- 1Department of Geological Sciences, University of Colorado Boulder, Boulder, CO, United States
- 2Department of Earth System Science, Stanford University, Stanford, CA, United States
- 3MARUM Center for Marine Environmental Sciences, University of Bremen, Bremen, Germany
Sphingolipids have long been of interest to the scientific community for their roles in eukaryotic cell structuring and disease pathology. Less is known about the occurrence and function of these diverse compounds in the bacterial domain of life, with most studies on bacterial sphingolipids focused on eukaryotic disease research and host-pathogen or host-symbiont interactions. Thus, bacterial contributions to environmental sphingolipid pools are poorly understood and the function of these lipids outside of pathogenicity remains largely unexplored. This report marks the first instance of sphingolipid production in a member of the phylum Acidobacteria, a globally ubiquitous phylum of soil bacteria. The occurrence of core- and intact-ceramides is reported for the Acidobacterium Solibacter usitatus under various environmentally relevant conditions. Shifts in the production of ceramides across temperature, pH, and oxygen gradients in this organism suggest that these compounds play a role in the physiological adaptation to environmental fluctuations. Additionally, the genetic basis of bacterial ceramide biosynthesis in this species is assessed and used to explore the potential for ceramide biosynthesis across the bacterial domain of life. The extent of the biosynthetic potential for Acidobacteria to produce ceramides coupled to the abundance of their genes in soil metagenomes suggests that soil sphingolipids should not be solely attributed to eukaryotic production.
1 Introduction
Sphingolipids are a diverse class of lipids characterized by their sphingoid bases, also referred to as long chain bases (LCBs), which are composed of a variety of aliphatic amino alcohols. Ceramides are a type of sphingolipid in which the LCB is N-acylated with any of a structurally diverse group of fatty acids. With vast structural varieties observed for both their LCBs and fatty acid components, ceramides are classically named after the combination of their LCBs and fatty acids. Ceramides are well documented components of eukaryotic cell membranes and structures (e.g., Harrison et al., 2018 and references therein for mammalian and fungal ceramides). While nearly ubiquitous in eukaryotes, the production of ceramides has only been reported in a few bacterial taxa to date (e.g., Harrison et al., 2018). Initially exclusive to the Sphingomonadaceae, bacterial sphingolipids have been found in members of the Bacteroidetes, Chlorobi, and other Proteobacteria over the past decade (An et al., 2011; Stankeviciute et al., 2019; Johnson et al., 2020; Couvillion et al., 2023).
Despite their significance in eukaryotes, the physiological role of sphingolipids in bacterial membranes is not well understood. While various studies have investigated the role of bacterial sphingolipids in the context of eukaryotic pathogenicity (Heaver et al., 2018), far less is known about the role of these compounds in environmental systems. This knowledge gap is in part a consequence of the previously unresolved biosynthetic pathway of ceramides in bacteria. Recently, the bacterial ceramide biosynthetic pathway was partially predicted (Olea-Ozuna et al., 2021) and independently confirmed (Stankeviciute et al., 2022) in the Proteobacterium C. crescentus. The bacterial pathway consists of three essential steps involving the condensation of L-serine and fatty-acyl CoA (Serine Palmitoyl Transferase, Spt) and subsequent addition of a second acyl chain (Bacterial Ceramide Synthase, bCerS) and reduction of the LCB (Bacterial Ceramide Reductase, CerR) (Figure 1) (Stankeviciute et al., 2022). While a combination of these three steps is necessary for producing a bacterial ceramide, other structural modifications to bacterial ceramides have been observed. Bacterial ceramide hydroxylase (CerH) was demonstrated to facilitate the addition of a hydroxyl group at the C2 position of the acyl chain in a ceramide from Caulobacter crescentus (Figure 1) (Stankeviciute et al., 2022).
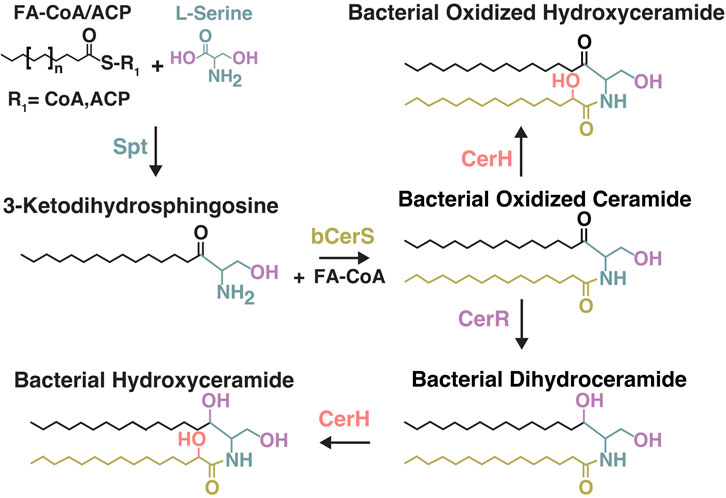
Figure 1. Overview of the bacterial ceramide biosynthesis pathway from Stankeviciute et al., 2022. Formation of 3-Ketodihydrosphingosine (Serine Palmitoyl Transferase, Spt). Formation of Bacterial Oxidized Ceramides (Bacterial Ceramide Synthase, bCerS). Formation of Bacterial Dihydroceramides ((Bacterial Ceramide Reductase, CerR)). Formation of Bacterial Hydroxy-Ceramides (Bacterial ceramide hydroxylase, CerH). Spt can use either acyl-CoA or acyl-ACP (acyl carrier protein) as a substrate. CerH was shown to use either oxidized- or dihydro-ceramides as substrates in C. crescentus.
In addition to reporting the enzymatic basis for bacterial ceramide biosynthesis, Stankeviciute et al. (2022) conducted bioinformatic analyses of the genetic potential for bacterial ceramide biosynthesis, predicting bacterial ceramide production in environmental niches such as soil and aquatic ecosystems. Ceramides have been widely reported as components of soils, although their production is often attributed to eukaryotic members of these ecosystems due to their abundance in plants (e.g., Markham et al., 2013) and occurrence in fungi (Harrison et al., 2018). While little is known about sphingolipid production by soil bacteria, the work presented here documents the production of ceramides by the Acidobacterium Solibacter usitatus, a bacterium isolated from a temperate, Australian pasture soil (Joseph et al., 2003). The occurrence of ceramides has not been previously reported within the Acidobacteria, a bacterial phylum that is an abundant and diverse component of global soil environments. Acidobacteria represent on average 20% of the microbial communities of global soils and are metabolically diverse heterotrophs (Eichorst et al., 2018). This work aims to explore the occurrence of ceramides in the bacterium Solibacter usitatus in the context of 1.) The physiological role of sphingolipids in soil bacteria and 2.) The unknown contribution of bacterial sphingolipids to soil ecosystems.
2 Materials and methods
2.1 Microbial culturing
Solibacter usitatus strain Ellin6076 (Joseph et al., 2003; Ward et al., 2009) was grown in triplicate in a modified DSMZ 1266 medium (see Halamka et al., 2023) at different temperature (15°C, 20°C, 25°C, 30°C), pH (5.5, 6.5), and oxygen (5%, 10%, 21% O2) conditions (Table 1). In brief, modifications to the DSMZ 1266 medium included the addition of 0.67 g/L yeast extract (YE) and 2.5 mM glucose (for full list of modifications, see Halamka et al., 2023). Media pH was buffered by MES (pKa 6.15) and adjusted with 5M NaOH to the reported pH values. One experimental condition was conducted with excess phosphate (10 mM) added to the base medium (0.2 mM). All aerobic culture experiments were conducted in standard yellow-capped 25-mL culture tubes (18 mm diameter) with 10 mL of media shaken at 250 rotations per minute (rpm) in atmosphere (21% O2). Suboxic culturing experiments (5% O2, 10% O2) were conducted in 100-mL media bottles with 60 mL of media and gasket-sealed screw-cap lids. Suboxic headspace was maintained by continuously flushing the culture headspace through gas-impermeable 1/8” PTFE tubing connected to in/out ports with standard ¼-28 liquid chromatography compression fittings at a rate of 100 mL/min with high purity N2 blended with compressed air using digital mass flow controllers (Alicat Scientific, MC-Series). All suboxic cultures were stirred continuously with a magnetic stir bar at 625 rpm to ensure gas equilibration between headspace and media. Cultures were harvested in stationary phase by centrifugation (5,000 rpm for 3 min), followed by freeze-drying prior to lipid extraction.
2.2 Lipid extraction
Before intact polar lipid (IPL) extraction, freeze-dried cell pellets were subjected to a pre-treatment procedure involving the freeze-thaw cycling of biomass in hexadecyltrimethylammonium bromide (CTAB). This technique has previously been reported by Evans et al. (2022) to increase the yields of extracted IPLs from archaeal biomass. The CTAB pre-treatment and subsequent lipid extraction technique described in Evans et al. (2022) was followed without modification for freeze dried cell pellets of S. usitatus. In brief, 1 g of CTAB was dissolved in 100 mL of MilliQ (MQ) H2O. Once fully dissolved, 3 mL of the CTAB solution was added to the freeze-dried cell pellets in organic clean Teflon tubes. Sample tubes were immediately transferred to a 50°C incubator for 20 min. After 20 min at 50°C, sample tubes were transferred to a −70°C freezer for 30 min. This process was repeated for a total of three freeze-thaw cycles before freeze drying overnight. After freeze-drying, the samples underwent an IPL extraction procedure modified from Bligh and Dyer (1959). Samples were extracted via the addition of 15 mL of buffer solution followed by centrifugation and addition of the subsequent supernatant to glass separatory funnels (this process conducted with a phosphate buffer (2x) followed by trichloroacetic acid (TCA) buffer (2x)). Following the buffer extractions, 15 mL of dichloromethane (DCM) was added to the funnels to induce phase separation of the organic layer. This process was repeated an additional time with DCM followed by a final addition of 15 mL of MQ H2O. The collected total lipid extract (TLE) was then evaporated under a stream of N2 gas for transfer and storage for subsequent analyses.
2.3 Lipid analysis
Total lipid extracts (TLEs) were analyzed on a high-performance liquid chromatography mass spectrometer (HPLC-MS) instrument consisting of a Dionex UltiMate 3000 UHPLC (Ultra High Performance Liquid Chromatography) and a Bruker maXis ultra-high resolution orthogonal acceleration quadrupole-time-of-flight (qTOF) mass spectrometer equipped with an electrospray ionization (ESI) source and operated in positive ion mode (Bruker Daltonik, Bremen, Germany). Chromatographic separation of IPLs and core lipids was achieved using the reverse phase (RP) method described in Zhu et al. (2013). IPLs were subsequently identified based on data dependent MS-MS (ddMS2) fragmentation patterns (e.g., Sturt et al., 2004; Schubotz et al., 2018).
2.4 Proteomic analyses
3 mL of cell culture was spun down at 17,000x g for 1 minute, the supernatant was removed, and the pellets were frozen until proteomic analysis. Protein concentrations were determined using tryptophan fluorescence (Wiśniewski and Gaugaz, 2015). Protein sample preparation, trypsin digest, and label-free quantitative proteomics were performed as described in Bassett et al. (2022). Briefly, after digestion, tryptic peptide samples were separated with an acetonitrile gradient from 2% to 20% on a reverse-phase C18 1.7 μm 130 Å, 75 mm x 250 mm M-class column (Waters) using an Ultimate 3000 UPLC and analyzed using a Q-Exactive HF-X mass spectrometer (Bassett et al., 2022). MaxQuant (Tyanova et al., 2016) was used to search the raw data against all proteins of S. usitatus strain Ellin6076 available on Uniprot (https://www.uniprot.org/taxonomy/234267). All peptides and proteins were thresholded at a 1% false discovery rate and intensity-based absolute quantitation values (iBAQs) were used to calculate relative protein abundances and abundance percentiles.
2.5 Phylogenetic analyses
Acidobacteria genomes in the Joint Genome Institute (JGI) and National Center for Biotechnology Information (NCBI) databases were searched for the presence of bCerS proteins using ACID_5449 as the BLAST query. Sequences with an e-value ≤ 1e-60 and a length = 350–500 amino acids were classified as bCerS proteins. JGI grassland, peat, and tundra metagenomes were searched for bCerS proteins with the same parameters. To assess the taxonomic affiliation of the metagenomic bCerS sequences, a phylogenetic tree was constructed containing: 1) the metagenomic bCerS sequences, 2) the Acidobacteria bCerS sequences, and 3) non-Acidobacterial bCerS sequences obtained from the NCBI non-redundant protein database (these sequences were retrieved by using ACID_5449 as the query with the same parameters as before except all genomes in the database were searched for bCerS proteins and then clustered at 70% sequence identity using CD-hit) (Fu et al., 2012). The sequences were aligned using MAFFT on XSEDE (7.505) with default parameters (BLOSUM matrix) (Katoh and Toh, 2010). The alignment was used to construct an initial phylogenetic tree with FastTreeMP on XSEDE (2.1.10) with default parameters (JTT+CAT substitution model) (Price et al., 2010). The tree was then visualized with the Interactive Tree of Life (iTOL) (Letunic and Bork, 2021). Metagenomic bCerS sequences clustering with Acidobacteria bCerS proteins were identified and selected for further analysis. A final phylogenetic tree was constructed containing the 1) metagenomic bCerS sequences clustering with Acidobacteria, 2) Acidobacteria bCerS sequences, and 3) non-Acidobacteria bCerS sequences. The sequences were aligned using MAFFT as before and a phylogenetic tree was constructed with the alignment using IQtree (2.2.0) with ModelFinder (best fit model chosen: Q. pfam+R10) and with 1,000 ultrafast bootstraps (Kalyaanamoorthy et al., 2017; Hoang et al., 2018; Minh et al., 2020). The resultant tree was visualized with iTOL.
3 Results
3.1 Identification of sphingolipids
The polar lipid extracts of S. usitatus revealed a diversity of glycerol- and sphingo-lipids. Previous studies have shown that S. usitatus is a bacterium capable of producing both ester and ether bound glycerol lipids (Sinninghe Damsté et al., 2018; Halamka et al., 2023; Chen et al., 2022), including a series of bacterial branched glycerol dialkyl glycerol tetraethers (brGDGTs) (Halamka et al., 2023, Chen et al., 2022). While the glycerol lipid and fatty acid contents of this organism have been discussed in previous works, sphingolipid production has not been detailed within this species. Thus, the occurrence of both core and intact ceramides in the polar lipid profile of S. usitatus prompted further investigation into the structural composition of sphingolipids in this organism.
Two varieties of core ceramides were identified, with the most abundant being ceramide NDS (non-hydroxy fatty acid dihydrosphingosine ceramides) (Figure 2A). NDS ceramides are the same configuration as the structure referred to as bacterial dihydroceramide shown in Figure 1C. The second variety of core ceramide identified in S. usitatus is referred to as Cer-HHDS in this text. The MS2 analysis of this compound indicates that the structure of Cer-HHDS contains a hydroxylated fatty acid component and a hydroxylated dihydrosphingosine LCB (Figure 3). The positions of the additional hydroxylations on the fatty acid and LCB could not be confirmed from the available mass spectra, therefore a traditional ceramide naming convention was not applied to this structure.
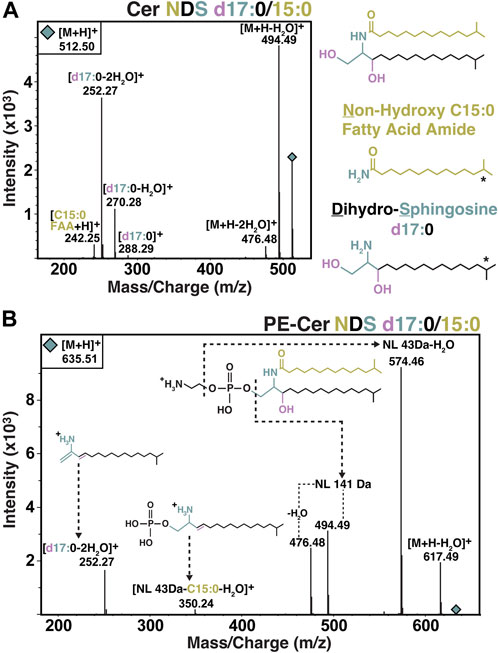
Figure 2. MS2 spectra and structural overview of Cer NDS and PE-Cer NDS observed in S. usitatus. MS-MS fragmentation and structure of Cer NDS d17:0/15:0 shown in panel (A). MS-MS fragmentation and structure of PE-Cer NDS d17:0/15:0 with neutral losses (NL) and key fragments shown in panel (B). Following conventional ceramide nomenclature, the prefix d (e.g., d17:0) refers to the number (di-) of hydroxyl groups on the LCB. * HPLC-MS-MS analysis did not determine the iso-branching of the displayed LCB and fatty acid. See discussion for further detail.
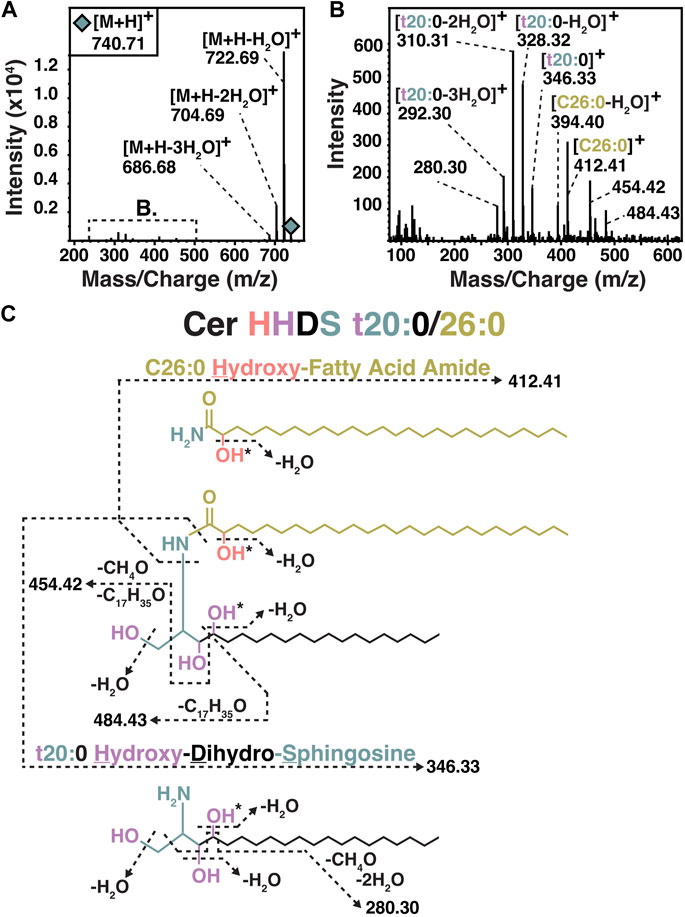
Figure 3. MS-MS (A, B) and structural overview (C) of Cer HHDS observed in S. usitatus. Following conventional ceramide nomenclature, the prefix t (e.g., t20:0) refers to the number (tri-) of hydroxyl groups on the LCB. * Position of hydroxyl group not confirmed by MS-MS.
The overall IPL composition of S. usitatus, including both glycerol lipids and sphingolipids, comprised four major polar headgroups: phosphohexose (PH), phosphatidylethanolamine (PE), phosphatidylcholine (PC), and di-phosphatidylglycerol (DPG, also referred to as cardiolipin). Of these main polar lipid classes, only PE head groups were detected in association with NDS ceramides (Figure 2B). No polar head groups were detected for Cer-HHDS.
3.2 Biosynthesis genes and protein abundances
Following the detection of sphingolipids in the lipid profile of S. usitatus, the genome was assessed for any of the known ceramide biosynthesis pathways. Homologs for the key genes involved in eukaryotic ceramide biosynthesis were not detected (data not shown) with the exception of Serine Palmitoyltransferase (Spt), which is the only shared step between the eukaryotic and bacterial pathways (Ikushiro et al., 2001; Yard et al., 2007; Stankeviciute et al., 2022). Following the exclusion of any known eukaryotic mechanisms for the biosynthesis of ceramides in S. usitatus, the genome was analyzed for homology to the bacterial ceramide biosynthesis pathway described from C. crescentus (Stankeviciute et al., 2022) (Figure 4A–D) (Supplementary Figure S1).
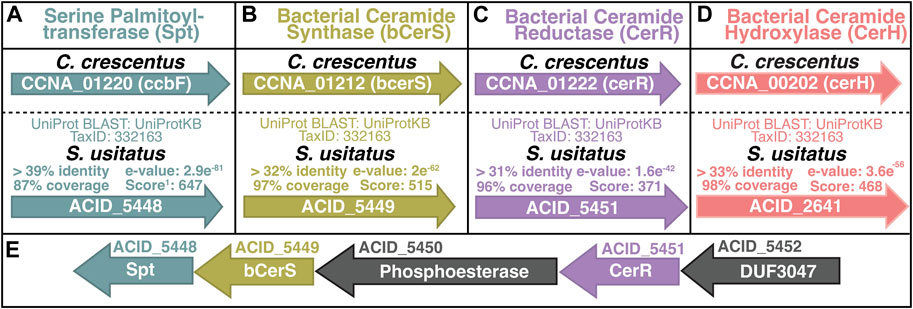
Figure 4. Overview of Solibacter usitatus homologs of bacterial ceramide biosynthesis proteins from Caulobacter crescentus (A–D) (Stankeviciute et al., 2022). Genes are represented by arrows inscribed with gene loci. (E) Predicted biosynthetic operon containing Spt, bCerS, and CerR. Gene loci ACID_XXXX denoted above arrows, predicted enzyme names listed inside each arrow. DUF - Domain of Unknown Function. Genes are color-coded for clarity following the scheme introduced in Figure 1.
The proposed homologs of the proteins encoded by spt (gene ACID_5448), bcerS (gene ACID_5449), and cerR (gene ACID_5451) in S. usitatus occur in the same predicted biosynthetic operon, further suggesting a shared functional relationship between these three homologs (Price et al., 2005) (Figure 4E). Protein BLAST results for CerH homologs in S. usitatus identified protein Q024E6 (gene ACID_2641), with the same annotation of fatty acid desaturase activity as CerH (Supplementary Figure S1). However, the gene ACID_2641 does not occur in the same predicted operon as Spt, bCerS, and CerR in S. usitatus and no further evidence of its function was found. Proteomic data generated from a subset of the culturing experiments from this study (5 samples in total) indicate average to above average expression levels for spt, bcerS, and cerR whereas the protein predicted by ACID_2641 was not detected (Figure 5).
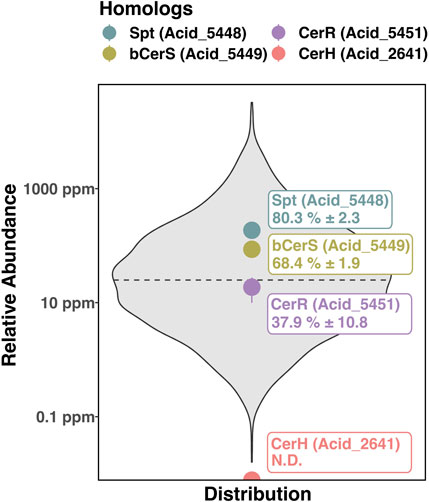
Figure 5. Averaged protein abundance data from 5 samples across the temperature and oxygen experiments of S. usitatus discussed in this study. Relative abundances of the ceramide biosynthesis enzymes are plotted relative to the distribution of the averaged proteomes. Spt, bCerS, CerR, and CerH are indicated by colored circles and labeled with their respective abundance percentiles. Dashed line represents the 50th percentile of relative protein abundance. Reported errors indicate 1 standard deviation of the 5 samples and demonstrate the relatively consistent expression of these proteins across experimental conditions. N.D. – Not detected (CerH).
3.3 Ceramide response to temperature, pH, and oxygen limitation
The most abundant PE ceramide detected across all analyzed temperature, pH, and oxygen culturing conditions was PE-Cer NDS d17:0/15:0 ([M+H]+ = 635.51, PE-Cer m/z 635). To compare the production of PE-Cer m/z 635 across all tested conditions, the response units of the compound were normalized for each sample by the amount of biomass extracted. Averages of biological triplicates were calculated for each temperature, oxygen, pH, and phosphate concentration (Figure 6). PE ceramides other than PE-Cer m/z 635 were detected in S. usitatus, but at intensities that were at least one order of magnitude lower than PE-Cer m/z 635 irrespective of culturing condition (Figure 7). Signal intensities for the minor PE ceramides in the excess phosphate experiment were too low for reliable quantification.
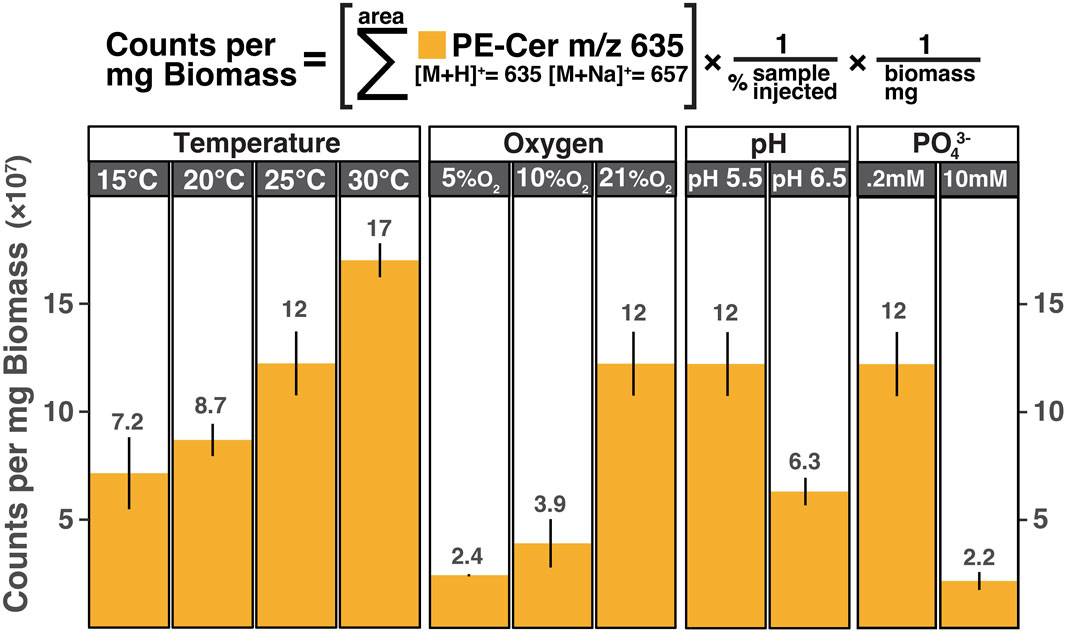
Figure 6. PE-Cer NDS d17:0/15:0 distributions from biological triplicates of S. usitatus. All temperature experiments were conducted at 21% O2 and pH 5.5. All oxygen experiments were conducted at pH 5.5 and 25°C. Both pH experiments were conducted at 21% O2 and 25°C. The 10 mM phosphate addition experiment was conducted at 25°C, 21% O2, and pH 5.5 and compared above to the corresponding temperature/oxygen/pH experiment that contained 0.2 mM phosphate from the base medium.
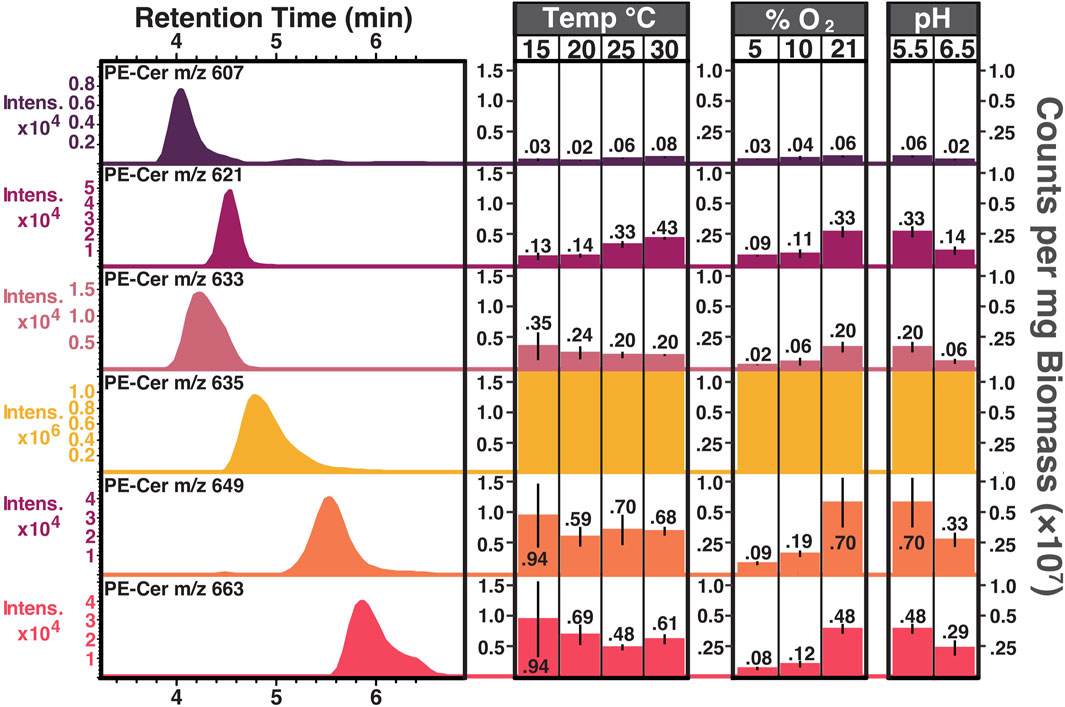
Figure 7. PE ceramide chromatography and counts per mg biomass distributions from biological triplicates of S. usitatus. Extracted ion chromatograms (EICs) of PE ceramides (left) shown with retention times (minutes, x-axis) and signal intensities (y-axis, note that intensity scales vary between chromatograms). EIC values are the same as m/z value listed for each PE ceramide. Amounts (right) calculated for each PE ceramide across various culturing conditions. Note that the y-axis scale (counts per mg biomass) is constant for each PE ceramide and the values for PE-Cer m/z 635 all exceed the upper scale limit (see Fig, 6 for PE-Cer m/z 635 amounts) All temperature experiments were conducted at 21% O2 and pH 5.5. Both pH experiments were conducted at 21% O2 and 25°C. All oxygen experiments were conducted at pH 5.5 and 25°C. PE-cers abundances (other than m/z 635) were too low for quantification in the phosphate experiments.
3.4 Phylogenetic assessment of ceramide biosynthesis potential in acidobacteria
To further elucidate the extent of Acidobacterial ceramide production in global soils, genomes available for the phylum from JGI and NCBI were assessed for similarity to the bCerS homolog from S. usitatus (ACID_5449). Genomes were assessed for the occurrence of bCerS due to the role of this enzyme as the first committed step in bacterial Cer-NDS biosynthesis. Homology to Spt alone is not a robust indicator of ceramide biosynthesis potential due to its high sequence similarity to synthases of unrelated biosynthetic pathways (Stankeviciute et al., 2022). BLAST results identified 359 bCerS sequences belonging to various classes of Acidobacteria including: Blastocatellia (n = 36), Holophagae (n = 17), Terriglobia (n = 72), Thermoanaerobaculia (n=79), Vicinamibacteria (n = 37), and unclassified Acidobacteria (n = 118).
After establishing the ceramide biosynthetic potential of the phylum, the occurrence of Acidobacterial bCerS genes in the environment was explored. Soil metagenomes from JGI were queried for ACID_5449, yielding a total of 16,477 bCerS sequences from grasslands (n = 4,593), tundras (n = 447), and peats (n = 11,437). To further assess which of these environmental bCerS sequences could be attributed back to Acidobacteria, a gene phylogeny for bCerS was constructed from the NCBI non redundant protein database using bCerS sequences from non-Acidobacteria (n = 2,656) and Acidobacteria (n = 319). A total of 2,963 bCerS genes from the soil metagenomes clustered within the resultant clades of Acidobacterial bCerS proteins: 74 belonging to tundra, 187 to grasslands, and strikingly, 2,702 to peats (Figure 8).

Figure 8. bCerS gene tree. Clades of Acidobacterial bCerS proteins are shown in purple, and S. usitatus’ bCerS gene is indicated by a purple circle. Outer ring colors delineate the environment that metagenomic hits came from: pink indicates peat soil hits, orange indicates grassland soil hits, and blue indicates tundra soil hits. A purple outer ring color indicates that the sequence belongs to an Acidobacterial genome or MAG from NCBI or JGI. Bootstrap percentages ≥900/1,000 are shown with an open circle. Branch lengths are shown to scale. Tree is rooted arbitrarily for visualization.
4 Discussion
4.1 Ceramide structures in S. usitatus
The dominant species of ceramide structure detected in S. usitatus (found as both a core ceramide and functionalized P/PE ceramide) was ceramide NDS d17:0/15:0. Formation of the LCB component of this compound, d17:0, is presumably facilitated by the enzyme Spt using the substrates L-serine and a C15:0 fatty acyl component. Activity and substrate preferences for Spt have been described for a variety of eukaryotes and some bacteria, with varying acyl-CoA/ACP carbon chain lengths and amino acids other than L-serine (e.g., Ikushiro et al., 2001; Ikushiro et al., 2023). Although the carbon chain length preference of the Spt from S. usitatus cannot be determined without further analyses, the dominance of a d17:0 LCB aligns well with the most abundant fatty acid components of this organism including saturated C15 compounds (Sinninghe Damsté et al., 2018; Chen et al., 2022; Halamka et al., 2023). Both straight chain and iso-branched C15:0 fatty acids occur in S. usitatus, but the MS2 analyses performed for this study did not clarify which of these features occurred in the d17:0 LCB. The ceramide structures depicted in Figure 2 contain iso-branched LCBs and fatty acyl components due to average relative abundance of iC15:0 (11.0 ± 13.2) in S. usitatus being approximately double that of nC15:0 (5.3 ± 3.2) (Halamka et al., 2023).
While the enzymes required for Cer-NDS biosynthesis in bacteria are known (Spt, bCerS, CerR) and can be discussed in context for the production of the major ceramide species in S. usitatus, the occurrence of Cer-HHDS presents unknowns in the enzymatic pathway. The Cer-HHDS structure proposed in Figure 3 contains an additional hydroxyl group on both the LCB (t20:0) and fatty acyl (C26:0) components of the ceramide. Additionally hydroxylated ceramides are commonly occurring compounds in many plants and fungi. Enzymes responsible for many of the site-specific hydroxylations on the LCBs and acyl chains of eukaryotic ceramides have been characterized, in addition to the acyl chain modifying bacterial ceramide hydroxylase (CerH). At present, mechanisms for LCB hydroxylation in bacterial ceramides are unknown. Both the acyl chain and LCB positions of the additional hydroxylations in the Cer-HHDS of S. usitatus are unconfirmed. Due to these uncertainties, the genome of S. usitatus was assessed for the occurrence of homologs to the known acyl chain bacterial ceramide hydroxylase CerH and to any described LCB hydroxylases from eukaryotes.
4.1.1 Ceramide acyl chain hydroxylation
The enzyme CerH was reported to facilitate C-2 hydroxylation on the ceramide fatty acyl chain in C. crescentus (gene CCNA_00202), and while a homolog was proposed for S. usitatus (gene ACID_2641) the associated protein (Q024E6) was not detected in the proteome. If Q024E6 is performing C-2 hydroxylation in Cer-HHDS, the lack of its detection in the proteome could be due to the low relative abundance of the compound. However, despite the predicted similarities in the protein classifications of CerH and Q024E6 (Supplementary Figure S1) BLAST results using a fatty acid hydroxylase or desaturase as a query (such as CerH) have the potential to find homologs of genes encoding for similar lipid modification enzymes that are unrelated to sphingolipids. Without further work, it is not possible to determine whether the protein Q024E6 plays any role in ceramide modification in S. usitatus.
4.1.2 Ceramide LCB hydroxylation
A C-4 hydroxylation on the LCB component of ceramides is the defining feature of phytoceramides and phytosphingosines in eukaryotes. Mammalian phytoceramide biosynthesis (protein DES2) was first characterized in Mus musculus (Ternes et al., 2002; Omae et al., 2004). Fungal phytosphingosine biosynthesis (protein SUR2/SYR2) was described in Saccharomyces cerevisiae (Haak et al., 1997; Grilley et al., 1998; Bae et al., 2004). Plant phytosphingosine biosynthesis (protein Sbh1/Sbh2) was identified in Arabidopsis thaliana from its homology to SUR2/SYR2 in S. cerevisiae (Sperling et al., 2001). However, BLAST searches in S. usitatus yielded no homologs for the mammalian, fungal, or plant phyto-ceramide/sphingosine enzymes (data not shown).
4.2 Physiological role of sphingolipids in bacteria
Previous microbial culturing work has provided some insights into the physiological role of sphingolipids in bacteria. The bacterial phylum Bacteroidetes contains a variety of sphingolipid producing genera identified from mammalian gut and oral microbiomes (LaBach and White, 1969; Ogawa et al., 2010; Kato et al., 1995; Nichols et al., 2004). Studies within the Bacteroidetes have established the role of ceramides in mediating prostaglandin secretion in oral fibroblasts (Nichols et al., 2004) and signaling functions for survival in mammalian intestines (An et al., 2011). Sphingolipid producing members of the Bacteroidetes phylum are also found in soil ecosystems (e.g., Sphingobacterium), but the physiology of these organisms is understudied.
The IPL profiles from S. usitatus generated for this study demonstrate that the production of PE ceramides in this organism responds to shifting temperature, oxygen, pH, and phosphate concentrations. While multiple varieties of PE ceramides were detected in S. usitatus, PE-Cer m/z 635 was the dominant species and was at least one order of magnitude higher intensity than other PE ceramides regardless of culturing condition. For this reason, trends observed in PE-Cer m/z 635 can be generalized to overall ceramide production at any given culturing condition irrespective of trends observed within the individual minor PE ceramide species.
The highest production of PE-Cer m/z 635 across all tested conditions was at 30°C, pH 5.5, 21% O2, grown in the base medium. At the same pH, O2, and media conditions, PE-Cer m/z 635 production positively correlated with increasing culturing temperatures. Efforts to culture S. usitatus below 15°C and above 30°C resulted in little or zero cell growth, suggesting that the observed temperature trend applies across the growth range of this strain and that increased production of ceramides is a temperature adaptation mechanism. Evidence of bacterial ceramide production in response to elevated temperature stress has previously been reported for the acetic acid bacterium Acetobacter malorum and C. crescentus (Ogawa et al., 2010; Olea-Ozuna et al., 2021). The same study of A. malorum by Ogawa et al., 2010 also identified low pH stress as a trigger for ceramide production. Although only two pH conditions were tested in this study, a decrease in the production of PE-Cer m/z 635 was found when culturing pH was increased. Further pH conditions would need to be assessed to confirm a trend in ceramide production for S. usitatus, but the present results suggest higher production of ceramides at lower pH values.
While some culturing variables in this study have more than two experiments from which a trend can be extrapolated, it is important to note that physiological implications derived from trends in absolute abundances (e.g., counts/mg biomass) can have limitations. If a given culturing variable increases the production of other cellular components (thus increasing the biomass yield) while ceramide production remains constant, the resultant trend in abundance could be misinterpreted as a decrease in ceramide production despite the cellular concentration remaining constant. Taking this limitation into consideration is particularly important when considering results from the oxygen limited cultures of S. usitatus. Cellular clumping and an excess of extracellular materials is often observed in liquid cultures of S. usitatus grown at suboxic conditions. These observations suggest that additional cellular components, not observed in the fully oxygenated cultures of S. usitatus, could be contributing to the amount of biomass generated in the suboxic experiments. Thus, the reported decrease in counts/mg biomass of PE-Cer m/z 635 at 10% and 5% O2 may be reflective of the increase in cellular components other than ceramides rather than a physiological shift in ceramide production.
Interestingly, the lowest production of PE-Cer m/z 635 was observed for the culturing experiment that was given excess phosphate. Phosphate starvation was previously found to trigger the production of a novel glycosphingolipid in C. crescentus, which was attributed to a lack of available phosphate for the biosynthesis of phospho-glycerol or sphingolipids (Stankeviciute et al., 2019). Data from S. usitatus suggests that a reduction in phosphate availability correlates to an increase in the production of PE ceramides. However, neither of the experimental phosphate conditions under which S. usitatus was grown for this study would be considered phosphate-limited for this organism, suggesting that the availability of phosphate for the biosynthesis of phospholipids is not an important factor in this instance. While the mechanism underpinning the decrease in PE ceramides at 10 mM phosphate remains unclear, it should be noted that this concentration of phosphate represents a bioavailable phosphorus quantity that is not representative of most soils.
4.3 Bacterial sphingolipids in soils
Ceramides and other sphingolipid derivatives are commonly reported in soil lipidomes. The provenance of these soil-derived ceramides is often attributed to eukaryotic members of soil ecosystems without the consideration of bacterial inputs. The significance of whether this assumption is accurate depends in part on the absolute amount of soil sphingolipids derived from bacterial producers relative to eukaryotic producers. Although the proportion of total soil ceramides produced from bacterial sources cannot be assessed by the methods employed in this study, the findings presented here suggest that the Acidobacteria are an understudied source of soil sphingolipids. Genomic analyses of the Acidobacteria found that five different classes of the phylum (and 118 unclassified members) have the genetic potential for ceramide biosynthesis. Acidobacteria are the third most abundant bacterial phylum in global soils, further underscoring the importance of their contributions to global soil lipidomes (Delgado-Baquerizo et al., 2018).
While the Acidobacteria are a major component of soils, other bacterial phyla undoubtedly have the potential to contribute to soil sphingolipid pools. The phylum Bacteroidetes contains multiple sphingolipid producing genera, and though many of these genera are not associated with soils, the genus Sphingobacterium is predominantly found in and isolated from soils. Indeed, phylogenetic analyses identified the class containing the genus Sphingobacterium as possessing members with homologs for all three essential bacterial ceramide biosynthesis genes (Stankeviciute et al., 2022). These same analyses also identified a class of Acidobacteria, providing the first genomic evidence suggesting that Acidobacteria can produce ceramides. In addition to these recent biosynthetic insights, evidence suggesting the occurrence of ceramides in the phylum Acidobacteria was reported in a recent study investigating shifts in the environmental lipidomes of grassland soils during wetting and drying cycles (Couvillion et al., 2023). In this study, a close association was found between increases in cluster amplicon sequence variants of Acidobacteria and the increased occurrence of sphingolipids in dry soils. Without previous evidence of ceramide biosynthesis in Acidobacteria, Couvillion et al. discussed the underexplored potential of bacterial ceramide production and caution against the assumption that soil sphingolipids are eukaryotic biomarkers.
The distribution of bCerS within available genomes of the Acidobacteria provides compelling evidence for their contribution to soil ceramide pools, but is limited in terms of predicting the extent of these contributions in the environment. To address this disconnect from environmental data, the distribution of bCerS in soil metagenomes was surveyed. Of the over 16,000 bCerS sequences recovered from the environmental metagenomes, nearly one-fifth (n= 2,963, 17.9%) of these sequences were grouped to Acidobacterial bCerS gene clades. When these environmental bCerS sequences are grouped by their source environment, the proportion of Acidobacterial biosynthetic potential in the surveyed soil subtypes can be assessed. Only 4% of the bCerS sequences from grassland soils clustered with Acidobacteria, whereas 16.5% and 23.6% of the sequences for tundra and peat, respectively, clustered within Acidobacterial bCerS gene clades. This suggests that while Acidobacteria may not be significant drivers of bacterial sphingolipid production in grassland soils, they represent nearly a quarter of the genetic potential for bacterial ceramide biosynthesis in peats.
5 Conclusion
This study combines some of the first insights into the physiological role of ceramides in a cultured soil bacterium with a more broad investigation of how the phylum Acidobacteria may influence environmental sphingolipid pools. Additional physiology-based studies of Acidobacterial sphingolipid producers should be conducted to further constrain the role of ceramides in Acidobacteria and thus their subsequent role in soil ecosystems. Although the culturing results presented here come from only one member of the phylum, 84% of the environmental peat bCerS sequences associated with Acidobacteria were found in the same clade as the bCerS gene from S. usitatus. This suggests that investigations of ceramide biosynthesis and physiology in S. usitatus may be particularly relevant for understanding bacterial sphingolipid dynamics in peat soil environments.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
TH: Writing–original draft, Writing–review and editing, Conceptualization, Investigation. AG: Writing–original draft, Formal analysis. TE: Writing–original draft, Investigation. SS: Writing–original draft, Investigation. AY: Writing–original draft. K-UH: Writing–original draft. SK: Writing–original draft.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was supported by a European Association of Organic Geochemists Research Award to TH. Lipid analyses at the University of Bremen were supported through the DFG und Germany’s Excellence Strategy (no. EXC-2077-390741603). This research was supported by an NSF grant to SHK (EAR1945484) and by the University of Colorado Boulder via start-up funds.
Acknowledgments
We further thank the CU Boulder Organic Geochemistry Lab (OGL) and the CU Boulder Earth Systems Stable Isotope Lab (CUBES-SIL) Core Facility (RRID: SCR_019300) for the analytical infrastructure that enabled this work.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgeoc.2024.1400278/full#supplementary-material
References
An, D., Na, C., Bielawski, J., Hannun, Y. A., and Kasper, D. L. (2011). Membrane sphingolipids as essential molecular signals for Bacteroides survival in the intestine. Proc. Natl. Acad. Sci. 108 (1), 4666–4671. doi:10.1073/pnas.1001501107
Bassett, J., Rimel, J. K., Basu, S., Basnet, P., Luo, J., Engel, K. L., et al. (2022). Systematic mutagenesis of TFIIH subunit p52/Tfb2 identifies residues required for XPB/Ssl2 subunit function and genetic interactions with TFB6. J. Biol. Chem. 298 (10), 102433. doi:10.1016/j.jbc.2022.102433
Bae, J.-H., Sohn, J.-H., Park, C.-S., Rhee, J.-S., and Choi, E.-S. (2004). Cloning and functional characterization of the SUR2/SYR2 gene encoding sphinganine hydroxylase in Pichia ciferrii. Yeast 21, 437–443. doi:10.1002/yea.1082
Bligh, E. G., and Dyer, W. J. (1959). A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiology 37 (8), 911–917. doi:10.1139/o59-099
Chen, Y., Zheng, F., Yang, H., Yang, W., Wu, R., Liu, X., et al. (2022). The production of diverse brGDGTs by an Acidobacterium providing a physiological basis for paleoclimate proxies. Geochim. Cosmochim. Acta. 337, 155–165. doi:10.1016/j.gca.2022.08.033
Couvillion, S. P., Danczak, R. E., Naylor, D., Smith, M. L., Stratton, K. G., Paurus, V. L., et al. (2023). Rapid remodeling of the soil lipidome in response to a drying-rewetting event. Microbiome 11 (1), 34. doi:10.1186/s40168-022-01427-4
Delgado-Baquerizo, M., Oliverio, A. M., Brewer, T. E., Benavent-González, A., Eldridge, D. J., Bardgett, R. D., et al. (2018). A global atlas of the dominant bacteria found in soil. Sci. (New York, N.Y.) 359 (6373), 320–325. doi:10.1126/science.aap9516
Eichorst, S. A., Trojan, D., Roux, S., Herbold, C., Rattei, T., and Woebken, D. (2018). Genomic insights into the Acidobacteriareveal strategies for their success in terrestrial environments. Environ. Microbiol. 20, 1041–1063. doi:10.1111/1462-2920.14043
Evans, T. W., Elling, F. J., Li, Y., Pearson, A., and Summons, R. E. (2022). A new and improved protocol for extraction of intact polar membrane lipids from archaea. Org. Geochem. 165, 104353. doi:10.1016/j.orggeochem.2021.104353
Fu, L., Niu, B., Zhu, Z., Wu, S., and Li, W. (2012). CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 28 (24), 3150–3152. doi:10.1093/bioinformatics/bts565
Grilley, M. M., Stock, S. D., Dickson, R. C., Lester, R. L., and Takemoto, J. Y. (1998). Syringomycin action gene SYR2 is essential for sphingolipid 4-hydroxylation in Saccharomyces cerevisiae. J. Biol. Chem. 273 (18), 11062–11068. doi:10.1074/jbc.273.18.11062
Haak, D., Gable, K., Beeler, T., and Dunn, T. (1997). Hydroxylation of Saccharomyces cerevisiae ceramides requires Sur2p and Scs7p. J. Biol. Chem. 272 (47), 529704–529710. doi:10.1074/jbc.272.47.29704
Halamka, T. A., Raberg, J. H., McFarlin, J. M., Younkin, A. D., Mulligan, C., Liu, X.-L., et al. (2023). Production of diverse brGDGTs by Acidobacterium solibacter usitatus in response to temperature, PH, and O 2 provides a culturing perspective on Br GDGT proxies and biosynthesis. Geobiology 21 (1), 102–118. doi:10.1111/gbi.12525
Harrison, P. J., Dunn, T. M., and Campopiano, D. J. (2018). Sphingolipid biosynthesis in man and microbes. Nat. Product. Rep. 35 (9), 921–954. doi:10.1039/C8NP00019K
Heaver, S. L., Johnson, E. L., and Ley, R. E. (2018). Sphingolipids in host–microbial interactions. Curr. Opin. Microbiol. 43, 92–99. doi:10.1016/j.mib.2017.12.011
Hoang, D. T., Chernomor, O., von Haeseler, A., Minh, B. Q., and Vinh, L. S. (2018). UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35 (2), 518–522. doi:10.1093/molbev/msx281
Holmes, B., Owen, R. J., and Hollis, D. G. (1982). “Flavobacterium spiritivorum, a new species isolated from human clinical specimens,” in International journal of systematic and evolutionary microbiology (Microbiology Society). doi:10.1099/00207713-32-2-157
Ikushiro, H., Hayashi, H., and Kagamiyama, H. (2001). A water-soluble homodimeric serine Palmitoyltransferase fromSphingomonas paucimobilis EY2395T strain. J. Biol. Chem. 276 (21), 18249–18256. doi:10.1074/jbc.M101550200
Ikushiro, H., Murakami, T., Takahashi, A., Katayama, A., Sawai, T., Goto, H., et al. (2023). Structural insights into the substrate recognition of serine Palmitoyltransferase from Sphingobacterium multivorum. J. Biol. Chem. 299 (5), 104684. doi:10.1016/j.jbc.2023.104684
Johnson, E. L., Heaver, S. L., Waters, J. L., Kim, B. I., Bretin, A., Goodman, A. L., et al. (2020). Sphingolipids produced by gut bacteria enter host metabolic pathways impacting ceramide levels. Nat. Commun. 11 (1), 2471. doi:10.1038/s41467-020-16274-w
Joseph, S. J., Hugenholtz, P., Sangwan, P., Osborne, C. A., and Janssen, P. H. (2003). Laboratory cultivation of widespread and PreviouslyUncultured SoilBacteria. Appl. Environ. Microbiol. 69 (12), 7210–7215. doi:10.1128/AEM.69.12.7210-7215.2003
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A., and Jermiin, L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14 (6), 587–589. doi:10.1038/nmeth.4285
Kato, M., Muto, Y., Tanaka-Bandoh, K., Watanabe, K., and Ueno, K. (1995). Sphingolipid composition in Bacteroides species. Anaerobe 1 (2), 135–139. doi:10.1006/anae.1995.1009
Katoh, K., and Toh, H. (2010). Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics 26 (15), 1899–1900. doi:10.1093/bioinformatics/btq224
LaBach, J. P., and White, D. C. (1969). Identification of ceramide phosphorylethanolamine and ceramide phosphorylglycerol in the lipids of an anaerobic bacterium. J. Lipid Res. 10 (5), 528–534. doi:10.1016/s0022-2275(20)43045-7
Letunic, I., and Bork, P. (2021). Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49 (W1), W293–W296. doi:10.1093/nar/gkab301
Markham, J. E., Lynch, D. V., Napier, J. A., Dunn, T. M., and Cahoon, E. B. (2013). Plant sphingolipids: function follows form. Curr. Opin. Plant Biol. 16 (3), 350–357. doi:10.1016/j.pbi.2013.02.009
Minh, B. Q., Schmidt, H. A., Chernomor, O., Schrempf, D., Woodhams, M. D., von Haeseler, A., et al. (2020). IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37 (5), 1530–1534. doi:10.1093/molbev/msaa015
Naka, T., Fujiwara, N., Yano, I., Maeda, S., Doe, M., Minamino, M., et al. (2003). Structural analysis of sphingophospholipids derived from Sphingobacterium spiritivorum, the type species of genus Sphingobacterium. Biochimica Biophysica Acta (BBA) - Mol. Cell. Biol. Lipids 1635 (2), 83–92. doi:10.1016/j.bbalip.2003.10.010
Nichols, F. C., Riep, B., Mun, J. Y., Morton, M. D., Bojarski, M. T., Dewhirst, F. E., et al. (2004). Structures and biological activity of phosphorylated dihydroceramides of porphyromonas gingivalis. J. Lipid Res. 45 (12), 2317–2330. doi:10.1194/jlr.M400278-JLR200
Ogawa, S., Tachimoto, H., and Kaga, T. (2010). Elevation of ceramide in acetobacter malorum S24 by low PH stress and high temperature stress. J. Biosci. Bioeng. 109 (1), 32–36. doi:10.1016/j.jbiosc.2009.07.007
Olea-Ozuna, R. J., Poggio, S., Bergström, E., Quiroz-Rocha, E., García-Soriano, D. A., Sahonero-Canavesi, D. X., et al. (2021). Five structural genes required for ceramide synthesis in Caulobacter and for bacterial survival. Environ. Microbiol. 23, 143–159. doi:10.1111/1462-2920.15280
Omae, F., Miyazaki, M., Enomoto, A., Suzuki, M., Suzuki, Y., and Suzuki, A. (2004). DES2 protein is responsible for phytoceramide biosynthesis in the mouse small intestine. Biochem. J. 379 (Pt 3), 687–695. doi:10.1042/BJ20031425
Price, M. N., Dehal, P. S., and Arkin, A. P. (2010). FastTree 2 – approximately maximum-likelihood trees for large alignments. PLOS ONE 5 (3), e9490. doi:10.1371/journal.pone.0009490
Price, M. N., Huang, K. H., Alm, E. J., and Arkin, A. P. (2005). A novel method for accurate operon predictions in all sequenced prokaryotes. Nucleic Acids Res. 33 (3), 880–892. doi:10.1093/nar/gki232
Schubotz, F., Xie, S., Lipp, J. S., Hinrichs, K.-U., and Wakeham, S. G. (2018). Intact polar lipids in the water column of the eastern tropical north pacific: abundance and structural variety of non-phosphorus lipids. Biogeosciences 15 (21), 6481–6501. doi:10.5194/bg-15-6481-2018
Sinninghe Damsté, J. S., Rijpstra, W. I. C., Foesel, B. U., Huber, K. J., Overmann, J., Nakagawa, S., et al. (2018). An overview of the occurrence of ether-and ester-linked iso-diabolic acid membrane lipids in microbial cultures of the Acidobacteria: implications for brGDGT paleoproxies for temperature and pH. Org. Geochem. 124, 63–76. doi:10.1016/j.orgge.ochem.2018.07.006
Stankeviciute, G., Tang, P., Ashley, B., Chamberlain, J. D., Hansen, M. E. B., Coleman, A., et al. (2022). Convergent evolution of bacterial ceramide synthesis. Nat. Chem. Biol. 18 (3), 305–312. doi:10.1038/s41589-021-00948-7
Stankeviciute, G., Ziqiang, G., Goldfine, H., and Klein, E. A. (2019). Caulobacter crescentus adapts to phosphate starvation by synthesizing anionic glycoglycerolipids and a novel glycosphingolipid. MBio 10 (2), e00107-e00119. doi:10.1128/mBio.00107-19
Sturt, H. F., Summons, R. E., Smith, K., Elvert, M., and Hinrichs, K.-U. (2004). Intact polar membrane lipids in prokaryotes and sediments deciphered by high-performance liquid chromatography/electrospray ionization multistage mass spectrometry—new biomarkers for biogeochemistry and microbial ecology. Rapid Commun. Mass Spectrom. 18 (6), 617–628. doi:10.1002/rcm.1378
Sperling, P., Ternes, P., Moll, H., Franke, S., Zähringer, U., and Heinz, E. (2001). Functional characterization of sphingolipid C4-hydroxylase genes from Arabidopsis thaliana. FEBS Lett. 494(1–2), 90–94. doi:10.1016/s0014-5793(01)02332-8
Ternes, P., Franke, S., Zähringer, U., Sperling, P., and Heinz, E. (2002). Identification and characterization of a sphingolipid delta 4-desaturase family. J. Biol. Chem. 277 (28), 25512–25518. doi:10.1074/jbc.M202947200
Tyanova, S., Temu, T., and Cox, T. (2016). The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 11, 2301–2319. doi:10.1038/nprot.2016.136
Ward, N. L., Challacombe, J. F., Janssen, P. H., Henrissat, B., Coutinho, P. M., Wu, M., et al. (2009). Three genomes from the phylum Acidobacteria provide insight into the lifestyles of these microorganisms in soils. Appl. Environ. Microbiol. 75 (7), 2046–2056. doi:10.1128/AEM.02294-08
Wiśniewski, J. R., and Gaugaz, F. Z. (2015). Fast and sensitive total protein and peptide assays for proteomic analysis. Anal. Chem. 87 (8), 4110–4116. doi:10.1021/ac504689z
Yard, B. A., Carter, L. G., Johnson, K. A., Overton, I. M., Dorward, M., Liu, H., et al. (2007). The structure of serine Palmitoyltransferase; gateway to sphingolipid biosynthesis. J. Mol. Biol. 370 (5), 870–886. doi:10.1016/j.jmb.2007.04.086
Keywords: ceramides, acidobacteria, sphingolipids, soil bacteria, biosynthesis
Citation: Halamka TA, Garcia A, Evans TW, Schubert S, Younkin A, Hinrichs K-U and Kopf S (2024) Occurrence of ceramides in the Acidobacterium Solibacter usitatus: implications for bacterial physiology and sphingolipids in soils. Front. Geochem. 2:1400278. doi: 10.3389/fgeoc.2024.1400278
Received: 13 March 2024; Accepted: 10 June 2024;
Published: 10 July 2024.
Edited by:
Manuel Miguel Jordán, Miguel Hernández University of Elche, SpainReviewed by:
Sabine Mehay, Schlumberger, GermanyChristian Sohlenkamp, National Autonomous University of Mexico, Mexico
Copyright © 2024 Halamka, Garcia, Evans, Schubert, Younkin, Hinrichs and Kopf. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Toby A. Halamka, dG9ieS5oYWxhbWthQGJyaXN0b2wuYWMudWs=
†Present address: Thomas W. Evans, Shell Global Solutions International B.V., Amsterdam, Netherlands
 Toby A. Halamka
Toby A. Halamka Andy Garcia2
Andy Garcia2 Thomas W. Evans
Thomas W. Evans Kai-Uwe Hinrichs
Kai-Uwe Hinrichs Sebastian Kopf
Sebastian Kopf