
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 06 March 2025
Sec. Genetics of Common and Rare Diseases
Volume 16 - 2025 | https://doi.org/10.3389/fgene.2025.1519108
Background: High altitude polycythemia (HAPC) is a disease with high morbidity and great harm in high altitude populations. It has been shown that Single Nucleotide Polymorphisms (SNPs) correlate with the genetic basis of adaptation to plateau hypoxia in Tibetan populations. The EPAS1 and PPARA genes are involved in hypoxia adaptation by encoding transcription factors in Tibetan populations at high altitude. The aim of this study was to investigate the association of EPAS1 and PPARA gene locus polymorphisms with genetic susceptibility to HAPC in the Chinese Tibetan population.
Methods: We included 78 HAPC patients and 84 healthy controls, and genotyped the EPAS1 gene SNP loci (rs6735530, rs6756667, rs7583392, and rs12467821) and PPARA rs6520015 by using TaqMan polymerase chain reaction. Logistic regression was used to analyze the association between these SNPs and HAPC; interactions between SNPs were also predicted by multifactorial dimensionality reduction (MDR) analysis.
Results: We found that the PPARA rs6520015 polymorphism was not associated with the risk of HAPC in the Chinese Tibetan population; EPAS1 rs6735530, rs6756667, rs7583392, and rs12467821 increased the risk of HAPC in some models. Haplotype TCAGC decreases the risk of HAPC; Haplotype TTGAT increases the risk of HAPC; and EPAS1 rs7583392 is in complete linkage disequilibrium with rs12467821. The best prediction model was the EPAS1 rs6756667 unit point model, but the P value was greater than 0.05 in all three models, which was not statistically significant.
Conclusion: The present findings suggest that among the Tibetan population in China, There is an association between EPAS1 rs6735530, rs6756667, rs7583392, and rs12467821 and the risk of HAPC, and that there is no significant correlation between PPARA rs6520015 and the risk of HAPC.
High altitude polycythemia (HAPC) is a chronic plateau disease in which the organism lives for a long period of time on a plateau at an altitude of more than 2,500 m above sea level, and the lack of oxygen in the tissues leads to the compensatory overproliferation of erythrocytes (women’s hemoglobin ≥19 g/dL, men’s hemoglobin ≥ 21 g/dL), increased blood viscosity, and clinical symptoms such as dizziness, shortness of breath, and chest tightness (León-Velarde et al., 2005; Beall and Goldstein, 1987; Wu, 2005). HAPC is the most prevalent and harmful chronic high altitude disease among high altitude residents. It is estimated that up to 5%–10% of high altitude residents may develop HAPC(1). There are differences in the prevalence of HAPC by race, gender, age, altitude of residence, and duration of residence (Meyer et al., 2017). The risk of HAPC is associated with genetic, environmental and physiological factors. In the low oxygen environment of plateau, lowlanders are more likely to suffer from HAPC than highlanders (Garrido et al., 1996). It has been reported that the prevalence of HAPC is lower in Tibetan long-established populations at the same altitude compared to migrated Han Chinese populations (Wu et al., 2005). Genome-wide association studies (GWAS) have confirmed that the differences in genetic background between lowlanders and highlanders are the result of natural selection of genomic loci over hundreds of generations in highlanders. SNPs in genes such as EPAS1, PPARA, EGLN1, EDNRA, and PTEN were found to correlate with hemoglobin (Hb) concentrations in high altitude Tibetan populations, and they found that most of these genes play a role in the HIF-1 pathway (Simonson et al., 2010). In addition, Mallik N suggests that gain-of-function mutations in EPAS1 and loss-of-function mutations in EGLN1 lead to constitutive activation of EPO signaling and thus to HAPC. Strong and significant associations between Hb concentrations and haplotypic variation in PPARA and EGLN1 provide evidence of a genetic mechanism for the form of high-altitude acclimation that characterizes Tibetan populations (Simonson et al., 2010; Mallik et al., 2021). Polymorphisms in the CYP17A1 and CYP2E1 gene loci have been shown to play a role in mediating susceptibility to HAPC (Xu et al., 2015a).
Endothelial-type PAS structural domain protein 1 (EPAS1), also known as hypoxia-inducible factor 2 (HIF-2ɑ), is encoded by the EPAS1 gene located at 2p21, and is a protein composed of 870 amino acids (Karaghiannis et al., 2023). Numerous studies have shown that polymorphisms in EPAS1 are associated with the pathogenesis, pathologic staging, progression, and prognosis of cancers such as colorectal cancer, hepatocellular carcinoma, esophageal squamous cell carcinoma, non-small cell lung cancer, and papillary thyroid carcinoma (Baba et al., 2010; Mohammed et al., 2011; Bangoura et al., 2004; Islam et al., 2020; Zhen et al., 2021; Zhang et al., 2023). EPAS1 rs7583392, rs6756667, and rs12467821 have been reported to be closely associated with plateau diseases and are thought to be related to high altitude acclimatization in Tibetans (Li et al., 2024; Basang et al., 2015; Bhandari et al., 2017; Pan et al., 2019). The hypoxia-inducible factor (HIF) oxygen signaling pathway plays an important role in hypoxia acclimatization in Tibetan populations (Simonson et al., 2010; Wang et al., 2011; Beall et al., 2010; Peng et al., 2011), but whether there is an association with HAPC is not yet known. The protein stability of HIF-2ɑ increases when the body is in a hypoxic state, and accumulated HIF-2ɑ continues to stimulate the HIF pathway, promoting transcription of downstream genes involved in erythropoiesis, hypoxic metabolism, inflammation, angiogenesis, and tumorigenesis (Wang et al., 2023; van Patot and Gassmann, 2011; Davis et al., 2022).
The peroxisome proliferator-activated receptor α gene (PPARA) is located on chromosome 22 and encodes peroxisome proliferator-activated receptor α (PPARα), a member of the nuclear receptor family of transcription factors, which regulates physiological processes, such as lipid metabolism, gluconeogenesis, and immune responses, particularly fatty acid β-oxidation in mitochondria and peroxisomes (Standage et al., 2017; Jay and Ren, 2007). Polymorphisms in the PPARA have been reported to be associated with the risk of liver disease, celiac disease, type 2 diabetes, and hyperlipidemia, among others (Jay and Ren, 2007; Li et al., 2014; Mostowy et al., 2016; Eurlings et al., 2002). A variant in the “C” allele of PPARA rs6520015 affects cardiac ejection function after acute plateau exposure (Standage et al., 2017; Yang et al., 2019). The PPARA promoter is associated with vascular function, target gene transcription and hemoglobin concentration during hypoxia in a Tibetan population (Shen et al., 2020).
In Tibetan populations at high altitude, EPAS1 and PPARA genes are involved in hypoxia adaptation by encoding transcription factors, and they play important roles in hypoxia-responsive pathways and oxidative stress (Simonson et al., 2010). However, whether the polymorphism of these genes is related to the incidence rate of HAPC in Chinese Tibetan population still needs to be investigated. Therefore, the aim of this study was to investigate the genetic relationship between polymorphisms in the EPAS1 and PPARA loci and HAPC in the Chinese Tibetan population.
This study was approved by the Research Ethics Committee of the First Affiliated Hospital of Dali University (approval number: DFY20171210002, date: 10 December 2017). We selected 162 participants from the Tibetan population in the Lhasa area of Tibet, all of whom were residents of high altitude areas. Among them, 78 cases were HAPC patients in the experimental group and 84 cases were healthy people in the control group. The inclusion criteria of the experimental group were: (León-Velarde et al., 2005): Hb ≥21 g/dL for men and ≥19 g/dL for women; (Beall and Goldstein, 1987); long-term residence in the plateau area with an elevation of more than 3000 m above sea level; and (Wu, 2005) having three or more of the following symptoms: headache, dizziness, fatigue, cyanosis, sleep disturbance, conjunctival congestion, and purplish skin. Patients with true cytosis and other secondary erythrocytosis; cardiovascular and cerebrovascular diseases, autoimmune diseases, malignant tumors, immune system diseases, and neurological diseases; and incomplete relevant data were excluded from the study. Clinical data: red blood cell count, hemoglobin, and hematocrit were also collected from the 2 groups.
The flow chart of this study is shown in Figure 1. Fasting venous blood (2 mL) was taken into EDTA tubes, and the collected blood samples were stored in −20°C refrigerator for subsequent use, genomic DNA was extracted using DNA extraction kit, the integrity of genomic DNA was detected by agarose gel electrophoresis, and DNA concentration was detected by microspectrophotometer.
The National Center for Biotechnology Information (NCBI, USA, http://www.ncbi.nlm.nih.gov/) database was used to obtain reference information of SNPs of EPAS1 and PPARA genes. PPARA rs6520015 (C/T) and four loci of the EPAS1 gene, rs6735530 (C/T), rs6756667 (A/G), rs7583392 (A/G), and rs12467821 (C/T), were selected.
TaqMan fluorescent probe technology was used for genotyping detection of polymorphic loci, and different fluorescence signals were detected by real-time fluorescence quantitative PCR. The reaction conditions were as follows: pre-denaturation at 95°C for 5 min, 40 cycles (denaturation at 95°C for 10 s, annealing at 60°C for 30 s, and extension at 72°C for 2 min), and finally extension at 16°C for 5 min. The primer probe sequences for the five SNP loci (rs6520015, rs6735530, rs6756667, rs7583392, and rs12467821) are shown in Table 1.
SPSS 27.0 software was used for statistical analysis (P < 0.05 is statistically different), and t-test and chi-square test were used for numerical and categorical data, respectively. Logistic regression was used to assess the association between genotyping of polymorphisms of EPAS1 and PPARA gene locus and the risk of occurrence of HAPC in the present study by constructing multiple genetic models (allele, dominant model, recessive model, codominant model, overdominant model). The HWE (Hardy-Weinberg equilibrium) test was used to assess the population genetics of the control group, and if P > 0.05, it indicated that the data of this study followed the law of population genetics and was representative of the population.
Linkage disequilibrium determination and haplotype analysis were performed using online SHEsis (http://analysis.bio-x.cn/myAnalysis.php) to analyze the association between gene haplotypes and the risk of developing HAPC. Multi-factor dimensionality reduction software MDR3.0.2 was used to analyze the interactions among the five gene loci studied, and the Cross-validation consistency and Testing accuracy of each model were calculated, and P < 0.05 was statistically significant.
Table 2 shows the demographic and clinical characteristics of these participants. The mean age of the control group and cases was 45.63 ± 18.208 years and 48.41 ± 14.671, respectively (P > 0.05). There were significant differences in gender, red blood cell count, hemoglobin, and hematocrit between the two groups (P < 0.05 statistically significant).
The HWE-P of five SNPs (rs6520015, rs6735530, rs6756667, rs7583392, and rs12467821) in the control and experimental groups were all >0.05, indicating that the gene frequencies observed in the HAPC study population were representative of the gene distributions observed in the general population, and the results are shown in Table 3.
To assess the association of SNPs in the EPAS1 and PPARA gene with the effect of HAPC disease, we analyzed the association of rs6520015, rs6735530, rs6756667, rs7583392, and rs12467821 with the risk of developing HAPC in 162 participants by constructing multiple genetic model analyses, including codominant, dominant, recessive, and overdominant models, and by using binary logistic regression analysis. The genotyping and allele frequency distributions of the healthy and HAPC groups are summarized in Table 4. The results showed that no association was found between PPARA rs6520015 and the risk of HAPC disease in the Tibetan population (P > 0.05).
In EPAS1 rs6735530, the genotyping distribution was as follows: 33.3% CC, 42.3% CT, and 24.4% TT in the HAPC group, while it was 42.9% CC, 46.4% CT, and 10.7% TT in the control group. Using the C allele as a reference, allele T was correlated with the risk of developing HAPC [OR = 1.627, CI = (1.039–2.548), P = 0.034]. Under the codominant model, TT was associated with susceptibility to HAPC compared to the CC genotype [OR = 2.923, CI = (1.142–7.482), P = 0.025], and under the recessive model, individuals with the TT genotype had an elevated risk of developing the disease [OR = 2.684, CI = (1.132–6.363), P = 0.025].
In EPAS1 rs6756667, the genotyping distribution was as follows: 48.7% AA, 38.5% AG, and 12.8% GG in HAPC group, while it was 77.4% AA, 20.2% AG, and 2.4% GG in control group. The frequency of allele G was elevated in HAPC patients compared to allele A [OR = 3.302, CI = (1.872–5.824), P = 0.000]. Under the codominant model, using the AA genotype as a reference, genotypes AG and GG both seemed to raise the risk of HAPC [AG:OR = 3.019, CI = (1.474–6.183), P = 0.003; GG:OR = 8.553, CI = (1.779–41.112), P = 0.007]. Individuals carrying the G allele (AG + GG) under the dominant model {OR = 3.601, CI = [(1.830–7.087], P = 0.000}, the GG genotype under the recessive model [OR = 6.029, CI = (1.277–28.460), P = 0.023], and carrying the heterozygous AG genotype under the overdominant model [OR = 2.463, CI = (1.222–4.965), P = 0.012] were all more likely to have HAPC.
In EPAS1 rs7583392, the genotyping distribution was as follows: 50.0% GG, 37.2% AG, and 12.8% AA in HAPC group; while it was 72.6% GG, 25.0% AG, and 2.4% AA in control group. rs7583392 allele G/A [A vs. G:OR = 2.619, CI = (1.522–4.508), P = 0.001], codominant model [AG vs. GG: OR = 2.160, CI = (1.083–4.309), P = 0.029; AA vs. GG:OR = 7.821, CI = (1.626–37.608), P = 0.010], dominant model [AG-AA vs. GG:OR = 2.652,CI = (1.380–5.098), P = 0.003], and recessive model [AA vs. GG-AG:OR = 6.029, CI = (1.277–28.460), P = 0.023] were risk factors for HAPC.
In EPAS1 rs12467821, the genotyping distribution was as follows: 50.0% CC, 38.5% CT, and 11.5% TT in the HAPC group; while it was 73.8% CC, 23.8% CT, and 2.4% TT in the control group. The susceptibility to HAPC was found in allele C/T [T vs. C:OR = 2.667, CI = (1.539–4.621), P = 0.000], codominant model [CT vs. CC:OR = 2.385, CI = (1.192–4.770), P = 0.014; TT vs. CC:OR = 7.154, CI = (1.468–34.859), P = 0.015], dominant model [CT-TT vs. CC:OR = 2.818, CI = (1.459–5.444), P = 0.002], recessive model [TT vs. CC-CT:OR = 5.348,CI = (1.118–25.584), P = 0.036], and overdominant models [CT vs. CC-TT:OR = 2.000,CI = (1.015–3.941), P = 0.045] would be significantly higher.
Five SNPs loci of this study were analyzed using online SHEsis. The SNP order of haolotype was rs6520015, rs6735530, rs6756667, rs7583392, and rs12467821. Based on the analysis results, haplotypes with frequencies below 3% were removed, and a total of six haplotypes were obtained. In descending order of frequency, they were TCAGC, TTGAT, TTAGC, CCAGC, CTGAT, and CTAGC; among them, two haplotypes, TCAGC and TTGAT, differed between the HAPC group and the control group (P < 0.05), and TCAGC appeared in HAPC cases with a significantly lower frequency than that of the control group, so the TCAGC haplotype had a protective effect against HAPC had a protective effect, while TTGAT appeared significantly more frequently in HAPC cases than in the control group, so the TTGAT haplotype increased the risk of HAPC; the specific results of the analyses are shown in Table 5. In Figure 2, it can be seen that there was a complete linkage disequilibrium between EPAS1 rs7583392 and rs12467821 (D’ = 1.00, r2 = 0.96).

Figure 2. Linkage disequilibrium (LD) of 5 SNPs. (A) The numbers inside the diamonds indicate the D’ for pairwise analyses. (B) The numbers inside the diamonds indicate the r2 for pairwise analyses.
MDR analysis was used to analyze the association between interactions among the five SNP loci and high plateau erythrocytosis. Figure 3 shows a dendrogram of the interactions among the three loci obtained by MDR analysis. The blue line indicates that the SNPs have a redundancy effect in regulating the risk of HAPC, and the gold line represents the intermediate point between synergistic and redundancy effects. The closer the loci are, the stronger the interactions are. In this way, we investigated whether there were significant interactions between different loci to further explore the effect of SNP-SNP interactions on HAPC disease. The results showed that the cross-consistency was high in both the EPAS1 rs6756667 model and the PPARA rs6520015, EPAS1 rs6735530, and EPAS1 rs6756667 model, which were 10 and 9, respectively, and the best prediction model was the EPAS1 rs6756667 unit-point model (CVC: 10/10, test balance accuracy: 0.6433, P = 0.231), and in all three models the P-value was greater than 0.05, which was not statistically significant. It indicates that there is a strong interaction between gene loci, but there is no significant effect on the development of HAPC, and the results are shown in Table 6, Figures 3, 4.

Figure 3. Dendrogram of SNP-SNP interactions. The blue line indicates that the SNPs have a redundancy effect in regulating the risk of HAPC, and the gold line represents the intermediate point between synergistic and redundancy effects. The closer the loci are, the stronger the interactions are.
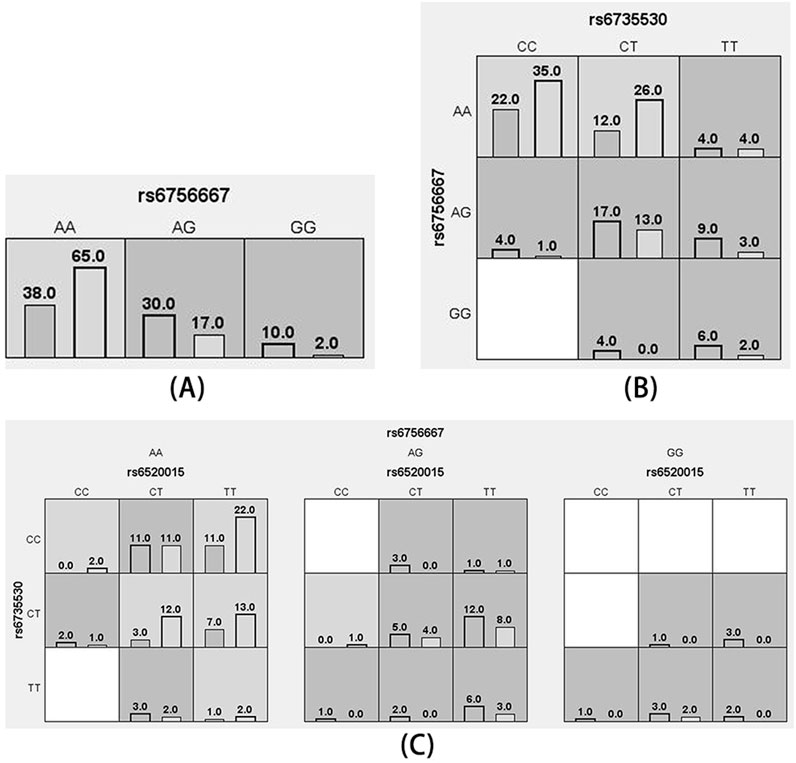
Figure 4. Cell diagram of the optimal model. (A): rs6756667 model; (B): rs6735530, rs6756667 model; (C): rs6520015, rs6735530, rs6756667 model (black bars on the left side indicate the case group, black bars on the right side indicate the control group, one cell represents one interaction combination, light gray cells indicate that the ratio of the combination does not exceed the ratio threshold and is low-risk, dark gray indicates that the ratio threshold is exceeded and is high-risk, and white indicates that there is no data on the combination).
More and more studies have suggested that genetic factors are one of the important factors contributing to the development of HAPC in Tibetan populations. EPAS1 rs12619696, rs13419896, and rs4953354 were significantly associated with the risk of HAPC disease in Tibetan populations on the Tibetan Plateau (Xu et al., 2015b). HAPC is an adverse consequence of adaptation to the high altitude environment and is closely related to the low-pressure environment of plateau hypoxia. The increasing number of erythrocytes and hemoglobin concentration poses a great threat to the health status of the population at high altitude (Wang et al., 2019). Therefore, it is important to explore the risk of HAPC genetically to improve the knowledge and diagnosis of this disease.
One of the hotspots of previous studies on the correlation between hypoxia adaptation and genetic polymorphisms is the EPAS1 gene, which encodes HIF-2α. Hypoxia-inducible factor (HIF) is a transcription factor that activates the adaptive hypoxia response at low levels of oxygen, and the two proteins, HIF-1 and HIF-2, consist of an unstable ɑ-chain and a stabilized β-chain (Jaśkiewicz et al., 2022; Yi et al., 2010); under normoxic conditions, the hydroxylation of HIF-1ɑ and HIF-2ɑ are post-translationally regulated by hydroxylation of specific proline residues within the oxygen-dependent degradation structural domain, which is carried out by prolyl hydrolase structural domain (PHD) proteins; hydroxylated HIF-α is recognized by von Hippel-Lindau (VHL) proteins, whose target HIFs are degraded via the ubiquitin-proteasomal pathway; in a hypoxic state, HIF-2ɑ hydroxylation is reduced, preventing the binding of HIF-2ɑ to VHL proteins, and the accumulated HIF-2ɑ continuously activates the HIF pathway, leading to a possible impact on the expression of downstream genes (Pamenter et al., 2020; Semenza, 2012; Sergi, 2019). Beall et al. found that EPAS1 variants downregulate its transcript levels, while its encoded transcription factor HIF-2α stimulates erythropoiesis, resulting in an increase in blood Hb concentration (Beall et al., 2010). Gruber et al. find that postnatal EPAS1 deficiency in mice causes anemia (Gruber et al., 2007). Tan et al. found that the EPAS1 G536W missense mutation causes erythrocytosis (Tan et al., 2013). In this study, we found that the rs6735530, rs6756667, rs7583392, and rs12467821 polymorphisms of the EPAS1 gene were significantly associated with HAPC in the Chinese Tibetan population by analysis. Among them, allele “G” in EPAS1 rs6756667 and allele “T” in rs12467821 were both significantly associated with increased risk of HAPC, and the analysis of codominant, dominant, recessive and overdominant models for rs6756667, rs12467821 showed an increased risk of HAPC. The allele “T” in EPAS1 rs6735530 is significantly associated with an increased risk of HAPC, and its codominant and recessive model analyses show an increased risk of HAPC. The allele “A” in EPAS1 rs7583392 is significantly associated with an increased risk of HAPC, and its codominant, dominant, and recessive models also show an increased risk of HAPC.
Under hypoxia, PPARα encoded by the PPARA gene enhances the regulation of anaerobic glycolysis and lactate accumulation in the venous blood, thereby increasing oxygen utilization and obtaining sufficient ATP for the organism (Kinota et al., 2023). In Tibetans, the expression of the putative favorable PPARA haplotype and PPARα was significantly and positively correlated with fatty acid oxidative capacity (FAO), and reduced expression of PPARα could enhance the body’s efficiency in utilizing oxygen, suggesting that PPARA is associated with hypoxic metabolic adaptation (Murray et al., 2018; Horscroft et al., 2017). We found no correlation between PPARA rs6520015 and the risk of HAPC in the Chinese Tibetan population. This is consistent with the results of previous studies. It has been reported that HIF can regulate PPARA by repressing its promoter during hypoxia, which reduces hemoglobin concentration in Tibetan population (Bhandari and Cavalleri, 2019; Bai et al., 2012).
In addition, the results of haplotype testing in this study showed that the TCAGC haplotype at the rs6520015, rs6735530, rs6756667, rs7583392, and rs12467821 loci decreases the risk of HAPC in Tibetans, and the TTGAT haplotype increases the risk of HAPC in Tibetans. And rs7583392 is in complete linkage disequilibrium with rs12467821. It indicates that rs7583392 and rs12467821 are highly interlinked, and the chance of their alleles appearing together on the same chromosome is higher than the frequency of random appearance. We also predicted the interactions between these SNPs by MDR analysis and found that there was no effect on the development of HAPC in the EPAS1 rs6756667 model and the PPARA rs6520015, EPAS1 rs6735530, EPAS1 rs6756667 model, although there was a strong interaction. This may have occurred because the MDR analysis could not adjust for covariates; and it could only analyze dichotomous phenotypes. It can only find interactions but does not identify the main effectors among the many factors. When we study fewer factors, there are limitations in using MDR analysis to accurately detect the effect of interactions on disease due to reduced dimensionality.
In this experiment, there are still some limitations. First, the sample size was small and limited to Tibetan patients in the Lhasa region of Tibet, so it is not possible to extrapolate the significance of the findings to other populations, and therefore the practical significance of the genetic polymorphisms needs to be investigated with larger sample sizes and different populations; second, we studied fewer loci, and in the future, we should investigate the correlation between other polymorphic loci in the EPAS1 and PPARA gene and the prevalence of HAPC.
In the Chinese Tibetan population, EPAS1 rs6735530, rs6756667, rs7583392, and rs12467821 polymorphisms were associated with the risk of developing HAPC, while PPARA rs6520015 was not significantly associated with HAPC.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
The studies involving humans were approved by the study was conducted in accordance with the Declaration of Helsinki, and approved by the Research Ethics Committee of the First Affiliated Hospital of Dali University (approval number: DFY20171210002, date: 10 December 2017). The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
ZC: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Software, Supervision, Validation, Visualization, Writing–original draft, Writing–review and editing. ZD: Investigation, Resources, Supervision, Visualization, Writing–original draft. RZ: Conceptualization, Data curation, Visualization, Writing–original draft. MX: Data curation, Visualization, Writing–original draft. YZ: Data curation, Supervision, Visualization, Writing–review and editing. QD: Conceptualization, Investigation, Resources, Supervision, Writing–original draft. GW: Conceptualization, Funding acquisition, Project administration, Supervision, Writing–review and editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the following grants: The National Natural Science Foundation of China. No: 82160244.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Baba, Y., Nosho, K., Shima, K., Irahara, N., Chan, A. T., Meyerhardt, J. A., et al. (2010). Hif1a overexpression is associated with poor prognosis in a cohort of 731 colorectal cancers. Am. J. Pathol. 176 (5), 2292–2301. doi:10.2353/ajpath.2010.090972
Bai, Z. Z., Jin, G. E., Wu-Ren, T., Ga, Q., and Ge, R. L. (2012). Energy power in mountains: difference in metabolism pattern results in different adaption traits in Tibetans. Zhongguo Ying Yong Sheng Li Xue Za Zhi 28 (6), 488–493.
Bangoura, G., Yang, L. Y., Huang, G. W., and Wang, W. (2004). Expression of Hif-2alpha/Epas1 in hepatocellular carcinoma. World J. Gastroenterol. 10 (4), 525–530. doi:10.3748/wjg.v10.i4.525
Basang, Z., Wang, B., Li, L., Yang, L., Liu, L., Cui, C., et al. (2015). Hif2a variants were associated with different levels of high-altitude hypoxia among native Tibetans. PLoS One 10 (9), e0137956. doi:10.1371/journal.pone.0137956
Beall, C. M., Cavalleri, G. L., Deng, L., Elston, R. C., Gao, Y., Knight, J., et al. (2010). Natural selection on Epas1 (Hif2alpha) associated with low hemoglobin concentration in Tibetan highlanders. Proc. Natl. Acad. Sci. U. S. A. 107 (25), 11459–11464. doi:10.1073/pnas.1002443107
Beall, C. M., and Goldstein, M. C. (1987). Hemoglobin concentration of pastoral nomads permanently resident at 4,850-5,450 meters in Tibet. Am. J. Phys. Anthropol. 73 (4), 433–438. doi:10.1002/ajpa.1330730404
Bhandari, S., and Cavalleri, G. L. (2019). Population history and altitude-related adaptation in the sherpa. Front. Physiol. 10, 1116. doi:10.3389/fphys.2019.01116
Bhandari, S., Zhang, X., Cui, C., Yangla, L. L., Ouzhuluobu, , Ouzhuluobu, , et al. (2017). Sherpas share genetic variations with Tibetans for high-altitude adaptation. Mol. Genet. Genomic Med. 5 (1), 76–84. doi:10.1002/mgg3.264
Davis, L., Recktenwald, M., Hutt, E., Fuller, S., Briggs, M., Goel, A., et al. (2022). Targeting Hif-2α in the tumor microenvironment: redefining the role of Hif-2α for solid cancer therapy. Cancers (Basel) 14 (5), 1259. doi:10.3390/cancers14051259
Eurlings, P. M., van der Kallen, C. J., Geurts, J. M., Flavell, D. M., and de Bruin, T. W. (2002). Identification of the Ppara locus on chromosome 22q13.3 as a modifier gene in familial combined hyperlipidemia. Mol. Genet. Metab. 77 (4), 274–281. doi:10.1016/s1096-7192(02)00174-9
Garrido, E., Segura, R., Capdevila, A., Pujol, J., Javierre, C., and Ventura, J. L. (1996). Are himalayan sherpas better protected against brain damage associated with extreme altitude climbs? Clin. Sci. (Lond) 90 (1), 81–85. doi:10.1042/cs0900081
Gruber, M., Hu, C. J., Johnson, R. S., Brown, E. J., Keith, B., and Simon, M. C. (2007). Acute postnatal ablation of Hif-2alpha results in anemia. Proc. Natl. Acad. Sci. U. S. A. 104 (7), 2301–2306. doi:10.1073/pnas.0608382104
Horscroft, J. A., Kotwica, A. O., Laner, V., West, J. A., Hennis, P. J., Levett, D. Z. H., et al. (2017). Metabolic basis to sherpa altitude adaptation. Proc. Natl. Acad. Sci. U. S. A. 114 (24), 6382–6387. doi:10.1073/pnas.1700527114
Islam, F., Gopalan, V., Law, S., Lam, A. K., and Pillai, S. (2020). Molecular deregulation of Epas1 in the pathogenesis of esophageal squamous cell carcinoma. Front. Oncol. 10, 1534. doi:10.3389/fonc.2020.01534
Jaśkiewicz, M., Moszyńska, A., Króliczewski, J., Cabaj, A., Bartoszewska, S., Charzyńska, A., et al. (2022). The transition from Hif-1 to Hif-2 during prolonged hypoxia results from reactivation of Phds and Hif1a Mrna instability. Cell Mol. Biol. Lett. 27 (1), 109. doi:10.1186/s11658-022-00408-7
Jay, M. A., and Ren, J. (2007). Peroxisome proliferator-activated receptor (Ppar) in metabolic syndrome and type 2 diabetes mellitus. Curr. Diabetes Rev. 3 (1), 33–39. doi:10.2174/157339907779802067
Karaghiannis, V., Maric, D., Garrec, C., Maaziz, N., Buffet, A., Schmitt, L., et al. (2023). Comprehensive in silico and functional studies for classification of Epas1/Hif2a genetic variants identified in patients with erythrocytosis. Haematologica 108 (6), 1652–1666. doi:10.3324/haematol.2022.281698
Kinota, F., Droma, Y., Kobayashi, N., Horiuchi, T., Kitaguchi, Y., Yasuo, M., et al. (2023). The contribution of genetic variants of the peroxisome proliferator-activated receptor-alpha gene to high-altitude hypoxia adaptation in sherpa highlanders. High. Alt. Med. Biol. 24 (3), 186–192. doi:10.1089/ham.2018.0052
León-Velarde, F., Maggiorini, M., Reeves, J. T., Aldashev, A., Asmus, I., Bernardi, L., et al. (2005). Consensus statement on chronic and subacute high altitude diseases. High. Alt. Med. Biol. 6 (2), 147–157. doi:10.1089/ham.2005.6.147
Li, H. H., Tyburski, J. B., Wang, Y. W., Strawn, S., Moon, B. H., Kallakury, B. V., et al. (2014). Modulation of fatty acid and bile acid metabolism by peroxisome proliferator-activated receptor α protects against alcoholic liver disease. Alcohol Clin. Exp. Res. 38 (6), 1520–1531. doi:10.1111/acer.12424
Li, X., Xu, S., Li, X., Wang, Y., Sheng, Y., Zhang, H., et al. (2024). Novel insight into the genetic signatures of altitude adaptation related body composition in Tibetans. Front. Public Health 12, 1355659. doi:10.3389/fpubh.2024.1355659
Mallik, N., Das, R., Malhotra, P., and Sharma, P. (2021). Congenital erythrocytosis. Eur. J. Haematol. 107 (1), 29–37. doi:10.1111/ejh.13632
Meyer, M. C., Aldenderfer, M. S., Wang, Z., Hoffmann, D. L., Dahl, J. A., Degering, D., et al. (2017). Permanent human occupation of the central Tibetan plateau in the early holocene. Science 355 (6320), 64–67. doi:10.1126/science.aag0357
Mohammed, N., Rodriguez, M., Garcia, V., Garcia, J. M., Dominguez, G., Peña, C., et al. (2011). Epas1 Mrna in plasma from colorectal cancer patients is associated with poor outcome in advanced stages. Oncol. Lett. 2 (4), 719–724. doi:10.3892/ol.2011.294
Mostowy, J., Montén, C., Gudjonsdottir, A. H., Arnell, H., Browaldh, L., Nilsson, S., et al. (2016). Shared genetic factors involved in celiac disease, type 2 diabetes and anorexia nervosa suggest common molecular pathways for chronic diseases. PLoS One 11 (8), e0159593. doi:10.1371/journal.pone.0159593
Murray, A. J., Montgomery, H. E., Feelisch, M., Grocott, M. P. W., and Martin, D. S. (2018). Metabolic adjustment to high-altitude hypoxia: from genetic signals to physiological implications. Biochem. Soc. Trans. 46 (3), 599–607. doi:10.1042/bst20170502
Pamenter, M. E., Hall, J. E., Tanabe, Y., and Simonson, T. S. (2020). Cross-species insights into genomic adaptations to hypoxia. Front. Genet. 11, 743. doi:10.3389/fgene.2020.00743
Pan, W., Liu, C., Zhang, J., Gao, X., Yu, S., Tan, H., et al. (2019). Association between Single nucleotide polymorphisms in Ppara and Epas1 genes and high-altitude appetite loss in Chinese young men. Front. Physiol. 10, 59. doi:10.3389/fphys.2019.00059
Peng, Y., Yang, Z., Zhang, H., Cui, C., Qi, X., Luo, X., et al. (2011). Genetic variations in Tibetan populations and high-altitude adaptation at the himalayas. Mol. Biol. Evol. 28 (2), 1075–1081. doi:10.1093/molbev/msq290
Semenza, G. L. (2012). Hypoxia-inducible factors in physiology and medicine. Cell 148 (3), 399–408. doi:10.1016/j.cell.2012.01.021
Sergi, C. (2019). Epas 1, congenital heart disease, and high altitude: disclosures by genetics, bioinformatics, and experimental embryology. Biosci. Rep. 39 (5). doi:10.1042/bsr20182197
Shen, Y., Zhang, J., Yang, J., Liu, C., Bian, S., Zhang, C., et al. (2020). Association of Epas1 and Ppara gene polymorphisms with high-altitude headache in Chinese han population. Biomed. Res. Int. 2020, 1593068. doi:10.1155/2020/1593068
Simonson, T. S., Yang, Y., Huff, C. D., Yun, H., Qin, G., Witherspoon, D. J., et al. (2010). Genetic evidence for high-altitude adaptation in Tibet. Science 329 (5987), 72–75. doi:10.1126/science.1189406
Standage, S. W., Bennion, B. G., Knowles, T. O., Ledee, D. R., Portman, M. A., McGuire, J. K., et al. (2017). Pparα augments heart function and cardiac fatty acid oxidation in early experimental polymicrobial sepsis. Am. J. Physiol. Heart Circ. Physiol. 312 (2), H239–H249. doi:10.1152/ajpheart.00457.2016
Tan, Q., Kerestes, H., Percy, M. J., Pietrofesa, R., Chen, L., Khurana, T. S., et al. (2013). Erythrocytosis and pulmonary hypertension in a mouse model of human Hif2a gain of function mutation. J. Biol. Chem. 288 (24), 17134–17144. doi:10.1074/jbc.M112.444059
van Patot, M. C., and Gassmann, M. (2011). Hypoxia: adapting to high altitude by mutating Epas-1, the gene encoding Hif-2α. High. Alt. Med. Biol. 12 (2), 157–167. doi:10.1089/ham.2010.1099
Wang, B., Zhang, Y. B., Zhang, F., Lin, H., Wang, X., Wan, N., et al. (2011). On the origin of Tibetans and their genetic basis in adapting high-altitude environments. PLoS One 6 (2), e17002. doi:10.1371/journal.pone.0017002
Wang, Y., Lin, A., He, R., Chen, C., Zeng, X., Pan, Y., et al. (2023). The role of Epas1 polymorphisms on Copd susceptibility in Southern Chinese. Heliyon 9 (10), e20226. doi:10.1016/j.heliyon.2023.e20226
Wang, Z., Liu, F., Ye, S., Jiang, P., Yu, X., Xu, J., et al. (2019). Plasma proteome profiling of high-altitude polycythemia using Tmt-based quantitative proteomics approach. J. Proteomics 194, 60–69. doi:10.1016/j.jprot.2018.12.031
Wu, T., Wang, X., Wei, C., Cheng, H., Wang, X., Li, Y., et al. (2005). Hemoglobin levels in Qinghai-Tibet: different effects of gender for Tibetans Vs. Han. J. Appl. Physiol. (1985) 98 (2), 598–604. doi:10.1152/japplphysiol.01034.2002
Wu, T. Y. (2005). Chronic Mountain sickness on the Qinghai-Tibetan plateau. Chin. Med. J. Engl. 118 (2), 161–168.
Xu, J., Yang, Y. Z., Tang, F., Ga, Q., Tana, W., and Ge, R. L. (2015b). Epas1 gene polymorphisms are associated with high altitude polycythemia in Tibetans at the Qinghai-Tibetan plateau. Wilderness Environ. Med. 26 (3), 288–294. doi:10.1016/j.wem.2015.01.002
Xu, J., Yang, Y. Z., Tang, F., Ga, Q., Wuren, T., Wang, Z., et al. (2015a). Cyp17a1 and Cyp2e1 Variants Associated with High Altitude Polycythemia in Tibetans at the Qinghai-Tibetan Plateau. Gene 566 (2), 257–263. doi:10.1016/j.gene.2015.04.056
Yang, J., Liu, C., Jihang, Z., Yu, J., Dai, L., Ding, X., et al. (2019). Ppara genetic variants increase the risk for cardiac pumping function reductions following acute high-altitude exposure: a self-controlled study. Mol. Genet. Genomic Med. 7 (10), e00919. doi:10.1002/mgg3.919
Yi, X., Liang, Y., Huerta-Sanchez, E., Jin, X., Cuo, Z. X., Pool, J. E., et al. (2010). Sequencing of 50 human exomes reveals adaptation to high altitude. Science 329 (5987), 75–78. doi:10.1126/science.1190371
Zhang, R., Zhao, J., and Zhao, L. (2023). Epas1/Hif-2α acts as an unanticipated tumor-suppressive role in papillary thyroid carcinoma. Int. J. Gen. Med. 16, 2165–2174. doi:10.2147/ijgm.S409874
Keywords: EPAS1, PPARA, high altitude polycythemia, single nucleotide polymorphisms, Tibetans
Citation: Chen Z, Dong Z, Zeng R, Xu M, Zhang Y, Dan Q and Wang G (2025) Association between single nucleotide polymorphisms in EPAS1 and PPARA genes and high altitude polycythemia in Chinese Tibetan population. Front. Genet. 16:1519108. doi: 10.3389/fgene.2025.1519108
Received: 30 October 2024; Accepted: 13 February 2025;
Published: 06 March 2025.
Edited by:
Pilar Giraldo, University of Zaragoza, SpainReviewed by:
Cagri Gulec, Istanbul University, TürkiyeCopyright © 2025 Chen, Dong, Zeng, Xu, Zhang, Dan and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qu Dan, ZGFuZWlsd2luZEAxMjYuY29t; Guangming Wang, d2dtMTk5MUBkYWxpLmVkdS5jbg==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.