
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 18 December 2024
Sec. Genomics of Plants and the Phytoecosystem
Volume 15 - 2024 | https://doi.org/10.3389/fgene.2024.1510864
This article is part of the Research Topic Responses and Adaptation of Plants to Abiotic Stress: Genetics, Evolution and Molecular Insights View all 3 articles
 Hao Yuan1,2‡
Hao Yuan1,2‡ Baoxia Liu1,2‡
Baoxia Liu1,2‡ Guwen Zhang2
Guwen Zhang2 Zhijuan Feng2
Zhijuan Feng2 Bin Wang2
Bin Wang2 Yuanpeng Bu2
Yuanpeng Bu2 Yu Xu2
Yu Xu2 Yaming Gong2
Yaming Gong2 Zhihong Sun1†*
Zhihong Sun1†* Na Liu1,2*†
Na Liu1,2*†Peas (Pisum sativum L.) serve as a vital model for plant development and stress research. The kinesin (KIN) gene family, encoding essential motor proteins, remains understudied in peas. Our research conducted a comprehensive genomic analysis of the KIN genes in peas, identifying 105 genes categorized into seven subfamilies based on evolutionary relationships, gene structures, conserved motifs, and interaction networks. A comparative analysis with Arabidopsis and soybean KIN gene families showed a non-uniform distribution of PsKIN genes across subfamilies. Homology analysis revealed that the PsKIN family has undergone segmental duplication and is under negative selection pressures, with conserved genes on chromosomes Ps5, Ps6, and Ps7 playing a significant role in pea evolution. Transcriptomics revealed 38 PsKIN genes with distinct tissue-specific expression, with PsKIN76, PsKIN96, PsKIN82, and PsKIN103 showing significant levels in roots, lateral roots, stems, petals, and seeds, respectively. Differential expression under drought and saline stress was observed, with PsKIN8, PsKIN11, PsKIN54 upregulated under drought, and PsKIN47 and PsKIN51 under saline stress. These genes are potential candidates for improving plant stress tolerance. This study offers insights into the pea KIN gene family, highlighting their potential in enhancing plant stress tolerance and setting a stage for future research.
Peas (Pisum sativum L.) are integral to the global agricultural and nutritional sectors. Despite their importance, the intricate nature of their genome has posed challenges for in-depth study. However, recent scientific progress has paved the way for a better understanding of their genetic diversity and its potential applications. By leveraging high-throughput genotyping methods, comprehensive genome-wide association studies have been performed, shedding light on the genetic makeup of pea germplasm resources (Aubert et al., 2022).
Peas are notable for their rich content of essential macronutrients and bioactive substances, positioning them as strong contenders for the development of functional foods with a range of health benefits (Wu et al., 2023). The unique chemical profile of pea protein isolate also presents significant opportunities for innovation in the food industry (Golovko et al., 2023). The introduction of cutting-edge genetic manipulation techniques has revolutionized pea breeding, allowing for precise genetic modifications that have a substantial impact on crop enhancement (Li et al., 2023). Despite these advancements, there are still hurdles in fully harnessing the genetic variability for nutritional traits and in the construction of genomic resources. Nonetheless, the application of omics-based strategies offers considerable promise for deciphering the genetic pathways that control nutritional attributes, while biotechnological approaches provide new avenues for enhancing the nutraceutical potential of peas. Kinesin proteins is indispensable ATP-driven microtubule motor enzymes and play a multifaceted role in cellular functions, are essential for photosynthesis in plants, and lay the foundation for further exploration of the pea gene family.
They participate in microtubule dynamics, chromosome segregation, organelle transport, and developmental processes such as cell division, expansion, and hormonal signaling (Solon et al., 2021). The interaction between kinesins and KIFBP modulates their binding to microtubules, thereby influencing a range of cellular activities (Seneviratne et al., 2023; Solon et al., 2021). Research across various plant species has identified and characterized the kinesin gene family, highlighting their regulatory roles, especially in early fruit development and cytoskeletal reorganization during cell division (Tian et al., 2021; Galindo-Trigo et al., 2020). The expansion of plant kinesin families underscores their critical role in facilitating plant-specific cytoskeletal rearrangements. The study of specific kinesin members, such as AtKRP125b in Arabidopsis thaliana, has provided insights into their involvement in mitosis and potential extranuclear functions, (Strauß et al., 2021). Gaining insights into the functions and regulatory mechanisms of plant kinesins is crucial for understanding the molecular basis of plant growth and development.
In our study, we employed the most current genomic data and search methodologies to identify 105 PsKIN (Kinesin) genes in pea. The evolutionary and structural characteristics of these PsKINs were elucidated through phylogenetic analysis, gene structure examination, identification of conserved domains, and interaction network predictions. Additionally, we systematically investigated the expression patterns of PsKINs across 20 RNA-seq samples, and a subset of hormone response candidates was further validated through transcriptome sequencing analysis. This research establishes a foundation for future functional studies of PsKINs and uncovers their roles in hormone response and plant growth regulation.
The complete genomic sequence of the pea plant was sourced from the National Center for Biotechnology Information (NCBI) database. A hidden Markov model (HMM) was crafted using a variety of kinesin (KIN) protein sequences from different species, which are accessible on the NCBI (GCA_024323335.1) platform. The HMMER software package (version 3.3.2) was then applied to perform searches against the local protein database of the pea plant, with an E-value threshold set at 10^-5 to filter results (Supplementary Table S1). This process resulted in the identification of a preliminary list of KIN candidate genes, which was subsequently refined by eliminating any duplicates. This method has been reported in the literature (Potter et al., 2018). In rice, Arabidopsis, and soybean, the HMMER model construction was also used (Guo et al., 2008; Chen et al., 2014).
To substantiate the authenticity of these candidate sequences, the NCBI Conserved Domain Database (CDD) and the SMART tool were employed. These tools were used with an E-value threshold of 10^-5 to filter the sequences based on the presence of the characteristic KIN domain. The KIN genes that passed this validation were systematically renamed according to their respective chromosomal locations within the pea genome.
For the prediction of subcellular localization, the Cell-PLoc software (version 2.0) was engaged. Additionally, the ExPASy ProtParam tool was leveraged to forecast various physicochemical attributes of the proteins, encompassing molecular weight (MW), isoelectric point (pI), instability index, and grand average of hydropathicity (GRAVY).
To elucidate the phylogenetic connections among the kinesin (KIN) genes of the pea plant, we conducted multiple sequence alignments involving the identified pea KIN proteins and those from Manihot esculenta, Populus trichocarpa, and A. thaliana. The sequence alignment was executed using the ClustalW tool, and the resultant data were utilized to construct a maximum likelihood (ML) phylogenetic tree with IQ-TREE 2 software, applying a bootstrap value of 1,000 to ensure the robustness of the tree topology.
The chromosomal positions and the exon-intron structures of the KIN genes were extracted from the genome annotation files. These files were sourced from the Ensembl Plants database (http://plants.ensembl.org/index.html). The physical locations of the KIN genes on the chromosomes were visualized using the MapGene2Chromosome v2.0 (http://mg2c.iask.in/mg2c_v2.0/). The exon-intron structures were graphically represented with the aid of the Gene Structure Display Server (GSDS2.0) (http://gsds.cbi.pku.edu.cn/).
Furthermore, to identify the conserved domains within each KIN protein sequence, we employed the MEME tool (http://meme-suite.org/). The search for motifs was configured to identify up to 10 distinct motifs, with all other parameters left at their default settings to ensure a thorough and unbiased search for conserved elements.
The genomic coordinates and structural attributes of genes on the chromosomes of pea were extracted from the GFF3 file, which was sourced from the pea genome database (htKIN://www.peagdb.com/download/). Subsequently, the MapGene2Chrom web v2 (http://mg2c.iask.in/mg2c_v2.0/) was applied to graphically represent the PsKIN genes on their respective chromosomes. The analysis of synteny among the KIN genes across pea, A. thaliana, and Glycine max was conducted using the Multiple Collinearity Scan Toolkit (MCScanX) and TBtools. The genomic datasets for A. thaliana and G. max were obtained from the Phytozome12 database (htKIN://phytozome.jgi.doe.gov/pz/portal.html).
The analytical capabilities of MCScanX were leveraged to determine the Ka (non-synonymous substitution rate) and Ks (synonymous substitution rate) for gene pairs involved in both segmental and tandem duplications. The Ks values were used to calculate the dates of duplication events (T) according to the following equation: T = Ks/2λ, with λ = 1.5 × 10−8 s for dicots (Bian et al., 2020). The Ka/Ks value was further employed to identify the selection mode of the PsKINs. STRING (htKIN://string-db.org/) was used to construct the functional interaction network of the proteins.
The propagation of the ‘Zhewan No.1’ (ZW1) TM-1 seeds of variety was initiated in a substrate composed of nutrient-enriched soil within an artificially regulated greenhouse. The conditions within the greenhouse were tailored to provide a 16 h period of light exposure at a steady temperature of 27°C, followed by an 8 h interval of darkness maintained at 22°C, with the atmospheric humidity consistently maintained between 60% and 80%. Peas seedlings of the TM-1 variety, which had reached a developmental stage characterized by the presence of three leaves and demonstrated signs of robust health and stable growth, were subsequently introduced to treatments designed to induce drought and salt stress. For the induction of salt stress, the root systems of the seedlings were treated with an irrigation solution containing a concentration of 300 mM sodium chloride (NaCl). In contrast, to mimic the effects of drought stress, a 20% solution of polyethylene glycol 6,000 (PEG6000) was applied to the root systems. Following the application of these stressors, leaf samples were harvested at several time points post-treatment: immediately post-treatment (designated as 0 h), as well as at 3 h and 24 h intervals. Upon collection, each leaf sample was rapidly immersed in liquid nitrogen to achieve rapid freezing, and then securely stored at a temperature of −80°C for the purpose of RNA extraction in subsequent experimental procedures.
Post-treatment at 3 h and 24 h time points, the fresh pea seedling leaves from the untreated control group and those subjected to 300 mmol NaCl for both time intervals, as well as the leaves treated with PEG6000, were flash-frozen in liquid nitrogen and dispatched to Tianjin Jizhi Gene Technology Co., Ltd. For further analysis. In order to validate the uniformity across our experiments, triplicate biological samples were analyzed. RNA integrity was determined by agarose gel electrophoresis, revealing clear 28S and 18 S rRNA bands in the total RNA, with the 28 S intensity of band being roughly double that of the 18 S band. For the preparation of the RNA-seq library, a multi-step protocol was adhered to, encompassing total RNA isolation, mRNA purification, fragmentation, cDNA generation, end-repair of double-stranded cDNA, adapter attachment, and PCR amplification. Additionally, the RNA-seq dataset was subjected to Weighted Gene Co-expression Network Analysis (WGCNA) to assess and secure the precision and dependability of the transcriptome data.
The resulting raw sequencing reads underwent a stringent quality control process and were subsequently cleaned. These high-quality reads were aligned to the reference genome, accessible via the provided link, to facilitate the assembly of transcripts and the quantification of gene expression levels. The quality of the RNA-seq alignments was meticulously evaluated. The subsequent analysis of the transcriptome was performed utilizing a proprietary cloud-based platform developed by Tianjin Jizhi Gene Technology Co., Ltd., ensuring a comprehensive and in-depth examination of the data.
In our research, we initiated the investigation by employing a hidden Markov model (HMM) specific to plant kinesin (KIN) proteins, utilizing this model to conduct a comprehensive search across the entire proteome of pea. Subsequently, we proceeded to authenticate the credibility of the identified candidate KIN proteins. A total of 105 KIN proteins, characterized by the presence of a conserved KIN domain, were identified and designated as constituents of the pea KIN family, with each member named PsKIN1 through PsKIN105, corresponding to their chromosomal positions (Figure 1; Supplementary Table S2).
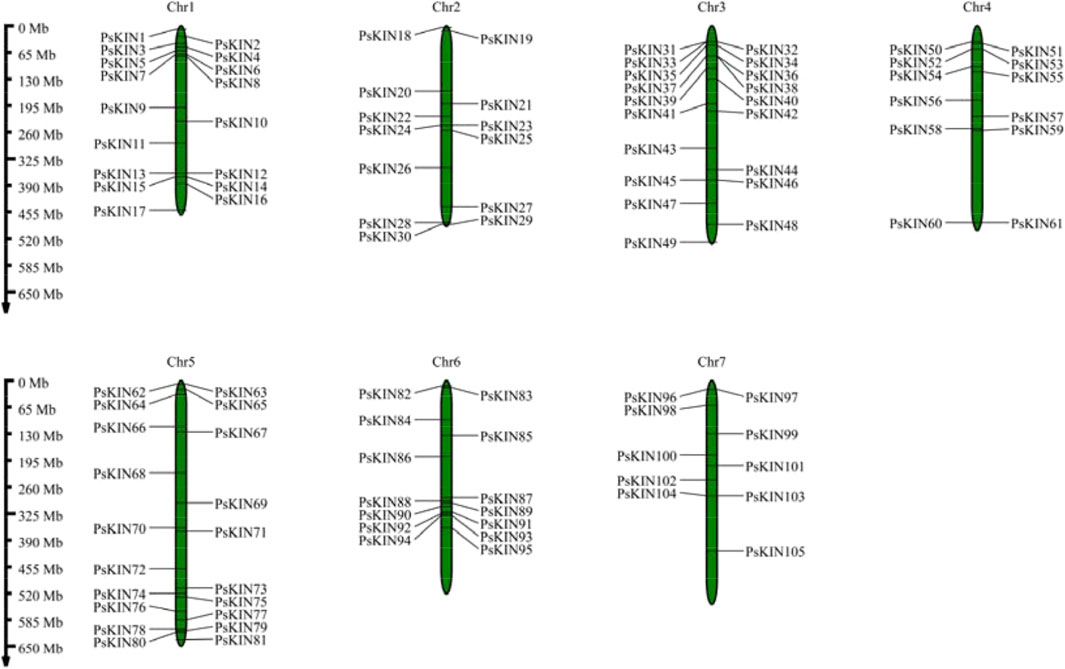
Figure 1. Chromosomal locations of the PsKIN genes on the eighteen pea chromosomes. The number of PsKIN genes is relatively low, and they are not distributed on every chromosome. The highest distribution of PsKIN genes is observed on Chr5, with seven genes present.
Further analysis was conducted to elucidate the fundamental characteristics of the PsKIN genes, encompassing parameters such as the length of the protein in amino acids (aa), molecular weight (MW), isoelectric point (PI), instability index, and grand average of hydropathy (GRAVY). The length of the PsKIN proteins varied significantly, extending from 70 aa for PsKIN60 to a substantial 2,980 aa for PsKIN101, with an overall mean length of 541.2 aa. The molecular weight of these proteins also exhibited a broad range, from 7.66 kD for PsKIN60 to a notably higher value of 341.67 kD for PsKIN101, with an average MW of 76.50 kD. Notably, both the length and MW of PsKIN101 exceeded those of the other PsKIN genes by a considerable margin. Additionally, the isoelectric points of the PsKIN genes spanned from 4.5 to 9.94, the instability indices ranged from 34.84 to 58.89, and the GRAVY values varied from −0.867 to 0.088.
To elucidate the evolutionary connections among the KIN gene family members in pea (comprising 105 PsKINs), Arabidopsis (encompassing 65 AtKINs), and G. max (consisting of 150 GmKINs), we developed a maximum likelihood (ML) phylogenetic framework. This framework was utilized to delineate the phylogenetic affinities among the species in question (Figure 2). Within the ML phylogenetic tree, the 320 KIN genes were categorized into seven distinct subfamilies, following the taxonomic and topological criteria established by prior research. The distribution of the 105 PsKINs across these subfamilies was found to be non-uniform, as detailed in Supplementary (Supplementary Table S2). Among the subfamilies, Subfamily III was identified as the most populous, housing 25 PsKIN genes. In contrast, Subfamily I was the least represented, with only 5 PsKIN genes. This differential distribution underscores the diversity and evolutionary complexity within the KIN gene family across the examined species.
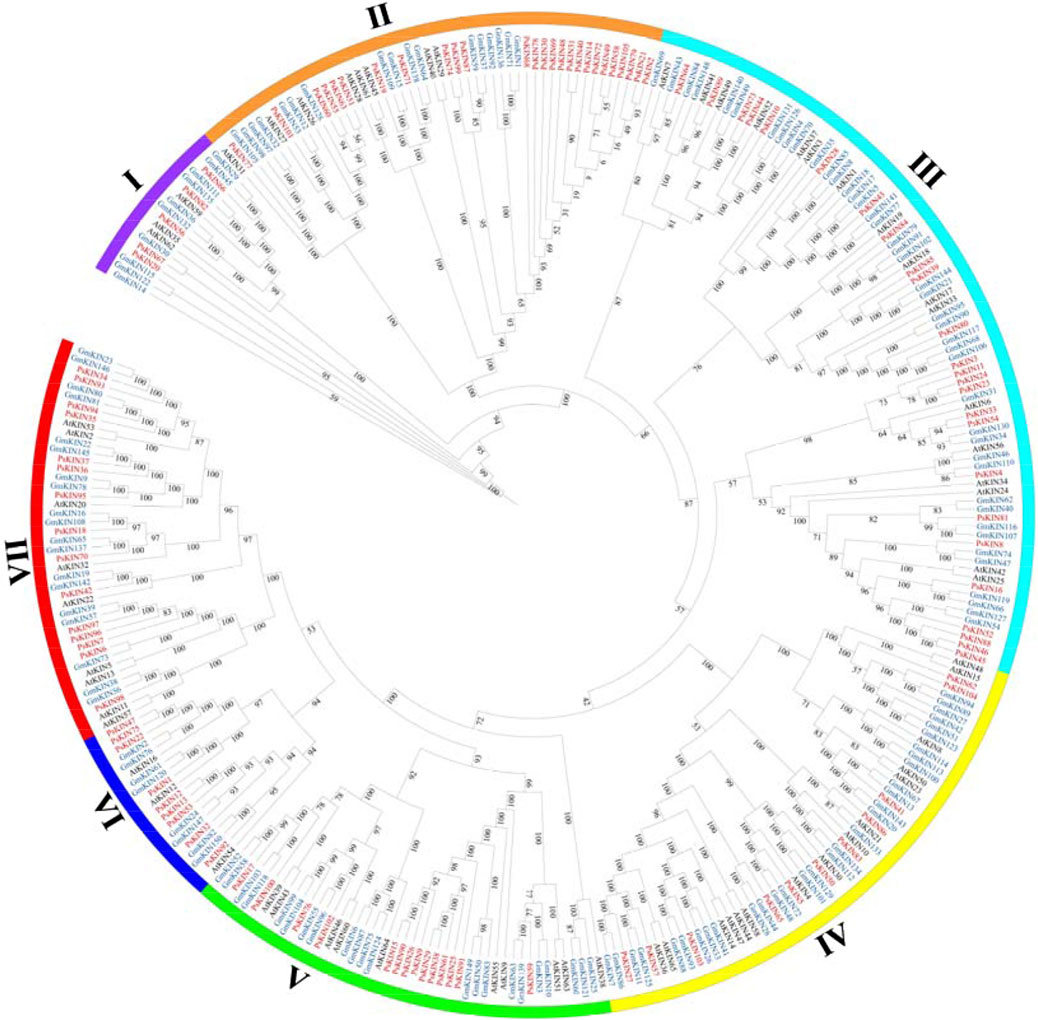
Figure 2. Phylogenetic tree incorporating KIN proteins from Pisum sativum, Arabidopsis and Glycine max. A phylogenetic tree of the KIN gene family was constructed by the IQ-TREE 2 software using the maximum likelihood (ML) option with 1,000 bootstrap replicates. The color of the outer ring and branches highlights different subfamilies.
In our investigation, we conducted a comprehensive analysis of the gene structure and the conserved motifs present within the 105 PsKIN proteins, correlating these findings with their phylogenetic relationships (Figures 3A–D). Utilizing the MEME online tool, we identified 10 conserved motifs (motifs 1–10; Figure 3B) across the PsKIN proteins. The analysis highlighted variations in the composition of these motifs, with a general trend showing that the motif distribution aligns well with the classification into subfamilies. Notably, motifs 5, 6, and 7 emerged as the predominant motifs across all PsKIN genes.

Figure 3. The phylogenetic relationship, conserved motifs and gene structure of PsKINs. (A) The maximum likelihood (ML) phylogenetic tree of PsKIN proteins was constructed using full-length sequence with 1,000 bootstrap replicates; (B) Distribution of conserved motifs in PsKIN proteins. A total of 10 motifs were predicted, and the scale bar represents 100 aa; (C) Distribution of KIN domain of PsKINs; (D) The gene structures of the PsKINs, include introns (black lines) and exons (green rectangles). The scale bar indicates 1,000 bp.
Concurrently, we examined the gene domains of the PsKINs, presenting a visual representation of the conserved KIN domains mapped directly onto the genetic structure of the pea (Figure 3C). Our findings indicated the presence of 12 distinct types of KIN domains, with the majority of genes anticipated to encompass the “Kinesin motor domain”.
Further analysis of the exon count within the 105 PsKINs, as per the pea GFF annotation file, revealed a range from 1 to 36 exons (Figure 3D). A significant number of PsKINs were found to possess more than 10 coding sequences (CDSs). PsKINs from related subfamilies displayed a tendency towards similar exon/intron structures, indicating a strong correlation between phylogenetic relationships and gene structure within the gene family. This correlation underscores the evolutionary significance of gene structure in shaping the diversity and function of the KIN gene family in pea.
Cis-regulatory elements (CREs), which are non-coding DNA sequences located within the promoter region of genes, play a pivotal role in gene expression and are integral to the regulation of a multitude of biological processes. In our study, we harnessed the 2000 base pair (bp) promoter sequences upstream of the identified PsKIN genes to forecast the presence of CREs, utilizing the PlantCARE database as a predictive tool. A total of 2,111 CREs sites, deemed representative, were culled from the predictive outcomes and are presented in Figure 4A. Within this set, elements associated with growth and developmental processes were found to be the most prevalent, closely trailed by those linked to hormone responsiveness, as illustrated in Figure 4B.
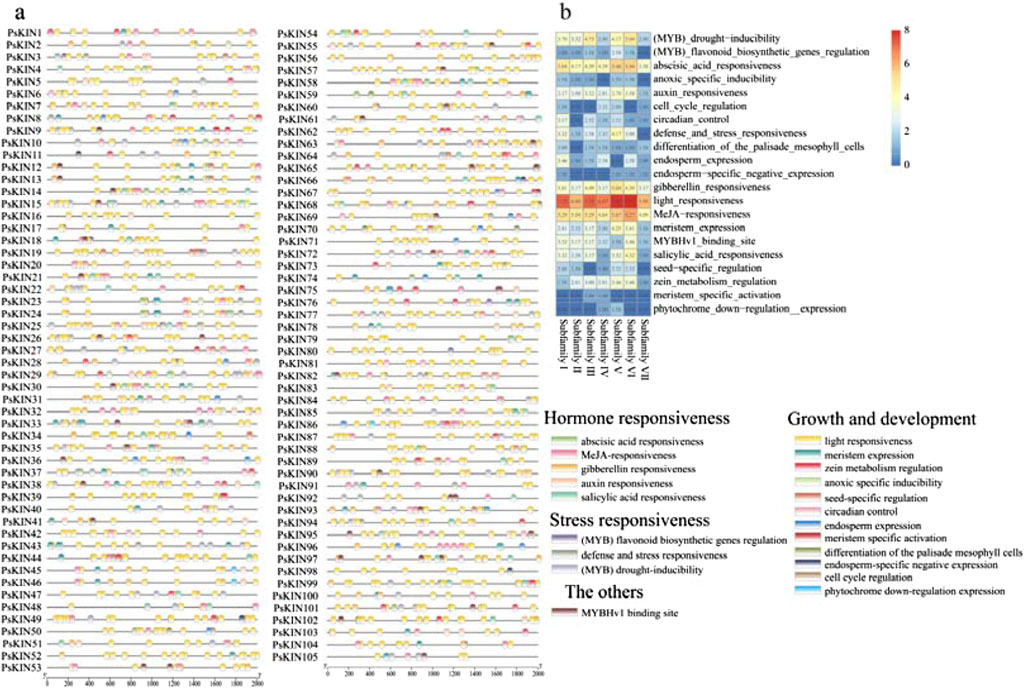
Figure 4. The CREs on the putative promoter of the PsKINs. (A) Distribution of CREs identified in the 2000 bp upstream promoter region of PsKINs; (B) The number of CREs on putative promoters of PsKINs. The numbers in the heatmap represent the quantity of elements.
This analysis underscores the complexity of gene regulation within the PsKIN family, highlighting the significance of promoter-associated CREs in modulating gene expression in response to various developmental cues and hormonal signals. The abundance of growth and development elements, as well as hormone responsiveness elements, suggests a dynamic interplay between these regulatory factors and the expression of PsKIN genes, potentially influencing their roles in plant physiology and adaptation.
The CREs on the putative promoter of the PsKINs. (a) Distribution of CREs identified in the 2000 bp upstream promoter region of PsKINs; (b) The number of CREs on putative promoters of PsKINs. The numbers in the heatmap represent the quantity of elements.
To delve into the genomic expansion mechanisms of the PsKINs gene family within pea, an examination of the syntenic relationships among the PsKINs was conducted. Out of the 105 PsKINs, 37 exhibited syntenic relationships, with 21 of these forming syntenic pairs that have undergone segmental duplication events within the pea genome, as detailed in Supplementary Table S4. The ratio of the rate of non-synonymous substitutions to the rate of synonymous substitutions, denoted as Ka/Ks, serves as an indicator of selective pressures acting on genes post-duplication. For the 21 segmental duplication gene pairs, the Ka/Ks ratios were found to be less than 1, suggesting that these pairs have experienced strong negative selection throughout their evolutionary trajectory.
In an effort to enhance our comprehension of genetic divergence, gene duplication, and evolutionary patterns between the KIN gene families of P. sativum, A. thaliana, and G. max, syntenic relationships were scrutinized to pinpoint orthologous KIN genes among these species using the MCScanX tool. A total of 56 and 184 pairs of orthologous KIN genes were identified across three comparative analyses (P. sativum versus A. thaliana, P. sativum versus G. max), as depicted in Figure 5. It is noteworthy that certain genes exhibit multiple homologous relationships across the different species. Notably, a significant concentration of syntenic genes was observed on chromosomes Ps5, Ps6, and Ps7.
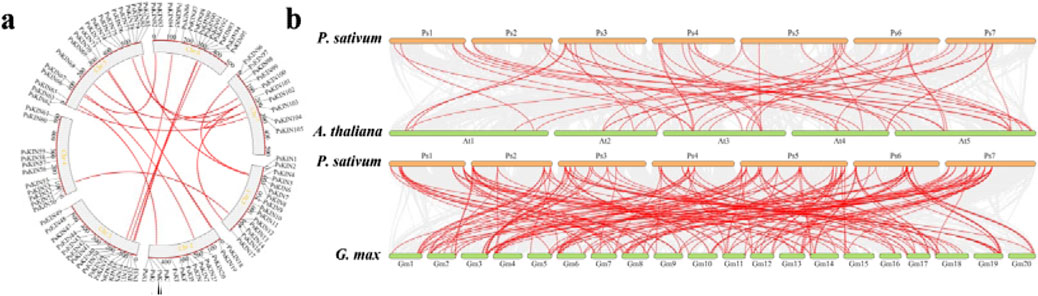
Figure 5. Syntenic analyses of KIN genes in P. mume, Arabidopsis, and Glycine max. (A) Seven chromosomes from P. sativum (Ps1 – Ps7) are mapped. The unit of chromosome length is Mb. Lines denote syntenic KIN gene pairs on the chromosomes. (B) The seven chromosomes of P. sativum (Ps1–7), five chromosomes of Arabidopsis thaliana (At1–5), and twenty chromosomes of Glycine max (Gm1–20) are mapped. The lines represent syntenic KIN gene pairs.
To gain a deeper insight into the biological roles and the intricate regulatory networks associated with PsKINs, the potential protein-protein interactions (PPIs) among these proteins were forecasted employing an orthology-based approach. The outcomes revealed that 43 of the PsKIN proteins shared orthologous connections with their counterparts in Arabidopsis. Intriguingly, this analysis identified the presence of both physical interactions and co-expression patterns among these 43 PsKIN proteins (Figure 6, Supplementary Table S5). It is hypothesized that these interactions are instrumental in orchestrating plant growth and development through the modulation of protein networks.
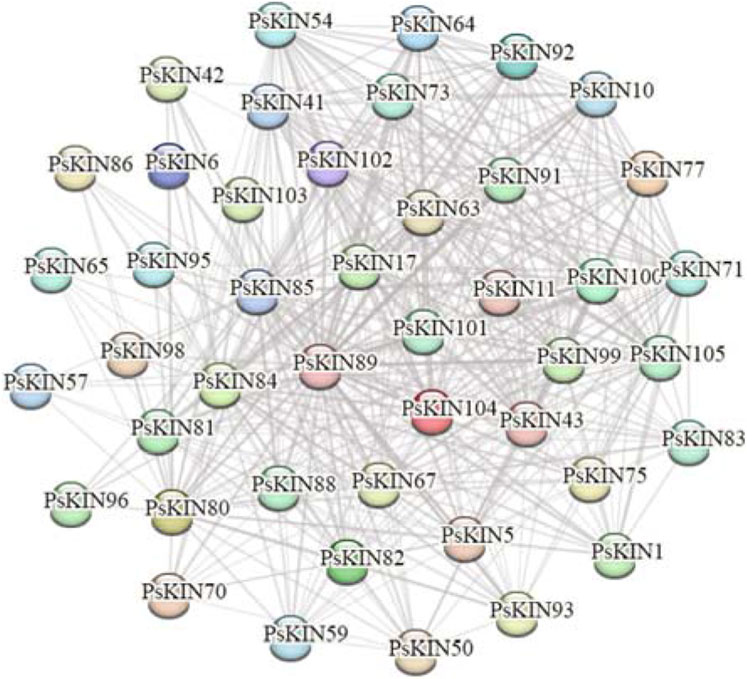
Figure 6. Predicted protein-protein interaction networks of PsKINs proteins with other proteins using STRING tool. The two circles connected by the gray line represent the interaction between the proteins.
To gain a comprehensive understanding of the expression profiles of the PSKIN gene family across various tissues and to anticipate their possible biological roles, an expression heatmap was developed, encompassing 38 PSKIN genes (Figure 7). These were meticulously picked from a collection of 105 genes, showing distinct patterns in a range of tissues including primary and lateral roots, nodules, young and mature stems, tendrils, young and mature leaves, sepals, petals, pods, and seeds.
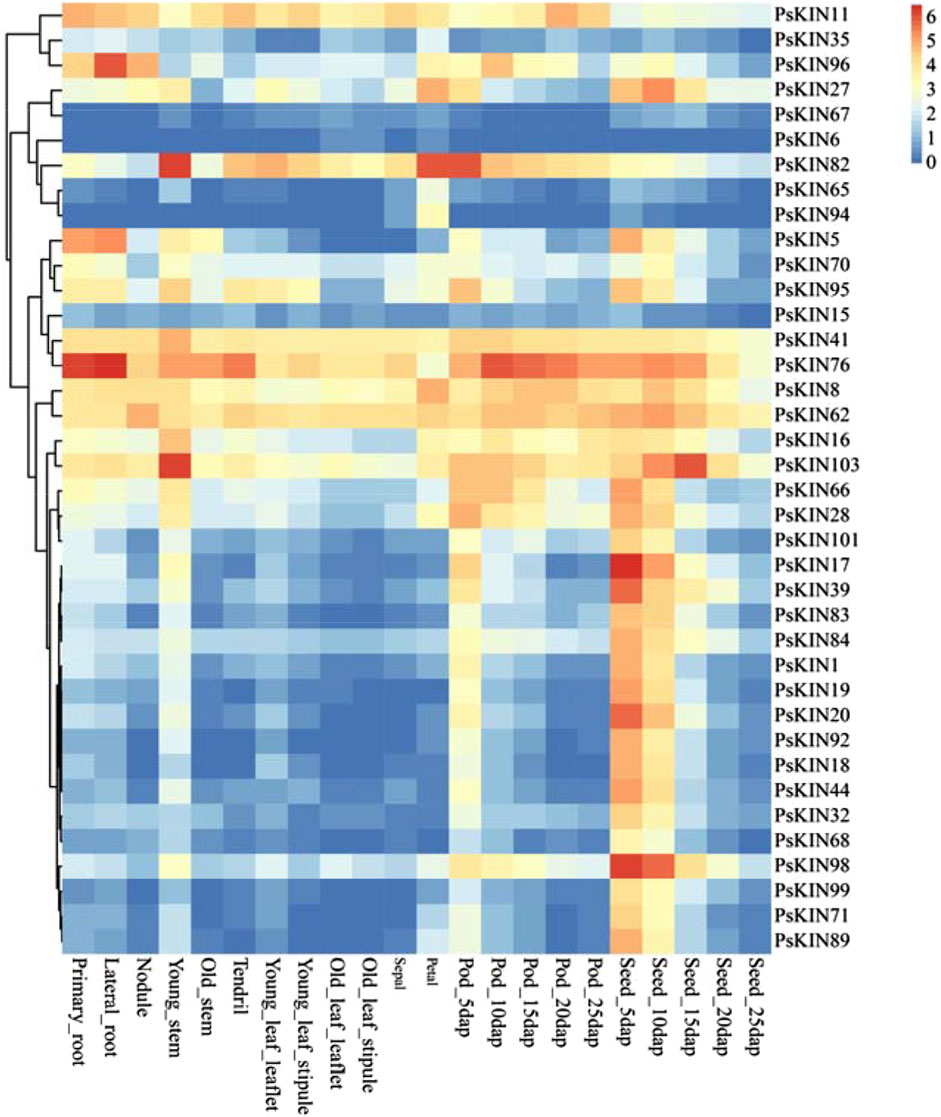
Figure 7. Expression profiles of the 38 PsKIN genes. The color scale from blue to red indicates increasing log2-transformed FPKM values.
We identified that PSKIN76 is markedly expressed in the roots, with PSKIN96 showing elevated levels specifically in lateral roots. Conversely, expression peaks of PSKIN82 in young stems, petals, and pods. Additionally, there is a notable upregulation of PSKIN103 in young stems and seeds, while PSKIN17 and PSKIN98 are highly expressed in seeds.
Our in-depth analysis indicated that 20 genes are predominantly expressed in seeds, hinting at their involvement in seed maturation and dormancy. Meanwhile, 11 genes displayed high expression specificity in pod tissues, potentially linked to pod development and seed safeguarding mechanisms.
To explore the reaction of the KIN gene family under drought conditions, pea seedlings were placed in an environment with a 20% concentration PEG6000 solution. Leaf tissues were harvested at 3 h and 24 h intervals post-treatment for subsequent RNA extraction and transcriptome profiling. As illustrated in eight of picture, a notable fluctuation in the expression levels of 47 PsKIN genes was discerned, including PsKIN8, PsKIN11, PsKIN36, PsKIN37, PsKIN54, PsKIN102, and PsKIN104, following exposure to PEG6000 (Figure 8). This biphasic response, characterized by an initial upregulation followed by a downregulation, could be indicative of acute reaction of the plant to water scarcity and the subsequent acclimation phase. Preliminary functional analyses hint that these genes might partake in signal transduction and metabolic pathways that are instrumental in the plant’s drought stress management.
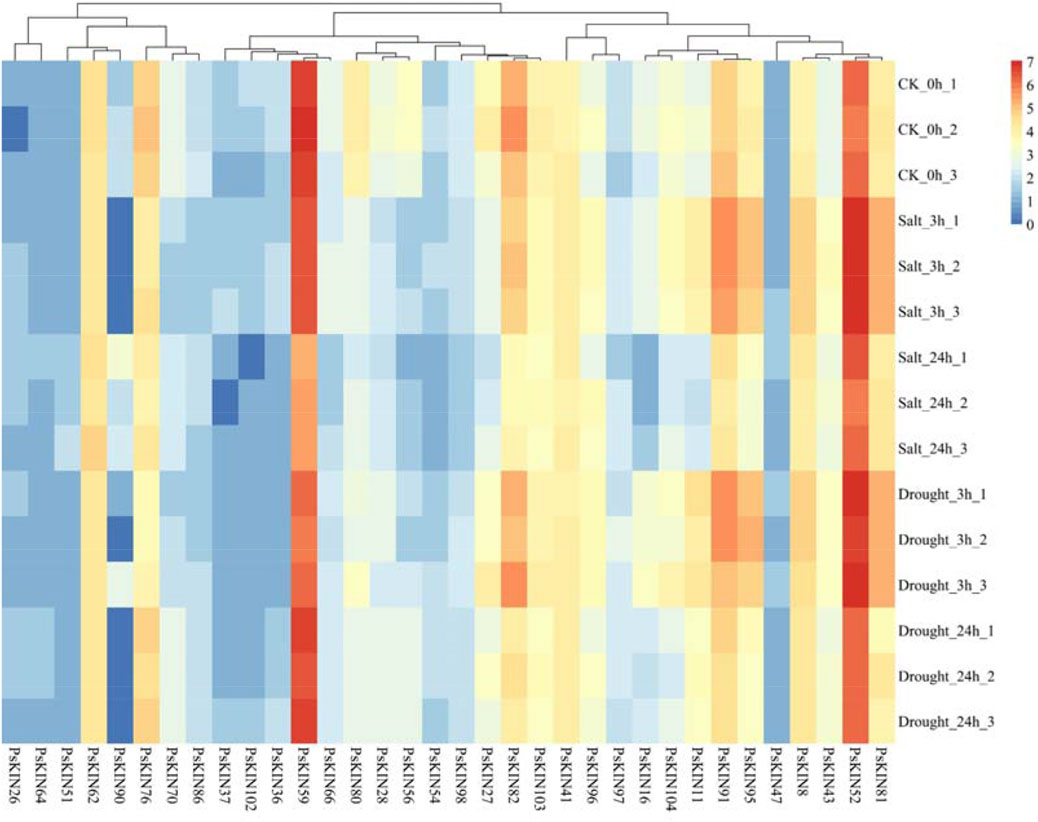
Figure 8. Transcriptome analysis describes the expression of 34 PsKIN genes in peas under drought stress induced by 20% PEG6000 and salt stress induced by 300 mM NaCl. Each experiment was conducted independently with a minimum of three replicates. Error bars indicate the standard deviation among replicates. “CK_0h” denotes the control group.
Conversely, a subset of genes, namely, PsKIN26, PsKIN47, PsKIN54, and PsKIN64, exhibited a sustained decrease in expression, potentially highlighting their involvement in long-term stress endurance mechanisms. This sustained response may be crucial for the preservation of cellular hydration and the reinforcement of antioxidant systems.
Expanding our inquiry to the realm of saline stress, seedlings were treated with escalating concentrations of NaCl, 100, 200, and 300 mM. Post-treatment analysis revealed pronounced alterations in the expression profiles of numerous PsKIN genes, with PsKIN47, PsKIN51, and PsKIN90 standing out as the most significantly affected. These changes are presumed to be intimately linked to the mechanisms of the plant of acclimatization to high salinity, possibly implicating these genes in osmotic adjustment and ionic equilibrium.
The Kinesin gene family in plants is tasked with the production of crucial motor proteins that are instrumental in intracellular transport and cell division. These proteins facilitate directional movement along microtubules, powered by ATP hydrolysis. The family is characterized by a functional diversity; while most kinesins are directed towards the plus ends of microtubules, the kinesin-14 subclass is unique in moving towards the minus ends, thus showcasing the wide-ranging capabilities of this gene family (Yamagishi et al., 2022). In upland cotton, 159 KIN genes have been identified, which belong to the kinesin motor protein gene family and may play a crucial role in the development of cotton fibers and bolls (Zhu et al., 2024). A total of 48 KIN gene family members have been identified in watermelon, highlighting their significant role in early fruit development (Tian et al., 2021). Studies on a range of plant species, such as A. thaliana and Physcomitrium patens, underscore the evolutionary conservation and essential roles that Kinesin proteins play in plant biology (Herrmann et al., 2021; Yoshida et al., 2023). Phylogenetic analyses have identified around 17 distinct Kinesin families, each with roles varying from organelle transport to spindle pole organization, which are vital for deciphering the mechanisms of plant growth, development, and adaptation (Ali and Yang, 2020; Shen et al., 2012; Ye et al., 2022).
This investigation provides an in-depth analysis of the structural and functional diversity of the Kinesin (KIN) gene family in pea, emphasizing the interplay between conserved and divergent features across species. The study identified 105 Kinesin genes in the pea genome, which were classified into 7 subfamilies based on phylogenetic analysis. This classification highlights the evolutionary adaptability and complexity of the Kinesin gene family and its significance in various organisms. Comparative analysis of Kinesin gene numbers in different species—such as 48 in watermelon (Zhou et al., 2019) and 71 in moss (Wickstead andGull, 2006) —demonstrates variability in gene counts within the KIN family, suggesting differences in roles and classifications of these subfamilies. This variability suggests evolutionary divergence and specialization within the KIN gene family, reflecting the unique biological needs of each species. Further analysis of the motif composition and exon/intron structures of PsKINs revealed that genes within the same subfamily often share similar structural features, with a significant variation in the number of exons/introns, ranging from 1 to 36. Protein modeling results echo this pattern, showing that genes within the same subfamily have comparable secondary and 3-D structures, while those from different subfamilies exhibit diverse structural characteristics. These findings support the phylogenetic classifications and suggest that while structural features within each subfamily are conserved, the diversity among different PsKINs likely contributes to the functional versatility of the KIN gene family.
The study also explores the mechanisms behind the expansion of the KIN gene family in pea, identifying segmental duplicates within 21 KIN gene families. This indicates that gene duplication, particularly segmental duplication, plays a significant role in the evolution and expansion of the KIN gene family in pea. Homologous genes identified between pea and other species were predominantly traced to chromosomes Ps5, Ps6, and Ps7, indicating strong conservation of these chromosomes throughout pea evolution and suggesting broader implications of segmental duplications on species evolution. Such duplications are known to be instrumental in the rapid evolution of primate genes and are associated with chromosome abnormalities and genetic diseases (Lawce, 2022).
The promoter regions of PsKIN genes contain cis-regulatory elements essential for the regulation of plant physiological processes. These elements can be broadly categorized into those associated with plant growth and development, hormonal responses, and reactions to environmental stressors. Among these, light-responsive elements are most frequently observed, with a significant number also responding to hormones like methyl jasmonate (MeJA) and gibberellins. Light significantly impacts plant growth and development by influencing processes such as seedling establishment, leaf expansion, and photosynthesis, activating specific photoreceptors and signal transduction pathways that regulate plant physiological and developmental responses, thus modulating gene expression and plant hormone levels throughout the plant’s lifecycle (Singh et al., 2023; Yadukrishnan et al., 2021). MeJA positively affects plant growth and development by promoting seed enlargement, improving plant resistance to adversity, regulating defense mechanisms, and influencing the synthesis of metabolic products, thereby enhancing plant growth parameters and increasing its nutritional and medicinal value (Song et al., 2023; Đurić et al., 2023; Wang et al., 2022). Gibberellins affect plant performance and metabolic product composition by regulating stem elongation, germination, flowering, as well as impacting nutritional growth, sex expression, and yield (Wang et al., 2022).
Transcriptome sequencing in our study has unveiled the distinct expression profiles of 47 PsKIN gene family members across various plant tissues, offering critical insights into their roles in the developmental processes of plants. Notably, PsKIN76 exhibited pronounced expression in primary and lateral roots, hinting at its potential involvement in the development of the root system. This is reminiscent of the role of DEEPER ROOTING 1 (DRO1), which is known to manipulate root growth angles and enhance rice yields under drought conditions (Uga Y et al., 2013). Drawing parallels, we hypothesize that PsKIN76 might regulate root morphogenesis through analogous mechanisms.
The significant expression of PsKIN82 in young stems and petals raises the possibility of its contribution to the reproductive development of plants. The ectopic expression of the KNOX gene SHOOT MERISTEMLESS (STM) in Arabidopsis is known to induce carpel formation and promote the transformation of ovules into carpels, essential for reproductive meristem development (Scofield S et al., 2007). We anticipate that PsKIN82 might perform a comparable regulatory function in the reproductive development of plants.
Furthermore, Elevated expression of PsKIN103 in young stems and seeds points towards a potential role in seed development and seedling growth. The large family of receptor-like protein kinases (RLKs) participates in the regulation of plant growth and development and is capable of adapting to stress (Zhu et al., 2024). We, therefore, speculate that PsKIN103 might serve similar functions, exerting a substantial influence on early plant development and seed maturation.
In response to abiotic stress, our study revealed that the expression of 14 PsKIN genes was significantly modulated, casting light on the plant’s adaptive mechanisms under harsh conditions. Under drought stress, an upregulation of genes like PsKIN8, PsKIN11, PsKIN36, PsKIN37, PsKIN54, PsKIN102, and PsKIN104 was observed, suggesting their participation in the drought response of plant. The GhKLCR1 gene, a kinesin light chain-related gene homologous to AtKLCR1, is upregulated under drought stress. Overexpression of GhKLCR1 in transgenic Arabidopsis plants leads to increased sensitivity to drought stress (Li et al., 2019). GmCAMTA12 Arabidopsis AtAnnexin2 AtWRKY1 PsKIN8. These findings suggest that these genes may play a key role in the response of plant to drought stress.
Conversely, the downregulation of PsKIN26, PsKIN47, PsKIN54, and PsKIN64 under drought stress may be linked to the drought adaptation of plant, akin to the improved drought resistance in grapevines with silenced glutathione S-transferase genes (Nerva L et al., 2022). Under salt stress, the expression changes of PsKIN47, PsKIN51, and PsKIN90 are the most significant. Transgenic rice overexpressing OsASR6 also exhibits adaptability to salt stress (Zhang Q et al., 2022). The OsTUB1 gene in rice interacts with Kinesin13A to stabilize microtubules and ion transporters, thereby conferring salt stress insensitivity to the plant (Chen et al., 2022). These findings propose that these genes might be pivotal in the plant’s response to salt stress.
In essence, our findings underscore the multifaceted roles of the PsKIN gene family in both plant development and stress response.
In our study, we identified the PsKIN gene family in the pea genome, consisting of 115 PsKIN genes that underwent in-depth characterization. This included sequence analysis, phylogenetics, assessment of gene structures and identification of conserved motifs, mapping of their chromosomal locations, analysis of their homologs, and prediction of their potential protein-protein interaction networks. To further our research, we explored the tissue-specific expression profiles of PsKIN genes, with particular attention to their expression dynamics under drought and salt stress conditions. The findings of this study provide profound insights and lay a solid theoretical foundation for future research aimed at uncovering the exact roles of PsKIN genes, especially in the context of abiotic stress.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
HY: Writing–original draft. BL: Writing–original draft. GZ: Writing–original draft. ZF: Writing–original draft. BW: Writing–original draft. YB: Writing–original draft. YX: Writing–original draft. YG: Writing–original draft. ZS: Writing–review and editing. NL: Investigation, Resources, Writing–review and editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. The research was supported by the Zhejiang Provincial Important Science and Technology Specific Projects of Vegetable Breeding (2021C02065) and National Natural Science Foundation of China (2045210652).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2024.1510864/full#supplementary-material
Ali, I., and Yang, W. C. (2020). Why are ATP-driven microtubule minus-end directed motors critical to plants? An overview of plant multifunctional kinesins. Funct. Plant Biol. 47, 524–536. doi:10.1071/FP19177
Aubert, G., Kreplak, J., Leveugle, M., Duborjal, H., Klein, A., Boucherot, K., et al. (2022). SNP discovery by exome capture and resequencing in a 1 pea genetic resource collection. Peer J. 3. doi:10.24072/pcjournal.332
Bian, S., Jin, D., Sun, G., Shan, B., Zhou, H., Wang, J., et al. (2020). Characterization of the soybean R2R3-MYB transcription factor GmMYB81 and its functional roles under abiotic stresses. Gene 753, 144803. doi:10.1016/j.gene.2020.144803
Chen, G., Xuan, W., Zhao, P., Yao, X., Peng, C., Tian, Y., et al. (2022). OsTUB1 confers salt insensitivity by interacting with Kinesin13A to stabilize microtubules and ion transporters in rice. New Phytol. 235, 1836–1852. doi:10.1111/nph.18282
Chen, X., Chen, Z., Zhao, H., Zhao, Y., Cheng, B., and Xiang, Y. (2014). Genome-wide analysis of soybean HD-Zip gene family and expression profiling under salinity and drought treatments. PLoS One 9, e87156. doi:10.1371/journal.pone.0087156
Đurić, M., Subotić, A., Prokić, L., Trifunović-Momčilov, M., and Milošević, S. (2023). Foliar application of methyl jasmonate affects impatiens walleriana growth and leaf physiology under drought stress. Plant Signal Behav. 18, 2219936. doi:10.1080/15592324.2023.2219936
Galindo-Trigo, S., Grand, T. M., Voigt, C. A., and Smith, L. M. (2020). A malectin domain kinesin functions in pollen and seed development in Arabidopsis. J. Exp. Bot. 71, 1828–1841. doi:10.1093/jxb/eraa023
Golovko, T., Golovko, M., Vasilenko, O., Pertsevoi, F., Bolgova, N., Tischenko, V. I., et al. (2023). Technology of protein isolate from peas (Pisum sativum var. arvense). Technol. audit prod. Reserves 2, 37–40. doi:10.15587/2706-5448.2023.278118
Guo, J., Wu, J., Ji, Q., Wang, C., Luo, L., Yuan, Y., et al. (2008). Genome-wide analysis of heat shock transcription factor families in rice and Arabidopsis. J. Genet. Genomics 35, 105–118. doi:10.1016/S1673-8527(08)60016-8
Herrmann, A., Livanos, P., Zimmermann, S., Berendzen, K., Rohr, L., Lipka, E., et al. (2021). KINESIN-12E regulates metaphase spindle flux and helps control spindle size in Arabidopsis. Plant Cell 33, 27–43. doi:10.1093/plcell/koaa003
Lawce, H. (2022). The role of repeat DNA sequences in human evolution and disease. J. Assoc. Genet. 48, 54–61.
Li, G., Liu, R., Xu, R., Varshney, R. K., Ding, H., Li, M., et al. (2023). Development of an Agrobacterium-mediated CRISPR/Cas9 system in pea (Pisum sativum L.). Crop J. 11, 132–139. doi:10.1016/j.cj.2022.04.011
Li, J., Yu, D., Qanmber, G., Lu, L., Wang, L., Zheng, L., et al. (2019). GhKLCR1, a kinesin light chain-related gene, induces drought-stress sensitivity in Arabidopsis. Sci. China Life Sci. 62, 63–75. doi:10.1007/s11427-018-9307-y
Nerva, L., Guaschino, M., Pagliarani, C., De Rosso, M., Lovisolo, C., and Chitarra, W. (2022). Spray-induced gene silencing targeting a glutathione S-transferase gene improves resilience to drought in grapevine. Plant Cell Environ. 45, 347–361. doi:10.1111/pce.14228
Potter, S. C., Luciani, A., Eddy, S. R., Park, Y., Lopez, R., and Finn, R. D. (2018). HMMER web server. Nucleic Acids Res. 2, 46.
Scofield, S., Dewitte, W., and Murray, J. A. (2007). The KNOX gene SHOOT MERISTEMLESS is required for the development of reproductive meristematic tissues in Arabidopsis. Plant J. 50, 767–781. doi:10.1111/j.1365-313X.2007.03095.x
Seneviratne, A. M. P. B., Lidagoster, S., Valbuena-Castor, S., Lashley, K., Saha, S., Alimova, A., et al. (2023). Kinesins modify ERR1-dependent transcription using a conserved nuclear receptor box motif. Int. J. Mol. Sci. 24, 3795. doi:10.3390/ijms24043795
Shen, Z., Collatos, A. R., Bibeau, J. P., Furt, F., and Vidali, L. (2012). Phylogenetic analysis of the Kinesin superfamily from physcomitrella. Front. Plant Sci. 3, 230. doi:10.3389/fpls.2012.00230
Singh, D., Ravindran, N., Job, N., Rahul, P. V., Bhagavatula, L., and Datta, S. (2023). “Plant transcription factors in light-regulated development and UV-B protection,” in Plant transcription factors Editors V. Srivastava, S. Mishra, S. Mehrotra, and S. K. Upadhyay 4th ed. (New York: Academic Press), 139–157.
Solon, A. L., Tan, Z., Schutt, K. L., Jepsen, L., Haynes, S. E., Alexey, I., et al. (2021). Kinesin-binding protein remodels the kinesin motor to prevent microtubule binding. Sci. Adv. 7, eabj9812. doi:10.1126/sciadv.abj9812
Song, Q., Gong, W., Yu, X., Ji, K., Jiang, Y., Chang, Y., et al. (2023). Transcriptome and anatomical comparisons reveal the effects of methyl jasmonate on the seed development of camellia oleifera. J. Agric. Food Chem. 71, 6747–6762. doi:10.1021/acs.jafc.3c00059
Strauß, T., Schattner, S., Hoth, S., and Walter, W. J. (2021). The Arabidopsis thaliana kinesin-5 AtKRP125b is a processive, microtubule-sliding motor protein with putative plant-specific functions. Int. J. Mol. Sci. 22, 11361. doi:10.3390/ijms222111361
Tian, S., Jiang, J., Xu, G. Q., Wang, T., Liu, Q., Chen, X., et al. (2021). Genome wide analysis of kinesin gene family in Citrullus lanatus reveals an essential role in early fruit development. BMC Plant Biol. 21, 210–216. doi:10.1186/s12870-021-02988-6
Uga, Y., Sugimoto, K., Ogawa, S., Rane, J., Ishitani, M., Hara, N., et al. (2013). Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat. Genet. 45, 1097–1102. doi:10.1038/ng.2725
Wang, C., Zhang, J., Lv, J., Li, J., Gao, Y., Patience, B. E., et al. (2022). Effect of methyl jasmonate treatment on primary and secondary metabolites and antioxidant capacity of the substrate and hydroponically grown Chinese chives. Front. Nutr. 9, 859035. doi:10.3389/fnut.2022.859035
Wickstead, B., and Gull, K. (2006). A “holistic” kinesin phylogeny reveals new kinesin families and predicts protein functions. Mol. Biol. Cell. 17, 1734–1743. doi:10.1091/mbc.e05-11-1090
Wu, D. T., Li, W. X., Wan, J. J., Hu, Y. C., Gan, R. Y., and Zou, L. (2023). A comprehensive review of pea (Pisum sativum L.): chemical composition, processing, health benefits, and food applications. Foods 12, 2527. doi:10.3390/foods12132527
Yadukrishnan, P., Singh, D., Ravindran, N., Kushwaha, A. K., Job, N., Rahul, P. V., et al. (2021). “Hormones and light-regulated seedling development,” in Hormones and plant response, plant in challenging environments. Editors D. K. Gupta,, and F. J. Corpas 2nd ed. (Cham: Springer), 91–116.
Yamagishi, M., Sumiyoshi, R., Drummond, D. R., and Yajima, J. (2022). Anchoring geometry is a significant factor in determining the direction of kinesin-14 motility on microtubules. Sci. Rep. 12, 15417. doi:10.1038/s41598-022-19589-4
Ye, Z., Yuan, Z., Xu, H., Pan, L., Chen, J., Gatera, A., et al. (2022). Genome-wide identification and expression analysis of kinesin family in barley (Hordeum vulgare). Genes (Basel) 13, 2376. doi:10.3390/genes13122376
Yoshida, M. W., Hakozaki, M., and Goshima, G. (2023). Armadillo repeat-containing kinesin represents the versatile plus-end-directed transporter in Physcomitrella. Nat. Plants 9, 733–748. doi:10.1038/s41477-023-01397-x
Zhang, Q., Liu, Y., Jiang, Y., Li, A., Cheng, B., and Wu, J. (2022). OsASR6 enhances salt stress tolerance in rice. Int. J. Mol. Sci. 23, 9340. doi:10.3390/ijms23169340
Zhou, L., Ouyang, L., Chen, K., and Wang, X. (2019). Research progress on KIF3B and related diseases. Ann. Transl. Med. 7, 492. doi:10.21037/atm.2019.08.47
Keywords: Pisum sativum L., PsKIN, gene expression, drought stress, salt stress
Citation: Yuan H, Liu B, Zhang G, Feng Z, Wang B, Bu Y, Xu Y, Gong Y, Sun Z and Liu N (2024) Genome-wide identification and expression analysis of the PsKIN gene family in pea. Front. Genet. 15:1510864. doi: 10.3389/fgene.2024.1510864
Received: 14 October 2024; Accepted: 21 November 2024;
Published: 18 December 2024.
Edited by:
Zhiqiang Wang, Chengdu Agriculture College, ChinaReviewed by:
Xiefeng Yao, Jiangsu Academy of Agricultural Sciences (JAAS), ChinaCopyright © 2024 Yuan, Liu, Zhang, Feng, Wang, Bu, Xu, Gong, Sun and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Na Liu, bG4yMDA4MTFAMTYzLmNvbQ==; Zhihong Sun, emguc3VuQHphZnUuZWR1LmNu
‡These authors share first authorship
†ORCID: Zhihong Sun, orcid.org/0000-0002-9172-2062; Na Liu, orcid.org/0000-0002-8154-7037
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.