- 1 Emory Vaccine Center, Emory University of School of Medicine, Atlanta, GA, USA
- 2 Department of Microbiology and Immunology, Emory University of School of Medicine, Atlanta, GA, USA
The emergence of a novel influenza A virus strain into humans poses a continuous public health threat. Vaccination is the most effective means of protection against influenza. The generation of memory B cells and long-lived plasma cells that can maintain protective levels of influenza-specific antibodies for protracted periods of time is the foundation for the success of such vaccines. Influenza vaccines elicit an antibody response that is primarily targeting viral surface glycoproteins. However, frequent amino acid mutations within the immunodominant epitopes allow the virus to efficiently escape neutralization by pre-existing antibodies and consequently cause annual epidemics and occasional pandemics. Recently, monoclonal antibodies (mAbs) that target subdominant influenza epitopes have been extensively characterized. These epitopes are immunogenic, can mediate virus neutralization, and most importantly are conserved among different influenza strains. It remains puzzling, however, that despite being repeatedly exposed to such conserved domains of influenza hemagglutinin (HA) either in the form of vaccination or natural infection, most humans do not develop immunological memory that can provide broad protection against emerging virus strains. Here we will discuss the conditions that may be required for engaging such cross-reactive memory B cells in the immune response to influenza infection and vaccination in humans.
Introduction
Since the isolation of the first human influenza virus in 1933, controlling infections mediated by this respiratory pathogen has represented a formidable challenge (Webby and Webster, 2003). The impediment stems from the ever-evolving nature of influenza viruses, which enables the viruses to escape pre-existing immune surveillance (Kilbourne, 1978). Moreover, effective vaccines against influenza viruses work by eliciting antibody responses that primarily target the hemagglutinin (HA) molecule, which is the most variable among virus proteins (Kilbourne, 1978). Vaccination remains, however, the most successful intervention to prevent influenza virus infection and transmission (Ellebedy and Webby, 2009). The need for a broadly protective influenza vaccine is clearly demonstrated by the failure of seasonal influenza vaccines to control the annual epidemics of influenza viruses, which result in about three to five million cases of severe illness, and up to 500,000 deaths worldwide (http://www.who.int/mediacentre/factsheets/fs211/en/). In addition, influenza viruses cause occasional pandemics when an antigenically novel virus spills into the human population or when, as in the case of the 2009 pandemic H1N1 virus, an influenza virus that has ceased to circulate among humans for decades reemerges (Smith et al., 2009). The vast majority of adults possess pre-existing immunity to influenza viruses because of prior infection and/or vaccination (Sasaki et al., 2007). Thus, influenza represents a direct and unique challenge to the human immunological memory.
Immunological memory is typically established following immunization or infections, and it is the corner-stone for the success of all prophylactic vaccines (Ahmed and Gray, 1996). Immunological memory is established in the cellular (CD4 and CD8 T cells) and humoral (B cells) immune compartments. Two B cell populations are responsible for sustaining the humoral immune memory: the long-lived plasma cells (PCs) and memory B cells (Slifka and Ahmed, 1998; Yoshida et al., 2010). Long-lived PCs are mainly concentrated in the bone marrow where stromal and other cell types provide ligands that promote their long-term survival. Long-lived PCs secrete antigen-specific antibodies for protracted periods of time (Slifka and Ahmed, 1998; Manz et al., 2002; Yoshida et al., 2010). Whether in blood or on mucosal surfaces, these antibodies represent the first line of pathogen-specific defense against re-infection. Unlike long-lived PCs, significant proportions of memory B cells can be isolated from peripheral blood in humans. Memory B cells rapidly proliferate and differentiate into antibody-secreting PCs upon re-encountering the immunizing antigen or pathogen.
Although protective vaccines against influenza viruses have been introduced several decades ago, a thorough understanding of the role of memory B cells in mediating protection against influenza virus challenge remains poorly defined. Here, we will discuss recent advances, and yet to be answered questions, regarding the role of cross-reactive memory B cell responses elicited by either infection or vaccination in protection against influenza. Cross-reactive or broad immunity to influenza viruses has been described using several terms such as heterotypic, heterosubtypic, and cross-neutralizing interchangeably. Here, we used the term cross-reactive in its broadest sense that is to describe B cell targeting of epitopes broadly conserved not only among different influenza virus subtypes but also among different viruses within the same subtype.
Antibody Responses to Influenza Virus HA: Catching a Constantly Moving Target
Preformed, neutralizing antibodies in serum or, preferably, on the airway mucosal surface are the best protective measure against influenza infection. Viral surface glycoproteins are the prime targets of protective antibodies. Binding to these glycoproteins can block initial viral attachment and the subsequent establishment of infection, thus providing “sterilizing protection” (Gerhard, 2001). The major two viral surface glycoproteins are the HA and the neuraminidase (NA). HA is the primary target of infection- and immunization-induced protective antibodies generated against influenza viruses (Wilson and Cox, 1990; Skehel and Wiley, 2000). HA is a homotrimer that mediates influenza virus binding to its sialic acid receptors on lung epithelial cells leading to virus entry via receptor-mediated endocytosis. In addition, once inside the endocytic vesicle, HA mediates a pH-dependent virus membrane fusion with the endosomal membrane, releasing viral genomic materials into the cytosol (Wilson and Cox, 1990; Skehel and Wiley, 2000).
To date, 16 antigenically distinct HA subtypes have been characterized. These subtypes are phylogenetically classified into group 1 (H1, H2, H5, H6, H8, H9, H11, H12, H13, and H16) and group 2 (H3, H4, H7, H10, H14, and H15) HA molecules (Figure 1; Nabel and Fauci, 2010; Sui et al., 2011). Screening viral escape mutants have revealed that anti-HA antibodies primarily target few immunodominant epitopes within the globular head region of the HA molecule. For example, in the influenza H1 HA five immunodominant epitopes (Sa, Sb, Ca1, Ca2, and Cb) were mapped (Figure 2; Caton et al., 1982; Wilson and Cox, 1990). Expectedly, these epitopes are located around the receptor-binding domain within the HA globular head. While antibodies targeting such epitopes are potentially neutralizing and protective, they are strain-specific, and thus lack the much-desired broad cross-neutralizing activity to different HA subtypes. Moreover, single amino acid mutations that accumulate over time within these immunodominant epitopes allow influenza viruses to escape existing immunity. This process is known as the antigenic drift. On the other hand, human monoclonal antibodies (mAbs) that recognize the globular head immunodominant epitopes across drift variants of the same influenza HA subtype and even across different HA subtypes have been characterized (Table 1; Corti et al., 2010; Krause et al., 2011; Whittle et al., 2011; Wrammert et al., 2011). This indicates that even within these hypervariable globular head epitopes there are residues that are conserved (possibly due to the structural and functional requirements for interaction with the sialic acid receptors), and thus constitute an important target for broad neutralizing antibodies. It can be speculated that changes within these conserved residues significantly affect virus fitness.
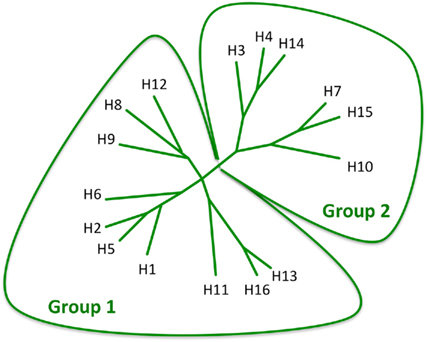
Figure 1. Phylogenetic analysis of the 16 influenza HA subtypes. The diagram was adapted with permission from Nabel and Fauci (2010).
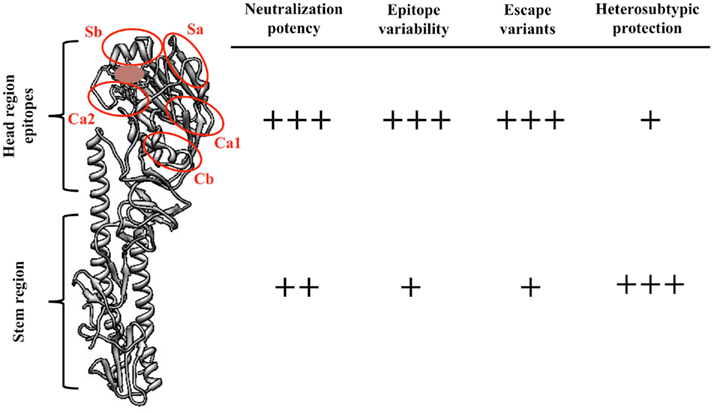
Figure 2. Comparisons between the features of influenza HA globular head epitopes and HA stem region epitopes. A structural view of the H1 HA molecule (monomer, A/PR8/34) showing the different globular head epitopes (red circles) and their localization close to the receptor-binding domain (brown). The number of (+) signs was arbitrarily determined. Figure was prepared using Chimera (Pettersen et al., 2004).
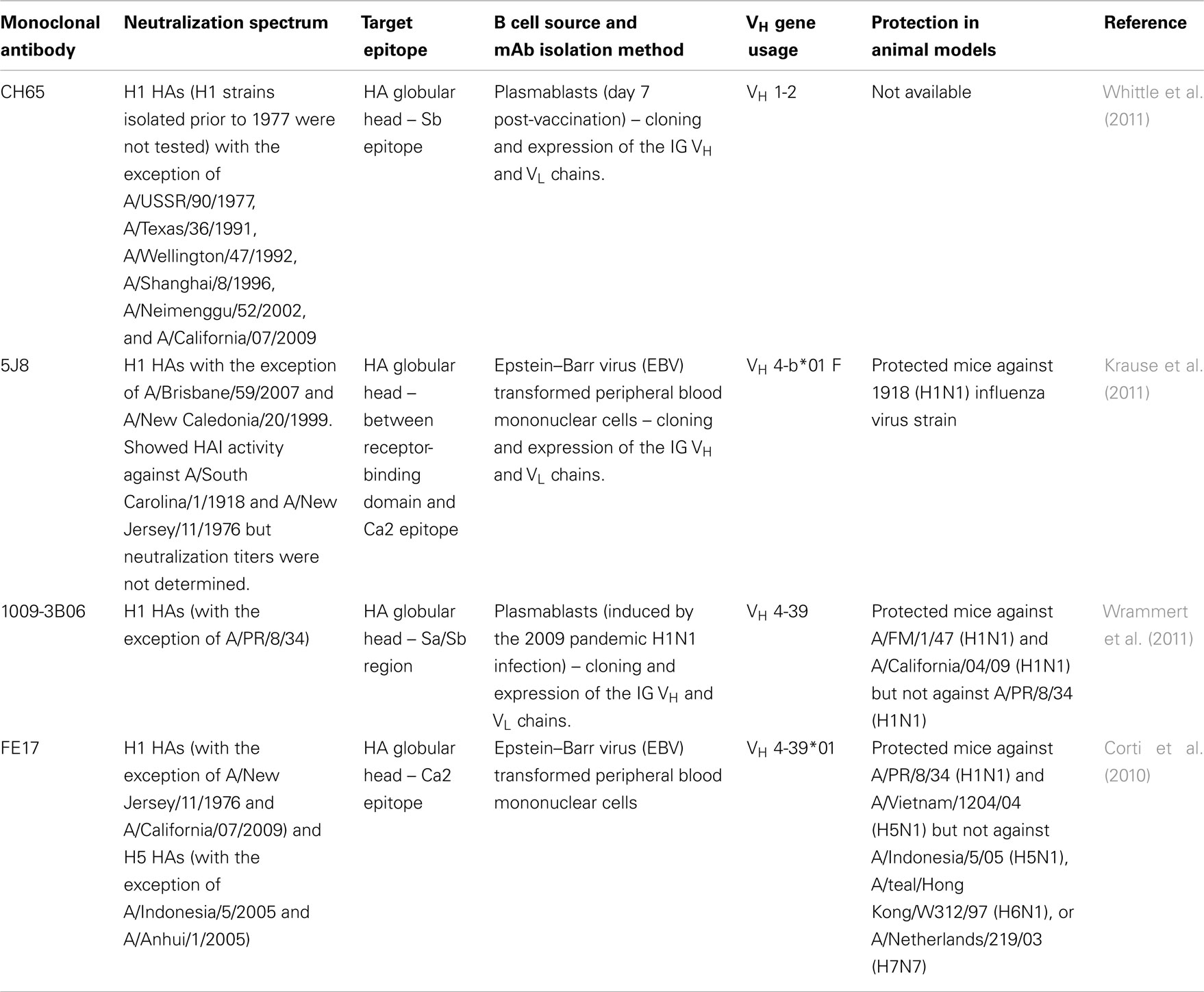
Table 1. Examples of cross-reactive human monoclonal antibodies that bind to HA globular head epitopes.
Apart from the immunodominant HA head epitopes, at least two epitopes within the HA stalk (or HA2) region have been identified as potential targets for an interesting group of murine and human mAbs (Okuno et al., 1993; Throsby et al., 2008; Ekiert et al., 2009; Sui et al., 2009; Corti et al., 2010; Wrammert et al., 2011). The striking advantage of such epitopes is that they are more conserved across different influenza HA subtypes compared to HA head ones. Therefore, antibodies targeting HA stalk region can be broadly neutralizing. Indeed, some mAbs in this group (CR6261 and F10) showed a remarkable broad neutralizing activity against influenza viruses belonging to all group 1 HAs (Ekiert et al., 2009; Sui et al., 2009). Ekiert et al. (2011) have recently demonstrated the existence of a similarly conserved epitope among group 2 HAs as well. The latter epitope is the target of CR8020, a human mAb that can neutralize all group 2 influenza viruses (Ekiert et al., 2011). Corti et al. (2011) were able to isolate a human mAb that is able to neutralize all viruses from group A and group B HAs. This pan-neutralizing mAb or FI6 was isolated through extensive screening of mAbs derived from recently generated antibody-secreting cells (ASCs) that produced antibodies cross-reacted with H1, H5, and H7 HAs (Corti et al., 2011). It is currently difficult to precisely determine the frequency of HA stem-targeting B cells among influenza-specific memory B cell pool, but the fact that out of more than 13,000 mAbs (over 90% of which is influenza-specific) screened from one donor only a few FI6-like mAbs were detected gives a glimpse of how rare such specificity can be in humans (Corti et al., 2011). Structural studies have revealed that similarly to CR6261 and F10, FI6 bound to an epitope in the F subdomain within the HA stem region. However, and in contrast to CR6261 and F10, the binding of FI6 to HA is mediated by its remarkably long heavy chain complementarity determining region 3 or HCDR3 (out of the three HCDRs) and that light chain CDR1 (LCDR1) also contributes to the binding (Corti et al., 2011). The mechanism by which anti-stem antibodies neutralize influenza viruses is not entirely clear, but the observation that CR6261 and FI6 can inhibit virus-mediated syncytia formation but not hemagglutination suggest that they interfere with the virus membrane fusion step (Corti et al., 2011).
The first evidence of the protective potential of HA stem-targeting antibodies dates back to 1993 when Okuno et al. (1993, 1994) described C179, a murine mAb that cross-reacted with H1 and H2, but not H3, HA subtypes. C179 protected mice from a lethal H1 challenge when administered 1 day prior, or 2 days after, virus challenge (Okuno et al., 1993, 1994). Recently, another murine mAb, 12D1, was generated following an immunization schedule designed to boost B cells targeting conserved epitopes on the H3 HA (Wang et al., 2010a). The same group then later showed that immunizing mice with the peptide suggested to enclose the epitope targeted by 12D1 afforded protection for mice against H3N2, H1N1, and H5N1 influenza viruses’ challenge (Wang et al., 2010b). Finally, Wei et al. reported a prime–boost influenza immunization strategy that enriched the anti-HA-antibody responses with those targeting the conserved stem epitopes. The immunization provided protection against a variety of H1N1 viruses in mice and ferrets (Wei et al., 2010). The same strategy showed promising results when used to elicit antibodies against H5 HA in humans (Ledgerwood et al., 2011).
Memory B Cell Responses to Influenza Virus Vaccination in Humans
Human influenza vaccines are designed to elicit antibody responses to HA and NA and, more importantly, to induce the generation of persistent antigen-specific memory B cells. However, due to influenza antigenic drift, it is necessary to perform an annual revision of the antigens included in human seasonal vaccines to ensure that they match the circulating influenza viruses. Currently, human annual vaccines contain antigens from three different influenza viruses: H1N1, and H3N2 influenza A viruses and an influenza B virus. The vaccines are provided in two forms; the inactivated, subunit form and the live attenuated form. Studying primary B cell responses to influenza vaccines in humans is difficult due to the wide prevalence of pre-existing immunity. Several studies have already demonstrated that the vast majority of adults and even older children possess low but consistent base line levels of influenza virus-specific IgG memory B cells (0.1–0.5% of total IgG memory B cells; Sasaki et al., 2007). Using the 2005/2006 and 2006/2007 seasonal vaccine formulations, we demonstrated that, at least in adults, the influenza-specific ASCs response to seasonal vaccines peaks around 1 week post-immunization, and is characterized by several features that are reminiscent of memory B cell responses such as the enhanced kinetics of the ASCs generation, the predominance of isotype switched, highly mutated IgG secreting cells among responding ASCs (Wrammert et al., 2008). Measuring the frequency, kinetics, and specificity of antigen-specific memory B cells post-vaccination has been technically challenging. Tracking memory B cells by flow cytometry using fluorescent antigen has been difficult mainly due to the complexity of the HA and NA molecules. Using an ex vivo ELISPOT assay in which memory B cells are polyclonally activated, we have shown that Influenza-specific IgG memory B cells are transiently boosted after immunization (peaking at 2–4 weeks post-immunizations). Pinna et al. (2009) developed a similar assay in which they selectively activate – using the TLR7/8 agonist R848 and IL-2 – and clone memory B cells from total peripheral blood monocytes (PBMCs). Using this method, the authors measured the kinetics of influenza-specific memory B cells generation following immunization and demonstrated that it peaks at day 14 post-immunization. Several aspects of the memory B cell responses to influenza immunization in humans remain largely undefined. For example, what is the fine specificity (also cross-reactivity and neutralizing activity) of the antibodies secreted by reactivated memory B cells? How does influenza infection affect the reactivity spectrum of the memory B cell pool? Other important aspects include the differences in kinetics, magnitude, and quality of immune responses to inactivated subunit vs. live attenuated influenza vaccines in influenza-naïve individuals.
Role of Memory B Cells in Cross-Protection Against Influenza: The 2009 H1N1 Pandemic Example
Apart from the annual epidemics caused by drift variants of seasonal influenza viruses, influenza viruses cause occasional pandemics. While influenza drift variants usually emerge as a result of few amino acid mutations within the immunodominant head epitopes, the 2009 pandemic H1 HA had a remarkable 10-fold increase in HA variability compared to H1 HAs from earlier seasons (Han and Marasco, 2011). The bulk of these changes are concentrated within the HA globular head (Han and Marasco, 2011). The HA stem region remained relatively conserved between the pandemic and H1 HA from prior seasonal strains. This begged the obvious question: if the broadly neutralizing HA stem-specific antibodies exist in humans and can be boosted following influenza seasonal immunization, why did the 2009 H1N1 influenza pandemic show such high infectivity among young adults with history of previous influenza exposure? And why, in general, influenza is still a serious public health problem?
One possible explanation for this puzzle is that the titers of HA stem-specific antibodies are too low to provide protection, which in turn might be due to the low frequency of HA stem-specific B cells participating in the primary response to influenza immunization or infection compared to the immunodominant HA head-specific ones resulting in a lower frequency of precursor memory B cells with such specificity. And thus upon re-exposure, it becomes difficult to boost HA stem-specific memory B cells to differentiate into ASCs, as they are outcompeted by HA head-specific memory B cells for the antigen, which in turn result in waning of the former cells over time. Surprisingly, in individuals infected with the 2009 pandemic H1N1, out of the 11 neutralizing, anti-HA mAbs retrieved from peripheral ASCs, 9 mAbs were cross-reactive targeting the conserved epitopes in the HA stem region (Wrammert et al., 2011). We hypothesized that the significant changes in the immunodominant globular head epitopes of the pandemic H1 HA compared to the contemporary seasonal H1 HAs lead to decreased competition for antigen among pre-existing memory B cells, which in turn allowed for the expansion of the subdominant stem-specific memory B cells.
Interestingly, the incidence of infection and mortality from the 2009 influenza pandemic was lowest among the elderly especially those aged >65 years (Chowell et al., 2009; Dawood et al., 2009; Jain et al., 2009). While such protection can be attributed to the accumulation of memory B cells or serum antibodies that are specific to the conserved cross-reactive epitopes, there are no data that either support or contradict this possibility. Another explanation for the observed protection would be the close antigenic relatedness between the HA of the 2009 pandemic H1N1 viruses and that of H1N1 viruses circulated after the 1918 pandemic and prior to 1950 or that of the swine-origin H1N1 (A/NJ/76; Hancock et al., 2009; Skowronski et al., 2011). Results from a recent study support the latter explanation showing that the elderly elicited superior responses to the 2009 H1N1 monovalent immunization in comparison to young adults. These responses were largely directed to the globular head epitopes (Skowronski et al., 2011). There are still open questions that are yet to be answered. For example, what is the frequency of HA stem-specific memory B cells? Can such memory B cells be efficiently boosted with the regular influenza vaccines?
Can Adjuvants Help?
Adjuvants have been used with human influenza vaccines for decades. For split or subunit influenza vaccines, the use of adjuvants (especially with antigens derived from avian influenza viruses) is recommended to enhance their intrinsic immunogenicity (Treanor et al., 2006). Currently, two main classes of adjuvants are being used with human influenza vaccines; the insoluble aluminum salts or alum and the oil-in-water emulsion adjuvants such as MF59 and AS03. These adjuvants were empirically developed and the mechanisms through which they potentiate immune responses are not fully understood. Assuming that the frequency of memory B cells that are specific to conserved HA epitopes are relatively low due to the intrinsic poor immunogenicity of the epitopes or due to the immunodominance of the HA globular head epitopes, can adjuvants help in re-engaging these cells in response to influenza immunization. Data from immunization studies in combination with oil-in-water emulsion based adjuvants have been promising in this respect. For example, MF59-adjuvanted influenza vaccines elicited not only a significantly higher antibody titers but also greater cross-reactive antibody responses (Stephenson et al., 2005; Banzhoff et al., 2008). Moreover, Golding and colleagues have shown that MF59 expands the diversity of the antibody responses to human influenza vaccines including H5N1 and the 2009 H1N1 pandemic monovalent influenza vaccines (Khurana et al., 2010, 2011). The authors examined the binding patterns of sera obtained from individuals immunized with MF59-adjuvanted vs. unadjuvanted influenza vaccine to whole influenza genome fragment phage display libraries (Khurana et al., 2010, 2011). In contrast to prevaccination and unadjuvanted vaccination sera that predominantly bound to linear epitopes within the HA2 region, MF59 increased the number of recognized fragments mapped to the HA1 region and NA protein. These findings are important as they shed light on the impact of adjuvants on the breadth of antibody responses to influenza vaccines. However, the question remains whether the “broadening” of the antibody responses observed with adjuvanted vaccines is a result of activation of a naïve B cell population that targeted novel epitopes within the immunizing antigen or simply due to more efficient recruitment of a broader repertoire of the influenza-specific memory B cells.
Concluding Remarks
Recent advances in techniques used in the isolation of human mAbs together with the high resolution three-dimensional structures of the HA-antibody complexes have greatly expanded our understanding of the neutralization potential of anti-influenza antibodies. This was exemplified by the characterization of the human mAbs that target conserved influenza HA stem epitopes as well as conserved epitopes within the HA globular head. More importantly, recent data suggest that antibodies that target these conserved sites can elicited by immunization in humans (Ledgerwood et al., 2011). It is important that we next focus on understanding the factors that determine the immunodominance hierarchy within the influenza-specific memory B cell pool, and how to enrich the B cell responses to immunization with those that target conserved and neutralizing epitopes on HA, and thus provide broader cross-protection.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was funded in part by National Institutes of Health (NIH)/National Institute of Allergy and Infectious Diseases (NIAID) U19-AI057266 with ARRA supplement funding U19 AI057266-06S2 (R. Ahmed), by NIH/NIAID HHSN266200700006C Center of Excellence for Influenza Research and Surveillance (R. Ahmed), by the Northeast Biodefense Center U54-AI057158-Lipkin (R. Ahmed).
References
Ahmed, R., and Gray, D. (1996). Immunological memory and protective immunity: understanding their relation. Science 272, 54–60.
Banzhoff, A., Pellegrini, M., Del Giudice, G., Fragapane, E., Groth, N., and Podda, A. (2008). MF59-adjuvanted vaccines for seasonal and pandemic influenza prophylaxis. Influenza Other Respi. Viruses 2, 243–249.
Caton, A. J., Brownlee, G. G., Yewdell, J. W., and Gerhard, W. (1982). The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype). Cell 31, 417–427.
Chowell, G., Bertozzi, S. M., Colchero, M. A., Lopez-Gatell, H., Alpuche-Aranda, C., Hernandez, M., and Miller, M. A. (2009). Severe respiratory disease concurrent with the circulation of H1N1 influenza. N. Engl. J. Med. 361, 674–679.
Corti, D., Suguitan, A. L. Jr., Pinna, D., Silacci, C., Fernandez-Rodriguez, B. M., Vanzetta, F., Santos, C., Luke, C. J., Torres-Velez, F. J., Temperton, N. J., Weiss, R. A., Sallusto, F., Subbarao, K., and Lanzavecchia, A. (2010). Heterosubtypic neutralizing antibodies are produced by individuals immunized with a seasonal influenza vaccine. J. Clin. Invest. 120, 1663–1673.
Corti, D., Voss, J., Gamblin, S. J., Codoni, G., Macagno, A., Jarrossay, D., Vachieri, S. G., Pinna, D., Minola, A., Vanzetta, F., Silacci, C., Fernandez-Rodriguez, B. M., Agatic, G., Bianchi, S., Giacchetto-Sasselli, I., Calder, L., Sallusto, F., Collins, P., Haire, L. F., Temperton, N., Langedijk, J. P., Skehel, J. J., and Lanzavecchia, A. (2011). A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 333, 850–856.
Dawood, F. S., Jain, S., Finelli, L., Shaw, M. W., Lindstrom, S., Garten, R. J., Gubareva, L. V., Xu, X., Bridges, C. B., and Uyeki, T. M. (2009). Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N. Engl. J. Med. 360, 2605–2615.
Ekiert, D. C., Bhabha, G., Elsliger, M. A., Friesen, R. H., Jongeneelen, M., Throsby, M., Goudsmit, J., and Wilson, I. A. (2009). Antibody recognition of a highly conserved influenza virus epitope. Science 324, 246–251.
Ekiert, D. C., Friesen, R. H., Bhabha, G., Kwaks, T., Jongeneelen, M., Yu, W., Ophorst, C., Cox, F., Korse, H. J., Brandenburg, B., Vogels, R., Brakenhoff, J. P., Kompier, R., Koldijk, M. H., Cornelissen, L. A., Poon, L. L., Peiris, M., Koudstaal, W., Wilson, I. A., and Goudsmit, J. (2011). A highly conserved neutralizing epitope on group 2 influenza A viruses. Science 333, 843–850.
Gerhard, W. (2001). The role of the antibody response in influenza virus infection. Curr. Top. Microbiol. Immunol. 260, 171–190.
Han, T., and Marasco, W. A. (2011). Structural basis of influenza virus neutralization. Ann. N. Y. Acad. Sci. 1217, 178–190.
Hancock, K., Veguilla, V., Lu, X., Zhong, W., Butler, E. N., Sun, H., Liu, F., Dong, L., DeVos, J. R., Gargiullo, P. M., Brammer, T. L., Cox, N. J., Tumpey, T. M., and Katz, J. M. (2009). Cross-reactive antibody responses to the 2009 pandemic H1N1 influenza virus. N. Engl. J. Med. 361, 1945–1952.
Jain, S., Kamimoto, L., Bramley, A. M., Schmitz, A. M., Benoit, S. R., Louie, J., Sugerman, D. E., Druckenmiller, J. K., Ritger, K. A., Chugh, R., Jasuja, S., Deutscher, M., Chen, S., Walker, J. D., Duchin, J. S., Lett, S., Soliva, S., Wells, E. V., Swerdlow, D., Uyeki, T. M., Fiore, A. E., Olsen, S. J., Fry, A. M., Bridges, C. B., and Finelli, L. (2009). Hospitalized patients with 2009 H1N1 influenza in the United States. N. Engl. J. Med. 361, 1935–1944.
Khurana, S., Chearwae, W., Castellino, F., Manischewitz, J., King, L. R., Honorkiewicz, A., Rock, M. T., Edwards, K. M., Del Giudice, G., Rappuoli, R., and Golding, H. (2010). Vaccines with MF59 adjuvant expand the antibody repertoire to target protective sites of pandemic avian H5N1 influenza virus. Sci. Transl. Med. 2, 15ra15.
Khurana, S., Verma, N., Yewdell, J. W., Hilbert, A. K., Castellino, F., Lattanzi, M., Del Giudice, G., Rappuoli, R., and Golding, H. (2011). MF59 adjuvant enhances diversity and affinity of antibody-mediated immune response to pandemic influenza vaccines. Sci. Transl. Med. 3, 85ra48.
Krause, J. C., Tsibane, T., Tumpey, T. M., Huffman, C. J., Basler, C. F., and Crowe, J. E. Jr. (2011). A broadly neutralizing human monoclonal antibody that recognizes a conserved, novel epitope on the globular head of the influenza H1N1 virus hemagglutinin. J. Virol. 85, 10905–10908.
Ledgerwood, J. E., Wei, C. J., Hu, Z., Gordon, I. J., Enama, M. E., Hendel, C. S., McTamney, P. M., Pearce, M. B., Yassine, H. M., Boyington, J. C., Bailer, R., Tumpey, T. M., Koup, R. A., Mascola, J. R., Nabel, G. J., and Graham, B. S. (2011). DNA priming and influenza vaccine immunogenicity: two phase 1 open label randomised clinical trials. Lancet. Infect. Dis. 11, 916–924.
Manz, R. A., Arce, S., Cassese, G., Hauser, A. E., Hiepe, F., and Radbruch, A. (2002). Humoral immunity and long-lived plasma cells. Curr. Opin. Immunol. 14, 517–521.
Nabel, G. J., and Fauci, A. S. (2010). Induction of unnatural immunity: prospects for a broadly protective universal influenza vaccine. Nat. Med. 16, 1389–1391.
Okuno, Y., Isegawa, Y., Sasao, F., and Ueda, S. (1993). A common neutralizing epitope conserved between the hemagglutinins of influenza A virus H1 and H2 strains. J. Virol. 67, 2552–2558.
Okuno, Y., Matsumoto, K., Isegawa, Y., and Ueda, S. (1994). Protection against the mouse-adapted A/FM/1/47 strain of influenza A virus in mice by a monoclonal antibody with cross-neutralizing activity among H1 and H2 strains. J. Virol. 68, 517–520.
Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., and Ferrin, T. E. (2004). UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612.
Pinna, D., Corti, D., Jarrossay, D., Sallusto, F., and Lanzavecchia, A. (2009). Clonal dissection of the human memory B-cell repertoire following infection and vaccination. Eur. J. Immunol. 39, 1260–1270.
Sasaki, S., Jaimes, M. C., Holmes, T. H., Dekker, C. L., Mahmood, K., Kemble, G. W., Arvin, A. M., and Greenberg, H. B. (2007). Comparison of the influenza virus-specific effector and memory B-cell responses to immunization of children and adults with live attenuated or inactivated influenza virus vaccines. J. Virol. 81, 215–228.
Skehel, J. J., and Wiley, D. C. (2000). Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69, 531–569.
Skowronski, D. M., Hottes, T. S., McElhaney, J. E., Janjua, N. Z., Sabaiduc, S., Chan, T., Gentleman, B., Purych, D., Gardy, J., Patrick, D. M., Brunham, R. C., De Serres, G., and Petric, M. (2011). Immuno-epidemiologic correlates of pandemic H1N1 surveillance observations: higher antibody and lower cell-mediated immune responses with advanced age. J. Infect. Dis. 203, 158–167.
Slifka, M. K., and Ahmed, R. (1998). Long-lived plasma cells: a mechanism for maintaining persistent antibody production. Curr. Opin. Immunol. 10, 252–258.
Smith, G. J., Bahl, J., Vijaykrishna, D., Zhang, J., Poon, L. L., Chen, H., Webster, R. G., Peiris, J. S., and Guan, Y. (2009). Dating the emergence of pandemic influenza viruses. Proc. Natl. Acad. Sci. U.S.A. 106, 11709–11712.
Stephenson, I., Bugarini, R., Nicholson, K. G., Podda, A., Wood, J. M., Zambon, M. C., and Katz, J. M. (2005). Cross-reactivity to highly pathogenic avian influenza H5N1 viruses after vaccination with nonadjuvanted and MF59-adjuvanted influenza A/Duck/Singapore/97 (H5N3) vaccine: a potential priming strategy. J. Infect. Dis. 191, 1210–1215.
Sui, J., Hwang, W. C., Perez, S., Wei, G., Aird, D., Chen, L. M., Santelli, E., Stec, B., Cadwell, G., Ali, M., Wan, H., Murakami, A., Yammanuru, A., Han, T., Cox, N. J., Bankston, L. A., Donis, R. O., Liddington, R. C., and Marasco, W. A. (2009). Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat. Struct. Mol. Biol. 16, 265–273.
Sui, J., Sheehan, J., Hwang, W. C., Bankston, L. A., Burchett, S. K., Huang, C. Y., Liddington, R. C., Beigel, J. H., and Marasco, W. A. (2011). Wide prevalence of heterosubtypic broadly neutralizing human anti-influenza A antibodies. Clin. Infect. Dis. 52, 1003–1009.
Throsby, M., van den Brink, E., Jongeneelen, M., Poon, L. L., Alard, P., Cornelissen, L., Bakker, A., Cox, F., van Deventer, E., Guan, Y., Cinatl, J., ter Meulen, J., Lasters, I., Carsetti, R., Peiris, M., de Kruif, J., and Goudsmit, J. (2008). Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS ONE 3, e3942. doi:10.1371/journal.pone.0003942
Treanor, J. J., Campbell, J. D., Zangwill, K. M., Rowe, T., and Wolff, M. (2006). Safety and immunogenicity of an inactivated subvirion influenza A (H5N1) vaccine. N. Engl. J. Med. 354, 1343–1351.
Wang, T. T., Tan, G. S., Hai, R., Pica, N., Petersen, E., Moran, T. M., and Palese, P. (2010a). Broadly protective monoclonal antibodies against H3 influenza viruses following sequential immunization with different hemagglutinins. PLoS Pathog. 6, e1000796. doi:10.1371/journal.ppat.1000796
Wang, T. T., Tan, G. S., Hai, R., Pica, N., Ngai, L., Ekiert, D. C., Wilson, I. A., Garcia-Sastre, A., Moran, T. M., and Palese, P. (2010b). Vaccination with a synthetic peptide from the influenza virus hemagglutinin provides protection against distinct viral subtypes. Proc. Natl. Acad. Sci. U.S.A. 107, 18979–18984.
Webby, R. J., and Webster, R. G. (2003). Are we ready for pandemic influenza? Science 302, 1519–1522.
Wei, C. J., Boyington, J. C., McTamney, P. M., Kong, W. P., Pearce, M. B., Xu, L., Andersen, H., Rao, S., Tumpey, T. M., Yang, Z. Y., and Nabel, G. J. (2010). Induction of broadly neutralizing H1N1 influenza antibodies by vaccination. Science 329, 1060–1064.
Whittle, J. R., Zhang, R., Khurana, S., King, L. R., Manischewitz, J., Golding, H., Dormitzer, P. R., Haynes, B. F., Walter, E. B., Moody, M. A., Kepler, T. B., Liao, H. X., and Harrison, S. C. (2011). Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc. Natl. Acad. Sci. U.S.A. 108, 14216–14221.
Wilson, I. A., and Cox, N. J. (1990). Structural basis of immune recognition of influenza virus hemagglutinin. Annu. Rev. Immunol. 8, 737–771.
Wrammert, J., Koutsonanos, D., Li, G. M., Edupuganti, S., Sui, J., Morrissey, M., McCausland, M., Skountzou, I., Hornig, M., Lipkin, W. I., Mehta, A., Razavi, B., Del Rio, C., Zheng, N. Y., Lee, J. H., Huang, M., Ali, Z., Kaur, K., Andrews, S., Amara, R. R., Wang, Y., Das, S. R., O’Donnell, C. D., Yewdell, J. W., Subbarao, K., Marasco, W. A., Mulligan, M. J., Compans, R., Ahmed, R., and Wilson, P. C. (2011). Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J. Exp. Med. 208, 181–193.
Wrammert, J., Smith, K., Miller, J., Langley, W. A., Kokko, K., Larsen, C., Zheng, N. Y., Mays, I., Garman, L., Helms, C., James, J., Air, G. M., Capra, J. D., Ahmed, R., and Wilson, P. C. (2008). Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature 453, 667–671.
Keywords: memory, B cells, cross-reactive, influenza, epitopes
Citation: Ellebedy AH and Ahmed R (2012) Re-engaging cross-reactive memory B cells: the influenza puzzle. Front. Immun. 3:53. doi: 10.3389/fimmu.2012.00053
Received: 11 January 2012; Accepted: 02 March 2012;
Published online: 30 March 2012.
Edited by:
Katja Fink, Agency for Science, Technology and Research A*Star, SingaporeReviewed by:
Katja Fink, Agency for Science, Technology and Research A*Star, SingaporeFrances E. Lee, University of Rochester Medical Center, USA
Copyright: © 2012 Ellebedy and Ahmed. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Rafi Ahmed, Emory University, G211 Rollins Research Building, 1510 Clifton Road, Atlanta, GA 30322, USA. e-mail:cmFobWVkQGVtb3J5LmVkdQ==
