
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Immunol. , 02 February 2012
Sec. Antigen Presenting Cell Biology
Volume 3 - 2012 | https://doi.org/10.3389/fimmu.2012.00009
This article is part of the Research Topic Eat Thyself, Heal Thyself: Autophagy in pathogen recognition and antigen processing View all 13 articles
T cells recognize proteolytic fragments of antigens that are presented to them on major histocompatibility complex (MHC) molecules. MHC class I molecules present primarily products of proteasomal proteolysis to CD8+ T cells, while MHC class II molecules display mainly degradation products of lysosomes for stimulation of CD4+ T cells. Macroautophagy delivers intracellular proteins to lysosomal degradation, and contributes in this fashion to the pool of MHC class II displayed peptides. Both self- and pathogen-derived MHC class II ligands are generated by this pathway. In addition, however, recent evidence points also to regulation of extracellular antigen processing by macroautophagy. In this review, I will discuss these two aspects of antigen processing for MHC class II presentation via macroautophagy, namely its influence on intracellular and extracellular antigen presentation to CD4+ T cells.
T cells recognize antigenic fragments presented to them by major histocompatibility complex (MHC) molecules. CD4+ T helper cells, which orchestrate adaptive humoral and cell-mediated immune responses, are stimulated by MHC class II molecules. These are present in the steady-state on antigen presenting cells (APCs), like B cells, macrophages and dendritic cells, leukocytes that are thought to initiate immune responses, but they can be up-regulated on most human cells upon inflammation or activation (Neefjes et al., 2011). Antigenic fragments and self-peptides are loaded onto MHC class II molecules in late endosomes or MHC class II containing compartments (MIICs). Most likely proteins that are transported to MIICs bind to MHC class II and are then trimmed by lysosomal proteolysis to yield peptides of at least nine amino acids length, but often 15-mers or longer, which then stabilize MHC class II for export from MIICs to the cell surface for T cell stimulation (Trombetta and Mellman, 2005). In addition to proteases, the oxidoreductase GILT (gamma-IFN-inducible lysosomal thiol reductase) participates in the unfolding of antigens by reducing disulfide bonds (Maric et al., 2001). MHC class II molecules reach MIICs with the help of the invariant chain (Ii), which associates with them in the endoplasmic reticulum, where it prevents premature peptide loading onto MHC class II and guides MHC class II transport to MIICs via its cytoplasmic tail. Ii is degraded in MIIC by lysosomal hydrolysis, and its last remnant the CLIP peptide (class II associated invariant chain peptide) is then released from the MHC class II peptide binding groove under the influence of the HLA-DM chaperone, which is negatively regulated by HLA-DO in some cell types. Some self-protein ligands of MHC class II might reach the MIICs via a similar route as MHC class II molecules themselves. Indeed, membrane proteins including MHC class I and II constitute around 40% of natural MHC class II ligands (Dengjel et al., 2005). Classically, endocytosis delivers non-self antigens to MIICs, but might also account for some surface receptor delivery for MHC class II loading. However, in B cell lines only around 10% of natural MHC class II ligands are derived from bona fide extracellular proteins. In addition, a substantial amount of MHC class II ligands originates from cytosolic and nuclear antigens (Dengjel et al., 2005). This fraction makes up 20–30% of natural MHC class II ligands. In this review I will discuss how macroautophagy, a pathway that engulfs cytoplasmic constituents with a double-membrane surrounded autophagosome and delivers them to lysosomes for degradation (Figure 1), might contribute not only to the transport of cytosolic and nuclear antigens to MIICs, but also how it might facilitate the delivery of endocytosed cargo to this compartment.
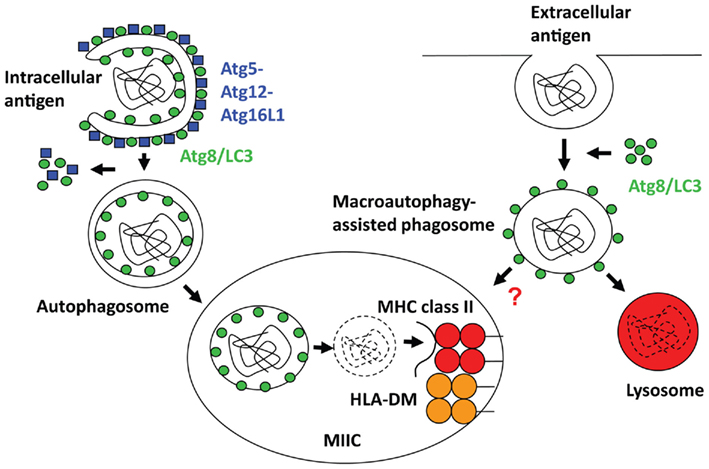
Figure 1. Regulation of intra- and extracellular antigen processing for MHC class II presentation by macroautophagy. Left side: Autophagosomes, which recruit their cargo through binding to Atg8/LC3, fuse with MHC class II containing compartments (MIICs), in which limited lysosomal hydrolysis breaks down these antigens for MHC class II loading with the assistance of HLA-DM (H2-M in mice). Right side: phagosomes, especially those carrying Toll-like receptor (TLR) ligands or apoptotic cells, get decorated with Atg8/LC3, which enhances fusion with lysosomes, and might also increase fusion with MIICs for antigen loading onto MHC class II molecules.
Indeed, when macroautophagy was induced in B cell lines by starvation, cytosolic, and nuclear antigen presentation was increased by 50%, while membrane bound antigen presentation remained largely unaffected (Dengjel et al., 2005). For individual ligands the increase even exceeded 130%. This increase in intracellular antigen presentation correlated with macroautophagy induction as determined by autophagic vacuole content. Among the cytosolic MHC class II ligands also two components of the macroautophagic machinery could be found, namely LC3 and GABARAP (Dengjel et al., 2005; Suri et al., 2008). Both of them are mammalian homologs of the autophagy related gene (atg) product Atg8, which are coupled to the autophagosome membrane and involved in the extension of the double-membrane around the cargo and closure of the final autophagosomes, as well as substrate recruitment into the autophagosome (Weidberg et al., 2010, 2011a,b). Interestingly, Atg8 homologs are also the only essential macroautophagy proteins that remain with the completed autophagosome on its inner membrane and get degraded with the autophagosome cargo by lysosomal proteolysis. Therefore, their turn-over can be monitored to analyze macroautophagy. These findings suggest that autophagosome cargo, including LC3 and GABARAP proteins gain access to MHC class II presentation and, therefore, autophagosomes might frequently fuse with MIICs. Indeed, such fusion events were observed in epithelial cells that expressed MHC class II upon IFN-γ treatment and also B cell lines and dendritic cells, as classical APCs (Schmid et al., 2007). The fusion vesicles contained hallmarks of MIICs with expression of HLA-DM and the lysosome associated membrane protein (LAMP) 2. Furthermore they resemble multivesicular bodies (MVBs) with both MHC class II and LC3 molecules primarily on the intravesicular membranes. Such autophagosome fusion delivers also melanosomes and tumor differentiation antigens to MIICs in melanoma cells (van den Boorn et al., 2011). Thus, autophagosomes fuse quite frequently with MIICs in APCs and some of their cargo is loaded onto MHC class II. This self-protein presentation on MHC class II molecules after macroautophagy seems to be especially important during thymic CD4+ T cell education. Indeed, also in thymic epithelial cells (TECs), which constitutively express MHC class II molecules, autophagosomes fuse with MIICs (Kasai et al., 2009). This fusion seems to deliver self-proteins for both positive CD4+ T cell selection in cortical TECs and negative selection in medullary TECs (Nedjic et al., 2008). Only some, but not other CD4+ T cell specificities were selected by cortical TECs deficient in the essential autophagy protein Atg5, which is involved in Atg8/LC3 coupling to the autophagosome membrane. In contrast, CD8+ T cells were correctly educated through Atg5 deficient thymic cortex. In addition, defective negative selection through Atg5 deficient thymii has been suggested to cause autoimmune inflammation in the intestines and other organs. This suggests that a substantial portion of self-protein derived ligands originates from macroautophagy cargo and that these are required to positively select some T cell specificities through the thymus and to induce central tolerance in this organ.
In addition to self-proteins, some viral and bacterial antigens are presented on MHC class II molecules after macroautophagy. The first viral example for this pathway was the nuclear antigen 1 (EBNA1) of the human tumor virus Epstein Barr virus (EBV; Paludan et al., 2005). This viral protein, which limits both its translation and its proteasomal degradation via a glycine–alanine (GA) repeat domain, is turned over by lysosomal degradation after macroautophagy. The engulfment by autophagosomes leads to MHC class II presentation on EBV transformed B cells to EBNA1 specific CD4+ T cell clones, a T cell specificity that is consistently found in healthy EBV carriers (Münz et al., 2000). Its nuclear localization, however, limits antigen processing via macroautophagy, and EBNA1 that lacks its nuclear localization domain is more efficiently presented to CD4+ T cells (Leung et al., 2010). More efficient processing of cytosolic EBNA1 via macroautophagy leads to CD4+ T cell recognition of more EBNA1 derived epitopes on EBV transformed B cells.
In addition to EBNA1, processing of bacterial antigens has been reported to require macroautophagy. The bacterial transposon-derived neomycin phosphotransferase II (NeoR) is presented to CD4+ T cells after macroautophagic processing for MHC class II presentation (Nimmerjahn et al., 2003; Comber et al., 2011). Interestingly and in contrast to EBNA1, forced nuclear localization of this after transfection cytosolic protein resulted in similar or even slightly enhanced presentation on MHC class II molecules (Riedel et al., 2008). This presentation was still dependent on macroautophagy and lysosomal degradation. While NeoR was in these studies introduced into the cytosol by transfection, some bacteria inject antigens via secretion systems into the same cellular compartment. Among these is the mycobacterial Ag85B antigen, whose MHC class II presentation by DCs is enhanced upon macroautophagy stimulation (Jagannath et al., 2009). DCs with macroautophagically increased Ag85B presentation elicit then more efficiently protective immune responses after vaccination by adoptive transfer. Thus, macroautophagy might enhance antigen presentation during mycobacterial infection. Furthermore, Yersinia outer proteins YopE and H block MHC class II loading via endocytosis. In the absence of this extracellular antigen processing, YopE fusion proteins get endogenously processed for MHC class II presentation (Russmann et al., 2010). This antigen processing is sensitive to lysosomal and macroautophagy inhibition. Thus bacterial cytosolic, and maybe even nuclear antigens are degraded via macroautophagy and this leads to MHC class II presentation to CD4+ T cells.
The above discussed evidence suggests that cytosolic and nuclear antigens get processed for MHC class II processing via macroautophagy. But how are these substrates recruited to autophagosomes? In higher eukaryotes two pathways of substrate recruitment to macroautophagy have been described. Both rely on anchoring of cytoplasmic constituents to Atg8/LC3, presumably on the inner autophagosomal membrane and deliver cell organelles like mitochondria as well as protein aggregates to autophagosomes. One pathway uses integral organelle proteins, like NIX for mitochondria (Schweers et al., 2007; Sandoval et al., 2008), to recruit them to forming autophagosomes or autophagosomal membranes to their cargo. The second mechanism relies on protein adaptors that link polyubiquitinylated substrates to Atg8/LC3 via ubiquitin-binding domains (UBA or UBZ) and LC3 interacting domains (LIRs). The four identified members of this class of proteins are p62/sequestosome 1, NBR1, NDP52, and optineurin (Bjorkoy et al., 2005; Pankiv et al., 2007; Kirkin et al., 2009; Thurston et al., 2009; Wild et al., 2011). They recruit protein aggregates, mitochondria, and bacterial pathogens, like Salmonella, to autophagosomes. Because they anchor these substrates to the inner autophagosomal membrane, they end up in the completed autophagosomes and are degraded with their content. Thus, they can be used to monitor autophagosome turn-over or macroautophagic flux. While none of these adaptor proteins has so far been directly linked to antigen processing for MHC class II presentation, covalent coupling of antigens to the N-terminus of Atg8/LC3 enhances their presentation on MHC class II molecules to CD4+ T cells by epithelial cells, B cells, and dendritic cells up to 20-fold. Such increase has been observed for the influenza A virus antigens matrix protein 1 (MP1; Schmid et al., 2007) and hemagglutinin (HA; Comber et al., 2011), as well as the tumor antigen NY-ESO-1 (unpublished data). Delivery of these fusion constructs to MIICs was dependent on the macroautophagy machinery, namely Atg7 and 12, and mutating the C-terminal glycine residue of Atg8/LC3, which is used to couple this protein to the autophagosomal membrane, abolishes enhanced MHC class II presentation of these fusion constructs (Schmid and Münz, 2007; Comber et al., 2011). These data argue that Atg8/LC3 can recruit antigens for MHC class II presentation to autophagosomes, which then frequently fuse with MIICs. To which extent the UBA/LIR anchor proteins and organelle specific LIR containing proteins contribute to self- and foreign-protein recruitment for MHC class II presentation remains to be determined.
Major histocompatibility complex class II containing compartments are usually characterized as late endosomal compartments with intravesicular membranes that morphologically appear either as MVBs, multilamellar (MLBs), or electron dense bodies (EDBs; Stern et al., 2006; Neefjes et al., 2011). Rarely, they have a clear lysosomal appearance, which suggests that their hydrolytic potential is controlled, and does not readily degrade antigens and MHC class II molecules all the way to amino acids. This notion is also supported by recent studies on limited acidification of endosomes in dendritic cells by alkalinization via for example reactive oxygen species (ROS) production by phagosomal NADPH oxidase 2 (NOX2; Savina et al., 2006). Elevated pH causes less efficient hydrolysis by for example lysosomal cathepsins. Along these lines MHC class II antigen presentation of macroautophagy substrates can be enhanced by slightly neutralizing the endolysosomal compartment with pharmacological reagents in macrophages (Brazil et al., 1997). In support of these cell biological studies, dendritic cells, and macrophages handle antigen also differently in vivo. While macrophages, which have a decreased ability to initiate immune responses, degrade injected antigen rapidly within 1 day, dendritic cells, superior in activation of adaptive immune compartments, retain injected antigens even 2 days after injection (Delamarre et al., 2005). Accordingly, antigen formulations, which are highly sensitive to lysosomal degradation are less well presented to CD4+ T cells and induce weaker immune responses than antigens with some resistance to hydrolysis (Delamarre et al., 2006). These data suggest that extracellular antigen is more efficiently presented on MHC class II molecules, when it is degraded less efficiently, and that rapid fusion with lysosomes would rather be detrimental for CD4+ T cell stimulation. This has to be kept in mind when we now discuss what the macroautophagic machinery contributes to phagocytosis and phagosome maturation.
Fusion of autophagosomes and endosomes is a frequent process in higher eukaryotic cells and the resulting vesicles were termed amphisomes (Berg et al., 1998). While Rab7 seems to be involved in direct fusion of autophagosomes with lysosomes, endosome, and MVB fusion is mediated by Rab11 (Gutierrez et al., 2004b; Fader et al., 2008). Amphisomes are then also delivered to lysosomes. Thus, autophagosomes might modify phagosomes via delivering additional cargo to amphisomes. Along these lines macroautophagy has been described to transport source proteins of microbial peptides to phagosomes (Alonso et al., 2007; Ponpuak et al., 2010). These were found to be derived from ubiquitin itself or proteins with ubiquitin-like domains, that were imported into autophagosomes by some of the anchor proteins described above. In part due to this cargo delivery autophagosome fusion with phagosomes renders the resulting amphisomes more degradative for endocytosed bacterial pathogens (Gutierrez et al., 2004a; Birmingham et al., 2006). ROS production by NOX2 and diacylglycerol (DAG) formation at the phagosomal membrane was proposed to enhance autophagosome fusion with bacteria containing phagosomes (Huang et al., 2009; Shahnazari et al., 2010). Thus, amphisome formation can enhance bactericidal activity of phagosomes.
In addition, macroautophagy has been described to accelerate phagosome fusion with lysosomes (Sanjuan et al., 2007; Florey et al., 2011). However, this pathway might not require autophagosome fusion with lysosomes, but coupling of Atg8/LC3 to the phagosomal membrane (Figure 1). Accordingly, Atg8/LC3 co-localization to phagosomes and enhanced fusion of phagosomes with lysosomes is dependent on the core machinery of Atg8/LC3 lipidation, for example Atg5 and Atg7, but not on factors that are required for starvation induced macroautophagy, like mTOR and Atg1/ULK1. Toll-like receptor (TLR) agonist carrying particles and apoptotic cells have been identified in Atg8/LC3 positive phagosomes. How Atg8/LC3 on the outside of the phagosome, however, mediates enhanced fusion with lysosomes remains unclear. It is tempting to speculate that Atg8/LC3, which is involved in autophagosome membrane extension by possibly mediating fusion with additional membranes (Nakatogawa et al., 2007; Weidberg et al., 2011a), could also promote other vesicular fusion events directly, like phagosome fusion with lysosomes. Irrespective of the mechanism, the macroautophagy machinery seems to modify phagocytosis by delivering precursors of bactericidal peptides to phagosomes and accelerating their fusion with lysosomes.
Although rapid degradation of phagosomal content rather destroys antigens than leads to their presentation and phagosomes have been described to mature faster with the help of the macroautophagy machinery, it was found that mice with macroautophagy deficiency in their dendritic cell compartment were less able to prime CD4+ T cell responses after herpes simplex virus (HSV) infection (Lee et al., 2010). Similarly, HSV lacking its ICP34.5 antigen, which blocks autophagosome formation (Orvedahl et al., 2007), stimulates stronger CD4+ T cell responses than wild-type HSV infection (Leib et al., 2009). Even so these data could still suggest intracellular HSV antigen processing by macroautophagy and cytosolic ovalbumin was also less efficiently presented on MHC class II molecules by Atg5 deficient dendritic cells, in addition, extracellular ovalbumin was less efficiently presented to CD4+ T cells by macroautophagy deficient dendritic cells (Lee et al., 2010). This was correlated with less efficient recruitment of lysosomal hydrolases, particularly cathepsins, to phagosomes in the absence of macroautophagy. However, to which extend this more rapid phagosome maturation with the help of the macroautophagy machinery contributes to extracellular antigen presentation on MHC class II molecules remains unknown, and influenza antigen processing for MHC class II presentation to CD4+ T cells was unaffected by macroautophagy inhibition (Comber et al., 2011). Therefore, CD4+ T cell responses only to some, but not other pathogens might be dependent on macroautophagy in dendritic cells for their induction, and the mechanism of macroautophagic assistance for extracellular antigen processing onto MHC class II molecules remains poorly defined.
Macroautophagy delivers cytoplasmic constituents for lysosomal degradation. This machinery is also used to visualize intracellular antigens to CD4+ T cells by processing them for MHC class II presentation. However, in the process of studying dependency of antigen processing on macroautophagy an additional pathway was revealed, by which macroautophagy influences extracellular antigen processing for MHC class II presentation to CD4+ T cells. How coupling of Atg8/LC3 to the phagosomal membrane, however, modifies phagosome maturation, potentially accelerating fusion with lysosomes remains unknown. Furthermore, the role of this accelerated phagosome maturation for MHC class II presentation remains to be determined. A better understanding of these molecular processes during antigen presentation should allow us to design antigens that are then more efficiently processed for CD4+ T cell stimulation and choose adjuvants, like TLR agonists, that utilize macroautophagy to optimize antigen presentation.
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Research in my laboratory is supported by grants from the National Cancer Institute (R01CA108609), the Sassella Foundation (10/02), Cancer Research Switzerland (KFS-02652-08-2010), the Association for International Cancer Research (11-0516), the Vontobel Foundation, Novartis, and the Swiss National Science Foundation (310030_126995).
Alonso, S., Pethe, K., Russell, D. G., and Purdy, G. E. (2007). Lysosomal killing of Mycobacterium mediated by ubiquitin-derived peptides is enhanced by autophagy. Proc. Natl. Acad. Sci. U.S.A. 104, 6031–6036.
Berg, T. O., Fengsrud, M., Stromhaug, P. E., Berg, T., and Seglen, P. O. (1998). Isolation and characterization of rat liver amphisomes. Evidence for fusion of autophagosomes with both early and late endosomes. J. Biol. Chem. 273, 21883–21892.
Birmingham, C. L., Smith, A. C., Bakowski, M. A., Yoshimori, T., and Brumell, J. H. (2006). Autophagy controls Salmonella infection in response to damage to the Salmonella-containing vacuole. J. Biol. Chem. 281, 11374–11383.
Bjorkoy, G., Lamark, T., Brech, A., Outzen, H., Perander, M., Overvatn, A., Stenmark, H., and Johansen, T. (2005). p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 171, 603–614.
Brazil, M. I., Weiss, S., and Stockinger, B. (1997). Excessive degradation of intracellular protein in macrophages prevents presentation in the context of major histocompatibility complex class II molecules. Eur. J. Immunol. 27, 1506–1514.
Comber, J. D., Robinson, T. M., Siciliano, N. A., Snook, A. E., and Eisenlohr, L. C. (2011). Functional macroautophagy induction by influenza A virus without a contribution to MHC-class II restricted presentation. J. Virol. 85, 6453–6463.
Delamarre, L., Couture, R., Mellman, I., and Trombetta, E. S. (2006). Enhancing immunogenicity by limiting susceptibility to lysosomal proteolysis. J. Exp. Med. 203, 2049–2055.
Delamarre, L., Pack, M., Chang, H., Mellman, I., and Trombetta, E. S. (2005). Differential lysosomal proteolysis in antigen-presenting cells determines antigen fate. Science 307, 1630–1634.
Dengjel, J., Schoor, O., Fischer, R., Reich, M., Kraus, M., Muller, M., Kreymborg, K., Altenberend, F., Brandenburg, J., Kalbacher, H., Brock, R., Driessen, C., Rammensee, H. G., and Stevanovic, S. (2005). Autophagy promotes MHC class II presentation of peptides from intracellular source proteins. Proc. Natl. Acad. Sci. U.S.A. 102, 7922–7927.
Fader, C. M., Sanchez, D., Furlan, M., and Colombo, M. I. (2008). Induction of autophagy promotes fusion of multivesicular bodies with autophagic vacuoles in K562 cells. Traffic 9, 230–250.
Florey, O., Kim, S. E., Sandoval, C. P., Haynes, C. M., and Overholtzer, M. (2011). Autophagy machinery mediates macroendocytic processing and entotic cell death by targeting single membranes. Nat. Cell Biol. 13, 1335–1343.
Gutierrez, M. G., Master, S. S., Singh, S. B., Taylor, G. A., Colombo, M. I., and Deretic, V. (2004a). Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 119, 753–766.
Gutierrez, M. G., Munafo, D. B., Beron, W., and Colombo, M. I. (2004b). Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J. Cell. Sci. 117, 2687–2697.
Huang, J., Canadien, V., Lam, G. Y., Steinberg, B. E., Dinauer, M. C., Magalhaes, M. A., Glogauer, M., Grinstein, S., and Brumell, J. H. (2009). Activation of antibacterial autophagy by NADPH oxidases. Proc. Natl. Acad. Sci. U.S.A. 106, 6226–6231.
Jagannath, C., Lindsey, D. R., Dhandayuthapani, S., Xu, Y., Hunter, R. L. Jr., and Eissa, N. T. (2009). Autophagy enhances the efficacy of BCG vaccine by increasing peptide presentation in mouse dendritic cells. Nat. Med. 15, 267–276.
Kasai, M., Tanida, I., Ueno, T., Kominami, E., Seki, S., Ikeda, T., and Mizuochi, T. (2009). Autophagic compartments gain access to the MHC class II compartments in thymic epithelium. J. Immunol. 183, 7278–7285.
Kirkin, V., Lamark, T., Sou, Y. S., Bjorkoy, G., Nunn, J. L., Bruun, J. A., Shvets, E., Mcewan, D. G., Clausen, T. H., Wild, P., Bilusic, I., Theurillat, J. P., Overvatn, A., Ishii, T., Elazar, Z., Komatsu, M., Dikic, I., and Johansen, T. (2009). A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell 33, 505–516.
Lee, H. K., Mattei, L. M., Steinberg, B. E., Alberts, P., Lee, Y. H., Chervonsky, A., Mizushima, N., Grinstein, S., and Iwasaki, A. (2010). In vivo requirement for Atg5 in antigen presentation by dendritic cells. Immunity 32, 227–239.
Leib, D. A., Alexander, D. E., Cox, D., Yin, J., and Ferguson, T. A. (2009). Interaction of ICP34.5 with beclin 1 modulates herpes simplex virus type 1 pathogenesis through control of CD4+ T-cell responses. J. Virol. 83, 12164–12171.
Leung, C. S., Haigh, T. A., Mackay, L. K., Rickinson, A. B., and Taylor, G. S. (2010). Nuclear location of an endogenously expressed antigen, EBNA1, restricts access to macroautophagy and the range of CD4 epitope display. Proc. Natl. Acad. Sci. U.S.A. 107, 2165–2170.
Maric, M., Arunachalam, B., Phan, U. T., Dong, C., Garrett, W. S., Cannon, K. S., Alfonso, C., Karlsson, L., Flavell, R. A., and Cresswell, P. (2001). Defective antigen processing in GILT-free mice. Science 294, 1361–1365.
Münz, C., Bickham, K. L., Subklewe, M., Tsang, M. L., Chahroudi, A., Kurilla, M. G., Zhang, D., O’donnell, M., and Steinman, R. M. (2000). Human CD4+ T lymphocytes consistently respond to the latent Epstein-Barr virus nuclear antigen EBNA1. J. Exp. Med. 191, 1649–1660.
Nakatogawa, H., Ichimura, Y., and Ohsumi, Y. (2007). Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 130, 165–178.
Nedjic, J., Aichinger, M., Emmerich, J., Mizushima, N., and Klein, L. (2008). Autophagy in thymic epithelium shapes the T-cell repertoire and is essential for tolerance. Nature 455, 396–400.
Neefjes, J., Jongsma, M. L., Paul, P., and Bakke, O. (2011). Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat. Rev. Immunol. 11, 823–836.
Nimmerjahn, F., Milosevic, S., Behrends, U., Jaffee, E. M., Pardoll, D. M., Bornkamm, G. W., and Mautner, J. (2003). Major histocompatibility complex class II-restricted presentation of a cytosolic antigen by autophagy. Eur. J. Immunol. 33, 1250–1259.
Orvedahl, A., Alexander, D., Talloczy, Z., Sun, Q., Wei, Y., Zhang, W., Burns, D., Leib, D., and Levine, B. (2007). HSV-1 ICP34.5 confers neurovirulence by targeting the beclin 1 autophagy protein. Cell Host Microbe 1, 23–35.
Paludan, C., Schmid, D., Landthaler, M., Vockerodt, M., Kube, D., Tuschl, T., and Münz, C. (2005). Endogenous MHC class II processing of a viral nuclear antigen after autophagy. Science 307, 593–596.
Pankiv, S., Clausen, T. H., Lamark, T., Brech, A., Bruun, J. A., Outzen, H., Overvatn, A., Bjorkoy, G., and Johansen, T. (2007). p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282, 24131–24145.
Ponpuak, M., Davis, A. S., Roberts, E. A., Delgado, M. A., Dinkins, C., Zhao, Z., Virgin, H. W. T., Kyei, G. B., Johansen, T., Vergne, I., and Deretic, V. (2010). Delivery of cytosolic components by autophagic adaptor protein p62 endows autophagosomes with unique antimicrobial properties. Immunity 32, 329–341.
Riedel, A., Nimmerjahn, F., Burdach, S., Behrends, U., Bornkamm, G. W., and Mautner, J. (2008). Endogenous presentation of a nuclear antigen on MHC class II by autophagy in the absence of CRM1-mediated nuclear export. Eur. J. Immunol. 38, 2090–2095.
Russmann, H., Panthel, K., Kohn, B., Jellbauer, S., Winter, S. E., Garbom, S., Wolf-Watz, H., Hoffmann, S., Grauling-Halama, S., and Geginat, G. (2010). Alternative endogenous protein processing via an autophagy-dependent pathway compensates for Yersinia-mediated inhibition of endosomal major histocompatibility complex class II antigen presentation. Infect. Immun. 78, 5138–5150.
Sandoval, H., Thiagarajan, P., Dasgupta, S. K., Schumacher, A., Prchal, J. T., Chen, M., and Wang, J. (2008). Essential role for Nix in autophagic maturation of erythroid cells. Nature 454, 232–235.
Sanjuan, M. A., Dillon, C. P., Tait, S. W., Moshiach, S., Dorsey, F., Connell, S., Komatsu, M., Tanaka, K., Cleveland, J. L., Withoff, S., and Green, D. R. (2007). Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature 450, 1253–1257.
Savina, A., Jancic, C., Hugues, S., Guermonprez, P., Vargas, P., Moura, I. C., Lennon-Dumenil, A. M., Seabra, M. C., Raposo, G., and Amigorena, S. (2006). NOX2 controls phagosomal pH to regulate antigen processing during crosspresentation by dendritic cells. Cell 126, 205–218.
Schmid, D., and Münz, C. (2007). Innate and adaptive immunity through autophagy. Immunity 26, 11–21.
Schmid, D., Pypaert, M., and Münz, C. (2007). MHC class II antigen loading compartments continuously receive input from autophagosomes. Immunity 26, 79–92.
Schweers, R. L., Zhang, J., Randall, M. S., Loyd, M. R., Li, W., Dorsey, F. C., Kundu, M., Opferman, J. T., Cleveland, J. L., Miller, J. L., and Ney, P. A. (2007). NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc. Natl. Acad. Sci. U.S.A. 104, 19500–19505.
Shahnazari, S., Yen, W. L., Birmingham, C. L., Shiu, J., Namolovan, A., Zheng, Y. T., Nakayama, K., Klionsky, D. J., and Brumell, J. H. (2010). A diacylglycerol-dependent signaling pathway contributes to regulation of antibacterial autophagy. Cell Host Microbe 8, 137–146.
Stern, L. J., Potolicchio, I., and Santambrogio, L. (2006). MHC class II compartment subtypes: structure and function. Curr. Opin. Immunol. 18, 64–69.
Suri, A., Walters, J. J., Rohrs, H. W., Gross, M. L., and Unanue, E. R. (2008). First signature of islet {beta}-cell-derived naturally processed peptides selected by diabetogenic class II MHC molecules. J. Immunol. 180, 3849–3856.
Thurston, T. L., Ryzhakov, G., Bloor, S., Von Muhlinen, N., and Randow, F. (2009). The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat. Immunol. 10, 1215–1221.
Trombetta, E. S., and Mellman, I. (2005). Cell biology of antigen processing in vitro and in vivo. Annu. Rev. Immunol. 23, 975–1028.
van den Boorn, J. G., Picavet, D. I., Van Swieten, P. F., Van Veen, H. A., Konijnenberg, D., Van Veelen, P. A., Van Capel, T., Jong, E. C., Reits, E. A., Drijfhout, J. W., Bos, J. D., Melief, C. J., and Luiten, R. M. (2011). Skin-depigmenting agent monobenzone induces potent T-cell autoimmunity toward pigmented cells by tyrosinase haptenation and melanosome autophagy. J. Invest. Dermatol. 131, 1240–1251.
Weidberg, H., Shpilka, T., Shvets, E., Abada, A., Shimron, F., and Elazar, Z. (2011a). LC3 and GATE-16 N termini mediate membrane fusion processes required for autophagosome biogenesis. Dev. Cell 20, 444–454.
Weidberg, H., Shvets, E., and Elazar, Z. (2011b). Biogenesis and cargo selectivity of autophagosomes. Annu. Rev. Biochem. 80, 125–156.
Weidberg, H., Shvets, E., Shpilka, T., Shimron, F., Shinder, V., and Elazar, Z. (2010). LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. 29, 1792–1802.
Keywords: autophagosome, CD4+ T cell, amphisome, MHC class II containing compartment, lysosome
Citation: Münz C (2012) Antigen processing for MHC class II presentation via autophagy. Front. Immun. 3:9. doi: 10.3389/fimmu.2012.00009
Received: 08 December 2011;
Paper pending published: 03 January 2012;
Accepted: 16 January 2012;
Published online: 02 February 2012.
Edited by:
Irina Caminschi, Walter and Eliza Hall Institute, AustraliaReviewed by:
Ludger Klein, Ludwig-Maximilians-University, GermanyCopyright: © 2012 Münz. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Christian Münz, Viral Immunobiology, Institute of Experimental Immunology, University of Zürich, Winterthurerstrasse 190, CH-8057 Zürich, Switzerland. e-mail:Y2hyaXN0aWFuLm11ZW56QHV6aC5jaA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.