- 1 Michael Smith Laboratories, University of British Columbia, Vancouver, BC, Canada
- 2 Department of Microbiology and Immunology, University of British Columbia, Vancouver, BC, Canada
- 3 Department of Biochemistry and Molecular Biology, University of British Columbia, Vancouver, BC, Canada
For millions of years, microbes have coexisted with eukaryotic cells at the mucosal surfaces of vertebrates in a complex, yet usually harmonious symbiosis. An ever-expanding number of reports describe how eliminating or shifting the intestinal microbiota has profound effects on the development and functionality of the mucosal and systemic immune systems. Here, we examine some of the mechanisms by which bacterial signals affect immune homeostasis. Focusing on the strategies that microbes use to keep our immune system healthy, as opposed to trying to correct the immune imbalances caused by dysbiosis, may prove to be a more astute and efficient way of treating immune-mediated disease.
Introduction
Higher organisms benefit from their microbiota, which offers nutritional and physiological advantages to the host in exchange for a nutrient-rich habitat. Microbes (bacteria, archaea, viruses, fungi, protozoans, etc.) reside on all mucosal surfaces of the body, with the gut harboring by far the most numerous population. Separating the microbial cells from the host is the intestinal barrier, which serves as an interface for many complex interactions. Two major components of the intestinal barrier are the intestinal epithelium and the gut associated lymphoid tissue (GALT). This tissue has the challenging dual task of selectively absorbing nutrients from the intestinal lumen, while preventing microbial entry and infection. The majority of microbes that reside in the intestine are commensals and do not pose a threat to the host under normal conditions. Thus, the host’s immune system must learn to tolerate this enormous commensal antigenic load. One of the strategies that the host utilizes to avoid an inflammatory response against the microbiota is to remain “ignorant” to a portion of the microbial antigens, by using the intestinal barrier, including the mucous layer and immunoglobulin (Ig) A, to maintain these antigens at a distance to the mucosal surface (Hansson and Johansson, 2010; Hooper and Macpherson, 2010). However, it is now clear that there is also constant communication between microbial cells or signals and host cell receptors, and that many of these interactions lead to the development of immune tolerance toward bacterial antigens (Hooper and Macpherson, 2010; Geuking et al., 2011).
Traditionally, it has been accepted that the evolution of the mammalian immune system has been driven by selective pressures imposed by pathogenic bacteria (Matsunaga and Rahman, 1998). For example, the emergence of jaws in cartilaginous fish matches the appearance of antigen receptor specificity and immunological memory in B and T cells, as well as the major histocompatibility complexes (MHC) I and II, all characteristic of the adaptive immune system (Agrawal et al., 1998). Immunologists have speculated that the jaw gave organisms the ability to ingest harder food sources that could damage the gut mucosa, likely increasing their risk for infections. The nutritional advantages of having a jaw allowed organisms to augment their body size and life expectancy, increasing their risk for repeated microbial attack and the necessity to develop immunological memory (Matsunaga and Rahman, 1998). While this may be true for the emergence of some features of the adaptive immune system, this view may not apply to many other components of the immune system. First, it does not take into account that the vast majority of interactions that occur between the microbiota and the mammalian immune system involve non-pathogenic bacteria. More importantly, commensals, as opposed to pathogens, do not limit the host’s lifespan, reproducibility, and consequent vertical gene transfer, and actually probably enhance it. Thus, it is more likely that commensal microorganisms pose a stronger evolutionary pressure on the host’s immune system than pathogens, a view that we share with others (McFall-Ngai, 2007; Lee and Mazmanian, 2010).
The goal of this minireview is to describe some of the diverse mechanisms of commensal–host communication in the mammalian intestine and to explore the effect that these have in immune homeostasis within the intestine and systemically.
Immune Consequences of an Axenic Life
The generation of germ-free mice has given us great insight into the immune effects of a microbe-free life, the development of the immune system during early microbial colonization, and the effects of specific bacterial groups and species on the local and systemic immune system.
The immune system is poorly developed in the absence of microbes. Structurally, the aggregated lymphoid structures of the GALT: mesenteric lymph nodes (MLNs), isolated lymphoid follicles, cryptopatches, and Peyer’s patches are all underdeveloped in germ-free mice (Mazmanian et al., 2005; Bouskra et al., 2008). These mice exhibit decreased amounts of most cytokines, CD4+ T helper cells, FoxP3+ Tregs, B cells, Th17 cells, IgA, antimicrobial peptides, MHC class II, etc. (Moreau et al., 1978; Dobber et al., 1992; Strauch et al., 2005; Ivanov et al., 2008; Niess et al., 2008; Gaboriau-Routhiau et al., 2009; Round and Mazmanian, 2009), and inoculating them with a single species of bacteria can revert many of these defects (Umesaki et al., 1995; Mazmanian et al., 2005). Gut bacteria also have critical effects on the development of intestinal epithelial cells (IECs). IECs are the first cells to encounter the microbiota and are able to integrate most microbial signals and dictate the tone of the immune response of the underlying lamina propria immune cells (Rescigno, 2011). IECs express microbial pattern recognition receptors (PRR) that bind to microbial signals and upregulate the expression of mucus, cytokines, and other immune components (Rescigno, 2011). In the absence of bacteria, not only do these cells have a low expression of PRRs (Willing and Van Kessel, 2007), they also fail to develop their typical microvilli pattern and show a slow cell turnover rate (Abrams et al., 1963).
Not surprisingly, the functionality of the immune system in axenic mice is very poor as well. Germ-free and even mono-colonized mice do not develop oral tolerance (Rask et al., 2005). Germ-free mice and antibiotic-treated mice are significantly more susceptible to infectious disease from several intestinal pathogens, including Shigella flexneri (Sprinz et al., 1961), Citrobacter rodentium (Wlodarska et al., 2011), Listeria monocytogenes (Zachar and Savage, 1979), and Salmonella typhimurium (Nardi et al., 1989; Sekirov et al., 2008; Ferreira et al., 2011). The microbiota can prevent or ameliorate infection by direct microbial antagonism or by indirectly promoting appropriate host immune defenses. Direct microbial effects, also defined as “colonization resistance” (van der Waaij et al., 1971), include host receptor and nutrient competition, and secretion of antimicrobial substances (Sekirov and Finlay, 2009). The importance of these effects were demonstrated in a study where transfer of a normal microflora, in the absence of T and B cells, cleared infection with S. typhimurium (Endt et al., 2010). This study highlights the importance of the competitive pressures exerted by the microbiota as well as the role of the innate immune response in enteric infections. In another study, a fecal transplant from a mouse strain resistant to C. rodentium was sufficient to delay pathogen colonization and reduce mortality in a mouse strain lethally susceptible to infection. In addition, antibiotic therapy increased disease severity in resistance mice, suggesting that the difference in infection susceptibility between the two mice strains can be explained, at least in part, by differences in microbiota composition (Willing et al., 2011).
A normal microbiota has also been shown to help clear pathogens by stimulating several host immune effectors. In one study, oral treatment with peptidoglycan from the intestinal microbiota was effective in preventing pneumococcal sepsis in mice. Levels of this bacterial membrane component in serum correlated with systemic neutrophil killing capacity and activation of the innate immune receptor NOD1, indicating that signals from the microbiota translocate across the intestinal barrier and interact with systemic immune cells (Clarke et al., 2010). Another example of how microbial-derived signals regulate the systemic immune response comes from a study that showed that administration of the TLR5 ligand, flagellin, to antibiotic-treated mice inhibited colonization with vancomycin resistant Enterococcus (VRE). Intraperitoneal injection of flagellin upregulated the expression of the bactericidal lectin REGIIIγ in IECs and Paneth cells in the small intestine via activation of TLR5 in hematopoietic cells and secretion of IL-22 (Kinnebrew et al., 2010). Flagellin stimulation has also been shown to induce differentiation of non-specific IgA+ plasma cells via activation of TLR5 in intestinal dendritic cells (Uematsu et al., 2008). Colonization with the microorganism Bacteroides fragilis is sufficient to prevent murine experimental colitis induced by Helicobacter hepaticus. B. fragilis is coated with polysaccharide A (PSA) and this capsular component was shown to suppress IL-17, increase IL-10 production and ameliorate disease in this model (Mazmanian et al., 2008).
The Commensal Microbiota Balances Pro and Anti-Inflammatory Immune Mechanisms
The Th17 immune response provides a good example of how the microbiota mediates the balance between immune homeostasis and uncontrolled inflammation. Overproduction of Th17 cytokines IL-17 and IL-23 is associated with colitis (Atarashi et al., 2008; Buonocore et al., 2010) and autoimmunity (Wen et al., 2008; Lee et al., 2010; Wu et al., 2010); however, Th17 cells are critical in controlling extracellular bacterial and fungal infections (Smith et al., 2007; Wu et al., 2010). Induction of Th17 immunity is crucial to prevent infection with C. rodentium, a murine pathogen that induces similar inflammation to enteropathogenic E. coli in humans (Wu et al., 2010). Thus, the right intensity of Th17 response must be achieved in order to prevent microbial attack and avoid uncontrolled inflammation. Segmented filamentous bacteria (SFB) are an unclassified species that resides in the murine ileum and has been shown to be a potent stimulator of the Th17 response (Gaboriau-Routhiau et al., 2009; Wu et al., 2010). Colonization with SFB induced the upregulation of cytokines, antimicrobial peptides, and serum amyloid A (SAA), an acute phase protein secreted during inflammation. SAA is believed to promote the Th17 differentiation of CD4+ T cells (Ivanov et al., 2009; Ather et al., 2011). Another mechanism that leads to Th17 differentiation is the production of commensal-derived adenosine 5′ triphosphate (ATP). ATP in the lamina propria stimulates dendritic cells to produce IL-6 and TGF-β, two cytokines necessary for Th17 cell expansion (Atarashi et al., 2008). Not surprisingly, microbial signals are also involved in limiting the Th17 immune response. IECs produce IL-25, which inhibits IL-23 production and prevents Th17 differentiation in response to the commensal flora (Zaph et al., 2008). In addition, oral administration of purified PSA from B. fragilis downregulated the Th17 response and induced propagation of Tregs in mice that develop autoimmune encephalomyelitis. This treatment was sufficient to prevent disease in this animal model (Ochoa-Reparaz et al., 2010). Collectively, these findings show how the microbiota produces inflammatory and tolerogenic signals in order to achieve immune homeostasis, a state that is favorable for both the microbiota and the host.
It is still unknown how certain microbes induce a pro-inflammatory response and others a tolerogenic one, but due to the central role that Treg cells have in regulating inflammatory responses (Figure 1), one could speculate that microbes manipulate the Treg cell response as a strategy to regulate immune responses towards them. But how do Treg cells perform such task? These cells are selected in the thymus for their ability to suppress T cells with a high affinity to self-MHC molecules, thus preventing autoimmune responses, but it is still unknown how these cells differentiate between pathogens or commensal pathogen signals. One explanation may be that the antigens from the commensal microbiota are used to train naïve T cells to differentiate into Treg cells. A recent study provides evidence that this may in fact occur. Analysis of several repertoires of T cell receptor (TCR) α chain revealed that the TCRs from FoxP3+ Treg cells within the colonic lamina propria are highly heterogeneous compared to Treg cells from effector/memory T cells in the lamina propria and from Treg cells from secondary lymphoid organs. Moreover, the study showed that the mouse microbiota was essential for the induction of this particular population of colonic Treg cells from naïve T cells, implying that there may be post-thymic mechanisms of immune cell education that occur peripherally via interactions with the commensal microbiota (Lathrop et al., 2011). This implies that the immune system may have evolved to rely on the microbiota to complete the training of a specific population of immune cells.
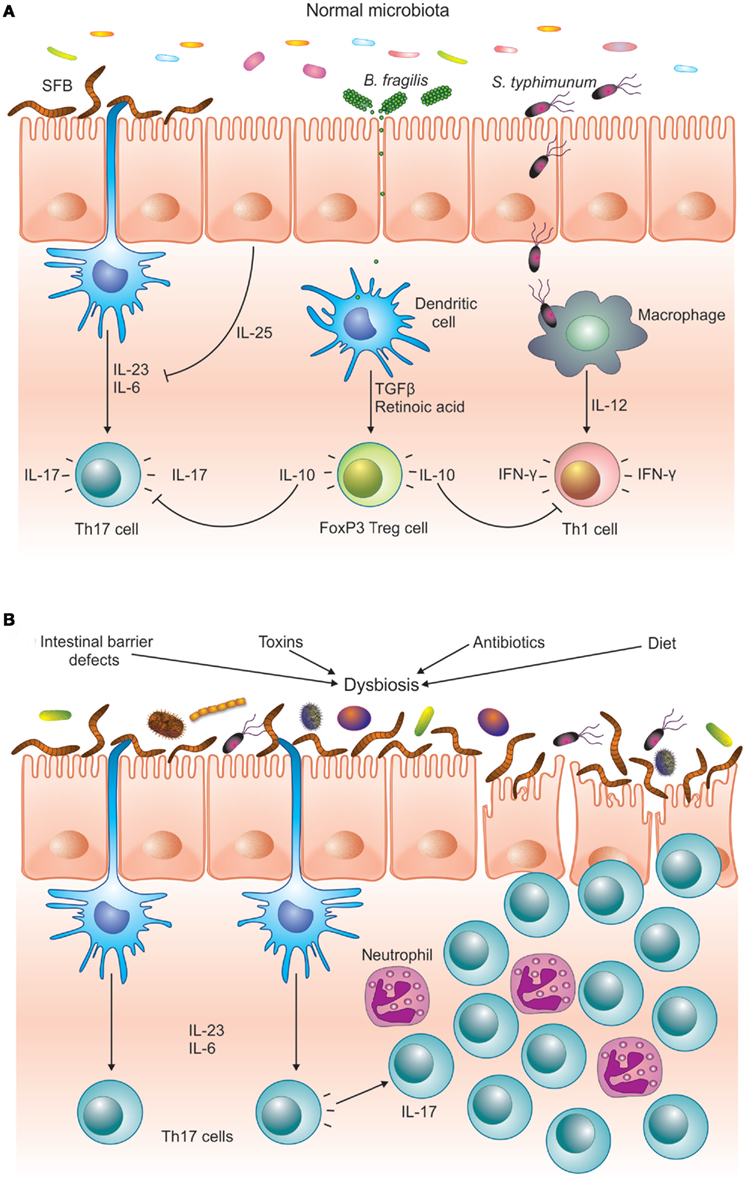
Figure 1. The microbiota helps to maintain immune homeostasis by stimulating different arms of the T cell response. (A) SFB is a potent inducer of Th17 cells, whereas pathogens like S. typhimurium induce an effector Th1 response. PSA from B. fragilis stimulates the differentiation of FoxP3+ T cells, which downregulate the pro-inflammatory Th1 and Th17 responses. (B) Different environmental and host-derived factors are known to cause dysbiosis. Imbalances in the intestinal microbiota can favor Th17 pro-inflammatory T cells over regulatory FoxP3+ T cells, inducing inflammatory response that can damage the intestinal epithelium. In a genetically predisposed host this can lead to chronic inflammatory disease, such as IBD or autoimmunity.
Dissecting the Gut Microbiota
Although our knowledge of the immune effects caused by the microbiota is growing, much less is known about what constitutes a normal microbiota and which groups of microorganisms are beneficial or detrimental to immune homeostasis. Sequencing-based studies have substantially increased our knowledge of the ecology of the mammalian intestine, although it is safe to assume that we are likely at the early stages of comprehending this complex population.
Analyses of the human gut microbiota have yielded several important findings. First, although there is great inter-individual variability at the species level, there appears to be low variability between individuals at the phylum level (Eckburg et al., 2005; Turnbaugh et al., 2009). The human gut is colonized by mainly three phyla: Firmicutes, Bacteroidetes, and Actinobacteria. The most predominant bacterial families are Ruminococcaceae, Lachnospiraceae, Eubacteriaceae, and Bacteroidaceae, in what has been defined as the “core members of the microbiota” (Tap et al., 2009). Genetics and environmental factors, such as birth delivery method, diet, and antibiotic use have an important role in establishing the composition of the microbiota (Ley et al., 2006; Penders et al., 2006; Adlerberth, 2008; Benson et al., 2010; Willing et al., 2010). Compositional studies have also shown that certain members of the flora may be indicative of disease states. For example, the bacterium Faecalibacterium prausnitzii, a member of the Ruminococcaceae family is absent in samples from Crohn’s disease (CD) patients (Sokol et al., 2008; Willing et al., 2009). Moreover, amelioration of disease with the anti TNF antibody, infliximab, allows F. prausnitzii to recolonize (Swidsinski et al., 2008). Furthermore, F. prausnitzii levels do not change in patients with ulcerative colitis, a disease characterized by Th2-type inflammation, suggesting that this microorganism is particularly susceptible to Th1 (predominant in CD) and not Th2-type inflammation.
A recent study analyzed the human fecal microbiota of 39 individuals from 6 different developed countries and found that there are three distinct ecosystems in the human gut, or “enterotypes.” The genus Bacteroides is highly abundant in enterotype 1, Prevotella is very common for enterotype type 2 and Ruminococcus in enterotype 3. Although the functional implications of all enterotypes are still under study, it seems that each enterotype favors the synthesis of certain metabolic products. Enterotype 1 ecosystem favors the production of enzymes involved in biotin synthesis, whereas enterotype 2 species produce more enzymes for thiamine synthesis (Arumugam et al., 2011). Another study examined the fecal microbiota of 98 individuals and showed that samples clustered into two of the previously described enterotypes: Bacteroides-rich and Prevotella-rich ecosystems. Interestingly, the authors were able to correlate these enterotypes with long-term diets. Individuals with high animal fat in their diet clustered in the Bacteroides enterotype, whereas diets high in carbohydrate clustered in the Prevotella enterotype. A short-term intervention with a carbohydrate-rich diet did not cause a switch from Bacteroides into Prevotella enterotypes, suggesting that they reflect long-term diet conditions (Wu et al., 2011). There are still many unanswered questions regarding the correlation between intestinal enterotypes and the immune status of the host, as well as the possibility that there are more than three enterotypes or subenterotypes. However, the compositional classification of the human gut microbiota into functional ecosystems has enormous potential as a marker of disease and as a treatment.
Final Remarks
The immune environment of the gut is characterized by tolerance. To achieve this state, host immune cells must learn to exhibit a tolerogenic behavior toward most microbial antigens. Thus, besides protecting the host from microbial attack, post-natal bacterial colonization likely serves the immune system with another important function: to be used as the second “training ground” for some naïve immune cells. Although this view poses a caveat to the traditional view of the main function of immune system – to distinguish between self and non-self – it may explain why the majority of the bacterial antigens in the gut are perceived as self-antigens.
More mechanisms on how and what components of the commensal microbiota affect mucosal and systemic immunity will undoubtedly continue to be found. This will not only expand our view of our microbiota as an integral part of our bodies (Gill et al., 2010), but it also opens the real possibility of manipulating specific functions of the microbiota to tune immune defects that cause infectious, inflammatory, or autoimmune disease. Learning from the strategies that the microbiota use to balance the different arms of the immune response and using them to overcome disease may prove to be more efficient than modifying the immune imbalances that originate from defective genes and microbial dysbiosis.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Marta Wlodarska, Dr. Benjamin Willing, and Dr. Jerôme Tourret for the critical revision of this manuscript, and Fern Ness for the artwork for Figure 1. The Finlay laboratory is supported by operating grants from Canadian Institutes of Heath Research (CIHR), the Crohn and Colitis Foundation of Canada (CCFC), and the Howard Hughes Medical Institute (HHMI).
References
Abrams, G. D., Bauer, H., and Sprinz, H. (1963). Influence of the normal flora on mucosal morphology and cellular renewal in the ileum. A comparison of germ-free and conventional mice. Lab. Invest. 12, 355–364.
Adlerberth, I. (2008). Factors influencing the establishment of the intestinal microbiota in infancy. Nestle Nutr. Workshop Ser. Pediatr. Program. 62, 13–29; discussion 29–33.
Agrawal, A., Eastman, Q. M., and Schatz, D. G. (1998). Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature 394, 744–751.
Arumugam, M., Raes, J., Pelletier, E., Le Paslier, D., Yamada, T., Mende, D. R., Fernandes, G. R., Tap, J., Bruls, T., Batto, J. M., Bertalan, M., Borruel, N., Casellas, F., Fernandez, L., Gautier, L., Hansen, T., Hattori, M., Hayashi, T., Kleerebezem, M., Kurokawa, K., Leclerc, M., Levenez, F., Manichanh, C., Nielsen, H. B., Nielsen, T., Pons, N., Poulain, J., Qin, J., Sicheritz-Ponten, T., Tims, S., Torrents, D., Ugarte, E., Zoetendal, E. G., Wang, J., Guarner, F., Pedersen, O., De Vos, W. M., Brunak, S., Dore, J., Antolin, M., Artiguenave, F., Blottiere, H. M., Almeida, M., Brechot, C., Cara, C., Chervaux, C., Cultrone, A., Delorme, C., Denariaz, G., Dervyn, R., Foerstner, K. U., Friss, C., Van De Guchte, M., Guedon, E., Haimet, F., Huber, W., Van Hylckama-Vlieg, J., Jamet, A., Juste, C., Kaci, G., Knol, J., Lakhdari, O., Layec, S., Le Roux, K., Maguin, E., Merieux, A., Melo Minardi, R., M’Rini, C., Muller, J., Oozeer, R., Parkhill, J., Renault, P., Rescigno, M., Sanchez, N., Sunagawa, S., Torrejon, A., Turner, K., Vandemeulebrouck, G., Varela, E., Winogradsky, Y., Zeller, G., Weissenbach, J., Ehrlich, S. D., and Bork, P. (2011). Enterotypes of the human gut microbiome. Nature 473, 174–180.
Atarashi, K., Nishimura, J., Shima, T., Umesaki, Y., Yamamoto, M., Onoue, M., Yagita, H., Ishii, N., Evans, R., Honda, K., and Takeda, K. (2008). ATP drives lamina propria T(H)17 cell differentiation. Nature 455, 808–812.
Ather, J. L., Ckless, K., Martin, R., Foley, K. L., Suratt, B. T., Boyson, J. E., Fitzgerald, K. A., Flavell, R. A., Eisenbarth, S. C., and Poynter, M. E. (2011). Serum amyloid A activates the NLRP3 inflammasome and promotes Th17 allergic asthma in mice. J. Immunol. 187, 64–73.
Benson, A. K., Kelly, S. A., Legge, R., Ma, F., Low, S. J., Kim, J., Zhang, M., Oh, P. L., Nehrenberg, D., Hua, K., Kachman, S. D., Moriyama, E. N., Walter, J., Peterson, D. A., and Pomp, D. (2010). Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc. Natl. Acad. Sci. U.S.A. 107, 18933–18938.
Bouskra, D., Brezillon, C., Berard, M., Werts, C., Varona, R., Boneca, I. G., and Eberl, G. (2008). Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature 456, 507–510.
Buonocore, S., Ahern, P. P., Uhlig, H. H., Ivanov, Ii., Littman, D. R., Maloy, K. J., and Powrie, F. (2010). Innate lymphoid cells drive interleukin-23-dependent innate intestinal pathology. Nature 464, 1371–1375.
Clarke, T. B., Davis, K. M., Lysenko, E. S., Zhou, A. Y., Yu, Y., and Weiser, J. N. (2010). Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat. Med. 16, 228–231.
Dobber, R., Hertogh-Huijbregts, A., Rozing, J., Bottomly, K., and Nagelkerken, L. (1992). The involvement of the intestinal microflora in the expansion of CD4+ T cells with a naive phenotype in the periphery. Dev. Immunol. 2, 141–150.
Eckburg, P. B., Bik, E. M., Bernstein, C. N., Purdom, E., Dethlefsen, L., Sargent, M., Gill, S. R., Nelson, K. E., and Relman, D. A. (2005). Diversity of the human intestinal microbial flora. Science 308, 1635–1638.
Endt, K., Stecher, B., Chaffron, S., Slack, E., Tchitchek, N., Benecke, A., Van Maele, L., Sirard, J. C., Mueller, A. J., Heikenwalder, M., Macpherson, A. J., Strugnell, R., Von Mering, C., and Hardt, W. D. (2010). The microbiota mediates pathogen clearance from the gut lumen after non-typhoidal Salmonella diarrhea. PLoS Pathog. 6, e1001097. doi:10.1371/journal.ppat.1001097
Ferreira, R. B., Gill, N., Willing, B. P., Antunes, L. C., Russell, S. L., Croxen, M. A., and Finlay, B. B. (2011). The intestinal microbiota plays a role in Salmonella-induced colitis independent of pathogen colonization. PLoS ONE 6, e20338. doi:10.1371/journal.pone.0020338
Gaboriau-Routhiau, V., Rakotobe, S., Lecuyer, E., Mulder, I., Lan, A., Bridonneau, C., Rochet, V., Pisi, A., De Paepe, M., Brandi, G., Eberl, G., Snel, J., Kelly, D., and Cerf-Bensussan, N. (2009). The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity 31, 677–689.
Geuking, M. B., Cahenzli, J., Lawson, M. A., Ng, D. C., Slack, E., Hapfelmeier, S., Mccoy, K. D., and Macpherson, A. J. (2011). Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity 34, 794–806.
Gill, N., Wlodarska, M., and Finlay, B. B. (2010). The future of mucosal immunology: studying an integrated system-wide organ. Nat. Immunol. 11, 558–560.
Hansson, G. C., and Johansson, M. E. (2010). The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Gut Microbes 1, 51–54.
Hooper, L. V., and Macpherson, A. J. (2010). Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat. Rev. Immunol. 10, 159–169.
Ivanov, I. I., Atarashi, K., Manel, N., Brodie, E. L., Shima, T., Karaoz, U., Wei, D., Goldfarb, K. C., Santee, C. A., Lynch, S. V., Tanoue, T., Imaoka, A., Itoh, K., Takeda, K., Umesaki, Y., Honda, K., and Littman, D. R. (2009). Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498.
Ivanov, I. I., Frutos Rde, L., Manel, N., Yoshinaga, K., Rifkin, D. B., Sartor, R. B., Finlay, B. B., and Littman, D. R. (2008). Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe 4, 337–349.
Kinnebrew, M. A., Ubeda, C., Zenewicz, L. A., Smith, N., Flavell, R. A., and Pamer, E. G. (2010). Bacterial flagellin stimulates Toll-like receptor 5-dependent defense against vancomycin-resistant Enterococcus infection. J. Infect. Dis. 201, 534–543.
Lathrop, S. K., Bloom, S. M., Rao, S. M., Nutsch, K., Lio, C. W., Santacruz, N., Peterson, D. A., Stappenbeck, T. S., and Hsieh, C. S. (2011). Peripheral education of the immune system by colonic commensal microbiota. Nature 478, 250–254.
Lee, Y. K., and Mazmanian, S. K. (2010). Has the microbiota played a critical role in the evolution of the adaptive immune system? Science 330, 1768–1773.
Lee, Y. K., Menezes, J. S., Umesaki, Y., and Mazmanian, S. K. (2010). Microbes and health Sackler colloquium: proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc. Natl. Acad. Sci. U.S.A. 108(Suppl. 1), 4615–4622.
Ley, R. E., Peterson, D. A., and Gordon, J. I. (2006). Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124, 837–848.
Matsunaga, T., and Rahman, A. (1998). What brought the adaptive immune system to vertebrates? The jaw hypothesis and the seahorse. Immunol. Rev. 166, 177–186.
Mazmanian, S. K., Liu, C. H., Tzianabos, A. O., and Kasper, D. L. (2005). An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122, 107–118.
Mazmanian, S. K., Round, J. L., and Kasper, D. L. (2008). A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453, 620–625.
Moreau, M. C., Ducluzeau, R., Guy-Grand, D., and Muller, M. C. (1978). Increase in the population of duodenal immunoglobulin A plasmocytes in axenic mice associated with different living or dead bacterial strains of intestinal origin. Infect. Immun. 21, 532–539.
Nardi, R. M., Silva, M. E., Vieira, E. C., Bambirra, E. A., and Nicoli, J. R. (1989). Intragastric infection of germfree and conventional mice with Salmonella typhimurium. Braz. J. Med. Biol. Res. 22, 1389–1392.
Niess, J. H., Leithauser, F., Adler, G., and Reimann, J. (2008). Commensal gut flora drives the expansion of proinflammatory CD4 T cells in the colonic lamina propria under normal and inflammatory conditions. J. Immunol. 180, 559–568.
Ochoa-Reparaz, J., Mielcarz, D. W., Wang, Y., Begum-Haque, S., Dasgupta, S., Kasper, D. L., and Kasper, L. H. (2010). A polysaccharide from the human commensal Bacteroides fragilis protects against CNS demyelinating disease. Mucosal Immunol. 3, 487–495.
Penders, J., Thijs, C., Vink, C., Stelma, F. F., Snijders, B., Kummeling, I., Van Den Brandt, P. A., and Stobberingh, E. E. (2006). Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics 118, 511–521.
Rask, C., Evertsson, S., Telemo, E., and Wold, A. E. (2005). A full flora, but not monocolonization by Escherichia coli or lactobacilli, supports tolerogenic processing of a fed antigen. Scand. J. Immunol. 61, 529–535.
Rescigno, M. (2011). The intestinal epithelial barrier in the control of homeostasis and immunity. Trends Immunol. 32, 256–264.
Round, J. L., and Mazmanian, S. K. (2009). The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 9, 313–323.
Sekirov, I., and Finlay, B. B. (2009). The role of the intestinal microbiota in enteric infection. J. Physiol. (Lond.) 587, 4159–4167.
Sekirov, I., Tam, N. M., Jogova, M., Robertson, M. L., Li, Y. L., Lupp, C., and Finlay, B. B. (2008). Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect. Immun. 76, 4726–4736.
Smith, K., Mccoy, K. D., and Macpherson, A. J. (2007). Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin. Immunol. 19, 59–69.
Sokol, H., Pigneur, B., Watterlot, L., Lakhdari, O., Bermudez-Humaran, L. G., Gratadoux, J. J., Blugeon, S., Bridonneau, C., Furet, J. P., Corthier, G., Grangette, C., Vasquez, N., Pochart, P., Trugnan, G., Thomas, G., Blottiere, H. M., Dore, J., Marteau, P., Seksik, P., and Langella, P. (2008). Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. U.S.A. 105, 16731–16736.
Sprinz, H., Kundel, D. W., Dammin, G. J., Horowitz, R. E., Schneider, H., and Formal, S. B. (1961). The response of the germfree guinea pig to oral bacterial challenge with Escherichia coli and Shigella flexneri. Am. J. Pathol. 39, 681–695.
Strauch, U. G., Obermeier, F., Grunwald, N., Gurster, S., Dunger, N., Schultz, M., Griese, D. P., Mahler, M., Scholmerich, J., and Rath, H. C. (2005). Influence of intestinal bacteria on induction of regulatory T cells: lessons from a transfer model of colitis. Gut 54, 1546–1552.
Swidsinski, A., Loening-Baucke, V., Vaneechoutte, M., and Doerffel, Y. (2008). Active Crohn’s disease and ulcerative colitis can be specifically diagnosed and monitored based on the biostructure of the fecal flora. Inflamm. Bowel Dis. 14, 147–161.
Tap, J., Mondot, S., Levenez, F., Pelletier, E., Caron, C., Furet, J. P., Ugarte, E., Munoz-Tamayo, R., Paslier, D. L., Nalin, R., Dore, J., and Leclerc, M. (2009). Towards the human intestinal microbiota phylogenetic core. Environ. Microbiol. 11, 2574–2584.
Turnbaugh, P. J., Hamady, M., Yatsunenko, T., Cantarel, B. L., Duncan, A., Ley, R. E., Sogin, M. L., Jones, W. J., Roe, B. A., Affourtit, J. P., Egholm, M., Henrissat, B., Heath, A. C., Knight, R., and Gordon, J. I. (2009). A core gut microbiome in obese and lean twins. Nature 457, 480–484.
Uematsu, S., Fujimoto, K., Jang, M. H., Yang, B. G., Jung, Y. J., Nishiyama, M., Sato, S., Tsujimura, T., Yamamoto, M., Yokota, Y., Kiyono, H., Miyasaka, M., Ishii, K. J., and Akira, S. (2008). Regulation of humoral and cellular gut immunity by lamina propria dendritic cells expressing Toll-like receptor 5. Nat. Immunol. 9, 769–776.
Umesaki, Y., Okada, Y., Matsumoto, S., Imaoka, A., and Setoyama, H. (1995). Segmented filamentous bacteria are indigenous intestinal bacteria that activate intraepithelial lymphocytes and induce MHC class-II molecules and fucosyl asialo Gm1 glycolipids on the small-intestinal epithelial-cells in the ex-germ-free mouse. Microbiol. Immunol. 39, 555–562.
van der Waaij, D., Berghuis-De Vries, J. M., and Lekkerkerk, L.-V. (1971). Colonization resistance of the digestive tract in conventional and antibiotic-treated mice. J. Hyg. (Lond.) 69, 405–411.
Wen, L., Ley, R. E., Volchkov, P. Y., Stranges, P. B., Avanesyan, L., Stonebraker, A. C., Hu, C. Y., Wong, F. S., Szot, G. L., Bluestone, J. A., Gordon, J. I., and Chervonsky, A. V. (2008). Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature 455, 1109–1113.
Willing, B., Halfvarson, J., Dicksved, J., Rosenquist, M., Jarnerot, G., Engstrand, L., Tysk, C., and Jansson, J. K. (2009). Twin studies reveal specific imbalances in the mucosa-associated microbiota of patients with ileal Crohn’s disease. Inflamm. Bowel Dis. 15, 653–660.
Willing, B. P., Gill, N., and Finlay, B. B. (2010). The role of the immune system in regulating the microbiota. Gut Microbes 1, 213–223.
Willing, B. P., Vacharaksa, A., Croxen, M., Thanachayanont, T., and Finlay, B. B. (2011). Altering host resistance to infections through microbial transplantation. PLoS ONE 6, e26988. doi:10.1371/journal.pone.0026988
Willing, B. P., and Van Kessel, A. G. (2007). Enterocyte proliferation and apoptosis in the caudal small intestine is influenced by the composition of colonizing commensal bacteria in the neonatal gnotobiotic pig. J. Anim. Sci. 85, 3256–3266.
Wlodarska, M., Willing, B., Keeney, K. M., Menendez, A., Bergstrom, K. S., Gill, N., Russell, S. L., Vallance, B. A., and Finlay, B. B. (2011). Antibiotic treatment alters the colonic mucus layer and predisposes the host to exacerbated Citrobacter rodentium-induced colitis. Infect. Immun. 79, 1536–1545.
Wu, G. D., Chen, J., Hoffmann, C., Bittinger, K., Chen, Y. Y., Keilbaugh, S. A., Bewtra, M., Knights, D., Walters, W. A., Knight, R., Sinha, R., Gilroy, E., Gupta, K., Baldassano, R., Nessel, L., Li, H., Bushman, F. D., and Lewis, J. D. (2011). Linking long-term dietary patterns with gut microbial enterotypes. Science 334, 105–108.
Wu, H. J., Ivanov, Ii., Darce, J., Hattori, K., Shima, T., Umesaki, Y., Littman, D. R., Benoist, C., and Mathis, D. (2010). Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity 32, 815–827.
Zachar, Z., and Savage, D. C. (1979). Microbial interference and colonization of the murine gastrointestinal tract by Listeria monocytogenes. Infect. Immun. 23, 168–174.
Keywords: intestinal microbiota, immune homeostasis
Citation: Arrieta M-C and Finlay BB (2012) The commensal microbiota drives immune homeostasis. Front. Immun. 3:33. doi: 10.3389/fimmu.2012.00033
Received: 01 December 2011; Paper pending published: 22 December 2011;
Accepted: 14 February 2012; Published online: 05 March 2012.
Edited by:
Larry J. Dishaw, University of South Florida, USAReviewed by:
Michael Bailey, The Ohio State University, USAJohn Rawls, University of North Carolina at Chapel Hill, USA
Copyright: © 2012 Arrieta and Finlay. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Barton Brett Finlay, Michael Smith Laboratories, University of British Columbia, #301 – 2185 East Mall, Vancouver, BC, Canada V6T 1Z4. e-mail:YmZpbmxheUBpbnRlcmNoYW5nZS51YmMuY2E=
