- 1Institute of Cardiovascular Sciences, Guangxi Academy of Medical Sciences, Nanning, China
- 2Department of Blood Transfusion, The People’s Hospital of Guangxi Zhuang Autonomous Region, Nanning, China
- 3Department of Radiation Oncology, Renji Hospital, School of Medicine, Shanghai Jiao Tong University, Shanghai, China
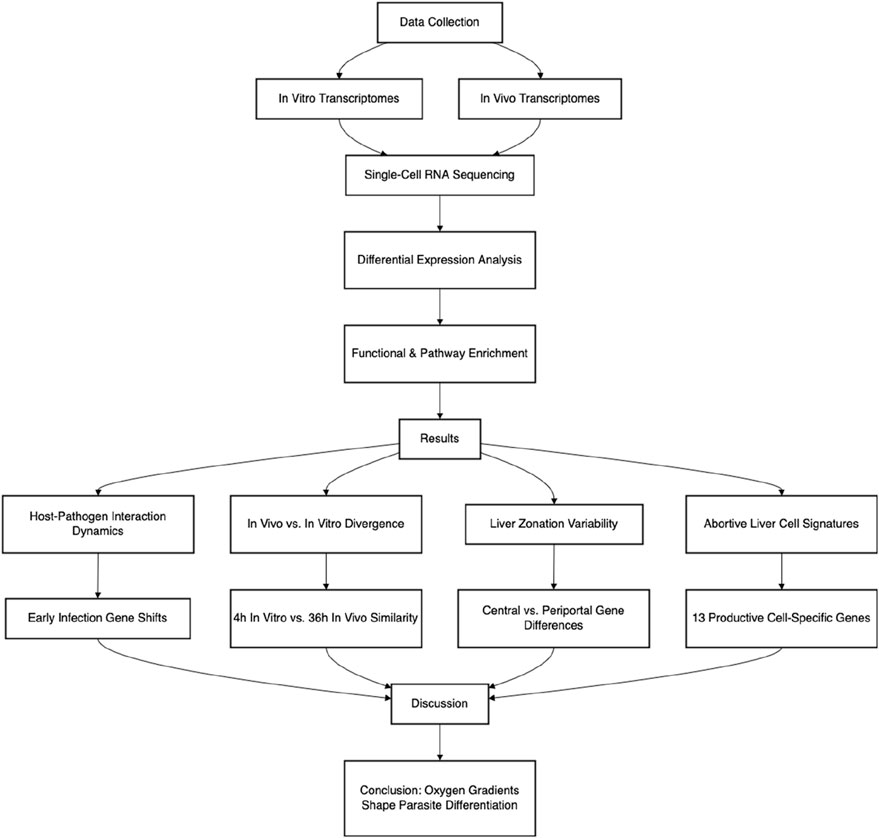
Intrahepatocytic development is a key stage in human Plasmodium infection, in which sporozoites replicate and transform into merozoites. Due to technological limitations, however, previous gene expression studies on malaria parasite liver infection were mostly conducted in vitro. In order to bridge these gaps, our current study compared the gene expression of in vitro-infected parasites at different time points with that of in vivo-infected parasites and revealed distinct patterns between parasite subpopulations in vitro and in vivo. A joint investigation of the Plasmodium transcriptome and their host transcriptome was carried out to confer a comprehensive analysis of gene expression in the liver stage of Plasmodium infection in vivo, which is similar to the disease setting, and therefore deepen our understanding of parasite and host transcriptional dynamics during intrahepatocytic infection.
1 Introduction
Malaria is a vector-borne infectious disease caused by Plasmodium parasites that have complex life cycles with various life stages in mosquito vectors and human hosts. Key stages in the human host include the liver stage and the intraerythrocytic development (IDE) period (Vaughan and Kappe, 2017). After transmission by mosquitoes, sporozoites enter the bloodstream and migrate to the liver. Within liver cells, sporozoites transform into merozoites, replicate extensively, and release these merozoites into the bloodstream (Lopes da Silva et al., 2012). In the liver stage, Plasmodium employ various mechanisms to escape the host immune reactions, taking advantage of the immunoprivileged nature of liver cells, which limits immune surveillance (Little et al., 2021; Real et al., 2021). Parasites modulate host cell signals, suppressing the activation of immune responses, including interference with interferon signaling and other crucial immune activation pathways (Liehl et al., 2014; Inacio et al., 2015; Ribot et al., 2019). Parasite proteins may also disrupt antigen presentation pathways, reducing immune cell visibility of infected liver cells (Hodgson et al., 2019; Mancio-Silva et al., 2022). Parasites strive to minimize antigen expression recognizable by the immune system, evading detection (Toro-Moreno et al., 2020; Hirako et al., 2022). Understanding these evasion mechanisms is crucial for developing strategies targeting the liver stage and preventing infection. Therefore, it is important not only to study the transformation of Plasmodium in the liver stage but also to know the ways in which liver cells respond to the parasites.
Understanding the complex life cycle of Plasmodium, especially its liver stage, is crucial for developing effective preventive and therapeutic strategies. With the advent of molecular biology techniques, gene expression studies on the Plasmodium liver stage emerged. Although these studies provided valuable information about the heterogeneity of host cells during infection, they often focused solely on the host side and did not jointly investigate the transcriptomes of both the parasite and the host in a comprehensive manner. In addition, as most of the previous molecular mechanism studies were based on the in vitro infection of the parasite because of its availability, it is important to integrate the in vitro information into the emerging in vivo studies (Howick et al., 2019).
Recent studies using the combination of single-cell sequencing and dual RNA sequencing (RNA-seq) allowed the investigation of in vivo stages (Afriat et al., 2022; Yang et al., 2023; Zanghi et al., 2023). In order to bridge these gaps, we jointly investigated the transcriptome profiles of intrahepatocytic Plasmodium and their host hepatocytes by using deposited RNA-seq data and single-cell sequencing data. To map these recent results to the vast amount of information from previous studies, we established a method based on gene set enrichment analysis (GSEA) and compared the gene expression of in vitro-infected parasites at different time points with that of in vivo-infected parasites, which revealed distinct patterns. This approach allows us to gain a more comprehensive understanding of the infection dynamics and gene expression patterns during the Plasmodium liver stage. It can potentially uncover novel molecular targets for anti-malarial drugs and provide new insights into the complex interactions between the parasite and the host at the molecular level.
2 Methods
2.1 Data sources
The bulk transcriptome data were derived from the previous datasets (Toro-Moreno et al., 2020; Caldelari et al., 2019; LaMonte et al., 2019), comprising multiple Plasmodium datasets collected at various times after infection in vitro. Furthermore, data for single-cell transcriptional dynamics during different phases of Plasmodium in vitro were sourced via the interactive Malaria Cell Atlas website (www.sanger.ac.uk/science/tools/mca/mca/). Single-cell transcriptome information on Plasmodium and their liver cell host at different infection times was collected from GEO databases (GSE181725).
2.2 Single-cell RNA sequencing (scRNA-seq)
The Seurat (version 4.2) package was used to analyze the scRNA-seq data. Cells with less than 300 or with more than 20% mitochondrial genes were excluded. After filtering, the data were normalized using the LogNormalize method. Uniform Manifold Approximation and Projection (UMAP) was established utilizing the primary components. The cluster analysis of single-cell data was performed using the graph-based clustering method in Seurat. The resolution of the FindClusters feature was set to 0.1.
2.3 Differential expression analyses
Differential expression analyses between the two groups were conducted using the limma package, and the filter condition for differentially expressed genes (DEGs) was set to the adjusted p-value < 0.05. The heatmap plot was built to visualize DEGs using the heatmap package. For the generation of volcano plots, we used ggplot2 in the R software package.
Subsequently, triwise plots and rose diagrams were generated with the log-transformed data using the triwise R software package (van de Laar et al., 2016).
2.4 Functional and pathway enrichment analyses
The clusterProfiler package served to target genes for functional and pathway enrichment analyses (Yu et al., 2012). Gene set enrichment analysis (www.gsea-msigdb.org/gsea/index.jsp) was executed for the respective DEGs (transcriptome-defined) to investigate whether these genes were enriched in the compared gene profiles of the Plasmodium or host transcriptome. In GSEA, we first ranked all the genes in our dataset based on their differential expression fold change. Then we tested whether the genes in a pre-defined gene set were preferentially located at the top or bottom of this ranked list. The software calculates an enrichment score (ES) for each gene set. A positive ES indicates that the genes in the gene set are upregulated under a particular condition, whereas a negative ES indicates downregulation.
The enrichment levels of the gene signatures determined from 4-h infection by Plasmodium berghei were then used to perform hierarchical clustering. Finally, the samples were divided into two directional groups based on the requirement of different time points or in vitro/in vivo comparison.
Furthermore, Gene Ontology (GO) functional enrichment analyses were performed. The results were visualized using the ggplot2 R software package.
3 Results
3.1 Host–pathogen interaction dynamics
When comparing the gene expression profile of Plasmodium in three major states before and after malaria parasite liver infection, distinct differences were observed. We identified 713 DEGs between sporozoites and Plasmodium berghei that infected liver cells, with a fold change of more than 10 and all having an adjusted p-value < 0.001. The roseplot in Figure 1A indicates the major upregulation of Plasmodium genes toward the direction of the intrahepatocytic phase when compared with those in sporozoites and merozoites (Figure 1A). This indicates that the gene expression of Plasmodium undergoes significant changes during its development within liver cells. The differences in gene expression profiles are likely related to the different functions and survival strategies of the parasite at these different life stages. For example, sporozoites are in the stage of invading liver cells, sporozoite replication within liver cells results in merozoite release, and the Plasmodium that infected liver cells are in an intermediate developmental stage.
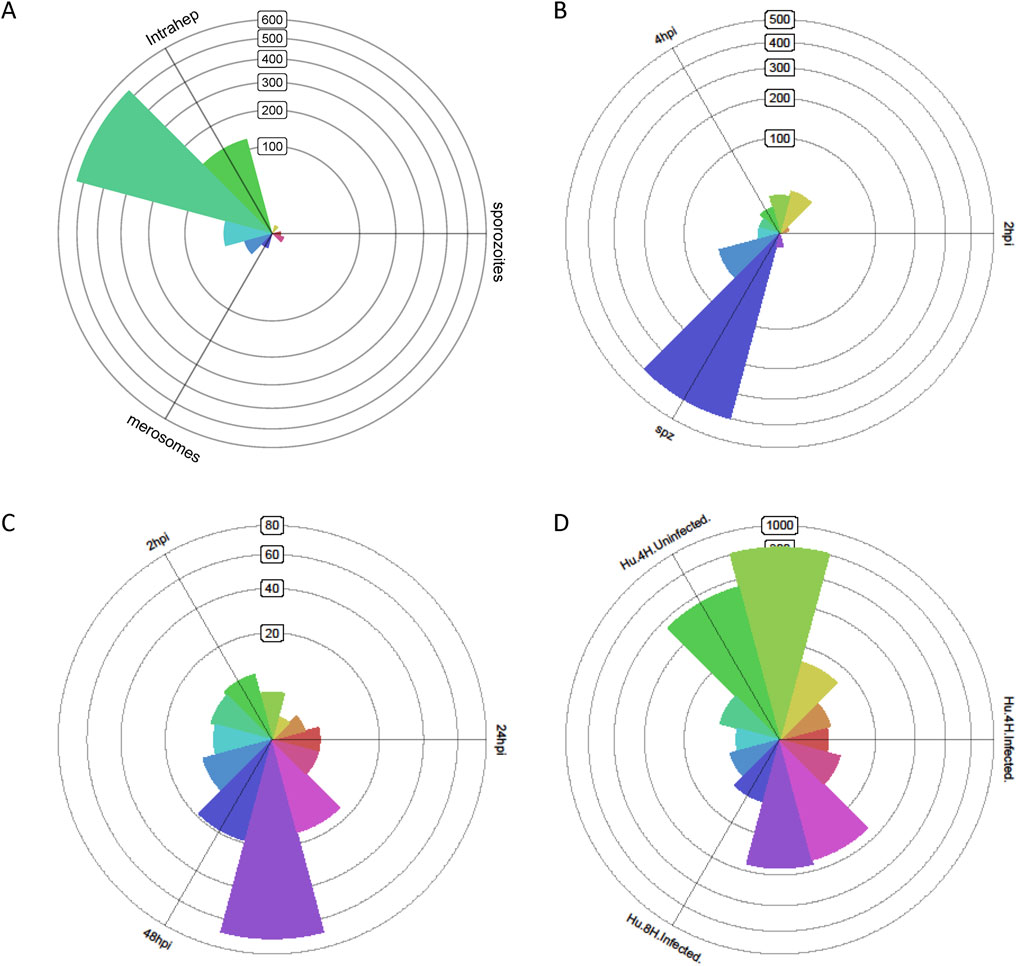
Figure 1. Transcriptome of Plasmodium that infected liver cells, and the infected liver cells demonstrate distinct transcriptional profiles at different stages. (A) Triwise analysis shows that the transcriptome of Plasmodium berghei that infected liver cells is distinctly different from that of sporozoites and merozoites. (B) Triwise analysis shows that the transcriptome of sporozoites is distinctly different from that of Plasmodium that infected liver cells. (C) Triwise analysis shows that the transcriptome profile of Plasmodium berghei at the late-stage liver infection is markedly different from the transcriptome profile of those at the early-stage liver infection. (D) The transcriptome of hepatocytes infected with Plasmodium berghei is distinct from that of hepatocytes not infected with Plasmodium berghei.
Compared to early stages of liver infection, gene expression of parasites differed markedly before entering liver cells. The 502 most differentially expressed genes at the early stage of liver infection demonstrated a 10-fold upregulation (p < 0.001) compared to pre-infection sporozoites. The roseplot in Figure 1B indicates the major upregulation of the Plasmodium genes toward the direction of the sporozoite phase when compared with the intrahepatocytic phase at 2 h and 4 h of infection (Figure 1B). This further emphasizes the dramatic transformation in the parasite’s gene expression once it enters the liver cells. Sporozoites have specific genes that involve in migration and invasion, whereas Plasmodium in the infected liver express genes related to replication and interaction with the host cell.
As the infection progresses, the parasite modifies its gene expression to adapt to the host environment, evade the immune system, and continue its replication. There were 149 significantly differentially expressed Plasmodium genes between infected and non-infected hepatocytes, showing a 10-fold increase in infected cells (p-value < 0.001). The roseplot in Figure 1C indicates the major upregulation of Plasmodium genes toward the direction of the intrahepatocytic phase at 48 h of infection when compared with the intrahepatocytic phase at 2 h and 24 h of infection (Figure 1C). In contrast, there was some similarity in the transcriptome of hepatocytes after 4 h of infection (Figure 1C).
On the other hand, dramatic differences were also found in the transcriptome of host cells. To study the host–pathogen interaction during malaria parasite liver infection, a dual RNA sequencing study was conducted, comparing the parasites and their host at different stages. Infected liver cells demonstrated 2,666 genes with a fold change of 10 and p-value <0.001 compared to non-infected hepatocytes (Figure 1D). This indicates that the infection has a significant impact on the host hepatocyte’s gene expression, altering its normal functions. The infected hepatocytes may respond to the parasite by changing their metabolic pathways, immune-related gene expression, and cell-cycle regulation.
3.2 In vivo vs. in vitro transcriptional divergence
The combination of single-cell sequencing and dual RNA sequencing allowed the investigation of in vivo stages. Comparing gene expression of in vitro-infected parasites at different time points with that of in vivo-infected parasites revealed distinct patterns. Notably, the in vitro-infected parasites exhibited a plateau in gene expression at 4 h, followed by a significant decline after 48 h, with the most specific gene expression observed at 4 h after infection (Figure 2A). This pattern indicates that the in vitro culture conditions led to a unique gene expression dynamic, which may not fully represent the in vivo situation. The specific gene expression at 4 h might be related to the initial adaptation of the parasite to the in vitro environment. In vitro parasites exhibit transient gene activation, whereas in vivo parasites maintain sustained expression (Figure 2A). This is probably because of oxygen gradients and host immune evasion mechanisms (HIF-1α signaling in pericentral zones), which could act as drivers of divergence between in vitro and in vivo environments.
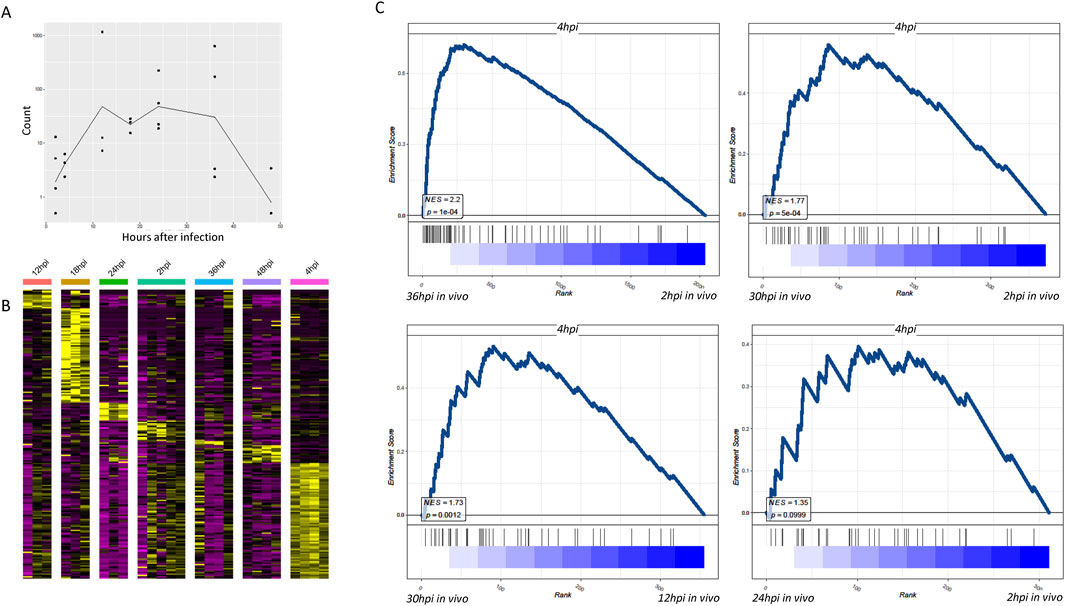
Figure 2. Dual RNA sequencing findings indicate that the transcriptomes of Plasmodium that infected the liver and their host cells are distinctly different from that of Plasmodium infected in vitro. (A) Line graph showing temporal profiles of gene expression in parasites infected in vitro. The curves in the figure represent the average of total counts of DEGs with adjusted p-values <0.01. DEGs are determined across different time points and samples, which have therefore been normalized. (B) Heatmap showing that relative to other time periods, Plasmodium demonstrate the most specific gene expression in hepatocytes 4 h after in vitro infection. Time hours in the figure represent the characteristic genes of Plasmodium after the respective time in the liver cells. Compared to other time hours, 4-h infection confers the most pronounced number of DEGs. (C) Using the transcriptome profile of in vitro-infected Plasmodium at 4 h, we compared the transcriptomes of hepatocytes infected at different times in vivo. Both normalized enrichment score and p-values of GSEA scoring indicated the similarity in the Plasmodium gene profiling between the in vivo group and 4-h in vitro infection group.
When comparing in vivo infected liver cells at 36 h, their gene transcription characteristics were more identical to those of in vitro-infected parasites at 4 h, indicating significant differences between in vitro cell culture and in vivo infection (Figure 2B). This further supports the finding in Figure 2A and suggests that 4 h is a critical time point for the parasite’s gene expression in vitro. It could be a time when the parasite is actively interacting with the in vitro cultured hepatocytes and adjusting its gene expression accordingly.
Using the transcriptome profile of 4-h in vitro-infected Plasmodium, we compared the transcriptomes of hepatocytes infected at different times in vivo. Both normalized enrichment score (NES) and p-values of GSEA scoring indicated the similarity of Plasmodium gene profiling between the in vivo group and the 4-h in vitro infection group (Figure 2C). The analysis of transcriptional profile differences between in vitro and in vivo was challenging before 24 h of in vivo infection, suggesting different transformation times for parasites within liver cells in vivo. This implies that the transformation time of parasites within liver cells in vivo is different from that in the in vitro environment, and it takes time for the differences in gene expression to become apparent. The in vivo environment, with its complex immune system and physiological factors, may influence the parasite’s gene expression in a way that is distinct from that in the in vitro culture.
3.3 Regional variability in liver zonation and infection outcomes
Apart from the timeliness of malaria parasite infection in liver cells, the complex liver environment may influence the differentiation status of the parasites. Studies have shown significant differences in gene expression between parasites infecting the cells in the pericentral region and those around the periportal region of the liver (Afriat et al., 2022). The central region had higher infection abundance, whereas the survival rate of parasites infecting liver cells around the portal vein was lower, possibly related to uneven oxygen distribution in liver lobules (Ben-Moshe and Itzkovitz, 2019). We found that although parasite-infected liver cells exhibited a distinct gene profiling based on their infected time (Supplementary Figure S1A), almost all the temporal clusters contain clear spatial segregation based on the expression of periportally or pericentrally zonated genes (Supplementary Figure S1B–D). GSEA indicated that parasites infecting the cells in the pericentral region of the liver closely resembled parasites that infected in vitro at 4 h (Figure 3A), suggesting that in vitro culture primarily simulates parasites infecting the cells in the central region of the liver in an oxygen-rich state. However, further research is needed to understand the in vitro model of parasites infecting liver cells around the portal vein. The genes associated with parasites infecting the cells in the pericentral region of the liver at 36 h in vivo were mainly related to synthesis (Figure 3B). These differential genes can provide insights into the different adaptation mechanisms of the parasite while infecting cells in different regions of the liver. For example, genes related to metabolism, immune evasion, and cell adhesion may be differentially expressed to cope with the different microenvironments in the pericentral and periportal regions. To study the host cells infected in the pericentral and portal vein regions, differential genes between the infected cells in the pericentral and portal regions of the liver, including uninfected and 36-h in vivo infected cells, were compared. Venn diagrams delineated unique differential genes for uninfected and 36-h in vivo infected cells (Figure 3C), indicating detoxification and transformation functions for uninfected cells and associations with alcohol and cholesterol metabolism for 36-h in vivo infected cells (Figures 3D–F). The common genes were associated with oxygen levels.
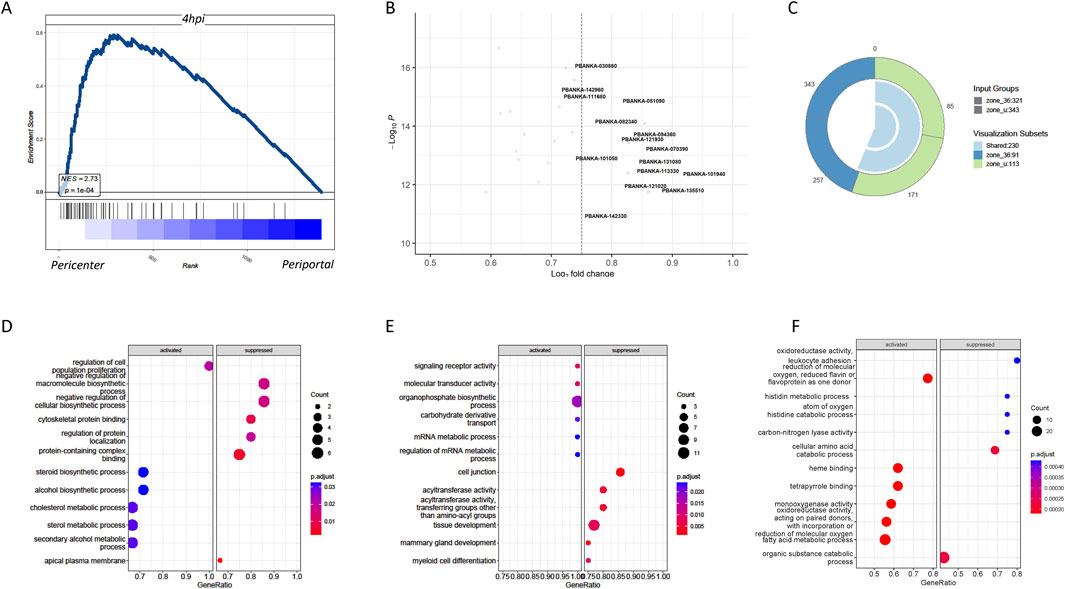
Figure 3. Significant differential transcriptome between Plasmodium in hepatocytes around the central vein and the portal vein as well as their host transcriptome. (A) GSEA showed that compared to the periportal infection, the gene expression of pericentrally infected Plasmodium significantly resembled the gene expression of those infected in vitro after 4 h. (B) Volcano graph comparing the gene expression of Plasmodium in the pericentral and periportal regions of the liver. (C) Venn diagram of host DEGs in pericentral (PC) versus periportal (PP) zones. Zone_u: 250 unique DEGs in uninfected cells; zone_36: 180 unique DEGs in infected cells; overlap: 45 shared genes. (D) GO enrichment analyses showed the host differential genes specific to no infection in pericentric versus periportal hepatocytes. (E) GO enrichment analyses showed the host differential genes specific to 36-h infection in vivo in pericentral versus periportal hepatocytes. (F) GO enrichment analyses showed the shared host differential genes by both no infection and 36-h infection in pericentral versus periportal hepatocytes.
3.4 Significance of abortive liver cells
The most notable discovery in in vivo malaria parasite liver infection was the identification of abortive liver cells, which were not previously observed in in vitro experiments. To contrast with in vitro cultured parasites, the differential genes of abortive liver cells at 36 h in vivo and those of productive cells were compared with the gene features of parasites infected in vitro at 4 h. It was found that the gene features of productive cells closely resembled those of parasites infected in vitro at 4 h, with genes primarily associated with parasite synthesis (Figure 4A). This suggests the difficulty of simulating abortive liver cell in parasites in the in vitro culture. Productive cells, which are more identical to in vitro-infected parasites at 4 h, may have a different set of gene expression patterns related to successful replication and development, whereas abortive cells have a distinct gene expression pattern that makes their replication complex in vitro. Genes related to DNA replication, protein synthesis, and cell-cycle regulation may be differentially expressed between abortive and productive cells (Figure 4B). Although there is an overlap, productive cells also contain unique genes, indicating that they have specific characteristics that are not fully captured by the pericentral infection model (Figure 4C). The volcano graph shows the 13 related genes that are specific to the productive cells (Figure 4D). These genes are likely to play important roles in the successful development of Plasmodium in hepatocytes and may be potential targets for future studies on malaria treatment or prevention. The data not only indicate similarity between the gene features of productive cells and parasites cultured in vitro, similar to those infecting cells in the pericentral region of the liver in an oxygen-rich state (Hirako et al., 2022; Ben-Moshe and Itzkovitz, 2019; Ben-Moshe et al., 2019) but also suggest that these are the major genes shared between liver cells in the pericentral and portal regions.
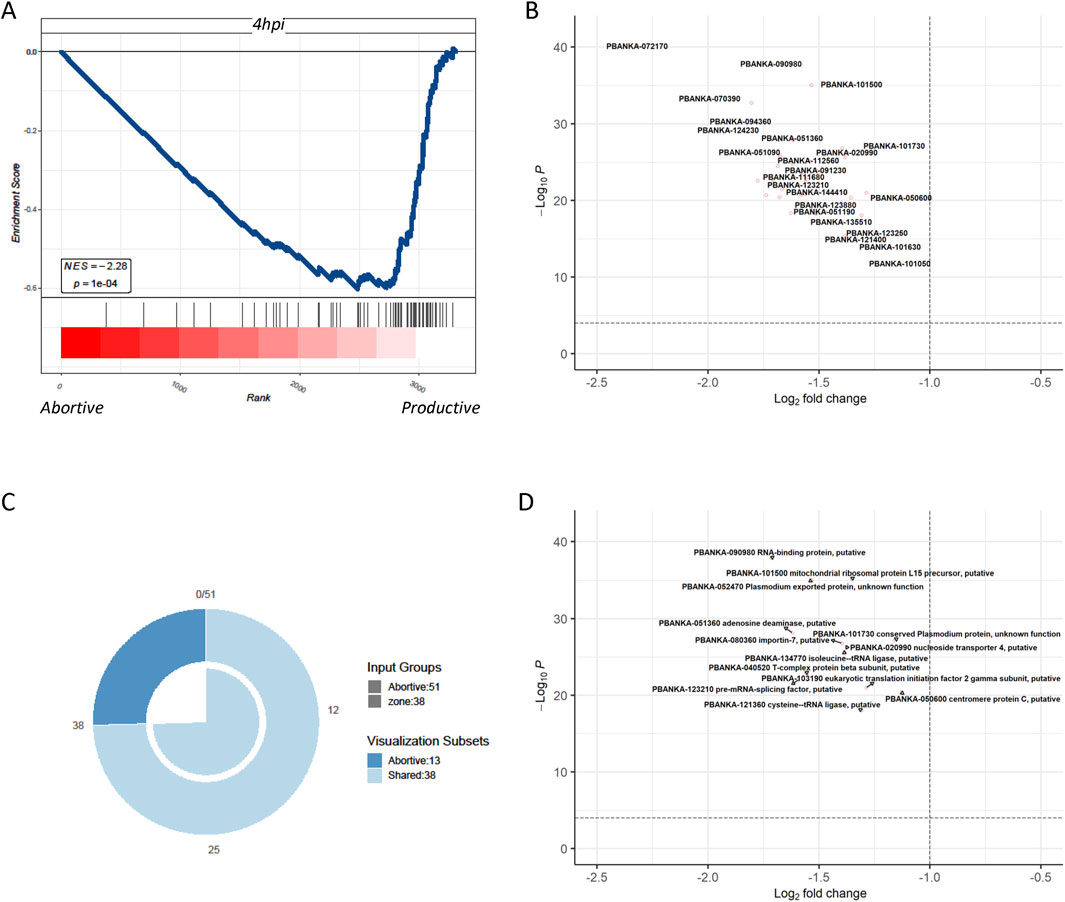
Figure 4. Comparison of the differential genes of Plasmodium in abortive hepatocytes and in productive cells with the genetic profile of Plasmodium after 4 h of in vitro infection (A) GSEA shows that compared to the transcriptome of Plasmodium in abortive hepatocytes, the transcriptome of Plasmodium in productive cells significantly resembled the transcriptome of those infected in vitro after 4 h. (B) Differential genes in Plasmodium in abortive hepatocytes versus productive cells at 4 hours post-infection in Plasmodium in abortive hepatocytes versus those in Plasmodium in productive cells. (C) Venn diagrams define, respectively, the Plasmodium-specific genes of the productive cells, Plasmodium-specific genes of pericentral infection, and their shared genes. (D) Volcano graph showing the 13 related genes that were specific to the productive cells.
4 Discussion
Our data on gene expression during Plasmodium liver infection emphasize the importance of the early stages of Plasmodium infection. In-depth analyses of single-cell profiles of Plasmodium-infected hepatocytes in vivo revealed differences between parasite subpopulations in vitro and in vivo. A joint investigation of the Plasmodium transcriptome and their host transcriptome provided a comprehensive analysis of gene expression in the liver stage of Plasmodium in vivo, deepening our understanding of parasite and host transcriptional dynamics during liver infection.
To study host–pathogen interactions in Plasmodium liver infections, we have taken advantage of the fact that single-cell sequencing technology can compare the transcriptomes of the host and the pathogen in a cell. We performed a dual RNA sequencing study and compared the transcriptome profiles of not only infected hepatocytes at different times but also those at different locations in the physiological and infected states, which were compared with the corresponding cellular pathogen transcriptomes. This comparison allowed us to find that the pathogen transcriptome of pericentric hepatocytes infected with Plasmodium berghei was more identical to that of Plasmodium berghei infection in vitro, which may be related to the oxygen content.
The bile ducts in the hepatic lobules carry bile acids secreted by the hepatocytes outward in a direction opposite to that of blood flow to the portal node. In contrast, blood enters the lobules from the portal node and flows inward through the hepatic blood sinusoids to the draining central vein (Hoehme et al., 2010). Hepatocyte zonation facilitates the allocation of high-energy-consuming tasks to high-oxygen areas and spatial recycling of materials (Ben-Moshe and Itzkovitz, 2019). The host transcriptome profiles of infected pericentric hepatocytes and uninfected pericentric hepatocytes were mostly identical, suggesting the positional conservation of the hepatocyte transcriptome. Interestingly, the host transcriptome after infection not only has a portion of the unique genes but also loses a portion of the site-specific genes of the hepatocyte transcriptome, suggesting that these genes were originally associated with the stress response and that pericentric and portal hepatocytes not only differ in metabolic function but also play different roles related to infection.
Hepatocytes exhibit significant variability in their transcriptome depending on their location along the central axis of the lobular portal vein. This heterogeneity is a consequence of the intrinsic epigenetic features inherent in the partitioning of oxygen, nutrient, and hormone gradients. The study of Plasmodium infection in hepatocytes requires systematic characterization of these heterogeneous layers to better understand Plasmodium differentiation and liver dysfunction during Plasmodium infection.
In summary, the current studies contribute to our understanding of malaria parasite liver infection dynamics, uncovering crucial gene expression patterns and molecular interactions at various stages.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding author.
Author contributions
ZM: Methodology. Conceptualization, Investigation, Writing – original draft, Writing – review and editing. YC: Methodology. Conceptualization, Investigation, Writing – original draft, Writing – review and editing. and YQ: investigation, writing – original draft, and writing – review and editing. JS: conceptualization, data curation, formal analysis, funding acquisition, investigation, methodology, project administration, resources, software, supervision, validation, visualization, writing – original draft, and writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study would not be possible without the funding from the National Natural Science Foundation of China (32360176, 82060036).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2025.1548487/full#supplementary-material
References
Afriat, A., Zuzarte-Luis, V., Bahar Halpern, K., Buchauer, L., Marques, S., Chora, A. F., et al. (2022). A spatiotemporally resolved single-cell atlas of the Plasmodium liver stage. Nature 611 (7936), 563–569. doi:10.1038/s41586-022-05406-5
Ben-Moshe, S., and Itzkovitz, S. (2019). Spatial heterogeneity in the mammalian liver. Nat. Rev. Gastroenterol. Hepatol. 16 (7), 395–410. doi:10.1038/s41575-019-0134-x
Ben-Moshe, S., Shapira, Y., Moor, A. E., Manco, R., Veg, T., Bahar Halpern, K., et al. (2019). Spatial sorting enables comprehensive characterization of liver zonation. Nat. Metab. 1 (9), 899–911. doi:10.1038/s42255-019-0109-9
Caldelari, R., Dogga, S., Schmid, M. W., Franke-Fayard, B., Janse, C. J., Soldati-Favre, D., et al. (2019). Transcriptome analysis of Plasmodium berghei during exo-erythrocytic development. Malar. J. 18 (1), 330. doi:10.1186/s12936-019-2968-7
Hirako, I. C., Antunes, M. M., Rezende, R. M., Hojo-Souza, N. S., Figueiredo, M. M., Dias, T., et al. (2022). Uptake of Plasmodium chabaudi hemozoin drives Kupffer cell death and fuels superinfections. Sci. Rep. 12 (1), 19805. doi:10.1038/s41598-022-23858-7
Hodgson, S. H., Muller, J., Lockstone, H. E., Hill, A. V. S., Marsh, K., Draper, S. J., et al. (2019). Use of gene expression studies to investigate the human immunological response to malaria infection. Malar. J. 18 (1), 418. doi:10.1186/s12936-019-3035-0
Hoehme, S., Brulport, M., Bauer, A., Bedawy, E., Schormann, W., Hermes, M., et al. (2010). Prediction and validation of cell alignment along microvessels as order principle to restore tissue architecture in liver regeneration. Proc. Natl. Acad. Sci. U. S. A. 107 (23), 10371–10376. doi:10.1073/pnas.0909374107
Howick, V. M., Russell, A. J. C., Andrews, T., Heaton, H., Reid, A. J., Natarajan, K., et al. (2019). The Malaria Cell Atlas: Single parasite transcriptomes across the complete Plasmodium life cycle. Science 365 (6455), eaaw2619. doi:10.1126/science.aaw2619
Inacio, P., Zuzarte-Luis, V., Ruivo, M. T., Falkard, B., Nagaraj, N., Rooijers, K., et al. (2015). Parasite-induced ER stress response in hepatocytes facilitates Plasmodium liver stage infection. EMBO Rep. 16 (8), 955–964. doi:10.15252/embr.201439979
LaMonte, G. M., Orjuela-Sanchez, P., Calla, J., Wang, L. T., Li, S., Swann, J., et al. (2019). Dual RNA-seq identifies human mucosal immunity protein Mucin-13 as a hallmark of Plasmodium exoerythrocytic infection. Nat. Commun. 10 (1), 488. doi:10.1038/s41467-019-08349-0
Liehl, P., Zuzarte-Luis, V., Chan, J., Zillinger, T., Baptista, F., Carapau, D., et al. (2014). Host-cell sensors for Plasmodium activate innate immunity against liver-stage infection. Nat. Med. 20 (1), 47–53. doi:10.1038/nm.3424
Little, T. S., Cunningham, D. A., Vandomme, A., Lopez, C. T., Amis, S., Alder, C., et al. (2021). Analysis of pir gene expression across the Plasmodium life cycle. Malar. J. 20 (1), 445. doi:10.1186/s12936-021-03979-6
Lopes da Silva, M., Thieleke-Matos, C., Cabrita-Santos, L., Ramalho, J. S., Wavre-Shapton, S. T., Futter, C. E., et al. (2012). The host endocytic pathway is essential for Plasmodium berghei late liver stage development. Traffic 13 (10), 1351–1363. doi:10.1111/j.1600-0854.2012.01398.x
Mancio-Silva, L., Gural, N., Real, E., Wadsworth, M. H., Butty, V. L., March, S., et al. (2022). A single-cell liver atlas of Plasmodium vivax infection. Cell Host Microbe 30 (7), 1048–1060.e5. doi:10.1016/j.chom.2022.03.034
Real, E., Howick, V. M., Dahalan, F. A., Witmer, K., Cudini, J., Andradi-Brown, C., et al. (2021). A single-cell atlas of Plasmodium falciparum transmission through the mosquito. Nat. Commun. 12 (1), 3196. doi:10.1038/s41467-021-23434-z
Ribot, J. C., Neres, R., Zuzarte-Luis, V., Gomes, A. Q., Mancio-Silva, L., Mensurado, S., et al. (2019). γδ-T cells promote IFN-γ–dependentPlasmodiumpathogenesis upon liver-stage infection. Proc. Natl. Acad. Sci. U. S. A. 116 (20), 9979–9988. doi:10.1073/pnas.1814440116
Toro-Moreno, M., Sylvester, K., Srivastava, T., Posfai, D., and Derbyshire, E. R. (2020). RNA-seq analysis illuminates the early stages of Plasmodium liver infection. mBio 11 (1), 032344–19. doi:10.1128/mBio.03234-19
van de Laar, L., Saelens, W., De Prijck, S., Martens, L., Scott, C. L., Van Isterdael, G., et al. (2016). Yolk sac macrophages, fetal liver, and adult monocytes can colonize an empty niche and develop into functional tissue-resident macrophages. Immunity 44 (4), 755–768. doi:10.1016/j.immuni.2016.02.017
Vaughan, A. M., and Kappe, S. H. I. (2017). Malaria parasite liver infection and exoerythrocytic biology. Cold Spring Harb. Perspect. Med. 7 (6), a025486. doi:10.1101/cshperspect.a025486
Yang, A. S. P., Dutta, D., Kretzschmar, K., Hendriks, D., Puschhof, J., Hu, H., et al. (2023). Development of Plasmodium falciparum liver-stages in hepatocytes derived from human fetal liver organoid cultures. Nat. Commun. 14 (1), 4631. doi:10.1038/s41467-023-40298-7
Yu, G., Wang, L. G., Han, Y., and He, Q. Y. (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16 (5), 284–287. doi:10.1089/omi.2011.0118
Keywords: Plasmodium, intraerythrocytic stage, dual transcriptome, spatiotemporal analyses, contrast between in vivo and in vitro
Citation: Mo Z, Chen Y, Qin Y and Song J (2025) Transcriptome profiling of intrahepatocytic Plasmodium and their host hepatocytes based on the infection phase and the zonation of the liver. Front. Genet. 16:1548487. doi: 10.3389/fgene.2025.1548487
Received: 19 December 2024; Accepted: 19 March 2025;
Published: 07 April 2025.
Edited by:
Tizhen Yan, Dongguan Maternal and Child Health Hospital, ChinaReviewed by:
Chenqi Wang, University of South Florida, United StatesRui Guo, Fujian Agriculture and Forestry University, China
Copyright © 2025 Mo, Chen, Qin and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian Song, anNvbmdAZ3hhbXMub3JnLmNu
†These authors have contributed equally to this work
 Zhuning Mo
Zhuning Mo Yuhong Chen
Yuhong Chen Yujuan Qin
Yujuan Qin Jian Song
Jian Song