
25. Tollner TL, Yudin AI, Treece CA, Overstreet JW, Cherr GN. Macaque sperm coating protein DEFB126 facilitates sperm penetration of cervical mucus. Hum Reprod (2008) 23:2523–34. doi:10.1093/humrep/den276
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
26. Zhou YS, Webb S, Lettice L, Tardif S, Kilanowski F, Tyrrell C, et al. Partial deletion of chromosome 8 β-defensin cluster confers sperm dysfunction and infertility in male mice. PLoS Genet (2013) 9:e1003826. doi:10.1371/journal.pgen.1003826
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
27. Tollner TL, Venners SA, Hollox EJ, Yudin AI, Liu X, Tang G, et al. A common mutation in the defensin DEFB126 causes impaired sperm function and subfertility. Sci Transl Med (2011) 3:92ra65. doi:10.1126/scitranslmed.3002289
28. Candille SI, Kaelin CB, Cattanach BM, Yu B, Thompson DA, Nix MA, et al. A -defensin mutation causes black coat color in domestic dogs. Science (2007) 318:1418–23. doi:10.1126/science.1147880
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
29. Swope VB, Jameson JA, McFarland KL, Supp DM, Miller WE, McGraw DW, et al. Defining MC1R regulation in human melanocytes by its agonist α-melanocortin and antagonists agouti signaling protein and β-defensin 3. J Invest Dermatol (2012) 132:2255–62. doi:10.1038/jid.2012.135
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
30. Maaser C, Kannengiesser K, Kucharzik T. Role of the melanocortin system in inflammation. Ann N Y Acad Sci (2006) 1072:123–34. doi:10.1196/annals.1326.016
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
31. Mathews M, Jia HP, Guthmiller JM, Losh G, Graham S, Johnson GK, et al. Production of β-defensin antimicrobial peptides by the oral mucosa and salivary glands. Infect Immun (1999) 67:2740–5.
32. Chen P-H, Fang S-Y. Expression of human β-defensin 2 in human nasal mucosa. Eur Arch Otorhinolaryngol (2004) 261:238–41. doi:10.1007/s00405-003-0682-z
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
33. Hiratsuka T, Nakazato M, Date Y, Ashitani J, Minematsu T, Chino N, et al. Identification of human β-defensin-2 in respiratory tract and plasma and its increase in bacterial pneumonia. Biochem Biophys Res Commun (1998) 249:943–7. doi:10.1006/bbrc.1998.9239
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
34. O’Neil DA, Porter EM, Elewaut D, Anderson GM, Eckmann L, Ganz T, et al. Expression and regulation of the human β-defensins hBD-1 and hBD-2 in intestinal epithelium. J Immunol (1999) 163:6718–24.
35. Hamanaka Y, Nakashima M, Wada A, Ito M, Kurazono H, Hojo H, et al. Expression of human β-defensin 2 (hBD-2) in Helicobacter pylori induced gastritis: antibacterial effect of hBD-2 against Helicobacter pylori. Gut (2001) 49:481–7. doi:10.1136/gut.49.4.481
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
36. Otri AM, Mohammed I, Al-Aqaba MA, Fares U, Peng C, Hopkinson A, et al. Variable expression of human Beta defensins 3 and 9 at the human ocular surface in infectious keratitis. Invest Ophthalmol Vis Sci (2012) 53:757–61. doi:10.1167/iovs.11-8467
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
37. Harder J, Bartels J, Christophers E, Schroder JM. A peptide antibiotic from human skin. Nature (1997) 387:861. doi:10.1038/43088
38. Lehmann J, Retz M, Harder J, Krams M, Kellner U, Hartmann J, et al. Expression of human beta-defensins 1 and 2 in kidneys with chronic bacterial infection. BMC Infect Dis (2002) 2:20. doi:10.1186/1471-2334-2-20
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
39. Sun L, Finnegan CM, Kish-catalone T, Blumenthal R, Garzino-demo P, La GM, et al. Human beta-defensins suppress human immunodeficiency virus infection: potential role in mucosal protection. J Virol (2005) 79:14318–29. doi:10.1128/JVI.79.22.14318-14329.2005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
40. Diamond G, Russell JP, Bevins CL. Inducible expression of an antibiotic peptide gene in lipopolysaccharide-challenged tracheal epithelial cells. Proc Natl Acad Sci U S A (1996) 93:5156–60. doi:10.1073/pnas.93.10.5156
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
41. Kumar A, Zhang J, Fu-Shin XY. Innate immune response of corneal epithelial cells to Staphylococcus aureus infection: role of peptidoglycan in stimulating proinflammatory cytokine secretion. Invest Ophthalmol Vis Sci (2004) 45:3513–22. doi:10.1167/iovs.04-0467
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
42. Birchler T, Seibl R, Büchner K, Loeliger S, Seger R, Hossle JP, et al. Human toll-like receptor 2 mediates induction of the antimicrobial peptide human beta-defensin 2 in response to bacterial lipoprotein. Eur J Immunol (2001) 31:3131–7. doi:10.1002/1521-4141(200111)31:11<3131::AID-IMMU3131>3.0.CO;2-G
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
43. McDermott AM, Redfern RL, Zhang B, Pei Y, Huang L, Proske RJ. Defensin expression by the cornea: multiple signalling pathways mediate IL-1β stimulation of hBD-2 expression by human corneal epithelial cells. Invest Ophthalmol Vis Sci (2003) 44:1859–65. doi:10.1167/iovs.02-0787
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
44. Harder J, Meyer-Hoffert U, Teran LM, Schwichtenberg L, Bartels J, Maune S, et al. Mucoid Pseudomonas aeruginosa, TNF-α, and IL-1 β, but Not IL-6, induce human β-defensin-2 in respiratory epithelia. Am J Respir Cell Mol Biol (2000) 22:714–21. doi:10.1165/ajrcmb.22.6.4023
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
45. Vankeerberghen A, Nuytten H, Dierickx K, Quirynen M, Cassiman J-J, Cuppens H. Differential induction of human beta-defensin expression by periodontal commensals and pathogens in periodontal pocket epithelial cells. J Periodontol (2005) 76:1293–303. doi:10.1902/jop.2005.76.8.1293
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
46. Kumar A, Zhang J, Yu F-SX. Toll-like receptor 2-mediated expression of β-defensin-2 in human corneal epithelial cells. Microbes Infect (2006) 8:380–9. doi:10.1016/j.micinf.2005.07.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
47. Semlali A, Witoled C, Alanazi M, Rouabhia M. Whole cigarette smoke increased the expression of TLRs, HBDs, and proinflammory cytokines by human gingival epithelial cells through different signaling pathways. PLoS One (2012) 7:e52614. doi:10.1371/journal.pone.0052614
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
48. García J-R, Jaumann F, Schulz S, Krause A, Rodríguez-Jiménez J, Forssmann U, et al. Identification of a novel, multifunctional β-defensin (human β-defensin 3) with specific antimicrobial activity. Cell Tissue Res (2001) 306:257–64. doi:10.1007/s004410100433
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
49. Lüthje P, Hirschberg AL, Brauner A. Estrogenic action on innate defense mechanisms in the urinary tract. Maturitas (2014) 77:32–6. doi:10.1016/j.maturitas.2013.10.018
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
50. Premratanachai P, Joly S, Johnson GK, McCray PB, Jia HP, Guthmiller JM. Expression and regulation of novel human β-defensins in gingival keratinocytes. Oral Microbiol Immunol (2004) 19:111–7. doi:10.1111/j.0902-0055.2002.00127.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
51. Huang L, Ching CB, Jiang R, Leong SSJ. Production of bioactive human beta-defensin 5 and 6 in Escherichia coli by soluble fusion expression. Protein Expr Purif (2008) 61:168–74. doi:10.1016/j.pep.2008.05.016
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
52. Kao CY, Chen Y, Zhao YH, Wu R. ORFeome-based search of airway epithelial cell-specific novel human [beta]-defensin genes. Am J Respir Cell Mol Biol (2003) 29:71–80. doi:10.1165/rcmb.2002-0205OC
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
53. Bals R, Wang X, Wu Z, Freeman T, Bafna V, Zasloff M, et al. Human beta-defensin 2 is a salt-sensitive peptide antibiotic expressed in human lung. J Clin Invest (1998) 102:874–80. doi:10.1172/JCI2410
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
54. García JR, Krause A, Schulz S, Rodríguez-Jiménez FJ, Klüver E, Adermann K, et al. Human beta-defensin 4: a novel inducible peptide with a specific salt-sensitive spectrum of antimicrobial activity. FASEB J (2001) 15:1819–21. doi:10.1096/fj.00-0865fje
55. Harder J, Bartels J, Christophers E, Schroder JM. Isolation and characterization of human beta -defensin-3, a novel human inducible peptide antibiotic. J Biol Chem (2001) 276:5707–13. doi:10.1074/jbc.M008557200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
56. Goldman MJ, Anderson GM, Stolzenberg ED, Kari UP, Zasloff M, Wilson JM. Human beta-defensin-1 is a salt-sensitive antibiotic in lung that is inactivated in cystic fibrosis. Cell (1997) 88:553–60. doi:10.1016/S0092-8674(00)81895-4
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
57. Singh PK, Jia HP, Wiles K, Hesselberth J, Liu L, Conway BA, et al. Production of beta-defensins by human airway epithelia. Proc Natl Acad Sci U S A (1998) 95:14961–6. doi:10.1073/pnas.95.25.14961
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
58. Selsted ME, Ouellette AJ. Mammalian defensins in the antimicrobial immune response. Nat Immunol (2005) 6:551–7. doi:10.1038/ni1206
59. Sørensen OE, Thapa DR, Rosenthal A, Liu L, Roberts AA, Ganz T. Differential regulation of beta-defensin expression in human skin by microbial stimuli. J Immunol (2005) 174:4870–9. doi:10.4049/jimmunol.174.8.4870
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
60. Doss M, White MR, Tecle T, Hartshorn KL. Human defensins and LL-37 in mucosal immunity. J Leukoc Biol (2010) 87:79–92. doi:10.1189/jlb.0609382
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
61. Vylkova S, Nayyar N, Li W, Edgerton M. Human beta-defensins kill Candida albicans in an energy-dependent and salt-sensitive manner without causing membrane disruption. Antimicrob Agents Chemother (2007) 51:154–61. doi:10.1128/AAC.00478-06
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
62. Sun L, Finnegan CM, Kish-Catalone T, Blumenthal R, Garzino-Demo P, La Terra Maggiore GM, et al. Human beta-defensins suppress human immunodeficiency virus infection: potential role in mucosal protection. J Virol (2005) 79:14318–29. doi:10.1128/JVI.79.22.14318-14329.2005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
63. Quiñones-Mateu ME, Lederman MM, Feng Z, Chakraborty B, Weber J, Rangel HR, et al. Human epithelial beta-defensins 2 and 3 inhibit HIV-1 replication. AIDS (2003) 17:F39–48. doi:10.1097/00002030-200311070-00001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
64. Weinberg A, Quiñones-Mateu ME, Lederman MM. Role of human beta-defensins in HIV infection. Adv Dent Res (2006) 19:42–8. doi:10.1177/154407370601900109
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
65. Leikina E, Delanoe-Ayari H, Melikov K, Cho M-S, Chen A, Waring AJ, et al. Carbohydrate-binding molecules inhibit viral fusion and entry by crosslinking membrane glycoproteins. Nat Immunol (2005) 6:995–1001. doi:10.1038/ni1248
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
66. Lacerda AF, Vasconcelos EAR, Pelegrini PB, Grossi de Sa MF. Antifungal defensins and their role in plant defense. Front Microbiol (2014) 5:116. doi:10.3389/fmicb.2014.00116
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
67. Colilla FJ, Rocher A, Mendez E. gamma-Purothionins: amino acid sequence of two polypeptides of a new family of thionins from wheat endosperm. FEBS Lett (1990) 270:191–4. doi:10.1016/0014-5793(90)81265-P
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
68. Mendez E, Moreno A, Colilla F, Pelaez F, Limas GG, Mendez R, et al. Primary structure and inhibition of protein synthesis in eukaryotic cell-free system of a novel thionin, gamma-hordothionin, from barley endosperm. Eur J Biochem (1990) 194:533–9. doi:10.1111/j.1432-1033.1990.tb15649.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
69. Semple F, Dorin JR. β-defensins: multifunctional modulators of infection, inflammation and more? J Innate Immun (2012) 4:337–48. doi:10.1159/000336619
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
70. Liang SC, Tan X-Y, Luxenberg DP, Karim R, Dunussi-Joannopoulos K, Collins M, et al. Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J Exp Med (2006) 203:2271–9. doi:10.1084/jem.20061308
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
71. Matusevicius D, Kivisäkk P, He B, Kostulas N, Ozenci V, Fredrikson S, et al. Interleukin-17 mRNA expression in blood and CSF mononuclear cells is augmented in multiple sclerosis. Mult Scler (1999) 5:101–4. doi:10.1191/135245899678847275
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
72. Aarvak T, Chabaud M, Miossec P, Natvig JB. IL-17 is produced by some proinflammatory Th1/Th0 cells but not by Th2 cells. J Immunol (1999) 162:1246–51.
73. Albanesi C, Scarponi C, Cavani A, Federici M, Nasorri F, Girolomoni G. Interleukin-17 is produced by both Th1 and Th2 lymphocytes, and modulates interferon-gamma- and interleukin-4-induced activation of human keratinocytes. J Invest Dermatol (2000) 115:81–7. doi:10.1046/j.1523-1747.2000.00041.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
74. Röhrl J, Yang D, Oppenheim JJ, Hehlgans T. Human beta-defensin 2 and 3 and their mouse orthologs induce chemotaxis through interaction with CCR2. J Immunol (2010) 184:6688–94. doi:10.4049/jimmunol.0903984
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
75. Funderburg NT, Jadlowsky JK, Lederman MM, Feng Z, Weinberg A, Sieg SF. The toll-like receptor 1/2 agonists Pam(3) CSK(4) and human β-defensin-3 differentially induce interleukin-10 and nuclear factor-κB signalling patterns in human monocytes. Immunology (2011) 134:151–60. doi:10.1111/j.1365-2567.2011.03475.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
76. Funderburg N, Lederman MM, Feng Z, Drage MG, Jadlowsky J, Harding CV, et al. Human -defensin-3 activates professional antigen-presenting cells via toll-like receptors 1 and 2. Proc Natl Acad Sci U S A (2007) 104:18631–5. doi:10.1073/pnas.0702130104
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
77. Biragyn A, Ruffini PA, Leifer CA, Klyushnenkova E, Shakhov A, Chertov O, et al. Toll-like receptor 4-dependent activation of dendritic cells by beta-defensin 2. Science (2002) 298:1025–9. doi:10.1126/science.1075565
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
78. Röhrl J, Yang D, Oppenheim JJ, Hehlgans T. Identification and biological characterization of mouse beta-defensin 14, the orthologue of human beta-defensin 3. J Biol Chem (2008) 283:5414–9. doi:10.1074/jbc.M709103200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
79. Barabas N, Röhrl J, Holler E, Hehlgans T. Beta-defensins activate macrophages and synergize in pro-inflammatory cytokine expression induced by TLR ligands. Immunobiology (2013) 218:1005–11. doi:10.1016/j.imbio.2012.11.007
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
80. Ganz T. Defensins: antimicrobial peptides of innate immunity. Nat Rev Immunol (2003) 3:710–20. doi:10.1038/nri1180
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
81. Abu Bakar S, Hollox EJ, Armour JAL. Allelic recombination between distinct genomic locations generates copy number diversity in human beta- defensins. Proc Natl Acad Sci U S A (2009) 106:853–8. doi:10.1073/pnas.0809073106
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
82. Aldhous MC, Bakar SA, Prescott NJ, Palla R, Soo K, Mansfield JC, et al. Measurement methods and accuracy in copy number variation: failure to replicate associations of beta-defensin copy number with Crohn’s disease. Hum Mol Genet (2010) 19(24):4930–8. doi:10.1093/hmg/ddq411
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
83. Hardwick RJ, Machado LR, Zuccherato LW, Antolinos S, Xue Y, Shawa N, et al. A worldwide analysis of beta-defensin copy number variation suggests recent selection of a high-expressing DEFB103 gene copy in East Asia. Hum Mutat (2011) 32(7):743–50. doi:10.1002/humu.21491
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
84. Barber JC, Joyce CA, Collinson MN, Nicholson JC, Willatt LR, Dyson HM, et al. Duplication of 8p23.1: a cytogenetic anomaly with no established clinical significance. J Med Genet (1998) 35:491–6. doi:10.1136/jmg.35.6.491
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
85. Hollox EJ, Barber JCK, Brookes AJ, Armour JAL. Defensins and the dynamic genome: what we can learn from structural variation at human chromosome band 8p23.1. Genome Res (2008) 18:1686–97. doi:10.1101/gr.080945.108
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
86. Hollox E, Hardwick R, Machado L, Zuccherato L, Antolinos S, Xue Y, et al. A worldwide analysis of beta-defensin copy number variation suggests recent selection of a high-expressing DEFB103 gene copy in East Asia. Hum Mutat (2011) 32(7):743–50. doi:10.1002/humu.21491
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
87. Jansen PAM, Rodijk-Olthuis D, Hollox EJ, Kamsteeg M, Tjabringa GS, de Jongh GJ, et al. Beta-defensin-2 protein is a serum biomarker for disease activity in psoriasis and reaches biologically relevant concentrations in lesional skin. PLoS One (2009) 4:e4725. doi:10.1371/journal.pone.0004725
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
88. Hollox EJ, Hoh B-P. Human gene copy number variation and infectious disease. Hum Genet (2014) 133:1217–33. doi:10.1007/s00439-014-1457-x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
89. Hollox EJ, Armour JAL, Barber JCK. Extensive normal copy number variation of a beta-defensin antimicrobial-gene cluster. Am J Hum Genet (2003) 73:591–600. doi:10.1086/378157
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
90. Linzmeier RM, Ganz T. Human defensin gene copy number polymorphisms: comprehensive analysis of independent variation in alpha- and beta-defensin regions at 8p22-p23. Genomics (2005) 86:423–30. doi:10.1016/j.ygeno.2005.06.003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
91. Hollox EJ, Davies J, Griesenbach U, Burgess J, Alton EWFW, Armour JAL. Beta-defensin genomic copy number is not a modifier locus for cystic fibrosis. J Negat Results Biomed (2005) 4:9. doi:10.1186/1477-5751-4-9
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
92. Chen Q, Book M, Fang X, Hoeft A, Stuber F. Screening of copy number polymorphisms in human beta-defensin genes using modified real-time quantitative PCR. J Immunol Methods (2006) 308:231–40. doi:10.1016/j.jim.2005.11.001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
93. Fellermann K, Stange DE, Schaeffeler E, Schmalzl H, Wehkamp J, Bevins CL, et al. A chromosome 8 gene-cluster polymorphism with low human beta-defensin 2 gene copy number predisposes to Crohn disease of the colon. Am J Hum Genet (2006) 79:439–48. doi:10.1086/505915
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
94. Hollox EJ, Huffmeier U, Zeeuwen PLJM, Palla R, Lascorz J, Rodijk-Olthuis D, et al. Psoriasis is associated with increased beta-defensin genomic copy number. Nat Genet (2008) 40:23–5. doi:10.1038/ng.2007.48
95. Armour JAL, Palla R, Zeeuwen PLJM, den Heijer M, Schalkwijk J, Hollox EJ. Accurate, high-throughput typing of copy number variation using paralogue ratios from dispersed repeats. Nucleic Acids Res (2007) 35:e19. doi:10.1093/nar/gkl1089
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
96. Groth M, Szafranski K, Taudien S, Huse K, Mueller O, Rosenstiel P, et al. High-resolution mapping of the 8p23.1 beta-defensin cluster reveals strictly concordant copy number variation of all genes. Hum Mutat (2008) 29:1247–54. doi:10.1002/humu.20751
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
97. Bentley RW, Pearson J, Gearry RB, Barclay ML, McKinney C, Merriman TR, et al. Association of higher DEFB4 genomic copy number with Crohn’s disease. Am J Gastroenterol (2010) 105:354–9. doi:10.1038/ajg.2009.582
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
98. Hardwick RJ, Amogne W, Mugusi S, Yimer G, Ngaimisi E, Habtewold A, et al. β-defensin genomic copy number is associated with HIV load and immune reconstitution in Sub-Saharan Africans. J Infect Dis (2012) 206:1012–9. doi:10.1093/infdis/jis448
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
99. Zhou X-J, Cheng F-J, Lv J-C, Luo H, Yu F, Chen M, et al. Higher DEFB4 genomic copy number in SLE and ANCA-associated small vasculitis. Rheumatology (Oxford) (2012) 51:992–5. doi:10.1093/rheumatology/ker419
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
100. Taudien S, Gäbel G, Kuss O, Groth M, Grützmann R, Huse K, et al. Association studies of the copy-number variable ß-defensin cluster on 8p23.1 in adenocarcinoma and chronic pancreatitis. BMC Res Notes (2012) 5:629. doi:10.1186/1756-0500-5-629
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
101. Wain LV, Odenthal-Hesse L, Abujaber R, Sayers I, Beardsmore C, Gaillard EA, et al. Copy number variation of the beta-defensin genes in Europeans: no supporting evidence for association with lung function, chronic obstructive pulmonary disease or asthma. PLoS One (2014) 9:e84192. doi:10.1371/journal.pone.0084192
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
102. Jones EA, Kananurak A, Bevins CL, Hollox EJ, Bakaletz LO. Copy number variation of the beta defensin gene cluster on chromosome 8p influences the bacterial microbiota within the nasopharynx of otitis-prone children. PLoS One (2014) 9:e98269. doi:10.1371/journal.pone.0098269
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
103. Taudien S, Huse K, Groth M, Platzer M. Narrowing down the distal border of the copy number variable beta-defensin gene cluster on human 8p23. BMC Res Notes (2014) 7:93. doi:10.1186/1756-0500-7-93
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
104. Barber JCK, Maloney V, Hollox EJ, Stuke-Sontheimer A, du Bois G, Daumiller E, et al. Duplications and copy number variants of 8p23.1 are cytogenetically indistinguishable but distinct at the molecular level. Eur J Hum Genet (2005) 13:1131–6. doi:10.1038/sj.ejhg.5201475
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
105. Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res (2002) 30:e57. doi:10.1093/nar/gnf056
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
106. Field SF, Howson JMM, Maier LM, Walker S, Walker NM, Smyth DJ, et al. Experimental aspects of copy number variant assays at CCL3L1. Nat Med (2009) 15:1115–7. doi:10.1038/nm1009-1115
107. He W, Kulkarni H, Castiblanco J, Shimizu C, Aluyen U, Maldonado R, et al. Reply to: “experimental aspects of copy number variant assays at CCL3L1”. Nat Med (2009) 15:1117–20. doi:10.1038/nm1009-1117
108. Aklillu E, Odenthal-Hesse L, Bowdrey J, Habtewold A, Ngaimisi E, Yimer G, et al. CCL3L1 copy number, HIV load, and immune reconstitution in sub-Saharan Africans. BMC Infect Dis (2013) 13:536. doi:10.1186/1471-2334-13-536
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
109. Stuart PE, Hüffmeier U, Nair RP, Palla R, Tejasvi T, Schalkwijk J, et al. Association of β-defensin copy number and psoriasis in three cohorts of European origin. J Invest Dermatol (2012) 132:2407–13. doi:10.1038/jid.2012.191
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
110. Harder J, Schröder J-M. Psoriatic scales: a promising source for the isolation of human skin-derived antimicrobial proteins. J Leukoc Biol (2005) 77:476–86. doi:10.1189/jlb.0704409
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
111. Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature (2007) 447:661–78. doi:10.1038/nature05911
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
112. Peyrin-Biroulet L, Beisner J, Wang G, Nuding S, Oommen ST, Kelly D, et al. Peroxisome proliferator-activated receptor gamma activation is required for maintenance of innate antimicrobial immunity in the colon. Proc Natl Acad Sci U S A (2010) 107:8772–7. doi:10.1073/pnas.0905745107
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
113. Jespersgaard C, Fode P, Dybdahl M, Vind I, Nielsen OH, Csillag C, et al. Alpha-defensin DEFA1A3 gene copy number elevation in Danish Crohn’s disease patients. Dig Dis Sci (2011) 56:3517–24. doi:10.1007/s10620-011-1794-8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
114. Khan FF, Carpenter D, Mitchell L, Mansouri O, Black HA, Tyson J, et al. Accurate measurement of gene copy number for human alpha-defensin DEFA1A3. BMC Genomics (2013) 14:719. doi:10.1186/1471-2164-14-719
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
115. Milanese M, Segat L, Arraes LC, Garzino-Demo A, Crovella S. Copy number variation of defensin genes and HIV infection in Brazilian children. J Acquir Immune Defic Syndr (2009) 50:331–3. doi:10.1097/QAI.0b013e3181945f39
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
116. Mehlotra RK, Zimmerman PA, Weinberg A, Jurevic RJ. Variation in human β-defensin genes: new insights from a multi-population study. Int J Immunogenet (2013) 40:261–9. doi:10.1111/iji.12021
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: copy number variation, defensins, HIV, psoriasis, Crohn’s disease
Citation: Machado LR and Ottolini B (2015) An evolutionary history of defensins: a role for copy number variation in maximizing host innate and adaptive immune responses. Front. Immunol. 6:115. doi: 10.3389/fimmu.2015.00115
Received: 22 December 2014; Accepted: 01 March 2015;
Published online: 18 March 2015.
Edited by:
Uday Kishore, Brunel University, UKReviewed by:
Kenneth Reid, University of Oxford, UKSilvia Bulfone-Paus, University of Manchester, UK
Copyright: © 2015 Machado and Ottolini. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lee R. Machado, Institute of Health and Wellbeing, School of Health, University of Northampton, Boughton Green Road, Northampton NN2 7AL, UK e-mail:bGVlLm1hY2hhZG9Abm9ydGhhbXB0b24uYWMudWs=
 Lee R. Machado
Lee R. Machado Barbara Ottolini
Barbara Ottolini