- 1Center for Clinical Molecular Medicine, National Clinical Research Center for Child Health and Disorders, Ministry of Education Key Laboratory of Child Development and Disorders, China International Science and Technology Cooperation Base of Child Development and Critical Disorders, Chongqing Key Laboratory of Pediatrics, Children’s Hospital of Chongqing Medical University, Chongqing, China
- 2Department of Neonatology, National Clinical Research Center for Child Health and Disorders, Ministry of Education Key Laboratory of Child Development and Disorders, Chongqing Key Laboratory of Pediatrics, Children’s Hospital of Chongqing Medical University, Chongqing, China
A Corrigendum on
Disease spectrum, prevalence, genetic characteristics of inborn errors of metabolism in 21,840 hospitalized infants in Chongqing, China, 2017–2022
by Wang D, Zhang J, Yang R, Zhang D, Wang M, Yu C, Yang J, Huang W, Liu S, Tang S and He X (2024). Front. Genet. 15:1395988. doi: 10.3389/fgene.2024.1395988
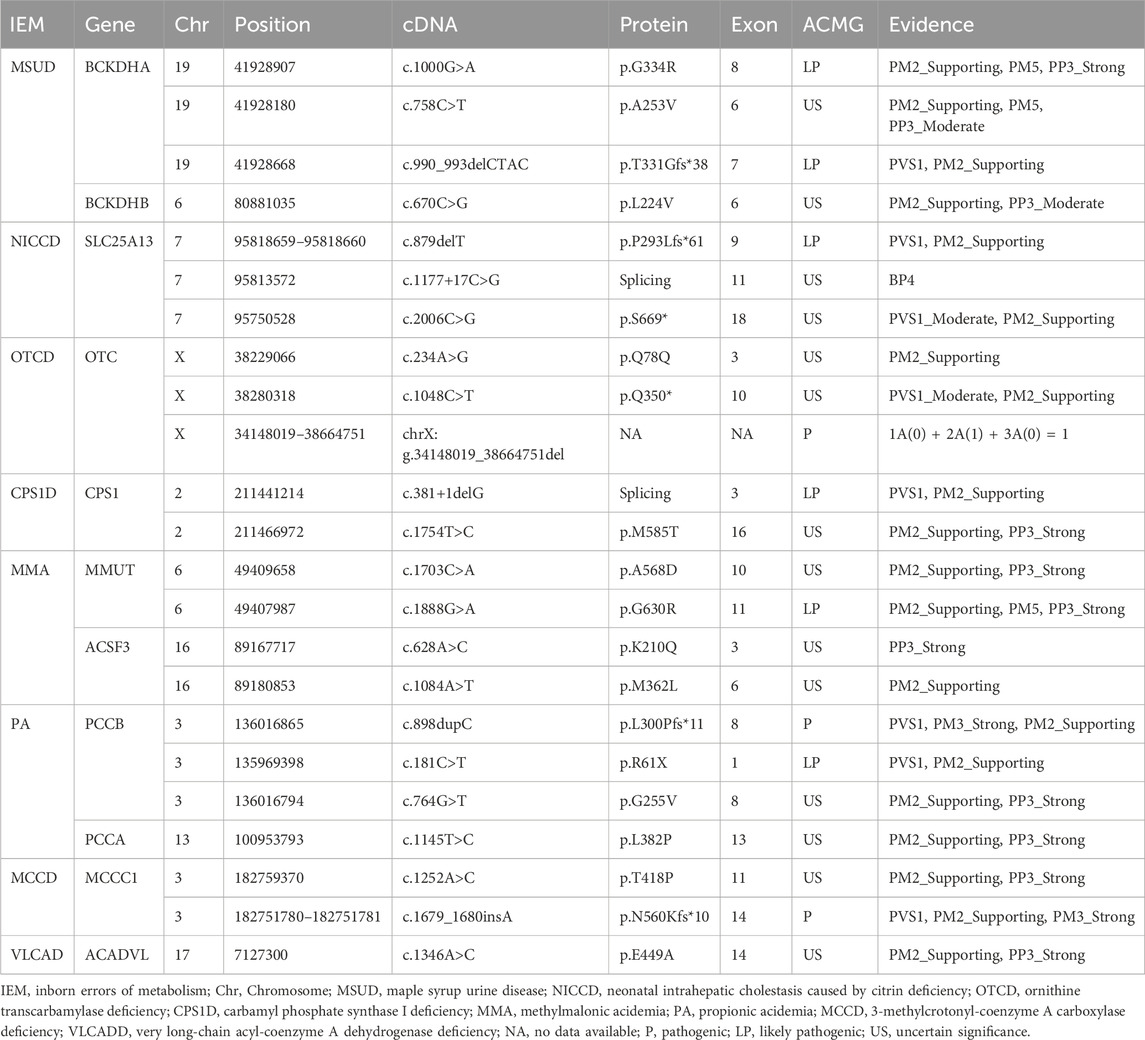
In the published article, there were four errors in Table 3 as published.
(1) For BCKDHA gene variant c.990_993delCTAC, the “ACMG” classification was erroneously listed as “US,” and it should be corrected to “LP”; the “Evidence” for the variant was incorrectly stated as “PM2_Supporting,” and it should be updated to “PVS1, PM2_Supporting.”
(2) For CPS1 gene variant c.381+1delG, the “ACMG” classification was erroneously listed as “US,” and it should be corrected to “LP”; the “Evidence” for the variant was incorrectly stated as “PM2_Supporting,” and it should be updated to “PVS1, PM2_Supporting.”
(3) For OTC gene variant chrX:g.34148019_38664751del, the “Evidence” was incorrectly stated as “1A(0) + 2A(1) + 3A(0) + 4L(0.45) = 1.45,” and it should be updated to “1A(0) + 2A(1) + 3A(0) = 1.”
(4) For PCCB gene variant c.898dupC, the “ACMG” classification was erroneously listed as “US,” and it should be corrected to “P”; the “Evidence” for the variant was incorrectly stated as “PM3_Strong, PM2_Supporting,” and it should be updated to “PVS1, PM3_Strong, PM2_Supporting.”
The correct Table 3 and its caption appear below.
In the published article, there were two errors. The text in the Results and Discussion sections referencing Table 3 requires updates to align with the corrected information of Table 3.
1. A correction has been made to Results Section, Gene detection in patients with inherited metabolic disorders, Paragraph 3.
The sentences previously stated:
“The pathogenicity of the 23 previously unreported variants mentioned above was analyzed using the ACMG rating system. Two mutations [a large 4.52 Mb hemizygous deletion containing the OTC gene (seq[hg19]del(X)(p21.1p11.4)chrX:g.34148019_38664751del) and c.1679_c.1680insA in the MCCC1 gene] were identified as pathogenic and 4 mutations (c.1000G>A in the BCKDHA gene, c.879delT in the SLC25A13 gene, c.1888G>A in the MMUT gene, and c.181C>T in the PCCB gene) were identified as likely pathogenic. The remainder were of uncertain significance, of which four were reported with different nucleotide changes at the same position, three variants were indexed in the Clinvar database but lacked relevant literature, and the remaining ten unreported variants of unknown significance included two frame-shift mutations, two termination mutations, two shear mutations and one intronic variant. The data were shown in Table 3.”
The corrected sentence appears below:
“The pathogenicity of the 23 previously unreported variants mentioned above was analyzed using the ACMG rating system. Three mutations [a large 4.52 Mb hemizygous deletion containing the OTC gene (seq[hg19]del(X)(p21.1p11.4)chrX:g.34148019_38664751del), c.898dupC in the PCCB gene and c.1679_c.1680insA in the MCCC1 gene] were identified as pathogenic and 6 mutations (c.1000G>A and c.990_993del CTAC in the BCKDHA gene, c.879delT in the SLC25A13 gene, c.381+1delG in the CPS1 gene, c.1888G>A in the MMUT gene, and c.181C>T in the PCCB gene) were identified as likely pathogenic. The remainder were of uncertain significance, of which four were reported with different amino acid substitutions at the same position, three variants were indexed in the Clinvar database lacked relevant literature, and the remaining seven unreported variants of unknown significance included a synonymous mutation, two termination mutations, three missense mutations and one intronic variant. The data were shown in Table 3.”
2. A correction has been made to Discussion Section, Paragraph 9.
The sentences previously stated:
“In addition, we analyzed 23 previously unreported genetic variants and evaluated their pathogenicity using the ACMG rating system. Among these, 2 variants were classified as pathogenic: a large 4.52 Mb hemizygous deletion containing the OTC gene (seq[hg19]del(X)(p21.1p11.4)chrX:g.34148019_38664751del) and the c.1679_c.1680 insA mutation in the MCCC1 gene. Furthermore, 4 variants were rated as likely pathogenic: c.1000G>A in the BCKDHA gene, c.879delT in the SLC25A13 gene, c.1888G>A in the MMUT gene, and c.181C>T in the PCCB gene. The remaining 17 unreported variants were categorized as uncertain significance by the ACMG system.”
The corrected sentence appears below:
“In addition, we analyzed 23 previously unreported genetic variants and evaluated their pathogenicity using the ACMG rating system. Among these, 3 variants were classified as pathogenic: a large 4.52 Mb hemizygous deletion containing the OTC gene (seq[hg19]del(X) (p21.1p11.4)chrX:g.34148019_38664751del), c.898dupC in the PCCB gene and the c.1679_c.1680insA mutation in the MCCC1 gene. Furthermore, 6 variants were rated as likely pathogenic: c.1000G>A and c.990_993delCTAC in the BCKDHA gene, c.879delT in the SLC25A13 gene, c.381+1delG in the CPS1D, c.1888G>A in the MMUT gene, and c.181C>T in the PCCB gene. The remaining 14 unreported variants were categorized as uncertain significance by the ACMG system.”
The authors apologize for these errors and state that these do not change the scientific conclusions of the article in any way. The original article has been updated.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Keywords: inborn errors of metabolism, newborn screening, disease spectrum, genetic characteristics, tandem mass spectrometry
Citation: Wang D, Zhang J, Yang R, Zhang D, Wang M, Yu C, Yang J, Huang W, Liu S, Tang S and He X (2024) Corrigendum: Disease spectrum, prevalence, genetic characteristics of inborn errors of metabolism in 21,840 hospitalized infants in Chongqing, China, 2017–2022. Front. Genet. 15:1449534. doi: 10.3389/fgene.2024.1449534
Received: 15 June 2024; Accepted: 24 June 2024;
Published: 10 July 2024.
Edited and reviewed by:
Madelyn Gillentine, Seattle Children’s Hospital, United StatesCopyright © 2024 Wang, Zhang, Yang, Zhang, Wang, Yu, Yang, Huang, Liu, Tang and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoyan He, aGV4aWFveWFuQGhvc3BpdGFsLmNxbXUuZWR1LmNu
 Dongjuan Wang
Dongjuan Wang Juan Zhang1
Juan Zhang1