
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Genet. , 02 July 2024
Sec. Stem Cell Research
Volume 15 - 2024 | https://doi.org/10.3389/fgene.2024.1429844
This article is part of the Research Topic Tissue Repair Modeling of Locomotor Apparatus View all 7 articles
Mesenchymal stem cells (MSCs) have promising potential for bone tissue engineering in bone healing and regeneration. They are regarded as such due to their capacity for self-renewal, multiple differentiation, and their ability to modulate the immune response. However, changes in the molecular pathways and transcription factors of MSCs in osteogenesis can lead to bone defects and metabolic bone diseases. DNA methylation is an epigenetic process that plays an important role in the osteogenic differentiation of MSCs by regulating gene expression. An increasing number of studies have demonstrated the significance of DNA methyltransferases (DNMTs), Ten-eleven translocation family proteins (TETs), and MSCs signaling pathways about osteogenic differentiation in MSCs. This review focuses on the progress of research in these areas.
Friedenstein et al. first discovered mesenchymal stem cells (MSCs) in bone marrow (Friedenstein et al., 1970). MSCs originate from embryonic stem cells and can differentiate into various cell types, including osteoblasts, chondrocytes, adipocytes, and other cell lineages (da Silva Meirelles et al., 2006; Rastegar et al., 2010). It can be extracted from adult tissues, including bone marrow, umbilical cord blood, adipose tissue, and tooth tissue (Hu et al., 2018). In an aging society, severe bone defects caused by trauma, infection, tumors, and skeletal developmental abnormalities are increasingly challenging treatments in orthopedics and trauma surgery. The development of bone healing and regeneration strategies is of great significance for improving patient function and quality of life (Cancedda et al., 2007). MSCs are considered promising seed cells for bone tissue engineering in bone healing and regeneration due to their self-renewal, multiple differentiation, and immunomodulatory properties (Seong et al., 2010). In normal bones, the differentiation of MSCs into osteoblasts is regulated by multiple signaling pathways, such as bone morphogenetic proteins (BMPs), Notch, Hedgehog, neuroepidermal growth factor like protein 1 (NELL-1), wnt signaling, growth factors, and MAPK signaling pathways (Montecino et al., 2020; Ahmadi et al., 2022; Thomas and Jaganathan, 2022). The transcription factors Runx2 and Osterix (Osx) are the main regulatory factors of osteogenesis, inducing osteogenic lineage commitment and differentiation of MSCs through interactions with various signaling pathways (Yang et al., 2004). There is evidence to suggest that DNA methylation plays an important role in regulating the molecular pathways involved in the osteogenesis of MSCs (Levi and Longaker, 2011).
DNA methylation is a part of epigenetics, controlled by enzymes from the DNA methyltransferases (DNMTs) family. It modifies DNA without changing its sequence, which affects gene expression (Moore et al., 2013). DNA methylation plays an important role in genomic imprinting, tissue-specific gene expression, and X chromosome inactivation (Walewska et al., 2023). It also regulates stem cell maintenance and differentiation by activating or inhibiting certain genes (Kouzarides, 2007). Research has shown that DNA methylation status is associated with the osteogenic ability of MSCs. DNA demethylation has been shown to play a beneficial role in the osteogenesis of MSCs (Yan et al., 2014). During the osteogenic differentiation process of MSCs, the expression of osteogenic specific transcription factor genes, RUNX2 and OSX, increases, accompanied by a decrease in their DNA methylation (Farshdousti Hagh et al., 2012; Xu et al., 2017). However, there are also opposing reports indicating that DNA demethylation impairs the osteogenic potential of MSCs (Xin et al., 2020). The role of DNA methylation in regulating the osteogenic differentiation of MSCs is not fully understood. Further research on the mechanism of DNA methylation regulating the osteogenic differentiation of MSCs can help us gain a more comprehensive understanding of the basic biological processes involved in stem cell differentiation and cell fate determination. This can allow us to fully utilize the characteristics of MSCs as seed cells to treat related diseases. This article reviews the progress made in understanding the mechanism of DNA methylation on the osteogenic differentiation of MSCs from the viewpoint of enzymes mediating DNA methylation and their impact on downstream signaling pathways related to osteogenic differentiation.
Epigenetic phenomena play a crucial role in cellular regulation and are significant factors in comprehending complex human diseases. Among the extensively studied epigenetic mechanisms, DNA methylation holds a prominent position (Kriukienė et al., 2024). DNA methylation, in the field of epigenetics, usually refers to 5’methylcytosine (5mC) where the fifth carbon atom of cytosine is added with a methyl group to form 5-methylcytosine (Patil et al., 2014). Most 5-methylcytosine is located directly in front of the 5′guanine residue, known as CpG methylation, in most mammalian cells. Non-CpG methylation can occur on CHG (H = A, C, or T) and CHH sites (Sarkies, 2022). CpG islands (CGIs) are high CpG density regions spread throughout the mammalian genome, with a length of 300-3000 bp, primarily located in the promoter and first exon/5′- untranslated region of the gene. Approximately 70% of human genes have promoter regions that overlap with these regions, making DNA methylation capable of affecting mRNA levels without changing the gene sequence (Luczak and Jagodziński, 2006; Saxonov et al., 2006). DNA methylation is closely related to gene transcriptional inhibition. It is generally believed that DNA methylation achieves negative regulation of gene expression through two pathways: direct inhibition of gene transcription by interfering with transcription factors binding to gene promoters (or other cis-regulatory sequences) and indirect inhibition of gene transcription by recruiting transcription inhibitors through 5mC binding proteins (Greenberg and Bourc’his, 2019). However, DNA methylation often occurs within the gene body, between the transcription start and end sites. In contrast to the effect of promoter methylation, genomic methylation is usually positively correlated with gene expression (Neri et al., 2017). The primary purpose of DNA methylation is genome defense, which maintains the silence of transposable elements to prevent transposition from affecting the stability of the host genome. As species evolve, DNA methylation gradually participates in other biological processes, making its functions increasingly complex (Jones, 2012).
DNA methylation is catalyzed by DNMTs, using S-adenosylmethionine as a methyl donor to transfer the methyl group to the fifth carbon atom (C5) of cytosine (Figure 1). According to the different modes of action of methyltransferases, DNMTs can be mainly divided into two categories: one is initiating DNA methyltransferases (de novo DNMTs), and the other is maintenance DNA methyltransferases (maintenance DNMTs) (Goll and Bestor, 2005). DNMT1 is responsible for maintaining DNA methylation. It binds to partially methylated CpG sites and spreads the methylation pattern to newly synthesized DNA. DNMT1’s activity is higher at partially methylated sites compared to unmethylated sites, indicating its role in maintenance methylation (Pradhan et al., 1999). DNMT1 is expressed in cells that are actively dividing and is associated with DNA replication. When the DNMT1 gene is knocked out in mouse embryonic stem cells, the genome-wide DNA methylation decreases significantly (Leonhardt et al., 1992; Li et al., 1992; Lei et al., 1996). DNMT3 family members, DNMT3A, DNMT3B, and DNMT3L are responsible for de novo methylation, which establishes new methylation patterns on unmethylated DNA (Okano et al., 1998). DNMT3A and DNMT3B are expressed during embryonic development, consistent with the timing of de novo methylation (Okano et al., 1998). In mouse embryonic stem cells and embryos, DNMT3A and DNMT3B double knockout prevents de novo methylation without affecting maintenance methylation (Okano et al., 1999). DNMT3L lacks catalytic activity but binds to DNMT3A and DNMT3B, enhancing their catalytic activity (Veland et al., 2019). DNMTs are essential for maintaining genomic integrity and silencing specific sequences, including imprinted genes, inactive X-chromosome genes, and transposons. Loss of DNMTs function may lead to chromosomal instability and tumor progression (Robertson, 2005).
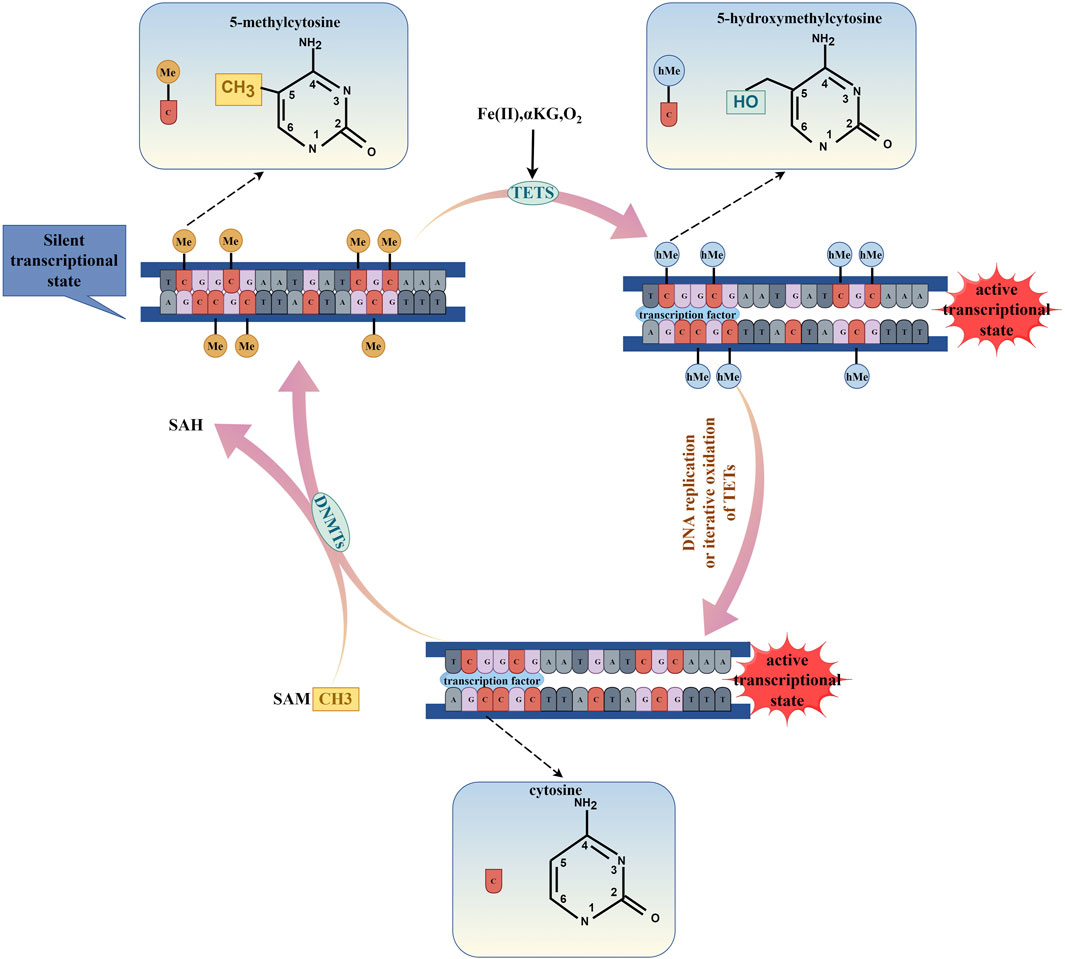
Figure 1. The process of DNA methylation and DNA demethylation. (By Figdraw.). DNA methylation is enzymatically catalyzed by DNMTs, which utilize S-adenosylmethionine (SAM) as a methyl donor to transfer a methyl group to the fifth carbon atom (C5) of the cytosine ring, thereby leading to the transcriptional repression of genes. Conversely, DNA demethylation is facilitated by the TETs enzymes through a process of iterative oxidation, which requires the presence of Fe(II), α-KG, and O2. Additionally, DNA demethylation can occur during DNA replication. This demethylation process results in the transcriptional activation of genes.
DNA demethylation is a process that is mediated by proteins belonging to the Ten Eleven Translation Family (TETs), which include TET1, TET2, and TET3 in mammals. The process involves oxidative modification of 5-methylcytosine (5mC) to produce 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC) that ultimately lead to DNA demethylation and promote gene transcription (Ooi and Bestor, 2008; Wang L. et al., 2022). TETs are crucial for stem cell differentiation, and the loss of TET1/2/3 can impair the differentiation of mouse embryonic stem cells (Dawlaty et al., 2014). Active demethylation intermediates can act as heritable epigenetic markers. 5hmC is involved in gene regulation and is predominantly present in actively transcribed genes. It binds to specific regulatory proteins capable of transmitting regulatory functions (Bachman et al., 2014). 5fC and 5caC interact with many proteins, including TDG, p53, DNA repair factors, chromatin remodeling factors, and forkhead box TFs. The presence of 5fC and 5caC in template DNA strands can temporarily suspend and decrease the elongation of RNA polymerase II (pol II) in vivo and in vitro. Controlled deposition of 5fC and 5caC can fine-tune the elongation of specific genes and indirectly affect gene expression (Rasmussen and Helin, 2016).
There is increasing evidence that DNA methylation, facilitated by various DNMTs, plays a crucial role in bone biology (Husain and Jeffries, 2017). Studies have shown that abnormal expression of DNMTs can influence the expression of genes and pathways that are related to osteoblasts and osteoclasts, thereby affecting bone remodeling (Liu et al., 2016). The process of MSCs differentiation requires the gradual loss of stem cell features and the acquisition of specific cell line or tissue-specific cellular identity, which is achieved through regulatory mechanisms such as epigenetic modification. The methylation of osteogenic-related gene promoters or enhancers regulates the osteogenic process of MSCs, while the DNMTs family mediates DNA methylation (Weissbein et al., 2017; Wang X. et al., 2023). In fact, OCT4 enhances the transcription of DNMTs genes, leading to an increase in p21 promoter methylation, promoting the self-renewal ability of human hair follicular mesenchymal stem cells (hHFMSCs), and reversing aging and osteogenic differentiation ability (Lu et al., 2019). Moreover, the expression of DNMT3A and DNMT3B increased in three-dimensional spherical osteogenic differentiation cultures, and the significant downregulation of DNMT3B in two-dimensional monolayer osteogenic differentiation culture affected the osteogenic potential of Dental pulp stem cells (DPSCs). However, the expression of DNMT1 remained unchanged in both two-dimensional monolayer and three-dimensional spherical osteogenic differentiation cultures (Raik et al., 2022). By gaining a deeper understanding of DNMTs that regulate gene expression, we can better comprehend the structure and osteogenic differentiation functional behavior of MSCs, and improve treatment methods based on MSCs.
DNMT1 is a vital component in both developmental and adult somatic cells, which ensures cell proliferation and survival (Jurkowska et al., 2011). It plays a crucial role in maintaining CpG methylation, regulating chromatin modification and remodeling in stem cell renewal and differentiation (Robert et al., 2003; Tsumura et al., 2006). Additionally, the downregulation of DNMT1 can lead to passive DNA demethylation, which can enhance the spontaneous differentiation of MSCs (Zhang et al., 2016). Many studies have reported the role of DNMT1 in regulating the directional osteogenic lineage differentiation of MSCs (Table 1).
DNMT1 plays a crucial role in the process of osteogenic differentiation of MSCs (Figure 2). Studies have shown that the expression of DNMT1 is reduced during the differentiation of bone marrow mesenchymal stem cells (BMSCs) or osteoprogenitor cells into osteoblasts. However, in humans and mice with age-related bone loss, the expression of DNMT1 increases in osteoprogenitor cells (Tao et al., 2022). Similarly, another study found that the expression level of DNMT1 decreased after osteogenic differentiation of human amniotic fluid-derived mesenchymal stem cells (AF-MSCs) (Glemžaitė and Navakauskienė, 2016). During the process of osteogenic differentiation of MSCs, DNMT1 is downregulated, which may be related to the upregulation of osteogenic lineage differentiation genes. On the other hand, DNMT1 methylates osteogenic differentiation-related genes and inhibits their expression (Cakouros and Gronthos, 2020).
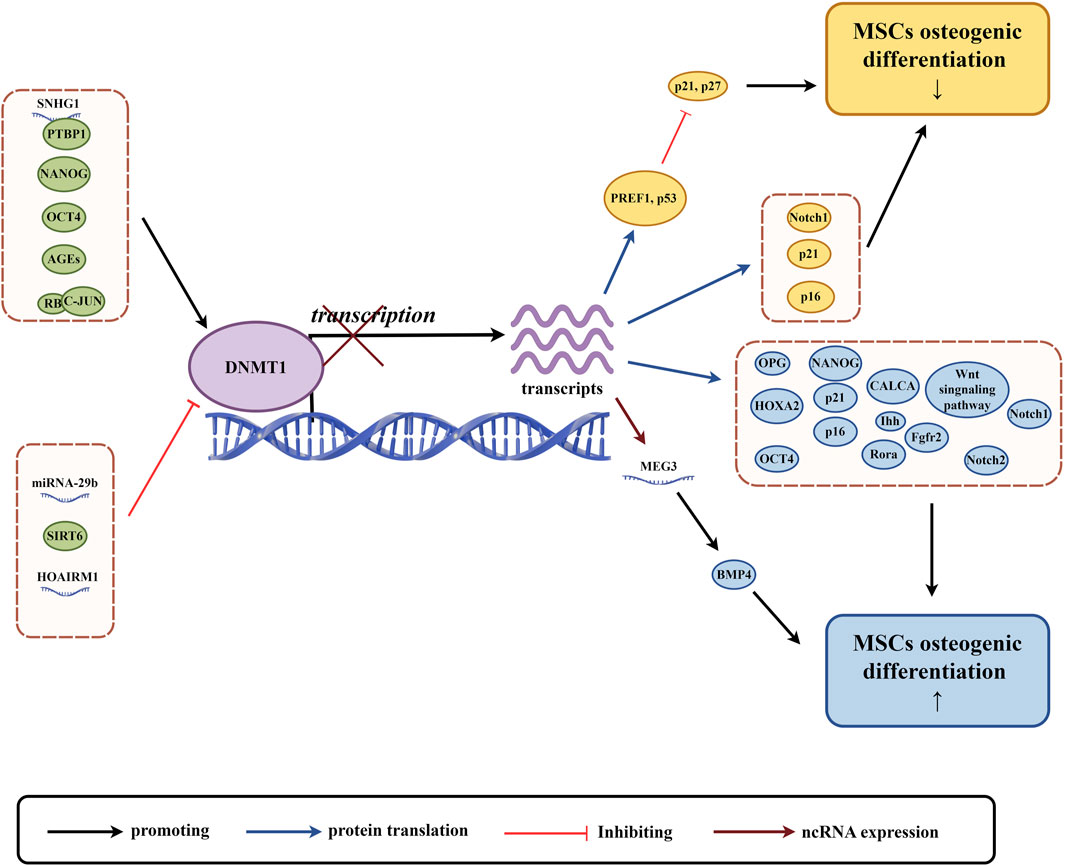
Figure 2. The role of DNMT1 in osteogenic differentiation of MSCs. (By Figdraw.). The expression and activity of DNMT1 are regulated by various factors, including non-coding RNAs and proteins (highlighted in green circles). These factors influence DNA methylation and subsequent gene transcription and translation. This regulation affects the expression of proteins related to osteogenic differentiation of MSCs, the Wnt signaling pathway, and non-coding RNAs, with yellow circles indicating factors associated with the downregulation of MSCs osteogenic differentiation, and blue circles indicating factors associated with the upregulation of MSCs osteogenic differentiation.
There is a relationship between DNMT1 (a protein that helps modify DNA) and non-coding RNA (RNA that does not produce proteins), which influences the way in which genomic methylation patterns are established and abnormal DNA methylation is regulated (Di Ruscio et al., 2013). This relationship can affect the process of creating bone cells from adult stem cells in various ways. For example, a type of non-coding RNA called LncRNA SNHG1 can activate the expression of DNMT1, which in turn can lead to the hypermethylation of a gene called osteoprotegerin (OPG) and the inhibition of OPG expression. This can damage the process of creating bone cells from mouse BMSCs (Yu X. et al., 2022). Conversely, another type of non-coding RNA known as LncRNA HOTAIRM1 can inhibit DNMT1 expression, which leads to low methylation of a gene called HOXA2 and an increase in HOXA2 expression. This promotes the process of creating bone cells from human dental follicle stem cells (hDFSCs) (Chen et al., 2020). In mice with lupus disease, a deficiency in a protein called Fas can lead to higher levels of miR-29b, a type of RNA. This can decrease the expression of DNMT1 in MRL/lpr BMSCs and contribute to low methylation of the Notch1 promoter, which can impair the process of creating bone cells (Liu et al., 2015). LncRNA, like any other protein coding gene, is regulated by the same epigenetic mechanism, and DNMT1 can regulate its expression (Lee, 2012).Finally, the expression of a non-coding RNA called LncRNA MEG3 is inhibited in children with aplastic anemia (AA) due to high levels of DNMT1 expression, which can lead to lower levels of bone morphogenetic protein 4 (BMP4) transcriptional activity and impair the process of creating bone cells from BMSCs in AA patients (Li H. et al., 2021). Overall, the relationship between DNMT1 and non-coding RNA may play an important role in regulating the process of creating bone cells from MSCs, and further research is necessary to understand this relationship more completely.
The OCT4, SOX2, and NANOG genes are responsible for regulating the self-renewal and pluripotency of stem cells. They form a transcriptional network that is essential for the differentiation potential of MSCs (Hyslop et al., 2005; Liu et al., 2009). DNMT1 is a protein that can affect the methylation of OCT4 and NANOG promoters and regulate their expression. This regulation can alter the tripartite differentiation of pig BMSCs and human BMSCs, making DNMT1 an early indicator and regulator of stem cell potential (Li K. et al., 2020). NANOG and OCT4, in turn, can bind to the promoter of DNMT1 and enhance its expression, indirectly regulating gene expression (Tsai et al., 2012). Recent research suggests that upregulation of DNMT1 can maintain DNA methylation of p16 and p21, as well as genes related to development and lineage differentiation. This maintenance inhibits gene expression, maintains cell proliferation, and undifferentiated status, and decreases osteogenic differentiation ability of human BMSCs (hBMSCs) (Tsai et al., 2012). On the other hand, downregulation of NANOG in human adipose-derived stem cells (hADSCs) can cause a decrease in DNMT1 protein expression, leading to decreased methylation of PREF1, p21, and p27. This decrease results in an upregulation of p21 and p27, causing hADSCs to stagnate in the G0/G1 phase. This stagnation ultimately results in decreased differentiation ability and impaired osteogenic differentiation function (Pitrone et al., 2019).
Advanced glycation end products (AGEs) are harmful pathogenic factors that are closely related to complications of diabetes. AGEs cause oxidative stress by cross-linking extracellular matrix, which damages the proliferation and osteogenic differentiation of MSCs and increases osteoclast production (Asadipooya and Uy, 2019). During the process of osteogenic induction of human periodontal ligament mesenchymal stem cells (hPDLSCs), AGEs upregulate the expression of advanced glycosylation end product-specific receptor (RAGE) and DNMT1, and increase the expression of calcitonin-related peptides α (CALCA). The methylation level of the CALCA promoter is increased, and the downregulation of CALCA expression impairs the osteogenic differentiation ability of hDPLSCs (Wang Q. N. et al., 2022). When rat ADSCs and mouse ADSCs were cultivated in media containing AGEs, high levels of DNMTs (DNMT1, DNMT3A, and DNMT3B) in cells led to an increase in global 5-mC. The classical Wnt signaling pathway and osteogenic differentiation were inhibited as a result (Zhang et al., 2018; Li Y. et al., 2020).
In addition to the research reports mentioned above, DNMT1 can also be influenced by other factors that affect the osteogenic differentiation of MSCs. The nicotinamide adenine dinucleotide (NAD) dependent deacetylase SIRT6 deacetylates DNMT1, causing protein instability and preventing abnormal DNA methylation of Notch1 and Notch2, resulting in their transcriptional upregulation and promoting osteogenic differentiation of hADSCs (Jia et al., 2021). Retinoblastoma (RB) protein binds to c-JUN to increase the expression of DNMT1, methylate the promoter of p16/p21 cyclin dependent kinase inhibitor gene, reduce its expression, inhibit aging of hBMSCs, increase differentiation potential and bone repair ability (Lin et al., 2014).
DNMT3A and DNMT3B are genes that are highly active in the early stages of mammalian embryos. However, their activity decreases as cells differentiate (Zhang and Xu, 2017). These genes are responsible for adding methyl groups to DNA, a process known as DNA methylation. This process plays a significant role in the differentiation of MSCs (Table 2) (Figure 3) (Busque et al., 2018).
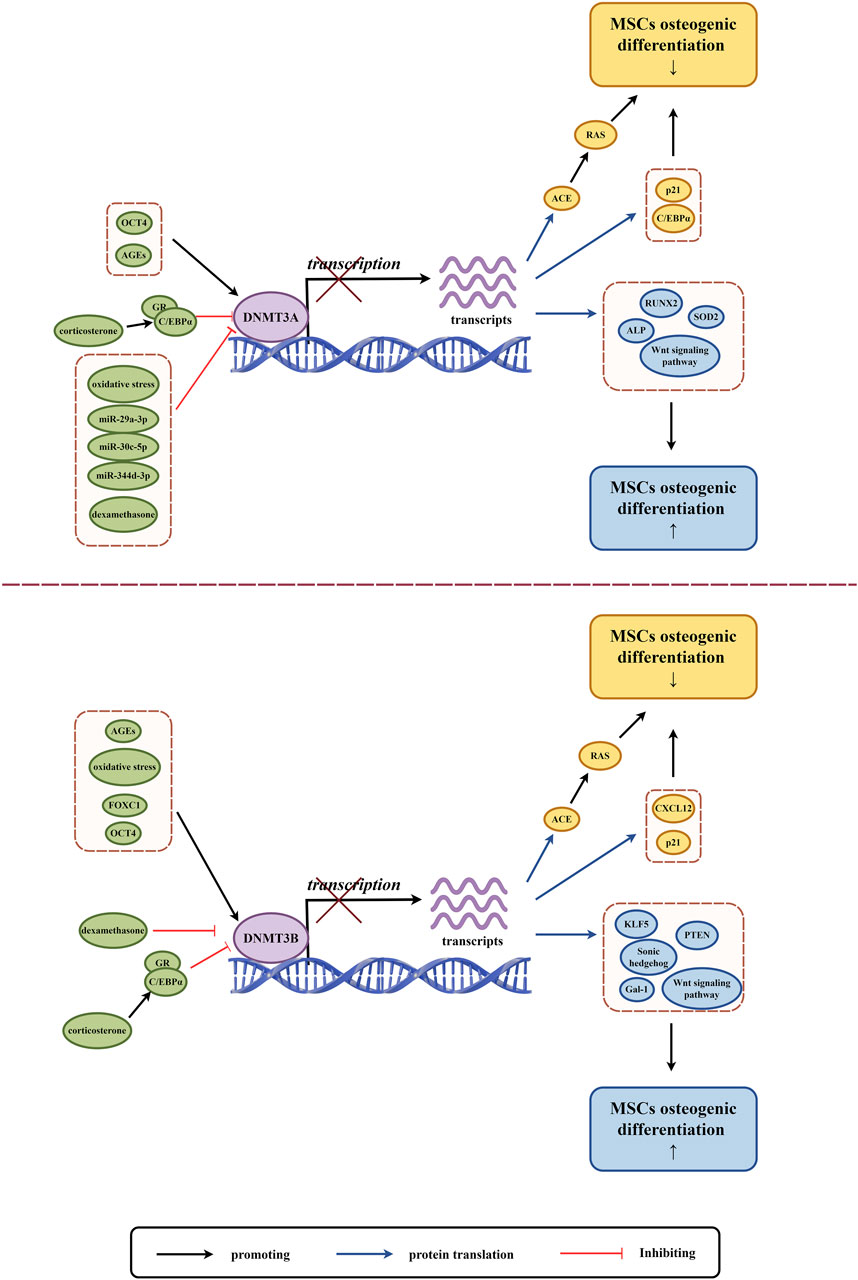
Figure 3. The role of DNMT3A and DNMT3B in osteogenic differentiation of MSCs. (By Figdraw.). The expression and activity of DNMT3A and DNMT3B are regulated by various factors (highlighted in green circles), which influence DNA methylation and subsequent gene transcription and translation. This regulation impacts the expression of proteins and the Wnt signaling pathway related to MSCs osteogenic differentiation, where yellow circles indicate factors associated with the downregulation of MSCs osteogenic differentiation, and blue circles indicate factors associated with the upregulation of MSCs osteogenic differentiation.
MSCs with varying osteogenic differentiation potentials exhibit different levels of de novo DNA methyltransferases. MSCs with low osteogenic potential in the PDLCs express higher levels of DNMT3A and lower levels of RUNX2 compared to high osteogenic potential PDLCs (Ferreira et al., 2022). As a stem gene of human induced pluripotent stem cell-derived mesenchymal stem cells (iPSC-MSCs), DNMT3B expression level is lower than that of BMSCs, and it also shows insufficient osteogenic differentiation ability (Kang et al., 2015). MSCs derived from the subchondral bone of the rat condyle (SMSCs) have higher expression of DNMT3B and significantly higher mineralization and osteogenic differentiation potential compared to MSCs derived from the tibia (TMSCs) and mandibular ramus bone marrow (MMSCs) (Wang Y. et al., 2023). Osteogenesis imperfecta (OI) mice with ADSCs express high levels of DNMT3A compared to wild-type mice with ADSCs, and their osteogenic differentiation ability and collagen secretion are significantly impaired (Shao et al., 2023). Differentially expressed DNMTs may reflect the differences in epigenetic regulation between different subgroups of MSCs, and this regulation may be one of the key factors in maintaining the characteristics of different MSCs subgroups.
Oxidative stress (OS) refers to an imbalance between the creation and removal of reactive oxygen species (ROS), which leads to a build-up of ROS in the body. This condition is caused by various factors such as ischemia, hypoxia, and inflammation (Muinos-López et al., 2016). Excessive ROS can harm nucleic acids, proteins, and lipids. Severe oxidative stress can inhibit cell survival, proliferation, and differentiation, and it can also inhibit osteogenic differentiation (Feng et al., 2013). Studies have shown that OS can induce changes in DNMTs and regulate genomic DNA methylation (O'Hagan et al., 2011; Li et al., 2014). OS mediates high methylation of zinc finger transcription factor krüppel-like factor 5 (KLF5) through DNMT3B, leading to a decrease in KLF5 expression and a decrease in association with β-catenin. This impairs the osteogenic differentiation of mouse BMSCs (Li L. et al., 2021). Liangping Li et al. established a new 3D cell culture model in which OS mediated low methylation of (Alkaline Phosphatase) ALP and RUNX2 promoters through DNMT3A downregulation, and upregulation of ALP and RUNX2 expression improved osteogenic differentiation of hBMSCs (Li L. et al., 2019).
As discussed previously, microRNAs (miRNAs) can affect DNA methylation and interfere with epigenetic mechanisms by altering the expression of DNA methylase or its co-proteins, which are intricately linked to osteoblast differentiation (Kim E. J. et al., 2013; Saviana et al., 2023). MiR-29a-3p and miR-30c-5p have an inhibitory function by binding directly to the DNMT3A 3′UTR, which reduces the methylation of CpG islands in the upstream regulatory region of the antioxidant gene superoxide dismutase 2 (SOD2). This leads to the restoration of SOD2 expression and alleviation of oxidative stress in hADSCs, improving mitochondrial function, cellular aging status, multi-directional differentiation potential of hADSCs and enhancing osteogenic differentiation ability (Jung et al., 2020). In the bone microenvironment of postmenopausal osteoporosis, the expression of miR-344d-3p in mouse mandibular BMSCs decreases. Due to the presence of two binding sites of miR-344d-3p in the 3′-UTR region of DNMT3A, an increase in DNMT3A leads to downregulation of osteogenic differentiation and upregulation of adipogenic differentiation in mouse mandibular BMSCs (Cao et al., 2023).
Glucocorticoids in normal concentrations promote osteoblast growth and stimulate the differentiation of MSCs into bone cells. However, at drug concentrations, glucocorticoids can cause apoptosis (cell death) of osteoblasts and bone cells, as well as hinder the differentiation and growth of bone progenitor cells (Han et al., 2019). Dexamethasone, for instance, blocks the binding of DNMT3A and DNMT3B to C/EBP alpha promoters, which can lead to an increase in CCAAT/enhancer α (C/EBP alpha) mRNA and protein levels in vivo and in vitro. High levels of C/EBP alpha can shift the fate of rat BMSCs towards adipocytes instead of osteoblasts (Li et al., 2013). In a study on prenatal exposure to caffeine (PCE), corticosterone was found to act through GR-C/EBP alpha, which in turn led to low methylation of angiotensin-converting enzyme (ACE). As a result, ACE expression increased and led to sustained activation of the RAS pathway, which inhibited osteogenic differentiation of rat BMSCs (Wen et al., 2019).
Wharton’s jelly derived MSCs (WJ-MSCs) obtained from human umbilical cord were treated with graphene oxide (GO) and were found to exhibit a decrease in DNMT3B expression after undergoing osteogenic induction (Hiew and Teoh, 2024). Interestingly, RNA interference was used to inhibit endogenous DNMT3B expression in human umbilical cord mesenchymal stem cells (hUMSCs), which resulted in impaired osteogenic differentiation of hUMSCs (Zhu et al., 2014). Depending on the intracellular and extracellular environment, as well as the interaction of regulatory factors, DNMT3B can play both a promoting and inhibitory role in the osteogenic differentiation of MSCs. DNMT3B mediated hypermethylation of the Galactin-1 gene (Gal-1) promoter plays an important role in the downregulation of Gal-1 in elderly mouse BMSCs. This inhibits β-catenin binding to the Gal-1 promoter gene, reduces Gal-1 expression, impairs the function of mouse BMSCs, and inhibits their osteogenic differentiation (Tang et al., 2024). During mechanical unloading in mice, the protein level of DNMT3B increases. This leads to the methylation of the Sonic hedgehog (Shh) gene promoter, which causes the downregulation of Shh. This process inhibits the Hedgehog (Hh) signaling pathway, impairs the osteogenic differentiation ability of mouse BMSCs (Wang et al., 2017). Fork shaped box C1 (FOXC1) can target DNMT3B and promote its expression. DNMT3B methylates the promoter of C-X-C chemokine ligand 12 (CXCL12), which inhibits the expression of CXCL12. As a result, this process promotes osteogenic differentiation of hBMSCs (Zhang et al., 2024). hBMSCs exhibit lower phosphatase and tensin homolog (PTEN) expression and lower osteogenic potential compared to human dental pulp mesenchymal stem cells (hDPMSCs) due to the high methylation of DNMT3B-mediated PTEN gene promoters (Shen et al., 2019).
DNMT3L is a protein that does not have enzymatic activity but plays a crucial role in promoting de novo DNA methylation. It interacts with DNMT3A and DNMT3B to affect gene expression. However, its role in regulating MSCs osteogenic differentiation is not well studied. In a study by Chih Yi Yang et al., it was found that DNMT3L protein is not expressed in cultured BMSCs. They also discovered that knockout mice derived BMSCs (DNMT3L-KO-MSCs) have impaired in vitro osteogenesis and reduced colony forming ability. It was observed that the osteogenic differentiation ability is linked to the pre-deposition epigenetic characteristics of DNMT3L expressing progenitor cells (Yang et al., 2021).
Mammalian TETs such as TET1, TET2, and TET3, are responsible for DNA demethylation, which can regulate gene expression by opening up chromatin for transcription regulators (Chakraborty et al., 2022). TETs are expressed in various cell lineages and play a crucial role in maintaining MSCs homeostasis and regulating their lineage differentiation (Yu et al., 2019; Alicka et al., 2020) (Table 3). TETs mediated DNA demethylation is essential for maintaining embryonic bone formation. Knocking out TET1/2/3 in mesenchymal stem cells under Prx1 Cre conditions can lead to severe defects in bone development, including delayed closure of the fontanelle, clavicular hypoplasia, and limb shortening. However, any allele that retains the TETs gene can maintain bone formation, indicating the functional redundancy of TETs in bone development (Wang L. et al., 2022).
TETs are molecules that play a crucial role in promoting the process of osteogenic differentiation of MSCs. They are especially important in promoting the differentiation of MSCs into bone cells. The osteogenic potential of hADSCs from elderly donors is improved by an increase in 5hmC levels and increased expression of TET2 and TET3 genes (Yan et al., 2014). During the process of osteogenic differentiation of hADSCs, TET2 is upregulated. Inhibition of TET2 may decrease the expression of osteogenic-related genes and downregulate 5hmC, which can ultimately have a negative effect on osteoporosis (Feng et al., 2020). In patients with aplastic anemia, low expression of TET2 in hBMSCs leads to a decrease in overall 5hmC levels, inhibiting osteogenic differentiation (Li N. et al., 2020). However, hDPSCs derived from three-dimensional sphere culture (3D) are more inclined to differentiate into osteogenic lineage than two-dimensional monolayer culture (2D) hDPSCs. TET1 mRNA transcripts are significantly upregulated in hDPSCs cultured in three-dimensional spheres, and TET1 plays a crucial role in the osteogenic differentiation of hDPSCs cultured in three-dimensional spheres (Raik et al., 2022). The demethylation of the P2rX7 promoter in BMSCs of TET1 and TET2 knockout mice is inhibited, resulting in impaired osteogenic differentiation of mouse BMSCs (Yang et al., 2018). Finally, the hydrogel system developed by Tingting Yu et al. showed an upregulation of TET2 in PDLSCs. TET2 combined with histone deacetylase 1(HDAC1) to inhibit the transcriptional expression of E-cadherin and promote the osteogenic differentiation of PDLSCs (Yu T. et al., 2022).
Some studies have suggested that TETs may impair the osteogenesis process of MSCs. For instance, TET2 knockout mouse BMSCs have shown dysregulation of 5mC hydroxymethylation and altered expression of key genes related to osteogenic differentiation, thereby exhibiting enhanced osteogenic differentiation potential, cell proliferation, and self-renewal (Li et al., 2018). However, other research has suggested that TETs can both inhibit and promote osteogenic differentiation of MSCs. Research results have shown that TET1 in hBMSCs inhibits osteogenic gene transcription by recruiting SIN3A and EZH2, thereby inhibiting osteogenic differentiation, while TET2 leads to low methylation of osteogenic genes and upregulation, promoting osteogenic differentiation. TET1 also plays a role in this process (Cakouros et al., 2019). Although TETs have some inhibitory effect on the osteogenic differentiation of MSCs, they mainly exhibit a promoting effect. However, this situation may be influenced by factors such as the gene regulatory network it participates in, epigenetic modifications, and intracellular environment.
The process of MSCs differentiation involves two steps: lineage shaping (from MSCs to lineage-specific progenitor cells) and maturation (from progenitor cells to specific cell types) (Chen Y. S. et al., 2016). Before directly differentiating into bone cells, MSCs tend to differentiate into osteoblasts. Pre-osteoblasts then develop into mature osteoblasts, which synthesize bone matrix and embed bone cells into it (James, 2013). Multiple signaling pathways play a role in regulating the osteogenic differentiation of MSCs, such as Notch (Fiddes et al., 2018), Hedgehog signaling (Carballo et al., 2018), Bone Morphogenetic Protein (BMP)—Smad (Wang et al., 2014), Wnt signaling (Monroe et al., 2012), Mitogen-activated protein kinase (MAPK) (Shukla et al., 2007), Fibroblast growth factor (FGF) (Ornitz and Marie, 2002), Ras homologous gene family member A/Rho kinase (RhoA/ROCK) (McBeath et al., 2004), and Neuroepidermal growth factor like protein 1 (NELL-1) (Li C. et al., 2019). These signaling pathways interact to form a complex regulatory network that regulates the osteogenic differentiation of MSCs. As targets of these signaling pathways, Runx2 and Osterix are important transcription factors that play a crucial role in the process of osteogenic differentiation of MSCs. In addition to Runx2 and Osterix, there are other transcription factors such as Dlx5, Dlx3, FRA1, Twist1, ZBTB16, and ATF4 that also participate in osteoblast differentiation (Harada and Rodan, 2003; Yang et al., 2004). Once osteogenic signals are activated in pre-osteoblasts, these signaling pathways regulate the expression and function of major transcription factors in osteoblasts. This helps control the expression of downstream bone phenotype genes, ultimately establishing the osteoblast components of mammalian bones (Maroni et al., 2012).
The impact of DNA methylation on the differentiation of MSCs towards bone-forming cells is complex and multi-faceted (Figure 4). Studies have shown that DNA methylation can regulate the expression, activity, and interaction of certain transcription factors, which in turn affect MSCs differentiation (Farshdousti Hagh et al., 2015; Marofi et al., 2019a; Marofi et al., 2019b; Fang et al., 2019; Marofi et al., 2020). Additionally, DNA methylation can also influence the activity of signaling pathways by regulating the expression or methylation status of genes involved in these pathways, ultimately affecting the osteogenic differentiation of MSCs (Table 4).
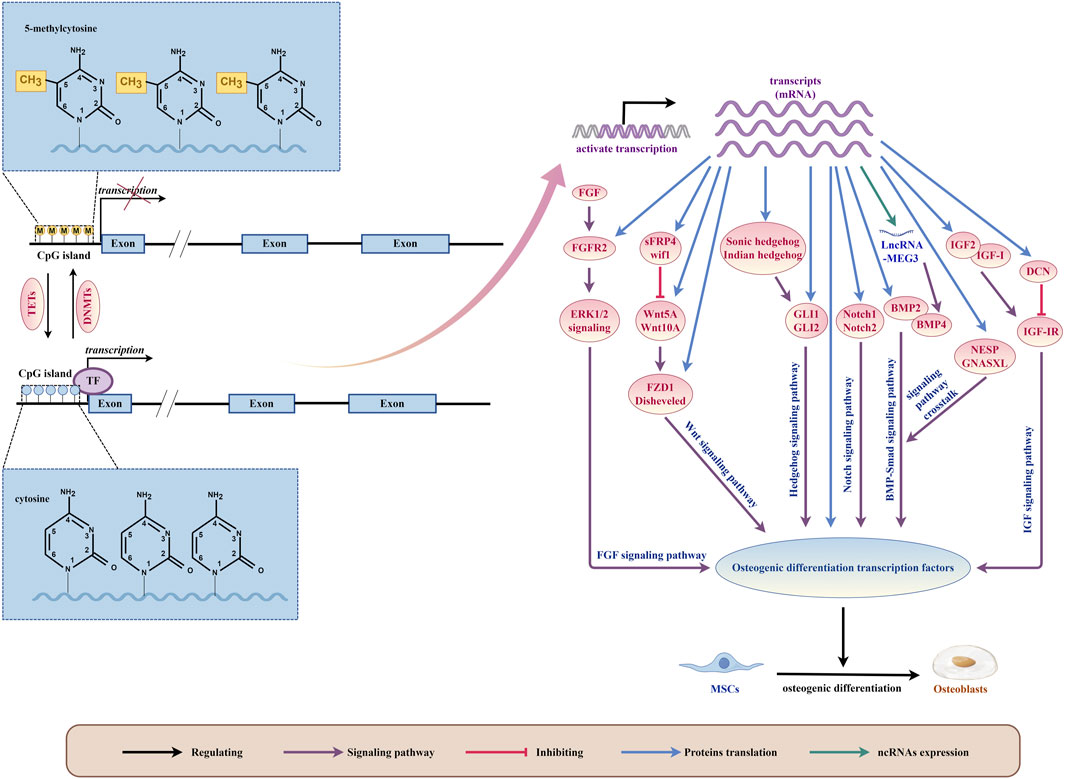
Figure 4. DNA methylation plays a critical role as a regulatory mechanism in orchestrating the osteogenic differentiation of bone marrow mesenchymal stem cells (MSCs) by modulating intricate signaling pathways and osteogenic differentiation transcription factors. (By Figdraw.). This biochemical phenomenon occurs specifically at the 5mC site on the DNA strand, catalyzed by DNA methyltransferases (DNMTs). Conversely, the action of Ten-eleven translocation family proteins (TETs) leads to the oxidation of 5mC to 5hmC, thereby facilitating active demethylation of DNA. This demethylation process dynamically influences gene transcription, thereby enabling the translation of diverse signaling pathway components pivotal for regulating osteogenic transcription factors essential for MSCs’ osteogenic differentiation. Furthermore, transcripts themselves can directly translate osteogenic differentiation transcription factors, further intricately fine-tuning the regulatory machinery governing MSCs’ osteogenic fate determination.
The Wnt signaling pathway is a crucial factor in promoting the osteogenic differentiation of MSCs. The classical Wnt pathway inhibits the primary lipid inducer, PPAR γ Binding protein, with C/EBP alpha. This inhibition prevents adipogenic differentiation and upregulates osteogenic regulatory factors such as Runx2, Dlx5, and Osterix, which promotes osteogenic differentiation. Furthermore, non-classical Wnt pathways induce osteogenic differentiation through different mechanisms (Kim J. H. et al., 2013). During the osteogenic differentiation process of MSCs, a whole-genome DNA methylation analysis revealed the enrichment of the Wnt signaling pathway, demonstrating its critical role in the osteogenic differentiation process of MSCs (Del Real et al., 2017; Cao et al., 2018; Lyu et al., 2019).
As discussed in the previous section of this review, AGEs can negatively impact the ability of MSCs to differentiate into bone cells and promote the formation of bone-dissolving cells. When rat and mouse ADSCs were grown in a medium containing AGEs, it caused an increase in global 5-mC, inhibited the classic Wnt signaling pathway, and hindered osteogenic differentiation (Zhang et al., 2018; Li Y. et al., 2020). In mice with diabetes-induced osteoporosis, the mesenchymal stem cells derived from adipose tissue (DOP ADSCs) had hypermethylation of genes related to osteogenesis and the Wnt/β-catenin signaling pathway, which reduced the activity of the pathway and impaired the osteogenic differentiation of DOP ADSCs (Zhang M. et al., 2022).
DNA methylation plays a significant role in regulating the Wnt signaling pathway during the osteogenic differentiation process of MSCs. This process affects the expression and activity of the components of the Wnt signaling pathway. When isolated from spinal ligaments with ectopic ossification, human mesenchymal stem cells (hMSCs) tend to have an osteogenic lineage, which is associated with low methylation of the Wnt5A gene (Chiba et al., 2015). The Wnt10A gene is a crucial factor that determines the fate of mesenchymal stem cells towards osteoblasts. Its 5′-region is rich in CpG sites, and its expression can be influenced by methylation regulation of these sites, which in turn regulates the lineage determination of MSCs, such as adipogenesis and osteoblast generation (Chen Q. et al., 2016). As the differentiation of osteoblasts increases, the expression of Disheveled also increases. The regulation of the degree of methylation in the Disheveled gene promoter region can affect the expression of Disheveled, which can then affect the expression of Wnt signaling molecules and, thus, affect the osteogenic differentiation of BMSCs (Han et al., 2017). Patients with femoral head necrosis have hBMSCs and Wnt signaling pathway receptor FZD1 with abnormal CpG island hypermethylation, leading to low transcription and translation levels. As a result, the Wnt/β-catenin signaling pathway is inhibited, and the osteogenic ability of hBMSCs is weakened, while their ability to generate fat is enhanced (Wu et al., 2019).
The Wnt antagonist family, which includes sFRP (secreted curl related protein), WIF-1 (Wnt inhibitor 1), Cerberus, and Dickkopf (Dkk), affect the transmission of the Wnt signaling pathway in various ways (Urakami et al., 2006). DNA methylation indirectly influences the Wnt signaling pathway by regulating the expression, activity, and interaction of regulatory factors with Wnt signaling pathway antagonists, thereby affecting the osteogenic differentiation of MSCs. During different stages of osteogenic differentiation in hBMSCs, dynamic downregulation of the Wif1 gene is related to the maintenance of methylation of the gene promoter (Mashhadikhan et al., 2020). Histone lysine demethylase 4A (KDM4A) directly binds to sFRP4 and C/EBP alpha, reducing the DNA methylation level of CpG in the promoter region of the C/EBP alpha and sFRP4 promoters, increasing the expression of C/EBP alpha and sFRP4, inactivating classical Wnt signaling, promoting adipogenic differentiation of mouse BMSCs, and blocking osteogenic differentiation (Qi et al., 2020). Oxidative stress-induced guanine modification relieves the inhibition of the sFRP4 gene by isolating MeCP2 and instead recruiting TBP at the TATA box. This inhibits Wnt signaling and impairs osteogenic differentiation of mouse bone marrow stromal derived ST2 cells (Mori et al., 2014).
The Notch signaling pathway is a pathway that has been conserved throughout evolution and is responsible for regulating cell proliferation and fate determination in both embryonic and adult organs. This pathway plays an important role in maintaining the stemness of multiple stem cells and guiding differentiation (Boni et al., 2008). The Notch signaling pathway has been extensively studied in bone formation and remodeling. It helps to maintain the bone marrow mesenchymal progenitor cell bank and regulates osteoclastogenesis by directly or indirectly regulating osteoblasts (Engin et al., 2008).
In vitro cell culture experiments have shown that activating Notch signaling can have both promoting and inhibiting effects on osteoblast differentiation and mineralization, while inhibiting Notch signaling can also have promoting or inhibiting effects (Ji et al., 2017). However, the contradictory role of the Notch signaling pathway in bone formation appears to be influenced by the DNA methylation process that regulates the osteogenic differentiation of MSCs. Abnormal DNA methylation of Notch1 and Notch2 can lead to transcriptional upregulation, promoting osteogenic differentiation of hADSCs (Jia et al., 2021). On the other hand, increased methylation levels of the Notch1 promoter result in a decrease in Notch1 expression, which can lead to poor multi-lineage differentiation potential of hBMSCs in patients with cyanotic congenital heart disease (CCHD) and impaired osteogenic differentiation of hBMSCs (Wang et al., 2020). In lupus disease mice, low methylation levels of the Notch1 promoter and activation of Notch signaling have been found to lead to impaired osteogenic differentiation of BMSCs (Liu et al., 2015). The contradictory role of Notch in osteogenesis can be explained by considering the relationship between the Notch signaling pathway effect in bones and the cellular environment. Specifically, when Notch is expressed in immature osteoblasts, it can prevent their differentiation, leading to bone loss. Conversely, when Notch is expressed in bone cells, it can initially inhibit bone resorption and increase bone mass (Canalis et al., 2013; Shao et al., 2018).
The Hedgehog signaling (Hh) pathway is highly evolved and plays important roles in embryonic and bone development. Research has shown that Hh signaling positively regulates the differentiation of MSCs into osteoblasts (James et al., 2010; Kim et al., 2010; Jiang et al., 2019). This is achieved by affecting the expression of two important transcription factors, RUNX2 and Osterix (Oliveira et al., 2012).
Hedgehog proteins, which include Sonic Hedgehog (SHH), Indian Hedgehog (IHH), and Desert Hedgehog (DHH), exist in mammals. When the extracellular Hedgehog protein binds to the transmembrane receptor (Ptch), it releases the inhibition of a specific labeled smooth receptor (Smo) and further phosphorylates it. This activation of Smo releases the Hedgehog pathway transcriptional effector Ci/Gli from Cos2 and transfers it to the nucleus, activating the expression of the corresponding downstream target genes (Petrova and Joyner, 2014). Overexpression of SHH significantly increases the osteogenic ability of rat BMSCs, both in vitro and in vivo (Jiang et al., 2019). However, mechanical unloading in mice leads to methylation of the SHH gene promoter and downregulation of SHH, inhibiting the Hh pathway and impairing the osteogenic differentiation ability of mouse BMSCs (Wang et al., 2017). IHH upregulates the expression of RUNX2 and osteogenesis through GLI2, thereby stimulating the differentiation of BMSCs into osteoblasts (Shimoyama et al., 2007). Overexpression of GLI1 can reverse oxidative stress-mediated reduction in the osteogenic differentiation of mouse MSCs (Kim et al., 2010). Studies have shown that demethylation of the IHH promoter in mouse C3H10T1/2 mesenchymal stem cells improves their osteogenic differentiation ability (Tao et al., 2022). The carcinogenic metabolite R-2HG induces a significant DNA hypermethylation state in hBMSCs, inducing high methylation and reducing mRNA levels of SHH, GLI1, and GLI2. Consequently, SHH signaling is inhibited, blocking the osteogenic differentiation of hBMSCs while promoting adipose differentiation of hBMSCs (Liu et al., 2021).
BMP is a type of pleiotropic cytokine that belongs to the transforming growth factor-β (TGF-β) superfamily. BMP signal transduction occurs through type I and type II BMP receptors. When BMP ligands bind to these receptors, homodimers of type II receptors form a tetrameric complex with homodimers of type I receptors. This leads to the phosphorylation of type I receptors, resulting in signal transduction through Smads. This dynamic interaction activates the transcription of specific target genes involved in osteoblast differentiation and bone formation (Wu et al., 2016).
Overexpression of bone morphogenetic protein 2 (BMP2) promotes the differentiation of BMSCs into osteogenic cells. The osteogenic cell line differentiation is believed to be stimulated by the BMP-2 receptor IB, which subsequently activates various transcription factors such as Runx2 and Osterix (Kato et al., 2009; Davis et al., 2011). Lovastatin treatment induces demethylation of the BMP2 promoter, which increases the expression of BMP2 and RUNX2, and promotes the osteogenic lineage differentiation of human endometrial mesenchymal stem cells (eMSCs) (Taghizadeh and Noruzinia, 2017). In human periodontal ligament CD105+ MSCs, 5-aza-dC demethylates BMP2 by increasing the level of hydroxymethylation, which in turn increases BMP2 transcriptional expression. BMP2 signaling molecules activate the phosphorylation and nuclear translocation of Smad protein, which interacts with RUNX2, leading to upregulation of ALP and OCN transcripts. This results in osteoblast differentiation (Sacramento et al., 2022). BMP4 has the potential to regulate the osteogenic differentiation of MSCs. After transfection of BMP4 gene into MSCs of patients with aplastic anemia (AA), the adipogenic differentiation ability is reduced, while the osteogenic differentiation is enhanced (Cheng et al., 2015). High methylation of the promoter of the LncRNA MEG3 gene in children with aplastic anemia (AA) inhibits the expression of LncRNA MEG3. Subsequently, low expression of LncRNA MEG3 leads to downregulation of BMP4 transcriptional activity, impairing the osteogenic differentiation of BMSCs in AA patients (Li H. et al., 2021).
Studies have demonstrated that the dynamic regulation of GNAS and cAMP levels, along with the crosstalk with BMP signaling pathways, are also associated with the initiation and progression of osteoblast differentiation (Zhang et al., 2012). The application of mechanical stress leads to the demethylation and upregulation of critical CpG sites in the GNAS subtype NESP and GNASXL of hADSCs, which promotes early osteogenic differentiation (Vlaikou et al., 2017). This demethylation and upregulation of key CpG sites may be linked to crosstalk in the BMP signaling pathway.
Insulin-like growth factors (IGFs), including insulin-like growth factor 1 (IGF-1) and insulin-like growth factor 2 (IGF-2), have the ability to stimulate and promote the differentiation of stem cells into various lineages in all three embryonic layers. This includes the differentiation of osteoblasts and bone development in vivo. IGF-1, IGF-2, and their receptor type 1 insulin-like growth factor receptor (IGF-1R) are closely associated with osteogenesis (Youssef and Han, 2017).
The ability of BMSCs to stimulate cell growth (mitogenic activity) through IGF-I is reduced in elderly donors due to aging. However, the overexpression of IGF-I can boost the proliferation and bone formation ability of BMSCs from elderly donors (Chen et al., 2017). During the aging process of hBMSCs, the expression level of the decorin (DCN) gene is positively related to the CpG methylation in the second intron of the gene. This methylation inhibits the IGF-I signaling through IGF-IR, which impairs the osteogenic differentiation potential of hBMSCs (Wong et al., 2020).
The imprinting region of the IGF2 gene is controlled by the H19DMR (differentially methylated region) gene, which plays a crucial role in parental-specific silencing of the adjacent H19 and IGF2 genes (Reik et al., 2001). IGF2 is crucial for the differentiation of MSCs into osteoblasts. During the process of osteogenic differentiation, IGF2 and IGF binding protein 2 (IGFBP2) are upregulated (Karasik et al., 2002), which also occurs in dental mesenchymal stem cells (Morsczeck et al., 2010). The methylation of the imprint control region within the IGF2-H19 locus in human deciduous teeth mesenchymal stem cells (SHED) increases by 4%. The upregulation of IGF2 in SHED is associated with the loss of imprints, which gives it higher osteogenic potential compared to hADSCs (Fanganiello et al., 2015).
Fibroblast Growth Factor (FGF) works by binding to fibroblast growth factor receptors (FGFRs) present on cell surfaces. This binding triggers a series of complex downstream signaling pathways, including the PLCγ pathway, the RAS-MAP kinase pathway involving ERK1/2, p38, and JNK kinases, and the PI3K/AKT pathway (Moosa and Wollnik, 2016). FGF signaling plays a crucial role in maintaining bone homeostasis, and the level of FGF receptor 2 (FGFR2) is linked to the osteogenic potential of hMSCs in vitro (Zhang Y. et al., 2022).
Mutations in any of the three members of the FGFRs family (FGFR1, FGFR2, and FGFR3) can lead to an imbalance in FGF signaling and cause premature closure of cranial sutures (Azoury et al., 2017). Studies suggest that nickel exposure at an early age can reduce the methylation of the FGFR2 promoter, which enhances its affinity for the transcription factor Sp1. This leads to the activation of FGFR2, which plays a vital role in regulating ERK1/2 signaling. Consequently, osteogenic differentiation of hMSCs is promoted through ERK1/2 signaling, which results in cranial suture fusion (Weng et al., 2024).
DNA methylation is a crucial biological process that regulates gene transcription and plays a vital role in various cellular activities, including stem cell fate determination, and cell differentiation. MSCs are self-renewing cells with the potential for multiple types of cell differentiation, making them a highly promising candidate for tissue engineering and regenerative medicine applications.
Current research suggests that DNA methylation plays a critical role in the osteogenic differentiation of MSCs. However, directing the osteogenic lineage differentiation of MSCs and utilizing their unique properties for bone tissue engineering represents a significant challenge. Due to variations in their osteogenic differentiation potential among different tissue sources, MSCs derived from diverse sources cannot be used for therapeutic purposes. Although some MSCs, such as BMSCs, have higher osteogenic efficiency, their limited availability and discomfort associated with obtaining them limit their widespread use. Future research should focus on enhancing the osteogenic differentiation potential of MSCs through DNA methylation mechanisms or developing novel strategies for osteogenic differentiation therapy of bone defect-related MSCs based on DNA methylation.
MSCs have been studied extensively and used successfully in bone tissue engineering and clinical therapy under in vitro culture conditions. However, various factors in vivo could impair their osteogenic differentiation, and MSCs need to be guided to the corresponding sites in vivo to exert their effects. Therefore, the study of DNA methylation in the in vivo osteogenic differentiation of MSCs and their directional migration in vivo is particularly crucial.
By identifying the target genes regulated by DNA methylation during MSCs osteogenic differentiation, we can develop novel epigenetic drugs using new drug screening techniques and drug delivery methods. The development of such epigenetic drugs is expected to bring new therapeutic strategies and drug choices to the field of orthopedics, potentially improving bone tissue regeneration and treatment outcomes for patients.
ZL: Supervision, Writing–original draft, Writing–review and editing. QS: Funding acquisition, Resources, Supervision, Writing–review and editing. YS: Validation, Writing–original draft. AT: Validation, Writing–original draft. JT: Funding acquisition, Resources, Supervision, Writing–review and editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by grants from the General Program of National Natural Science Foundation of China (No. 82174494) and the Hubei Natural Science Foundation project (2022CFB192).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Ahmadi, A., Mazloomnejad, R., Kasravi, M., Gholamine, B., Bahrami, S., Sarzaeem, M. M., et al. (2022). Recent advances on small molecules in osteogenic differentiation of stem cells and the underlying signaling pathways. Stem Cell Res. Ther. 13 (1), 518. doi:10.1186/s13287-022-03204-4
Alicka, M., Kornicka-Garbowska, K., Kucharczyk, K., Kępska, M., Rӧcken, M., and Marycz, K. (2020). Age-dependent impairment of adipose-derived stem cells isolated from horses. Stem Cell Res. Ther. 11 (1), 4. doi:10.1186/s13287-019-1512-6
Asadipooya, K., and Uy, E. M. (2019). Advanced glycation end products (AGEs), receptor for AGEs, diabetes, and bone: review of the literature. J. Endocr. Soc. 3 (10), 1799–1818. doi:10.1210/js.2019-00160
Azoury, S. C., Reddy, S., Shukla, V., and Deng, C. X. (2017). Fibroblast growth factor receptor 2 (FGFR2) mutation related syndromic craniosynostosis. Int. J. Biol. Sci. 13 (12), 1479–1488. doi:10.7150/ijbs.22373
Bachman, M., Uribe-Lewis, S., Yang, X., Williams, M., Murrell, A., and Balasubramanian, S. (2014). 5-Hydroxymethylcytosine is a predominantly stable DNA modification. Nat. Chem. 6 (12), 1049–1055. doi:10.1038/nchem.2064
Boni, A., Urbanek, K., Nascimbene, A., Hosoda, T., Zheng, H., Delucchi, F., et al. (2008). Notch1 regulates the fate of cardiac progenitor cells. Proc. Natl. Acad. Sci. U. S. A. 105 (40), 15529–15534. doi:10.1073/pnas.0808357105
Busque, L., Buscarlet, M., Mollica, L., and Levine, R. L. (2018). Concise review: age-related clonal hematopoiesis: stem cells tempting the devil. Stem Cells 36 (9), 1287–1294. doi:10.1002/stem.2845
Cakouros, D., and Gronthos, S. (2020). Epigenetic regulators of mesenchymal stem/stromal cell lineage determination. Curr. Osteoporos. Rep. 18 (5), 597–605. doi:10.1007/s11914-020-00616-0
Cakouros, D., Hemming, S., Gronthos, K., Liu, R., Zannettino, A., Shi, S., et al. (2019). Specific functions of TET1 and TET2 in regulating mesenchymal cell lineage determination. Curr. Osteoporos. Rep. 12 (1), 3. doi:10.1186/s13072-018-0247-4
Canalis, E., Parker, K., Feng, J. Q., and Zanotti, S. (2013). Osteoblast lineage-specific effects of notch activation in the skeleton. Endocrinology 154 (2), 623–634. doi:10.1210/en.2012-1732
Cancedda, R., Giannoni, P., and Mastrogiacomo, M. (2007). A tissue engineering approach to bone repair in large animal models and in clinical practice. Biomaterials 28 (29), 4240–4250. doi:10.1016/j.biomaterials.2007.06.023
Cao, W., Yang, X., Hu, X. H., Li, J., Tian, J., OuYang, R., et al. (2023). miR-344d-3p regulates osteogenic and adipogenic differentiation of mouse mandibular bone marrow mesenchymal stem cells. PeerJ 11, e14838. doi:10.7717/peerj.14838
Cao, Y., Yang, H., Jin, L., Du, J., and Fan, Z. (2018). Genome-wide DNA methylation analysis during osteogenic differentiation of human bone marrow mesenchymal stem cells. Stem Cells 2018, 8238496. doi:10.1155/2018/8238496
Carballo, G. B., Honorato, J. R., de Lopes, G. P. F., and Spohr, T. (2018). A highlight on Sonic hedgehog pathway. Cell Commun. Signal 16 (1), 11. doi:10.1186/s12964-018-0220-7
Chakraborty, R., Ostriker, A. C., Xie, Y., Dave, J. M., Gamez-Mendez, A., Chatterjee, P., et al. (2022). Histone acetyltransferases p300 and CBP coordinate distinct chromatin remodeling programs in vascular smooth muscle plasticity. Circulation, 145(23), 1720–1737. doi:10.1161/circulationaha.121.057599
Chen, C. Y., Tseng, K. Y., Lai, Y. L., Chen, Y. S., Lin, F. H., and Lin, S. (2017). Overexpression of insulin-like growth factor 1 enhanced the osteogenic capability of aging bone marrow mesenchymal stem cells. Theranostics 7 (6), 1598–1611. doi:10.7150/thno.16637
Chen, Q., Shou, P., Zheng, C., Jiang, M., Cao, G., Yang, Q., et al. (2016a). Fate decision of mesenchymal stem cells: adipocytes or osteoblasts? Cell Death Differ. 23 (7), 1128–1139. doi:10.1038/cdd.2015.168
Chen, Y. S., Wu, R., Yang, X., Kou, S., MacDougald, O. A., Yu, L., et al. (2016b). Inhibiting DNA methylation switches adipogenesis to osteoblastogenesis by activating Wnt10a. Sci. Rep. 6, 25283. doi:10.1038/srep25283
Chen, Z., Zheng, J., Hong, H., Chen, D., Deng, L., Zhang, X., et al. (2020). lncRNA HOTAIRM1 promotes osteogenesis of hDFSCs by epigenetically regulating HOXA2 via DNMT1 in vitro. J. Cell Physiol. 235 (11), 8507–8519. doi:10.1002/jcp.29695
Cheng, H. C., Liu, S. W., Li, W., Zhao, X. F., Zhao, X., Cheng, M., et al. (2015). Arsenic trioxide regulates adipogenic and osteogenic differentiation in bone marrow MSCs of aplastic anemia patients through BMP4 gene. Acta Biochim. Biophys. Sin. (Shanghai) 47 (9), 673–679. doi:10.1093/abbs/gmv065
Chiba, N., Furukawa, K., Takayama, S., Asari, T., Chin, S., Harada, Y., et al. (2015). Decreased DNA methylation in the promoter region of the WNT5A and GDNF genes may promote the osteogenicity of mesenchymal stem cells from patients with ossified spinal ligaments. J. Pharmacol. Sci. 127 (4), 467–473. doi:10.1016/j.jphs.2015.03.008
da Silva Meirelles, L., Chagastelles, P. C., and Nardi, N. B. (2006). Mesenchymal stem cells reside in virtually all post-natal organs and tissues. J. Cell Sci. 119 (Pt 11), 2204–2213. doi:10.1242/jcs.02932
Davis, H. E., Case, E. M., Miller, S. L., Genetos, D. C., and Leach, J. K. (2011). Osteogenic response to BMP-2 of hMSCs grown on apatite-coated scaffolds. Biotechnol. Bioeng. 108 (11), 2727–2735. doi:10.1002/bit.23227
Dawlaty, M. M., Breiling, A., Le, T., Barrasa, M. I., Raddatz, G., Gao, Q., et al. (2014). Loss of Tet enzymes compromises proper differentiation of embryonic stem cells. Dev. Cell 29 (1), 102–111. doi:10.1016/j.devcel.2014.03.003
Del Real, A., Pérez-Campo, F. M., Fernández, A. F., Sañudo, C., Ibarbia, C. G., Pérez-Núñez, M. I., et al. (2017). Differential analysis of genome-wide methylation and gene expression in mesenchymal stem cells of patients with fractures and osteoarthritis. Epigenetics 12 (2), 113–122. doi:10.1080/15592294.2016.1271854
Di Ruscio, A., Ebralidze, A. K., Benoukraf, T., Amabile, G., Goff, L. A., Terragni, J., et al. (2013). DNMT1-interacting RNAs block gene-specific DNA methylation. Nature 503 (7476), 371–376. doi:10.1038/nature12598
Engin, F., Yao, Z., Yang, T., Zhou, G., Bertin, T., Jiang, M. M., et al. (2008). Dimorphic effects of Notch signaling in bone homeostasis. Nat. Med. 14 (3), 299–305. doi:10.1038/nm1712
Fang, B., Wang, D., Zheng, J., Wei, Q., Zhan, D., Liu, Y., et al. (2019). Involvement of tumor necrosis factor alpha in steroid-associated osteonecrosis of the femoral head: friend or foe? Stem Cell Res. Ther. 10 (1), 5. doi:10.1186/s13287-018-1112-x
Fanganiello, R. D., Ishiy, F. A., Kobayashi, G. S., Alvizi, L., Sunaga, D. Y., and Passos-Bueno, M. R. (2015). Increased in vitro osteopotential in SHED associated with higher IGF2 expression when compared with hASCs. Stem Cell Rev. Rep. 11 (4), 635–644. doi:10.1007/s12015-015-9592-x
Farshdousti Hagh, M., Noruzinia, M., Mortazavi, Y., Soleimani, M., Kaviani, S., Abroun, S., et al. (2015). Different methylation patterns of RUNX2, OSX, DLX5 and BSP in osteoblastic differentiation of mesenchymal stem cells. Cell J. 17 (1), 71–82. doi:10.22074/cellj.2015.513
Farshdousti Hagh, M., Noruzinia, M., Mortazavi, Y., Soleimani, M., Kaviani, S., and Mahmodinia Maymand, M. (2012). Zoledrinic acid induces steoblastic differentiation of mesenchymal stem cells without change in hypomethylation status of OSTERIX promoter. Cell J. 14 (2), 90–97.
Feng, L., Zhou, J., Xia, B., and Tian, B. F. (2020). The positive effect of TET2 on the osteogenic differentiation of human adipose-derived mesenchymal stem cells. Cell Reprogr. 22 (1), 3–13. doi:10.1089/cell.2019.0045
Feng, Y. F., Wang, L., Zhang, Y., Li, X., Ma, Z. S., Zou, J. W., et al. (2013). Effect of reactive oxygen species overproduction on osteogenesis of porous titanium implant in the present of diabetes mellitus. Biomaterials 34 (9), 2234–2243. doi:10.1016/j.biomaterials.2012.12.023
Ferreira, R. S., Assis, R. I. F., Feltran, G. d. S., do Rosário Palma, I. C., Françoso, B. G., Zambuzzi, W. F., et al. (2022). Genome-wide DNA (hydroxy) methylation reveals the individual epigenetic landscape importance on osteogenic phenotype acquisition in periodontal ligament cells. J. Periodontol. 93 (3), 435–448. doi:10.1002/jper.21-0218
Fiddes, I. T., Lodewijk, G. A., Mooring, M., Bosworth, C. M., Ewing, A. D., Mantalas, G. L., et al. (2018). Human-specific NOTCH2NL genes affect notch signaling and cortical neurogenesis. Cell 173 (6), 1356–1369. doi:10.1016/j.cell.2018.03.051
Friedenstein, A. J., Chailakhjan, R. K., and Lalykina, K. S. (1970). The development of fibroblast colonies in monolayer cultures of Guinea-pig bone marrow and spleen cells. Cell Tissue Kinet. 3 (4), 393–403. doi:10.1111/j.1365-2184.1970.tb00347.x
Glemžaitė, M., and Navakauskienė, R. (2016). Osteogenic differentiation of human amniotic fluid mesenchymal stem cells is determined by epigenetic changes. Stem Cells Int. 2016, 6465307. doi:10.1155/2016/6465307
Goll, M. G., and Bestor, T. H. (2005). Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 74, 481–514. doi:10.1146/annurev.biochem.74.010904.153721
Greenberg, M. V. C., and Bourc'his, D. (2019). The diverse roles of DNA methylation in mammalian development and disease. Nat. Rev. Mol. Cell Biol. 20 (10), 590–607. doi:10.1038/s41580-019-0159-6
Han, L., Wang, B., Wang, R., Gong, S., Chen, G., and Xu, W. (2019). The shift in the balance between osteoblastogenesis and adipogenesis of mesenchymal stem cells mediated by glucocorticoid receptor. Stem Cell Res. Ther. 10 (1), 377. doi:10.1186/s13287-019-1498-0
Han, X., Li, X., Zhong, G., and Liu, Z. (2017). Regulation of osteogenic differentiation by DNA methylation of the dishevelled gene in bone marrow mesenchymal stem cells. Am. J. Transl. Res. 9 (11), 4848–4855.
Harada, S., and Rodan, G. A. (2003). Control of osteoblast function and regulation of bone mass. Nature 423 (6937), 349–355. doi:10.1038/nature01660
Hiew, V. V., and Teoh, P. L. (2024). Differential gene expression of Wharton's jelly-derived mesenchymal cells mediated by graphene oxide in basal and osteo-induced media. Mol. Biol. Rep. 51 (1), 383. doi:10.1007/s11033-024-09324-9
Hu, L., Yin, C., Zhao, F., Ali, A., Ma, J., and Qian, A. (2018). Mesenchymal stem cells: cell fate decision to osteoblast or adipocyte and application in osteoporosis treatment. Int. J. Mol. Sci. 19 (2), 360. doi:10.3390/ijms19020360
Husain, A., and Jeffries, M. A. (2017). Epigenetics and bone remodeling. Curr. Osteoporos. Rep. 15 (5), 450–458. doi:10.1007/s11914-017-0391-y
Hyslop, L., Stojkovic, M., Armstrong, L., Walter, T., Stojkovic, P., Przyborski, S., et al. (2005). Downregulation of NANOG induces differentiation of human embryonic stem cells to extraembryonic lineages. Stem Cells 23 (8), 1035–1043. doi:10.1634/stemcells.2005-0080
James, A. W. (2013). Review of signaling pathways governing MSC osteogenic and adipogenic differentiation. Sci. (Cairo) 2013, 684736. doi:10.1155/2013/684736
James, A. W., Leucht, P., Levi, B., Carre, A. L., Xu, Y., Helms, J. A., et al. (2010). Sonic Hedgehog influences the balance of osteogenesis and adipogenesis in mouse adipose-derived stromal cells. Tissue Eng. Part A 16 (8), 2605–2616. doi:10.1089/ten.TEA.2010.0048
Ji, Y., Ke, Y., and Gao, S. (2017). Intermittent activation of notch signaling promotes bone formation. Am. J. Transl. Res. 9 (6), 2933–2944.
Jia, B., Chen, J., Wang, Q., Sun, X., Han, J., Guastaldi, F., et al. (2021). SIRT6 promotes osteogenic differentiation of adipose-derived mesenchymal stem cells through antagonizing DNMT1. Front. Cell Dev. Biol. 9, 648627. doi:10.3389/fcell.2021.648627
Jiang, Z. L., Jin, H., Liu, Z. S., Liu, M. Y., Cao, X. F., Jiang, Y. Y., et al. (2019). Lentiviral-mediated Shh reverses the adverse effects of high glucose on osteoblast function and promotes bone formation via Sonic hedgehog signaling. Mol. Med. Rep. 20 (4), 3265–3275. doi:10.3892/mmr.2019.10540
Jones, P. A. (2012). Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 13 (7), 484–492. doi:10.1038/nrg3230
Jung, Y. D., Park, S. K., Kang, D., Hwang, S., Kang, M. H., Hong, S. W., et al. (2020). Epigenetic regulation of miR-29a/miR-30c/DNMT3A axis controls SOD2 and mitochondrial oxidative stress in human mesenchymal stem cells. Redox Biol. 37, 101716. doi:10.1016/j.redox.2020.101716
Jurkowska, R. Z., Jurkowski, T. P., and Jeltsch, A. (2011). Structure and function of mammalian DNA methyltransferases. Chembiochem 12 (2), 206–222. doi:10.1002/cbic.201000195
Kang, R., Zhou, Y., Tan, S., Zhou, G., Aagaard, L., Xie, L., et al. (2015). Mesenchymal stem cells derived from human induced pluripotent stem cells retain adequate osteogenicity and chondrogenicity but less adipogenicity. Stem Cell Res. Ther. 6 (1), 144. doi:10.1186/s13287-015-0137-7
Karasik, D., Rosen, C. J., Hannan, M. T., Broe, K. E., Dawson-Hughes, B., Gagnon, D. R., et al. (2002). Insulin-like growth factor binding proteins 4 and 5 and bone mineral density in elderly men and women. Calcif. Tissue Int. 71 (4), 323–328. doi:10.1007/s00223-002-1002-0
Kato, S., Kawabata, N., Suzuki, N., Ohmura, M., and Takagi, M. (2009). Bone morphogenetic protein-2 induces the differentiation of a mesenchymal progenitor cell line, ROB-C26, into mature osteoblasts and adipocytes. Life Sci. 84 (9-10), 302–310. doi:10.1016/j.lfs.2008.12.011
Kim, E. J., Kang, I. H., Lee, J. W., Jang, W. G., and Koh, J. T. (2013a). MiR-433 mediates ERRγ-suppressed osteoblast differentiation via direct targeting to Runx2 mRNA in C3H10T1/2 cells. Life Sci. 92 (10), 562–568. doi:10.1016/j.lfs.2013.01.015
Kim, J. H., Liu, X., Wang, J., Chen, X., Zhang, H., Kim, S. H., et al. (2013b). Wnt signaling in bone formation and its therapeutic potential for bone diseases. Ther. Adv. Musculoskelet. Dis. 5 (1), 13–31. doi:10.1177/1759720x12466608
Kim, W. K., Meliton, V., Bourquard, N., Hahn, T. J., and Parhami, F. (2010). Hedgehog signaling and osteogenic differentiation in multipotent bone marrow stromal cells are inhibited by oxidative stress. J. Cell Biochem. 111 (5), 1199–1209. doi:10.1002/jcb.22846
Kouzarides, T. (2007). Chromatin modifications and their function. Cell 128 (4), 693–705. doi:10.1016/j.cell.2007.02.005
Kriukienė, E., Tomkuvienė, M., and Klimašauskas, S. (2024). 5-Hydroxymethylcytosine: the many faces of the sixth base of mammalian DNA. DNA 53 (5), 2264–2283. doi:10.1039/d3cs00858d
Lee, J. T. (2012). Epigenetic regulation by long noncoding RNAs. Science 338 (6113), 1435–1439. doi:10.1126/science.1231776
Lei, H., Oh, S. P., Okano, M., Jüttermann, R., Goss, K. A., Jaenisch, R., et al. (1996). De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development 122 (10), 3195–3205. doi:10.1242/dev.122.10.3195
Leonhardt, H., Page, A. W., Weier, H. U., and Bestor, T. H. (1992). A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell 71(5), 865–873. doi:10.1016/0092-8674(92)90561-p
Levi, B., and Longaker, M. T. (2011). Concise review: adipose-derived stromal cells for skeletal regenerative medicine. Stem Cells 29 (4), 576–582. doi:10.1002/stem.612
Li, C., Zhang, X., Zheng, Z., Nguyen, A., Ting, K., and Soo, C. (2019a). Nell-1 is a key functional modulator in osteochondrogenesis and beyond. J. Dent. Res. 98 (13), 1458–1468. doi:10.1177/0022034519882000
Li, E., Bestor, T. H., and Jaenisch, R. (1992). Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69 (6), 915–926. doi:10.1016/0092-8674(92)90611-f
Li, H., Xu, X., Wang, D., Zhang, Y., Chen, J., Li, B., et al. (2021a). Hypermethylation-mediated downregulation of long non-coding RNA MEG3 inhibits osteogenic differentiation of bone marrow mesenchymal stem cells and promotes pediatric aplastic anemia. Int. Immunopharmacol. 93, 107292. doi:10.1016/j.intimp.2020.107292
Li, J., Zhang, N., Huang, X., Xu, J., Fernandes, J. C., Dai, K., et al. (2013). Dexamethasone shifts bone marrow stromal cells from osteoblasts to adipocytes by C/EBPalpha promoter methylation. Cell Death Dis. 4 (10), e832. doi:10.1038/cddis.2013.348
Li, K., Ning, T., Wang, H., Jiang, Y., Zhang, J., and Ge, Z. (2020a). Nanosecond pulsed electric fields enhance mesenchymal stem cells differentiation via DNMT1-regulated OCT4/NANOG gene expression. Stem Cell Res. Ther. 11 (1), 308. doi:10.1186/s13287-020-01821-5
Li, L., Ling, Z., Dong, W., Chen, X., Vater, C., Liao, H., et al. (2019b). Dnmt3a-Mediated DNA methylation changes regulate osteogenic differentiation of hMSCs cultivated in the 3D scaffolds under oxidative stress. Oxid. Med. Cell. Longev. 2019, 4824209. doi:10.1155/2019/4824209
Li, L., Wang, H., Chen, X., Li, X., Wang, G., Jie, Z., et al. (2021b). Oxidative stress-induced hypermethylation of KLF5 promoter mediated by DNMT3B impairs osteogenesis by diminishing the interaction with β-catenin. Antioxid. Redox Signal 35 (1), 1–20. doi:10.1089/ars.2020.8200
Li, N., Liu, L., Liu, Y., Luo, S., Song, Y., and Fang, B. (2020b). miR-144-3p suppresses osteogenic differentiation of BMSCs from patients with aplastic anemia through repression of TET2. Mol. Ther. Nucleic Acids 19, 619–626. doi:10.1016/j.omtn.2019.12.017
Li, R., Zhou, Y., Cao, Z., Liu, L., Wang, J., Chen, Z., et al. (2018). TET2 loss dysregulates the behavior of bone marrow mesenchymal stromal cells and accelerates tet2(-/-)-driven myeloid malignancy progression. Stem Cell Rep. 10 (1), 166–179. doi:10.1016/j.stemcr.2017.11.019
Li, Y., Gorelik, G., Strickland, F. M., and Richardson, B. C. (2014). Oxidative stress, T cell DNA methylation, and lupus. Arthritis Rheumatol. 66 (6), 1574–1582. doi:10.1002/art.38427
Li, Y., Wang, L., Zhang, M., Huang, K., Yao, Z., Rao, P., et al. (2020c). Advanced glycation end products inhibit the osteogenic differentiation potential of adipose-derived stem cells by modulating Wnt/β-catenin signalling pathway via DNA methylation. Cell Prolif. 53 (6), e12834. doi:10.1111/cpr.12834
Lin, S. P., Chiu, F. Y., Wang, Y., Yen, M. L., Kao, S. Y., and Hung, S. C. (2014). RB maintains quiescence and prevents premature senescence through upregulation of DNMT1 in mesenchymal stromal cells. Stem Cell Rep. 3 (6), 975–986. doi:10.1016/j.stemcr.2014.10.002
Liu, H., Liu, Z., Du, J., He, J., Lin, P., Amini, B., et al. (2016). Thymidine phosphorylase exerts complex effects on bone resorption and formation in myeloma. Sci. Transl. Med. 8 (353), 353ra113. doi:10.1126/scitranslmed.aad8949
Liu, L., Hu, K., Feng, J., Wang, H., Fu, S., Wang, B., et al. (2021). The oncometabolite R-2-hydroxyglutarate dysregulates the differentiation of human mesenchymal stromal cells via inducing DNA hypermethylation. DNA hypermethylation 21 (1), 36. doi:10.1186/s12885-020-07744-x
Liu, S., Liu, D., Chen, C., Hamamura, K., Moshaverinia, A., Yang, R., et al. (2015). MSC transplantation improves osteopenia via epigenetic regulation of notch signaling in lupus. Cell Metab. 22 (4), 606–618. doi:10.1016/j.cmet.2015.08.018
Liu, T. M., Wu, Y. N., Guo, X. M., Hui, J. H., Lee, E. H., and Lim, B. (2009). Effects of ectopic Nanog and Oct4 overexpression on mesenchymal stem cells. Stem Cells Dev. 18 (7), 1013–1022. doi:10.1089/scd.2008.0335
Lu, Y., Qu, H., Qi, D., Xu, W., Liu, S., Jin, X., et al. (2019). OCT4 maintains self-renewal and reverses senescence in human hair follicle mesenchymal stem cells through the downregulation of p21 by DNA methyltransferases. Stem Cell Res. Ther. 10 (1), 28. doi:10.1186/s13287-018-1120-x
Luczak, M. W., and Jagodziński, P. P. (2006). The role of DNA methylation in cancer development. Folia Histochem Cytobiol. 44 (3), 143–154.
Lyu, M., Zheng, Y., Jia, L., Zheng, Y., Liu, Y., Lin, Y., et al. (2019). Genome-wide DNA-methylation profiles in human bone marrow mesenchymal stem cells on titanium surfaces. Eur. J. Oral Sci. 127 (3), 196–209. doi:10.1111/eos.12607
Marofi, F., Choupani, J., Solali, S., Vahedi, G., Hassanzadeh, A., Gharibi, T., et al. (2020). ATF4, DLX3, FRA1, MSX2, C/EBP-ζ, and C/EBP-α shape the molecular basis of therapeutic effects of zoledronic acid in bone disorders. Anticancer Agents Med. Chem. 20 (18), 2274–2284. doi:10.2174/1871520620666200721101904
Marofi, F., Hassanzadeh, A., Solali, S., Vahedi, G., Mousavi Ardehaie, R., Salarinasab, S., et al. (2019a). Epigenetic mechanisms are behind the regulation of the key genes associated with the osteoblastic differentiation of the mesenchymal stem cells: the role of zoledronic acid on tuning the epigenetic changes. J. Cell Physiol. 234 (9), 15108–15122. doi:10.1002/jcp.28152
Marofi, F., Vahedi, G., Solali, S., Alivand, M., Salarinasab, S., Zadi Heydarabad, M., et al. (2019b). Gene expression of TWIST1 and ZBTB16 is regulated by methylation modifications during the osteoblastic differentiation of mesenchymal stem cells. stem cells 234 (5), 6230–6243. doi:10.1002/jcp.27352
Maroni, P., Brini, A. T., Arrigoni, E., de Girolamo, L., Niada, S., Matteucci, E., et al. (2012). Chemical and genetic blockade of HDACs enhances osteogenic differentiation of human adipose tissue-derived stem cells by oppositely affecting osteogenic and adipogenic transcription factors. Biochem. Biophys. Res. Commun. 428 (2), 271–277. doi:10.1016/j.bbrc.2012.10.044
Mashhadikhan, M., Kheiri, H., and Dehghanifard, A. (2020). DNA methylation and gene expression of sFRP2, sFRP4, Dkk 1, and Wif1 during osteoblastic differentiation of bone marrow derived mesenchymal stem cells. J. Oral Biosci. 62 (4), 349–356. doi:10.1016/j.job.2020.08.001
McBeath, R., Pirone, D. M., Nelson, C. M., Bhadriraju, K., and Chen, C. S. (2004). Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev. Cell 6 (4), 483–495. doi:10.1016/s1534-5807(04)00075-9
Monroe, D. G., McGee-Lawrence, M. E., Oursler, M. J., and Westendorf, J. J. (2012). Update on Wnt signaling in bone cell biology and bone disease. Gene 492 (1), 1–18. doi:10.1016/j.gene.2011.10.044
Montecino, M., Carrasco, M. E., and Nardocci, G. (2020). Epigenetic control of osteogenic lineage commitment. Front. Cell Dev. Biol. 8, 611197. doi:10.3389/fcell.2020.611197
Moore, L. D., Le, T., and Fan, G. (2013). DNA methylation and its basic function. Neuropsychopharmacology 38 (1), 23–38. doi:10.1038/npp.2012.112
Moosa, S., and Wollnik, B. (2016). Altered FGF signalling in congenital craniofacial and skeletal disorders. Semin. Cell Dev. Biol. 53, 115–125. doi:10.1016/j.semcdb.2015.12.005
Mori, K., Kitazawa, R., Kondo, T., Mori, M., Hamada, Y., Nishida, M., et al. (2014). Diabetic osteopenia by decreased β-catenin signaling is partly induced by epigenetic derepression of sFRP-4 gene. PLoS One 9 (7), e102797. doi:10.1371/journal.pone.0102797
Morsczeck, C., Völlner, F., Saugspier, M., Brandl, C., Reichert, T. E., Driemel, O., et al. (2010). Comparison of human dental follicle cells (DFCs) and stem cells from human exfoliated deciduous teeth (SHED) after neural differentiation in vitro. Clin. Oral Investig. 14 (4), 433–440. doi:10.1007/s00784-009-0310-4
Muinos-López, E., Ripalda-Cemboráin, P., López-Martínez, T., González-Gil, A. B., Lamo-Espinosa, J. M., Valentí, A., et al. (2016). Hypoxia and reactive oxygen species homeostasis in mesenchymal progenitor cells define a molecular mechanism for fracture nonunion. Stem Cells 34 (9), 2342–2353. doi:10.1002/stem.2399
Neri, F., Rapelli, S., Krepelova, A., Incarnato, D., Parlato, C., Basile, G., et al. (2017). Intragenic DNA methylation prevents spurious transcription initiation. Nature 543 (7643), 72–77. doi:10.1038/nature21373
O'Hagan, H. M., Wang, W., Sen, S., Destefano Shields, C., Lee, S. S., Zhang, Y. W., et al. (2011). Oxidative damage targets complexes containing DNA methyltransferases, SIRT1, and polycomb members to promoter CpG Islands. Cancer Cell 20 (5), 606–619. doi:10.1016/j.ccr.2011.09.012
Okano, M., Bell, D. W., Haber, D. A., and Li, E. (1999). DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99 (3), 247–257. doi:10.1016/s0092-8674(00)81656-6
Okano, M., Xie, S., and Li, E. (1998). Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat. Genet. 19 (3), 219–220. doi:10.1038/890
Oliveira, F. S., Bellesini, L. S., Defino, H. L., da Silva Herrero, C. F., Beloti, M. M., and Rosa, A. L. (2012). Hedgehog signaling and osteoblast gene expression are regulated by purmorphamine in human mesenchymal stem cells. J. Cell Biochem. 113 (1), 204–208. doi:10.1002/jcb.23345
Ooi, S. K., and Bestor, T. H. (2008). The colorful history of active DNA demethylation. Cell 133 (7), 1145–1148. doi:10.1016/j.cell.2008.06.009
Ornitz, D. M., and Marie, P. J. (2002). FGF signaling pathways in endochondral and intramembranous bone development and human genetic disease. Genes Dev. 16 (12), 1446–1465. doi:10.1101/gad.990702
Patil, V., Ward, R. L., and Hesson, L. B. (2014). The evidence for functional non-CpG methylation in mammalian cells. Epigenetics 9 (6), 823–828. doi:10.4161/epi.28741
Petrova, R., and Joyner, A. L. (2014). Roles for Hedgehog signaling in adult organ homeostasis and repair. Development 141 (18), 3445–3457. doi:10.1242/dev.083691
Pitrone, M., Pizzolanti, G., Coppola, A., Tomasello, L., Martorana, S., Pantuso, G., et al. (2019). Knockdown of NANOG reduces cell proliferation and induces G0/G1 cell cycle arrest in human adipose stem cells. Stem Cells 20 (10), 2580. doi:10.3390/ijms20102580
Pradhan, S., Bacolla, A., Wells, R. D., and Roberts, R. J. (1999). Recombinant human DNA (cytosine-5) methyltransferase. I. Expression, purification, and comparison of de novo and maintenance methylation. J. Biol. Chem. 274 (46), 33002–33010. doi:10.1074/jbc.274.46.33002
Qi, Q., Wang, Y., Wang, X., Yang, J., Xie, Y., Zhou, J., et al. (2020). Histone demethylase KDM4A regulates adipogenic and osteogenic differentiation via epigenetic regulation of C/EBPα and canonical Wnt signaling. Cell Mol. Life Sci. 77 (12), 2407–2421. doi:10.1007/s00018-019-03289-w
Raik, S., Thakur, R., Rattan, V., Kumar, N., Pal, A., and Bhattacharyya, S. (2022). Temporal modulation of DNA methylation and gene expression in monolayer and 3D spheroids of dental pulp stem cells during osteogenic differentiation: a comparative study. Study 19 (6), 1267–1282. doi:10.1007/s13770-022-00485-x
Rasmussen, K. D., and Helin, K. (2016). Role of TET enzymes in DNA methylation, development, and cancer. Dev. cancer 30 (7), 733–750. doi:10.1101/gad.276568.115
Rastegar, F., Shenaq, D., Huang, J., Zhang, W., Zhang, B. Q., He, B. C., et al. (2010). Mesenchymal stem cells: molecular characteristics and clinical applications. World J. Stem Cells 2 (4), 67–80. doi:10.4252/wjsc.v2.i4.67
Reik, W., Dean, W., and Walter, J. (2001). Epigenetic reprogramming in mammalian development. Science 293 (5532), 1089–1093. doi:10.1126/science.1063443
Robert, M. F., Morin, S., Beaulieu, N., Gauthier, F., Chute, I. C., Barsalou, A., et al. (2003). DNMT1 is required to maintain CpG methylation and aberrant gene silencing in human cancer cells. Nat. Genet. 33 (1), 61–65. doi:10.1038/ng1068
Robertson, K. D. (2005). DNA methylation and human disease. Nat. Rev. Genet. 6 (8), 597–610. doi:10.1038/nrg1655
Sacramento, C. M., Assis, R. I. F., Saito, M. T., Coletta, R. D., da Rocha Dourado, M., Sallum, E. A., et al. (2022). BMP-2 and asporin expression regulate 5-aza-dC-mediated osteoblast/cementoblast differentiation of periodontal dental ligament mesenchymal progenitor cells. Differentiation 124, 17–27. doi:10.1016/j.diff.2022.02.003
Sarkies, P. (2022). Encyclopaedia of eukaryotic DNA methylation: from patterns to mechanisms and functions. Biochem. Soc. Trans. 50 (3), 1179–1190. doi:10.1042/bst20210725
Saviana, M., Le, P., Micalo, L., Del Valle-Morales, D., Romano, G., Acunzo, M., et al. (2023). Crosstalk between miRNAs and DNA methylation in cancer. Genes (Basel). 14 (5), 1075. doi:10.3390/genes14051075
Saxonov, S., Berg, P., and Brutlag, D. L. (2006). A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc. Natl. Acad. Sci. U. S. A. 103 (5), 1412–1417. doi:10.1073/pnas.0510310103
Seong, J. M., Kim, B. C., Park, J. H., Kwon, I. K., Mantalaris, A., and Hwang, Y. S. (2010). Stem cells in bone tissue engineering. Biomed. Mater 5 (6), 062001. doi:10.1088/1748-6041/5/6/062001
Shao, C., Liu, Y., Zhao, Y., Jing, Y., Li, J., Lv, Z., et al. (2023). DNA methyltransferases inhibitor azacitidine improves the skeletal phenotype of mild osteogenesis imperfecta by reversing the impaired osteogenesis and excessive osteoclastogenesis. Bone 170, 116706. doi:10.1016/j.bone.2023.116706
Shao, J., Zhou, Y., Lin, J., Nguyen, T. D., Huang, R., Gu, Y., et al. (2018). Notch expressed by osteocytes plays a critical role in mineralisation. J. Mol. Med. Berl. 96 (3-4), 333–347. doi:10.1007/s00109-018-1625-x
Shen, W. C., Lai, Y. C., Li, L. H., Liao, K., Lai, H. C., Kao, S. Y., et al. (2019). Methylation and PTEN activation in dental pulp mesenchymal stem cells promotes osteogenesis and reduces oncogenesis. oncogenesis 10 (1), 2226. doi:10.1038/s41467-019-10197-x
Shimoyama, A., Wada, M., Ikeda, F., Hata, K., Matsubara, T., Nifuji, A., et al. (2007). Ihh/Gli2 signaling promotes osteoblast differentiation by regulating Runx2 expression and function. Mol. Biol. Cell 18 (7), 2411–2418. doi:10.1091/mbc.e06-08-0743
Shukla, V., Coumoul, X., Wang, R. H., Kim, H. S., and Deng, C. X. (2007). RNA interference and inhibition of MEK-ERK signaling prevent abnormal skeletal phenotypes in a mouse model of craniosynostosis. Nat. Genet. 39 (9), 1145–1150. doi:10.1038/ng2096
Taghizadeh, M., and Noruzinia, M. (2017). Lovastatin reduces stemness via epigenetic reprograming of BMP2 and GATA2 in human endometrium and endometriosis. Cell J. 19 (1), 50–64. doi:10.22074/cellj.2016.3894
Tang, L., Zhang, Y. Y., Liu, W. J., Fu, Q., Zhao, J., and Liu, Y. B. (2024). DNA methylation of promoter region inhibits galectin-1 expression in BMSCs of aged mice. Am. J. Physiol. Cell Physiol. 326 (2), C429–c441. doi:10.1152/ajpcell.00334.2023
Tao, C., Liu, J., Li, Z., Lai, P., Zhang, S., Qu, J., et al. (2022). DNMT1 is a negative regulator of osteogenesis. Biol. Open 11 (3), bio058534. doi:10.1242/bio.058534
Thomas, S., and Jaganathan, B. G. (2022). Signaling network regulating osteogenesis in mesenchymal stem cells. stem cells 16 (1), 47–61. doi:10.1007/s12079-021-00635-1
Tsai, C. C., Su, P. F., Huang, Y. F., Yew, T. L., and Hung, S. C. (2012). Oct4 and Nanog directly regulate Dnmt1 to maintain self-renewal and undifferentiated state in mesenchymal stem cells. Mol. Cell 47 (2), 169–182. doi:10.1016/j.molcel.2012.06.020
Tsumura, A., Hayakawa, T., Kumaki, Y., Takebayashi, S., Sakaue, M., Matsuoka, C., et al. (2006). Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes cells. 11 (7), 805–814. doi:10.1111/j.1365-2443.2006.00984.x
Urakami, S., Shiina, H., Enokida, H., Hirata, H., Kawamoto, K., Kawakami, T., et al. (2006). Wnt antagonist family genes as biomarkers for diagnosis, staging, and prognosis of renal cell carcinoma using tumor and serum DNA. Clin. Cancer Res. 12 (23), 6989–6997. doi:10.1158/1078-0432.ccr-06-1194
Veland, N., Lu, Y., Hardikar, S., Gaddis, S., Zeng, Y., Liu, B., et al. (2019). DNMT3L facilitates DNA methylation partly by maintaining DNMT3A stability in mouse embryonic stem cells. Nucleic Acids Res. 47 (1), 152–167. doi:10.1093/nar/gky947
Vlaikou, A. M., Kouroupis, D., Sgourou, A., Markopoulos, G. S., Bagli, E., Markou, M., et al. (2017). Mechanical stress affects methylation pattern of GNAS isoforms and osteogenic differentiation of hAT-MSCs. Biochim. Biophys. Acta Mol. Cell Res. 1864 (8), 1371–1381. doi:10.1016/j.bbamcr.2017.05.005
Walewska, A., Janucik, A., Tynecka, M., Moniuszko, M., and Eljaszewicz, A. (2023). Mesenchymal stem cells under epigenetic control - the role of epigenetic machinery in fate decision and functional properties. Cell Death Dis. 14 (11), 720. doi:10.1038/s41419-023-06239-4
Wang, C., Shan, S., Wang, C., Wang, J., Li, J., Hu, G., et al. (2017). Mechanical stimulation promote the osteogenic differentiation of bone marrow stromal cells through epigenetic regulation of Sonic Hedgehog. Exp. Cell Res. 352 (2), 346–356. doi:10.1016/j.yexcr.2017.02.021
Wang, L., You, X., Ruan, D., Shao, R., Dai, H. Q., Shen, W., et al. (2022a). TET enzymes regulate skeletal development through increasing chromatin accessibility of RUNX2 target genes. Nat. Commun. 13 (1), 4709. doi:10.1038/s41467-022-32138-x
Wang, Q. N., Yan, Y. Z., Zhang, X. Z., Lv, J. X., Nie, H. P., Wu, J., et al. (2022b). Rescuing effects of periostin in advanced glycation end-products (AGEs) caused osteogenic and oxidative damage through AGE receptor mediation and DNA methylation of the CALCA promoter. Chem. Biol. Interact. 354, 109835. doi:10.1016/j.cbi.2022.109835
Wang, R. N., Green, J., Wang, Z., Deng, Y., Qiao, M., Peabody, M., et al. (2014). Bone Morphogenetic Protein (BMP) signaling in development and human diseases. Genes Dis. 1 (1), 87–105. doi:10.1016/j.gendis.2014.07.005
Wang, T., Xing, J., Ying, Y., Tang, H., Li, J., Wei, Y., et al. (2020). Notch1 signaling mediated dysfunction of bone marrow mesenchymal stem cells derived from cyanotic congenital heart disease. Biochem. Biophys. Res. Commun. 527 (4), 847–853. doi:10.1016/j.bbrc.2020.04.129
Wang, X., Yu, F., and Ye, L. (2023a). Epigenetic control of mesenchymal stem cells orchestrates bone regeneration. Front. Endocrinol. (Lausanne) 14, 1126787. doi:10.3389/fendo.2023.1126787
Wang, Y., Li, H. Y., Guan, S. Y., Yu, S. H., Zhou, Y., Zheng, L. W., et al. (2023b). Different sources of bone marrow mesenchymal stem cells: a comparison of subchondral, mandibular, and tibia bone-derived mesenchymal stem cells. Stem Cells 19, 1029–1041. doi:10.2174/011574888x260686231023091127
Weissbein, U., Plotnik, O., Vershkov, D., and Benvenisty, N. (2017). Culture-induced recurrent epigenetic aberrations in human pluripotent stem cells. stem cells 13 (8), e1006979. doi:10.1371/journal.pgen.1006979
Wen, Y., Shangguan, Y., Pan, Z., Hu, H., Magdalou, J., Chen, L., et al. (2019). Activation of local bone RAS by maternal excessive glucocorticoid participated in the fetal programing of adult osteopenia induced by prenatal caffeine exposure. Toxicol. Appl. Pharmacol. 363, 1–10. doi:10.1016/j.taap.2018.11.003
Weng, Z., Xu, C., Chen, X., Yan, Q., Fu, Z., Jiao, J., et al. (2024). Sp1-activated FGFR2 is involved in early-life exposure to nickel-induced craniosynostosis by regulating the ERK1/2 signaling pathway. Environ. Int. 184, 108477. doi:10.1016/j.envint.2024.108477
Wong, T. H., Chen, T. Y., Tseng, K. Y., Chen, Z. Y., Chen, C. H., Lin, F. H., et al. (2020). Decorin inhibits the insulin-like growth factor I signaling in bone marrow mesenchymal stem cells of aged humans. Aging (Albany NY) 13 (1), 578–597. doi:10.18632/aging.202166
Wu, F., Jiao, J., Liu, F., Yang, Y., Zhang, S., Fang, Z., et al. (2019). Hypermethylation of Frizzled1 is associated with Wnt/β-catenin signaling inactivation in mesenchymal stem cells of patients with steroid-associated osteonecrosis. Exp. Mol. Med. 51 (2), 1–9. doi:10.1038/s12276-019-0220-8
Wu, M., Chen, G., and Li, Y. P. (2016). TGF-β and BMP signaling in osteoblast, skeletal development, and bone formation, homeostasis and disease. Bone Res. 4, 16009. doi:10.1038/boneres.2016.9
Xin, T. Y., Yu, T. T., and Yang, R. L. (2020). DNA methylation and demethylation link the properties of mesenchymal stem cells: regeneration and immunomodulation. World J. Stem Cells 12 (5), 351–358. doi:10.4252/wjsc.v12.i5.351
Xu, L., Liu, Y., Sun, Y., Wang, B., Xiong, Y., Lin, W., et al. (2017). Tissue source determines the differentiation potentials of mesenchymal stem cells: a comparative study of human mesenchymal stem cells from bone marrow and adipose tissue. Stem Cell Res. Ther. 8 (1), 275. doi:10.1186/s13287-017-0716-x
Yan, X., Ehnert, S., Culmes, M., Bachmann, A., Seeliger, C., Schyschka, L., et al. (2014). 5-azacytidine improves the osteogenic differentiation potential of aged human adipose-derived mesenchymal stem cells by DNA demethylation. PLoS One 9 (6), e90846. doi:10.1371/journal.pone.0090846
Yang, C. Y., Lu, R. J., Lee, M. K., Hsiao, F. S., Yen, Y. P., Cheng, C. C., et al. (2021). Transcriptome analysis of Dnmt3l knock-out mice derived multipotent mesenchymal stem/stromal cells during osteogenic differentiation. Front. Cell Dev. Biol. 9, 615098. doi:10.3389/fcell.2021.615098
Yang, R., Yu, T., Kou, X., Gao, X., Chen, C., Liu, D., et al. (2018). Tet1 and Tet2 maintain mesenchymal stem cell homeostasis via demethylation of the P2rX7 promoter. Nat. Commun. 9 (1), 2143. doi:10.1038/s41467-018-04464-6
Yang, X., Matsuda, K., Bialek, P., Jacquot, S., Masuoka, H. C., Schinke, T., et al. (2004). ATF4 is a substrate of RSK2 and an essential regulator of osteoblast biology; implication for Coffin-Lowry Syndrome. Cell 117 (3), 387–398. doi:10.1016/s0092-8674(04)00344-7
Youssef, A., and Han, V. K. M. (2017). Regulation of osteogenic differentiation of placental-derived mesenchymal stem cells by insulin-like growth factors and low oxygen tension. Stem Cells Int. 2017, 4576327. doi:10.1155/2017/4576327
Yu, T., Liu, D., Zhang, T., Zhou, Y., Shi, S., and Yang, R. (2019). Inhibition of Tet1-and Tet2-mediated DNA demethylation promotes immunomodulation of periodontal ligament stem cells. Cell Death Dis. 10 (10), 780. doi:10.1038/s41419-019-2025-z
Yu, T., Zhang, L., Dou, X., Bai, R., Wang, H., Deng, J., et al. (2022a). Mechanically robust hydrogels facilitating bone regeneration through epigenetic modulation. Adv. Sci. 9 (32), e2203734. doi:10.1002/advs.202203734
Yu, X., Song, M. S., Rong, P. Z., Chen, X. J., Shi, L., Wang, C. H., et al. (2022b). LncRNA SNHG1 modulates adipogenic differentiation of BMSCs by promoting DNMT1 mediated Opg hypermethylation via interacting with PTBP1. J. Cell. Mol. Med. 26 (1), 60–74. doi:10.1111/jcmm.16982
Zhang, M., Gao, Y., Li, Q., Cao, H., Yang, J., Cai, X., et al. (2022a). Downregulation of DNA methyltransferase-3a ameliorates the osteogenic differentiation ability of adipose-derived stem cells in diabetic osteoporosis via Wnt/β-catenin signaling pathway. Stem Cell Res. Ther. 13 (1), 397. doi:10.1186/s13287-022-03088-4
Zhang, M., Li, Y., Rao, P., Huang, K., Luo, D., Cai, X., et al. (2018). Blockade of receptors of advanced glycation end products ameliorates diabetic osteogenesis of adipose-derived stem cells through DNA methylation and Wnt signalling pathway. Cell Prolif. 51 (5), e12471. doi:10.1111/cpr.12471
Zhang, P., Feng, B., Dai, G., Niu, K., and Zhang, L. (2024). FOXC1 promotes osteoblastic differentiation of bone marrow mesenchymal stem cells via the dnmt3b/CXCL12 Axis. Biochem. Genet. 62 (1), 176–192. doi:10.1007/s10528-023-10403-y
Zhang, R., Wang, N., Zhang, L. N., Huang, N., Song, T. F., Li, Z. Z., et al. (2016). Knockdown of DNMT1 and DNMT3a promotes the angiogenesis of human mesenchymal stem cells leading to arterial specific differentiation. Stem Cells 34 (5), 1273–1283. doi:10.1002/stem.2288
Zhang, S., Kaplan, F. S., and Shore, E. M. (2012). Different roles of GNAS and cAMP signaling during early and late stages of osteogenic differentiation. Horm. Metab. Res. 44 (10), 724–731. doi:10.1055/s-0032-1321845
Zhang, W., and Xu, J. (2017). DNA methyltransferases and their roles in tumorigenesis. Biomark. Res. 5, 1. doi:10.1186/s40364-017-0081-z
Zhang, Y., Ling, L., Ajay, D. O. A. A., Eio, Y. M., van Wijnen, A. J., Nurcombe, V., et al. (2022b). FGFR2 accommodates osteogenic cell fate determination in human mesenchymal stem cells. Gene 818, 146199. doi:10.1016/j.gene.2022.146199
Keywords: mesenchymal stem cells (MSCs), DNA methylation, DNA methyltransferases (DNMTs), Ten-eleven translocation family proteins (TETs), osteogenic differentiation, signaling pathway
Citation: Lai Z, Shu Q, Song Y, Tang A and Tian J (2024) Effect of DNA methylation on the osteogenic differentiation of mesenchymal stem cells: concise review. Front. Genet. 15:1429844. doi: 10.3389/fgene.2024.1429844
Received: 08 May 2024; Accepted: 10 June 2024;
Published: 02 July 2024.
Edited by:
Serena Duchi, The University of Melbourne @ St. Vincent’s Hospital, AustraliaReviewed by:
Xiaolei Li, University of Pennsylvania, United StatesCopyright © 2024 Lai, Shu, Song, Tang and Tian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun Tian, dGlhbmp1bkB6bmhvc3BpdGFsLmNu
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.