
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Genet., 11 February 2025
Sec. Genomics of Plants and the Phytoecosystem
Volume 16 - 2025 | https://doi.org/10.3389/fgene.2025.1539311
This article is part of the Research TopicSurvival Strategies and Biotechnological Applications of Plants in Extreme EnvironmentsView all articles
 Huan Zhang1,2†
Huan Zhang1,2† Qilin Yang1,2†
Qilin Yang1,2† Leyi Wang1,2
Leyi Wang1,2 Huawei Liu1,3
Huawei Liu1,3 Daoyuan Zhang1,3
Daoyuan Zhang1,3 Cheng-Guo Duan4*
Cheng-Guo Duan4* Xiaoshuang Li1,3*
Xiaoshuang Li1,3*In complex and diverse environments, plants face constant challenges from various pathogens, including fungi, bacteria, and viruses, which can severely impact their growth, development, and survival. Mosses, representing early divergent lineages of land plants, lack traditional vascular systems yet demonstrate remarkable adaptability across diverse habitats. While sharing the fundamental innate immune systems common to all land plants, mosses have evolved distinct chemical and physical defense mechanisms. Notably, they exhibit resistance to many pathogens that typically affect vascular plants. Their evolutionary significance, relatively simple morphology, and well-conserved defense mechanisms make mosses excellent model organisms for studying plant-pathogen interactions. This article reviews current research on moss-pathogen interactions, examining host-pathogen specificity, characterizing infection phenotypes and physiological responses, and comparing pathogen susceptibility and defense mechanisms between mosses and angiosperms. Through this analysis, we aim to deepen our understanding of plant immune system evolution and potentially inform innovative approaches to enhancing crop disease resistance.
Mosses are characterized by their diminutive stature and relatively simple organization. Their gametophytes and sporophytes predominantly comprise a single layer of cells and lack true vascular tissue, placing them in the category of non-vascular plants. Bryophytes, which include mosses, liverworts, and hornworts, are important representatives of early terrestrial plants that evolved from aquatic plants around 450 million years ago (Clarke et al., 2011; Delaux et al., 2019; Resemann et al., 2019). These mosses represent an ancient lineage in plant evolution, having diversified over at least 300 million years (Dobbeler, 1997). Mosses hold a foundational position in the evolution of terrestrial plants and play a crucial role in the transition from aquatic plants to terrestrial plants, linking single-celled green algae to vascular plants (De León and Montesano, 2013; De León and Montesano, 2017). Additionally, they serve as an effective model organism (De León, 2011) due to their compact size, simple structure, strong regenerative ability, and short cultivation cycle (Schaefer, 2002; Cove et al., 2006). Furthermore, mosses are valuable for molecular biology and genetic studies because they respond similarly to environmental signals and plant growth factors as other vascular plants, and their life cycle is dominated by the haploid gametophyte stage (Schaefer and Zryd, 1997; Schaefer, 2002).
Mosses, like other plants, can be infected by various pathogens including fungi, bacteria, oomycetes, and viruses. Among these, several key model pathogens have been instrumental in understanding moss-pathogen interactions. Botrytis cinerea, one of the most extensively studied fungal pathogens, serves as a primary model system due to its ability to directly penetrate the cell wall and invade intercellular spaces of Physcomitrella patens, causing tissue browning and death (Yan et al., 2018). Similarly, the bacterial pathogen Pseudomonas syringae pv. tomato DC3000 has emerged as another important model organism, as it can effectively colonize P. patens and induce characteristic disease symptoms, providing valuable insights into the conservation of immune responses between mosses and vascular plants (Yan et al., 2023). Other significant pathogens include Pectobacterium carotovorum, which causes tissue maceration in P. patens (Alvarez et al., 2016), and the oomycetes Pythium irregulare and P. debaryanum, which infect multiple tissue types and cause browning of the stem, midrib, and leaf base (Oliver et al., 2009). While less studied, viral infections have also been documented, with Tobacco Mosaic Virus (TMV) and Cucumber Green Mottle Mosaic Virus (CGMMV) detected in moss species such as Polytrichum commune (Polischuk et al., 2007). Understanding these diverse pathosystems is crucial for elucidating the evolution of plant-pathogen relationships and may provide insights into developing disease resistance strategies in both non-vascular and vascular plants. Due to their significant evolutionary position and relatively simple morphological structure, mosses are regarded as an ideal model system for studying plant-pathogen interactions. Research on mosses allows for a clear understanding of the mechanisms behind plant disease resistance responses, as well as insights into the defense strategies of early land plants and their evolutionary trajectories. This knowledge contributes to better understanding of how pathogens threaten major crops and how plants activate their complex defense mechanism, which could aid in managing or preventing crop diseases. This review aims to provide a comprehensive overview of studies on moss-pathogen interactions. It examines the susceptibility of mosses compared to angiosperms to various pathogens and highlights the similarities and differences in the defense mechanisms activated after infection. Additionally, it discusses the interactions between various hosts and pathogen and the physiological phenotypes in mosses. Investigating these interactions is crucial for identifying key factors of early land plant defense systems and for elucidating the molecular mechanisms of plant-pathogen interactions from an evolutionary perspective. This review will definitely provide valuable insights for molecular breeding of plant disease resistance.
Fungal pathogens have been identified in moss populations, and the associated disease symptoms have been documented for decades (Tsuneda et al., 2001a; De León, 2011). Various fungi, including Thyronectria hyperantartica, Teprocybe palustris, Bryoscyphus dicrani, Scleroconidioma sphagnicola, Acrospermum adenum, Ardapia retitruga, Lizonia baldinii, and Atradidymella muscivora, cause necrotic lesions and the death of moss gametophytes (Davey and Currah, 2006; Davey et al., 2009). Some of these pathogens have been studied for their ability to penetrate bryophyte tissues, destroy cells, and elicit host responses (Martínez-Abaigar et al., 2005; Davey and Currah, 2006; Davey et al., 2009). The invasion of moss pathogens into host cells typically involves vegetative hyphae, osmotic plugs, and sometimes appressoria, along with the enzymatic digestion of plant cell walls (Tsuneda et al., 2001a; Davey and Currah, 2009; Davey et al., 2009). Research has particularly focused on the interaction between P. patens and B. cinerea. B. cinerea can directly penetrate the cell wall of P. patens and invade the intercellular space, which can cause brown spots in moss leaves. With the extension of time, the number of brown spots increases and expands, spreading from the base and tip of the leaves to the middle, eventually leading to leaf decay and plant death, showing wilting symptoms (Yan et al., 2018). A strain isolated from Leucobryum glaucum, when introduced to P. patens, produced symptoms similar to L. glaucum blight disease. This fungus was closely related to Mucor racemosus based on sequencing, indicating that M. racemosus can infect both P. patens and L. glaucum, causing wilting, chlorosis, and other symptoms, although the detailed infection mechanisms are not fully understood (Liu, 2011). In contrast, M. racemosus does not infect Haplocladium microphyllum or Mnium hornum (Liu, 2011). Colletotrichum gloeosporioides can infect P. patens by directly penetrating the host cell wall, primarily damaging the leaves and resulting in tissue maceration, necrosis, etc., (Reboledo et al., 2015; Otero-Blanca et al., 2021). Additionally, Verticillium dahliae can also infect P. patens, Bryum argenteum, and Syntrichia caninervis, causing tissue browning and chloroplast degradation, with leaves and stems being the primary sites of infection. Infected mosses exhibit localized lesions and browning at the leaf edges. Sphagnum fuscum can be infected by Lyophyllum palustre, S. sphagnicola, Oidiodendron maius, Acremonium cf. curvulum, Arrhenia retiruga and Pochonia bulbillosa, where the hyphae selectively degrade the moss cell wall, leading to wavy deformations as they grow both outside and inside the cell wall, resulting in wavy deformation of the cell wall (Untiedt and Muller, 1985; Tsuneda et al., 2001b; Davey and Currah, 2006). Rozellopsidalean fungi can penetrate protonema and rhizoid cells to infect the Funaria hygrometrica, Bryum pseudotriquetrum and Bryum capillare (Martínez-Abaigar et al., 2005), resulting in compression of moss cell tissues, the release of numerous lipid droplets, necrosis of cortex cells in stems and leaves, and chloroplast detachment. Nectri mnii has also been observed to spread throughout the stem tissues of Plagiomnium medium (Döbbeler, 1988; Lawton and Saidasan, 2009). In contrast, the interaction between mosses and fungi has not been thoroughly examined. For instance, Phyllosticta tetraplodontis Lebedeva causes browning and chlorosis of the sporophytic tissues of Tetraplodon, ultimately resulting in the loss of the capsules. The process of cell disruption and degradation for this organism remains unidentified (Davey and Currah, 2006). Detailed descriptions of fungal infections in mosses were listed in Table 1.
There have been few studies on the interaction between mosses and oomycetes. Two oomycetes, P. irregulare and P. debaryanum, can develop appressoria to penetrate moss tissues, affecting all tissue types including leaves, protonema, rhizoids and stems. This leads to the browning of the stem, midrib and leaf base in P. patens Appressoria are visible during the early stages of infection. When several cells are infected, multi-digital haustoria-like structures appear in moss-infected tissues. The penetration of host cell walls into adjacent cells occurs through constricted hyphae. As the duration of infection increases, hyphal colonization becomes more extensive, leading to tissue rot in P. patens, shrinkage of cytoplasm, relocation of chloroplasts, and browning (Oliver et al., 2009). The oomycete Pythium ultimum cause the formation of areas of dying and dead moss gametophytes, while symptoms such as chlorosis and necrosis, followed by the death of gametophyte, are typical of all known mosses pathogens (fungi, oomycetes, and bacteria). (Redhead and Spicer, 1981; Untiedt and Muller, 1985; Döbbeler, 2003; Davey and Currah, 2006; De León, 2011), The mechanisms of penetration and destruction of moss cells by different pathogens, the causes of disease, and the host‘s response to infection are different (Hühns et al., 2003; Oliver et al., 2009; Alvarez et al., 2016).
There are currently few reports on the interactions between moss and bacteria. Erwinia carotovora can infect P. patens (De León et al., 2007) by entering through wounds, resulting in damage in moss tissue, browning of gametophytes, and gradual rotting. This bacterium can also cause cytoplasmic shrinkage, accumulation of autofluorescent substances, changes in chloroplast structure, and pigments in P. patens. P. carotovorum produces a high concentrations of enzymes that break down plant cell walls, including cellulases, proteases and pectinases (Toth and Birch, 2005). These enzymes work synergistically with other virulence factors to dissolve host tissues and facilitate host cell death (Davidsson et al., 2013). P. patens is susceptible to P. carotovorum infection (Alvarez et al., 2016), and after infection, P. patens exhibits basal tissue browning and partial wilting within 24 h (Alvarez et al., 2016). The proteases and cell wall-degrading enzymes degrade moss tissues leading to cell death. Furthermore, P. syringae is recognized as one of the most destructive agricultural pathogens (Liu et al., 2020). P. syringae pv. tomato DC3000 (Pst DC3000) can also infect P. patens, B. argenteum and S. caninervis, causing typical disease symptoms such as wilting and browning of the stems and leaves, etc., (Yan et al., 2023).
Viruses represent a major class of pathogens on earth that utilize their own viral suppressors of RNA silencing (VSRs) (Duan et al., 2012; Qiao et al., 2013; Yin et al., 2019) to overcome plant defenses and infect all types of organisms (Retel et al., 2019). Although the small RNA pathways in mosses and vascular plants share high similarity, little is known about the infection of mosses with viral pathogens (Marttinen et al., 2022). Researchers detected infections by TMV and CGMMV in Antarctic P. commune, as well as TMV infection in Barbilophozia attenuate (Polischuk et al., 2007). These two viruses typically infect dicotyledonous plants, and it remains unclear how they naturally infect mosses cells in the field. Additionally, viruses have been detected in the phyllosphere of Sphagnum, which serves as an important source of virus diversity and activity (Marttinen et al., 2022). Furthermore, it has been demonstrated that P. patens is infected with Tomato Bushy Stunt Virus (TBSV) and Cucumber Mosaic Virus (CMV) (Marttinen et al., 2022), which primarily damaging its gametophyte. This could facilitate the investigation of virus-moss host interactions using P. patens as a laboratory viral host.
In summary, mosses can interact with pathogens, particularly fungi, throughout their life cycles. The infection sites are varied, and there is no clear preference for specific tissues. P. patens is the main moss model used in current research, with a notable focus on the Physcomitrium-moss relationship. However, studies on other moss species are still quite limited. It is necessary to strength the research that elucidate the mechanisms behind bacterial and viral infections, as these pose major challenges in comprehending moss disease resistance.
We compared the symptoms and incidence rates of representative moss P. patens against the vascular plants Arabidopsis thaliana or tobacco after pathogen infection. It discovered that in most cases, the disease symptoms in infected mosses were similar to those in vascular plants, but the disease development rate was often faster than that in vascular plants (Table 2). Lesions can be seen on the tissue surface within 24 h of B. cinerea infecting P. patens and the moss dies after 5 days infection. Similarly, when B. cinerea infects A. thaliana, after 1-day, necrotic lesions appeared on the leaves. The symptoms are similar in the two hosts, mainly manifesting as lesions on leaves, wilting and eventual death (González et al., 2006; Jiang et al., 2013; Yan et al., 2018). For B. cinerea, the onset time of the two plants is not much different. The leaves of P. patens dipped with spore suspension of V. dahliae showed typical V-shaped necrotic spots within 15 days, while the leaves of A. thaliana dipped with spore suspension of V. dahliae showed rosette yellowing and premature senescence within 21 days (Fradin and Thomma, 2006; Reusche et al., 2014). Symptoms like tissue maceration occurred in P. patens within 24 h after C. gloeosporioides inoculation (Otero-Blanca et al., 2021), while the same concentration of conidia suspension droplets were added to the leaves of A. thaliana, the leaves wilted, chlorotic and exhibited water--soaked lesions after 3 days (Gao et al., 2021). Oomycetes P. irregulare and P. debaryanum were inoculated with P. patens by agar block, stem browning and tissue maceration was observed 1 day post inoculation, with increasing rotting and eventual death around 2 days as infection progressed (Toth and Birch, 2005; Oliver et al., 2009). However, after 2 days inoculation, A. thaliana with P. irregulare agar block, causing leaf wilting and brown, watery lesions symptoms (Castro et al., 2016). When P. patens is spray-inoculated with P. carotovorum, browning in stems and rhizoids can be observed within 1 day (Alvarez et al., 2016), whereas around 2 days are required to observe disease symptoms of water-soaked rotting leaves when A. thaliana was dipped in the bacterial suspension. The leaves of P. patens were soaked in the suspension of P. syringae at a concentration of 1 × 108 cfu/mL. After 3 days, the leaves appeared bacterial spots, yellowing, wilting and other phenomena (Yan et al., 2023), while the higher concentration of bacteria was used to soak A. thaliana, the symptoms were observed after 4 days (Summermatter et al., 1995). Soft rot E. carotovora also displayed faster pathogenesis in P. patens compared to A. thaliana (Kariola et al., 2005; De León et al., 2007). The same is true for viruses. For example, Tomato spotted wilt virus (TSWV) inoculation of simultaneously in P. patens and A. thaliana, and the structural protein of the virus could be detected in P. patens after 11 days. In contrast, A. thaliana could be detected after 21 days, indicating that the incidence of TSWV in P. patens was faster than that in A. thaliana (Hühns et al., 2003).
Current research reports indicate that several pathogens have been primarily isolated and identified from mosses, highlighting the unique host-pathogen interactions in bryophyte systems. Maybe these findings represent only a fraction of the potential moss-specific pathogens yet to be discovered. For example, after infecting P. patens, M. racemosus caused symptoms of tissue browning and leaf wilting around 3 days post inoculation, with gradual moss death starting after 1 week (Liu, 2011). S. sphagnicola is a potentially destructive necrotrophic pathogen capable of infecting S. fuscum. When S. fuscum tips were placed upside down on hyphae to inoculate the pathogen, yellowing was observed, and after 12–15 days, leaf necrosis occurred, causing the plants to become wrinkly and fragile (Tsuneda et al., 2001b). After artificially inoculating healthy S. fuscum, diseased plants showed brown lesions similar to those naturally infected. Detailed electron microscopic examination revealed the penetration of S. fuscum cell contents and walls by S. sphagnicola hyphae (Koukol and Kovarova, 2007). A. muscivora with F. hygrometrica as primary host can also infect Hylocomium splendens, Aulacomnium palustre and Polytrichum juniperinum, causing necrosis and wilting in all hosts (Davey et al., 2009). Rozellopsidalean fungi was only found in bryophytes, infecting F. hygrometrica, B. pseudotriquetrum and B. capillare, with browning observed at the penetration site and lateral shoots turning brown (Martínez-Abaigar et al., 2005). These findings collectively underscore the importance of studying moss-specific pathogens to better understand the unique aspects of plant-microbe interactions in non-vascular plants and their potential implications for broader plant pathology research.
Over a long period, plants and pathogens have evolved together. Plants utilize their cell walls as the first layer of defense against pathogenic microbial diseases, and modifications to these walls play a crucial role in their defense mechanisms (De León, 2011). Plant cell walls are composed of cellulose, hemicelluloses, pectin, xyloglucan, and hydroxyproline-rich proteins. In contrast, moss cell walls are thinner, lacking a clear distinction between primary and secondary walls. Additionally, mosses do not contain lignin but have polymers that are similar to lignans or lignin (lignans) (Ligrone et al., 2002; Popper, 2008). When pathogens invade, plants enhance their cell wall defense by incorporating phenolic compounds, depositing callose, and activating Dirigent (DIR) protein-coding genes involved in the synthesis of similar lignin compounds (De León et al., 2012; Reboledo et al., 2015; Alvarez et al., 2016). Moss can also release phenolic compounds from their thallus to prevent the germination of fungal spores and produce secondary metabolites to mitigate biological stress (Dixon, 2001; Commisso et al., 2021).
Plants have evolved intricate signaling systems for perception, transduction, and response as a means of defending against pathogen invasion (Gabriel and Rolfe, 1990; Boller and Felix, 2009; Takken and Tameling, 2009). The pattern recognition receptors (PRRs) positioned on the plasma membrane of angiosperms are responsible for detecting conserved pathogen-associated molecular patterns (PAMPs) and inducing PAMP-triggered immunity (PTI), which offers protection against non-adapted pathogens (Jones and Dangl, 2006; Zipfel, 2009). Pathogens that are well-adapted to their host plants deliver effector molecules into plant cells to target key PTI components and suppress plant defenses (Boller and Felix, 2009; Boller and He, 2009). In response, plants possess a second layer of immune receptors encoded by resistance (R) genes. These receptors can detect effectors either directly or indirectly, leading to effector-triggered immunity (ETI). ETI is a highly specific immune response that is frequently accompanied by a hypersensitive response (HR) and systemic acquired resistance (SAR). Several PAMPs and their corresponding PRRs have been identified so far in angiosperms. In A. thaliana, the receptors FLS2 (Flagellin-Sensing 2), EFR1 (Elongation Factor Tu Receptor 1), and LYK1/CERK1 (LysM-containing Receptor-like Kinase1/chitin Elicitor Receptor Kinase1) recognize bacterial flagellin, Elongation Factor Tu, and fungal chitin, respectively (Gómez-Gómez and Boller, 2000; Zipfel et al., 2006; Miya et al., 2007), with LYK4 and LYK5 implicated as crucial for chitin signaling and immunity in A. thaliana (Wan et al., 2012; Cao et al., 2014). LYK5 has been identified as the chitin receptor in A. thaliana, forming chitin-induced receptor complexes with CERK1 to activate plant immunity (Cao et al., 2014). P. patens lacks close relatives of the FLS2 and EFR receptors (Boller and He, 2009), which aligns with findings that moss cells do not respond to flagellin (flg22) and Elongation Factor Tu (Bressendorff et al., 2016). A functional CERK1 receptor has recently been found in P. patens that can perceive PAMPs such as fungal chitin and bacterial peptidoglycan (Bressendorff et al., 2016). Mutation in PpCERK1 led to reduced defenses, including lower expression of defense genes and reduced levels of cell wall-associated phenolic compounds, suggesting that PTI is an ancient plant defense response (Bressendorff et al., 2016). Further studies are needed to understand chitin perception in P. patens, including the analysis of molecular complexes associated with PpCERK1 and a potential LYK5-like receptor, to better understand the immune response in moss.
Studies show that P. patens utilizes mechanisms for pathogen recognition that are similar to those found in vascular plants, as evidenced by the presence of typical R genes in its genome (De León, 2011; De León and Montesano, 2013). It also activates additional defense responses, such as the production of reactive oxygen species (ROS), programmed cell death (PCD), cell wall reinforcement, and increased expression of defense-related genes, which mirror the responses observed in angiosperms upon pathogen attack (De León et al., 2007; De León and Montesano, 2017). One of the initial plant responses following pathogen recognition is the production of ROS, which directly damages pathogens and signals cell wall strengthening, HR induction and gene expression changes (Torres et al., 2006). Conversely, necrotrophic pathogens appear to enhance the production of ROS by damaging host cells, leading to their death (Govrin and Levine, 2000). Typical hallmarks of PCD observed in pathogen-infected moss tissues include chloroplast degradation, accumulation of autofluorescent compounds, cytoplasmic shrinkage, nuclear fragmentation and nuclease activity (De León et al., 2007; De León et al., 2012; Wang et al., 2015). Several canonical intracellular receptor genes have been identified in P. patens, including kinase-Nucleotide Binding Site (NBS)-Leucine Rich Repeat (LRR) receptors and Toll/interleukin-1 like Receptor (TIR)-NBS-LRR (Akita and Valkonen, 2002; Xue et al., 2012; Tanigaki et al., 2014). Previous studies have shown that algae lack the homologous genes of NBS-LRR, TIR-NBS-LRR or TIR-LRR (Sarris et al., 2016), but recent studies have found that algae may have the original form of NBS-LRR or TIR-NBS-LRR genes (Andolfo et al., 2019; Andolfo et al., 2020). It is speculated that the origin and differentiation of RNL subclass are earlier than the separation of green algae and charophytes (Shao et al., 2019), and the emergence of NLR genes is related to the origin of terrestrial plants. This suggests that the evolution of receptor-like genes may represent an adaptive strategy for sensing pathogens and triggering defenses, which plays an important role in plant adaptation to terrestrial environments. The NBS-LRR protein family in terrestrial plants has expanded in vascular plants, which may be caused by polyploidization or paleopolyploidization events. The NBS-LRR protein family has expanded in vascular plants, likely due to polyploidy or ancient polyploidization events. Intriguingly, Selaginella tamariscina possesses a much smaller number of NBS-LRRs and other potential receptor genes compared to P. patens (Sarris et al., 2016). Further research is needed to explore whether pathogen effectors can inhibit moss defenses and if certain receptor-like proteins in activated P. patens can detect these effectors, either directly or indirectly, to initiate ETI.
In angiosperm, calcium ions (Ca2+) play a vital role as a second messenger in PTI and ETI responses, in the process of plant-microorganism interaction, the change of intracellular Ca2+concentration is one of the earliest biochemical characteristics detected after microbial recognition. leading to the production of ROS and (nitric oxide) NO, as well as the activation of defense gene expression (Seybold et al., 2014; Zhang et al., 2014). The signaling of biotic stress in plants is regulated by calcium-dependent protein kinases (CDPKs) and mitogen-activated protein kinase (MAPK) pathways, both of which elevate cytoplasmic Ca2+ levels upon perception of pathogens (Seybold et al., 2014). In P. patens, chitin oligosaccharide treatment resulted in Ca2+ oscillations, and calcium carriers could stimulate expression of defense-related genes. The influx of Ca2+ alone was enough to provoke defense responses similar to those observed in angiosperms (Galotto et al., 2020).
A comprehensive analysis of immune-related gene families across plant lineages has revealed intriguing patterns of conservation and innovation (Figure 1), particularly in mosses. The examination of key immune components, including PRRs, Nucleotide-binding Leucine-rich Repeat (NLR) proteins, Receptor-like Kinases (RLKs), and other plant immune-related proteins across various plant groups (Wu and Zhou, 2013), has uncovered significant insights into the evolution of plant immunity.
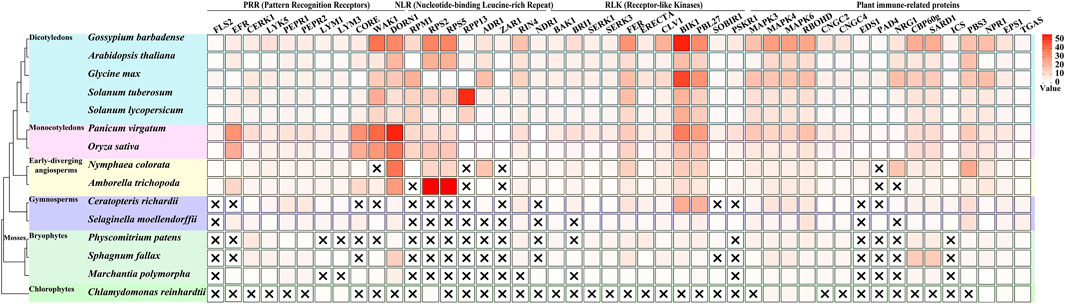
Figure 1. Evolutionary distribution of immune-related genes across plant lineages. This figure illustrates the presence and copy number of key immune-related genes across 15 representative plant species, spanning from chlorophytes to angiosperms, with a focus on mosses. The heatmap represents the number of orthologous genes for each immune component (columns) in each species (rows), with darker colors indicating higher copy numbers. Data acquisition and analysis: Genome sequences were obtained from Phytozome (https://phytozome-next.jgi.doe.gov/). The following genome versions were used: Chlamydomonas reinhardtii CC-4532 v6.1, Marchantia polymorpha v3.1, Sphagnum fallax v1.1, Physcomitrium patens v6.1, Selaginella moellendorffii v1.0, Ceratopteris richardii v2.1, Amborella trichopoda v1.0, Nymphaea colorata v1.2, Oryza sativa Kitaake v3.1, Panicum virgatum v5.1, Solanum lycopersicum ITAG5.0, Solanum tuberosum v6.1, Glycine max Wm82. a6. v1, Arabidopsis thaliana Araport11, and Gossypium barbadense v1.1. Orthologous genes were identified using OrthoFinder (Emms and Kelly, 2019).
In the realm of Pattern Recognition Receptors, a clear evolutionary trend in their diversification is observed. Notably, the bacterial PAMP receptors FLS2 and EFR are absent in chlorophytes and mosses, only emerging in lycophytes and becoming more diverse in angiosperms (Wu and Zhou, 2013; de Vries et al., 2018; Delaux and Schornack, 2021). This suggests that these specific bacterial recognition mechanisms evolved after the divergence of mosses from the plant lineage leading to vascular plants. In contrast, mosses possess multiple copies of the chitin-sensing receptors CERK1 and LYK5, with numbers comparable to or exceeding those found in many angiosperms. This indicates an early evolution and potential importance of fungal pathogen recognition in mosses. The presence of PEPR1 and PEPR2, receptors for damage-associated molecular patterns (DAMPs), in mosses but with lower copy numbers compared to vascular plants, suggests a more ancient origin of DAMP recognition systems (Engelsdorf et al., 2018; Liu et al., 2024). One of the most striking observations is the complete absence of canonical NLR genes (such as RPM1, RPS2, RPS4, RPS5, RPP13) in mosses and other non-vascular plants. This finding suggests that the complex NLR-mediated immunity, crucial for effector-triggered immunity, evolved after the divergence of vascular plants, representing a major innovation in plant immune systems. This absence implies that mosses must rely on alternative strategies for pathogen resistance. The analysis of RLKs revealed a mixed pattern of conservation and innovation. Co-receptors such as BAK1, SERK1, and SERK3 are present in mosses, indicating an early evolution of these signaling components (Ngou et al., 2024).
The examination of other immune-related proteins yielded several noteworthy findings. The MAPK cascade components (MAPK3, MAPK4, MAPK6) are present in similar numbers in mosses and vascular plants, suggesting early evolution and conservation of this signaling module (Ghelis, 2011; Ramírez-Zavaleta et al., 2022; Singh et al., 2024). RBOHD, involved in ROS production, is also present in mosses, indicating an ancient origin of this defense mechanism (Marchetti et al., 2024). However, key regulators of basal immunity and systemic acquired resistance, EDS1 and PAD4, are absent in mosses, correlating with the lack of canonical NLRs (Ramírez-Zavaleta et al., 2022). These observations collectively paint a picture of moss immunity as a unique system that combines ancient, conserved elements with lineage-specific innovations. The absence of certain immune components (e.g., FLS2, EFR, NLRs) coupled with the expansion of others (e.g., CERK1, CBP60g) suggests that mosses have evolved alternative strategies to cope with pathogens. The expansion of chitin recognition receptors implies an enhanced capacity for fungal pathogen detection, possibly reflecting the importance of fungal interactions in their evolutionary history. The absence of canonical NLRs suggests a heavier reliance on PTI rather than ETI for pathogen resistance in mosses. Furthermore, the expansion of salicylic acid-related transcription factors could indicate a novel regulatory mechanism for defense responses, potentially compensating for the lack of NLR-mediated immunity.
In conclusion, the unique immune profile of mosses, characterized by both the absence of seemingly crucial components and the expansion of others, challenges conventional understanding of plant immunity. It suggests that there may be alternative, yet undiscovered mechanisms of pathogen resistance in these ancient plant lineages, which could potentially inform new strategies for crop protection and disease resistance in the future.
In angiosperms, AP2/ERFs (APETALA2/Ethylene Responsive Factors) play important regulatory roles in plant defenses against pathogens and abiotic stresses by controlling the expression of their target genes (Xu et al., 2011). On the other hand, a number of ERF family members are induced in P. patens during pathogen infection, indicating their role as immune modulators in mosses (Reboledo et al., 2022a). Researchers studied a pathogen-induced ERF called PpERF24 in P. patens and found that its direct orthologs exist only in other mosses, being absent in the bryophytes Marchantia polymorpha and Anthoceros agrestis, the vascular plant S. tamariscina, or angiosperms (Reboledo et al., 2022b). This demonstrates that PpERF24 belongs to a moss-specific clade with unique amino acid features in the AP2 DNA-binding domain. Interestingly, all members of the PpERF24 sub-clade are induced by fungal pathogens, making PpERF24 a unique pathogen-responsive gene in moss (Reboledo et al., 2022b). Among the upregulated differentially expressed genes during B. cinerea infection, 216 were specifically expressed in P. patens, whereas showing minimal or no expression in other vascular plants (Reboledo et al., 2021). Additionally, 22 genes encoding putative fungal major facilitator superfamily transporters were also upregulated in P. patens, but typically exhibited no expression or reduced expression in vascular plants (Reboledo et al., 2021). Other specifically upregulated genes in P. patens included carboxylic ester hydrolases, cellobiose dehydrogenase, β-glucosidases, peroxidase, polyketide synthases and numerous hypothetical proteins. Additionally, Bcaba2, which encodes a cytochrome P450 monooxygenase involved in abscisic acid (ABA) biosynthesis, was only upregulated in P. patens tissues after a sustained 24-h infection and was absent in vascular plants (Reboledo et al., 2021).
The activation of defense responses upon pathogen infection involves the induction of various host genes (Wise et al., 2007). Some pathogen-induced genes encode enzymes involved in synthesizing antimicrobial compounds, oxidant stress protection enzymes, tissue repair enzymes, and cell wall fortification enzymes, while others encode proteins with regulatory functions in defense signaling pathways. When B. cinerea infects P. patens, there is an increased expression of phenylalanine ammonia lyase (PAL), chalcone synthase (CHS), lipoxygenase (LOX) and the classic vascular plant host defense marker pathogenesis-related protein PR-1 (Oliver et al., 2009; De León and Montesano, 2013). The enzymes encoded by these genes contribute to the synthesis of phenylpropanoids, flavonoids, and oxygenated fatty acids, which play various roles in defense responses (Feussner and Wasternack, 2002; De León and Montesano, 2013). Additionally, genes associated with PCD are also upregulated, encoding proteases, nucleases and Bax Inhibitor-1 proteins that regulate PCD (De León and Montesano, 2013). The genome of P. patens contains many gene family members involved in phenylpropanoid metabolism (Rensing et al., 2008). P. patens has a higher number of members composing PAL and CHS multigene family members as compared to vascular plants (Koduri et al., 2010; Wolf et al., 2010), and some of these are induced upon pathogen attack (De León et al., 2007; Oliver et al., 2009). These enzymes can produce novel metabolites that may function in impeding pathogen infection. Moreover, the presence of oxylipids (produced by LOX) in P. patens, which are not found in vascular plants, makes it a valuable model for identifying new defense related compounds and defense mechanisms that may have evolved or been lost in plants over time (Oliver et al., 2009).
Plants resist pathogenic microbes by altering hormone levels and expression of defense genes. Salicylic acid (SA), jasmonic acid (JA), and ethylene (ET) play important roles in host responses to pathogen infections (Glazebrook, 2005; Lorenzo and Solano, 2005; Van Loon et al., 2006; Loake and Grant, 2007; Oliver et al., 2009). SA is primarily associated with resistance against biotrophic pathogens and serves as an important endogenous signal for triggering HR and SAR (Saleem et al., 2021). In contrast, JA and ET are typically involved in responses to necrotrophic pathogens, with their signaling pathways working synergistically to produce induced systemic resistance (ISR) (Jiang et al., 2019). These hormones also influence susceptible responses. Generally, the SA and JA pathways are antagonistic, while the JA and ET pathways are synergistic (Xie and Duan, 2023). JA also confers resistance against (hemi)biotrophic pathogens in rice (De Vleesschauwer et al., 2014). And studies have shown that JA activation while abscisic acid signal inhibition may be a key factor in activating the basic response to fungal resistance (Amoroso et al., 2023). Additionally, plant hormones such as abscisic acid and auxin are also involved in PAMP-triggered immune responses. Upon pathogen invasion, JA-isoleucine rapidly accumulates in angiosperms, promoting the interaction between the receptor COI1 (Coronatine-insensitive protein 1) and inhibitory protein JAZ (Jasmonate-ZIM domain) (Chini et al., 2007; Jeong et al., 2023). Although P. patens encodes orthologs of all JA signaling components, the role of JA in moss disease resistance remains unclear (De León et al., 2015). Instead, its precursor cis-oxophytodienoic acid (OPDA) accumulates in moss tissues after pathogen attack (Oliver et al., 2009; De León et al., 2012). Both methyl jasmonate and OPDA can induce the expression of the defense gene PAL. Similar to A. thaliana, OPDA can act as a signaling molecule in mosses, resulting in the induction of defense-related genes (Taki et al., 2005). In mosses, SA treatment also induces expression of PAL. ABA contributes to the synthesis of papillae upon pathogen perception and also promotes the expression of certain defense genes (Adie et al., 2007). In P. patens, ABA induces the synthesis of defense proteins, including the RPM1-related R protein, LOX, an intracellular pathogenesis-related protein, a N-hydroxycinnamoyl/benzoyl transferase involved in phytoalexin production, a hydroxyproline-rich glycoprotein, and a proline-rich protein that aids in cell wall reinforcement, as well as ascorbate peroxidase and peroxiredoxin, indicating ABA may play a role in moss defenses against pathogens (Wang et al., 2010; De León and Montesano, 2017). While auxin can promote wound healing tissue formation in plants, it has a negative regulatory effect during biotrophic pathogen infections. Auxin plays significant roles in resistance against B. cinerea and P. carotovorum (Llorente et al., 2008). It is also implicated in moss defenses, with auxin signaling responding to infection by P. irregulare, P. debaryanum, and C. gloeosporioides (Mittag et al., 2015; Reboledo et al., 2015; Alvarez et al., 2016), and can induce PAL expression (Reboledo et al., 2015). The roles of cytokinin and brassinosteroid in moss defenses have yet to be reported. Overall, these phytohormone signaling pathways have played critical roles in the adaptation of land plants to microbial pathogens (De León and Montesano, 2017).
Gene mining in mosses can provide promising insights for stress-resistant breeding. Currently, a number of disease-resistant genes have been identified in mosses and successfully utilized in vascular plants. Soloist gene, which encodes a unique subfamily member of the AP2/ERF transcription factor family, plays an important role in plant responses to both biotic and abiotic stresses. Overexpression the Soloist gene (ScAPD1-like gene) found in S. caninervis significantly increased the ability of transgenic A. thaliana and S. caninervis to combat V. dahliae, reduced ROS accumulation, and improved their scavenging ability (Li et al., 2023). Additionally, the ectopic expression of PpBURP2 from P. patens in rice enhanced tolerance to osmotic and salt stresses, as well as drought stress. Overexpressing PpBURP2 in rice can enhance resistance to bacterial leaf blight (Yu et al., 2022). Cinnamyl alcohol dehydrogenase (CAD) is a key enzyme in the final step of lignin biosynthesis. PpCAD1 can inhibit the invasion of B. cinerea by boosting phenylpropanoid levels in the cell wall. Studies have shown that plants overexpressing PpCAD1 in P. patens exhibit enhanced resistance to B. cinerea, while knockout mutants show reduced resistance (Jiang et al., 2022). In the vascular plant A. thaliana, AtCAD1 is also essential for lignification (Eudes et al., 2006), and the AtCAD1 gene negatively regulates the cell death aspect of the plant immune response by SA signaling pathway (Morita-Yamamuro et al., 2005). However, ectopic expression of PpCAD1 in A. thaliana significantly enhanced its tolerance to B. cinerea (Jiang et al., 2022). In addition, ectopic expression of the pathogenesis-related (PR) −10 gene (PpPR-10) from P. patens could enhance the resistance of A. thaliana to P. irregulare, which was proved by smaller lesions and less cell damage compared with wild-type plants (Castro et al., 2016). These findings suggest that the discovery of moss genes can be effectively applied to other vascular plants, indicating that studying moss-pathogen interaction can inspire new strategies for molecular breeding aimed at enhancing plant disease resistance.
The study of plant pathology has traditionally centered on the interaction between pathogens and plants. Recent findings indicate that fungi, bacteria, and viruses can also infect mosses. Due to their small size, simple structure, and short life cycles, mosses present an excellent model systems for studying plant-pathogen interactions. Mosses are more suitable for genetic manipulation, allowing for extensive gene deletion and overexpression studies compared to vascular plants (Zhou et al., 2024). Despite the fact that vascular plants and mosses share similar pathogen sensing mechanisms and downstream pathways, many issues in this field remain unresolved. Moreover, mosses like P. patens also have a telomere-to-telomere (T2T) genome (Bi et al., 2024) and extensive data from multiple histological studies, making them ideal for in-depth research on plant-pathogen interactions. Although there have been numerous recent publications on moss-pathogen interactions, further investigation is necessary to fully understand the underlying molecular mechanisms. Firstly, future research could focus on expanding the scope of studies to interactions between different moss species and significant pathogens. As mentioned above, current knowledge of moss-pathogen interactions is limited, primarily concentrating on a few model mosses and typical pathogens. Different moss species may exhibit varying responses and interacting mechanisms against the same pathogen. Therefore, more systematic studies are needed to explore the interactions between diverse moss species and important pathogens, which is crucial for understanding disease resistance mechanisms in mosses.
Secondly, improving the investigations of the molecular pathways that govern how mosses detect pathogens and trigger immune response is essential. Our current understanding of the morphological, physiological, invasion strategies, sensing mechanisms, and downstream signaling pathways in moss-pathogen interactions is quite limited. According to the existing reports, it is found that P. patens lack many receptors that already exist in A. thaliana, such as CREK1 and LYK5, while retaining some basic modules of the immune system (Figure 2), which reveals that bryophytes may adopt different immune strategies from angiosperms. At present, the research on moss immune pathway is still relatively scarce. Subsequent investigations may employ multi-omics and bioinformatic approaches to comprehensively analyze regulatory changes at various levels in pathogen-infected mosses, thereby identifying key immune-related genes and signals associated with immunity. To gain deeper insights into the specificity of immune regulation in plants and the conservation of evolutionary traits, it is important to enhance comparisons between mosses and vascular plants.
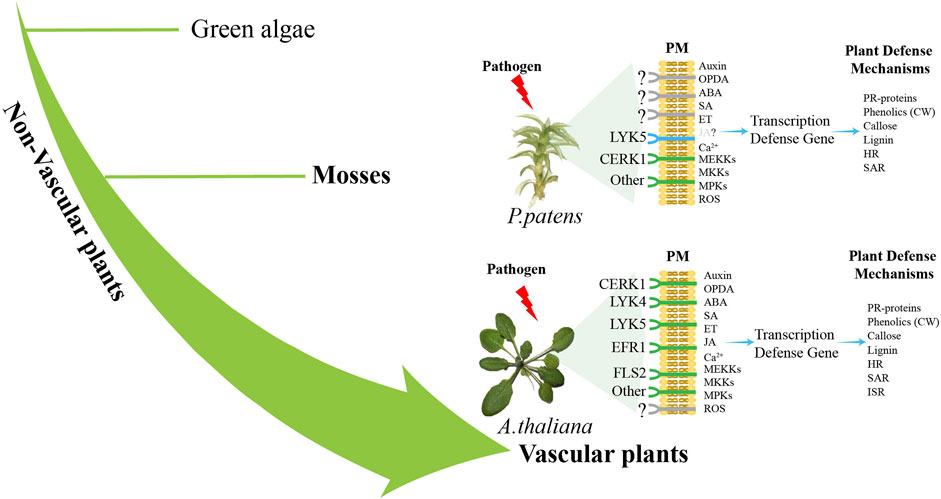
Figure 2. Difference pathways of pathogen sensing between vascular plants and non-vascular plants. Plants sense pathogen-associated molecular patterns (PAMPs) such as FLS2, EFR1, CERK1, and LYK5 through plasma membrane (PM) PRRs (Couto and Zipfel, 2016). Pathogen recognition triggers Ca2+production and activates the MAPK cascade (Seybold et al., 2014). P. patens lacks the close relatives of receptors FLS2 and EFR, while a functional CERK1 receptor senses fungal chitin and bacterial peptidoglycan (Bressendorff et al., 2016). Subsequently, at least one MAP kinase kinase (MEKKs), one MAP kinase (MKKs) and two MAP kinases (MPKs) are activated to participate in the defense response of moss to fungal chitin (Bressendorff et al., 2016). ROS, SA and auxin activate the expression of defense genes, leading to the activation of defense mechanisms (De León et al., 2012; De León and Montesano, 2013), including the expression of genes encoding PR proteins, the entry of phenols into cell wall (CW), callose deposition and the accumulation of pre-lignin compounds (Overdijk et al., 2016; De León and Montesano, 2017). HR-like reaction and SAR were also activated in infected mosses (De León et al., 2012). A. thaliana and other angiosperms have receptors such as LYK5, LYK4, CERK1, EFR1 and FLS2 (Couto and Zipfel, 2016), which activate MEKK, MKK and MPKs, leading to the production of ROS and the expression of defense genes (Meng and Zhang, 2013). Hormones SA, JA and OPDA, ABA, auxin and ET activate the expression of defense genes, leading to the activation of defense mechanisms, including PR proteins, phenolic substances into the CW, callose and lignin deposition, HR, SAR and ISR activation (Glazebrook, 2005). The green color is the identified immune-related receptor, and the blue color is the homologous gene of the receptor identified by genomic comparison.
Thirdly, creating a high-throughput screening platform based on moss mutants is also recommended. Mosses are ideal for high-throughput gene knockout or overexpression due to their small size, short cultivation cycle, and high frequency of homologous recombination. These characteristics allow for large-scale mutant screening under pathogen infections, facilitating the identification of important genes or mechanisms that influence disease resistance.
Additionally, developing a rapid gene screening system in mosses for mining disease resistance genes is crucial. Mosses can serve as ideal platforms for validating plant resistance genes because they are easy to cultivate and support rapid pathogen growth. This allows for the efficient heterologous expression of both known and unknown resistance genes from vascular plants, enabling quick functional verification and high-quality gene selection, which can yield promising candidates for molecular plant breeding. In addition, with advancements in gene editing technology and crop genetic transformation systems (Lu et al., 2020; Cao et al., 2023; Tian et al., 2024), we can look forward to exciting breakthroughs in gene applications that will drive progress and innovation in agriculture and related fields.
The advancement of high-throughput omics technologies opens up opportunities for future research to conduct in-depth analyses of important activated genes, signals, and metabolites in pathogen-infected mosses. By combining gene editing and transgenic techniques to eliminate or overexpress key immune genes in mosses, researchers can gain a better understanding of their specific roles in plant immune responses. Furthermore, comparing how mosses react to various pathogens may reveal conserved immune mechanisms. Mosses are also excellent models for validating plant resistance genes. As our knowledge in this field expands, mosses will play a crucial role in elucidating the evolution of immune mechanisms in plants and will provide the theoretical foundations and technological support for developing disease-resistant crops.
HZ: Data curation, Formal Analysis, Writing–original draft. QY: Data curation, Formal Analysis, Writing–original draft. LW: Investigation, Writing–review and editing. HL: Writing–review and editing. DZ: Writing–review and editing. C-GD: Supervision, Writing–review and editing. XL: Supervision, Writing–review and editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was supported by the Key Research Program of Frontier Sciences and Chinese Academy of Sciences (ZDBS-LY-SM009), Leading Talents in Technological Innovation program (2022TSYCLJ0049) and Third Xinjiang Scientific Expedition Program (Grant No. 2021xjkk0500).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Adie, B. A. T., Pérez-Pérez, J., Pérez-Pérez, M. M., Godoy, M., Sánchez-Serrano, J. J., Schmelz, E. A., et al. (2007). ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis. Plant Cell 19, 1665–1681. doi:10.1105/tpc.106.048041
Akita, M., and Valkonen, J. P. T. (2002). A novel gene family in moss (Physcomitrella patens) shows sequence homology and a phylogenetic relationship with the TIR-NBS class of plant disease resistance genes. J. Mol. Evol. 55, 595–605. doi:10.1007/s00239-002-2355-8
Alvarez, A., Montesano, M., Schmelz, E., and de León, I. P. (2016). Activation of shikimate, phenylpropanoid, oxylipins, and auxin pathways in Pectobacterium carotovorum elicitors-treated moss. Front. Plant Sci. 7, 328. doi:10.3389/fpls.2016.00328
Amoroso, C. G., D'Esposito, D., Cigliano, R. A., and Ercolano, M. R. (2023). Comparison of tomato transcriptomic profiles reveals overlapping patterns in abiotic and biotic stress responses. Int. J. Mol. Sci. 24, 23. doi:10.3390/ijms24044061
Andolfo, G., Iovieno, P., Ricciardi, L., Lotti, C., Filippone, E., Pavan, S., et al. (2019). Evolutionary conservation of MLO gene promoter signatures. BMC Plant Biol. 19, 150. doi:10.1186/s12870-019-1749-3
Andolfo, G., Villano, C., Errico, A., Frusciante, L., Carputo, D., Aversano, R., et al. (2020). Inferring RPW8-NLRs's evolution patterns in seed plants: case study in Vitis vinifera. Planta 251, 32. doi:10.1007/s00425-019-03324-x
Bi, G. Q., Zhao, S. J., Yao, J. W., Wang, H., Zhao, M. K., Sun, Y. Y., et al. (2024). Near telomere-to-telomere genome of the model plant Physcomitrium patens. Nat. Plants 10, 327–343. doi:10.1038/s41477-023-01614-7
Boller, T., and Felix, G. (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60, 379–406. doi:10.1146/annurev.arplant.57.032905.105346
Boller, T., and He, S. Y. (2009). Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science 324, 742–744. doi:10.1126/science.1171647
Bressendorff, S., Azevedo, R., Kenchappa, C. S., de León, I. P., Olsen, J. V., Rasmussen, M. W., et al. (2016). An innate immunity pathway in the moss Physcomitrella patens. Plant Cell 28, 1328–1342. doi:10.1105/tpc.15.00774
Cao, X. S., Xie, H. T., Song, M. L., Zhao, L. H., Deng, S., Tian, Y. F., et al. (2023). Extremely simplified cut-dip-budding method for genetic transformation and gene editing in Taraxacum kok-saghyz. Innovation Life 1, 100040. doi:10.59717/j.xinn-life.2023.100040
Cao, Y. R., Liang, Y., Tanaka, K., Nguyen, C. T., Jedrzejczak, R. P., Joachimiak, A., et al. (2014). The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin-induced complex with related kinase CERK1. eLife 3, e03766. doi:10.7554/eLife.03766
Castro, A., Vidal, S., and de León, I. P. (2016). Moss pathogenesis-related-10 protein enhances resistance to Pythium irregulare in physcomitrella patens and Arabidopsis thaliana. Front. Plant Sci. 7, 17. doi:10.3389/fpls.2016.00580
Chau, R. (1979). Conidial ultrastructure and taxonomic affinity of a fungal parasite of Sphagnum. Mich Bot. 18, 15–18.
Chini, A., Fonseca, S., Fernández, G., Adie, B., Chico, J. M., Lorenzo, O., et al. (2007) The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448, 666–671. doi:10.1038/nature06006
Clarke, J. T., Warnock, R. C. M., and Donoghue, P. C. J. (2011). Establishing a time-scale for plant evolution. New Phytol. 192, 266–301. doi:10.1111/j.1469-8137.2011.03794.x
Commisso, M., Guarino, F., Marchi, L., Muto, A., Piro, A., and Degola, F. (2021). Bryo-Activities: a review on how bryophytes are contributing to the arsenal of natural bioactive compounds against fungi. Plants-Basel 10, 203. doi:10.3390/plants10020203
Couto, D., and Zipfel, C. (2016). Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol. 16, 537–552. doi:10.1038/nri.2016.77
Cove, D., Bezanilla, M., Harries, P., and Quatrano, R. (2006). Mosses as model systems for the study of metabolism and development. Annu. Rev. Plant Biol. 57, 497–520. doi:10.1146/annurev.arplant.57.032905.105338
Davey, M. L., and Currah, R. S. (2006). Interactions between mosses (Bryophyta) and fungi. Can. J. Bot-Rev Can. Bot. 84, 1509–1519. doi:10.1139/b06-120
Davey, M. L., and Currah, R. S. (2009). Atradidymella muscivora gen. et sp. nov. (Pleosporales) and its anamorph Phoma muscivora sp. nov.: A new pleomorphic pathogen of boreal bryophytes. Am. J. Bot. 96, 1281–1288. doi:10.3732/ajb.0900010
Davey, M. L., Tsuneda, A., and Currah, R. S. (2009). Pathogenesis of bryophyte hosts by the ascomycete Atradidymella muscivora. Am. J. Bot. 96, 1274–1280. doi:10.3732/ajb.0800239
Davidsson, P. R., Kariola, T., Niemi, O., and Palva, E. T. (2013). Pathogenicity of and plant immunity to soft rot pectobacteria. Front. Plant Sci. 4, 191. doi:10.3389/fpls.2013.00191
Delaux, P. M., Hetherington, A. J., Coudert, Y., Delwiche, C., Dunand, C., Gould, S., et al. (2019). Reconstructing trait evolution in plant evo-devo studies. Curr. Biol. 29, R1110–R1118. doi:10.1016/j.cub.2019.09.044
Delaux, P. M., and Schornack, S. (2021). Plant evolution driven by interactions with symbiotic and pathogenic microbes. Science 371, eaba6605. doi:10.1126/science.aba6605
De León, I. P. (2011). The moss Physcomitrella patens as a model system to study interactions between plants and phytopathogenic fungi and oomycetes. J. Pathog. 2011, 719873. doi:10.4061/2011/719873
De León, I. P., Hamberg, M., and Castresana, C. (2015). Oxylipins in moss development and defense. Front. Plant Sci. 6, 12. doi:10.3389/fpls.2015.00483
De León, I. P., and Montesano, M. (2013). Activation of defense mechanisms against pathogens in mosses and flowering plants. Int. J. Mol. Sci. 14, 3178–3200. doi:10.3390/ijms14023178
De León, I. P., and Montesano, M. (2017). Adaptation mechanisms in the evolution of moss defenses to microbes. Front. Plant Sci. 8, 14. doi:10.3389/fpls.2017.00366
De León, I. P., Oliver, J. P., Castro, A., Gaggero, C., Bentancor, M., and Vidal, S. (2007). Erwinia carotovora elicitors and Botrytis cinerea activate defense responses in Physcomitrella patens. BMC Plant Biol. 7, 52. doi:10.1186/1471-2229-7-52
De León, I. P., Schmelz, E. A., Gaggero, C., Castro, A., Alvarez, A., and Montesano, M. (2012). Physcomitrella patens activates reinforcement of the cell wall, programmed cell death and accumulation of evolutionary conserved defence signals, such as salicylic acid and 12-oxo-phytodienoic acid, but not jasmonic acid, upon Botrytis cinerea infection. Mol. Plant Pathol. 13, 960–974. doi:10.1111/j.1364-3703.2012.00806.x
De Vleesschauwer, D., Xu, J., and Höfte, M. (2014). Making sense of hormone-mediated defense networking: from rice to Arabidopsis. Front. Plant Sci. 5, 611. doi:10.3389/fpls.2014.00611
de Vries, S., de Vries, J., von Dahlen, J. K., Gould, S. B., Archibald, J. M., Rose, L. E., et al. (2018). On plant defense signaling networks and early land plant evolution. Commun. and Integr. Biol. 11, 1–14. doi:10.1080/19420889.2018.1486168
Dixon, R. A. (2001). Natural products and plant disease resistance. Nature 411, 843–847. doi:10.1038/35081178
Döbbeler, P. (1988). Drei neue moosebewohnende ascomyceten. Plant Syst. Evol. 158, 329–340. doi:10.1007/BF00936354
Dobbeler, P. (1997). Biodiversity of bryophilous ascomycetes. Biodivers. Conserv. 6, 721–738. doi:10.1023/a:1018370304090
Döbbeler, P. (2003). Ascomycetes on dendroligotrichum (musci). Nova Hedwig. 76, 1–44. doi:10.1127/0029-5035/2003/0076-0001
Duan, C. G., Fang, Y. Y., Zhou, B. J., Zhao, J. H., Hou, W. N., Zhu, H., et al. (2012). Suppression of Arabidopsis ARGONAUTE1-mediated slicing, transgene-induced RNA silencing, and DNA methylation by distinct domains of the cucumber mosaic virus 2b protein. Plant Cell 24, 259–274. doi:10.1105/tpc.111.092718
During, H. J., and Vantooren, B. F. (1990). Bryophyte interactions with other plants. J. Linn. Soc. 104, 79–98. doi:10.1111/j.1095-8339.1990.tb02212.x
Emms, D. M., and Kelly, S. (2019). OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20, 238. doi:10.1186/s13059-019-1832-y
Engelsdorf, T., Gigli-Bisceglia, N., Veerabagu, M., McKenna, J. F., Vaahtera, L., Augstein, F., et al. (2018). The plant cell wall integrity maintenance and immune signaling systems cooperate to control stress responses in Arabidopsis thaliana. Sci. Signal 11, eaao3070. doi:10.1126/scisignal.aao3070
Eudes, A., Pollet, B., Sibout, R., Do, C. T., Seguin, A., Lapierre, C., et al. (2006). Evidence for a role of AtCAD 1 in lignification of elongating stems of Arabidopsis thaliana. Planta 225, 23–39. doi:10.1007/s00425-006-0326-9
Feussner, I., and Wasternack, C. (2002). The lipoxygenase pathway. Annu. Rev. Plant Biol. 53, 275–297. doi:10.1146/annurev.arplant.53.100301.135248
Fradin, E. F., and Thomma, B. (2006). Physiology and molecular aspects of Verticillium wilt diseases caused by V-dahliae and V-albo-atrum. Mol. Plant Pathol. 7, 71–86. doi:10.1111/j.1364-3703.2006.00323.X
Gabriel, D. W., and Rolfe, B. G. (1990). Working models of specific recognition in plant-microbe interactions. Annu. Rev. Phytopathol. 28, 365–391. doi:10.1146/annurev.py.28.090190.002053
Galotto, G., Abreu, I., Sherman, C., Liu, B. Y., Gonzalez-Guerrero, M., and Vidali, L. (2020). Chitin triggers calcium-mediated immune response in the plant model physcomitrella patens. Mol. Plant-Microbe Interact. 33, 911–920. doi:10.1094/mpmi-03-20-0064-r
Gao, M. Z., Wan, M. T., Yang, L. Y., Zhao, M., Liu, X. J., Chen, J. J., et al. (2021). Molecular and physiological characterization of Arabidopsis Colletotrichum gloeosporioides pathosystem. Plant Pathol. 70, 1168–1179. doi:10.1111/ppa.13364
Ghelis, T. (2011). Signal processing by protein tyrosine phosphorylation in plants. Plant Signal. Behav. 6, 942–951. doi:10.4161/psb.6.7.15261
Glazebrook, J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227. doi:10.1146/annurev.phyto.43.040204.135923
Gómez-Gómez, L., and Boller, T. (2000). FLS2:: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5, 1003–1011. doi:10.1016/s1097-2765(00)80265-8
González, J., Reyes, F., Salas, C., Santiago, M., Codriansky, Y., Colihueque, N., et al. (2006). Arabidopsis thaliana:: a model host plant to study plant-pathogen interaction using Chilean field isolates of Botrytis cinerea. Biol. Res. 39, 221–228. doi:10.4067/s0716-97602006000200004
Govrin, E. M., and Levine, A. (2000). The hypersensitive response facilitates plant infection by the necrotrophic pathogen Botrytis cinerea. Curr. Biol. 10, 751–757. doi:10.1016/s0960-9822(00)00560-1
Hühns, S., Bauer, C., Buhlmann, S., Heinze, C., von Bargen, S., Paape, M., et al. (2003). Tomato spotted wilt virus (TSWV) infection of Physcomitrella patens gametophores. Plant Cell Tissue Organ Cult. 75, 183–187. doi:10.1023/a:1025070722420
Jeong, H. M., Patterson, H., and Carella, P. (2023). Bryo-FIGHTs: emerging insights and principles acquired from non-vascular plant-pathogen interactions. Curr. Opin. Plant Biol. 76, 102484. doi:10.1016/j.pbi.2023.102484
Jiang, N. H., Fan, X. Y., Lin, W. P., Wang, G. P., and Cai, K. Z. (2019). Transcriptome analysis reveals new insights into the bacterial wilt resistance mechanism mediated by silicon in tomato. Int. J. Mol. Sci. 20, 761. doi:10.3390/ijms20030761
Jiang, S., Tian, X., Huang, X. L., Xin, J. K., and Yan, H. Q. (2022). Physcomitrium patens CAD1 has distinct roles in growth and resistance to biotic stress. BMC Plant Biol. 22, 518. doi:10.1186/s12870-022-03892-3
Jiang, S., Yu, Z. J., and Lan, S. C. (2013). The changes on metabolism of reactive oxygen species and expressionof pathogenesis related protein 1 during Interaction of Physcomitrella patens with Botrytis cinerea. Chin. J. Aeronaut. 37, 349–354. doi:10.16357/j.cnki.issn1000-58622013.04.005
Jones, J. D. G., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi:10.1038/nature05286
Kariola, T., Brader, G., Li, J., and Palva, E. T. (2005). Chlorophyllase 1, a damage control enzyme, affects the balance between defense pathways in plants. Plant Cell 17, 282–294. doi:10.1105/tpc.104.025817
Koduri, P. K. H., Gordon, G. S., Barker, E. I., Colpitts, C. C., Ashton, N. W., and Suh, D. Y. (2010). Genome-wide analysis of the chalcone synthase superfamily genes of Physcomitrella patens. Plant MolBiol 72, 247–263. doi:10.1007/s11103-009-9565-z
Koukol, O., and Kovarova, M. (2007). Autecology of Scleroconidioma sphagnicola particularly in šumava national park (Czech republic). Natl. Park Czech Mycol. 59, 111–123. doi:10.33585/cmy.59113
Li, X. S., Yang, R. R., Liang, Y. Q., Gao, B., Li, S. M., Bai, W. W., et al. (2023). The ScAPD1-like gene from the desert moss Syntrichia caninervis enhances resistance to Verticillium dahliae via phenylpropanoid gene regulation. Plant J. 113, 75–91. doi:10.1111/tpj.16035
Ligrone, R., Vaughn, K. C., Renzaglia, K. S., Knox, J. P., and Duckett, J. G. (2002). Diversity in the distribution of polysaccharide and glycoprotein epitopes in the cell walls of bryophytes: new evidence for the multiple evolution of water-conducting cells. New Phytol. 156, 491–508. doi:10.1046/j.1469-8137.2002.00538.x
Liu, J., Li, W. J., Wu, G., and Ali, K. (2024). An update on evolutionary, structural, and functional studies of receptor-like kinases in plants. Front. Plant Sci. 15, 1305599. doi:10.3389/fpls.2024.1305599
Liu, J. J., Zhang, C. G., Jia, X. C., Wang, W. X., and Yin, H. (2020). Comparative analysis of RNA-binding proteomes under Arabidopsis thaliana-Pst DC3000-PAMP interaction by orthogonal organic phase separation. Int. J. Biol. Macromol. 160, 47–54. doi:10.1016/j.ijbiomac.2020.05.164
Liu, Y. (2011). Pathogenicity of GFP transformed Botrytis cinerea and its application inestablishment of Physcomifrella patens disease interaetion system, Master. Shanghai, China: East China Normal University.
Llorente, F., Muskett, P., Sánchez-Vallet, A., López, G., Ramos, B., Sánchez-Rodríguez, C., et al. (2008). Repression of the auxin response pathway increases Arabidopsis susceptibility to necrotrophic fungi. Mol. Plant 1, 496–509. doi:10.1093/mp/ssn025
Loake, G., and Grant, M. (2007). Salicylic acid in plant defence-the players and protagonists. Curr. Opin. Plant Biol. 10, 466–472. doi:10.1016/j.pbi.2007.08.008
Lorenzo, O., and Solano, R. (2005). Molecular players regulating the jasmonate signalling network. Curr. Opin. Plant Biol. 8, 532–540. doi:10.1016/j.pbi.2005.07.003
Lu, Y. M., Tian, Y. F., Shen, R. D., Yao, Q., Wang, M. G., Chen, M., et al. (2020). Targeted, efficient sequence insertion and replacement in rice. Nat. Biotechnol. 38, 1402–1407. doi:10.1038/s41587-020-0581-5
Marchetti, F., Distéfano, A. M., Cainzos, M., Setzes, N., Cascallares, M., López, G. A., et al. (2024). Cell death in bryophytes: emerging models to study core regulatory modules and conserved pathways. Ann. Bot. 134, 367–384. doi:10.1093/aob/mcae081
Martínez-Abaigar, J., Núñez-Olivera, E., Matcham, H. W., and Duckett, J. G. (2005). Interactions between parasitic fungi and mosses:: pegged and swollen-tipped rhizoids in Funaria and Bryum. J. Bryol. 27, 47–53. doi:10.1179/174328205x40572
Marttinen, E. M., Lehtonen, M. T., van Gessel, N., Reski, R., and Valkonen, J. P. T. (2022). Viral suppressor of RNA silencing in vascular plants also interferes with the development of the bryophyte Physcomitrella patens. Plant Cell Environ. 45, 220–235. doi:10.1111/pce.14194
Meng, X. Z., and Zhang, S. Q. (2013). “MAPK cascades in plant disease resistance signaling,” in Annual review of phytopathology. Editor N. K. VanAlfen (Palo Alto: Annual Reviews) 51, 245–266. doi:10.1146/annurev-phyto-082712-102314
Mittag, J., Sola, I., Rusak, G., and Ludwig-Müller, J. (2015). Physcomitrella patens auxin conjugate synthetase (GH3) double knockout mutants are more resistant to Pythium infection than wild type. J. Plant Physiol. 183, 75–83. doi:10.1016/j.jplph.2015.05.015
Miya, A., Albert, P., Shinya, T., Desaki, Y., Ichimura, K., Shirasu, K., et al. (2007). CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 104, 19613–19618. doi:10.1073/pnas.0705147104
Morita-Yamamuro, C., Tsutsui, T., Sato, M., Yoshioka, H., Tamaoki, M., Ogawa, D., et al. (2005). The Arabidopsis gene CAD1 controls programmed cell death in the plant immune system and encodes a protein containing a MACPF domain. Plant Cell Physiol. 46, 902–912. doi:10.1093/pcp/pci095
Ngou, B. P. M., Wyler, M., Schmid, M. W., Kadota, Y., and Shirasu, K. (2024). Evolutionary trajectory of pattern recognition receptors in plants. Nat. Commun. 15, 308. doi:10.1038/s41467-023-44408-3
Oliver, J. P., Castro, A., Gaggero, C., Cascón, T., Schmelz, E. A., Castresana, C., et al. (2009). Pythium infection activates conserved plant defense responses in mosses. Planta 230, 569–579. doi:10.1007/s00425-009-0969-4
Otero-Blanca, A., Pérez-Llano, Y., Reboledo-Blanco, G., Lira-Ruan, V., Padilla-Chacon, D., Folch-Mallol, J. L., et al. (2021). Physcomitrium patens Infection by Colletotrichum gloeosporioides: understanding the fungal-bryophyte interaction by microscopy, phenomics and RNA sequencing. J. Fungi 7, 677. doi:10.3390/jof7080677
Overdijk, E. J. R., De Keijzer, J., De Groot, D., Schoina, C., Bouwmeester, K., Ketelaar, T., et al. (2016). Interaction between the moss Physcomitrella patens and Phytophthora: a novel pathosystem for live-cell imaging of subcellular defence. J. Microsc. 263, 171–180. doi:10.1111/jmi.12395
Polischuk, V., Budzanivska, I., Shevchenko, T., and Oliynik, S. (2007). Evidence for plant viruses in the region of Argentina Islands, Antarctica. FEMS Microbiol. Ecol. 59, 409–417. doi:10.1111/j.1574-6941.2006.00242.x
Popper, Z. A. (2008). Evolution and diversity of green plant cell walls. Curr. Opin. Plant Biol. 11, 286–292. doi:10.1016/j.pbi.2008.02.012
Qiao, Y. L., Liu, L., Xiong, Q., Flores, C., Wong, J., Shi, J. X., et al. (2013). Oomycete pathogens encode RNA silencing suppressors. Nat. Genet. 45, 330–333. doi:10.1038/ng.2525
Ramírez-Zavaleta, C. Y., García-Barrera, L. J., Rodríguez-Verástegui, L. L., Arrieta-Flores, D., and Gregorio-Jorge, J. (2022). An overview of PRR- and NLR-mediated immunities: conserved signaling components across the plant kingdom that communicate both pathways. Int. J. Mol. Sci. 23, 12974. doi:10.3390/ijms232112974
Reboledo, G., Agorio, A., and De León, I. P. (2022a). Moss transcription factors regulating development and defense responses to stress. J. Exp. Bot. 73, 4546–4561. doi:10.1093/jxb/erac055
Reboledo, G., Agorio, A., Vignale, L., Alvarez, A., and De Leon, I. P. (2022b). The moss-specific transcription factor PpERF24 positively modulates immunity against fungal pathogens in Physcomitrium patens. Front. Plant Sci. 13, 908682. doi:10.3389/fpls.2022.908682
Reboledo, G., Agorio, A., Vignale, L., Batista-García, R. A., and De León, I. P. (2021). Botrytis cinerea transcriptome during the infection process of the bryophyte Physcomitrium patens and angiosperms. J. Fungi 7, 11. doi:10.3390/jof7010011
Reboledo, G., del Campo, R., Alvarez, A., Montesano, M., Mara, H., and de León, I. P. (2015). Physcomitrella patens activates defense responses against the pathogen Colletotrichum gloeosporioides. Int. J. Mol. Sci. 16, 22280–22298. doi:10.3390/ijms160922280
Redhead, S. A., and Spicer, K. W. (1981). Discinella schimperi, a circumpolar parasite of Sphagnum squarrosum, and notes on bryophytomyces sphagni. Mycologia 73, 904–913. doi:10.2307/3759801
Rensing, S. A., Lang, D., Zimmer, A. D., Terry, A., Salamov, A., Shapiro, H., et al. (2008). The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319, 64–69. doi:10.1126/science.1150646
Resemann, H. C., Lewandowska, M., Gömann, J., and Feussner, I. (2019). Membrane lipids, waxes and oxylipins in the moss model organism Physcomitrella patens. Plant Cell Physiol. 60, 1166–1175. doi:10.1093/pcp/pcz006
Retel, C., Märkle, H., Becks, L., and Feulner, P. G. D. (2019). Ecological and evolutionary processes shaping viral genetic diversity. Viruses-Basel 11, 220. doi:10.3390/v11030220
Reusche, M., Truskina, J., Thole, K., Nagel, L., Rindfleisch, S., Tran, V. T., et al. (2014). Infections with the vascular pathogens Verticillium longisporum and Verticillium dahliae induce distinct disease symptoms and differentially affect drought stress tolerance of Arabidopsis thaliana. Environ. Exp. Bot. 108, 23–37. doi:10.1016/j.envexpbot.2013.12.009
Rice, A. V., Tsuneda, A., and Currah, R. S. (2006). In vitro decomposition of Sphagnum by some microfungi resembles white rot of wood. FEMS Microbiol. Ecol. 56, 372–382. doi:10.1111/j.1574-6941.2006.00071.x
Saleem, M., Fariduddin, Q., and Castroverde, C. D. M. (2021). Salicylic acid: a key regulator of redox signalling and plant immunity. Plant Physiol. Biochem. 168, 381–397. doi:10.1016/j.plaphy.2021.10.011
Sarris, P. F., Cevik, V., Dagdas, G., Jones, J. D. G., and Krasileva, K. V. (2016). Comparative analysis of plant immune receptor architectures uncovers host proteins likely targeted by pathogens. BMC Biol. 14, 18. doi:10.1186/s12915-016-0228-7
Schaefer, D. G. (2002). A newmoss genetics:: targeted mutagenesis in Physcomitrella patens. Annu. Rev. Plant Biol. 53, 477–501. doi:10.1146/annurev.arplant.53.100301.135202
Schaefer, D. G., and Zryd, J. P. (1997). Efficient gene targeting in the moss Physcomitrella patens. Plant J. 11, 1195–1206. doi:10.1046/j.1365-313X.1997.11061195.x
Seybold, H., Trempel, F., Ranf, S., Scheel, D., Romeis, T., and Lee, J. (2014). Ca2+ signalling in plant immune response: from pattern recognition receptors to Ca2+ decoding mechanisms. New Phytol. 204, 782–790. doi:10.1111/nph.13031
Shao, Z. Q., Xue, J. Y., Wang, Q., Wang, B., and Chen, J. Q. (2019). Revisiting the origin of plant NBS-LRR genes. Trends Plant Sci. 24, 9–12. doi:10.1016/j.tplants.2018.10.015
Singh, V. P., Jaiswal, S., Wang, Y. Y., Feng, S. L., Tripathi, D. K., Singh, S., et al. (2024). Evolution of reactive oxygen species cellular targets for plant development. Trends Plant Sci. 29, 865–877. doi:10.1016/j.tplants.2024.03.005
Sivaranjani, M., Krishnan, S. R., Kannappan, A., Ramesh, M., and Ravi, A. V. (2016). Curcumin from Curcuma longa affects the virulence of Pectobacterium wasabiae and P. carotovorum subsp carotovorum via quorum sensing regulation. Eur. J. Plant Pathol. 146, 793–806. doi:10.1007/s10658-016-0957-z
Summermatter, K., Sticher, L., and Metraux, J. P. (1995). Systemic responses in Arabidopsis thaliana infected and challenged with Pseudomonas syringae pv syringae. Plant Physiol. 108, 1379–1385. doi:10.1104/pp.108.4.1379
Taki, N., Sasaki-Sekimoto, Y., Obayashi, T., Kikuta, A., Kobayashi, K., Ainai, T., et al. (2005). 12-oxo-phytodienoic acid triggers expression of a distinct set of genes and plays a role in wound-induced gene expression in Arabidopsis. Plant Physiol. 139, 1268–1283. doi:10.1104/pp.105.067058
Takken, F. L. W., and Tameling, W. I. L. (2009). To nibble at plant resistance proteins. Science 324, 744–746. doi:10.1126/science.1171666
Tanigaki, Y., Ito, K., Obuchi, Y., Kosaka, A., Yamato, K. T., Okanami, M., et al. (2014). Physcomitrella patens has kinase-LRR RGene homologs and interacting proteins. PLoS One 9, e95118. doi:10.1371/journal.pone.0095118
Tian, Y. F., Li, X. B., Xie, J. Y., Zheng, Z., Shen, R. D., Cao, X. S., et al. (2024). Targeted G-to-T base editing for generation of novel herbicide-resistance gene alleles in rice. J. Integr. Plant Biol. 66, 1048–1051. doi:10.1111/jipb.13657
Torres, M. A., Jones, J. D. G., and Dangl, J. L. (2006). Reactive oxygen species signaling in response to pathogens. Plant Physiol. 141, 373–378. doi:10.1104/pp.106.079467
Toth, I. K., and Birch, P. R. J. (2005). Rotting softly and stealthily. Curr. Opin. Plant Biol. 8, 424–429. doi:10.1016/j.pbi.2005.04.001
Tsuneda, A., Chen, M. H., and Currah, R. S. (2001b). Characteristics of a disease of Sphagnum fuscum caused by Scleroconidioma sphagnicola. Can. J. Bot-Rev Can. Bot. 79, 1217–1224. doi:10.1139/b01-102
Tsuneda, A., Thormann, M. N., and Currah, R. S. (2001a). Modes of cell-wall degradation of Sphagnum fuscum by Acremonium cf. curvulum and Oidiodendron maius. Can. J. Bot-Rev Can. Bot. 79, 93–100. doi:10.1139/b00-149
Untiedt, E., and Muller, K. (1985). Colonization of Sphagnum cells by Lyophyllum palustre. Can. J. Bot-Rev Can. Bot. 63, 757–761. doi:10.1139/b85-095
Van Loon, L. C., Geraats, B. P. J., and Linthorst, H. J. M. (2006). Ethylene as a modulator of disease resistance in plants. Trends Plant Sci. 11, 184–191. doi:10.1016/j.tplants.2006.02.005
Wan, J. R., Tanaka, K., Zhang, X. C., Son, G. H., Brechenmacher, L., Tran, H. N. N., et al. (2012). LYK4, a lysin motif receptor-like kinase, Is important for chitin signaling and plant innate immunity in Arabidopsis. Plant Physiol. 160, 396–406. doi:10.1104/pp.112.201699
Wang, C. Y., Liu, Y., Li, S. S., and Han, G. Z. (2015). Insights into the origin and evolution of the plant hormone signaling machinery. Plant Physiol. 167, 872–886. doi:10.1104/pp.114.247403
Wang, X. Q., Kuang, T. Y., and He, Y. K. (2010). Conservation between higher plants and the moss Physcomitrella patens in response to the phytohormone abscisic acid: a proteomics analysis. BMC Plant Biol. 10, 192. doi:10.1186/1471-2229-10-192
Wise, R. P., Moscou, M. J., Bogdanove, A. J., and Whitham, S. A. (2007). Transcript profiling in host-pathogen interactions. Annu. Rev. Phytopathol. 45, 329–369. doi:10.1146/annurev.phyto.45.011107.143944
Wolf, L., Rizzini, L., Stracke, R., Ulm, R., and Rensing, S. A. (2010). The molecular and physiological responses of Physcomitrella patens to ultraviolet-B radiation. Plant Physiol. 153, 1123–1134. doi:10.1104/pp.110.154658
Wu, Y., and Zhou, J. M. (2013). Receptor-like kinases in plant innate immunity. J. Integr. Plant Biol. 55, 1271–1286. doi:10.1111/jipb.12123
Xie, S. S., and Duan, C. G. (2023). Epigenetic regulation of plant immunity: from chromatin codes to plant disease resistance. aBIOTECH 4, 124–139. doi:10.1007/s42994-023-00101-z
Xu, Z. S., Chen, M., Li, L. C., and Ma, Y. Z. (2011). Functions and application of the AP2/ERF transcription factor family in crop improvement. J. Integr. Plant Biol. 53, 570–585. doi:10.1111/j.1744-7909.2011.01062.x
Xue, J. Y., Wang, Y., Wu, P., Wang, Q., Yang, L. T., Pan, X. H., et al. (2012). A primary survey on bryophyte species reveals two novel classes of nucleotide-ninding site (NBS) genes. PLoS One 7, 7. doi:10.1371/journal.pone.0036700
Yan, H. Q., Zhang, T. T., Lan, S. C., and Jiang, S. (2018). Ultrastructural study on the interaction between Physcomitrella patens and Botrytis cinerea. Plant Pathol. 67, 42–50. doi:10.1111/ppa.12720
Yan, Z., Haxim, Y., Liu, X. J., Li, X. S., Zhang, D. Y., and Li, J. (2023). Research on the pathogenicity of Pseudomonas syringae to three moss species. J. Yunnan Agric. Univ. 38, 383–391. doi:10.12101/j.issn.1004-390X(n).202111051
Yin, C. T., Ramachandran, S. R., Zhai, Y., Bu, C. Y., Pappu, H. R., and Hulbert, S. H. (2019). A novel fungal effector from Puccinia graminis suppressing RNA silencing and plant defense responses. New Phytol. 222, 1561–1572. doi:10.1111/nph.15676
Yu, S. W., Yang, F. W., Zou, Y. Q., Yang, Y. A., Li, T. F., Chen, S. J., et al. (2022). Overexpressing PpBURP2 in rice increases plant defense to abiotic stress and bacterial leaf blight. Front. Plant Sci. 13, 812279. doi:10.3389/fpls.2022.812279
Zhang, L., Du, L., and Poovaiah, B. W. (2014). Calcium signaling and biotic defense responses in plants. Plant Signal. Behav. 9, e973818. doi:10.4161/15592324.2014.973818
Zhou, P., Liu, X. J., Liang, Y. Q., Zhang, Y., Li, X. S., and Zhang, D. Y. (2024). A simple, highly efficient Agrobacterium tumefaciens-mediated moss transformation system with broad applications. aBIOTECH 22, 476–487. doi:10.1007/s42994-024-00174-4
Zipfel, C. (2009). Early molecular events in PAMP-triggered immunity. Curr. Opin. Plant Biol. 12, 414–420. doi:10.1016/j.pbi.2009.06.003
Keywords: moss, pathogen, interaction, disease resistance mechanism, plant immune receptors
Citation: Zhang H, Yang Q, Wang L, Liu H, Zhang D, Duan C-G and Li X (2025) Moss-pathogen interactions: a review of the current status and future opportunities. Front. Genet. 16:1539311. doi: 10.3389/fgene.2025.1539311
Received: 04 December 2024; Accepted: 21 January 2025;
Published: 11 February 2025.
Edited by:
Tao Xu, Jiangsu Normal University, ChinaReviewed by:
Aroldo Cisneros, National Polytechnic Institute (IPN), MexicoCopyright © 2025 Zhang, Yang, Wang, Liu, Zhang, Duan and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cheng-Guo Duan, Y2dkdWFuQGNlbXBzLmFjLmNu; Xiaoshuang Li, bGl4c0Btcy54amIuYWMuY24=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.