- 1Department of Infectious Disease Control and Prevention, Changshu Center for Disease Control and Prevention, Suzhou, China
- 2Department of Epidemiology and Biostatistics, School of Public Health, Nanjing Medical University, Nanjing, China
- 3Department of Tuberculosis, The Second People’s Hospital of Changshu, Changshu, China
- 4Department of Infectious Disease, The People’s Hospital of Taixing, Taixing, China
- 5Department of Tuberculosis, The Third People’s Hospital of Zhenjiang Affiliated to Jiangsu University, Zhenjiang, China
- 6Department of Infectious Disease, The Jurong Hospital Affiliated to Jiangsu University, Jurong, China
Objective: The pathogenesis of antituberculosis drug-induced liver injury (AT-DILI) remains largely unknown. The current investigation aimed to determine the genetic contribution of the nuclear receptor subfamily 1 Group I member 3 (NR1I3) and nuclear receptor subfamily 1 Group H member 4 (NR1H4) genes to the risk of AT-DILI in the Chinese population.
Methods: A 1:4 matched case‒control study was conducted, and five single nucleotide polymorphisms (SNPs) in the NR1I3 and NR1H4 genes were detected and assessed. Utilizing a multivariate conditional logistic regression model, the effects of haplotype and genotype on the risk of AT-DILI were examined. Extended subgroup analysis was carried out based on sex. The distribution of the peak value of serum liver enzymes also compared among different genotypes.
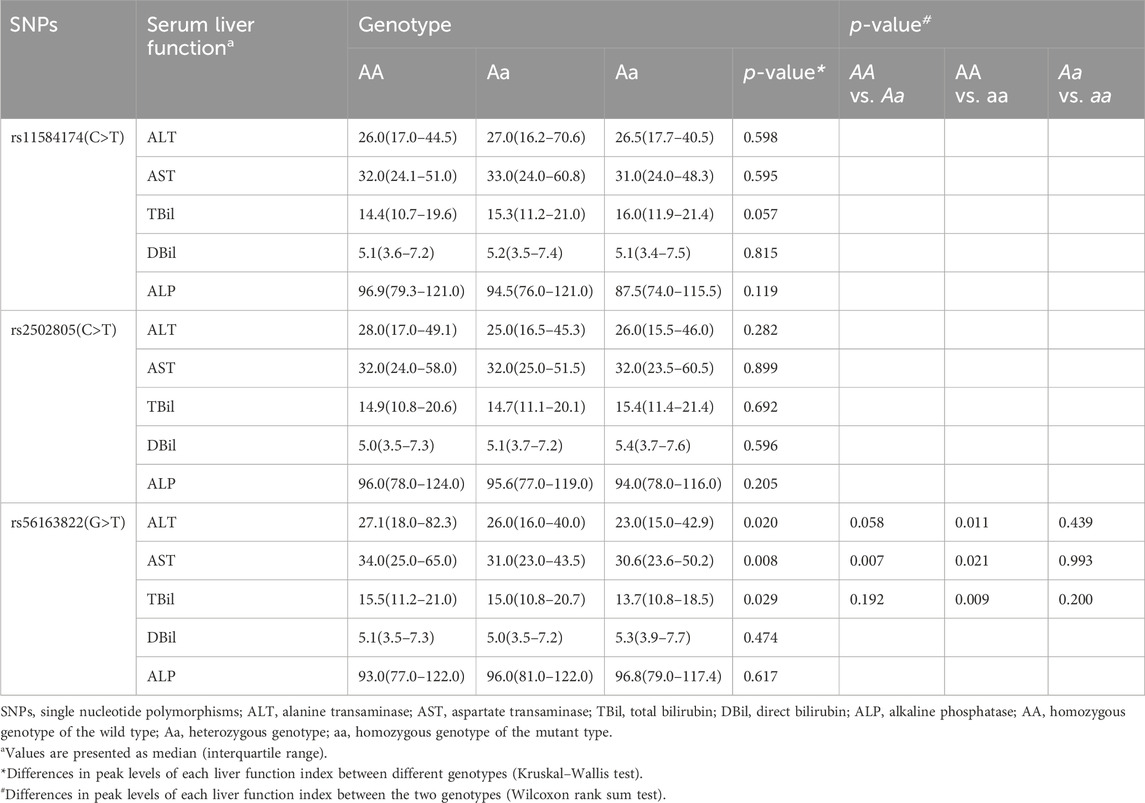
Results: 224 AT-DILI cases and 896 controls were included in this study. No significant difference was observed in genotypes or haplotypes frequencies between AT-DILI cases and controls. However, comparisons of liver function indicators revealed significant differences in the peak values of alanine aminotransferase (ALT), aspartate aminotransferase (AST) and total bilirubin (TBil) among patients with different genotypes of NR1H4 rs56163822 (GG vs. GT vs. TT, 27.1 U/L vs. 26.0 U/L vs. 23.0 U/L, p = 0.020; 34.0 U/L vs. 31.0 U/L vs. 30.6 U/L, p = 0.008; 15.5 μmol/L vs. 15.0 μmol/L vs. 13.7 μmol/L, p = 0.029, respectively), as well as in the peak values of ALT and AST among male patients with different genotypes of NR1H4 rs56163822 (29.0 U/L vs. 26.9 U/L vs. 22.6 U/L, p = 0.002; 34.0 U/L vs. 32.0 U/L vs. 30.5 U/L, p = 0.019, respectively).
Conclusion: Based on this 1:4 individual-matched case‒control study, the SNP rs56163822 in the NR1H4 gene may be linked to the susceptibility to AT-DILI in Chinese patients receiving anti-TB treatment. Further studies in larger varied populations are needed to validate our findings.
Introduction
Tuberculosis (TB) is treated in China with the regimen recommended by the World Health Organization (WHO), which includes a combined therapy of isoniazid (INH), rifampicin (RIF), pyrazinamide, ethambutol and/or streptomycin for 6 months (WHO, 2017). This is the most commonly used standard anti-TB treatment; however, long-term use of multidrug combination therapy may lead to a variety of adverse drug reactions (ADRs) in patients (Arbex et al., 2010). Among all kinds of ADRs, anti-TB drug-induced liver injury (AT-DILI) exhibits a comparatively higher occurrence rate (Wang et al., 2022). At present, the exact mechanism of AT-DILI is not fully understood, and there are many factors affecting the occurrence of AT-DILI, including social, environmental, and genetic factors, such as age, female sex, concomitant hepatitis C, and gene polymorphisms (Gaude et al., 2015; Ding et al., 2017; Zheng et al., 2020; Yang et al., 2019a).
In recent years, animal experiments have shown that the accumulation of protoporphyrin Ⅸ (PPIX) in hepatocytes may be a potential new mechanism of AT-DILI (Sachar et al., 2016a). Pregnane X receptor (PXR), a significant member of the nuclear receptor family, functions as a transcriptional regulator of cytochrome P450 (CYP450) 3A4 (CYP3A4). It binds to the response element of the CYP3A4 gene’s promoter and operates in a heterodimeric form with the retinoid X receptor (RXR). RIF-mediated activation of PXR induces aminolevulinic synthase-1 (ALAS1) expression by upregulating CYP450 through the PXR pathway (Sachar et al., 2016b; Lyoumi et al., 2013; He et al., 2017). Animal experimental studies have shown that in mice, human-derived PXR can upregulate ALAS1 expression by binding to the drug response enhancer sequence in the flanking region of ALAS1 (Podvinec et al., 2004; Fraser et al., 2003). When exposed to agents that stimulate CYP450 and other drug-metabolizing enzymes, ALAS1 transcription is upregulated, the rate of haem biosynthesis is limited, and PPIX synthesis is increased, leading to hepatic accumulation and liver injury. In addition, studies have suggested that the metabolites of INH, hydrazine and acetyl hydrazine, are related to drug-induced liver injury (DILI). Recent animal experiments have suggested that INH can induce the expression of ALAS1 and reduce that of ferrochelatase (FECH) in mice, resulting in a decrease in the synthesis of haem from PPIX and Fe2+ and an increase in PPIX levels in vivo (Sachar et al., 2016b).
In the PXR pathway, heat shock protein 90 (HSP90) serves as a molecular chaperone. The cytosolic constitutive androstane receptor (CAR, encoded by the NR1I3 gene) interacts with PXR (Squires et al., 2004; Moffatt et al., 2008) and is also involved in regulating HSP90 activity (Brychzy et al., 2003). The CAR is highly expressed in the liver and shows an overlapping set of target genes with PXR in response to potentially harmful chemicals (Swales and Negishi, 2004; Timsit and Negishi, 2007). Overexpression of CAR in the presence of PXR leads to an elevation in the cytoplasmic concentrations of the PXR-CAR-HSP90 complex, which is retained in the cytoplasm of HepG2 cells (Squires et al., 2004). Upon binding to its ligand, PXR undergoes a conformational alteration, prompting its association with RXR to generate PXR/RXR heterodimers. These heterodimers then transfer to the nucleus and recruit transcriptional coactivators or corepressors to form complexes. These complexes specifically recognize and bind to response elements situated within the proximal promoter regions of target genes, thereby modulating their transcriptional activity and expression patterns (Tian, 2013). However, PXR/RXR heterodimers activate the xenobiotic-responsive enhancer module in response to RIF (Goodwin et al., 1999), thereby affecting the body function. Normally, the PXR-HSP90-CAR complex is located in the cytoplasm, however, the RIF-INH combination triggers the dissociation of this complex, the translocation of PXR to mitochondria via interaction with RXR. Subsequently, the ligand-bound PXR-RXR complex binds to specific DNA response elements, leading to the transcription of ALAS1 and CYP450. Direct transcription of ALAS1 can be activated by RIF or INH, or it can be downregulated by negative feedback disinhibition triggered by the incorporation of haem into CYP450 apolipoproteins (Lyoumi et al., 2013). This dual mechanism may be the cause for the accumulation of the endogenous hepatotoxicant PPIX, which leads to AT-DILI. However, few studies have investigated the association between NR1I3 gene polymorphisms and AT-DILI, and the results are negative (Wang et al., 2019). Therefore, we hoped to verify this result by expanding the sample size.
Research suggest that RIF-induced liver injury, steatosis, and cholestasis are associated with farnesoid X receptor (FXR, encoded by the NR1H4 gene) dysfunction and altered bile acid (BA) metabolism, and that the JNK signaling pathway is partially implicated in this injury. Therefore, FXR may represent a novel therapeutic target for addressing DILI (Zhou et al., 2024; Ezhilarasan, 2023). Wen et al. found that HRZE may upregulate FXR with short-term administration, but more prolonged treatment appears to suppress FXR function, resulting in hepatic BA accumulation, suggestive of hepatotoxicity (Wen et al., 2022). INH + RIF administration decreased the expression of BA transporters such as the bile salt export pump and multidrug resistance-associated protein 2, and induced liver injury through sirtuin 1 and FXR pathway (Ezhilarasan, 2023). PZA induces cholestatic liver injury by FXR inhibition, leading to dysfunction in BA synthesis and transport (Guo et al., 2016). Pharmacological studies have provided compelling evidence that FXR exerts protective effects against diverse etiologies of DILI through its anticholestatic, anti-adipogenic, antioxidant, anti-inflammatory, and antifibrotic effects. Further research has demonstrated that FXR agonists potently mitigate hepatotoxicity and foster liver regeneration in mice models with liver injury (Carino et al., 2018). However, the association between FXR gene polymorphism and AT-DILI has not been studied.
Therefore, the current study endeavors to elucidate the genetic predisposition of NR1I3 and NR1H4 genes in the risk of AT-DILI among the Chinese population, employing single-nucleotide polymorphisms (SNPs) as markers and adopting an individual-based matched case-control study design.
Methods
Study design and setting
The Ethics Committee of Nanjing Medical University granted approval for this research. Between May 2016 and December 2021, patients receiving anti-TB treatment were recruited from the outpatient departments of four designated infectious disease hospitals (Jurong People’s Hospital, Taixing People’s Hospital, The Second People’s Hospital of Changshu and The Third People’s Hospital of Zhenjiang) in Jiangsu Province, covering both urban and rural areas. This study was conducted according to STROBE checklist for case-control studies (Supplementary Table S1).
Patients and clinical criteria
The inclusion criteria for this study encompassed patients who: (1) had a newly diagnosed TB; (2) were poised to undergo anti-TB therapy; (3) could actively undergo regular liver function tests during the first 2 months of treatment and had at least 3 cycles of liver function test information available; and (4) provided voluntary informed consent. The exclusion criteria for this study: (1) TB patients exhibiting abnormal baseline liver function indicators, such as alanine aminotransferase (ALT), aspartate aminotransferase (AST), and total bilirubin (TBil) levels; (2) with mental illness; (3) with severe comorbidities.
AT-DILI case and control definition and selection
The diagnosis of AT-DILI was confirmed in accordance with the Chinese guidelines (Yu et al., 2017), ALT is greater than or equal to 3 upper limit of normal (ULN) and/or TBil is greater than or equal to 2 ULN; or AST, ALP and TBil increased at the same time, and at least one item is greater than or equal to 2 ULN. Three distinct patterns of injury were delineated, namely, ‘hepatocellular’, ‘mixed’, or ‘cholestatic’, based on the R ratio. The R ratio was determined by employing the ratio of the ALT level to the alkaline phosphatase (ALP) level, both values being normalized by their respective local upper limits of normal (ULN). Specifically, the calculation was conducted as follows: R = (ALT value/ALT ULN)/(ALP value/ALP ULN). An R ratio > 5 indicated hepatocellular injury; that of < 2 indicated cholestatic injury; and an R ratio between 2 and 5 indicated mixed injury. Additionally, causality assessment was conducted in accordance with the International Consensus Criteria (Roussel Uclaf Causality Assessment Method, RUCAM) (Danan and Teschke, 2015), and all patients with possible (score 3–5), probable (score 6–8), or highly probable (score > 8) were included in the study. Furthermore, the severity of liver injury was categorized as mild (ALT < 5 ULN), moderate (ALT 5–10 ULN), or severe (ALT > 10 ULN), adhering to the WHO toxicity classification standards (Tostmann et al., 2008).
Patients who met the diagnostic criteria for AT-DILI were categorized into the case group, while those whose serum liver function tests remained consistently normal throughout anti-TB treatment were deemed candidate controls. For each AT-DILI patient, four control individuals of the same sex and within a 5-year age range were randomly selected.
Variables
In this study, the prevalence of AT-DILI was selected as the outcome variable. Liver disease history is defined as having had a liver condition in the past. Similarly, smoking history is defined as having smoked at least two cigarettes per day for the past 2 weeks. In relation to alcohol intake, drink history is defined as having drunk at least 50 mL of alcohol per day for the past 2 weeks. Furthermore, hepatoprotectant use refers to the use of hepatoprotectant during the course of treatment.
Data sources/measurement
Prior to the initiation of anti-TB treatment, liver function tests (LFTs) were administered. During the initial 2 months of anti-TB therapy, patients underwent scheduled outpatient follow-ups and underwent LFTs every 2 weeks at the designated hospitals, adhering to the local free-TB service policy. In the event that patients exhibited symptoms indicative of potential hepatitis (such as anorexia, nausea, vomiting, malaise, or tea-coloured urine), they were instructed to promptly visit the designated hospital for immediate LFTs (Chen et al., 2019; Yang et al., 2019b). Despite the absence of any manifestations suggesting drug-induced hepatitis, patients were counseled to undergo regular LFTs at a biweekly interval. Residual blood samples from the LFTs of each patient were retained for further analysis. All recruited TB patients were closely monitored throughout their treatment regimen until completion.
SNP selection and genotyping
The leftover blood sample from an LFT was utilized for the extraction of genomic DNA. The Han Chinese individuals in Beijing (CHB) dataset of the 1000 Genomes Project database served as the source for downloading all eligible SNPs in the NR1I3 and NR1H4 gene regions, along with regions 2 kb upstream and downstream of the genes. Utilizing the Haploview software version 4.2 from Broad Institute in Cambridge, MA, SNPs were chosen based on specific criteria: (1) a minor allele frequency (MAF) ≥ 10% and (2) an r2 of pairwise linkage disequilibrium ≥ 0.8 (Eberle et al., 2006). Furthermore, HaploReg version 4.1 was used to maximally select potentially functional SNPs. Finally, a total of five SNPs, rs2307424, rs2502805, rs10157822, and rs11584174 in the NR1I3 gene and rs56163822 in the NR1H4 gene, were identified for the genotyping using TaqMan allelic discrimination technology (Supplementary Table S2). To ensure unbiased genotyping, the case or control status was blinded during the process. Additionally, over 10% of the samples were randomly selected for duplicate genotyping using the same assay, resulting in a 100% accuracy rate.
Statistical analysis
Continuous variables are described as the means ± standard deviations or as medians with interquartile ranges (IQRs), and the independent-samples t-test or the Wilcoxon rank-sum test were used to analyse differences between groups. Categorized variables are represented as the count of patients and a percentage and were analysed using a conditional logistic regression model. The chi-square goodness-of-fit test was conducted to assess Hardy-Weinberg Equilibrium (HWE) in the control group. Utilizing the multivariate conditional logistic regression model, odds ratios (ORs) were computed, along with their respective 95% confidence intervals (CIs). The liver disease history and smoking history were adjusted. Three genetic models (dominant, recessive, and additive) were constructed to comprehensively analyse the effects of the SNPs on the risk of AT-DILI. Haplotype blocks were constructed using the HaploView 4.2 software, considering the linkage disequilibrium (LD) among SNPs. The haplo.stats package in the R software was employed to estimate the haplotype of each individual based on the observations. Subgroup analyses were performed, each according to sex. For the association analysis of gene polymorphisms, false discovery rate (FDR) correction was implemented to correct for multiple comparisons. The genetic variants that could affect different gene expression were identified via online expression quantitative trait loci (eQTL) analysis from GETx-Portal (https://www.gtexportal.org/home/). All the analyses were conducted using the R software for Windows version 4.2.2. Statistical significance was inferred when the two-tailed p-value < 0.05.
Results
Baseline and clinical characteristics
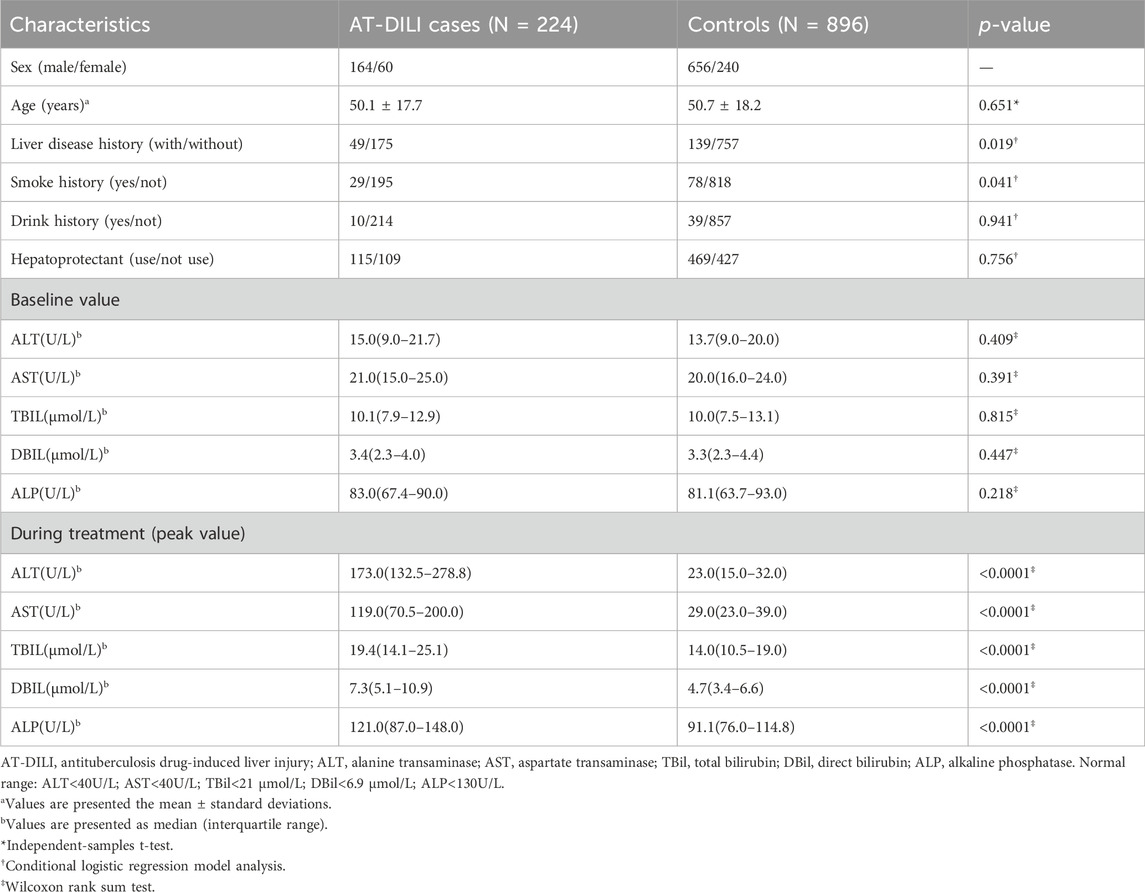
A total of 1120 TB patients (224 AT-DILI cases and 896 controls) were included in the study. There were 749 patients treated with the 2HREZ/4HR regimen, 149 with the 3HREZ/9HR regimen, and 222 with other treatment regimens. The chi-square test results show that there was no difference in treatment regimens between the two groups (p > 0.05). Among the 224 AT-DILI cases, 124 (55.4%) were judged as probable, 131 (58.5%) had mild liver injury, and 147 (65.6%) were classified as the hepatocellular type of liver injury. Table 1 summarized the baseline demographic and clinical characteristics of the patients and matched controls. No significant differences were observed in the distributions of age, sex, drinking history, hepatoprotectant use or baseline hepatic biochemical parameters between the case groups and control groups (p > 0.05). The proportions of AT-DILI cases with liver disease or smoking history exceeded those of controls. During the course of anti-TB therapy, the peak levels of AST, ALT, TBil, direct bilirubin (DBil) and ALP in the AT-DILI cases were significantly elevated compared to those in the controls (p < 0.0001).
Genotype analysis
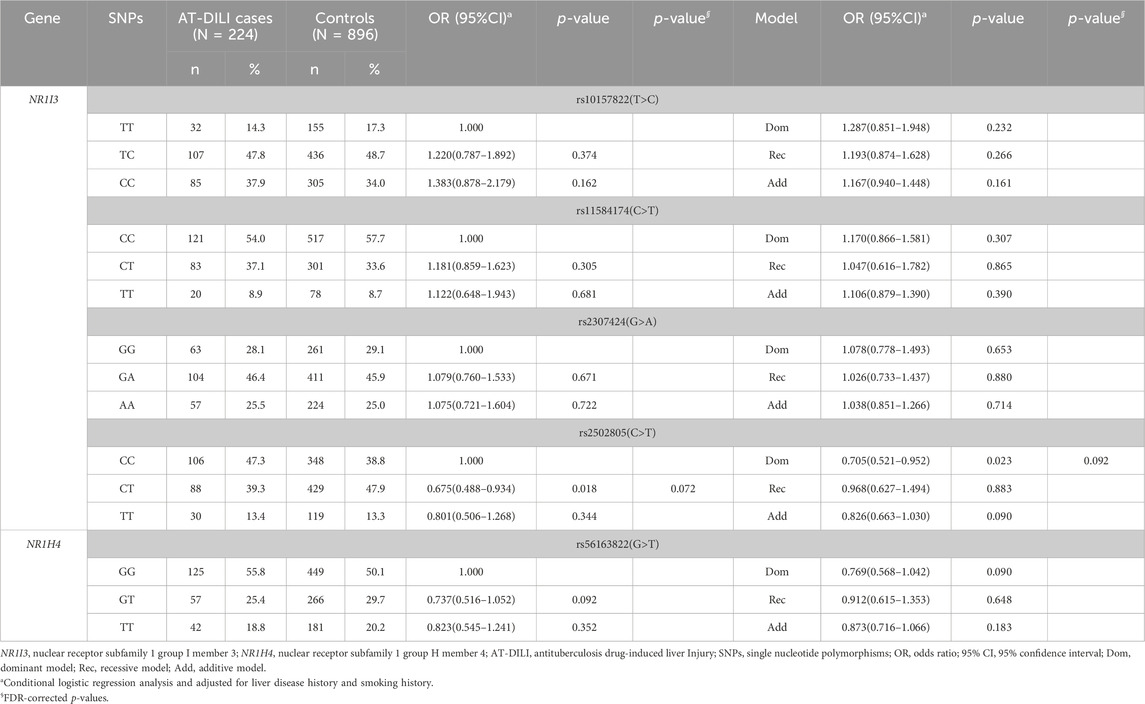
Multivariate conditional logistic regression analysis exhibited that patients carrying the CT genotype of rs2502805 were at a lower risk of AT-DILI than those with the CC genotype (adjusted OR = 0.675, 95% CI: 0.488–0.934, p = 0.018), and the risk of AT-DILI in patients with CT + TT genotypes was lower than that in patients with the CC genotype (adjusted OR = 0.705, 95% CI: 0.521–0.952, p = 0.023) according to the dominant model. However, after FDR correction, the difference between the groups was no longer statistically significant (p > 0.05) (Table 2).
Haplotype analysis
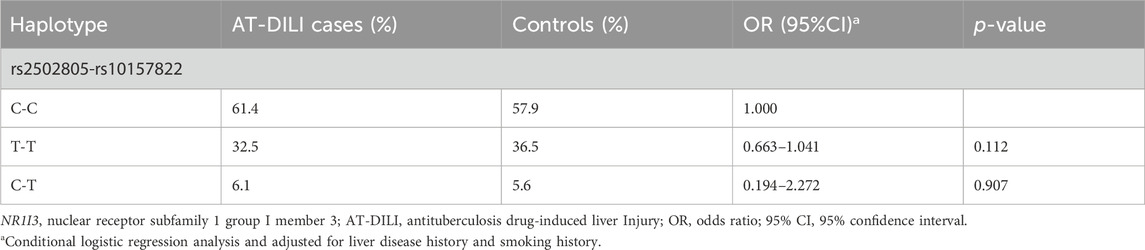
A reconstructed LD plot of the NR1I3 gene, which has been generated utilizing the Haploview 4.2 software was shown in Supplementary Figure S1, and the results indicate that SNPs rs2502805 and rs10157822 were in linkage. According to the frequency distribution of haplotypes, the haplotypes with frequencies that were too low were excluded, and multivariate conditional logistic regression analysis was conducted on the haplotypes with frequencies greater than 2%. There was no significant difference in the haplotype distribution between the case and control groups (p > 0.05) (Table 3).
Subgroup analysis
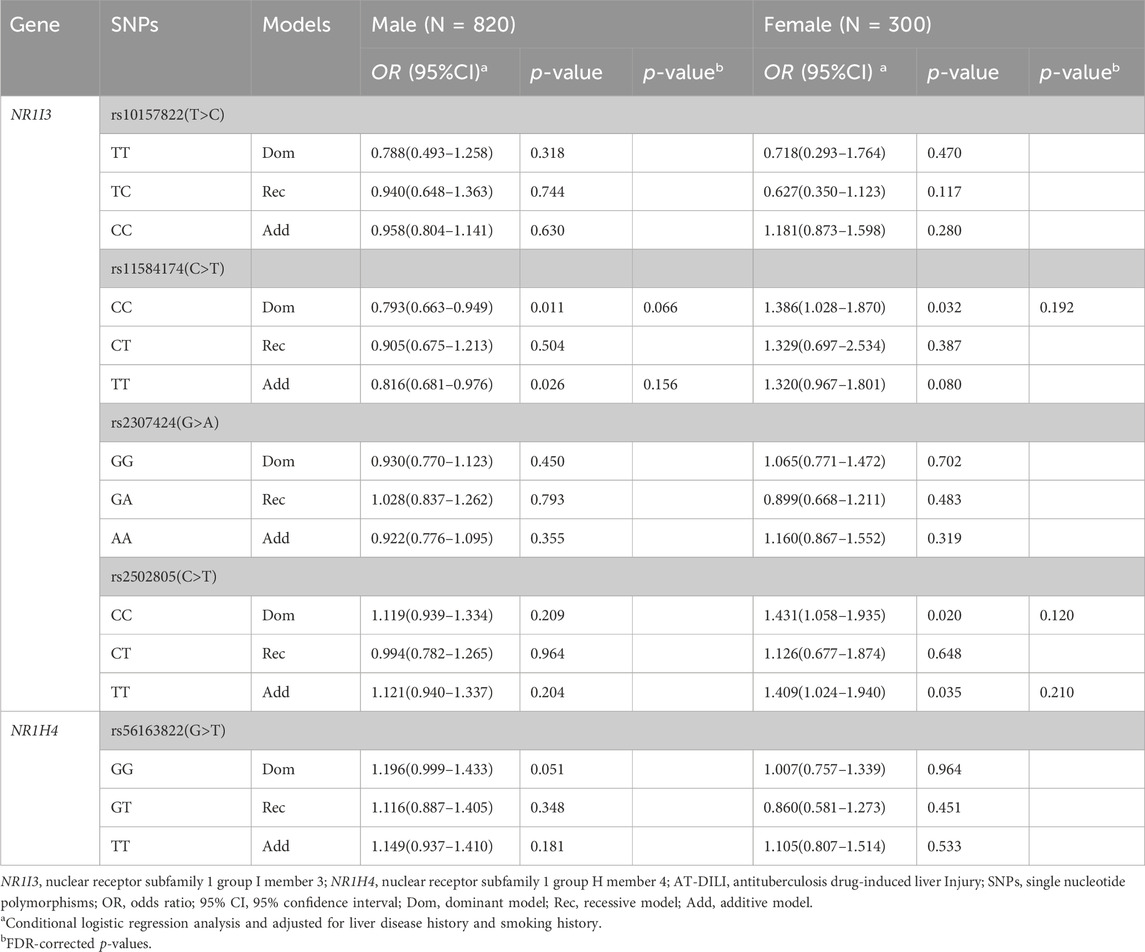
The associations between SNPs and AT-DILI by sex are shown in Table 4. Utilizing multivariate conditional logistic regression analysis, we found that female patients carrying the T alleles of rs11584174 and rs2502805 in the NR1I3 gene exhibited an elevated risk of AT-DILI (dominant model, adjusted OR = 1.386, 95% CI: 1.028–1.870, p = 0.032; dominant model, adjusted OR = 1.431, 95% CI: 1.058–1.935, p = 0.020, respectively). In addition, female patients harboring the TT allele of rs2502805 in the NR1I3 gene displayed a heightened risk of AT-DILI (additive model, adjusted OR = 1.409, 95% CI: 1.024–1.940, p = 0.035). Marginally significant differences were also found in male patients carrying the GT/TT alleles of rs56163822 in the NR1H4 gene (dominant model, adjusted OR = 1.196, 95% CI: 0.999–1.433, p = 0.051). Additionally, male patients carrying the T allele of rs11584174 in the NR1I3 gene displayed a decreased risk of AT-DILI (dominant model, adjusted OR = 0.793, 95% CI: 0.663–0.949, p = 0.011). However, after FDR correction, the difference was insignificant (p > 0.05).
Comparisons of liver function indicators among patients with different genotypes
The correlations between liver function indicators of participants and different genotypes are shown in Table 5, Supplementary Table S3. The peak levels of ALT, AST and TBil in all patients with the NR1H4-rs56163822-GG genotype were notably greater than those in patients with the TT genotype (p = 0.011, p = 0.021, and p = 0.009, respectively). Additionally, this analysis revealed a noteworthy significant difference in the peak value of AST between the GG and GT genotype groups (34.0 U/L vs. 31.0 U/L, p = 0.007). Further subgroup analysis of rs56163822 in NR1H4 also revealed significant differences in the peak values of ALT and AST among male patients (p = 0.002 and p = 0.019, respectively).
Online eQTL analysis
From the GTEx portal, further online eQTL analysis of NR1H4 (SNP rs56163822) and NR1I3 (SNP rs10157822, rs11584174, rs2307424 and rs2502805) genes was conducted with the gene expression in 208 liver samples, and the result revealed that significant differences in the expression levels of the three genotypes of NR1H4 rs56163822 in liver samples (p = 0.022), whereas no significant differences were observed in the expression levels of the three genotypes of NR1I3 in liver samples (p > 0.05) in Supplementary Figure S2.
Discussion
This research indicated that genetic variants in NR1H4 might affect the susceptibility to AT-DILI, which further corroborates the hypothesis that perturbation of BA homeostasis causes liver injury.
The NR1I3 gene, situated at the 1q23.3 locus on the human chromosome, comprises nine exons and encodes a member of the nuclear receptor superfamily and regulates ectobiotic and endobiotic metabolism. Distinct from many nuclear receptors, this transcriptional regulator remains constitutively active in the absence of a ligand, yet its activity is modulated by both agonists and inverse agonists. Upon ligand binding, this protein undergoes translocation to the nucleus to activate or repress the transcription of target genes. The ligands encompass bilirubin, various foreign compounds, steroid hormones, and prescribed medications like RIF and efavirenz (Petros et al., 2022). CAR and Th17 cells may be potential targets for novel therapeutic strategies for xenobiotic-induced liver inflammation (Shi et al., 2022). The indirect activation of the CAR/PXR, FXR, and Nrf2 pathways provides important molecular mechanisms in PFOA-induced human hepatotoxicity (Murase et al., 2023). Furthermore, the PXR and CAR pathways were activated in the PPARα−/− mice, which may explain the lipid accumulation and hepatomegaly observed in the livers of mice exposed to PFDMO2HpA/PFDMO2OA (Ren et al., 2024).
Studies have illuminated the involvement of rs2307424 in the metabolism of efavirenz (Wyen et al., 2011). A meta-analysis revealed that the TT genotype of rs2307424 is associated with a lower concentration of efavirenz compared to the CC genotype. The TT genotype may exhibit linkage disequilibrium, which can lead to increased CAR and CYP2B6 activities. It increases the efficiency of metabolic enzymes and transporters, thereby reducing efavirenz concentrations (Ayuso et al., 2019), and rs2307424 is also associated with increased virological response (de Almeida Velozo et al., 2022). The homozygous rs10157822 variant genotype is correlated with a decreased risk of hyperbilirubinemia among female neonates (OR = 0.44, 95% CI: 0.20–0.95, p = 0.034) (Cheung et al., 2015), which may be due to acceleration of bilirubin transport by the homozygous variant. However, one case-control study failed to detect any association between six SNPs of CAR and risk of AT-DILI (Wang et al., 2019). Rs2502805 and rs11584174 are transcription factor-binding sites, and the Regulome DB score of rs11584174 is less than 1. However, no notable linkage was detected between rs11584174 and AT-DILI susceptibility, which may be due to differences in the distribution of gene frequencies in the population. The distribution of rs11584174 in the control population of this study was not consistent with the HWE, and there is no study on these two SNPs and related diseases, which can be further explored in different populations and larger sample sizes.
The NR1H4 gene, which is situated on human chromosome 12q23.1 and comprises 17 exons, encodes FXR, a member of the nuclear receptor superfamily and a ligand-induced transcription factor. As a receptor for BAs, FXR binds to DNA and modulates the expression of genes that are integral to the synthesis and transportation of BAs (Yang et al., 2007; Attinkara et al., 2012; Kim et al., 2017). The suppression of the BA receptor FXR by pathogenic influences disrupts the downstream expression of BA synthesis and transport proteins, leading to abnormal functioning (He et al., 2024). In turn, this ultimately leads to elevated BA levels within the liver and intestine. Certain BA are toxic and induce inflammation at specific concentrations; whereas others can induce oxidative damage by interacting with cell membranes and fostering the production of reactive oxygen species (He et al., 2024). FXR reportedly participates in liver regeneration, which is mediated by hepatocyte proliferation, by regulating cell cycle progression, and NR1H4 gene expression is significantly upregulated in severe liver injury (Cai et al., 2021). FXR also functions as a proviral host factor for hepatitis B virus (Mouzannar et al., 2019) and hinders liver fibrosis (Wang et al., 2018). Studies have shown that FXR agonists have antifibrotic and portal pressure-lowering effects in animal models; therefore, genotypes that lead to the suppression of FXR (such as rs56163822) expression elevate the risk of hepatic decompensation and death; however, no significant correlation was observed between the rs56163822 genetic polymorphism and the development of advanced chronic liver disease (Semmler et al., 2019). A study has shown that the rs56163822 GT genotype enhances the likelihood of developing spontaneous bacterial peritonitis among cirrhotic patients exhibiting ascites (Lutz et al., 2014).
This is the first study to explore the potential linkage between NR1H4 and AT-DILI. This study revealed significant differences in the peak values of the serum liver function indicators ALT, AST and TBil among all patients with the GG and GT/TT genotypes of rs56163822 (27.1 U/L vs. 26.0 U/L vs. 23.0 U/L, p = 0.020; 34.0 U/L vs. 31.0 U/L vs. 30.6 U/L, p = 0.008; 15.5 μmol/L vs. 15.0 μmol/L vs. 13.7 μmol/L, p = 0.029, respectively). The present study also revealed significant differences in the peak values of ALT and AST among male patients with the three genotypes of rs56163822 (29.0 U/L vs. 26.9 U/L vs. 22.6 U/L, p = 0.002; 34.0 U/L vs. 32.0 U/L vs. 30.5 U/L, p = 0.019, respectively). Previous studies have revealed that median plasma BA concentrations are greater in men than in women (Frommherz et al., 2016; Xiang et al., 2012). The present results suggested that male patients carrying the NR1H4-rs56163822-GG may be more prone to liver injury, possibly because the G allele can affect the function of NR1H4, resulting in the disruption of BA homeostasis and increasing the risk of liver injury. Furthermore, the eQTL online database showed that the gene expression of SNP rs56163822-GG genotype was significantly higher than that of other genotypes (p = 0.022), further confirming the research findings at the mRNA level. If due to certain specific conditions or reasons of the patient, the NR1H4 gene site can be tested to determine whether there is a genetic variation. If the test finds that the patient carries this variation, the doctor can advise the patient to use other anti-tuberculosis drugs instead of those that may cause liver damage, so as to avoid the increased risk of liver injury when the patient uses certain drugs. However, a larger cohort study is needed to verify this conclusion.
The pivotal strength of this research is that it was a 1:4 individual-matched case‒control design with a considerable sample size in a Chinese population. This approach, coupled with comprehensive factor matching and multivariate logistic regression, effectively mitigates the influence of confounding variables potentially associated with AT-DILI. The substantial sample size further enhances statistical power and lays a solid foundation for in-depth subgroup analysis. Additionally, each AT-DILI patient strictly underwent a stringent causality assessment, minimizing the risk of misclassification bias. However, there are several limitations that ought to be acknowledged. Primarily, the rs2307424, rs11584174 and rs56163822 SNPs did not adhere to the HWE, which may be attributed to the nature of the study’s participant pool, where all controls were TB patients. Secondly, our participants were recruited from outpatient departments of four hospitals designated in Jiangsu Province, China, potentially limiting the generalizability of our findings to the entire Chinese population. Furthermore, we did not collect data on patients with previous hepatitis C infection or HIV infection. While the prevalence of these infections is relatively low in China, they are well-documented risk factors for hepatotoxicity.
Conclusion
The present study is the first to undertake the initial evaluation of the potential correlations between genetic variations in NR1I3 and NRIH4 and the risk of AT-DILI among Chinese population. Our 1:4 case‒control design, with individual matching, suggests that the SNP rs56163822 in the NR1H4 gene could be implicated in the vulnerability to AT-DILI in Chinese patients receiving anti-TB therapy. However, to substantiate these initial observations, additional research encompassing larger and more diverse populations is imperative.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The studies involving humans were approved by Ethics Committee of Nanjing Medical University. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s), and minor(s)’ legal guardian/next of kin, for the publication of any potentially identifiable images or data included in this article.
Author contributions
XX: Writing–review and editing, Funding acquisition, Investigation, Data curation, Formal Analysis, Resources. RC: Investigation, Methodology, Writing–original draft, Validation. LL: Investigation, Writing–original draft, Data curation. JC: Investigation, Methodology, Writing–original draft. XH: Investigation, Writing–original draft, Data curation. Hongqiu Pan: Investigation, Writing–original draft, Data curation. MZ: Investigation, Writing–original draft, Supervision. HY: Methodology, Writing–review and editing, Validation. ST: Supervision, Writing–review and editing, Funding acquisition, Validation.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was funded by a grant from the National Natural Science Foundation of China (grant no. 82073614) and Changshu Science and Technology Development Plan Project (grant no. CS202230).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2024.1428319/full#supplementary-material
References
Arbex, M. A., Varella, M. d. C. L., Siqueira, H. R. d., and Mello, F. A. F. d. (2010). Antituberculosis drugs: drug interactions, adverse effects, and use in special situations. Part 1: first-line drugs. J. Bras. Pneumol. 36 (5), 626–640. doi:10.1590/s1806-37132010000500016
Attinkara, R., Mwinyi, J., Truninger, K., Regula, J., Gaj, P., Rogler, G., et al. (2012). Association of genetic variation in the NR1H4 gene, encoding the nuclear bile acid receptor FXR, with inflammatory bowel disease. BMC Res. Notes 5, 461. doi:10.1186/1756-0500-5-461
Ayuso, P., Neary, M., Chiong, J., and Owen, A. (2019). Meta-analysis of the effect of CYP2B6, CYP2A6, UGT2B7 and CAR polymorphisms on efavirenz plasma concentrations. J. Antimicrob. Chemother. 74 (11), 3281–3290. doi:10.1093/jac/dkz329
Brychzy, A., Rein, T., Winklhofer, K. F., Hartl, F. U., Young, J. C., and Obermann, W. M. J. (2003). Cofactor Tpr2 combines two TPR domains and a J domain to regulate the Hsp70/Hsp90 chaperone system. EMBO J. 22 (14), 3613–3623. doi:10.1093/emboj/cdg362
Cai, P., Mao, X., Zhao, J., Nie, L., Jiang, Y., Yang, Q., et al. (2021). Farnesoid X receptor is required for the redifferentiation of bipotential progenitor cells during biliary-mediated zebrafish liver regeneration. Hepatology 74 (6), 3345–3361. doi:10.1002/hep.32076
Carino, A., Biagioli, M., Marchianò, S., Scarpelli, P., Zampella, A., Limongelli, V., et al. (2018). Disruption of TFGβ-SMAD3 pathway by the nuclear receptor SHP mediates the antifibrotic activities of BAR704, a novel highly selective FXR ligand. Pharmacol. Res. 131, 17–31. doi:10.1016/j.phrs.2018.02.033
Chen, S., Pan, H., Chen, Y., Lu, L., He, X., Chen, H., et al. (2019). Association between genetic polymorphisms of NRF2, KEAP1, MAFF, MAFK and anti-tuberculosis drug-induced liver injury: a nested case-control study. Sci. Rep. 9 (1), 14311. doi:10.1038/s41598-019-50706-y
Cheung, T. P., Van Rostenberghe, H., Ismail, R., Nawawi, N. N., Abdullah, N. A., Ramli, N., et al. (2015). High resolution melting analysis of the NR1I3 genetic variants: is there an association with neonatal hyperbilirubinemia? Gene 573 (2), 198–204. doi:10.1016/j.gene.2015.07.045
Danan, G., and Teschke, R. (2015). RUCAM in drug and herb induced liver injury: the update. Int. J. Mol. Sci. 17 (1), 14. doi:10.3390/ijms17010014
de Almeida Velozo, C.A.-O., de Almeida, T. B., de Azevedo, M. C. V. M., Espasandin, I., da Cunha Pinto, J. F., López, S., et al. (2022). Polymorphisms at CYP enzymes, NR1I2 and NR1I3 in association with virologic response to antiretroviral therapy in Brazilian HIV-positive individuals. Pharmacogenomics J. 22 (1), 33–38. doi:10.1038/s41397-021-00254-4
Ding, C., Zhang, Y., Yang, Z., Wang, J., Jin, A., Wang, W., et al. (2017). Incidence, temporal trend and factors associated with ventilator-associated pneumonia in mainland China: a systematic review and meta-analysis. BMC Infect. Dis. 17 (1), 468. doi:10.1186/s12879-017-2566-7
Eberle, M. A., Rieder, M. J., Kruglyak, L., and Nickerson, D. A. (2006). Allele frequency matching between SNPs reveals an excess of linkage disequilibrium in genic regions of the human genome. PLoS Genet. 2 (9), e142. doi:10.1371/journal.pgen.0020142
Ezhilarasan, D. (2023). Antitubercular drugs induced liver injury: an updated insight into molecular mechanisms. Drug Metab. Rev. 55 (3), 239–253. doi:10.1080/03602532.2023.2215478
Fraser, D. J., Fau - Meyer, U. A. Z. A., and Meyer, U. A. (2003). Nuclear receptors constitutive androstane receptor and pregnane X receptor activate a drug-responsive enhancer of the murine 5-aminolevulinic acid synthase gene. J. Biol. Chem. 278 (41), 39392–39401. doi:10.1074/jbc.M306148200
Frommherz, L., Bub, A., Hummel, E., Rist, M. J., Roth, A., Watzl, B., et al. (2016). Age-related changes of plasma bile acid concentrations in healthy adults--results from the cross-sectional KarMeN study. PLoS One 11 (4), e0153959. doi:10.1371/journal.pone.0153959
Gaude, G. S., Chaudhury, A., and Hattiholi, J. (2015). Drug-induced hepatitis and the risk factors for liver injury in pulmonary tuberculosis patients. J. Fam. Med. Prim. Care 4 (2), 238–243. doi:10.4103/2249-4863.154661
Goodwin, B., Hodgson, C., Fau - Liddle, E., and Liddle, C. (1999). The orphan human pregnane X receptor mediates the transcriptional activation of CYP3A4 by rifampicin through a distal enhancer module. Mol. Pharmacol. 56 (6), 1329–1339. doi:10.1124/mol.56.6.1329
Guo, H. L., Hassan, H. M., Zhang, Y., Dong, S. Z., Ding, P. P., Wang, T., et al. (2016). Pyrazinamide induced rat cholestatic liver injury through inhibition of FXR regulatory effect on bile acid synthesis and transport. Toxicol. Sci. 152 (2), 417–428. doi:10.1093/toxsci/kfw098
He, L., Guo, Y., Deng, Y., Zuo, C., and Peng, W. (2017). Involvement of protoporphyrin IX accumulation in the pathogenesis of isoniazid/rifampicin-induced liver injury: the prevention of curcumin. Xenobiotica 47 (2), 154–163. doi:10.3109/00498254.2016.1160159
He, X., Zhou, Y., Yu, J., Huang, Q., Chen, Z., Xiao, R., et al. (2024). JiaGaSongTang improves chronic cholestasis via enhancing FXR-mediated bile acid metabolism. Phytomedicine 128, 155347. doi:10.1016/j.phymed.2024.155347
Kim, K. H., Choi, S., Zhou, Y., Kim, E. Y., Lee, J. M., Saha, P. K., et al. (2017). Hepatic FXR/SHP axis modulates systemic glucose and fatty acid homeostasis in aged mice. Hepatology 66 (2), 498–509. doi:10.1002/hep.29199
Lutz, P., Berger, C., Langhans, B., Grünhage, F., Appenrodt, B., Nattermann, J., et al. (2014). A farnesoid X receptor polymorphism predisposes to spontaneous bacterial peritonitis. Dig. Liver Dis. 46 (11), 1047–1050. doi:10.1016/j.dld.2014.07.008
Lyoumi, S., Lefebvre, T., Karim, Z., Gouya, L., and Puy, H. (2013). PXR-ALAS1: a key regulatory pathway in liver toxicity induced by isoniazid-rifampicin antituberculosis treatment. Clin. Res. Hepatol. Gastroenterol. 37 (5), 439–441. doi:10.1016/j.clinre.2013.06.010
Moffatt, N. S., Bruinsma, E., Uhl, C., Obermann, W. M. J., and Toft, D. (2008). Role of the cochaperone Tpr2 in Hsp90 chaperoning. Biochemistry 47 (31), 8203–8213. doi:10.1021/bi800770g
Mouzannar, K., Fusil, F., Lacombe, B., Ollivier, A., Ménard, C., Lotteau, V., et al. (2019). Farnesoid X receptor-α is a proviral host factor for hepatitis B virus that is inhibited by ligands in vitro and in vivo. FASEB J. 33 (2), 2472–2483. doi:10.1096/fj.201801181R
Murase, W., Kubota, A., Ikeda-Araki, A., Terasaki, M., Nakagawa, K., Shizu, R., et al. (2023). Effects of perfluorooctanoic acid (PFOA) on gene expression profiles via nuclear receptors in HepaRG cells: comparative study with in vitro transactivation assays. Toxicology 494, 153577. doi:10.1016/j.tox.2023.153577
Petros, Z., Habtewold, A., Makonnen, E., and Aklillu, E. (2022). Constitutive androstane receptor and pregnane X receptor genotype influence efavirenz plasma concentration and CYP2B6 enzyme activity. Sci. Rep. 12 (1), 9698. doi:10.1038/s41598-022-14032-0
Podvinec, M., Handschin, C., Looser, R., and Meyer, U. A. (2004). Identification of the xenosensors regulating human 5-aminolevulinate synthase. Proc. Natl. Acad. Sci. U. S. A. 101 (24), 9127–9132. doi:10.1073/pnas.0401845101
Ren, W., Wang, Z., Guo, H., Gou, Y., Dai, J., Zhou, X., et al. (2024). GenX analogs exposure induced greater hepatotoxicity than GenX mainly via activation of PPARα pathway while caused hepatomegaly in the absence of PPARα in female mice. Environ. Pollut. 344, 123314. doi:10.1016/j.envpol.2024.123314
Sachar, M., Anderson, K. E., and Ma, X. (2016a). Protoporphyrin IX: the good, the bad, and the ugly. J. Pharmacol. Exp. Ther. 356 (2), 267–275. doi:10.1124/jpet.115.228130
Sachar, M., Li, F., Liu, K., Wang, P., Lu, J., and Ma, X. (2016b). Chronic treatment with isoniazid causes protoporphyrin IX accumulation in mouse liver. Chem. Res. Toxicol. 29 (8), 1293–1297. doi:10.1021/acs.chemrestox.6b00121
Semmler, G.A.-O. X., Simbrunner, B., Scheiner, B., Schwabl, P., Paternostro, R., Bucsics, T., et al. (2019). Impact of farnesoid X receptor single nucleotide polymorphisms on hepatic decompensation and mortality in cirrhotic patients with portal hypertension. J. Gastroenterol. Hepatol. 34 (12), 2164–2172. doi:10.1111/jgh.14700
Shi, Z., Li, X., Zhang, Y. M., Zhou, Y. Y., Gan, X. F., Fan, Q. Y., et al. (2022). Constitutive androstane receptor (CAR) mediates pyrene-induced inflammatory responses in mouse liver, with increased serum amyloid A proteins and Th17 cells. Br. J. Pharmacol. 179 (23), 5209–5221. doi:10.1111/bph.15934
Squires, E. J., Sueyoshi, M., Fau - Negishi, T., and Negishi, M. (2004). Cytoplasmic localization of pregnane X receptor and ligand-dependent nuclear translocation in mouse liver. J. Biol. Chem. 279 (47), 49307–49314. doi:10.1074/jbc.M407281200
Swales, K., and Negishi, M. (2004). CAR, driving into the future. Mol. Endocrinol. 18 (7), 1589–1598. doi:10.1210/me.2003-0397
Tian, Y. (2013). Epigenetic regulation of pregnane X receptor activity. Drug Metab. Rev. 45 (2), 166–172. doi:10.3109/03602532.2012.756012
Timsit, Y. E., and Negishi, M. (2007). CAR and PXR: the xenobiotic-sensing receptors. Steroids 72 (3), 231–246. doi:10.1016/j.steroids.2006.12.006
Tostmann, A., Boeree, M. J., Aarnoutse, R. E., de Lange, W. C. M., van der Ven, A. J. A. M., and Dekhuijzen, R. (2008). Antituberculosis drug-induced hepatotoxicity: concise up-to-date review. J. Gastroenterol. Hepatol. 23 (2), 192–202. doi:10.1111/j.1440-1746.2007.05207.x
Wang, H., Ge, C., Zhou, J., Guo, Y., Cui, S., Huang, N., et al. (2018). Noncanonical farnesoid X receptor signaling inhibits apoptosis and impedes liver fibrosis. EBioMedicine 37, 322–333. doi:10.1016/j.ebiom.2018.10.028
Wang, N., Chen, X., Hao, Z., Guo, J., Wang, X., Zhu, X., et al. (2022). Incidence and temporal trend of antituberculosis drug-induced liver injury: a systematic review and meta-analysis. J. Trop. Med. 2022, 8266878. doi:10.1155/2022/8266878
Wang, Y., Xiang, X., Huang, W. W., Sandford, A. J., Wu, S. Q., Zhang, M. M., et al. (2019). Association of PXR and CAR polymorphisms and antituberculosis drug-induced hepatotoxicity. Sci. Rep. 9 (1), 2217. doi:10.1038/s41598-018-38452-z
Wen, Y., Zhang, G., and Wu, X. (2022). The role of the farnesoid X receptor in quadruple anti-tuberculosis drug-induced liver injury. Toxicology 476, 153256. doi:10.1016/j.tox.2022.153256
WHO (2017). Guidelines for treatment of drug-susceptible tuberculosis and patient care. Geneva: World Health Organization.
Wyen, C., Hendra, H., Siccardi, M., Platten, M., Jaeger, H., Harrer, T., et al. (2011). Cytochrome P450 2B6 (CYP2B6) and constitutive androstane receptor (CAR) polymorphisms are associated with early discontinuation of efavirenz-containing regimens. J. Antimicrob. Chemother. 66 (9), 2092–2098. doi:10.1093/jac/dkr272
Xiang, X., Backman, J. T., Neuvonen, P. J., and Niemi, M. (2012). Gender, but not CYP7A1 or SLCO1B1 polymorphism, affects the fasting plasma concentrations of bile acids in human beings. Basic Clin. Pharmacol. Toxicol. 110 (3), 245–252. doi:10.1111/j.1742-7843.2011.00792.x
Yang, F., Huang, X., Yi, T., Yen, Y., Moore, D. D., and Huang, W. (2007). Spontaneous development of liver tumors in the absence of the bile acid receptor farnesoid X receptor. Cancer Res. 67 (3), 863–867. doi:10.1158/0008-5472.CAN-06-1078
Yang, M., Pan, H., Lu, L., He, X., Chen, H., Tao, B., et al. (2019b). Home-based Anti-Tuberculosis Treatment Adverse Reactions (HATTAR) study: a protocol for a prospective observational study. BMJ Open 9 (3), e027321. doi:10.1136/bmjopen-2018-027321
Yang, S., Hwang, S. J., Park, J. Y., Chung, E. K., and Lee, J. I. (2019a). Association of genetic polymorphisms of CYP2E1, NAT2, GST and SLCO1B1 with the risk of anti-tuberculosis drug-induced liver injury: a systematic review and meta-analysis. BMJ Open 9 (8), e027940. doi:10.1136/bmjopen-2018-027940
Yu, Y. C., Mao, Y. M., Chen, C. W., Chen, J. J., Chen, J., Cong, W. M., et al. (2017). CSH guidelines for the diagnosis and treatment of drug-induced liver injury. Hepatol. Int. 11 (3), 221–241. doi:10.1007/s12072-017-9793-2
Zheng, J.A.-O., Guo, M. H., Peng, H. W., Cai, X. L., Wu, Y. L., and Peng, X. E. (2020). The role of hepatitis B infection in anti-tuberculosis drug-induced liver injury: a meta-analysis of cohort studies. Epidemiol. Infect. 148, e290. doi:10.1017/S0950268820002861
Keywords: antituberculosis drug-induced liver injury, nuclear receptor subfamily 1 group I member 3, nuclear receptor subfamily 1 group H member 4, genetic polymorphisms, case-control study
Citation: Xu X, Chen R, Lu L, Cheng J, He X, Pan H, Zhang M, Yi H and Tang S (2024) Roles of NR1I3 and NR1H4 polymorphisms in the susceptibility to antituberculosis drug-induced liver injury in China: a case‒control study. Front. Genet. 15:1428319. doi: 10.3389/fgene.2024.1428319
Received: 06 May 2024; Accepted: 08 October 2024;
Published: 23 October 2024.
Edited by:
Paulo Caleb J. L. Santos, Federal University of São Paulo, BrazilReviewed by:
Shicheng Fan, Southern Medical University, ChinaCarolina Dagli Hernandez, State University of Campinas, Brazil
Copyright © 2024 Xu, Chen, Lu, Cheng, He, Pan, Zhang, Yi and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shaowen Tang, dG9tc3dlbkBuam11LmVkdS5jbg==
 Xiaoyan Xu1
Xiaoyan Xu1 Ruina Chen
Ruina Chen Honggang Yi
Honggang Yi Shaowen Tang
Shaowen Tang