- Department of Genetics, University Medical Center Groningen, University of Groningen, Groningen, Netherlands
Cellular differentiation is regulated on the level of gene expression, and it is known that dysregulation of gene expression can lead to deficiencies in differentiation that contribute to a variety of diseases, particularly of the immune system. Until recently, it was thought that the dysregulation was governed by changes in the binding or activity of a class of proteins called transcription factors. However, the discovery of micro-RNAs and recent descriptions of long non-coding RNAs (lncRNAs) have given enormous momentum to a whole new field of biology: the regulatory RNAs. In this review, we describe these two classes of regulatory RNAs and summarize what is known about how they regulate aspects of the adaptive and innate immune systems. Finally, we describe what is known about the involvement of micro-RNAs and lncRNAs in three different autoimmune diseases (celiac disease, inflammatory bowel disease, and multiple sclerosis).
Introduction
The discovery of the first micro-RNA (miRNA) in 1993 (1, 2) was the start of research that has led to the understanding that gene regulation is not only controlled by proteins (transcription factors) but also RNA molecules. Since then, thousands of novel non-coding RNAs, which can be subdivided into dozens of families (3), have been identified. Two of the most widely studied classes of non-coding RNAs, miRNAs and long non-coding RNAs (lncRNAs), are now recognized as important regulators of gene expression. These molecules are also designated as (small or long) regulatory RNAs. At the time of writing this review, the authorative miRNA database miRBase (release 21) describes 1,881 human miRNA precursors and 2,588 human mature miRNA sequences (4), whereas the GENCODE compendium (V19) mentions 13,870 human lncRNA genes (5). MiRNAs are thought to affect gene expression by inhibiting target mRNA translation (which leads indirectly to degradation of the target) or they can directly induce target mRNA degradation. Many lncRNAs are thought to be involved in chromatin modification processes that, in turn, affect gene expression levels (Figure 1). The role of miRNAs in homeostasis and the deregulation of miRNAs in human disease have been well established, but the role of lncRNAs in these processes is not yet fully appreciated. Here, we will review what is known about the role of miRNAs and lncRNAs in the development and activation of the adaptive and innate immune systems in health and disease.
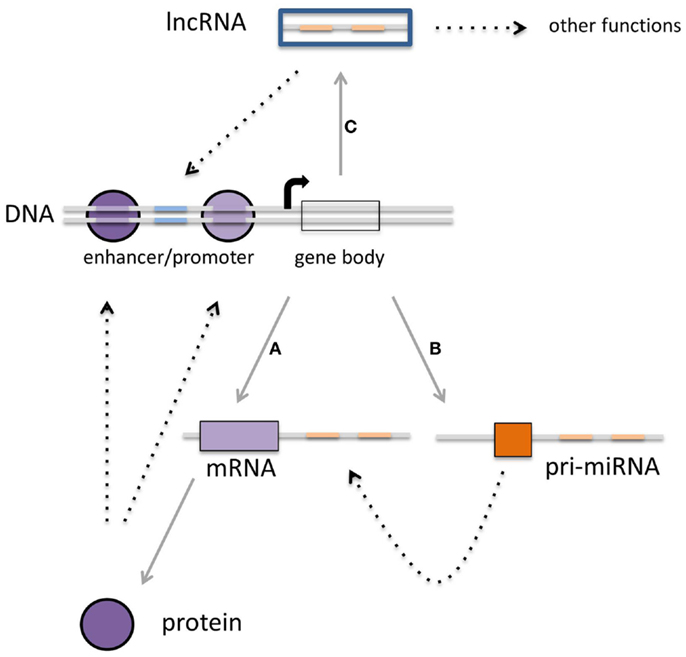
Figure 1. Multiple layers of gene expression controlled by transcription factors, miRNAs, and lncRNAs. (A) Protein-coding genes are transcribed into mRNA, which subsequently are translated into proteins. These proteins can function as the classical transcription factors. (B) There is a second class of RNAs that is not translated into protein but rather is regulating the expression of other transcripts. The third class of transcripts described in this review (C) is the long non-coding RNAs that can regulate gene expression as well, although other functions for these transcripts have been described (see Figure 3). It is becoming clear that there is interaction within each class, but also between these three classes, which can converge on transcriptional outcome (see text for details).
MiRNAs
Micro-RNAs are short (19–24 nt), single-stranded, RNAs that are involved in the post-transcriptional regulation of gene expression. Their sequences are evolutionary strongly conserved. miRNA expression profiles and target mRNA sites are also conserved, allowing the translation of findings in mouse models to human physiology.
Micro-RNAs are transcribed by RNA polymerase II into longer (several hundred to several thousand nucleotides) primary miRNAs (pri-miRNAs), containing a cap as well as a poly-A tail. The pri-miRNA is processed in the nucleus by a microprocessor, a complex composed of Drosha (a class III RNase) and the DiGeorge syndrome critical region gene 8 (DGCR8), into a ~60 nt precursor-miRNA (pre-miRNA). In this step, the double-stranded stem-loop structures are specifically recognized by the microprocessor, which catalyzes the cleavage of the pri-miRNA near the base of the stem (6, 7). Then, the double-stranded pre-miRNA stem-loop structure is transported into the cytoplasm by a complex containing exportin 5 (XPO5), where it is recognized and further processed by a class III RNase named Dicer into a double-stranded RNA duplex of ~19–24 nt in length. Next, only one of the strands is incorporated into the RNA-induced silencing complex (RISC) composed of Argonaute (AGO) and GW182. The RISC complex is guided to the 3′-untranslated region (3′-UTR) of target mRNA molecules. This leads successively to a decrease in target stability, resulting in accelerated uncapping and deadenylation (8) and/or inhibition of translation (9) (Figure 2). It has been suggested that the translational repression of mRNAs takes place in specialized compartments called processing bodies (P-bodies), compartments in the cytoplasm involved in the storage, and degradation of repressed mRNAs (10). To make things more complex, miRNAs were shown to be transported to the nucleus, where they can affect their own expression or the expression of other miRNAs (11). Moreover, lncRNAs can act as sponges for miRNAs. It was demonstrated that the lncRNA phosphatase and tensin homolog (PTEN) pseudogene1 (PTENpg1) sequesters various PTEN-targeting miRNAs, thereby indirectly regulating the PTEN mRNA level (12).
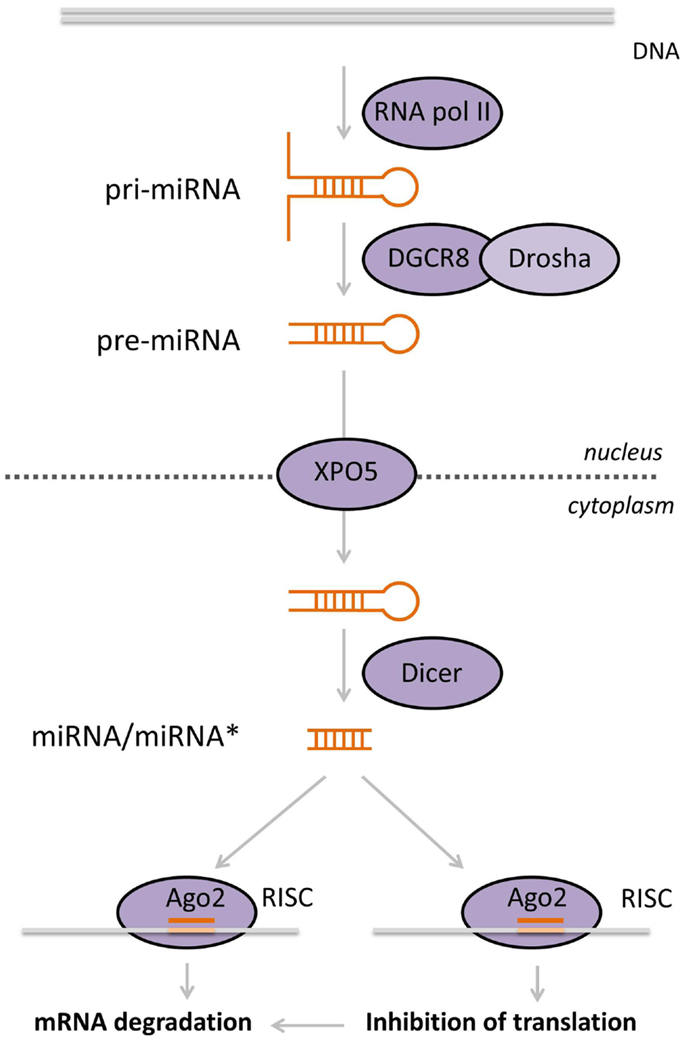
Figure 2. Biogenesis of miRNAs. MiRNAs are transcribed by RNA pol II in the nucleus. Double-stranded miRNA hairpins in the pri-miRNA transcript are recognized and cleaved by the microprocessor complex, composed of Drosha and DGCR8, producing the pre-miRNA. These are subsequently exported into the cytoplasm by XPO5, where they are recognized by Dicer, which cuts off the loop of the hairpin yielding a small double-stranded RNA molecule. One of these strands, the mature miRNA, is loaded into the RISC complex that contains AGO2. This complex is guided to the target mRNA, based on sequence homology between the miRNA and the target. Ultimately, this leads to diminished mRNA translation and/or degradation of the target.
Reports describing that miRNAs can originate from other regulatory RNAs, like tRNAs (13) or pre-ribosomal RNAs (14), complicate the “canonical pathway of miRNA production.” MiRNAs can also be derived from introns, which mimic the structural features of pre-miRNAs and these miRNAs can, therefore, enter the miRNA processing pathway independent of Drosha (15). This group of miRNAs can even be subdivided into two groups: (a) splicing-dependent miRNAs (mirtrons, e.g., human miR-877 and miR-1226) or (b) splicing-independent simtrons (human miR-1225 and miR-1228). The processing of mirtrons requires the spliceosome but not Drosha or DGCR8, whereas the generation of simtrons depends on Drosha (but not on DGCR8). Mirtrons are exported from the nucleus by XPO5, cleaved by Dicer, and subsequently enter the RISC complex, similarly to canonical miRNAs. We do not yet know what factors regulate simtron export from the nucleus, but simtrons also enter the RISC complex in the cytoplasm (16).
Interest in miRNAs grew when it was found that miRNAs can be detected in many body fluids such as serum, cerebrospinal fluid (CSF), saliva, and urine (17) and that miRNA profiles are remarkably stable (e.g., resistant to RNases, freeze-thaw cycles). This protection from degradation is probably conferred by one or more mechanisms: (1) miRNAs can be bound to protein components of the RISC complex (AGO2), (2) they can be bound to high density lipoproteins, or (3) they can be packaged in exosomes (18). It was exciting to discover that miRNA profiles in circulation can be disease or even disease stage specific (19). Moreover, they can be useful in predicting treatment response (20). Research into the biological role of circulating miRNAs is still in its infancy, but recent papers describe the intriguing possibility that miRNA can be secreted by one cell type and can then exert its function on or in other cell types (21–23). Exosomes have been shown to participate in various processes that are crucial for immune system function and they can be released by various immune cell types, e.g., T-cells, B-cells, and dendritic cells (DCs). Importantly, these exosomes contain miRNAs, some of which are cell-type specific, while others are present in exosomes of various cell types. Moreover, some miRNAs are more highly expressed in exosomes than in the cells that excrete them, implying that a subset of miRNAs is specifically packaged (24). The selection of miRNA for packaging into exosomes has been described based on two specific motifs in the miRNA sequence; these are recognized by sumoylated heterogeneous nuclear ribonucleoprotein A2/B1 (hnRNPA2B1), a protein controlling miRNA loading into exosomes (25).
Long Non-Coding RNAs
Long non-coding RNAs are a heterogeneous group of non-coding transcripts longer than 200 nucleotides (26, 27) and they constitute the major class of regulatory RNA genes (28, 29). Thousands of mammalian lncRNAs have been identified since the first genome-wide discovery studies in the early 2000s and it has become clear that they play important roles in regulating several biological processes, such as gene expression, chromatin remodeling, and protein transport. Although many lncRNAs have been identified, little is known about either their general characteristics or their possible mechanisms of action in health and disease. They can be detected both in the nucleus and in the cytosol, and can be polyadenylated or not. Compared to protein-coding genes, lncRNAs have fewer but longer exons, which are poorly conserved across species (26, 30). In general, the expression of lncRNAs is lower than that of protein-coding genes, although in a cell-type-specific context the expression can be just as high (26). There is growing evidence pointing to changes in lncRNA expression being associated with the etiopathology of diseases, for instance in cancer and autoimmune disease (18, 31). Expression profiling of specific immune cell subsets has revealed an enrichment of long intergenic non-coding RNAs (lincRNAs) that are expressed in immune cells in autoimmune disease-associated loci, thereby implying that these non-coding RNAs play a role in the etiology of autoimmune disease (Barbara Hrdlickova, personal communication). Furthermore, expression quantitative trait locus (eQTL) analysis has demonstrated that disease-associated single nucleotide polymorphisms (SNPs) can affect the expression of lncRNAs, relating lncRNAs to disease susceptibility (32).
Long non-coding RNAs are a structurally and functionally heterogeneous group of transcripts. One approach classifies them into four different subclasses based on their location with respect to the closest protein-coding gene (5, 18). The largest subclass consists of the lincRNAs, which do not overlap with protein-coding genes. Of the remaining “genic” lncRNAs (the second largest subclass), the antisense lncRNA group contains transcripts that overlap with exons of protein-coding genes on the opposite strand (natural antisense transcripts, or NATs) or transcripts that reside in an intron of the protein-coding gene on the opposite strand (antisense intronic transcripts). The antisense and sense transcripts are often co-expressed. The third subclass of lncRNAs encompasses the sense lncRNAs. These transcripts can contain coding genes within an intron on the same strand (sense overlapping transcripts), or they can be located within an intron of a protein-coding gene on the same strand (sense intronic transcripts). The fourth subclass comprises the bi-directional or divergent lncRNA transcripts. These are antisense transcripts that co-transcribe in the opposite direction to the protein-coding gene.
The GENCODE (V7) compendium has annotated over 13,000 human lncRNAs, of which only a fraction, however, has a known function (26). LncRNAs can have diverse molecular functions relayed by the molecules they interact with: mRNA, protein, miRNAs, or DNA (Figure 3) (33). These interactions can affect processes like transcription, translation, splicing, translation, or RNA degradation (34, 35). Chang and Rinn classified lncRNAs into four subclasses by their different functions (34). For example, lncRNAs can function as molecular scaffolds to bring proteins together in a complex, but they can also act as a signal for a specific biological condition or state, for instance cellular stress or temperature. The signal can subsequently activate or repress the expression of other genes. Another function lncRNAs can exhibit is that of being a decoy, in which they bind to other RNAs or proteins and interfere with their function. Finally, lncRNAs can guide protein complexes to targets, where they can act as activators or repressors of other genes. In addition to these four main functions, some lncRNAs can inhibit the function of miRNAs, thereby alleviating the downregulating effect of the miRNA on the gene expression (36–38). Note that it is also possible for lncRNAs to exert multiple of these functions.
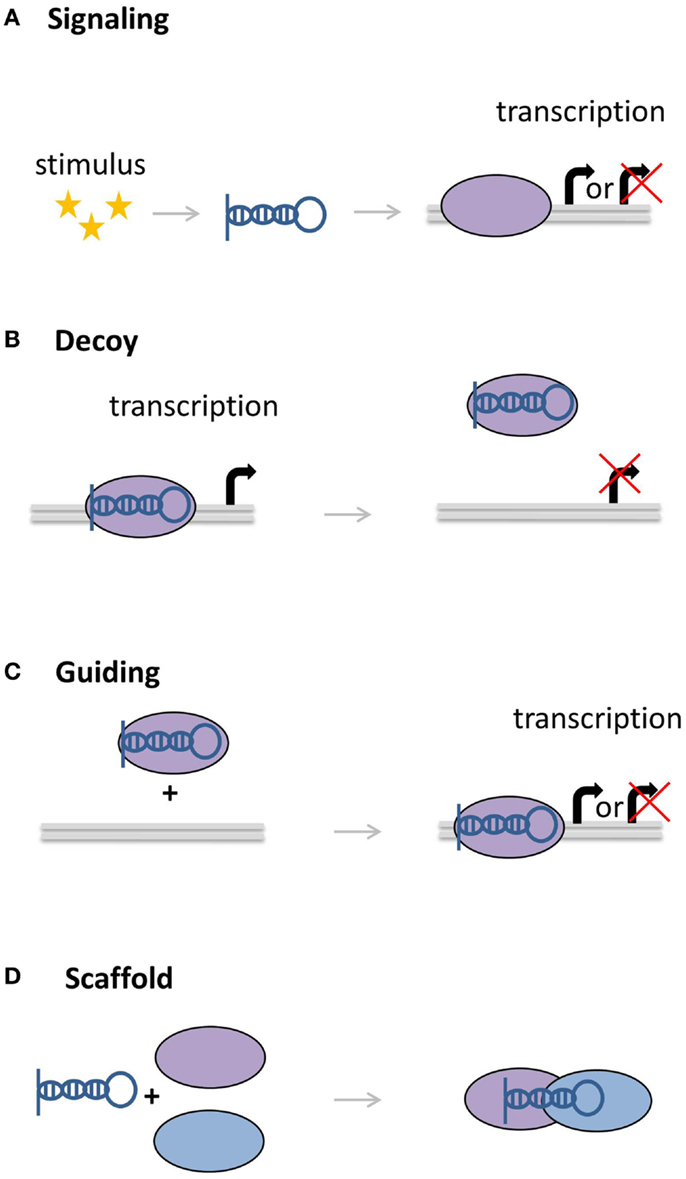
Figure 3. Molecular functions of lncRNAs. (A) LncRNAs can act as signaling molecules, affecting the expression of genes in response to a stimulus. (B) LncRNAs can divert transcription factors or other proteins away from the DNA. (C) Other lncRNAs can recruit proteins, bringing them closer to target genes. (D) As scaffolds, lncRNAs can bring together multiple proteins to form complexes.
The Role of miRNAs and lncRNAs in the Immune System
It has been proposed that miRNA emerged as a primitive immune response against viral infection. The fact that they show remarkable conservation in animals and plants suggests they hold important biological functions. MiRNAs play important roles in cell physiology, as clearly demonstrated by the fact that Dicer knockout in mouse embryos is incompatible with life. Deleting or overexpressing individual miRNAs in mice offers the opportunity to study their roles in the immune response (39). Lineage-specific knockout (KO) of miRNAs in specific immune cell types results in severe perturbation of immune cell numbers, their composition, and function. These all points to miRNAs being essential for immune cell development, differentiation, function, and homeostasis (39–43).
Dicer1 deletion in granulocyte-macrophage progenitors (derived from a myeloid-specific CCAAT/enhancer binding protein-α (Cebpa)-Cre-driven Dicer1-deleter mouse strain) resulted in changes in gene expression profiles, increased self-renewal ability of precursors in the bone marrow (BM), monocyte depletion, and myeloid dysplasia, underlining the essential contribution of miRNAs to myeloid development (44). The role of Dicer and miRNAs has also been demonstrated in natural killer (NK) cells. By using mice with conditional deletion of Dicer1 or Dgcr8 in NK cells, a reduced cellularity in the spleen was observed with a concomitant reduced frequency of splenic NK cells, but without alterations in T- and B-cell frequencies. Dicer and Dgcr8 deficiency was associated with an increase in NK apoptosis and an impairment in NK activation, suggesting that miRNAs are required for NK homeostasis and function (45).
As discussed above, miRNAs can be packaged into exosomes that are subsequently secreted from the cell. It has been suggested that circulating miRNAs could act in intercellular communication, also in the immune system. For example, it was suggested that T-cells communicate with antigen-presenting cells (APCs) by a unidirectional transfer of exosomal miRNA. Another example is the transfer of miR-335 (downregulating SOX4, a progenitor cell transcription factor), which was correlated with the transfer of CD63 upon formation of the immune synapse (24). Furthermore, exosomes released from mature BM-derived DCs contain more miR-125-5p, miR-146a, and miR-148, which are negative regulators of pro-inflammatory factors in myeloid cells and DCs. Exosomes released by both immature and by mature BM-derived DCs contain miR-34a and miR-21 (known to regulate the differentiation of hematopoietic precursors into myeloid DCs), as well as miR-221 and miR-222 (that prevent differentiation into plasmacytoid DCs). Such exosomes can be taken up by recipient DCs, and the packaged miRNAs can then be released to target known binding sites, as shown by 3′-UTR-luciferase experiments (46). Exosome-derived miRNAs have also been implicated in the progression of Epstein–Barr virus (EBV) infection. Exosomes containing EBV-derived miRNAs are released from infected B-cells and taken up by DCs, where the miRNAs can then downregulate the expression of genes encoding immune-stimulation factors (47). Together, these limited but suggestive data point to a role for miRNA-based intercellular communication, mediated by exosomes, in the immune system, which has implications for health and disease.
Because lncRNAs are not produced via a lncRNA-specific biochemical pathway, it is not feasible to generate general or lineage-specific lncRNA-knockout mice. The lack of evolutionary conservation of lncRNAs across species further complicates the study of their individual function. Nevertheless, several mouse knockouts have been generated for single lncRNAs. A landmark study described the generation of 18 mouse strains, all with one lncRNA deleted (48). Although the lncRNA candidates were not selected for immune cell specificity, the study revealed key roles for several individual lincRNAs in the viability and developmental processes of the mice and it also highlighted the importance of using in vivo models to reveal the biological significance and functional diversity of lncRNAs (48).
In the next section, we give examples of how key miRNAs play regulatory roles in the development and activation of the immune system and we summarize the much smaller body of evidence implicating lncRNAs in these processes.
The Role of MiRNAs in the Development of Innate Immune Cells
The innate immune system includes myeloid cells derived from hematopoietic stem cells (HSCs) and myeloid progenitors. These cells give rise to monocytes, which can develop into macrophages and DCs, and to granulocytes (neutrophils, eosinophils, basophils) through a series of developmental stages (myeloblast, promyelocyte, myelocyte, metamyelocyte, band cell or monoblast, and promonocyte) (Figure 4). One of the first studies of miRNA expression in normal human granulocytes reported sets of miRNAs that were subject to upregulation or downregulation at discrete maturation stages in neutrophil development. Although the majority of miRNA family members showed coordinated expression patterns, the expression of some miRNAs in the same cluster is not always synchronized. For example, the miR-17-92/oncomir-1 cluster encompasses six miRNAs (miR-17, -18a, -19a, -20a, -19b-1, -92a-1). Among the cluster’s targets are antitumor, pro-apoptotic, and tumor suppressor proteins. HSCs and early progenitors in the BM express high levels of miRNAs from this cluster, whereas their expression is reduced during myeloid and lymphoid differentiation (49, 50). Of this cluster, miR-20a and miR-92 are downregulated in metamyelocytes, miR-18a, miR-19a and miR-19b are downregulated in neutrophils, while miR17-5p gradually decreased from myeloblasts in the subsequent stages of development (51). In miR-223 KO mice, it was shown that miR-223 deletion leads to an increase in the number of granulocyte progenitors and neutrophil hyperactivity, suggesting that miR-223 acts as a crucial regulator of granulocyte production and the inflammatory response (52).
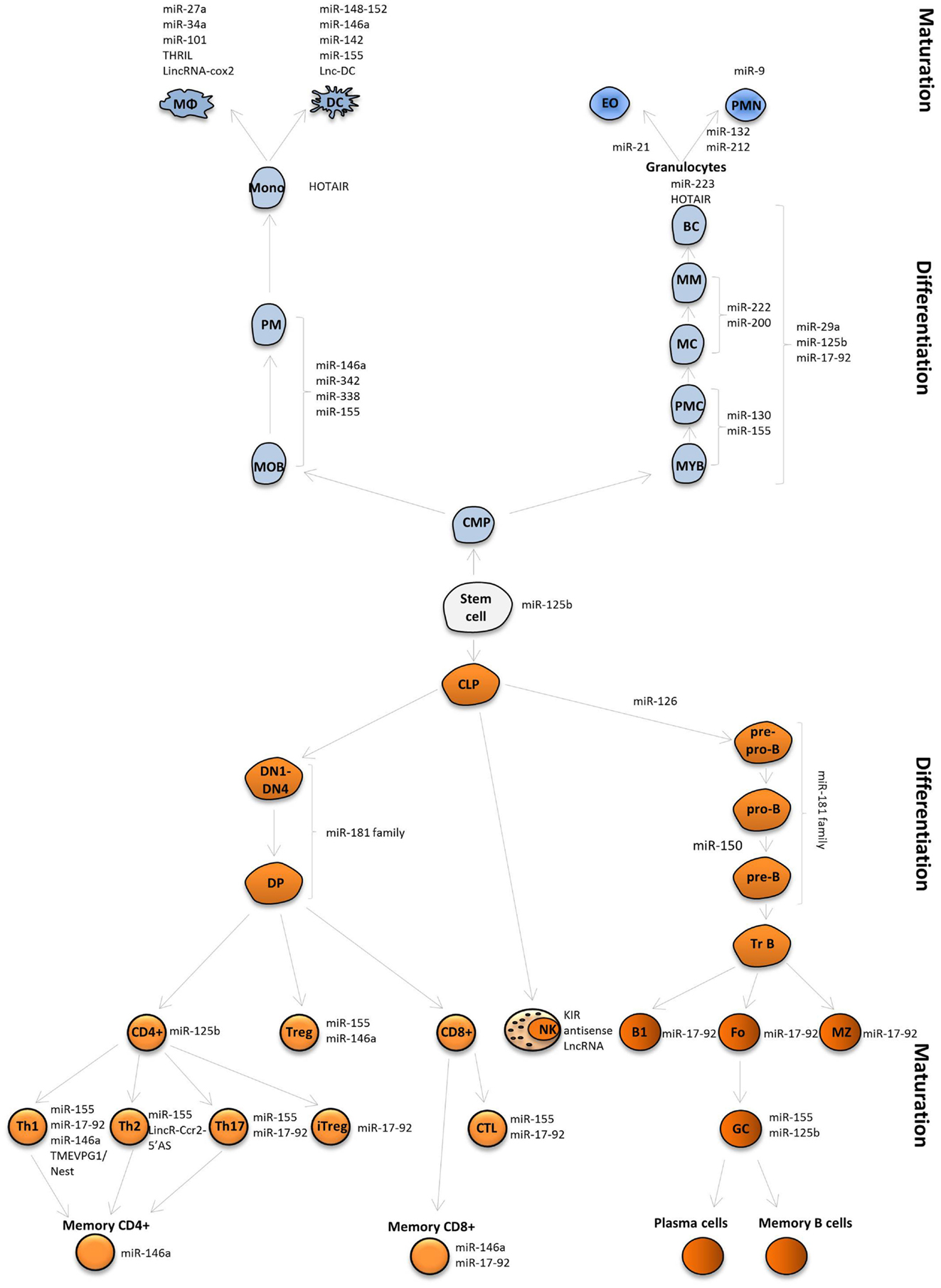
Figure 4. MiRNAs and lncRNAs influence immune cell fate and function. MiRNAs and lncRNAs were shown to modulate development and function the immune system. LncRNAs and selected miRNAs that are discussed in this review are depicted.
In vitro overexpression or knockdown experiments of miR-29a or miR-142-3p in human leukemia cell lines showed that the miRNA overexpression promoted monocytic and myelocytic maturation, while blockage with antisense inhibitors promoted not only the expression of early progenitor markers but also reduced cell maturation, indicating these miRNAs play roles as regulators of normal myeloid differentiation. MiR-29a and miR-142-3p were both shown to target cyclin T2 (CCNT2), while they were also specifically targeting the cyclin-dependent kinase 6 (CDK6) gene (miR-29a) and the transforming growth factor β (TGFβ) activated kinase 1/MAP3K7 binding protein 2 (TAB2) gene (miR-142-3p) (41).
An important group of miRNAs in myeloid biology is the miR-125 family, consisting of miR-125a, -125b1, and -125b2. The family members target crucial factors involved in HSC survival and apoptosis (42). MiR-125b overexpression, instigated by transplanting fetal liver cells ectopically expressing miR-125b in mice, caused a lethal myeloproliferative disorder (43). In addition, enforced expression of miR-125b in BM chimeric mice promoted myelopoiesis. B-cell lymphoma-2 (Bcl-2) homologous antagonist/killer 1 (Bak1) and the signal transducer and activator of transcription 3 (Stat3) were proposed as possible target genes (53). Moreover, using a miRNA-sponge approach, it was shown that miR-125b can also regulate myelopoiesis in mice by targeting Lin28A, an important regulator of hematopoiesis (54).
In PU.1-deficient mice, the development of macrophages, granulocytes, and B-lymphocytes is impaired, revealing that the PU.1 transcription factor is involved in myeloid and lymphoid development (55). Several miRNAs, including the miR-17-92 cluster, are activated by PU.1 to modulate macrophage development. In PUER cells (murine myeloid progenitors in which macrophage development can be supported on inducing a tamoxifen-inducible PU.1 transgene), it was demonstrated that macrophage differentiation requires downregulation of miR-17-92 (49). Moreover, PU.1 may also regulate macrophage development by inducing miR-146a, miR-342, miR-338, and miR-155 (56). miR-142 is another miRNA involved in myeloid development. In miR-142-deficient mice, a reduction of CD4+ DCs is accompanied by a severe defect in their ability to prime CD4+ T-cells (57). MiRNA expression profiling during human monocyte differentiation has shown a decrease in levels of miRNAs, the miR-17-92 cluster (miR-17-5p and miR-20a), as well as of miR-106a (a member of miR-106a-363, a paralog of the miR-17-92 cluster), compared to early progenitors. One of the shared targets of miR-17-5p, miR-20a, and miR-106a is the runt-related transcription factor 1 (RUNX-1) gene, an important regulator of hematopoiesis (58).
Analysis of miRNA profiles in human BM precursors and neutrophils revealed that 135 miRNAs were differentially expressed between the myeloid developmental stages. For instance, high levels of miR-130a, miR-155, and miR-146a were observed in myeloblasts and promyelocytes, followed by a decrease in expression in more mature cells. Potential targets for these miRNAs include transcripts encoding members of the TGFβ signaling pathway, such as TGFβ-receptor 1 (TGFβR1) and TGFβR2, SMAD2, SMAD4, and SMAD5 (miR-130). Some miRNAs clustered with the intermediate stages of development (miR-222, miR-200, miR 29a), while others were associated with mature neutrophils (miR-132, miR-212). Among the predicted targets of the miRNAs listed above are transcripts encoding cell cycle regulators, such as CDK2 (miR-155), or proteins associated with apoptosis such as apoptotic protease activating factor 1 (APAF1), CASP8, and Fas-associated death domain (FADD), which are targeted by miR-132, miR-212 (59).
Another miRNA important in the development of the innate immune system is miR-21. It has been identified as one of the most highly upregulated miRNAs in allergic diseases and this is associated with high numbers of eosinophils, the main effector cells in allergic responses. In a report by Lu et al. (60), the role of miR-21 was evaluated in a murine ex vivo culture system. By using RT-PCR, it was shown that during eosinophil differentiation miR-21 was upregulated threefold from day 4 to day 14 in culture. Cultures derived from miR-21−/− eosinophil progenitor cells showed higher apoptosis than cultures from miR-21+/+ progenitor cells, suggesting that miR-21 regulates the development of eosinophils by modulating eosinophil progenitor cell growth. In agreement with these findings, miR-21−/−mice showed reduced blood eosinophil levels, concomitant with a reduced capacity to produce eosinophils in the BM. Microarray analysis revealed the differential expression of genes involved in cell proliferation, cell cycle control, and the immune response (60).
MiRNAs Involved in Innate Immune Cell Activation
The best-known example of a miRNA involved in the activation of innate and adaptive immune cells is miR-155. It has also been implicated as a general and conserved feature of mouse and human DC activation by various Toll-like receptor (TLR) ligands (61, 62). Analyses of miR-155-deficient mice demonstrated that although the development of DCs was unaffected, miR-155 is required for DC maturation and the ability to promote antigen-specific T-cell activation (61). DC maturation is also affected by miR-150, miR-34a, and let-7i by mechanisms that involve silencing c-Fos (miR-155), Csf1r (which controls M-CSF receptor expression; miR-34a), and suppressor of cytokine signaling-1 (SOCS1) (let-7i) expression (61–64). Another group of miRNAs, including miR-146a, miR-148, and miR-142, have been associated with downregulation of inflammatory pathways and modulation of DC maturation (65–67). MiR-146a controls DC cross-priming (by suppressing Notch1 expression and IL-12p70 production) and DC activation (by targeting TLR9, TLR2, interleukin-1 receptor-associated kinase 1 (IRAK1), and tumor necrosis factor (TNF) receptor-associated factor 6 (TRAF6) signaling) (65–68). Another important regulator of DC maturation is calcium/calmodulin-dependent protein kinase II (CaMKII), which is a target of three members of the miR-148/152 family (miR-148a, miR-148b, and miR-152). These miRNAs downregulate CaMKII leading to reduced expression of MHCII, reduced cytokine production (IL-12, IL-6, TNF-α), and a reduced antigen-presenting capacity of DCs (66).
Micro-RNA profiling of macrophages stimulated with TLR ligands and cytokines has shown the involvement of several miRNAs in inflammation. After incubation of murine macrophages with lipopolysaccharide (LPS), poly I:C, or interferon β (IFN-β), the expression of miR-9, miR-101, miR-155 was upregulated, while miR-34 and miR-27a were downregulated to modulate the levels of important regulators of inflammation. The upregulated miRNAs target nuclear factor-κB1 (NFκB1, miR-9), mitogen-activated protein kinase 1 (MAPK1, miR-101), and JUN N-terminal kinase (JNK, miR-155), while the downregulated miRNAs target NOTCH1 (miR-34a) and STAT3 (miR-27a), and the production of pro-inflammatory cytokines such as TNF-α, interleukin-6 (IL-6), and IL-10 (69–73).
MiR-9 was also associated with the response of polymorphonuclear cells (PMN) to TLR stimulation. It was interesting that out of the 365 miRNAs tested, miR-9 was the only one upregulated in both human macrophages and PMN after LPS activation (69). In contrast, miR-155, miR-146a, miR-146b, miR-187, miR-125a, miR-99b, and let-7e appeared to be macrophage-specific, while miR-196a was PMN-specific. This underscores how some miRNAs are involved in the activation of multiple lineages of innate immune cells, while others play a more lineage-specific role (69).
Recent studies have shown that microorganisms can modulate miRNA expression and thus the immune response during infection, as a mechanism of immune evasion. Mycobacterium tuberculosis induces miR-21 expression in macrophages and DCs. It was suggested that by targeting IL-12, miR-21 modulates the Th1 immune response (74). Leishmania has also developed strategies to subvert the host macrophage response. On Leishmania infection of human macrophages in vitro, approximately 64 out of 365 analyzed miRNAs were found to be modulated. Enrichment analyses have revealed that several of these differentially expressed miRNAs are involved in the regulation of TLR and pro-apoptotic pathways (75). By using the murine model of Toxoplasma infection, an increase in the levels of the immune-miRNAs miR-146a and miR-155 was observed in the brain of chronically infected mice compared with non-infected controls. Further assays in miR-146 KO mice demonstrated that miR-146 ablation promotes parasite control, resulting in long-term survival (76). MiRNA profile expression analyses of human macrophages infected with Toxoplasma showed that the miR-17-92 cluster expression was significantly upregulated and that the levels of miR-17-92 were closely related with a decrease in expression of the pro-apoptotic regulator Bim. Interestingly, the Bim 3’-UTR contains predicted binding sites for multiple miRNAs derived from the miR-17-92 family. All the above evidence suggests miRNAs are involved in parasite persistence and modulation of apoptosis (77).
Long Non-Coding RNAs in Innate Immunity
So far, most lncRNA studies have been performed in normal cellular development or in cancer (78–82), although the role of lncRNAs in hematopoiesis and the immune system is slowly starting to emerge. The lncRNA HOTAIRM1, located antisense to homeobox A1 (HOXA1) and HOXA2 of the HOXA gene cluster, is expressed specifically in the myeloid lineage (83). HOTAIRM1 is upregulated during retinoic acid-driven granulocytic differentiation in NB4 promyelocytic leukemia cells, which are a model for granulocytic differentiation. Knockdown of HOTAIRM1 prevents the expression of HOXA1, HOXA4, CD11b, and CD18, but not of the more distal HOXA genes and decreased myeloid differentiation.
The KIR antisense lncRNA was found to be expressed only in human embryonic stem cells and other cell types with stem cell properties (84). KIR genes encode class-I MHC receptors expressed on human NK cells. KIR antisense lncRNA overlaps with exons 1 and 2 of the protein-coding KIR gene, as well as with an upstream proximal promoter region of the KIR genes. Overexpression of the lncRNA in NK cells was found to decrease the expression of the KIR protein-coding gene. Wright et al. speculated that the KIR genes are silenced in NK progenitors so that they are not able to influence the process of NK cell differentiation (84). As the KIR distal antisense promoter contains myeloid zinc finger-1 (MZF-1)-binding sites, it is assumed that this transcription factor regulates the expression of the KIR antisense lncRNA. MZF-1 is a transcriptional regulator that is able to activate transcription in cells of hematopoietic origins, whereas it can repress transcription in other cell types. However, the precise mechanism of the regulation of KIR antisense lncRNA expression is unknown.
Lnc-DC, exclusively expressed in conventional human DCs (85), was found to induce the nuclear translocation of STAT3. The proposed mechanism of action for this lncRNA is to prevent SHP1 from binding to phosphorylated STAT3 and dephosphorylating it, thereby preventing its dimerization and translocation to the nucleus. This is an example of a lncRNA affecting cellular differentiation by a mechanism that takes place in the cytoplasm.
In another study, 54 mouse pseudogene lncRNAs were found to be induced by TNF-α (86). One of these, Lethe, functions as a negative feedback signal that inhibits NF-κB. Its expression is increased when TNF-α activates NF-κB, after which Lethe binds to NF-κB and prevents it from binding to DNA, thereby inhibiting the expression of inflammatory proteins, such as IL-6, IL-8, and superoxide dismutase 2 (SOD2).
A whole-transcriptome profiling of mouse macrophages stimulated with different TLR ligands uncovered dozens of expressed lncRNAs (87). Activation by the synthetic bacterial lipopeptide Pam3CSK4, a TLR2 ligand, resulted in the expression of 62 lncRNAs. One of them, lincRNA-Cox2, acts as a key regulator of the inflammatory response by mediating both activation and repression of several immune genes. In response to TLR2-stimulation, lincRNA-Cox2 induces the expression of hundreds of genes, including Tlr1, Il-6, and Il-23a. LincRNA-Cox2-mediated repression of target gene expression was found to require the interaction of lincRNA-Cox2 with hnRNPA/B and hnRNPA2/B1, repressing the transcription of immune cells.
Stimulation of human THP1 macrophage cells by a synthetic lipopeptide ligand of TLR2 induced 159 lincRNAs (88). One of these, TNFα and hnRNPL-related immunoregulatory lincRNA (THRIL), form a complex with hnRNPL. This complex can bind the promoter of TNFα and regulate its transcription. Microarray analysis showed that THRIL is required for the expression of various immune genes, including cytokines and other regulators of TNFα expression, including IL-8, C-X-C motif chemokine 10 (CXCL10), chemokine (C-C motif) ligand 1 (CCL1), and the colony stimulating factor 1 (CSF1). THRIL expression was also reported to be correlated with the severity of symptoms in patients with Kawasaki disease, an autoimmune disease mostly seen in children.
NEAT1 (nuclear enriched abundant transcript 1) is a lncRNA that was shown to be essential for the formation of paraspeckles. Paraspeckles are nuclear bodies found in mammalian cell nuclei and it has been proposed that they play a role in several biological processes, including cellular differentiation and the stress response (89). It has been shown that NEAT1 is induced by viral infection as well as by poly I:C stimulation and that, in response to such a stimulus, NEAT1 binds to paraspeckle protein splicing factor proline/glutamine-rich (SFPQ). This complex binds to and regulates the expression of several antiviral genes, including IL-8, which induces the formation of paraspeckles (90).
The Role of miRNA in the Development of Cells of the Adaptive Immune System
MiRNAs in T-Cell Development and Activation
Micro-RNAs have been shown to be crucial for both immune system development and its functioning. MiRNAs that are characteristically enriched in HSCs and progenitor cells are miR-125a-5p, miR-125b-5p, miR-155, miR-130a, miR-196b, miR-99a, miR-126-3p, miR-181c, miR-193b, miR-542-5p, and let-7e (91). Their expression changes during immune cell development.
Some miRNAs are selectively expressed in specific stages of immune cell development, whereas others are more broadly expressed. Profiling studies showed that there are miRNAs, which are preferentially upregulated in lymphocytes. The miR-181 family is abundant expressed in developing BM B-cells and thymocytes.
The importance of miRNAs in T-cell biology has been extensively studied in mice with conditional Dicer1 deletion. Conditional deletion of Dicer1 in T-cell precursors using Lck-Cre demonstrated that Dicer is necessary for the generation and survival of normal numbers of αβ T-cells (92). Cd4-Cre-mediated deletion did not affect the viability of Cd4+ T-cells, but the numbers of Th1 and Th2 cells were significantly reduced, resulting from both decreased proliferation as well as from increased apoptosis. Dicer-deficient Cd4+ cells have been described as more prone to differentiate into Th1 cells and Dicer-deficient cells cannot repress Inf-γ upon Th2 stimulation (93).
Profiling of different stages of T-cell development, starting from the double negative 1 (DN1) thymocyte stage, reveals that miRNA profiles are similar for cells with similar developmental status (94). DN3 and DN4 populations cluster together based on their miRNA expression profiles, as do mature single positive Cd4+ and Cd8+ cells. DN1 cells are more similar to DN3 and DN4 cells. Nevertheless, expression of individual miRNAs changes depending on their developmental stage. Each of the stages is characterized by elevated expression of at least one miRNA or miRNA family. In DN1 cells, miR-21, miR-29b, miR-342, miR-221, and miR-223 are elevated and miR-16, miR-181a, and miR-15b are decreased. MiR-191 is upregulated and miR-142-3p is downregulated in DN3 cells. MiR-142-5p, miR-20a, miR-16, and miR-128b are increased, whereas miR-150 is decreased in DN4 cells. In double positive (DP) cells, expression of miR-92, miR-181a, miR-181b, and miR-350 are enhanced, while in Cd4+ cells, miR-669c and miR-297 are elevated. In Cd4+ and Cd8+ cells, miR-128 is abundant.
It is interesting that, on activation of Cd4+ T-cells, Ago2 ubiquitination and consequentially its proteosomal degradation is induced, leading to global miRNA downregulation. Moreover, naive T-cells display reduced levels of Ago2 and differentiate more rapidly. These findings led to the hypothesis that the decrease in the miRNA pool on T-cell activation allows the expression of genes regulating CD4+ T-cell differentiation and facilitates the gain of T-cell effector functions (95). Although these are global effects on the miRNA pool, some miRNAs can be picked out that play key roles in T-cell biology. For example, the miR-181 family is upregulated in DP cells and its family member miR-181a decreases the expression of Cd69, T-cell receptor α (Tcrα), and Bcl-2 (94).
Another key player is miR-125b. This miRNA is part of the miR-99a/100~125b tricistrons, located on human chromosomes 11, 19, and 21. The tricistron on chromosome 21, encompassing miR-99a/let-7c/miR-125b-2, is highly expressed in HSCs and is responsible for maintaining stem cell properties (96). In human naive CD4+ T-cells, miR-125b downregulates proteins that are critically involved in T-cell differentiation: IFN-γ, IL-2RB, IL-10RA, and PR domain zinc finger protein 1 (PRDM1, encoding B lymphocyte-induced maturation protein-1, BLIMP-1). Overexpression of miR-125b inhibits the differentiation of naive T-cells into effector cells (97). This miRNA is an example of one that affects various stages of immune cell differentiation in different immune cell lineages.
T-cell activation leads to highly elevated expression of miR-155 (98). Experiments conducted on Cd4+ T-cells isolated from mice deficient for Bic, the primary transcript encoding miR-155, uncovered elevated Th2 polarization, and Th2 cytokine production (Il-4, Il-5, and Il-10) in these cells. This effect is mediated by upregulation of c-Maf, a Th2-specific transcription factor known to induce the expression of Il-4/5/10 (99). Th1 and Th17 responses are also regulated by miR-155. Transfection with miR-155 promotes, whereas miR-155 inhibition decreases, the number of Th1 and Th17 cells in mice with experimental autoimmune encephalomyelitis (EAE) (100). Mice lacking Bic also display decreased levels of regulatory T-cells (Tregs) in the thymus and in the periphery while the function of these cells in vitro is not affected. This indicates that miR-155 is required for Treg development (101). Characteristically, miR-155 expression is induced in Tregs by forkhead box P3 (Foxp3), while one of the main targets of miR-155 in Tregs is Socs1. When miR-155 is high, Socs1 is low, which contributes to maintaining the competitive fitness and proliferative potential of Tregs (102).
The miR-17-92 cluster is a master switch involved in the differentiation into Th1 and Th17 cells. Experiments conducted in cd4-cre-driven miR-17-92 conditional KO mice demonstrated that Th1 development is critically controlled by miR-17 and miR-19b, which target TgfβrII and cyclic AMP-responsive element binding protein 1 (Creb1, miR-17), and Pten (miR-19b). Together, these two miRNAs enhance T-cell proliferation and Inf-γ production, protect from activation-induced cell death, and repress induced Treg (iTreg) differentiation. Interestingly, miR-18a of the same cluster antagonizes the pro-Th1 effect of miR-17 and miR-19b through elevation of activation-induced cell death and inhibition of proliferation (103). Subsequent experiments conducted on T-cells isolated from conditional miR-17-92-depleted mice (that had been retrovirally transduced with selected miRNAs from the miR-17-92 cluster), showed that miR-17 and miR-19b were also the miRNAs promoting Th17 differentiation. This is mediated by miR-17-induced downregulation of Ikaros family zinc finger 4 (Ikzf4) and the downregulation of Pten by miR-19b (104).
Another miRNA regulating the adaptive response is miR-146a. Profiling studies in mice showed that miR-146a expression is high in Th1 cells and low in naive T-cells and Th2 cells (105), but very high in Tregs (106). Level of miR-146a is also elevated in human memory cells (both in CD4 and CD8 memory cells). MiR-146 expression is induced on TCR stimulation and is regulated by NF-κB and the c-E26 transformation specific (c-ETS) transcription factor. It was suggested that miR-146a exerts its regulatory function by targeting FADD, leading to a decrease in apoptosis. On TCR stimulation, activator protein-1 (AP-1) activity and IL-2 production are induced, but miR-146 targets both of them, thereby enabling miR-146a to affect the duration of T-cell activation phases (107).
As can be expected, the importance of miRNAs in CD8+ biology has been well studied. Cd4-Cre-induced Dicer deletion in mice leads to reduced development of peripheral Cd8+ cells due to decreased cell survival and defective migration out of the primary lymphoid compartment (93). As was also the case for CD4+ cells, a decrease in Dicer (and therefore in the miRNA pool) leads to differentiation, characterized by increased levels of perforin, granzymes, and effector cytokines that are usually targeted by miR-139 (targets Eomes and perforin) and miR-150 (targets Cd25) (108). Another study reported that miR-15b, miR-150, miR-24, and miR-27a were increased in Cd8+ cells (94).
MiR-155 expression also plays an important regulatory role in CD8+ cells. It is most highly expressed in primary effector Cd8+, shows intermediate expression in effector memory Cd8+ T-cells, and low expression in naive Cd8+ and central memory Cd8+ cells. Its role in the antiviral response of Cd8+ cells was demonstrated in miR-155-deficient mice, which are characterized by an attenuated antiviral response due to a diminished response of Cd8+ cells. On the contrary, overexpression of miR-155 in mice enhanced the antiviral Cd8+ response (109).
In short-lived effector CD8+ cells, miR-17-92 is upregulated in contrast to memory cells. Experiments on mice with a conditional gain or loss of miR-17-92 expression in mature Cd8+ cells after activation (controlled by the human Granzyme B promoter) revealed that miR-17-92 regulates Cd8+ expansion and the balance between effector and memory differentiation. MiR-17-92 overexpression elevates the differentiation into terminal effector cells and concomitantly decreases the formation of polyfunctional lymphoid memory cells. As miR-17-92 overexpression correlates with downregulation of Pten, and in consequence induces the PI3K (phosphoinositide 3-kinase)-Akt-mTor (mammalian target of rapamycin) pathway, here too it was suggested that this could be the main pathway involved in regulating cellular proliferation. In contrast, conditional deletion of miR-17-92 leads to attenuated proliferation of antigen-specific cells, increased Il-7Rα and Bck-2 expression, and faster acquisition of memory cell properties (110).
miRNAs in B-Cell Biology
Experiments with conditional Dicer-KO mice in early B-cell progenitors (Mb1-Cre drives deletion starting from the pro-B-cell stage) resulted in a block in B-cell development at the transition from pro- to pre-B-cell stage (111). Conditional deletion of Dicer-1 in later stages of B-cell development showed that miRNAs are also critically important for the transition from transitional B-cells to germinal center (GC) or follicular (Fo) B-cells (112). Conditional deletion of Dicer in activated B-cells [activation-induced cytidine deaminase (Aicda-cre)] confirmed that Dicer is essential for GC B-cell generation. The ablation of Bim partially rescued this effect (113).
The analysis of different cell subtypes during B-cell development in mice underscored the highly regulated, developmental stage specific, expression of miRNAs. Hierarchical clustering of BM and spleen B populations leads to a perfect recapitulation of the B-cell developmental pathway. This study found the population with the most distinct miRNA profile was that of fraction A (FrA) B-cells, characterized by expression of miR-2138, -542-3p, -500, -1959, -221, -1965, -1900, -1893, -501-5p, and let-7f*. FrA cells have been reported to still retain the capacity to differentiate into T-cells, while Pax5 and Cd19 expression is induced in the next developmental stage leading to FrB/C B-cells. Among the miRNAs not expressed in the BM are miR-150 and miR-155. These miRNAs start to be expressed in transitional B-cells and are most highly expressed in mature B-cells in the spleen (50).
In this B-cell lineage, specific or more broadly expressed miRNAs also control the differentiation and activation. MiR-126 expression decreases during B-cell maturation. Injecting miR-126-overexpressing HSC/progenitors cells into lethally irradiated mice showed that miR-126 induces the differentiation of B-cell myeloid progenitors. One of the genes regulated by miR-126 in this process is insulin regulatory subunit-1 (Irs-1) (114).
MiR-181 is preferentially expressed in the B-cells in the BM. Its overexpression in HSCs and progenitor cells leads to increased levels of B-cells. Moreover, ectopic expression of miR-181 in Lin- BM cells transplanted into sublethally irradiated mice leads to an increase in B-cells, with a concomitant decrease of T-lymphoid cells (115).
Mice without miR-17-92 (a Cre-deletor mouse strain with Cre controlled by the human β-actin promoter) die shortly after birth. Analysis of fetal liver cells from these mice showed that the frequency of HSCs and the number of early progenitors was not affected, but that the number of pre-B-cells was significantly reduced. In miR-17-92-deficient adult mice, marginal zone (MZ), Fo, and newly formed B-cells in spleen (as well as peritoneal B1a and B1b cells) are reduced, whereas the relative number of transitional B-cells was not altered. Moreover, in these mice, the frequency of red blood cells, granulocytes, and monocytes was also not altered (116). In contrast, another study showed that B-cell-specific miR-17-92 overexpressing mice develop B-cell lymphomas due to the downregulation of negative regulators of the PI3K pathway [Pten and PH domain and leucine-rich repeat protein phosphatase 2 (Phlpp2)] and the NF-κB pathway (Cyld, A20, Itch, Rnfl1, and Tax1bp1), as well as due to the downregulation pro-apoptotic protein Bim and cell cycle regulator E2F3. Together, this results in constitutive activation of pro-survival pathways (117).
MiR-150 is expressed in mature B-cells but not in BM B-cells. Premature overexpression of miR-150 in HSCs showed that miR-150 blocks the generation of mature B-cells by preventing the transition of pro-B-cells to pre-B-cells, but not the development of T-cells, granulocytes, or macrophages. The main target involved in this mechanism is c-Myb (118, 119).
MiR-155 controls the GC response at least partially via regulation of cytokine production. Mature miR-155−/− cells isolated from spleens are deficient in Tnf and lymphotoxin-α (Lt-α) production (120). MiR-155 was also shown to be critically involved in isotype switching, as reduced extrafollicular and GC responses, and a concomitant lack of high-affinity IgG1, were observed in the absence of miR-155 (121). MiR-155 is often overexpressed in B-cell lymphomas including DLBCL (122), and Eμ-enhancer driven miR-155 overexpression leads to lympho-proliferative disease followed by B-cell malignancy (123).
Finally, miR-125b appears to inhibit GC B-cell differentiation by targeting BLIMP-1 and IFN regulatory factor 4 (IRF4); it is essential for the post-GC plasma B-cell differentiation (124).
The Role of Long Non-Coding RNA in the Adaptive Immune System
Mouse CD8 + T-cells were found to specifically express hundreds of lncRNA genes. Many of these are specific for lymphoid cells and their expression was dynamically changed during lymphocyte differentiation or activation (125). A subset of 39 lncRNAs appear to be precursor transcripts to small regulatory RNAs (miRNAs and small interference RNAs, siRNAs), suggesting that some lncRNAs function via smaller RNA species.
The dynamic nature and cell-specific lncRNA expression during mouse T-cell differentiation was demonstrated by RNA-seq analysis of 42 T-cell subsets (from early T-cell progenitors to terminally differentiated T helper subsets, at multiple time points during differentiation) (126). This led to the identification of 1,524 lincRNAs, most of which are located adjacent to key proteins that regulate the immune system. Knockdown of one of these lncRNAs, LincR-Ccr2-5’AS, led to deregulation of its neighboring chemokine receptor genes and prevented Th2 migration into the lung tissue (126).
In 2003, the Tmevpg1 gene was shown to control the persistence of Theiler’s virus in the mouse central nervous system (127). Both the mouse gene and its human ortholog, TMEVPG1/NeST, encode a non-coding RNA located in a cluster of cytokine genes, including the IFN-γ gene, and it was suggested to be involved in controlling IFN-γ expression (128). Both the mouse and human lncRNAs are expressed in Th1 cells and depend on Stat4 and T-box expressed in T-cells (T-bet), two transcription factors regulating Th1 differentiation (128). Comparison of mouse strains with and without the capacity to clear Theiler’s virus revealed that mice that cannot clear the infection express Tmevpg1 to a higher level, concomitantly with increased IFN-γ synthesis and enhanced resistance to Salmonella enterica infection (129). These results indicate that lncRNA TMEVPG1/NeST regulates IFN-γ expression and plays an important role in the susceptibility to viral and bacterial infections.
The Role of miRNAs and lncRNAs in Autoimmune Diseases
Celiac Disease
Celiac disease (CeD) is characterized by a severe inflammatory reaction to gluten peptides derived from grain storage proteins; it occurs in patients with a susceptibility genotype. Besides sharing a number of phenotypic characteristics with inflammatory bowel disease (IBD), CeD also shares multiple genetic susceptibility loci with IBD (130). So far, there are limited data on the involvement of miRNAs in CeD and there are no publications on the role of lncRNAs in this autoimmune disease. The lack of a suitable animal model for CeD makes it impossible to study the role of specific miRNAs in vivo. However, profiling of miRNA expression in small intestinal biopsies from patients with active CeD versus controls showed that miR-449, -492, -644, -503, -196a, -504, -500, and -330 were differentially expressed in CeD patients, with miR-449 as the most upregulated miRNA. Putative targets of miR-449 include mRNAs encoding proteins involved in the NOTCH signaling pathway. In agreement with this, the expression of the inflammation regulator, NOTCH1, was found to be decreased in the small intestine of CeD patients, suggesting that miRNAs can also control inflammation in CeD (131). Indirect evidence for the involvement of particular miRNAs in CeD was found by analyzing genome-wide association study (GWAS) data. Kumar et al. have described how CeD-associated SNPs may actually affect the 3′-UTR of IRF4, PTPRK, and ICOSLG and suggested that these might change miRNA-binding sites (132).
Inflammatory Bowel Disease
Inflammatory bowel disease includes Crohn’s disease (CD) and ulcerative colitis (UC) (133). Recent GWAS and meta-analyses have identified 163 common risk loci for IBD and 47 unique risk loci associated with UC (134). Although the cause of IBD is unknown, there is evidence to suggest that an abnormal immune response to intestinal flora leads to this disease in genetically susceptible individuals. There have been various miRNA profiling studies published on IBD and the miRNA profiles in tissues or serum of UC and CD patients at different stages underscore the importance of miRNA as key regulators of the immune response in this disease (130, 131). Circulating miRNAs in serum were suggested as useful biomarkers for CD diagnosis. MiRNA RT-PCR revealed a set of 11 miRNAs that were significantly elevated in CD patients, but not in the serum of controls or in the serum of patients with active CeD (135).
The first report of miRNA expression in colonic mucosa samples from IBD patients identified 11 miRNA differentially expressed in active UC patients versus controls (136). Since then, the number of miRNAs linked with IBD has increased gradually (137). Several reports have demonstrated the alterations in expression of miRNAs involved in modulating different aspects of the innate and adaptive immunity, such as miR-21, miR-29a, miR-150, and miR-155 (136, 138, 139). Recently, by using microarray-based miRNA profiling of colonic mucosal biopsies, five miRNAs were shown to be upregulated in patients with active UC compared to quiescent UC, CD patients, and controls. In addition, expression of two miRNAs, miR-125b-1 and let-7e*, was enhanced in patients with quiescent UC compared with active UC, CD patients, and controls, supporting the utility of miRNAs as biomarkers to distinguish the different IBD stages (140). An interesting point is that SNP rs2910164, which has been associated with susceptibility to CD, has also been linked to miR-146a (141). Subsequent reports have identified other miRNAs that may affect the control of inflammation during IBD. MiRNA-155, which has been associated with T-cell, B-cell, and innate cell function, was detected in the blood of CD and UC patients, but not in that of healthy controls (142). RT-PCR has revealed an upregulation in miR-21 levels in mucosal tissue and serum and UC patients (143). Further, in vitro analyses demonstrated that overexpression of miR-21 in mucosa from UC patients and in the Caco-2 cell model resulted in impaired tight junction formation and decreased barrier function, suggesting a pathogenic role for miR-21 in UC (143). Using the murine model of dextran sulfate sodium (DSS)-induced colitis, it was found that overexpression of miR-146b (by expression vector) or ablation of miR-21 (miR-21 KO mice) reduces intestinal inflammation and restores epithelial barrier function by activating NF-κB (miR-146b) or by negatively regulating RhoB (miR- 21) (144, 145).
The involvement of other inflammatory pathways and processes regulated by miRNAs was also proven to be important in IBD pathogenesis. Several studies have shown that the pattern recognition receptor NOD 2 is upregulated by miR-146 or downregulated by miR-122, while the colonic leukocytic trafficking is regulated by miR-141 (146–148).
More studies have led to an understanding of the role of miRNAs on carcinogenesis, since IBD has been well established to be a predisposing condition for colorectal carcinoma (CRC). For instance, the levels of miR-143 and miR-145 were downregulated in UC patients compared with normal controls. Among the putative targets of these miRNAs are proteins associated with cell cycle regulation, such as K-RAS, API5, MAPK kinase-2 (MEK-2), and IRS-1 (149). A recent study identified miR-224 as one of the most upregulated miRNAs during the transition from IBD to IBD-associated CRC. In silico analysis and functional assays confirmed that miR-224 targets the cell cycle regulator p21, which could suggest the involvement of miR-224 in IBD-associated carcinogenesis (150). Other studies have demonstrated a dysregulation of miR-21 and miR-155 during active IBD in IBD-dysplastic lesions (151, 152).
In conclusion, these inflammation-related miRNAs target important regulators of carcinogenesis, such as programed cell death 4 and mismatch repair elements, which could provide a biochemical link from IBD to cancer development (151, 152).
Because of their specific expression profiles, miRNAs are considered useful biomarkers for IBD diagnosis and as predictors of disease progression (153). MiR-122, miR-17, and let-7e were found to be altered during the progression of IBD (147), while a set of studies on immune-mediated diseases (including IBD) highlighted miRNAs as promising indicators of response to immunosuppressor treatment (20).
However, the role of lncRNA in the pathogenesis of CD remains elusive. A GWAS study identified leucine-rich repeat kinase-2 (LLRK2), which is part of a complex including the large non-coding RNA repressor of NFAT, as associated with CD. In line with this, when wild-type mice were sublethally irradiated and reconstituted with Lrrk2-deficient hematopoietic cells, they were more susceptible to DSS-induced colitis. This suggests that LLRK2 deficiency increases UC severity (154). In addition, high levels of lncRNA DQ786243 were found in the blood of patients with CD. Subsequent overexpression of DQ786243 in Jurkat cells showed a correlation between the lncRNA and the expression of the Foxp3 regulator, CREB, suggesting that DQ786243 is involved in inflammation control and CD pathogenesis (155).
Multiple Sclerosis
Multiple sclerosis (MS) is an autoimmune, demyelinating neurodegenerative disorder, mostly affecting adults. Most often, the disease first manifests itself as relapsing-remitting MS (RR-MS), which typically progresses into secondary progressive MS (SP-MS) after 10–25 years. It is thought that RR-MS is more pro-inflammatory in nature, while SP-MS is mostly associated with neurodegeneration, although there is a small percentage of patients who present with neurodegenerative symptoms, without any evidence for former pro-inflammatory episodes. They have an aggressive form of MS called primary-progressive MS (PP-MS) (156). Multiple studies suggest the involvement of miRNAs in MS and changes in miRNAs have been detected in circulation, in brain tissue, in immune cells, and in CSF. Several miRNAs associated with the immune response have also been linked with MS, including miRNAs regulating the Th1 response, such as miR-155 (100) and miR-29 (157), but also Th17-associated miRNAs [miR-155 (100), miR-17-92 (104), miR-132 (158), and miR-326 (159)]. Of interest is that the level of miR-326 in peripheral blood lymphocytes was suggested to distinguish between the relapsing and remitting phases of MS (160).
One of the miRNAs important in MS pathology is miR-155, the expression of which is elevated in RR-MS patients during relapse, as well as in the murine EAE model. In EAE mice, delivery of miR-155 stimulates the development of inflammatory Th17 and Th1 cells, whereas applying miR-155 inhibitors reduces these processes (100).
The immune response in MS in the brain may be also regulated indirectly by changes in the blood–brain barrier (BBB) permeability. Elevated levels of miR-155 in the neurovascular part of the brain of MS patients as well as in EAE mice are linked with a decreased function of the BBB. The potential targets of miR-155 suggested to be involved in BBB function are focal adhesion proteins (DOCK-1, dedicator of cytokinesis-1, and Syntenin-1) and proteins involved in cell–cell interaction (Annexin-2 and Claudin-1) (161).
Another miRNA that plays a crucial role in regulating the function of the BBB, and consequently in regulating the immune cell influx in the brain, is miR-125a-5p. Overexpression of this miRNA elevates barrier tightness, with thicker and more continuous tight junctions formed by vascular endothelial-cadherin (VE-cadherin) and zona-occludens-1 (162).
MiR-92a-1* is reported to be increased specifically in the plasma of RR-MS patients compared to healthy controls or SP-MS patients. Furthermore, its level correlates negatively with disease duration and disability score. Elevated levels of miR-92 can also distinguish RR-MS from another neuronal disease, amyotrophic lateral sclerosis. Using ingenuity pathway analysis (IPA), it was predicted that miR-92 affects CD40 signaling by targeting CD40 directly (163).
Let-7a expression is diminished in SP-MS patients compared to healthy controls. IPA analysis led to the suggestion that let-7a targets TLR4 and TLR9, as well as IL-12RB2 and TGF-βR1 (163). Let-7a also regulates neuronal differentiation (164).
MiR-145 appears to decrease during the transition from RR-MS to SP-MS and is negatively correlated with disability score (163). This miRNA is known to regulate insulin-like growth factor 1 (IGF-1) signaling (IGF-1R, IRS1, and IRS2) (165).
Long non-coding RNAs have been shown to be involved in the development of neurodegenerative disorders, like β-secretase-1 (BACE1)-AS, the brain cytoplasmic RNA (BC200 RNA) in Alzheimer’s disease, and the brain-derived neurothrophic factor (BDNF)-AS in Huntington’s disease (166). BACE1-AS elevates the protein level of BACE1, an enzyme involved in generating β-amyloid, by binding and stabilizing BACE1 mRNA (167). The level of BC200 RNA is elevated in the brains of Alzheimer’s patients (168) and this lncRNA was shown to interact with proteins participating in mRNA transport in neurons (166). BDNF-AS inhibits the expression of BDNF, a growth factor promoting neuron maturation and survival (169, 170). It is not yet known whether these lncRNAs are also involved in MS.
Impaired remyelination and neuron apoptosis are two hallmarks of the neurodegenerative phase of SP-MS. Interestingly, the level of the small non-coding RNA 715 (sncRNA715), which inhibits oligodendrogial myelin basic protein (MBP) translation, is elevated in demyelinated chronic lesions in MS patients (171). In contrast, remyelination can be affected by exosomes released by DCs. Stimulating DCs with low levels of INF-γ causes the release of exosomes enriched in miRNA involved in the regulation of myelin production (miR-219) and anti-inflammatory response (miR-181a, -451, -532-5p, and -665) compared to exosomes released from unstimulated DCs. This, in turn, increases myelination and reduces oxidative stress in hippocampal slice cultures. Moreover, intranasal delivery of these exosomes elevates the central nervous system myelination in vivo (172). LncRNAs have also been implicated in the regulation of apoptosis, and because neuron apoptosis is important in MS, the role of lncRNA in apoptosis in MS should be investigated further (173).
Two MS SNPs are associated with miRNA genes (miR-223; rs1044165 and miR-23; rs3745453) and the expression of these miRNAs was investigated in MS patients (174). MiR-223 is important for the innate immune system, e.g., affecting the non-canonical NF-κB pathway during macrophage differentiation (175), whereas miR-23a promotes myelination by elevating oligodendrocyte differentiation and myelin synthesis (176). Both miR-223 and miR-23a are decreased in serum from RR-MS patients. However, the expression of miR-233 is increased in peripheral blood mononuclear cells (PBMCs) from both RR-MS as well as PP-MS patients, whereas miRNA-23a is upregulated only in PBMCs from RR-MS patients (174). Thus far, no eQTL-effect of both SNPs on the respective miRNAs has been found.
Another study predicted that SNP rs28366, which is in moderate linkage disequilibrium (r2 = 0.4) with MS risk variant rs17066096, might affect the binding of miR-2278 and miR-411-5p- to the 3′-UTR of IL22RA2. However, although these miRNAs did indeed bind to the 3′-UTR in an in vitro assay, the SNP did not affect the binding (177).
Perspectives
Integration of autoimmune disease-associated SNPs with data on the functional regions in the genome shows that ~90% of these SNPs intersect with regulatory regions in the DNA (enhancers, promoters) rather than with protein-coding regions (exons) (29, 130). Autoimmune disease-associated loci often encompass genes known to play critical roles in immunological diseases. In these common illnesses, characteristically dozens of disease-associated SNPs have been identified, each predisposing moderately to disease. The general consensus for these immune-related diseases is that many SNPs with modest effects converge on a limited number of biological pathways and it is the sum of these effects that influences the phenotype to a large extent.
In the last two decades, thousands of regulatory RNA genes have been identified that also regulate gene expression. In fact, the total number of known miRNA genes plus lncRNA genes is now more than 15,000 and this is starting to approach the number of known protein-coding genes (~22,000). There is convincing evidence that autoimmune SNPs also affect miRNA and lncRNA biology. SNPs have been identified that locate to miRNA-binding sites and some SNPs appear to regulate the expression of lncRNAs (32, 132, 174, 177). This means that to fully understand the changes to transcriptional programs that contribute to disease, we need to take into account multiple gene-regulatory pathways, involving classical transcription factors, miRNAs, and lncRNAs. This situation is further complicated by the fact that these pathways, and the molecules involved in them, not only regulate other molecules within their own class, but also molecules of other classes. In the case of miRNAs and lncRNAs, for instance, it has already been reported that lncRNAs can be targeted by miRNAs, that lncRNAs can encode miRNAs, that lncRNAs and miRNAs can compete for targets, and that some lncRNAs can function as miRNA sponges (178).
With advances in next generation sequencing technology and the anticipated reduction in the cost of using these techniques, many more regulatory RNAs (and perhaps even novel classes of regulatory RNAs) are likely to be discovered. If we focus on discovering novel miRNAs and lncRNAs, we should take into account that the expression of both classes of regulatory RNAs is more cell-type- and stimulus-dependent than the expression of protein-coding genes. This has two consequences: (1) for regulatory RNAs affecting other RNA species, both the regulator and the target need to be expressed in the same cell type and (2) to better understand the molecular basis of human disease, it means that the tissues that specifically contribute to a disease need to be identified, isolated, and profiled. Subsequently, the targets of the known and novel regulatory RNAs need to be identified; this is currently a major challenge. It is known that miRNAs can bind dozens of targets and that single mRNAs can be regulated by multiple miRNAs. Canonical miRNA action is based on complementarity between the seed sequence of the miRNA and a binding sequence in the 3′-UTR of its target. This “rule” has been used to design algorithms that predict miRNA-binding sites in the 3′-UTRs of target mRNAs [see review by Hrdlickova et al. (18) for listing of web-based tools]. The field of lncRNA research is quite new and the functions of lncRNAs are still poorly understood. Although characteristics such as cell-type specificity and their role in epigenetic regulation also make lncRNAs interesting therapeutic targets, much more basic research is required to fully understand and appreciate the potential importance of lncRNAs in diagnostic work and treatment. There are currently no target prediction algorithms for lncRNAs, primarily because lncRNAs represent a heterogeneous group of regulatory RNAs that exhibit many different mechanisms of action. Moreover, their interaction with specific targets, which can be DNA or protein, appears to be determined by their structure rather than by their sequence. A factor that complicates lncRNA research is that they exhibit little sequence conservation throughout evolution. It is, therefore, not easy to identify mouse homologs of human lncRNAs for instance. However, it is possible that functional structures – independent of sequence – are more conserved, but there is not yet enough data available to prove definitely that this is a general feature. In contrast, miRNAs and their targets are well conserved throughout evolution, which makes it relatively easy to extrapolate mouse miRNA results to the human system.
Importantly, in silico predictions of targets and functions for regulatory RNAs need to be followed up by in vitro validation. Recently, advanced assays have been developed based on cross-linking of regulatory RNAs to their targets followed by analysis of their binding partners. By using such assays, miRNA targets can be identified by RNA sequencing [e.g., (179)] and lncRNA interacting partners can be determined by DNA sequencing or mass spectrometry [e.g., (180)]. These technically challenging assays are currently considered to be the state-of-the-art in pinpointing the targets of regulatory RNAs.
Much attention in the miRNA field is currently focused on the potential of using circulating miRNAs as biomarkers for disease. Tissue-specific expression profiles of miRNAs and lncRNAs are one of the features that make them attractive biomarker candidates. MiRNAs are of particular interest because it has been shown that they exhibit stability in plasma and serum, a feature that is not expected of lncRNAs. MiRNA (and lncRNA) levels can be measured by PCR, array technology, or by next generation sequencing applications, with each technique having its own advantages and disadvantages (181). Unfortunately, there are no simple tests (for instance dip-sticks) available to detect RNAs. Additionally, miRNAs are attractive targets for therapy. For instance, decoys can be applied to prevent miRNAs or lncRNAs from binding to their targets. Miravirsen, an inhibitor of miR-122, was shown to reduce HCV viremia in monkeys and is currently the most advanced miRNA-based drug in the clinical testing pipeline (Phase II). For more details on the potential of miRNAs or lncRNAs in the clinic and on the challenges associated with their delivery to target tissues, we recommend the reviews by Ling et al (182) and Li and Rana (183). It is clear, however, that the elucidation of more complete transcriptional networks, containing regulatory RNAs and protein-coding genes, will soon provide several more potential targets for therapeutic options.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Jackie Senior for editing the manuscript. The authors are supported by the European Research Council (ERC) under the European Union’s Seventh Framework Program (FP/2007-2013; ERC Grant Agreement no. 2012-322698 to C. Wijmenga), the Dutch Multiple Sclerosis Foundation (11-752 to Sebo Withoff), NWO-VENI grant 863.09.007 to J. Fu, and the Systems Biology Centre for Metabolism and Ageing (SBC-EBA).
Abbreviations
3′-UTR, 3′-untranslated region; Aicda, activation-induced cytidine deaminase; BACE1, gene encoding β-secretase-1; AGO, Argonaute; AP-1, activator protein-1; APAF1, apoptotic protease activating factor 1; APC, antigen-presenting cell; Bak1, Bcl-2 homologus antagonist/killer 1; BBB, blood–brain barrier; Bcl-2, B-cell lymphoma-2; BDNF, brain-derived neurothrophic factor; Bim, pro-apoptotic factor; BLIMP-1, B lymphocyte-induced maturation protein-1; BM, bone marrow; CaMKII, calcium/calmodulin-dependent protein kinase II; Cepba, CCAAT/enhancer binding protein-α; CCL1, chemokine (C-C motif) ligand 1; CD, Crohn’s disease; CDK, cyclin-dependent kinase; CeD, celiac disease; C-ETS, c-E26 transformation specific transcription factor; CLP, common lymphoid progenitor; CMP, common myeloid progenitor; CRC, colorectal carcinoma; CREB, cyclic AMP-responsive element binding protein; CSF, cerebrospinal fluid; CSF1, colony stimulating factor 1; CXCL10, C-X-C motif chemokine 10; DC, dendritic cell; DGCR8, DiGeorge syndrome critical region gene 8; DN, double negative; DOCK-1, dedicator of cytokinesis-1; DP, double positive (CD4+CD8+); DSS, dextran sulfate sodium; EAE, autoimmune encephalomyelitis; EBV, Epstein–Barr virus; eQTL, expression quantitative trait locus; FADD, Fas-associated death domain; Fo, follicular B-cell; Foxp3, forkhead box P3; GC, germinal center; GWAS, genome-wide association study; hnRNP, heterogeneous nuclear ribonucleoprotein; HOX, homeobox; HSC, hematopoietic stem cell; IBD, inflammatory bowel disease; IFN, interferon; IGF-1, insulin-like growth factor 1; Ikzf4, Ikaros family zinc finger 4; IL, interleukin; IPA, ingenuity pathway analysis; IRAK1, interleukin-1 receptor-associated kinase 1; IRF4, IFN regulatory factor 4; IRS-1, insulin regulatory subunit-1; iTreg, induced regulatory T-cell; JNK, JUN N-terminal kinase; KO, knockout; lincRNA, long intergenic ncRNA; LLRK2, leucine-rich repeat kinase-2; lncRNA, long non-coding RNA; LPS, lipopolysaccharide; Lt-α, lymphotoxin-α; MAPK, mitogen-activated protein kinase; MBP, oligodendrogial myelin basic protein; MEK, MAPK kinase; miRNA, micro-RNA; mirtron, intron-derived splicing-dependent miRNA; MS, multiple sclerosis; mTOR, mammalian target of rapamycin; MZ, marginal zone; MZF-1, myeloid zinc finger-1; NATs, natural antisense transcripts; ncRNA, non-coding RNA; NEAT1, nuclear enriched abundant transcript 1; NF-κB, nuclear factor-κB; NK, natural killer; PBMC, peripheral blood mononuclear cell; Phlpp2, PH domain and leucine-rich repeat protein phosphatase 2; PI3K, phosphoinositide 3-kinase; PP-MS, primary-progressive multiple sclerosis; PMN, polymorphonuclear cells; PRDM1, PR domain zinc finger protein 1; pre-miRNA, precursor-miRNA; pri-miRNA, primary miRNA; PTEN, phosphatase and tensin homolog; RISC, RNA-induced silencing complex; RR-MS, relapsing-remitting multiple sclerosis; RUNX-1, runt-related transcription factor 1; siRNA, small interference RNA; SFPQ, splicing factor proline/glutamine-rich; SOCS1, suppressor of cytokine signaling-1; simtron, intron-derived splicing-independent miRNA; sncRNA715, small non-coding RNA 715; SNP, single nucleotide polymorphism; SOD2, superoxide dismutase 2; SP-MS, secondary progressive multiple sclerosis; STAT, signal transducer and activator of transcription; T-bet, T-box expressed in T-cells; TCR, T-cell receptor; TGFβ, transforming growth factor β; TGFβR, TGFβ-receptor; Th, T helper; THRIL, TNFα- and hnRNPL-related immunoregulatory lincRNA; TLR, Toll-like receptor; TNF, tumor necrosis factor; TRAF, TNF receptor-associated factor; Treg, regulatory T-cell; UC, ulcerative colitis; VE-cadherin, vascular endothelial-cadherin; XPO5, exportin 5.
References
1. Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell (1993) 75:843–54. doi:10.1016/0092-8674(93)90529-Y
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
2. Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell (1993) 75:855–62. doi:10.1016/0092-8674(93)90530-4
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
3. Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Non-coding RNAs: regulators of disease. J Pathol (2010) 220:126–39. doi:10.1002/path.2638
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
4. Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res (2014) 42:D68–73. doi:10.1093/nar/gkt1181
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
5. Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, et al. GENCODE: the reference human genome annotation for the ENCODE project. Genome Res (2012) 22:1760–74. doi:10.1101/gr.135350.111
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
6. Denli AM, Tops BBJ, Plasterk RHA, Ketting RF, Hannon GJ. Processing of primary microRNAs by the microprocessor complex. Nature (2004) 432:231–5. doi:10.1038/nature03049
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
7. Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature (2003) 425:415–9. doi:10.1038/nature01957
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
8. Chen C-YA, Zheng D, Xia Z, Shyu A-B. Ago-TNRC6 triggers microRNA-mediated decay by promoting two deadenylation steps. Nat Struct Mol Biol (2009) 16:1160–6. doi:10.1038/nsmb.1709
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
9. Takimoto K, Wakiyama M, Yokoyama S. Mammalian GW182 contains multiple argonaute-binding sites and functions in microRNA-mediated translational repression. RNA (2009) 15:1078–89. doi:10.1261/rna.1363109
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
10. Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol (2005) 7:719–23. doi:10.1038/ncb1274
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
11. Wei Y, Li L, Wang D, Zhang C-Y, Zen K. Importin 8 regulates the transport of mature microRNAs into the cell nucleus. J Biol Chem (2014) 289:10270–5. doi:10.1074/jbc.C113.541417
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
12. Johnsson P, Ackley A, Vidarsdottir L, Lui W-O, Corcoran M, Grandér D, et al. A pseudogene long-noncoding-RNA network regulates PTEN transcription and translation in human cells. Nat Struct Mol Biol (2013) 20:440–6. doi:10.1038/nsmb.2516
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
13. Cole C, Sobala A, Lu C, Thatcher SR, Bowman A, Brown JWS, et al. Filtering of deep sequencing data reveals the existence of abundant dicer-dependent small RNAs derived from tRNAs. RNA (2009) 15:2147–60. doi:10.1261/rna.1738409
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
14. Son DJ, Kumar S, Takabe W, Kim CW, Ni C-W, Alberts-Grill N, et al. The atypical mechanosensitive microRNA-712 derived from pre-ribosomal RNA induces endothelial inflammation and atherosclerosis. Nat Commun (2013) 4:3000. doi:10.1038/ncomms4000
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
15. Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature (2007) 448:83–6. doi:10.1038/nature05983
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
16. Havens MA, Reich AA, Duelli DM, Hastings ML. Biogenesis of mammalian microRNAs by a non-canonical processing pathway. Nucleic Acids Res (2012) 40:4626–40. doi:10.1093/nar/gks026
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
17. Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, et al. The microRNA spectrum in 12 body fluids. Clin Chem (2010) 56:1733–41. doi:10.1373/clinchem.2010.147405
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
18. Hrdlickova B, de Almeida RC, Borek Z, Withoff S. Genetic variation in the non-coding genome: involvement of micro-RNAs and long non-coding RNAs in disease. Biochim Biophys Acta (2014) 1842:1910–22. doi:10.1016/j.bbadis.2014.03.011
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
19. Iborra M, Bernuzzi F, Correale C, Vetrano S, Fiorino G, Beltrán B, et al. Identification of serum and tissue micro-RNA expression profiles in different stages of inflammatory bowel disease. Clin Exp Immunol (2013) 173:250–8. doi:10.1111/cei.12104
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
20. De Iudicibus S, Lucafò M, Martelossi S, Pierobon C, Ventura A, Decorti G. MicroRNAs as tools to predict glucocorticoid response in inflammatory bowel diseases. World J Gastroenterol (2013) 19:7947–54. doi:10.3748/wjg.v19.i44.7947
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
21. Liu Y, Zhao L, Li D, Yin Y, Zhang C-Y, Li J, et al. Microvesicle-delivery miR-150 promotes tumorigenesis by up-regulating VEGF, and the neutralization of miR-150 attenuate tumor development. Protein Cell (2013) 4:932–41. doi:10.1007/s13238-013-3092-z
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
22. Lehmann SM, Krüger C, Park B, Derkow K, Rosenberger K, Baumgart J, et al. An unconventional role for miRNA: let-7 activates toll-like receptor 7 and causes neurodegeneration. Nat Neurosci (2012) 15:827–35. doi:10.1038/nn.3113
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
23. Park C-K, Xu Z-Z, Berta T, Han Q, Chen G, Liu X-J, et al. Extracellular microRNAs activate nociceptor neurons to elicit pain via TLR7 and TRPA1. Neuron (2014) 82:47–54. doi:10.1016/j.neuron.2014.02.011
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
24. Mittelbrunn M, Gutiérrez-Vázquez C, Villarroya-Beltri C, González S, Sánchez-Cabo F, González MÁ, et al. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Commun (2011) 2:282. doi:10.1038/ncomms1285
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
25. Villarroya-Beltri C, Gutiérrez-Vázquez C, Sánchez-Cabo F, Pérez-Hernández D, Vázquez J, Martin-Cofreces N, et al. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun (2013) 4:2980. doi:10.1038/ncomms3980
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
26. Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res (2012) 22:1775–89. doi:10.1101/gr.132159.111
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
27. Guttman M, Russell P, Ingolia NT, Weissman JS, Lander ES. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell (2013) 154:240–51. doi:10.1016/j.cell.2013.06.009
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
28. Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, et al. Landscape of transcription in human cells. Nature (2012) 489:101–8. doi:10.1038/nature11233
29. Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature (2012) 489:57–74. doi:10.1038/nature11247
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
30. Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature (2009) 458:223–7. doi:10.1038/nature07672
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
31. Li J, Xuan Z, Liu C. Long non-coding RNAs and complex human diseases. Int J Mol Sci (2013) 14:18790–808. doi:10.3390/ijms140918790
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
32. Kumar V, Westra H-J, Karjalainen J, Zhernakova DV, Esko T, Hrdlickova B, et al. Human disease-associated genetic variation impacts large intergenic non-noding RNA expression. PLoS Genet (2013) 9:e1003201. doi:10.1371/journal.pgen.1003201
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
33. Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell (2011) 43:904–14. doi:10.1016/j.molcel.2011.08.018
34. Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem (2012) 81:145–66. doi:10.1146/annurev-biochem-051410-092902
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
35. Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature (2012) 482:339–46. doi:10.1038/nature10887
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
36. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell (2011) 146:353–8. doi:10.1016/j.cell.2011.07.014
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
37. Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell (2011) 147:358–69. doi:10.1016/j.cell.2011.09.028
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
38. Karreth FA, Tay Y, Perna D, Ala U, Tan SM, Rust AG, et al. In vivo identification of tumor- suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell (2011) 147:382–95. doi:10.1016/j.cell.2011.09.032
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
39. Contreras J, Rao DS. MicroRNAs in inflammation and immune responses. Leukemia (2012) 26:404–13. doi:10.1038/leu.2011.356
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
40. O’Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol (2010) 10:111–22. doi:10.1038/nri2708
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
41. Wang XS, Gong JN, Yu J, Wang F, Zhang XH, Yin XL, et al. MicroRNA-29a and microRNA-142-3p are regulators of myeloid differentiation and acute myeloid leukemia. Blood (2012) 119:4992–5004. doi:10.1182/blood-2011-10-385716
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
42. O’Connell RM, Zhao JL, Rao DS. MicroRNA function in myeloid biology. Blood (2011) 118:2960–9. doi:10.1182/blood-2011-03-291971
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
43. Bousquet M, Harris MH, Zhou B, Lodish HF. MicroRNA miR-125b causes leukemia. Proc Natl Acad Sci U S A (2010) 107:21558–63. doi:10.1073/pnas.1016611107
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
44. Alemdehy MF, van Boxtel NGJA, de Looper HWJ, van den Berge IJ, Sanders MA, Cupedo T, et al. Dicer1 deletion in myeloid-committed progenitors causes neutrophil dysplasia and blocks macrophage/dendritic cell development in mice. Blood (2012) 119:4723–30. doi:10.1182/blood-2011-10-386359
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
45. Bezman NA, Cedars E, Steiner DF, Blelloch R, Hesslein DGT, Lanier LL. Distinct requirements of microRNAs in NK cell activation, survival, and function. J Immunol (2010) 185:3835–46. doi:10.4049/jimmunol.1000980
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
46. Montecalvo A, Larregina AT, Shufesky WJ, Stolz DB, Sullivan MLG, Karlsson JM, et al. Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood (2012) 119:756–66. doi:10.1182/blood-2011-02-338004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
47. Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, van Eijndhoven MAJ, Hopmans ES, Lindenberg JL, et al. Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci U S A (2010) 107:6328–33. doi:10.1073/pnas.0914843107
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
48. Sauvageau M, Goff LA, Lodato S, Bonev B, Groff AF, Gerhardinger C, et al. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. Elife (2013) 2:e01749. doi:10.7554/eLife.01749
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
49. Pospisil V, Vargova K, Kokavec J, Rybarova J, Savvulidi F, Jonasova A, et al. Epigenetic silencing of the oncogenic miR-17-92 cluster during PU.1-directed macrophage differentiation. EMBO J (2011) 30:4450–64. doi:10.1038/emboj.2011.317
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
50. Spierings DC, McGoldrick D, Hamilton-Easton AM, Neale G, Murchison EP, Hannon GJ, et al. Ordered progression of stage-specific miRNA profiles in the mouse B2 B-cell lineage. Blood (2011) 117:5340–9. doi:10.1182/blood-2010-10-316034
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
51. Sun SM, Dijkstra MK, Bijkerk AC, Brooimans RA, Valk PJM, Erkeland SJ, et al. Transition of highly specific microRNA expression patterns in association with discrete maturation stages of human granulopoiesis. Br J Haematol (2011) 155:395–8. doi:10.1111/j.1365-2141.2011.08682.x
52. Johnnidis JB, Harris MH, Wheeler RT, Stehling-Sun S, Lam MH, Kirak O, et al. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature (2008) 451:1125–9. doi:10.1038/nature06607
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
53. Surdziel E, Cabanski M, Dallmann I, Lyszkiewicz M, Krueger A, Ganser A, et al. Enforced expression of miR-125b affects myelopoiesis by targeting multiple signaling pathways. Blood (2011) 117:4338–48. doi:10.1182/blood-2010-06-289058
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
54. Chaudhuri AA, So AY-L, Mehta A, Minisandram A, Sinha N, Jonsson VD, et al. Oncomir miR-125b regulates hematopoiesis by targeting the gene Lin28A. Proc Natl Acad Sci U S A (2012) 109:4233–8. doi:10.1073/pnas.1200677109
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
55. Scott EW, Simon MC, Anastasi J, Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science (1994) 265:1573–7. doi:10.1126/science.8079170
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
56. Ghani S, Riemke P, Schönheit J, Lenze D, Stumm J, Hoogenkamp M, et al. Macrophage development from HSCs requires PU.1-coordinated microRNA expression. Blood (2011) 118:2275–84. doi:10.1182/blood-2011-02-335141
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
57. Mildner A, Chapnik E, Manor O, Yona S, Kim KW, Aychek T, et al. Mononuclear phagocyte miRNome analysis identifies miR-142 as critical regulator of murine dendritic cell homeostasis. Blood (2013) 121:1016–27. doi:10.1182/blood-2012-07-445999
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
58. Fontana L, Pelosi E, Greco P, Racanicchi S, Testa U, Liuzzi F, et al. MicroRNAs 17-5p-20a-106a control monocytopoiesis through AML1 targeting and M-CSF receptor upregulation. Nat Cell Biol (2007) 9:775–87. doi:10.1038/ncb1613
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
59. Larsen MT, Hother C, Häger M, Pedersen CC, Theilgaard-Mönch K, Borregaard N, et al. MicroRNA profiling in human neutrophils during bone marrow granulopoiesis and in vivo exudation. PLoS One (2013) 8:e58454. doi:10.1371/journal.pone.0058454
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
60. Lu TX, Lim EJ, Itskovich S, Besse JA, Plassard AJ, Mingler MK, et al. Targeted ablation of miR-21 decreases murine eosinophil progenitor cell growth. PLoS One (2013) 8:e59397. doi:10.1371/journal.pone.0059397
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
61. Dunand-Sauthier I, Santiago-Raber ML, Capponi L, Vejnar CE, Schaad O, Irla M, et al. Silencing of c-Fos expression by microRNA-155 is critical for dendritic cell maturation and function. Blood (2011) 117:4490–500. doi:10.1182/blood-2010-09-308064
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
62. Riepsaame J, van Oudenaren A, den Broeder BJH, van Ijcken WFJ, Pothof J, Leenen PJM. MicroRNA-mediated down-regulation of M-CSF receptor contributes to maturation of mouse monocyte-derived dendritic cells. Front Immunol (2013) 4:353. doi:10.3389/fimmu.2013.00353
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
63. Mi QS, Xu YP, Qi RQ, Shi YL, Zhou L. Lack of microRNA miR-150 reduces the capacity of epidermal langerhans cell cross-presentation. Exp Dermatol (2012) 21:876–7. doi:10.1111/exd.12008
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
64. Zhang M, Liu F, Jia H, Zhang Q, Yin L, Liu W. Inhibition of microRNA let-7i depresses maturation and functional state of dendritic cells in response to lipopolysaccharide stimulation via targeting suppressor of cytokine signaling 1. J Immunol (2011) 187:1674–83. doi:10.4049/jimmunol.1001937
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
65. Jurkin J, Schichl YM, Koeffel R, Bauer T, Richter S, Konradi S, et al. miR-146a is differentially expressed by myeloid dendritic cell subsets and desensitizes cells to TLR2-dependent activation. J Immunol (2010) 184:4955–65. doi:10.4049/jimmunol.0903021
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
66. Liu X, Zhan Z, Xu L, Ma F, Li D, Guo Z, et al. MicroRNA-148/152 impair innate response and antigen presentation of TLR-triggered dendritic cells by targeting CaMKIIα. J Immunol (2010) 185:7244–51. doi:10.4049/jimmunol.1001573
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
67. Bai Y, Qian C, Qian L, Ma F, Hou J, Chen Y, et al. Integrin CD11b negatively regulates TLR9-triggered dendritic cell cross-priming by upregulating microRNA-146a. J Immunol (2012) 188:5293–302. doi:10.4049/jimmunol.1102371
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
68. Hua C, Sun L, Yang Y, Tan R, Hou Y. Mechanisms of CpG-induced CD40 expression on murine bone marrow-derived dendritic cells. Autoimmunity (2013) 46:177–87. doi:10.3109/08916934.2012.751980
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
69. Bazzoni F, Rossato M, Fabbri M, Gaudiosi D, Mirolo M, Mori L, et al. Induction and regulatory function of miR-9 in human monocytes and neutrophils exposed to proinflammatory signals. Proc Natl Acad Sci U S A (2009) 106:5282–7. doi:10.1073/pnas.0810909106
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
70. Jiang P, Liu R, Zheng Y, Liu X, Chang L, Xiong S, et al. MiR-34a inhibits lipopolysaccharide-induced inflammatory response through targeting notch1 in murine macrophages. Exp Cell Res (2012) 318:1175–84. doi:10.1016/j.yexcr.2012.03.018
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
71. Zhu Q-Y, Liu Q, Chen J-X, Lan K, Ge B-X. MicroRNA-101 targets MAPK phosphatase-1 to regulate the activation of MAPKs in macrophages. J Immunol (2010) 185:7435–42. doi:10.4049/jimmunol.1000798
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
72. O’Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci U S A (2007) 104:1604–9. doi:10.1073/pnas.0610731104
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
73. Xie N, Cui H, Banerjee S, Tan Z, Salomao R, Fu M, et al. miR-27a regulates inflammatory response of macrophages by targeting IL-10. J Immunol (2014) 193:327–34. doi:10.4049/jimmunol.1400203
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
74. Wu Z, Lu H, Sheng J, Li L. Inductive microRNA-21 impairs anti-mycobacterial responses by targeting IL-12 and Bcl-2. FEBS Lett (2012) 586:2459–67. doi:10.1016/j.febslet.2012.06.004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
75. Lemaire J, Mkannez G, Guerfali FZ, Gustin C, Attia H, Sghaier RM, et al. MicroRNA expression profile in human macrophages in response to Leishmania major infection. PLoS Negl Trop Dis (2013) 7:e2478. doi:10.1371/journal.pntd.0002478
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
76. Cannella D, Brenier-Pinchart M-P, Braun L, van Rooyen JM, Bougdour A, Bastien O, et al. miR-146a and miR-155 delineate a microRNA fingerprint associated with Toxoplasma persistence in the host brain. Cell Rep (2014) 6:928–37. doi:10.1016/j.celrep.2014.02.002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
77. Cai Y, Chen H, Mo X, Tang Y, Xu X, Zhang A, et al. Toxoplasma gondii inhibits apoptosis via a novel STAT3-miR-17-92-Bim pathway in macrophages. Cell Signal (2014) 26:1204–12. doi:10.1016/j.cellsig.2014.02.013
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
78. Gibb EA, Vucic EA, Enfield KSS, Stewart GL, Lonergan KM, Kennett JY, et al. Human cancer long non-coding RNA transcriptomes. PLoS One (2011) 6:e25915. doi:10.1371/journal.pone.0025915
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
79. Gutschner T, Hämmerle M, Diederichs S. MALAT1 – a paradigm for long noncoding RNA function in cancer. J Mol Med (Berl) (2013) 91:791–801. doi:10.1007/s00109-013-1028-y
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
80. Kim K, Jutooru I, Chadalapaka G, Johnson G, Frank J, Burghardt R, et al. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene (2013) 32:1616–25. doi:10.1038/onc.2012.193
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
81. Hu W, Alvarez-Dominguez JR, Lodish HF. Regulation of mammalian cell differentiation by long non-coding RNAs. EMBO Rep (2012) 13:971–83. doi:10.1038/embor.2012.145
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
82. Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet (2014) 15:7–21. doi:10.1038/nrg3606
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
83. Zhang X, Lian Z, Padden C, Gerstein MB, Rozowsky J, Snyder M, et al. A myelopoiesis-associated regulatory intergenic noncoding RNA transcript within the human HOXA cluster. Blood (2009) 113:2526–34. doi:10.1182/blood-2008-06-162164
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
84. Wright PW, Huehn A, Cichocki F, Li H, Sharma N, Dang H, et al. Identification of a KIR antisense lncRNA expressed by progenitor cells. Genes Immun (2013) 14:427–33. doi:10.1038/gene.2013.36
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
85. Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S, et al. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science (2014) 344:310–3. doi:10.1126/science.1251456
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
86. Rapicavoli NA, Qu K, Zhang J, Mikhail M, Laberge R-M, Chang HY. A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. Elife (2013) 2:e00762. doi:10.7554/eLife.00762
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
87. Carpenter S, Aiello D, Atianand MK, Ricci EP, Gandhi P, Hall LL, et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science (2013) 341:789–92. doi:10.1126/science.1240925
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
88. Li Z, Chao T-C, Chang K-Y, Lin N, Patil VS, Shimizu C, et al. The long noncoding RNA THRIL regulates TNFα expression through its interaction with hnRNPL. Proc Natl Acad Sci U S A (2014) 111:1002–7. doi:10.1073/pnas.1313768111
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
89. Mao YS, Zhang B, Spector DL. Biogenesis and function of nuclear bodies. Trends Genet (2011) 27:295–306. doi:10.1016/j.tig.2011.05.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
90. Imamura K, Imamachi N, Akizuki G, Kumakura M, Kawaguchi A, Nagata K, et al. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol Cell (2014) 53:393–406. doi:10.1016/j.molcel.2014.01.009
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
91. O’Connell RM, Chaudhuri AA, Rao DS, Gibson WSJ, Balazs AB, Baltimore D. MicroRNAs enriched in hematopoietic stem cells differentially regulate long-term hematopoietic output. Proc Natl Acad Sci U S A (2010) 107:14235–40. doi:10.1073/pnas.1009798107
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
92. Cobb BS, Nesterova TB, Thompson E, Hertweck A, O’Connor E, Godwin J, et al. T cell lineage choice and differentiation in the absence of the RNase III enzyme Dicer. J Exp Med (2005) 201:1367–73. doi:10.1084/jem.20050572
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
93. Muljo SA, Ansel KM, Kanellopoulou C, Livingston DM, Rao A, Rajewsky K. Aberrant T cell differentiation in the absence of Dicer. J Exp Med (2005) 202:261–9. doi:10.1084/jem.20050678
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
94. Neilson JR, Zheng GXY, Burge CB, Sharp PA. Dynamic regulation of miRNA expression in ordered stages of cellular development. Genes Dev (2007) 21:578–89. doi:10.1101/gad.1522907
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
95. Bronevetsky Y, Villarino AV, Eisley CJ, Barbeau R, Barczak AJ, Heinz GA, et al. T cell activation induces proteasomal degradation of argonaute and rapid remodeling of the microRNA repertoire. J Exp Med (2013) 210:417–32. doi:10.1084/jem.20111717
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
96. Ooi AGL, Sahoo D, Adorno M, Wang Y, Weissman IL, Park CY. MicroRNA-125b expands hematopoietic stem cells and enriches for the lymphoid-balanced and lymphoid-biased subsets. Proc Natl Acad Sci U S A (2010) 107:21505–10. doi:10.1073/pnas.1016218107
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
97. Rossi RL, Rossetti G, Wenandy L, Curti S, Ripamonti A, Bonnal RJP, et al. Distinct microRNA signatures in human lymphocyte subsets and enforcement of the naive state in CD4+ T cells by the microRNA miR-125b. Nat Immunol (2011) 12:796–803. doi:10.1038/ni.2057
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
98. Kuchen S, Resch W, Yamane A, Kuo N, Li Z, Chakraborty T, et al. Regulation of microRNA expression and abundance during lymphopoiesis. Immunity (2010) 32:828–39. doi:10.1016/j.immuni.2010.05.009
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
99. Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, et al. Requirement of bic/microRNA-155 for normal immune function. Science (2007) 316:608–11. doi:10.1126/science.1139253
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
100. Zhang J, Cheng Y, Cui W, Li M, Li B, Guo L. MicroRNA-155 modulates Th1 and Th17 cell differentiation and is associated with multiple sclerosis and experimental autoimmune encephalomyelitis. J Neuroimmunol (2014) 266:56–63. doi:10.1016/j.jneuroim.2013.09.019
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
101. Kohlhaas S, Garden OA, Scudamore C, Turner M, Okkenhaug K, Vigorito E. Cutting edge: the Foxp3 target miR-155 contributes to the development of regulatory T cells. J Immunol (2009) 182:2578–82. doi:10.4049/jimmunol.0803162
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
102. Lu L-F, Thai T-H, Calado DP, Chaudhry A, Kubo M, Tanaka K, et al. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity (2009) 30:80–91. doi:10.1016/j.immuni.2008.11.010
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
103. Jiang S, Li C, Olive V, Lykken E, Feng F, Sevilla J, et al. Molecular dissection of the miR-17-92 cluster’s critical dual roles in promoting Th1 responses and preventing inducible Treg differentiation. (2011). Blood (2011) 118:5487–97. doi:10.1182/blood-2011-05-355644
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
104. Liu SQ, Jiang S, Li C, Zhang B, Li QJ. miR-17-92 cluster targets phosphatase and tensin homology and ikaros family zinc finger 4 to promote TH17-mediated inflammation. (2014). J Biol Chem (2014) 289:12446–56. doi:10.1074/jbc.M114.550723
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
105. Monticelli S, Ansel KM, Xiao C, Socci ND, Krichevsky AM, Thai TH, et al. MicroRNA profiling of the murine hematopoietic system. Genome Biol (2005) 6:R71. doi:10.1186/gb-2005-6-8-r71
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
106. Cobb BS, Hertweck A, Smith J, O’Connor E, Graf D, Cook T, et al. A role for dicer in immune regulation. J Exp Med (2006) 203:2519–27. doi:10.1084/jem.20061692
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
107. Curtale G, Citarella F, Carissimi C, Goldoni M, Carucci N, Fulci V, et al. An emerging player in the adaptive immune response: microRNA-146a is a modulator of IL-2 expression and activation-induced cell death in T lymphocytes. Blood (2010) 115:265–73. doi:10.1182/blood-2009-06-225987
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
108. Trifari S, Pipkin ME, Bandukwala HS, Äijö T, Bassein J, Chen R, et al. MicroRNA-directed program of cytotoxic CD8+ T-cell differentiation. Proc Natl Acad Sci U S A (2013) 110:18608–13. doi:10.1073/pnas.1317191110
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
109. Gracias DT, Stelekati E, Hope JL, Boesteanu AC, Doering TA, Norton J, et al. The microRNA miR-155 controls CD8(+) T cell responses by regulating interferon signaling. Nat Immunol (2013) 14:593–602. doi:10.1038/ni.2576
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
110. Khann AA, Penny LA, Yuzefpolskiy Y, Sarkar S, Kalia V. MicroRNA-17 92 regulates effector and memory CD8 T-cell fates by modulating proliferation in response to infections. (2013). Blood (2013) 121:4473–83. doi:10.1182/blood-2012-06-435412
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
111. Koralov SB, Muljo SA, Galler GR, Krek A, Chakraborty T, Kanellopoulou C, et al. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell (2008) 132:860–74. doi:10.1016/j.cell.2008.02.020
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
112. Belver L, de Yébenes VG, Ramiro AR. MicroRNAs prevent the generation of autoreactive antibodies. Immunity (2010) 33:713–22. doi:10.1016/j.immuni.2010.11.010
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
113. Xu S, Guo K, Zeng Q, Huo J, Lam K-P. The RNase III enzyme dicer is essential for germinal center B-cell formation. Blood (2012) 119:767–76. doi:10.1182/blood-2011-05-355412
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
114. Okuyama K, Ikawa T, Gentner B, Hozumi K, Harnprasopwat R, Lu J, et al. MicroRNA-126-mediated control of cell fate in B-cell myeloid progenitors as a potential alternative to transcriptional factors. Proc Natl Acad Sci U S A (2013) 110:13410–5. doi:10.1073/pnas.1220710110
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
115. Chen C-Z, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science (2004) 303:83–6. doi:10.1126/science.1091903
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
116. Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell (2008) 132:875–86. doi:10.1016/j.cell.2008.02.019
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
117. Jin HY, Oda H, Lai M, Skalsky RL, Bethel K, Shepherd J, et al. MicroRNA-17 92 plays a causative role in lymphomagenesis by coordinating multiple oncogenic pathways. EMBO J (2013) 32:2377–91. doi:10.1038/emboj.2013.178
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
118. Zhou B, Wang S, Mayr C, Bartel DP, Lodish HF. miR-150, a microRNA expressed in mature B and T cells, blocks early B cell development when expressed prematurely. Proc Natl Acad Sci U S A (2007) 104:7080–5. doi:10.1073/pnas.0702409104
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
119. Xiao C, Calado DP, Galler G, Thai T-H, Patterson HC, Wang J, et al. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell (2007) 131:146–59. doi:10.1016/j.cell.2007.07.021
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
120. Thai T-H, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, et al. Regulation of the germinal center response by microRNA-155. Science (2007) 316:604–8. doi:10.1126/science.1141229
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
121. Vigorito E, Perks KL, Abreu-Goodger C, Bunting S, Xiang Z, Kohlhaas S, et al. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity (2007) 27:847–59. doi:10.1016/j.immuni.2007.10.009
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
122. Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF, et al. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci U S A (2005) 102:3627–32. doi:10.1073/pnas.0500613102
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
123. Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N, et al. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci U S A (2006) 103:7024–9. doi:10.1073/pnas.0602266103
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
124. Gururajan M, Haga CL, Das S, Leu C-M, Hodson D, Josson S, et al. MicroRNA 125b inhibition of B cell differentiation in germinal centers. Int Immunol (2010) 22:583–92. doi:10.1093/intimm/dxq042
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
125. Pang KC, Dinger ME, Mercer TR, Malquori L, Grimmond SM, Chen W, et al. Genome-wide identification of long noncoding RNAs in CD8+ T cells. J Immunol (2009) 182:7738–48. doi:10.4049/jimmunol.0900603
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
126. Hu G, Tang Q, Sharma S, Yu F, Escobar TM, Muljo SA. Expression and regulation of intergenic long noncoding RNAs during T cell development and differentiation. Nat Immunol (2013) 14:1190–8. doi:10.1038/ni.2712
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
127. Vigneau S, Rohrlich P-S, Brahic M, Bureau J-F. Tmevpg1, a candidate gene for the control of Theiler’s virus persistence, could be implicated in the regulation of gamma interferon. J Virol (2003) 77:5632–8. doi:10.1128/JVI.77.10.5632-5638.2003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
128. Collier SP, Collins PL, Williams CL, Boothby MR, Aune TM. Cutting edge: influence of Tmevpg1, a long intergenic noncoding RNA, on the expression of Ifng by Th1 cells. J Immunol (2012) 189:2084–8. doi:10.4049/jimmunol.1200774
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
129. Gomez JA, Wapinski OL, Yang YW, Bureau J-F, Gopinath S, Monack DM, et al. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-γ locus. Cell (2013) 152:743–54. doi:10.1016/j.cell.2013.01.015
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
130. Ricaño-Ponce I, Wijmenga C. Mapping of immune-mediated disease genes. Annu Rev Genomics Hum Genet (2013) 14:325–53. doi:10.1146/annurev-genom-091212-153450
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
131. Capuano M, Iaffaldano L, Tinto N, Montanaro D, Capobianco V, Izzo V, et al. MicroRNA-449a overexpression, reduced NOTCH1 signals and scarce goblet cells characterize the small intestine of celiac patients. PLoS One (2011) 6:e29094. doi:10.1371/journal.pone.0029094
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
132. Kumar V, Wijmenga C, Withoff S. From genome-wide association studies to disease mechanisms: celiac disease as a model for autoimmune diseases. Semin Immunopathol (2012) 34:567–80. doi:10.1007/s00281-012-0312-1
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
133. Geremia A, Biancheri P, Allan P, Corazza GR, Di Sabatino A. Innate and adaptive immunity in inflammatory bowel disease. Autoimmun Rev (2014) 13:3–10. doi:10.1016/j.autrev.2013.06.004
134. Beaudoin M, Goyette P, Boucher G, Lo KS, Rivas MA, Stevens C, et al. Deep resequencing of GWAS loci identifies rare variants in CARD9, IL23R and RNF186 that are associated with ulcerative colitis. PLoS Genet (2013) 9:e1003723. doi:10.1371/journal.pgen.1003723
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
135. Zahm AM, Thayu M, Hand NJ, Horner A, Leonard MB, Friedman JR. Circulating MicroRNA is a biomarker of pediatric Crohn disease. J Pediatr Gastroenterol Nutr (2011) 53:26–33. doi:10.1097/MPG.0b013e31822200cc
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
136. Wu F, Zikusoka M, Trindade A, Dassopoulos T, Harris ML, Bayless T, et al. MicroRNAs are differentially expressed in ulcerative colitis and alter expression of macrophage inflammatory peptide-2 alpha. Gastroenterology (2008) 135:1624–35. doi:10.1053/j.gastro.2008.07.068
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
137. Coskun M, Bjerrum JT, Seidelin JB, Nielsen OH. MicroRNAs in inflammatory bowel disease – pathogenesis, diagnostics and therapeutics. World J Gastroenterol (2012) 18:4629–34. doi:10.3748/wjg.v18.i34.4629
138. Takagi T, Naito Y, Mizushima K, Hirata I, Yagi N, Tomatsuri N, et al. Increased expression of microRNA in the inflamed colonic mucosa of patients with active ulcerative colitis. J Gastroenterol Hepatol (2010) 25(Suppl 1):S129–33. doi:10.1111/j.1440-1746.2009.06216.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
139. Fasseu M, Tréton X, Guichard C, Pedruzzi E, Cazals-Hatem D, Richard C, et al. Identification of restricted subsets of mature microRNA abnormally expressed in inactive colonic mucosa of patients with inflammatory bowel disease. PLoS One (2010) 5(pii):e13160. doi:10.1371/journal.pone.0013160
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
140. Coskun M, Bjerrum JT, Seidelin JB, Troelsen JT, Olsen J, Nielsen OH. miR-20b, miR-98, miR-125b-1*, and let-7e* as new potential diagnostic biomarkers in ulcerative colitis. World J Gastroenterol (2013) 19:4289–99. doi:10.3748/wjg.v19.i27.4289
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
141. Gazouli M, Papaconstantinou I, Stamatis K, Vaiopoulou A, Zeglinas C, Vassiliou I, et al. Association study of genetic variants in miRNAs in patients with inflammatory bowel disease: preliminary results. Dig Dis Sci (2013) 58:2324–8. doi:10.1007/s10620-013-2640-y
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
142. Paraskevi A, Theodoropoulos G, Papaconstantinou I, Mantzaris G, Nikiteas N, Gazouli M. Circulating microRNA in inflammatory bowel disease. J Crohns Colitis (2012) 6:900–4. doi:10.1016/j.crohns.2012.02.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
143. Yang Y, Ma Y, Shi C, Chen H, Zhang H, Chen N, et al. Overexpression of miR-21 in patients with ulcerative colitis impairs intestinal epithelial barrier function through targeting the Rho GTPase RhoB. Biochem Biophys Res Commun (2013) 434:746–52. doi:10.1016/j.bbrc.2013.03.122
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
144. Nata T, Fujiya M, Ueno N, Moriichi K, Konishi H, Tanabe H, et al. MicroRNA-146b improves intestinal injury in mouse colitis by activating nuclear factor-κB and improving epithelial barrier function. J Gene Med (2013) 15:249–60. doi:10.1002/jgm.2717
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
145. Shi C, Liang Y, Yang J, Xia Y, Chen H, Han H, et al. MicroRNA-21 knockout improve the survival rate in DSS induced fatal colitis through protecting against inflammation and tissue injury. PLoS One (2013) 8:e66814. doi:10.1371/journal.pone.0066814
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
146. Ghorpade DS, Sinha AY, Holla S, Singh V, Balaji KN. NOD2-nitric oxide-responsive microRNA-146a activates Sonic hedgehog signaling to orchestrate inflammatory responses in murine model of inflammatory bowel disease. J Biol Chem (2013) 288:33037–48. doi:10.1074/jbc.M113.492496
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
147. Chen Y, Wang C, Liu Y, Tang L, Zheng M, Xu C, et al. MiR-122 targets NOD2 to decrease intestinal epithelial cell injury in Crohn’s disease. Biochem Biophys Res Commun (2013) 438:133–9. doi:10.1016/j.bbrc.2013.07.040
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
148. Huang Z, Shi T, Zhou Q, Shi S, Zhao R, Shi H, et al. miR-141 regulates colonic leukocytic trafficking by targeting CXCL12β during murine colitis and human Crohn’s disease. Gut (2013):1–11. doi:10.1136/gutjnl-2012-304213
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
149. Pekow JR, Dougherty U, Mustafi R, Zhu H, Kocherginsky M, Rubin DT, et al. miR-143 and miR-145 are downregulated in ulcerative colitis: putative regulators of inflammation and protooncogenes. Inflamm Bowel Dis (2012) 18:94–100. doi:10.1002/ibd.21742
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
150. Olaru AV, Yamanaka S, Vazquez C, Mori Y, Cheng Y, Abraham JM, et al. MicroRNA-224 negatively regulates p21 expression during late neoplastic progression in inflammatory bowel disease. Inflamm Bowel Dis (2013) 19:471–80. doi:10.1097/MIB.0b013e31827e78eb
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
151. Ludwig K, Fassan M, Mescoli C, Pizzi M, Balistreri M, Albertoni L, et al. PDCD4/miR-21 dysregulation in inflammatory bowel disease-associated carcinogenesis. Virchows Arch (2013) 462:57–63. doi:10.1007/s00428-012-1345-5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
152. Svrcek M, El-Murr N, Wanherdrick K, Dumont S, Beaugerie L, Cosnes J, et al. Overexpression of microRNAs-155 and 21 targeting mismatch repair proteins in inflammatory bowel diseases. Carcinogenesis (2013) 34:828–34. doi:10.1093/carcin/bgs408
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
153. Kanaan Z, Rai SN, Eichenberger MR, Barnes C, Dworkin AM, Weller C, et al. Differential microRNA expression tracks neoplastic progression in inflammatory bowel disease-associated colorectal cancer. Hum Mutat (2012) 33:551–60. doi:10.1002/humu.22021
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
154. Liu Z, Lee J, Krummey S, Lu W, Cai H, Lenardo MJ. The kinase LRRK2 is a regulator of the transcription factor NFAT that modulates the severity of inflammatory bowel disease. Nat Immunol (2011) 12:1063–70. doi:10.1038/ni.2113
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
155. Qiao YQ, Huang ML, Xu AT, Zhao D, Ran ZH, Shen J. LncRNA DQ786243 affects Treg related CREB and Foxp3 expression in Crohn’s disease. J Biomed Sci (2013) 20:87. doi:10.1186/1423-0127-20-87
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
156. Vukusic S, Confavreux C. Prognostic factors for progression of disability in the secondary progressive phase of multiple sclerosis. J Neurol Sci (2003) 206:135–7. doi:10.1016/S0022-510X(02)00426-4
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
157. Smith KM, Guerau-de-Arellano M, Costinean S, Williams JL, Bottoni A, Mavrikis Cox G, et al. miR-29ab1 deficiency identifies a negative feedback loop controlling Th1 bias that is dysregulated in multiple sclerosis. J Immunol (2012) 189:1567–76. doi:10.4049/jimmunol.1103171
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
158. Nakahama T, Hanieh H, Nquyen NT, Chinen I, Ripley B, Millrine D, et al. Aryl hydrocarbon receptor-mediated induction of the microRNA-132/212 cluster promotes interleukin-17-producing T-helper cell differentiation. (2013). Proc Natl Acad Sci U S A (2013) 110:11964–9. doi:10.1073/pnas.1311087110
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
159. Du C, Liu C, Kang J, Zhao G, Ye Z, Huang S, et al. MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. (2009). Nat Immunol (2009) 10:1252–9. doi:10.1038/ni.1798
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
160. Honardoost MA, Kiani-Esfahani A, Ghaedi K, Etemadifar M, Salehi M. miR-326 and miR-26a, two potential markers for diagnosis of relapse and remission phases in patient with relapsing-remitting multiple sclerosis. Gene (2014) 544:128–33. doi:10.1016/j.gene.2014.04.069
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
161. Lopez-Ramirez MA, Wu D, Pryce G, Simpson JE, Reijerkerk A, King-Robson J, et al. MicroRNA-155 negatively affects blood-brain barrier function during neuroinflammation. FASEB J (2014) 28:2551–65. doi:10.1096/fj.13-248880
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
162. Reijerkerk A, Lopez-Ramirez MA, van Het Hof B, Drexhage JAR, Kamphuis WW, Kooij G, et al. MicroRNAs regulate human brain endothelial cell-barrier function in inflammation: implications for multiple sclerosis. J Neurosci (2013) 33:6857–63. doi:10.1523/JNEUROSCI.3965-12.2013
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
163. Gandhi R, Healy B, Gholipour T, Egorova S, Musallam A, Hussain MS, et al. Circulating microRNAs as biomarkers for disease staging in multiple sclerosis. Ann Neurol (2013) 73:729–40. doi:10.1002/ana.23880
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
164. Schwamborn JC, Berezikov E, Knoblich JA. The TRIM-NHL protein TRIM32 activates microRNAs and prevents self-renewal in mouse neural progenitors. Cell (2009) 136:913–25. doi:10.1016/j.cell.2008.12.024
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
165. Li JJ, Dolios G, Wang R, Liao F-F. Soluble beta-amyloid peptides, but not insoluble fibrils, have specific effect on neuronal microRNA expression. PLoS One (2014) 9:e90770. doi:10.1371/journal.pone.0090770
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
166. Ng S-Y, Lin L, Soh BS, Stanton LW. Long noncoding RNAs in development and disease of the central nervous system. Trends Genet (2013) 29:461–8. doi:10.1016/j.tig.2013.03.002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
167. Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, et al. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat Med (2008) 14:723–30. doi:10.1038/nm1784
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
168. Mus E, Hof PR, Tiedge H. Dendritic BC200 RNA in aging and in Alzheimer’s disease. Proc Natl Acad Sci U S A (2007) 104:10679–84. doi:10.1073/pnas.0701532104
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
169. Modarresi F, Faghihi MA, Lopez-Toledano MA, Fatemi RP, Magistri M, Brothers SP, et al. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat Biotechnol (2012) 30:453–9. doi:10.1038/nbt.2158
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
170. Xie Y, Hayden MR, Xu B. BDNF overexpression in the forebrain rescues Huntington’s disease phenotypes in YAC128 mice. J Neurosci (2010) 30:14708–18. doi:10.1523/JNEUROSCI.1637-10.2010
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
171. Bauer NM, Moos C, van Horssen J, Witte M, van der Valk P, Altenhein B, et al. Myelin basic protein synthesis is regulated by small non-coding RNA 715. EMBO Rep (2012) 13:827–34. doi:10.1038/embor.2012.97
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
172. Pusic AD, Pusic KM, Clayton BLL, Kraig RP. IFNγ-stimulated dendritic cell exosomes as a potential therapeutic for remyelination. J Neuroimmunol (2014) 266:12–23. doi:10.1016/j.jneuroim.2013.10.014
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
173. Rossi MN, Antonangeli F. LncRNAs: new players in apoptosis control. Int J Cell Biol (2014) 2014:473857. doi:10.1155/2014/473857
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
174. Ridolfi E, Fenoglio C, Cantoni C, Calvi A, De Riz M, Pietroboni A, et al. Expression and genetic analysis of microRNAs involved in multiple sclerosis. Int J Mol Sci (2013) 14:4375–84. doi:10.3390/ijms14034375
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
175. Li T, Morgan MJ, Choksi S, Zhang Y, Kim Y-S, Liu Z. MicroRNAs modulate the noncanonical transcription factor NF-kappaB pathway by regulating expression of the kinase IKKalpha during macrophage differentiation. Nat Immunol (2010) 11:799–805. doi:10.1038/ni.1918
176. Lin S-T, Huang Y, Zhang L, Heng MY, Ptácek LJ, Fu Y-H. MicroRNA-23a promotes myelination in the central nervous system. Proc Natl Acad Sci U S A (2013) 110:17468–73. doi:10.1073/pnas.1317182110
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
177. Lill CM, Schilling M, Ansaloni S, Schröder J, Jaedicke M, Luessi F, et al. Assessment of microRNA-related SNP effects in the 3′ untranslated region of the IL22RA2 risk locus in multiple sclerosis. Neurogenetics (2014) 15:129–34. doi:10.1007/s10048-014-0396-y
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
178. Yoon J-H, Abdelmohsen K, Gorospe M. Functional interactions among microRNAs and long noncoding RNAs. Semin Cell Dev Biol (2014) 34C:9–14. doi:10.1016/j.semcdb.2014.05.015
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
179. Grosswendt S, Filipchyk A, Manzano M, Klironomos F, Schilling M, Herzog M, et al. Unambiguous identification of miRNA:target site interactions by different types of ligation reactions. Mol Cell (2014) 54:1042–54. doi:10.1016/j.molcel.2014.03.049
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
180. Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell (2011) 44:667–78. doi:10.1016/j.molcel.2011.08.027
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
181. Mestdagh P, Hartmann N, Baeriswyl L, Andreasen D, Bernard N, Chen C, et al. Evaluation of quantitative miRNA expression platforms in the microRNA quality control (miRQC) study. Nat Methods (2014) 11:809–15. doi:10.1038/nmeth.3014
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
182. Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov (2013) 12:847–65. doi:10.1038/nrd4140
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
183. Li Z, Rana TM. Therapeutic targeting of microRNAs: current status and future challenges. Nat Rev Drug Discov (2014) 13:622–38. doi:10.1038/nrd4359
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: regulatory RNAs, miRNAs, long non-coding RNAs, immune system, autoimmune diseases
Citation: Stachurska A, Zorro MM, van der Sijde MR and Withoff S (2014) Small and long regulatory RNAs in the immune system and immune diseases. Front. Immunol. 5:513. doi: 10.3389/fimmu.2014.00513
Received: 24 July 2014; Accepted: 02 October 2014;
Published online: 20 October 2014.
Edited by:
Susan Carpenter, University of Massachusetts Medical School, USACopyright: © 2014 Stachurska, Zorro, van der Sijde and Withoff. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sebo Withoff, University Medical Center Groningen, Hanzeplein 1, 9713 GZ, Groningen, Netherlands e-mail:cy53aXRob2ZmQHVtY2cubmw=
†Anna Stachurska, Maria M. Zorro and Marijke R. van der Sijde have contributed equally to this work.
 Anna Stachurska
Anna Stachurska Maria M. Zorro
Maria M. Zorro Marijke R. van der Sijde
Marijke R. van der Sijde Sebo Withoff
Sebo Withoff