- 1College of Horticulture Science and Technology, Hebei Normal University of Science and Technology, Qinhuangdao, China
- 2Hebei Key Laboratory of Horticultural Germplasm Excavation and Innovative Utilization, Qinhuangdao, China
- 3Hebei Higher Institute Application Technology Research and Development Center of Horticultural Plant Biological Breeding, Hebei Normal University of Science and Technology, Qinhuangdao, China
Background: Low temperature pose significant challenges to peach cultivation, causing severe damage to peach buds and restricting production and distribution. Ethylene, an important phytohormone, plays a critical role in enhancing plant cold resistance. Structural genes and transcription factors involved in ethylene biosynthesis and signal transduction pathways are associated with cold resistance. However, no research has specifically addressed their roles in peach cold resistance.
Methods: In this study, we aimed for cold-resistance gene discovery in cold-sensitive peach cultivar “21Shiji” (21SJ) and cold-resistance cultivar “Shijizhixing” (SJZX) using RNA-seq and gas chromatography.
Results: The findings revealed that under cold stress conditions, ethylene biosynthesis in “SJZX” was significantly induced. Subsequently, a structural gene, PpACO1-1, involved in ethylene biosynthesis in peach buds was significantly upregulated and showed a higher correlation with ethylene release rate. To identify potential transcription factors associated with PpACO1-1 expression and ethylene signal transduction, weighted gene co-expression network analysis was conducted using RNA-seq data. Four transcription factors: PpERF2, PpNAC078, PpWRKY65 and PpbHLH112, were identified.
Conclusion: These findings provide valuable theoretical insights for investigating the regulatory mechanisms of peach cold resistance and guiding breeding strategies.
Introduction
Peach (Prunus persica L.) originates in China. It is widely cultivated in the North China Plain and the Yangtze River Basin, where it holds the top rank globally in both yield and cultivation areas. However, the low temperature (<0°C) during winter and early spring in Northern China significantly restricts peach growth and geographical distribution. Most peach cultivars are susceptible to the environments, and those with higher cold resistance are insufficient to meet the market demand. This scarcity significantly limits the high-quality development of the peach industry.
Two regulatory pathways are recognized in plant cold resistance research: the abscisic acid (ABA)-dependent and ABA-independent pathway. Among them, the ABA-independent pathway can be further categorized into C-repeat binding transcription factor (CBF)-dependent regulatory and CBF-independent regulatory pathways (Park et al., 2015; Lim and Lee, 2020). Multiple studies have reported that ABA signaling significantly induces cold resistance gene expression in plants (Ju et al., 2020; Guo et al., 2021; Lee et al., 2021; Song et al., 2022; Yu et al., 2023). The ABA receptor protein (PYR/PYL/RCAR), 2C protein phosphatase (PP2C), and sucrose nonfermenting-1-related protein kinase 2 (SnRK2) constitute the primary components of the ABA signal transduction pathway. ABA presence inactivates PP2C and releases SnRK2. Activated SnRK2 then binds to ABA response elements (ABRE) or ABA response binding factors (ABF) in downstream cold gene promoters, thereby activating their expression (Ma et al., 2009; Nakashima et al., 2009; Klingler et al., 2010; Gonzalez-Guzman et al., 2012; Soon et al., 2012). Recent studies indicate that several transcription factors, such as b-ZIPs, LEAs, MYBs, WRKYs, and NACs, can be regulated by ABA signaling under cold stress (Sun et al., 2019; Ju et al., 2020; Shu Y. et al., 2023; Mei et al., 2023; Shen et al., 2023).
CBFs belong to the APETALA2/ETHYLENE RESPONSE FACTOR (AP/ERF) family. In the ABA-independent pathway, CBFs play a central role in regulating plant cold resistance. Under cold stress, the CBFs can be induced by Inducer of CBF Expression (ICE). The induced CBFs then bind to the C-repeat/Dehydration Responsive Element (CRT/DRE), a cis-acting element in the promoter region of Cold Regulated (COR) genes, thereby activating their transcription (Thomashow, 1999; Chinnusamy et al., 2007). Several CBFs involved in plant cold resistance have been identified (Park et al., 2015; Jia et al., 2016; Wang H. et al., 2021; Wang et al., 2021). Among these, the ICE-CBFs-COR signaling cascade stands out as a typical cod-resistant pathway, extensively documented in the literature (Hwarari et al., 2022; Kopeć et al., 2022; Ma et al., 2023; Wang et al., 2023). Furthermore, several transcription factors can directly regulate the expression of CBF genes, thereby improving plant cold resistance, such as MdNAC104, MdHYL1, MdMYB88, and MdMYB124 in apples (Mei et al., 2023; Shen et al., 2023).
In CBF-independent pathways, structural genes and transcription factors involved in ethylene signal transduction are pivotal for cold resistance (Shu P. et al., 2023). Ethylene biosynthesis begins with methionine conversion to S-adenosyl-methionine (SAM) by SAM synthetases. SAM is further converted to the ethylene precursor 1-aminocyclopropane-1-carboxylic acid (ACC) by ACC synthetases. Ultimately, ACC is converted to ethylene by ACC oxidases (ACO) (Adams and Yang, 1977; Adams and Yang, 1979; Hamilton et al., 1991; Pattyn et al., 2021). The ethylene receptors and CONSTITUTIVE TRIPLE RESPONSE1 (CTR1) downstream of ethylene synthesis negatively regulate the ethylene-signaling pathway. The presence of ethylene inactivates the ethylene receptors and CTR1, activating ETHYLENE INSENSITIVE 2 (EIN2) expression. EIN2, downstream of CTR1, promotes the activity of ETHYLENE INSENSITIVE 3 (EIN3) and EIN3-LIKE 1 (EIL1) (Chen et al., 2010; Shakeel et al., 2015; Zhao et al., 2021), which controls the expression of numerous ethylene-responsive genes, including ETHYLENE RESPONSE FACTORs (ERFs) (Lorenzo et al., 2003; Cheng et al., 2013; Li et al., 2019; Hu et al., 2020). In plants, ERFs contribute positively to cold resistance. For instance, in apples, overexpressing MdERF1B enhances cold tolerance by interacting with MdACO1 and MdERF3, key components in ethylene biosynthesis (Wang et al., 2021a). In Vitis amurensis, overexpressing the VaERF092 gene in the ethylene signal transduction pathway enhances cold resistance of Arabidopsis. Additionally, VaERF092 interacts with the cis-acting element (GCC-box) in the VaWRKY33 promoter, indirectly enhancing the cold resistance of Arabidopsis (Sun et al., 2019). In bermudagrass, CdERF1 positively regulates plant cold response by activating the expression of PODs, CBF2 and LTPs (Hu et al., 2020). ERF41 and ERF180 in kiwi fruit are significantly induced under low temperatures (Gunaseelan et al., 2019).
Several studies have explored the chilling response of postharvest peach fruit and the cold response of peach shoots (Pons et al., 2014; Wang et al., 2017; Yu et al., 2020; Guo et al., 2023; Li et al., 2023). Peach buds are notably more susceptible to cold stress compared to the trunk and shoots. In Northern China, prolonged winter low temperatures can lead to peach bud ossification, significantly reducing peach orchard yield. However, their studies regarding the cold resistance of peach buds remain unexplored. Therefore, in this study, we aimed to investigate the essential structural genes and transcription factors associated with resistance in peach buds using the cold-sensitive peach cultivar “21Shiji” (21SJ) and the cold-resistant cultivar “Shijizhixing” (SJZX), commonly cultivated in Northern China. “21SJ” came from the hybridization of peach cultivar “Dangui” × “Xuetao” and “SJZX” came from the hybridization of “21Shiji” × “Jiucui”. The approach involves transcriptome analysis and weighted gene co-expression network analysis (WGCNA). The findings could serve as a valuable reference for future research on cold resistance research in peach buds.
Materials and methods
Plant material
Peach cultivars “21SJ” and “SJZX” were cultivated in the Peach Experimental Garden of Hebei Normal University of Science and Technology (39°42′N, 119°10′E). Dormant buds from “21SJ” and “SJZX” were utilized to identify cold resistance. Peach shoots (1 year old) containing dormant buds were randomly collected in 20 November 2022, the temperature at that time was ranged from 1°C to 5°C. The collected buds were then subjected to cold storage in a programmable incubator set at −4°C for 0, 12, 24, 48, and 72 h. For ethylene release rates assessment, dormant bud samples of these two cultivars were collected and randomly assigned to 5 sets of 10 buds each (in total 100 buds per time point). Dormant bud samples which used for RNA-seq were randomly collected at 12, 24, 48 h, 3 sets of 10 buds were used at each time point.
Determination of electrolyte leakage and ethylene release rate
The full buds were excised from peach shoots using flat cuts and subsequently placed in a dish with moist blotting paper and cut into small pieces. Approximately 0.2 g of each sample was incubated in 30 mL of ddH2O for 2 h at 25°C with shaking at 200 rpm. The first electrolyte (C1) and second electrolyte leakage were measured using a digital conductivity meter (DDS−307, Rex, China). C2 was achieved after boiling the bud samples at 100°C for 30 min and subsequently cooled down to 25°C with shaking. Relative electrolyte leakage (REL) was calculated as (C1/C2) × 100%. 10 buds of each set were sampled and placed in sealed tube (5 mL), 1 mL of air from the headspace of each tube was withdrawn with a syringe and manually injected into a gas chromatograph (7890A, Agilent Technology, United States). The ethylene release rate in the buds was determined according to the method described by Tian et al. (2013).
Analysis of peach bud transcriptomics at different cold stress times
RNA concentration and purity were determined using the NanoDrop 2000 (Thermo Fisher Scientific, Wilmington, DE, United States). RNA integrity was assessed with the RNA Nano 6000 Assay Kit on the Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, United States). Sequencing libraries were prepared with 1 μg RNA per sample using the NEBNext Ultra TM RNA Library Prep Kit for Illumina (NEB, United States) following the instructions of the manufacturer. The prepared libraries were sequenced on an Illumina platform, yielding clean reads after adapter, poly-N sequence, and low-quality read removal. The high-quality, paired-end clean reads from each sample were aligned to the Prunus_persica.Chinese_Cling_v1.0.genome [Prunus persica genome assembly ASM1834083v1 - NCBI - NLM (nih.gov)] using Hisat2 software. Subsequently, differential gene expression analysis was performed using DESeq2.
Identification of candidate genes using qRT-PCR
Total RNA was extracted from “21SJ” and “SJZX” bud samples at 0, 12, 24, 48, and 72 h using the Plant Total RNA Isolation Kit (SK8631; Sangon Biotech, Shanghai, China), following the instruction of the manufacturer. Subsequently, cDNA synthesis was conducted using the PrimeScript™ RT-PCR Kit (RR047A; TaKaRa Bio, Kusatsu, Japan) and diluted fivefold. Quantitative real-time PCR (qRT-PCR) was performed using ABI QuantStudio™ 6 Flex System (Applied Biosystems). Gene expression levels were calculated using the 2−ΔΔCT method, with three biological replicates per reaction. Primer sequences for the candidate genes are provided in Supplementary Table S5.
Results
Cold resistance and ethylene release rate assessment in “21SJ” and “SJZX” buds
The cold resistance of “21SJ” and “SJZX” buds was measured at different time points under stress using their electrolyte leakage rate (ELR) (Figure 1A). Both cultivars showed increased ELR under cold stress conditions. “21SJ” exhibited an increase from 21.25% to 56.17%, while “SJZX” increased from 18.43% to 45.42%. “21SJ” consistently had significantly higher ELR values compared to “SJZX” from 12 h to 72 h. No significant difference in ELR between “SJZX” at 12 h and 24 h was observed.
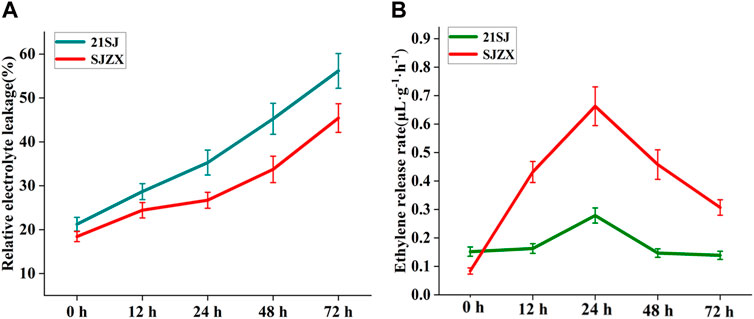
Figure 1. Cold resistance identification and ethylene release rate determination. (A) Electrolyte leakage rates determination for “21SJ” and “SJZX” buds at different time. (B) Ethylene release rate determination for “21SJ” and “SJZX” buds at different time.
To investigate the relationship between ethylene biosynthesis and the different cold resistance of “21SJ” and “SJZX”, ELR was analyzed in these cultivars at different stages of cold stress using gas chromatography (Figure 1B). The findings showed that both cultivars experienced an increase in ELR from 0 h to 24 h during cold stress, reaching a peak at 24 h, followed by a decrease from 24 h to 72 h “SJZX” exhibited a significantly higher ELR compared to “21SJ” from 12 h to 72 h, suggesting a strong link between ethylene biosynthesis and cold resistance in peach buds.
Differential gene expression analysis between “21SJ” and “SJZX” using transcriptomics
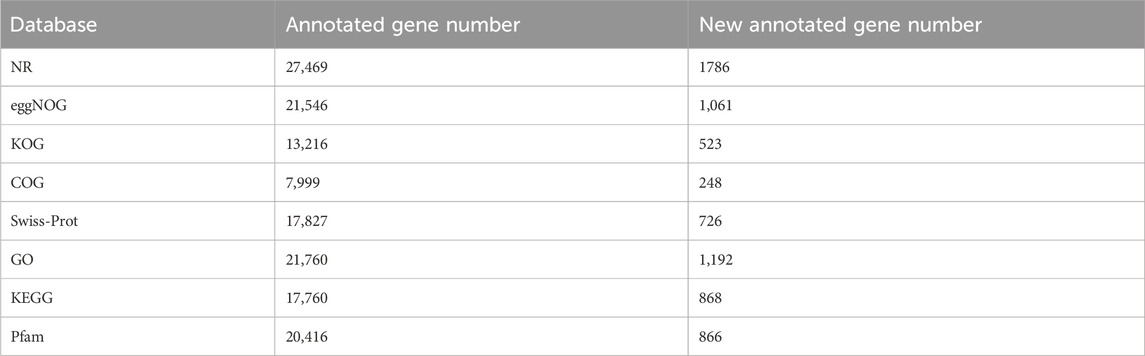
In this study, peach buds subjected to cold stress treatments for 12, 24, and 48 h underwent RNA-Seq to identify candidate genes. After filtering out low-quality raw reads, a total of 123.91 Gb clean data were obtained and deposited in the NCBI Sequence Read Archive (SRA) with accession number PRJNA1071065 and Q30 (%) was 93.58%–95.29% (Supplementary Table S1). These clean reads were aligned to the reference genome sequence using Hisat2 tools, resulting in the identification of 27,506 annotated unigenes through alignment with Nr, eggNOG, KOG, COG, Swiss-Prot, GO, KEGG, and Pfam databases (Table 1; Supplementary Table S2), including 1,807 novel genes (Supplementary Table S3). Expression levels of each unigene are presented in Supplementary Table S4. Correlation coefficients were calculated to assess gene expression consistency (Figure 2A). After this, differential expression analysis revealed significant differences: 526 genes were differentially expressed in 21SJ-12 vs. SJZX-12 (|[log2(fold change)]| >1 and adjusted p < 0.05), with 168 upregulated and 358 downregulated genes. Moreover, 489 genes displayed differential expression in 21SJ-24 vs. SJZX-24, including 293 upregulated and 196 downregulated genes. Furthermore, 489 genes exhibited differential expression in 21SJ-48 vs. SJZX-48, with 197 upregulated and 292 downregulated genes (Figure 2B).
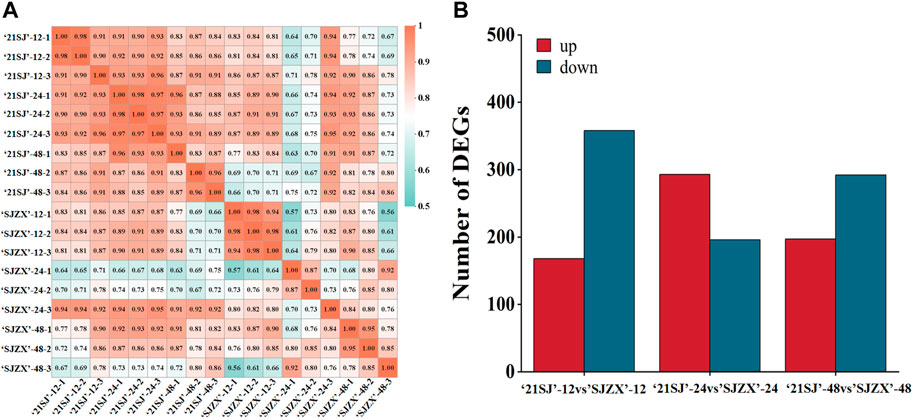
Figure 2. Pearson’s correlation coefficients analysis for all expressed genes and different expressed genes statistics between each group. (A) reliability and rationality between samples based on Pearson’s correlation coefficients for all gene expression levels between each sample. (B) Values plotted in red represent up-regulated genes in “21SJ” vs. “SJZX”; those plotted in blue represent down-regulated genes in “21SJ” vs. “SJZX” and red plot represent up-regulated genes in “21SJ” vs. “SJZX”.
Discovery of candidate structural genes in ethylene signal transduction
Ethylene biosynthesis and signal transduction in plants are complex processes involving several structural genes (Figure 3A). 14 candidate genes were identified from the RNA-Seq data based on their annotation and expression levels in 21SJ-12 vs. SJZX-12, 21SJ-24 vs. SJZX-24, and 21SJ-48 vs. SJZX-48 (Figure 3B).
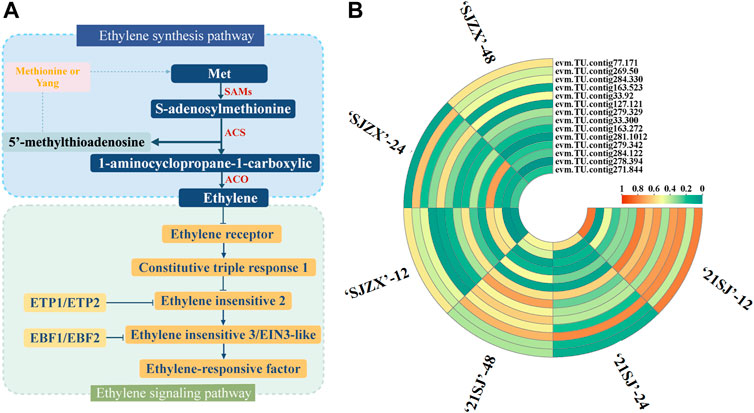
Figure 3. Candidate structural genes in ethylene biosynthesis and signal transduction pathway. (A) Structural genes involved in ethylene biosynthesis and signal transduction pathway in plants. (B) Candidate genes filtration in ethylene biosynthesis and signal transduction pathway based on RNA-seq data. Orange corresponds to highly expressed and blue to poorly expressed.
To further identify candidate genes associated with ethylene biosynthesis and signal transduction during cold stress in peach buds, Pearson correlation coefficients (p < 0.05) were calculated for these 14 candidate genes and ELR (Figure 4A). Subsequently, one structural gene significantly correlated with ELR was identified and designated as PpACO1-1 (evm.TU.contig279.342). The expression of PpACO1-1 was significantly induced under cold stress in “SJZX”, peaking at 24 h. Moreover, the expression level of PpACO1-1 was significantly higher in “SJZX” compared to “21SJ” (Figure 4B).
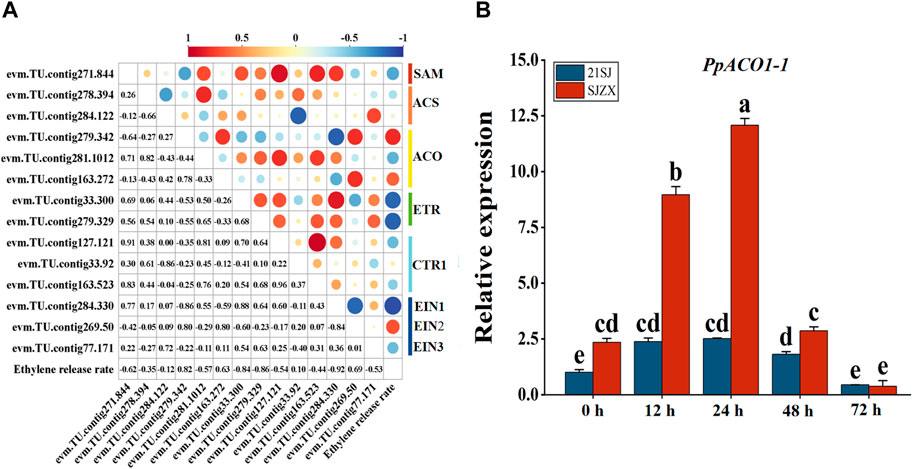
Figure 4. Candidate structural genes discovery and qRT-PCR verification based on Pearson correlation coefficients analysis. Blue bars represent cultivar “21SJ” and red bars represent cultivar “21SJ”. (A) Pearson correlation coefficients analysis for candidate genes and ELR. (B) qRT-PCR verification for PpACO1-1. Error bars represent the standard deviation of three biological replicates. Lowercase letters on the bar chart represent significant differences between the two cultivars at different cold stress stages according to Duncan’s multiple range test at p < 0.05.
Transcription factors influencing ethylene biosynthesis and the PpACO1-1 gene were identified using WGCNA. Different modules represented clusters of genes with high correlation (Figure 5A). Initially, 17 transcription factors linked to ERFs, WRKYs, NACs, and bHLHs were identified (Figure 5B). Subsequently, four candidate transcription factors PpERF2 (evm.TU.contig268.88), PpNAC078 (evm.TU.contig277.535), PpWRKY65 (evm.TU.contig38.293), and PpbHLH112 (evm.TU.contig38.614) were selected based on qRT-PCR (Figures 5C–F). PpERF2, PpNAC078, and PpbHLH112 exhibited upregulation in both “21SJ” and “SJZX” peach buds under cold stress, with significantly higher expression levels in “SJZX” than in “21SJ”. Furthermore, PpWRKY65 showed upregulation in “SJZX” and downregulation in “21SJ,” with its expression level significantly higher in “SJZX” than in “21SJ”.

Figure 5. Transcription factors discovery related to peach buds cold resistance based on weighted gene co-expression network analysis and qRT-PCR validation. (A) Weighted gene co-expression network analysis based on the expression of PpACO1-1 and ethylene release rate. (B) Cluster heat map of differentially expressed transcription factors in ERF, WRKY, NAC and bHLH family. Genes are shown horizontally; samples are represented by columns. Red corresponds to highly expressed genes and green to poorly expressed genes. (C–F) qRT-PCR analysis of selected transcription factors involved in peach buds cold resistance. Blue bars represent cultivar “21SJ” and red bars represent cultivar “SJZX”. Error bars represent the standard deviation of three biological replicates. Lowercase letters on the bar chart represent significant differences between the two cultivars and different developmental stages according to Duncan’s multiple range test at p < 0.05.
Discussion
Relationship between ethylene biosynthesis and plant cold resistance
Ethylene, an important phytohormone, influences plant growth and development, especially in fruit ripening (McMurchie et al., 1972; Iqbal et al., 2017). Low temperatures causing freezing stress significantly affect plant distribution, growth, and yield, including in peach cultivation (Li and Wang, 2020). Several studies have explored the deep mechanisms of plant resistance to cold stress (Ding et al., 2020; Hwarari et al., 2022; Ma et al., 2022). Phytohormones such as ABA, ET, Jasmonic Acid (JA), and Salicylic acid (SA) are pivotal in plant cold resistance (Huang et al., 2017; Yang et al., 2019; Huang et al., 2023; Zhang et al., 2023). Studies have also highlighted their role in peach cold resistance, including ABA, JA, and SA (Zhang et al., 2009; Zhao et al., 2021b; Zhao et al., 2021c). Ethylene enhances cold resistance in various fruit tree species, such as apples, pears, and grapevine (Hershkovitz et al., 2009; Sun et al., 2016; Wang Y. et al., 2021). However, until now, no research has investigated the relationship between ethylene biosynthesis and cold resistance in peaches. The findings in this study identified a positive correlation between ethylene biosynthesis and cold resistance in peach buds.
Plants synthesize ethylene in response to various biotic and abiotic stresses, either inducing or repressing structural genes in the ethylene biosynthesis and signal transduction pathway (Chen et al., 2009; Boutrot et al., 2010; Xu et al., 2019; Zhao Z. X. et al., 2021; Hartman et al., 2021; Wei et al., 2022). Structural genes such as SlACS1A, SlACS1B, SlACO1, and SlACO4 in tomato (Dong et al., 2022), PaACS1, PaACS2, PaACO, and PaCTR1 in pear (Hershkovitz et al., 2009), and VvACO and MdACO1 in apple and grapevine (Sun et al., 2016; Wang Y. et al., 2021), are known for their roles in ethylene biosynthesis under cold stress. However, the key structural gene involved in peach ethylene biosynthesis under cold stress remains unreported. The findings of this study reveal PpACO1-1 (evm.TU.contig279.342) as a key structural gene responding significantly to cold stress in peach buds.
Transcription factors involved in ethylene signaling pathway associated with cold resistance
Extensive literature exists on ethylene signaling pathway genes and transcription factors associated with cold resistance compared to the genes involved in ethylene biosynthesis. ERFs, important transcription factors regulated by ET, play a significant role in plant cold resistance. For instance, PtrERF108 in trifoliate orange regulates raffinose synthesis by modulating PtrRafS expression, PtrERF109 positively regulates POD-encoding genes to scavenge reactive oxygen species (ROS), and PtrERF9 positively modulates ROS homeostasis by regulating PtrGSTU17 expression under cold stress (Wang M. et al., 2019; Khan et al., 2021; Zhang et al., 2022). Additionally, ThERF5, ThERF31, ThERF46, and ThERF55 in Tetrastigma hemsleyanum exhibit a sensitive response to cold stress (Xie et al., 2022). Furthermore, MfERF1 from Medicago falcata enhances cold tolerance through upregulation of polyamine turnover, antioxidant protection, and proline accumulation (Zhuo et al., 2018). MdERF1B in apples enhances cold tolerance by upregulating the expression of the cold-responsive gene MdCBF1 and ethylene biosynthesis gene MdACO1 (Wang Y. et al., 2021). VaERF092 in Amur grape induces cold tolerance calli by regulating VaWRKY33 expression (Sun et al., 2019). The investigation in this study identified an ERF designated PpERF2, which exhibits significantly induced expression in the buds of the cold-resistant cultivar “SJZX”, suggesting its potential function in peach cold resistance.
Transcription factors involved in CBF-COR cold resistance pathway
NACs, WRKYs, and bHLHs represent three major TF families in plants, acting as key regulators that transmit upstream stress signals to downstream stress responses. Transcription factors from these families are primarily involved in the CBF-COR cold resistance pathway, directly binding to the promoter regions of CBFs to enhance their expression. For instance, GmNAC20 in soybean (Hao et al., 2011), PbeNAC1 in pear (Jin et al., 2017), and MdNAC104 in apple (Mei et al., 2023), MdCIbHLH1/MdICE1, MdICE1L and MdbHLH4 in apple (Feng et al., 2012; An et al., 2021; An et al., 2022; Yang et al., 2023), PavbHLH106 and PavbHLH28 in sweet cherry (Cao et al., 2023; Hou et al., 2023), VaWRKY33 in grapevine (Sun et al., 2019), VbWRKY32 in Verbena bonariensis (Wang M. Q. et al., 2019), KoWRKY40 in Kandelia obovate (Fei et al., 2022), CdWRKY2 in bermudagrass (Huang et al., 2022). To date, no research has reported candidate transcription factors from these families involved in peach cold resistance. Three transcription factors, PpNAC078, PpWRKY65, and PpbHLH112, were preliminarily identified in this study. However, further investigation is needed to understand the precise cold resistance mechanisms mediated by these transcription factors.
Conclusion
In the study, cold-sensitive peach cultivar “21SJ” and cold-resistance cultivar “SJZX” were used to discovering peach bud cold resistant genes by using RNA-seq and gas chromatography. In total of 123.91 Gb clean data were achieved based on RNA-seq and 526, 489 and 489 genes were differentially expressed in 21SJ-12 vs. SJZX-12, 21SJ-24 vs. SJZX-24 and 21SJ-48 vs. SJZX-48, respectively. Finally, the ethylene biosynthesis gene, PpACO1-1, was discovered as pivotal in peach bud cold resistance, given its significant response to cold stress in resistant cultivars. Subsequently, four transcription factors PpERF2, PpNAC078, PpWRKY65, and PpbHLH112 were selected based on WGCNA, as they potentially regulate PpACO1-1 expression and ethylene biosynthesis. These findings provide crucial insights for future research and breeding endeavors aimed at bolstering peach cold resistance.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
WX: Investigation, Methodology, Writing–review and editing, Writing–original draft. YY: Formal Analysis, Writing–original draft. CZ: Methodology, Software, Writing–review and editing. CL: Data curation, Formal Analysis, Writing–review and editing. KX: Formal Analysis, Writing–original draft. XX: Funding acquisition, Writing–original draft. JW: Funding acquisition, Writing–original draft. YS: Writing–original draft. LZ: Methodology, Resources, Writing–original draft, Investigation. KS: Writing–original draft, Funding acquisition, Methodology, Resources, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was funded by Science and Technology Project of Hebei Education Department (grant numbers QN2022011); Hebei Natural Science Foundation (grant number C2022407048); Central Guiding Local Science and Technology Development Fund Projects (grant number 236Z6401G); and Modern Agricultural Industry Technology System in Hebei Province (grant number HBCT2024160205, HBCT2024160203). The content of the manuscript have not previously appeared online. Funding bodies were not involved in the design of the study and collection, analysis, interpretation of data and in writing the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2024.1438276/full#supplementary-material
References
Adams, D. O., and Yang, S. F. (1977). Methionine metabolism in apple tissue: implication of s-adenosylmethionine as an intermediate in the conversion of methionine to ethylene. Plant Physiol. 60 (6), 892–896. doi:10.1104/pp.60.6.892
Adams, D. O., and Yang, S. F. (1979). Ethylene biosynthesis: identification of 1-aminocyclopropane-1-carboxylic acid as an intermediate in the conversion of methionine to ethylene. Proc. Natl. Acad. Sci. U. S. A. 76 (1), 170–174. doi:10.1073/pnas.76.1.170
An, J. P., Wang, X. F., Zhang, X. W., You, C. X., and Hao, Y. J. (2021). Apple B-box protein BBX37 regulates jasmonic acid mediated cold tolerance through the JAZ-BBX37-ICE1-CBF pathway and undergoes MIEL1-mediated ubiquitination and degradation. New Phytol. 229 (5), 2707–2729. doi:10.1111/nph.17050
An, J. P., Xu, R. R., Liu, X., Su, L., Yang, K., Wang, X. F., et al. (2022). Abscisic acid insensitive 4 interacts with ICE1 and JAZ proteins to regulate ABA signaling-mediated cold tolerance in apple. J. Exp. Bot. 73 (3), 980–997. doi:10.1093/jxb/erab433
Boutrot, F., Segonzac, C., Chang, K. N., Qiao, H., Ecker, J. R., Zipfel, C., et al. (2010). Direct transcriptional control of the Arabidopsis immune receptor FLS2 by the ethylene-dependent transcription factors EIN3 and EIL1. Proc. Natl. Acad. Sci. U. S. A. 107 (32), 14502–14507. doi:10.1073/pnas.1003347107
Cao, X., Wen, Z., Shen, T., Cai, X., Hou, Q., Shang, C., et al. (2023). Overexpression of PavbHLH28 from Prunus avium enhances tolerance to cold stress in transgenic Arabidopsis. BMC Plant Biol. 23 (1), 652. doi:10.1186/s12870-023-04666-1
Chen, H., Xue, L., Chintamanani, S., Germain, H., Lin, H., Cui, H., et al. (2009). ETHYLENE INSENSITIVE3 and ETHYLENE INSENSITIVE3-LIKE1 repress SALICYLIC ACID INDUCTION DEFICIENT2 expression to negatively regulate plant innate immunity in Arabidopsis. Plant Cell 21 (8), 2527–2540. doi:10.1105/tpc.108.065193
Chen, Y. F., Gao, Z., Kerris, R. J., Wang, W., Binder, B. M., and Schaller, G. E. (2010). Ethylene receptors function as components of high-molecular-mass protein complexes in Arabidopsis. PLoS One 5 (1), e8640. doi:10.1371/journal.pone.0008640
Cheng, M. C., Liao, P. M., Kuo, W. W., and Lin, T. P. (2013). The Arabidopsis ETHYLENE RESPONSE FACTOR1 regulates abiotic stress-responsive gene expression by binding to different cis-acting elements in response to different stress signals. Plant Physiol. 162 (3), 1566–1582. doi:10.1104/pp.113.221911
Chinnusamy, V., Zhu, J., and Zhu, J. K. (2007). Cold stress regulation of gene expression in plants. Trends Plant Sci. 12 (10), 444–451. doi:10.1016/j.tplants.2007.07.002
Ding, Y., Shi, Y., and Yang, S. (2020). Molecular regulation of plant responses to environmental temperatures. Mol. Plant 13 (4), 544–564. doi:10.1016/j.molp.2020.02.004
Dong, Y., Tang, M., Huang, Z., Song, J., Xu, J., Ahammed, G. J., et al. (2022). The miR164a-NAM3 module confers cold tolerance by inducing ethylene production in tomato. Plant J. 111 (2), 440–456. doi:10.1111/tpj.15807
Fei, J., Wang, Y. S., Cheng, H., Su, Y. B., Zhong, Y. J., and Zheng, L. (2022). The Kandelia obovata transcription factor KoWRKY40 enhances cold tolerance in transgenic Arabidopsis. BMC Plant Biol. 22 (1), 274. doi:10.1186/s12870-022-03661-2
Feng, X. M., Zhao, Q., Zhao, L. L., Qiao, Y., Xie, X. B., Li, H. F., et al. (2012). The cold-induced basic helix-loop-helix transcription factor gene MdCIbHLH1 encodes an ICE-like protein in apple. BMC Plant Biol. 12, 22. doi:10.1186/1471-2229-12-22
Gonzalez-Guzman, M., Pizzio, G. A., Antoni, R., Vera-Sirera, F., Merilo, E., Bassel, G. W., et al. (2012). Arabidopsis PYR/PYL/RCAR receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid. Plant Cell 24 (6), 2483–2496. doi:10.1105/tpc.112.098574
Gunaseelan, K., McAtee, P. A., Nardozza, S., Pidakala, P., Wang, R., David, K., et al. (2019). Copy number variants in kiwifruit ETHYLENE RESPONSE FACTOR/APETALA2 (ERF/AP2)-like genes show divergence in fruit ripening associated cold and ethylene responses in C-REPEAT/DRE BINDING FACTOR-like genes. PLoS One 14 (5), e0216120. doi:10.1371/journal.pone.0216120
Guo, Q., Li, X., Niu, L., Jameson, P. E., and Zhou, W. (2021). Transcription-associated metabolomic adjustments in maize occur during combined drought and cold stress. Plant Physiol. 186 (1), 677–695. doi:10.1093/plphys/kiab050
Guo, X., Huang, D., Jing, G., Feng, J., and Zhu, S. (2023). Nitric oxide-mediated DNA methylation enhances cold resistance in postharvest peach fruit. Food Chem. 404 (1), 134660. doi:10.1016/j.foodchem.2022.134660
Hamilton, A. J., Bouzayen, M., and Grierson, D. (1991). Identification of a tomato gene for the ethylene-forming enzyme by expression in yeast. Proc. Natl. Acad. Sci. U. S. A. 88 (16), 7434–7437. doi:10.1073/pnas.88.16.7434
Hao, Y. J., Wei, W., Song, Q. X., Chen, H. W., Zhang, Y. Q., Wang, F., et al. (2011). Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants. Plant J. 68 (2), 302–313. doi:10.1111/j.1365-313X.2011.04687.x
Hartman, S., Sasidharan, R., and Voesenek, L. (2021). The role of ethylene in metabolic acclimations to low oxygen. New Phytol. 229 (1), 64–70. doi:10.1111/nph.16378
Hershkovitz, V., Friedman, H., Goldschmidt, E. E., Feygenberg, O., and Pesis, E. (2009). Induction of ethylene in avocado fruit in response to chilling stress on tree. J. Plant Physiol. 166 (17), 1855–1862. doi:10.1016/j.jplph.2009.05.012
Hou, Q., Shen, T., Yu, R., Deng, H., Wen, X., and Qiao, G. (2023). Functional analysis of sweet cherry PavbHLH106 in the regulation of cold stress. Plant Cell Rep. 43 (1), 7. doi:10.1007/s00299-023-03115-5
Hu, Z., Huang, X., Amombo, E., Liu, A., Fan, J., Bi, A., et al. (2020). The ethylene responsive factor CdERF1 from bermudagrass (Cynodon dactylon) positively regulates cold tolerance. Plant Sci. 294, 110432. doi:10.1016/j.plantsci.2020.110432
Huang, J., Zhao, X., Bürger, M., Chory, J., and Wang, X. (2023). The role of ethylene in plant temperature stress response. Trends Plant Sci. 28 (7), 808–824. doi:10.1016/j.tplants.2023.03.001
Huang, X., Cao, L., Fan, J., Ma, G., and Chen, L. (2022). CdWRKY2-mediated sucrose biosynthesis and CBF-signalling pathways coordinately contribute to cold tolerance in bermudagrass. Plant Biotechnol. J. 20 (4), 660–675. doi:10.1111/pbi.13745
Huang, X., Shi, H., Hu, Z., Liu, A., Amombo, E., Chen, L., et al. (2017). ABA is involved in regulation of cold stress response in bermudagrass. Front. Plant Sci. 8, 1613. doi:10.3389/fpls.2017.01613
Hwarari, D., Guan, Y., Ahmad, B., Movahedi, A., Min, T., Hao, Z., et al. (2022). ICE-CBF-COR signaling cascade and its regulation in plants responding to cold stress. Int. J. Mol. Sci. 23 (3), 1549. doi:10.3390/ijms23031549
Iqbal, N., Khan, N. A., Ferrante, A., Trivellini, A., Francini, A., and Khan, M. I. R. (2017). Ethylene role in plant growth, development and senescence: interaction with other phytohormones. Front. Plant Sci. 8, 475. doi:10.3389/fpls.2017.00475
Jia, Y., Ding, Y., Shi, Y., Zhang, X., Gong, Z., and Yang, S. (2016). The cbfs triple mutants reveal the essential functions of CBFs in cold acclimation and allow the definition of CBF regulons in Arabidopsis. New Phytol. 212 (2), 345–353. doi:10.1111/nph.14088
Jin, C., Li, K. Q., Xu, X. Y., Zhang, H. P., Chen, H. X., Chen, Y. H., et al. (2017). A novel NAC transcription factor, PbeNAC1, of pyrus betulifolia confers cold and drought tolerance via interacting with PbeDREBs and activating the expression of stress-responsive genes. Front. Plant Sci. 8, 1049. doi:10.3389/fpls.2017.01049
Ju, Y. L., Yue, X. F., Min, Z., Wang, X. H., Fang, Y. L., and Zhang, J. X. (2020). VvNAC17, a novel stress-responsive grapevine (Vitis vinifera L.) NAC transcription factor, increases sensitivity to abscisic acid and enhances salinity, freezing, and drought tolerance in transgenic Arabidopsis. Plant Physiol. Biochem. 146, 98–111. doi:10.1016/j.plaphy.2019.11.002
Khan, M., Hu, J., Dahro, B., Ming, R., Zhang, Y., Wang, Y., et al. (2021). ERF108 from Poncirus trifoliata (L.) Raf. functions in cold tolerance by modulating raffinose synthesis through transcriptional regulation of PtrRafS. Plant J. 108 (3), 705–724. doi:10.1111/tpj.15465
Klingler, J. P., Batelli, G., and Zhu, J. K. (2010). ABA receptors: the START of a new paradigm in phytohormone signalling. J. Exp. Bot. 61 (12), 3199–3210. doi:10.1093/jxb/erq151
Kopeć, P., Rapacz, M., and Arora, R. (2022). Post-translational activation of CBF for inducing freezing tolerance. Trends Plant Sci. 27 (5), 415–417. doi:10.1016/j.tplants.2022.01.003
Lee, Y., Do, V. G., Kim, S., Kweon, H., and McGhie, T. K. (2021). Cold stress triggers premature fruit abscission through ABA-dependent signal transduction in early developing apple. PLoS One 16 (4), e0249975. doi:10.1371/journal.pone.0249975
Li, M. Y., Liu, J. X., Hao, J. N., Feng, K., Duan, A. Q., Yang, Q. Q., et al. (2019). Genomic identification of AP2/ERF transcription factors and functional characterization of two cold resistance-related AP2/ERF genes in celery (Apium graveolens L.). Planta 250 (4), 1265–1280. doi:10.1007/s00425-019-03222-2
Li, Y., Tian, Q., Wang, Z., Li, J., Liu, S., Chang, R., et al. (2023). Integrated analysis of transcriptomics and metabolomics of peach under cold stress. Front. Plant Sci. 14, 1153902. doi:10.3389/fpls.2023.1153902
Li, Y., and Wang, L. (2020). Genetic Resources, breeding programs in China, and gene mining of peach: a review. Hortic. Plant J. 6 (4), 205–215. doi:10.1016/j.hpj.2020.06.001
Lim, C. W., and Lee, S. C. (2020). ABA-dependent and ABA-independent functions of RCAR5/PYL11 in response to cold stress. Front. Plant Sci. 11, 587620. doi:10.3389/fpls.2020.587620
Lorenzo, O., Piqueras, R., Sánchez-Serrano, J. J., and Solano, R. (2003). ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15 (1), 165–178. doi:10.1105/tpc.007468
Ma, T., Wang, S., Sun, C., Tian, J., Guo, H., Cui, S., et al. (2023). Arabidopsis LFR, a SWI/SNF complex component, interacts with ICE1 and activates ICE1 and CBF3 expression in cold acclimation. Front. Plant Sci. 14, 1097158. doi:10.3389/fpls.2023.1097158
Ma, X., Zhao, F., Su, K., Lin, H., and Guo, Y. (2022). Discovery of cold-resistance genes in Vitis amurensis using bud-based quantitative trait locus mapping and RNA-seq. BMC Genomics 23 (1), 551. doi:10.1186/s12864-022-08788-y
Ma, Y., Szostkiewicz, I., Korte, A., Moes, D., Yang, Y., Christmann, A., et al. (2009). Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324 (5930), 1064–1068. doi:10.1126/science.1172408
McMurchie, E. J., McGlasson, W. B., and Eaks, I. L. (1972). Treatment of fruit with propylene gives information about the biogenesis of ethylene. Nature 237 (5352), 235–236. doi:10.1038/237235a0
Mei, C., Yang, J., Mei, Q., Jia, D., Yan, P., Feng, B., et al. (2023). MdNAC104 positively regulates apple cold tolerance via CBF-dependent and CBF-independent pathways. Plant Biotechnol. J. 21 (10), 2057–2073. doi:10.1111/pbi.14112
Nakashima, K., Fujita, Y., Kanamori, N., Katagiri, T., Umezawa, T., Kidokoro, S., et al. (2009). Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 50 (7), 1345–1363. doi:10.1093/pcp/pcp083
Park, S., Lee, C. M., Doherty, C. J., Gilmour, S. J., Kim, Y., and Thomashow, M. F. (2015). Regulation of the Arabidopsis CBF regulon by a complex low-temperature regulatory network. Plant J. 82 (2), 193–207. doi:10.1111/tpj.12796
Pattyn, J., Vaughan-Hirsch, J., and Van de Poel, B. (2021). The regulation of ethylene biosynthesis: a complex multilevel control circuitry. New Phytol. 229 (2), 770–782. doi:10.1111/nph.16873
Pons, C., Martí, C., Forment, J., Crisosto, C. H., Dandekar, A. M., and Granell, A. (2014). A bulk segregant gene expression analysis of a peach population reveals components of the underlying mechanism of the fruit cold response. PLoS One 9 (3), e90706. doi:10.1371/journal.pone.0090706
Shakeel, S. N., Gao, Z., Amir, M., Chen, Y. F., Rai, M. I., Haq, N. U., et al. (2015). Ethylene regulates levels of ethylene receptor/CTR1 signaling complexes in Arabidopsis thaliana. J. Biol. Chem. 290 (19), 12415–12424. doi:10.1074/jbc.M115.652503
Shen, X., Song, Y., Ping, Y., He, J., Xie, Y., Ma, F., et al. (2023). The RNA-binding protein MdHYL1 modulates cold tolerance and disease resistance in apple. Plant Physiol. 192 (3), 2143–2160. doi:10.1093/plphys/kiad187
Shu, P., Li, Y., Sheng, J., and Shen, L. (2023). SlMAPK3 positively regulates the ethylene production of postharvest tomato fruits and is involved in ethylene-mediated cold tolerance. J. Agric. Food Chem. doi:10.1021/acs.jafc.2c08723
Shu, Y., Zhang, W., Tang, L., Li, Z., Liu, X., Liu, X., et al. (2023). ABF1 positively regulates rice chilling tolerance via inducing trehalose biosynthesis. Int. J. Mol. Sci. 24 (13), 11082. doi:10.3390/ijms241311082
Song, Z., Lai, X., Chen, H., Wang, L., Pang, X., Hao, Y., et al. (2022). Role of MaABI5-like in abscisic acid-induced cold tolerance of 'Fenjiao' banana fruit. Hortic. Res. 9, uhac130. doi:10.1093/hr/uhac130
Soon, F. F., Ng, L. M., Zhou, X. E., West, G. M., Kovach, A., Tan, M. H., et al. (2012). Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases. Science 335 (6064), 85–88. doi:10.1126/science.1215106
Sun, X., Zhang, L., Wong, D. C. J., Wang, Y., Zhu, Z., Xu, G., et al. (2019). The ethylene response factor VaERF092 from Amur grape regulates the transcription factor VaWRKY33, improving cold tolerance. Plant J. 99 (5), 988–1002. doi:10.1111/tpj.14378
Sun, X., Zhao, T., Gan, S., Ren, X., Fang, L., Karungo, S. K., et al. (2016). Ethylene positively regulates cold tolerance in grapevine by modulating the expression of ETHYLENE RESPONSE FACTOR 057. Sci. Rep. 6, 24066. doi:10.1038/srep24066
Tan, D., Li, T., and Wang, A. (2013). Apple 1-aminocyclopropane-1-carboxylic acid synthase genes, MdACS1 and MdACS3a, are expressed in different systems of ethylene biosynthesis. Plant Mol. Biol. Rep. 31 (1), 204–209. doi:10.1007/s11105-012-0490-y
Thomashow, M. F. (1999). PLANT COLD ACCLIMATION: freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 571–599. doi:10.1146/annurev.arplant.50.1.571
Wang, K., Yin, X. R., Zhang, B., Grierson, D., Xu, C. J., and Chen, K. S. (2017). Transcriptomic and metabolic analyses provide new insights into chilling injury in peach fruit. Plant Cell Environ. 40 (8), 1531–1551. doi:10.1111/pce.12951
Wang, M., Dai, W., Du, J., Ming, R., Dahro, B., and Liu, J. H. (2019). ERF109 of trifoliate orange (Poncirus trifoliata (L.) Raf.) contributes to cold tolerance by directly regulating expression of Prx1 involved in antioxidative process. Plant Biotechnol. J. 17 (7), 1316–1332. doi:10.1111/pbi.13056
Wang, M. Q., Huang, Q. X., Lin, P., Zeng, Q. H., Li, Y., Liu, Q. L., et al. (2019). The overexpression of a transcription factor gene VbWRKY32 enhances the cold tolerance in Verbena bonariensis. Front. Plant Sci. 10, 1746. doi:10.3389/fpls.2019.01746
Wang, X., Song, Q., Guo, H., Liu, Y., Brestic, M., and Yang, X. (2023). StICE1 enhances plant cold tolerance by directly upregulating StLTI6A expression. Plant Cell Rep. 42 (1), 197–210. doi:10.1007/s00299-022-02949-9
Wang, Y., Jiang, H., Mao, Z., Liu, W., Jiang, S., Xu, H., et al. (2021). Ethylene increases the cold tolerance of apple via the MdERF1B-MdCIbHLH1 regulatory module. Plant J. 106 (2), 379–393. doi:10.1111/tpj.15170
Wang, Y., Xin, H., Fan, P., Zhang, J., Liu, Y., Dong, Y., et al. (2021). The genome of Shanputao (Vitis amurensis) provides a new insight into cold tolerance of grapevine. Plant J. 105 (6), 1495–1506. doi:10.1111/tpj.15127
Wang Z., , Wong, D. C. J., Wang, Y., Xu, G., Ren, C., Liu, Y., et al. (2021). GRAS-domain transcription factor PAT1 regulates jasmonic acid biosynthesis in grape cold stress response. Plant Physiol. 186 (3), 1660–1678. doi:10.1093/plphys/kiab142
Wei, Y., Zhu, B., Ma, G., Shao, X., Xie, H., Cheng, X., et al. (2022). The coordination of melatonin and anti-bacterial activity by EIL5 underlies ethylene-induced disease resistance in cassava. Plant J. 111 (3), 683–697. doi:10.1111/tpj.15843
Xie, Z., Yang, C., Liu, S., Li, M., Gu, L., Peng, X., et al. (2022). Identification of AP2/ERF transcription factors in Tetrastigma hemsleyanum revealed the specific roles of ERF46 under cold stress. Front. Plant Sci. 13, 936602. doi:10.3389/fpls.2022.936602
Xu, L., Xiang, G., Sun, Q., Ni, Y., Jin, Z., Gao, S., et al. (2019). Melatonin enhances salt tolerance by promoting MYB108A-mediated ethylene biosynthesis in grapevines. Hortic. Res. 6, 114. doi:10.1038/s41438-019-0197-4
Yang, J., Duan, G., Li, C., Liu, L., Han, G., Zhang, Y., et al. (2019). The crosstalks between jasmonic acid and other plant hormone signaling highlight the involvement of jasmonic acid as a core component in plant response to biotic and abiotic stresses. Front. Plant Sci. 10, 1349. doi:10.3389/fpls.2019.01349
Yang, J., Guo, X., Mei, Q., Qiu, L., Chen, P., Li, W., et al. (2023). MdbHLH4 negatively regulates apple cold tolerance by inhibiting MdCBF1/3 expression and promoting MdCAX3L-2 expression. Plant Physiol. 191 (1), 789–806. doi:10.1093/plphys/kiac512
Yu, D. J., Jun, S. H., Park, J., Kwon, J. H., and Lee, H. J. (2020). Transcriptome analysis of genes involved in cold hardiness of peach tree (Prunus persica) shoots during cold acclimation and deacclimation. Genes (Basel) 11 (6), 611. doi:10.3390/genes11060611
Yu, W., Ma, P., Sheng, J., and Shen, L. (2023). Postharvest fruit quality of tomatoes influenced by an ethylene signaling component during long-term cold storage. Food Chem. 422, 136087. doi:10.1016/j.foodchem.2023.136087
Zhang, M., Leng, P., Zhang, G., and Li, X. (2009). Cloning and functional analysis of 9-cis-epoxycarotenoid dioxygenase (NCED) genes encoding a key enzyme during abscisic acid biosynthesis from peach and grape fruits. J. Plant Physiol. 166 (12), 1241–1252. doi:10.1016/j.jplph.2009.01.013
Zhang, M., Zhang, M., Wang, J., Dai, S., Zhang, M., Meng, Q., et al. (2023). Salicylic acid regulates two photosystem II protection pathways in tomato under chilling stress mediated by ETHYLENE INSENSITIVE 3-like proteins. Plant J. 114 (6), 1385–1404. doi:10.1111/tpj.16199
Zhang, Y., Ming, R., Khan, M., Wang, Y., Dahro, B., Xiao, W., et al. (2022). ERF9 of Poncirus trifoliata (L.) Raf. undergoes feedback regulation by ethylene and modulates cold tolerance via regulating a glutathione S-transferase U17 gene. Plant Biotechnol. J. 20 (1), 183–200. doi:10.1111/pbi.13705
Zhao, H., Yin, C. C., Ma, B., Chen, S. Y., and Zhang, J. S. (2021a). Ethylene signaling in rice and Arabidopsis: new regulators and mechanisms. J. Integr. Plant Biol. 63 (1), 102–125. doi:10.1111/jipb.13028
Zhao, Y., Song, C., Brummell, D. A., Qi, S., Lin, Q., and Duan, Y. (2021b). Jasmonic acid treatment alleviates chilling injury in peach fruit by promoting sugar and ethylene metabolism. Food Chem. 338, 128005. doi:10.1016/j.foodchem.2020.128005
Zhao, Y., Song, C., Qi, S., Lin, Q., and Duan, Y. (2021c). Jasmonic acid and salicylic acid induce the accumulation of sucrose and increase resistance to chilling injury in peach fruit. J. Sci. Food Agric. 101 (10), 4250–4255. doi:10.1002/jsfa.11064
Zhao, Z. X., Feng, Q., Liu, P. Q., He, X. R., Zhao, J. H., Xu, Y. J., et al. (2021). RPW8.1 enhances the ethylene-signaling pathway to feedback-attenuate its mediated cell death and disease resistance in Arabidopsis. New Phytol. 229 (1), 516–531. doi:10.1111/nph.16857
Keywords: peach buds, cold resistance, ethylene, transcription factor, regulatory mechanism, candidate gene
Citation: Xia W, Yang Y, Zhang C, Liu C, Xiao K, Xiao X, Wu J, Shen Y, Zhang L and Su K (2024) Discovery of candidate genes involved in ethylene biosynthesis and signal transduction pathways related to peach bud cold resistance. Front. Genet. 15:1438276. doi: 10.3389/fgene.2024.1438276
Received: 25 May 2024; Accepted: 08 July 2024;
Published: 18 July 2024.
Edited by:
Xiyang Zhao, Jilin Agricultural University, ChinaReviewed by:
Tangren Cheng, Beijing Forestry University, ChinaBa Liangjie, Guiyang University, China
Copyright © 2024 Xia, Yang, Zhang, Liu, Xiao, Xiao, Wu, Shen, Zhang and Su. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kai Su, Z3JhcGVzdWthaUAxMjYuY29t
 Wenqian Xia1
Wenqian Xia1 Chenguang Zhang
Chenguang Zhang Kun Xiao
Kun Xiao Xiao Xiao
Xiao Xiao Kai Su
Kai Su