
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 18 July 2024
Sec. Genomics of Plants and the Phytoecosystem
Volume 15 - 2024 | https://doi.org/10.3389/fgene.2024.1432376
This article is part of the Research Topic Genetic Dissection and Improvement of Crop Quality and Stress Adaptation View all 3 articles
The Epidermal Patterning Factor/EPF-like (EPF/EPFL) family encodes a specific type of secreted protein in plants and plays an important role in plant growth and development, especially in the process of morphogenesis. To investigate the characteristics of EPF/EPFL gene family members and their regulatory functions in stomatal development of Populus trichocarpa, a total of 15 EPF/EPFL family genes were identified. Then the gene structure, chromosome location, phylogenetic relationship, protein conserved domain and gene expression profile were analyzed. According to phylogenetic analysis, PtEPF/EPFL can be classified into four groups. The gene structure and protein conservation motifs within the EPF family indicate the high conservation of the PtEPF/EPFL sequence. The promoter region of PtEPF/EPFL was found to contain cis-elements in response to stress and plant hormones. In addition, RT-qPCR results indicated that the PtEPF/EPFL have a differentially expressed in different tissues. Under drought stress treatment, a substantial upregulation was observed in the majority of PtEPF/EPFL members, suggesting their potential involvement in drought response. These results provide a theoretical basis for future exploration of the characteristics and functions of more PtEPF/EPFL genes.
Drought stress is the most important environmental constraint affecting plant growth and distribution (Zhu, 2002; Kissoudis et al., 2016). The majority of water absorbed by plants is lost to the air via stomata and reducing the transpiration water loss through stomata is one of the effective strategy to improve plant drought resistance (Lawson and Blatt, 2014). Stomata play a crucial role in regulating both carbon dioxide intake and water loss in plants, which determines the productivity and water use efficiency (WUE) of plants (Wang et al., 2016; Buckley et al., 2019). By changing their stomata, plants could optimize the absorption of carbon dioxide by photosynthesis while minimizing water loss, thus drought tolerance and WUE of plants were improved (Bertolino et al., 2019; Zhao et al., 2020).
The development of stomata is a complex process, which involving the regulation of signal transduction, cell fate transformation and asymmetric division. Researches have revealed that the EPIDERMAL PATTERNING FACTOR (EPF) family consists of a group of cysteine-rich secreted peptides and plays a prominent role in stomatal development (Kondo et al., 2010; Katsir et al., 2011). The EPF family in Arabidopsis consists of 11 members, including EPF1, EPF2, and EPFL1-9. Mature EPF peptides typically exhibit conserved six to eight cysteine residues at the C-terminal, which form intramolecular disulfide bonds that are important for maintaining their spatial structure (Ohki et al., 2011). Some members of the EPF family are involved in regulating stomatal density and patterning within the plant epidermis. AtEPF1 and AtEPF2 have been identified as negative regulators of stomatal development. Although the amino acid sequences of AtEPF1 and AtEPF2 are highly similar, they show different functions in stomatal development. AtEPF2 regulates the differentiation of pro-epidermal cells into meristemoid mother cell (MMC), while AtEPF1 regulates the direction of spacing division and produces satellite meristem (Hara et al., 2007; Hara et al., 2009; Hunt and Gray, 2009). AtEPFL9, also known as Stomagen, functions as a positive regulator of leaf stomatal density while exerting an antagonistic effect on AtEPF1 and AtEPF2 (Hunt et al., 2010; Sugano et al., 2010). As an important member of the EPF family, the CHAL subfamily has similar functions to EPF1 and EPF2 and is a negative regulator of stomatal development (Abrash and Bergmann, 2010). In addition, EPF genes are also involved in the regulation of plant inflorescence structure, leaf morphology, pollen tube elongation and awn development (Abrash et al., 2011; Uchida et al., 2012; Xiong et al., 2022).
Genetic and physiological analysis demonstrated that EPF genes have great potential in improving plant WUE and drought tolerance by regulating stomata development. It was found that overexpression EPF1 reduced stomatal density and improved WUE and drought tolerance of plants in barley, rice, and wheat (Hughes et al., 2017; Caine et al., 2019; Dunn et al., 2019). Overexpression of StEPF2 and PdEPF2 in Arabidopsis leads to a significant reduction in stomatal density. However, under drought stress conditions, these plants maintain higher photosynthetic rates, photosystem II efficiency, and instantaneous WUE compared to wild-type (WT) plants (Wang et al., 2016; Wang et al., 2020). Similar results were also obtained in Arabidopsis (Yu et al., 2008; Franks et al., 2015), maize (Xiang et al., 2021), tomato (Li et al., 2019; Li et al., 2020) and poplar (Wang et al., 2016; Li et al., 2021). Recent studies have revealed the crucial functions performed by members of the EPF gene family in the growth and development of plants. Overexpression of TaEPFL1 can shorten the filament of transgenic Arabidopsis thaliana and cause abnormal development of horn fruit, thus affecting the development of male stamens (Sun et al., 2019). EPFL2 and EPFL9 contribute to the coordination of ovule patterning, influencing seed number alongside gynoecium and fruit growth via the ERECTA family receptors (Kawamoto et al., 2020). These studies suggest that genes regulating stomatal development are potential candidate genes for improving plants WUE by reducing stomatal density.
Poplar is one of the most important plantation species in temperate regions of the world. It has the advantages of fast growth rate, strong adaptability, early timber formation and large wood accumulation, etc. With the completion of whole genome sequencing of Populus trichocarpa and Populus euphratica (Ma et al., 2013; Zhang et al., 2014), poplar has become an ideal woody model plant. EPF gene family is an important factor in the regulation of stomatal development. In this study, the identification of the poplar EPF/EPFL gene family, covering aspects such as physicochemical property analysis, chromosomal location, phylogenetic analysis, gene structure, conserved domains, and cis-regulatory elements in the promoter. Furthermore, the study explored the tissue-specific expression patterns and transcriptional responses to drought stress and abscisic acid (ABA) of this gene family. Identification and analysis of EPF/EPFL family gene phenotype in poplar are helpful to explore its role in stomatal development, which provided a theoretical basis for breeding poplar varieties with high WUE and fast growth rate.
P. trichocarpa genome and annotation information were downloaded from the Ensembl Plants database (https://plants.ensembl.org). The genome data for the 11 EPF family in A. thaliana were downloaded from TAIR (https://www.arabidopsis.org). Download genome data from the Phytozome (v4.1) database (https://phytozome-next.jgi.doe.gov/), which includes genomes of Physcomitrella patens, Selaginella moellendorffii, Sorghum bicolor, Medicago truncatula, Carica papaya, Picea abies, and Oryza sativa.
The EPF Hidden Markov Model (HMM) profile of the EPF domain (pfam17181) was used to blast against the P. trichocarpa reference genome to identify EPF genes. A total of 15 putative EPF-encoding genes were identified. Physicochemical properties such as CDS and protein lengths, molecular weights (MW), isoelectric points (IP), and hydrophilicity analysis were forecasted using ExPasy ProtParam (https://web.expasy.org/protparam/). SingnalP-4.1 was used to predict the signal peptide of PtEPF/EPFL protein. Using the online website WoLFPOSRT (https://wolfpsort.hgc.jp/) for subcellular localization prediction. According to the gff3 format annotation file downloaded from the poplar genome database, the size of each member chromosome and the gene location information of each member were obtained. Then the chromosomal localization and syntenic analysis were performed using TBtools software (Chen et al., 2020).
To identify EPF genes across various species, TBLASTN searches were conducted against genomic databases using amino acid sequences encoded by AtEPF/EPFL genes as references. Sequence comparisons were performed using MEGA7, and a phylogenetic tree was constructed using the Neighbor-Joining (NJ) method with 1,000 repetitions. The gene structure was mapped in TBTools based on the downloaded genome annotation information. The protein conserved motifs of PtEPF/EPFL gene family were predicted using the MEME online program (https://meme-suite.org), with default algorithm parameters (Bailey et al., 2015). The maximum number of motifs was set to 10 and visualized using TBtools software (Chen et al., 2020)
Based on the identification of the obtained gene IDs, the promoter sequences (2,000 bp upstream of the start codon) of PtEPF/EPFL were extracted by TBtools software. The promoter cis-acting elements of PtEPF/EPFL genes were predicted by PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) to analyze the cis-elements species, number and function.
Uniformly growing one-year-old P. trichocarpa trees were selected for this study. Plants grown in the nursery at Beijing Forestry University, Beijing, China (40°000N, 116°200E; 49 m above sea level), in loamy sandy soil (pH = 6.0). Three seedlings with good growth were selected for biological replicates in each pot. For drought treatment, water was withheld from plants for 0, 3, 6, 9, or 12 days. The control group was watered once every 3 days. And young leaves (the first one to three leaves from the shoot apex) were harvested. ABA treatment involved spraying the leaves once with a 250 µM ABA solution, followed by the collection of young leaves at 0, 3, 6, 9, or 12 h post-treatment. Various tissues of one-year-old Populus L., including young leaves, mature leaves, senescent leaves, stems, and roots, were collected. All collected samples were promptly frozen in liquid nitrogen and stored at −80°C for subsequent RNA isolation.
Total RNA was extracted using CTAB method, and reverse transcription was carried out according to the instructions of the FastKing RT kit (Tiangen, Beijing, China). The cDNA was used as a template for quantitative real-time polymerase chain reaction (qRT-PCR). Quantitative (R) RT-PCRs were conducted in 96-well plates, each containing 100 ng of template (1 µL), 0.6 µL of forward primer (10 µM), 0.6 µL of reverse primer (10 µM), 10 µL of SYBR Green Master Mix (Tiangen, Beijing, China), 2 µL of ROXTM Reference Dye, and 5.8 µL of RNase-free ddH2O, resulting in a total reaction volume of 20 µL. The amplification was carried out for 40 cycles, with denaturation at 95°C for 10 s, annealing at 55°C for 30 s, and extension at 72°C for 30 s. The reactions were performed in biological triplicates using RNA samples extracted from three independent plant materials, and all experiments were repeated three times. PtUBQ was used as the internal reference gene. Transcript levels of all candidate genes were determined using the 2-△△CT method, and heatmap visualization was conducted using TBtools (Chen et al., 2020). Primers were designed using Primer6 software, as listed in Supplementary Table S1.
All statistical analyses and plots were conducted using SPSS (IBM Corporation, Armonk, NY, United States). Statistical comparisons were performed using one-way analysis of variance (ANOVA) in SPSS. Different letters denote significant differences at a significance level of α = 0.05 (one-way ANOVA). Data were presented as mean ± standard error (SE).
The gene family was discovered through a search using information probes comprising 11 members of the EPF/EPFL family in Arabidopsis, along with the conserved domain of the EPF family (pfam17181), resulting in the identification of 15 EPF/EPFL members in poplar. Detailed information on the gene ID, CDs size, amino acid length, isoelectric point (PI), molecular weight, signal peptide, prediction of subcellular location, and other characteristics of each PtEPF/EPFL family member is provided. As shown in Table 1, all EPF/EPFL family proteins are relatively short and are classified as secretory polypeptides, with lengths ranging from 107 aa to 177 aa. Their molecular weights vary from 11.98 to 19.80 kDa, and their isoelectric points range from 4.65 to 10.06. The nomenclature of the PtEPF/EPFL gene family was based on the sequences of 11 members of the EPF/EPFL family genes in Arabidopsis.
The chromosomal localization analysis revealed that the 15 PtEPF/EPFL genes were dispersed across ten chromosomes of P. trichocarpa, as depicted in Figure 1. The distribution of PtEPF/EPFL genes across chromosomes varied, with an uneven range from one to three genes per chromosome. Chromosome three exhibited the highest distribution with three genes, followed by chromosomes 2, 3, and 9, each containing two genes. Additionally, chromosomes 5, 7, 8, 10, and 18 harbored one PtEPF/EPFL gene each. To characterize the expansion patterns and replication relationships of the PtEPF/EPFL genes, a syntenic analysis of the family genes was performed using the TBtools software. A total of seven colinear gene pairs were identified, including two pairs on chromosome 1/3, one pair on 5/7, one pair on 8/10, one pair on 11/13, and two pairs on 13/19 (Figure 2).
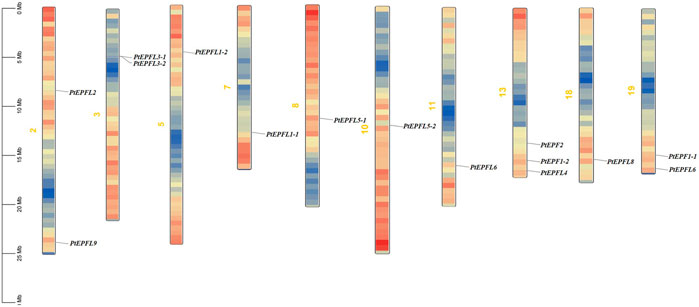
Figure 1. Chromosomal localization of PtEPF/EPFL family in poplar. Size of each chromosome and gene position can be estimated according to the scale on the left of the figure. Chromosome colors represent gene abundance.
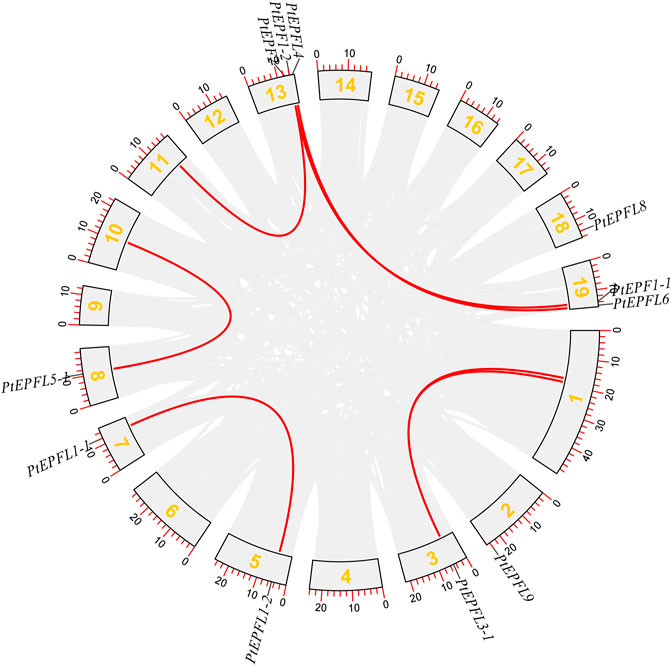
Figure 2. Syntenic analysis of PtEPF/EPFL genes. Gray lines indicate all collinearity blocks in the poplar genome, red lines indicates segmental duplicated PtEPF/EPFL gene pairs.
To investigate the evolutionary relationship of EPF family genes, we selected various representative plant EPF family genes to construct an evolutionary tree. There are 11 AtEPF family members in Arabidopsis, and 9, 8, 7, 13, 10, 8, 10, and 15 EPF family members in P. patens, S. moellendorffii, P. abies, O. sativa, S. bicolor, C. papaya, and P. trichocarpa, respectively. Using MEGA7 and the Nei M. truncatula ghbor-Joining (NJ) method, we conducted multiple sequence alignment of full-length EPF proteins from nine species, and a phylogenetic tree was constructed. The evolutionary tree showed that the EPF gene family can be divided into four groups: EPFL9/Stomagen branch (yellow line), EPF1-EPF2-EPFL7 branch (blue line), EPFL4-6 branch (red line), and EPFL1-3-EPFL8 branch (green line) (Figure 3). Among them, P. patens exists only in the branches of EPF1-EPF2-EPFL7 and EPFL4-6, while there were no members of the branches of EPFL9/Stomagen and EPFL1-3-EPFL8, suggesting that these two branches may have originated after pteridophytes.
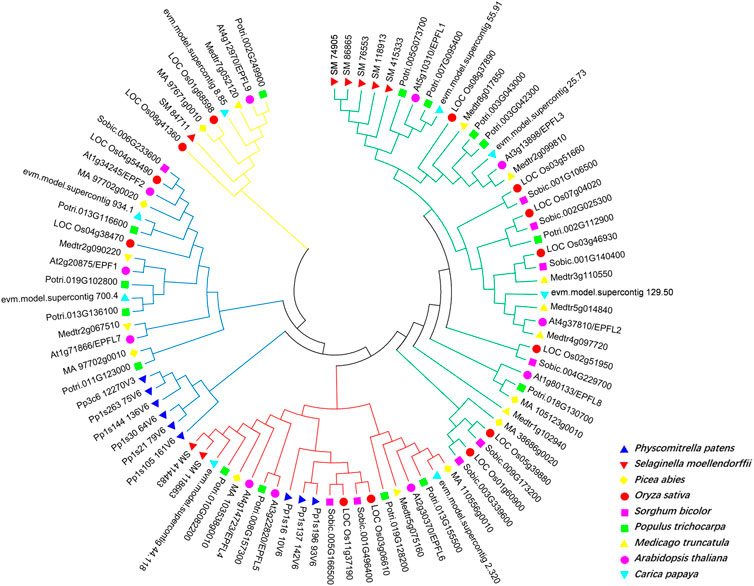
Figure 3. Phylogenetic analysis of EPF family genes in Physcomitrella patens, Selaginella moellendorffii, Picea abies, Oryza sativa, Sorghum bicolor, Populus trichocarpa, Medicago truncatula, A. thaliana and Carica papaya. Different species are indicated by different colors.
Phylogenetic tree analysis showed that EPF1-EPF2-EPFL7 was closely related to EPFL9/Stomagen, and amino acid sequences of the primary protein structure of the two branches were compared. As shown in Figure 4, both the EPF1-EPF2-EPFL7 branch and the EPFL9/Stomagen branch have six conserved cysteine residues in the C-terminal region of the mature peptide, and in the EPF1-EPF2-EPFL7 branch, in addition to the P. patens, other species also have two additional conserved cysteine residues.
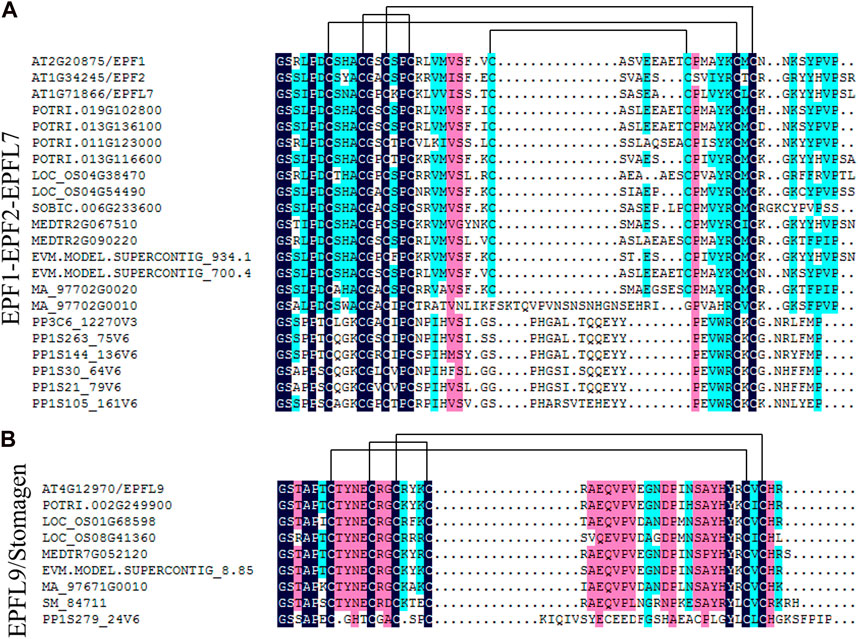
Figure 4. Analysis of conserved of EPF1-EPF2-EPFL7 clade (A) and EPFL9/Stomagen clade (B) in land plants. Sequence alignment was generated by the DNAMAN. Pairs of cysteine residues forming disulfide bonds predicted for EPF/EPFL family genes are indicated by lines.
Gene structure analysis revealed that the PtEPF/EPFL gene family exhibits a range of exon numbers, from 2 to 5. And the PtEPFL3-1 had the highest number of exons. Moreover, the lengths of the 5′-UTR or 3′-UTR varied among different PtEPF/EPFL members (Figure 5A). The results of conserved motif analysis (Figure 5B) showed that PtEPF/EPFL proteins all contained five motif structures, some motif structures were relatively conserved in family genes, and some motifs showed varying degrees of specificity among family members. The PtEPFL9, which correspond to the only positive regulator of stomatal development in Arabidopsis, AtEPFL9, exhibited the lowest conservation compared to other PtEPF/EPFL family members. The difference of protein conserved motifs may be the cause of functional diversity of poplar EPF/EPFL family.
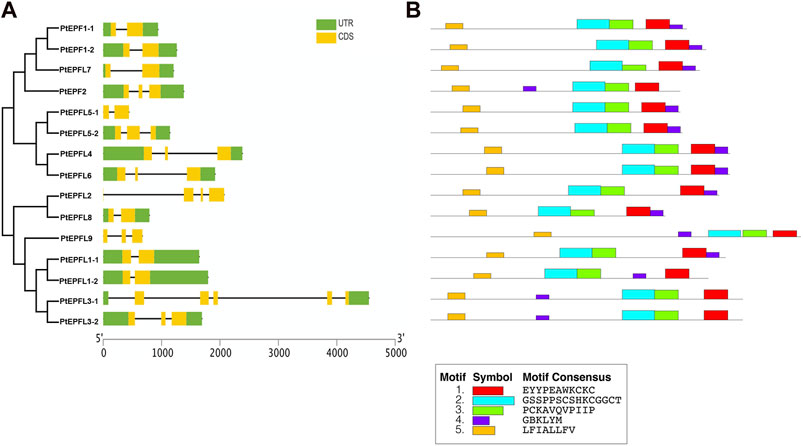
Figure 5. Gene structure and conserved protein motifs analysis of PtEPF/EPFL. (A) Exon–intron structures of PtEPF/EPFL. (B) Conserved motifs of PtEPF/EPFL.
To predict the cis-acting elements among the PtEPF/EPFL, the 2,000 bp upstream promoter sequence of EFP genes was collected and analyzed using PlantCARE. As shown in Figure 6, the cis-acting elements associated with hormonal response, stress response elements, light response, growth and development-related elements, site binding and other functional categories. The hormonal response elements include salicylic acid response elements (SARE and TCA-element), the ABA response element (ABRE), the auxin response elements (AuxRR-core and TGA-element), the P-box and the gibberellin response elements (GARE-motif), CGTCA-motif and TGACG-motif related to jasmonic acid (MeJA). Additionally, the GC-motif, low temperature response element (LTR), the defense and stress response element (TC-rich repeats), the anaerobic induction element (ARE) and the MYB binding site involved in light responsiveness (MBS) were associated with stress responsiveness. Growth and development-related elements comprise the GCN4_motif related to endosperm expression, the CAT-box associated with meristem expression, and the zein metabolism regulation (O2-site). Light responsive elements include G-box, 3-AF1 binding site and GT1-motif.
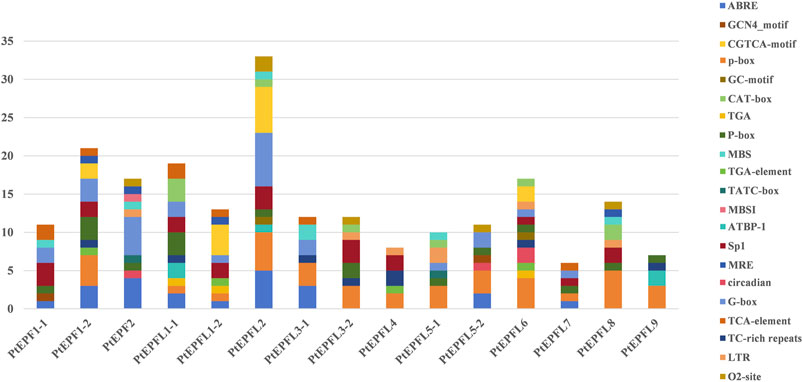
Figure 6. Cis-acting elements were predicted within the 2,000 bp promoter regions of PtEPF/EPFL, with different cis-acting elements indicated by differently coloured boxes. Names of cis-acting elements are shown on the right.
To explore the potential biological roles of PtEPF/EPFL, qRT-PCR was employed to assess the relative transcription levels of PtEPF/EPFL in different tissues of poplar. The result showed that the expression of PtEPF/EPFL has obvious tissue specificity (Figure 7A). Among them, PtEPF1-1, PtEPF1-2, PtEPF2, PtEPF3-1, PtEPF3-2, PtEPFL6 and PtEPFL7 were mainly expressed in young leaves, PtEPFL1-1 was mainly expressed in young leaves and stems, PtEPFL1-2, PtEPFL5-1, PtEPFL5-2 and PtEPFL8 were mainly expressed in roots, PtEPFL4 and PtEPFL6 was mainly expressed in stems, and PtEPFL9 was highly expressed in young and mature leaves.
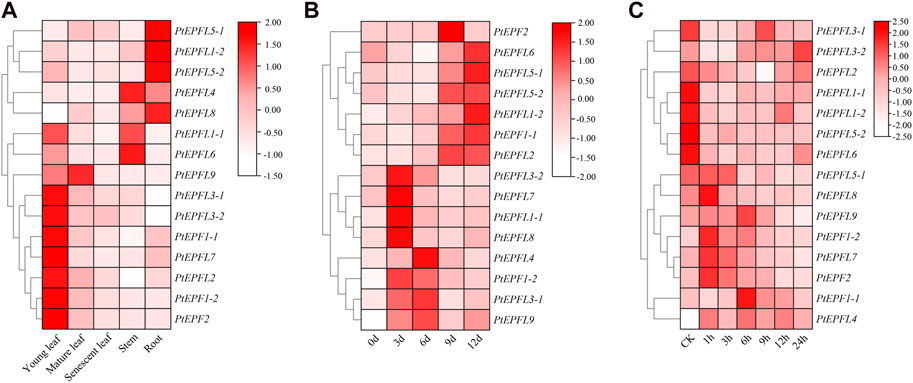
Figure 7. The expression pattern of PtEPF/EPFL. (A) Heatmap showing the tissue-specific expression of PtEPF/EPFL genes. “Young leaf”, the first one to three leaves from the shoot apex; ‘‘mature leaf”, a fully expanded leaf; “senescent leaf”, basal most two to three leaves above the root system. (B) Expression pattern of PtEPF/EPFL genes under drought stress. (C) Expression pattern of PtEPF/EPFL genes under ABA treatment. Data represent mean ± SE (n = 3).
In order to identify the response of PtEPF/EPFL family genes to stress treatment, poplar plants were subjected to drought and ABA stress treatment. Then the expression of these 15 genes was quantitatively analyzed. The expression pattern of PtEPF/EPFL gene was different between drought stress and ABA treatment. As can be seen from Figure 7B, the expression of PtEPF/EPFL family genes were basically increased after drought stress treatment, but the space and time of the increase were different. The expression levels of PtEPF1-2, PtEPFL1-1, PtEPFL3-2, PtEPFL7 and PtEPFL8 reached their peak at 3 days of drought stress, and the expression of PtEPF1-2 was the highest. The expression levels of PtEPFL3-1, PtEPFL4 and PtEPFL9 were highest at 6 days after drought stress. The expression levels of PtEPF2 and PtEPFL2 were highest at 9 days after drought stress. PtEPF1-1, PtEPFL1-2, PtEPFL5-1, PtEPFL5-2, and PtEPFL6 expressed highest at 12 days after drought stress. ABA is an important stress hormone in plants, which can regulate plant stress adaptation to drought and other environmental factors. As can be seen from Figure 7C, some PtEPF/EPFL family genes were upregulated and some were downregulated after ABA spraying treatment. The expression levels of PtEPF1-1, PtEPF1-2, PtEPF2, PtEPFL7 and PtEPFL8 were rapidly upregulated after spraying ABA, while the expression levels of PtEPFL1-1, PtEPFL1-2, PtEPFL5-2 and PtEPFL6 were significantly downregulated. The expression levels of other genes were not significantly responsive to ABA.
As an important channel for gas exchange (CO2 and water vapor) between the external environment and plants, stomata play a crucial role in regulating plant photosynthesis and transpiration (Bertolino et al., 2019; Buckley et al., 2019). As a kind of epidermal model factor, EPF gene family can encode a specific secretory polypeptide in plants and play an important role in stomatal development (Takata et al., 2013; Uchida and Tasaka, 2013). Through identification and analysis of poplar EPF gene family, the characteristics and functions of poplar EPF gene were revealed.
In this study, 15 EPF genes were identified in poplar and the PtEPF/EPFL family members are characterized by relatively short protein lengths and similar molecular weights (Table 1). According to the phylogenetic tree shown in Figure 3, these results support the classification of 15 PtEPF members into four subfamilies (I, II, III, and IV). The analysis of the phylogenetic tree of plants of different evolutionary levels showed that the stomata and vascular tissues had not evolved in the lower bryophytes and ferns, but members of the EPF family had appeared. EPF1-EPF2-EPFL7 and EPFL4-6 branches contain genes from bryophytes and genes from vascular plants, while EPFL1-3-EPFL8 and EPFL9/Stomagen branches contain no genes from bryophytes and are composed of genes from vascular plants. EPF family members have increased during evolution possibly due to the duplication of genes or genomes. EPFs have six conserved cysteine residues in the C-terminal region of the mature peptide, in addition to two additional conserved cysteine residues in the EPF1-EPF2-EPFL7 branch (Figure 4). Disulfide bonds can be formed between cysteines. Shimada et al. speculated that this may be the site where the proteins of this family interact with downstream protein kinase receptors (Shimada et al., 2011). The promoter region of the PtEPF/EPFL family gene has been analyzed predictively, revealing the presence of various elements responsible for hormonal response, stress response, growth and development, light response, site binding, and other functional categories. This suggests that the PtEPF/EPFL gene family exhibits functional diversity due to the presence of these diverse regulatory elements.
The results of tissue expression revealed that most PtEPF genes exhibited expression across all organs (Figure 7A). Specifically, PtEPF1-1, PtEPF1-2, PtEPF2, and PtEPFL9, which are homologous to the AtEFPL9, AtEFP1 and AtEFP2 genes that regulate stomatal development in Arabidopsis (Hara et al., 2007; Hara et al., 2009; Hunt et al., 2010), showed high expression levels in leaves. The expression levels of PtEPFL4, PtEPFL5-1, PtEPFL5-2, and PtEPFL6 were notably elevated in both roots and stems. This observation aligns with the known function of the CHAL subfamily, which primarily revolves around promoting plant growth (Abrash et al., 2011; Uchida et al., 2012). The expression of EPF family genes were basically increased after drought stress treatment (Figure 7B). It is speculated that these genes play an important role in drought resistance. Drought stress induced the accumulation of ABA in plant cells, in response to the expression of ABA regulatory genes, resulting in stomatal closure, thereby improving the drought resistance of plants (Wang et al., 2021). The signal pathways of plants responding to drought stress can be divided into ABA-dependent signaling pathways and ABA-independent signaling pathways (Park et al., 2016). Further analysis of PtEPF genes response to ABA stress showed that the PtEPF1-1, PtEPF1-2, PtEPF2, PtEPFL7 and PtEPFL8 genes were induced to increase expression after ABA spraying treatment, while the PtEPFL1-1, PtEPFL1-2, PtEPFL5-2 and PtEPFL6 were downregulated of gene expression (Figure 7C). The expression patterns of the genes were different under drought or ABA treatment, suggesting that these PtEPF/EPFL genes may have regulatory functions in the tolerance to drought and ABA stress and deserve further investigation.
In populus genome, we identified 15 EPF genes and analyzed the gene structure, genome location, phylogenetic relationship, conserved domain of proteins, promoter elements and gene expression using bioinformatics methods. Our results provided important information on the functional role of EPF genes in populus. Generally, studying the molecular characteristics of PtEPF/EPFL contributes to the understanding of their specific biological functions, which provided a theoretical basis for breeding poplar varieties with high water use efficiency and fast growth rate.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
SL: Data curation, Formal Analysis, Supervision, Writing–original draft, Writing–review and editing. TC: Visualization, Writing–review and editing. XL: Writing–review and editing. JC: Writing–review and editing. YT: Formal Analysis, Supervision, Visualization, Writing–review and editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was funded by Hebei Natural Science Foundation (C2022402026).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2024.1432376/full#supplementary-material
Abrash, E. B., and Bergmann, D. C. (2010). Regional specification of stomatal production by the putative ligand CHALLAH. Development 137, 447–455. doi:10.1242/dev.040931
Abrash, E. B., Davies, K. A., and Bergmann, D. C. (2011). Generation of signaling specificity in Arabidopsis by spatially restricted buffering of ligand-receptor interactions. Plant Cell. 23, 2864–2879. doi:10.1105/tpc.111.086637
Bailey, T. L., Johnson, J., Grant, C. E., and Noble, W. S. (2015). The MEME suite. Nucleic acids Res. 43, W39–W49. doi:10.1093/nar/gkv416
Bertolino, L. T., Caine, R. S., and Gray, J. E. (2019). Impact of stomatal density and morphology on water-use efficiency in a changing world. Front. Plant Sci. 10, 225. doi:10.3389/fpls.2019.00225
Buckley, C. R., Caine, R. S., and Gray, J. E. (2019). Pores for thought: can genetic manipulation of stomatal density protect future rice yields? Front. Plant Sci. 10, 1783. doi:10.3389/fpls.2019.01783
Caine, R. S., Yin, X., Sloan, J., Harrison, E. L., Mohammed, U., Fulton, T., et al. (2019). Rice with reduced stomatal density conserves water and has improved drought tolerance under future climate conditions. New Phytol. 221, 371–384. doi:10.1111/nph.15344
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 13, 1194–1202. doi:10.1016/j.molp.2020.06.009
Dunn, J., Hunt, L., Afsharinafar, M., Meselmani, M. A., Mitchell, A., Howells, R., et al. (2019). Reduced stomatal density in bread wheat leads to increased water-use efficiency. J. Exp. Bot. 70, 4737–4748. doi:10.1093/jxb/erz248
Franks, P. J., T, W.D.-A., Britton-Harper, Z. J., and Gray, J. E. (2015). Increasing water-use efficiency directly through genetic manipulation of stomatal density. New Phytol. 207, 188–195. doi:10.1111/nph.13347
Hara, K., Kajita, R., Torii, K. U., Bergmann, D. C., and Kakimoto, T. (2007). The secretory peptide gene EPF1 enforces the stomatal one-cell-spacing rule. Genes. Dev. 21, 1720–1725. doi:10.1101/gad.1550707
Hara, K., Yokoo, T., Kajita, R., Onishi, T., Yahata, S., Peterson, K. M., et al. (2009). Epidermal cell density is autoregulated via a secretory peptide, EPIDERMAL PATTERNING FACTOR 2 in Arabidopsis leaves. Plant Cell Physiol. 50, 1019–1031. doi:10.1093/pcp/pcp068
Hughes, J., Hepworth, C., Dutton, C., Dunn, J. A., Hunt, L., Stephens, J., et al. (2017). Reducing stomatal density in barley improves drought tolerance without impacting on yield. Plant Physiol. 174, 776–787. doi:10.1104/pp.16.01844
Hunt, L., Bailey, K. J., and Gray, J. E. (2010). The signalling peptide EPFL9 is a positive regulator of stomatal development. New Phytol. 186, 609–614. doi:10.1111/j.1469-8137.2010.03200.x
Hunt, L., and Gray, J. E. (2009). The signaling peptide EPF2 controls asymmetric cell divisions during stomatal development. Curr. Biol. 19, 864–869. doi:10.1016/j.cub.2009.03.069
Katsir, L., Davies, K. A., Bergmann, D. C., and Laux, T. (2011). Peptide signaling in plant development. Curr. Biol. 21, R356–R364. doi:10.1016/j.cub.2011.03.012
Kawamoto, N., Del Carpio, D. P., Hofmann, A., Mizuta, Y., Kurihara, D., Higashiyama, T., et al. (2020). A peptide pair coordinates regular ovule initiation patterns with seed number and fruit size. Curr. Biol. 30, 4352–4361. doi:10.1016/j.cub.2020.08.050
Kissoudis, C., van de Wiel, C., Visser, R. G., and van der Linden, G. (2016). Future-proof crops: challenges and strategies for climate resilience improvement. Curr. Opin. Plant Biol. 30, 47–56. doi:10.1016/j.pbi.2016.01.005
Kondo, T., Kajita, R., Miyazaki, A., Hokoyama, M., Nakamura-Miura, T., Mizuno, S., et al. (2010). Stomatal density is controlled by a mesophyll-derived signaling molecule. Plant Cell Physiol. 51, 1–8. doi:10.1093/pcp/pcp180
Lawson, T., and Blatt, M. R. (2014). Stomatal size, speed, and responsiveness impact on photosynthesis and water use efficiency. Plant Physiol. 164, 1556–1570. doi:10.1104/pp.114.237107
Li, H., Yang, Y., Wang, H., Liu, S., Jia, F., Su, Y., et al. (2021). The receptor-like kinase ERECTA confers improved water use efficiency and drought tolerance to poplar via modulating stomatal density. Int. J. Mol. Sci. 22, 7245. doi:10.3390/ijms22147245
Li, S., Zhang, J., Liu, L., Wang, Z., Li, Y., Guo, L., et al. (2020). SlTLFP8 reduces water loss to improve water-use efficiency by modulating cell size and stomatal density via endoreduplication. Plant, Cell Environ. 43, 2666–2679. doi:10.1111/pce.13867
Li, X., Ye, J., Munir, S., Yang, T., Chen, W., Liu, G., et al. (2019). Biosynthetic gene pyramiding leads to ascorbate accumulation with enhanced oxidative stress tolerance in tomato. Int. J. Mol. Sci. 20, 1558. doi:10.3390/ijms20071558
Ma, T., Wang, J., Zhou, G., Yue, Z., Hu, Q., Chen, Y., et al. (2013). Genomic insights into salt adaptation in a desert poplar. Nat. Commun. 4, 2797. doi:10.1038/ncomms3797
Ohki, S., Takeuchi, M., and Mori, M. (2011). The NMR structure of stomagen reveals the basis of stomatal density regulation by plant peptide hormones. Nat. Commun. 2, 512. doi:10.1038/ncomms1520
Park, Y., Xu, Z. Y., Kim, S. Y., Lee, J., Choi, B., Lee, J., et al. (2016). Spatial regulation of ABCG25, an ABA exporter, is an important component of the mechanism controlling cellular ABA levels. Plant Cell 28, 2528–2544. doi:10.1105/tpc.16.00359
Shimada, T., Sugano, S. S., and Hara-Nishimura, I. (2011). Positive and negative peptide signals control stomatal density. Cell. Mol. life Sci. 68, 2081–2088. doi:10.1007/s00018-011-0685-7
Sugano, S. S., Shimada, T., Imai, Y., Okawa, K., Tamai, A., Mori, M., et al. (2010). Stomagen positively regulates stomatal density in Arabidopsis. Nature 463, 241–244. doi:10.1038/nature08682
Sun, Q., Qu, J., Yu, Y., Yang, Z., Wei, S., Wu, Y., et al. (2019). TaEPFL1, an EPIDERMAL PATTERNING FACTOR-LIKE (EPFL) secreted peptide gene, is required for stamen development in wheat. Genetica. 147, 121–130. doi:10.1007/s10709-019-00061-7
Takata, N., Yokota, K., Ohki, S., Mori, M., Taniguchi, T., and Kurita, M. (2013). Evolutionary relationship and structural characterization of the EPF/EPFL gene family. PloS one 8, e65183. doi:10.1371/journal.pone.0065183
Uchida, N., Lee, J. S., Horst, R. J., Lai, H. H., Kajita, R., Kakimoto, T., et al. (2012). Regulation of inflorescence architecture by intertissue layer ligand-receptor communication between endodermis and phloem. Proc. Natl. Acad. Sci. U. S. A. 109, 6337–6342. doi:10.1073/pnas.1117537109
Uchida, N., and Tasaka, M. (2013). Regulation of plant vascular stem cells by endodermis-derived EPFL-family peptide hormones and phloem-expressed ERECTA-family receptor kinases. J. Exp. Bot. 64, 5335–5343. doi:10.1093/jxb/ert196
Wang, C., Liu, S., Dong, Y., Zhao, Y., Geng, A., Xia, X., et al. (2016). PdEPF1 regulates water-use efficiency and drought tolerance by modulating stomatal density in poplar. Plant Biotechnol. J. 14, 849–860. doi:10.1111/pbi.12434
Wang, Y. H., Que, F., Li, T., Zhang, R. R., Khadr, A., Xu, Z. S., et al. (2021). DcABF3, an ABF transcription factor from carrot, alters stomatal density and reduces ABA sensitivity in transgenic Arabidopsis. Plant Sci. 302, 110699. doi:10.1016/j.plantsci.2020.110699
Wang, Y., Que, F., Xie, T., and Zhang, C. (2020). Overexpression of the potato StEPF2 gene confers enhanced drought tolerance in arabidopsis. Plan. Biotech. Rep. 14, 479–490.
Xiang, Y., Sun, X., Bian, X., Wei, T., Han, T., Yan, J., et al. (2021). The transcription factor ZmNAC49 reduces stomatal density and improves drought tolerance in maize. J. Exp. Bot. 72, 1399–1410. doi:10.1093/jxb/eraa507
Xiong, L., Huang, Y., Liu, Z., Li, C., Yu, H., Shahid, M. Q., et al. (2022). Small EPIDERMAL PATTERNING FACTOR-LIKE2 peptides regulate awn development in rice. Plant physiol. 190, 516–531. doi:10.1093/plphys/kiac278
Yu, H., Chen, X., Hong, Y. Y., Wang, Y., Xu, P., Ke, S. D., et al. (2008). Activated expression of an Arabidopsis HD-START protein confers drought tolerance with improved root system and reduced stomatal density. Plant Cell 20, 1134–1151. doi:10.1105/tpc.108.058263
Zhang, J., Feng, J., Lu, J., Yang, Y., Zhang, X., Wan, D., et al. (2014). Transcriptome differences between two sister desert poplar species under salt stress. BMC Genomics 15, 337. doi:10.1186/1471-2164-15-337
Zhao, P. X., Miao, Z. Q., Zhang, J., Chen, S. Y., Liu, Q. Q., and Xiang, C. B. (2020). Arabidopsis MADS-box factor AGL16 negatively regulates drought resistance via stomatal density and stomatal movement. J. Exp. Bot. 71, 6092–6106. doi:10.1093/jxb/eraa303
Keywords: EPF/EPFL gene family, poplar, gene expression, drought, stomata
Citation: Liu S, Chen T, Li X, Cui J and Tian Y (2024) Genome-wide identification and expression analysis of EPF/EPFL gene family in Populus trichocarpa. Front. Genet. 15:1432376. doi: 10.3389/fgene.2024.1432376
Received: 14 May 2024; Accepted: 01 July 2024;
Published: 18 July 2024.
Edited by:
Liwei Zheng, Zhengzhou University, ChinaReviewed by:
Chenguang Zhang, Hebei Normal University of Science and Technology, ChinaCopyright © 2024 Liu, Chen, Li, Cui and Tian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yinshuai Tian, dGlhbnlpbnNodWFpQGhlYmV1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.