
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Imaging , 18 May 2023
Sec. Imaging Applications
Volume 2 - 2023 | https://doi.org/10.3389/fimag.2023.1175860
Non-invasive, label-free hyperspectral imaging technologies are powerful tools that can provide critical insights into the spatiotemporal dynamics of evolving systems. They are already integrated into autonomous adaptive surveillance workflows for precision agriculture, remote sensing, environmental monitoring, and food analysis. However, these technologies are not yet routinely used in medicine. With these technologies applied to the biomedical sciences, we anticipate landmark advances in the physicochemical behavioral refinement of high signal-to-noise probes and in the rapid screening of patient exosome populations. To implement label-free spatiochemical imaging techniques while meeting the practical needs of clinicians, technology, workflow design and development must be tailored to the specific medical field while considering existing visualization modalities. This perspective merges basic, translational, and clinical views of label-free, non-invasive hyperspectral imaging. We explore its potential role in positively impacting cancer-related precision medicine and early diagnostics to support clinically implemented point-of-care technologies.
Spatiochemical comprehension of biological systems is important for early diagnostics and precision medicine in oncology (Li et al., 2021), especially since these systems are dynamic (Biswas and Drivers, 2021) while being chemically and structurally complex. Throughout the early 2000s, synchrotron-based FTIR spectromicroscopy was shown to be non-invasive and non-destructive (Martin et al., 2001), even when observing living biological systems (Holman et al., 2010) in real-time over the course of multiple days (Loutherback et al., 2015). Research performed as early as 2003 showed the potential of FTIR spectromicroscopy's use in biomedical applications (Holman et al., 2003). By 2016, FTIR spectromicroscopy's application to cancer research had progressed to the level of identifying spectroscopic biomarker regions relating to identification and characterization of cancer drug resistance (Junhom et al., 2016) and cancer cell reactivity to potentially therapeutic agents (Al-Jorani et al., 2018). As new advancements in quantum cascade laser (Razeghi et al., 2015), optics (Prater et al., 2023), detector (Verma et al., 2021), and software technologies (Toplak et al., 2017; Holman et al., 2020) are commercially implemented, FTIR spectromicroscopy with acceptable spatiochemical resolution and computationally efficient autonomous adaptive data acquisition (Holman et al., 2020, 2023) will become more accessible to the general biomedical science community (Großerueschkamp and Gerwert, 2020).
In this perspective, we select colorectal cancer as our specific medical field of conversation for two reasons. Firstly, it has one of the highest cancer incidence and cancer-related mortality rates (Bray et al., 2018; Siegel et al., 2023); hence, there is an unmet clinical need for precision medicine and early diagnostics. Secondly, colorectal cancer screening, treatment, and surveillance involve endoscopic technologies that have progressed rapidly in the past decade to incorporate multimodal real-time imaging; this implies that the endoscopic hardware is at a point where the imaging technology can benefit from advances in supportive fields such as probe design and performance optimization. By developing these supportive fields, we can improve the quality of patient care using existing imaging technologies. Herein, we will discuss near-future applications of non-invasive, label-free hyperspectral imaging for translational research in probe characterization and development as well as early diagnostics via exosome screening. It is important to note that our near-future applications were chosen for their conserved generalizability and modularity to the overall cancer management framework, despite our decision to narrow the medical field scope for succinct discussion.
Over the past two decades, endoscopic technologies for gastrointestinal diagnostic and therapeutic procedures (Figures 1A, B) (Yim et al., 2020) have improved drastically to present clinicians with a functional variety of endoscopes ranging from multimodal (Lombardini et al., 2018), smart (Schembre, 2004; Zhou et al., 2017; Erendgenova et al., 2018), intraoperative optical guidance (Mieog et al., 2010; Boni et al., 2015; Garai et al., 2015), and wireless (Iddan et al., 2000) capabilities. However, a key challenge to improving the clinical performance of multimodal intraoperative guidance systems emerges from the need to understand the underlying mechanisms behind the physicochemical behavior of the used medical imaging contrast agents. A major reason for incomplete removal of cancerous tissue in potentially resectable lesions stems from difficulties in completely identifying tumor-positive margins during surgery (Macdonald, 1999). Although many factors contribute to inaccurate identification of lesions, there exists a conserved difficulty when using contrast agents due to the altered mucosal appearance. Specifically, clinical and experimental visualization modalities such as standard white light endoscopy and near-infrared imaging have difficulty in accurately differentiating between tumor and inflammatory colonic tissue (Holt et al., 2014; Feuerstein et al., 2019). Further studies and characterization of existing contrast agents are necessary in order to reduce the persistent background retention (PBR) of the imaging probes which potentially are related to non-specific probe binding, non-specific cellular uptake of the probe, inadequate probe clearance from non-target tissue, and non-target tissue metabolic rates (Frangioni, 2008). By increasing our mechanistic understanding of probe-tissue interactions that drive an imaging probe's PBR, we believe that the research community can not only improve and optimize probe chemistry for the specific target system of interest but also obtain better comprehension of the diverse, heterogenous tumor microenvironments and their physicochemical relationship to contrast agent performance.
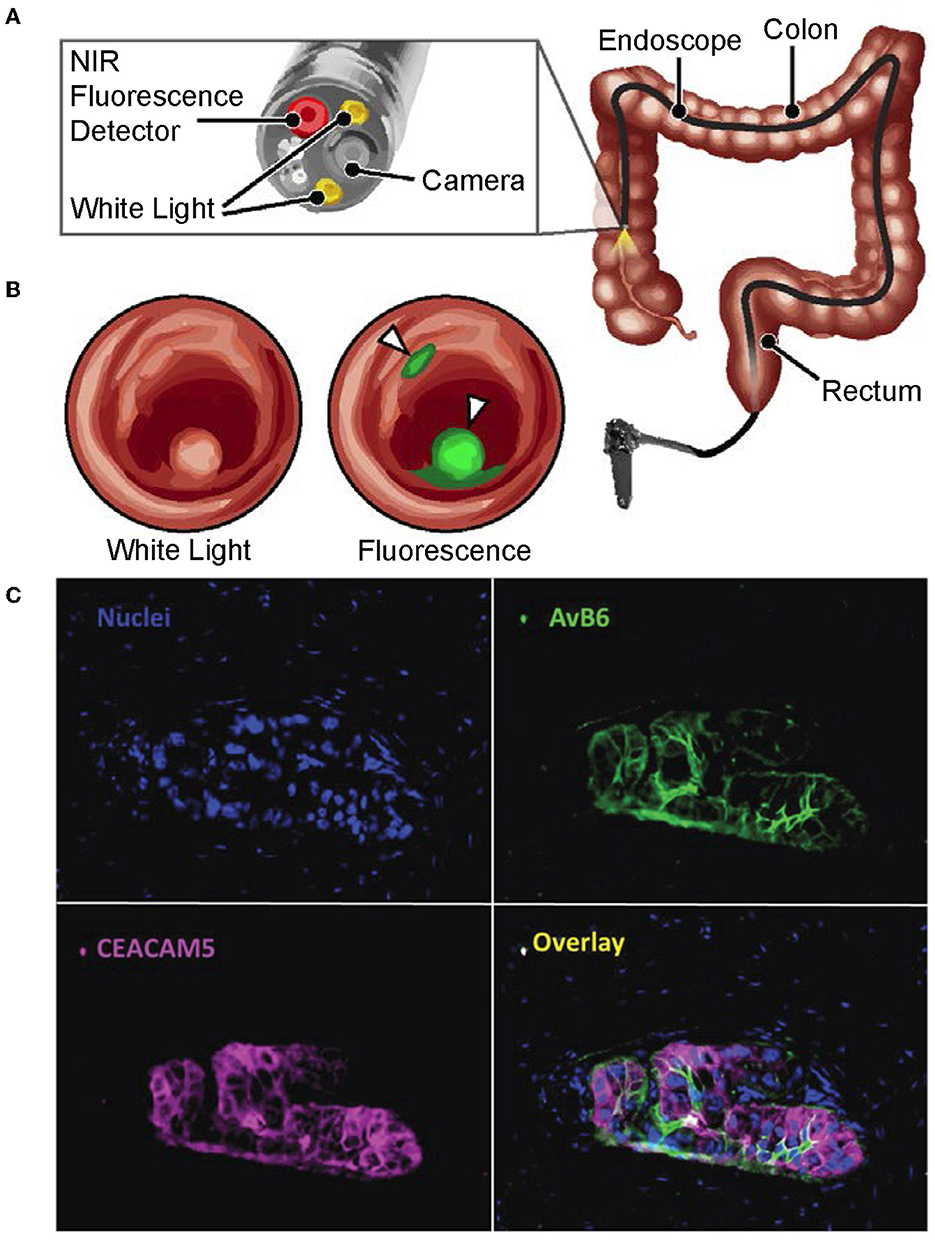
Figure 1. Multimodal endoscopic technologies for cancer surveillance. (A, B) Reproduced with permission from Yim et al. (2020). Copyright 2020 National Academy of Sciences. (A) Schematic of a near-infrared fluorescence endoscope with white-light capabilities that is used in the clinic. (B) Conceptual image illustrating how a fluorescent dye can be used by multimodal image-guidance systems in clinic to identify cancerous lesions, with these results being overlayed onto a white-light image. (C) Reproduced with permission from Tummers et al. (2017). Copyright 2017 Authors licensed under a Creative Commons Attribution (CC BY) License. The visualization of advantages made possible by using image probe multiplexing. Nuclei of normal and tumor-positive tissue are overlayed with results from simultaneous immunofluorescent staining of αvβ6 and CEACAM5 on the same sample. The image overlay highlights the value of using multiple biomarkers to accurately identify total tumor volumes.
Tummers et al. (2017) emphasized and visually characterized the differential physicochemical behavior of cancer-specific diagnostic agents that exhibited differential physicochemical behavior across a tumor. As shown in Figure 1C (Tummers et al., 2017), the tumor tissue retention of two imaging probes together are able to completely identify the full tumor volume with its true tumor-positive margins whereas one alone is insufficient. These results suggest that accurate and comprehensive identification of tumor-positive tissue by image-guided systems in the clinic can be achieved through the combined use of multiple imaging probes, which can be performed either simultaneously or sequentially as needed. Although imaging probe, panel-based approaches relevant to clinical implementation are discussed later, we foresee the need of non-invasive, label-free imaging and chemometrics to characterize each probe's behavior in varying tumor microenvironments. We believe that this can be achieved by hyperspectral imaging modalities that provide spatially resolved, physicochemical characterization. By being able to describe the spatiochemical microenvironments that lead to clinically acceptable signal-to-background (SBR) levels to image-guided systems, we can:
(1) Identify which probes should be used in tandem in order to confidently label tumor-positive tissue as well as its margins and
(2) Begin to study and design probes for tumor microenvironments that do not have an acceptable SBR in clinical practice.
Our proposed approach that utilizes non-invasive label-free hyperspectral imaging is inspired by FTIR spectromicroscopy approaches that have been used for decades in the space and geological sciences (Ventura, 2015). Similar to endoscopic practices relating to early colorectal cancer diagnostics (Davnall et al., 2012), these fields characterize the physical aspect of their geological samples with metrics that assess the physical structure of the material, such as the level of structural order, pore density, and metrics describing pore shape (Selvadurai and Suvorov, 2020). These metrics together aid in elucidating the relationship between a material's microstructure and its poroelasticity; in the medical context, this would correspond to the tumor matrix's microstructure and how the fluid-saturated porous microstructure can influence the flow of fluids through, within, and eternal to the tumor.
We propose to combine non-invasive, non-destructive and label-free hyperspectral imaging approaches such as FTIR spectromicroscopy with already existing analytical techniques for assessing physical structure to chemical function correlations. This experimental workflow would use hyperspectral imaging upstream of the more destructive X-ray based techniques (Gianoncelli et al., 2015; Petruzzellis et al., 2018) to assess the same sample for spatiochemical information and high-resolution physical microstructural data. Since these datasets are obtained from the same sample with maintained sample integrity for their relevant imaging methods, they are directly comparable to one another. We believe that elucidating these relationships through experimental design and careful execution can lead to a better understanding of how microstructure-driven compartmentalized chemistries could affect dynamic intratumor phenotypic heterogeneity and its phenotypic plasticity (Biswas and Drivers, 2021).
By performing physicochemical probe-based ex vivo studies on tumor-positive colonic tissue and contrasting its spatiochemically-defined tissue architecture to normal colonic tissue, we can begin to construct the necessary foundation of knowledge that can link genetic (Hoorn et al., 2022) and non-genetic characterizations of cancers through the spatiochemical domain of function-based physicochemical microstructures and their subsequent functional effect on probe performance. As an example, non-targeted probes such as indocyanine green (ICG) (Fox, 1960) or indigo carmine dye (Kudo et al., 1997) have been used predominantly in molecular surgery and endoscopy prior to the emergence of targeted probes (Stibbe et al., 2022). However, ICG has been shown in intraoperative fluorescence-based studies to have variable levels of success depending upon its cancer application (Schmidt et al., 2017). Some clinical trials highlight a lack of significant difference in tumor-to-background (TBR) ratios between malignant (TBR = 2.0) and benign lesions (TBR = 2.0) in their specific cancer type (Tummers et al., 2015). This suggests at least two alternative approaches to address the TBR challenge. First, one can use target-specific contrast agents such as enzyme-based probes (Ofori et al., 2015; Fernandez et al., 2019; Widen et al., 2021; Liang et al., 2022) and design the molecular targets with a specific tumor region in mind (leading edge vs. the tumor core). Second, one can design non-specific environmental probes (Walke et al., 2023) that follow spatiochemical AND-gate logic when used in combination with other non-specific probes such as ICG—phrased alternatively, if these two microstructure-based spatial chemistries (e.g. acidity) exist in proximity to one another, we are relatively confident in classifying the labeled tissue as tumor-positive. These spatial relationships to compartmentalized tumor chemistries and their local microbial and chemical environments can be characterized by FTIR spectromicroscopy, an analytical technique that already has been used to study microbial chemistries (Holman, 2010) and to characterize the relationships between mineral, organic, and fluid phases in porous geologic materials (Pisapia et al., 2018).
The emergence and subsequent growth of this knowledge domain will increase our scientific and clinical understanding of the relationships between tumor microstructure, its microenvironments (Walke et al., 2023), and its effect on diagnostic and therapeutic agents. These relationships also can aid the deconvolution of multilevel genetic and non-genetic tumor heterogeneity information (Biswas and Drivers, 2021), which can build toward eventual in vivo dynamic combinatorial labeling for intraoperative probe-based fluorescence endoscopy (Vandebriel et al., 2023). We believe that advances in these areas will lead to the potential identification of novel and practical avenues of diagnostic and therapeutic practices in precision medicine for cancer management.
Surgical or endoscopic resection and identification of target lesions is solely dependent on the experience of the performing physician. Visual alterations such as different tissue textures and consistencies are used to identify the tumor margins and biopsy locations are then selected based on the endoscopist's domain knowledge (Harmsen et al., 2017; Hernot et al., 2019; Stibbe et al., 2022). Long-term outcomes depend on accuracy of resection; unfortunately, residual positive cancerous margins appear in up to 70% of the cases (Hernot et al., 2019). More complex procedures such as endoscopic submucosal dissection can assist with the piece-meal resection of polyps that otherwise show a high recurrence rate; however, medical specialists are required for these treatments (Jacques et al., 2022). Thus, medical innovation in readily available imaging agents and technologies is desperately needed to enhance the contrast between normal and tumor-positive tissue to assist the clinician during image-guided medical procedures. Currently, there are very few fluorescent dyes that allow for optical surgical navigation and are fully FDA-approved (Harmsen et al., 2017; Hernot et al., 2019; Stibbe et al., 2022). Of these available FDA-approved agents, all are classified as non-targeted imaging probes. Finding new cancer-specific microenvironments and molecules to target is crucial in visualizing the entire tumor—with an ideal visualization to completeness including not only the tumor-positive tissue but also the dormant malignant and precancerous cells in the periphery (Harmsen et al., 2017; Tummers et al., 2017). However, new agents and technologies face significant hurdles when designing and implementing these findings into a clinical workflow. In addition, this opens a large opportunity to omics-technologies to identify new markers or characteristics inside tissue (Mayer et al., 2023).
The physicochemical properties of an agent, its route of administration, and its feasibility for procedural use must always be considered (Stibbe et al., 2022). Topical applications reduce risk of systemic reactions but can only be applied during the medical procedure and may have insufficient tissue penetration within a clinically relevant timeframe. Intravenous applications allow for good penetration depth but increases the risk of systemic reaction and toxicity. Furthermore, its administration typically requires the patient to be present at the facility well in advance of the procedure (Stibbe et al., 2022). In addition, procedure-relevant hardware must physically fit in a crowded clinical workspace, be safe to use, sterilizable, and cost-effective for medical insurance reimbursement. The new technology can neither increase procedural time nor require extensive training for integration into the clinical workflow (Harmsen et al., 2017). Moreover, the system must be compatible and optimized for the imaging agent's properties and for tissue autofluorescence. These requirements lead to only a handful systems that are currently used in experimental clinical settings (Harmsen et al., 2017). In summary, the agent and hardware must successfully integrate chemistry, hardware, and software as well as address medical needs without interrupting the clinical workflow (Harmsen et al., 2017).
The early detection of cancer is critical for improving treatment outcomes and patient survival (Schiffman et al., 2022). However, achieving early detection can be difficult, especially when cancer diagnosis requires an invasive procedure as with colonoscopies for colorectal cancer. Even when the diagnostic procedure is performed, early manifestations of cancer such as flat pre-cancerous dysplastic lesions may be difficult to differentiate from normal colonic tissue (Kaltenbach et al., 2008). Consequently, there is tremendous interest in generating personalized health and disease profiles through a patient's regular submission of liquid biopsies such as saliva and blood.
Exosomes are extracellular vesicles that are released by most cell types and found in all body fluids (Yáñez et al., 2015). Importantly, exosomes produced by cancer cells are chemically and biologically distinct from those produced by normal tissue (De Toro et al., 2015). Therefore, the routine screening of exosomes from liquid biopsies is a promising solution since exosomes serve as membrane-enclosed information capsules that are representative of their cells of origin. However, these samples in practice are highly complex. Exosomes that contribute to a patient's background populations may be derived from cells existing across a spectrum of different aging phenotypes, injuries, or sites of inflammation (Borges et al., 2013; Kulshreshtha et al., 2013; Zhang et al., 2017). For diagnostic purposes, these exosomes effectively form a patient-specific “background”. Thus, early diagnosis of cancer in a minimally invasive manner will require high-throughput, rapid screens of both background and disease associated exosomes with a preference for single-exosome characterization.
We believe that FTIR spectromicroscopy is well-suited for this application because it is label-free, noninvasive, and non-destructive (Martin et al., 2001; Holman et al., 2010). Furthermore, exosome sample preparations are often dried, negating the impact of water absorption bands while enabling the use of FTIR spectromicroscopy over other vibrational spectroscopy methods for increased chemical sensitivity per unit of time (Palonpon et al., 2013). Therefore, we primarily discuss exosome screening in the context of FTIR spectromicroscopy, though many of our outlined considerations can be generalized to other hyperspectral imaging technologies.
Numerous publications identify cancer-associated spectral shifts that can reliably differentiate among cancer cell lines as well as between cancer-derived and normal exosomes under carefully controlled laboratory conditions (Zlotogorski-Hurvitz et al., 2019; Romanò et al., 2020, 2022). However, these spectral changes are often subtle and difficult to interpret (Uthamacumaran et al., 2022). The subtlety of spectral changes may be further complicated by the site-to-site variability that has adversely affected applying machine learning to clinical imaging analysis (Willemink et al., 2020). Due to these considerations, we anticipate that the earliest clinically relevant implementations of FTIR spectroscopy for routine screening will serve as general indicators of disease presence. Diagnostic refinement would then be achieved via more focused and established technologies such as those detecting cancer-subtype specific exosome surface markers (Liu et al., 2018) and miRNAs (Taylor and Gercel-Taylor, 2008). Later implementations may eventually achieve broad, multi-functional diagnostics, but the practical performance of integrated classifiers in complex exosome backgrounds must first be determined with large numbers of patient samples. Broadly, FTIR spectral characterization of exosomes can be classified into two categories, and this categorization provides the important context for accurate sample analysis.
The first category is bulk exosome characterization. A purified exosome sample is often immobilized onto a slide by drying then assessed either in transmission mode, in reflectance mode, or by using an ATR objective for improved signal. In all the above cases, the incident beam is at best diffraction limited (2–10 μm) (Holman et al., 2010) and is considerably larger than the ~120 nm diameter of exosomes (Yu et al., 2018). Furthermore, depending on how rapidly sample drying is achieved, the process may result in a ‘coffee ring' effect (Choi and Birarda, 2017); exosomes may be concentrated in certain regions while depleted in others. This implies that each successful measurement will likely represent the properties of multiple exosomes. If disease-associated spectral changes are significant and present in a large fraction of exosomes, this method can be viable for the diagnosis and monitoring of disease progression (Blat et al., 2019). However, this approach's effectiveness may be greatly reduced for early diagnostic applications, as the fraction of disease-associated exosomes would be relatively small when compared to normal background exosomes.
The second category is single-exosome characterization, in which the signal is obtained on a per-exosome basis. The resolution improvements necessary for single-exosome sampling are achieved via scanning probe-based technologies. For these technologies, resolution is no longer diffraction-limited; it is instead a function of utilized probe-tip diameter. Two FTIR-dependent infrared nanospectroscopy methods are currently in use: AFM-IR (Dazzi et al., 2005) and SINS (Bechtel et al., 2020). As with bulk FTIR measurements, spectra acquired using these modalities are reported to be able to distinguish between cancer-derived and normal exosomes (Kim et al., 2018). While this single-exosome strategy has potential for establishing a per-patient normal background, the characterization in practice is limited to a small number of exosomes due to long data acquisition times. Although long acquisition times are not ideal for large-scale screening that potentially examines millions of exosomes, recent advances in infrared sources (Weida and Yee, 2011), signal enhancement methods (Dixon et al., 2020), and array-based strategies can bring high-throughput single-exosome characterization into the realm of feasibility.
Hyperspectral imaging has great potential for clinical use. However, due to being a relatively recent technology in the medical field that is still undergoing rapid advancement, it has yet to be substantively incorporated into clinical workflows. In this perspective, we present several biomedical applications of non-invasive label-free hyperspectral imaging that take into consideration the clinical requirements for practical implementation as well as the areas in which we believe that technological advances will likely occur. Each of these potential application cases is addressed in a dedicated section and has the potential for contributing directly or indirectly to early cancer diagnostics and precision medicine.
As shown in Figure 2, the general cycle of cancer patient diagnosis and treatment is a multi-step process requiring affordable and accessible point-of-care technologies; the applications we present directly address critical steps in this cycle. Our first proposed use of FTIR spectromicroscopy is for creating a relational database of physicochemical-microstructural information to clinical cancer subtype. This database would be used in optimizing combinatorial imaging probe selection for potential diagnostic and theragnostic improvement by the clinician. Our second proposed use is for the routine screening of exosomes, providing an information-rich yet minimally invasive method for early cancer diagnostics that could be performed outside of the clinic by specialized laboratories.
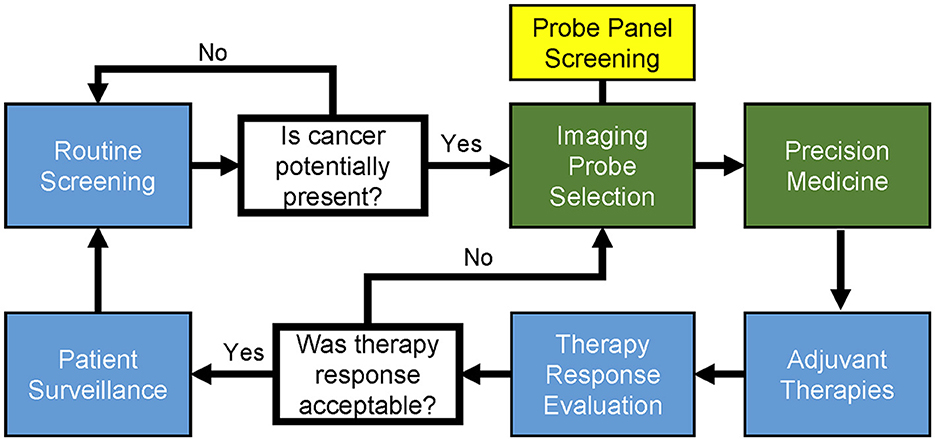
Figure 2. General workflow and cycle of patient diagnosis and therapy in precision medicine for cancer. Professional societies use statistically powerful, evidence-based medicine to establish and update the clinical guidelines (blue boxes). The diagnostic and precision medicine stages (green boxes) can be clinician-specific, often being the stages in which experimental procedures are performed. Patients enter the cycle at routine screening and proceed through the workflow as indicated. Once cancer presence is detected, identification of cancerous tissue can be performed with the visual aid of imaging probes. Probe screening and the subsequent selection of complementary imaging probe combinations is indicated (yellow box) as a modular component of general image probe selection. Notably, there exist imaging probes that contain theragnostic capabilities, such as those that can be used to image and photothermally ablate tumor-positive tissue.
By taking practical considerations of clinical implementation into account, we emphasize approaches that do not necessitate direct integration of broadband hyperspectral imaging at the point-of-care level but instead can improve a clinician's ability to perform early diagnostics and precision medicine for their patients through the efficient and effective use of existing imaging and treatment modalities.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
EH structured the manuscript, discussed each section with all authors, gathered materials from all authors, and wrote the introduction, biomedical research application, and discussion sections. DH wrote the exosome-based rapid screening section. SR wrote the practical considerations for clinical implementation section. EH, DH, and SR edited the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by NIH/NCI R01CA222836 (SR).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer FB declared a shared affiliation with the authors to the handling editor at the time of review.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Al-Jorani, K., Rüther, A., Haputhanthri, R., Deacon, G. B. L. i. H. L., Cullinane, C., et al. (2018). ATR-FTIR spectroscopy shows changes in ovarian cancer cells after incubation with novel organoamidoplatinum (ii) complexes. Analyst. 143, 6087–6094. doi: 10.1039/C8AN01558A
Bechtel, H. A., Johnson, S. C., Khatib, O., Muller, E.A., Raschke, M.B., et al. (2020). Synchrotron infrared nano-spectroscopy and –imaging. Surface Science Reports. 75, 100493. doi: 10.1016/j.surfrep.2020.100493
Biswas, A., and Drivers, D. e. S. (2021). Drivers of dynamic intratumor heterogeneity and phenotypic plasticity. Am. J. Physiol. Cell Physiol. 320, C750–C760. doi: 10.1152/ajpcell.00575.2020
Blat, A., Wiercigroch, E., Smeda, M., Wislocka, A., Chlopicki, S., Malek, K., et al. (2019). Fourier transform infrared spectroscopic signature of blood plasma in the progression of breast cancer with simultaneous metastasis to lungs. J. Biophotonics. 12, e201900067. doi: 10.1002/jbio.201900067
Boni, L., David, G., Mangano, A., Dionigi, G., Rausei, S., Spampatti, S., et al. (2015). Clinical applications of indocyanine green (ICG) enhanced fluorescence in laparoscopic surgery. Surg. Endosc. 29, 2046–2055. doi: 10.1007/s00464-014-3895-x
Borges, F. T., Melo, S. A., Özdemir, B. C., Kato, N., Revuelta, I., Miller, C. A., et al. (2013). TGF-beta1-containing exosomes from injured epithelial cells activate fibroblasts to initiate tissue regenerative responses and fibrosis. J. Am. Soc. Nephrol. 24, 385–392. doi: 10.1681/ASN.2012101031
Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R. L., Torre, L. A., Jemal, A., et al. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424. doi: 10.3322/caac.21492
Choi, S., and Birarda, G. (2017). Protein Mixture Segregation at Coffee-Ring: Real-Time Imaging of Protein Ring Precipitation by FTIR Spectromicroscopy. J. Phys. Chem. B. 121, 7359–7365. doi: 10.1021/acs.jpcb.7b05131
Davnall, F., Yip, C. S. P., Ljungqvist, G., Selmi, M., Ng, F., Sanghera, B., et al. (2012). Assessment of tumor heterogeneity: an emerging imaging tool for clinical practice? Insights Imaging. 3, 573–589. doi: 10.1007/s13244-012-0196-6
Dazzi, A., Prazeres, R., Glotin, F., and Ortega, J. M. (2005). Local infrared microspectroscopy with subwavelength spatial resolution with an atomic force microscope tip used as a photothermal sensor. Opt. Lett. 30, 2388–2390. doi: 10.1364/OL.30.002388
De Toro, J., Herschlik, L., Waldner, C., and Mongini, C. (2015). Emerging roles of exosomes in normal and pathological conditions: new insights for diagnosis and therapeutic applications. Front. Immunol. 6, 203. doi: 10.3389/fimmu.2015.00203
Dixon, K., Montazeri, A. O., Shayegannia, M., Barnard, E. S., Cabrini, S., Matsuura, N., et al. (2020). Tunable rainbow light trapping in ultrathin resonator arrays. Light Sci. Appl. 9, 194. doi: 10.1038/s41377-020-00428-y
Erendgenova, K. U., Fedorov, E. D., Kadushnikov, R. M., and Kulagina, O. (2018). A Smart Endoscope—Firmware Complex for Real-Time Analysis and Recognition of Endoscopic Videos. Cham: Springer International Publishing. doi: 10.1007/978-3-319-62870-7_3
Fernandez, A., Thompson, E. J., Pollard, J. W., Kitamura, T., and Vendrell, M. A. (2019). Fluorescent activatable AND-gate chemokine CCL2 enables in vivo detection of metastasis-associated macrophages. Angew Chem Int Ed Engl. 58, 16894–16898. doi: 10.1002/anie.201910955
Feuerstein, J. D., Rakowsky, S., Sattler, L., Yadav, A., Foromera, J., Grossberg, L., et al. (2019). Meta-analysis of dye-based chromoendoscopy compared with standard- and high-definition white-light endoscopy in patients with inflammatory bowel disease at increased risk of colon cancer. Gastrointestinal Gastrointest. Endoscopy. 90, 186–195.e1. doi: 10.1016/j.gie.2019.04.219
Fox, I. J. (1960). Indocyanine green: physical and physiological properties. Proc. Staff. Meet Mayo. Clin. 35, 732–744.
Frangioni, J. V. (2008). New technologies for human cancer imaging. J. Clin. Oncol. 26, 4012–4021. doi: 10.1200/JCO.2007.14.3065
Garai, E., Sensarn, S., Zavaleta, C. L., Loewke, N. O., Rogalla, S., Mandella, M. J., et al. (2015). A real-time clinical endoscopic system for intraluminal, multiplexed imaging of surface-enhanced Raman scattering nanoparticles. PLoS ONE. 10, e0123185. doi: 10.1371/journal.pone.0123185
Gianoncelli, A., Vaccari, L., Kourousias, G., Cassese, D., Bedolla, D. E., Kenig, S., et al. (2015). Soft X-ray microscopy radiation damage on fixed cells investigated with synchrotron radiation FTIR microscopy. Sci Rep. 5, 1–11. doi: 10.1038/srep10250
Großerueschkamp, F., and Gerwert, K. (2020). Label-free digital pathology by infrared imaging. Biomedical Biomed. Spectroscopy and. ImagingImag.. 9, 5–12. doi: 10.3233/BSI-200196
Harmsen, S., Teraphongphom, N., Tweedle, M. F., Basilion, J. P., and Rosenthal, E. L. (2017). Optical surgical navigation for precision in tumor resections. Mol. Imaging Biol. 19, 357–362. doi: 10.1007/s11307-017-1054-1
Hernot, S., van Manen, L., Debie, P., Mieog, J. S. D., and Vahrmeijer, A. L. (2019). Latest developments in molecular tracers for fluorescence image-guided cancer surgery. Lancet Oncol. 20, e354–e367. doi: 10.1016/S1470-2045(19)30317-1
Holman, E. A., Fang, Y. S., Chen, L., DeWeese, M., Holman, H. N., Sternberg, P. W., et al. (2020). Autonomous adaptive data acquisition for scanning hyperspectral imaging. Commun Biol. 3, 684. doi: 10.1038/s42003-020-01385-3
Holman, E. A., Fang, Y. S., Chen, L., DeWeese, M., Holman, H. N., Sternberg, P. W., et al. (2023). Toward implementing autonomous adaptive data acquisition for scanning hyperspectral imaging of biological systems. Applied Appl. Physics Phy. Reviews Rev. 10, 011319. doi: 10.1063/5.0123278
Holman, H.- .Y. N., Bechtel, H. A., Hao, Z., and Martin, M. C. (2010). Synchrotron IR spectromicroscopy: chemistry of living cells. Anal Chem. 82, 8757–8765. doi: 10.1021/ac100991d
Holman, H.-. Y. N., Martin, M. C., and McKinney, W. R. (2003). Tracking chemical changes in a live cell: biomedical applications of SR-FTIR spectromicroscopy. Spectroscopy. 17, 486940. doi: 10.1155/2003/486940
Holman, H.-Y. N. (2010). Chapter 4 - Synchrotron Infrared Spectromicroscopy for Studying Chemistry of Microbial Activity in Geologic, M.aterials, in Developments in Soil Science, B., and Singh, and, M., Gräfe, Editors. Elsevier. 103–130. doi: 10.1016/S0166-2481(10)34004-9
Holt, D., Okusanya, O., Judy, R., Venegas, O., Jiang, J., DeJesus, E., et al. (2014). Intraoperative near-infrared imaging can distinguish cancer from normal tissue but not inflammation. PLoS One. 9, e103342. doi: 10.1371/journal.pone.0103342
Hoorn, S. T., de Back, T. R., Sommeijer, D. W., and Vermeulen, L. (2022). Clinical value of consensus molecular subtypes in colorectal cancer: a systematic review and meta-analysis. J. Natl. Cancer Inst. 114, 503–516. doi: 10.1093/jnci/djab106
Iddan, G., Meron, G., Glukhovsky, A., Swain, P., Wang, A., Banerjee, S., et al. (2000). Wireless capsule endoscopy. Nature 405, 417. doi: 10.1038/35013140
Jacques, J., Wallenhorst, T., Chevaux, J-. B., Lépilliez, V., Chaussade, S., Rivory, J., et al. (2022). Endoscopic submucosal dissection (ESD) versus piece-meal endoscopic mucosal resection (PM-EMR) for large laterally spreading lesions: french randomized controlled trial resect-colon. Gastrointest. Endosc. 95, AB138. doi: 10.1016/j.gie.2022.04.384
Junhom, C., Weerapreeyakul, N., Tanthanuch, W., and Thumanu, K. (2016). FTIR microspectroscopy defines early drug resistant human hepatocellular carcinoma (HepG2) cells. Exp. Cell. Res. 340, 71–80. doi: 10.1016/j.yexcr.2015.12.007
Kaltenbach, T., Friedland, S., and Soetikno, R. A. (2008). randomised tandem colonoscopy trial of narrow band imaging versus white light examination to compare neoplasia miss rates. Gut. 57, 1406–1412. doi: 10.1136/gut.2007.137984
Kim, S. Y., Khanal, D., Tharkar, P., Kalionis, B., and Chrzanowski, W. (2018). None of us is the same as all of us: resolving the heterogeneity of extracellular vesicles using single-vesicle, nanoscale characterization with resonance enhanced atomic force microscope infrared spectroscopy (AFM-IR). Nanoscale Horiz. 3, 430–438. doi: 10.1039/C8NH00048D
Kudo, S., Kashida, H., Nakajima, T., Tamura, S., and Nakajo, K. (1997). Endoscopic diagnosis and treatment of early colorectal cancer. World J. Surg. 21, 694–701. doi: 10.1007/s002689900293
Kulshreshtha, A., Ahmad, T., Agrawal, A., and Ghosh, B. (2013). Proinflammatory role of epithelial cell-derived exosomes in allergic airway inflammation. J. Allergy Clin. Immunol. 131, 1194–203. 1203:e1–14. doi: 10.1016/j.jaci.2012.12.1565
Li, L., Wu, J., Yang, L., Wang, H., Xu, Y., Shen, K., et al. (2021). Fourier transform infrared spectroscopy: an innovative method for the diagnosis of ovarian cancer. Cancer Manag. Res. 13, 2389–2399. doi: 10.2147/CMAR.S291906
Liang, D., Yu, C., Qin, X., Yang, X., Dong, X., Hu, M., et al. (2022). Discovery of small-molecule fluorescent probes for C-Met. Eur. J. Med. Chem. 230, 114114. doi: 10.1016/j.ejmech.2022.114114
Liu, C., Zeng, X., An, Z., Yang, Y., Eisenbaum, M., Gu, X., et al. (2018). Sensitive detection of exosomal proteins via a compact surface plasmon resonance biosensor for cancer diagnosis. ACS Sens. 3, 1471–1479. doi: 10.1021/acssensors.8b00230
Lombardini, A., Mytskaniuk, V., Sivankutty, S., Andresen, E. R., Chen, X., Wenger, J., et al. (2018). High-resolution multimodal flexible coherent Raman endoscope. Light Sci. Appl. 7, 10. doi: 10.1038/s41377-018-0003-3
Loutherback, K., Chen, L., and Holman, H. Y. (2015). Open-channel microfluidic membrane device for long-term FT-IR spectromicroscopy of live adherent cells. Anal Chem. 87, 4601–4606. doi: 10.1021/acs.analchem.5b00524
Macdonald, J. S. (1999). Adjuvant therapy of colon cancer. CA Cancer J. Clin. 49, 202–219. doi: 10.3322/canjclin.49.4.202
Martin, M. C., Tsvetkova, N. M., Crowe, J. H., and McKinney, W. R. (2001). Negligible sample heating from synchrotron infrared beam. Appl Spectrosc. 55, 111–113. doi: 10.1366/0003702011951551
Mayer, A. T., Holman, D. R., Sood, A., Tandon, U., Bhate, S. S., Bodapati, S., et al. (2023). A tissue atlas of ulcerative colitis revealing evidence of sex-dependent differences in disease-driving inflammatory cell types and resistance to TNF inhibitor therapy. Sci. Adv. 9, eadd1166. doi: 10.1126/sciadv.add1166
Mieog, J. S. D., Vahrmeijer, A. L., Hutteman, M., van der Vorst, J. R., van Hooff, M. D., Dijkstra, J., et al. (2010). Novel intraoperative near-infrared fluorescence camera system for optical image-guided cancer surgery. Mol. Imaging. 9, 223–231. doi: 10.2310/7290.2010.00014
Ofori, L. O., Withana, N. P., Prestwood, T. R., Verdoes, M., Brady, J. J., Winslow, M. M., et al. (2015). Design of protease activated optical contrast agents that exploit a latent lysosomotropic effect for use in fluorescence-guided surgery. ACS Chem. Biol. 10, 1977–1988. doi: 10.1021/acschembio.5b00205
Palonpon, A. F., Ando, J., Yamakoshi, H., Dodo, K., Sodeoka, M., Kawata, S., et al. (2013). Raman and SERS microscopy for molecular imaging of live cells. Nat. Protoc. 8, 677–692. doi: 10.1038/nprot.2013.030
Petruzzellis, F., Pagliarani, C., Savi, T., Losso, A., Cavalletto, S., Tromba, G., et al. (2018). The pitfalls of in vivo imaging techniques: evidence for cellular damage caused by synchrotron X-ray computed micro-tomography. New Phytol. 220, 104–110. doi: 10.1111/nph.15368
Pisapia, C., Jamme, F., Duponchel, L., and Ménez, B. (2018). Tracking hidden organic carbon in rocks using chemometrics and hyperspectral imaging. Sci. Rep. 8, 2396. doi: 10.1038/s41598-018-20890-4
Prater, C., Bai, Y., Konings, S. C., Martinsson, I., Swaminathan, V. S., Nordenfelt, P., et al. (2023). Fluorescently guided optical photothermal infrared microspectroscopy for protein-specific bioimaging at subcellular level. J. Med. Chem. 66, 2542–2549. doi: 10.1021/acs.jmedchem.2c01359
Razeghi, M., Lu, Q. Y., Bandyopadhyay, N., Zhou, W., Heydari, D., Bai, Y., et al. (2015). Quantum cascade lasers: from tool to product. Opt. Express. 23, 8462–8475. doi: 10.1364/OE.23.008462
Romanò, S., Di Giacinto, F., Primiano, A., Gervasoni, J., Mazzini, A., Papi, M., et al. (2022). Label-free spectroscopic characterization of exosomes reveals cancer cell differentiation. Anal Chim. Acta. 1192, 339359. doi: 10.1016/j.aca.2021.339359
Romanò, S., Di Giacinto, F., Primiano, A., Mazzini, A., Panzetta, C., Papi, M., et al. (2020). Fourier Transform Infrared Spectroscopy as a useful tool for the automated classification of cancer cell-derived exosomes obtained under different culture conditions. Anal. Chim. Acta. 1140, 219–227. doi: 10.1016/j.aca.2020.09.037
Schembre, D. (2004). Smart endoscopes. Gastrointest Endosc. Clin. N. Am., 14, 709–16. ix. doi: 10.1016/j.giec.2004.04.010
Schiffman, J. D., Fisher, P. G., and Gibbs, P. (2022). Early detection of cancer. Science, 375:eaay9040. doi: 10.14694/EdBook_AM.2015.35.57
Schmidt, F., Dittberner, A., Koscielny, S., Petersen, I., and Guntinas-Lichius, O. (2017). Feasibility of real-time near-infrared indocyanine green fluorescence endoscopy for the evaluation of mucosal head and neck lesions. Head Neck. 39, 234–240. doi: 10.1002/hed.24570
Selvadurai, A. P. S., and Suvorov, A. P. (2020). The influence of the pore shape on the bulk modulus and the Biot coefficient of fluid-saturated porous rocks. Sci Rep. 10, 18959. doi: 10.1038/s41598-020-75979-6
Siegel, R. L., Miller, K. D., Wagle, N. S., and Jemal, A. (2023). Cancer statistics, 2023. CA Cancer J. Clin. 73, 17–48. doi: 10.3322/caac.21763
Stibbe, J. A., Hoogland, P., Achterberg, F. B., Holman, D. R., Sojwal, R. S., Burggraaf, J., et al. (2022). Highlighting the undetectable — fluorescence molecular imaging in gastrointestinal endoscopy. Mol. Imaging Biol. 25, 18–35. doi: 10.1007/s11307-022-01741-1
Taylor, D. D., and Gercel-Taylor, C. (2008). MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 110, 13–21. doi: 10.1016/j.ygyno.2008.04.033
Toplak, M., Birarda, G., Read, S., and Sandt, C. (2017). Infrared orange: connecting hyperspectral data with machine learning. Synchrotron Radiat News. 30, 40–45. doi: 10.1080/08940886.2017.1338424
Tummers, Q. R. J. G., Hoogstins, C. E. S., Peters, A. A. W., de Kroon, C. D., Trimbos, J. B. M. Z., van de Velde, C. J. H., et al. (2015). The value of intraoperative near-infrared fluorescence imaging based on enhanced permeability and retention of indocyanine green: feasibility and false-positives in ovarian cancer. PLoS ONE 10, e0129766. doi: 10.1371/journal.pone.0129766
Tummers, W. S., Farina-Sarasqueta, A., Boonstra, M. C., Prevoo, H. A., Sier, C. F., Mieog, J. S., et al. (2017). Selection of optimal molecular targets for tumor-specific imaging in pancreatic ductal adenocarcinoma. Oncotarget 8, 56816–56828. doi: 10.18632/oncotarget.18232
Uthamacumaran, A., Elouatik, S., Abdouh, M., and Berteau-Rainville, M. (2022). Machine learning characterization of cancer patients-derived extracellular vesicles using vibrational spectroscopies: results from a pilot study. Appl. Intel. 52, 12737–12753. doi: 10.1007/s10489-022-03203-1
Vandebriel, R., Luthman, S., Vunckx, K., Jayapala, M., Charle, W., Solie, L., et al. (2023). Integrating hyperspectral imaging in an existing intra-operative environment for detection of intrinsic brain tumors. SPIE BiOS. 12368, XXI. doi: 10.1117/12.2647690
Ventura, G. D. (2015). FTIR imaging in diffusion studies: CO2 and H2O in a synthetic sector-zoned beryl. Front. Earth Sci. 3, 1–11. doi: 10.3389/feart.2015.00033
Verma, V. B., Korzh, B., Walter, A. B., and Lita, A. (2021). Single-photon detection in the mid-infrared up to 10 μm wavelength using tungsten silicide superconducting nanowire detectors. APL Photonics 6, 056101. doi: 10.1063/5.0048049
Walke, A., Black, D., Valdes, P. A., Stummer, W., König, S., Suero-Molina, E., et al. (2023). Challenges in, and recommendations for, hyperspectral imaging in ex vivo malignant glioma biopsy measurements. Sci. Rep. 13, 3829. doi: 10.1038/s41598-023-30680-2
Weida, M., and Yee, B. (2011). Quantum Cascade Laser-Based Replacement for FTIR Microscopy. Bellingham, DC: SPIE. p. 7902.
Widen, J. C., Tholen, M., Yim, J. J., Antaris, A., Casey, K. M., Rogalla, S., et al. (2021). AND-gate contrast agents for enhanced fluorescence-guided surgery. Nat. Biomed. Eng. 5, 264–277. doi: 10.1038/s41551-020-00616-6
Willemink, M. J., Koszek, W. A., Hardell, C., Wu, J., Fleischmann, D., Harvey, H., et al. (2020). Preparing medical imaging data for machine learning. Radiology 295, 4–15. doi: 10.1148/radiol.2020192224
Yáñez-Mó, M., Siljander, P. R-.M., Andreu, Z., Zavec, A. B., Borràs, F. E., Buzas, E. I., et al. (2015). Biological properties of extracellular vesicles and their physiological functions. J. Extracell Vesicles. 4, 27066. doi: 10.3402/jev.v4.27066
Yim, J. J., Harmsen, S., Flisikowski, K., and Slisikowska, T. (2020). A protease-activated, near-infrared fluorescent probe for early endoscopic detection of premalignant gastrointestinal lesions. Proc. Natl. Acad. Sci. U. S. A. 118, e2008072118. doi: 10.1073/pnas.2008072118
Yu, L. L., Zhu, J., Liu, J. X., Jiang, F., Ni, W. K., Qu, L. S., et al. (2018). A comparison of traditional and novel methods for the separation of exosomes from human samples. Biomed. Res. Int. 2018, 3634563. doi: 10.1155/2018/3634563
Zhang, Y., Kim, M. S., Jia, B., Yan, J., Zuniga-Hertz, J. P., Han, C., et al. (2017). Hypothalamic stem cells control ageing speed partly through exosomal miRNAs. Nature 548, 52–57. doi: 10.1038/nature23282
Zhou, T., Han, G., Li, B. N., Lin, Z., Ciaccio, E. J., Green, P. H., et al. (2017). Quantitative analysis of patients with celiac disease by video capsule endoscopy: a deep learning method. Comput. Biol. Med. 85, 1–6. doi: 10.1016/j.compbiomed.2017.03.031
Keywords: hyperspectral imaging (HSI), precision medicine, early diagnostics, contrast agents (CAs), label-free, infrared spectromicroscopy, cancer
Citation: Holman EA, Holman DR and Rogalla S (2023) Expanding hyperspectral imaging applications to the clinical scene: non-invasive, label-free approaches for early diagnostics and precision medicine. Front. Imaging. 2:1175860. doi: 10.3389/fimag.2023.1175860
Received: 28 February 2023; Accepted: 02 May 2023;
Published: 18 May 2023.
Edited by:
Marcin Wozniak, Silesian University of Technology, PolandReviewed by:
Sankar Raju Narayanasamy, Lawrence Livermore National Laboratory (DOE), United StatesCopyright © 2023 Holman, Holman and Rogalla. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Elizabeth A. Holman, ZWhvbG1hbkBzdGFuZm9yZC5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.