
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Geochem. , 21 August 2024
Sec. Biogeochemistry
Volume 2 - 2024 | https://doi.org/10.3389/fgeoc.2024.1410338
This article is part of the Research Topic The Biome of Glaciers and Ice Sheets and their Downstream Ecosystems View all articles
 Jared J. Clance1†
Jared J. Clance1† Jacob M. C. Shaffer1†
Jacob M. C. Shaffer1† Morgan L. Cable2
Morgan L. Cable2 Christian Stenner3
Christian Stenner3 Glyn Williams-Jones4
Glyn Williams-Jones4 Anna Szynkiewicz5
Anna Szynkiewicz5 Michael Paton2
Michael Paton2 Kathleen Graham3
Kathleen Graham3 Olivia Vinnes1
Olivia Vinnes1 Jill A. Mikucki1*
Jill A. Mikucki1*The Mount Meager Volcanic Complex (Q̓welq̓welústen) is an active glacier-capped volcanic massif in the Garibaldi Volcanic Belt (British Columbia) and the only known glaciovolcanic cave system in North America steadily releasing sulfur-rich gases. In September 2022, leveraging specialized cave explorer expertise, the fumarole-carved ice cave at the Job Glacier on Mt. Meager was surveyed. Direct measurements of fumarolic gas concentrations were taken at the source, with H2S >200 ppm, SO2 >100 ppm, CO2 ∼5,200 ppm, and CO ∼230 ppm. Snowpack and fumarole-associated sediments were characterized for microbial diversity, functional potential, and biogeochemistry including measurements of nutrients, major ions, dissolved organic and inorganic carbon concentrations as well as the stable isotope compositions of carbon, sulfur, hydrogen and oxygen. Green algae (Chlorophyta) dominated the snowpack, consistent with other Pacific Northwest glaciers. Representatives of Firmicutes were the most abundant bacterial sequences detected in our samples, contrasting with other glacier and snowpack samples which harbor abundant Sphingobacteria, Betaproteobacteria, and Alphaproteobacteria. Sediments and water collected inside the cave were mostly high in SO42- (5.3–185.2 mg/L) and acidic (pH = 3.6–6.0), while most other major anions and cations were below detection of the method used. Snow at the cave entrance had more SO42- (0.08 mg/L) and lower pH (5.9) than snow collected at a distance (SO42- undetectable, pH 7.6), suggesting influence by fumarole exhalations. Negative δ13C values of organic matter (−29.0‰ to −26.1‰, respectively) in sediments suggest in-situ microbial carbon transformations, findings that are supported by the presence of genes encoding complete heterotrophic and autotrophic carbon transformation pathways. The δ34S value of H2S was ∼0‰, suggesting a deep magmatic origin; however, both sulfur-oxidizing and sulfate-reducing microbial phyla were present in the sediment samples as were genes encoding both dissimilatory sulfur-oxidizing and sulfate-reducing pathways. Metagenomic data suggest diverse chemosynthetic lifestyles in the cave microbial community. This study provides insight on the microbiomes associated with a sulfidic glaciovolcanic system and identifies unique analog features for icy celestial bodies like Saturn’s moon Enceladus, where cryovolcanic activity may carry biomarkers from the subsurface and deposit them on surface ice.
Glaciovolcanism is a collection of processes by which volcanic heat interacts with glacial ice at the Earth’s surface (Smellie and Edwards, 2016). One such interaction is the formation of void spaces underneath ice, termed glaciovolcanic cave systems (Sobolewski et al., 2022; Unnsteinsson et al., 2024). Although glaciovolcanic caves are distributed globally in volcanic settings, they remain understudied. These cave systems provide access points to glacial interior environments, including the interfaces between basal ice and the underlying rock, referred to as the subglacial realm. This is a critical zone in the glacier ecosystem where microbial processes such as chemosynthesis and oxidation-reduction reactions including denitrification, methane oxidation, sulfur transformations, and methanogenesis appear to be enhanced (e.g., Hotaling et al., 2017; Livingstone et al., 2022). Geothermal heat from fumaroles on glaciated volcano slopes can cause melting and form dramatic glaciovolcanic features. For example, on Mt. Erebus in Antarctica, volcanic heat and steam outgassing rapidly cool near the surface, forming extensive caves and towers (Curtis and Kyle, 2011). Mount Rainier, USA, has well-documented glaciovolcanic cave systems formed by subglacial fumarolic heat in the glaciated summit craters (Florea et al., 2021; Stenner et al., 2023). Measured volcanogenic gases within glaciovolcanic caves are typically dominated by CO2 rather than sulfidic gases (Stenner et al., 2022).
Numerous investigations of cave geomicrobiology in karst settings show how functionally diverse microorganisms, including sulfur-, iron- and manganese-oxidizing bacteria shape the ecosystem (e.g., Northup and Lavoie, 2001), while the microbiology of surface fumarolic systems is less studied. Volcanic tephra heated by fumarolic gases in the Atacama Desert (Chile/Argentina) contained viable microorganisms, including photoautotrophs (Costello et al., 2009; Solon et al., 2018), while molecular studies of soils and mineral surfaces associated with fumaroles in the Galápagos Islands found that pH controlled community composition selecting for abundant acidophiles (e.g., Sulfobacillus, Acidobacteria; Mayhew et al., 2007). At Mt. Erebus, microbial community structure correlates with pH, and amplicon sequence data suggest chemolithotrophy is an important metabolic process (Noell et al., 2022). To date, one microbiological study has examined temperate fumarole steam directly, detecting DNA and viable microorganisms related to halophilic archaea (Ellis et al., 2008). Glaciovolcanic caves combine processes observed in caves, fumaroles, and subglacial settings (Sobolewski et al., 2022). However, little is known about the microbial ecology of these systems with existing studies of glaciovolcanic cave microbiology limited to Mt. St. Helens in Washington, USA (Anitori et al., 2011; Connell and Staudigel, 2013; Tebo et al., 2015; Fraser et al., 2018; Sobolewski et al., 2022).
Studies of the terrestrial cryosphere can inform our understanding of the potential ecology of astrobiological targets, such as ocean worlds, and can help us develop and test tools and operational concepts for future extraterrestrial missions (Howell et al., 2021; Glass et al., 2022). Ocean worlds are planetary bodies, such as Jupiter’s moon Europa and Saturn’s moon Enceladus, that harbor liquid water oceans below an icy shell (Hand et al., 2020) and are considered prime targets for astrobiological investigations (Hendrix et al., 2019; Cable et al., 2021; Vance et al., 2023; Cockell et al., 2024). Glaciovolcanic systems feature diverse extreme icy terrains including fissures, crevasses, and voids, providing features analogous to what may be expected on icy ocean world surfaces. In most terrestrial glacial settings, meltwater generally migrates from the glacier surface to the base through cracks and fissures known as moulins. There are very few cases where subsurface materials transit from the base to the surface (Badgeley et al., 2017). Glaciovolcanic fumaroles provide a unique ocean world analog feature where reduced gases vent to the surface and a photic and oxidizing environment is encountered. This can serve as a potential analog for cryovolcanic plumes on ocean worlds, where venting delivers ocean fluids, and possibly biomarkers such as organic molecules (Postberg et al., 2018) or even cells (Perera and Cockell, 2023) from the deeper subsurface; cryovolcanic plumes have been observed on Enceladus and possibly Europa (Dougherty et al., 2006; Hansen et al., 2006; Porco et al., 2006; Spahn et al., 2006; Tokar et al., 2006; Waite et al., 2006; Roth et al., 2014; Arnold et al., 2019).
Sub-ice realms are difficult to sample due to the logistical challenges of drilling through tens to hundreds of meters of ice, as well as the potential for contamination (Schuler and Mikucki, 2023). Glacier cave structures can provide access into the subglacial realm enabling the investigation of structural features and the direct collection of fumarole gases, sediments and ice for biogeochemical analyses. As such, access to subglacial realms via glaciovolcanic caves is operationally simpler and can facilitate the study of this poorly understood ecosystem.
The Mount Meager Volcano Complex (MMVC) is known as Q̓welq̓welústen in the local Líĺwat First Nation Language. In Ucwalmícwts, Q̓welq̓welústen (Qual-qual-OSH-tin) means “cooked face place” or “cooked fire place; ” Qwelqwel is a repetition, meaning “very cooked,” the suffix -us is used for “face,” “hill,” or “fire” (also suggesting “personality” or “spirit”), and the suffix -tn indicates a location, place, instrument or thing; mention of heat, cooking, hot springs, and landscape changes recur as key elements in Lílwat oral traditions about this massif (Wilson et al., 2024).
As part of the Mount Meager Volcano Project, we endeavored to explore, map, and sample glacier caves associated with a fumarole system in order to use these subsurface access points to 1) test robotic mapping and sensing capabilities, 2) take in situ measurements of the cave ecosystem, and 3) collect samples of subglacial sediments, glacial melt water, and snow that surrounds the fumarole for geochemical and microbiological analyses. The MMVC is a potentially valuable astrobiological analog feature in which to study the distribution and evolution of discharged subsurface materials, including nutrients and biosignatures, that may accumulate on surface ice and snow, interacting with the surficial chemistry and biota.
Mt. Meager is a large, long-lived (>1.9 Ma), and active glacier-clad stratovolcano complex in the Garibaldi Volcanic Belt in the northern portion of the Cascade Volcanic Arc in southwestern British Columbia, Canada (e.g., Read, 1990; Russell et al., 2021). The last major explosive eruption at the MMVC occurred approximately 2,360 years BP (Clague et al., 1995; Hickson et al., 1999). Currently, the MMVC experiences frequent landslides, ranging from small events almost annually to less frequent large slope failures (e.g., eight historical events, 105–5.3 x 107 m3), which may be exacerbated by glacial movement (Friele et al., 2008; Roberti et al., 2018; Connelly et al., 2024). At Job Glacier on the north side of Mt. Meager (50°37′54″N, 123°32′26″W), glacial ice overlies an active fumarole field with glaciovolcanic cave openings (near horizontal, lateral conduits and near vertical chimneys; Figure 1A; Figure 2A) which have been observed since 2015 (Roberti et al., 2018; Unnsteinsson et al., 2024). Due in part to this fumarolic activity, Mt. Meager has been classified as a Very High threat volcano in Canada (Kelman and Wilson, 2024). Job Glacier has experienced rapid loss of ice since 1987, downwasting by up to 50 m and retreating at an average rate of 11 m/yr between 1987 and 2016 (Roberti, 2018). Thinning of ice over the active fumarole field is a major contributing factor in the formation of these caves and release of fumarole-derived gases to the surface (Unnsteinsson et al., 2024).
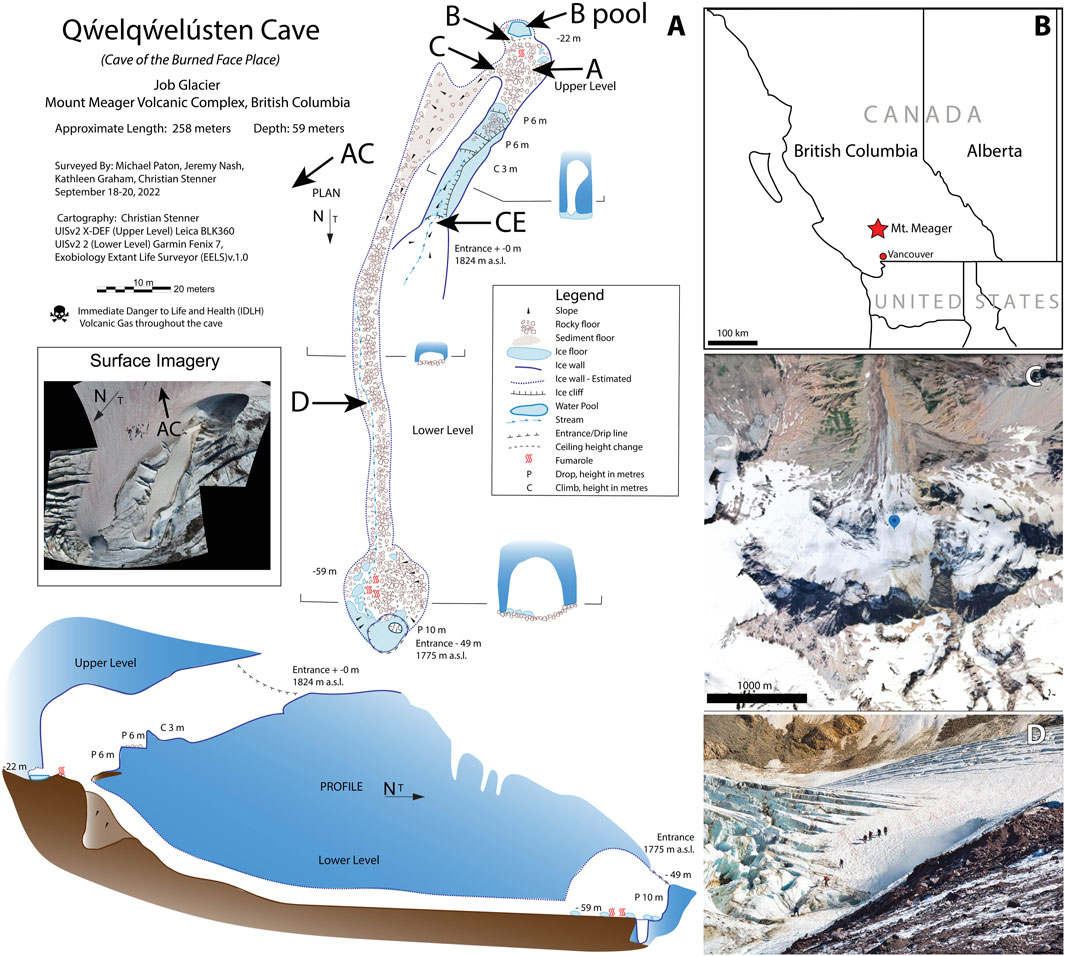
Figure 1. The Q̓welq̓welústen/Mount. Meager Volcano Complex system. (A) Plan-view and profile-view maps of the cave system at Job Glacier in September 2022. Approximate locations of sites sampled in this study are indicated. AC = Above Cave, CE = Cave Entrance. Approximate location of site AC is also shown on the inset surface image. (B) Location of Mt. Meager in British Columbia, Canada. (C) Satellite image of Job Glacier, MMVC. Marker indicates location of the September 2022 field survey. Image retrieved from Google Earth. (D) September 2022 survey team members at cave entrance on Job Glacier.

Figure 2. Sample collection at the Mt. Meager Glaciovolcanic cave system. (A) Aerial view of the cave system at Mt. Meager; (B) visibly pigmented snow surrounding the fumarole cave entrance with collection Above Cave (AC); (C) Cave Entrance (CE); (D) sample acquisition inside the ice cave with visible H2S vapors; (E) VNIR/SWIR reflectance measurement collection.
A 3-day field survey of the glaciovolcanic cave system at Job Glacier was conducted in September 2022, which included mapping of the cave interior (Figure 1A), in-situ measurements of select physical and geochemical parameters, and collection of sediment, water, and snow samples for biological and geochemical analyses. Sample sites, types, and their designated nomenclature are briefly described below with additional details in Table 1; Figure 1. Snow on and surrounding the west slope of the cave entrance featured visibly red streaks presumed to be snow algae (Figures 2A, B). An area approximately 50 m NW uphill of the southern cave entrance, with a prominent red streak, was designated Above Cave (AC) snow and selected for sampling (Figure 2B). The cave entrance formed an approximately 300-m2 depression with both standing water and deposited sediments (Figures 2A, C). Snow within this area had a brown-yellow coloration. Snow with a yellow tint was collected from the wall of the depression designated Cave Entrance (CE) snow. Sediments inside the cave were sampled at sites selected to represent a diversity of environmental conditions such as apparent water saturation (or lack thereof) in sediments, concentration of volcanic gases, and sediment temperatures (Table 1). Two sites close to an active fumarole near the southern cave entrance were designated sites A and B; a pool of water near this fumarole was designated site B pool. A site downslope from this fumarole was designated site C, and a site further downslope in a lower level of the cave was designated site D (Figure 1).
The glaciovolcanic caves at Job Glacier were mapped and sampled by expert cave explorers which required the use of a specialized self-contained breathing apparatus. At each site, ground surface temperature was measured using a Seek Thermal Pro camera (instrument range = −40°C–330°C ± 0.3°C) and/or an Omega HH11B digital thermometer (instrument range −20°C–1,372°C ± 0.1°C). Ambient air temperature and gas concentrations including H2S, CO2, O2, CO, SO2, and CH4 were measured in-situ using body worn, calibrated RKI GX-6000 and GX-3R monitors with an upper detection limit of 200 ppm (H2S), 10 vol% (CO2), 2000 ppm (CO), 99.9 ppm (SO2), 40% (O2), and 5% CH4 (CH4 expressed as LEL, lower explosive limit). RKI GX-6000 and GX-3R recorded gas and temperature measurements at 10 s intervals.
Snow, glacier cave and fumarole materials including sediment, rocks, and ice were collected at each site using individually wrapped sterile scoops (Bel-Art) and transferred to sterile polyethylene bags (Whirl-Pak). All sediment samples were surficial except at site C where a sample from ∼5 cm depth was also taken. Materials were stored in a cooler and transported to our camp laboratory (∼2–4 h) where they were aseptically subsampled with sterile tools for additional analyses or stabilized. Collected sediment subsamples were designated for either molecular analysis, geochemistry, or direct counts of microbial cells and were stored at −80°C, −20°C, or +4°C, respectively and shipped at temperature to the University of Tennessee (Knoxville, Tennessee, USA).
Chemical parameters of streams and melted snow samples were analyzed at our camp laboratory using an AquaTROLL 500 Multiparameter Sonde (In-Situ Inc.; PN: 0050770) outfitted with sensors for pH and oxidation-reduction potential (pH/ORP; PN: 0063470), conductivity (PN: 0063460) and rugged dissolved oxygen (RDO; PN: 0063450). Briefly, ∼10 mL of water or melted snow was transferred to a clean polypropylene sample container (Corning) and the sonde was submerged until all sensors were covered. pH was measured in a 2:1 water:sediment solution (10 mL of ultrapure water added to 5 g of sediment, stirred and equilibrated for 10 min prior to measurements). Measurements were taken in Live mode and data points were averaged.
Adenosine triphosphate (ATP) concentrations in liquid and melted snow samples were determined using a Hygiena EnSURE Luminometer (V.2) and AquaSnap Total ATP Water Tests. Samples were transferred to ATP-free microcentrifuge tubes and measured following the manufacturer’s protocol. All measurements were performed in triplicate and blank measurements were made using UltraPure water (Intermountain Life Sciences). Concentrations were recorded as relative light units (RLU) and converted to pmol/mL via a standard curve of an ATP standard (Thermo Scientific; R2 = 0.9962). Chlorophyll a (CHLa) and colored dissolved organic matter (CDOM) were measured using a Turner Trilogy Fluorometer with a CHLa in vivo module (460 nm) and a CDOM/FDOM module (365 nm), respectively. CHLa relative fluorescence units (RFU) were converted to μg/L using a CHLa standard curve (Turner; R2 = 0.9999). CDOM RFU were converted to mg/mL using an IHSS Suwannee River Fulvic Acid Standard II (www.humic-substances.org; Averett et al., 1994; R2 = 0.9963).
The composition of various snow and rock deposits, as well as rock samples collected from within the fumarole, was analyzed using an ASD TerraSpec Halo Mineral Identifier (Malvern Panalytical; see Figure 2E). This handheld field instrument contains an internal broad-spectrum halogen light source and measures the visible to short wave infrared reflectance (350–2,500 nm, spectral resolution 3 nm at 700 nm, 9.8 nm at 1,400 nm, and 8.1 nm at 2,100 nm) of a sample surface. A white reference calibration was performed with Spectralon® prior to sample measurements, after which spectra were collected in triplicate. Each spectrum collected was an average of 50 scans for analyses performed at the field site, or 100 scans for analyses of samples returned to the camp laboratory. Spectra were splice corrected (splice correction gap = 5, no experimental slope correction) and converted from relative to absolute reflectance in ViewSpec Pro (ver. 6.2) using a laboratory reference spectrum obtained with the same spectrometer (Unit 30241). Minerals identified by the TerraSpec Halo onboard spectral library (HaloStandard ver. 2.3) were cross-referenced against the USGS Spectral Library (Ver. 7; Kokaly et al., 2017).
Sediment extracts for soluble ion analyses (1:5 sediment:ultrapure water) were prepared from 10 g of sediments by shaking in 50 mL ultrapure water for 1 h, then centrifugation at 26,200 x g for 15 min. Sediment leachate, water and snowmelt samples were filtered through a 0.45 μm polycarbonate filter for ion analysis; cations were acidified (5% HNO3 final concentration). Major cation and anion concentrations in the water/snow samples were measured using Dionex ICS-2100 and Dionex ICS-2000 ion chromatographs (IC) in the Stable Isotope Laboratory of University of Tennessee with analytical precision of ±5%. Oxygen (O) and hydrogen (H) isotope compositions of water (δ18O and δ2H, respectively) were measured using a Los Gatos Research DLT-100 with analytical precision of ± 0.2‰ for δ18O and 0.7‰ for δ2H.
Samples were passed through a pre-combusted (4 h at 450°C) glass fiber filter (grade GF/F) into combusted amber glass vials for dissolved organic carbon (DOC) quantification and stored at −20°C until analyses. Concentrations and C isotope compositions (δ13C) of dissolved inorganic carbon (DIC) and DOC in the water/snow samples were measured using the Thermo-Scientific Gas Bench II coupled with a Delta Plus XL isotope ratio mass spectrometer. In the first step, CO2 gas headspace liberated by phosphoric acid was measured to determine the concentration and δ13C of DIC. In the second step, the remaining water was bubbled with He gas for 5 min at the rate of 100 mL/min to remove any remaining DIC/CO2. The water was then oxidized to convert DOC into CO2 using 0.8 mL of 1 N sodium persulfate. The water was heated in metal sleeves up to 90°C–100°C for 1 h, allowed to cool, and the concentration and δ13C of DOC was determined. The analytical precision for δ13C and wt% of DIC and DOC were ± 0.2‰ and 0.2%, and ± 0.2‰ and 0.6%, respectively.
Fumarolic H2S was reacted in-situ with a concentrated solution of zinc acetate and precipitated to zinc sulfide (ZnS). Sediments collected in the cave were air dried in the lab and ground using an agate mortar. The δ34S of ZnS and bulk δ13C of sediments were analyzed using a Costech EA coupled with a Delta Plus XL. Analytical precisions were ± 0.2‰ for δ34S and 0.1‰ for δ13C. δ2H and δ18O values are reported in units of per mil with respect to Standard Mean Ocean Water (V-SMOW), the δ13C values to Pee Dee Belemnite (V-PDB), and the δ34S values to Vienna Cañon Diablo Troilite (V-CDT).
Aliquots for cell enumeration were fixed with paraformaldehyde (final concentration 4%) and stored at +4°C until analysis. Direct counts of DNA-containing cells in both fixed snowmelt and sediment supernatant were conducted via microscopy (Mikucki and Priscu, 2007; Purcell et al., 2014). Approximately 1–2 g of sediment was suspended in sterile 1X Tris-borate-EDTA buffer and fixed with paraformaldehyde (final concentration 4%). The suspension was shaken on an orbital shaker at 100 rpm and 10°C–15°C for 30 min, then centrifuged at 50 g for 1 h, and the resulting supernatant was collected for staining. Three technical replicates were prepared for each sample. 1–2 mL of the fixed sample were stained with SYBR Gold (Invitrogen) for 15 min, then vacuum-filtered through a 25-mm black polycarbonate 0.2-μm membrane filter (Sterlitech) and observed by fluorescence on a Zeiss AXIO Imager two microscope. At least 30 100-μm square fields of view were counted for each sample, biovolume was calculated for select cells in each field. Direct counts were used to calculate cells/g of sediment.
Snow samples were melted at ambient temperature (∼21°C) before subsampling. Aliquots of snow or water designated for nucleic acid sequencing (120–600 mL) were filtered through Sterivex filters (Millipore-Sigma), shipped frozen, and stored at −80°C until extraction. DNA was extracted from samples using the Dneasy PowerLyzer PowerSoil kit (Qiagen) following manufacturer recommendations with minor modifications; Lysing Matrix E bead-beating tubes (MP Biomedicals) were used instead of the provided bead-beating tubes. DNA was quantified with a Qubit 4 fluorometer (Invitrogen).
DNA samples (≥200 ng) were sequenced by Novogene Corporation, Inc. (Sacramento, CA). Amplicon sequencing targeted the V3-V4 regions of the 16S rRNA gene (primers 341F–CCTAYGGGRBGCASCAG, 806R–GGACTACNNGGGTATCTAAT; Caporaso et al., 2011; Klindworth et al., 2012) and the V4 region of the 18S rRNA gene (primers 528F–GCGGTAATTCCAGCTCCAA, 706R–AATCCRAGAATTTCACCTCT; Xu et al., 2020) using the NovaSeq 6000 platform (Illumina) with 250-bp paired-end reads at 100,000 raw reads per sample. Shotgun metagenomic sequencing was also performed by Novogene on the NovaSeq 6000 platform with 150-bp paired-end reads at ≥20 million raw read pairs per sample.
Initial amplicon read filtering was performed using a Novogene in-house pipeline. Sequencing adapters and barcodes were trimmed for all samples, and paired-end reads were merged using FLASH (ver. 1.2.11; Magoč and Salzberg, 2011) and quality filtered using fastp (ver. 0.23.1; Bokulich et al., 2013). rRNA gene amplicon sequence reads were further processed using the R (ver. 4.3.2) package DADA2 (ver. 1.29.0; Callahan et al., 2016). Reads were filtered to remove all sequences containing Ns and with expected errors >2. Forward and reverse reads were paired to generate amplicon sequence variants (ASVs) and chimeric sequences were removed. Taxonomic identities were assigned to ASVs according to the Silva database (ver. 132; Quast et al., 2013), and analyses were performed using the R package phyloseq (ver. 1.44.0; McMurdie and Holmes, 2013). Abundances of microbial families present as ≥1% of reads at each site were plotted as heat maps using the R package ampvis2 (Albertsen et al., 2015). Alpha diversity was calculated as Hill numbers using the “renyi” function in the R package vegan (ver. 2.6–4; Roswell et al., 2021; Oksanen et al., 2023). Using the R package ggdendro (ver. 0.1.20; de Vries et al., 2016), hierarchical clustering of sampling sites was performed and plotted as dendrograms. Relative abundances were plotted using the R packages phyloseq and ggplot2 (Wickham, 2016).
ASVs were compared to sequences from Mt. Erebus surficial geothermal soils amplicon libraries (Noell et al., 2022) and fumarole cave clone libraries (Tebo et al., 2015) using BLAST (ver. 2.15.0; Altschul et al., 1990). Sequences were considered to belong to the same species if sequences had greater than 97% similarity (Stackebrandt and Goebel, 1994). Before comparison, raw reads from Noell et al. (2022) were assigned taxonomically using the above DADA2 pipeline, following additional trimming and error processing recommendations for Ion Torrent reads (Callahan et al., 2016).
Shotgun metagenomic reads were quality filtered by a Novogene in-house pipeline; raw reads containing adapter sequences, with >10% Ns, and/or more than 50% of bases with quality scores ≤5 were removed before sequence return. Metagenomic reads were further processed using the Kbase online platform (Arkin et al., 2018). Read quality was assessed with FastQC (ver. 0.12.1; http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Reads were assembled into contigs using metaSPAdes (ver. 3.15.3; Nurk et al., 2017) and putative population genomes (bins) were generated using MaxBin2 (ver. 2.2.4; Wu et al., 2016). Bins were quality-checked using CheckM (ver. 1.0.18; Parks et al., 2015) and Bowtie2 (ver. 2.3.2; Parks et al., 2015). Metagenomic reads were taxonomically classified with Kaiju (ver. 1.9.0; Menzel et al., 2016), and classifications of bins were assigned by GTDB-Tk (ver. 2.3.2; Chaumeil et al., 2022). Both the assembled metagenome and high-quality bins were annotated using Prokka (ver. 1.14.6; Seemann, 2014), in addition to KEGG annotation assigned via GhostKOALA (ver. 2.0; Kanehisa and Goto, 2000; Kanehisa et al., 2016).
The physical exploration and survey conducted via LiDAR (Light Detection and Ranging), tacheometric measurements and subsequent detailed sketches (Figure 1A) revealed a single system of subglacial passages connected via two entrances and a depth range of 59 m from the top entrance to the lowest measured point. Two distinct fumarolic areas characterized by visible sulfur deposition (Figure 3) or visible degassing (Figure 2D) and multiple areas of heated bedrock and sediment were located, along with a subglacial stream and pooled standing water. Fumarolic areas and resulting dome shaped morphologies were connected by a lateral conduit at the rock-ice margin in the lowest part of the cave. In the upper level of the cave, the lateral conduit was connected to the surface via an inclined sloping chimney composed of ice.
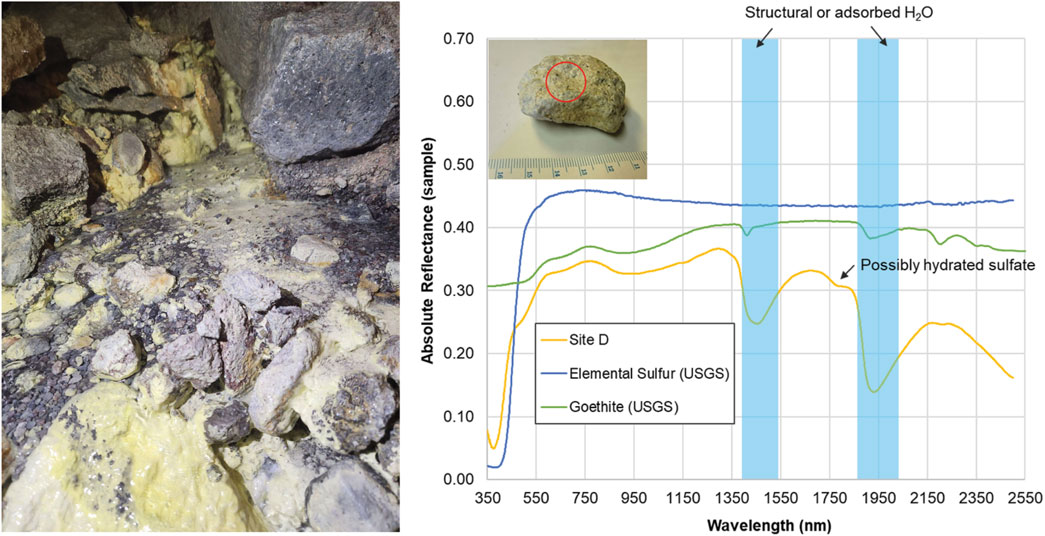
Figure 3. Spectral analyses of sulfur deposits collected within the glacier cave at site D. (Left) Photo of Site D at time of sample collection. (Right) VNIR/SWIR absolute reflectance of a rock sample collected from Site D. Image (inset, top left) shows the sample with a circle indicating the approximate placement of the TerraSpec Halo collection window (1.3 cm diameter). The steep absorption edge at 450 nm is characteristic of elemental sulfur. Other absorptions are consistent with goethite, an iron oxyhydroxide formed through hydrothermal alteration of iron-rich minerals, and structural or adsorbed H2O (blue). Elemental sulfur (USGS s07_ASD record 10973) and goethite (USGS splib07a, record 4,390) reference spectra are shifted and scaled for clarity.
Fumarole temperatures within the glaciovolcanic cave measured by thermal camera ranged from 28°C to 90°C with the maximum temperature of >90°C measured with the Omega thermometer inserted into the ground (Table 2; Supplementary Figure S1). Surrounding cave ice walls ranged from −3 to +2°C. Ambient air temperatures measured with the portable gas detectors were 10°C–17.3°C near the fumarole. Gas concentrations close to the fumaroles reached maxima of 5200 ppm CO2, >200 ppm H2S, >100 ppm SO2, 230 ppm CO, and 0% CH4. The O2 minimum was 20.4% at site B.
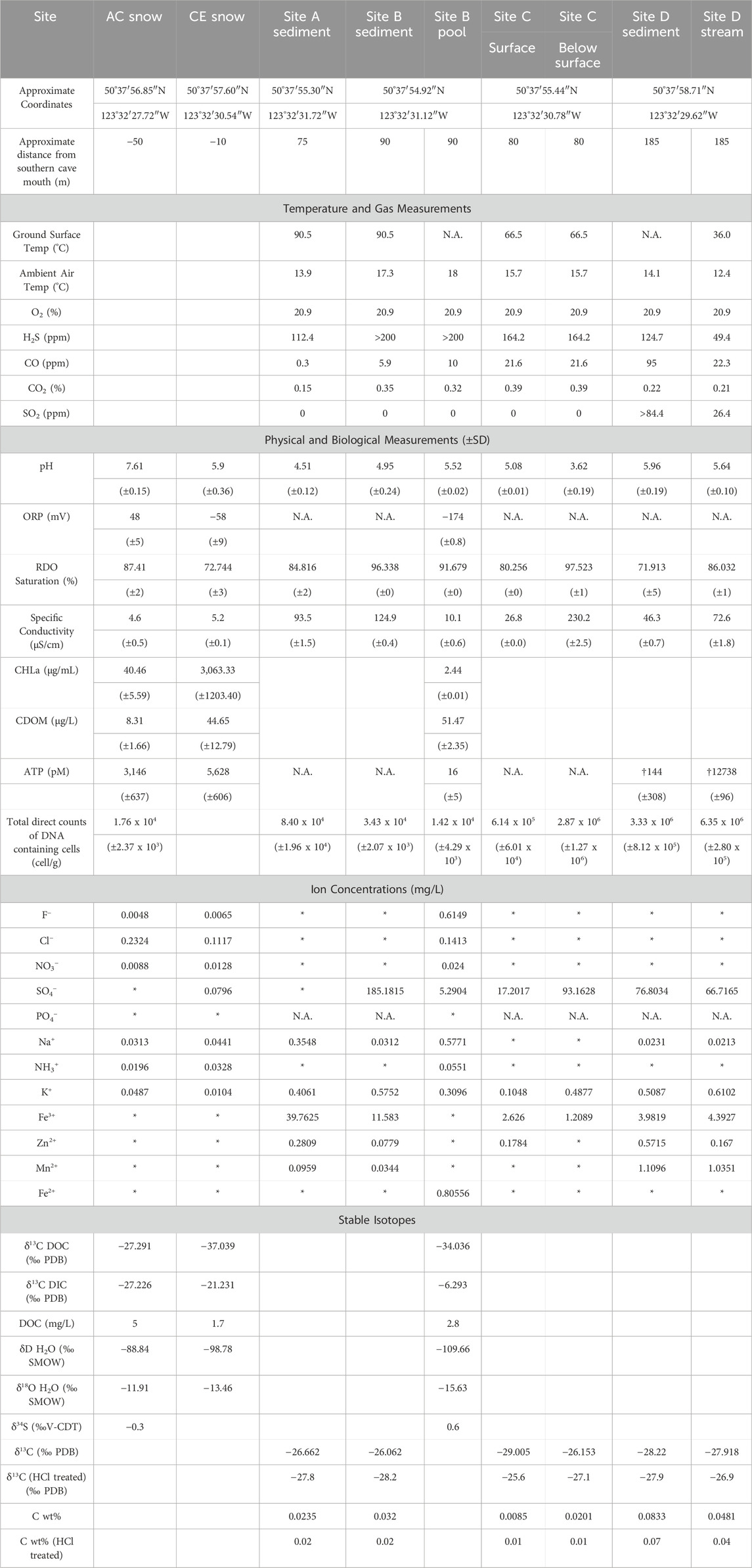
Table 2. Summary of geochemical and biological findings in collected Mt. Meager materials. N.A = not assessed. * = below detection. † = water collected with sediments. Br−, Li+, Ni2+, Co2+, Cu+, and NO2− were below detection in all samples. PDB = Pee Dee Belemnite Standard; SMOW = Standard Mean Ocean Water; V-CDT = Vienna- Cañon Diablo Troilite.
Snow from site AC had a pH of 7.61, while the snow from site CE, where plumes of vapor were visible (Figure 2C), had a pH of 5.90, which was closer to the pH of water collected from the pool at site B (pH = 5.52; Table 2). Oxidation-reduction potential (ORP) measurements in snowmelt and water ranged from −174 mV (site B pool) to 48 mV (AC snow). Specific conductivity for AC snow, CE snow, and the site B pool was 4.6 μS/cm, 5.2 μS/cm, and 10.1 μS/cm, respectively. Sediment pH ranged from 3.62 (site C below surface) to 5.96 (site D), while pH of sediments collected in the upper cave (sites A, B) was between 4.51 and 4.95 (Table 2). Specific conductivity of sediments was variable, with the highest and lowest values also recorded at site C (surface 27 μS/cm; below surface 230 μS/cm).
Chlorophyll a (CHLa) measurements (Table 2) were almost two orders of magnitude greater in CE snow, which was green/yellow in appearance (3,060.33 μg/mL) compared to the visibly red snow collected from AC (40.46 μg/mL; Figure 2B). CHLa concentrations from site B pool were within detection limits (2.44 μg/mL). Colored dissolved organic matter (CDOM) was highest in the site B pool (51.47 μg/L), compared to CE snow (44.65 μg/L) and AC snow (8.34 μg/L). Concentrations of total ATP in snow samples averaged 3,146 pM in AC snow and 5,628 pM in CE snow, while only 16 pM ATP was detected in site B pool water. ATP was detectable within a small amount of water contained in site D sediments which averaged 144 pM ATP. The site D stream had the highest ATP concentration of all samples with an average of 12739 pM. ATP was not detectable in any of the blanks.
Visible to short wavelength infrared (VNIR/SWIR) spectral analyses of samples from the cave entrance and surrounding area indicated the presence of minerals associated with volcanic fumaroles and hydrothermal alteration (illite, epidote, goethite, heulandite; Supplementary Figures S2-S4). Elemental (native) sulfur, characterized by a steep absorption edge at 450 nm, and ferric oxyhydroxides (features at ∼670 nm and 900 nm) were observed on the exteriors of samples collected throughout the fumarole (Supplementary Figures S5-S6). Features consistent with goethite were observed in rocks collected near the heated Site D stream, indicating hydrothermal alteration (Figure 3).
Snow samples contained carotenoid (550 nm) and chlorophyll (680 nm) absorption features (Figure 4) associated with red microalgae detected in surface snow deposits at depths up to 14 cm. The 438 nm absorption feature identified at depths up to 45 cm could be due to octahedrally coordinated Fe3+ as in jarosite (KFe3(SO4)2(OH)6), although no other longer-wavelength features of this mineral were observed. Spectral absorption features were observed at 550 nm (possible carotenoid) that disappeared below 14 cm depth, but a 438 nm feature persisted. Neither were present in pure snow standard (Painter et al., 2001).
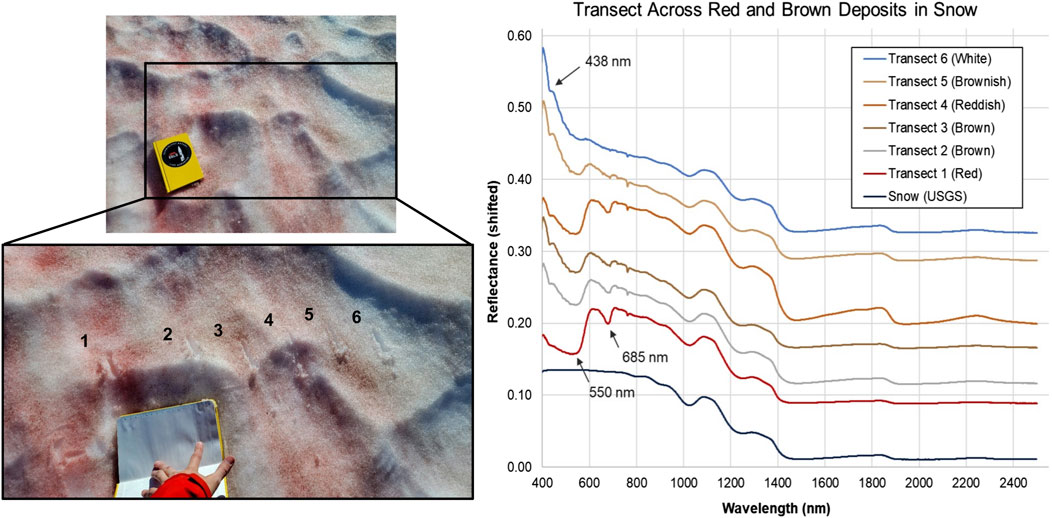
Figure 4. Spectral analyses of visible snow algae communities in the Mt. Meager snowpack. (Left) Photos of transect across snow located at Above Cave site. Spacings between transect points range from 7 to 15 cm. (Right) VNIR/SWIR reflectance (shifted for clarity) of transect points. Absorptions at 550 nm and 685 nm are consistent with carotenoid and chlorophyll, respectively. The 438 nm absorption feature could be due to octahedrally coordinated Fe3+ as in jarosite, although no other longer-wavelength features of this mineral are observed. Ice absorptions occur at approximately 810, 900, 1,030, 1,260, 1,500 and 2000 nm (Painter et al., 2001). USGS reference spectrum for melting snow (s07_ASD record 14508) also shifted and scaled for clarity.
Most major anions (F−, Cl−, NO3−, and NH3+) were below detection limits (<0.1 mg/L) in the sampled sediments except for SO42-, which ranged from <0.1 mg/L at site A to 185.2 mg/L at site B (Table 2). K+ was present in all sediment samples (0.11–0.61 mg/L), and Na+ was present in most sediments (0.02–0.88 mg/L) except site C (Table 2). Iron was present as Fe3+ in all sediment samples (1.21–39.76 mg/L), while Fe2+ was not detected in any sediment samples. Most sediments also contained Zn2+ and Mn2+ at concentrations of 0.08–0.57 and 0.03–1.11 mg/L, respectively (Table 2). F− (0.005–0.615 mg/L), Cl− (0.112–0.232 mg/L), NO3− (0.009–0.024 mg/L), and NH3+ (0.020–0.055 mg/L) were present in snowmelt and water samples from sites AC, CE, and site B pool; with the highest Cl− at site AC and highest F−, NO3−, and NH3+ in the site B pool (Table 2). Fe, Zn, and Mn were below detection in snowmelt and water samples except for Fe2+ in the site B pool (0.806 mg/L; Table 2). NO2− and Br− were below detection in all samples.
Bulk carbon content of sediments was low, ranging from 0.01% to 0.08% by weight (Table 2). δ13C of organic matter in sediments ranged from −28.2‰ to −25.6‰ with lowest and highest values at site B and site C (surface), respectively; organic carbon content ranged from 0.01% to 0.07% by weight (Table 2) with lowest and highest values at site C and site D. δ13C of DOC in snowmelt ranged from −37.0‰ to −27.3‰ with lowest and highest values at site CE and site AC, respectively (Table 2). δ13C of DIC was more variable, ranging from −27.2‰ to −6.3‰ with lowest and highest values at site AC and the site B pool (Table 2). DOC concentration in snowmelt ranged from 1.7 to 5.0 mg/L, and was highest at site AC. δD and δ18O of snowmelt ranged from −109.7‰ to −88.8‰ and −15.6‰ to −11.9‰, respectively, with lowest and highest values at the site B pool and site AC (Table 2). δ34S of H2S from site AC and the site B pool were −0.3‰ and +0.6‰, respectively (Table 2).
Direct counts of cells (Table 2) in snow and sediment samples ranged from 3.43 ± 0.12 x 10⁴ to 6.35 ± 0.16 x 10⁶ cells/g of sediment and 1.42 ± 0.42 x 10⁴ to 1.76 ± 0.23 x 10⁴ cells/mL of snowmelt water. Calculated biovolume ranged between 0.47 μm³ ± 0.01 to 1.065 μm³ ± 0.12. Diverse morphologies were observed consisting of primarily very small bacilli and cocci as well as a few endospores. Site C sediments varied by an order of magnitude between the surficial (top) sample and sediments collected at approximately 5 cm depth (6.14 ± 0.35 x 10⁵ cells/g and 2.87 ± 0.73 x 10⁶ cells/g, respectively). Additionally, morphologies varied between two Site C collections with the sample from ∼5 cm hosting larger cells, the majority of which were bacilli, and a larger cellular biovolume (1.21 μm³ ± 0.04) compared to 0.838 μm³ ± 0.04 in the surficial sample. Cells were the most abundant in site D stream sediments (6.35 ± 0.16 x 10⁶ cells/g) with the most diverse morphologies including large cocci and bacilli, some of which were segmented, as well as a small number of filaments and possible vibrio. Algal cells were not observed microscopically for this study.
Concentrations of DNA used for analyses ranged from 0.76 to 1.89 ng/μL. High-quality, non-chimeric rRNA gene amplicon sequences across five sampling sites totaled 428,999 reads for 16S and 583,639 reads for 18S rRNA gene analyses (average quality score 35.5 before filtering). Sequences sorted into 5,716 non-chloroplast 16S rRNA gene and 588 18S rRNA gene. In the prokaryotic communities, sequences matching Firmicutes were the most abundant at the phylum level across all samples (38.0%), followed by Proteobacteria (32.2%) and Bacteroidetes (19.3%). The most abundant taxa varied by site; at sites A and B, Firmicutes were the most abundant (62.4% and 98.7%, respectively) while sequences matching Proteobacteria were most abundant at sites D (48.7%) and CE (63.2%), Bacteroidetes were most abundant at site AC (49.9%). The most abundant prokaryotic family was different at each site; Bacillaceae at site A (44.8%), Planococcaceae at site B (82.2%), Acidithiobacillaceae at site D (36.1%), Chitinophagaceae at site AC (26.4%), and Burkholderiaceae at site CE (24.2%; Figure 5).
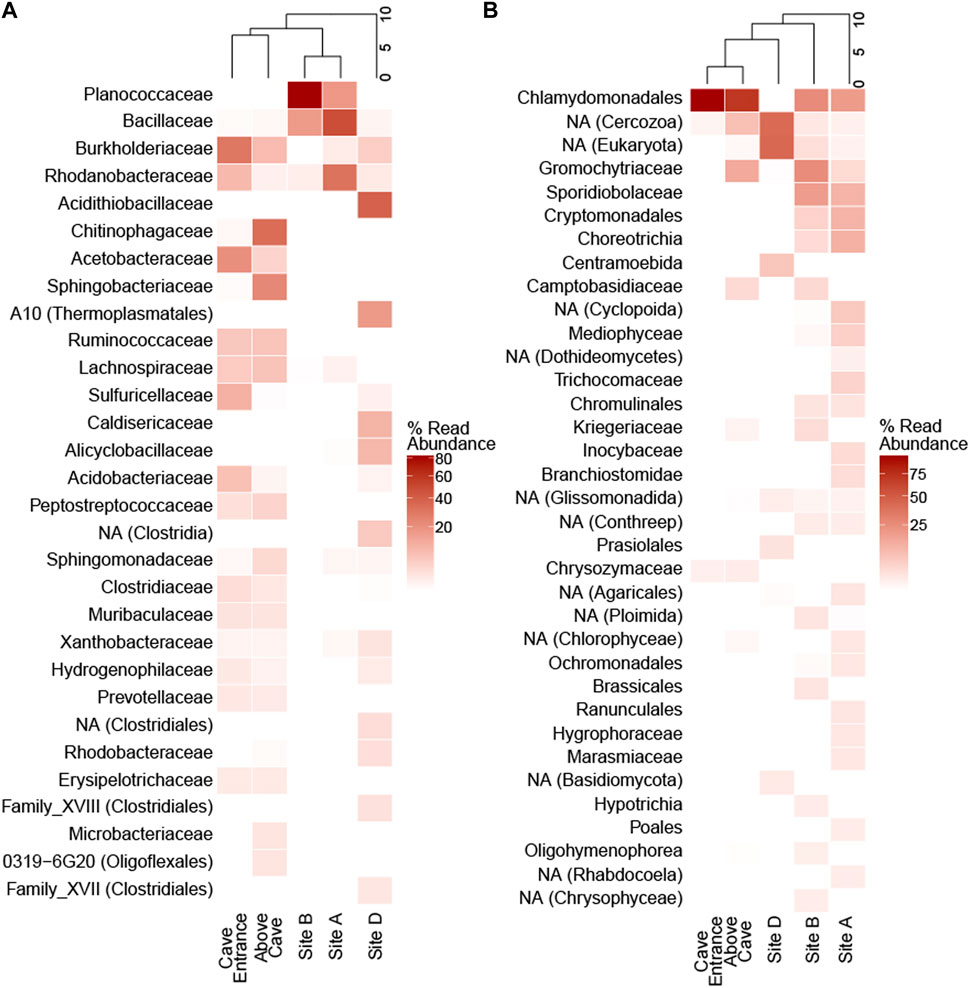
Figure 5. Family-level community diversity in ice cave sediments and surface snow. Heatmaps show percent abundance of amplicon sequence variants that are present as >1% of reads in (A) 16S and (B) 18S rRNA gene amplicon libraries at the family level. Dendrograms show hierarchical clustering of sampling sites. NA = not assigned at family level; the lowest assigned level of taxonomic classification is given in parentheses when family is not assigned.
Microbial taxa with known chemosynthetic lifestyles (Wirsen and Jannasch, 1978; Kelly and Wood, 2014; Kuever, 2014; Oren, 2014; Orlygsson and Kristjansson, 2014; Prosser et al., 2014; Pujalte et al., 2014; Stackebrandt, 2014; Watanabe et al., 2015) were present at ≥1% relative abundance in amplicon libraries from all five sampling sites. Site D contained a high abundance of Acidithiobacillaceae (36.1% of reads) and Sulfuricellaceae was abundant in site CE (8.7% of reads). Seven families identified as chemosynthetic belonged to Proteobacteria; Thermoanaerobacteraceae which falls within the Firmicutes and Thiovulaceae a member of the Epsilonbacteriota were also detected (Supplementary Figures S7).
Across all samples, the most abundant eukaryotic phylum was Chlorophyta (42.0% of 18S amplicon reads) comprising the majority of eukaryotic reads at sites AC (70.0%) and CE (98.5%; Supplementary Figures S8). Chlorophyta were present, but less abundant at sites A (19.7%), B (25.1%), and D (3.8%); mixtures of fungi and other eukaryotes made up the majority of the eukaryotic communities at these three sites. Chlamydomonadales was the most abundant eukaryotic family at sites A (17.1%), B (25.1%), AC (69.6%), and CE (98.4%); Centramoebida was the most abundant eukaryotic ASV assigned at the family level at site D (6.7%; Figure 5). Snow algae of the genera Chloromonas and Chlamydomonas were the most abundant eukaryotes in surface snow (Supplementary Figures S8). Chlamydomonas was more abundant than Chloromonas at site AC, where red-pigmented snow was dense.
Prokaryotic diversity indices were one to two orders of magnitude higher than eukaryotic diversity indices at each site (Supplementary Table S1). Based on the exponential Shannon entropy (Hill q = 1) and the inverse Simpson (Hill q = 2) indices, site AC had the highest diversity of prokaryotes (Hill q = 1: 693.1; Hill q = 2: 435.2). Eukaryotic diversity was highest at site B (Hill q = 1: 83.0; Hill q = 2: 50.6), while site CE showed the lowest diversity for both prokaryotes and eukaryotes (Supplementary Table S1).
Metagenomic sequencing of sediments from site D yielded 35,755,611 total sequences with average quality scores of 36.1 for forward reads (97.9% reads with quality score ≥30) and 35.9 for reverse reads (97.9% reads with quality score ≥30). Both forward and reverse reads had average GC content of 58.4%. The assembled metagenome consisted of 15,838 contigs (N50 = 18,509 and L50 = 1,259). Average miscalled bases per 100 kbp was 72.52 in the final assembly. Annotation by Prokka identified features with 132,212 putative coding sequences. Of these coding sequences, 49.9% were annotated by the KEGG database.
Prior to binning, 47.4% of metagenomic reads were assigned taxonomic classifications (Supplementary Figures S9). The most abundant phylum in the assigned reads was Proteobacteria (81.3%), followed by Firmicutes (7.3%) and Candidatus Thermoplasmatota (2.2%). Acidithiobacillaceae was the most abundant family in the assigned reads (24.8%), which agree with amplicon sequencing results (Figure 5A). Several complete chemosynthetic carbon fixation pathways (Hügler and Sievert, 2011) were annotated including the Calvin-Benson-Bassham (CBB) cycle, Wood-Ljungdahl (WL), and Arnon-Buchanan (AB) pathways (Supplementary Table S4). Genes encoding the Crassulacean acid metabolism (CAM) pathway are also present; however, because CAM includes light-dependent reactions and overlying ice excludes sunlight at site D, these CAM-related genes are more likely involved in anaplerotic reactions or alternative processes of carbohydrate metabolism (e.g., Smith et al., 2021). The assembled metagenome contained complete pathways for energy-conserving metabolisms including dissimilatory sulfate reduction (DSR), sulfur oxidation via the Sox system, and dissimilatory nitrate reduction (DNR).
Binning of reads from the site D metagenome yielded 40 bins with 14 considered high-quality (≥90% completion and <5% contamination; Supplementary Table S3); only high-quality bins are discussed further. Of these 14 high-quality bins, nine were assigned to taxa present in the amplicon sequences from site D including Firmicutes, Thermoplasmata, Anaerolineae, Rhodobacteraceae, Acidithiobacillus, Dissulfurimicrobium, Sulfuricurvum, Thermithiobacillus, and Xanthobacter. The remaining bins, assigned to Acidobacteriota, Armatimonadota, Symbiobacteriia, Limnocylindrales, and Rectinemataceae, were not detected by amplicon sequencing, likely reflecting differences in taxonomic assignment between the SILVA and GTDB databases (as described by Robeson et al., 2021). Binned metagenomic reads averaged 293 coding sequences with 54% annotation and 44 complete pathways identified per bin. The complete WL pathway was found in Bin011, which was taxonomically assigned as a Firmicutes sp.; the complete AB pathway was found in Bin018, which was assigned as a Sulfuricurvum sp.; Bin005 was assigned as a Rhodobacteraceae sp. and contained complete pathways for CBB and Sox; Bin006 was assigned as a Xanthobacter sp. and contained complete pathways for CAM, CBB, and Sox; Bin036 was assigned as a Rectinemataceae sp. and contained complete CAM and DSR pathways (Supplementary Table S4).
The MMVC cave system is one of only six described glaciovolcanic cave areas on Earth (Sobolewski et al., 2022) and its sulfidic gas emissions are unique among these. Other examples reviewed in Stenner et al. (2022) included reference to historical observations of H2S at ice caves on Mt. Hood and Mt. Baker, however these caves have not been subject to further detailed study. Studies of fumarole-associated ice cave sediments at Mt. Erebus are also relevant, though not only were concentrations of H2S below detection limits, but sediment temperatures at Mt. Erebus (0.1–18.5°C; Tebo et al., 2015) were much cooler than temperatures of sampled MMVC sediments (36–90.5°C). Fumarolic gas compositions at other glaciovolcanic systems including Mt. Rainier, Mt. St. Helens (both in Washington, USA) and Mt. Erebus are dominated by water vapor, CO2, and H2 (Gerlach and Casadevall, 1986; Zimbelman et al., 2000; Oppenheimer and Kyle, 2008); H2S was undetected or very low in these caves, with the highest reported concentration (2.16 mol %) present at Mt. St. Helens in 1980–81 (Gerlach and Casadevall, 1986), although sulfate mineral deposits around a dormant fumarole vent at Mt. Rainier suggests the presence of H2S or SO2 in fumarole gases in the past (Zimbelman et al., 2000). Thus, while there have been several studies on the biogeochemistry and microbial ecology of fumarole-associated subglacial cave environments (Anitori et al., 2011; Connell and Staudigel, 2013; Tebo et al., 2015; Fraser et al., 2018), none of these systems share the sulfidic nature of the MMVC caves. Thus, the MMVC represents a valuable addition to the sparse global inventory of glaciovolcanic cave systems, as well as a novel type of subglacial ecosystem whose biogeochemistry has not been previously described. The true diversity in terms of physical setting, geochemistry and biome diversity remain poorly constrained for glacier ecosystems, and subglacial environments in particular (Stibal et al., 2020). In general, direct analyses of subglacial materials are limited with studies of mountain glacier subglacial environments primarily focused on the water that emanates from beneath glaciers into proglacial outflow streams (Anesio et al., 2017; Hotaling et al., 2017). Glacier beds are difficult to access due to logistical challenges, and a limited inventory of subglacial samples have been collected to date; samples from the beds of temperate glaciers are particularly lacking. Exceptions include Hamilton et al. (2013), who collected subglacial sediments through an ice cave access at the terminus of the Robertson Glacier in the Canadian Rockies, Alberta, that was carved by subglacial meltwater, and an acidic, sulfidic volcanic lake formed below the Vatnajökull ice cap in Iceland was sampled using a hot water drill (Gaidos et al., 2004). The MMVC provides a rare subglacial access point for direct observations, measurements and sample collection. It is worth noting that despite our ability to access the MMVC caves directly, sample volumes and replicates remained limited because time in the caves was restricted due to hazardous conditions such as darkness, technical glaciated terrain, icefall, and dangerously toxic concentrations of volcanic gases.
Our measurements of MMVC indicate that volcanic processes influence snow, water, and sediment geochemistry in and around the cave complex. Chloride was the most abundant ion in our snow samples, suggesting either condensation of volcanic gas or meteoric precipitation. Typically, atmospheric pollution contributes to increased nitrate in meteoric precipitation (Galloway et al., 2004). However, the MMVC is relatively remote from large urban or industrial centers. Winds blow from west to east along the MMVC, and the closest large urban area (Vancouver) is ∼150 km to the southeast (Warwick et al., 2022). Since nitrate was low or below detection in our samples, we infer that the chloride in snow and water samples mainly originated from fumarolic exhalations. Water inside the cave likely originated from snowmelt, as suggested by negative δD and δ18O values typical in meteoric precipitation, and most major dissolved ions being below detection limits. Elevated SO42- concentrations in site B sediments (Table 2) were likely due to oxidation of fumarolic H2S. The measured δ34S of H2S was ∼0‰ in the snowmelt and site B pool water (Table 2) suggesting a deep, magmatic origin for the sulfide (e.g., Oppenheimer et al., 2011) consistent with fumarolic exhalation. VNIR spectra taken from yellow crystal-like and amorphous deposits observed at multiple sites within the cave were consistent with elemental sulfur (Figure 3; Supplementary Figures S6), suggesting deposition of elemental sulfur from the sulfur-rich fumarole gases.
Bulk δ13C of organic carbon in MMVC cave sediments is typical for biomass on Earth (e.g., Dijkstra et al., 2006). Bulk sediment δ13C (average −27‰) was higher than for the bulk DOC fraction (that passed through a grade GF/F glass fiber filter) in the Site B pool (−34‰), suggesting different organic endmembers are likely present in this setting. Given the high elevation and snow cover in the MMVC system, biological inputs through surface soil processes are limited, thus negative δ13C DOC values may represent in-situ microbial processes. Metabolic processes likely also affect the δ13C of inorganic carbon (DIC) in this system. In snow at site AC, the δ13C value of DIC was depleted (−27‰), which may result from decomposition of organic-rich material transported by wind, as this value was similar to DIC from organic matter decomposition in soils of forested areas (Wynn et al., 2006; Spencer et al., 2009). Alternatively, the δ13C-depleted value of DIC may indicate remineralization, decomposition of organic matter or methane oxidation, as DIC produced from methane oxidation is depleted in 13C relative to the source methane (Barker and Fritz, 1981). Although methane was not detected in our gas measurements, it is possible that any methane present is rapidly oxidized by chemolithotrophic microbes. Methane oxidizers were detected at low (<0.2%) relative abundance in the amplicon libraries from sites AC and CE, but not in the cave sediments. In site B pool, the δ13C value of DIC was significantly higher (−6‰) suggesting dissolution of atmospheric CO2 in the standing water (Graven et al., 2020). However, several lines of evidence support the presence of an active microbial community in the MMVC cave sediments including direct counts of microbial cells, measurable ATP concentrations, microbial community composition reflective of both heterotrophic and chemosynthetic taxa, and the presence of genes encoding pathways for inorganic carbon fixation.
ATP is used by all cells to transport and convert energy for metabolism, and has been used to assess viability in diverse environmental samples (Hammes et al., 2010; Tan et al., 2022). ATP was detectable in all snowmelt and water samples (Table 2) indicating that the MMVC system hosts a viable microbial community. Snow samples outside the cave contained more ATP (3146–5628 pM) than melt water inside at the site B pool (16 pM). However, the highest ATP concentration measured in this study was in water collected from inside the cave at the site D stream (12738 pM), while water collected with site D sediments was significantly lower (144 pM ATP) despite similar concentrations of cells (6.35 x 106 and 3.33 x 106 cells/g, respectively) and carbon (0.08% and 0.05% by weight, respectively; Table 2). While the cause of variation in ATP at site D is unclear, we hypothesize the stream provides more amenable conditions for metabolically active cells than the surrounding sediments. While direct temperature measurements of site D sediments were not collected, the thermal camera recorded temperatures of 36°C for the site D stream and concentrations of CO and H2S were much higher where sediments were collected compared to the stream (Table 2).
Snow algae, including members of the family Chlamydomonadales, are known to produce pigments such as the carotenoid astaxanthin as protection against UV light-induced damage (Gorton and Vogelmann, 2003; Hoham and Remias, 2020) which results in bright red swaths of snow globally (Lutz et al., 2016; Hoham and Remias, 2020). Densely red-pigmented snow was observed in the snow surrounding the cave entrance at the MMVC in September 2022 (Figure 2B) and corresponded with high CHLa concentrations (40.5–3063 μg/mL; Table 2). In addition, VNIR spectra of red-pigmented snow taken in-situ showed broad absorption features typical of astaxanthin (Buchwald and Jencks, 1968), which were absent in white snow (Figure 4). Site CE, which is directly influenced by fumarole exhalation and had a lower pH (5.9) than our more distant snow sample at site AC (7.6), also had a significantly higher CHLa concentration (CE: 3,063 μg/mL; AC: 40.5 μg/mL). Stressed algal cells producing copious amounts of astaxanthin have shown decreased photosynthetic efficiency (Zhang et al., 2016); however, lower pH can reduce stress (Hoham and Mohn, 1985) leading to higher photosynthetic activity. Thus, interplay between pH and UV stress at the MMVC may affect primary production and biogeochemical cycles in-situ and in downstream environments distinct from glacier terrain and without fumarolic inputs.
Microbial community assessments of snowpack associated with peaks along the Cascade Volcanic Arc, but not associated with fumaroles, at Mt. Adams (Washington, USA), Mt. Hood, and North Sister (both Oregon, USA; Hamilton and Havig, 2017; Hamilton and Havig, 2020; Havig and Hamilton, 2019) offer reference for assessing the potential influence of fumarolic exhalations on MMVC surface snow ecology. Additionally, snowpack studies at Mt. Baker (Washington, USA), a glacier-clad volcanic peak in the Cascade Volcanic Arc with fumarolic activity (Werner et al., 2009), were conducted outside of the influence of any fumaroles (Schuler and Mikucki, 2023). CHLa concentrations in the MMVC snowpack (40.46–3063.3 μg/mL; Table 2) were several orders of magnitude higher than those reported for snowpack at Mt. Baker (0.01–1.9 μg/mL; Schuler and Mikucki, 2023). In contrast, CDOM at the MMVC (8.3–51.5 μg/L; Table 2) was much lower than at Mt. Baker (2.1 x 104–1.9 x 106 μg/L; Schuler and Mikucki, 2023). Chlamydomonas and Chloromonas were the most abundant eukaryotic organisms in snowpack associated with Mt. Adams, Mt. Hood, North Sister, and Mt. Baker, (Hamilton and Havig, 2017; 2020; Havig and Hamilton, 2019; Schuler and Mikucki, 2023), which is similar to the snowpack communities at the MMVC. Direct counts of algal cells and measurements of in-situ primary production are necessary to accurately draw comparisons between snowpack microbial communities at the MMVC and other sites.
Bacterial community structure in the MMVC snowpack differs from that of other sites in the Cascade Volcanic Arc. At Mt. Adams and Mt. Hood, Sphingobacteriaceae comprised nearly 50% of the prokaryotic communities at sites where Chlamydomonadaceae dominated the eukaryotic communities (Hamilton and Havig, 2020). At Mt. Baker, Chloromonas-rich snowpack was associated with an abundance of Chitinophagaceae and Comamonadaceae, while other sites had abundant Sphingobacteriacea and Oxalobacteraceae (Schuler and Mikucki, 2023). At MMVC, Chitinophagaceae (26.4% of 16S reads) and Sphingobacteriaceae (18.1% of 16S reads) were abundant at site AC where Chlamydomonas algae dominate the eukaryotic community. However, at site CE, where fumarole exhalations alter snow chemistry and Chloromonas algae are abundant (98% of the eukaryotic community), reads of Chitinophagaceae and Sphingobacteriaceae were relatively low (0.29% and 0.17% of prokaryotes, respectively). Firmicutes, major constituents of the bacterial communities at MMVC, were not detected at Mt. Adams, Mt. Hood, or North Sister (Hamilton and Havig, 2017; Havig and Hamilton, 2019).
The MMVC system is largely distinct when compared to other glaciovolcanic and subglacial features. The MMVC prokaryotic communities showed higher abundance of several phyla, including Bacteroidetes, Firmicutes, and Proteobacteria, compared to Mt. Erebus soils (Figure 6; Noell et al., 2022). Caldiserica and Euryarchaeota both make up a significant portion of reads in MMVC site D sediments though, were not detected at Mt. Erebus. The phylum Caldiserica contains a single cultured species, C. exile, originally isolated from a hot spring in Japan (Mori et al., 2008; 2009). However, members of the proposed Caldiserica superphylum have been detected globally in cryosphere lakes and permafrost (Martinez et al., 2019) including in the subglacial lakes beneath the Vatnajökull ice cap (Marteinsson et al., 2013). C. exile strain AZM16c01 has been shown in culture to reduce both elemental sulfur and sulfite, though not sulfate. Clone libraries from two caves on Mt. Erebus, Harry’s Dream and Warren Cave, had substantial populations of Chloroflexi and Cyanobacteria, neither of which were present at greater than 1% in MMVC amplicon libraries. The Hubert’s Nightmare cave on Mt. Erebus contained a greater percentage of phyla present in MMVC sediments, though Firmicutes were not detected. When compared at the species level, several ASVs from site D had high similarity (>97%) to Thiobacillus clone HD11 and Sulfuriferula clone HD13, both from Harry’s Dream. Many Thiobacillus spp. are sulfur oxidizing chemolithoautotrophs (Boden et al., 2012), and have been shown to contribute to primary production in sulfidic cave systems, such as Movile Cave, Romania (Chen et al., 2009). Similarly, Sulfuriferula spp. are known sulfur oxidizers largely isolated from freshwater environments (Watanabe et al., 2016).
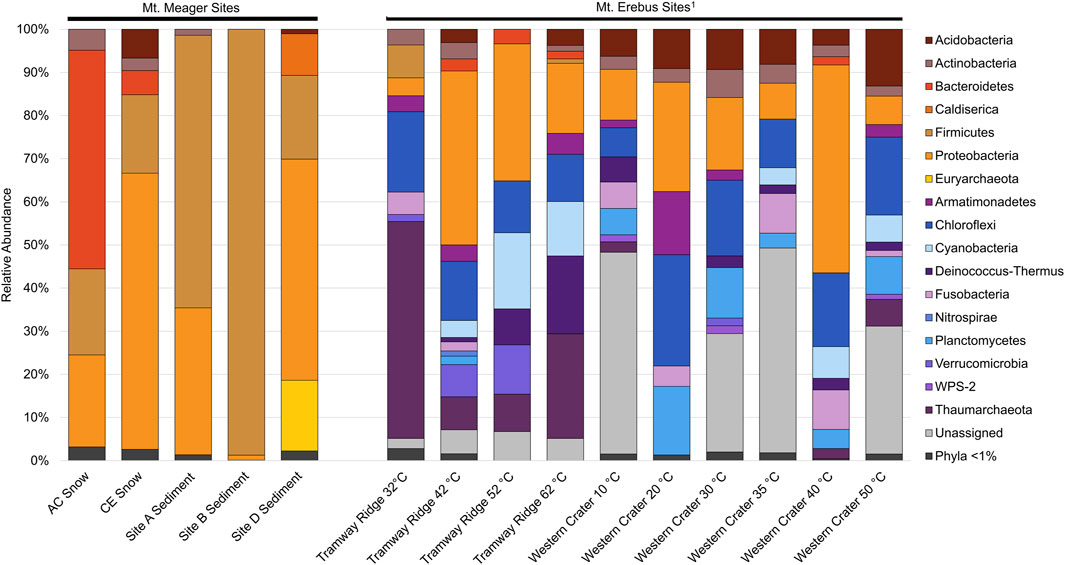
Figure 6. Phylum level comparison between 16S rRNA gene amplicon sequence libraries from Mt. Meager and Mt. Erebus, Antarctica. Phyla in warm colors are present within at least 1 Mt. Meager site at ≥ 1% abundance. Mt. Erebus sequences from Noell et al. (2022).
Euryarchaeota ASVs were identified in all sediment samples except for site A, and site B had fewer than 40 reads assigned to archaea. Site D Euryarchaeota contained a total of 71 ASVs, all assigned to the uncultured Thermoplasmatales group A10, except for one ASV assigned to the phylum Crenarchaeota. Putative thermophilic members of the class Thermoplasmata were detected in cave sediments but not in snowpack. Thermoplasmatales group A10 has been detected in several acidic and/or geothermal environments, including hot springs and acid mine drainage (Merkel et al., 2017). In hot springs at the Uzon Caldera and near Mutnovsky Volcano (Kamchatka Krai, Russia), this group was present in high abundance (up to 52%) and were hypothesized to function as sulfur oxidizers (Merkel et al., 2017). Sediments collected at site D were mildly acidic (5.96), and the stream at the site was geothermally heated (36°C; Table 2). Both sulfide and sulfate were detectable at site D (gas and sediment measurements, respectively), and noticeable yellow deposits were documented near and around the stream. The presence of both Caldiserica and Thermoplasmatales group A10 suggest that both sulfur oxidation and reduction reactions may be occurring within the lower cave sediments. Sulfur-oxidizing taxa are not present at Mt. Erebus (Tebo et al., 2015), which is unsurprising as H2S has only been detected in trace amounts in Mt. Erebus plumes. Microbial communities in snow at the MMVC are largely distinct from the communities at Mt. Erebus, with five cloned sequences from Mt. Erebus (HD10, HD6, NH2, HN6, and WC4) making up a total of ∼0.1% of AC reads and ∼0.6% of CE reads.
Comparison to the subglacial volcanic Skaftá lake beneath the Vatnajökull ice cap, Iceland) yields few similarities. The bacterial community in the anoxic, sulfide-rich portion of the water column is dominated by members of the genus Acetobacterium (63.7%), Thermus (9.7%), and Paludibacter (8.5%; Gaidos et al., 2009). No MMVC amplicon sequences were assigned to Acetobacterium or Thermus, and only three reads in the MMVC sediments were assigned to Paludibacter. Sulfuricurvum, the fourth-most abundant genus detected in the Skaftá lake water (2.8%), was present in MMVC sediments, although at lower relative abundance (0.3%). In subglacial sediments collected from beneath the Robertson Glacier (Alberta, Canada), the bacterial community was dominated at the order level by Burkholderiales (Hamilton et al., 2013), which was not detected in any MMVC sediment samples. The orders Bacillales, Desulfuromonadales, and Rhodobacterales were shared between Robertson Glacier and MMVC sediments, albeit at very different relative abundances. For example, 61.7% of 16S rRNA gene reads in MMVC sediments were assigned to Bacillales, while the same order makes up <15% of reads in Robertson Glacier sediments (Hamilton et al., 2013). Archaeal communities between MMVC and Robertson Glacier sediments also differ, Methanomicrobiales and Methanosarcinales were abundant at Robertson Glacier (Hamilton et al., 2013), but absent in MMVC cave sediments. The only eukaryotic orders shared between MMVC and Robertson Glacier sediments were Hypocreales, Chlamydomonadales, and Chaetophorales, though Hypocreales and Chaetophorales were represented by only 54 and 26 reads, respectively, at the MMVC while Chlamydomonadales was far more abundant in MMVC sediments than in Robertson Glacier sediments (Hamilton et al., 2013). With the exception of these few similarities, the microbial communities in the MMVC sediments are distinct from those documented in other subglacial environments.
Overall, we detected microbial communities typical of high-temperature, acidic fumarolic systems with numerous sequences related to strains that utilize sulfur for chemosynthetic metabolism. All samples contained taxa capable of sulfur oxidation (e.g., Acidithiobacillus, Sulfuriferula, Thermithiobacillus, Thiomonas, Thiobacillus, etc.). While metabolic properties cannot be directly derived from 16S rRNA gene diversity, these data, in combination with metagenomic analyses and geochemistry, support a significant role of sulfur metabolism and chemosynthesis in the MMVC. Sulfur is required for cellular components, such as the amino acids cysteine and methionine; however, some microorganisms utilize sulfur compounds in dissimilatory, energy-yielding metabolic processes. For example, sulfur-oxidizing prokaryotes, which are metabolically and phylogenetically diverse (Friedrich et al., 2001; Friedrich et al., 2005), can mediate the fixation of inorganic C by coupling sulfur oxidation to the reduction of O2, NO3−, Mn3+/4+, and Fe3+ (Mattes et al., 2013). Sulfate-reducing prokaryotes respire organic material for energy using sulfate as an electron acceptor (Jorgensen, 1982; Jorgensen and Postgate, 1982). Previous studies of glaciovolcanic cave systems have implicated reduced inorganic molecules (e.g., H2, H2S) as important energy sources for chemosynthetic lifestyles of resident microbial communities (Tebo et al., 2015; Sobolewski et al., 2022). Given the high concentrations of H2S in the fumarole-associated gases at the MMVC, we expect sulfur oxidation to be a prominent form of energy metabolism in this system. The Sox system is a gene cluster encoding a number of enzymes involved in dissimilatory sulfur oxidation (Meyer et al., 2007) that can oxidize H2S to SO42- with elemental sulfur (S0) and/or SO32- as intermediates (Wu et al., 2021). The metagenome generated from site D sediments had 95 genes related to the Sox system, and contained the complete gene cluster (i.e., soxABCDXYZ). Additionally, the metagenome contains the complete dissimilatory sulfate reduction (Dsr) pathway, which reduces sulfate to sulfide and can act in reverse to oxidize sulfide (Neukirchen et al., 2023). Genes encoding adenosine-5′-phosphosulfate (APS) reductase (Apr) are also present in the metagenome; Apr reduces APS to sulfide and can operate in reverse to oxidize sulfite to APS and possibly APS to SO42- (Meyer and Kuever, 2007). Sulfur-oxidizing bacteria (e.g., Acidithiobacillaceae) were abundant in the site D 16S rRNA gene amplicon library and several bins were assigned to Acidithiobacillaceae, including Bin002 (Acidithiobacillus sp.) which contained 5 of the 7 Sox system genes (soxABXYZ). Bin005 (Rhodobacteraceae sp.) and Bin006 (Xanthobacter sp.) each contained the complete Sox system (Supplementary Table S4), with partial Sox systems noted in several other bins. The presence of sulfur oxidizers and multiple dissimilatory sulfur oxidation pathways supports the likelihood that MMVC sediment microbial communities utilize H2S from fumarolic gases as an energy source. Dissimilatory sulfur oxidation and reduction by microbes may cause measurable S isotope fractionation (Bradley et al., 2016; Pellerin et al., 2019). Our 34S of H2S data do not show clear evidence of biological influence; however, additional 34S measurements of sulfate from this system, including from the fumarole exhalations and sediments, are necessary in order to properly assess biological effects.
Carbon isotope composition of DIC and DOC in cave sediments indicate likely microbial transformations of the carbon pool, while measurable ATP indicates the cells in our samples are metabolically active. Metagenomic data from site D sediments further supports active microbial carbon transformations by revealing genes encoding complete heterotrophic and autotrophic pathways. Thus both heterotrophy and autotrophy appear to contribute to carbon cycling. In MMVC sediments, genes encoding photosystems I and II are absent, while the complete autotrophic Wood-Ljungdahl (WL), Arnon-Buchanan (AB), and Calvin-Benson-Bassham (CBB) pathways are present. Complete versions of these pathways were annotated in six bins; the WL pathway was present in Bin011 (Firmicutes sp.), the AB pathway in Bin018 (Sulfuricurvum sp.), the CBB pathway in Bin005 and Bin006 (Rhodobacteraceae and Xanthobacter spp., respectively). The WL pathway is prevalent among Firmicutes (Ragsdale and Pierce, 2008), which are abundant in the MMVC amplicon libraries (Figure 6). The AB pathway has been largely described in lithotrophic sulfur-oxidizing bacteria (Chlorobium; Buchanan et al., 2017), aligning with the metabolic lifestyle of Sulfuricurvum. Members of Rhodobacteraceae and Xanthobacter have been documented to perform steps of the CBB cycle (e.g., van den Bergh et al., 1996; Brinkmann et al., 2018). The abundance of carbon fixation pathways in the metagenome suggest that the MMVC cave sediment microbial community is sustained by chemosynthesis. The presence of several carbon fixation pathways suggest different strategies that may be shaped by available energy or geochemistry. For example, the WL (1 ATP) and the AB pathway (2 ATP) require significantly less ATP per mole of carbon fixed than the CBB (7 ATP) pathway (Bar-Even et al., 2012). The site D metagenome contains more genes related to the AB pathway (727) than the CBB (451), suggesting the lower energy requirements of the AB pathway may provide an advantage in this environment. The variety of carbon fixation pathways present in the site D sediments may also indicate niche partitioning around electron donor availability and energetic costs of macromolecule biosynthesis, as has been suggested for other subglacial systems (Mikucki et al., 2016).
Binning of sequences from the MMVC fumarole sediments enable more detailed examination of the metabolic potential of specific microbial community members. Bin014 from the site D metagenome was assigned to the class Symbiobacteriia (Supplementary Table S3) and contains genes comprising a pathway for dissimilatory nitrate reduction to nitrous oxide. Members of Symbiobacteriia have been reported to perform anaerobic oxidation of propane coupled to nitrate reduction (Wu et al., 2022). We did not measure propane in the September 2022 survey, but other fumaroles emitting propane have been documented (e.g., Mariner et al., 2008; Fiebig et al., 2013). Nitrate was below detection at site D; however, sulfate can also function as the electron acceptor in this reaction. Bin008 was assigned to the genus Dissulfurimicrobium (Supplementary Table S3) a group shown to perform sulfur disproportionation, i.e., utilizing sulfur (as S0, sulfite, or thiosulfate) as both an electron donor and acceptor (Slobodkin et al., 2016). While no single pathway has been described for sulfur disproportionation, most sulfur disproportionators contain genes encoding the Apr and Dsr pathways (Slobodkin and Slobodkina, 2019). Bin008 contains these genes as well as genes for sulfate adenylyltransferase (Sat), which reduces sulfate to APS, and SoxB, which is involved in oxidation of thiosulfate to sulfate. While we did not measure sulfite or thiosulfate, VNIR spectra suggest S0 is present in yellow mineral deposits at site D (Figure 3) and may provide a source of sulfur for disproportionation. Bin021 was assigned to the order Limnocylindrales, a member of the phylum Chloroflexota (Supplementary Table S3) and contained genes encoding an anaerobic carbon monoxide dehydrogenase, indicating this organism may be able to utilize carbon monoxide as an electron donor for anaerobic respiration of sulfate, hydrogen or CO2 (Oelgeschläger and Rother, 2008). The pathways identified in these bins demonstrate a subset of the diverse metabolic strategies employed by the microbial communities in MMVC sediments and emphasize the prevalence of chemosynthesis as a microbial lifestyle in glaciovolcanic cave environments. The sulfur-rich nature of the MMVC cave system provides a suite of potential metabolic strategies that are likely rare or absent in previously documented glaciovolcanic cave systems, positioning the MMVC glacier caves as a novel cryosphere microbial ecosystem.
Using a combination of exploration and mapping, in-situ and laboratory-based geochemical measurements, and environmental nucleic acid sequencing analyses, we present an initial characterization of a novel sulfidic glaciovolcanic cave system at the MMVC. Bacterial and archaeal community compositions appear largely distinct from other Pacific Northwest glacial systems, glaciovolcanic caves, temperate subglacial systems and a sulfidic subglacial caldera lake. Community structure and function is consistent with high-temperature acidic systems, suggesting that volcanic activity at the MMVC plays a substantial role in structuring microbial communities. The fumarolic soils support a chemosynthetic community that employs energy-yielding sulfur metabolism and utilizes several carbon fixation strategies. Our findings show that volcanic sulfidic gases influence the microbial ecology and geochemistry of the ice cave system and its surrounding snowpack. Further studies are needed to better understand the influence of volcanic degassing on snowpack microbial community structure and function and how the presence of dynamic glaciovolcanic cave systems may influence downstream ecology, for example, through the generation of acidic and solute-rich meltwater distinct from typical glacial melt.
Further geobiological study of the MMVC will improve understanding of subglacial microbial ecology and may provide opportunities to test operational concepts and technology for future life detection missions to icy ocean worlds, such as Saturn’s moon Enceladus. The MMVC system provides a rare access point into a unique subglacial realm, facilitating the study of fumarole biogeochemistry beneath glacial ice. This adds to our limited number of subglacial samples and expands our understanding of the diversity of subglacial ecosystem structure and function. The attributes of the MMVC also provide a potential astrobiological analog feature. Cryovolcanism, plume eruptions, or other mechanisms on Enceladus and Europa may transport constituents of these moons’ sub-ice oceans or near-surface reservoirs to the outer surface of their ice shells, likely depositing material on the surface (Roth et al., 2014; Porco et al., 2017; Quick and Hedman, 2020; Cable et al., 2021); if life exists in the sub-ice oceans, cryovolcanic plumes and deposits may contain biosignatures (Hendrix et al., 2019; NASEM, 2023; Perera and Cockell, 2023). While cryovolcanism does not occur on Earth, glaciovolcanism provides a terrestrial analog in the release of fumarole vapors from subglacial cave systems. Understanding the composition, transport and depositional characteristics of the vapor phase in a habitable system may provide important insight. For example, measurements of molecular hydrogen and methane in the vapor of the Enceladus plume by the Ion and Neutral Mass Spectrometer (INMS) aboard Cassini provided evidence in support of hydrothermal activity at the seafloor (Bouquet et al., 2015; Waite et al., 2017). Thorough characterization of glaciovolcanic activity at the MMVC for terrestrial studies provides important planetary analog context and can support the development and testing of mission concepts, methods, and instrumentation which may be applied to future space-based missions, such as an orbiter or lander deployed to an icy ocean world (MacKenzie et al., 2021; Hand et al., 2017; MacKenzie et al., 2022; Mousis et al., 2022).
The sequencing data presented in this study are deposited in the NCBI Sequence Read Archive; BioProject number PRJNA1144430, accession numbers SRR30131605 and SRR30196498 through SRR30196521.
JC: Investigation, Writing–original draft, Conceptualization, Formal Analysis, Visualization, Data curation. JS: Investigation, Writing–original draft, Formal Analysis, Data curation, Validation, Visualization. MC: Writing–original draft, Writing–review and editing, Investigation, Formal Analysis, Funding acquisition, Visualization. CS: Writing–original draft, Writing–review and editing, Investigation, Conceptualization, Funding acquisition, Visualization. GW-J: Writing–review and editing, Conceptualization, Funding acquisition. AS: Writing–original draft, Investigation. MP: Writing–review and editing, Funding acquisition, Investigation. OV: Writing–original draft, Investigation. KG: Writing–review and editing, Investigation. JM: Conceptualization, Funding acquisition, Investigation, Project administration, Writing–original draft, Writing–review and editing, Formal Analysis, Visualization.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. Partial funding to support the project was provided by the Trebek Initiative, a partnership between the Royal Canadian Geographical Society and National Geographic, a Natural Sciences and Engineering Research Council of Canada Discovery grant (RGPIN-2016-03953), and the University of Tennessee’s Office of Research, Innovation and Economic Development. Some of this work was also funded by an internal grant (Exobiology Extant Life Surveyor Project) supported by the JPL NEXT Program.
The authors respectfully acknowledge that the area which inspired this research is within the traditional unceded territories of the Líĺwat First Nation. Fieldwork was conducted under BC Parks permit 109950 and BC Ministry of Forests permit S26564. Some of this work was conducted at the Jet Propulsion Laboratory, California Institute of Technology, under a contract with the National Aeronautics and Space Administration (80NM0018D0004). Reference herein to any specific commercial product, process, or service by trade name, trademark, manufacturer, or otherwise, does not constitute or imply its endorsement by the United States Government or the Jet Propulsion laboratory, California Institute of Technology. E. Cartaya, T. Gall, S. Linn, B. Swerdlow and A. Walker provided safety and surface support. Aaron Messinger and Special Projects Operations provided specialized self-contained breathing apparatus for cave exploration. Meager Creek Development Corporation, Innergex Renewable Energy, and No Limits Helicopter Adventures provided invaluable logistical support and assistance in the field.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgeoc.2024.1410338/full#supplementary-material
Albertsen, M., Karst, S. M., Ziegler, A. S., Kirkegaard, R. H., and Nielsen, P. H. (2015). Back to basics – the influence of DNA extraction and primer choice on phylogenetic analysis of activated sludge communities. PloS ONE 10 (7), e0132783. doi:10.1371/journal.pone.0132783
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215 (3), 403–410. doi:10.1016/S0022-2836(05)80360-2
Anesio, A. M., Lutz, S., Chrismas, N. A. M., and Benning, L. G. (2017). The microbiome of glaciers and ice sheets. npj Biofilms Microbiomes 3 (10), 10. doi:10.1038/s41522-017-0019-0
Anitori, R., Davis, R., Connell, L., Kelley, M., Staudigel, H., and Tebo, B. (2011). Targeting autotrophic and lithotrophic microorganisms from fumarolic ice caves of Mt. Erebus. AGU Fall Meet. Abstr.
Arkin, A. P., Cottingham, R. W., Henry, C. S., Harris, N. L., Stevens, R. L., Maslov, S., et al. (2018). Kbase: the United States department of energy systems biology knowledgebase. Nat. Biotechnol. 36, 566–569. doi:10.1038/nbt.4163
Arnold, H., Liuzzo, L., and Simon, S. (2019). Magnetic signatures of a plume at Europa during the galileo E26 flyby. Geophys. Res. Lett. 46 (3), 1149–1157. doi:10.1029/2018GL081544
Averett, R. C., Leenheer, J. A., McKnight, D. M., and Thorn, K. A. (1994). Humic substances in the Suwannee River, Georgia: interactions, properties, and proposed structures. Denver, CO: US Government Printing Office. No. 2373. doi:10.3133/Wsp2373
Badgeley, J. A., Pettit, E. C., Carr, C. G., Tulaczyk, S., Mikucki, J. A., Lyons, W. B., et al. (2017). An englacial hydrologic system of brine within a cold glacier: blood Falls, McMurdo Dry Valleys, Antarctica. J. Glaciol. 63 (239), 387–400. doi:10.1017/jog.2017.16
Bar-Even, A., Noor, E., and Milo, R. (2012). A survey of carbon fixation pathways through a quantitative lens. J. Exp. Bot. 63 (6), 2325–2342. doi:10.1093/jxb/err417
Barker, J. F., and Fritz, P. (1981). Carbon isotope fractionation during microbial methane oxidation. Nature 293, 289–291. doi:10.1038/293289a0
Boden, R., Cleland, D., Green, P. N., Katayama, Y., Uchino, Y., Murrell, J. C., et al. (2012). Phylogenetic assessment of culture collection strains of Thiobacillus thioparus, and definitive 16S rRNA gene sequences for T. thioparus, T. denitrificans, and Halothiobacillus neapolitanus. Arch. Microbiol. 194, 187–195. doi:10.1007/s00203-011-0747-0
Bokulich, N. A., Subramanian, S., Faith, J. J., Gevers, D., Gordon, J. I., Knight, R., et al. (2013). Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 10, 57–59. doi:10.1038/nmeth.2276
Bouquet, A., Mousis, O., Waite, J. H., and Picaud, S. (2015). Possible evidence for a methane source in Enceladus’ ocean. Geophys. Res. Lett. 42 (5), 1334–1339. doi:10.1002/2014GL063013
Bradley, A. S., Leavitt, W. D., Schmidt, M., Knoll, A. H., Girguis, P. R., and Johnston, D. T. (2016). Patterns of sulfur isotope fractionation during microbial sulfate reduction. Geobiology 14, 91–101. doi:10.1111/gbi.12149
Brinkmann, H., Gӧker, M., Koblížek, M., Wagner-Dӧbler, I., and Petersen, J. (2018). Horizontal operon transfer, plasmids, and the evolution of photosynthesis in Rhodobacteraceae. ISME J. ISME J. 12 (8), 1994–2010. doi:10.1038/s41396-018-0150-9
Buchanan, B. B., Sirevåg, R., Fuchs, G., Ivanovsky, R. N., Igarashi, Y., Ishii, M., et al. (2017). The Arnon-Buchanan cycle: a retrospective, 1966-2016. Photosynth Res. 134, 117–131. doi:10.1007/s11120-017-0429-0
Buchwald, M., and Jencks, W. (1968). Optical properties of astaxanthin solutions and aggregates. Biochemistry 7 (2), 834–843. doi:10.1021/bi00842a042
Cable, M. L., Porco, C., Glein, C. R., German, C. R., MacKenzie, S. M., Neveu, M., et al. (2021). The science case for a return to Enceladus. Planet Sci. J. 2 (132), 132. doi:10.3847/PSJ/abfb7a
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13 (7), 581–583. doi:10.1038/nmeth.3869
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Lozupone, C. A., Turnbaugh, P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. 108, 4516–4522. doi:10.1073/pnas.1000080107
Chaumeil, P., Mussig, A. J., Hugenholtz, P., and Parks, D. H. (2022). GTDB-Tk v2: memory friendly classification with the genome taxonomy database. Bioinformatics 38 (23), 5315–5316. doi:10.1093/bioinformatics/btac672
Chen, Y., Wu, L., Boden, R., Hillebrand, A., Kumaresan, D., Moussard, H., et al. (2009). Life without light: microbial diversity and evidence of sulfur- and ammonium-based chemolithotrophy in Movile Cave. ISME J. 3, 1093–1104. doi:10.1038/ismej.2009.57
Clague, J. J., Evans, S. G., Rampton, V. N., and Woodsworth, G. J. (1995). Improved age estimates for the white river and bridge river tephras, western Canada. Can. J. Earth Sci. 32 (8), 1172–1179. doi:10.1139/e95-096
Cockell, C. S., Simons, M., Castillo-Rogez, J., Higgins, P. M., Kaltenegger, L., Keane, J. T., et al. (2024). Sustained and comparative habitability beyond Earth. Nat. Astron. 8, 30–38. doi:10.1038/s41550-023-02158-8
Connell, L., and Staudigel, H. (2013). Fungal diversity in a dark oligotrophic volcanic ecosystem (DOVE) on mount erebus, Antarctica. Biology 2, 798–809. doi:10.3390/biology2020798
Connelly, J. P., Sepúlveda, S. A., and Williams-Jones, G. (2024). Qwelqwelústen Mount Meager Volcanic Complex, British Columbia: inter-eruptive landslide susceptibility assessment using statistical machine learning techniques. Volcanica.
Costello, E. K., Halloy, S. R. P., Reed, S. C., Sowell, P., and Schmidt, S. K. (2009). Fumarole-Supported Islands of Biodiversity within a Hyperarid, High-Elevation Landscape on Socompa Volcano, Puna de Atacama, Andes. Appl. Environ. Microbiol. 75 (3), 735–747. doi:10.1128/AEM.01469-08
Curtis, A., and Kyle, P. (2011). Geothermal point sources identified in a fumarolic ice cave on Erebus volcano, Antarctica using fiber optic distributed temperature sensing. Geophys. Res. Lett. 38 (16). doi:10.1029/2011GL048272
de Vries, A., Klevtsov, A., and Gilliland, J. D. A. (2016). ggdendro. Available at: https://github.com/andrie/ggdendro.
Dijkstra, P., Ishizu, A., Doucett, R., Hart, S. C., Schwartz, E., Menyailo, O. V., et al. (2006). 13C and 15N natural abundance of the soil microbial biomass. Soil Biol. Biochem. 38, 3257–3266. doi:10.1016/j.soilbio.2006.04.005
Dougherty, M. K., Khurana, K. K., Neubauer, F. M., Russell, C. T., Saur, J., Leisner, J. S., et al. (2006). Identification of a dynamic atmosphere at Enceladus with the Cassini magnetometer. Science 311, 1406–1409. doi:10.1126/science.1120985
Ellis, D. G., Bizzoco, R. W., and Kelley, S. T. (2008). Halophilic Archaea determined from geothermal steam vent aerosols. Environ. Microbiol. 10 (6), 1582–1590. doi:10.1111/j.1462-2920.2008.01574.x
Fiebig, J., Tassi, F., D’Alessandro, W., Vaselli, O., and Woodland, A. B. (2013). Carbon-bearing gas geothermometers for volcanic-hydrothermal systems. Chem. Geol. 351, 66–75. doi:10.1016/j.chemgeo.2013.05.006
Florea, L., Pflitsch, A., Cartaya, E., and Stenner, C. (2021). Microclimates in fumarole ice caves on volcanic edifices—Mount Rainier, Washington, USA. J. Geophys. Res. Atmos. 126 (4). doi:10.1029/2020JD033565
Fraser, C. I., Connell, L., Lee, C. K., and Cary, S. C. (2018). Evidence of plant and animal communities at exposed and subglacial (cave) geothermal sites in Antarctica. Polar Biol. 41 (3), 417–421. doi:10.1007/s00300-017-2198-9
Friedrich, C. G., Bardischewsky, F., Rother, D., Quentmeier, A., and Fischer, J. (2005). Prokaryotic sulfur oxidation. Curr. Opin. Microbiol. 8, 253–259. doi:10.1016/j.mib.2005.04.005
Friedrich, C. G., Rother, D., Bardischewsky, F., Quentmeier, A., and Fischer, J. (2001). Oxidation of reduced inorganic sulfur compounds by bacteria: emergence of a common mechanism?. Appl. Environ. Microbiol. 67, 2873–2882. doi:10.1128/AEM.67.7.2873-2882.2001
Friele, P., Jakob, M., and Clague, J. (2008). Hazard and risk from large landslides from Mount Meager volcano, British Columbia, Canada. Georisk Assess. Manag. Risk Eng. Syst. Geohazards 2, 48–64. doi:10.1080/17499510801958711
Gaidos, E., Lanoil, B., Thorsteinsson, T., Graham, A., Skidmore, M., Han, S. K., et al. (2004). A viable microbial community in a subglacial volcanic crater lake, Iceland. Astrobiology 4 (3), 327–344. doi:10.1089/ast.2004.4.327
Gaidos, E., Marteinsson, V., Thorsteinsson, T., Jóhannesson, T., Rúnarsson, A. R., Stefansson, A., et al. (2009). An oligarchic microbial assemblage in the anoxic bottom waters of a volcanic subglacial lake. ISME J. 3, 486–497. doi:10.1038/ismej.2008.124
Galloway, J. N., Dentener, F. J., Capone, D. G., Boyer, E. W., Howarth, R. W., Seitzinger, S. P., et al. (2004). Nitrogen cycles: past, present, and future. Biogeochemistry 70, 153–226. doi:10.1007/s10533-004-0370-0
Gerlach, T. M., and Casadevall, T. J. (1986). Evaluation of gas data from high-temperature fumaroles at Mount St. Helens, 1980-1982. J. Volcanol. Geotherm. Res. 28, 107–140. doi:10.1016/0377-0273(86)90008-9
Glass, J. B., Dierssen, H. M., Glein, C. R., Schmidt, B. E., and Winebrenner, D. P. (2022). Defining and characterizing habitable environments in ocean world systems. Oceanography 35 (1), 30–38. doi:10.5670/oceanog.2021.414
Gorton, H., and Vogelmann, T. (2003). Ultraviolet radiation and the snow alga Chlamydomonas nivalis (bauer) wille. Photochem. Photobiol. 77 (6), 608–615. doi:10.1562/0031-8655(2003)0770608URATSA2.0.CO2
Graven, H., Keeling, R., and Rogelj, J. (2020). Changes to carbon isotopes in atmospheric CO2 over the industrial era and into the future. Glob. Biogeochem. Cycles 34 (11), e2019GB006170. doi:10.1029/2019GB006170
Hamilton, T. L., and Havig, J. R. (2017). Primary productivity of snow algae communities on stratovolcanoes of the Pacific Northwest. Geobiology 15, 280–295. doi:10.1111/gbi.12219
Hamilton, T. L., and Havig, J. R. (2020). Inorganic carbon addition stimulates snow algae primary productivity. ISME J. 14, 857–860. doi:10.1038/s41396-018-0048-6
Hamilton, T. L., Peters, J. W., Skidmore, M. L., and Boyd, E. S. (2013). Molecular evidence for an active endogenous microbiome beneath glacial ice. ISME J. 7 (7), 1402–1412. doi:10.1038/ismej.2013.31
Hammes, F., Goldschmidt, F., Vital, M., Wang, Y., and Egli, T. (2010). Measurement and interpretation of microbial adenosine tri-phosphate (ATP) in aquatic environments. Water Res. 44 (13), 3915–3923. doi:10.1016/j.watres.2010.04.015
Hand, K. P., Chyba, C. F., Priscu, J. C., Carlson, R. W., and Nealson, K. H. (2017). “Astrobiology and the potential for life on Europa, in Europa. Editors R. T. Pappalardo, W. B. McKinnon, and K. Khurana (Tucson, AZ: University of Arizona Press), 589–630.
Hand, K. P., Sotin, C., Hayes, A., and Coustenis, A. (2020). On the habitability and future exploration of ocean worlds. Space Sci. Rev. 216, 95–24. doi:10.1007/s11214-020-00713-7
Hansen, C. J., Esposito, L., Stewart, A. I. F., Colwell, J., Hendrix, A., Pryor, W., et al. (2006). Enceladus’ water vapor plume. Science 311, 1422–1425. doi:10.1126/science.1121254
Havig, J. R., and Hamilton, T. L. (2019). Snow algae drive productivity and weathering at volcanic rock-hosted glaciers. Geochimica Cosmochimica Acta 247, 220–242. doi:10.1016/j.gca.2018.12.024
Hendrix, A. R., Hurford, T. A., Barge, L. M., Bland, M. T., Bowman, J. S., Brinckerhoff, W., et al. (2019). The NASA roadmap to ocean worlds. Astrobiology 19 (1), 1–27. doi:10.1089/ast.2018.1955
Hickson, C. J., Russell, J. K., and Stasiuk, M. V. (1999). Volcanology of the 2350 B.P. Eruption of Mount Meager volcanic complex, British Columbia, Canada: implications for hazards from eruptions in topographically complex terrain. Bull. Volcanol. 60 (7), 489–507. doi:10.1007/s004450050247
Hoham, R. W., and Mohn, W. W. (1985). The optimum pH of four strains of acidophilic snow algae in the genus Chloromonas (Chlorophyta) and possible effects of acid precipitation. J. Phycol. 21, 603–609. doi:10.1111/j.0022-3646.1985.00603.x
Hoham, R. W., and Remias, D. (2020). Snow and glacial algae: a review. J. Phycol. 56, 264–282. doi:10.1111/jpy.12952
Hotaling, S., Hood, E., and Hamilton, T. L. (2017). Microbial ecology of mountain glacier ecosystems: biodiversity, ecological connections and implications of a warming climate. Environ. Microbiol. 19 (8), 2935–2948. doi:10.1111/1462-2920.13766
Howell, S., Stone, W. C., Craft, K., German, C., Murray, A., Rhoden, A., et al. (2021). Ocean worlds exploration and the search for life. Bull. AAS 53 (4). doi:10.3847/25c2cfeb.8920f9ae
Hügler, M., and Sievert, S. M. (2011). Beyond the Calvin cycle: autotrophic carbon fixation in the ocean. Annu. Rev. Mar. Sci. 3, 261–289. doi:10.1146/annurev-marine-120709-142712
Jørgensen, B. B. (1982). Mineralization of organic matter in the sea bed - the role of sulphate reduction. Nature 296, 643–645. doi:10.1038/296643a0
Jorgensen, B. B., and Postgate, J. R. (1982). Ecology of the bacteria of the sulphur cycle with special reference to anoxicoxic interface environments. Philos. T R Soc. B 298 (1093), 543–561. doi:10.1098/rstb.1982.0096
Kanehisa, M., and Goto, S. (2000). KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. doi:10.1093/nar/28.1.27
Kanehisa, M., Sato, Y., and Morishima, K. (2016). BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 428, 726–731. doi:10.1016/j.jmb.2015.11.006
Kelly, D. P., and Wood, A. P. (2014). “The family Acidithiobacillaceae,” in The prokaryotes. 4th Edn, Editors E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson doi:10.1007/978-3-642-38922-1_250
Kelman, M. C., and Wilson, A. M. (2024). Assessing the relative threats from Canadian volcanoes. Can. J. Earth Sci. 61 (3), 408–430. doi:10.1139/cjes-2023-0074
Klindworth, A., Pruesse, E., Schweer, T., Peplies, J., Quast, C., Horn, M., et al. (2012). Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41 (1), e1. doi:10.1093/nar/gks808
Kokaly, R. F., Clark, R. N., Swayze, G. A., Livo, K. E., Hoefen, T. M., Pearson, N. C., et al. (2017) USGS spectral library version 7. Denver, CO: U.S. Geological Survey Data Series, Vol. 1035, 61. doi:10.3133/ds1035
Kuever, J. (2014). “The family desulfobulbaceae,” in The prokaryotes. 4th Edn, Editors E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson doi:10.1007/978-3-642-39044-9_267
Livingstone, S. J., Li, Y., Rutishauser, A., Sanderson, R. J., Winter, K., Mikucki, J. A., et al. (2022). Subglacial lakes and their changing role in a warming climate. Nat. Rev. Earth and Environ. 3, 106–124. doi:10.1038/s43017-021-00246-9
Lutz, S., Anesio, A. M., Raiswell, R., Edwards, A., Newton, R. J., Gill, F., et al. (2016). The biogeography of red snow microbiomes and their role in melting arctic glaciers. Nat. Commun. 7, 11968. doi:10.1038/ncomms11968
MacKenzie, S. M., Neveu, M., Davila, A. F., Lunine, J. I., Cable, M. L., Phillips-Lander, C. M., et al. (2022). Science objectives for flagship-class mission concepts for the search for evidence of life at Enceladus. Astrobiology 22 (6), 685–712. doi:10.1089/ast.2020.2425
MacKenzie, S. M., Neveu, M., Davila, A. F., Lunine, J. I., Craft, K. L., Cable, M. L., et al. (2021). The Enceladus orbilander mission concept: balancing return and resources in the search for life. Planet Sci. J. 2 (77), 77. doi:10.3847/PSJ/abe4da
Magoč, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi:10.1093/bioinformatics/btr507
Mariner, R. H., Minor, S. A., King, A. P., Boles, J. R., Kellogg, K. S., Evans, W. C., et al. (2008). A landslide in Tertiary marine shale with superheated fumaroles, Coast Ranges, California. Geology 36 (12), 959–962. doi:10.1130/G25285A.1
Marteinsson, V. T., Rúnarsson, Á., Stefánsson, A., Thorsteinsson, T., Jóhannesson, T., Magnússon, S. H., et al. (2013). Microbial communities in the subglacial waters of the Vatnajӧkull ice cap, Iceland. ISME J. 7, 427–437. doi:10.1038/ismej.2012.97
Martinez, M. A., Woodcroft, B. J., Ignacio Espinoza, J. C., Zayed, A. A., Singleton, C. M., Boyd, J. A., et al. (2019). Discovery and ecogenomic context of a global Caldiserica-related phylum active in thawing permafrost, Candidatus Cryosericota phylum nov., Ca. Cryosericia class nov., Ca. Cryosericales ord. nov., Ca. Cryosericaceae fam. Nov., comprising the four species Cryosericum septentrionale gen. nov. sp. nov., Ca. C. hinesii sp. nov., Ca. C. odellii sp. nov., Ca. C. terrychapinii sp. nov. Syst. Appl. Microbiol. 42, 54–66. doi:10.1016/j.syapm.2018.12.003
Mattes, T. E., Nunn, B. L., Marshall, K. T., Proskurowski, G., Kelley, D. S., Kawka, O. E., et al. (2013). Sulfur oxidizers dominate carbon fixation at a biogeochemical hot spot in the dark ocean. ISME J. 7 (12), 2349–2360. doi:10.1038/ismej.2013.113
Mayhew, L. E., Geist, D. J., Childers, S. E., and Pierson, J. D. (2007). Microbial community comparisons as a function of the physical and geochemical conditions of galapagos island fumaroles. Geomicrobiol. J. 24 (7-8), 615–625. doi:10.1080/01490450701672133
McMurdie, P. J., and Holmes, S. (2013). Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PloS ONE 8 (4), e61217. doi:10.1371/journal.pone.0061217
Menzel, P., Ng, K., and Krogh, A. (2016). Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nat. Commun. 7, 11257. doi:10.1038/ncomms11257
Merkel, A.Yu., Pimenov, N. V., Rusanov, I. I., Slobodkin, A. I., Slobodkina, G. B., Tarnovetckii, I. Y., et al. (2017). Microbial diversity and autotrophic activity in Kamchatka hot springs. Extremophiles 21, 307–317. doi:10.1007/s00792-016-0903-1
Meyer, B., Imhoff, J. F., and Kuever, J. (2007). Molecular analysis of the distribution and phylogeny of the soxB gene among sulfur-oxidizing bacteria – evolution of the Sox sulfur oxidation enzyme system. Environ. Microbiol. 9 (12), 2957–2977. doi:10.1111/j.1462-2920.2007.01407.x
Meyer, B., and Kuever, J. (2007). Molecular analysis of the distribution and phylogeny of dissimilatory adenosine-5’-phosphosulfate reductase-encoding genes (aprBA) among sulfur-oxidizing prokaryotes. Microbiology 153, 3478–3498. doi:10.1099/mic.0.2007/008250-0
Mikucki, J. A., and Priscu, J. C. (2007). Bacterial diversity associated with blood falls, a subglacial outflow from the taylor glacier, antarctica. Appl. Environ. Microb. 73 (12), 4029–4039. doi:10.1128/AEM.01396-06
Mikucki, J. A., Lee, P. A., Ghosh, D., Purcell, A. M., Mitchell, A. C., Mankoff, K. D., et al. (2016). Subglacial Lake Whillans microbial biogeochemistry: a synthesis of current knowledge. Philos. T. R. Soc. A. 374. doi:10.1098/rsta.2014.0290
Mori, K., Sunamura, M., Yanagawa, K., Ishibashi, J., Miyoshi, Y., Iino, T., et al. (2008). First cultivation and ecological investigation of a bacterium affiliated with the candidate phylum OP5 from hot springs. Appl. Environ. Microbiol. 74, 6223–6229. doi:10.1128/AEM.01351-08
Mori, K., Yamaguchi, K., Sakiyama, Y., Urabe, T., and Suzuki, K. (2009). Caldisericum exile gen. nov., sp. nov., an anaerobic, thermophilic, filamentous bacterium of a novel bacterial phylum, Caldiserica phyl. Nov., originally called the candidate phylum OP5, and description of Caldisericaceae fam. Nov., Caldisericales ord. nov. and Caldisericia classis nov. Int. J. Syst. Evol. Micr 59, 2894–2898. doi:10.1099/ijs.0.010033-0
Mousis, O., Bouquet, A., Langevin, Y., André, N., Boithias, H., Durry, G., et al. (2022). Moonraker: Enceladus multiple flyby mission. Planet Sci. J. 3 (268), 268. doi:10.3847/PSJ/ac9c03
National Academies of Sciences, Engineering, and Medicine (NASEM) (2023). Origins, worlds, and life: a decadal strategy for planetary science and astrobiology 2023–2032. Washington, DC: The National Academies Press.
Neukirchen, S., Pereira, I. A. C., and Sousa, F. L. (2023). Stepwise pathway for early evolutionary assembly of dissimilatory sulfite and sulfate reduction. ISME J. 17 (10), 1680–1692. doi:10.1038/s41396-023-01477-y
Noell, S. E., Baptista, M. S., Smith, E., McDonald, I. R., Lee, C. K., Stott, M. B., et al. (2022). Unique geothermal chemistry shapes microbial communities on Mt. Erebus, Antarctica. Front. Microbiol. 13, 836943. doi:10.3389/fmicb.2022.836943
Northup, D. E., and Lavoie, K. H. (2001). Geomicrobiology of caves: a review. Geomicrobiol. J. 18 (3), 199–222. doi:10.1080/01490450152467750
Nurk, S., Meleshko, D., Korobeynikov, A., and Pevzner, P. A. (2017). metaSPAdes: a new versatile metagenomic assembler. Genome Res. 27 (5), 824–834. doi:10.1101/gr.213959.116
Oelgeschläger, E., and Rother, M. (2008). Carbon monoxide-dependent energy metabolism in anaerobic bacteria and archaea. Arch. Microbiol. 190, 257–269. doi:10.1007/s00203-008-0382-6
Oksanen, J., Simpson, G., Blanchet, F., Kindt, R., Legendre, P., Minchin, P., et al. (2023). Vegan: community ecology package. R. package version 2, 6–5.
Oppenheimer, C., and Kyle, P. R. (2008). Probing the magma plumbing of Erebus volcano, Antarctica, by open-path FTIR spectroscopy of gas emissions. J. Volcanol. Geotherm. Res. 177, 743–754. doi:10.1016/j.jvolgeores.2007.08.022
Oppenheimer, C., Scaillet, B., and Martin, R. S. (2011). Sulfur degassing from volcanoes: source conditions, surveillance, plume chemistry and earth system impacts. Rev. Mineralogy Geochem. 73 (1), 363–421. doi:10.2138/rmg.2011.73.13
Oren, A. (2014). “The family xanthobacteraceae,” in The prokaryotes. 4th Edn, Editors Editors E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson doi:10.1007/978-3-642-30197-1_258
Orlygsson, J., and Kristjansson, J. K. (2014). “The family hydrogenophilaceae,” in The prokaryotes. 4th Edn, Editors E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson doi:10.1007/978-3-642-30197-1_244
Painter, T. H., Duval, B., Thomas, W. H., Mendez, M., Heintzelman, S., and Dozier, J. (2001). Detection and quantification of snow algae with an airborne imaging spectrometer. Appl. Environ. Microbiol. 67 (11), 5267–5272. doi:10.1128/AEM.67.11.5267-5272.2001
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P., and Tyson, G. W. (2015). CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055. doi:10.1101/gr.186072.114
Pellerin, A., Antler, G., Holm, S. A., Findlay, A. J., Crockford, P. W., Turchyn, A. V., et al. (2019). Large sulfur isotope fractionation by bacterial sulfide oxidation. Sci. Adv. 5 (7), eaaw1480. doi:10.1126/sciadv.aaw1480
Perera, L. J., and Cockell, C. S. (2023). Dispersion of bacteria by low-pressure boiling: life detection in Enceladus’ plume material. Astrobiology 23 (3), 269–279. doi:10.1089/ast.2022.0009
Porco, C. C., Dones, L., and Mitchell, C. (2017). Could it Be snowing microbes on Enceladus? Assessing conditions in its plume and implications for future missions. Astrobiology 17 (9), 876–901. doi:10.1089/ast.2017.1665
Porco, C. C., Helfenstein, P., Thomas, P. C., Ingersoll, A. P., Wisdom, J., West, R., et al. (2006). Cassini observes the active south Pole of Enceladus. Science 311 (5776), 1393–1401. doi:10.1126/science.1123013
Postberg, F., Khawaja, N., Abel, B., Choblet, G., Glein, C. R., Gudipati, M. S., et al. (2018). Macromolecular organic compounds from the depths of Enceladus. Nature 558, 564–568. doi:10.1038/s41586-018-0246-4
Prosser, J. I., Head, I. M., and Stein, L. Y. (2014). “The family nitrosomonadaceae,” in The prokaryotes. 4th Edn, Editors E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson doi:10.1007/978-3-642-30197-1_372
Pujalte, M. J., Lucena, T., Ruvira, M. A., Arahal, D. R., and Macián, M. C. (2014). “The family Rhodobacteraceae,” in The prokaryotes. 4th Edn, Editors E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson doi:10.1007/978-3-642-30197-1_377
Purcell, A. M., Mikucki, J. A., Achberger, A. M., Alekhina, I. A., Barbante, C., Christner, B. C., et al. (2014). Microbial sulfur transformations in sediments from subglacial lake whillans. Front. Microbiol. 5, 594. doi:10.3389/fmicb.2014.00594
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, 590–596. doi:10.1093/nar/gks1219
Quick, L. C., and Hedman, M. M. (2020). Characterizing deposits emplaced by cryovolcanic plumes on Europa. Icarus 343, 113667. doi:10.1016/j.icarus.2020.113667
Ragsdale, S. W., and Pierce, E. (2008). Acetogenesis and the Wood-Ljungdahl pathway of CO2 fixation. Biochimica Biophysica Acta (BBA) - Proteins Proteomics 1784, 1873–1898. doi:10.1016/j.bbapap.2008.08.012
Read, P. B. (1990). Mount meager complex, Garibaldi Belt, southwestern British Columbia. Geosci. Can. 17 (3), 167–170.
Roberti, G. (2018). Mount Meager, a glaciated volcano in a changing cryosphere: hazard and risk challenges. PhD Dissertation. Vancouver (BC). Burnaby: Simon Fraser University. Available at: https://summit.sfu.ca/item/18653.
Roberti, G., Ward, B., van Wyk de Vries, B., Falorni, G., Menounos, B., Friele, P., et al. (2018) “Landslides and glacier retreat at Mt. Meager volcano: hazard and risk challenges,” in 7th Canadian Geohazards Conference.
Robeson, M. S. I. I., O’Rourke, D. R., Kaehler, B. D., Ziemski, M., Dillon, M. R., Foster, J. T., et al. (2021). RESCRIPt: reproducible sequence taxonomy reference database management. PLOS Comput. Biol. 17 (11), e1009581. doi:10.1371/journal.pcbi.1009581
Roswell, M., Dushoff, J., and Winfree, R. (2021). A conceptual guide to measuring species diversity. Oikos 130, 321–338. doi:10.1111/oik.07202
Roth, L., Saur, J., Retherford, K., Strobel, D., Feldman, P., McGrath, M., et al. (2014). Transient water vapor at europa’s south Pole. Science 343 (6167), 171–174. doi:10.1126/science.1247051
Russell, J. K., Stewart, M., Wilson, A., and Williams-Jones, G. (2021). Eruption of Mount Meager, British Columbia, during the early fraser glaciation. Can. J. Earth Sci. 58 (10), 1146–1154. doi:10.1139/cjes-2021-0023
Schuler, C. G., and Mikucki, J. A. (2023). Microbial ecology and activity of snow algae within a Pacific Northwest snowpack. Arct. Antarct. Alp. Res. 55 (1). doi:10.1080/15230430.2023.2233785
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30 (14), 2068–2069. doi:10.1093/bioinformatics/btu153
Slobodkin, A. I., and Slobodkina, G. B. (2019). Diversity of sulfur-disproportionating microorganisms. Microbiology 88 (5), 509–522. doi:10.1134/S0026261719050138
Slobodkin, A. I., Slobodkina, G. B., Panteleeva, A. N., Chernyh, N. A., Novikov, A. A., and Bonch-Osmolovskaya, E. A. (2016). Dissulfurimicrobium hydrothermale gen. nov., sp. nov., a thermophilic, autotrophic, sulfur-disproportionating deltaproteobacterium isolated from a hydrothermal pond. Int. J. Syst. Evol. Microbiol. 66, 1022–1026. doi:10.1099/ijsem.0.000828
Smellie, J. L., and Edwards, B. R. (2016). Glaciovolcanism on Earth and mars: products, processes, and palaeoenvironmental significance. Cambridge, United Kingdom: Cambridge University Press.
Smith, A. R., Mueller, R., Fisk, M. R., and Colwell, F. S. (2021). Ancient metabolisms of a thermophilic subseafloor bacterium. Front. Microbiol. 12, 764631. doi:10.3389/fmicb.2021.764631
Sobolewski, L., Stenner, C., Williams-Jones, G., Anitori, R., Davis, R. E., and Pflitsch, A. (2022). Implications of the study of subglacial volcanism and glaciovolcanic cave systems. Bull. Volcanol. 84, 21. doi:10.1007/s00445-022-01525-z
Solon, A. J., Vimercati, L., Darcy, J. L., Arán, P., Porazinska, D., Dorador, C., et al. (2018). Microbial Communities of High-Elevation Fumaroles, Penitentes, and Dry Tephra “Soils” of the Puna de Atacama Volcanic Zone. Microb. Ecol. 76 (2), 340–351. doi:10.1007/s00248-017-1129-1
Spahn, F., Schmidt, J., Albers, N., Horning, M., Makuch, M., Seiß, M., et al. (2006). Cassini dust measurements at Enceladus and implications for the origin of the E ring. Science 311 (5766), 1416–1418. doi:10.1126/science.1121375
Spencer, C. N., Matzner, S. L., Smalley, J., Bukrey, M., Onberg, J., Chapman, M., et al. (2009). Forest expansion and soil carbon changes in the loess hills of eastern south Dakota. Am. Midl. Nat. 161, 273–285. doi:10.1674/0003-0031-161.2.273
Stackebrandt, E. (2014). “The family Thermoanaerobacteraceae,” in The prokaryotes. 4th Edn, Editors E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson doi:10.1007/978-3-642-30120-9_367
Stackebrandt, E., and Goebel, B. M. (1994). Taxonomic note: a place for DNA–DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Evol. Microbiol. 44, 846–849. doi:10.1099/00207713-44-4-846
Stenner, C., Florea, L., Pflitsch, A., Cartaya, E., and Riggs, D. (2023). Morphodynamics of glaciovolcanic caves—mount rainier, Washington, USA. J. Cave Karst Stud. 85 (3-4), 65–85. doi:10.4311/2021EX0131
Stenner, C., Pflitsch, A., Florea, L., Graham, K., and Cartaya, E. (2022). Development and persistence of hazardous atmospheres in a glaciovolcanic cave system—Mount Rainier, Washington, USA. J. Cave Karst Stud. 84 (2), 66–82. doi:10.4311/2021EX0102
Stibal, M., Bradley, J. A., Edwards, A., Hotaling, S., Zawierucha, K., Rosvold, J., et al. (2020). Glacial ecosystems are essential to understanding biodiversity responses to glacier retreat. Nat. Ecol. Evol. 4, 686–687. doi:10.1038/s41559-019-1042-8
Tan, G. K., Simpson, A., Holtzen, S., Amador, E., Cable, M. L., Cantrell, T., et al. (2022). Spatial variation in results of biosignature analyses of apparently homogeneous samples from mars analogue environments in Iceland. ACS Earth Space Chem. 6 (6), 1472–1481. doi:10.1021/acsearthspacechem.1c00390
Tebo, B. M., Davis, R. E., Anitori, R. P., Connell, L. B., Schiffman, P., and Staudigel, H. (2015). Microbial communities in dark oligotrophic volcanic ice cave ecosystems of Mt. Erebus, Antarctica. Front. Microbiol. 6, 179. doi:10.3389/fmicb.2015.00179
Tokar, R. L., Johnson, R. E., Hill, T. W., Pontius, D. H., Kurth, W. S., Crary, F. J., et al. (2006). The interaction of the atmosphere of Enceladus with Saturn’s plasma. Science 311 (5776), 1409–1412. doi:10.1126/science.1121061
Unnsteinsson, T., Flowers, G., and Williams-Jones, G. (2024). Formation and persistence of glaciovolcanic voids explored with analytical and numerical models. J. Glaciol., 1–15. doi:10.1017/jog.2024.8
Vance, S. D., Craft, K. L., Shock, E., Schmidt, B. E., Lunine, J., Hand, K. P., et al. (2023). Investigating europa’s habitability with the europa clipper. Space Sci. Rev. 219. doi:10.1007/s11214-023-01025-2
van den Bergh, E. R. E., Baker, S. C., Raggers, R. J., Terpstra, P., Woudstra, E. C., Dijkhuizen, L., et al. (1996). Primary structure and phylogeny of the Calvin cycle enzymes transketolase and fructosbisphosphate aldolase of Xanthobacter flavus. J bacteriol. J. Bacteriol. 178 (3), 888–893. doi:10.1128/jb.178.3.888-893.1996
Waite, J. H., Combi, M. R., Ip, W., Cravens, T. E., McNutt, R. L., Kasprzak, W., et al. (2006). Cassini ion and neutral mass spectrometer: Enceladus plume composition and structure. Science 311 (5776), 1419–1422. doi:10.1126/science.1121290
Waite, J. H., Glein, C. R., Perryman, R. S., Teolis, B. D., Magee, B. A., Miller, G., et al. (2017). Cassini finds molecular hydrogen in the Enceladus plume: evidence for hydrothermal processes. Science 356 (6334), 155–159. doi:10.1126/science.aai8703
Warwick, R., Williams-Jones, G., Kelman, M., and Witter, J. B. (2022). A scenario-based volcanic hazard assessment for the Mount Meager volcanic complex, British Columbia. J. Appl. Volcanol. 11 (5), 5–22. doi:10.1186/s13617-022-00114-1
Watanabe, T., Kojima, H., and Fukui, M. (2015). Sulfuriferula multivorans gen. nov., sp. nov., isolated from a freshwater lake, reclassification of ‘Thiobacillus plumbophilus’ as Sulfuriferula plumbophilus sp. nov., and description of Sulfuricellaceae fam. Nov. and Sulfuricellales ord. nov. Int. J. Syst. Evol. Microbiol. 65, 1504–1508. doi:10.1099/ijs.0.000129
Watanabe, T., Kojima, H., and Fukui, M. (2016). Sulfuriferula thiophila sp. nov., a chemolithoautotrophic sulfur-oxidizing bacterium, and correction of the name Sulfuriferula plumbophilus to Sulfuriferula plumbiphila corrig. Int. J. Syst. Evol. Microbiol. 66, 2041–2045. doi:10.1099/ijsem.0.000988
Werner, C., Evans, W. C., Poland, M., Tucker, D. S., and Doukas, M. P. (2009). Long-term changes in quiescent degassing at Mount Baker Volcano, Washington, USA; Evidence for a stalled intrusion in 1975 and connection to a deep magma source. J. Volcanol. Geotherm. Res. 186 (3-4), 379–386. doi:10.1016/j.jvolgeores.2009.07.006
Wilson, M. C., Angelbeck, B., and Jones Yaqalatqa, J. (2024). Lílwat oral traditions of Qwelqwelústen (Mount Meager): indigenous records of volcanic eruption, outburst flood, and landscape change in southwest British Columbia. Can. J. Earth Sci. 61 (6), 661–677. doi:10.1139/cjes-2023-0098
Wirsen, C. O., and Jannasch, H. W. (1978). Physiological and morphological observations on thiovulum sp. J. Bacteriol. 136 (2), 765–774. doi:10.1128/jb.136.2.765-774.1978
Wu, B., Liu, F., Fang, W., Yang, T., Chen, G., He, Z., et al. (2021). Microbial sulfur metabolism and environmental implications. Sci. Total Environ. 778, 146085. doi:10.1016/j.scitotenv.2021.146085
Wu, M., Li, J., Leu, A. O., Erler, D. V., Stark, T., Tyson, G. W., et al. (2022). Anaerobic oxidation of propane coupled to nitrate reduction by a lineage within the class Symbiobacteriia. Nat. Commun. 13, 6115. doi:10.1038/s41467-022-33872-y
Wu, Y., Simmons, B. A., and Singer, S. W. (2016). MaxBin 2.0: an automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics 32 (4), 605–607. doi:10.1093/bioinformatics/btv638
Wynn, J. G., Harden, J. W., and Fries, T. L. (2006). Stable carbon isotope depth profiles and soil organic carbon dynamics in the lower Mississippi Basin. Geoderma 131 (1-2), 89–109. doi:10.1016/j.geoderma.2005.03.005
Xu, Z., Li, Y., Lu, Y., Li, Y., Yuan, Z., Dai, M., et al. (2020). Impacts of the Zhe-Min Coastal Current on the biogeographic pattern of microbial eukaryotic communities. Prog. Oceanogr. 183, 102309. doi:10.1016/j.pocean.2020.102309
Zhang, L., Su, F., Zhang, C., Gong, F., and Liu, J. (2016). Changes of photosynthetic behaviors and photoprotection during cell transformation and astaxanthin accumulation in Haematococcus pluvialis grown outdoors in tubular photobioreactors. Int. J. Mol. Sci. 18 (1), 33. doi:10.3390/ijms18010033
Keywords: glaciovolcanism, glacier biogeochemistry, glacier caves, astrobiology, ocean worlds, subglacial microbiology
Citation: Clance JJ, Shaffer JMC, Cable ML, Stenner C, Williams-Jones G, Szynkiewicz A, Paton M, Graham K, Vinnes O and Mikucki JA (2024) Biogeochemistry of the rare sulfidic glaciovolcanic cave system on Mount Meager, British Columbia, Canada. Front. Geochem. 2:1410338. doi: 10.3389/fgeoc.2024.1410338
Received: 01 April 2024; Accepted: 29 July 2024;
Published: 21 August 2024.
Edited by:
William Berry Lyons, The Ohio State University, United StatesReviewed by:
Mohammed Abdul Rasheed, Gujarat Energy Research and Management Institute (GERMI), IndiaCopyright © 2024 Clance, Shaffer, Cable, Stenner, Williams-Jones, Szynkiewicz, Paton, Graham, Vinnes and Mikucki. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jill A. Mikucki, am1pa3Vja2lAdXRrLmVkdQ==
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.