
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
HYPOTHESIS AND THEORY article
Front. Immunol. , 03 May 2012
Sec. Molecular Innate Immunity
Volume 3 - 2012 | https://doi.org/10.3389/fimmu.2012.00096
This article is part of the Research Topic Changing Views of the Evolution of Immunity View all 11 articles
 Larry J. Dishaw1*
Larry J. Dishaw1* Jaime A. Flores-Torres2
Jaime A. Flores-Torres2 M. Gail Mueller1,3
M. Gail Mueller1,3 Charlotte R. Karrer3
Charlotte R. Karrer3 Diana P. Skapura3
Diana P. Skapura3 Daniela Melillo4
Daniela Melillo4 Ivana Zucchetti4
Ivana Zucchetti4 Rosaria De Santis4
Rosaria De Santis4 Maria Rosaria Pinto4
Maria Rosaria Pinto4 Gary W. Litman1,3
Gary W. Litman1,3
Complex symbiotic interactions at the surface of host epithelia govern most encounters between host and microbe. The epithelium of the gut is a physiologically ancient structure that is comprised of a single layer of cells and is thought to possess fully developed immunological capabilities. Ciona intestinalis (sea squirt), which is a descendant of the last common ancestor of all vertebrates, is a potentially valuable model for studying barrier defenses and gut microbial immune interactions. A variety of innate immunological phenomena have been well characterized in Ciona, of which many are active in the gut tissues. Interactions with gut microbiota likely involve surface epithelium, secreted immune molecules including variable region-containing chitin-binding proteins, and hemocytes from a densely populated laminar tissue space. The microbial composition of representative gut luminal contents has been characterized by molecular screening and a potentially relevant, reproducible, dysbiosis can be induced via starvation. The dialog between host and microbe in the gut can be investigated in Ciona against the background of a competent innate immune system and in the absence of the integral elements and processes that are characteristic of vertebrate adaptive immunity.
Because of their phylogenetic position relative to vertebrates, deuterostome invertebrates (Figure 1) are compelling models for studies of immunity. Of the representative deuterostome invertebrates, echinoderms, which include sea urchins and starfish, diverged prior to chordates and have proven to be highly informative models in which to examine innate immunity (Rast and Messier-Solek, 2008; Messier-Solek et al., 2010). Protochordates are comprised of invertebrate chordates such as sea squirts (Urochordata or Tunicata) and amphioxus (Cephalochordata). These species, which share certain developmental features with vertebrates and possess competent innate immunity, diverged prior to the origins of the vertebrate adaptive immune system.
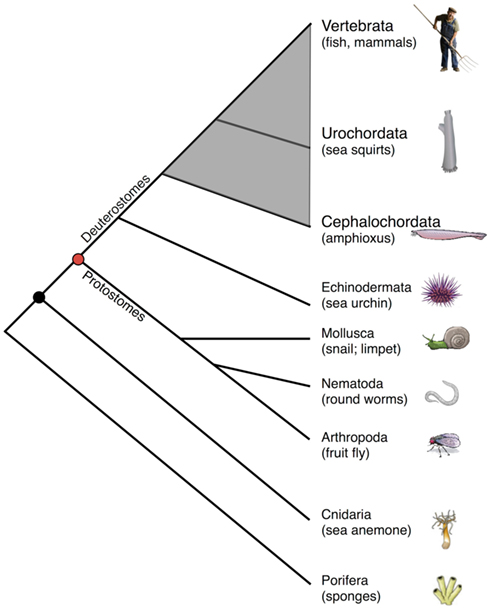
Figure 1. Simplified cladogram of animal phylogeny emphasizing chordates (shaded gray) relative to other animal forms. Note that only two protochordate taxa are shown, and all are exclusively marine animals.
Ciona intestinalis (sea squirt), which has been the focus of our recent efforts, is relatively easy to maintain and propagate (Figure 2) at room temperature and continues to serve as a highly informative model for studies of animal development (Katz, 1983; Meinertzhagen and Okamura, 2001; Canestro et al., 2003; Shi et al., 2005; Baghdiguian et al., 2007; Davidson, 2007; Sasakura et al., 2009) and immune defense (Fujita et al., 2004; Melillo et al., 2006; Parrinello, 2009; Sasaki et al., 2009; Zucchetti et al., 2009). As a model of animal development (Satoh and Levine, 2005; Lemaire et al., 2008; Christiaen et al., 2009) Ciona has proven invaluable in: (1) unraveling details of Hox-gene influences on development (Ikuta et al., 2010), (2) mapping pathways in cardiac development (Davidson, 2007), (3) defining the roles of cis-regulatory networks (Kubo et al., 2010), (4) modeling the effects of maternally derived epigenetic silencing (Sasakura et al., 2010), and (5) defining the evolution of the cell death machinery (Terajima et al., 2003; Baghdiguian et al., 2007). Many of the involved processes utilize signaling pathways that are relevant to studies concerning immunity and immune homeostasis.
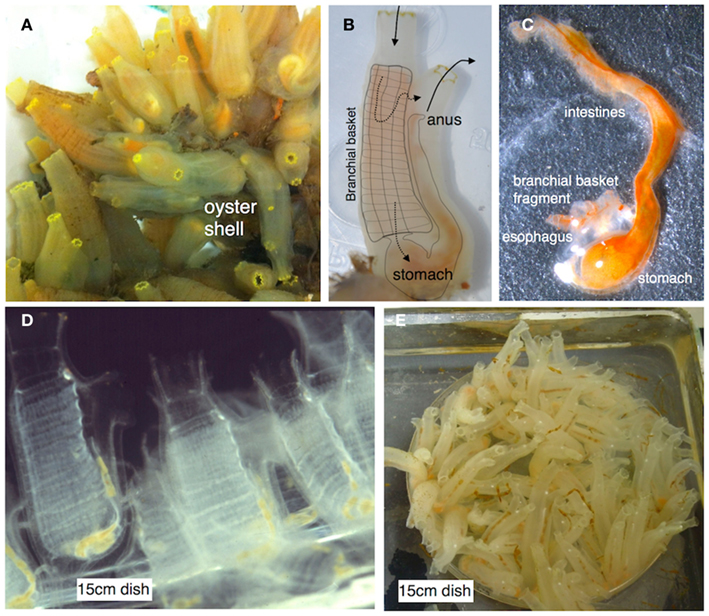
Figure 2. (A) Ciona intestinalis is a solitary urochordate that typically grows in close proximity on suitable substrates. (B) Ciona is relatively translucent and the gut can be visualized through the tunic (overlay depicts the branchial basket and gut; solid arrows depict water flow; dotted arrows indicate two routes of flow). (C) The entire gut can be readily dissected. (D) Juveniles are attracted and attach to standard tissue culture plastic plates. (E) Ciona can be reared to adulthood in a laboratory environment (Cirino et al., 2002; Joly et al., 2007). Ciona adults are hermaphrodites and release sperm and egg into the water column. Typically, fertilization leads to rapid development and hatching of swimming tadpole larva. After settlement, the animal undergoes metamorphosis into a permanent, sessile, filter-feeding lifestyle; colonization of the gut by microbes is likely immediate. Images (D,E) are courtesy Dr. Paola Cirino.
Deuterostome invertebrates possess homologs of a large number of vertebrate innate immune receptors, effectors, and their corresponding regulatory elements (Rast and Messier-Solek, 2008; Messier-Solek et al., 2010). The most surprising finding regarding immunity in sea urchin and amphioxus is the expansion of multigene families encoding homologs of different innate immune pattern recognition receptors (PRRs), including: Toll-like receptors (TLRs), scavenger receptors, and leucine-rich receptor (LRR)-containing intracellular sensors such as nucleotide-binding oligomerization domain-like receptors (NLRs; Rast et al., 2006; Holland et al., 2008). Novel evolutionary constraints most likely led to the lineage-specific expansions and functional divergence of the various gene families encoding these molecules in amphioxus and sea urchin. Detailed sequence analyses have revealed examples of parallel or convergent evolution relating to the expansions (Leulier and Lemaitre, 2008). Very little sequence similarity among presumed homologs (i.e., lack of one-to-one orthology) is evident between these receptors in invertebrate deuterostomes and their vertebrate counterparts. Innate immune genes in Ciona have not undergone the expansions seen in amphioxus and sea urchin (Dehal et al., 2002; Hughes and Friedman, 2005).
Comparative biological studies have revealed that most multicellular life is protected by conserved mechanisms of innate immunity which include: barrier defenses [i.e., host (mucosal) epithelium] and the associated non-specific secretory components (e.g., antibacterial peptides), PRRs, which are present on the surface and within the cytoplasm of phagocytes and other host cells, various phagocyte effector mechanisms (e.g., reactive oxygen species, induced cell death), and different enzymatically catalyzed cascades involved in clotting, melanization, and complement activation (Figure 3; Lavelle et al., 2010; Maldonado-Contreras and McCormick, 2011). A variety of innate immune genes and mechanisms have been identified in Ciona (Parrinello, 2009), which based on histological and molecular characterizations have evolved complex barrier defense strategies. Challenge with microbe-associated molecular patterns (MAMPs) such as the Gram-negative lipopolysaccharide (LPS) induces inflammatory-like reactions that are characterized, in part, by tumor-necrosis factor (TNF)-like gene expression and cell recruitment to various body compartments (Di Bella and De Leo, 2000; Melillo et al., 2006; Parrinello et al., 2007, 2008, 2010; Cammarata et al., 2008; Bonura et al., 2009). These responses can recruit a variety of cell types (Parrinello et al., 1996; Arizza and Parrinello, 2009; Arizza et al., 2011) and induce a number of immunological phenomena (Smith and Peddie, 1992; Melillo et al., 2006; Parrinello et al., 2007), including the expression of characteristic innate immune receptors (Shida et al., 2003; Parrinello, 2009; Sasaki et al., 2009; Dishaw et al., 2011). Although a variety of different MAMPs induce Ciona TLRs, LPS does not activate TLR expression directly, as with TLR4 in vertebrates, suggesting the presence of alternative LPS sensors (Sasaki et al., 2009). A repertoire of innate effectors [e.g., TLRs, TNF, complement components, and the protochordate-specific variable region-containing chitin-binding proteins (VCBPs)] are expressed in the gut of Ciona (Marino et al., 2002; Sasaki et al., 2009; Skjoedt et al., 2010; Dishaw et al., 2011) and could play essential roles in the stable maintenance of host–microbial interactions (see below).
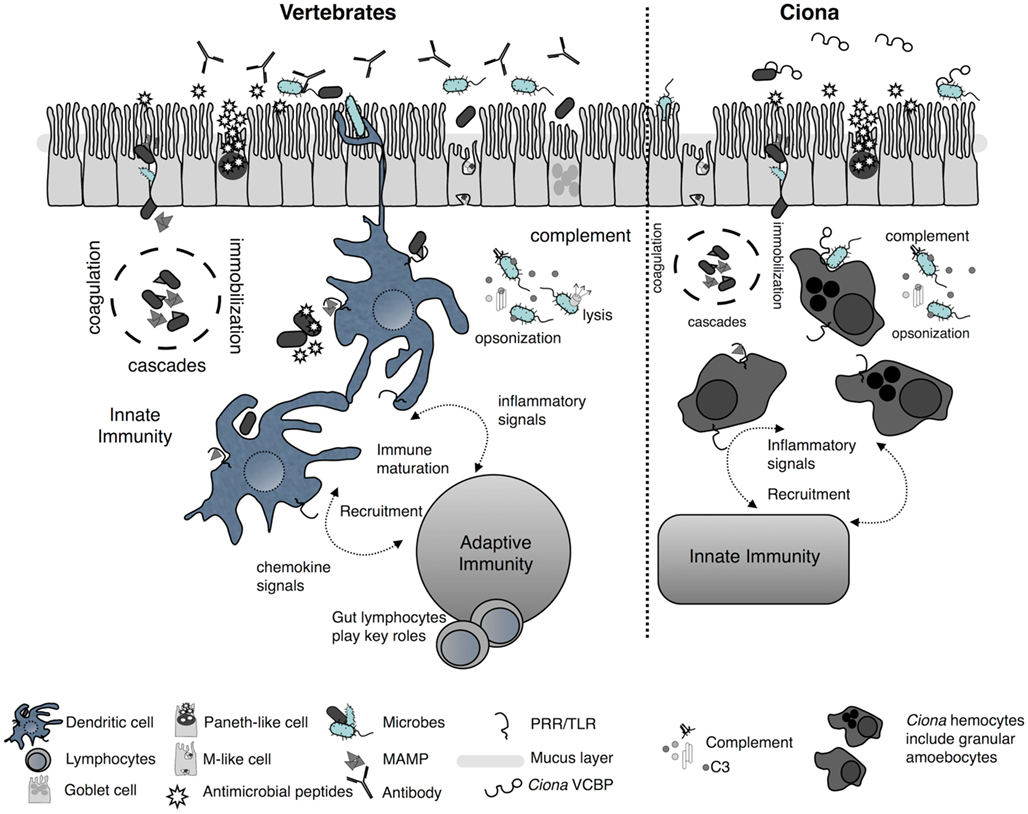
Figure 3. Simplified schematic of mucosal immunity emphasizing barrier defense strategies of vertebrates and Ciona. Barrier defenses (across the epithelium) are governed by innate immune phenomena characterized by the secretion of mucus, antimicrobials, and soluble immune molecules. On the basolateral surface and deeper, host innate immunity consists of various proteolytic-coagulation cascades for wound healing and microbe trapping, as well as complement defense pathways. Distinct phagocytes populate this area as well other cell types [e.g., dendritic cells (DC) in vertebrates]. Gut DCs sample luminal antigens and present them to the adaptive immune system which includes gut-specific lymphocytes of both T and B cell lineages. This results in the maturation of immunity and the recruitment of additional cell types. A parallel, more simplified, system in Ciona has evolved to also include barrier defenses across distinct epithelial lineages in the gut and the secretion of immune mediators, including antimicrobial peptides, into the lumen. In the basolateral side, distinct populations of hemocytes, including granular amoebocytes, populate the laminar connective tissues. Immunological competence in this area is mediated by coagulation/immobilization cascades and microbe trapping, complement defenses, distinct antigenic sampling via PRRs, secretion of pro-inflammatory signals, and recruitment of specialized hemocytes. However, as opposed to the vertebrate gut, the Ciona gut generates immune amplification within an innate immune context and does not couple to a distinct, more specialized, system such as adaptive immunity.
Documentation of these types of functional effects is critical to the design of experimental approaches for characterizing the interactions of immune receptors with gut microbiota. However, enumerating gene homologs and defining their expression patterns is one thing, understanding their dynamic interactions in a cellular and molecular context is a daunting challenge in invertebrate systems, particularly those that inhabit marine and aquatic environments. Despite a number of potential shortcomings, Ciona has the potential to reveal conserved mechanisms sustaining the evolution of host–microbial interactions (see below). Experimental manipulation of host–microbial interactions at the gut epithelial interface is critical to such studies and in order to approach these questions, it is essential to first understand the Ciona gut as both a physical barrier and as an immunological organ.
Ciona is a highly successful, cosmopolitan solitary tunicate that has adapted to living in diverse marine environments (Caputi et al., 2007). Other tunicates include both colonial and solitary forms and spend their adult lives as sessile, filter-feeding, organisms. In addition, a few pelagic forms have been identified (Denoeud et al., 2010; Nishida et al., 2010). Because the tunicate feeding strategy involves siphoning seawater, the gut in Ciona is in continuous contact with both dietary and seawater microbiota where in addition to its essential physiological role, it acts as both a physical barrier and site of continuous immunological interaction. An immunocompetent gut, which includes mucosal immunity mediated by surface epithelium, is present even in the simplest metazoans (Bosch et al., 2009). In mammals, proper development of gut mucosal immune tissues is dependent on the proper timing and colonization of the gut by microbial communities (see below); however, details governing these events remain to be defined (Cebra et al., 1998; Cebra, 1999; Hooper and Gordon, 2001; Mazmanian et al., 2005; Edelman and Kasper, 2008; Kanther and Rawls, 2010). Intensive studies in comparative models, such as Ciona, have the potential for shedding light on the basic biology of gut immune homeostasis and in turn, may reveal basic mechanisms of dysfunctional gut immunity [e.g., inflammatory bowel diseases (IBD) in mammals]. As an initial step in adapting a urochordate as a new model system for understanding gut immune homeostasis and mucosal immunity, we have combined cellular, molecular, and microbiological approaches to characterize the Ciona gut and its microbiota. At this preliminary stage of investigation, basic aspects of complex microbial community dynamics can be identified that mirror many of the core features and symbiotic intricacies of host–microbe interactions that are recognized in the mammalian gut ecosystem (Savage, 1977; Hooper and Gordon, 2001; Willing et al., 2011). The interactions between host and gut microflora are not simple, but instead involve complex mutualisms (Bischoff, 2011) that ultimately help govern immune development, and the establishment and maintenance of immune homeostasis (Artis, 2008; Chung and Kasper, 2010; Hooper and Macpherson, 2010).
Ciona filters microbe-rich seawater through a modified pharynx where ciliated cells push food particles into a gut, which is divided into esophagus, stomach, mid-gut, and hind-gut; the latter two are referred to collectively as intestines. The esophagus connects the branchial basket to the stomach in which both cilia and mucous glands are highly developed for the efficient transfer of foods. The stomach epithelium forms many cilia-rich ridges and grooves and is composed of at least two major epithelial cell types, as well as an undifferentiated cell population (Burighel and Cloney, 1977). The stomach epithelium, which is presumed to be the site of most digestive enzyme secretion, contains secretory cells, and is coated by a thin layer of mucus. The mid-gut is distinguished by an interior typhlosole that runs the entire length and is rich in testicular acini. Three types of largely granular epithelial cells, absorptive, mucous, and large round (or elliptical), define the mid-gut. Glycogen stores are concentrated in the mid-gut and to a lesser degree in the stomach. Energy is stored as both fat and glycogen (Yonge, 1925). The sexual ducts exit the atrial siphon adjacent to the hind-gut. Although absorption is most prominent in the mid-gut, diffusion of dissolved substances occurs throughout the alimentary track. The Ciona gut demonstrates complex epithelial cell renewal traits (Ermak, 1981), which are of particular importance to several aspects of mucosal physiology, including immune function. The highly developed and compartmentalized stomach and distinct intestinal region in Ciona morphologically resemble that of more recently diverged chordates (Millar, 1953; Burighel and Cloney, 1977).
Details surrounding the relationships between filter-feeding invertebrates and the microbial communities colonizing their guts are lacking but may be broadly relevant to gut immunity in vertebrates for determining: (1) the role of diet in gut microbial ecology, (2) the nature of host selection of gut microbiota, and (3) the role of microbiota in gut immune homeostasis. These features of gut physiology are essential to our defining of homeostasis, even if a model organism such as Ciona does not share all the innate receptor orthologs found in mammals. Because the composition of gut microbiota is relevant to host physiology for a variety of reasons not limited to their metabolic output, we suspect that the gut microbial communities in Ciona will not simply reflect diet and/or environmental availability, but will reveal species-specific communities that determine or influence various aspects of overall immune competence.
Gut bacteria from four Ciona populations (Woods Hole, MA, USA; San Diego and Monterey Bay, CA, USA; and Naples, Italy) have been partially sampled using PCR-amplified 16S ribosomal genes recovered in small clone libraries derived from whole gut homogenates (and/or recovered fecal matter). The 16S products were then characterized by sequencing of individual clones as well as by screening of restriction fragment length polymorphisms (Figure 4). A fraction of the recovered microflora (∼25 bacterial species) was cultured successfully. The Ciona gut revealed distinct communities of bacteria that were affected by both diet and environment, including species of metabolic significance (e.g., Chitinophaga). Starvation induced reproducible dysbiosis (i.e., disruption or displacement of microbial communities) and revealed bacterial families and genera that were conserved across populations and between two Ciona species (e.g., Gammaproteobacteria such as Vibrio sp. and Shewanella sp., as well as various Oceanospirillales genera; Dishaw, unpublished observations).
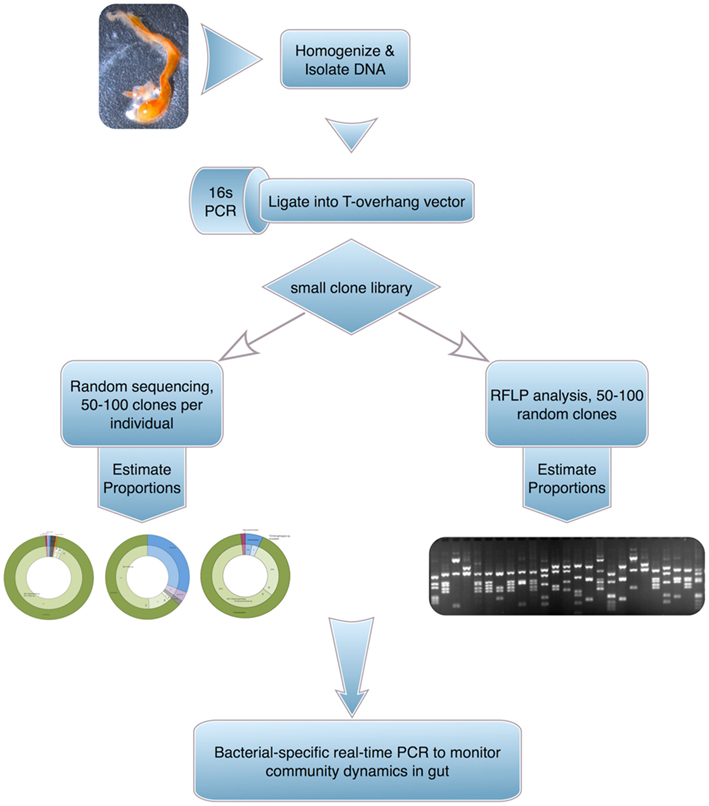
Figure 4. A general approach to assess gut microbial communities in Ciona. The technique, albeit labor intensive, provides high-grade sequence-based identifications.
The successful recovery, identification, and growth of native gut bacteria from wild Ciona adults provides the basic background to inoculate the developing gut of juveniles grown in controlled laboratory environments (e.g., under semi- or sterile conditions or colonized by complex mixtures of microbes). Changes in community structure subsequent to experimental manipulation can be monitored using real time quantitative PCR. These experiments, which currently are in progress, have the potential to define the onset and normal timing of microbial colonization in the development of the Ciona gut in general, and more specifically, characterize how this interaction affects the maturation of the hemocyte-rich laminar spaces (i.e., immune tissues). Determining how interactions with microbiota affect maturation of the gut immune tissues will utilize dysbiosis techniques including antibiotic treatments and/or development of juveniles under germ-free or semi-germ-free conditions. In this regard, two features of the Ciona model are particularly attractive: (1) it is relatively easy to produce and maintain hundreds of Ciona juveniles and (2) transparency of tissues makes it feasible to visually characterize gut development and gauge luminal content (Figure 2) and make possible the tracking of host–microbe interactions (e.g., the use of labeled bacteria) from early in development through adulthood.
The circulatory system of Ciona is open and continuous with histologically defined blood lacunae. Gut-associated lacunae, which we have termed the Ciona gut lamina propria (Dishaw et al., 2011), share blood cells between the various tissue spaces and are richly infiltrated by a variety of hemocyte types. A complex developmental maturation of this subepithelial lamina that likely is influenced by luminal antigen exposure is indicated. Notably, the hemocytes are not restricted to the gut as morphologically indistinguishable cell types can be detected in other parts of the body. Adult Ciona injected with MAMPs (e.g., in the tunic) can be induced to generate localized inflammatory responses, which include an active recruitment of these and other immunocompetent cells (Di Bella and De Leo, 2000; Pinto et al., 2003; Bonura et al., 2009; Parrinello et al., 2010). In this regard, variable (V) region-containing chitin-binding proteins (VCBPs), which are expressed by distinct gut epithelial cells in amphioxus (Cannon et al., 2002), also have been identified in Ciona (Figure 5). VCBPs have been shown to be secreted by discrete cells of the stomach and intestinal epithelium into gut lumen where they interact with bacteria via their V-type immunoglobulin domains (Dishaw et al., 2011). The functional relevance of the VCBP chitin-binding domain remains unclear. Ciona granular amoebocytes, which also express VCBPs, are present in both blood and the laminar spaces of the gut. In vitro experiments have demonstrated that granular amoebocytes, recovered from blood, recognize (phagocytose) bacteria coated with VCBPs (Dishaw et al., 2011) in vitro. We have hypothesized that the morphologically indistinguishable cells found in the lamina propria function in an equivalent manner and play a major role in the dynamics of gut immunity in both Ciona and amphioxus. This hypothesis is supported further by the finding that native VCBPs bind luminal bacteria (Dishaw et al., 2011) and it is entirely likely that VCBPs enhance phagocytic recognition of gut bacteria coated with VCBPs that traverse epithelial barriers (e.g., in instances of epithelial damage). An emerging functional role for VCBPs places them in the broader context of our hypothesis by suggesting that some innate receptors, secreted into the gut lumen, may be serving still undefined roles as symbiosis factors. Penetration of the mucosal barrier would trigger an immunological event by the tagged (i.e., opsonized) microbe to protect the integrity of self (i.e., phagocytosis, inflammation, and cell recruitment). Broad analogies can be drawn between this process and immune recognition in the vertebrate mucosal environment, albeit involving different effectors. MAMP challenge across the gut mucosal barriers also may recruit cells from distant tissue spaces; however, this may be unnecessary since the subepithelial lamina is densely populated with many hemocyte types along the gut length (Figure 5). Several preliminary observations (Dishaw, De Stantis, and Pinto, unpublished) suggest that lamina-associated gut hemocytes from Ciona may be exposed to luminal contents (e.g., through injury of epithelium or exposure to factors that affect epithelial tight junctions). Electron microscopic analysis will be critical to demonstrate if hemocytes actively cross the epithelium and interact with gut microbiota and if luminal antigens enter the laminar spaces.
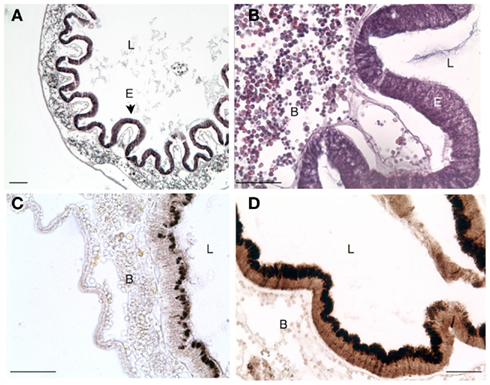
Figure 5. Transverse section of stomach in Ciona depicting. (A) Characteristic folding of epithelium that is not seen in the intestines. (B) Higher magnification view [of (A)] emphasizing the abundance of laminar tissue blood hemocytes. (C) Antibody staining for VCBP-C in untreated Ciona. (D) Extensive upregulation of VCBP expression in animals challenged with E. coli lysates. Scale bar: 100 μm in (A) and 50 μm in (B–D); Hematoxylin and eosin staining in (A,B) and 3,3′ diaminobenzidine (DAB) substrate detection in (C,D); L, lumen; B, blood; and E, epithelium.
The extent to which innate receptors are expressed by gut epithelium and the functional implications of their interaction with symbiotic or pathogenic gut microbes, as well as virulence factors, are of central interest. Innate immune receptors in vertebrates, primarily TLRs, are expressed selectively in a polarized fashion on intestinal epithelial cells (IECs) of the small and large intestines (Abreu, 2010). TLRs provide indirect signals to the adaptive immune system by first providing innate immune signals to the underlying (basal lamina) tissues. In Ciona, the expression, function, and regulation of a variety of immune molecules (Azumi et al., 2003; Shida et al., 2003), including antimicrobial peptides (Fedders and Leippe, 2008), TLRs, Gram-negative binding proteins (GNBPs), lipopolysaccharide binding protein (LBP), TNF, MBL, complement protein C3, as well as VCBPs, are of potential interest. Ciona responds to bacterial ligands in a manner consistent with the patterns of expression of TLRs and other PRRs in gut tissues, resulting in the induction of pro-inflammatory molecules, e.g., TNF (Sasaki et al., 2009). Whereas Ciona only has two TLRs, both are expressed in distinct locations along the gut and along with other innate immune molecules likely play a significant role in discriminating among gut commensals and sensing pathogens (Sasaki et al., 2009; Abreu, 2010). The presence of only two TLRs, which interact with more than one ligand, could be seen as a disadvantage of the Ciona system over mammals, in which multiple TLRs discriminate among distinct ligands. Multi-functional PRRs, such as the TLRs, may be coupled to downstream pathways which may serve to better discriminate ligands; such pathways could differ significantly in Ciona and investigations in this system will help reveal how various organisms discriminate among TLR ligands. Under normal conditions, the gut in Ciona likely maintains a state of balance (i.e., homeostasis) between tolerance and protection through host epithelial–microbe interactions, as has been seen in vertebrates. However, most such interactions are designed to sustain ancient symbiotic relationships and are not necessarily immune-restricted (Hooper and Macpherson, 2010). Innate receptors play significant roles in these basic processes that govern homeostasis and studies that define conserved mechanisms governing host–microbial interactions in the gut are of fundamental biomedical interest.
The animal gut is host to a massively populated dynamic ecosystem of microbes (Savage, 1977) with enormously complex antigenic diversity. Such gut microbial communities in mammals are linked intimately to the maturation and development of mucosal immunity and represent an important determinant of health and disease (Mazmanian et al., 2005; Fujimura et al., 2010; Hooper and Macpherson, 2010). A particularly complex physiological challenge is posed to the mucosal immune system of the host, which must differentiate distinct populations of commensal (i.e., possibly useful) microorganisms from pathogenic communities. Specifically, some symbionts are recognized and tolerated, and subsequently form a cooperative system in the gut, whereas pathogens, which in many cases are invasive, are not well tolerated and are cleared. Traditional views of gut immunity are complicated further by numerous commensals that although beneficial, can induce a pathogenic state in the host (i.e., pathobionts, Round and Mazmanian, 2009). Breakdown of commensal-immune suppression and tolerance mechanisms can lead to disruptions of homeostasis and in turn to inflammation, resulting in a range of distinct clinical phenotypes that define acute and/or chronic IBD (Chung and Kasper, 2010; Nell et al., 2010). Details of how and why such changes in the gut ecosystem can lead to disease manifestations remain largely unknown (Bischoff, 2011). Although chronic inflammation in mammals involves a variety of gut-specific lymphocytes (i.e., adaptive immunity), gut microbial recognition, and immune homeostasis largely represent an innate immune phenomenon, for which the epithelium is primarily responsible (Artis, 2008). Details of the microbial and innate immune dialog within the gut (e.g., across the epithelium), defined exclusively in terms of innate immunity (i.e., in the absence of the adaptive immune system), remain largely unknown and underscore the unique potential of Ciona as a model system for investigating gut microbial–immune interactions.
Gut microbiota also appear to play a major role beyond the gut immune environment extending into a range of host physiological responses (Bischoff, 2011; Maslowski and Mackay, 2011), which include but are not limited to metabolomics and behavior (Sekirov et al., 2010). In addition to their roles in host nutritional physiology, disease protection, and pathogenesis, symbiotic microbial interactions in the gut may be linked to the phylogenetic origins of vertebrate adaptive immunity, which evolved through the recruitment of pre-existing genetic elements (Flajnik and Kasahara, 2010; Litman et al., 2010) and the spontaneous acquisition of specialized components of the recombination machinery (Dreyfus, 1992; Agrawal et al., 1998). The selective pressures that drive these events appear to have led to more than one form of molecular innovation (Cooper and Alder, 2006) and remain elusive (Matsunaga and Rahman, 1998). It has been proposed that selective pressures were maintained by an increasing need to reduce collateral damage caused by persistent or chronic infections (i.e., the toxic index hypothesis; Usharauli, 2010), as well as by innovations of strategies to manage increasingly complex symbiotic communities of the gut (McFall-Ngai, 2007; Lee and Mazmanian, 2010), which in turn also drove innovation in highly specialized cell types (Rescigno and Di, 2009; Chow and Mazmanian, 2010). The two theories may not be mutually exclusive if both phenomena intersect in the gut, which evolved as a major factor in adaptive immune maturation (Cebra, 1999; Ivanov et al., 2009; Atarashi et al., 2011). A variety of gut-specific adaptive immune mechanisms help maintain microbial–immune homeostasis (Hooper and Macpherson, 2010; Feng and Elson, 2011), yet the effects on host physiology and immunity in distant parts of the body by the resident gut microbiota (Clarke et al., 2010; Lathrop et al., 2011; Maslowski and Mackay, 2011) support the existence of an ancient mutualism.
The animal gut has evolved to tolerate the presence of dense communities of commensal residents; however, the majority of this microbiota remain invisible to the majority of the gut immune system on the basal laminar surface, in part due to physical barriers such as mucus (Figure 3). The epithelium has evolved to become a fully functional immune system that expresses various innate receptors and secretes immune-related molecules into the gut lumen (Muller et al., 2005; Duerkop et al., 2009; Marchiando et al., 2010). However, details regarding epithelial recognition and tolerance of luminal bacteria remain largely unknown (Rautava and Walker, 2007; Marchiando et al., 2010), e.g., tolerance to endotoxin is likely governed at the earliest exposure in development (Lotz et al., 2006). It has been suggested that immune systems, as defined currently, evolved first to manage complex symbiotic relationships, while the preservation of “self” became an inevitable adaptation (Bosch and McFall-Ngai, 2011). By this account, mechanisms (later acquired by innate immunity) that govern host–microbial interactions are of ancient phylogenetic origins.
Investigations in Ciona, which are focused on the interaction of microbes at the epithelial surface, may help reveal: (1) if certain bacterial species effect barrier function (Lyczak, 2003; Ohland and Macnaughton, 2010); (2) if the degree of microbiome complexity influences epithelial response to infections (Mans et al., 2009); (3) how polarized expression of PRRs help maintain microbial recognition, immune integrity, and homeostasis; (4) how bacteria modulate intercellular tight junctions, which are key to barrier integrity (Turner, 2009); and (5) how secretory pro-inflammatory molecules, like TNF, affect barrier integrity by increased intercellular permeability. Much like mammals, rich microbial communities of distinct phyla and genera are selected in Ciona from an exceptionally vast range of choices of dietary and seawater microbes. Various commensal bacteria likely have co-evolved with the species and may have become integral to nutritional acquisition and gut homeostasis. As such, Ciona offers a unique opportunity to study and characterize not only the molecular events surrounding gut microbial engagement with mucosal immunity but to define the symbiotic ecosystem required for gut homeostasis and thereby host well-being in a controlled, systematic manner using a novel, tractable model system.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors would like to also acknowledge Mr. Alberto Macina (Aqua-Culture of Marine Organisms facility) and Rita Graziano and Franco Iamunno (Electron Microscopy facility) of the Stazione Zoologica Anton Dohrn for providing technical assistance. This work was supported by NIH grant AI23338 (Gary W. Litman) and startup funds from the All Children’s Hospital Foundation and USF Sponsored Research (Larry J. Dishaw).
Abreu, M. T. (2010). Toll-like receptor signalling in the intestinal epithelium: how bacterial recognition shapes intestinal function. Nat. Rev. Immunol. 10, 131–144.
Agrawal, A., Eastman, Q. M., and Schatz, D. G. (1998). Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system Nature 394, 744–751.
Arizza, V., and Parrinello, D. (2009). Inflammatory hemocytes in Ciona intestinalis innate immune response. Invertebrate Surviv. J. 6, S58–S66.
Arizza, V., Parrinello, D., Cammarata, M., Vazzana, M., Vizzini, A., Giaramita, F. T., and Parrinello, N. (2011). A lytic mechanism based on soluble phospholipases A2 (sPLA2) and beta-galactoside specific lectins is exerted by Ciona intestinalis (ascidian) unilocular refractile hemocytes against K562 cell line and mammalian erythrocytes. Fish Shellfish Immunol. 30, 1014–1023.
Artis, D. (2008). Epithelial-cell recognition of commensal bacteria and maintenance of immune homeostasis in the gut. Nat. Rev. Immunol. 8, 411–420.
Atarashi, K., Tanoue, T., Shima, T., Imaoka, A., Kuwahara, T., Momose, Y., Cheng, G., Yamasaki, S., Saito, T., Ohba, Y., Taniguchi, T., Takeda, K., Hori, S., Ivanov, I. I., Umesaki, Y., Itoh, K., and Honda, K. (2011). Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331, 337–341.
Azumi, K., De Santis, R., De Tomaso, A., Rigoutsos, I., Yoshizaki, F., Pinto, M. R., Marino, R., Shida, K., Ikeda, M., Ikeda, M., Arai, M., Inoue, Y., Shimizu, T., Satoh, N., Rokhsar, D. S., Du Pasquier, L., Kasahara, M., Satake, M., and Nonaka, M. (2003). Genomic analysis of immunity in a Urochordate and the emergence of the vertebrate immune system: “waiting for Godot” Immunogenetics 55, 570–581.
Baghdiguian, S., Martinand-Mari, C., and Mangeat, P. (2007). Using Ciona to study developmental programmed cell death. Semin. Cancer Biol. 17, 147–153.
Bischoff, S. C. (2011). “Gut health”: a new objective in medicine? BMC Med. 9, 24. doi: 10.1186/1741-7015-9-24
Bonura, A., Vizzini, A., Salerno, G., Parrinello, N., Longo, V., and Colombo, P. (2009). Isolation and expression of a novel MBL-like collectin cDNA enhanced by LPS injection in the body wall of the ascidian Ciona intestinalis. Mol. Immunol. 46, 2389–2394.
Bosch, T. C., Augustin, R., Anton-Erxleben, F., Fraune, S., Hemmrich, G., Zill, H., Rosenstiel, P., Jacobs, G., Schreiber, S., Leippe, M., Stanisak, M., Grotzinger, J., Jung, S., Podschun, R., Bartels, J., Harder, J., and Schroder, J. M. (2009). Uncovering the evolutionary history of innate immunity: the simple metazoan Hydra uses epithelial cells for host defence. Dev. Comp. Immunol. 33, 559–569.
Bosch, T. C., and McFall-Ngai, M. J. (2011). Metaorganisms as the new frontier. Zoology (Jena) 114, 185–190.
Burighel, P., and Cloney, R. A. (1977). “Urochordata: Ascidiacea,” in Microscopic Anatomy of Invertebrates, Vol. 15: Hemichordata, Chaetognatha and the Invertebrate Chordates, eds F. W. Harrison, and E. E. Ruppert 221–347.
Cammarata, M., Arizza, V., Cianciolo, C., Parrinello, D., Vazzana, M., Vizzini, A., Salerno, G., and Parrinello, N. (2008). The prophenoloxidase system is activated during the tunic inflammatory reaction of Ciona intestinalis. Cell Tissue Res. 333, 481–492.
Canestro, C., Bassham, S., and Postlethwait, J. H. (2003). Seeing chordate evolution through the Ciona genome sequence. Genome Biol. 4, 208.
Cannon, J. P., Haire, R. N., and Litman, G. W. (2002). Identification of diversified genes that contain immunoglobulin-like variable regions in a protochordate. Nat. Immunol. 3, 1200–1207.
Caputi, L., Andreakis, N., Mastrototaro, F., Cirino, P., Vassillo, M., and Sordino, P. (2007). Cryptic speciation in a model invertebrate chordate. Proc. Natl. Acad. Sci. U.S.A. 104, 9364–9369.
Cebra, J. J. (1999). Influences of microbiota on intestinal immune system development. Am. J. Clin. Nutr. 69, 1046S–1051S.
Cebra, J. J., Periwal, S. B., Lee, G., Lee, F., and Shroff, K. E. (1998). Development and maintenance of the gut-associated lymphoid tissue (GALT): the roles of enteric bacteria and viruses. Dev. Immunol. 6, 13–18.
Chow, J., and Mazmanian, S. K. (2010). A pathobiont of the microbiota balances host colonization and intestinal inflammation. Cell Host Microbe 7, 265–276.
Christiaen, L., Wagner, E., Shi, W., and Levine, M. (2009). Isolation of individual cells and tissues from electroporated sea squirt (Ciona) embryos by fluorescence-activated cell sorting (FACS). Cold Spring Harb. Protoc. 2009, pdb.prot5349.
Chung, H., and Kasper, D. L. (2010). Microbiota-stimulated immune mechanisms to maintain gut homeostasis. Curr. Opin. Immunol. 22, 455–460.
Cirino, P., Toscano, A., Caramiello, D., Macina, A., Miraglia, V., and Monte, A. (2002). Laboratory culture of the ascidian Ciona intestinalis (L.): a model system for molecular developmental biology research. Mar. Mod. Elec. Rec. [serial online]. Available at: http://www.mbl.edu/html/BB/MMER/CIR/CirTit.html
Clarke, T. B., Davis, K. M., Lysenko, E. S., Zhou, A. Y., Yu, Y., and Weiser, J. N. (2010). Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat. Med. 16, 228–231.
Cooper, M. D., and Alder, M. N. (2006). The evolution of adaptive immune systems. Cell 124, 815–822.
Davidson, B. (2007). Ciona intestinalis as a model for cardiac development. Semin. Cell Dev. Biol. 18, 16–26.
Dehal, P., Satou, Y., Campbell, R. K., Chapman, J., Degnan, B., De Tomaso, A., Davidson, B., Di Gregorio, A., Gelpke, M., Goodstein, D. M., Harafuji, N., Hastings, K. E. M., Ho, I., Hotta, K., Huang, W., Kawashima, T., Lamaire, P., Martinez, D., Meinertzhagan, I. A., Necula, S., Nonaka, M., Putnam, N., Rash, S., Salga, H., Sataka, M., Terry, A., Yamada, L., Wang, H.-G., Awazu, S., Azumi, K., Boore, J., Branno, M., Chin-bow, S., DeSantis, R., Doyle, S., Francino, P., Keys, D. N., Haga, S., Hayashi, H., Hino, K., Imai, K. S., Inaba, K., Kano, S., Kobayashi, K., Kobayashi, M., Lee, B.-I., Makabe, K. W., Manohar, C., Matassi, G., Medina, M., Mochizuki, Y., Mount, S., Morishita, T., Miura, S., Nakayama, A., Nishizaka, S., Nomoto, H., Ohta, F., Oishi, K., Rigoutsos, I., Sano, M., Sasaki, A., Sasakura, Y., Shoguchi, E., Shin-i, T., Spagnuolo, A., Stainier, D., Suzuki, M. M., Tassy, O., Takatori, N., Tokuoka, M., Yagi, K., Yoshizaki, F., Wada, S., Zhang, C., Hyatt, P. D., Larimer, F., Detter, C., Doggett, N., Glavina, T., Hawkins, T., Richardson, P., Lucas, S., Kohara, Y., Levine, M., Satoh, N., and Rokhsar, D. S. (2002). The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science 298, 2157–2160.
Denoeud, F., Henriet, S., Mungpakdee, S., Aury, J. M., Da Silva, C., Brinkmann, H., Mikhaleva, J., Olsen, L. C., Jubin, C., Canestro, C., Bouquet, J. M., Danks, G., Poulain, J., Campsteijn, C., Adamski, M., Cross, I., Yadetie, F., Muffato, M., Louis, A., Butcher, S., Tsagkogeorga, G., Konrad, A., Singh, S., Jensen, M. F., Cong, E. H., Eikeseth-Otteraa, H., Noel, B., Anthouard, V., Porcel, B. M., Kachouri-Lafond, R., Nishino, A., Ugolini, M., Chourrout, P., Nishida, H., Aasland, R., Huzurbazar, S., Westhof, E., Delsuc, F., Lehrach, H., Reinhardt, R., Weissenbach, J., Roy, S. W., Artiguenave, F., Postlethwait, J. H., Manak, J. R., Thompson, E. M., Jaillon, O., Du Pasquier, L., Boudinot, P., Liberles, D. A., Volff, J. N., Philippe, H., Lenhard, B., Crollius, H. R., Wincker, P., and Chourrout, D. (2010). Plasticity of animal genome architecture unmasked by rapid evolution of a pelagic tunicate. Science 330, 1381–1385.
Di Bella, M. A., and De Leo, G. (2000). Hemocyte migration during inflammatory-like reaction of Ciona intestinalis (Tunicata, Ascidiacea). J Invertebr. Pathol. 76, 105–111.
Dishaw, L. J., Giacomelli, S., Melillo, D., Zucchetti, I., Haire, R. N., Natale, L., Russo, N. A., De Santis, R., Litman, G. W., and Pinto, M. R. (2011). A role for variable region-containing chitin-binding proteins (VCBPs) in host gut-bacteria interactions. Proc. Natl. Acad. Sci. U.S.A. 108, 16747–16752.
Dreyfus, D. H. (1992). Evidence suggesting an evolutionary relationship between transposable elements and immune system recombination sequences. Mol. Immunol. 29, 807–810.
Duerkop, B. A., Vaishnava, S., and Hooper, L. V. (2009). Immune responses to the microbiota at the intestinal mucosal surface. Immunity 31, 368–376.
Edelman, S. M., and Kasper, D. L. (2008). Symbiotic commensal bacteria direct maturation of the host immune system. Curr. Opin. Gastroenterol. 24, 720–724.
Ermak, T. H. (1981). A comparison of cell proliferation patterns in the digestive track of ascidians. J. Exp. Zool. 217, 325–339.
Fedders, H., and Leippe, M. (2008). A reverse search for antimicrobial peptides in Ciona intestinalis: identification of a gene family expressed in hemocytes and evaluation of activity. Dev. Comp. Immunol. 32, 286–298.
Feng, T., and Elson, C. O. (2011). Adaptive immunity in the host-microbiota dialog. Mucosal Immunol. 4, 15–21.
Flajnik, M. F., and Kasahara, M. (2010). Origin and evolution of the adaptive immune system: genetic events and selective pressures. Nat. Rev. Genet. 11, 47–59.
Fujimura, K. E., Slusher, N. A., Cabana, M. D., and Lynch, S. V. (2010). Role of the gut microbiota in defining human health. Expert Rev. Anti Infect. Ther. 8, 435–454.
Fujita, T., Endo, Y., and Nonaka, M. (2004). Primitive complement system–recognition and activation. Mol. Immunol. 41, 103–111.
Holland, L. Z., Albalat, R., Azumi, K., Benito-Gutierrez, E., Blow, M. J., Bronner-Fraser, M., Brunet, F., Butts, T., Candiani, S., Dishaw, L. J., Ferrier, D. E., Garcia-Fernandez, J., Gibson-Brown, J. J., Gissi, C., Godzik, A., Hallbook, F., Hirose, D., Hosomichi, K., Ikuta, T., Inoko, H., Kasahara, M., Kasamatsu, J., Kawashima, T., Kimura, A., Kobayashi, M., Kozmik, Z., Kubokawa, K., Laudet, V., Litman, G. W., McHardy, A. C., Meulemans, D., Nonaka, M., Olinski, R. P., Pancer, Z., Pennacchio, L. A., Pestarino, M., Rast, J. P., Rigoutsos, I., Robinson-Rechavi, M., Roch, G., Saiga, H., Sasakura, Y., Satake, M., Satou, Y., Schubert, M., Sherwood, N., Shiina, T., Takatori, N., Tello, J., Vopalensky, P., Wada, S., Xu, A., Ye, Y., Yoshida, K., Yoshizaki, F., Yu, J. K., Zhang, Q., Zmasek, C. M., de Jong, P. J., Osoegawa, K., Putnam, N. H., Rokhsar, D. S., Satoh, N., and Holland, P. W. (2008). The amphioxus genome illuminates vertebrate origins and cephalochordate biology. Genome Res. 18, 1100–1111.
Hooper, L. V., and Gordon, J. I. (2001). Commensal host-bacterial relationships in the gut. Science 292, 1115–1118.
Hooper, L. V., and Macpherson, A. J. (2010). Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat. Rev. Immunol. 10, 159–169.
Hughes, A. L., and Friedman, R. (2005). Loss of ancestral genes in the genomic evolution of Ciona intestinalis. Evol. Dev. 7, 196–200.
Ikuta, T., Satoh, N., and Saiga, H. (2010). Limited functions of Hox genes in the larval development of the ascidian Ciona intestinalis. Development 137, 1505–1513.
Ivanov, I. I., Atarashi, K., Manel, N., Brodie, E. L., Shima, T., Karaoz, U., Wei, D., Goldfarb, K. C., Santee, C. A., Lynch, S. V., Tanoue, T., Imaoka, A., Itoh, K., Takeda, K., Umesaki, Y., Honda, K., and Littman, D. R. (2009). Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498.
Joly, J., Kano, S., Matsuoka, T., Auger, H., Hirayama, K., Satoh, N., Awazu, S., Legendre, L., and Sasakura, Y. (2007). Culture of Ciona intestinalis in closed systems. Dev. Dyn. 236, 1832–1840.
Kanther, M., and Rawls, J. F. (2010). Host-microbe interactions in the developing zebrafish. Curr. Opin. Immunol. 22, 10–19.
Katz, M. J. (1983). Comparative anatomy of the tunicate tadpole, Ciona intestinalis. Biol. Bull. 164, 1–27.
Kubo, A., Suzuki, N., Yuan, X., Nakai, K., Satoh, N., Imai, K. S., and Satou, Y. (2010). Genomic cis-regulatory networks in the early Ciona intestinalis embryo. Development 137, 1613–1623.
Lathrop, S. K., Bloom, S. M., Rao, S. M., Nutsch, K., Lio, C. W., Santacruz, N., Peterson, D. A., Stappenbeck, T. S., and Hsieh, C. S. (2011). Peripheral education of the immune system by colonic commensal microbiota. Nature 478, 250–254.
Lavelle, E. C., Murphy, C., O’Neill, L. A., and Creagh, E. M. (2010). The role of TLRs, NLRs, and RLRs in mucosal innate immunity and homeostasis. Mucosal Immunol. 3, 17–28.
Lee, Y. K., and Mazmanian, S. K. (2010). Has the microbiota played a critical role in the evolution of the adaptive immune system? Science 330, 1768–1773.
Lemaire, P., Smith, W. C., and Nishida, H. (2008). Ascidians and the plasticity of the chordate developmental program. Curr. Biol. 18, R620–R631.
Leulier, F., and Lemaitre, B. (2008). Toll-like receptors – taking an evolutionary approach. Nat. Rev. Genet. 9, 165–178.
Litman, G. W., Rast, J. P., and Fugmann, S. D. (2010). Origins of vertebrate adaptive immunity. Nat. Rev. Immunol. 10, 543–553.
Lotz, M., Gutle, D., Walther, S., Menard, S., Bogdan, C., and Hornef, M. W. (2006). Postnatal acquisition of endotoxin tolerance in intestinal epithelial cells. J. Exp. Med. 203, 973–984.
Lyczak, J. B. (2003). Commensal bacteria increase invasion of intestinal epithelium by Salmonella enterica serovar Typhi. Infect. Immun. 71, 6610–6614.
Maldonado-Contreras, A. L., and McCormick, B. A. (2011). Intestinal epithelial cells and their role in innate mucosal immunity. Cell Tissue Res. 343, 5–12.
Mans, J. J., von Lackum, K., Dorsey, C., Willis, S., Wallet, S. M., Baker, H. V., Lamont, R. J., and Handfield, M. (2009). The degree of microbiome complexity influences the epithelial response to infection. BMC Genomics 10, 380. doi: 10.1186/1471-2164-10-380
Marchiando, A. M., Graham, W. V., and Turner, J. R. (2010). Epithelial barriers in homeostasis and disease. Annu. Rev. Pathol. 5, 119–144.
Marino, R., Kimura, Y., De Santis, R., Lambris, J. D., and Pinto, M. R. (2002). Complement in urochordates: cloning and characterization of two C3-like genes in the ascidian Ciona intestinalis. Immunogenetics 53, 1055–1064.
Maslowski, K. M., and Mackay, C. R. (2011). Diet, gut microbiota and immune responses. Nat. Immunol. 12, 5–9.
Matsunaga, T., and Rahman, A. (1998). What brought the adaptive immune system to vertebrates? – The jaw hypothesis and the seahorse. Immunol. Rev. 166, 177–186.
Mazmanian, S. K., Liu, C. H., Tzianabos, A. O., and Kasper, D. L. (2005). An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122, 107–118.
Meinertzhagen, I. A., and Okamura, Y. (2001). The larval ascidian nervous system: the chordate brain from its small beginnings. Trends Neurosci. 24, 401–410.
Melillo, D., Sfyroera, G., De Santis, R., Graziano, R., Marino, R., Lambris, J. D., and Pinto, M. R. (2006). First identification of a chemotactic receptor in an invertebrate species: structural and functional characterization of Ciona intestinalis C3a receptor. J. Immunol. 177, 4132–4140.
Messier-Solek, C., Buckley, K. M., and Rast, J. P. (2010). Highly diversified innate receptor systems and new forms of animal immunity. Semin. Immunol. 22, 39–47.
Muller, C. A., Autenrieth, I. B., and Peschel, A. (2005). Innate defenses of the intestinal epithelial barrier. Cell. Mol. Life Sci. 62, 1297–1307.
Nell, S., Suerbaum, S., and Josenhans, C. (2010). The impact of the microbiota on the pathogenesis of IBD: lessons from mouse infection models. Nat. Rev. Microbiol. 8, 564–577.
Nishida, H., Satoh, N., and Hirose, E. (2010). More diversity and more convergence in tunicate biology. Zool. Sci. 27, 67–68.
Ohland, C. L., and Macnaughton, W. K. (2010). Probiotic bacteria and intestinal epithelial barrier function. Am. J. Physiol. Gastrointest. Liver Physiol. 298, G807–G819.
Parrinello, N. (2009). Focusing on Ciona intestinalis (tunicate) innate immune system. Evolutionary implications. Invertebrate Surviv. J. 6, S46–S57.
Parrinello, N., Arizza, V., Cammarata, M., Giaramita, F. T., Pergolizzi, M., Vazzana, M., Vizzini, A., and Parrinello, D. (2007). Inducible lectins with galectin properties and human IL1alpha epitopes opsonize yeast during the inflammatory response of the ascidian Ciona intestinalis. Cell Tissue Res. 329, 379–390.
Parrinello, N., Cammarata, M., and Arizza, V. (1996). Univacuolar refractile hemocytes from the tunicate Ciona intestinalis are cytotoxic for mammalian erythrocytes in vitro. Biol. Bull. 190, 418–425.
Parrinello, N., Vizzini, A., Arizza, V., Salerno, G., Parrinello, D., Cammarata, M., Giaramita, F. T., and Vazzana, M. (2008). Enhanced expression of a cloned and sequenced Ciona intestinalis TNFalpha-like (CiTNF alpha) gene during the LPS-induced inflammatory response. Cell Tissue Res. 334, 305–317.
Parrinello, N., Vizzini, A., Salerno, G., Sanfratello, M. A., Cammarata, M., Arizza, V., Vazzana, M., and Parrinello, D. (2010). Inflamed adult pharynx tissues and swimming larva of Ciona intestinalis share CiTNFalpha-producing cells. Cell Tissue Res. 341, 299–311.
Pinto, M. R., Chinnici, C. M., Kimura, Y., Melillo, D., Marino, R., Spruce, L. A., De Santis, R., Parrinello, N., and Lambris, J. D. (2003). CiC3-1a-mediated chemotaxis in the deuterostome invertebrate Ciona intestinalis (Urochordata). J. Immunol. 171, 5521–5528.
Rast, J. P., and Messier-Solek, C. (2008). Marine invertebrate genome sequences and our evolving understanding of animal immunity. Biol. Bull. 214, 274–283.
Rast, J. P., Smith, L. C., Loza-Coll, M., Hibino, T., and Litman, G. W. (2006). Genomic insights into the immune system of the sea urchin. Science 314, 952–956.
Rautava, S., and Walker, W. A. (2007). Commensal bacteria and epithelial cross talk in the developing intestine. Curr. Gastroenterol. Rep. 9, 385–392.
Rescigno, M., and Di Sabatino, A. (2009). Dendritic cells in intestinal homeostasis and disease. J. Clin. Invest. 119, 2441–2450.
Round, J. L., and Mazmanian, S. K. (2009). The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 9, 313–323.
Sasaki, N., Ogasawara, M., Sekiguchi, T., Kusumoto, S., and Satake, H. (2009). Toll-like receptors of the ascidian Ciona intestinalis: prototypes with hybrid functionalities of vertebrate Toll-like receptors. J. Biol. Chem. 284, 27336–27343.
Sasakura, Y., Inaba, K., Satoh, N., Kondo, M., and Akasaka, K. (2009). Ciona intestinalis and Oxycomanthus japonicus, representatives of marine invertebrates. Exp. Anim. 58, 459–469.
Sasakura, Y., Suzuki, M. M., Hozumi, A., Inaba, K., and Satoh, N. (2010). Maternal factor-mediated epigenetic gene silencing in the ascidian Ciona intestinalis. Mol. Genet. Genomics 283, 99–110.
Satoh, N., and Levine, M. (2005). Surfing with the tunicates into the post-genome era. Genes Dev. 19, 2407–2411.
Savage, D. C. (1977). Microbial ecology of the gastrointestinal tract. Annu. Rev. Microbiol. 31, 107–133.
Sekirov, I., Russell, S. L., Antunes, L. C., and Finlay, B. B. (2010). Gut microbiota in health and disease. Physiol. Rev. 90, 859–904.
Shi, W., Levine, M., and Davidson, B. (2005). Unraveling genomic regulatory networks in the simple chordate, Ciona intestinalis. Genome Res. 15, 1668–1674.
Shida, K., Terajima, D., Uchino, R., Ikawa, S., Ikeda, M., Asano, K., Watanabe, T., Azumi, K., Nonaka, M., Satou, Y., Satoh, N., Satake, M., Kawazoe, Y., and Kasuya, A. (2003). Hemocytes of Ciona intestinalis express multiple genes involved in innate immune host defense. Biochem. Biophys. Res. Commun. 302, 207–218.
Skjoedt, M. O., Palarasah, Y., Rasmussen, K., Vitved, L., Salomonsen, J., Kliem, A., Hansen, S., Koch, C., and Skjodt, K. (2010). Two mannose-binding lectin homologues and an MBL-associated serine protease are expressed in the gut epithelia of the urochordate species Ciona intestinalis. Dev. Comp. Immunol. 34, 59–68.
Smith, V. J., and Peddie, C. M. (1992). Cell cooperation during host defense in the solitary tunicate Ciona intestinalis (L). Biol. Bull. 183, 211–219.
Terajima, D., Shida, K., Takada, N., Kasuya, A., Rokhsar, D., Satoh, N., Satake, M., and Wang, H. G. (2003). Identification of candidate genes encoding the core components of the cell death machinery in the Ciona intestinalis genome. Cell Death Differ. 10, 749–753.
Turner, J. R. (2009). Intestinal mucosal barrier function in health and disease. Nat. Rev. Immunol. 9, 799–809.
Usharauli, D. (2010). Chronic infection and the origin of adaptive immune system. Med. Hypotheses 75, 241–243.
Willing, B. P., Antunes, L. C., Keeney, K. M., Ferreira, R. B., and Finlay, B. B. (2011). Harvesting the biological potential of the human gut microbiome. Bioessays 33, 414–418.
Yonge, C. M. (1925). Studies on the comparative physiology of digestion. III. Secretion, digestion, and assimilation in the gut of Ciona intestinalis. J. Exp. Biol. 2, 373–388.
Keywords: protochordate, innate immunity, Ciona, microbiota, gut epithelium
Citation: Dishaw LJ, Flores-Torres JA, Mueller MG, Karrer CR, Skapura DP, Melillo D, Zucchetti I, De Santis R, Pinto MR and Litman GW (2012) A basal chordate model for studies of gut microbial immune interactions. Front. Immun. 3:96. doi: 10.3389/fimmu.2012.00096
Received: 17 January 2012; Accepted: 12 April 2012;
Published online: 03 May 2012.
Edited by:
Maria Yazdanbakhsh, Leiden University Medical Center, NetherlandsCopyright: © 2012 Dishaw, Flores-Torres, Mueller, Karrer, Skapura, Melillo, Zucchetti, De Santis, Pinto and Litman. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Larry J. Dishaw, Department of Pediatrics, University of South Florida, 140 Seventh Avenue South, CRI 3008, St. Petersburg, FL 33701, USA. e-mail:bGRpc2hhd0BoZWFsdGgudXNmLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.