- 1Department of Biology, University of Maryland, College Park, MD, United States
- 2Institute of Biology, University of Graz, Graz, Austria
Advances in genome sequencing have greatly accelerated the identification of sex chromosomes in a variety of species. Many of these species have experienced structural rearrangements that reduce recombination between the sex chromosomes, allowing the accumulation of sequence differences over many megabases. Identification of the genes that are responsible for sex determination within these sometimes large regions has proved difficult. Here, we identify an XY sex chromosome system on LG19 in the West African cichlid fish Chromidotilapia guntheri in which the region of differentiation extends over less than 400 kb. We develop high-quality male and female genome assemblies for this species, which confirm the absence of structural variants, and which facilitate the annotation of genes in the region. The peak of differentiation lies within rin3, which has experienced several debilitating mutations on the Y chromosome. We suggest two hypotheses about how these mutations might disrupt endocytosis, leading to Mendelian effects on sexual development.
1 Introduction
Three classes of genes are frequently identified in studies of sex-determination in teleosts (Kitano et al., 2024). The first class of genes are elements of the TGFß pathway, which transduces extracellular signals to the nucleus. A duplication of the ligand amh is a sex determiner in Nile tilapia (Oreochromis niloticus) (Li et al., 2015) and Northern pike (Esox lucius) (Pan et al., 2019). Variations of the amh receptor are implicated in sex determination in several orders, including Cichliformes, Siluriformes, and Tetraodontiformes (Kamiya et al., 2012; Nacif et al., 2023; Wen et al., 2023). Polymorphisms of the ligand gsdf are associated with sex determination in the Philippine medaka (Oryzias luzonensis) (Myosho et al., 2012) and in cichlids from the Malawi basin (Munby et al., 2021). A recent study associated polymorphisms in bone morphogenetic protein receptor type 1b (bmprt1) with sex determination in Atlantic herring (Clupea harengus) (Rafati et al., 2020). The second class of genes are involved in steroid metabolism. Variation in steroid modification enzymes contribute to sex determination, including hsd17b1 in amberjacks (Seriola spp.) (Koyama et al., 2019) and sult1st6y in bluefin tunas (Thunnus maccoyii and T. orientalis) (Nakamura et al., 2021). The third class of genes include transcription factors such as dmrt and sox. Dmrt has taken on a role in sex determination in the Japanese ricefish (Oryzia curvinotus and O. latipes) (Matsuda et al., 2003) and a flatfish (Cynoglossus semilaevis) (Chen et al., 2014). Sox2 is implicated in sex determination in another flatfish (Scophthalmus maximus) (Martínez et al., 2021) and sox3 is associated with sex in the Indian ricefish (Oryzias dancena) (Takehana et al., 2014).
While most Mendelian factors associated with sex determination are part of these well-studied pathways, there are exceptions. A surprising role for an immune gene (irf9/sdY) was identified in salmonids (Yano et al., 2012; Yano et al., 2013). In Solea senegalensis, a species of flatfish, an allele of the receptor of the follicle stimulating hormone (fshr) inhibits the action of the follicle stimulating hormone tipping the gonad into testis (de la Herrán et al., 2023). Finally, in the Atlantic cod (Gadus morhua), a copy of the zinc knuckle gene (zkY) on the Y chromosomes leads to male development (Kirubakaran et al., 2019). So, while some genes have been evolved to become Mendelian factors in sex determination convergently in multiple lineages, this does not preclude the evolution of sex-determiners from other types of genes. Sex-determiners have been mapped in many other species, but the sex-linked regions do not appear to contain candidate genes in any of the three gene classes already identified. The true number of genes capable of becoming sex determiners is still unknown, and it seems likely that more genes will be implicated as we investigate sex determination in more species (Tao et al., 2021b; Behrens et al., 2024a).
There are multiple methodologies for identifying sex chromosomes, the effectiveness of which are dependent on properties of the sex chromosomes themselves (Palmer et al., 2019). Highly divergent sex chromosomes such as those of mammals (Hughes and Page, 2015) or birds (Zhou et al., 2014) are often visibly different when karyotyped and can be detected by looking for differences in DNA sequence coverage between the sexes (Vicoso et al., 2013; Darolti et al., 2019). Detection of homomorphic sex chromosomes, which are not detectable by karyotyping, is much more challenging. Many fish, including cichlids, have homomorphic sex chromosomes (Kitano and Peichel, 2012), which can be identified by examining the distribution of sex-specific SNPs along the chromosomes (Gammerdinger et al., 2018; Behrens et al., 2024a).
Characterization of the sex-determining genes on these sex chromosomes is critical to understanding the developmental mechanisms of sex determination. However, identification of a causative gene or variant within the sex-determining region has been challenging, especially when they are contained within structural variants that sequester differentiation across large genomic regions encompassing dozens to hundreds of genes. An example of this is the discovery of the Y-linked figla-like gene, proposed to have a role in sex determination in Nile tilapia, that is not present in the current assembly of an XX O. niloticus genome, but which was identified in an assembly of the Oreochromis aureus genome (Curzon et al., 2022). Another challenge in identifying candidate genes is the distance of the closest quality reference genome from the organism of interest (Darolti et al., 2022). Distantly related species may have accumulated structural differences, such as inversions, gene insertions, or gene duplications that can obscure the gene content of the sex-determining region.
Sex chromosomes often feature highly repetitive or degenerated regions that are difficult to assemble. To avoid this complication, assemblies were often generated only for the homogametic sex (Rhie et al., 2021; Carey et al., 2022). The advancement and reduction in cost of long read sequencing technologies such as PacBio HiFi and Oxford Nanopore now allow us to assemble sex chromosomes from long reads that can span these repetitive regions. An additional challenge with sex chromosomes is that while assemblers like hifiasm (Cheng et al., 2021) are able to phase autosomes, they may struggle to determine haplotypes in sex chromosomes due to potential changes in structure, gene content, and repeat content (Carey et al., 2022). Despite this, assembly of sex chromosomes is becoming increasingly successful. There were several early attempts to assemble the human Y chromosome from telomere to telomere (Skaletsky et al., 2003; Jain et al., 2018), but a complete assembly was only accomplished recently (Rhie et al., 2023). Relatively complete assemblies of the sex chromosomes have been completed for several fish, including the stickleback (Peichel et al., 2020), the zig-zag eel (Xue et al., 2021), and the spotted knifejaw (Li et al., 2021).
Cichlid fishes (Cichlidae) are a large and incredibly diverse group, featuring upwards of 1,500 species in Africa alone, which makes them an ideal group for studying the mechanisms of speciation, morphological divergence, and other evolutionary processes (Burress, 2015; Santos et al., 2023). They have proven to be a good system in which to study the evolution of sex chromosomes (El Taher et al., 2021). The development of long read sequencing technologies has allowed for several high quality cichlid assemblies, including O. niloticus and Metriaclima (Maylandia) zebra (Conte and Kocher, 2015; Conte et al., 2019), and O. aureus and O. mossambicus (Tao et al., 2021c).
A great diversity of sex determination systems have been characterized in East African cichlids (Gammerdinger and Kocher, 2018; Behrens et al., 2024b). Chromidotilapia guntheri is a paternally mouthbrooding West African cichlid. It is a member of the tribe Chromidotilapiini, a species-rich early-branching lineage within the African cichlid tree (Greenwood, 1987; Schwarzer et al., 2015; Astudillo-Clavijo et al., 2023). This places it as an outgroup to all African cichlid species that have previously been investigated for sex chromosomes. The closest relatives of C. guntheri that have been studied for sex chromosomes are members of the tribe Oreochromini. Oreochromis niloticus has an XY-LG23 system in some strains (Conte et al., 2017), and an XY-LG1 system in a Japanese strain (Tao et al., 2021c). O. aureus has a ZW-LG3 system (Tao et al., 2021c), and O. mossambicus segregates both XY-LG14 and a ZW-LG3 systems (Tao et al., 2021a). The enormity of the cichlid radiation means genome assemblies for many individual clades may be needed to understand the evolution of sex chromosomes.
Karyotype work shows C. guntheri has a diploid chromosomal number of 48 (Ozouf-Costaz et al., 2017). The only other known karyotype for the Chromidotilapiini is Pelvicachromis pulcher, a species of cichlid with environmental sex determination driven by temperature and water pH (Reddon and Hurd, 2013; Renn and Hurd, 2021), which also has a diploid chromosomal number of 48 (Post, 1965). This diploid number is greater than the modal number for cichlids, which is 2n = 44 (Feldberg et al., 2003; Poletto et al., 2010), but is close to the ancestral number for teleost fish (2n = 50) (Nakatani et al., 2007). The reduction from 50 to 48 is proposed to result from a fusion of two acrocentric chromosomes, likely forming LG7 (Liu et al., 2013; Ozouf-Costaz et al., 2017; Conte et al., 2019) and creating what was likely the ancestral karyotype for most cichlids (2n = 48) (Poletto et al., 2010). LG23 was also formed by a fusion (Conte et al., 2017; Conte et al., 2019) however this occurred only in the African cichlid lineage.
The goals of this study were to 1) look for evidence of genetic sex determination in C. guntheri, 2) map the sex determining loci, and 3) characterize the genes within these regions that might affect sexual development.
2 Materials and methods
2.1 Pooled-sequencing
The Chromidotilapia guntheri used were full siblings reared at the University of Graz from aquarium stock. We sampled tissue from 23 males and 26 females, and individuals were dissected to verify sex by visual inspection of gonad morphology. Animal care and use were approved under animal care protocols BMWFW-66.007/004-WF/V/3b/2016 (University of Graz) and R-OCT-22-46 (University of Maryland) and this study was carried out with the approval of the ethics committee of the University of Graz (permit number GZ. 39/115/63 ex 2022/23). DNA was purified separately for each individual using phenol:chloroform extraction in phase-lock silica gel tubes (Quantabio, Beverly MA, United States). The DNA from each individual was quantified by Picogreen fluorescent assay (ThermoFisher Scientific, Waltham, Massachusetts, United States) and then equimolar pools were constructed for males and for females. Sequencing libraries were constructed for 150bp paired-end DNA sequencing on a NovaSeq6000 S4 (Illumina, San Diego CA, United States) by Maryland Genomics (Institute for Genome Sciences, Baltimore MD, United States).
2.2 Sex-specific SNP analysis
The main basis of our analyses is the identification and analysis of sex-specific SNPs. These SNPs were identified following our methods described previously (Behrens et al., 2022) using the sex-SNP-finder pipeline (Gammerdinger et al., 2018). Previously reported code from that study is available (https://github.com/Gammerdinger/sex-SNP-finder). Briefly, the sequence reads were aligned with BWA version 0.7.12 using the default parameters along with read group labels. We initially aligned all samples to the closest high-quality reference assembly, O. niloticus UMD_NMBU (RefSeq GCF_001858045.2), which was the best assembly available at the time of our initial analysis. At each variable nucleotide site, we calculated the FST statistic between the populations of male and female sequence reads. The resulting FST plots provide a first indication of the differentiation between male and female genomes. We further identified XY- and ZW- patterned SNPs as SNPs that were fixed (frequency less than 0.1) in one sex and polymorphic (frequency between 0.3 and 0.7) in the other sex. Separate plots of the allele frequency of XY- and ZW- patterned SNPs suggested the type of heterogametic system segregating (XY or ZW). This process was repeated against the new C. guntheri assembly (this study, JBDKXC000000000).
We used Bedtools make windows and coverage to calculate the density of sex-patterned SNPs in 100kbp windows across the genome. We identified the top 1% of windows (∼78 of 7,800 anchored windows) with the highest number of sex-patterned SNPs using the methodology described in Kocher et al. (2022). The log2(XY:ZW) ratio of SNP density was then calculated for each window. A Kruskal–Wallis (KW) test on the ranked data was conducted in R (v.2023.03.0+386) using kruskal.test from the stats package to determine if the log ratio differed among chromosomes. If the differences were statistically significant, the Dunn’s test from the rstatix R package was conducted post-hoc to determine which chromosomes differed significantly from one another with Benjamini–Hochberg correction for multiple tests. Regions of elevated sex-specific SNP density were visualized in IGV to identify candidate sex determining genes.
2.3 Genome sequencing
Additional C. guntheri were obtained from the aquarium trade and maintained in the Tropical Aquaculture Facility at the University of Maryland. Animal use was approved under the animal care protocol R-OCT-22-46 (U. Maryland) and all experiments were conducted in accordance with the Guide for Care and Use of Laboratory Animals. High-molecular weight DNA was prepared from a single male and a single female individual, which were sexed by visual inspection of gonad morphology. DNA was extracted from heart tissue using the Circulomics/PacBio Nanobind Tissue RT kit and the short-read elimination >25 kb enrichment kit was used to remove shorter reads from the sample. DNA concentrations were quantified by fluorescence spectroscopy using a Quant-iT PicoGreen assay (ThermoFisher, Waltham MA, United States). At the Maryland Genomics center samples were size selected on a BluePippin pulse-field gel system (Sage Science, Beverly MA, United States), sequencing libraries were constructed and extra-long PacBio HiFi sequencing was conducted on the PacBio Revio machine (Pacific Bioscience, Menlo Park CA, United States).
2.4 Genome assembly
Genome assemblies were constructed using hifiasm (Cheng et al., 2021; Wang, 2022) with the default purge_dups setting on. Bandage (Wick et al., 2015) was then used to make supported merges based on contiguity of contigs. Contigs for each sex were then aligned against the O. aureus (ZZ) assembly (GCF_013358895.1) (Tao et al., 2021c), which was the most complete reference available at the time of this analysis, using D-Genies (Cabanettes and Klopp, 2018) to determine placement of contigs (Supplementary Figure S1). This initial alignment was used to inform preliminary manual merges between contigs, where a string of 50 N were used between two contigs to indicate a gap. When necessary, contigs were reverse complemented with seqtk (https://github.com/lh3/seqtk). After this initial assignment of contigs, the male and female assembly were aligned against each other and visualized in D-Genies to further guide joins and identify false joins. The Telomere Identification Toolkit (Tidk, v0.2.31) (https://github.com/tolkit/telomeric-identifier) was used to detect repeats with a pattern consistent with a telomeric identity. Once the repeat was detected (AACCCT and its reverse complement), this was used to guide correct orientation of contigs when scaffolding. Potential false joins were visually inspected in IGV (Thorvaldsdóttir et al., 2013) to determine if the contig produced by hifiasm was supported by the reads.
To identify the Y chromosome, the male haplotigs, the female contig for LG19, and the male and female PacBio HiFi reads were aligned. These alignments were used to identify regions of elevated heterozygosity in the male that were not present in the female. The haplotig with greatest number of male-specific SNPs in this region was inferred to be the Y chromosome. The X and Y were then compared using ModDotPlot (Sweeten et al., 2024).
2.5 Quality assessment
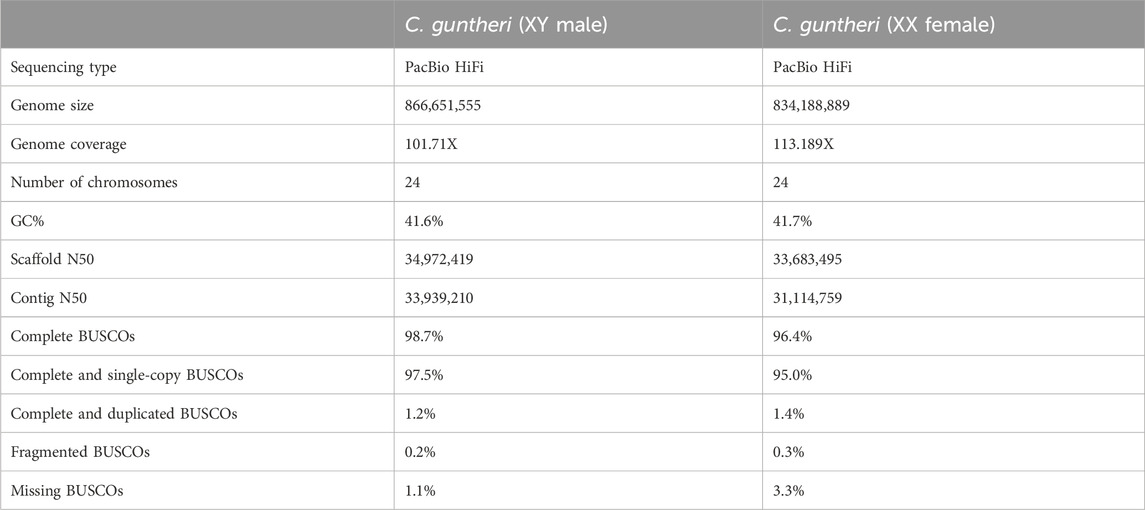
Genome assembly quality was assessed using the Genome Evaluation Pipeline (GEP) (https://git.imp.fu-berlin.de/cmazzoni/GEP), which contains the following programs: GenomeScope2 (https://github.com/tbenavi1/genomescope2.0), meryl (Miller et al., 2008; Koren et al., 2017), merqury (Rhie et al., 2020), BUSCOv5 (Manni et al., 2021), and a modified version of assembly_stats (v0.1.4) (http://doi.org/10.5281/zenodo.3968775). The O. aureus assembly (Tao et al., 2021c) was used as a guide for expected chromosome size. Coverage was calculated using samtools depth (Danecek et al., 2021).
2.6 Annotation of the sex-determining region
The sex-determining region on the new C. guntheri male assembly was annotated by aligning genes from the O. niloticus and O. aureus assemblies in that region against the new assembly using minimap2 (Li, 2018). Once oriented, we aligned the PacBio reads as well as the pooled sequencing reads for both sexes against the haplotigs identified as the Y. The genes of interest were extracted from the assembly fasta using samtools faidx and the region coordinates. This subset fasta was then aligned and annotated using the O. aureus annotation of rin3 in the EMSEMBL tool genewise (Madeira et al., 2022). The protein sequence called by genewise was then used in a multisequence alignment using ClustalOmega (Sievers et al., 2011; Madeira et al., 2022) with rin3 from both sexes of C. guntheri, and the annotations of rin3 on NCBI of for the outgroup species O. aureus, Haplochromis (Astatotilapia) burtoni, Simochromis diagramma, and Neolamprologus brichardi. This analysis was also conducted for the other genes in the region: adi1, tedc1, pth2, and dnal1. If the protein had multiple isoforms for a species, all potential isoforms were included in the multisequence alignment.
3 Results
3.1 Sex chromosome discovery
The analysis of sex-specific SNPs using the O. niloticus genome as a reference identified signal on LG19 in C. guntheri (Figure 1A). When the statistical methods were conducted based on this alignment, LG3 showed significant signal in the Dunn’s test. In the top 1% analysis, LG19 had 7 XY windows and 2 ZW windows and the top 100 kb window (124 XY SNPs) was on LG19 (Supplementary Table S1). The highest non-LG19 window was on LG1 with 59 XY SNPs. LG3 had 6 XY windows and 7 ZW windows, and the highest window contained 35 XY SNPs. The narrow signal on LG19, which against O. niloticus spans approximately 400 kb, was small enough that identifying a candidate sex-determining gene appeared feasible. Thus, we proceeded with genome assemblies of each sex.
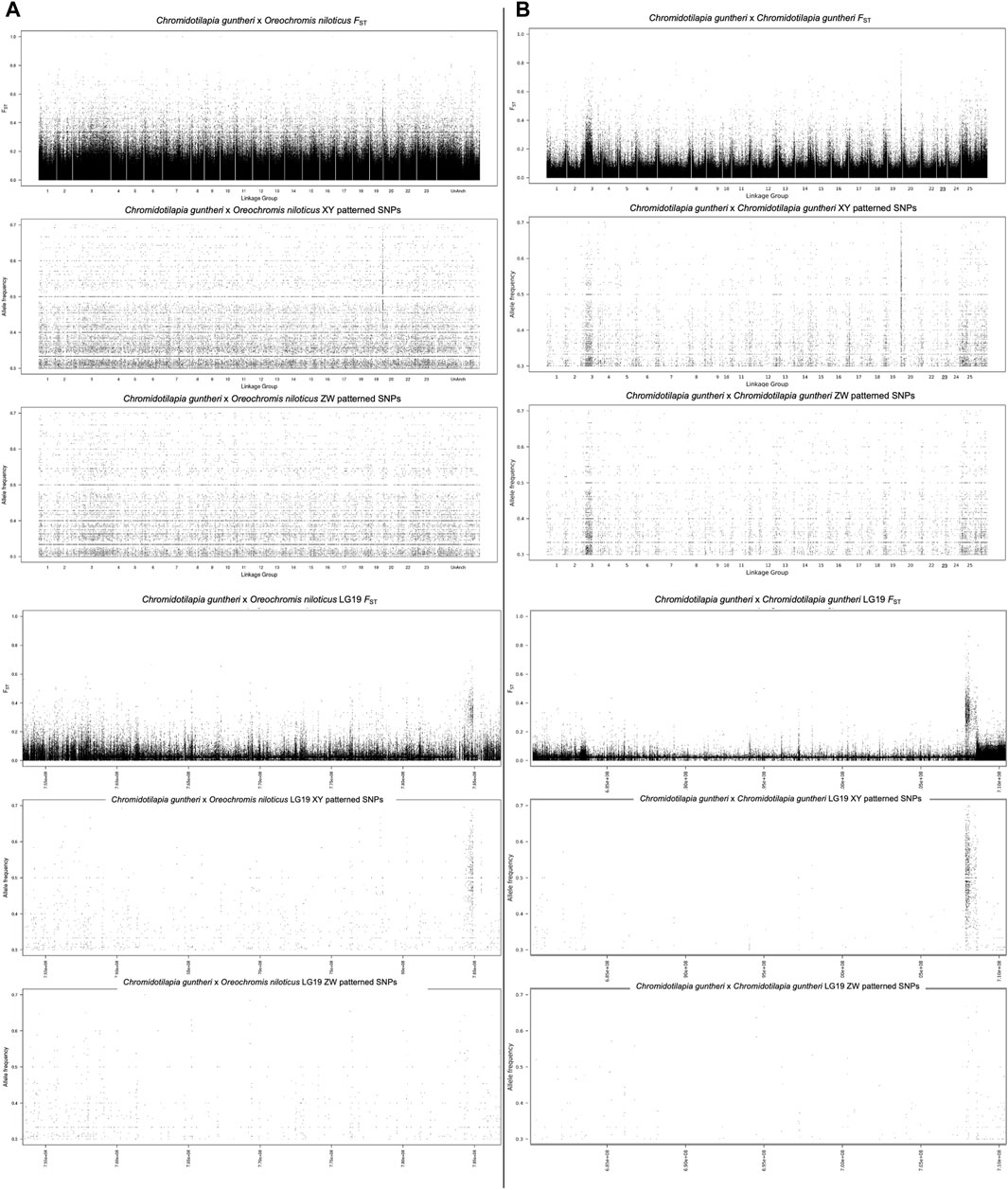
Figure 1. (A) Alignment of pooled-seq reads against O. niloticus (B) alignment of pooled-seq reads against C. guntheri.
3.2 Genome assemblies
We generated high quality genomes for each sex of C. guntheri. Summarized in Table 1, these genomes are on par with or better in quality than other recent cichlid genomes (Tao et al., 2021c). The contig N50 is high, indicating that hifiasm assembled chromosome length contigs for most of the chromosomes (Table 1). The metric for log-scaled probability of error for the consensus base calls, consensus quality (QV), was 65.86 in the male and 65.36 in the female, indicating a very accurate consensus. Additionally, k-mer completeness was 94.35% in the male, and 91.45% in the female. As the region sex-determining region on LG19 is small, and has not accumulated a large number of repeats, it assembled as a single contig in the female. A single join was made of two large contigs in the male LG19, but this breakpoint does not appear to be in the sex-determining region.
C. guntheri has 2n = 48 chromosomes (Ozouf-Costaz et al., 2017), and these additional two chromosomes assembled cleanly with clear telomeric regions (Supplementary Figure S2; Supplementary Tables S3–S5). Their homology to other cichlid genomes was confirmed via sequence alignments. LG24 aligned against parts of O. aureus LG23, and LG25 against parts of O. aureus LG3. LG3 is itself a result of a fusion of two chromosomes, which is thought to have occurred at the base of the African cichlid lineage (Poletto et al., 2010; Conte et al., 2017; Ozouf-Costaz et al., 2017). The LG3 fusion and further fusion with a B chromosome in the Oreochrominii has been previously characterized in cichlids (Conte et al., 2021), so we investigated which chromosome corresponded to each region of the fused LG3. The genes sarcs2 and adamsts1 of O. niloticus aligned to LG3 from the C. guntheri male assembly. The genes poln, nhs12, tec, and ccdc171 aligned exclusively to LG25. This suggests that the chromosome called LG25 in this assembly corresponds with the LG3a from the Conte et al., 2017 study, and LG3 in our assembly corresponds to O. niloticus LG3a’.
Two incorrect joins were made by hifiasm in the female assembly. The first was an incorrect assembly that was broken into LG16 and LG20, respectively using alignments comparing female contigs to the male assembly, and female reads to the female assembly. These were visually inspected in IGV (Thorvaldsdóttir et al., 2013) to determine that the contig produced by hifiasm was not supported by the reads. The same process was repeated for the second contig that broke into parts of LG11 and LG15 that required further manual joining to other contigs to form a more complete scaffold.
Once the assemblies were complete, we mapped the pool-seq reads to the new C. guntheri male assembly. The same signal was present on LG19, though it was stronger against the C. guntheri reference than against the O. niloticus reference (Figure 1B). In the top 1% analysis, LG19 had 9 XY windows and 1 ZW window and the top 100 kb window (340 XY SNPs) was on LG19. The highest non-LG19 window was on LG7 with 86 XY SNPs. LG14 showed significant signal in the Dunn’s test but did not appear in the top 1% windows. The signal on LG3 was weak and diffuse compared to LG19 and had both XY and ZW signal in equal strength. The highest window on LG3 contained 35 XY SNPs. Thus, we conclude that the sex chromosome in C. guntheri is LG19.
3.3 Structure of the sex determining region
To understand the structure of the sex-determining region, we aligned both the HiFi sequencing reads and the pool-seq reads against the new male genome. Additionally, we aligned the X and Y assemblies to each other. There is no evidence for a structural rearrangement in the sex determining region of LG19 (Figure 2). The percent identity between the X and the Y is high, with the exception of the region at the end of the chromosome. This sub-telomeric region has likely accumulated a large quantity of repetitive DNAs due to the low recombination rate at the ends of cichlid chromosomes (Conte et al., 2021).
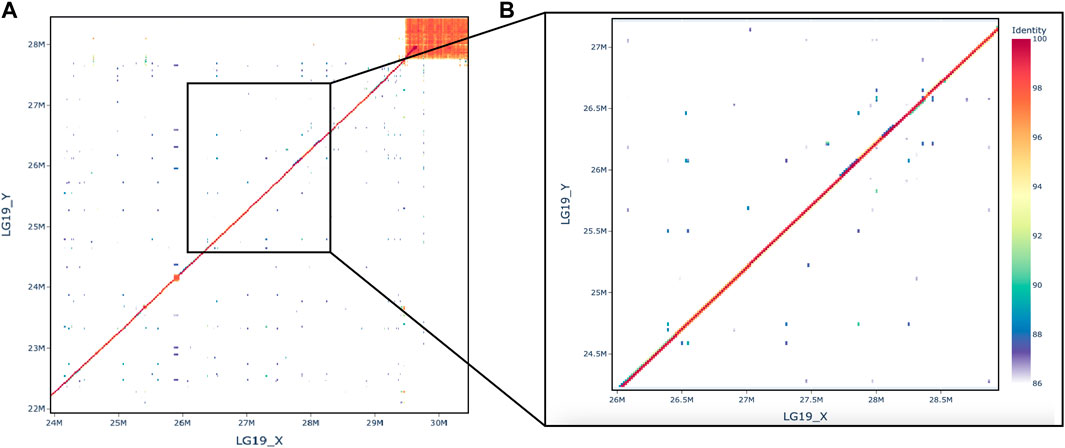
Figure 2. Dotplot comparing X and Y, (A) full chromosome and (B) the zoom on the region where rin3 is at 27.6 Mb.
In the narrow region of sex differentiation we found the following genes, listed in order from left to right: acireductone dioxygenase 1 (adi1), dynein axonemal light chain 1 (dnal1), tubulin epsilon and delta complex 1 (tedc1), parathyroid hormone 2 (pth2), ras and rab interactor 3 (rin3), and LOC120434950 which is uncharacterized and had no informative hits when BLASTed against the GenBank database. Perk2 was initially considered as a candidate gene based on the O. niloticus genome, however it became apparent that this gene is misannotated in the O. niloticus genome, likely because of the proline-rich domain. Alignments of the sex-determining region against the O. niloticus perk2 on LG19 were poor (<50% alignment), and the annotation is 7,143 bp with only two exons. When we searched the NCBI genome data viewer, the full copy of perk2 appears on LG3 and is 13,618 bp long with seven exons. Moving forward, we utilized the O. aureus annotation for this region.
While there is a high concentration of male-specific SNPs in this region, it is likely that many of these occur in introns or other non-coding regions. Identifying changes to enhancers and other regulatory regions is challenging without extensive functional annotation, so we chose to focus on SNPs that caused changes in the encoded proteins. Thus, we evaluated SNPs that resulted in a non-conserved amino acid change and were likely to impact the structure or function of the resultant protein. Only two genes from the list, adi1 and rin3, had a non-conserved amino acid change in C. guntheri that also differed from the reference cichlid species (Table 2). Any other amino acid changes to genes were conserved or shared with an outgroup species. Adi1 had a change at G185R, however this change was present in the female sequence (X) but not the male sequence which could result in a functional Y and a non-functional X.
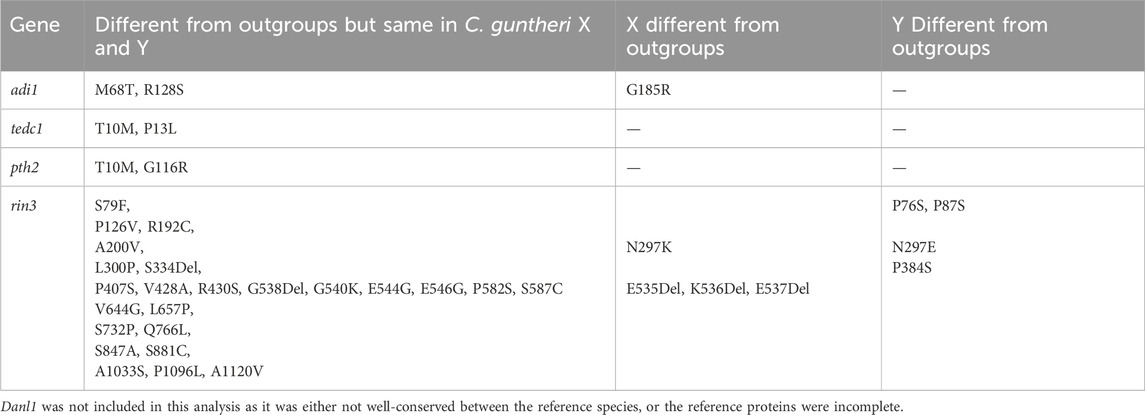
Table 2. Non-conserved amino acid changes in candidate sex-determining genes found in the region with the highest number of male sex-specific SNPs.
3.4 Candidate sex determining gene
We focused our analysis on rin3, as it features the highest density of heterozygous SNPs in the male (Figure 3, 27, 607, 824–27,629,697), which included several non-conserved amino acid changes. The multi-sequence alignment showed that the rin3 sequence on the Y is different from both the reference cichlid species and the X at three amino acid positions (Figure 4). All three of these are non-conserved. Two of these changes, P76S and P87S, occur just before the Src homology 2 (SH2) domain of the protein. The third, P384S, occurs in one of the three proline-rich domains (PRDs). Other changes include position 297, where the X sequence is N297K, and the Y is N297E. This is a conservative substitution, however both E to K are different from the outgroups. A four base pair deletion occurred after position 535 in the X. However, this is part of a repetitive string, and the Y also has a 1 base pair deletion in this region, suggesting that changes to this region have less impact on protein function.
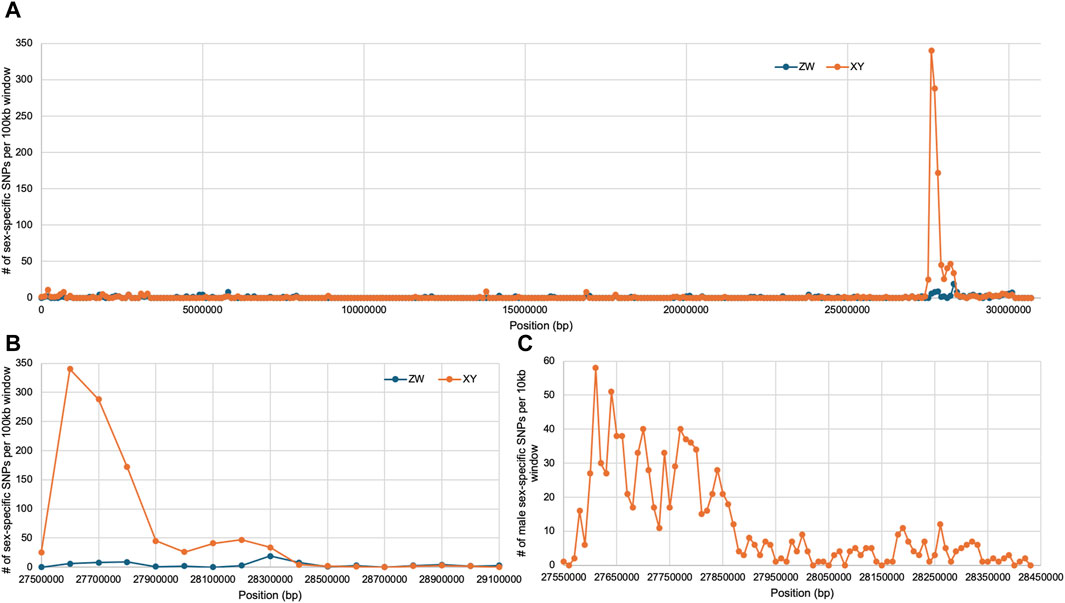
Figure 3. Sex-specific SNP density on LG19, rin3 is at 27.6 Mb. (A) Full chromosome with both XY and ZW SNPs plotted in 100 kb windows, (B) zoomed in plot on the sex-determining region in 100 kb windows, (C) zoomed in plot on sex-determining region in 10 kb windows.
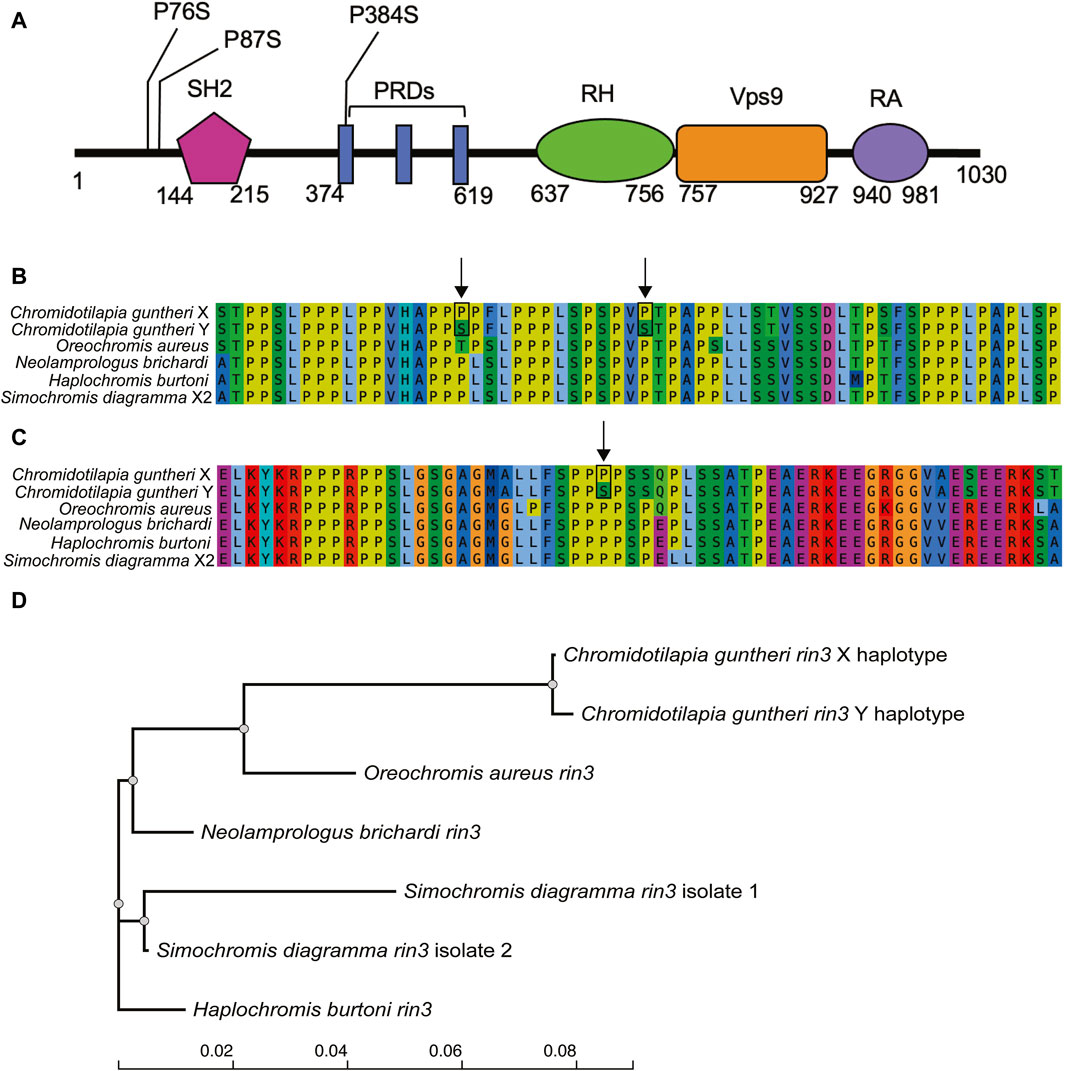
Figure 4. Variation in the candidate gene rin3. (A) domains of rin3 with positions of non-conserved amino acid changes (Kajiho et al., 2003), (B) alignment of C. guntheri and reference cichlid species corresponding to the first two amino acid changes which are indicated with an arrow, (C) alignment of C. guntheri and reference cichlid species corresponding to the amino acid change in the PR domain, (D) gene tree of rin3 in of C. guntheri and reference cichlid species.
4 Discussion
In this study, we used pooled-sequencing of males and females to identify an XY sex-determining locus on linkage group 19 in C. guntheri. We developed high-quality genome assemblies for a male and female of this species and used these assemblies to characterize a novel candidate gene for sex determination.
4.1 High quality genomes for studying sex chromosomes
Our assemblies of C. guntheri represent the first chromosome-level assemblies of a Chromidotilapiine cichlid. As C. guntheri is an outgroup to the hyper-diverse haplotilapiine cichlids, this is an important resource for understanding African cichlid genome evolution. These genomes assembled with a high N50 and minimal need for scaffolding, resulting in two highly contiguous reference assemblies that allow us to characterize any structural differences between the male and the female genomes. Hi-C might be the gold standard for scaffolding sex chromosomes (Carey et al., 2022), but for the purposes of generating the many reference genomes required for understanding cichlid evolution, our methodology assembling long high-coverage PacBio HiFi reads works well and is more affordable. As shown in Figure 1, the evolutionary distance to the reference sequence has a clear impact on sex chromosome detection. The signal identified on the C. guntheri reference is cleaner and stronger than in comparisons using the O. niloticus reference. The small sex-determining region means that our analyses were not affected by the usual challenges in phasing polymorphisms across the sex-determining region.
4.2 Lack of structural rearrangements
Many sex chromosomes are differentiated by one or more structural rearrangements that reduce recombination between the sex determiner and nearby genes (Jay et al., 2022; Charlesworth, 2023; Olito and Abbott, 2023). However, there is no evidence for an inversion or other structural rearrangement between the X and the Y in C. guntheri, and gene order in this region appears to be conserved in cichlids. The region of differentiation between the sex chromosomes is very narrow. This might suggest that the LG19 system is very young, however we do not have information on the sex chromosomes of any closely related species which would help date the origin of this sex chromosome system. It is likely that differentiation is maintained across this small region because of its position in near the end of the chromosome, which in cichlids usually has a low rate of recombination (Conte et al., 2021). The asymmetry of the peak on LG19 is consistent with the proximity of rin3 to the proximity of rin3 the region of low recombination near the telomere.
4.3 Rin3 as the candidate sex determining gene
Of the possible candidate genes in the narrow region on LG19, Ras and Rab Interactor 3 (rin3) stands out because it has the highest density of sex-specific substitutions of any genes in the region. The function of rin3 has been studied with respect to Alzheimer’s disease in humans, however the protein structure is not fully defined (Shen et al., 2022). The known domains include the Src homology 2 (SH2), proline rich regions (PR), RIN homology (RH), vacuolar protein sorting 9 (VPS9/GEF), and the Ub-like Ras association (RA) domain (Kajiho et al., 2003). Rin3 is distinguished from the rin1 and rin2 genes by featuring 3 PR domains (Kajiho et al., 2003). It is involved in the early stages of endocytosis via activation of Rab5 and Rab31, which bind to the VPS9 domain of the protein (Kajiho et al., 2003; Kajiho et al., 2011; Shen et al., 2022). Rin3 also activates Ras, which is involved in signaling pathways that regulate cellular function (Shen et al., 2022). The SH2 domain is involved in organizing cellular signaling network via protein tyrosine kinases (PTKs) (Pawson et al., 2001; Schlessinger and Lemmon, 2003). The PR domain of rin3 has been found to bind bridging integrator 1 (bin1) (Kajiho et al., 2003), bridging integrator 2 (bin2) (Janson et al., 2012), and CD2-associated protein (cd2ap) in humans (Rouka et al., 2015; Shen et al., 2020).
Interactions between the domains of rin3 make it somewhat difficult to discern how changes to a single domain might affect function. Previous work has found that if the SH2 domain is rendered inactive, rin3 had reduced colocalization with Rab5 indicating that the SH2 domain may self-inhibit the guanine-nucleotide exchange factor (GEF) activity of Rin proteins that are required for interaction with Rab5 (Yoshikawa et al., 2008). Other work has indicated that binding of the SH2 domain to the phosphotyrosine residue from the Ras/MAP signaling pathway inhibits Rab5 activity, while increased rin3 activity can result in an increase of Rab5 activation and formation of a complex of rin3-bin1-cd2ap in early endosomes that interrupts endocytic trafficking (Meshref et al., 2023). While the two amino acid changes fall just before the SH2 domain, cichlids seem to have a larger region before the SH2 domain than humans, which might induce conformational changes that impact binding.
Without functional data, we can only speculate on what effect the changes to the expression or structure of rin3 might have on the protein and the sex-determination network in C. guntheri males. Here, we present two possible mechanisms for how this might occur. The first hypothesis focuses on the role Rab5 plays in endocytosis via both the internalization of the clathrin-coated pits that lead to early endosomes and via the caveolin-mediated pathway (Shikanai et al., 2023). Both of these pathways are known to be involved in the endocytosis of TGFß receptors (Le Roy and Wrana, 2005; Chen, 2009; Heldin and Moustakas, 2016). Changes to Rab5 binding might impact the turnover and degradation of TGFß receptors, which has been shown for Rin1 (Hu et al., 2008). Thus, reduced ability of Rin3 to bind Rab5 may impact the TGFß pathway by reducing endocytic activity of these receptors, leaving more TGFß receptors available on the cell surface. An increased number of available receptors might be sufficient to tip the developmental pathway toward maleness. It is also important to note that bin2, which binds to Rin3 and plays a role in endocytosis via promotion of membrane bending (Janson et al., 2012), was found as a candidate gene for sex determination in the cichlid tribe Tropheini from Lake Tanganyika (Behrens et al., 2024a). A second hypothesis is less specific to TGFß receptors, but still relates to the role of rin3 in endocytosis. Endocytosis has been proposed to play a role in sex steroid regulation by enabling sex steroids to enter the cell, in contrast with previous theories that suggested sex steroids freely diffuse through the cell membrane (Hammes et al., 2005; Marko et al., 2022). Thus, reducing the function of rin3 in the endocytic pathway of males could affect female sex steroid uptake, also tipping the balance toward maleness. Evaluation of these hypotheses will require functional experiments, beginning with the development of CRISPR knockouts of rin3.
4.4 What genes can become sex-determining
Recent reviews have highlighted three major classes of genes that have become sex determiners in various vertebrate lineages (Curzon et al., 2023; Kitano et al., 2024). In fishes, genes in the TGFß pathway are the most frequently recruited to control sex determination. In the studies published to date, it is the genes for ligands (amh, gsdf, gdf6) or cell membrane receptors (amhr2, bmpr1) that are most often utilized (Pan et al., 2021; Yu et al., 2024). However, these signaling pathways extend deep within the cell, and it is possible that modification of other elements in the pathway might also become Mendelian factors in sex determination. A recent report proposed id2b as a candidate gene for sex determination in the arapaima (Adolfi et al., 2021). Id2 genes are downstream effectors of TGFß signaling, with effects primarily in the nucleus. Here we have proposed rin3 as a candidate gene for sex determination in C. guntheri. Rin3 may be a modifier of TGFß pathway function by modulating receptor turnover. These results suggest we may have to reconsider the hypothesis that sex determiners typically evolve at the top of biochemical pathways or gene regulatory networks (Wilkins, 1995).
5 Conclusion
This work furthers both our understanding of sex-determination in fish, as well as our understanding of the regulatory networks in which sex-determining genes function. Often, the differentiated regions of sex chromosomes span several megabases encompassing dozens of genes. Here we identified a narrow sex-determining region on LG19 in C. guntheri, which allowed us to identify rin3 as a strong candidate for the Mendelian factor regulating sex determination in this species. We also developed high quality male and female genome assemblies for C. guntheri, an outgroup to the extraordinary radiation of cichlids in East Africa, which will be an important resource for evolutionary studies of this group.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, JBDKXC000000000; https://www.ncbi.nlm.nih.gov/, JBDKXD000000000; https://www.ncbi.nlm.nih.gov/, PRJNA1100981.
Ethics statement
The animal study was approved by animal care protocols BMWFW-66.007/004-WF/V/3b/2016 (University of Graz) and the Institutional Animal Care and Use Committee protocol R-OCT-22-46 (University of Maryland) and this study was carried out with the approval of the ethics committee of the University of Graz (permit number GZ. 39/115/63 ex 2022/23). The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
KB: Writing–original draft, Writing–review and editing. SK: Writing–original draft, Writing–review and editing. TK: Writing–original draft, Writing–review and editing.
Funding
The authors declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by a grant from the National Science Foundation (DEB-1830753) to TK.
Acknowledgments
We thank the staff of Maryland Genomics in the Institute of Genome Science (University of Maryland School of Medicine) for their excellence in DNA sequencing, and the staff of the Division of Information Technology (University of Maryland, College Park) for their support of the Zaratan computer cluster.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2024.1447628/full#supplementary-material
References
Adolfi, M. C., Du, K., Kneitz, S., Cabau, C., Zahm, M., Klopp, C., et al. (2021). A duplicated copy of id2b is an unusual sex-determining candidate gene on the Y chromosome of arapaima (Arapaima gigas). Sci. Rep. 11 (1), 21544. doi:10.1038/s41598-021-01066-z
Astudillo-Clavijo, V., Stiassny, M. L. J., Ilves, K. L., Musilova, Z., Salzburger, W., and López-Fernández, H. (2023). Exon-based phylogenomics and the relationships of African cichlid fishes: tackling the challenges of reconstructing phylogenies with repeated rapid radiations. Syst. Biol. 72 (1), 134–149. doi:10.1093/sysbio/syac051
Behrens, K. A., Koblmüller, S., and Kocher, T. D. (2022). Sex chromosomes in the tribe cyprichromini (teleostei: cichlidae) of Lake Tanganyika. Sci. Rep. 12 (1), 17998–18013. doi:10.1038/s41598-022-23017-y
Behrens, K. A., Koblmüller, S., and Kocher, T. D. (2024b). Diversity of sex chromosomes in vertebrates: six novel sex chromosomes in basal haplochromines (Teleostei: Cichlidae). Genome Biol. Evol. 16 (7), evae152. doi:10.1093/gbe/evae152
Behrens, K. A., Zimmermann, H., Blažek, R., Reichard, M., Koblmüller, S., and Kocher, T. D. (2024a). Turnover of sex chromosomes in the Lake Tanganyika cichlid tribe Tropheini (Teleostei: Cichlidae). Sci. Rep. 14 (1), 2471. doi:10.1038/s41598-024-53021-3
Burress, E. D. (2015). Cichlid fishes as models of ecological diversification: patterns, mechanisms, and consequences. Hydrobiologia 748 (1), 7–27. doi:10.1007/s10750-014-1960-z
Cabanettes, F., and Klopp, C. (2018). D-GENIES: dot plot large genomes in an interactive, efficient and simple way. PeerJ 6, e4958. doi:10.7717/peerj.4958
Carey, S. B., Lovell, J. T., Jenkins, J., Leebens-Mack, J., Schmutz, J., Wilson, M. A., et al. (2022). Representing sex chromosomes in genome assemblies. Cell Genomics 2 (5), 100132. doi:10.1016/j.xgen.2022.100132
Charlesworth, D. (2023). Why and how do Y chromosome stop recombining? J. Evol. Biol. 36 (3), 632–636. doi:10.1111/jeb.14137
Chen, S., Zhang, G., Shao, C., Huang, Q., Liu, G., Zhang, P., et al. (2014). Whole-genome sequence of a flatfish provides insights into ZW sex chromosome evolution and adaptation to a benthic lifestyle. Nat. Genet. 46 (3), 253–260. doi:10.1038/ng.2890
Chen, Y.-G. (2009). Endocytic regulation of TGF-beta signaling. Cell Res. 19 (1), 58–70. doi:10.1038/cr.2008.315
Cheng, H., Concepcion, G. T., Feng, X., Zhang, H., and Li, H. (2021). Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat. Methods 18 (2), 170–175. doi:10.1038/s41592-020-01056-5
Conte, M. A., Clark, F. E., Roberts, R. B., Xu, L., Tao, W., Zhou, Q., et al. (2021). Origin of a giant sex chromosome. Mol. Biol. Evol. 38 (4), 1554–1569. doi:10.1093/molbev/msaa319
Conte, M. A., Gammerdinger, W. J., Bartie, K. L., Penman, D. J., and Kocher, T. D. (2017). A high quality assembly of the Nile Tilapia (Oreochromis niloticus) genome reveals the structure of two sex determination regions. BMC Genomics 18 (1), 341–419. doi:10.1186/s12864-017-3723-5
Conte, M. A., Joshi, R., Moore, E. C., Nandamuri, S. P., Gammerdinger, W. J., Roberts, R. B., et al. (2019). Chromosome-scale assemblies reveal the structural evolution of African cichlid genomes. GigaScience 8 (4), giz030. doi:10.1093/gigascience/giz030
Conte, M. A., and Kocher, T. D. (2015). An improved genome reference for the African cichlid, Metriaclima zebra. BMC Genomics 16 (1), 724. doi:10.1186/s12864-015-1930-5
Curzon, A. Y., Shirak, A., Benet-Perlberg, A., Naor, A., Low-Tanne, S. I., Sharkawi, H., et al. (2022). Absence of figla-like gene is concordant with femaleness in cichlids harboring the LG1 sex-determination system. Int. J. Mol. Sci. 23 (14), 7636. doi:10.3390/ijms23147636
Curzon, A. Y., Shirak, A., Ron, M., and Seroussi, E. (2023). Master-key regulators of sex determination in fish and other vertebrates—a review. Int. J. Mol. Sci. 24 (3), 2468. doi:10.3390/ijms24032468
Danecek, P., Bonfield, J. K., Liddle, J., Marshall, J., Ohan, V., Pollard, M. O., et al. (2021). Twelve years of SAMtools and BCFtools. GigaScience 10 (2), giab008. doi:10.1093/gigascience/giab008
Darolti, I., Almeida, P., Wright, A. E., and Mank, J. E. (2022). Comparison of methodological approaches to the study of young sex chromosomes: a case study in Poecilia. J. Evol. Biol. 35 (12), 1646–1658. doi:10.1111/jeb.14013
Darolti, I., Wright, A. E., Sandkam, B. A., Morris, J., Bloch, N. I., Farré, M., et al. (2019). Extreme heterogeneity in sex chromosome differentiation and dosage compensation in livebearers. Proc. Natl. Acad. Sci. 116 (38), 19031–19036. doi:10.1073/pnas.1905298116
de la Herrán, R., Hermida, M., Rubiolo, J. A., Gómez-Garrido, J., Cruz, F., Robles, F., et al. (2023). A chromosome-level genome assembly enables the identification of the follicule stimulating hormone receptor as the master sex-determining gene in the flatfish Solea senegalensis. Mol. Ecol. Resour. 23 (4), 886–904. doi:10.1111/1755-0998.13750
El Taher, A., Ronco, F., Matschiner, M., Salzburger, W., and Böhne, A. (2021). Dynamics of sex chromosome evolution in a rapid radiation of cichlid fishes. Sci. Adv. 7, eabe8215. doi:10.1126/sciadv.abe8215
Feldberg, E., Porto, J. I. R., and Bertollo, L. A. C. (2003). “Chromosomal changes and adaptation of cichlid fishes during evolution,” in Fish adaptations. Editors A. L. Val, and B. G. Kapoor (New Dehli and New York: Science Publishers), 285–308.
Gammerdinger, W. J., Conte, M. A., Sandkam, B. A., Ziegelbecker, A., Koblmüller, S., and Kocher, T. D. (2018). Novel sex chromosomes in 3 cichlid fishes from Lake Tanganyika. J. Hered. 109 (5), 489–500. doi:10.1093/jhered/esy003
Gammerdinger, W. J., and Kocher, T. D. (2018). Unusual diversity of sex chromosomes in african cichlid fishes. Genes 9 (10), 480. doi:10.3390/genes9100480
Greenwood, P. H. (1987). The genera of pelmatochromine fishes (Teleostei, Cichlidae). A phylogenetic review. Bull. Br. Mus. Nat. Hist. Zoology 53, 139–203.
Hammes, A., Andreassen, T. K., Spoelgen, R., Raila, J., Hubner, N., Schulz, H., et al. (2005). Role of endocytosis in cellular uptake of sex steroids. Cell 122 (5), 751–762. doi:10.1016/j.cell.2005.06.032
Heldin, C.-H., and Moustakas, A. (2016). Signaling receptors for TGF-β family members. Cold Spring Harb. Perspect. Biol. 8 (8), a022053. doi:10.1101/cshperspect.a022053
Hu, H., Milstein, M., Bliss, J. M., Thai, M., Malhotra, G., Huynh, L. C., et al. (2008). Integration of transforming growth factor beta and RAS signaling silences a RAB5 guanine nucleotide exchange factor and enhances growth factor-directed cell migration. Mol. Cell. Biol. 28 (5), 1573–1583. doi:10.1128/MCB.01087-07
Hughes, J. F., and Page, D. C. (2015). The biology and evolution of mammalian Y chromosomes. Annu. Rev. Genet. 49, 507–527. doi:10.1146/annurev-genet-112414-055311
Jain, M., Olsen, H. E., Turner, D. J., Stoddart, D., Bulazel, K. V., Paten, B., et al. (2018). Linear assembly of a human centromere on the Y chromosome. Nat. Biotechnol. 36 (4), 321–323. doi:10.1038/nbt.4109
Janson, C., Kasahara, N., Prendergast, G. C., and Colicelli, J. (2012). RIN3 is a negative regulator of mast cell responses to SCF. PLoS ONE 7 (11), e49615. doi:10.1371/journal.pone.0049615
Jay, P., Tezenas, E., Véber, A., and Giraud, T. (2022). Sheltering of deleterious mutations explains the stepwise extension of recombination suppression on sex chromosomes and other supergenes. PLoS Biol. 20 (7), e3001698. doi:10.1371/journal.pbio.3001698
Kajiho, H., Saito, K., Tsujita, K., Kontani, K., Araki, Y., Kurosu, H., et al. (2003). RIN3: a novel Rab5 GEF interacting with amphiphysin II involved in the early endocytic pathway. J. Cell Sci. 116 (20), 4159–4168. doi:10.1242/jcs.00718
Kajiho, H., Sakurai, K., Minoda, T., Yoshikawa, M., Nakagawa, S., Fukushima, S., et al. (2011). Characterization of RIN3 as a guanine nucleotide exchange factor for the Rab5 subfamily GTPase Rab31. J. Biol. Chem. 286 (27), 24364–24373. doi:10.1074/jbc.M110.172445
Kamiya, T., Kai, W., Tasumi, S., Oka, A., Matsunaga, T., Mizuno, N., et al. (2012). A trans-species missense SNP in Amhr2 is associated with sex determination in the tiger pufferfish, Takifugu rubripes (Fugu). PLoS Genet. 8 (7), e1002798. doi:10.1371/journal.pgen.1002798
Kirubakaran, T. G., Andersen, Ø., De Rosa, M. C., Andersstuen, T., Hallan, K., Kent, M. P., et al. (2019). Characterization of a male specific region containing a candidate sex determining gene in Atlantic cod. Sci. Rep. 9 (1), 116. doi:10.1038/s41598-018-36748-8
Kitano, J., Ansai, S., Takehana, Y., and Yamamoto, Y. (2024). Diversity and convergence of sex-determination mechanisms in teleost fish. Annu. Rev. Animal Biosci. 12, 233–259. doi:10.1146/annurev-animal-021122-113935
Kitano, J., and Peichel, C. L. (2012). Turnover of sex chromosomes and speciation in fishes. Environ. Biol. Fishes 94 (3), 549–558. doi:10.1007/s10641-011-9853-8
Kocher, T. D., Behrens, K. A., Conte, M. A., Aibara, M., Mrosso, H. D. J., Green, E. C. J., et al. (2022). New sex chromosomes in lake victoria cichlid fishes (Cichlidae: Haplochromini). Genes 13, 804. doi:10.3390/genes13050804
Koren, S., Walenz, B. P., Berlin, K., Miller, J. R., Bergman, N. H., and Phillippy, A. M. (2017). Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27 (5), 722–736. doi:10.1101/gr.215087.116
Koyama, T., Nakamoto, M., Morishima, K., Yamashita, R., Yamashita, T., Sasaki, K., et al. (2019). A SNP in a steroidogenic enzyme is associated with phenotypic sex in Seriola fishes. Curr. Biol. 29 (11), 1901–1909. doi:10.1016/j.cub.2019.04.069
Le Roy, C., and Wrana, J. L. (2005). Clathrin- and non-clathrin-mediated endocytic regulation of cell signalling. Nat. Rev. Mol. Cell Biol. 6 (2), 112–126. doi:10.1038/nrm1571
Li, H. (2018). Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34 (18), 3094–3100. doi:10.1093/bioinformatics/bty191
Li, M., Sun, Y., Zhao, J., Shi, H., Zeng, S., Ye, K., et al. (2015). A tandem duplicate of anti-Müllerian hormone with a missense SNP on the Y chromosome is essential for male sex determination in Nile Tilapia, Oreochromis niloticus. PLoS Genet. 11 (11), e1005678. doi:10.1371/journal.pgen.1005678
Li, M., Zhang, R., Fan, G., Xu, W., Zhou, Q., Wang, L., et al. (2021). Reconstruction of the origin of a neo-Y sex chromosome and its evolution in the spotted knifejaw, oplegnathus punctatus. Mol. Biol. Evol. 38 (6), 2615–2626. doi:10.1093/molbev/msab056
Liu, F., Sun, F., Li, J., Xia, J. H., Lin, G., Tu, R. J., et al. (2013). A microsatellite-based linkage map of salt tolerant tilapia (Oreochromis mossambicus x Oreochromis spp.) and mapping of sex-determining loci. BMC Genomics 14 (1), 58. doi:10.1186/1471-2164-14-58
Madeira, F., Pearce, M., Tivey, A. R. N., Basutkar, P., Lee, J., Edbali, O., et al. (2022). Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic acids Res. 50 (W1), W276–W279. doi:10.1093/nar/gkac240
Manni, M., Berkeley, M. R., Seppey, M., Simão, F. A., and Zdobnov, E. M. (2021). BUSCO update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol. Biol. Evol. 38 (10), 4647–4654. doi:10.1093/molbev/msab199
Marko, H. L., Hornig, N. C., Betz, R. C., Holterhus, P. M., Altmüller, J., Thiele, H., et al. (2022). Genomic variants reducing expression of two endocytic receptors in 46,XY differences of sex development. Hum. Mutat. 43 (3), 420–433. doi:10.1002/humu.24325
Martínez, P., Robledo, D., Taboada, X., Blanco, A., Moser, M., Maroso, F., et al. (2021). A genome-wide association study, supported by a new chromosome-level genome assembly, suggests sox2 as a main driver of the undifferentiatiated ZZ/ZW sex determination of turbot (Scophthalmus maximus). Genomics 113 (4), 1705–1718. doi:10.1016/j.ygeno.2021.04.007
Matsuda, M., Sato, T., Toyazaki, Y., Nagahama, Y., Hamaguchi, S., and Sakaizumi, M. (2003). Oryzias curvinotus has DMY, a gene that is required for male development in the medaka, O. latipes. Zoological Sci. 20 (2), 159–161. doi:10.2108/zsj.20.159
Meshref, M., Ghaith, H. S., Hammad, M. A., Shalaby, M. M. M., Ayasra, F., Monib, F. A., et al. (2023). The role of RIN3 gene in Alzheimer’s disease pathogenesis: a comprehensive review. Mol. Neurobiol. 61, 3528–3544. doi:10.1007/s12035-023-03802-0
Miller, J. R., Delcher, A. L., Koren, S., Venter, E., Walenz, B. P., Brownley, A., et al. (2008). Aggressive assembly of pyrosequencing reads with mates. Bioinformatics 24 (24), 2818–2824. doi:10.1093/bioinformatics/btn548
Munby, H., Linderoth, T., Fischer, B., Du, M., and Vernaz, G. (2021). Differential use of multiple genetic sex determination systems in divergent ecomorphs of an African crater lake cichlid. bioRxiv. 2021.08.05.455235.
Myosho, T., Otake, H., Masuyama, H., Matsuda, M., Kuroki, Y., Fujiyama, A., et al. (2012). Tracing the emergence of a novel sex-determining gene in medaka, Oryzias luzonensis. Genetics 191 (1), 163–170. doi:10.1534/genetics.111.137497
Nacif, C. L., Kratochwil, C. F., Kautt, A. F., Nater, A., Machado-Schiaffino, G., Meyer, A., et al. (2023). Molecular parallelism in the evolution of a master sex-determining role for the anti-Mullerian hormone receptor 2 gene (amhr2) in Midas cichlids. Mol. Ecol. 32 (1398–1410), 1398–1410. doi:10.1111/mec.16466
Nakamura, Y., Higuchi, K., Kumon, K., Yasuike, M., Takashi, T., Gen, K., et al. (2021). Prediction of the sex-associated genomic region in tunas (Thunnus fishes). Int. J. Genomics, 7226353. doi:10.1155/2021/7226353
Nakatani, Y., Takeda, H., Kohara, Y., and Morishita, S. (2007). Reconstruction of the vertebrate ancestral genome reveals dynamic genome reorganization in early vertebrates. Genome Res. 17 (9), 1254–1265. doi:10.1101/gr.6316407
Olito, C., and Abbott, J. K. (2023). The evolution of suppressed recombination between sex chromosomes and the lengths of evolutionary strata. Evolution 77 (4), 1077–1090. doi:10.1093/evolut/qpad023
Ozouf-Costaz, C., Coutanceau, J., Bonillo, C., Mercot, H., Fermon, Y., and Guidi-Rontani, C. (2017). New insights into the chromosomal differentiation patterns among cichlids from Africa and Madagascar. Cybium Int. J. Ichthyology 41 (1), 35–43.
Palmer, D. H., Rogers, T. F., Dean, R., and Wright, A. E. (2019). How to identify sex chromosomes and their turnover. Mol. Ecol. 28 (21), 4709–4724. doi:10.1111/mec.15245
Pan, Q., Kay, T., Depincé, A., Adolfi, M., Schartl, M., Guiguen, Y., et al. (2021). Evolution of master sex determiners: TGF-β signalling pathways at regulatory crossroads. Philosophical Trans. R. Soc. B Biol. Sci. 376 (1832), 20200091. doi:10.1098/rstb.2020.0091
Pan, X., Cao, Y., Stucchi, R., Hooikaas, P. J., Portegies, S., Will, L., et al. (2019). MAP7D2 localizes to the proximal axon and locally promotes kinesin-1-mediated cargo transport into the axon. Cell Rep. 26, 1988–1999. doi:10.1016/j.celrep.2019.01.084
Pawson, T., Gish, G. D., and Nash, P. (2001). SH2 domains, interaction modules and cellular wiring. Trends Cell Biol. 11 (12), 504–511. doi:10.1016/s0962-8924(01)02154-7
Peichel, C. L., McCann, S. R., Ross, J. A., Naftaly, A. F. S., Urton, J. R., Cech, J. N., et al. (2020). Assembly of the threespine stickleback y chromosome reveals convergent signatures of sex chromosome evolution. Genome Biol. 21 (1), 177. doi:10.1186/s13059-020-02097-x
Poletto, A. B., Ferreira, I. A., Cabral-de-Mello, D. C., Nakajima, R. T., Mazzuchelli, J., Ribeiro, H. B., et al. (2010). Chromosome differentiation patterns during cichlid fish evolution. BMC Genet. 11, 50. doi:10.1186/1471-2156-11-50
Post, V. A. (1965). Vergleichende untersuchungen der chromosomenzahlen hei Süßwasser-Teleosteern. J. Zoological Syst. Evol. Res. 3, 47–93. doi:10.1111/j.1439-0469.1965.tb00426.x
Rafati, N., Chen, J., Herpin, A., Pettersson, M. E., Han, F., Feng, C., et al. (2020). Reconstruction of the birth of a male sex chromosome present in Atlantic herring. Proc. Natl. Acad. Sci. 117 (39), 24359–24368. doi:10.1073/pnas.2009925117
Reddon, A. R., and Hurd, P. L. (2013). Water pH during early development influences sex ratio and male morph in a West African cichlid fish, Pelvicachromis pulcher. Zoology 116 (3), 139–143. doi:10.1016/j.zool.2012.11.001
Renn, S. C. P., and Hurd, P. L. (2021). Epigenetic regulation and environmental sex determination in cichlid fishes. Sex. Dev. 15 (1–3), 93–107. doi:10.1159/000517197
Rhie, A., McCarthy, S. A., Fedrigo, O., Damas, J., Formenti, G., Koren, S., et al. (2021). Towards complete and error-free genome assemblies of all vertebrate species. Nature 592 (7856), 737–746. doi:10.1038/s41586-021-03451-0
Rhie, A., Nurk, S., Cechova, M., Hoyt, S. J., Taylor, D. J., Altemose, N., et al. (2023). The complete sequence of a human Y chromosome. Nature 621 (7978), 344–354. doi:10.1038/s41586-023-06457-y
Rhie, A., Walenz, B. P., Koren, S., and Phillippy, A. M. (2020). Merqury: reference-free quality, completeness, and phasing assessment for genome assemblies. Genome Biol. 21 (1), 245. doi:10.1186/s13059-020-02134-9
Rouka, E., Simister, P. C., Janning, M., Kumbrink, J., Konstantinou, T., Muniz, J. R. C., et al. (2015). Differential recognition preferences of the three Src homology 3 (SH3) domains from the adaptor CD2-associated protein (CD2AP) and direct association with ras and rab interactor 3 (RIN3). J. Biol. Chem. 290 (42), 25275–25292. doi:10.1074/jbc.M115.637207
Santos, M. E., Lopes, J. F., and Kratochwil, C. F. (2023). East African cichlid fishes. EvoDevo 14 (1), 1. doi:10.1186/s13227-022-00205-5
Schlessinger, J., and Lemmon, M. A. (2003). SH2 and PTB domains in tyrosine kinase signaling. Science’s STKE signal Transduct. Knowl. Environ. 2003 (191), RE12. doi:10.1126/stke.2003.191.re12
Schwarzer, J., Lamboj, A., Langen, K., Misof, B., and Schliewen, U. K. (2015). Phylogeny and age of chromidotilapiine cichlids (Teleostei: cichlidae). Hydrobiologia 748 (1), 185–199. doi:10.1007/s10750-014-1918-1
Shen, R., Murphy, C. J., Xu, X., Ding, J., and Wu, C. (2022). Ras and rab interactor 3: from cellular mechanisms to human diseases. Front. Cell Dev. Biol. 10, 824961. doi:10.3389/fcell.2022.824961
Shen, R., Zhao, X., He, L., Ding, Y., and Xu, W. (2020). Upregulation of RIN3 induces endosomal dysfunction in Alzheimer’s disease. Transl. Neurodegener. 9, 26. doi:10.1186/s40035-020-00206-1
Shikanai, M., Ito, S., Nishimura, Y. V., Akagawa, R., Fukuda, M., Yuzaki, M., et al. (2023). Rab21 regulates caveolin-1-mediated endocytic trafficking to promote immature neurite pruning. EMBO Rep. 24 (3), e54701. doi:10.15252/embr.202254701
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539. doi:10.1038/msb.2011.75
Skaletsky, H., Kuroda-Kawaguchi, T., Minx, P. J., Cordum, H. S., Hillier, L., Brown, L. G., et al. (2003). The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 423 (6942), 825–837. doi:10.1038/nature01722
Sweeten, A. P., Schatz, M. C., and Phillippy, A. M. (2024). ModDotPlot—rapid and interactive visualization of complex repeats. bioRxiv. 2024.04.15.589623. doi:10.1101/2024.04.15.589623
Takehana, Y., Matsuda, M., Myosho, T., Suster, M. L., Kawakami, K., Shin-I, T., et al. (2014). Co-option of Sox3 as the male-determining factor on the Y chromosome in the fish Oryzias dancena. Nat. Commun. 5 (1), 4157. doi:10.1038/ncomms5157
Tao, W., Cao, J., Xiao, H., Zhu, X., Dong, J., Kocher, T. D., et al. (2021a). A chromosome-level genome assembly of Mozambique Tilapia (Oreochromis mossambicus) reveals the structure of sex determining regions. Front. Genet. 12, 796211. doi:10.3389/fgene.2021.796211
Tao, W., Conte, M. A., Wang, D., and Kocher, T. D. (2021b). Network architecture and sex chromosome turnovers. BioEssays 43 (3), 2000161. doi:10.1002/bies.202000161
Tao, W., Xu, L., Zhao, L., Zhu, Z., Wu, X., Min, Q., et al. (2021c). High-quality chromosome-level genomes of two tilapia species reveal their evolution of repeat sequences and sex chromosomes. Mol. Ecol. Resour. 21 (2), 543–560. doi:10.1111/1755-0998.13273
Thorvaldsdóttir, H., Robinson, J. T., and Mesirov, J. P. (2013). Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Briefings Bioinforma. 14 (2), 178–192. doi:10.1093/bib/bbs017
Vicoso, B., Emerson, J. J., Zektser, Y., Mahajan, S., and Bachtrog, D. (2013). Comparative sex chromosome genomics in snakes: differentiation, evolutionary strata, and lack of global dosage compensation. PLOS Biol. 11 (8), e1001643. doi:10.1371/journal.pbio.1001643
Wang, Y. (2022). “A comparative study of HiCanu and hifiasm,” in 2022 5th international conference on mathematics and statistics. ICoMS 2022, Paris, France, June 17–19, 2022 (New York, NY: ACM), 100–104. doi:10.1145/3545839.3545855
Wen, M., Pan, Q., Larson, W., Eché, C., and Guiguen, Y. (2023). Characterization of the sex determining region of channel catfish (Ictalurus punctatus) and development of a sex-genotyping test. Gene 850, 146933. doi:10.1016/j.gene.2022.146933
Wick, R. R., Schultz, M. B., Zobel, J., and Holt, K. E. (2015). Bandage: interactive visualization of de novo genome assemblies. Bioinformatics 31 (20), 3350–3352. doi:10.1093/bioinformatics/btv383
Wilkins, A. S. (1995). Moving up the hierarchy: a hypothesis on the evolution of a genetic sex determination pathway. BioEssays News Rev. Mol. Cell. Dev. Biol. 17 (1), 71–77. doi:10.1002/bies.950170113
Xue, L., Gao, Y., Wu, M., Tian, T., Fan, H., Huang, Y., et al. (2021). Telomere-to-telomere assembly of a fish Y chromosome reveals the origin of a young sex chromosome pair. Genome Biol. 22 (1), 203. doi:10.1186/s13059-021-02430-y
Yano, A., Guyomard, R., Nicol, B., Jouanno, E., Quillet, E., Klopp, C., et al. (2012). An immune-related gene evolved into the master sex-determining gene in rainbow trout, Oncorhynchus mykiss. Curr. Biol. 22 (15), 1423–1428. doi:10.1016/j.cub.2012.05.045
Yano, A., Nicol, B., Jouanno, E., Quillet, E., Fostier, A., Guyomard, R., et al. (2013). The sexually dimorphic on the Y-chromosome gene (sdY) is a conserved male-specific Y-chromosome sequence in many salmonids. Evol. Appl. 6 (3), 486–496. doi:10.1111/eva.12032
Yoshikawa, M., Kajiho, H., Sakurai, K., Minoda, T., Nakagawa, S., Kontani, K., et al. (2008). Tyr-phosphorylation signals translocate RIN3, the small GTPase Rab5-GEF, to early endocytic vesicles. Biochem. Biophysical Res. Commun. 372 (1), 168–172. doi:10.1016/j.bbrc.2008.05.027
Yu, H., Du, X., Chen, X., Liu, L., and Wang, X. (2024). Transforming growth factor-β (TGF-β): a master signal pathway in teleost sex determination. General Comp. Endocrinol. 355, 114561. doi:10.1016/j.ygcen.2024.114561
Keywords: sex chromosome, sex determination, African cichlids, genome assembly, RIN3
Citation: Behrens KA, Koblmüller S and Kocher TD (2024) Genome assemblies for Chromidotilapia guntheri (Teleostei: Cichlidae) identify a novel candidate gene for vertebrate sex determination, RIN3. Front. Genet. 15:1447628. doi: 10.3389/fgene.2024.1447628
Received: 11 June 2024; Accepted: 06 August 2024;
Published: 16 August 2024.
Edited by:
Diogo Teruo Hashimoto, São Paulo State University, BrazilReviewed by:
Gabriela Vanina Villanova, National University of Rosario, ArgentinaFei Liu, Chinese Academy of Sciences (CAS), China
Copyright © 2024 Behrens, Koblmüller and Kocher. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kristen A. Behrens, a2JlaHJlbjJAdW1kLmVkdQ==
 Kristen A. Behrens
Kristen A. Behrens Stephan Koblmüller
Stephan Koblmüller Thomas D. Kocher
Thomas D. Kocher