
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol., 06 January 2012
Sec. Molecular Innate Immunity
volume 2 - 2011 | https://doi.org/10.3389/fimmu.2011.00092
This article is part of the Research TopicState-of-the-Art research on C1q and the Classical Complement PathwayView all 11 articles
C1q, the ligand-binding unit of the C1 complex of complement, is a pattern recognition molecule with the unique ability to sense an amazing variety of targets, including a number of altered structures from self, such as apoptotic cells. The three-dimensional structure of its C-terminal globular domain, responsible for its recognition function, has been solved by X-ray crystallography, revealing a tightly packed heterotrimeric assembly with marked differences in the surface patterns of the subunits, and yielding insights into its versatile binding properties. In conjunction with other approaches, this same technique has been used recently to decipher the mechanisms that allow this domain to interact with various non-immune self ligands, including molecules known to provide eat-me signals on apoptotic cells, such as phosphatidylserine and DNA. These investigations provide evidence for a common binding area for these ligands located in subunit C of the C1q globular domain, and suggest that ligand recognition through this area down-regulates C1 activation, hence contributing to the control of the inflammatory reaction. The purpose of this article is to give an overview of these advances which represent a first step toward understanding the recognition mechanisms of C1q and their biological implications.
Triggering of the classical complement pathway is mediated by C1, a multimolecular complex resulting from the association of a recognition subunit, C1q, and a Ca2+-dependent tetramer comprising two copies of two serine proteases, C1r, and C1s (Cooper, 1985; Gaboriaud et al., 2004). C1q is a 460-kDa hexameric protein assembled from six heterotrimeric collagen-like fibers, each being prolonged by a C-terminal globular domain which mediates the recognition function of C1 (Gaboriaud et al., 2003, 2004; Kishore et al., 2004). A major characteristic of this domain lies in its ability to sense and engage an amazing variety of ligands (Cooper, 1985; Kishore et al., 2004). Thus, C1q is classically known for its ability to bind IgG- and IgM-containing immune complexes. In addition, it recognizes the lectin SIGN-R1, C-reactive protein, and other pentraxins bound to pathogens and other surfaces, as well as various molecular motifs on several Gram-negative bacteria and viruses (Cooper, 1985; Szalai et al., 1999; Thielens et al., 2002; Kishore et al., 2004; Kang et al., 2006). In most cases, recognition of these non-self ligands by C1q triggers activation of the classical complement pathway, thereby contributing to their elimination through enhanced phagocytosis, lysis, and inflammation.
This traditional view of the biological role of C1q should be reconsidered in light of recent studies providing evidence that C1q has the ability to sense many altered structures from self, including the pathological form of the prion protein (Klein et al., 2001; Erlich et al., 2010), β-amyloid fibrils (Tacnet-Delorme et al., 2001), modified forms of low-density lipoprotein (Biro et al., 2007, 2010), and apoptotic cells (Taylor et al., 2000; Navratil et al., 2001). Recognition by C1q triggers efficient clearance of apoptotic cells by phagocytes, but in this case lysis and inflammation are both inhibited, thus contributing to the maintenance of immune tolerance (Nauta et al., 2004; Fraser et al., 2009). Recent investigations on the C1q targets at the apoptotic cell surface have revealed that three molecules known to provide “eat-me” signals, phosphatidylserine, DNA, and calreticulin, are recognized by C1q, suggesting a multiligand-binding process (Païdassi et al., 2008a,b, 2011).
The three-dimensional (3-D) structure of the heterotrimeric globular domain of C1q, responsible for the recognition function of this protein, has been solved by X-ray crystallography, giving insights into its versatile binding properties (Gaboriaud et al., 2003). The same technique, as well as other approaches, have been recently applied to decipher the interaction of this domain with various non-immune self ligands, including molecules acting as eat-me signals on apoptotic cells, providing evidence for a common binding area (Païdassi et al., 2008a; Garlatti et al., 2010). The purpose of this article is to give an overview of these advances which shed light on the recognition of self ligands by C1q and reveal possible implications in the regulation of C1 activation.
The 3-D structure of the C-terminal globular domain of human C1q, obtained after digestion of the collagenous moiety of the protein with collagenase, has been solved by X-ray crystallography to a resolution of 1.9 Å (Gaboriaud et al., 2003). The structure revealed a globular, almost spherical heterotrimeric assembly, with a diameter of about 50 Å, the N- and C-termini of each subunit emerging at the base of the trimer, in close vicinity to one another (Figures 1A,B). A major information from the structure was that the gC1q subunits are arranged clockwise in the order A, B, C when the assembly is viewed from the top, this indication enabling us to derive a 3-D model of the collagen-like triple helix of C1q, and thereby to reconstruct the whole C1q molecule (Gaboriaud et al., 2003, 2004). In the resulting C1q model (Figure 1C), the B subunit of each globular domain lies on the outer part of the molecule, whereas A and C are positioned inside. As discussed later, it is very likely that this particular configuration has direct implications in terms of ligand recognition and C1 activation.
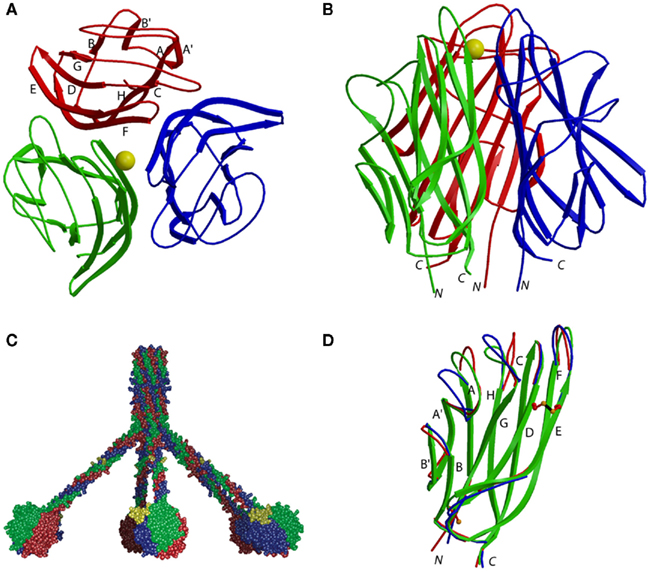
Figure 1. Structure of the C1q globular domain. (A) Top view and (B) side view of the heterotrimeric assembly. Subunits A, B, and C are shown in blue, green, and red, respectively. β-strands are labeled according to the tumor necrosis factor nomenclature, and the Ca2+ ion is represented as a golden sphere. N and C indicate the N- and C-terminal ends of each gC1q subunit. (C) Side view of the three-dimensional C1q model derived from the structure of the globular domain. The N-linked oligosaccharide attached to each globular domain is represented in yellow. (D) Structure of the gC1q subunits. Subunits A, B, and C are superimposed. Only the free cysteine and disulfide bond of subunit C are displayed for clarity. (Modified from Gaboriaud et al., 2003, 2004).
The three subunits each exhibit a 10-stranded β sandwich fold with a jelly roll topology homologous to the one described initially for tumor necrosis factor (Eck and Sprang, 1989; Jones et al., 1989) and subsequently for members of the gC1q family (Shapiro and Scherer, 1998; Bogin et al., 2002; Kvansakul et al., 2002), consisting of two five-stranded β-sheets (A′, A, H, C, F and B′, B, G, D, E), each made of anti-parallel strands (Figure 1D). Comparison of the three subunits indicates strong conservation of the β-strands and significant variability in the loops connecting them, a characteristic that also applies to other gC1q domains. In addition to a free cysteine conserved in all gC1q domains, which is essentially buried in the structure, each subunit of human C1q contains two other cysteines engaged in a disulfide bond (Figure 1D).
Assembly of the C1q heterotrimer involves a tight association of the subunits, as shown by a total buried surface of 5490 Å2 equally contributed by each subunit, this value being significantly less, however, than those in collagens VIII and X (Bogin et al., 2002). Trimerization involves a central interface as well as lateral interactions which in both cases are hydrophobic at the base of the trimer and become more polar when going to the apex. This results in the formation of a discontinuous central channel which is closed at both extremities. Interestingly, despite its heterotrimeric structure, the C1q globular domain assembles in the same way as its homotrimeric counterparts in ACRP30 and collagens VIII and X. Nevertheless, attempts to assemble gC1q subunits as homotrimers in silico were found to result in severe steric clashes, particularly at the level of lateral interactions, thus providing a structural basis for their natural propensity to associate only as heterotrimers (Gaboriaud et al., 2003). The structure of the C1q globular domain also reveals the presence of a Ca2+ ion bound at the top of the assembly (Figures 1A,B). The binding site is asymmetrical relative to the trimer, considering that Ca2+ is coordinated by oxygen ligands contributed by subunits A and B, but is not connected to subunit C. In contrast to the buried Ca2+ cluster observed in collagen X (Bogin et al., 2002), the single Ca2+ ion of C1q is exposed to the solvent and defines the upper end of the central channel. In addition to contributing to the stability of the heterotrimeric assembly, the Ca2+ ion may have a functional role. Thus, the loss of Ca2+ has been postulated to modify the direction of the electric moment of the C1q globular domain and thereby to influence the recognition of immune targets such as C-reactive protein and IgG (Roumenina et al., 2005). The fact that the Ca2+ ion is accessible to the solvent also opens the possibility of a direct implication in the recognition of certain charged targets (Gaboriaud et al., 2003).
A striking feature of the structure of the C1q globular domain lies in the fact that the three subunits exhibit marked differences in their surface patterns, with respect to both charged and hydrophobic residues (Figure 2). Thus, subunit A mainly shows a combination of arginine and acidic residues scattered on its surface (Figure 2A). Subunit C also shows a combination of basic and acidic residues spread over the surface (Figure 2C). In contrast, positively charged residues are predominant on the surface of module B, with in particular a cluster of three arginines ArgB101, ArgB114, and ArgB129. The latter two residues, which have been proposed to be involved in the interaction with IgG (Marqués et al., 1993), markedly protrude outside the structure (Figure 2B). Several hydrophobic residues are exposed to the solvent on the external face of each subunit, the most striking example being the IleB103, ValB105, ProB106 cluster lying over the ArgB101, ArgB114, ArgB129 triad (Figure 2B). In contrast, the only accessible aromatic residues (TyrC155, TrpC190) are found on the equatorial area of subunit C (Figure 2C). The top of the heterotrimer (Figure 2D) shows a predominance of positive charges mainly contributed by lysine residues. In each subunit, several hydrophobic patches and aromatic residues are also exposed.
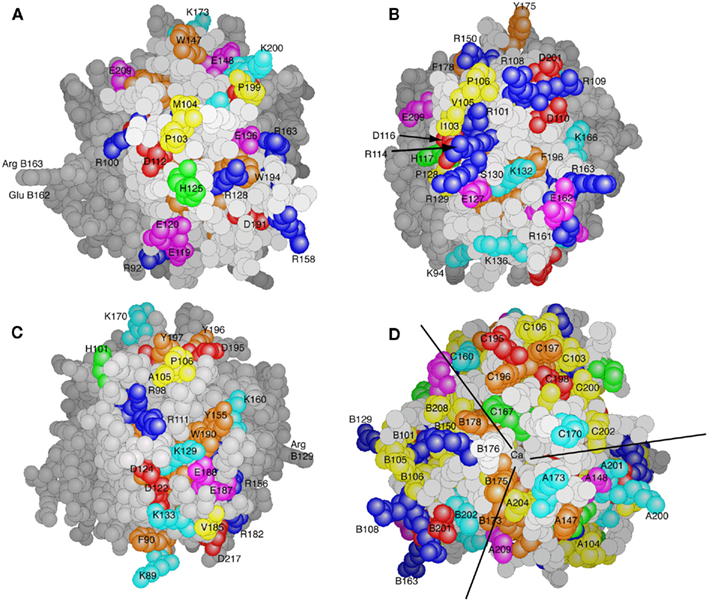
Figure 2. Surface properties of the C1q globular domain. (A–C) Side views of the heterotrimer seen from subunits A, B, and C, respectively. (D) Top view of the heterotrimer. The side chains of Arg, Lys, His, Asp, and Glu residues are shown in deep blue, light blue, green, red, and magenta, respectively. Hydrophobic residues are shown in yellow, and aromatic ones are in orange. The lines in (D) indicate the approximate subunit boundaries. (From Gaboriaud et al., 2003).
The heterotrimeric structure of the C1q globular domain is very likely a major determinant of its versatile recognition properties. Thus, because they display quite different surface patterns in terms of charged and hydrophobic residues, the three subunits are expected to mediate different individual binding properties. In addition, considering the tightly packed structure of the domain, it appears likely that recognition of certain ligands will involve residues contributed by several subunits, thereby considerably enlarging the spectrum of the C1q targets. This assumption appears consistent with the observation that many C1q ligands have been found to interact in a significant way with different subunits of its globular domain (Kishore et al., 2003). As discussed later, the diversity of the recognition modes of C1q is well illustrated by the models proposed for C-reactive protein and IgG1, where binding is thought to involve the apex of the C1q globular domain, and the equatorial region of subunit B, respectively (Gaboriaud et al., 2003).
Recent investigations on the targets recognized by C1q at the apoptotic cell surface have allowed us to identify several ligands (Païdassi et al., 2008a,b, 2011). In all cases, the C1q moiety responsible for binding has been shown to be the globular domain.
Phosphatidylserine is currently one of the best documented recognition signal required for apoptotic cell clearance. Its tethering is mediated directly by phagocyte receptors, with or without the help of soluble bridging molecules (Savill et al., 2002). We have recently shown that C1q is one of the molecules that bridge phosphatidylserine exposed at the surface of early apoptotic cells to the phagocyte (Païdassi et al., 2008a). The interaction is mediated by its globular domain, which recognizes the phosphoserine moiety of phosphatidylserine. Annexin V, which binds avidly to phosphatidylserine, inhibits the interaction, and C1q and its globular domain both co-localize with phosphatidylserine on the apoptotic cell surface. Surface plasmon resonance analysis of the interaction between the C1q globular domain and phosphatidylserine has revealed high-affinity (Figure 3A) and the phosphoserine binding site on C1q has been identified by X-ray crystallography (Figure 4).
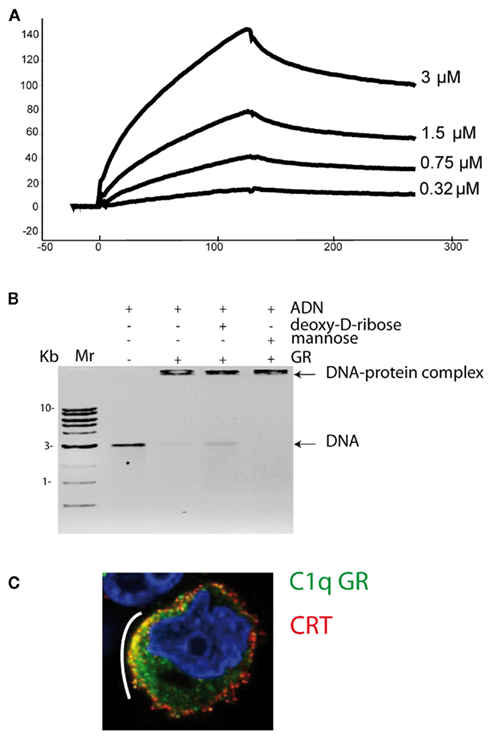
Figure 3. C1q binding to apoptotic cell ligands. (A) Surface plasmon resonance analysis of the binding of the C1q globular domain to immobilized phosphatidylserine. The concentrations of the C1q globular domain are indicated. (B) Electrophoretic mobility shift assay showing that complex formation between DNA and the C1q globular domain is partly inhibited by 100 mM deoxy-D-ribose. Mannose was used as a negative control. DNA molecular weight markers are shown on the left. (C) Early apoptotic HeLa cells were submitted to a double-immunofluorescence labeling for the C1q globular domain (C1q GR, green) and calreticulin (CRT, red) followed by confocal laser microscopy detection. Nuclei were labeled with Hoechst (blue). The white bow indicates areas where the C1q globular domain co-localizes with calreticulin. (From Païdassi et al., 2008a,b, 2011).
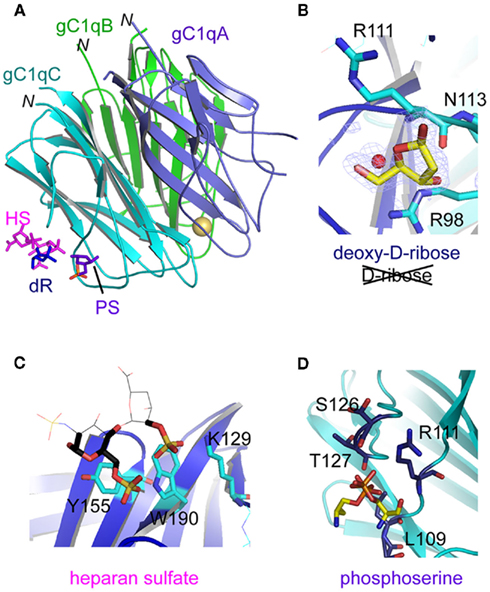
Figure 4. Location of three non-immune self ligand-binding sites in the C1q globular domain. (A) Overall view of the C1q globular domain indicating the relative positioning of the binding sites for heparan sulfate (HS), deoxy-D-ribose (dR), and phosphoserine (PS). (B–D) Detailed views of the binding sites for deoxy-D-ribose, heparan sulfate, and phosphoserine, respectively. Residues of the C subunit involved in the interaction are shown. In (C), the ligand portion displayed in sticks corresponds to the major interpretable extra electron-density. In (D), a double conformation was used to interpret the extra-density corresponding to phosphoserine. (Modified from Garlatti et al., 2010 and Païdassi et al., 2008a).
Nuclear splitting and DNA fragmentation are well-established features of apoptosis, and the generation of auto-antibodies directed against chromatin components is a characteristic of autoimmune diseases such as systemic lupus erythematosus and rheumatoid arthritis (Shoenfeld et al., 1987). It was recently demonstrated that nucleic acids are rapidly exposed during apoptosis and provide ligands for C1q binding (Elward et al., 2005). In an attempt to characterize C1q binding to DNA, we have shown for the first time that the globular region of C1q displays a lectin-like activity that contributes to DNA binding (Païdassi et al., 2008b). This observation is consistent with the observation that treatment of apoptotic cells with DNase decreases significantly binding to C1q. Studies at the molecular level have demonstrated that the C1q globular domain binds DNA through its pentose moiety, and that deoxy-D-ribose inhibits formation of complexes between DNA and the C1q globular domain (Figure 3B). The structure of the deoxy-D-ribose binding site on the C1q globular domain has been deciphered at high resolution by X-ray crystallography.
Calreticulin was first identified as a receptor for the collagenous moiety of C1q and the collectins at the surface of phagocytes. However, based on more recent studies, it is now widely accepted that calreticulin is a major recognition signal at the apoptotic cell surface, acting as an “eat-me” signal in the uptake of apoptotic cells by macrophages (Gardai et al., 2006; Obeid et al., 2007). This latter finding evokes a double play for this protein, as both a phagocyte receptor and an “eat-me” signal on the apoptotic cell surface, with the help of the bridging molecule C1q. In support of this hypothesis, we have recently demonstrated that the C1q globular domain, responsible for C1q binding to apoptotic cells, binds calreticulin with high-affinity and co-localizes with this protein on the apoptotic cell surface (Figure 3C; Païdassi et al., 2011).
To decipher the recognition properties of the C1q globular domain, its X-ray structure has been solved in the presence of three different small ligands corresponding to molecular determinants recognized by C1q in larger target molecules from self: (i) phosphoserine, the motif recognized in phosphatidylserine, (ii) deoxy-D-ribose, a specific determinant of DNA, and (iii) two repetitive units of heparan sulfate (Païdassi et al., 2008a; Garlatti et al., 2010). The three structures were solved using crystal soaking because attempts to generate co-crystals of the C1q globular domain in complex with a ligand were unsuccessful. The soaking technique introduces space constraints, and is more favorable to small ligands. The smallest ligand tested, deoxy-D-ribose, was observed at the highest resolution (1.25 Å) in a convincing binding site located in subunit C of the C1q globular domain (Garlatti et al., 2010). It is accommodated in a small pocket lying between ArgC98 and ArgC111 (Figure 4B). Two of its hydroxyl groups are stabilized by direct and water-mediated polar interactions with AsnC113, ArgC111, and ArgC118. The interaction is clearly specific to deoxy-D-ribose, since no ligand fixation could be detected when the same protocol was applied using D-ribose. Indeed, the additional hydroxyl group present in ribose would clash sterically with AsnC113 in the observed configuration, providing a structural basis for the strict specificity of C1q toward deoxy-D-ribose. The other two ligands investigated, phosphoserine and heparan sulfate, are both negatively charged, and interact approximately in the same area of the C subunit of the C1q globular domain (Figure 4A), although they differ in their precise interaction modes (Païdassi et al., 2008a; Garlatti et al., 2010). Thus, heparan sulfate is interacting with TyrC155, TrpC190, and LysC129 (Figure 4C), whereas phosphoserine binds ArgC111, SerC126, and ThrC127 (Figure 4D). The sulfate/phosphate binding propensity of ArgC98, ArgC111, TyrC155, and TrpC190 suggests that this area has the ability to bind larger polyanionic molecules. If this hypothesis is correct, then the larger polyanionic molecules DNA and heparin are expected to compete with each other because of the close proximity of their binding sites. This hypothesis has been tested using competition experiments, providing evidence that, indeed, DNA and heparan sulfate fragments each inhibit in a dose-dependent manner C1q binding to a HS-coated surface, in complete agreement with X-ray crystallography analyses (Garlatti et al., 2010).
According to the C1q model derived from the X-ray structure of its globular domain (Figure 1C), the binding sites for phosphoserine, deoxy-D-ribose, and heparan sulfate, all located in subunit C, would be positioned on the inner face of the C1q cone, and oriented toward the target (Figures 4A and 5A). In contrast, the IgG binding site has been proposed to lie on the equatorial region of subunit B, on the outer part of the C1q molecule (Gaboriaud et al., 2003; Garlatti et al., 2010). In light of our current knowledge of the structure and activation mechanism of the C1 complex (Budayova-Spano et al., 2002; Gaboriaud et al., 2003, 2004; Bally et al., 2009; Brier et al., 2010), these locations have direct functional implications in terms of C1 activation. Thus, the X-ray structure of the zymogen C1r catalytic domain has led to the conclusion that an outward movement of the C1q stems is necessary to disrupt the resting head-to-tail dimeric structure of C1r and thereby trigger C1 activation (Figures 5B–D). If this assumption is correct, any ligand recognized through the outer part of the C1q globular domain, as proposed for IgG (Figure 5A), is expected to trigger efficient C1 activation (Figure 5F). Conversely, given the inner positioning of their interaction sites, binding of ligands such as phosphoserine, deoxy-D-ribose, and heparan sulfate would be unable to generate the appropriate outward movement of the C1q stems, particularly if these ligands are clustered in dense surface patches, thus preventing or restraining activation of the C1 complex. In full agreement with this hypothesis, it has been shown that immune complexes trigger efficient activation of C1 in the presence of C1 inhibitor, in contrast to DNA and heparin (Figure 5E; Ziccardi, 1982; Garlatti et al., 2010).
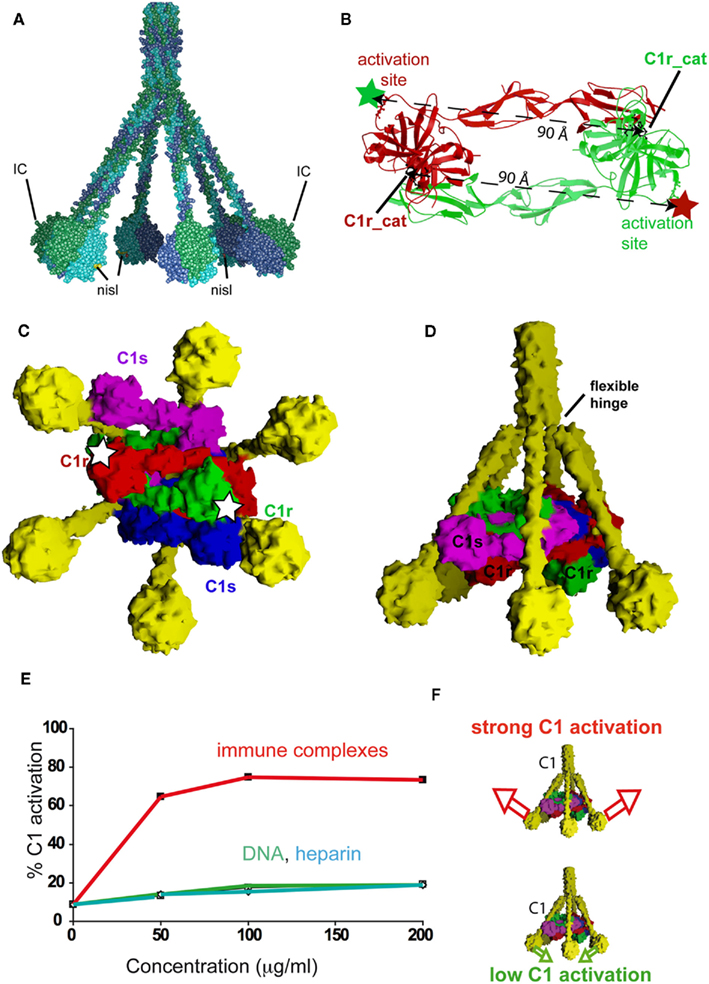
Figure 5. Implications on C1 activation of the location of ligand-binding sites in the C1q globular domain. (A) Location of the ligand-binding sites according to the current version of the C1q model. Nisl: non-immune self ligands binding area as described in Figure 4; IC: proposed binding sites for IgG-containing immune complexes. (B) Dimeric structure of the C1r catalytic domain in its “resting state.” In this configuration, a distance of 90 Å is observed between the catalytic site (C1r_cat) of one subunit and the activation site (colored star) to be cleaved in the other subunit. Such a distance prevents unwanted spontaneous C1r autoactivation, which requires transient disruption of the dimer. (C,D) Bottom and side views, respectively, of the current C1 model (Bally et al., 2009; Brier et al., 2010). In (C) white stars show the approximate positions of the C1r activation sites. (E) C1 activation by IgG-containing immune complexes, heparin and DNA. (F) Schematic interpretation of (E). According to our hypothesis, a strong outward movement of the C1q stems (red arrows) induced by binding to immune complexes is expected to disrupt the C1r catalytic dimer and thereby induce C1 activation. In contrast, binding to non-immune self ligands would generate little or no activation (green arrows). (Modified from Budayova-Spano et al., 2002 and Garlatti et al., 2010).
Uncontrolled activation of the classical complement pathway is involved in many inflammatory pathologies. Conversely, C1q deficiency is associated with major defaults in the uptake of apoptotic cells and correlates with autoimmune diseases such as systemic lupus erythematosus and glomerulonephritis, emphasizing the crucial role of C1q in the maintenance of immune tolerance (Botto and Walport, 2002). Whereas heparan sulfate and other sulfated molecules are known to inhibit complement activation, DNA, and phosphatidylserine are both eat-me signals involved in the removal of apoptotic cells by C1q. It is tempting to hypothesize that, because the binding sites for these ligands are all located within the same area of the C1q globular domain, on the inner edge of the C1q cone, their recognition by C1q only generates low C1 activation, hence contributing to the control of the inflammatory reaction, in addition to complement regulators (Zipfel and Skerka, 2009). Recently, another molecule from self, heme, was shown to bind C1q and inhibit C1 activation by immune complexes and C-reactive protein (Roumenina et al., 2011). Interestingly, one of the two heme binding sites postulated by these authors is located in subunit C of the C1q globular domain, in the vicinity of the sites identified for DNA, phosphatidylserine, and heparan sulfate, suggesting that heme-mediated inhibition of C1 activation could involve a similar mechanism.
It is clear, however, that this scheme does not apply to all known non-immune C1q self ligands. Thus, whereas the oxidized form of low-density lipoprotein (ox-LDL) does not trigger C1 activation despite its ability to bind C1q with high-affinity (Biro et al., 2007), the enzymatically modified species (E-LDL) is a potent C1 activator (Biro et al., 2007, 2010). In the same way, cardiolipin, β-amyloid fibrils, as well as different forms of the prion protein have been shown to bind C1q and activate C1 to various extents (Peitsch et al., 1988; Tacnet-Delorme et al., 2001; Dumestre-Pérard et al., 2007; Sim et al., 2007; Sjöberg et al., 2008). Further investigations at the molecular level will be necessary to generate a more detailed map of the ligand-binding sites of C1q and uncover all the secrets of this unique sensor molecule. Such advances would have important potential therapeutic applications, considering that ligand recognition by C1q may elicit both beneficial and deleterious effects, as exemplified in neurodegenerative diseases where both types of effects have been reported (van Beek et al., 2003; Veerhuis et al., 2011). Ideally, the aim would be to modulate the classical complement pathway by preventing or limiting its noxious effects while preserving its protective role. In the long term, this goal should be achievable through the design of inhibitory molecules able to specifically target the strong C1-activating binding sites located on the outer part of the C1q globular domain, while preserving the functionality of the inner binding area allowing recognition and efficient clearance of altered self ligands.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors greatly acknowledge the contributions to the studies referred to in this review of all past and present members of the laboratory, with particular attention to Claudine Darnault, Virginie Garlatti, Jordi Juanhuix, Monique Lacroix, Helena Païdassi, and Pascale Tacnet-Delorme.
Bally, I., Rossi, V., Lunardi, T., Thielens, N. M., Gaboriaud, C., and Arlaud, G. J. (2009). Identification of the C1q-binding sites of human C1r and C1s: a refined three-dimensional model of the C1 complex of complement. J. Biol. Chem. 284, 19340–19348.
Biro, A., Ling, W. L., and Arlaud, G. J. (2010). Complement protein C1q recognizes enzymatically modified low-density lipoprotein through unesterified fatty acids generated by cholesterol esterase. Biochemistry 49, 2167–2176.
Biro, A., Thielens, N. M., Cervenak, L., Prohaszka, Z., Füst, G., and Arlaud, G. J. (2007). Modified low density lipoproteins differentially bind and activate the C1 complex of complement. Mol. Immunol. 44, 1169–1177.
Bogin, O., Kvansakul, M., Rom, E., Singer, J., Yayon, A., and Hohenester, E. (2002). Insights into Schmid metaphyseal chondrodysplasia from the crystal structure of the collagen X NC1 domain trimer. Structure 10, 165–173.
Brier, S., Pflieger, D., Le Mignon, M., Bally, I., Gaboriaud, C., Arlaud, G. J., and Daniel, R. (2010). Mapping surface accessibility of the C1r/C1s tetramer by chemical modification and mass spectrometry provides new insights into assembly of the human C1 complex. J. Biol. Chem. 285, 32251–32263.
Budayova-Spano, M., Lacroix, M., Thielens, N. M., Arlaud, G. J., Fontecilla-Camps, J. C., and Gaboriaud, C. (2002). The crystal structure of the zymogen catalytic domain of complement protease C1r reveals that a disruptive mechanical stress is required to trigger activation of the C1 complex. EMBO J. 21, 231–239.
Cooper, N. R. (1985). The classical pathway of complement: activation and regulation of the first complement component. Adv. Immunol. 37, 151–216.
Dumestre-Pérard, C., Osmundson, J., Lemaire-Vieille, C., Thielens, N., Grives, A., Favier, B., Csopaki, F., Jamin, M., Gagnon, J., and Cesbron, J. Y. (2007). Activation of classical pathway of complement cascade by soluble oligomers of prion. Cell. Microbiol. 9, 2870–2879.
Eck, M. J., and Sprang, S. R. (1989). The structure of tumor necrosis factor-α at 2.6 Å resolution. Implications for receptor binding. J. Biol. Chem. 264, 17595–17605.
Elward, K., Griffiths, M., Mizuno, M., Harris, C. L., Neal, J. W., Morgan, B. P., and Gasque, P. (2005). CD46 plays a key role in tailoring innate immune recognition of apoptotic and necrotic cells. J. Biol. Chem. 280, 36342–36354.
Erlich, P., Dumestre-Pérard, C., Ling, W. L., Lemaire-Vieille, C., Schoehn, G., Arlaud, G. J., Thielens, N. M., Gagnon, J., and Cesbron, J. Y. (2010). Complement protein C1q forms a complex with cytotoxic prion protein oligomers. J. Biol. Chem. 285, 19267–19276.
Fraser, D. A., Laust, A. K., Nelson, E. L., and Tenner, A. J. (2009). C1q differentially modulates phagocytosis and cytokine responses during ingestion of apoptotic cells by human monocytes, macrophages, and dendritic cells. J. Immunol. 183, 6175–6185.
Gaboriaud, C., Juanhuix, J., Gruez, A., Lacroix, M., Darnault, C., Pignol, D., Verger, D., Fontecilla-Camps, J. C., and Arlaud, G. J. (2003). The crystal structure of the globular head of complement protein C1q provides a basis for its versatile recognition properties. J. Biol. Chem. 278, 46974–46982.
Gaboriaud, C., Thielens, N. M., Gregory, L. A., Rossi, V., Fontecilla-Camps, J. C., and Arlaud, G. J. (2004). Structure and activation of the C1 complex of complement: unravelling the puzzle. Trends Immunol. 25, 368–373.
Gardai, S. J., Bratton, D. L., Ogden, C. A., and Henson, P. M. (2006). Recognition ligands on apoptotic cells: a perspective. J. Leukoc. Biol. 79, 896–903.
Garlatti, V., Chouquet, A., Lunardi, T., Vivès, R., Païdassi, H., Lortat-Jacob, H., Thielens, N. M., Arlaud, G. J., and Gaboriaud, C. (2010). Cutting edge: C1q binds deoxyribose and heparan sulfate through neighboring sites of its recognition domain. J. Immunol. 185, 808–812.
Jones, E. Y., Stuart, D. I., and Walker, N. P. (1989). Structure of tumour necrosis factor. Nature 338, 225–228.
Kang, Y. S., Do, Y., Lee, H. K., Park, S. H., Cheong, C., Lynch, R. M., Loeffler, J. M., Steinman, R. M., and Park, C. G. (2006). A dominant complement fixation pathway for pneumococcal polysaccharides initiated by SIGN-R1 interacting with C1q. Cell 125, 47–58.
Kishore, U., Gaboriaud, C., Waters, P., Shrive, A. K., Greenhough, T. J., Reid, K. B., Sim, R. B., and Arlaud, G. J. (2004). C1q and tumor necrosis factor superfamily: modularity and versatility. Trends Immunol. 25, 551–561.
Kishore, U., Gupta, S. K., Perdikoulis, M. V., Kojouharova, M. S., Urban, B. C., and Reid, K. B. (2003). Modular organization of the carboxyl-terminal, globular head region of human C1q A, B, and C chains. J. Immunol. 171, 812–820.
Klein, M. A., Kaeser, P. S., Schwarz, P., Weyd, H., Xenarios, I., Zinkernagel, R. M., Carroll, M. C., Verbeek, J. S., Botto, M., Walport, M. J., Molina, H., Kalinke, U., Acha-Orbea, H., and Aguzzi, A. (2001). Complement facilitates early prion pathogenesis. Nat. Med. 7, 488–492.
Kvansakul, M., Bogin, O., Hohenester, E., and Yayon, A. (2002). Crystal structure of the collagen α1(VIII) NC1 trimer. Matrix Biol. 22, 145–152.
Marqués, G., Anton, L. C., Barrio, E., Sanchez, A., Ruiz, A., Gavilanes, F., and Vivanco, F. (1993). Arginine residues of the globular regions of human C1q involved in the interaction with immunoglobulin G. J. Biol. Chem. 268, 10393–10402.
Nauta, A. J., Castellano, G., Xu, W., Woltman, A. M., Borrias, M. C., Daha, M. R., van Kooten, C., and Roos, A. (2004). Opsonization with C1q and mannose-binding lectin targets apoptotic cells to dendritic cells. J. Immunol. 173, 3044–3050.
Navratil, J. S., Watkins, S. C., Wisnieski, J. J., and Ahearn, J. M. (2001). The globular heads of C1q specifically recognize surface blebs of apoptotic vascular endothelial cells. J. Immunol. 166, 3231–3239.
Obeid, M., Tesniere, A., Ghiringhelli, F., Fimia, G. M., Apetoh, L., Perfettini, J. L., Castedo, M., Mignot, G., Panaretakis, T., Casares, N., Metivier, D., Larochette, N., van Endert, P., Ciccosanti, F., Piacentini, M., Zitvogel, L., and Kroemer, G. (2007). Calreticulin exposure dictates the immunogenicity of cancer cell death. Nat. Med. 13, 54–61.
Païdassi, H., Tacnet-Delorme, P., Garlatti, V., Darnault, C., Ghebrehiwet, B., Gaboriaud, C., Arlaud, G. J., and Frachet, P. (2008a). C1q binds phosphatidylserine and likely acts as a multiligand bridging molecule in apoptotic cell recognition. J. Immunol. 180, 2329–2338.
Païdassi, H., Tacnet-Delorme, P., Lunardi, T., Arlaud, G. J., Thielens, N. M., and Frachet, P. (2008b). The lectin-like activity activity of human C1q and its implication in DNA and apoptotic cell recognition. FEBS Lett. 582, 3111–3116.
Païdassi, H., Tacnet-Delorme, P., Verneret, M., Gaboriaud, C., Houen, G., Duus, K., Ling, W. L., Arlaud, G. J., and Frachet, P. (2011). Investigations on the C1q-calreticulin-phosphatidylserine interactions yield new insights into apoptotic cell recognition. J. Mol. Biol. 408, 277–290.
Peitsch, M. C., Tschopp, J., Kress, A., and Isliker, H. (1988). Antibody-independent activation of the complement system by mitochondria is mediated by cardiolipin. Biochem. J. 249, 495–500.
Roumenina, L. T., Kantardjiev, A. A., Atanasov, B. P., Waters, P., Gadjeva, M., Reid, K. B., Mantovani, A., Kishore, U., and Kojouharova, M. S. (2005). Role of Ca2+ in the electrostatic stability and the functional activity of the globular domain of human C1q. Biochemistry 44, 14097–14109.
Roumenina, L. T., Radanova, M., Atanasov, B. P., Popov, K. T., Kaveri, S. V., Lacroix-Desmazes, S., Frémeaux-Bacchi, V., and Dimitrov, J. D. (2011). Heme interacts with C1q and inhibits the classical complement pathway. J. Biol. Chem. 286, 16459–16469.
Savill, J., Dransfield, I., Gregory, C., and Haslett, C. (2002). A blast from the past: clearance of apoptotic cells regulates immune responses. Nat. Rev. Immunol. 2, 965–975.
Shapiro, L., and Scherer, P. E. (1998). The crystal structure of a C1q family protein suggests an evolutionary link to tumor necrosis factor. Curr. Biol. 8, 335–338.
Shoenfeld, Y., Segol, G., Segol, O., Neary, B., Klajman, A., Stollar, B. D., and Isenberg, D. A. (1987). Detection of antibodies to total histones and their subfractions in systemic lupus erythematosus patients and their asymptomatic relatives. Arthritis Rheum. 30, 169–175.
Sim, R. B., Kishore, U., Villiers, C. L., Marche, P. N., and Mitchell, D. A. (2007). C1q binding and complement activation by prions and amyloids. Immunobiology 212, 355–362.
Sjöberg, A. P., Nyström, S., Hammarström, P., and Blom, A. M. (2008). Native, amyloid fibrils and beta-oligomers of the C-terminal domain of human prion protein display differential activation of complement and bind C1q, factor H and C4b-binding protein directly. Mol. Immunol. 45, 3213–3221.
Szalai, A. J., Agrawal, A., Greenhough, T. J., and Volanakis, J. E. (1999). C-reactive protein: structural biology and host defense function. Clin. Chem. Lab. Med. 37, 265–270.
Tacnet-Delorme, P., Chevallier, S., and Arlaud, G. J. (2001). Beta-amyloid fibrils activate the C1 complex of complement under physiological conditions: evidence for a binding site for A beta on the C1q globular regions. J. Immunol. 167, 6374–6381.
Taylor, P. R., Carugati, A., Fadok, V. A., Cook, H. T., Andrews, M., Carroll, M. C., Savill, J. S., Henson, P. M., Botto, M., and Walport, M. J. (2000). A hierarchical role for classical pathway complement proteins in the clearance of apoptotic cells in vivo. J. Exp. Med. 192, 359–366.
Thielens, N. M., Tacnet-Delorme, P., and Arlaud, G. J. (2002). Interaction of C1q and mannan-binding lectin with viruses. Immunobiology 205, 563–574.
van Beek, J., Elward, K., and Gasque, P. (2003). Activation of complement in the central nervous system: roles in neurodegeneration and neuroprotection. Ann. N. Y. Acad. Sci. 992, 56–71.
Veerhuis, R., Nielsen, H. M., and Tenner, A. J. (2011). Complement in the brain. Mol. Immunol. 48, 1592–1603.
Ziccardi, R. J. (1982). A new role for C1-inhibitor in homeostasis: control of activation of the first component of human complement. J. Immunol. 128, 2505–2508.
Keywords: C1q, complement, innate immunity, ligand recognition, X-ray crystallography
Citation: Gaboriaud C, Frachet P, Thielens NM and Arlaud GJ (2012) The human C1q globular domain: structure and recognition of non-immune self ligands. Front. Immun. 2:92. doi: 10.3389/fimmu.2011.00092
Received: 25 October 2011;
Paper pending published: 09 December 2011;
Accepted: 21 December 2011;
Published online: 06 January 2012.
Edited by:
Uday Kishore, Brunel University, UKReviewed by:
Uday Kishore, Brunel University, UKCopyright: © 2012 Gaboriaud, Frachet, Thielens and Arlaud. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Gérard J. Arlaud, Commissariat à l’énergie atomique, CNRS, Institut de Biologie Structurale, Université Joseph Fourier-Grenoble 1, 41 rue Jules Horowitz, 38027 Grenoble Cedex 1, France. e-mail:YXJsYXVkLmdlcmFyZEBvcmFuZ2UuZnI=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.