- 1Inria, Rennes, France
- 2Visages U746, INSERM, Rennes, France
- 3UMR 6074, CNRS, IRISA, University of Rennes I, Rennes, France
- 4Department of Neuroradiology, CHU Rennes, Rennes, France
Two of the major concerns of researchers and clinicians performing neuroimaging experiments are managing the huge quantity and diversity of data and the ability to compare their experiments and the programs they develop with those of their peers. In this context, we introduce Shanoir, which uses a type of cloud computing known as software as a service to manage neuroimaging data used in the clinical neurosciences. Thanks to a formal model of medical imaging data (an ontology), Shanoir provides an open source neuroinformatics environment designed to structure, manage, archive, visualize, and share neuroimaging data with an emphasis on managing multi-institutional, collaborative research projects. This article covers how images are accessed through the Shanoir Data Management System and describes the data repositories that are hosted and managed by the Shanoir environment in different contexts.
Introduction
Context
Two of the major concerns for researchers and clinicians performing neuroimaging experiments are managing the huge quantity and diversity of data and the ability to compare their experiments and the programs they develop with those of their peers. In practice, researchers and clinicians in the neuroimaging field are encouraged to set up large-scale experiments, but the inability to recruit sufficient local subjects who meet specific criteria results in the need for cooperation to gather the relevant imaging data. Pooling experimental results via the Internet and cooperative efforts by centers provide larger and more specific subject populations that expand the scope and value of scientific research.
Searches on distributed neuroimaging databases for similar results and images containing singularities (quirk, peculiarities, etc.) or the use of data mining techniques may highlight possible similarities. Such efforts also broaden the possible panel of people involved in neuroimaging studies while maintaining the quality of the work. Indeed, the explosion of data generated by the neurosciences community in the early 1990s has resulted in the need for innovative techniques for data and knowledge sharing and reuse (Roland and Zilles, 1994; Mazziotta et al., 1995; Shepherd et al., 1998). This has led to the emergence of large-scale projects on the human brain. A recent objective added to these initial issues is the application of data analysis and data processing software to various data repository systems for knowledge discovery and data mining, including its more recent extension to merging imaging and genetic data (Hibar et al., 2015). In parallel, the development of web applications has stimulated the interest of researchers and clinicians in distributed databases and information sharing.
Background
It is now commonly accepted in the neuroimaging community that sharing data and image processing services will play a crucial role in translational research (Barillot et al., 2003; Walport and Brest, 2011; Poline et al., 2012; Keator et al., 2013; Van Horn and Gazzaniga, 2013; Poldrack and Gorgolewski, 2014). Research funding agencies now clearly identify the sharing of scientific resources (data processing) as a top priority. International organizations such as the International Neuroinformatics Coordinating Facility (INCF)1 are now dedicated to promoting the field of neuroinformatics (Book et al., 2013; Kennedy et al., 2015). Sharing data and image processing services for translational research are needed for:
(1) the integration of large data sets for population-wide studies and construction of imaging cohorts (Shepherd et al., 1998; Van Horn et al., 2001; Barillot et al., 2006; Evans and Brain Development Cooperative Group, 2006; Jack et al., 2008; Hall et al., 2012; Weiner et al., 2012; Marcus et al., 2013; Van Essen et al., 2013),
(2) the validation of image processing tools on reference datasets for validation and quality control of image processing procedures (Styner et al., 2008; Menze et al., 2015),
(3) the reuse of image processing pipeline on different sets of data and different peers for sharing processing tools (Keator et al., 2009, 2013; Ooi et al., 2009; Dinov et al., 2010; Gorgolewski et al., 2011; Bellec et al., 2012; Glatard et al., 2014), and
(4) the validation of research results based on proofed control statistical analysis of images for validation and quality control of experimental research (Carp, 2012; Button et al., 2013; Ioannidis, 2014; Ioannidis et al., 2014).
This is particularly significant in the field of neuroimaging as several large recent multicenter initiatives have shown. These include Evans and Brain Development Cooperative Group (2006), which performed a study using magnetic resonance imaging (MRI) of normal brain maturation from birth to adulthood in approximately 500 children with behavior disorders, and the Alzheimer’s disease neuroimaging initiative (ADNI), which has assembled a very large variety of images for its work (Weiner et al., 2012). The Human Connectome Project (HCP), which worked with 1,200 healthy volunteers to investigate brain connectivity in the normal brain (Marcus et al., 2013; Van Essen et al., 2013), is another well-known example illustrating the importance of aggregate imaging data and relating data warehouses to image processing resources.
To provide archiving solutions for large or various multicenter projects, several architectures have already been proposed. The Biomedical Informatics Research Network (BIRN) has been a pioneer in launching brain imaging solutions (Gupta et al., 2003; Keator et al., 2008, 2009; Ashish et al., 2010). Another early initiative, the FMRIDC project sought to share task-based fMRI imaging data (Van Horn and Gazzaniga, 2013). @NeurIST set up a dedicated solution (funded by an Integrated European Project) to support research and treatment of cerebral aneurysms using heterogeneous data, computing, and complex processing services (Benkner et al., 2010). The LORIS/CBRAIN project is an initiative to develop a pan-Canadian platform for distributed processing, analysis, exchange, and visualization of brain imaging data (Das et al., 2011; Sherif et al., 2014). Finally, other generic data management systems have been proposed to offer shared solutions for managing multicenter studies. These include the Extensible Neuroimaging Archive Toolkit (XNAT) (Marcus et al., 2007), which has been successful due to its integration in the management of large projects (Marcus et al., 2013) and ability to communicate with data management servers via dedicated REST web services, and the Collaborative Informatics and Neuroimaging Suite (COINS), which provides a web-based neuroimaging and neuropsychology software suite (Scott et al., 2011). Although the extensibility of these platforms is part of the motivations, none of them are built on top of a formal semantic model or ontology that can guarantee the sustainability of any evolution of the original data scheme.
Significance
In this context, the Sharing Neuroimaging Resources (Shanoir) environment enables sharing between distributed sources of neuroimaging information over the Internet, whether the sources are located in various centers of experimentation, clinical departments of neurology, or research centers in cognitive neurosciences or image processing. A large variety of users can thus share, exchange, and have controlled access to neuroimaging information using the software as a service (SaaS) type of cloud computing (Rimal et al., 2009) almost as easily as if the data were stored locally.
In this paper, we introduce the Shanoir software environment for managing neuroimaging data production in the context of clinical neurosciences and show how the images are accessible through the Shanoir Data Management System. Shanoir is an open source neuroinformatics environment designed to structure, manage, archive, visualize, and share neuroimaging data with an emphasis on multi-institutional, collaborative research projects. The software offers features commonly found in neuroimaging data management systems along with research-oriented data organization capabilities and enhanced accessibility. It also provides user-friendly secure web access and an intuitive workflow that facilitates the collection and retrieval of neuroimaging data from multiple sources.
In Section “Shanoir Software Environment,” we provide a brief overview of the software environment including its core (web portal, Study Card, and quality control) and extensions for loading, querying, and processing data. Section “Data Repositories” describes the data repositories, while Section “Conclusion and Perspectives” covers the use of these repositories and potential evolution.
Shanoir Software Environment
General Description of the Software Environment
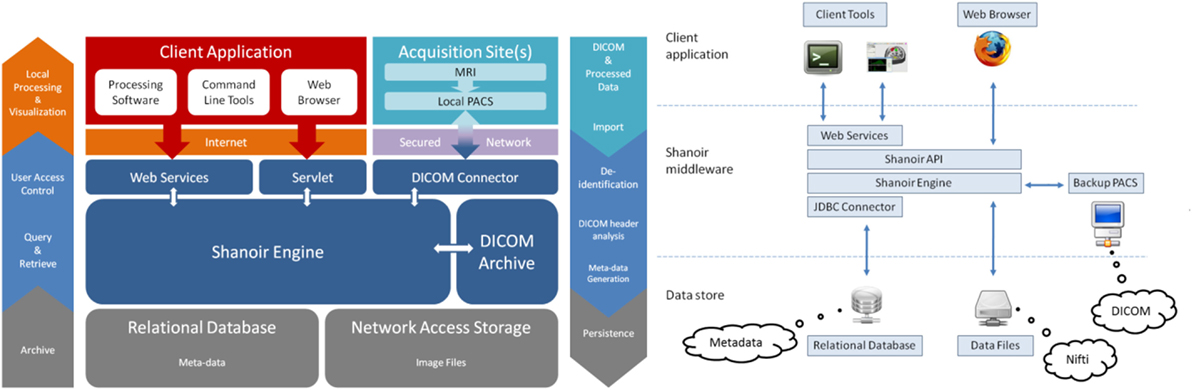
Shanoir is an open source software environment with QPL licensing designed to archive, structure, manage, visualize, and share neuroimaging data with an emphasis on managing collaborative research projects. It includes the common features of neuroimaging data management systems along with research-oriented data organization and enhanced accessibility. Shanoir is based on a secure J2EE application running on a JBoss server that is accessed via graphical interfaces in a browser or by third-party programs via web services using simple object access protocol (SOAP). It behaves like a repository of neuroimaging files coupled with a relational database containing metadata (Figure 1).
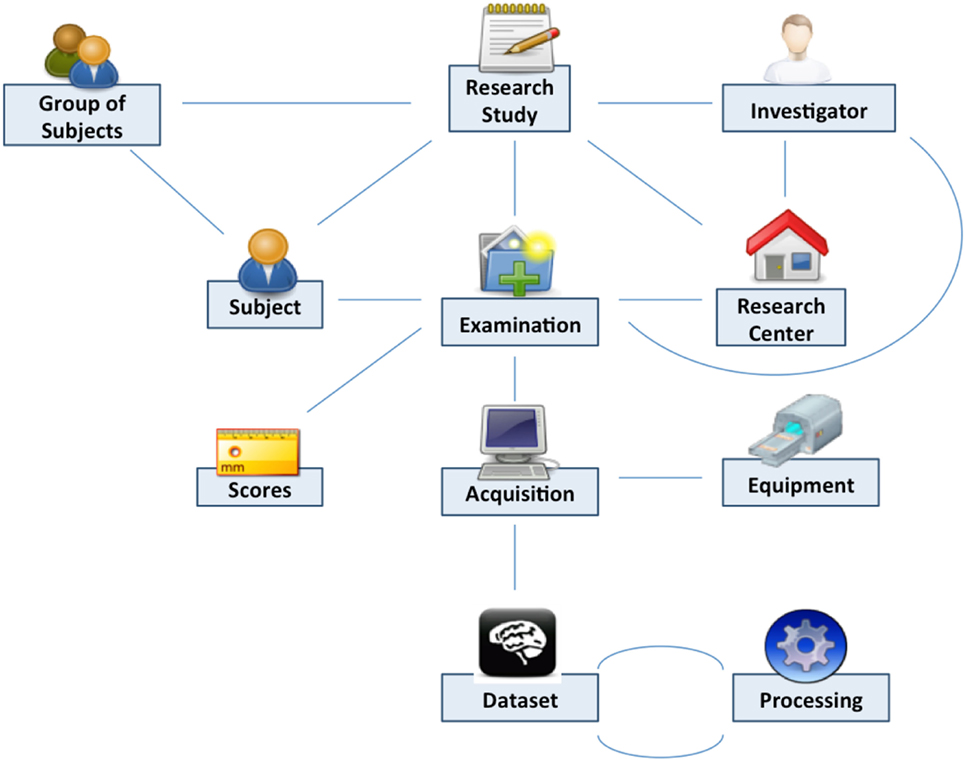
Shanoir uses semantics for structuring the concepts as defined by the OntoNeuroLOG2 ontology (Temal et al., 2008; Michel et al., 2010). OntoNeuroLOG reuses and extends the OntoNeuroBase ontology defined earlier (Barillot et al., 2006) (see Figure 2). Both were designed using the methodological framework (Temal et al., 2008) of the foundational Descriptive Ontology for Linguistic and Cognitive Engineering (DOLCE) (Masolo et al., 2003) and a number of core ontologies that provide generic, basic, and minimal concepts and relationships in specific fields such as artifacts, participant roles, information, and discourse acts. In Shanoir, the OWL-Lite implementation was manually derived from the OntoNeuroLOG initial expressive representation to Java classes. The data model based on this ontology is dedicated to the neuroimaging field and is structured around research studies in which patients are examined to produce image acquisitions or clinical scores. Each image acquisition is composed of datasets represented by acquisition parameters and image files. For security and legal reasons, all data on the system are anonymous by default, this can be customized with specific algorithm (e.g., defacing is not currently implemented but can easily be embedded in a specific anonymizer that Shanoir will call).
Raw as well as derived (i.e., post-processed) image files can also be imported into the system using medical imaging technology [e.g., media based on the Digital Imaging and Communications in Medicine (DICOM) standard, picture archiving and communication system (PACS), or image files in the Neuroimaging Informatics Technology Initiative (NIfTI)/Analyze-style data format] using online wizards, which complete related metadata, command line tools, or SOAP web services. Once identity information has been removed from raw data during the importation process, the DICOM header content is automatically extracted, enriched, and inserted into the database with the customizable “Study Card” feature. Shanoir can also record any execution process for retrieval of workflows applied to a particular dataset along with the derived data.
Clinical scores from instrument assessments (e.g., neuropsychological tests) can be recorded and easily retrieved and exported in different formats (Excel, CSV, and XML). The instrument database is scalable and new measures can be added in order to meet specific project needs (Figure 3). Scores, image acquisitions, and post-processed images are bound together, so that relationships can be analyzed. Using cross-data navigation and advanced search criteria, the user can quickly indicate a subset of data for download. Client-side applications have also been developed to locally access and exploit data though web services. The security features of the system require authentication with user rights set for each study. A study manager can define the users allowed to see, download, or import data into his/her study or simply make it public.
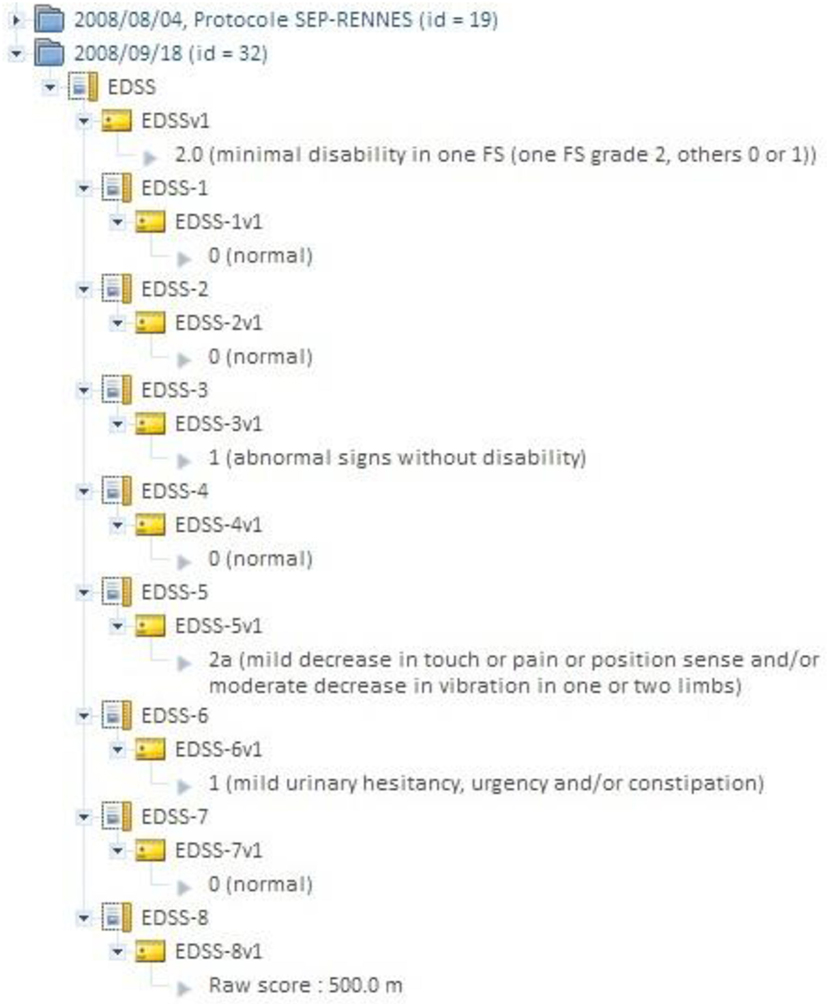
Figure 3. Shanoir “instrument” database can be used for attaching clinical scores to images (e.g., EDSS score in MS). An instrument can be any record where an alphanumerical value can be attached.
In practice, Shanoir serves neuroimaging researchers by efficiently organizing their studies while cooperating with other laboratories. By managing patient privacy, Shanoir offers the possibility of using clinical data in a research context. Finally, it is a handy solution for publishing and sharing data with a broader community.
Study Card and Quality Control Concepts
Images can be imported in Shanoir from various sources: DICOM media, PACS (with DICOM Query and Retrieve), and 3D/4D image files (in NIfTI/Analyze format). Users are guided step-by-step through online forms to perform imports. In addition to archiving DICOM files, NIfTI copies are automatically generated and saved. This is convenient since the NIfTI format is better suited to perform image processing (such as registration, segmentation, and statistical analysis) than the DICOM format.
The Study Card
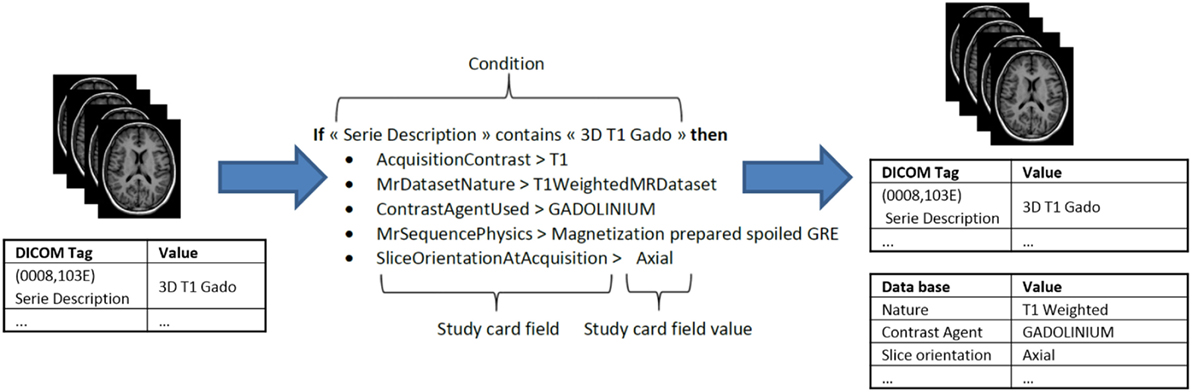
During archiving, the DICOM files are processed in two phases. The first phase de-identifies the images. The second phase populates the database with the new metadata items generated from the DICOM header and enriched with the Study Card, which enables online metadata wrapping between the local data to be imported (center, acquisition equipment, etc.) and the semantic concepts of the research study to which the data will be assigned. The actual DICOM metadata can thus be aligned with the ontology and also provides additional allocation of concepts to the stored images that are more closely related to the research study protocol (e.g., functional MRI, perfusion imaging, contrast agent, diffusion imaging, etc.). The mechanism behind this feature is based on a set of rules that the user predefines to associate specific acquisition equipment and a specific data production site to the desired research study. Each rule determines the specific value of a metadata item according to the value(s) of one or more specific DICOM tag(s) (e.g., Series Description, see Figure 4). This greatly facilitates the consistent recording and alignment to the ontology of metadata for all data in a research study without the need for tedious workflow during the online import of images. Due to the simplicity of the process, no specific skills are required to perform data import, and it only takes a few minutes over the Internet. The “Study Card” concept makes possible an automatic quality control of the imported data using their metadata. For instance, a conformal statement can be attached to the imported data according to a match score to the Study Card rules.
Quality Assessment
Shanoir’s next major functionality concerns the quality check of the images for conformity of the imported data with the predefined study protocol and ensures the integrity of the archived data. We have identified three levels of control:
• study protocol: controls the time interval between examinations (expected visits) as defined by the principal investigator (PI) of the study;
• acquisition protocol: controls the presence of all the sequences of the imaging protocol as defined by the PI of the study; and
• raw data:
○ the software automatically checks the range of parameters for a given protocol, experimental center, or acquisition scanner as defined in the Study Card by the PI’s technical representative;
○ visual inspection of the image quality and integrity can be reported and assigned to the imported data; however, the mechanism to detect the visual quality is not yet integrated in the Shanoir environment.
In the next release of Shanoir, quality assessment will be present as flags (flawless, acceptable, or inadmissible) in the database.
This QA capability does not address the control of image formation as, for instance, to check image artifacts (bias, motions, ghosting, etc.). This category of QA can be implemented in a dedicated image visualization and processing tools that interoperate with Shanoir through the dedicated web services.
Web Portal
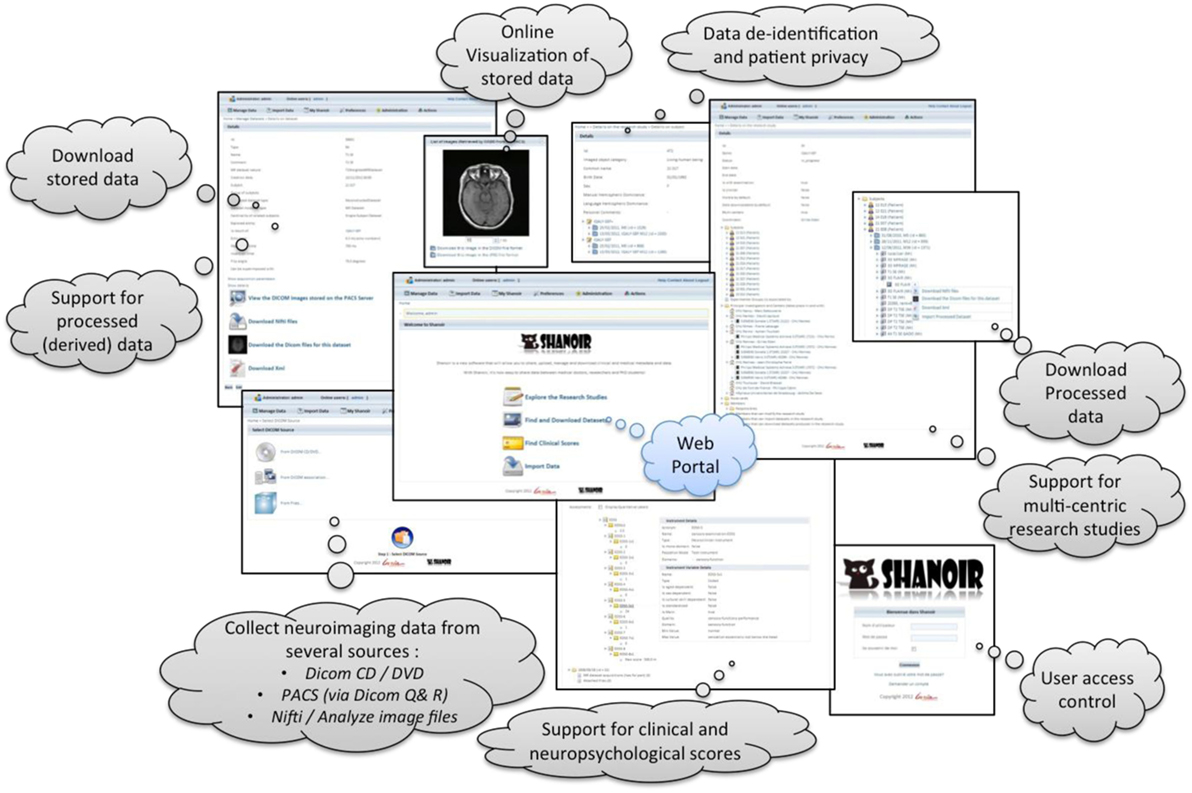
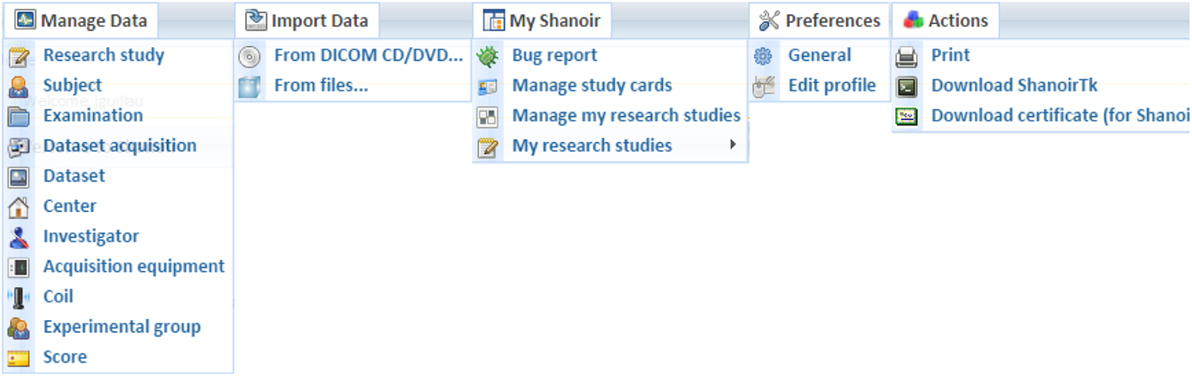
Shanoir provides user-friendly secure web access and offers an intuitive workflow to facilitate the collection and retrieval of neuroimaging data from multiple sources (Figure 5). On the home page, the user has direct access to the most frequent functionalities: Find and Download Datasets, Explore the Research Studies, Find Clinical Scores, and Import Data (Figure 6). On the top of all pages, the user always has a very complete navigation menu that leads to all services.
Interoperability
Interoperability is a very important concern for the Shanoir environment. Shanoir offers web services interface that is open to a large variety of clients. We already offer several dedicated interface that are already in used by different external applications. Hereafter, we described four of these external services that are currently available and run independently to each other: ShanoirUploader, QtShanoir, medInria, and iShanoir developed either in C++, Java, or Objective-C environments.
SOAP for Integration of Services
The Shanoir web services interface is based on the SOAP. Messages between clients and the server are exchanged using Extensible Markup Language (XML) with well-defined elements. The Hypertext Transfer Protocol (HTTP) is used with Transport Layer Security (TLS). Elements and services are described with the Web Service Description Language (WSDL). Based on this description, client stubs can be automatically generated to simplify the connection of new clients. The web service layer is implemented with the Java API for XML web services (JAX-WS). Shanoir offers numerous dedicated web services:
• “EntityCreator”: creates new entities, such as creating a new subject in the database
• “CredentialTester”: validates if username and password are correct
• “Downloader”: downloading files/datasets on base of dataset IDs
• “CenterFinder”: find center(s) based on different search criteria, i.e., study or investigator
• “DatasetAcquisitionFinder”: find acquisition(s) based on IDs or examinations
• “DatasetFinder”: find dataset(s) based on multiple search criteria/filters
• “DatasetProcessingFinder”: find dataset processing(s) based on IDs
• “ExaminationFinder”: find examination(s) based on multiple search criteria/filters
• “ExperimentalGroupOfSubjectsFinder”: find group of subjects based on multiple criteria
• “InvestigatorFinder”: find investigator(s) based on IDs or centers
• “MrDatasetFinder”: find MR dataset(s) based on multiple search criteria/filters
• “StudyFinder”: find study/-ies based on multiple search criteria/filters
• “SubjectFinder”: find subject(s) based on IDs with multiple filters
• “DatasetImporter”: import dataset files to already existing entities in the database
• “ReferenceLister”: shows list of reference strings stored in the database
• “FileUploader”: upload files in local archive for later import, used by ShanoirUploader
ShanoirUploader for Seamless Integration of Data
“ShanoirUploader” is a Java desktop application that transfers data securely between a PACS and a Shanoir server instance (e.g., within a hospital). It offers both a direct DICOM query/retrieve connection to search and download images from a local PACS and a DICOM CD upload facility. After retrieval, the DICOM files are locally anonymized and then uploaded to the Shanoir server (the anonymization algorithm can be customized according to specific operational/regulation constraints). The primary goals of the application are to enable mass data transfers between different remote server instances and reduce user waiting time when importing data into Shanoir. Most of the import time thus involves data transfer.
“ShanoirUploader” requires a local Java installation. For a simpler distribution and installation of the software, Java Web Start (JWS) can be used. The application can be installed with a simple web link that is opened in a web browser. Java takes care of the installation and, later, of automatic updates. Internal components are based on Java Swing for the graphical user interface (Figure 7), dcm4chee3 libraries to connect with a PACS and Java, and WebServices (JAX-WS) to transfer data to a Shanoir server.
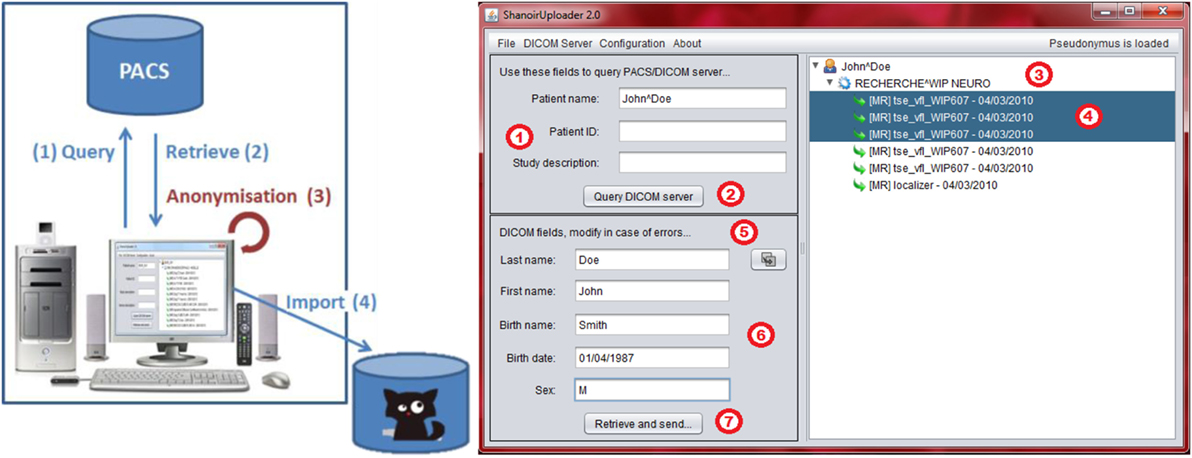
Figure 7. Shanoir Uploader architecture for secure transfer of local PACS data to a Shanoir Server (left) and user interface (right).
Apache Solr for Metadata Querying
Shanoir integrates the open source enterprise search platform Apache Solr,4 which provides users with a vast array of advanced features such as near real-time indexing and queries, full-text searching, faceted navigation, autosuggestion, and autocomplete.
One of the most important features of the Solr search is the faceted navigation. Facets correspond to properties of the Solr information elements and are derived by analyzing pre-existing metadata that are related to the ontology model used by Shanoir.
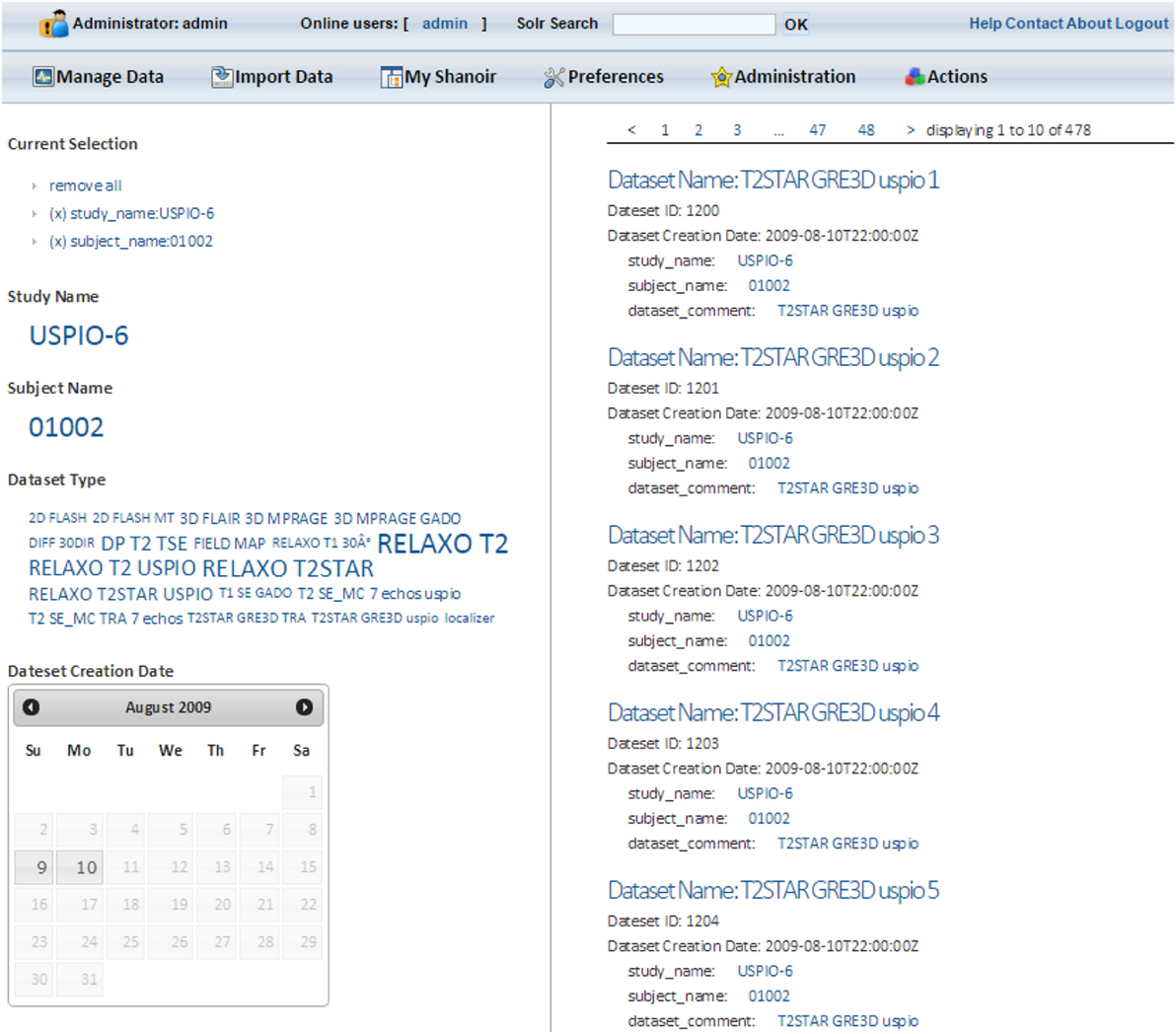
Shanoir users can access all metadata with a simple Solr search bar. After entering at least one character, a user will be automatically guided to complete his search. Data are sorted by categories and dynamically displayed once a facet is chosen. By clicking on Solr data results, users access all the additional information available in Shanoir corresponding to their search, and then use these queries for local downloading (Figure 8).
All metadata are indexed in a JBoss server that hosts the Solr servlets. A custom security post-filter has also been developed and implemented in Shanoir to control user access. This filter retrieves user identification and access rights in Shanoir and interacts with the Solr server to show relevant results that the user is allowed to access.
iShanoir for Mobile Data Access
An iOS application, iShanoir, has been developed for iPhones and iPads. It opens a secure connection with a Shanoir server and enables the user to access data stored on a Shanoir server. With iShanoir, the user can navigate within the Shanoir data tree structure on the server. After data are selected from the mobile app, the images can be downloaded to the local device, displayed, and analyzed with any local DICOM viewer or through cloud services (i.e., Dropbox, iCloud, Google, or OneDrive).
The iShanoir application has been developed with Xcode and implemented in Objective-C. For the graphical user interface, two storyboards have been developed to fit the different display sizes between iPhones and iPads (Figure 9). It uses the following iOS frameworks: Foundation, CoreFoundation, UIKit, and CFNetwork. For implementation of the SOAP web services client, the WSDL2ObjC utility has been used as it offers a client stub code generation based on the server WSDL document.
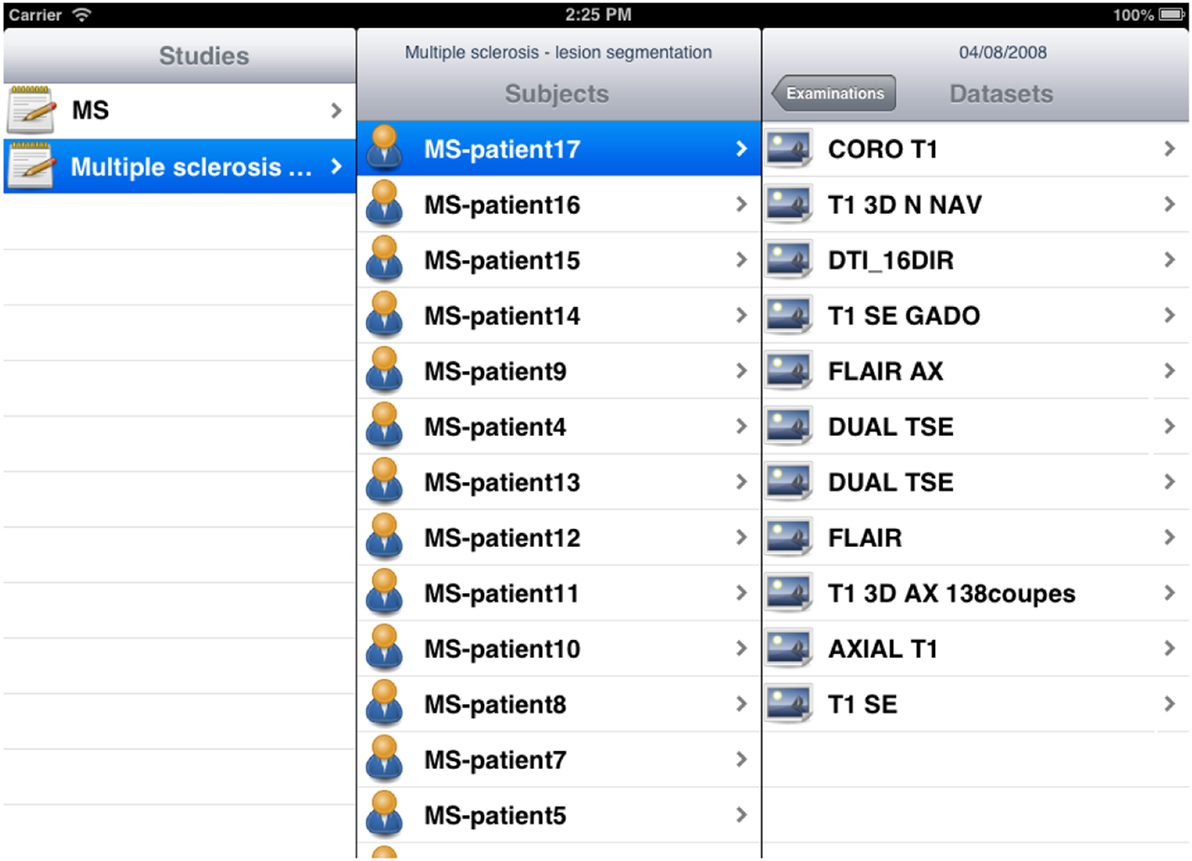
Figure 9. Example of storyboard interfaces under the iShanoir iOS mobile application connected to a Shanoir server.
QtShanoir for Image Processing
Shanoir web services may also be queried from standalone C++/Qt applications through the QtShanoir library,5 which uses SOAP web services provided by a Shanoir server to access and display studies, patients, and data with their associated metadata. In QtShanoir, a set of Qt widgets are defined that can be embedded in any Qt application. The library was used to implement a Shanoir query plugin inside the medInria visualization and processing software6 for interrogation and downloading of image data from Shanoir for processing within medInria, for example, using the available processing tools and then upload the processing results back to the Shanoir server with the correct metadata values (Figure 10).
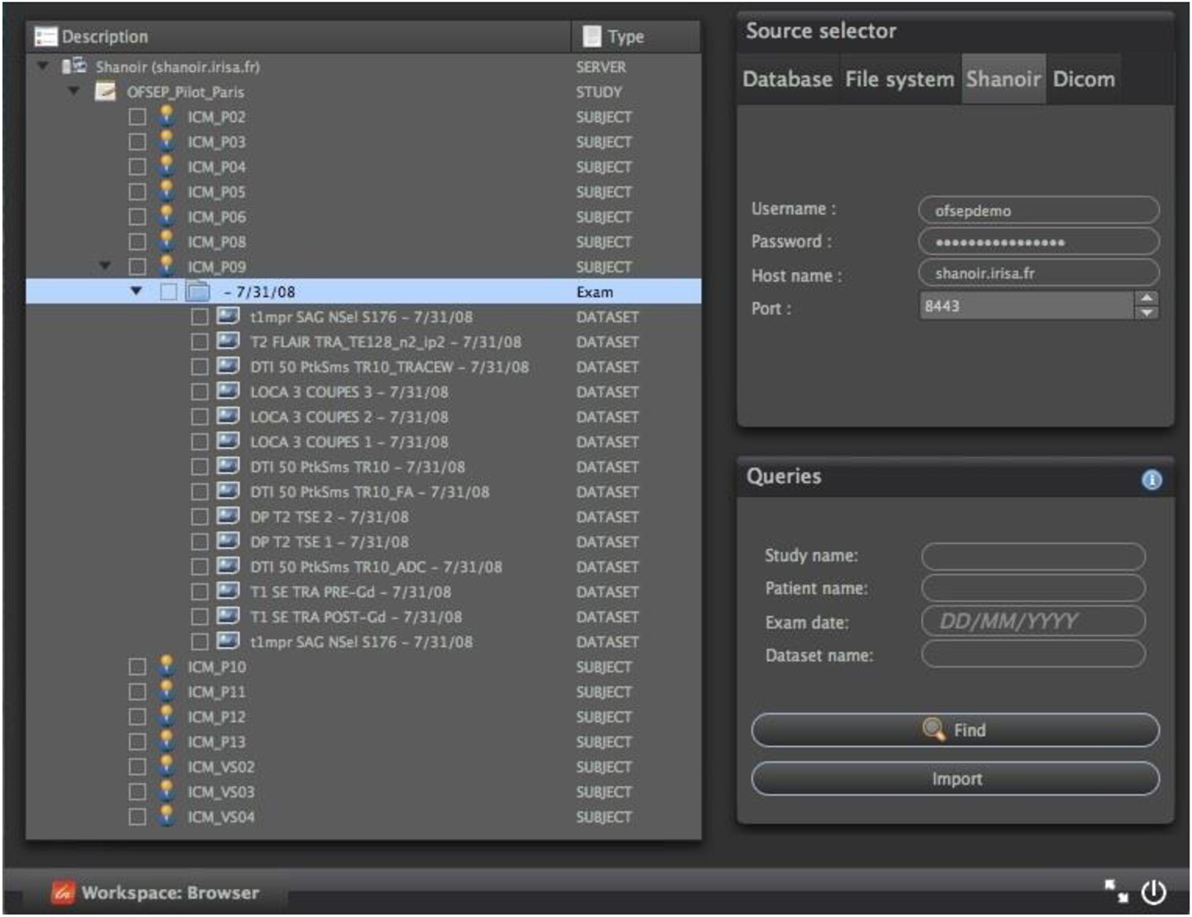
Figure 10. Example of a Shanoir query service within the medInria environment by using QtShanoir web services.
Distribution of Shanoir
The Shanoir server can be freely downloaded on request. It is currently deployed using Docker containers running on a Linux kernel. Linux containers are implemented using namespaces for locating each type of resource. Dockers are tools for managing lightweight method of virtualization (named containers) on Linux that are lighter than traditional virtual machines. The host and guest systems share the same kernel. The kernel is responsible for host ↔ guest and guest ↔ guest isolation (the result of system calls depends on the container in which the calling process is running). As described in Figure 11, a minimal Shanoir deployment consists of four servers running in at least four separate containers:
• “shanoir_container”: the actual Shanoir server. It relies on a mysql container (for the database) and on the PACS container (for archiving DICOM data),
• “pacs_container”: the DICOM PACS server, currently managed by dcm4chee,7
• “mysql_container”: the database server that is hosting two databases: shanoirdb and pacsdb,
• “nginx _container”: the web frontend server based on a nginx8 HTTP server configured as a reverse-proxy for reaching the Shanoir server. It is the only server that is publicly reachable. It provides TLS encryption and security filtering, and
• “smtpsink _container”: an optional SMTP server for outgoing e-mails.
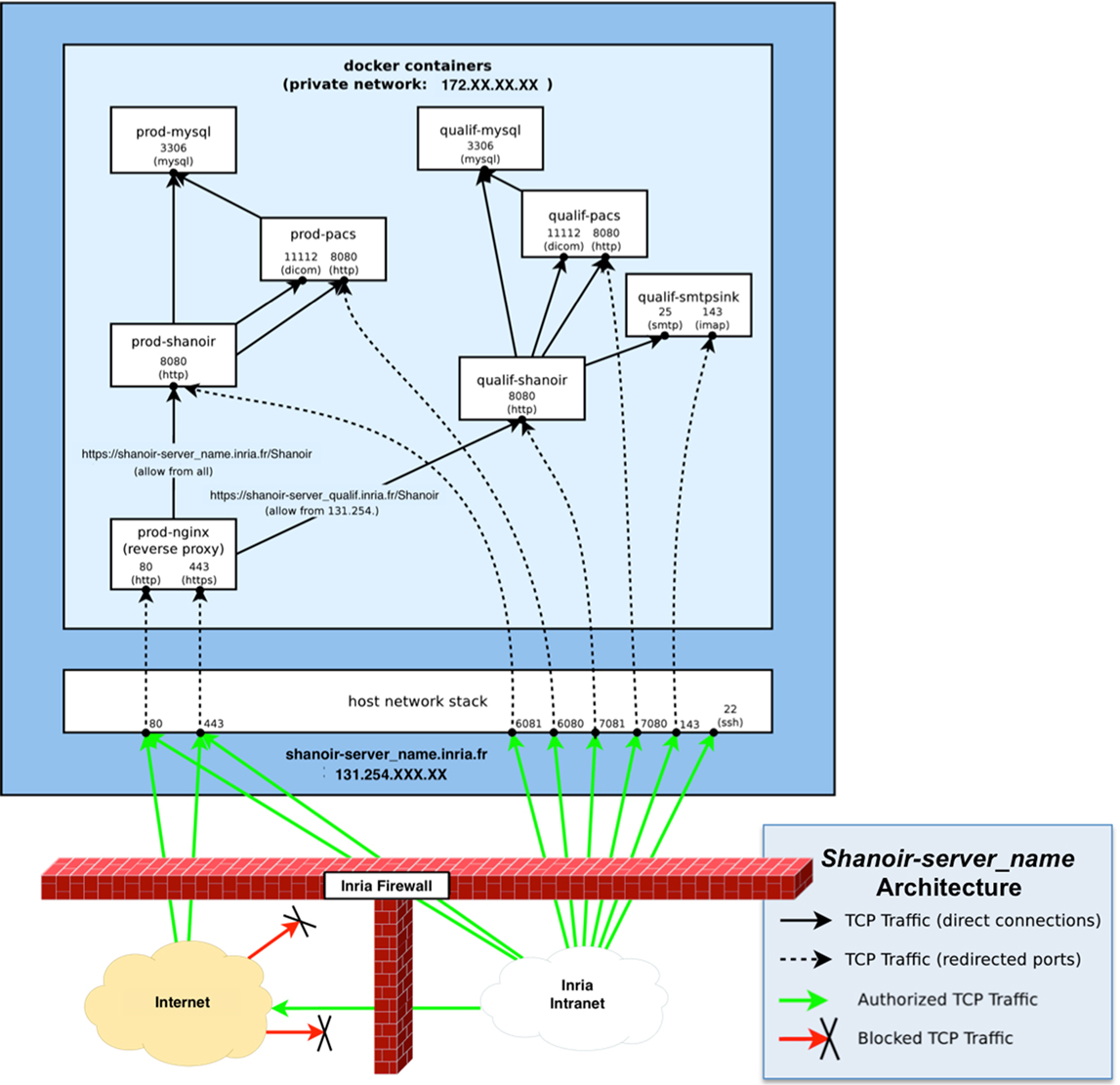
Figure 11. Schematic diagram of a Shanoir server. There is one set of containers for production: prod-Shanoir, prod-pacs, prod-mysql, and prod-nginx, and one set for qualification: qualif-shanoir, qualif-pacs, qualif-mysql, and qualif-nginx (not represented here). The arrows represent relationships between containers.
Data Repositories
Each Shanoir repository has an administrator that manages the access rights of the repository. Each user requests an account through a web-based form and specifies which study he/she wants to access, contact, role in the study, required level of expertise/access (guest, user, expert, and admin), etc. According to the information provided, the Shanoir administrator of the repository determines whether the user can access the system. Access to a specific study is granted by the person responsible for the study (i.e., the PI of the research study or the official representative). Depending on these settings, the new user will be able to see, download, and import datasets or even modify the study parameters. The corresponding rights are set for a limited time and must be renewed regularly. If requested, the user can receive a report by e-mail each time data are imported into the study.
The Shanoir@Neurinfo Repository
Started in 2009, the Neurinfo MRI research facility9 promotes translational clinical research and supports the development of clinical research, technological activity, and methodological activity. It offers resources for in vivo human imaging acquisition, image data analysis, and image data management. A large community of users, both clinicians and scientists, uses the resources as part of local, national, and international imaging-based research projects.
All data produced at Neurinfo for academic or clinical research purposes are managed through a dedicated Shanoir@Neurinfo repository (Figure 12) administered by the facility’s staff. The Shanoir@Neurinfo server also hosts data from imaging studies at multiple sites. In total, around 2To of data from 42 centers and 50 MR scanners are archived at this repository. The amount of data increases by 30 GB per month (see table in Figure 12).
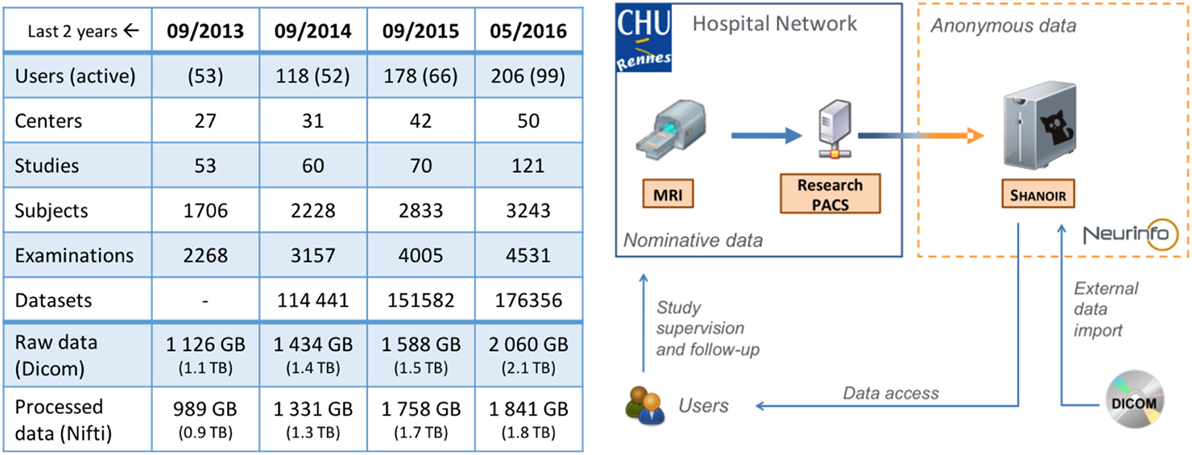
Figure 12. Evolution of the Shanoir@Neurinfo repository Global Statistics (left) and Service Infrastructure (right).
In daily practice, DICOM data are imported by a technician from either a local PACS, a CD/DVD, or a disk drive containing the DICOMDIR in its root directory and the DICOM files.
The clinical studies stored on the Shanoir@Neurinfo server concern the whole body (brain, spine, heart, lung, pelvis, vasculature, etc.) with a major focus on brain anatomy and function in normal control and pathological populations. Out of the 70 or so ongoing research studies on the Neurinfo platform, 75% relates to brain imaging, 15% concern abdominal imaging, and 10% concern heart imaging. Among the neuroimaging clinical studies, multiple sclerosis, dementia, tumors, stroke, and mood disorders are the most investigated pathologies.
Depending on the specific nature of the research study, typical neuroimaging protocols include structural imaging, functional BOLD MRI, Arterial Spin Labeling perfusion imaging, diffusion imaging, relaxometry sequences, pre- and post-gadolinium T1w sequences, or vascular sequences. The following studies are examples of research carried out with the Shanoir@Neurinfo service as well as the types of data that are managed by the Shanoir@Neurinfo repository.
Along with the MRI raw data, post-processed images can be stored for each dataset. For example, in a study on functional motor activation, the motor areas were delineated by a trained radiologist and associated with each 3D T1w image. Multiple sclerosis (MS) lesion segmentation masks can also be attached to the examination. In addition to image data, clinical scores can also be stored for each subject in the repository. Several MS clinical studies collect measurements such as the number of T2 new lesions, and number of T1w Gd enhancing lesions or clinical scores such as EDSS. These measurements are also included in the search engine and consequently easily accessible through requests. For more advanced clinical follow-ups, Shanoir can easily be interfaced with existing databases.
The general policy for the Shanoir@Neurinfo repository for dissemination of data related to a particular study is decided upon beforehand with the PI in compliance with the informed consent form approved by the ethics committee and signed by the participant. Any opening of the data to third parties is submitted to the approval of the PI prior to allowing (complete or partial) access to a third-party user. Nonetheless, to ensure dissemination and the best use of data acquired from public funding, the Neurinfo team strongly encourages investigators to share their data, which is usually done after an embargo period.
Shanoir@OFSEP Repository
The French Multiple Sclerosis Observatory (OFSEP),10 a major epidemiological tool on MS for the scientific community, was selected after a call for projects for Cohorts 2010, funded by France’s Investment in the Future Program. It is a collaborative project involving over 40 MS research centers in France. The aim of the project is to build and maintain a nationwide cohort of patients with MS and enrich the clinical data with biological samples, socio-economic data, and neuro-images.
A dedicated imaging working group is in charge of acquiring, processing, and integrating imaging and derived imaging data into a shared imaging resource center (IRC), and ensuring that the IRC is integrated with clinical databases. The consistent assessment of MRI-based measurements on a large scale requires robust and efficient image processing pipelines. A further goal of this project is to establish an information technology infrastructure enabling audited access to imaging data, as well as a virtual laboratory environment supporting the distributed, synergistic development, validation, and deployment of specialized image analysis procedures developed by different national and international research centers. To ensure easy access to the imaging data and allow modifications, queries, annotations, and access control, the Shanoir environment has been selected. It will also ensure interoperability and data management related to the imaging aspect of the cohort (the clinical part is managed by the EDMUS11 system). For this purpose, we have set up a specific Shanoir@OFSEP image repository that is currently in its pilot phase.
Begun in 2012, the Shanoir@OFSEP server was installed to store the imaging data of the OFSEP cohort, which will study the neuroimaging data of 40,000 MS patients over the next 10 years. A consensus has emerged concerning the acquisition protocol, which requires (at least) one brain MRI every 3 years, one spinal MRI every 6 years, i.e., 200,000 MRIs over 10 years. The Shanoir@OFSEP database will grow during this period and beyond (Cotton et al., 2015).
Since OFSEP is a nationwide project covering many patients, many IRCs, and many different kinds of MRI acquisition equipment, a national repository with nationwide access and uniform measures was therefore needed. The OFSEP imaging working group is continuously gathering new acquisition centers volunteering to take part to the cohort. In Shanoir@OFSEP, there are currently about 30 IRCs which include 31 pieces of MRI acquisition equipment representing 14 different MR scanner models from three MR manufacturers (Siemens, Philips, and GE). All the centers are importing data in one main study called the “Mother Cohort.” Each center follows the OFSEP protocol, which will be checked through the quality control module as described in Section “Quality Assessment.” If necessary, derived imaging data can then be imported back to the server in order to refer to potential post-processing information and MS-specific imaging biomarkers to make them available for authorized users.
Currently, the Shanoir@OFSEP repository is hosting five studies: the “Mother Cohort” (200,000 MRIs planned over the next 10 years) as well as four MS imaging clinical research projects. More of these “OFSEP-labeled” clinical research projects or nested cohorts will be integrated in coming years. Everyone can join the “Mother Cohort” study as long as they use the OFSEP protocol. One can also ask the OFSEP to contribute to the project through his study as soon as the PI presents his research study subject to the OFSEP scientific committee that can grant (or not) the hosting. Data hosted on Shanoir@OFSEP will remain confidential (private) throughout the duration of the study but can be made available to all researchers through a specific OFSEP application.
Conclusion and Perspectives
The Shanoir SaaS manages the sharing of distributed information sources in neuroimaging over the Internet, whether these resources are located in centers of experimentation, clinical departments in neurology, or research centers in cognitive neurosciences or image processing. Through the description of two repositories that administer a Shanoir environment (Neurinfo and OFSEP), we have illustrated how a large variety of users can diffuse, share, or access neuroimaging information between peers almost as easily as if the data were stored at their local hospital, research lab, or company. Through the description of the Shanoir software environment, we have illustrated how neuroimaging data can be structured, managed, archived, visualized, and shared.
In the medium term, we plan to integrate Shanoir’s resources and services with the open community through the French National Infrastructure’s “France Life Imaging” (FLI),12 and more specifically, the “Information Analysis and Management” (IAM) node that is dedicated to provide large scale IT infrastructure for in vivo imaging. For this purpose, the FLI–IAM node will build and operate an infrastructure to store, manage, and process in vivo imaging data from human or preclinical procedures. The main achievements of the IAM node will consist of a versatile software platform composed of several subcomponents that will connect hardware and software facilities to build:
• an archiving and management infrastructure of in vivo images as well as provide solutions to process and manage the acquired data through dedicated software and hardware solutions;
• versatile image analysis and data management solutions for in vivo imaging to facilitate interoperability between production sites and users and provide heterogeneous and distributed storage solutions for raw and metadata indexing (e.g., through the use of semantic models).
As such, we are under integrating Shanoir as one of the data management solutions of the FLI-IAM facilities along with a collection of companion data management software platforms such as CATI-DB13 and ArchiMed, or a collection of processing clients or high-performance computing workflow facilities such as medInria, BrainVisa,14 and the VIP platform.15 For this purpose, within FLI-IAM, we are setting up the “glue” between these platforms that will make it possible to connect and interoperate between them. In addition, through FLI-IAM, we will provide the necessary information for additional resources to join the FLI-IAM infrastructure by defining the basic conformal statement that will make the technology and scalability of FLI-IAM possible.
Nonetheless, as described in Section “Introduction,” there are a lot of similar initiatives going on recently in the medical imaging research field, such as Human Brain Project, ADNI, XNAT-based solutions, etc. For these initiatives, as well as for Shanoir, the goal is to share the data at a large extent. This cannot be done without a significant additional effort on standardization in the field and on interoperability between software platforms addressing similar services. This is what motivates the integration of Shanoir in the French FLI-IAM e-infrastructure initiative. This challenge of tomorrow is to continue this effort at the international level.
Author Contributions
All authors listed have made substantial, direct, and intellectual contribution to the work and approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work has been supported by Inria under a “technological development program” grant for the Neurinfo platform from the Brittany regional council and the EU-Feder program and two grants provided by the French government through the Agence Nationale de la Recherche as part of the Investment in the Future Program under references ANR-10-COHO-002 (for OFSEP) and ANR-11-INBS-006 (for FLI).
Abbreviations
DICOM, Digital Imaging and Communications in Medicine; DOLCE, Descriptive Ontology for Linguistic and Cognitive Engineering; IRC, imaging resource center; J2EE, Java platform enterprise edition; JAX-WS, Java API for XML web services; JWS, Java web start; NIfTI, neuroimaging informatics technology initiative; OFSEP, Observatoire Français de la Sclérose en Plaques (French multiple sclerosis observatory); OWL, web ontology language; PACS, picture archiving and communication system; PI, principal investigator; SaaS, software as a service; Shanoir, sharing neuroimaging resources; SOAP, simple object access protocol; WSDL, Web Service Description Language.
Footnotes
- ^http://www.incf.org/.
- ^OntoNeuroLOG: http://neurolog.i3s.unice.fr/public_namespace/ontology.
- ^http://www.dcm4che.org.
- ^http://lucene.apache.org/solr/.
- ^QtShanoir library: http://qtshanoir.gforge.inria.fr.
- ^medInria: http://med.inria.fr.
- ^http://www.dcm4che.org/.
- ^http://wiki.nginx.org/.
- ^http://www.neurinfo.org/.
- ^The OFSEP MS Cohort observatory: http://www.ofsep.org/en/.
- ^EDMUS: http://www.edmus.org.
- ^France Life Imaging https://www.francelifeimaging.fr) with the IAM node (https://project.inria.fr/fli/) is a national infrastructure for in vivo imaging.
- ^http://www.cati-neuroimaging.com.
- ^http://brainvisa.info.
- ^http://www.creatis.insa-lyon.fr/vip.
References
Ashish, N., Ambite, J. L., Muslea, M., and Turner, J. A. (2010). Neuroscience data integration through mediation: an (F)BIRN case study. Front. Neuroinformatics 4:12. doi:10.3389/fninf.2010.00118
Barillot, C., Amsaleg, L., Aubry, F., Bazin, J.-P., Benali, H., Cointepas, Y., et al. (2003). Neurobase: Management of Distributed Knowledge and Data Bases in Neuroimaging. Human Brain Mapping. New York, NY: Academic Press, 726.
Barillot, C., Benali, H., Dojat, M., Gaignard, A., Gibaud, B., Kinkingnehun, S., et al. (2006). Federating distributed and heterogeneous information sources in neuroimaging: the neurobase project. Stud. Health Technol. Inform. 120, 3–13.
Bellec, P., Lavoie-Courchesne, S., Dickinson, P., Lerch, J. P., Zijdenbos, A. P., and Evans, A. C. (2012). The pipeline system for Octave and Matlab (PSOM): a lightweight scripting framework and execution engine for scientific workflows. Front. Neuroinform. 6:7. doi:10.3389/fninf.2012.00007
Benkner, S., Arbona, A., Berti, G., Chiarini, A., Dunlop, R., Engelbrecht, G., et al. (2010). @neurIST: infrastructure for advanced disease management through integration of heterogeneous data, computing, and complex processing services. IEEE Trans. Inf. Technol. Biomed. 14, 1365–1377. doi:10.1109/TITB.2010.2049268
Book, G. A., Anderson, B. M., Stevens, M. C., Glahn, D. C., Assaf, M., and Pearlson, G. D. (2013). Neuroinformatics Database (NiDB) – a modular, portable database for the storage, analysis, and sharing of neuroimaging data. Neuroinformatics 11, 495–505. doi:10.1007/s12021-013-9194-1
Button, K. S., Ioannidis, J. P., Mokrysz, C., Nosek, B. A., Flint, J., Robinson, E. S., et al. (2013). Confidence and precision increase with high statistical power. Nat. Rev. Neurosci. 14, 585–586. doi:10.1038/nrn3475-c4
Carp, J. (2012). The secret lives of experiments: methods reporting in the fMRI literature. Neuroimage 63, 289–300. doi:10.1016/j.neuroimage.2012.07.004
Cotton, F., Kremer, S., Hannoun, S., Vukusic, S., Dousset, V., The Imaging Working Group of OFSEP. (2015). OFSEP, a nationwide cohort of people with multiple sclerosis: consensus minimal MRI protocol. J. Neuroradiol. 42, 133–140. doi:10.1016/j.neurad.2014.12.001
Das, S., Zijdenbos, A. P., Harlap, J., Vins, D., and Evans, A. C. (2011). LORIS: a web-based data management system for multi-center studies. Front. Neuroinform. 5:37. doi:10.3389/fninf.2011.00037
Dinov, I., Lozev, K., Petrosyan, P., Liu, Z., Eggert, P., Pierce, J., et al. (2010). Neuroimaging study designs, computational analyses and data provenance using the LONI pipeline. PLoS ONE 5:e13070. doi:10.1371/journal.pone.0013070
Evans, A. C., and Brain Development Cooperative Group. (2006). The NIH MRI study of normal brain development. Neuroimage 30, 184–202. doi:10.1016/j.neuroimage.2005.09.068
Glatard, T., Rousseau, M.-E., Camarasu-Pop, S., Rioux, P., Sherif, T., Beck, N., et al. (2014). Interoperability between the CBRAIN and VIP web platforms for neuroimage analysis. Front. Neuroinformatics. doi:10.3389/conf.fninf.2014.18.00070
Gorgolewski, K., Burns, C. D., Madison, C., Clark, D., Halchenko, Y. O., Waskom, M. L., et al. (2011). Nipype: a flexible, lightweight and extensible neuroimaging data processing framework in python. Front. Neuroinform. 5:13. doi:10.3389/fninf.2011.00013
Gupta, A., Ludaescher, B., Martone, M., and Rajasekar, A. (2003). BIRN-M: A Semantic Mediator for Solving Real-World Neuroscience Problems. ACM SIGMOD 2003. San Diego, CA: ACM, 678.
Hall, D., Huerta, M. F., McAuliffe, M. J., and Farber, G. K. (2012). Sharing heterogeneous data: the national database for autism research. Neuroinformatics 10, 331–339. doi:10.1007/s12021-012-9151-4
Hibar, D. P., Stein, J. L., Renteria, M. E., Arias-Vasquez, A., Desrivieres, S., Jahanshad, N., et al. (2015). Common genetic variants influence human subcortical brain structures. Nature 520, 224–229. doi:10.1038/nature14101
Ioannidis, J. P. (2014). How to make more published research true. PLoS Med. 11:e1001747. doi:10.1371/journal.pmed.1001747
Ioannidis, J. P., Munafo, M. R., Fusar-Poli, P., Nosek, B. A., and David, S. P. (2014). Publication and other reporting biases in cognitive sciences: detection, prevalence, and prevention. Trends Cogn. Sci. 18, 235–241. doi:10.1016/j.tics.2014.02.010
Jack, C. R. Jr., Bernstein, M. A., Fox, N. C., Thompson, P., Alexander, G., Harvey, D., et al. (2008). The Alzheimer’s disease neuroimaging initiative (ADNI): MRI methods. J. Magn. Reson. Imaging 27, 685–691. doi:10.1002/jmri.21049
Keator, D. B., Grethe, J. S., Marcus, D., Ozyurt, B., Gadde, S., Murphy, S., et al. (2008). A national human neuroimaging collaboratory enabled by the Biomedical Informatics Research Network (BIRN). IEEE Trans. Inf. Technol. Biomed. 12, 162–172. doi:10.1109/TITB.2008.917893
Keator, D. B., Helmer, K., Steffener, J., Turner, J. A., Van Erp, T. G., Gadde, S., et al. (2013). Towards structured sharing of raw and derived neuroimaging data across existing resources. Neuroimage 82, 647–661. doi:10.1016/j.neuroimage.2013.05.094
Keator, D. B., Wei, D., Gadde, S., Bockholt, J., Grethe, J. S., Marcus, D., et al. (2009). Derived data storage and exchange workflow for large-scale neuroimaging analyses on the BIRN grid. Front. Neuroinformatics 3:30. doi:10.3389/neuro.11.030.2009
Kennedy, D. N., Haselgrove, C., Riehl, J., Preuss, N., and Buccigrossi, R. (2015). The three NITRCs: a guide to neuroimaging neuroinformatics resources. Neuroinformatics 13, 383–386. doi:10.1007/s12021-015-9263-8
Marcus, D. S., Harms, M. P., Snyder, A. Z., Jenkinson, M., Wilson, J. A., Glasser, M. F., et al. (2013). Human Connectome Project informatics: quality control, database services, and data visualization. Neuroimage 80, 202–219. doi:10.1016/j.neuroimage.2013.05.077
Marcus, D. S., Olsen, T. R., Ramaratnam, M., and Buckner, R. L. (2007). The Extensible Neuroimaging Archive Toolkit: an informatics platform for managing, exploring, and sharing neuroimaging data. Neuroinformatics 5, 11–34. doi:10.1385/NI:5:1:11
Masolo, C., Borgo, S., Gangemi, A., Guarino, N., and Oltramari, A. (2003). WonderWeb Deliverable D18, Ontology Library (Final). Technical Report. Trento: LOA-ISTC, CNR.
Mazziotta, J. C., Toga, A. W., Evans, A., Fox, P., and Lancaster, J. (1995). A probabilistic atlas of the human brain: theory and rationale for its development. The International Consortium for Brain Mapping (ICBM). Neuroimage 2, 89–101. doi:10.1006/nimg.1995.1012
Menze, B. H., Jakab, A., Bauer, S., Kalpathy-Cramer, J., Farahani, K., Kirby, J., et al. (2015). The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans. Med. Imaging 34, 1993–2024. doi:10.1109/TMI.2014.2377694
Michel, F., Gaignard, A., Ahmad, F., Barillot, C., Batrancourt, B., Dojat, M., et al. (2010). Grid-wide neuroimaging data federation in the context of the NeuroLOG project. Stud. Health Technol. Inform. 159, 112–123.
Ooi, C., Bullmore, E. T., Wink, A.-M., Sendur, L., Barnes, A., Achard, S., et al. (2009). CamBAfx: workflow design, implementation and application for neuroimaging. Front. Neuroinformatics 3:27. doi:10.3389/neuro.11.027.2009
Poldrack, R. A., and Gorgolewski, K. J. (2014). Making big data open: data sharing in neuroimaging. Nat. Neurosci. 17, 1510–1517. doi:10.1038/nn.3818
Poline, J. B., Breeze, J. L., Ghosh, S., Gorgolewski, K., Halchenko, Y. O., Hanke, M., et al. (2012). Data sharing in neuroimaging research. Front. Neuroinform. 6:9. doi:10.3389/fninf.2012.00009
Rimal, B. P., Choi, E., and Lumb, I. (2009). “A taxonomy and survey of cloud computing systems. INC, IMS and IDC, 2009. NCM’09,” in Fifth International Joint Conference on IEEE (Seoul: IEEE), 44–51.
Roland, P. E., and Zilles, K. (1994). Brain atlases – a new research tool. Trends Neurosci. 17, 458–467. doi:10.1016/0166-2236(94)90131-7
Scott, A., Courtney, W., Wood, D., de la Garza, R., Lane, S., King, M., et al. (2011). COINS: an innovative informatics and neuroimaging tool suite built for large heterogeneous datasets. Front. Neuroinform. 5:33. doi:10.3389/fninf.2011.00033
Shepherd, G. M., Mirsky, J. S., Healy, M. D., Singer, M. S., Skoufos, E., Hines, M. S., et al. (1998). The Human Brain Project: neuroinformatics tools for integrating, searching and modeling multidisciplinary neuroscience data. Trends Neurosci. 21, 460–468. doi:10.1016/S0166-2236(98)01300-9
Sherif, T., Rioux, P., Rousseau, M. E., Kassis, N., Beck, N., Adalat, R., et al. (2014). CBRAIN: a web-based, distributed computing platform for collaborative neuroimaging research. Front. Neuroinform. 8:54. doi:10.3389/fninf.2014.00054
Styner, M., Lee, J., Chin, B., Chin, M. S., Commowick, O., Tran, H., et al. (2008). 3D segmentation in the clinic: a grand challenge II: MS lesion segmentation. Midas J. 11. Available at: http://www.midasjournal.org/browse/publication/638
Temal, L., Dojat, M., Kassel, G., and Gibaud, B. (2008). Towards an ontology for sharing medical images and regions of interest in neuroimaging. J. Biomed. Inform. 41, 766–778. doi:10.1016/j.jbi.2008.03.002
Van Essen, D. C., Smith, S. M., Barch, D. M., Behrens, T. E., Yacoub, E., Ugurbil, K., et al. (2013). The WU-Minn Human Connectome Project: an overview. Neuroimage 80, 62–79. doi:10.1016/j.neuroimage.2013.05.041
Van Horn, J. D., and Gazzaniga, M. S. (2013). Why share data? Lessons learned from the fMRIDC. Neuroimage 82, 677–682. doi:10.1016/j.neuroimage.2012.11.010
Van Horn, J. D., Grethe, J. S., Kostelec, P., Woodward, J. B., Aslam, J. A., Rus, D., et al. (2001). The Functional Magnetic Resonance Imaging Data Center (fMRIDC): the challenges and rewards of large-scale databasing of neuroimaging studies. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356, 1323–1339. doi:10.1098/rstb.2001.0916
Walport, M., and Brest, P. (2011). Sharing research data to improve public health. Lancet 377, 537–539. doi:10.1016/S0140-6736(10)62234-9
Keywords: neuroimaging, database, data sharing, neuroinformatics, software as a service, cloud computing, web application, web services
Citation: Barillot C, Bannier E, Commowick O, Corouge I, Baire A, Fakhfakh I, Guillaumont J, Yao Y and Kain M (2016) Shanoir: Applying the Software as a Service Distribution Model to Manage Brain Imaging Research Repositories. Front. ICT 3:25. doi: 10.3389/fict.2016.00025
Received: 08 April 2016; Accepted: 20 October 2016;
Published: 01 December 2016
Edited by:
Marleen De Bruijne, Erasmus MC, NetherlandsReviewed by:
Suyash P. Awate, Indian Institute of Technology Bombay, IndiaHenri Vrooman, Sophia Children’s Hospital, Netherlands
Copyright: © 2016 Barillot, Bannier, Commowick, Corouge, Baire, Fakhfakh, Guillaumont, Yao and Kain. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christian Barillot, Y2hyaXN0aWFuLmJhcmlsbG90QGlyaXNhLmZy
 Christian Barillot
Christian Barillot Elise Bannier
Elise Bannier Olivier Commowick1,2,3
Olivier Commowick1,2,3 Isabelle Corouge
Isabelle Corouge Ines Fakhfakh
Ines Fakhfakh