- 1Key Laboratory of Herbage & Endemic Crop Biology, Ministry of Education, School of Life Sciences, Inner Mongolia Normal University, Hohhot, China
- 2College of Agriculture and Bioengineering, Heze University, Heze, China
- 3College of Agriculture, Shanxi Agricultural University, Taigu, China
Among the largest transcription factor families in plants, bZIPs are crucial for various developmental and physiological processes, particularly abiotic stress resistance. Setaria italica has become a model for understanding stress resistance mechanisms. In this study, we identified 90 bZIP transcription factors in the Setaria italica genome. SibZIPs were classified into 13 groups based on references to Arabidopsis bZIPs. Members in the same group shared similar motifs and gene structure pattern. In addition, gene duplication analysis indenfied 37 pairs of segmental duplicated genes and none tandem duplicated genes in S. italica suggesting segmental duplication contributed to the expansion of the S. italica bZIP gene family. Moreover, the number of SibZIPs genes (39) exhibiting higher expression in roots was significantly more than that in other organs. Twelve SibZIP genes were upregulated in response to dehydration stress. In conclusion, our study advances the current understanding of SibZIP genes and provide a number of candidates for functional analysis of drought tolerance in S. italica.
1 Introduction
Drought stress is a major abiotic stress that affects global crop productivity, with almost all important crops being highly sensitive to drought (Zandalinas et al., 2018). To maintain normal growth and development under dehydration stress, plants have developed adaptive regulatory mechanisms to increase drought tolerance (Gupta et al., 2020). One such mechanism is the use of transcription factors (TFs) to regulate (inhibit or activate) specific protein expression to generate appropriate responses (Baillo et al., 2019; Hrmova and Hussain, 2021). The basic leucine zipper motif (bZIP) is a major TF family that actively responds to dehydration stress (Joo et al., 2021).
The bZIP TFs have 60–80 amino acid (aa) residues, with a basic structure comprising an alkaline binding domain and a leucine zipper dimerization motif (Hurst, 1995). The former contains a relatively conserved ACGT core motif in the form of DNA cis-elements, such as the G Box (CACGTG), C Box (CACGTC), and A Box (TACGTA) (Izawa et al., 1993; Foster et al., 1994). The latter is composed of two typical α-helices, each with at least four leucines (Leu) or another hydrophobic residue (e.g., isoleucine, valine, methionine) at every seventh position.
Members of the bZIP family play important regulatory roles in seed maturation (Alonso et al., 2009), flower development (Abe et al., 2005), carbon and nitrogen metabolism (Baena-González et al., 2007), and abiotic stress responses (Uno et al., 2000). Numerous studies have shown that plant could withstand drought stress by bZIP TFs through abscisic acid (ABA)-dependent pathways (Hrmova and Hussain, 2021; Guo et al., 2024). ABF1, AREB1/ABF2, ABF3, and AREB2/ABF4 could enhance drought stress tolerance in Arabidpsis (Yoshida et al., 2010; Yoshida et al., 2014). The manipulation of AREB1(AtbZIP36), which is involved in ABA response pathway, has been shown to improve drought tolerance in Arabidpsis (Fujita et al., 2005). Similarly, OsABF1 (OsbZIP42) acts as a positive regulator of drought tolerance in rice (Joo et al., 2019). Moreover, OsbZIP23, OsbZIP45, OsbZIP46 and OsbZIP72 also play important roles in drought tolerance in rice (Park et al., 2015; Tang et al., 2012; Lu et al., 2009). In addition, bZIP TFs in other plant species such as GmbZIP2 in soybean, ZmbZIP76 in maize, and PtrbZIP3 in Populus trichocarpa are also involved in drought tolerance (Yang et al., 2020; He et al., 2024; Zhou et al., 2023).
Initial genome-wide analyses of bZIP family members were made possible through the availability of genome sequences from the model plants Arabidopsis and rice (Oryza sativa). Arabidopsis bZIP proteins are classified into 10 groups and one unclassified group based on phylogeny and conserved motifs (Jakoby, 2002). In rice, the majority of phylogenetically related bZIP proteins were found to have similar DNA-binding properties based on binding site analyses (Nijhawan et al., 2008). As more plant genomes have been sequenced, bZIP families have been characterized in maize (Wei et al., 2012), sorghum (Wang et al., 2011), wheat (Agarwal et al., 2019), soybean (Zhang et al., 2018), barley (Pourabed et al., 2015), peanut (Wang et al., 2019), and peach (Aslam et al., 2023).
A draf genome of Setaria was developed over a decade ago, and the genome sequence has been updated to version 2.2 at Phytozome database (Zhang et al., 2012). Enomous trancriptomic and proteomic data of Setaria under drought stress has been generated (Pan et al., 2018; Zhang et al., 2022; Gao et al., 2023), and investigation of these data using bioinformatic and biotechnology approaches help us identify the possible candidate genes of drought tolerance. In this study, we used foxtail millet (Setaria italica L.) as a model species for examining the role of bZIP in drought tolerance. We performed a genome-wide analysis of the bZIP gene family in the S. italica genome to identify and classify S. italica bZIP (SibZIP) genes. We also analyzed SibZIP expression profiles within different organs and under drought treatment. The results from our study should offer valuable information for further understanding the role of SibZIPs in drought tolerance.
2 Materials and methods
2.1 Identification of the bZIP gene family in Setaria italica
To identify bZIP genes in S. italica, the bZIP domain (PF00170) was downloaded from the Pfam website (http://pfam.xfam.org/). HMMER software was then used to screen the protein sequences of S. italica, with a threshold set at an E-value <10−5. In addition, 78 Arabidosis AtbZIPs and 89 rice OsbZIP protein sequences, download from TAIR (http://arabidopsis.org) and TIGR (http://www.tigr.org) respectively, were used as queries to search S. italica protein sequences. Subsequently, the candidate proteins were further screened, and the conserved domains were validated using SMART (http://smart.embl-heidelberg.de/) in combination with the NCBI CDD online analysis website (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). The incomplete domain and redundant protein sequences were manually removed. Finally, a total of 90 SibZIP genes were identified from S. italica (Supplementary Table S1).
2.2 Chromosomal localization and syntenic analysis of SibZIP genes
The genome annotation file (Gene Transfer Format/General Feature Format version 3 [GTF/GFF3]) of S. italica was downloaded from the Phytozome database (https://phytozome-next.jgi.doe.gov/), and the chromosomal localization of each SibZIP gene was displayed using TBtools (Chen et al., 2020).
The gene duplication of the SibZIP genes in S. italica was predicted using MCScanX (Wang et al., 2012). The syntenic relationships between SibZIP genes and bZIP genes from A. thaliana and O. sativa were visualized by TBtools. The nonsynonymous substitution rate (Ka), synonymous substitution rate (Ks), and Ka/Ks ratio were determined using TBtools (Supplementary Table S2, S3, S4). The selection pressure of duplicated genes was evaluated using the Ka/Ks ratio. Ka/Ks > 1 suggests positive selection, Ka/Ks = 1 suggests neutral selection, and Ka/Ks < 1 meant negative selection (Krishnamurthy et al., 2015).
2.3 Phylogenetic tree construction of SibZIP proteins
The full bZIP protein sequences of 13 AtbZIP proteins from Arabidopsis, 89 OsbZIP proteins from rice and 90 SibZIP proteins were subjected to multiple sequence alignment using the MUSCLE wrapper and trimAL wrapper of TBtools (Supplementary Table S5). The results were used to construct a neighbor-joining phylogenetic tree in TBtools. Bootstrap values were calculated with 1,000 iterations. The phylogenetic tree was embellished using iTOL v6.7.6 (https://itol.embl.de/).
2.4 Conserved domains and gene structure analysis of the SibZIP genes
Protein motifs were identified using Multiple Expectation Maximization for Motif Elicitation (MEME) (http://meme.nbcr.net/meme/). The analysis was performed with the following settings: number of repetitions, any; maximum number of motifs, 20; and optimum width motifs, 10–60.
Full bZIP protein sequences from S. italica were subjected to the Conserved Domain Database (CDD) from the National Center for Biotechnology Information (NCBI), and the results were used to construct gene structure photographs using Gene Structure View in TBtools.
2.5 In silico expression profiling of SibZIP genes
The gene expression profiling data of SibZIP genes were retrieved from the Multi-omics Database for S. italica (MDSi) (http://foxtail-millet.biocloud.net/home). The reads per kilobase per million (RPKM) was downloaded (Supplementary Table S6) and a heatmap was generated in the HeatMap in TBtools.
In addition, transcriptomic data from roots of two drought-tolerant cultivars (Ci328 and Ci409) under normal condition (ERX5299071 and ERX5299091) and drought condition (ERX5987296 and ERX5299098) were retrieved from the European Nucleotide Archive (https://www.ebi.ac.uk/ena, PRJEB43702). The expression data of SibZIP genes were normalized as LogRPKM (Supplementary Table S7) and the expression levels of the SibZIP genes were visualized using HeatMap in TBtools.
2.6 Plant materials and drought treatment
Seeds of ‘Yugu1’ were obtained from Inner Mongolia Agriculture University, Hohhot, China, and grown in a greenhouse under the following conditions: 20 h of light (150 μmol⋅m-2⋅sec-1) at 26°C ± 2°C and 4 h of darkness at 22°C ± 2°C. eight pots containing 5-week-old seedlings were under drought stress for 10 days and re-watering afterwords. The pots with normal watering were used as controls. Roots from seedlings under drought stress and re-watering were collected and then stored at −80°C until RNA isolation. Three independent replicates were carried.
2.7 SibZIPs gene expression in response to dehydration stress
Total RNA was extracted from roots using RNAiso Plus (TaKaRa, T9108) according to the protocol. The integrity of the extracted RNA was determined through 1.5% agarose gel electrophoresis and the quantity was measured with a NanoDrop. The cDNA was synthesized using PrimeScriptTM Ⅱfirst Strand cDNA Synthesis Kit (TaKaRa, 6210A). RT-qPCR was carried out using PrimeScript RT reagent Kit with gDNA Eraser (Takara, RR047A) on ABI7500 (Applied Biosystems, USA). The primers of SibZIP genes were designed by Primer Premier 5.0 (Supplementary Table S8). The fold change of the expression levels of SibZIP genes was calculated via relative quantification (2-△△CT) and SiACTIN was used as internal reference gene (Zhang et al., 2017).
3 Results
3.1 Identification of SibZIP genes
An initial search of the Phytozome database yielded 92 SibZIP members. Of these, 2 were redundant transcripts, and the remainder contained 90 putative SibZIP genes, which were named SibZIP1-SibZIP90 based on their chromosomal position (Supplementary Table S1).
The predicted size of the SibZIP proteins was 319 aa on average, but they spanned a wide range (132–759 aa). Approximately 50.5% of the SibZIP proteins had predicted sizes between 200 and 400 aa, 27.5% had sizes less than 200 aa, and 23% had sizes greater than 400 aa. The longest predicted protein sequence (759 aa) was SibZIP62, whereas the shortest (132aa) was SibZIP69. The isoelectric point (pI) values (4.69–11.97) and molecular weights (15.260–80.305 kDa) of the SibZIPs varied widely. Supplementary Table S1 provides further characteristics of the SibZIP proteins.
3.2 Chromosome localization, gene duplication and syntenic analysis of SibZIP genes
The 90 SibZIP genes were unevenly distributed across the 9 chromosomes of S. italica (Figure 1). The number of genes on each chromosome was unrelated to chromosome size. Chromosome 2 had the greatest number of genes (15), accounting for 16.7% of all SibZIP genes, while chromosome 8 contained the least number of genes (4, 4.4%).
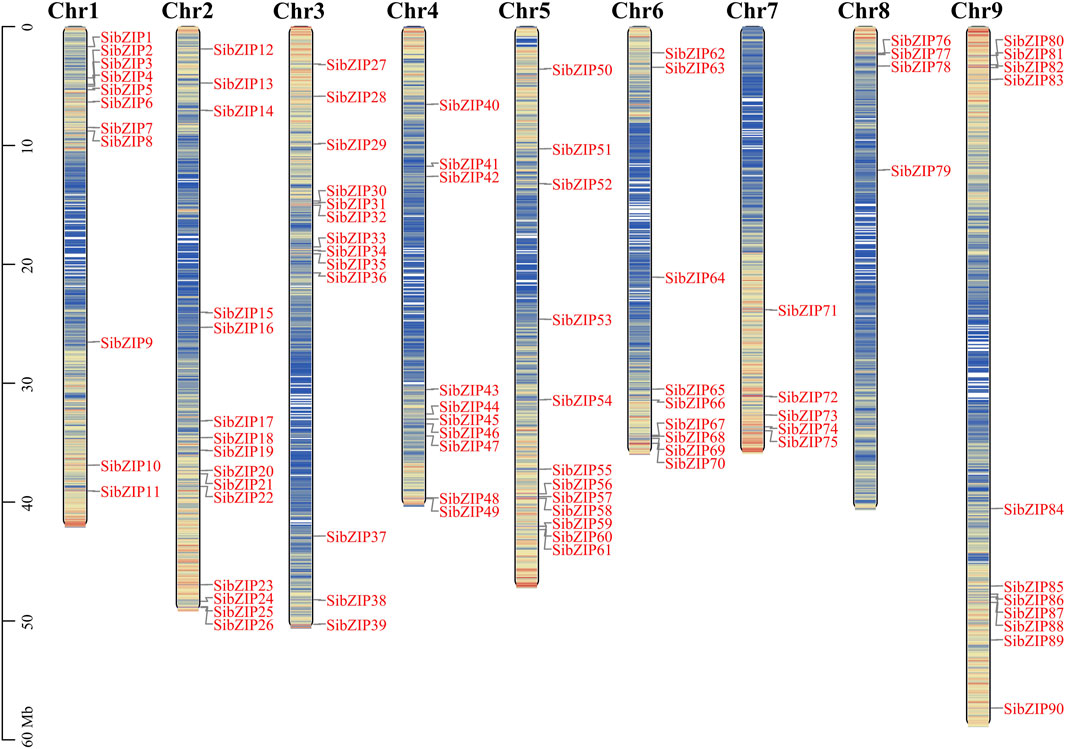
Figure 1. Distribution of 90 SibZIP genes onto nine Setaria italica chromosomes. Graphical representation of physical locations for each SibZIP gene on Setaria italica chromosomes (numbered Chr1–9). Chromosomal distances are given in Mb.
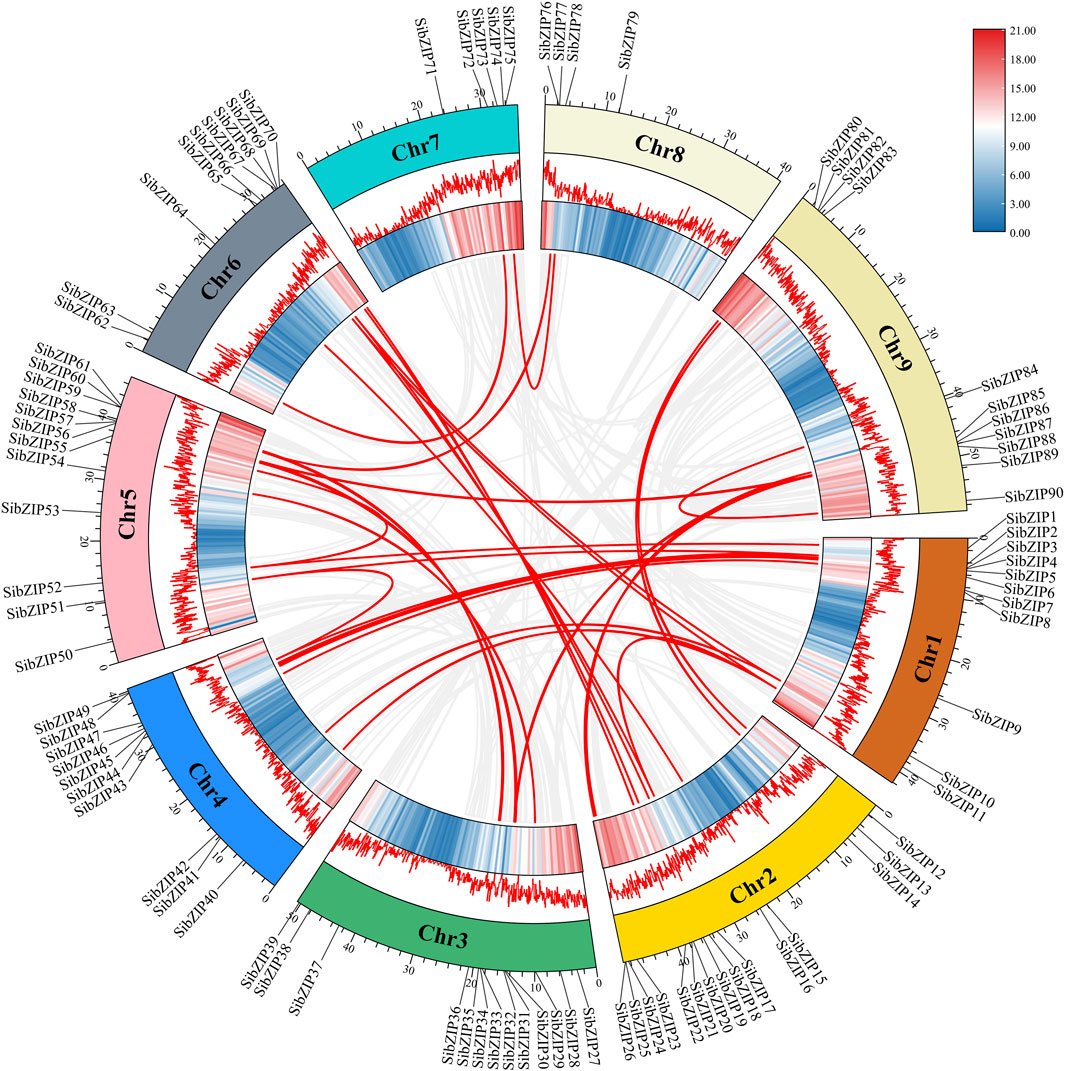
Moreover, gene duplication of the SibZIP genes was predicted using MCScanX. Thirty-seven segmental duplicated gene pairs were detected on different chromosomes of S. italica, and no tandemly duplicated genes were detected (Figure 2; Supplementary Table S2). This result indicates that segmental duplication contributed to the expansion of the S. italica bZIP gene family during evolution. The Ka/Ks ratio of 37 pairs of segmental duplicated genes were all lower than 1, varying from 0.07 to 0.90. The Ka/Ks value of SibZIP21 and SibZIP67 is larger than 0.9 and the ratios of the rest gene pairs were all less than 0.5.
In addition, to investigate the evolutionary relationships of bZIP genes from S. italica, A. thaliana and O. sativa, syntenic analysis was conducted using TBtools. The results revealed that 84 SibZIP genes exhibited collinear relationships with rice OsbZIP genes, while only 7 SibZIP genes exhibited collinear relationships with Arabidopsis AtbZIP genes (Figure 3; Supplementary Table S3, S4).
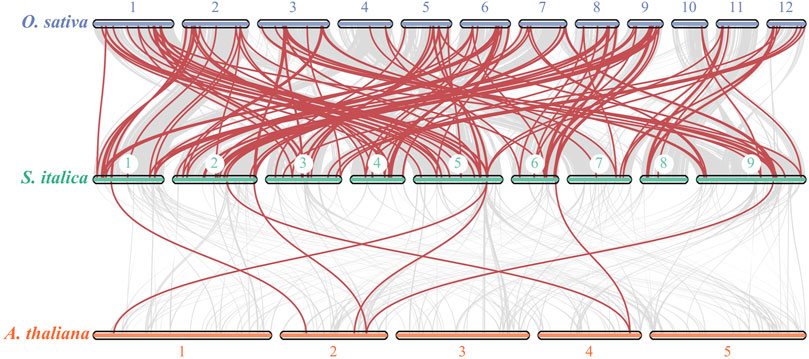
Figure 3. Synteny of bZIP gene family members between Setaria italica, A. thaliana, and Oryza sativa.
3.3 Phylogeny, motif and gene structure analysis of the SibZIP genes
Based on the multiple sequence alignment and the previously reported AtbZIP classification, the SibZIP genes and rice OsbZIP genes were assigned to 13 groups (A-M, and S) (Figure 4). The larger group was Group S containing 17 SibZIPs, followed by Group A (15) and Group D (15). The smallest groups were B and J with only two genes. In addition, no SibZIP and OsbZIP were classified wtih Arabidopsis AtbZIP72 (Group M).
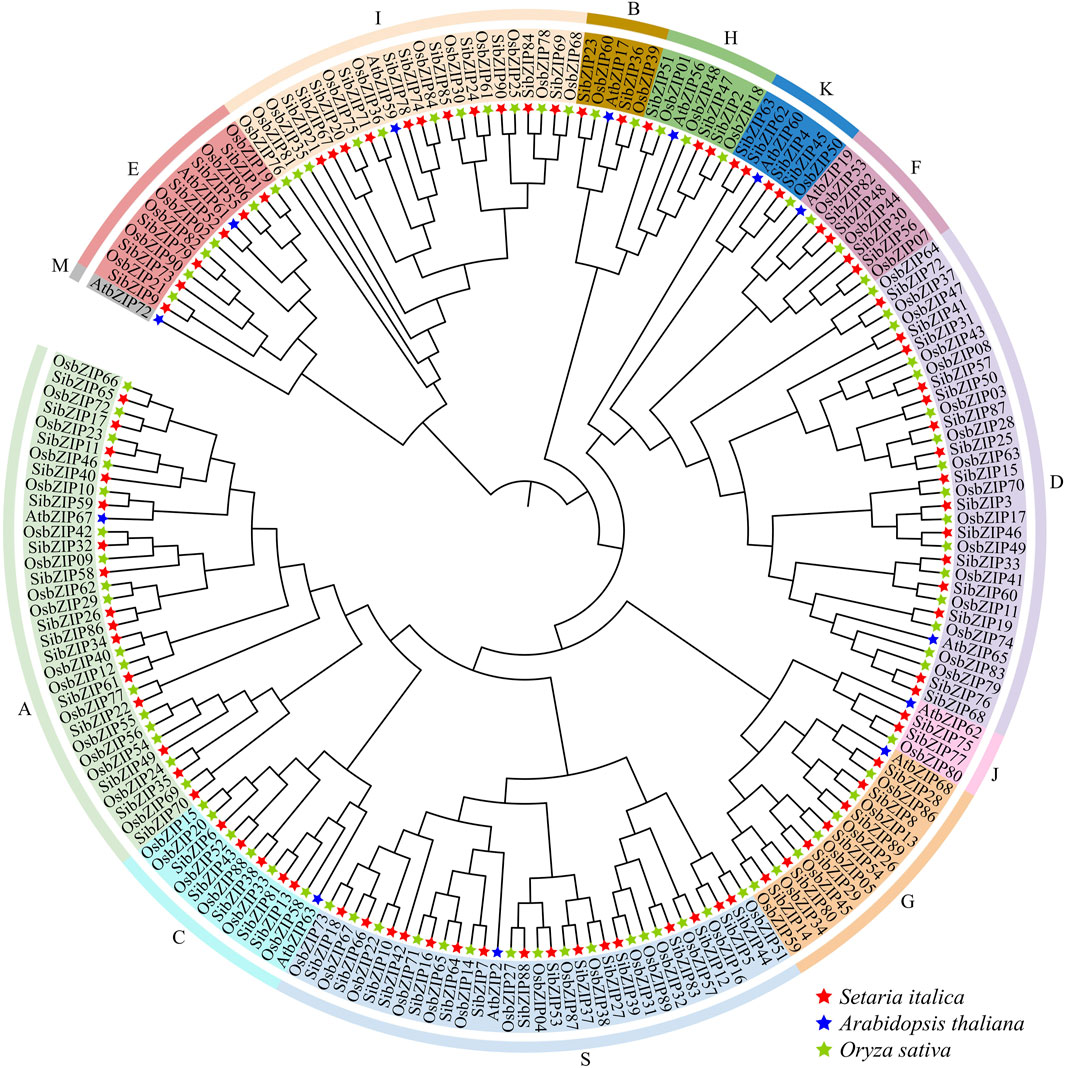
Figure 4. Phylogenetic tree of bZIP proteins from Setaria italica (SibZIPs) and Arabidopsis (AtbZIPs) and O. sativa (OsbZIPs). The proteins were classified into 13 distinct clusters. Each group was assigned a different color. The name of groups (A, B, C, D, E, F, G, H, I, J, K, M and S) were shown at the outside of the circle.
We identified 20 motifs in the 90 SibZIP genes and mapped their distribution to the phylogenetic tree (Figures 5A,B; Supplementary Table S9). All SibZIPs contained the basic leucine zipper domain (Motif 1; Supplementary Figure S1). Group A contained specific motif 8 (RQGSLGSLTLEEFLVRLGVVREDMGSD), which contains a phosphorylation site RXXS/T (Uno et al., 2000). Group D contained specific motif 3 and motif 6 which are glutamine rich (Q-rich) domains at C-terminus. Motif 13 was observed only in Group I, while motif 20 was only present in Group G. In addition, some motifs were present in multiple groups. Groups E and I, for instance, both contained motifs 4 and 18, whereas motif 10 was shared across most groups except D. Together, these observations demonstrate that most SibZIPs in the same group also tended to contain similar motifs.
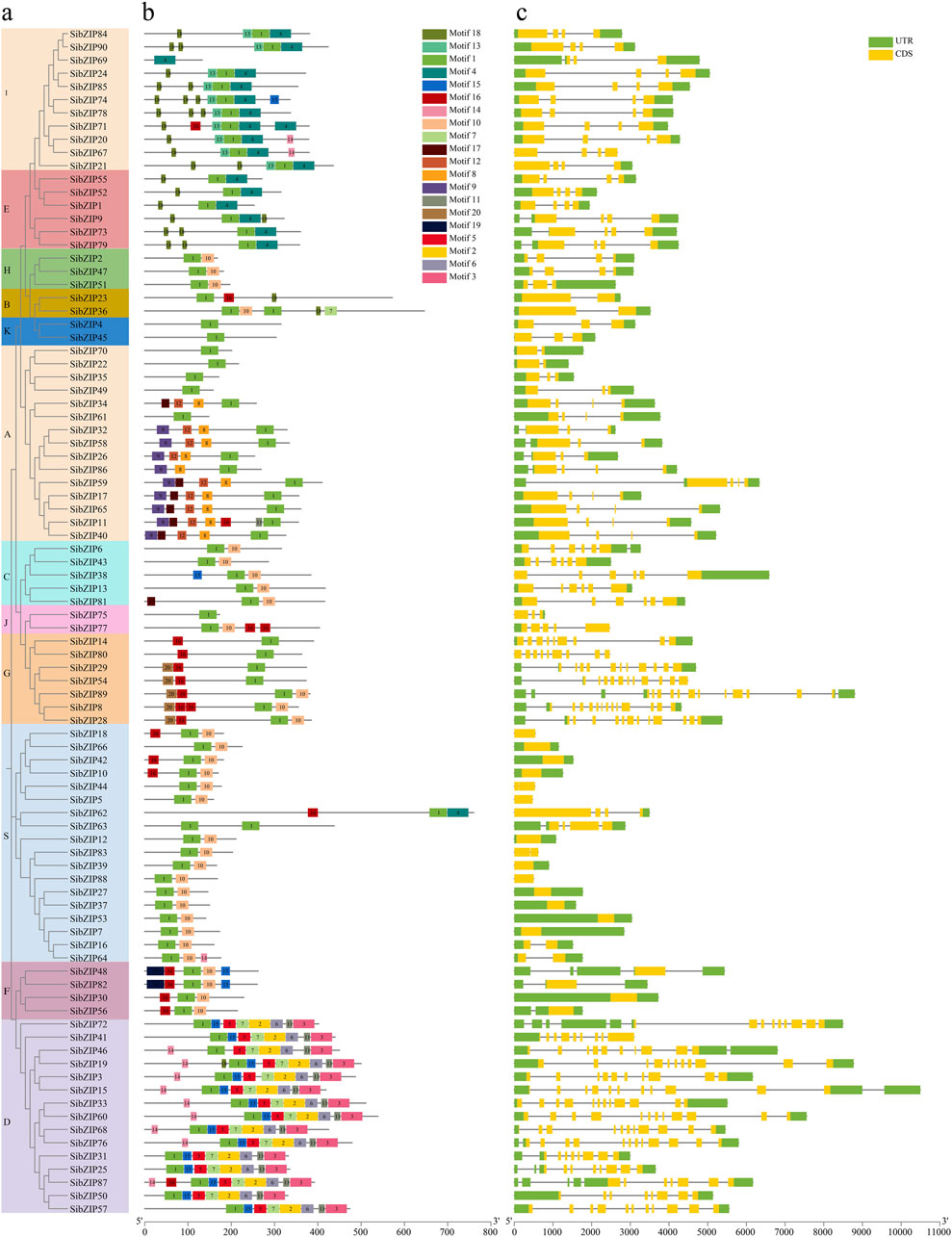
Figure 5. Schematic diagram of amino acid motifs and gene structure of SibZIP genes from different groups. (A) Phylogenetic tree of SibZIP genes. (B) Schematic diagram of amino acid motifs of SibZIP genes from different groups. (C) Schematic diagram of gene structure of SibZIP genes from different groups. Motif analysis was performed using Tbtools software as described in the Methods. The black solid line represents the corresponding SibZIP genes and its length. The different-colored boxes represent different motifs and their position in each SibZIP sequence.
The distributions of the coding sequences (CDSs), untranslated regions (UTRs), and introns of the SibZIPs are displayed in Figure 5C. The number of introns ranged from 1 to 14, and the number of introns in the same subgroup was similar. Most SibZIP genes from Group S were intronless.
3.4 Tissue-specific expression analysis of SibZIP genes
The gene expression profiling data of SibZIP genes from eight different organs (including seedling, young leaf, stem, flag leaf, root, panicle, immature seed and mature seed) were retrieved from the MDSi database (Supplementary Table S6). A heatmap was generated using TBtools (Figure 6), which showed that seven SibZIPs (SibZIP20, SibZIP57, SibZIP58, SibZIP65, SibZIP69, SibZIP73 and SibZIP74) were expressed in all the tissues, but many genes were specific to certain organs (Figure 6; Supplementary Table S6). For example, 39 SibZIP genes have highest expression in roots, and 13 and 14 genes were in panicle and stem. SibZIP12 were only expressed in mature seeds.
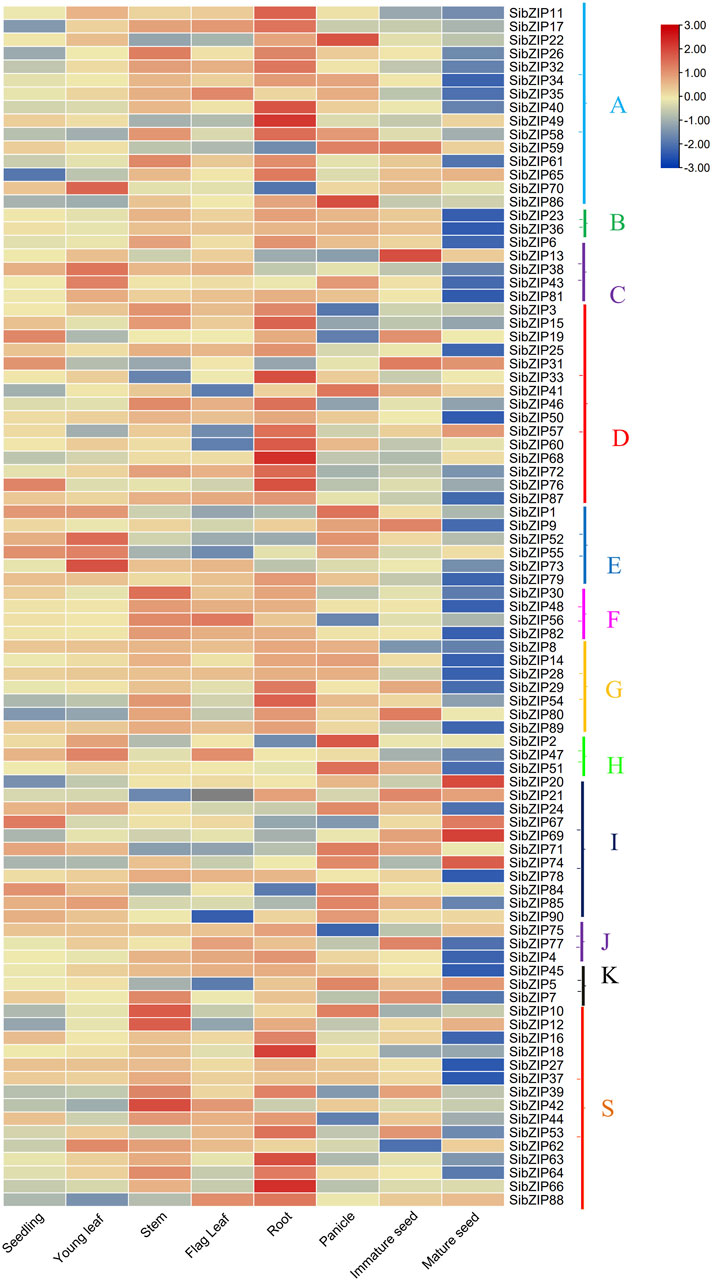
Figure 6. Heat-map showing the expression pattern of SibZIP genes in eight tissues namely seedlings, young leaves, stems, flag leaves, roots, panicles, immature seeds and mature seeds. The heat-map shows gene expression of SibZIP genes in different groups (A, B, C, D, E, F, G, H, I, J, K and S). The color scales for fold-change values are shown at the right. The figure showed that most SibZIP genes were highly expressed in at least one of the tested tissues. Note that expression values mapped to a color gradient from low (blue) to high expression (orange).
3.5 SibZIP gene expression patterns under dehydration stress
The transcriptome data of tolerance cultivars (Ci328 and Ci409) under drought stress was used to investigate SibZIP gene expression patterns under dehydration stress. The expression patterns of the SibZIP genes in roots under drought treatment and under normal conditions are shown in the heatmap (Figure 7; Supplementary Table S7). The results showed that 27 SibZIP genes were upregulated after drought stress in both Ci328 and Ci409 and marked with asterisks in Figure 7. These genes were from different groups. Group A had highest number of upregulated genes (5 genes), followed by Group D (4) and Group G (4).
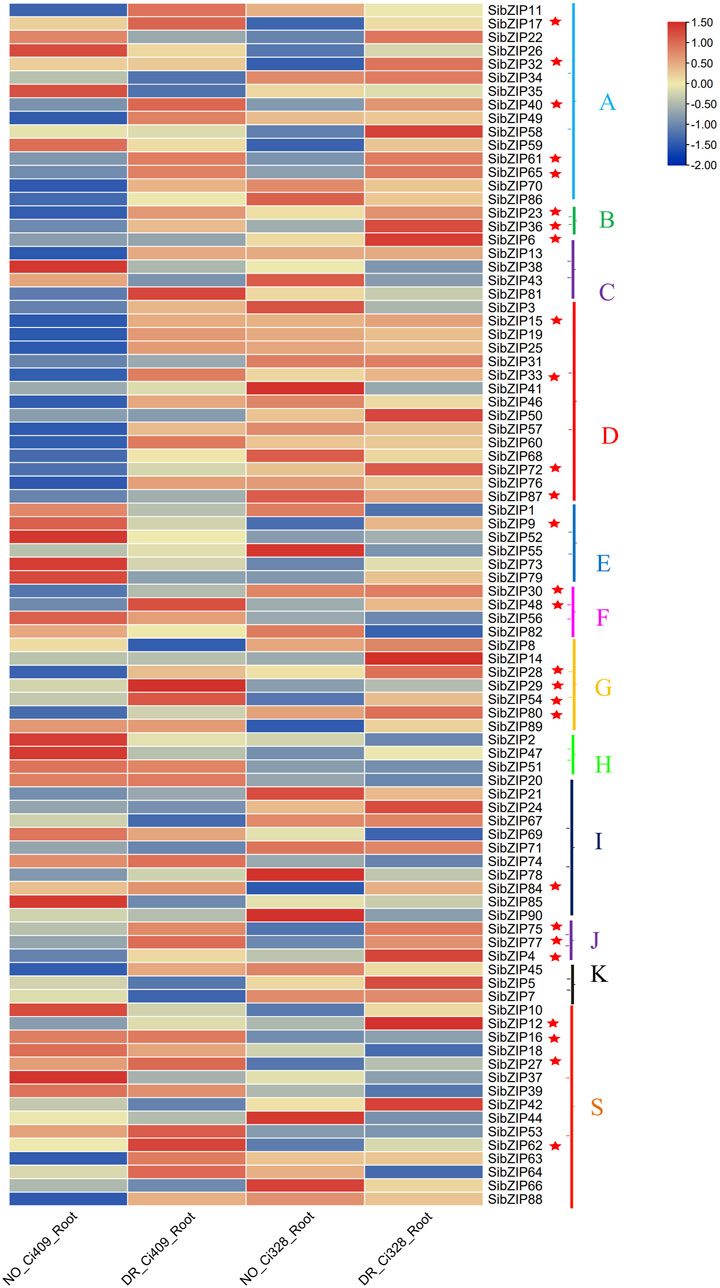
Figure 7. SibZIP genes expression patterns under dehydration stress. The transcriptome data of tolerance cultivars (Ci328 and Ci409) under normal condition and drought stress condition was downloaded to investigate SibZIP gene expression patterns under dehydration stress (Zhang et al., 2022).The heat-map shows gene expression of SibZIP genes in different groups (A, B, C, D, E, F, G, H, I, J, K and S).Twenty seven SibZIP genes were up-regulated after drought stress in both Ci328 and Ci409 and marked with asterisks.
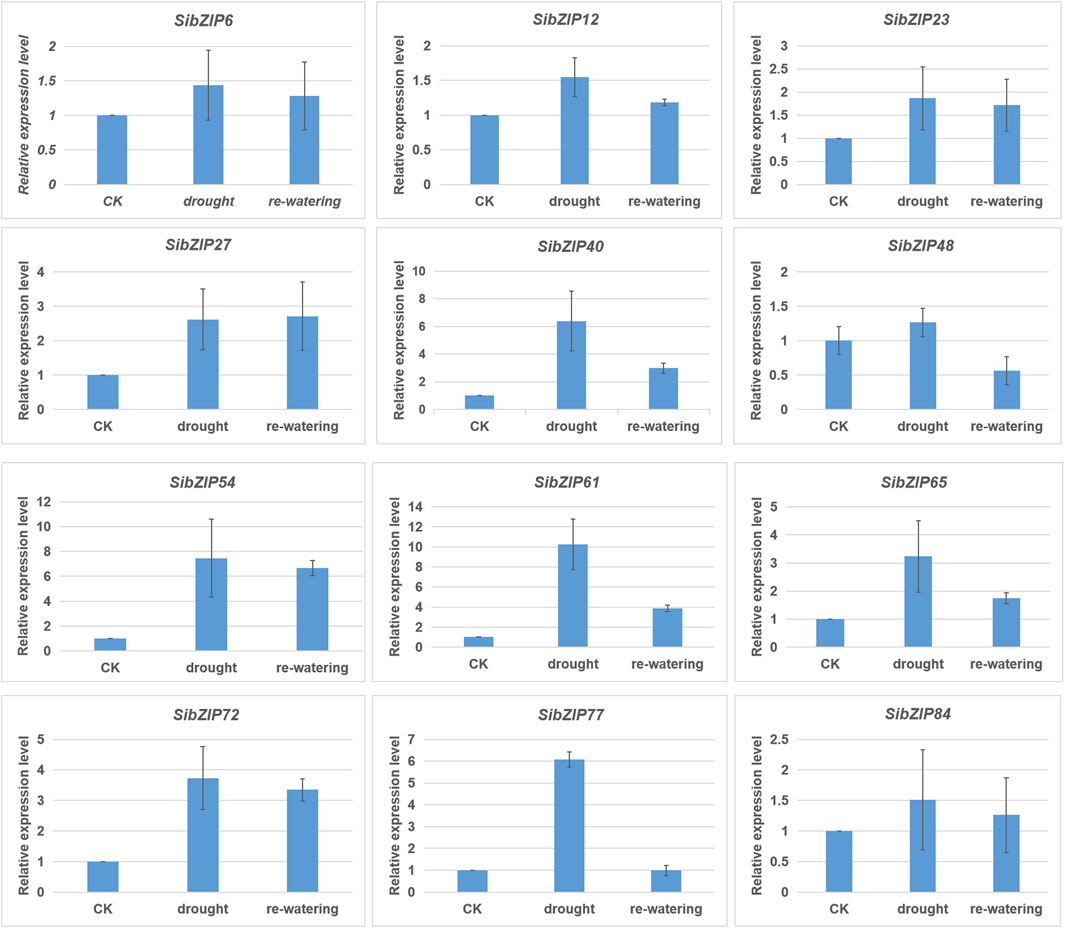
In addition, RT-qPCR were used to confirm the expression patterns of 12 genes (SibZIP6, SibZIP12, SibZIP23, SibZIP27, SibZIP40, SibZIP48, SibZIP54, SibZIP61, SibZIP65, SibZIP72, SibZIP77 and SibZIP84). The RT-qPCR profiles were shown in Figure 8. The results showed that 12 SibZIP genes were all upregulated after seedlings were subjected to drought stress, and decreased after re-watering (Figure 8). Additionally, over 5 fold-increase was observed for SibZIP40, SibZIP54, SibZIP61 and SibZIP77.
4 Discussion
S. italica is an important grain and forage crop in China, known for its excellent drought and barren soil tolerance (Muthamilarasan and Prasad 2015). In recent years, due to the impact of climate change, the cultivation of crops in arid regions has been receiving increasing attention (Zandalinas et al., 2018). Therefore, elucidation of the drought tolerance mechanisms in S. italica is of significant importance. Drought is a complex trait and regulated by many genes from different pathways, transcription factors including bZIP transcription factors actively respond to dehydration stress (Baillo et al., 2019; Joo et al., 2021).
In this study, we successfully identified 90 SibZIPs genes in S. italica using an updated S. italica genome at Phytozome database and added 17 more SibZIPs genes than previous identification of bZIP genes in S. italica (Liu et al., 2016). Phylogenetic analysis, conserved motif and gene structure analysis demonstrated that SibZIPs can be categorized into 13 distinct groups, with members of the same groups sharing similar motifs and gene structure pattern. Seventy five Arabidopsis bZIPs was first classified into 10 groups (A-I, and S) and Dröge-Laser proposed a updated classification of 13 groups (A-M, and S) (Jakoby et al., 2002; Dröge-Laser et al., 2018). The number of bZIP family members and classification in different species was diverse. For instance, 89 rice OsbZIP proteins were classified into 10 clades and 125 maize bZIPs were identified into 11 groups (Wei et al., 2012). Moreover, 98 pearl millet PgbZIPs into 12 subfamilies (Jha et al., 2024), and 86 poplar bZIP genes into 12 subfamilies (Zhao et al., 2021). Although the groups differed slightly in their the number of classification, subfamilies across different plants shared common 10 subfamilies (A-I, and S).
Gene duplication is the major force of bzip gene family expansion (Corrêa et al., 2008). We indenfied 37 pairs of segmental duplicated genes and none tandem duplicated genes in S. italica. This result indicates that segmental duplication contributed to the expansion of the S. italica bZIP gene family during evolution. Our result is generally in line with previous studies on the bZIP family in cucumber (Baloglu et al., 2014), legume (Wang et al., 2015), poplar (Zhao et al., 2021), tobacco (Li et al., 2021) Solanum tuberosum (Herath and Verchot, 2020), Perilla frutescens (Huang et al., 2024), ect. In addition, The Ka/Ks ratio of 37 pairs of segmental duplicated genes were all lower than 1, varying from 0.07 to 0.90. The ratios of most gene pairs were all less than 0.5. This result suggested that the duplicated genes was under negative selection and exhibited little functional divergence (Krishnamurthy et al., 2015).
In silico expression profiling in eight organs of S. italica revealed that seven SibZIP genes were ubiquitously expressed in all the tissues, suggesting transcriptional regulation of a broad gene set. Moreover the number of SibZIPs genes (39, 43%) exhibiting higher expression in roots was significantly more than that in other organs, which was also observed in cassava (Hu et al., 2016). In addition a number of transcriptome studies of Setaria italica in response to drought stress have generated large amount of genomic data (Yi et al., 2022; Wang et al., 2023; Zhang et al., 2022). The transcriptome data of tolerance cultivars (Ci328 and Ci409) under drought stress was used to investigate SibZIP gene expression patterns under dehydration stress in this study. Interestingly, most of SibZIPs genes showed response to drought stress in each drought tolerant cultivar, but only 27 genes showed response in two drought tolerant cultivars. In addition, 12 out of 27 genes were further investigated on their expression under drought stress using RT-qPCR and they were all upregulated after drought stress, indicating their possible function in drought tolerance. Numerous bZIP TFs have been identified as positive regulators of drought stress. For example, OsbZIP23 homologous genes of SibZIP40, are involved in ABA-dependent drought regulation (Park et al., 2015). OsbZIP72 overexpression significantly improved drought tolerance and ABA sensitivity in rice (Lu et al., 2009). Moreover, a role for GmTGA17 in the drought and salt tolerance was also suggested by the upregulation of GmTGA17 in both Arabidopsis and soybean (Li et al., 2019).
5 Conclusion
In this study, we identified and characterized bZIP TFs in S. italica using bioinformatics analysis and transcriptome sequencing data. Through a phylogenetic analysis, our genome-wide analysis revealed 90 SibZIP genes that were subsequently classified into 13 groups (with reference to Arabidopsis bZIP classification). The analysis of 20 conserved motifs and gene structure analysis supported this classification. Moreover, transcriptome data and RT-qPCR analysis revealed a number of SibZIP genes were upregulated under drought stress. This comprehensive study on S. italica bZIPs under drought stress provides useful information for further investigating the molecular mechanism of plant adaptation to drought stress.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author contributions
XY: Formal Analysis, Investigation, Writing–original draft. CG: Methodology, Validation, Writing–original draft. YH: Writing–original draft. QM: Writing–review and editing. ZeL: Writing–review and editing. JW: Writing–review and editing. ZhL: Writing–review and editing. LZ: Project administration, Writing–review and editing. DL: Funding acquisition, Project administration, Supervision, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by Natural Science Foundation of Inner Mongolia (2021ZD04, 2023QN03024), The Central Government Guiding Special Funds for the Development of Local Science and Technology (2020ZY0005), Research Program of science and technology at Universities of Inner Mongolia Autonomous Region (NJZY22342).
Acknowledgments
The authors would like to thank Shihua Guo, who provided ‘Yugu1’ seeds for our experiment.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2024.1466486/full#supplementary-material
References
Abe, M., Kobayashi, Y., Yamamoto, S., Daimon, Y., Yamaguchi, A., Ikeda, Y., et al. (2005). FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309, 1052–1056. doi:10.1126/science.1115983
Agarwal, P., Baranwal, V. K., and Khurana, P. (2019). Genome-wide analysis of bZIP transcription factors in wheat and functional characterization of a TabZIP under abiotic stress. Sci. Rep. 9, 4608. doi:10.1038/s41598-019-40659-7
Alonso, R., Oñate-Sánchez, L., Weltmeier, F., Ehlert, A., Diaz, I., Dietrich, K., et al. (2009). A pivotal role of the basic leucine zipper transcription factor bZIP53 in the regulation of Arabidopsis seed maturation gene expression based on heterodimerization and protein complex formation. Plant Cell 21, 1747–1761. doi:10.1105/tpc.108.062968
Aslam, M. M., Deng, L., Meng, J., Wang, Y., Pan, L., Niu, L., et al. (2023). Characterization and expression analysis of basic leucine zipper (bZIP) transcription factors responsive to chilling injury in peach fruit. Mol. Biol. Rep. 50, 361–376. doi:10.1007/s11033-022-08035-3
Baena-González, E., Rolland, F., Thevelein, J. M., and Sheen, J. (2007). A central integrator of transcription networks in plant stress and energy signalling. Nature 448, 938–942. doi:10.1038/nature06069
Baillo, E. H., Kimotho, R. N., Zhang, Z., and Xu, P. (2019). Transcription factors associated with abiotic and biotic stress tolerance and their potential for crops improvement. Genes (Basel) 10, 771. doi:10.3390/genes10100771
Baloglu, M. C., Eldem, V., Hajyzadeh, M., and Unver, T. (2014). Genome-wide analysis of the bZIP transcription factors in cucumber. PloS one, 9, e96014. doi:10.1371/journal.pone.0096014
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. plant, 13, 1194–1202. doi:10.1016/j.molp.2020.06.009
Dröge-Laser, W., Snoek, B. L., Snel, B., and Weiste, C. (2018). The Arabidopsis bZIP transcription factor family—an update. Curr. Opin. plant Biol. 45, 36–49. doi:10.1016/j.pbi.2018.05.001
Foster, R., Izawa, T., and Chua, N. H. (1994). Plant bZIP proteins gather at ACGT elements. FASEB J., 8, 192. 200. doi:10.1096/fasebj.8.2.8119490
Fujita, Y., Fujita, M., Satoh, R., Maruyama, K., Parvez, M. M., Seki, M., et al. (2005). AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17, 3470–3488. doi:10.1105/tpc.105.035659
Gao, H., Ge, W., Bai, L., Zhang, T., Zhao, L., Li, J., et al. (2023). Proteomic analysis of leaves and roots during drought stress and recovery in Setaria italica L. Front. Plant Sci. 14, 1240164. doi:10.3389/fpls.2023.1240164
Guo, Z., Dzinyela, R., Yang, L., and Hwarari, D. (2024). bZIP transcription factors: structure, modification, abiotic stress responses and application in plant improvement. Plants, 13, 2058. doi:10.3390/plants13152058
Gupta, A., Rico-Medina, A., and Caño-Delgado, A. I. (2020). The physiology of plant responses to drought. Science 368, 266–269. doi:10.1126/science.aaz7614
He, L., Wu, Z., Wang, X., Zhao, C., Cheng, D., Du, C., et al. (2024). A novel maize F-bZIP member, ZmbZIP76, functions as a positive regulator in ABA-mediated abiotic stress tolerance by binding to ACGT-containing elements. Plant Sci. 341, 111952. doi:10.1016/j.plantsci.2023.111952
Herath, V., and Verchot, J. (2020). Insight into the bZIP gene family in Solanum tuberosum: genome and transcriptome analysis to understand the roles of gene diversification in spatiotemporal gene expression and function. Int. J. Mol. Sci. 22, 253. doi:10.3390/ijms22010253
Hrmova, M., and Hussain, S. S. (2021). Plant transcription factors involved in drought and associated stresses. Int. J. Mol. Sci. 22, 5662. doi:10.3390/ijms22115662
Hu, W., Yang, H., Yan, Y., Wei, Y., Tie, W., Ding, Z., et al. (2016). Genome-wide characterization and analysis of bZIP transcription factor gene family related to abiotic stress in cassava. Sci. Rep. 6, 22783. doi:10.1038/srep22783
Huang, X., Zhou, Y., Shi, X., Wen, J., Sun, Y., Chen, S., et al. (2024). PfbZIP85 transcription factor mediates ω-3 fatty acid-enriched oil biosynthesis by down-regulating PfLPAT1B gene expression in plant tissues. Int. J. Mol. Sci., 25, 4375. doi:10.3390/ijms25084375
Izawa, T., Foster, R., and Chua, N. H. (1993). Plant bZIP protein DNA binding specificity. J. Mol. Biol. 230, 1131–1144. doi:10.1006/jmbi.1993.1230
Jakoby, M., Weisshaar, B., Dröge-Laser, W., Vicente-Carbajosa, J., Tiedemann, J., Kroj, T., et al. (2002). bZIP transcription factors in Arabidopsis. Trends Plant Sci. 7, 106–111. doi:10.1016/s1360-1385(01)02223-3
Jha, D. K., Chanwala, J., Barla, P., and Dey, N. (2024). Genome-wide identification of bZIP gene family in Pearl millet and transcriptional profiling under abiotic stress, phytohormonal treatments; and functional characterization of PgbZIP9. Front. Plant Sci. 15, 1352040. doi:10.3389/fpls.2024.1352040
Joo, J., Lee, Y. H., and Song, S. I. (2019). OsbZIP42 is a positive regulator of ABA signaling and confers drought tolerance to rice. Planta 249, 1521–1533. doi:10.1007/s00425-019-03104-7
Krishnamurthy, P., Hong, J. K., Kim, J. A., Jeong, M. J., Lee, Y. H., and Lee, S. I. (2015). Genome-wide analysis of the expansin gene superfamily reveals Brassica rapa-specific evolutionary dynamics upon whole genome triplication. Mol. Gen. Genomics. 290, 521-30. doi:10.1007/s00438-014-0935-0
Li, B., Liu, Y., Cui, X. Y., Fu, J. D., Zhou, Y. B., Zheng, W. J., et al. (2019). Genome-wide characterization and expression analysis of soybean TGA transcription factors identified a novel TGA gene involved in drought and salt tolerance. Front. Plant Sci. 10, 549. doi:10.3389/fpls.2019.00549
Li, Z., Chao, J., Li, X., Li, G., Song, D., Guo, Y., et al. (2021). Systematic analysis of the bZIP family in tobacco and functional characterization of NtbZIP62 involvement in salt stress. Agronomy 11, 148. doi:10.3390/agronomy11010148
Liu, B., Zhang, L., Sun, Y., Xue, J., Gao, C., Yuan, L., et al. (2016). Genome-wide characterization of bZIP transcription factors in foxtail millet and their expression profiles in response to drought and salt stresses. Chin. Bull. Bot. 51, 473. doi:10.11983/CBB15148
Lu, G., Gao, C., Zheng, X., and Han, B. (2009). Identification of OsbZIP72 as a positive regulator of ABA response and drought tolerance in rice. Planta 229, 605–615. doi:10.1007/s00425-008-0857-3
Nijhawan, A., Jain, M., Tyagi, A. K., and Khurana, J. P. (2008). Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol. 146, 333–350. doi:10.1104/pp.107.112821
Pan, J., Li, Z., Wang, Q., Garrell, A. K., Liu, M., Guan, Y., et al. (2018). Comparative proteomic investigation of drought responses in foxtail millet. BMC Plant Biol. 18, 315–319. doi:10.1186/s12870-018-1533-9
Park, S. H., Jeong, J. S., Lee, K. H., Kim, Y. S., Do Choi, Y., et al. (2015). OsbZIP23 and OsbZIP45, members of the rice basic leucine zipper transcription factor family, are involved in drought tolerance. Plant Biotechnol. Rep. 9, 89–96. doi:10.1007/s11816-015-0346-7
Pourabed, E., Ghane Golmohamadi, F., Soleymani Monfared, P., Razavi, S. M., and Shobbar, Z. S. (2015). Basic leucine zipper family in barley: genome-wide characterization of members and expression analysis. Mol. Biotechnol. 57, 12–26. doi:10.1007/s12033-014-9797-2
Tang, N., Zhang, H., Li, X., Xiao, J., and Xiong, L. (2012). Constitutive activation of transcription factor OsbZIP46 improves drought tolerance in rice. Plant Physiol. 158, 1755–1768. doi:10.1104/pp.111.190389
Uno, Y., Furihata, T., Abe, H., Yoshida, R., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2000). Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. PNAS 97, 11632–11637. doi:10.1073/pnas.190309197
Wang, J., Sun, Z., Wang, X., Tang, Y., Li, X., Ren, C., et al. (2023). Transcriptome-based analysis of key pathways relating to yield formation stage of foxtail millet under different drought stress conditions. Front. Plant Sci., 13, 1110910. doi:10.3389/fpls.2022.1110910
Wang, J., Zhou, J., Zhang, B., Vanitha, J., Ramachandran, S., and Jiang, S. Y. (2011). Genome-wide expansion and expression divergence of the basic leucine zipper transcription factors in higher plants with an emphasis on sorghum. J. Integr. Plant Biol. 53, 212–231. doi:10.1111/j.1744-7909.2010.01017.x
Wang, Z., Cheng, K., Wan, L., Yan, L., Jiang, H., Liu, S., et al. (2015). Genome-wide analysis of the basic leucine zipper (bZIP) transcription factor gene family in six legume genomes. BMC genomics 16, 1053. doi:10.1186/s12864-015-2258-x
Wang, Z., Yan, L., Wan, L., Huai, D., Kang, Y., Shi, L., et al. (2019). Genome-wide systematic characterization of bZIP transcription factors and their expression profiles during seed development and in response to salt stress in peanut. BMC Genom. 20, 51. doi:10.1186/s12864-019-5434-6
Wei, K., Chen, J., Wang, Y., Chen, Y., Chen, S., Lin, Y., et al. (2012). Genome-wide analysis of bZIP-encoding genes in maize. DNA Res. 19, 463–476. doi:10.1093/dnares/dss026
Yang, Y., Yu, T. F., Ma, J., Chen, J., Zhou, Y. B., et al. (2020).The soybean bZIP transcription factor gene GmbZIP2 confers drought and salt resistances in transgenic plants. Int. J. Mol. Sci. 21, 670. doi:10.3390/ijms21020670
Yi, F., Huo, M., Li, J., and Yu, J. (2022). Time-series transcriptomics reveals a drought-responsive temporal network and crosstalk between drought stress and the circadian clock in foxtail millet. Plant J. 110, 1213–1228. doi:10.1111/tpj.15725
Yoshida, T., Fujita, Y., Maruyama, K., Mogami, J., Todaka, D., Shinozaki, K., et al. (2014). Four Arabidopsis AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic acid signalling in response to osmotic stress. Plant Cell Environ. 38, 35–49. doi:10.1111/pce.12351
Yoshida, T., Fujita, Y., Sayama, H., Kidokoro, S., Maruyama, K., Mizoi, J., et al. (2010). AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 61, 672–685. doi:10.1111/j.1365-313X.2009.04092.x
Zandalinas, S. I., Mittler, R., Balfagón, D., Arbona, V., and Gómez-Cadenas, A. (2018). Plant adaptations to the combination of drought and high temperatures. Physiol. Plant. 162, 2–12. doi:10.1111/ppl.12540
Zhang, A., Zhang, L., Guo, E., Wang, R., Li, Q., Guo, S., et al. (2023). Setaria italica SiWRKY89 enhances drought tolerance in Arabidopsis. Plant Growth Regul. 9, 125–135. doi:10.1007/s10725-022-00916-8
Zhang, M., Liu, Y., Shi, H., Guo, M., Chai, M., He, Q., et al. (2018). Evolutionary and expression analyses of soybean basic Leucine zipper transcription factor family. BMC Genom. 19, 159. doi:10.1186/s12864-018-4511-6
Zhang, R., Zhi, H., Li, Y., Guo, E., Feng, G., Tang, S., et al. (2022). Response of multiple tissues to drought revealed by a weighted gene co-expression network analysis in foxtail millet [Setaria italica (L.) P. Beauv.]. Front. Plant Sci. 12, 746166. doi:10.3389/fpls.2021.746166
Zhao, K., Chen, S., Yao, W., Cheng, Z., Zhou, B., and Jiang, T. (2021). Genome-wide analysis and expression profile of the bZIP gene family in poplar. BMC Plant Biol. 21, 122. doi:10.1186/s12870-021-02879-w
Keywords: foxtail millet, bZIP transcription factor, dehydration stress, gene expression, transcriptome
Citation: Yang X, Gao C, Hu Y, Ma Q, Li Z, Wang J, Li Z, Zhang L and Li D (2024) Identification and expression analysis of bZIP transcription factors in Setaria italica in response to dehydration stress. Front. Genet. 15:1466486. doi: 10.3389/fgene.2024.1466486
Received: 18 July 2024; Accepted: 19 August 2024;
Published: 30 August 2024.
Edited by:
Jianbo Li, The University of Sydney, AustraliaReviewed by:
Cheng Liu, Shandong Academy of Agricultural Sciences, ChinaLinyi Qiao, Shanxi Agricultural University, China
Copyright © 2024 Yang, Gao, Hu, Ma, Li, Wang, Li, Zhang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dongming Li, bGlkb25nbWluZzAxMThAMTYzLmNvbQ==; Li Zhang, emhhbmdsaTc5MTJAMTYzLmNvbQ==
†These authors have contributed equally to this work
 Xuefei Yang
Xuefei Yang Changyong Gao2†
Changyong Gao2† Li Zhang
Li Zhang