
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Immunol., 22 April 2014
Sec. Vaccines and Molecular Therapeutics
Volume 5 - 2014 | https://doi.org/10.3389/fimmu.2014.00179
This article is part of the Research TopicAnimal allergens: Common protein characteristics featuring their allergenicityView all 10 articles
Fish is a common trigger of severe, food-allergic reactions. Only a limited number of proteins induce specific IgE-mediated immune reactions. The major fish allergens are the parvalbumins. They are members of the calcium-binding EF-hand protein family characterized by a conserved protein structure. They represent highly cross-reactive allergens for patients with specific IgE to conserved epitopes. These patients might experience clinical reactions with various fish species. On the other hand, some individuals have IgE antibodies directed against unique, species-specific parvalbumin epitopes, and these patients show clinical symptoms only with certain fish species. Furthermore, different parvalbumin isoforms and isoallergens are present in the same fish and might display variable allergenicity. This was shown for salmon homologs, where only a single parvalbumin (beta-1) isoform was identified as allergen in specific patients. In addition to the parvalbumins, several other fish proteins, enolases, aldolases, and fish gelatin, seem to be important allergens. New clinical and molecular insights advanced the knowledge and understanding of fish allergy in the last years. These findings were useful for the advancement of the IgE-based diagnosis and also for the management of fish allergies consisting of advice and treatment of fish-allergic patients.
In human diet, fish is a valuable source of essential amino acids, polyunsaturated fatty acids, and lipid-soluble vitamins. Although the fish consumption in European countries is quite stable, the global demand for fish and fish products is increasing still steadily (1). While many literature sources indicate that allergies to fish are on the rise, the actual prevalence is not well established as most studies are based on self-reported food allergies (2). It is estimated that up to 0.2% of the general population is affected by fish allergy (3, 4). However, the exposure to fish is an important factor determining the reported prevalence of fish allergy. Therefore, the prevalence of fish allergy is greater in countries with high fish consumption and fish-processing industries (5).
Fish allergy is a pathophysiological, IgE-mediated immune response to specific fish proteins. Patients become sensitized by allergen exposure via the gastro-intestinal tract during ingestion, which is the major route of sensitization, or via the respiratory system by fish aeroallergens or skin contact (6–9). Common clinical manifestations are oral allergy syndrome, rhinitis, abdominal pain, diarrhea, urticaria, angioedema, asthma, and in severe cases, even life-threatening anaphylaxis (10–12). Fish aeroallergens can be important triggers of atopic dermatitis (13). There are only a few clinical studies addressing minimal eliciting doses of fish allergy. However, already low milligram amounts of fish seem to be sufficient to trigger allergic symptoms in sensitized patients (14). Cross-reactivity among fish species has been commonly reported for fish allergy (11, 15, 16). This clinical cross-reactivity seems to be even more pronounced between closely related fishes. On the other hand, it has been shown that even highly fish-sensitized patients can tolerate certain fish species, such as tuna (17, 18). In addition, in case reports, clinical monosensitivity to single fishes has been proven for sole, swordfish, pangasius/tilapia, tuna/marlin, and more recently, for salmon and salmonid fishes (19–26). So far, no cross-reactivity has been shown for fish and other seafood allergens while in some cases, a co-sensitization to both allergen sources might occur (27).
The diagnostic procedure is based on four main pillars: the patient’s clinical history, in vivo analysis of skin reactivity, in vitro quantification of specific serum IgE and, in selected cases, oral provocation challenges. A broad diversity of fishes is globally consumed but only a limited number of commercial extracts are available for skin testing. Therefore, fresh or processed fish is commonly used for this in vivo analysis. However, the predictive value of skin tests is low (28). For in vitro analysis of specific IgE levels, the ImmunoCAP system (ThermoScientific) offers a wide panel of fish extracts. Meanwhile, two recombinant parvalbumins from carp and cod are available for this diagnostic assay. The predictive value of fish extract-specific IgE measurements is not well established, but a high titer of specific IgE (20 kUA/L) was reported to predict an allergy to cod with a likelihood of 95% (29).
Other adverse reactions might be misdiagnosed as fish allergy (30). Allergy-like symptoms occur upon ingestion of histamine-contaminated, spoiled fish (“scombroid fish poisoning”) (31). Also, consumption of fish contaminated with the parasite Anisakis simplex (herring worm) provokes acute allergic manifestations caused by IgE-mediated sensitization to Anisakis allergens (27, 32).
To avoid severe reactions, the management of fish allergy relies on the elimination of each fish product from the diet of the sensitized patient. In some cases, it has been reported that patients may lose their sensitivity upon eliminating diet (33, 34). The therapeutic desensitization to fish has been reported only for a single case (35). The development of specific immunotherapeutics based on hypoallergenic variants of parvalbumins, the major fish allergens, is the focus of ongoing studies (36, 37). Research on a primary strategy for the prevention of fish allergy is very limited. The current recommendations of the American Academy of Allergy, Asthma & Immunology also do not suggest a general, delayed introduction of fish in the diet of children (38).
The few foods that are responsible for causing most allergic reactions are milk, eggs, peanuts, tree nuts, fish, shellfish, soy, and wheat. They contain potent food allergens (39). Their allergenic potency has been related to specific protein features. These allergens are highly abundant in the food sources, and moreover, they possess a high stability toward food processing and digestion (40). The structural stability has been allocated to different protein characteristics such as intrinsic ligand binding and intramolecular disulfide bonds (41). Some food allergens form protein aggregates of high stability. Although some food allergens are sensitive to gastric and intestinal digestion, degradation fragments are still recognized by specific IgE antibodies (42). Food allergens of animal origin are mainly grouped into three protein superfamilies such as caseins, tropomyosins, and EF-hand proteins (43).
Most fish parvalbumins belong to the beta-subtype while the alpha-subtype is predominantly found in other organisms. Beta-parvalbumin has first been identified as fish allergen in Baltic cod (44). Later on, the importance of this protein as the fish panallergen was confirmed for a wide range of commonly consumed species such as salmon, carp, mackerel, tuna, and pilchard (45–49). Parvalbumins are highly stable, low-molecular-weight proteins (10–12 kDa), which are very common in fish muscle (5). The muscle of bony fishes is composed of two tissues, the light and dark muscle differing by their physiological function and composition (50). The parvalbumin expression is considerably higher in light than in dark muscle tissue (51). In contrast to most bony fishes, pelagic fishes such as tuna have mainly dark muscles of low parvalbumin content (48). This complies with the low allergenicity reported for tuna which is even used in canned preparations as matrix for food challenges (17). Overall, the parvalbumin content differs considerably in fish species and the different amount of parvalbumins correlates with the variable allergenicity of fishes (52–54). It has been shown that the parvalbumin level is up to 100-times higher in carp than in mackerel or tuna muscle. In addition, physical and chemical effects of food processing may alter the allergenicity of fish in food preparations by parvalbumin degradation or oligomerization which may decrease or increase the number of IgE epitopes (55).
Parvalbumins belong to the family of EF-hand proteins. As such, they contain specific EF-hand motifs composed of 12 residues of long loops which are involved in the binding of divalent metal ions. The N-terminal EF-hand site is a non-functional domain while the two other EF-hand motifs are binding calcium and magnesium ions (56). The physiological role of muscle parvalbumins is related to the regulation of the intracellular calcium concentration during muscle relaxation (57). Upon ion binding and ion release, the beta-parvalbumin structure is subjected to a global rearrangement indicating a general flexibility of the EF-hand domains (58). The apo-protein, which is calcium-depleted parvalbumin, has a reduced ability to bind IgE antibodies from fish-allergic patients (59). Consequently, these calcium-binding protein regions were attributed to conformational B-cell epitopes (60).
As of today, the official database of allergens contains 21 parvalbumins from 12 fish species1 (Table 1) (61). The Allergome database lists far more than 100 entries for fish parvalbumins and their isoforms/isoallergens2 (62). This much higher number arises from a compilation of homolog, potentially allergenic molecules, in addition to proteins of proven allergenicity. Parvalbumin has been defined as the major fish allergen as a majority of fish-allergic patients have IgE antibodies reacting to this muscle protein (63–65). However, the prevalence of parvalbumin-specific IgE antibodies seems to vary across different patient populations. For example, fish-allergic patients with sensitization to tropical fishes react mostly to allergens other than parvalbumins (66, 67). Generally, the high clinical cross-reactivity among fishes has been attributed to cross-reacting IgE antibodies recognizing parvalbumins from several species (68). Recently, clinical monosensitivity to salmonid fishes has been linked to salmonid parvalbumin-specific IgE antibodies suggesting that cross-reactivity among fish parvalbumins may be restricted to these closely related fishes (23, 25).
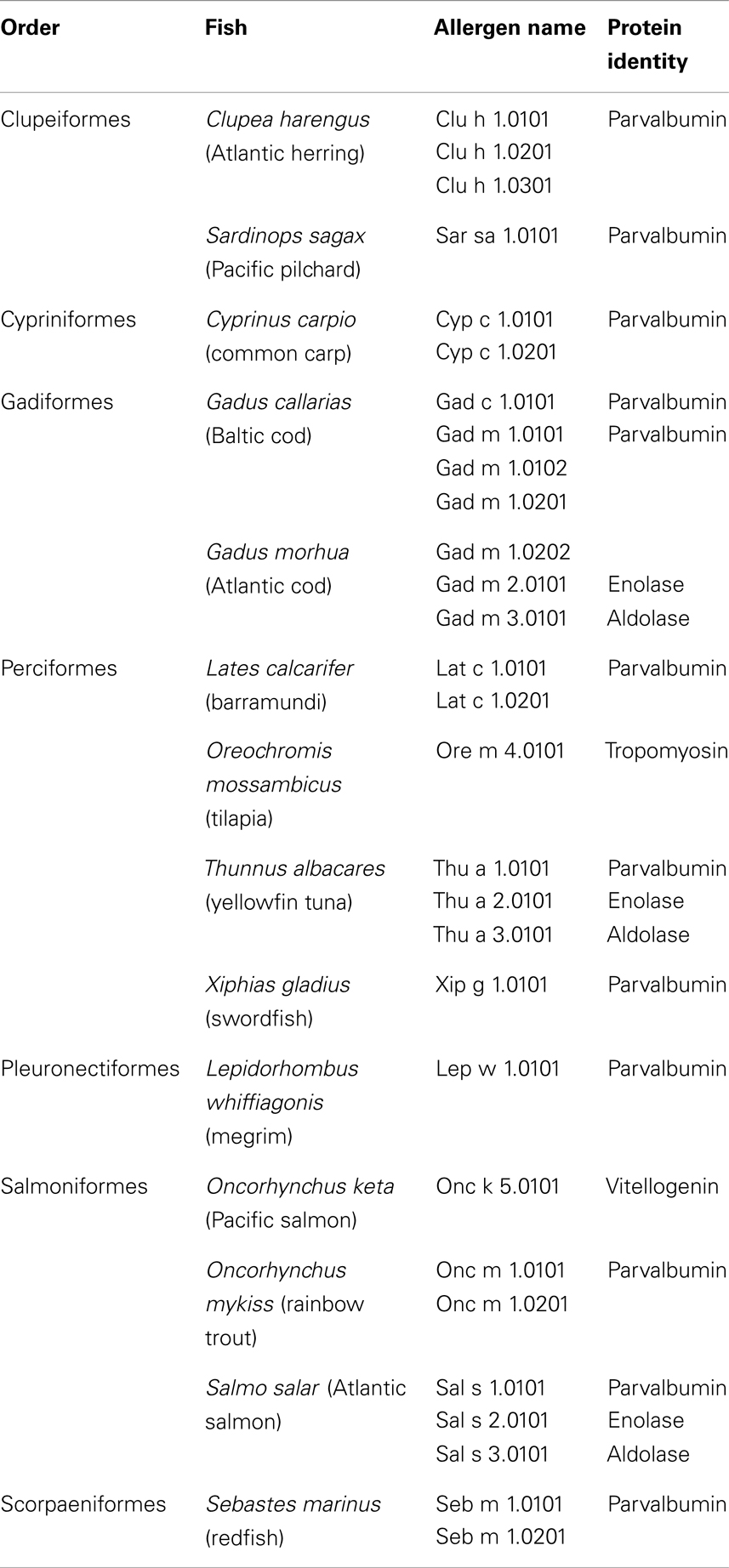
Table 1. Entries of official fish allergens by the International Union of Immunological Societies Allergen Nomenclature Subcommittee database (www.allergen.org).
Initially, IgE reactivity to fish beta-enolase and aldolase had been described in single fishes (69–71). In 2013, 50 kDa enolases and 40 kDa aldolases were identified as important fish allergens in cod, salmon, and tuna (Table 1) (72). Both enzymes are abundant in the fish muscle as they are involved in metabolic glycolysis, the sugar degradation during physiological production of energy.
In a recent study, IgE binding to enolases and aldolases was detected in a high number of fish-allergic patients (72). Most of the enolase/aldolase-positive patients also had IgE antibodies to parvalbumin. Both allergens were positive in the mediator release assay using rat basophilic leukemia cells expressing the human high-affinity IgE receptor passively sensitized with patient sera. They were found to be sensitive to heat treatment. Their relevance as food allergens is still not well understood especially in parvalbumin-negative patients.
The relevance of fish gelatin as a food allergen has been controversially discussed for many years (4). Fish gelatin is commonly used in food and pharmaceutical products replacing mammalian gelatins (73).
Originally, IgE binding to fish collagen had been shown in fish-allergic patients (74, 75). Later, the prevalence of fish gelatin allergy was addressed in single clinical studies (76, 77). In one study, skin prick tests were positive in 3 of 30 cod-allergic patients, while food challenges were positive in one of the three tested patients. Also, its potency as a food allergen was shown in a case report of severe anaphylaxis upon ingestion of sweets containing several grams of fish gelatin (78). No cross-reactivity of mammalian and fish gelatin has been reported so far. Another risk factor of fish gelatin is a potential contamination with fish parvalbumin as it is produced from fish as a natural source. In fact, parvalbumin traces were detected in isinglass, a fish collagen-based food supplement (79).
In the past decades, numerous studies reported allergens different from parvalbumin which were specified by their molecular weight or identified by other methods. Although most studies showed IgE reactivity to these proteins, the relevance of these potential allergens was not addressed. Several allergens (63 kDa protein, further IgE-reactive proteins) were described for cod while some were identified as parvalbumin oligomers (80). Furthermore, allergens were identified in swordfish (25 kDa), eel/eelpout (40 kDa), snapper (35–90 kDa), tuna/marlin (94–105 kDa), scad (46–50 kDa), tropical fishes (29–54 kDa), and pangasius/tilapia (18–45 kDa) (20–22, 64, 66, 81, 82). Potential allergens of known identity were found in cod (aldehyde phosphate isomerase), salmon (triose-phosphate isomerase, fructose-bis-phosphate isomerase, serum albumin), and tuna (creatine kinase, beta-enolase) (70, 83, 84).
Two further fish allergens have been registered by the IUIS-allergen database (Table 1). Food allergy to fish roe has been addressed in a few studies so far. Vitellogenin, a fish yolk protein, has been identified as a food allergen in caviar from different fishes (85, 86). In 2013, tropomyosin, a filamentous muscle protein, was purified, cloned, and identified as a fish allergen in tilapia-sensitized patients (87). This tilapia allergen showed high homology (88%) to the human homolog. It was suggested that it may be involved in autoimmune reaction of inflammatory bowel disease as a significant proportion of the patients with IgE to tropomyosin suffered from this autoimmune disease.
During last decades, parvalbumins, enolases, aldolases, and fish gelatin have been identified as allergenic fish proteins. Specific properties have been attributed to common food allergens such as resistance toward conditions of the gastrointestinal tract and influences by food processing conditions (88). So far, only the allergenicity of parvalbumins has been characterized in more detail as to their allergenicity while consolidated findings still need to be generated for the other allergens.
Parvalbumins have been classified into two phylogenetic lineages, alpha and beta subtype. Their isoelectric points (alpha, pI > 5.0; beta, pI < 4.5) and the other multiple characteristics of the primary protein structure are different (89, 90). Generally, parvalbumins occur in various organs such as the central nervous system and the muscles (56). In muscles, both subtypes have been detected in amphibian tissues while only the alpha-subtype has been reported in mammalian and avian muscles (91, 92). Cartilaginous fish muscles express alpha-parvalbumins whereas the beta-homolog is found in muscle tissue of bony fishes (93). So far, the potency as important food allergens has been shown only for fish beta-parvalbumins. Despite their overall structural similarity to beta-homologs, alpha-parvalbumins are generally considered as non-allergenic proteins (43). Indeed, parvalbumins of the alpha-lineage have been described only as frog meat allergens (94). However, IgE cross-reactivity has also been reported between homologs from fish and frog in a population of fish-sensitized patients (95). In 2011, chicken alpha-parvalbumin has been reported as a poultry allergen in chicken meat allergy (96). Interestingly, IgE recognition of both fish and avian homologs was found in one case study while cross-reactivity was not detected in another clinical case (97, 98). However, the sequence identity between fish parvalbumins and homologs from mammals and birds is low (<55%) so that IgE cross-reactivity seems to be unlikely.
Recent classification by structural, biochemical, and phylogenetic analysis assigned alpha-parvalbumins into a common cluster together with higher vertebrate, including human proteins while beta-subtypes form a separate, only remotely related cluster of parvalbumins which might explain the variable allergenicity of both lineages (99, 100).
The amino acid sequence identities of fish parvalbumins vary substantially (55–95%) but high structural similarity has been reported as a common protein characteristic (101). IgE-binding epitopes were suggested to be located in highly conserved, calcium-binding regions of the molecule. Indeed, reduced IgE binding was found for parvalbumin mutants with single amino acid modifications in calcium-binding motifs (60). The analysis of human B-cell epitopes has only been addressed so far in a few parvalbumins namely allergens from cod, carp, mackerel, and salmon (25, 102–104).
Both linear and conformational epitopes were found in these studies but overall, non-plausible peptide pattern was found to define a common IgE-binding region of fish parvalbumins. It seems that B-cell epitopes are distributed over the whole parvalbumin primary structure. A possible explanation might be the polyclonal B-cell response in fish-allergic patients, which is even more stimulated by individual’s eating habits (e.g., canned, fried, smoked fish) leading to a high variety of exposed allergen forms such as native, modified, or degraded parvalbumins from different species.
For most patients, clinical cross-reactivity among fish species was postulated as a specific feature of fish allergy. Although single clinical reports showed true monosensitivity to single fishes, there was no allergen-based explanation for these observations. Only recently, the species-specific sensitization was attributed to the presence of IgE antibodies selectively recognizing a single salmonid parvalbumin isoallergen (beta-1 parvalbumin) (25). These findings may allow two important conclusions concerning the allergenicity of parvalbumins.
First, parvalbumins from different fishes might vary by their allergenic potential. This might be explained by the specific sensitization of the individual patient, which results from the clinical history (eating habits, age of onset of fish allergy).
Second, the allergenicity of parvalbumin isoforms/isoallergens from the same fish might be variable, as reported for salmon allergens. In fact, fishes express often a high number of parvalbumins such as that reported for carp and catfish (105, 106). These isoforms seem to play a physiological role in the fish muscle adaptation to developmental and environmental changes. In the same fish, parvalbumins differing by sequence microheterogeneity (sequence identity >90%) have been reported (99). It is conceivable that these highly identical isoforms might be of variable allergenicity. For Bet v 1, the major birch pollen allergen, a number of isoforms with different allergenic properties were isolated (107). Several allergenic parvalbumins have been identified in commonly consumed fish, cod (beta-1, beta-2), salmon (beta-1, beta-2), and herring (beta-1, beta-2, beta-3) (5). During studies of monosensitivity to salmonid fishes, different antigenic regions were assumed but only a single epitope was defined as a species-specific allergy marker (23, 25). This unique epitope was localized on a single isoform, the beta-1 salmon parvalbumin. For a patient with documented monosensitivity to salmonid fishes, we could confirm a single region from salmon and trout as species-specific parvalbumin epitope (“allergenic peptide”; Figure 1). This antigenic region was located on the parvalbumin surface and was unique when compared with different homologs from other fish species. The allergenic region matched with the IgE-binding epitope identified by a subsequent study using a peptide-based microarray assay (25). Studies with further sera from patients with specific sensitizations are required to demonstrate the existence of antigenic parvalbumin regions linked to specific clinical phenotypes. A recent study showed that 9 out of 62 fish-allergic patients (15%) experienced clinical reactions with salmonid fishes only (72). This suggests the conclusions that the prevalence of clinically salmon-monosensitized patients is higher than previously assumed.
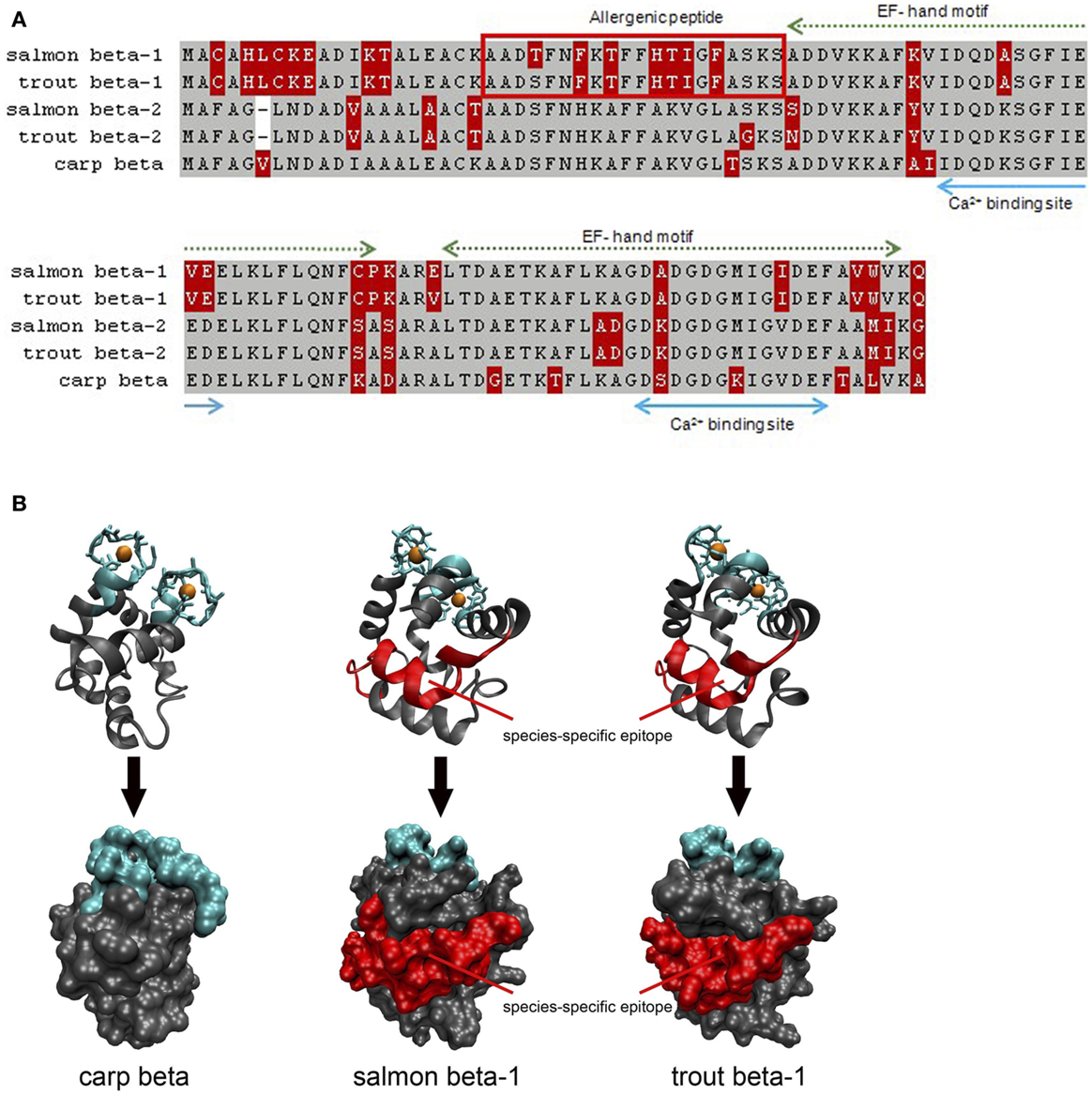
Figure 1. Comparison of salmon (X97824, X97825), trout (FN544258, FN544259), and carp (P02618; 4cpv) parvalbumins. (A) Both salmonid beta-1 parvalbumins sequences differ from their homologs in the N-terminal third of the protein which is not involved in calcium binding. The allergenic peptide is specifically recognized by IgE from a patient monosensitized to salmonid fishes. Gray, identical residues; red, variable residues. (B) Both Ribbon and surface models show that the allergenic peptide is localized on the surface of salmon and trout beta-1 parvalbumins. Blue, calcium-binding regions; red, IgE epitope.
During recent decades, important insights into clinical and allergen-based features were gained for fish allergy: the variable allergenicity among salmon parvalbumins (beta-1, beta-2) was shown, in addition, important new fish allergens (enolase, aldolase, fish gelatin) were identified. These findings will help to develop new immunotherapeutic strategies, but they have also shown that the clinical picture of fish allergy is more complex than anticipated. New molecules need to be implemented in IgE-based routine assays to advance patient diagnosis and advice.
All authors have contributed to the conception, design, and drafting of the paper.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Declaration of all sources of funding: this work has been granted by the Ministry of Research, Luxembourg.
1. Failler P, Van der Walle G, Lecrivain N, Himbes A, Lewins R. Future Prospects for Fish and Fishery Products, European Overview. FAO Fisheries Circular No. 972/4, Part 1 (2007). 204 p.
2. Cianferoni A, Spergel JM. Food allergy: review, classification and diagnosis. Allergol Int (2009) 58:457–66. doi:10.2332/allergolint.09-RAI-0138
3. Rona RJ, Keil T, Summer C, Gislason D, Zuidmeer L, Sodergren E, et al. The prevalence of food allergy: a meta-analysis. J Allergy Clin Immunol (2007) 120:638–46. doi:10.1016/j.jaci.2007.05.026
4. Taylor SL, Kabourek JL, Hefle SL. Fish allergy: fish and products thereof. J Food Sci (2004) 69:75–80. doi:10.1111/j.1750-3841.2004.tb18022.x
5. Sharp MF, Lopata AL. Fish allergy: in review. Clin Rev Allergy Immunol (2013). doi:10.1007/s12016-013-8363-1
6. Jeebhay MF, Robins TG, Seixas N, Baatjies R, George DA, Rusford E, et al. Environmental exposure characterization of fish processing workers. Ann Occup Hyg (2005) 49:423–37. doi:10.1093/annhyg/meh113
7. Pascual CY, Crespo JF, Dominguez Noche C, Ojeda I, Ortega N, Esteban MM. IgE-binding proteins in fish and fish steam. Monogr Allergy (1996) 32:174–80.
8. Seitz CS, Bröcker EB, Trautmann A. Occupational allergy due to seafood delivery: case report. J Occup Med Toxicol (2008) 3:11–3. doi:10.1186/1745-6673-3-11
9. Taylor SL, Gendel SM, Houben GF, Julien E. The key events dose-response framework: a foundation for examining variability in elicitation thresholds for food allergens. Crit Rev Food Sci Nutr (2009) 49:729–39. doi:10.1080/10408390903098707
10. Bock SA, Munoz-Furlong A, Sampson HA. Fatalities due to anaphylactic reactions to food. J Allergy Clin Immunol (2001) 107:191–3. doi:10.1067/mai.2001.112031
11. Helbling A, Haydet R, McCants ML, Musmand JJ, El-Dahr J, Lehrer SB. Fish allergy: is cross-reactivity among fish species relevant? Double-blind placebo-controlled food challenge studies of fish-allergic patients. Ann Allergy Asthma Immunol (1999) 83:517–23. doi:10.1016/S1081-1206(10)62862-1
12. Sicherer SH, Munoz-Furlong A, Sampson HA. Dose-response in double-blind, placebo-controlled oral food challenges in children with atopic dermatitis. J Allergy Clin Immunol (2000) 105:582–6. doi:10.1067/mai.2000.104941
13. Molkhou P. La dermatite atopique (DA) et l’allergie alimentaire (AA) en 2008. J Pediatr Pueric (2008) 22:5–13. doi:10.1016/j.jpp.2008.12.001
14. Untersmayr E, Vestergaard H, Malling HJ, Jensen LB, Platzer MH, Boltz-Nitulescu G, et al. Incomplete digestion of codfish represents a risk factor for anaphylaxis in patients with allergy. J Allergy Clin Immunol (2007) 119:711–7. doi:10.1016/j.jaci.2006.10.039
15. De Martino M, Novembre E, Galli L, de Marco A, Botarelli P, Marano E, et al. Allergy to different fish species in cod-allergic children: in vivo and in vitro studies. J Allergy Clin Immunol (1990) 86:909–14. doi:10.1016/S0091-6749(05)80154-X
16. Sicherer SH, Sampson HA. Food allergy. J Allergy Clin Immunol (2010) 125:116–25. doi:10.1016/j.jaci.2009.08.028
17. Bernhisel-Broadbent J, Strause D, Sampson HA. Fish hypersensitivity. II. Clinical relevance of altered fish allergenicity caused by various preparation methods. J Allergy Clin Immunol (1992) 90:622–9. doi:10.1016/0091-6749(92)90135-O
18. Pascual CY, Reche M, Fiandor A, Valbuena T, Cuevas T, Martin-Esteban MM. Fish allergy in childhood. Pediatr Allergy Immunol (2008) 19:573–9. doi:10.1111/j.1399-3038.2008.00822.x
19. Asero R, Mistrello G, Roncarolo D, Casarini M, Falagiani P. True monosensitivity to a tropical sole. Allergy (1999) 54:1228–9. doi:10.1034/j.1398-9995.1999.00359.x
20. Ebo DG, Kuehn A, Bridts CH, Hilger C, Hentges F, Stevens WJ. Monosensitivity to pangasius and tilapia caused by allergens other than parvalbumin. J Investig Allergol Clin Immunol (2010) 20:84–8.
21. Kelso JM, Jones RT, Yunginger JW. Monospecific allergy to swordfish. Ann Allergy Asthma Immunol (1996) 77:227–8. doi:10.1016/S1081-1206(10)63260-7
22. Kondo Y, Komatsubara R, Nakajima Y, Yasuda T, Kakami M, Tsuge I, et al. Parvalbumin is not responsible for cross-reactivity between tuna and marlin: a case report. J Allergy Clin Immunol (2006) 118:1382–3. doi:10.1016/j.jaci.2006.07.047
23. Kuehn A, Hutt-Kempf E, Hilger C, Hentges F. Clinical monosensitivity to salmonid fish linked to specific IgE-epitopes on salmon and trout beta-parvalbumins. Allergy (2011) 66:299–301. doi:10.1111/j.1398-9995.2010.02463.x
24. Peñas E, Uberti F, Baviera G, Di Lorenzo C, Restani P. Clinical monosensitivity to salmon and rainbow trout: a case report. Pediatr Allergy Immunol (2013) 25:98–100. doi:10.1111/pai.12150
25. Perez-Gordo M, Lin J, Bardina L, Pastor-Vargas C, Cases B, Vivanco F, et al. Epitope mapping of Atlantic salmon major allergen by peptide microarray immunoassay. Int Arch Allergy Immunol (2012) 157:31–40. doi:10.1159/000324677
26. Vázquez-Cortés S, Nuñez-Acevedo B, Jimeno-Nogales L, Ledesma A, Fernández-Rivas M. Selective allergy to the Salmonidae fish family: a selective parvalbumin epitope? Ann Allergy Asthma Immunol (2012) 108:62–3. doi:10.1016/j.anai.2011.10.006
27. Lopata AL, Lehrer SB. New insights into seafood allergy. Curr Opin Allergy Clin Immunol (2009) 9:270–7. doi:10.1097/ACI.0b013e32832b3e6f
28. Lee LA, Burks AW. Food allergies: prevalence, molecular characterization, and treatment/prevention strategies. Annu Rev Nutr (2006) 26:539–65. doi:10.1146/annurev.nutr.26.061505.111211
29. Sampson HA. Utility of food-specific IgE concentrations in predicting symptomatic food allergy. J Allergy Clin Immunol (2001) 107:891–6. doi:10.1067/mai.2001.114708
30. Attaran RR, Probst F. Histamine fish poisoning: a common but frequently misdiagnosed condition. Emerg Med J (2002) 19:474–5. doi:10.1136/emj.19.5.474
31. McLauchlin J, Little CL, Grant KA, Mithani V. Scombrotoxic fish poisoning. J Public Health (2006) 28:61–2. doi:10.1093/pubmed/fdi063
32. Valls A, Pascual CY, Martín Esteban M. Anisakis allergy: an update. Rev Fr Allergol (2005) 45:108–13. doi:10.1097/ACI.0b013e3283373bd0
33. Pite H, Prates S, Borrego LM, Matos V, Loureiro V, Leiria-Pinto P. Resolution of IgE-mediated fish allergy. Allergol Immunopathol (2012) 40:195–7. doi:10.1016/j.aller.2011.03.004
34. Solensky R. Resolution of fish allergy: a case report. Ann Allergy Asthma Immunol (2003) 91:411–2. doi:10.1016/S1081-1206(10)61690-0
35. Casimir G, Cuvelier P, Allard S, Duchateau J. Life-threatening fish allergy successfully treated with immunotherapy. Pediatr Allergy Immunol (1997) 8:103–5. doi:10.1111/j.1399-3038.1997.tb00152.x
36. Swoboda I, Balic N, Klug C, Focke M, Weber M, Spitzauer S, et al. A general strategy for the generation of hypoallergenic molecules for the immunotherapy of fish allergy. J Allergy Clin Immunol (2013) 132:979–81. doi:10.1016/j.jaci.2013.04.027
37. Zuidmeer-Jongejan L, Fernandez-Rivas M, Poulsen LK, Neubauer A, Asturias J, Blom L, et al. FAST: towards safe and effective subcutaneous immunotherapy of persistent life-threatening food allergies. Clin Transl Allergy (2012) 2:5. doi:10.1186/2045-7022-2-5
38. Fleischer DM, Spergel JM, Assa’ad AH, Pongracic JA. Primary prevention of allergic disease through nutritional interventions. J Allergy Clin Immunol Pract (2013) 1:29–36. doi:10.1016/j.jaip.2012.09.003
39. Sicherer SH, Sampson HA. 9. Food allergy. J Allergy Clin Immunol (2006) 117:470–5. doi:10.1016/j.jaci.2005.05.048
40. Bannon GA. What makes a food protein an allergen? Curr Allergy Asthma Rep (2004) 4:43–6. doi:10.1007/s11882-004-0042-0
41. Breiteneder H, Mills EN. Molecular properties of food allergens. J Allergy Clin Immunol (2005) 115:14–23. doi:10.1016/j.jaci.2004.10.022
42. Moreno FJ. Gastrointestinal digestion of food allergens: effect on their allergenicity. Biomed Pharmacother (2007) 61:50–60. doi:10.1016/j.biopha.2006.10.005
43. Radauer C, Bublin M, Wagner S, Mari A, Breiteneder H. Allergens are distributed into few protein families and possess a restricted number of biochemical functions. J Allergy Clin Immunol (2008) 121:847–52. doi:10.1016/j.jaci.2008.01.025
44. Aas K, Elsayed SM. Characterization of a major allergen (cod). Effect of enzymic hydrolysis on the allergenic activity. J Allergy (1969) 44:333–43. doi:10.1016/0021-8707(69)90025-2
45. Beale JE, Jeebhay MF, Lopata AL. Characterisation of purified parvalbumin from five fish species and nucleotide sequencing of this major allergen from Pacific pilchard, Sardinops sagax. Mol Immunol (2009) 46:2985–93. doi:10.1016/j.molimm.2009.06.018
46. Bugajska-Schretter A, Pastore A, Vangelista L, Rumpold H, Valenta R, Spitzauer S. Molecular and immunological characterization of carp parvalbumin, a major fish allergen. Int Arch Allergy Immunol (1999) 118:306–8. doi:10.1159/000024111
47. Hamada Y, Tanaka H, Ishizaki S, Ishida M, Nagashima Y, Shiomi K. Purification, reactivity with IgE and cDNA cloning of parvalbumin as the major allergen of mackerels. Food Chem Toxicol (2003) 41:1149–56. doi:10.1016/S0278-6915(03)00074-7
48. Lim DL, Neo KH, Goh DL, Shek LP, Lee BW. Missing parvalbumin: implications in diagnostic testing for tuna allergy. J Allergy Clin Immunol (2005) 115:874–5. doi:10.1016/j.jaci.2004.12.1117
49. Lindstrom CD, van Do T, Hordvik I, Endresen C, Elsayed S. Cloning of two distinct cDNAs encoding parvalbumin, the major allergen of Atlantic salmon (Salmo salar). Scand J Immunol (1996) 44:335–44. doi:10.1046/j.1365-3083.1996.d01-314.x
50. Rehbein H, Oehlenschlaeger J, editors. Fishery Products: Quality, Safety and Authenticity. Weinheim: Wiley-Blackwell (2009).
51. Kobayashi A, Tanaka H, Hamada Y, Ishizaki S, Nagashima Y, Shiomi K. Comparison of allergenicity and allergens between fish white and dark muscles. Allergy (2006) 61:357–63. doi:10.1111/j.1398-9995.2006.00966.x
52. Griesmeier U, Vázquez-Cortés S, Bublin M, Radauer C, Ma Y, Briza P, et al. Expression levels of parvalbumins determine allergenicity of fish species. Allergy (2010) 65:191–8. doi:10.1111/j.1398-9995.2009.02162.x
53. Kuehn A, Scheuermann T, Hilger C, Hentges F. Important variations in parvalbumin content in common fish species: a factor possibly contributing to variable allergenicity. Int Arch Allergy Immunol (2010) 153:359–66. doi:10.1159/000316346
54. Lee PW, Nordlee JA, Koppelman SJ, Baumert JL, Taylor SL. Measuring parvalbumin levels in fish muscle tissue: relevance of muscle locations and storage conditions. Food Chem (2012) 135:502–7. doi:10.1016/j.foodchem.2012.05.030
55. Sletten G, Van Do T, Lindvik H, Egaas E, Florvaag E. Effects of industrial processing on the immunogenicity of commonly ingested fish species. Int Arch Allergy Immunol (2010) 151:223–36. doi:10.1159/000242360
56. Arif SH. A Ca2+-binding protein with numerous roles and uses: parvalbumin in molecular biology and physiology. Bioessays (2009) 31:410–21. doi:10.1002/bies.200800170
57. Schwaller B. The continuing disappearance of “pure” Ca2+ buffers. Cell Mol Life Sci (2009) 66:275–300. doi:10.1007/s00018-008-8564-6
58. Permyakov SE, Bakunts AG, Denesyuk AI, Knyazeva EL, Uversky VN, Permyakov EA. Apo-parvalbumin as an intrinsically disordered protein. Proteins (2008) 72:822–36. doi:10.1002/prot.21974
59. Bugajska-Schretter A, Grote M, Vangelista L, Valent P, Sperr WR, Rumpold H, et al. Purification, biochemical, and immunological characterisation of a major food allergen: different immunoglobulin E recognition of the apo- and calcium-bound forms of carp parvalbumin. Gut (2000) 46:661–9. doi:10.1136/gut.46.5.661
60. Swoboda I, Bugajska-Schretter A, Linhart B, Verdino P, Keller W, Schulmeister U, et al. A recombinant hypoallergenic parvalbumin mutant for immunotherapy of IgE-mediated fish allergy. J Immunol (2007) 178:6290–6.
61. King TP, Hoffman D, Lowenstein H, Marsh DG, Platts-Mills TA, Thomas W. Allergen nomenclature. WHO/IUIS allergen nomenclature subcommittee. Int Arch Allergy Immunol (1994) 105:224–33. doi:10.1159/000236761
62. Mari A, Scala E, Palazzo P, Ridolfi S, Zennaro D, Carabella G. Bioinformatics applied to allergy: allergen databases, from collecting sequence information to data integration. The allergome platform as a model. Cell Immunol (2006) 244:97–100. doi:10.1016/j.cellimm.2007.02.012
63. Bugajska-Schretter A, Elfman L, Fuchs T, Kapiotis S, Rumpold H, Valenta R, et al. Parvalbumin, a cross-reactive fish allergen, contains IgE-binding epitopes sensitive to periodate treatment and Ca2+ depletion. J Allergy Clin Immunol (1998) 101:67–74. doi:10.1016/S0091-6749(98)70195-2
64. Lim DL, Neo KH, Yi FC, Chua KY, Goh DL, Shek LP, et al. Parvalbumin – the major tropical fish allergen. Pediatr Allergy Immunol (2008) 19:399–407. doi:10.1111/j.1399-3038.2007.00674.x
65. Swoboda I, Bugajska-Schretter A, Verdino P, Keller W, Sperr WR, Valent P, et al. Recombinant carp parvalbumin, the major cross-reactive fish allergen: a tool for diagnosis and therapy of fish allergy. J Immunol (2002) 168:4576–84.
66. Rosmilah M, Shahnaz M, Masita A, Noormalin A, Jamaludin M. Identification of major allergens of two species of local snappers: Lutjanus argentimaculatus (merah/red snapper) and Lutjanus johnii (jenahak/golden snapper). Trop Biomed (2005) 22:171–7.
67. Wang B, Li Z, Zheng L, Liu Y, Lin H. Identification and characterization of a new Ige-binding protein in mackerel (Scomber japonicus) by MALDI-TOF-MS. J Ocean Univ China (2011) 1:93–8. doi:10.1007/s11802-011-1793-6
68. Van Do T, Elsayed S, Florvaag E, Hordvik I, Endresen C. Allergy to fish parvalbumins: studies on the cross-reactivity of allergens from 9 commonly consumed fish. J Allergy Clin Immunol (2005) 116:1314–20. doi:10.1016/j.jaci.2005.07.033
69. Liu R, Krishnan HB, Xue W, Liu C. Characterization of allergens isolated from the freshwater fish blunt snout bream (Megalobrama amblycephala). J Agric Food Chem (2011) 59:458–63. doi:10.1021/jf103942p
70. Nakamura R, Satoh R, Nakajima Y, Kawasaki N, Yamaguchi T, Sawada J, et al. Comparative study of GH-transgenic and non-transgenic amago salmon (Oncorhynchus masou ishikawae) allergenicity and proteomic analysis of amago salmon allergens. Regul Toxicol Pharmacol (2009) 55:300–8. doi:10.1016/j.yrtph.2009.08.002
71. Tomm JM, van Do T, Jende C, Simon JC, Treudler R, von Bergen M, et al. Identification of new potential allergens from Nile perch (Lates niloticus) and cod (Gadus morhua). J Investig Allergol Clin Immunol (2013) 23:159–67.
72. Kuehn A, Hilger C, Lehners-Weber C, Codreanu-Morel F, Morisset M, Metz-Favre C, et al. Identification of enolases and aldolases as important fish allergens in cod, salmon and tuna: component resolved diagnosis using parvalbumin and the new allergens. Clin Exp Allergy (2013) 43:811–22. doi:10.1111/cea.12117
73. Karim AA, Bhat R. Fish gelatin: properties, challenges, and prospects as an alternative to mammalian gelatins. Food Hydrocoll (2009) 23:563–76. doi:10.1016/j.foodhyd.2008.07.002
74. Hamada Y, Nagashima Y, Shiomi K. Identification of collagen as a new fish allergen. Biosci Biotechnol Biochem (2001) 65:285–91. doi:10.1271/bbb.65.285
75. Sakaguchi M, Nakayama T, Inouye S. Food allergy to gelatin in children with systemic immediate-type reactions, including anaphylaxis, to vaccines. J Allergy Clin Immunol (1996) 98:1058–61. doi:10.1016/S0091-6749(96)80191-6
76. André F, Cavagna S, Andre C. Gelatin prepared from tuna skin: a risk factor for fish allergy or sensitization? Int Arch Allergy Immunol (2003) 130:17–24. doi:10.1159/000068370
77. Hansen TK, Poulsen LK, Stahl Skov P, Hefle SL, Hlywka JJ, Taylor SL, et al. A randomized, double-blinded, placebo-controlled oral challenge study to evaluate the allergenicity of commercial, food-grade fish gelatin. Food Chem Toxicol (2004) 42:2037–44. doi:10.1016/j.fct.2004.08.008
78. Kuehn A, Hilger C, Hentges F. Anaphylaxis provoked by ingestion of marshmallows containing fish gelatin. J Allergy Clin Immunol (2009) 123:708–9. doi:10.1016/j.jaci.2008.12.012
79. Weber P, Steinhart H, Paschke A. Competitive indirect ELISA for the determination of parvalbumins from various fish species in food grade fish gelatins and isinglass with PARV-19 anti-parvalbumin antibodies. J Agric Food Chem (2009) 57:11328–34. doi:10.1021/jf902470e
80. Dory D, Chopin C, Aimone-Gastin I, Gueant JL, Guerin L, Sainte-Laudy J, et al. Recognition of an extensive range of IgE-reactive proteins in cod extract. Allergy (1998) 53:42–50. doi:10.1111/j.1398-9995.1998.tb03772.x
81. Misnan R, Murad S, Jones M, Taylor G, Rahman D, Arip M, et al. Identification of the major allergens of Indian scad (Decapterus russelli). Asian Pac J Allergy Immunol (2008) 26:191–8.
82. Sten E, Hansen TK, Stahl Skov P, Andersen SB, Torp A, Bindslev-Jensen U, et al. Cross-reactivity to eel, eelpout and ocean pout in codfish-allergic patients. Allergy (2004) 59:1173–80. doi:10.1111/j.1398-9995.2004.00497.x
83. Das Dores S, Chopin C, Romano A, Galland-Irmouli AV, Quaratino D, Pascual C, et al. IgE-binding and cross-reactivity of a new 41 kDa allergen of codfish. Allergy (2002) 57:84–7. doi:10.1034/j.1398-9995.57.s72.6.x
84. Rosmilah M, Shahnaz M, Meinir J, Masita A, Noormalin A, Jamaluddin M. Identification of parvalbumin and two new thermolabile major allergens of Thunnus tonggol using a proteomics approach. Int Arch Allergy Immunol (2013) 162:299–309. doi:10.1159/000354544
85. Kondo Y, Kakami M, Koyama H, Yasuda T, Nakajima Y, Kawamura M, et al. IgE cross-reactivity between fish roe (salmon, herring and pollock) and chicken egg in patients anaphylactic to salmon roe. Allergol Int (2005) 54:317–23. doi:10.2332/allergolint.54.317
86. Perez-Gordo M, Sanchez-Garcia S, Cases B, Pastor C, Vivanco F, Cuesta-Herranz J. Identification of vitellogenin as an allergen in Beluga caviar allergy. Allergy (2008) 63:479–80. doi:10.1111/j.1398-9995.2007.01614.x
87. Liu R, Holck AL, Yang E, Liu C, Xue W. Tropomyosin from tilapia (Oreochromis mossambicus) as an allergen. Clin Exp Allergy (2013) 43:365–77. doi:10.1111/cea.12056
88. Weber P, Paschke A. Fish allergens. In: Jedrychowski L, Wichers HJ, editors. Chemical and Biological Properties of Food Allergens. Boca Raton: CRC Press (2010).
89. Moncrief ND, Kretsinger RH, Goodman M. Evolution of EF-hand calcium-modulated proteins. I. Relationships based on amino acid sequences. J Mol Evol (1990) 30:522–62. doi:10.1007/BF02101108
90. Nakayama S, Moncrief ND, Kretsinger RH. Evolution of EF-hand calcium-modulated proteins. II. Domains of several subfamilies have diverse evolutionary histories. J Mol Evol (1992) 34:416–48. doi:10.1007/BF00162998
91. Brewer JM, Arnold J, Beach GG, Ragland WL, Wunderlich JK. Comparison of the amino acid sequences of tissue-specific parvalbumins from chicken muscle and thymus and possible evolutionary significance. Biochem Biophys Res Commun (1991) 181:226–31. doi:10.1016/S0006-291X(05)81406-8
92. Föhr UG, Weber BR, Müntener M, Staudenmann W, Hughes GJ, Frutiger S, et al. Human alpha and beta parvalbumins. Structure and tissue-specific expression. Eur J Biochem (1993) 215:719–27. doi:10.1111/j.1432-1033.1993.tb18084.x
93. Goodman M, Pechère JF, Haiech J, Demaille JG. Evolutionary diversification of structure and function in the family of intracellular calcium-binding proteins. J Mol Evol (1979) 13:331–52. doi:10.1007/BF01731373
94. Hilger C, Grigioni F, Thill L, Mertens L, Hentges F. Severe IgE-mediated anaphylaxis following consumption of fried frog legs: definition of alpha-parvalbumin as the allergen in cause. Allergy (2002) 57:1053–8. doi:10.1034/j.1398-9995.2002.23677.x
95. Hilger C, Thill L, Grigioni F, Lehners C, Falagiani P, Ferrara A, et al. IgE antibodies of fish allergic patients cross-react with frog parvalbumin. Allergy (2004) 59:653–60. doi:10.1111/j.1398-9995.2004.00436.x
96. González-Mancebo E, Pastor C, González-de-Olano D, Gandolfo-Cano M, Melendez A, Cuesta J, et al. Identification of allergens in chicken meat allergy. J Investig Allergol Clin Immunol (2011) 21:326–7.
97. Kuehn A, Lehners C, Hilger C, Hentges F. Food allergy to chicken meat with IgE reactivity to muscle alpha-parvalbumin. Allergy (2009) 64:1557–8. doi:10.1111/j.1398-9995.2009.02094.x
98. González-de-Olano D, Bartolomé B, Maroto AS, Vivanco F, Pastor-Vargas C. Asthma after chicken consumption due to cross-reactivity between fish and chicken parvalbumin. J Investig Allergol Clin Immunol (2012) 22:227–8.
99. Lapteva YS, Uversky VN, Permyakov SE. Sequence microheterogeneity of parvalbumin, the major fish allergen. Biochim Biophys Acta (2013) 1834:1607–14. doi:10.1016/j.bbapap.2013.04.025
100. Saptarshi SR, Sharp MF, Kamath SD, Lopata AL. Antibody reactivity to the major fish allergen parvalbumin is determined by isoforms and impact of thermal processing. Food Chem (2014) 148:321–8. doi:10.1016/j.foodchem.2013.10.035
101. Jenkins JA, Breiteneder H, Mills EN. Evolutionary distance from human homologs reflects allergenicity of animal food proteins. J Allergy Clin Immunol (2007) 120:1399–405. doi:10.1016/j.jaci.2007.08.019
102. Elsayed S, Apold J. Immunochemical analysis of cod fish allergen M: locations of the immunoglobulin binding sites as demonstrated by the native and synthetic peptides. Allergy (1983) 38:449–59. doi:10.1111/j.1398-9995.1983.tb02353.x
103. Perez-Gordo M, Pastor-Vargas C, Lin J, Bardina L, Cases B, Ibáñez MD, et al. Epitope mapping of the major allergen from Atlantic cod in Spanish population reveals different IgE-binding patterns. Mol Nutr Food Res (2013) 57:1283–90. doi:10.1002/mnfr.201200332
104. Yoshida S, Ichimura A, Shiomi K. Elucidation of a major IgE epitope of Pacific mackerel parvalbumin. Food Chem (2008) 111:857–61. doi:10.1016/j.foodchem.2008.04.062
105. Brownridge P, de Mello LV, Peters M, McLean L, Claydon A, Cossins AR, et al. Regional variation in parvalbumin isoform expression correlates with muscle performance in common carp (Cyprinus carpio). J Exp Biol (2009) 212:184–93. doi:10.1242/jeb.021857
106. Huriaux F, Vandewalle P, Focant B. Immunological study of muscle parvalbumin isotypes in three African catfish during development. Comp Biochem Physiol B Biochem Mol Biol (2002) 132:579–84. doi:10.1016/S1096-4959(02)00071-4
Keywords: allergenicity, fish allergy, fish gelatin, food allergy, isoallergens, isoforms, monosensitivity, parvalbumin
Citation: Kuehn A, Swoboda I, Arumugam K, Hilger C and Hentges F (2014) Fish allergens at a glance: variable allergenicity of parvalbumins, the major fish allergens. Front. Immunol. 5:179. doi: 10.3389/fimmu.2014.00179
Received: 19 December 2013; Paper pending published: 13 February 2014;
Accepted: 05 April 2014; Published online: 22 April 2014.
Edited by:
Kendall A. Smith, Weill Medical College of Cornell University, USAReviewed by:
Richard E. Goodman, University of Nebraska-Lincoln, USACopyright: © 2014 Kuehn, Swoboda, Arumugam, Hilger and Hentges. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Annette Kuehn, Laboratory of Immunogenetics and Allergology, Public Research Centre for Health (CRP-Santé), 84 Val Fleuri, Luxembourg L-1526, Luxembourg e-mail:YW5uZXR0ZS5rdWVobkBjcnAtc2FudGUubHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.