
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 27 May 2024
Sec. Cancer Genetics and Oncogenomics
Volume 15 - 2024 | https://doi.org/10.3389/fgene.2024.1401100
This article is part of the Research Topic Emerging Relevance of Molecular Profiling in Global Cancer Research and Management View all 4 articles
 Aleksey Drobyshev1,2†
Aleksey Drobyshev1,2† Alexander Modestov2†
Alexander Modestov2† Maria Suntsova1,2
Maria Suntsova1,2 Elena Poddubskaya2,3
Elena Poddubskaya2,3 Alexander Seryakov4
Alexander Seryakov4 Aleksey Moisseev2
Aleksey Moisseev2 Maksim Sorokin1,2
Maksim Sorokin1,2 Victor Tkachev5
Victor Tkachev5 Galina Zakharova2
Galina Zakharova2 Aleksander Simonov2
Aleksander Simonov2 Marianna A. Zolotovskaia1,2,6
Marianna A. Zolotovskaia1,2,6 Anton Buzdin1,2,6,7*
Anton Buzdin1,2,6,7*The TERT gene encodes the reverse transcriptase subunit of telomerase and is normally transcriptionally suppressed in differentiated human cells but reactivated in cancers where its expression is frequently associated with poor survival prognosis. Here we experimentally assessed the RNA sequencing expression patterns associated with TERT transcription in 1039 human cancer samples of 27 tumor types. We observed a bimodal distribution of TERT expression where ∼27% of cancer samples did not express TERT and the rest showed a bell-shaped distribution. Expression of TERT strongly correlated with 1443 human genes including 103 encoding transcriptional factor proteins. Comparison of TERT- positive and negative cancers showed the differential activation of 496 genes and 1975 molecular pathways. Therein, 32/38 (84%) of DNA repair pathways were hyperactivated in TERT+ cancers which was also connected with accelerated replication, transcription, translation, and cell cycle progression. In contrast, the level of 40 positive cell cycle regulator proteins and a set of epithelial-to-mesenchymal transition pathways was specific for the TERT- group suggesting different proliferation strategies for both groups of cancer. Our pilot study showed that the TERT+ group had ∼13% of cancers with C228T or C250T mutated TERT promoter. However, the presence of promoter mutations was not associated with greater TERT expression compared with other TERT+ cancers, suggesting parallel mechanisms of its transcriptional activation in cancers. In addition, we detected a decreased expression of L1 retrotransposons in the TERT+ group, and further decreased L1 expression in promoter mutated TERT+ cancers. TERT expression was correlated with 17 genes encoding molecular targets of cancer therapeutics and may relate to differential survival patterns of TERT- positive and negative cancers.
The human telomerase reverse transcriptase gene TERT encodes a reverse transcriptase that maintains chromosome ends by adding hexameric repeats 5′-TTAGGG-3′ to the 3′termini using an RNA template encoded by the TERC gene (Wu et al., 2017). In the human body, telomerase activity could be found in few types of the somatic cells, such as activated B-lymphocytes and T-lymphocytes, endothelial cells and macrophages (Hsiao et al., 1997; Weng et al., 1998; Lin et al., 2010; Gizard et al., 2011). In general, telomerase expression is normally restricted to developing tissues, germ-line and stem cells but it can be also pathologically reactivated in cancers, thus supporting continuous proliferation and preventing senescence caused by shortened telomeres (Colebatch et al., 2019). Furthermore, expression of telomerase was reported as a prognostic factor of patient survival in some cancers, where an increased TERT expression is a negative prognostic biomarker (Gong et al., 2021; Kwon and Yi, 2021; Li et al., 2024). Thus, the analysis of expression patterns and structural changes of the TERT gene is of great interest. The increased expression of TERT was frequently reported to be associated with activating somatic mutations in its promoter region (e.g., (Huang et al., 2015)). However, the frequency of such promoter mutations differs significantly by cancer type (Blackburn et al., 2015). The highest frequency of TERT promoter mutations was found in melanomas and central nervous system tumors (in particular, over 70% in primary glioblastoma and oligodendroglioma cases) (Killela et al., 2013; Simon et al., 2015; Hugdahl et al., 2018; Colebatch et al., 2019; Olympios et al., 2021; Guo et al., 2022). TERT promoter mutations are also frequently observed in urothelial, thyroid, and hepatocellular carcinomas, and in squamous cell and basal cell carcinoma of the skin (Griewank et al., 2013; Scott et al., 2014; Liu and Xing, 2016; Nault and Zucman-Rossi, 2016; Colebatch et al., 2019; Hayashi et al., 2021; Zhang et al., 2022). In addition, mutations were found in most tumor types, but at a relatively low frequency (Vinagre et al., 2013; Bell et al., 2016; Gupta et al., 2021; El Zarif et al., 2023).
The significance of TERT promoter mutations in carcinogenesis has been assessed in many studies (Fredriksson et al., 2014; Barthel et al., 2017). It was hypothesized that it mainly occurs in tumors originating from tissues with a low self-renewal rate (Killela et al., 2013; Simon et al., 2015).
Two activating mutations in the promoter region of TERT: C228T (chr5:1,295,228 C>T on assembly GRCh37; NM_198253.3:c.-124C>T) and C250T (chr5:1,295,250 C>T on assembly GRCh37; NM_198253.3:c.-146C>T) are located at the positions −124 and −146 bp, respectively, upstream of the ATG site and are the most common reported mutations in the TERT gene (Colebatch et al., 2018; Powter et al., 2021; Kang et al., 2022). These mutations are typically heterozygous, rarely occur simultaneously and result in a 5′-CCCGGAAGGGG-3′ sequence of 11 bases that corresponds to a de novo binding site for the E26 transformation-specific transcriptional factor (ETS) protein (da Costa et al., 2021; Hasanau et al., 2022; Koleilat et al., 2023). In addition, TERT promoter mutations are associated with the switching of inactive chromatin marks to active ones in the TERT promoter (Stern et al., 2015; Salgado et al., 2020). It increases the transcription and thereby leads to enhanced activity of telomerase (Liu et al., 2016). It was also previously reported that in humans TERT is frequently expressed in the form of alternatively spliced transcripts in both normal and tumor cells, and only the longest transcripts possess the reverse transcriptase enzymatic activity (Hrdličková et al., 2012; Colebatch et al., 2019). Overexpression of some alternatively spliced variants could increase the proliferation rate without enhancing telomerase activity (Hrdličková et al., 2012).
In a number of studies, mutations in the promoter region of TERT were reported to correlate with its increased expression level. In melanomas, the increased telomerase expression was correlated with C228T/A and C250T/A TERT promoter mutations (Lee et al., 2016; Shaughnessy et al., 2020; Guo et al., 2022). Also, higher expression of TERT in tumors with promoter mutations was observed in urothelial carcinomas and in gliomas (Borah et al., 2015; Heidenreich et al., 2015).
However, these findings are contradictory as some researchers reported lack of statistical significance for such a phenomenon for patients diagnosed with thyroid cancer (Pestana et al., 2020), bladder cancer (Allory et al., 2014), and in melanoma cell lines (Salgado et al., 2020). Also, it was reported that the TERT promoter mutation status did not affect its mRNA level in hepatocellular carcinomas (Chen et al., 2014; Zhang et al., 2022). A recent pan-cancer study reported only a marginally significant association between promoter mutation status of TERT and its mRNA level (p = 0.0373, N = 503 cell lines) (Stern et al., 2020). These facts could be attributed to the structural variations of the TERT gene. For example, it was found that nearly 50% of glioblastoma patients with wild-type TERT promoter harbored structural rearrangements of TERT (Diplas et al., 2018). At the same time there was no significant difference in TERT expression between the rearranged and promoter-mutant glioblastoma subgroups. Moreover, it was reported that telomerase-expressing embryonic stem cells with an introduction of any of the most frequent TERT promoter mutations showed only a modest increase in this gene transcription with no detectable impact on telomerase functional activity (Chiba et al., 2015).
Interestingly, recent studies have identified several non-canonical functions of TERT that are not related to maintaining telomere stability (Ségal-Bendirdjian and Geli, 2019; Thompson and Wong, 2020; Akincilar et al., 2021). For example, TERT can act as a modulator in NF-kB and Wnt/β-catenin signaling pathways (Li and Tergaonkar, 2014). Other non-canonical functions of TERT include regulation of apoptosis, maintenance of the cellular redox homeostasis, cell proliferation, response to DNA damage, and formation of G-quadruplexes in the G-rich area of gene promoters, which can influence transcription by regulating binding of transcription factor proteins (Fleisig et al., 2016; Khattar et al., 2016; Yuan et al., 2019; Rosen et al., 2020; Gu et al., 2022; Pavlova et al., 2022; Palamarchuk et al., 2023). Thus, theoretically TERT promoter mutations may also affect these and probably other processes related to TERT.
In this study, we experimentally assessed the gene expression patterns associated with the status of TERT expression in 1039 human cancer samples of 27 tumor types investigated by RNA sequencing. On pan-cancer level we showed a bimodal distribution of TERT expression where ∼27% of cancer samples did not express TERT, whereas the rest showed a bell-shaped distribution. Expression of TERT was strongly correlated with 1443 human genes including 103 encoding transcriptional factor proteins. Comparison of TERT- positive and negative cancers showed the differential activation of 496 genes and 1975 molecular pathways. Among them, 32/38 (84%) of DNA repair pathways were hyperactivated in TERT+ cancers which was also connected with accelerated replication, transcription, translation, and cell cycle progression. Our pilot study showed that the TERT+ group included ∼13% of cancers with mutated TERT promoter (mutations C228T, C250T, or both). However, the presence of promoter mutations was not associated with a greater level of TERT expression compared with other TERT+ cancers. This evidences parallel mechanisms leading to the TERT gene transcriptional activation with only a rather minor impact related to promoter mutation.
The consent procedure and the design of the study were approved by the ethical committee of Clinical Center Vitamed, Moscow, protocol date 16.10.2017.
We enrolled 1039 adult 18–80 years old male and female patients with diagnosed tumors who were either eligible for Oncobox clinical trials NCT03521245 (Sorokin et al., 2020) and NCT03724097, or submitted their biomaterials for Oncobox molecular testing. For solid tumors the biomaterials were formalin-fixed paraffin embedded (FFPE) samples of surgically removed tumor tissue or core-needle biopsies. The materials were confirmed by a certified pathologist for diagnosis and content of cancer cells (no less than 50% of cancer cells in a biosample was acceptable). All the patients or their legal representatives signed written informed consents to participate in this study and to publish the results of RNA sequencing analysis without disclosure of personal genetic data.
Total RNA preps extracted from the tumor biomaterial were subjected to RNA sequencing as previously described (Suntsova et al., 2019). Solid tissue biosamples were either immediately stabilized in RNAlater (Qiagen, Germany) and then stored at −70 C or fixed in formalin and embedded in paraffin blocks. RNA extraction was performed immediately before the preparation of sequencing libraries using QIAGEN RNeasy Kit (Qiagen) or Direct-zol™ RNA MiniPrep (Zymo Research) with TRI Reagent (MRC) for tissues in RNAlater and the RecoverAll™ RNeasy FFPE Kit (Qiagen), following the manufacturer’s protocol. Also, KAPA RNA Hyper with RiboErase (KAPA Biosystem) Kit was used for further depletion of ribosomal RNA and library preparation (Suntsova et al., 2019).
RNAseq FASTQ files were processed with STAR aligner in “GeneCounts” mode with the Ensembl human transcriptome annotation (Dobin et al., 2013). Ensembl gene IDs were converted to HUGO Gene Nomenclature Committee (HGNC) gene symbols using the Complete HGNC dataset (https://www.genenames.org/, database version from 13 July 2017). Overall, expression levels were established for 35,126 annotated genes with the corresponding HGNC identifiers. For further assessments, only the samples with high-quality experimental RNA sequencing profiles reaching the threshold of 3.5*10^6 gene-mapped reads were selected (Suntsova et al., 2019), statistics shown on Table 1. By comparing the tumor and normal expression profiles, the case-to-normal ratios for the expression of individual genes were calculated, as well as the extent of differential activation of 2929 intracellular molecular pathways (Borisov et al., 2020) using OncoboxPD tool (Zolotovskaia et al., 2022). For these comparisons, the RNA sequencing profiles of healthy tissues previously obtained by us for human donors killed in road accidents using the same protocols, equipment, and reagents were used as the normal controls (Sorokin et al., 2023).
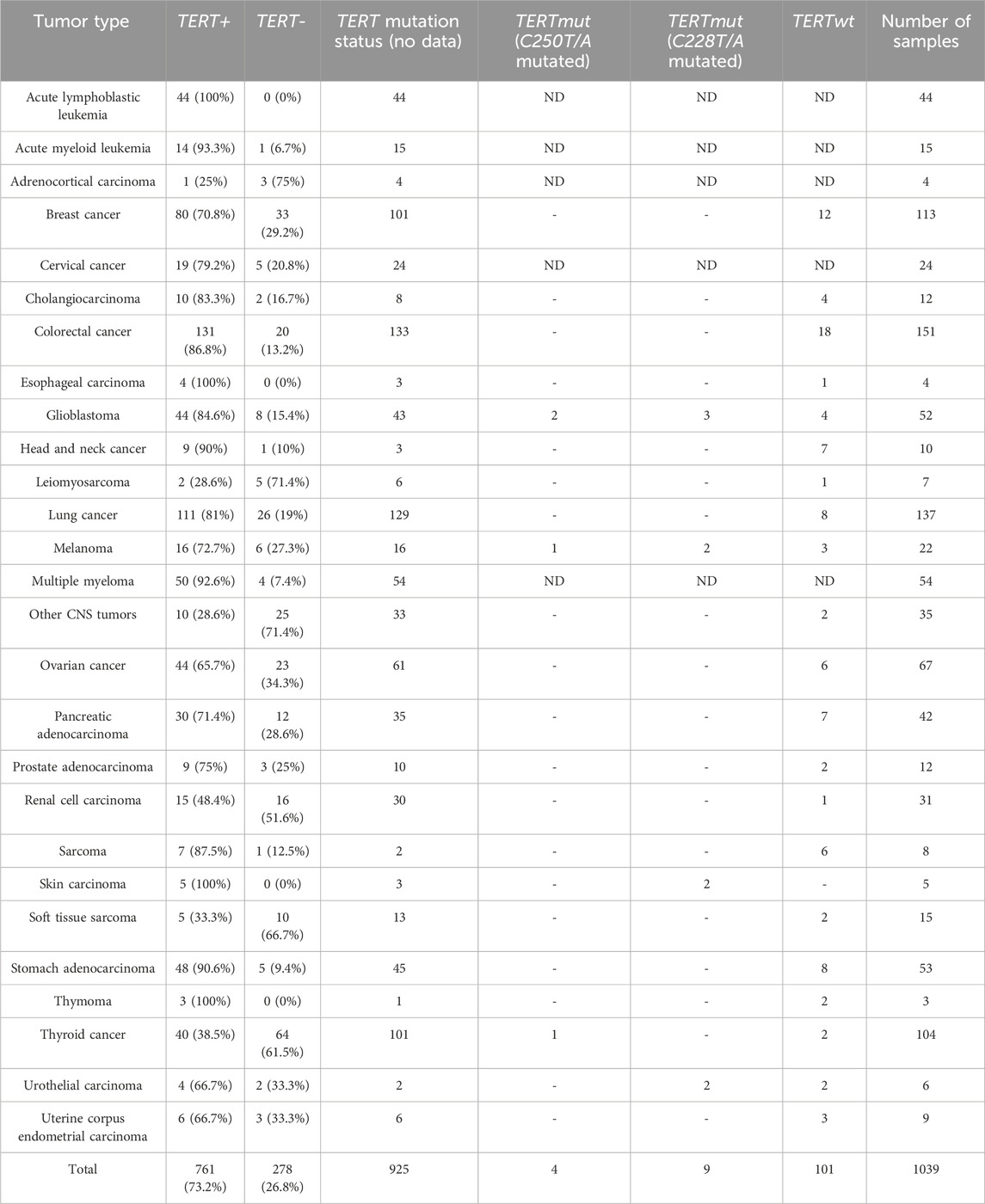
Table 1. Statistics for experimental cancer samples used in this study sorted according to cancer type diagnoses.
Pathway activation level (PAL) is an integral quantitative and qualitative characteristic of changes in the expression levels for genes participating in a molecular pathway (Aliper et al., 2017; Buzdin et al., 2018; Borisov et al., 2020; Zolotovskaia et al., 2023).
PAL values were calculated as follows:
where PALp is PAL for a pathway p, CNRn is the case-to-normal ratio for a gene n; ARR (activator/repressor role) is a Boolean value that depends on the function of this gene product in the pathway p (Zolotovskaia et al., 2023). ARRnp is a Boolean value defined as follows: −1 when the product of the gene n inhibits the pathway p; 1 when the product of n activates p; 0 when the product of n has an ambiguous role in p. The CNRn value is calculated as the ratio of a quantitative metric level for the gene n in a biosample under study to an average level for n in the control group. For each tumor type, we normalized the expression data using the group of 4-6 control biosamples of the corresponding healthy tissues obtained for either healthy donors (blood) or donors killed in road accidents (solid tissues) from the ANTE collection of RNA sequencing samples that was previously published by our group (Sorokin et al., 2023).
Principal component analysis was performed for the groups of biosamples stratified depending on the TERT expression (TERT+/TERT-) and TERT promoter mutation status (TERTwt/TERTmut).
Spearman correlation coefficients with p-values were calculated using the R “stats” package and visualized using the R “corrplot” package. We used a Benjamini-Hochberg adjusted p-value threshold 0.05 for the assessment of correlation significance.
Gene ontology (GO) enriched terms were defined for differential gene sets investigated using “clusterProfiler” R package, and p-values were adjusted according to Benjamini and Hochberg false discovery rate (FDR) correction procedure with the statistical threshold of 0.05. L1 retroelement quantification was calculated using the TEcount tool based on precomputed STAR alignments according to the recommendation of TEcount. Expression data of genes and L1 retroelements (of L1HS and L1PA2 subgroups) were then DESeq2 normalized. Comparisons of PAL for molecular pathways, gene expression, and L1 expression for the different groups of interest were performed using non-parametric Mann Whitney U-test in R (Benjamini-Hochberg adjusted p-value threshold 0.05). DESeq2 software was used to obtain the differentially expressed genes from raw counts. Genes with adjusted p-values < 0.05 and absolute value of fold change (FC) > 2 were considered as differentially expressed.
In this study we investigated TERT promoter mutation status for 114 independent FFPE solid cancer tissue samples representing 22 tumor types: Breast cancer (12 samples), Cholangiocarcinoma (4 samples), Colorectal cancer (18 samples), Esophageal carcinoma (1 sample), Glioblastoma (9 samples), Head and neck cancer (7 samples), Leiomyosarcoma (1 sample), Lung cancer (8 samples), Melanoma (6 samples), Other CNS tumors (2 samples), Ovarian cancer (6 samples), Pancreatic adenocarcinoma (7 samples), Prostate adenocarcinoma (2 samples), Renal cell carcinoma (1 sample), Sarcoma (6 samples), Skin carcinoma (2 samples), Soft tissue sarcoma (2 samples), Stomach adenocarcinoma (8 samples), Thymoma (2 samples), Thyroid cancer (3 samples), Urothelial carcinoma (4 samples), and Uterine corpus endometrial carcinoma (3 samples).
The TERT promoter genomic region potentially containing C250T/A and C228T/A mutations was amplified as described before (Colebatch et al., 2018) with minor modifications. The 30-µL reaction mixture contained 5 ng of tumor-isolated genomic DNA, 500 nM of each primer (forward: 5′-GCAGCGCTGCCTGAAACTCG-3′, reverse: 5′-CGTCCTGCCCCTTCACCTTC-3′), 200 nM of 7-deaza-dGTP and 1x PCR mastermix (HS-Sybr, Evrogen, Russia). Thermocycling conditions were as follows: 95°C for 5 min, 4 cycles of 98°C for 14 s, 66°C for 10 s, 75°C for 30 s, followed by 40 cycles of 95°C for 10 s, 66°C for 5 s, 75°C for 14 s, followed by melting curve recording from 95°C to 75°C with 0,1°C/s rate. Samples with clear melting peak at 88°C were used for further purification. The 60 µL of PCR product were purified with MDX041 NGS Clean and Select Beads (Meridian Bioscience, United States) according to manufacturer’s protocol and eluted with 10 µL of 10 mM Tris-HCl pH 8.0; 5 µL of purified product was sequenced with 3500xL Applied Biosystems sequencer using reverse primer.
In the TERT promoter mutation assessment process, the C228A mutation (chr5:1,295,228 C>A on assembly GRCh37; NM_198253.3:c.-124C>A) was detected in one biosample of the TERT+ group (Urothelial carcinoma), which creates a putative ETS motif (Spiegl-Kreinecker et al., 2018; Gutierrez-Rodrigues et al., 2019; Yu et al., 2021). The C250A mutation (chr5:1,295,250 C>A on assembly GRCh37; NM_198253.3:c.-146C>A) is also possible (Spiegl-Kreinecker et al., 2018; Gutierrez-Rodrigues et al., 2019; Yu et al., 2021) but was not detected in our study. For further analysis, C250T/A and C228T/A mutations were considered as the driver hotspot clinically relevant TERT promoter mutations.
Overall survival (OS) and progression-free survival (PFS) analysis and hazard ratio calculation were assessed with relation to extent of TERT expression for colorectal cancer (TCGA-COAD) and thyroid cancer (TCGA-THCA) patients using the TCGA database. Patient survival analysis and plotting were performed using R packages “survival,” “survminer,” “pheatmap,” and “ggplot2”. The statistical significance of differences was measured by log-rank test p-value. p-values were adjusted according to Benjamini and Hochberg false discovery rate (FDR) correction procedure. The statistical threshold for FDR-adjusted p-value was 0.05. The Kaplan–Meier plots were drawn to visualize survival differences.
In this study we aimed to identify transcriptomic patterns associated with cancer expression of the TERT gene in a high-throughput experimental assay. To this end, using the laboratory protocol and bioinformatic pipeline from (Suntsova et al., 2019), we obtained bulk RNA sequencing profiles for 1039 human cancer tissue biosamples representing 27 tumor types (Table 1). Cancer tissue specimens were obtained from the patients who were either included in clinical trials NCT03521245 (Sorokin et al., 2020) and NCT03724097, or who submitted their biomaterials for Oncobox molecular testing in 2018–2021 and agreed to participate in this study. Among them, for 114 patients with available biosamples and who gave their additional informed consent, the statuses of the TERT gene mutations at C228T and C250T sites were established later by Sanger sequencing. The experimental cancer RNA profiles were then bioinformatically assessed depending on the status of (i) TERT expression and, where available, (ii) TERT promoter mutation.
Our pan-cancer analysis clearly revealed a bimodal distribution of the RNA sequencing profiles sorted according to the expression of TERT (Figure 1A), where ∼73% of the samples expressed TERT, and ∼27% showed no detectable transcription (Table 1). We detected TERT expression—positive (TERT+) status in the majority of the samples in 21 (78%) tumor types. The biggest proportion of TERT+ samples was observed for the hematological cancers: in acute lymphoblastic leukemia (100%; n = 44), acute myeloid leukemia (93%; n = 15), and multiple myeloma (93%, n = 54), Table 1. Principal component analysis performed for all available expression profiles demonstrated that for most cancers, TERT expression status did not affect sample clustering. At the same time, a separate cluster was observed for a group of hematologic tumor samples, which were mostly TERT+ (Figure 1B).
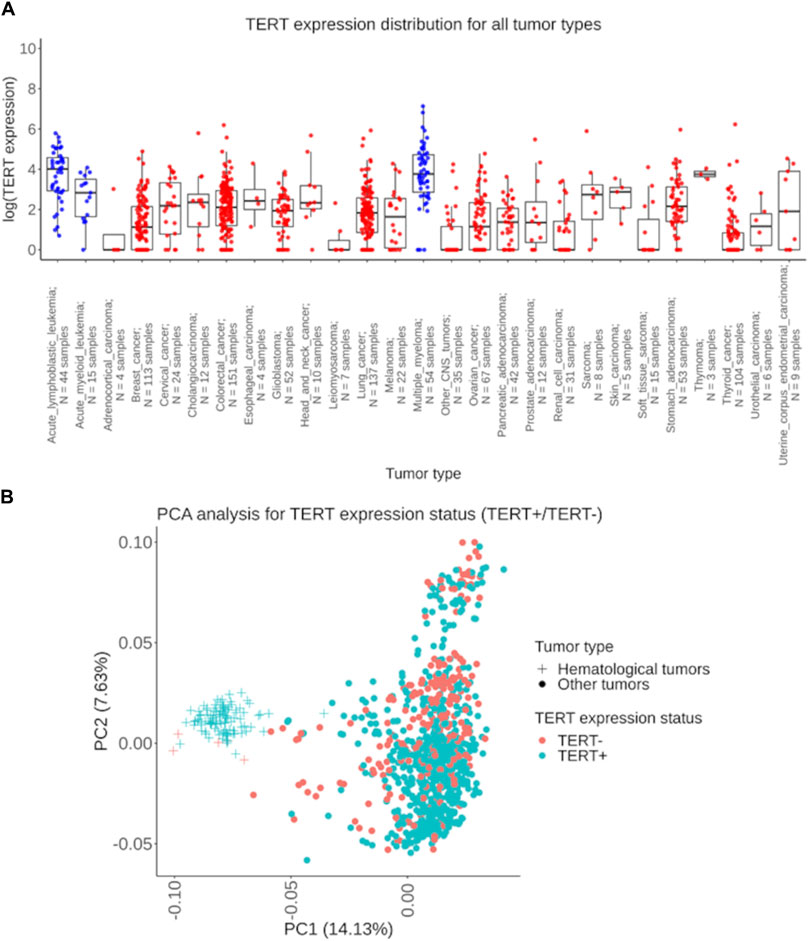
Figure 1. Assessment of TERT expression distribution in experimentally tested 1039 tumor samples. (A) The extent of TERT expression shown separately for 27 cancer types under analysis. The Y-axis indicates the natural logarithm of (TERT expression +1) value. Blue dots represent biosamples of the hematological cancers: acute lymphoblastic leukemia, acute myeloid leukemia, and multiple myeloma; red dots indicate biosamples of the non-hematological tumors. (B) Principal component analysis of TERT+ and TERT- tumors, hematologic and solid cancers are shown separately. Dots and crosses represent specific biosamples.
However, in six tumor types the proportion of TERT expression–negative (TERT-) samples was predominant: in adrenocortical carcinoma (75%; n = 4), in non-glioblastoma CNS tumors (71%; n = 35), in soft tissue sarcomas (67%; n = 15), in renal cell carcinoma (52%; n = 31), in leiomyosarcoma (71%; n = 7), and in thyroid cancer (62%; n = 104), Table 1. In 23 cancer types collectively representing 95% of the RNAseq profiles, both TERT+ and TERT- biosamples were detected, thus evidencing that the phenomenon of TERT- negative cancers is not restricted to specific tumor localizations.
TERT positive or negative expression status was not correlated with the fraction of cancer cells in the tissue section evaluated by a pathologist in the biosamples under analysis (data not shown). In addition, the representative microphotographs of hematoxylin/eosin stained lung cancer biosamples show no visible differences in numbers of viable tumor cells between TERT+ and TERT- tumor groups (Figure 2A).
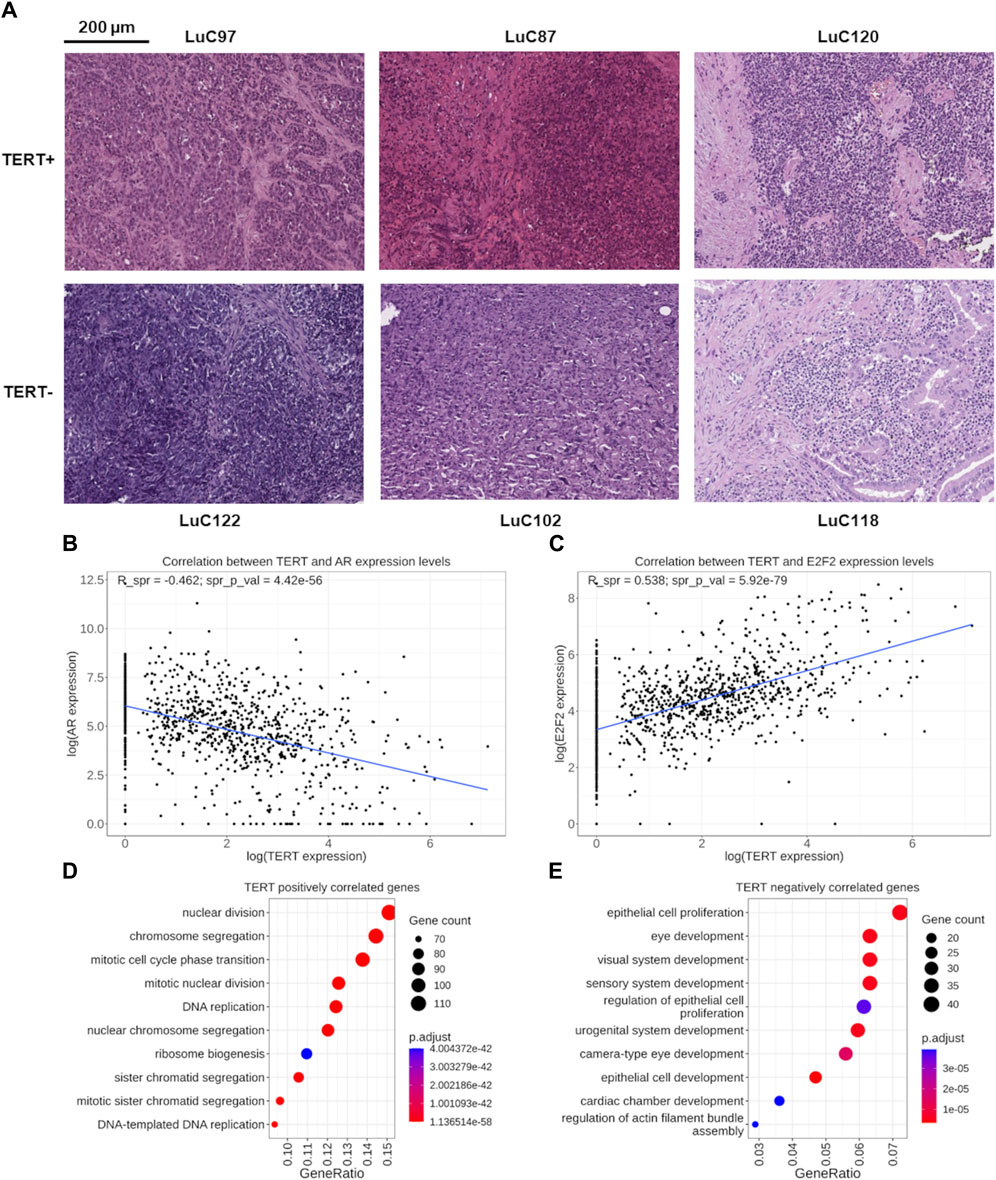
Figure 2. (A) Representative microphotographs of hematoxylin/eosin stained lung cancer biosamples. TERT- status was established for samples LuC122 (∼70% cancer cells), LuC102 (∼80% cancer cells), and LuC118 (∼60% cancer cells). TERT+ status was established for samples LuC97 (∼90% cancer cells), LuC87 (∼60% cancer cells), and LuC120 (∼80% cancer cells). (B) Functional assessment of genes correlated with TERT expression. Correlation of expression levels between the TERT and AR genes. Each dot represents an individual biosample. (C) Correlation of expression levels between the TERT and E2F2 genes. Each dot represents an individual biosample. (D) Gene Ontology (GO) analysis of 817 genes positively correlated with TERT expression. (E) GO analysis of 626 genes negatively correlated with TERT expression.
By correlating the expression of TERT with other 35,125 known human genes in all 1039 cancer samples under analysis using the threshold of FDR-adjusted p-value less than 0.05 and Spearman correlation with absolute value more than 0.3, we identified 817 genes positively and 626 genes negatively correlated with the transcription of TERT (Supplementary Table S1). Among them, 103 transcription factor (TF) genes were found: 48 positively and 55 negatively correlated with TERT (Supplementary Table S2). Nineteen most strongly correlated TF genes with absolute value of correlation coefficient starting from 0.4 included 11 positively and 8 negatively correlated genes (Table 2).
Interestingly, among these strongly correlated 19 TF genes, for six (32%) we found previous reports in the literature that they are involved in the regulation of TERT. Specifically, the AR gene (Figure 2B) encoding the androgen receptor regulates TERT through chromatin binding of TFs E2F1 and MYC (Dong et al., 2021). E2F1, E2F2, (Figure 2C), E2F8 genes are representatives of the E2F family of TFs which is known as the potent inducer of TERT (Suenaga et al., 2006; Kumari et al., 2009; Zhang et al., 2014). The FOXM1 (Tang et al., 2023) and HMGB1 (Kučírek et al., 2019) genes are other known regulators of TERT stability and activity. Other strongly correlating TF genes for which we found no mentioning in the literature on their association with TERT were: positively correlated—MTA2, MYBL2, TCF3, ZBED4, ZNF296, ZNF367; negatively correlated—CREBL2, NFIA, NFIC, PURA, ZFPM2, ZNF25, ZNF641. Thus, the strong connection of these 13 TF genes with telomerase expression is new and may be representing new regulatory mechanisms governing transcription of TERT. In addition, MYC is a well-known regulator of TERT transcription, and in this study it was found statistically significantly correlated with the expression of TERT with a correlation coefficient of 0.33.
We then performed Gene Ontology (GO) analysis separately for the genes identified as positively or negatively correlated with TERT expression. We found that the pool of 817 positively correlated genes was most strongly enriched among the terms dealing with protein synthesis, ribosome biogenesis and cell division: chromatid segregation, mitosis, nuclear division, DNA replication (Figure 2D). On the other hand, the most strongly enriched GO terms among the negatively correlated 626 genes dealt mostly with the cellular terminal differentiation and organogenesis, and also with the proliferation of epithelial cells (Figure 2E).
Interestingly, the latter finding means that many genes whose expression negatively correlates with TERT play a role in the regulation of epithelial cell proliferation. A more careful look on these terms identified 40 genes strongly negatively correlated with TERT, most of which are known as positive regulators of cell proliferation (Table 3). All of these genes were statistically significantly upregulated in the TERT- tumors compared to the TERT+ group (Supplementary Figure S1). These results may indicate more complex molecular mechanisms of proliferation in TERT- tumors, such as alternative lengthening of telomeres (ALT) which is observed in a subset of cancers, particularly in tumors with deficiencies in telomerase activity (Zhao et al., 2019). ALT is a telomerase-independent mechanism used by cancer cells to maintain their telomeres presumably by a recombination-based pathway which could promote cell proliferation even in the case of silent telomerase genes (Zhao et al., 2019; Ferreira et al., 2020).
In addition, we performed an analysis also at the level of molecular pathway activation. Here the pathway activation level (PAL) metric was used to assess the correlation of TERT expression with the activation levels of 2929 molecular pathways. Positive PAL for pathway indicates upregulation compared with the normal tissue, negative PAL means downregulation, and zero PAL shows no changes in gene expression between tumor and normal tissues, whereas the absolute value of PAL reflects the extent of up/downregulation of a pathway in a tumor sample under analysis (Buzdin et al., 2020). Meanwhile, PAL was shown to be a promising biomarker in tumor research that is more reliable than the expression levels of individual genes (Borisov et al., 2017; Buzdin et al., 2018). In this study, PAL scores were correlated with TERT expression, and positive and negative links were established. Using the same criteria of FDR-adjusted p-value less than 0.05 and absolute value of Spearman correlation exceeding 0.3, we totally found 241 positively and 24 negatively correlated pathways with the TERT gene expression (Supplementary Table S3), top correlated pathways shown on Table 4.
Overall, the figure of top correlated pathways was remarkably similar to what was observed on the gene level. Telomerase expression was shown to strongly correlate with the pathway of telomere extension by telomerase, with the pathways of ribosomal RNA transcription and ribosome biogenesis, nuclear export and import of RNAs, ribonucleotide biosynthesis, amino acid charging of transfer RNAs, transcription and processing of messenger RNAs, and mitochondrial transcription (Table 4).
In turn, the top negatively correlated pathways dealt with the processes of cytoskeleton and external matrix remodeling, SMAD signaling, JAK degradation regulation, PTEN dependent cell cycle arrest and apoptosis, and cell proliferation through ErbB2 and ErbB3 signaling, Table 5.
Among them, the activation profiles in TERT+ and in TERT- cancers can be seen in more detail for the pathways of Telomere extension by telomerase (Figure 3A), PTEN dependent cell cycle arrest and apoptosis (Figure 3B), and SMAD signaling pathway (Figure 3C). The group-averaged pathway activation charts are given relatively to the normal tissue, which was used to calculate pathway activation levels for every cancer sample under analysis. This provides a more detailed view of the differential activation of those pathway components in the TERT+ and TERT- cancers.
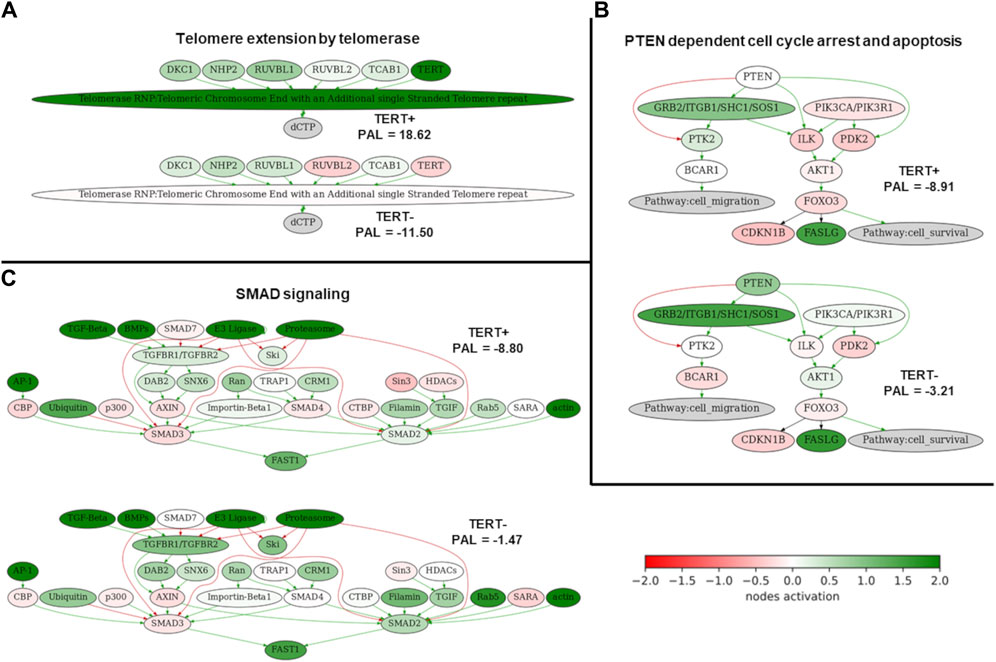
Figure 3. Activation profiles of three selected molecular pathways strongly correlated with the expression of TERT. (A) Telomere extension by telomerase pathway. (B) PTEN dependent cell cycle arrest and apoptosis pathway. (C) SMAD signaling pathway. Schemes for TERT+ samples are shown on the top of each panel, for TERT- samples– on the bottom of each panel. Color reflects the logarithm of the case-to-normal ratio (CNR) of the pathway nodes, color scale is given (green–upregulated, red–downregulated, white–intact). Arrows show molecular interactions within a pathway: green stands for activation, red for inhibition.
In order to investigate differential transcriptional patterns of TERT+ and TERT- cancers, we profiled differential activities of individual genes and molecular pathways where the TERT- group was taken as a reference. Using statistical criterion of FDR-adjusted p-value < 0.05 and |log2FoldChange| > 2, we identified 262 genes upregulated in TERT+ cancers and 234 genes upregulated in TERT- cancers (Figure 4A; Supplementary Table S4). At the level of molecular pathways, we found totally 1077 pathways upregulated in TERT+ and 898 pathways upregulated in TERT- cancers using non-parametric Mann Whitney U-test (Supplementary Table S5).
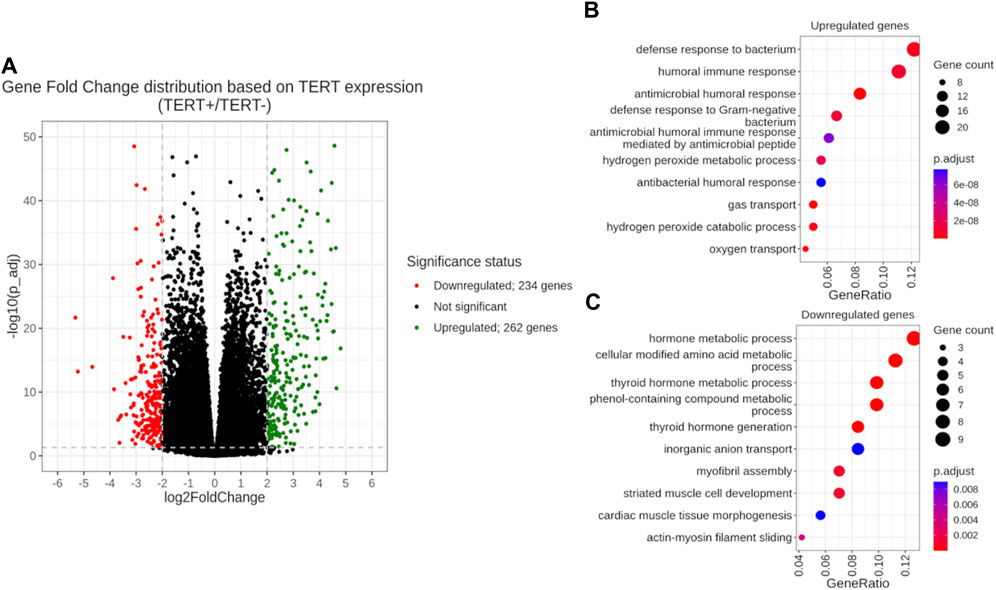
Figure 4. Differential gene expression analysis of TERT+ and TERT- groups with a TERT- group taken as the reference. (A) Volcano plot of gene fold changes. X-axis, logarithm of gene fold change; Y-axis, negative logarithm of Benjamini-Hochberg FDR-corrected p-value. (B) GO analysis of 262 genes upregulated in TERT + cancers. (C) GO analysis of 234 genes upregulated in TERT- cancers.
In TERT+ cancers, the group of upregulated genes was enriched in GO terms dealing with antimicrobial response, oxygen transport and hydrogen peroxide metabolism (Figure 4B). At the pathway level, in TERT+ cancers we observed most strongly upregulated pathways dealing with telomere extension, ribosomal RNA level and processing, ribosome biogenesis, transcription and processing of messenger RNA, nuclear transport of RNA and proteins, DNA replication and repair, and cell cycle progression (Table 6).
In TERT- cancers, upregulated genes gave GO terms for the amino acids modifications, metabolism of hormones and phenolic compounds, transfer of inorganic anions, and assembly of myofibrils (Figure 4C). The activated pathways were related to cell cycle progression, TGF beta and SMAD signaling, JAK degradation regulation, cytoskeleton and extracellular matrix remodeling, AP1 transcription factor network, AKT signaling, PDGFR, HGF and FGFR2 signaling, epithelial to mesenchymal transition (Table 7).
Note that the majority (32/38, 84%) of DNA repair pathways was statistically significantly upregulated in TERT+ compared to TERT- cancers (Supplementary Figure S2), except for the three pathways related to ATM mediated control of G2M cell cycle checkpoint and survival and three other pathways related to p53 control of DNA integrity and survival (Supplementary Figure S2). This trend fits previous figure of incremental increase in activation of the same 32/38 pathways in the range: healthy human normal tissue — tumor adjacent pathologically normal tissue — benign tumor — malignant tumor, with the opposite regulation for the six pathways related to ATM and p53 replication control (Vladimirova et al., 2021; Sorokin et al., 2023; Zolotovskaia et al., 2023).
Thus, we detected an overally differential trend where TERT+ cancers showed the increased transcription and translation, and DNA replication backed by enhanced DNA repair with disrupted quality checkpoints. In contrast, TERT- cancers showed the activation of peculiar set of positive cell cycle promoting genes and strong epithelial-to-mesenchymal transition marks (Table 7).
We then tried to characterize transcriptional patterns that could be associated with TERT promoter mutation status in the group of TERT- expressing (TERT+) cancers. To this end we collected where possible fragments of remaining biomaterials available after extraction of RNA for sequencing and proposed to the patients or their legal representatives to sign a new informed agreement form to participate in the investigation of molecular features associated with C228T/A and C250T/A mutations in TERT promoter. Both conditions were met for 114 patient cases, and our further mutation screening identified among them 101 cases with wild-type TERT promoter and 13 cases with the presence of either mutation (Table 1). TERT promoter mutations were found in 5 glioblastomas, 3 melanomas, 2 skin carcinomas, 2 urothelial carcinomas, and in one thyroid cancer sample (Figure 5A). For further analysis, we took only the samples showing detectable TERT expression, and selected totally 81 TERTwt and 12 TERTmut profiles. Since the sampling of tumor samples with known status of TERT promoter mutations was limited, our findings of comparing the TERTwt and TERTmut groups should be considered as the pilot study results that need to be validated on greater cohorts in the future.
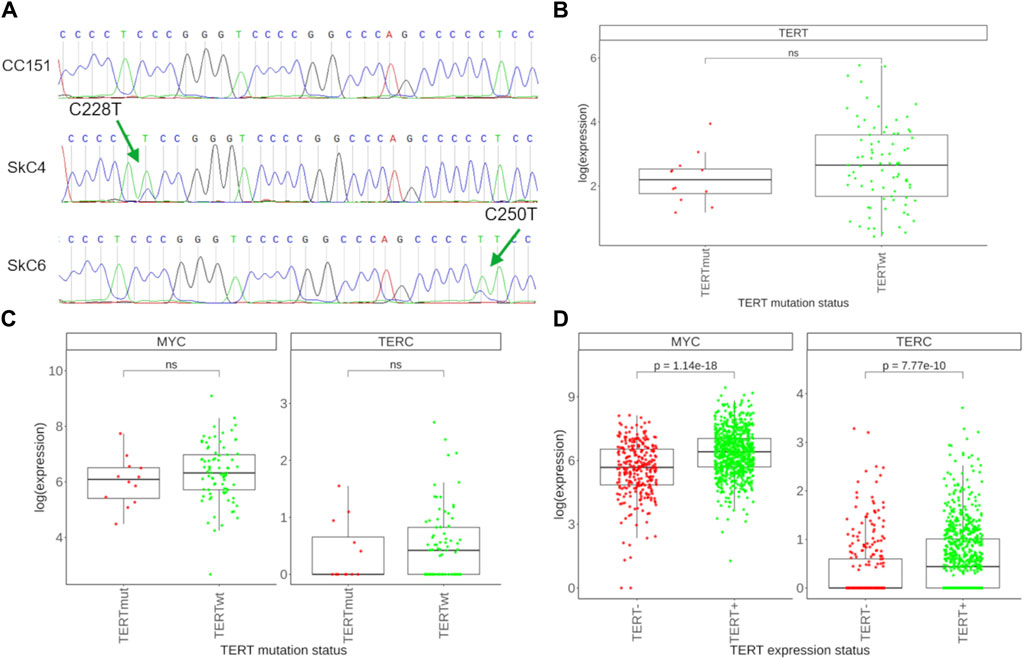
Figure 5. Assessment of TERT promoter mutations and associated transcriptional features. (A) Sanger-sequenced profile of the TERT gene promoter region spanning the putative C228T and C250T mutation sites. Examples are shown of no mutations found (top), of C228T mutation (center), and of C250T mutation identified (bottom). Green arrows indicate the position of the mutation sites. Tumor sample IDs are shown left to the respective tracks. (B) Expression of the TERT gene in 12 TERTmut and 81 TERTwt tumors. (C) Expression of the MYC and TERC genes in TERTmut and TERTwt tumors. (D) Expression of the MYC and TERC genes in 278 TERT- tumors and 761 TERT+ tumors.
We first compared expression of TERT in selected TERTmut and TERTwt tumors and found lack of detectable difference among the two groups (Figure 5B). This was also the case for the expression of TERC, a gene for the RNA component of telomerase (Figure 5C). We also found no difference for the expression levels of MYC, a known potent inducer of TERT in normal and malignant tissues (Figure 5C). At the same time, both TERC and MYC were found significantly upregulated in the group of TERT+ cancers compared to the TERT- group (Figure 5D). Thus, our results indicate that mutations in TERT promoter do not result in an overexpression of telomerase components compared to other TERT+ tumors.
In addition to the multiple important non-canonical roles of TERT including regulation of apoptosis, maintenance of the cellular redox homeostasis, the main function of active telomerase is adding specific DNA sequence monomers to chromosome ends thus preventing their pathologic shortening following a series of cell division cycles. However, in some organisms this role is played by the LINE retrotransposons which insert their DNA copies in the chromosome termini (Shpiz et al., 2007). In humans, L1 family of LINE retrotransposons occupies nearly 20% of the DNA (Suntsova and Buzdin, 2020) and is fully active thus generating new inserts through the genome (Terry and Devine, 2019). L1 retrotransposons can also mobilize other elements and may cause many kinds of DNA rearrangements (Schumann et al., 2010). Importantly, L1 retrotransposition may deal with the function of rescuing the chromosome ends in cancer independently of telomerase (Aschacher et al., 2020), as shown experimentally for glioblastoma. Furthermore, L1s are known to be transcriptionally reactivated in human cancers (Protasova et al., 2021; Mathavarajah and Dellaire, 2024). In the human genome, two subfamilies of L1 (L1HS and L1PA2) were reported to be able to transpose (Buzdin et al., 2003). Thus, we compared expression levels of these two L1 subfamilies in our experimental set of TERT-, TERT+, TERTmut and TERTwt tumors (Figure 6). Firstly, we detected a higher expression of L1 retrotransposons in TERT- compared to TERT+ tumors (statistically significant for L1HS subfamily, Figure 6A). Secondly, there was a lower expression of L1 elements in TERTmut compared to TERTwt TERT+ tumors (statistically significant for L1PA2; a trend is seen also for the L1HS elements, Figure 6B). Thus, we found that the factors known to be associated with telomerase reactivation (increased expression, activating promoter mutations) are also associated with the decrease of active L1 subfamilies expression, and vice versa. However, the ALT pathway could also be associated with a higher likelihood of genomic rearrangements, including the activation of L1 elements, which potentially causes genomic instability and contributes to tumorigenesis. Conversely, in TERT+ tumors with active telomerase, where telomere maintenance primarily occurs through the TERT function, the L1 activity is instead reduced (Aschacher et al., 2020).
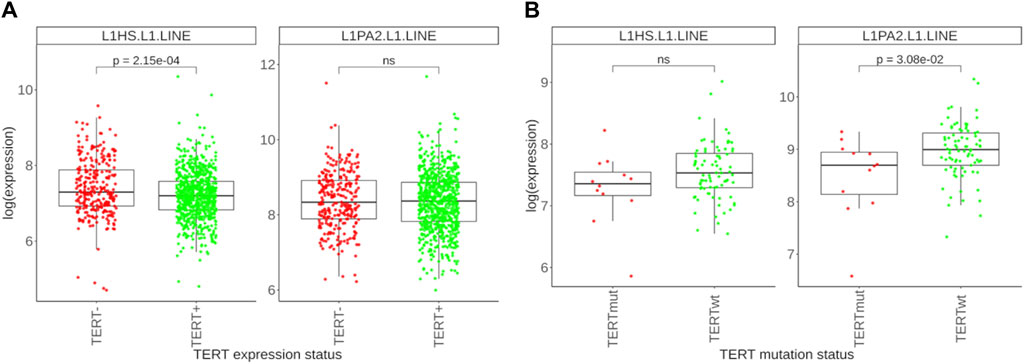
Figure 6. Differential expression profile of L1 retrotransposon subfamilies. (A) Expression levels of L1HS and L1PA2 retrotransposons in 278 TERT- tumors and 761 TERT+ tumors. (B) Expression levels of L1HS and L1PA2 retrotransposons in 12 TERTmut and 81 TERTwt tumors.
To summarize, in this study we experimentally investigated gene expression patterns associated with the status of TERT transcription on a pan-cancer level. We found a bimodal distribution of TERT expression where ∼27% of cancer samples did not express TERT. In turn, expression of TERT was strongly correlated with 1443 human genes and 265 molecular pathways. We also found differentially activated 496 genes and 1975 molecular pathways for the comparison of TERT- positive and negative cancers, which may reflect greater physiological impact of changes in the molecular pathways compared to the individual genes. Our pilot assay showed that the TERT+ group there was ∼13% of cancers with mutated TERT promoter, where the presence of promoter mutations was not associated with a greater level of TERT expression. Taken together, this evidences parallel mechanisms leading to the TERT gene transcriptional activation, and also parallel mechanisms of proliferation and survival in TERT- positive and negative cancers. Finally, we detected a decreased expression of L1 retrotransposons in the TERT+ group, and further decreased L1 expression in promoter mutated TERT+ cancers.
In over 80% of tumors, the increased TERT gene expression promotes abnormal telomerase activation and upregulated telomere maintenance mechanism (Lee et al., 2016; Dratwa et al., 2020; Liu et al., 2024). In the context of cancer development, TERT serves as a critical factor by contributing to the maintenance of chromosomal stability through its role in preserving telomere length (Colebatch et al., 2019). In cancer cells, it serves as a constraining element in the assembly of the telomerase complex (Dratwa et al., 2020).
However, the important roles of TERT in vital non-canonical processes other than telomerase activity should be also considered. For example, TERT can act as a modulator in NF-kB and Wnt/β-catenin signaling pathways, regulate gene expressions, proliferation, apoptosis, cell cycle, chromosomal rearrangements, epigenetic modifications (Li and Tergaonkar, 2014; Dratwa et al., 2020; Liu et al., 2024). At the same time, in tumor cells with deficiencies in telomerase activity, telomere elongation can occur due to the telomerase-independent ALT pathway (Zhao et al., 2019). Thus, alterations in TERT expression can lead to disruption of the above molecular mechanisms, which may have a role in tumorigenesis.
In this study, the pathway and differential gene analysis revealed the complex influence of TERT expression on multiple aspects of cancer cell physiology. For example, TERT expression was positively associated with many pathways mediating enhanced transcription, ribosome assembly, translation, replication, DNA repair, and cell division (Figure 2; Table 4). The upregulation of the ribosome biogenesis pathway could be due to various factors related to telomerase activity. One interpretation of this pathway being prominently upregulated in the context of telomerase reactivation could be related to the association of TERC with dyskerin, a component of the H/ACA ribonucleoprotein complex involved in ribosome biogenesis (Montanaro et al., 2008). Also, the protein encoded by the NAT10 gene is a part of the “ribosome biogenesis” GO term and participates in the acetylation of telomeric proteins (Lv et al., 2003; Fu and Collins, 2007). In general, the upregulation of ribosome biogenesis may reflect the increased need for ribosomes to facilitate the translation of mRNA into proteins during forced division of tumor cells (Gonzalez et al., 2014). Little or no expression of TERT was connected with the activation of pathways related to epithelial-to-mesenchymal transition (Figure 2; Table 5), and with a number of cell cycle and survival promoting gene products (Table 3). On Supplementary Figure S3 we schematized major cellular processes differentially regulated in TERT+/− tumors. However, the results obtained obviously reflect the complex genetic heterogeneity of the analyzed tumors in pan-cancer and the lack of possibility to investigate effects of TERT+/TERT- expression status in the same genetic background, as it could be implemented on experimental in vivo models.
Many tumors, such as gliomas, melanomas, and urothelial carcinomas show a high frequency of TERT promoter mutations, and TERT with mutations in its promoter can be highly expressed that can lead to abnormally enhanced telomerase activity (Borah et al., 2015; Heidenreich et al., 2015; Lee et al., 2016; Shaughnessy et al., 2020; Guo et al., 2022). For example, the increased TERT expression in mutant melanoma cell lines was associated with the MAPK signaling pathway, which constitutively activates the transcription factor ETS binding to the mutant TERT promoter and increasing its transcription (Vallarelli et al., 2016). However, several reports regarding thyroid cancer, bladder cancer, melanoma, and breast cancer indicate no connection between TERT promoter mutations and the extent of this gene expression (Allory et al., 2014; Pestana et al., 2020; Salgado et al., 2020). Our analysis on a pan-cancer level also revealed no statistically significant differences in the TERT gene expression between the groups of TERTmut and TERTwt tumors. These results may be associated with parallel TERT transcriptional regulation pathways, where methylation of the TERT promoter is one of the well-known regulatory mechanisms. For example, TERT promoter methylation is counterintuitively correlated with chromatin accessibility and TERT expression (Salgado et al., 2020; Liu et al., 2023). Also, the formation of G-quadruplexes in the G-rich region of the promoter has a major influence on the expression of TERT, and mutations in the G-rich region of the promoter may lead to disruption of their structure which in turn strongly affects transcription factor binding and enhances TERT expression (Gu et al., 2022; Pavlova et al., 2022).
We also found that the expression of active human retrotransposon family L1 is increased in the TERT- deficient group of cancers (Figure 6). Interestingly, connection between the transcription of L1 and TERT was previously reported for cancer cells (Aschacher et al., 2016). It has to be investigated in the future whether this may be somehow related to the hypothetical function of telomere extension by the L1 reverse transcriptase, or does it simply represent occasional transcriptional interplay between the processes. It is important to note that L1 elements are able to retrotranspose into telomere regions and may be a part of the alternative lengthening of telomeres (ALT) mechanism that operates independently of the telomerase and, therefore, potentially contributes to the elongation of telomeres in ALT-positive cells (Aschacher et al., 2020).
Finally, our analysis revealed 1443 genes whose expression is strongly correlated with the transcription of TERT. Among them, 17 genes represent molecular targets of cancer therapeutics, where 9 genes are positively and 8 negatively correlated with the transcription of TERT (Table 8).
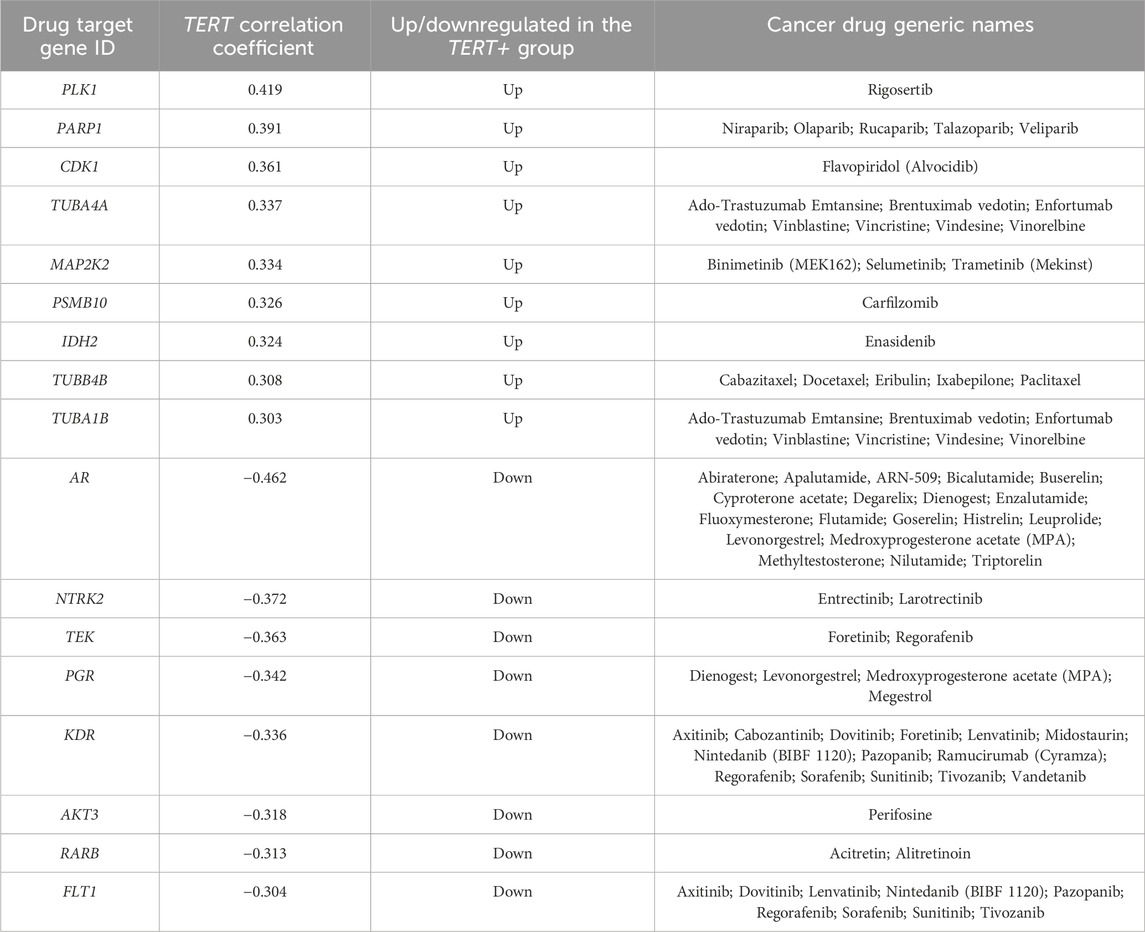
Table 8. Cancer drug target genes which are positively and negatively correlated with TERT expression.
Thus, our findings may shed light on the differential sensitivity of tumors on targeted therapies depending on the TERT expression status. TERT expression level is known to be a prognostic survival biomarker in many cancers. In agreement with this, our pilot assessment of overall survival (OS) and progression-free survival (PFS) data from the TCGA project for colorectal cancer (TCGA-COAD) and thyroid cancer (TCGA-THCA) revealed a strong association of lower expression of TERT with a favorable outcome (Figure 7). Thus, it has to be investigated in the future whether TERT expression typing of cancers may serve to personalize existing modes of therapy or even develop new approaches for the differential treatment of patients.
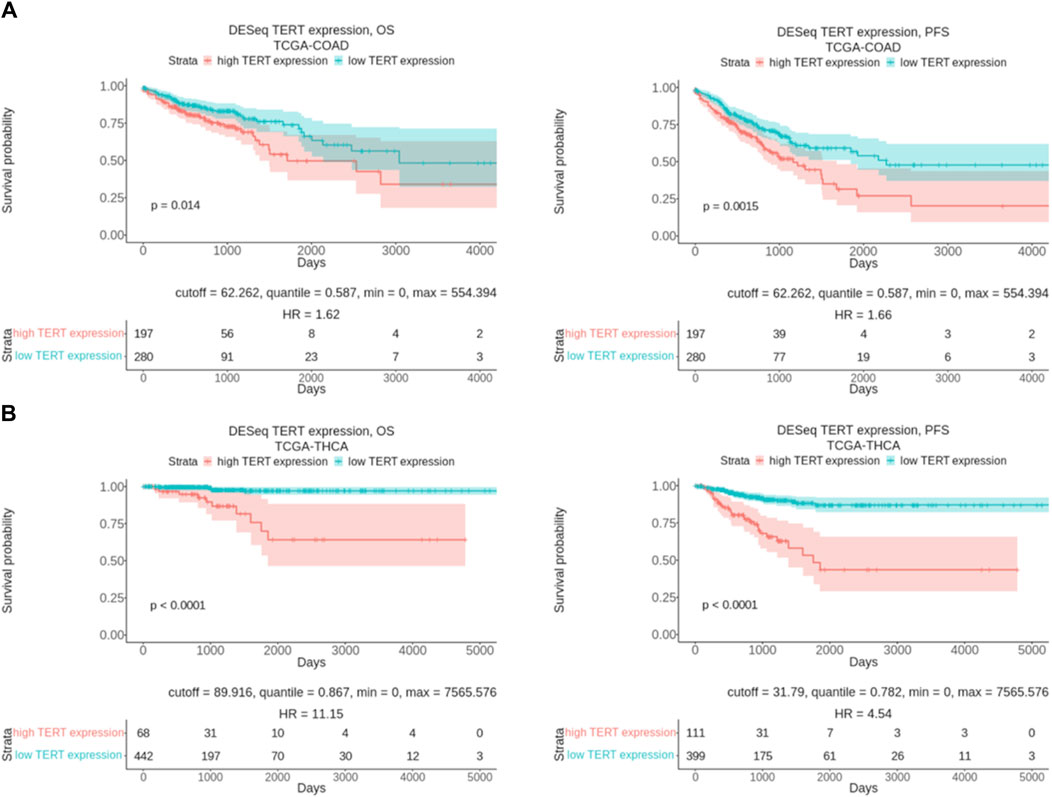
Figure 7. Overall and progression free survival analysis results in relation with TERT expression level. (A) Kaplan-Meier plots built for colorectal cancer patients from TCGA database (TCGA-COAD). (B) Kaplan-Meier plots built for thyroid cancer patients from TCGA database (TCGA-THCA). OS means overall survival and PFS means progression free survival.
For the first time, in this comprehensive study of a 1039-patient cohort we have tried to catalogue these and other molecular processes connected with the transcription of the TERT gene in cancers. We hope that the results communicated here may be useful to those interested in different ways of proliferation and survival of human cancer cells or working in the domain of advanced molecular diagnostics.
Here we experimentally assessed the gene expression patterns associated with the status of TERT transcription in 1039 human cancer samples of 27 tumor types investigated by RNA sequencing. We showed a bimodal distribution of TERT expression where ∼27% of cancer samples did not express TERT, whereas the rest showed a bell-shaped distribution. Expression of TERT was strongly correlated with 1443 human genes including 103 encoding transcriptional factor proteins. Comparison of TERT- positive and negative cancers showed the differential activation of 496 genes and 1975 molecular pathways. Among them, 32/38 (84%) of DNA repair pathways were hyperactivated in TERT+ cancers which was also connected with accelerated replication, transcription, translation, and cell cycle progression. In contrast, the level of 40 positive cell cycle regulator proteins was specific for the TERT- group which suggests different proliferation strategies for both groups of cancer. Our pilot study showed that the TERT+ group had ∼13% of cancers with mutated TERT promoter (mutations C228T, C250T, or both). However, the presence of promoter mutations was not associated with a greater level of TERT expression compared with other TERT+ cancers. This evidences parallel mechanisms leading to the TERT gene transcriptional activation. In addition, we detected a decreased expression of L1 retrotransposons in the TERT+ group, and further decreased L1 expression in promoter mutated TERT+ cancers. TERT expression was correlated with 17 target genes for cancer therapy which may relate to differential survival pattern of TERT- positive and negative cancers.
The data analyzed in this study is subject to the following licenses/restrictions: Patient raw genetic and gene expression data obtained in this study cannot be submitted to public databases due to patient written informed consent formulation. However, normalized gene expression and TERT promoter mutation data are available upon the request to the authors. Requests to access these datasets should be directed to AB, YnV6ZGluQG9uY29ib3guY29t.
The studies involving humans were approved by the Clinical Center Vitamed, Moscow, Russia. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
AD: Conceptualization, Investigation, Writing–original draft, Writing–review and editing. AxM: Data curation, Writing–original draft. MS: Formal Analysis, Investigation, Writing–review and editing. EP: Conceptualization, Formal Analysis, Writing–review and editing. AxS: Conceptualization, Formal Analysis, Writing–review and editing. AkM: Conceptualization, Formal Analysis, Writing–review and editing. MS: Conceptualization, Data curation, Software, Writing–review and editing. VT: Data curation, Software, Writing–review and editing. GZ: Conceptualization, Investigation, Writing–original draft, Writing–review and editing. AkS: Data curation, Software, Writing–review and editing. MZ: Data curation, Formal Analysis, Writing–review and editing. AB: Conceptualization, Writing–original draft, Writing–review and editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. Annotation of TERT- related gene expression and profiling of TERT promoter mutations was carried out with the financial support of a grant from the Ministry of Science and Higher Education of the Russian Federation (Agreement No. 075-15-2022-310 dated 20 April 2022). Differential analysis of molecular pathways in relation with the expression of TERT was supported by Russian Science Foundation grant 20-75-10071.
RNA sequencing profiles and partial clinical annotation of cancer cases were provided within the frameworks of Oncobox LLC (Russia) research initiative.
Author VT was employed by the Oncobox LLC.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2024.1401100/full#supplementary-material
Akincilar, S. C., Chan, C. H. T., Ng, Q. F., Fidan, K., and Tergaonkar, V. (2021). Non-canonical roles of canonical telomere binding proteins in cancers. Cell. Mol. Life Sci. 78, 4235–4257. doi:10.1007/s00018-021-03783-0
Aliper, A. M., Korzinkin, M. B., Kuzmina, N. B., Zenin, A. A., Venkova, L. S., Smirnov, P. Y., et al. (2017). Mathematical justification of expression-based pathway activation scoring (PAS). Methods Mol. Biol. Clifton N. J. 1613, 31–51. doi:10.1007/978-1-4939-7027-8_3
Allory, Y., Beukers, W., Sagrera, A., Flández, M., Marqués, M., Márquez, M., et al. (2014). Telomerase reverse transcriptase promoter mutations in bladder cancer: high frequency across stages, detection in urine, and lack of association with outcome. Eur. Urol. 65, 360–366. doi:10.1016/j.eururo.2013.08.052
Aschacher, T., Wolf, B., Aschacher, O., Enzmann, F., Laszlo, V., Messner, B., et al. (2020). Long interspersed element-1 ribonucleoprotein particles protect telomeric ends in alternative lengthening of telomeres dependent cells. Neoplasia N. Y. N. 22, 61–75. doi:10.1016/j.neo.2019.11.002
Aschacher, T., Wolf, B., Enzmann, F., Kienzl, P., Messner, B., Sampl, S., et al. (2016). LINE-1 induces hTERT and ensures telomere maintenance in tumour cell lines. Oncogene 35, 94–104. doi:10.1038/onc.2015.65
Barthel, F. P., Wei, W., Tang, M., Martinez-Ledesma, E., Hu, X., Amin, S. B., et al. (2017). Systematic analysis of telomere length and somatic alterations in 31 cancer types. Nat. Genet. 49, 349–357. doi:10.1038/ng.3781
Bell, R. J. A., Rube, H. T., Xavier-Magalhães, A., Costa, B. M., Mancini, A., Song, J. S., et al. (2016). Understanding TERT promoter mutations: a common path to immortality. Mol. Cancer Res. MCR 14, 315–323. doi:10.1158/1541-7786.MCR-16-0003
Blackburn, E. H., Epel, E. S., and Lin, J. (2015). Human telomere biology: a contributory and interactive factor in aging, disease risks, and protection. Science 350, 1193–1198. doi:10.1126/science.aab3389
Borah, S., Xi, L., Zaug, A. J., Powell, N. M., Dancik, G. M., Cohen, S. B., et al. (2015). Cancer. TERT promoter mutations and telomerase reactivation in urothelial cancer. Science 347, 1006–1010. doi:10.1126/science.1260200
Borisov, N., Sorokin, M., Garazha, A., and Buzdin, A. (2020). Quantitation of molecular pathway activation using RNA sequencing data. Methods Mol. Biol. Clifton N. J. 2063, 189–206. doi:10.1007/978-1-0716-0138-9_15
Borisov, N., Suntsova, M., Sorokin, M., Garazha, A., Kovalchuk, O., Aliper, A., et al. (2017). Data aggregation at the level of molecular pathways improves stability of experimental transcriptomic and proteomic data. Cell Cycle georget. Tex 16, 1810–1823. doi:10.1080/15384101.2017.1361068
Buzdin, A., Sorokin, M., Garazha, A., Glusker, A., Aleshin, A., Poddubskaya, E., et al. (2020). RNA sequencing for research and diagnostics in clinical oncology. Semin. Cancer Biol. 60, 311–323. doi:10.1016/j.semcancer.2019.07.010
Buzdin, A., Sorokin, M., Garazha, A., Sekacheva, M., Kim, E., Zhukov, N., et al. (2018). Molecular pathway activation - new type of biomarkers for tumor morphology and personalized selection of target drugs. Semin. Cancer Biol. 53, 110–124. doi:10.1016/j.semcancer.2018.06.003
Buzdin, A., Ustyugova, S., Gogvadze, E., Lebedev, Y., Hunsmann, G., and Sverdlov, E. (2003). Genome-wide targeted search for human specific and polymorphic L1 integrations. Hum. Genet. 112, 527–533. doi:10.1007/s00439-002-0904-2
Chen, Y.-L., Jeng, Y.-M., Chang, C.-N., Lee, H.-J., Hsu, H.-C., Lai, P.-L., et al. (2014). TERT promoter mutation in resectable hepatocellular carcinomas: a strong association with hepatitis C infection and absence of hepatitis B infection. Int. J. Surg. Lond. Engl. 12, 659–665. doi:10.1016/j.ijsu.2014.05.066
Chiba, K., Johnson, J. Z., Vogan, J. M., Wagner, T., Boyle, J. M., and Hockemeyer, D. (2015). Cancer-associated TERT promoter mutations abrogate telomerase silencing. eLife 4, e07918. doi:10.7554/eLife.07918
Colebatch, A. J., Dobrovic, A., and Cooper, W. A. (2019). TERT gene: its function and dysregulation in cancer. J. Clin. Pathol. 72, 281–284. doi:10.1136/jclinpath-2018-205653
Colebatch, A. J., Witkowski, T., Waring, P. M., McArthur, G. A., Wong, S. Q., and Dobrovic, A. (2018). Optimizing amplification of the GC-rich TERT promoter region using 7-Deaza-dGTP for droplet digital PCR quantification of TERT promoter mutations. Clin. Chem. 64, 745–747. doi:10.1373/clinchem.2017.284257
da Costa, V. R., Bim, L. V., Pacheco e Silva, L. D. P., Colloza-Gama, G. A., Bastos, A. U., Delcelo, R., et al. (2021). Advances in detecting low prevalence somatic TERT promoter mutations in papillary thyroid carcinoma. Front. Endocrinol. 12, 643151. doi:10.3389/fendo.2021.643151
Diplas, B. H., He, X., Brosnan-Cashman, J. A., Liu, H., Chen, L. H., Wang, Z., et al. (2018). The genomic landscape of TERT promoter wildtype-IDH wildtype glioblastoma. Nat. Commun. 9, 2087. doi:10.1038/s41467-018-04448-6
Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., et al. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinforma. Oxf. Engl. 29, 15–21. doi:10.1093/bioinformatics/bts635
Dong, X., Zhang, Q., Hao, J., Xie, Q., Xu, B., Zhang, P., et al. (2021). Large multicohort study reveals a prostate cancer susceptibility allele at 5p15 regulating TERT via androgen signaling-orchestrated chromatin binding of E2F1 and MYC. Front. Oncol. 11, 754206. doi:10.3389/fonc.2021.754206
Dratwa, M., Wysoczańska, B., Łacina, P., Kubik, T., and Bogunia-Kubik, K. (2020). TERT—regulation and roles in cancer formation. Front. Immunol. 11, 589929. doi:10.3389/fimmu.2020.589929
El Zarif, T., Machaalani, M., Nawfal, R., Nassar, A. H., Xie, W., Choueiri, T. K., et al. (2023). TERT promoter mutations frequency across race, sex, and cancer type. Oncol. 29, oyad208–14. doi:10.1093/oncolo/oyad208
Ferreira, M. S. V., Sørensen, M. D., Pusch, S., Beier, D., Bouillon, A.-S., Kristensen, B. W., et al. (2020). Alternative lengthening of telomeres is the major telomere maintenance mechanism in astrocytoma with isocitrate dehydrogenase 1 mutation. J. Neurooncol. 147, 1–14. doi:10.1007/s11060-020-03394-y
Fleisig, H. B., Hukezalie, K. R., Thompson, C. a. H., Au-Yeung, T. T. T., Ludlow, A. T., Zhao, C. R., et al. (2016). Telomerase reverse transcriptase expression protects transformed human cells against DNA-damaging agents, and increases tolerance to chromosomal instability. Oncogene 35, 218–227. doi:10.1038/onc.2015.75
Fredriksson, N. J., Ny, L., Nilsson, J. A., and Larsson, E. (2014). Systematic analysis of noncoding somatic mutations and gene expression alterations across 14 tumor types. Nat. Genet. 46, 1258–1263. doi:10.1038/ng.3141
Fu, D., and Collins, K. (2007). Purification of human telomerase complexes identifies factors involved in telomerase biogenesis and telomere length regulation. Mol. Cell 28, 773–785. doi:10.1016/j.molcel.2007.09.023
Gizard, F., Heywood, E. B., Findeisen, H. M., Zhao, Y., Jones, K. L., Cudejko, C., et al. (2011). Telomerase activation in atherosclerosis and induction of telomerase reverse transcriptase expression by inflammatory stimuli in macrophages. Arterioscler. Thromb. Vasc. Biol. 31, 245–252. doi:10.1161/ATVBAHA.110.219808
Gong, C., Yang, H., Wang, S., Liu, J., Li, Z., Hu, Y., et al. (2021). hTERT promotes CRC proliferation and migration by recruiting YBX1 to increase NRF2 expression. Front. Cell Dev. Biol. 9, 658101. doi:10.3389/fcell.2021.658101
Gonzalez, O. G., Assfalg, R., Koch, S., Schelling, A., Meena, J. K., Kraus, J., et al. (2014). Telomerase stimulates ribosomal DNA transcription under hyperproliferative conditions. Nat. Commun. 5, 4599. doi:10.1038/ncomms5599
Griewank, K. G., Murali, R., Schilling, B., Schimming, T., Möller, I., Moll, I., et al. (2013). TERT promoter mutations are frequent in cutaneous basal cell carcinoma and squamous cell carcinoma. PloS One 8, e80354. doi:10.1371/journal.pone.0080354
Gu, W., Lin, Z., Zhao, S., Wang, G., Shen, Z., Liu, W., et al. (2022). Research progress on G-quadruplexes in human telomeres and human telomerase reverse transcriptase (hTERT) promoter. Oxid. Med. Cell. Longev. 2022, 2905663. doi:10.1155/2022/2905663
Guo, Y., Chen, Y., Zhang, L., Ma, L., Jiang, K., Yao, G., et al. (2022). TERT promoter mutations and telomerase in melanoma. J. Oncol. 2022, 6300329. doi:10.1155/2022/6300329
Gupta, S., Vanderbilt, C. M., Lin, Y.-T., Benhamida, J. K., Jungbluth, A. A., Rana, S., et al. (2021). A pan-cancer study of somatic TERT promoter mutations and amplification in 30,773 tumors profiled by clinical genomic sequencing. J. Mol. Diagn. JMD 23, 253–263. doi:10.1016/j.jmoldx.2020.11.003
Gutierrez-Rodrigues, F., Donaires, F. S., Pinto, A., Vicente, A., Dillon, L. W., Clé, D. V., et al. (2019). Pathogenic TERT promoter variants in telomere diseases. Genet. Med. 21, 1594–1602. doi:10.1038/s41436-018-0385-x
Hasanau, T., Pisarev, E., Kisil, O., Nonoguchi, N., Le Calvez-Kelm, F., and Zvereva, M. (2022). Detection of TERT promoter mutations as a prognostic biomarker in gliomas: methodology, prospects, and advances. Biomedicines 10, 728. doi:10.3390/biomedicines10030728
Hayashi, Y., Fujita, K., Netto, G. J., and Nonomura, N. (2021). Clinical application of TERT promoter mutations in urothelial carcinoma. Front. Oncol. 11, 705440. doi:10.3389/fonc.2021.705440
Heidenreich, B., Rachakonda, P. S., Hosen, I., Volz, F., Hemminki, K., Weyerbrock, A., et al. (2015). TERT promoter mutations and telomere length in adult malignant gliomas and recurrences. Oncotarget 6, 10617–10633. doi:10.18632/oncotarget.3329
Hrdličková, R., Nehyba, J., and Bose, H. R. (2012). Alternatively spliced telomerase reverse transcriptase variants lacking telomerase activity stimulate cell proliferation. Mol. Cell. Biol. 32, 4283–4296. doi:10.1128/MCB.00550-12
Hsiao, R., Sharma, H. W., Ramakrishnan, S., Keith, E., and Narayanan, R. (1997). Telomerase activity in normal human endothelial cells. Anticancer Res. 17, 827–832.
Huang, D.-S., Wang, Z., He, X.-J., Diplas, B. H., Yang, R., Killela, P. J., et al. (2015). Recurrent TERT promoter mutations identified in a large-scale study of multiple tumour types are associated with increased TERT expression and telomerase activation. Eur. J. Cancer Oxf. Engl. 1990 51, 969–976. doi:10.1016/j.ejca.2015.03.010
Hugdahl, E., Kalvenes, M. B., Mannelqvist, M., Ladstein, R. G., and Akslen, L. A. (2018). Prognostic impact and concordance of TERT promoter mutation and protein expression in matched primary and metastatic cutaneous melanoma. Br. J. Cancer 118, 98–105. doi:10.1038/bjc.2017.384
Kang, S. Y., Kim, D. G., Kim, H., Cho, Y. A., Ha, S. Y., Kwon, G. Y., et al. (2022). Direct comparison of the next-generation sequencing and iTERT PCR methods for the diagnosis of TERT hotspot mutations in advanced solid cancers. BMC Med. Genomics 15, 25. doi:10.1186/s12920-022-01175-2
Khattar, E., Kumar, P., Liu, C. Y., Akıncılar, S. C., Raju, A., Lakshmanan, M., et al. (2016). Telomerase reverse transcriptase promotes cancer cell proliferation by augmenting tRNA expression. J. Clin. Invest. 126, 4045–4060. doi:10.1172/JCI86042
Killela, P. J., Reitman, Z. J., Jiao, Y., Bettegowda, C., Agrawal, N., Diaz, L. A., et al. (2013). TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc. Natl. Acad. Sci. U. S. A. 110, 6021–6026. doi:10.1073/pnas.1303607110
Koleilat, A., Kumar, V., Al Hmada, Y., Milosevic, D., Zheng, G., Lopes, M. B., et al. (2023). Concurrent TERT promoter C228T and C250T mutations in diffuse gliomas: rare occurrence of intratumoral heterogeneity. J. Neuropathol. Exp. Neurol. 82, 1044–1047. doi:10.1093/jnen/nlad081
Kučírek, M., Bagherpoor, A. J., Jaroš, J., Hampl, A., and Štros, M. (2019). HMGB2 is a negative regulator of telomerase activity in human embryonic stem and progenitor cells. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 33, 14307–14324. doi:10.1096/fj.201901465RRR
Kumari, A., Srinivasan, R., and Wig, J. D. (2009). Effect of c-MYC and E2F1 gene silencing and of 5-azacytidine treatment on telomerase activity in pancreatic cancer-derived cell lines. Pancreatol. Off. J. Int. Assoc. Pancreatol. IAP Al 9, 360–368. doi:10.1159/000212094
Kwon, J. H., and Yi, J. W. (2021). Correlation between telomerase reverse transcriptase messenger RNA expression and survival of patients with papillary thyroid carcinoma. Surgery 169, 43–49. doi:10.1016/j.surg.2020.04.054
Lee, S., Opresko, P., Pappo, A., Kirkwood, J. M., and Bahrami, A. (2016). Association of TERT promoter mutations with telomerase expression in melanoma. Pigment. Cell Melanoma Res. 29, 391–393. doi:10.1111/pcmr.12471
Li, S., Xue, J., Jiang, K., Chen, Y., Zhu, L., and Liu, R. (2024). TERT promoter methylation is associated with high expression of TERT and poor prognosis in papillary thyroid cancer. Front. Oncol. 14, 1325345. doi:10.3389/fonc.2024.1325345
Li, Y., and Tergaonkar, V. (2014). Noncanonical functions of telomerase: implications in telomerase-targeted cancer therapies. Cancer Res. 74, 1639–1644. doi:10.1158/0008-5472.CAN-13-3568
Lin, J., Epel, E., Cheon, J., Kroenke, C., Sinclair, E., Bigos, M., et al. (2010). Analyses and comparisons of telomerase activity and telomere length in human T and B cells: insights for epidemiology of telomere maintenance. J. Immunol. Methods 352, 71–80. doi:10.1016/j.jim.2009.09.012
Liu, M., Zhang, Y., Jian, Y., Gu, L., Zhang, D., Zhou, H., et al. (2024). The regulations of telomerase reverse transcriptase (TERT) in cancer. Cell Death Dis. 15, 90–12. doi:10.1038/s41419-024-06454-7
Liu, R., and Xing, M. (2016). TERT promoter mutations in thyroid cancer. Endocr. Relat. Cancer 23, R143–R155. doi:10.1530/ERC-15-0533
Liu, T., Li, S., Xia, C., and Xu, D. (2023). TERT promoter mutations and methylation for telomerase activation in urothelial carcinomas: new mechanistic insights and clinical significance. Front. Immunol. 13, 1071390. doi:10.3389/fimmu.2022.1071390
Liu, T., Yuan, X., and Xu, D. (2016). Cancer-specific telomerase reverse transcriptase (TERT) promoter mutations: biological and clinical implications. Genes 7, 38. doi:10.3390/genes7070038
Lv, J., Liu, H., Wang, Q., Tang, Z., Hou, L., and Zhang, B. (2003). Molecular cloning of a novel human gene encoding histone acetyltransferase-like protein involved in transcriptional activation of hTERT. Biochem. Biophys. Res. Commun. 311, 506–513. doi:10.1016/j.bbrc.2003.09.235
Mathavarajah, S., and Dellaire, G. (2024). LINE-1: an emerging initiator of cGAS-STING signalling and inflammation that is dysregulated in disease. Biochem. Cell Biol. Biochim. Biol. Cell. 102, 38–46. doi:10.1139/bcb-2023-0134
Montanaro, L., Calienni, M., Ceccarelli, C., Santini, D., Taffurelli, M., Pileri, S., et al. (2008). Relationship between dyskerin expression and telomerase activity in human breast cancer. Cell. Oncol. Off. J. Int. Soc. Cell. Oncol. 30, 483–490. doi:10.3233/CLO-2008-0436
Nault, J.-C., and Zucman-Rossi, J. (2016). TERT promoter mutations in primary liver tumors. Clin. Res. Hepatol. Gastroenterol. 40, 9–14. doi:10.1016/j.clinre.2015.07.006
Olympios, N., Gilard, V., Marguet, F., Clatot, F., Di Fiore, F., and Fontanilles, M. (2021). TERT promoter alterations in glioblastoma: a systematic review. Cancers 13, 1147. doi:10.3390/cancers13051147
Palamarchuk, A. I., Kovalenko, E. I., and Streltsova, M. A. (2023). Multiple actions of telomerase reverse transcriptase in cell death regulation. Biomedicines 11, 1091. doi:10.3390/biomedicines11041091
Pavlova, A. V., Savitskaya, V.Yu., Dolinnaya, N. G., Monakhova, M. V., Litvinova, A. V., Kubareva, E. A., et al. (2022). G-quadruplex formed by the promoter region of the hTERT gene: structure-driven effects on DNA mismatch repair functions. Biomedicines 10, 1871. doi:10.3390/biomedicines10081871
Pestana, A., Batista, R., Celestino, R., Canberk, S., Sobrinho-Simões, M., and Soares, P. (2020). Comprehensive assessment of TERT mRNA expression across a large cohort of benign and malignant thyroid tumours. Cancers 12, 1846. doi:10.3390/cancers12071846
Powter, B., Jeffreys, S. A., Sareen, H., Cooper, A., Brungs, D., Po, J., et al. (2021). Human TERT promoter mutations as a prognostic biomarker in glioma. J. Cancer Res. Clin. Oncol. 147, 1007–1017. doi:10.1007/s00432-021-03536-3
Protasova, M. S., Andreeva, T. V., and Rogaev, E. I. (2021). Factors regulating the activity of LINE1 retrotransposons. Genes 12, 1562. doi:10.3390/genes12101562
Rosen, J., Jakobs, P., Ale-Agha, N., Altschmied, J., and Haendeler, J. (2020). Non-canonical functions of telomerase reverse transcriptase – impact on redox homeostasis. Redox Biol. 34, 101543. doi:10.1016/j.redox.2020.101543
Salgado, C., Roelse, C., Nell, R., Gruis, N., Doorn, R. V., and Velden, P. van der (2020). Interplay between TERT promoter mutations and methylation culminates in chromatin accessibility and TERT expression. PLOS ONE 15, e0231418. doi:10.1371/journal.pone.0231418
Schumann, G. G., Gogvadze, E. V., Osanai-Futahashi, M., Kuroki, A., Münk, C., Fujiwara, H., et al. (2010). Unique functions of repetitive transcriptomes. Int. Rev. Cell Mol. Biol. 285, 115–188. doi:10.1016/B978-0-12-381047-2.00003-7
Scott, G. A., Laughlin, T. S., and Rothberg, P. G. (2014). Mutations of the TERT promoter are common in basal cell carcinoma and squamous cell carcinoma. Mod. Pathol. Off. J. U. S. Can. Acad. Pathol. Inc. 27, 516–523. doi:10.1038/modpathol.2013.167
Ségal-Bendirdjian, E., and Geli, V. (2019). Non-canonical roles of telomerase: unraveling the imbroglio. Front. Cell Dev. Biol. 7, 332. doi:10.3389/fcell.2019.00332
Shaughnessy, M., Njauw, C.-N., Artomov, M., and Tsao, H. (2020). Classifying melanoma by TERT promoter mutational status. J. Invest. Dermatol. 140, 390–394. doi:10.1016/j.jid.2019.06.149
Shpiz, S., Kwon, D., Uneva, A., Kim, M., Klenov, M., Rozovsky, Y., et al. (2007). Characterization of Drosophila telomeric retroelement TAHRE: transcription, transpositions, and RNAi-based regulation of expression. Mol. Biol. Evol. 24, 2535–2545. doi:10.1093/molbev/msm205
Simon, M., Hosen, I., Gousias, K., Rachakonda, S., Heidenreich, B., Gessi, M., et al. (2015). TERT promoter mutations: a novel independent prognostic factor in primary glioblastomas. Neuro-Oncol. 17, 45–52. doi:10.1093/neuonc/nou158
Sorokin, M., Buzdin, A. A., Guryanova, A., Efimov, V., Suntsova, M. V., Zolotovskaia, M. A., et al. (2023). Large-scale assessment of pros and cons of autopsy-derived or tumor-matched tissues as the norms for gene expression analysis in cancers. Comput. Struct. Biotechnol. J. 21, 3964–3986. doi:10.1016/j.csbj.2023.07.040
Sorokin, M., Ignatev, K., Barbara, V., Vladimirova, U., Muraveva, A., Suntsova, M., et al. (2020). Molecular pathway activation markers are associated with efficacy of trastuzumab therapy in metastatic HER2-positive breast cancer better than individual gene expression levels. Biochem. Biokhimiia 85, 758–772. doi:10.1134/S0006297920070044
Spiegl-Kreinecker, S., Lötsch, D., Neumayer, K., Kastler, L., Gojo, J., Pirker, C., et al. (2018). TERT promoter mutations are associated with poor prognosis and cell immortalization in meningioma. Neuro-Oncol. 20, 1584–1593. doi:10.1093/neuonc/noy104
Stern, J. L., Hibshman, G., Hu, K., Ferrara, S. E., Costello, J. C., Kim, W., et al. (2020). Mesenchymal and MAPK expression signatures associate with telomerase promoter mutations in multiple cancers. Mol. Cancer Res. MCR 18, 1050–1062. doi:10.1158/1541-7786.MCR-19-1244
Stern, J. L., Theodorescu, D., Vogelstein, B., Papadopoulos, N., and Cech, T. R. (2015). Mutation of the TERT promoter, switch to active chromatin, and monoallelic TERT expression in multiple cancers. Genes Dev. 29, 2219–2224. doi:10.1101/gad.269498.115
Suenaga, M., Yamaguchi, A., Soda, H., Orihara, K., Tokito, Y., Sakaki, Y., et al. (2006). Antiproliferative effects of gefitinib are associated with suppression of E2F-1 expression and telomerase activity. Anticancer Res. 26, 3387–3391.
Suntsova, M., Gaifullin, N., Allina, D., Reshetun, A., Li, X., Mendeleeva, L., et al. (2019). Atlas of RNA sequencing profiles for normal human tissues. Sci. Data 6, 36. doi:10.1038/s41597-019-0043-4
Suntsova, M. V., and Buzdin, A. A. (2020). Differences between human and chimpanzee genomes and their implications in gene expression, protein functions and biochemical properties of the two species. BMC Genomics 21, 535. doi:10.1186/s12864-020-06962-8
Tang, Q., Liu, C., Zhang, S., He, L., Liu, Y., Wang, J., et al. (2023). FOXM1 increases hTERT protein stability and indicates poor prognosis in gastric cancer. Neoplasia N. Y. N. 36, 100863. doi:10.1016/j.neo.2022.100863
Terry, D. M., and Devine, S. E. (2019). Aberrantly high levels of somatic LINE-1 expression and retrotransposition in human neurological disorders. Front. Genet. 10, 1244. doi:10.3389/fgene.2019.01244
Thompson, C. A. H., and Wong, J. M. Y. (2020). Non-canonical functions of telomerase reverse transcriptase: emerging roles and biological relevance. Curr. Top. Med. Chem. 20, 498–507. doi:10.2174/1568026620666200131125110
Vallarelli, A. F., Rachakonda, P. S., André, J., Heidenreich, B., Riffaud, L., Bensussan, A., et al. (2016). TERT promoter mutations in melanoma render TERT expression dependent on MAPK pathway activation. Oncotarget 7, 53127–53136. doi:10.18632/oncotarget.10634
Vinagre, J., Almeida, A., Pópulo, H., Batista, R., Lyra, J., Pinto, V., et al. (2013). Frequency of TERT promoter mutations in human cancers. Nat. Commun. 4, 2185. doi:10.1038/ncomms3185
Vladimirova, U., Rumiantsev, P., Zolotovskaia, M., Albert, E., Abrosimov, A., Slashchuk, K., et al. (2021). DNA repair pathway activation features in follicular and papillary thyroid tumors, interrogated using 95 experimental RNA sequencing profiles. Heliyon 7, e06408. doi:10.1016/j.heliyon.2021.e06408
Weng, N., Hathcock, K. S., and Hodes, R. J. (1998). Regulation of telomere length and telomerase in T and B cells: a mechanism for maintaining replicative potential. Immunity 9, 151–157. doi:10.1016/S1074-7613(00)80597-X
Wu, R. A., Upton, H. E., Vogan, J. M., and Collins, K. (2017). Telomerase mechanism of telomere synthesis. Annu. Rev. Biochem. 86, 439–460. doi:10.1146/annurev-biochem-061516-045019
Yu, Y., Fan, D., Song, X., Zakeri, K., Chen, L., Kang, J., et al. (2021). TERT promoter mutations are enriched in oral cavity cancers and associated with locoregional recurrence. JCO Precis. Oncol. 5, 1259–1269. doi:10.1200/PO.20.00515
Yuan, X., Larsson, C., and Xu, D. (2019). Mechanisms underlying the activation of TERT transcription and telomerase activity in human cancer: old actors and new players. Oncogene 38, 6172–6183. doi:10.1038/s41388-019-0872-9
Zhang, H., Zhang, X., and Yu, J. (2022). Integrated analysis of altered lncRNA, circRNA, microRNA, and mRNA expression in hepatocellular carcinoma carrying TERT promoter mutations. J. Hepatocell. Carcinoma 9, 1201–1215. doi:10.2147/JHC.S385026
Zhang, Y., Zhang, A., Shen, C., Zhang, B., Rao, Z., Wang, R., et al. (2014). E2F1 acts as a negative feedback regulator of c-Myc-induced hTERT transcription during tumorigenesis. Oncol. Rep. 32, 1273–1280. doi:10.3892/or.2014.3287
Zhao, S., Wang, F., and Liu, L. (2019). Alternative lengthening of telomeres (ALT) in tumors and pluripotent stem cells. Genes 10, 1030. doi:10.3390/genes10121030
Zolotovskaia, M. A., Modestov, A. A., Suntsova, M. V., Rachkova, A. A., Koroleva, E. V., Poddubskaya, E. V., et al. (2023). Pan-cancer antagonistic inhibition pattern of ATM-driven G2/M checkpoint pathway vs other DNA repair pathways. DNA Repair 123, 103448. doi:10.1016/j.dnarep.2023.103448
Zolotovskaia, M. A., Tkachev, V. S., Guryanova, A. A., Simonov, A. M., Raevskiy, M. M., Efimov, V. V., et al. (2022). OncoboxPD: human 51 672 molecular pathways database with tools for activity calculating and visualization. Comput. Struct. Biotechnol. J. 20, 2280–2291. doi:10.1016/j.csbj.2022.05.006
Keywords: TERT promoter C228T and C250T mutations, reverse transcriptase, differential gene expression analysis, RNA sequencing, pathway activation profiling, pan-cancer investigation
Citation: Drobyshev A, Modestov A, Suntsova M, Poddubskaya E, Seryakov A, Moisseev A, Sorokin M, Tkachev V, Zakharova G, Simonov A, Zolotovskaia MA and Buzdin A (2024) Pan-cancer experimental characteristic of human transcriptional patterns connected with telomerase reverse transcriptase (TERT) gene expression status. Front. Genet. 15:1401100. doi: 10.3389/fgene.2024.1401100
Received: 18 March 2024; Accepted: 08 May 2024;
Published: 27 May 2024.
Edited by:
Cosmeri Rizzato, University of Pisa, ItalyReviewed by:
Ismail Hosen, University of Dhaka, BangladeshCopyright © 2024 Drobyshev, Modestov, Suntsova, Poddubskaya, Seryakov, Moisseev, Sorokin, Tkachev, Zakharova, Simonov, Zolotovskaia and Buzdin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anton Buzdin, YnV6ZGluQG9uY29ib3guY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.