- 1Key Laboratory of Xinjiang Phytomedicine Resource and Utilization of Ministry of Education, Key Laboratory of Oasis Town and Mountain-Basin System Ecology of Xinjiang Production and Construction Corps, College of Life Sciences, Shihezi University, Shihezi, China
- 2Zhuhai College of Science and Technology, Zhuhai, China
- 3Business School of Xinjiang Normal University, Urumqi, China
- 4College of Life Sciences, Shaanxi Normal University, Xi’an, China
Excessive reactive oxygen species stress due to salinity poses a significant threat to the growth of Glycyrrhiza uralensis Fisch. To adapt to salt stress, G. uralensis engages in alternative splicing (AS) to generate a variety of proteins that help it withstand the effects of salt stress. While several studies have investigated the impact of alternative splicing on plants stress responses, the mechanisms by which AS interacts with transcriptional regulation to modulate the salt stress response in G. uralensis remain poorly understood. In this study, we utilized high-throughput RNA sequencing data to perform a comprehensive analysis of AS events at various time points in G. uralensis under salt stress, with exon skipping (SE) being the predominant AS type. KEGG enrichment analysis was performed on the different splicing genes (DSG), and pathways associated with AS were significantly enriched, including RNA transport, mRNA surveillance, and spliceosome. This indicated splicing regulation of genes, resulting in AS events under salt stress conditions. Moreover, plant response to salt stress pathways were also enriched, such as mitogen-activated protein kinase signaling pathway – plant, flavonoid biosynthesis, and oxidative phosphorylation. We focused on four differentially significant genes in the MAPK pathway by AS and qRT-PCR analysis. The alternative splicing type of MPK4 and SnRK2 was skipped exon (SE). ETR2 and RbohD were retained intron (RI) and alternative 5’splice site (A5SS), respectively. The expression levels of isoform1 of these four genes displayed different but significant increases in different tissue sites and salt stress treatment times. These findings suggest that MPK4, SnRK2, ETR2, and RbohD in G. uralensis activate the expression of isoform1, leading to the production of more isoform1 protein and thereby enhancing resistance to salt stress. These findings suggest that salt-responsive AS directly and indirectly governs G. uralensis salt response. Further investigations into AS function and mechanism during abiotic stresses may offer novel references for bolstering plant stress tolerance.
1 Introduction
Increased emissions of greenhouse gases have led to global warming, while high evapotranspiration rates have exacerbated soil salinization (Cicek et al., 2022). A lack of annual rainfall is also preventing salt from reaching the subsoil, resulting in serious ecological changes to soil (Khosravichenar et al., 2023). Xinjiang has a large area of saline-alkali land as a result of insufficient annual precipitation. In recent times, Glycyrrhiza uralensis Fisch, widely cultivated in Northwest China for medicinal and food purposes, has suffering from salt exposure throughout the year. For plants to prevent ROS proliferation, the antioxidant defense system must be activated. The SOS pathway, for instance, is activated by salt stress, which results in ROS accumulation and MAPK cascades (Yang et al., 2018).
Plants respond to salt stress through mitogen-activated protein kinase (MAPK) cascades. Three primary components comprise these cascades, each of which is involved in phosphorylation and activation in succession. In order for the second element to function, a MAP kinase (MAPKK or MKK) must be phosphorylated and activated by the first. As a result of the sequential phosphorylation process, the stress signal can be amplified, thus allowing for a more robust response. Salt-induced stress requires MAPK cascades to transduce signals and enable plants to adapt it (Mishra et al., 2006). The salt stress response of Arabidopsis is mediated by the MPK4 cascade. Null mutants of the gene exhibit hypersensitivity, while overexpression increases salt tolerance (Teige et al., 2004). Other plants also activate the MAPK cascade under salt stress; for example, alfalfa (Medicago sativa) activates the MAPK cascade under salt stress (Kiegerl et al., 2000). In rice, the MPK4 cascade regulates transcription factor gene expression in response to salt stress (F. Wang et al., 2014). When Arabidopsis is dehydrated, mutants of the MKK4 cascade are more salt sensitive and shed more water than the wild type (Kim et al., 2012).
When G. uralensis is exposed to salt stress, proteins that are involved in the synthesis of glycyrrhizic acid and liqueritin are activated, resulting in the cumulative production of glycyrrhizin and its components (C. Wang et al., 2020). Alternative splicing events can involve five different forms: skipped exon (SE), alternate 5-splice site (A5SS), alternate 3-splice site (A3SS), mutually exclusion exon (MXE), and retained intron (RI). Some AS events may be gene-specific and some species-specific, with SE and RI being the most and least prevalent, respectively, in animals. The majority of AS cases are caused by SE, while only 0.01% are caused by RI; in contrast, RI is the most common form of AS in Arabidopsis and maize (Daszkowska-Golec et al., 2017; Laloum et al., 2018; Thatcher et al., 2016). However, in recent years, SE and RI are the most prevalent AS in licorice and soybeans because of advances in sequencing technology and detection tools (Li et al., 2022; Song et al., 2020). The number of genes with introns that undergo alternative splicing in Arabidopsis thaliana (Marquez et al., 2012) will increase as more transcriptome data from plants in different developmental conditions and environmental conditions are collected. High-throughput analyses are becoming more sophisticated as more advanced tools are developed for splice variant identification.
Multiple studies have illustrated the important functions of alternative splicing in the cellular response to abiotic stresses—specifically, salt stress (Laloum et al., 2018). Recent research provides evidence that highlights the essential biological function of alternative splicing (AS) in increasing plant resilience to salt stress. These events have been implicated in regulating stress responses in plants. Spliceosomal components of Arabidopsis also affect response to stress in plants. There are over 6,000 Arabidopsis genes, for instance, that alter their alternative splicing patterns when exposed to salt stress (Feng et al., 2015). Mutations in SKIP protein interact physically with SR45, which modulates recognition or cleavage of splice sites 5′ and 3′ in alternative splicing in SNW/Ski-interacting protein (SKIP) (Wang et al., 2012), leading to increased salt and osmotic sensitivity in plants (Feng et al., 2015). In rice, OsNHX1-related gene undergoes AS under the salt stress, yielding three distinct transcripts. Notably, the transcript carrying the 3′UTR has been demonstrated to boost salt endurance the most (Amin et al., 2016). Three transcript isoforms of WDREB2 were produced by AS in wheat: WDREB2α, WDREB2β, and WDREB2γ. During a 24-hour stress treatment period, the expression of the non-functional isoform WDREB2β lacking the third exon remained relatively constant. In contrast, the functional isoforms WDREB2α and WDREB2γ exhibited temporary increases in transcript levels when subjected to drought, salt, and ABA stress treatment (Egawa et al., 2006; Liu et al., 2018).
Recent studies have demonstrated that alternative splicing plays a regulatory role in plants under salt stress (Cinatl et al., 2003). However, studies on how AS responds to salt in G. uralensis are rarely reported. Therefore, this study aims to investigate the regulatory mechanism of AS and expression profiles of different AS isoforms in response to salt stress by transcriptional regulation in G. uralensis. To gain valuable insights into the mechanisms underlying salt stress response in G. uralensis, we investigated the regulation of alternative splicing (AS) and expression patterns of various AS isoforms. Moreover, we aimed to examine the impact of AS on the development of G. uralensis under salt stress. In this study, we conducted RNA sequencing (RNA-seq) experiments on both aboveground parts (AP) and underground parts (UP) of G. uralensis. The plants were exposed to salt stress for durations of 0, 2, 6, and 12 h. We identified isoforms and quantified events of alternative splicing at diverse stages of the salt stress. Five AS types were identified at the AP and UP of salt-stressed G. uralensis. We identified differentially spliced genes (DSGs) that were then subjected to Gene Ontology (GO) term and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. The predominant AS events observed in our study were exon skipping (SE). In G. uralensis, we also identified several new AS events that greatly contributed to the response process to salt stress. Our results provide valuable insights into the stress response mechanism in G. uralensis. The findings illuminate the post-transcriptional regulation patterns of AS at different time points throughout the salt stress process.
2 Materials and methods
2.1 Plant material and salt stress treatments
The experimental materials were sourced from wild G. uralensis plants (Ding et al., 2014). Wild G. uralensis seeds were treated with 95% concentrated H2SO4 for 60 min and rinsed 8–10 times with sterile water and were then let dry. Later, these seeds were placed in a flowerpot containing nutrient soil and vermiculite 1:1. These flowerpots were allowed to germinate at 25°C in an illumination incubator (GXZ-430D) under dark conditions before being grown in a climate-controlled chamber. Licorice seedlings 60 days after emergence were transplanted into a hydroponic box (38 × 28 × 14 cm) with nutrient solution (Hogrland nutrient solution) for 5 days to adapt to the hydroponic conditions. NaCl was added to the hydroponic culture solution to a concentration of 150 mM/L. The culture media was changed once every 2 days. After 2, 6, and 12 h of NaCl stress, tissue samples of NaCl treatment for different times were collected. All samples were collected immediately, washed with distilled water, dried, frozen in liquid nitrogen, and stored at −80°C in three biological replicates of each treatment.
2.2 RNA-seq datasets and processing methods
The experiment involved all AS in the RNA-seq datasets which were detected by rMATs [(version 4.0.1) (http://rnaseq-mats.sourceforge.net/index.html)]. This incorporated data from 24 libraries, each representing different conditions (two tissue types × time points of salt stress treatment × three biological replicates).
2.3 Reference genome-based assembly of transcript structures, mapping reads
A stringent quality control procedure, which can ensure the accuracy of rMATs analysis results, was performed on the RNA-seq raw read datasets. Attaining high-quality raw sequencing data necessitates the elimination of low-quality reads, which contain sequences with high “N” and short reads. To screen out low-quality bases, SeqPrep (https://github.com/jstjohn/SeqPrep) and Sickle (https://github.com/najoshi/sickle) were used (Li et al., 2022). We used the reference genome for the species name: Glycyrrhiza_uralensis Reference genome Version: riken (Reference Genome Source: http://ngs-data-archive.psc.riken.jp/Gur-genome/index.pl). We employed TopHat2 (http://ccb.jhu.edu/software/tophat/index.shtml) to locate new splice sites mapped directly to pre-existing transcripts for accurate comparison. In addition, we used String Tie (http://ccb.jhu.edu/software/stringtie/) to assemble complex datasets into transcripts and compared them with known transcripts.
2.4 AS events identification in response to salt stress
We used rMATs to identify and analyze AS events between these samples. We detected AS events showing a significant salt stress response by comparing them to AS events with a false discovery rate (FDR) < 0.01. We identified five AS event types: SE, MXE, A5SS, A3SS, and RI.
2.5 DEGs, gene ontology, and pathway enrichment analysis
We used DESeq2V (http://bioconductor.org/packages/stats/bioc/DESeq2/) to analyze RNA-seq datasets in pair comparisons between the salt-stressed treatment and control group to identify DEGs. We identified genes as differentially expressed (DEGs) when their fold change was greater than or equal to 2 and their false discovery rate (FDR) was below 0.05. Transcripts per million (TPM) were utilized to quantify the gene expression levels in our study. DSGs were determined based on the FDR (p < 0.05) from exclusive nodal reads, and significant AS were mapped to the GO term (http://www.geneontology.org/). The significance of DSGs in KEGG pathways for genome KEGG background was tested (p < 0.05).
2.6 Genes of encoding salt stress regulators express patterns
To enable qRT-PCR analysis, cDNA synthesis was performed using total RNA extracted from G. uralensis as the template. This specific method is consistent with Li et al. (2022). The primers used are lists in Supplementary Table S1.
3 Results
3.1 Overview of RNA-seq datasets sequencing quality under salt stress conditions
We analyzed AS events in G. uralensis in response to salt stress by utilizing 24 high-throughput RNA-seq libraries. The seedlings were exposed to salt stress at different time points: 0 (APSSCK and UPDSCK), 2 (APSS_2h and UPSS_2h), 6 (APSS_6h and UPSS_6h), and 12 h (APSS_12h and UPSS_12h). The analysis was performed using TopHat2 software, which has been proven superior to previously used TopHat software for analyzing soybean and G. uralensis RNA-seq data (Li et al., 2020; Song et al., 2020). We analyzed each sample using over 4.2 million clean reads. The presence of high percentages of Q20 (>98%) and Q30 (>94%) confirmed high sequencing accuracy (Supplementary Table S2), enabling further data analyses. The distribution of filtered reads across the first 20 chromosomes of G. uralensis was indicated (Supplementary Figure S1A). There was no conspicuous preference in the sequence coverage (Supplementary Figure S1B), allowing us to perform further data analysis for AS event identification. Principal component analysis (PCA) was performed to validate sample repeatability (Figure 1A). Clusters of biological replicates were observed to have close distances, indicating acceptable variation between different time points. In addition, the biological replicates showed strong correlations with correlation coefficients (r^2) ranging from 0.77 to 1 in AP and 0.94 to 1 in UP (Figure 1B). This indicated that these RNA-seq datasets were high-quality and reliable.
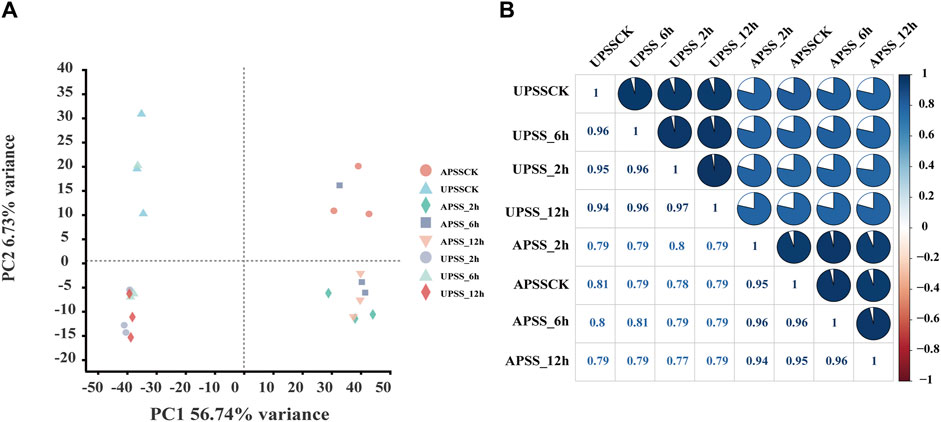
Figure 1. Assessment of RNA-seq dataset for identifying AS events. (A) Principal component analysis (PCA) of RNA-seq data. The sample replicates of the same group formed the same clusters and of different groups were classified into diverse clusters. (B) To assess the relationship between the samples, a correlation analysis of the RNA-seq was performed. Correlation matrices were generated by calculating the mean values of AP and UP at different time points under salt-treatment. Pearson correlation coefficients were computed using R scripts.
3.2 Identification of AS events in G. uralensis tissues under salt treatment
We used 24 RNA-Seq datasets to analyze the comprehensive landscape of AS events in salt-treated G. uralensis. After mapping the high-quality reads to the reference genome of G. uralensis (Mochida et al., 2017), we utilized the rMATS software with the junction count-only setting to identify and quantify AS events, and tested the quantity of the five AS types (SE, MXE, A5SS, A3SS, and RI) using the Kolmogorov–Smirnov method (Supplementary Table S3). After correction using one-way ANOVA, the Bonferroni method was applied to the mean values (Supplementary Table S4). The results showed that salt treatment for 2, 6, and 12 h significantly increased the amount of SE, A3SS, A5SS, and RI AS events in the AP compared to the UP of G. uralensis. In the UP of salt-treated G. uralensis, significant differences (p < 0.05) in SE and MXE AS events were detected without significant differences in A3SS, A5SS, and RI AS events (Figure 2). When comparing the mean differences of five AS events in AP and UP, significant differences were identified in the mean discrepancies among the five AS events, especially in the UP of G. uralensis. Furthermore, significant mean difference in SE and MXE but not in RI, A3SS, and A5SS AS events were observed in the UP of salt-treated G. uralensis (Supplementary Figure S2). These results suggest that AS events are common during salt stress, with the UP being the major tissue site in G. uralensis for the response to salt.
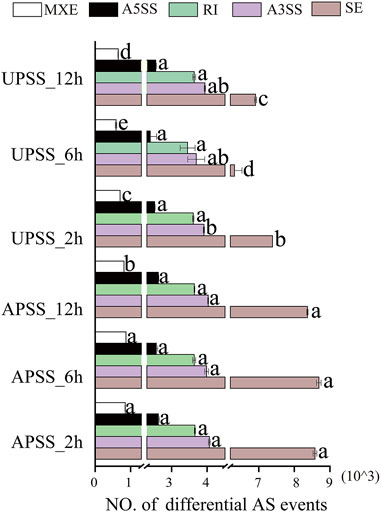
Figure 2. Quantities of various AS event types in salt-treated transcriptomes in Glycyrrhiza uralensis. Five AS event types of SE, MXE, A5SS, A3SS, and RI were analyzed, with the data of three biological replicates to be indicated as mean ± standard deviation. The mean values were evaluated by Bonferroni test, with distinct letters to represent significant differences among diverse treatments (p < 0.05).
3.3 Recognition of AS events responding to salt-treated Glycyrrhiza uralensis AP and UP sections
Reports indicated that AS events in wheat roots led to diverse spliced gene isoforms under salt stress. Under drought stress, the generation of variant splicing isoforms in G. uralensis genes is similar Li et al. (2022). It suggest that these events contribute to G. uralensis’ responsiveness to salt. In this study, a total of 1648, 1505, and 2329 AS events in AP and 2983, 1742, and 3086 AS events in UP were obtained in salt-treated G. uralensis for different time points of 2, 6, and 12 h, respectively (Figure 3A). SE was the most prevalent AS event, accounting for 34% of AP and 29% of UP. Furthermore, following 2 h and 12 h of salt stress exposure in G. uralensis of AP tissue, the SE events significantly tend towards the inclusion type. In contrast, the same events in UP leaned towards the exclusion type (Figure 3A). Hence, the abundance of diverse isoforms varies widely across different anatomical regions of G. uralensis plants.
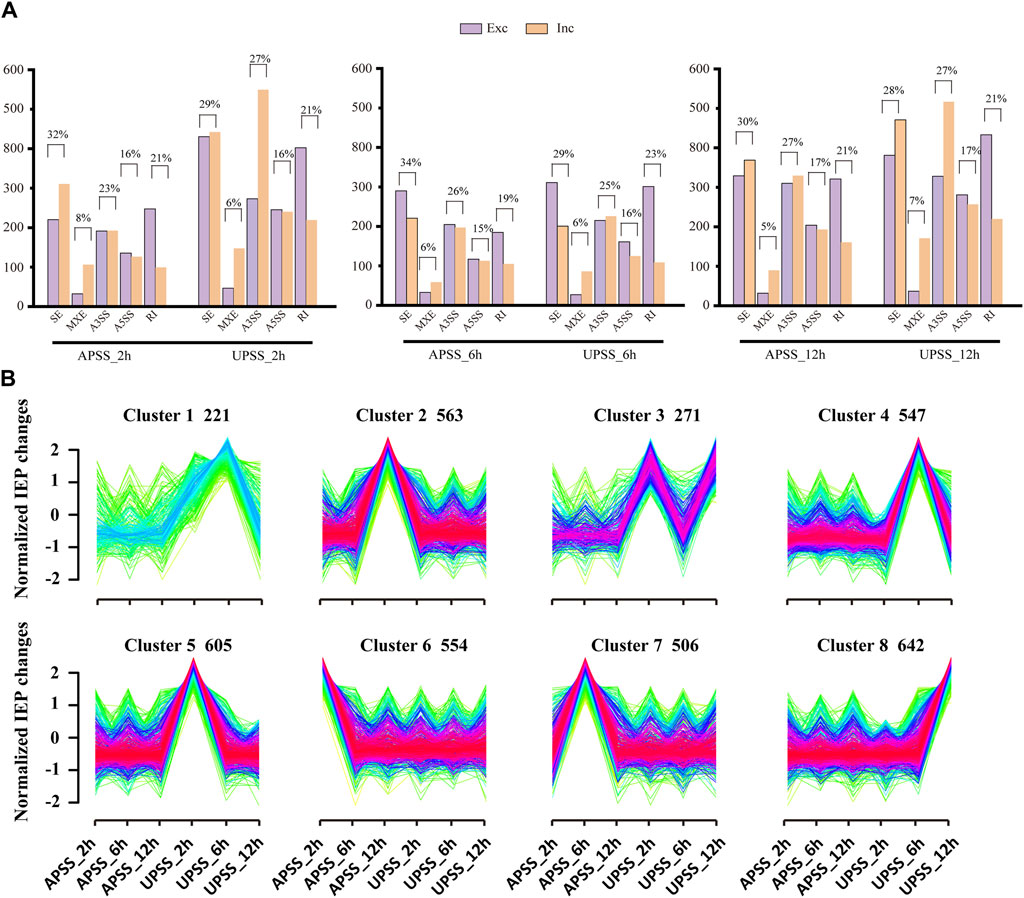
Figure 3. Identification and comparative analysis of the AS events in G. uralensis under salt stress. (A) Comparative analysis of the quantity of AS events in salt-treated G. uralensis UP and AP sections. (B) IEP-based cluster examination of AS events in G. uralensis in response to salt stress. The count of AS events in each cluster was indicated, with x-axis representing the different treatment time points in G. uralensis UP and AP sections and y-axis representing the IEP values. The red lines indicate the trend of IEP mean value for each AS event in different clusters.
Isoform expression percentage (IEP) alterations have been used to categorize AS events in wheat (Li et al., 2022) and soybean (Song et al., 2020) under salt stress. In G. uralensis AP and UP sections, SE-type AS events emerged as the primary manifestation of salt-related AS in response to salt stress. Consequently, we utilized Mfuzz software to perform the IEP cluster analysis in SE-type with the conditions of “FDR < 0.05” and “change in isoform expression percentage ≥30%” (Figure 3B; Supplementary Table S5). The results indicated that eight groups were classified representing 221, 563, 271, 547, 605, 554, 506, and 642 AS events, respectively. Group 3 exemplified changes in the AS pattern at the UPSS_2 h, UPSS_6 h, and UPSS_12h time points. Similarly, groups 1, 4, 5, and 8 showed variations in AS patterns in the salt-stressed G. uralensis UP section, revealing their sensitivity to salt stress. Conversely, Groups 2, 6, and 7 exhibited minimal adjustments in the AS pattern in the salt-stressed AP tissues of G. uralensis. Therefore, significant alterations in AS patterns in response to salt stress occurred mainly in G. uralensis UP tissues. These results demonstrated that the substantial AS pattern alterations in response to salt stress occurred predominantly in G. uralensis UP.
3.4 Comparative evaluation of differentially expressed and spliced genes
DSGs were generated due to AS events and exhibited significant variation under salt stress. Overall, 4,117 DSGs in G. uralensis were identified and distributed across various phases throughout the salt stress (Supplementary Table S6). Of these, 1,017 genes showed clear salt-responsive and AS regulatory expression patterns (Figure 4A). In order to better understand the AS events in response to salt treatment, we compared the levels of DSGs and DEGs in salt-stressed G. uralensis UP and AP tissues at various time intervals throughout the salt stress treatment period (Figures 4B–D; Supplementary Table S6). We observed 68, 11, and 72 genes evident in DSGs and DEGs in AP tissues at 2, 6, 12 h. Similarly, 120, 13, and 93 genes were identified in UP tissues as DSGs and DEGs at 2, 6, and 12 h, respectively (Figure 4B). These results showed that salt-responsive expression tendencies and AS regulation were more significant in 2-h and 12-h treated tissues. Furthermore, the newly discovered AS events were analyzed in G. uralensis under salt stress; the results indicated that 1,746, 356, and 150 novel AS events of the SE, MXE, and RI types were identified and had a significant response (Supplementary Table S7), highlighting the substantial involvement of SE-type AS in G. uralensis salt stress response.
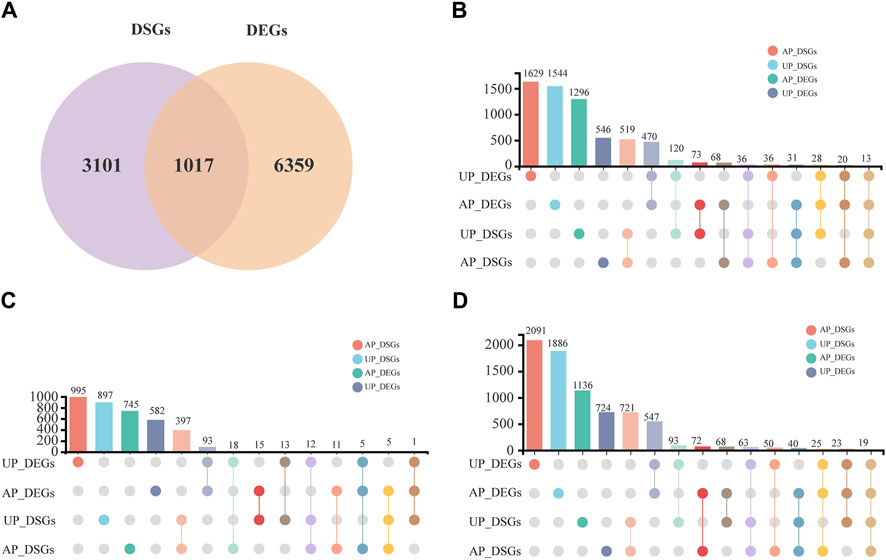
Figure 4. Analysis of DSGs and DEGs in salt-treated G. uralensis AP and UP sections. (A) Comparative analysis was performed on the overlapping DEGs and DSGs subjected to different salt stress treatments. The DSGs and DEGs in the G. uralensis AP and UP sections were measured at 2 h (B), 6 h (C), and 12 h (D) intervals during salt treatment.
3.5 GO analysis of DSGs in salt-treated G. uralensis AP and UP
In order to comprehend the impact of AS regulation on the biological functions of G. uralensis during the process of salt stress response, a GO enrichment analysis was performed on all DSGs. The results showed that numerous DSGs regulated by AS were related to metabolic, spliceosome, and oxidase activity (Figure 5). A total of 19 GO terms were significantly enriched in underground tissues, including cellular response to stress, DNA repair, and organonitrogen compound metabolism. The AP tissues exhibited significant enrichment of six GO terms, including metabolic processes for nitrogen compounds, phosphatase activity, and RNA processing. GO term enrichment in UP tissues is much higher than in AP tissues. Salt stress-related GO terms were only significantly enriched in UP tissues, not AP tissues, such as protoporphyrinogen oxidase activity, oxygen-dependent protoporphyrinogen oxidase activity, and oxidoreductase activity. It is possible that UP tissues first experience salt stress and then send signals to ground tissues. Furthermore, DEGs from UP and AP tissues in salt stress conditions were analyzed using GO to assess their functional significance. There was a significant enrichment in GO terms related to salt stress, such as oxidoreductase activity, flavonoid glucuronidation, and oxidoreductase activity, acting on CH–OH (Supplementary Figure S3). The enriched regulation of intracellular metabolic, spliceosome, and oxidase activity provides a comprehensive understanding of how AS regulates the salt stress response in G. uralensis.
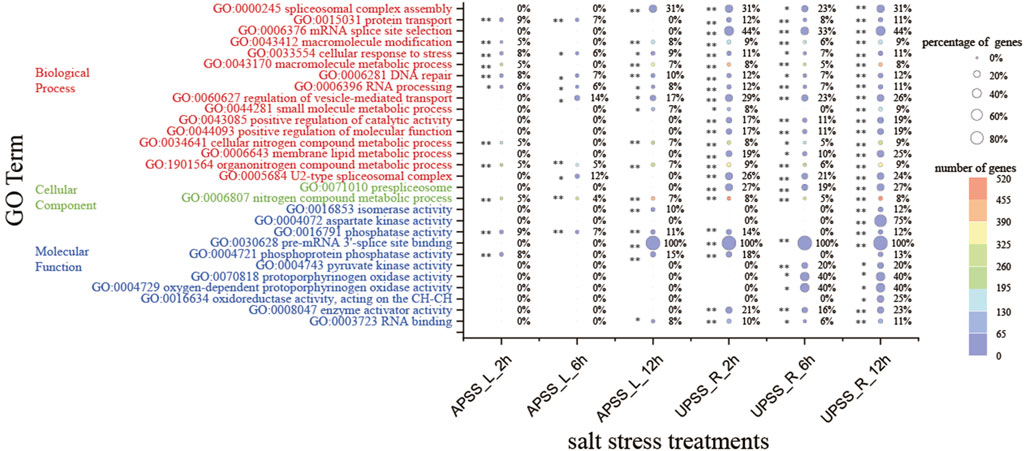
Figure 5. GO analyses of DSGs under salt stress. (A) GO enrichment analysis of DSGs. DSGs were subject to GO terms with the condition of FDR < 0.05. (B) Top 20 GO terms of the DSGs in the category of molecular function. **denotes p < 0.01 and * represents p < 0.05.
3.6 KEGG enrichment analysis of DEGs and DSGs that encode splicing regulators
Under various stress conditions, salt stress triggers the response of splicing regulatory factors (SPFs) and induces diverse patterns of splicing (Ding et al., 2014; Filichkin et al., 2010; Li et al., 2022; Palusa et al., 2007; Tanabe et al., 2007). In our work, KEGG enrichment analysis was performed on the difference splicing genes (DSGs). Pathways associated with AS, such as RNA transport, mRNA surveillance, and spliceosome, were significantly enriched, indicated splicing regulation of genes and resulting in AS events under salt stress conditions. These results indicated that the SPFs-encoding genes appeared to respond to salt stress in G. uralensis UP sections and exhibited variable splicing (Figure 6A). Moreover, plant response to salt stress pathways were also enriched, such as MAPK signaling pathway-plant, oxidative phosphorylation, and flavonoid biosynthesis. A total of 81 SPF-related genes and 45 genes related to salt stress that displayed significant splicing variation at diverse salt treatment time points were discovered (Supplementary Table S8). After salt stress treatment for 2 and 12 h, there were more upregulated than downregulated genes (Figure 6B). The main AS events were SE and RI in salt-treated G. uralensis (Figure 6C). Most SPF-related genes showed increased expressions in salt-treated G. uralensis UP section (UPSS_2h, UPSS_6h, and UPSS_12h) (Supplementary Figure S4). These results suggest that the SPF-encoding genes and genes related to salt stress are regulated by AS in G. uralensis UP under salt stress.
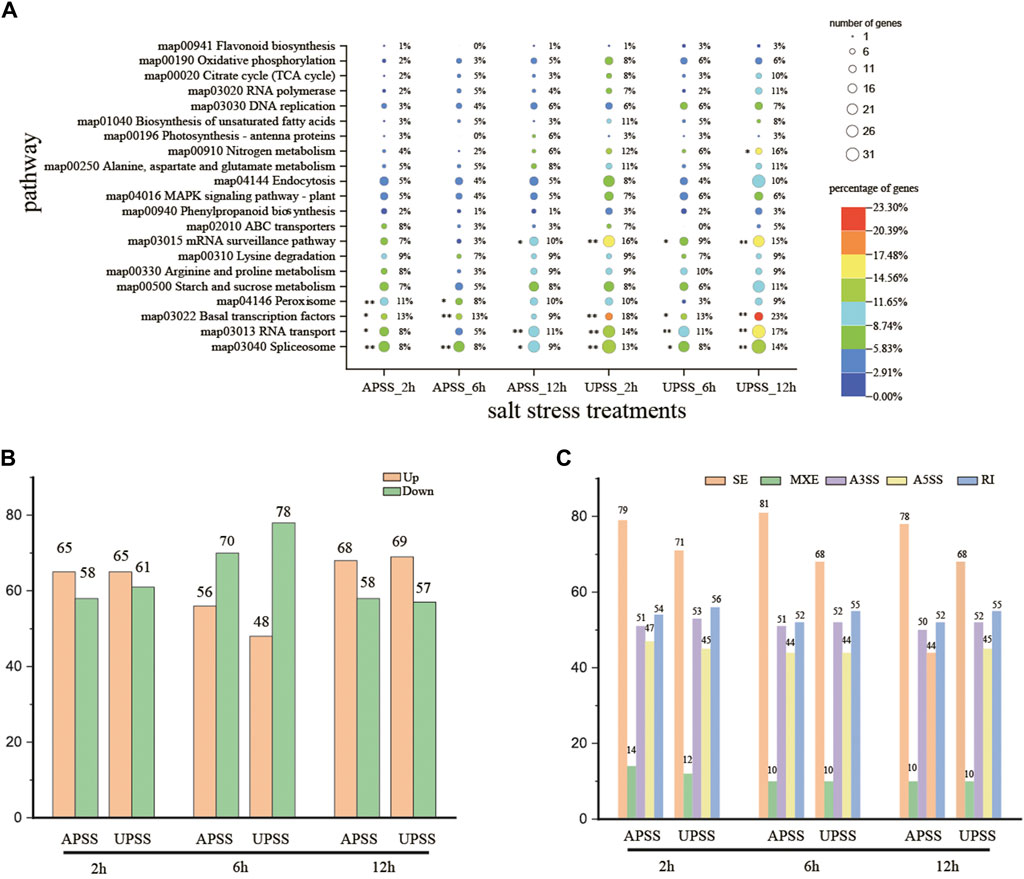
Figure 6. Analysis of KEGG enrichment and AS pattern of SPF-related genes during salt stress. (A) Top 20 pathways demonstrated enrichment of DSGs. (B) Notable differential splicing was observed within SPF-related genes under salt-stress conditions. (C) Significant differential expression was recorded within SPF-related genes under salt stress. **denotes p < 0.01 and * represents p < 0.05.
To comprehend how stress response genes in the root tissue of G. uralensis adapt to salt stress, we examined the expression levels of four genes encoding SPFs. As illustrated in Supplementary Figure S5, under salt stress at various time points, the four genes—Glyur000002s00000283 (similar to Glycine rich RNA-binding protein RZ1B), Glyur000834s00025309 (akin to Polyglutamine-binding protein 1), Glyur000659s00029682 (paralleling Serine/threonine protein phosphatase PP2A), and Glyur000136s00007936 (reminding of Polyadenylate binding protein 2)—exhibited high expressions as isoform 1 (spliced partial protein) and low expressions as isoform 2 (non-spliced intact protein) (Supplementary Figures 5A–D). These SPF-encoding genes displayed comparable responses to salt stress, implying their potential functions in modulating the salt-stress response in G. uralensis.
3.7 Identification of potential regulators in G. uralensis in response to salt stress
Many genes encoding the calcium ion and MAPK signal transduction pathway proteins showed notable AS alterations (Supplementary Figure S6). Glyur001147s00029003 (MPK4) encodes mitogen-activated protein kinase, with a significantly higher proportion of isoform 1 observed under salt stress 2, 6, and 12 h treatment. Under APSS_2h, APSS_12h, UPSS_2h, UPSS_6h, and UPSS_12h treatment conditions, there was a significant increase in isoform 1 of Glyur000143s00011203 (RbohD). Glyur000099s00014961(SnRK2) encodes erine/threonine–protein kinase, in which isoform 1 was detected in higher proportions under APSS_2h treatment conditions than control conditions. APSS_2h UPSS_2h, UPSS_6h, and UPSS_12h treatment conditions resulted in significantly higher proportions of isoform 1 than control conditions for Glyur000837s00027495 (ETR2) (Supplementary Figures S6C,E,F,G).
The expression patterns of the four genes in the above-mentioned MAPK pathway were also examined using qRT-PCR under salt stress. MPK4 had a significantly higher expression level observed under salt stress 2, 6 and 12 h treatment than control conditions. Under APSS_2h, APSS_12h, UPSS_2h, UPSS_6h, and UPSS_12h treatment conditions , there was a significant increase in expression level of RbohD compared to control conditions. SnRK2 expression level was higher under APSS_2h treatment conditions than control. APSS_2h, UPSS_2h, UPSS_6h, and UPSS_12h treatment conditions resulted in significantly higher proportions of expression level than control conditions for ETR2 (Figure 7). G. uralensis may be able to respond better to salt stress by increasing the proportion of isoform 1 of salt-stress-related genes. These findings indicate that significant alternative splicing events in the MPK4, RbohD, SnRK2, and ETR2 genes are especially generated in G. uralensis, which lead to increased protein isoform 1 production in response to salt stress.
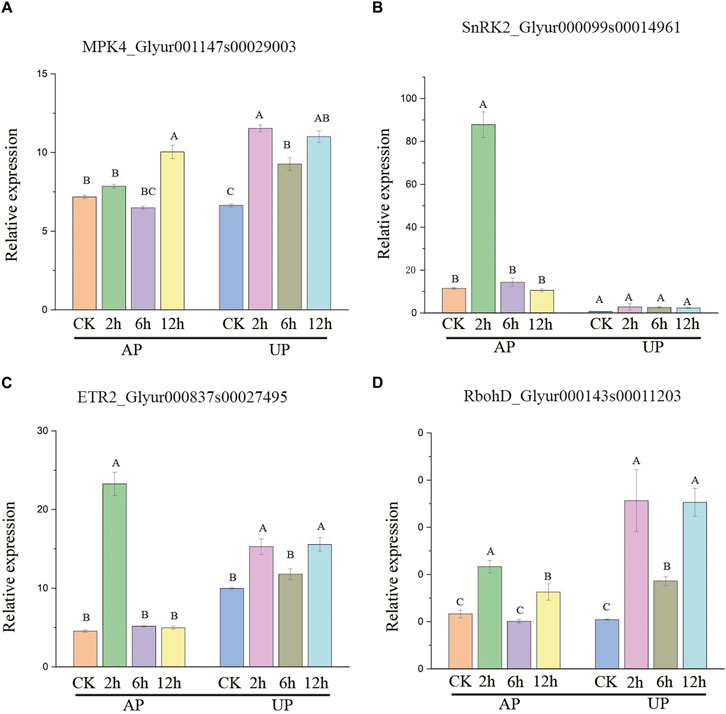
Figure 7. Expression levels of BAM1 (A), cyclin-dependent kinase B2-2 (CDKB2-2) (B), DRM2 (C), and PP2C12 (D) in salt-stressed G. uralensis AP and UP sections. Expression value was mean ± SD with three independent replicates. Bonferroni test used to analyze average value. Diverse letters denote significantly different salt stress treatments (p < 0.01).
4 Discussion
4.1 Salt stress significantly induced AS events in G. uralensis
AS events are an important factor for plants to respond to stress; RNA-seq studies-based analysis of abiotic stress-regulated AS events in G. uralensis is still limited (Calixto et al., 2018; Ding et al., 2014; Li et al., 2020; Liu et al., 2018; Thatcher et al., 2016). A total of 2591 and 3068 AS events were identified in the aboveground parts (AP) and underground parts (UP) of G. uralensis under salt stress, respectively, including SE, RI, A5SS, A3SS, and MXE. Under salt stress, the UP exhibited a higher occurrence of AS events related to stress response in comparison to the AP, indicating a more vigorous AS response. Some AS events may be gene-specific and some species-specific, with SE and RI being the most and least prevalent respectively in animals. The majority of AS cases are caused by SE, while only 0.01% are caused by RI; in contrast, RI is the most common form of AS in Arabidopsis and maize (Thatcher et al., 2016). In recent years, however, SE and RI have been the most prevalent AS in licorice and soybeans (Li et al., 2022; Song et al., 2020) but MXE the least. These results are in line with our study. This may be due to advances in sequencing technology and detection tools that allow for more detailed detection of AS. Among these, the SE-types of AS were the most prevalent. In AP tissue, SE-types accounted for 30%–34% of the responsive AS events, while in UP tissue they accounted for 28%–29% of the responsive AS occurrences in salt-stressed G. uralensis. Under salt stress, there was a higher occurrence of A3SS events than RI events (Figure 2), but the count of A3SS and RI events was not significantly different (Figure 2). This finding aligns with a previous study that reported SE and A3SS as the predominant AS models in soybeans and G. uralensis under drought stress. However, RI was the notable AS type in reaction to drought in soybeans. Under high temperatures and drought treatments in wheat, the predominant type of AS was found to be RI, whereas the primary model in maize of AS in coping with stress was also RI (Iniguez et al., 2017; Liu et al., 2018; Shen et al., 2014; L; Wang et al., 2014; Y; Wang et al., 2020). These differences in the major AS types could be attributed to variations in SRP-related genes. Consequently, our investigation revealed many AS events in G. uralensis under salt stress condition, underscoring their crucial contribution to the response of G. uralensis to saline environment pressures.
4.2 DSGs enriched in key GO terms and KEGG pathways associated with salt stress and AS
Global crop production is severely constrained by salt stress, which negatively impacts the growth of G. uralensis (Xiao et al., 2024). There are many genes that are responsive to salt stress, but the alternative splicing patterns of these are still unclear. In this study, we found that DEGs and DSGs mainly occurred in the root (Figures 4B–D). To investigate what functions these DEGs and DSGs have, we analyzed their GO enrichment separately. The results indicate that most of these GO terms are enriched in UP tissues. In DSGs, GO terms such as “pyruvate kinase activity”, “protoporphyrinogen oxidase activity”, and “protoporphyrinogen oxidase activity” were significantly enriched in UPSS_6h and UPSS_12h (Figure 5). In DEG, GO terms such as “isoflavonoid biosynthetic process”, ”flavonoid glucuronidation”, “oxidation-reduction process”, and “isoflavonoid metabolic process”, consistent with previous studies, all participated in salt stress response (Kalifa et al., 2004; Kononenko et al., 2023; Tavakkoli et al., 2011; Zhang et al., 2023). Meanwhile, in DSGs, some GO terms such as “spliceosomal complex assembly”,“U2-type spliceosomal complex”, “prespliceosome”, and “pre-mRNA 3′-splice site binding” were significantly enriched at UPSS_2h, UPSS_6h, and UPSS_12h especially, and “mRNA splice site selection” was enriched at UPSS_2h, UPSS_6h, and UPSS_12h in G. uralensis (Figure 5), which was known to be important for alternative splicing (Clayton et al., 2011; Schindler et al., 2008). Interestingly, these GO terms were not enriched in the DEG. These results indicate that the alternative splicing genes played important roles in the response to salt stress. Moreover, some previously identified stress-related proteins were also enriched, such as enzyme activator activity (Gao et al., 2017).
To further investigate on which signaling pathways these DSGs function, we performed KEGG enrichment. We observed remarkable enrichment of DSGs in the “RNA transport”, “mRNA surveillance”, and “spliceosome” pathways, which were significantly enriched in UPSS_2h, UPSS_6h, and UPSS_12h, which are known to be important for alternative splicing (Liu et al., 2018; Song et al., 2020). The regulation of alternative splicing represents an important means of fine-tuning gene expression that may save the time required for changes in transcriptional activation and pre-mRNA accumulation, thus allowing rapid plant adaptation to adverse environmental conditions. Ultimately, the effects of alternative splicing on mRNAs’ encoding effectors and modulators of abiotic stress responses are determined by the levels and/or activity of the splicing factors regulating this process. Pre-RNA undergoes maturation to become mature mRNA through the spliceosome process, making it an important splicing process (Syed et al., 2012). Marondedze et al. (2019) recently reported that spliceosome component modifications were consistent with changes in drought-stressed Arabidopsis proteome and transcriptional profiles. We also discovered that genes encoding SPFs proteins, which these are certain RNA-binding (Glyur000002s00000283, Glyur000834s00025309, and Glyur000136s00007936) were potentially regulated by AS. Spliceosomes have been reported in G. uralensis under drought stress (Li et al., 2022), indicating their involvement in the reaction of G. uralensis to salt stress.
The mRNA surveillance pathway detects and degrades abnormal mRNAs and plays an important role in maintaining accurate gene expression with salt stress in soybeans (Song et al., 2020). In soybeans, nonsense-mediated decay (NMD) is a crucial pathway responsible for mRNA surveillance and facilitates the degradation of mRNAs that contain premature termination codons (Lareau et al., 2007). According to Baena-Gonzalez et al. (2007), approximately 17.4% of multi-exonic and protein-coding genes in Arabidopsis are known to produce splicing variants aimed by NMD. Salt stress modulates AS in conjunction with NMD in Arabidopsis (Drechsel et al., 2013). In the present study, we identified one gene (Glyur000659s00029682) that encodes the serine/threonine PP2A regulated by salt stress and AS (Supplementary Figure S4D). Hence, the salt stress response in G. uralensis could potentially be regulated by the mRNA surveillance pathway. Pre-mRNAs and proteins abundant in Ser/Arg residues are key players in AS, helping maintain cellular and tissue homeostasis (Ding et al., 2014; Laloum et al., 2018; Li et al., 2022; Reddy et al., 2011). AS occurs in SRP-related genes in plants in a developmental and tissue-specific manner, responding to various hormonal and abiotic stresses (Ding et al., 2014; Li et al., 2022; Zhang et al., 2009; Zhu et al., 2017). BrSR45a was found by Muthusamy et al. (2020) to increase stress-inducible genes and influence the AS of target genes in Arabidopsis. Interestingly, our study discovered that AS during salt stress affected specific genes involved in encoding Ser/Arg-rich proteins, which are identified as participants in pre-mRNA splicing (Figure 6). Therefore, SRP-related genes in G. uralensis may play a significant role in coping with salt stress.
Some KEEG pathways such as “Peroxisome” were significantly enriched at APSS_2 h and APSS_6h, which was consistent with previous studies that all participated in salt stress response (Liu et al., 2018). Moreover, some previously identified stress-related signaling pathways were also enriched, such as MAPK signaling pathway – plant (Yang et al., 2018). Plants have collectively evolved specific responses at both transcriptional and AS levels to cope with salt stress.
4.3 Signaling pathways in response to salt stress
In addition to AS, salt stress can also give rise to oxidative stress by increasing the levels of reactive oxygen species (ROS) (Carillo, 2019). Plant antioxidant systems have been shown to effectively reduce the effects of oxidative stress and reduce ROS formation (Farhangi-Abriz et al., 2017). Long-term evolution has led to a variety of adaptive physiological and biochemical strategies for plants, including protecting against high-salt environments, restoring ROS equilibrium, and maintaining ion and osmotic homeostasis. Plants respond to salt stress by producing H2O2 from NADPH oxidase (Rboh) (Liu et al., 2020). RbohD under APSS 2,12, UPSS 2,6,12 elevated the proportion of isform 1 through A5SS events and then increased its expression. The excess H2O2 was then removed to improve salt stress response in plants.
Licorice increases antioxidant activity when exposed to salt stress conditions and reduces lipid peroxidation due to free radical damage (Xu et al., 2021). In response to salt stress, MAPK cascades are triggered, such as on the SOS pathway (Yang et al., 2018c). Plants can adapt to salt stress by utilizing MAPK cascades for transducing signals (Mishra et al., 2006). Salt stress is regulated by the MPK4 cascade in Arabidopsis. Plants with the MPK4 gene null showed hypersensitivity to salt stress, whereas plants with overexpressed MPK4 showed greater salt tolerance (Teige et al., 2004). There are other plants that also rely on the MAPK cascade for salt stress signal transduction. Alfalfa (M. sativa), for example, uses the MKK-MK cascade to trigger its response to salt stress (Kiegerl et al., 2000). The MPK4 cascade controls salt stress signal transduction in rice by regulating the expression of transcription factor genes (F. Wang et al., 2014). When Arabidopsis is dehydrated, mutants of the MKK4 cascade are more salt sensitive and shed more water than the wild type (Kim et al., 2012). We demonstrate that MPK 4 under salt stress increases isform 1 through SE events, increases its expression, and regulates downstream genes responding to salt stress to improve plant tolerance. When Asterochloris erici is subjected to hyperosmotic stresses such as high salt and desiccation, MAPK signaling cascades are activated (Gasulla et al., 2016). More studies are needed to better understand how salt/osmotic stress in plants affects MAPK signaling pathways. As a result of osmotic stress, all ten SnRK2 isoforms are activated except SnRK2.9. The signaling pathway is dependent on ABA in order to activate SnRK2.2/3/6/7/8 (Boudsocq et al., 2004; Nykiel et al., 2023). Osmotic stress activates transcription of downstream effectors through ABAactivated SnRK2.2/3/6-ABA-responsive element (ABRE)-binding protein and ABRE-binding factor (AREB/ABF) signaling (Soma et al., 2017). The moss Physcomitrella patens expresses an ABA-responsive Raf-like kinase (ARK) that activates SnRK2 in response to osmotic stress (Saruhashi et al., 2015). As part of our research, we found that SnRK2 enhanced isform 1 proportions by enhancing SE event expression, regulating downstream salt stress response genes, and improving salt stress tolerance in plants. Furthermore, many studies have shown that the ethylene (ET) synthesis genes ETR1 and ETR2 also regulate the synthesis of ABA and that ABA can interact with ET to regulate salt tolerance in plants (Zhang et al., 2022). ETR2 at APSS _ 2 h, UPS SS _ 2, and 12h increased the proportion of isform 1 by RI events to improve ET synthesis and then regulate downstream salt stress response genes to improve plant tolerance to salt stress. In this study, expression levels of MPK4, RbohD, SnRK2, and ETR2 were markedly elevated under CK treatment compared to salt stress, demonstrating a significant increase (Figure 7). In the root tissues of G. uralensis, these four genes exhibited significant AS events that produced additional proteins in response to salt stress. Furthermore, researchers can examine plant transcriptome datasets to analyze AS events and their variations across different species, tissues, and developmental stages. By examining AS patterns in various environments, researchers can uncover insights into the functions and mechanisms of AS in reaction to abiotic stress. Enhanced comprehension of this matter could potentially pave the way for the identification of novel strategies to bolster plant resistance against stress, thus advancing our knowledge of plant biology and augmenting our capacity to cultivate more resilient crops.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
HY: Writing–original draft, Investigation, Validation, Visualization. GL: Methodology, Software, Writing–original draft. GZ: Software, Writing–original draft. GF: Software, Writing–original draft. JF: Writing–original draft. GX: Writing–review and editing. HS: Funding acquisition, Project administration, Resources, Supervision, Writing–original draft, Writing–review and editing. HL: Writing–review and editing.
Funding
The authors declare that financial support was received for the research, authorship, and/or publication of this article. This research was funded by the Natural Science Foundation of China (grant number 32260083), the Science and Technology Project of Bingtuan (grant number 2023AB052, and 2023CB008-17), the Starting Fund for High-Scientific Study of Genetics (grant number RCZK2021B31), and the Special Project for New Variety Cultivation at Shihezi University (grant number YZZX202102), and the Science and Technology Project of Huyanghe City (grant number QS2023008), and the Department of Education of Guangdong Province (grant number 2022Z DZX2063) and Science and Technology Project of Shihezi University (CXPY202107), and Ministry Key Laboratory of Xinjiang Phytomedicine Resource and Utilization Project (XPRU202105, XPRU202202, XPRU202002).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2024.1397502/full#supplementary-material
References
Amin, U. S., Biswas, S., Elias, S. M., Razzaque, S., Haque, T., Malo, R., et al. (2016). Enhanced salt tolerance conferred by the complete 2.3 kb cdna of the rice vacuolar na(+)/h(+) antiporter gene compared to 1.9 kb coding region with 5' utr in transgenic lines of rice. Front. Plant Sci. 7, 14. doi:10.3389/fpls.2016.00014
Baena-Gonzalez, E., Rolland, F., Thevelein, J. M., and Sheen, J. (2007). A central integrator of transcription networks in plant stress and energy signalling. Nature 448 (7156), 938–942. doi:10.1038/nature06069
Boudsocq, M., Barbier-Brygoo, H., and Lauriere, C. (2004). Identification of nine sucrose nonfermenting 1-related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana. J. Biol. Chem. 279 (40), 41758–41766. doi:10.1074/jbc.M405259200
Calixto, C., Guo, W., James, A. B., Tzioutziou, N. A., Entizne, J. C., Panter, P. E., et al. (2018). Rapid and dynamic alternative splicing impacts the arabidopsis cold response transcriptome. Plant. Cell. 30 (7), 1424–1444. doi:10.1105/tpc.18.00177
Carillo, P. C. M. V., Cirillo, C., De Micco, V., Arena, C., De Pascale, S., and Rouphael, Y. (2019). Morpho-anatomical, physiological and biochemical adaptive responses to saline water of bougainvillea spectabilis willd. Trained to different canopy shapes. Agric. Water Manag. 212, 12–22. doi:10.1016/j.agwat.2018.08.037
Cinatl, J., Morgenstern, B., Bauer, G., Chandra, P., Rabenau, H., and Doerr, H. W. (2003). Glycyrrhizin, an active component of liquorice roots, and replication of SARS-associated coronavirus. Lancet 361 (9374), 2045–2046. doi:10.1016/s0140-6736(03)13615-x
Clayton, J. C., Phelan, M., Goult, B. T., Hautbergue, G. M., Wilson, S. A., and Lian, L. Y. (2011). The 1H, 13C and 15N backbone and side-chain assignment of the RRM domain of SC35, a regulator of pre-mRNA splicing. Biomol. NMR Assign. 5 (1), 7–10. doi:10.1007/s12104-010-9254-5
Daszkowska-Golec, A., Skubacz, A., Marzec, M., Slota, M., Kurowska, M., Gajecka, M., et al. (2017). Mutation in hvcbp20 (cap binding protein 20) adapts barley to drought stress at phenotypic and transcriptomic levels. Front. Plant Sci. 8, 942. doi:10.3389/fpls.2017.00942
Ding, F., Cui, P., Wang, Z., Zhang, S., Ali, S., and Xiong, L. (2014). Genome-wide analysis of alternative splicing of pre-mRNA under salt stress in Arabidopsis. BMC Genomics 15 (1), 431. doi:10.1186/1471-2164-15-431
Drechsel, G., Kahles, A., Kesarwani, A. K., Stauffer, E., Behr, J., Drewe, P., et al. (2013). Nonsense-mediated decay of alternative precursor mRNA splicing variants is a major determinant of the Arabidopsis steady state transcriptome. Plant. Cell. 25 (10), 3726–3742. doi:10.1105/tpc.113.115485
Egawa, C., Kobayashi, F., Ishibashi, M., Nakamura, T., Nakamura, C., and Takumi, S. (2006). Differential regulation of transcript accumulation and alternative splicing of a DREB2 homolog under abiotic stress conditions in common wheat. Genes. Genet. Syst. 81 (2), 77–91. doi:10.1266/ggs.81.77
Farhangi-Abriz, S., and Torabian, S. (2017). Antioxidant enzyme and osmotic adjustment changes in bean seedlings as affected by biochar under salt stress. Ecotoxicol. Environ. Saf. 137, 64–70. doi:10.1016/j.ecoenv.2016.11.029
Feng, J., Li, J., Gao, Z., Lu, Y., Yu, J., Zheng, Q., et al. (2015). SKIP confers osmotic tolerance during salt stress by controlling alternative gene splicing in arabidopsis. Mol. Plant. 8 (7), 1038–1052. doi:10.1016/j.molp.2015.01.011
Filichkin, S. A., Priest, H. D., Givan, S. A., Shen, R., Bryant, D. W., Fox, S. E., et al. (2010). Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 20 (1), 45–58. doi:10.1101/gr.093302.109
Gao, Y., Wang, Y., Xin, H., Li, S., and Liang, Z. (2017). Involvement of ubiquitin-conjugating enzyme (E2 gene family) in ripening process and response to cold and heat stress of Vitis vinifera. Sci. Rep. 7 (1), 13290. doi:10.1038/s41598-017-13513-x
Gasulla, F., Barreno, E., Parages, M. L., Camara, J., Jimenez, C., Dormann, P., et al. (2016). The role of phospholipase d and mapk signaling cascades in the adaption of lichen microalgae to desiccation: changes in membrane lipids and phosphoproteome. Plant Cell. Physiol. 57 (9), 1908–1920. doi:10.1093/pcp/pcw111
Iniguez, L. P., Ramirez, M., Barbazuk, W. B., and Hernandez, G. (2017). Identification and analysis of alternative splicing events in phaseolus vulgaris and glycine max. BMC Genomics 18 (1), 650. doi:10.1186/s12864-017-4054-2
Kalifa, Y., Gilad, A., Konrad, Z., Zaccai, M., Scolnik, P. A., and Bar-Zvi, D. (2004). The water- and salt-stress-regulated Asr1 (abscisic acid stress ripening) gene encodes a zinc-dependent DNA-binding protein. Biochem. J. 381 (Pt 2), 373–378. doi:10.1042/BJ20031800
Kiegerl, S., Cardinale, F., Siligan, C., Gross, A., Baudouin, E., Liwosz, A., et al. (2000). SIMKK, a mitogen-activated protein kinase (MAPK) kinase, is a specific activator of the salt stress-induced MAPK, SIMK. Plant. Cell. 12 (11), 2247–2258. doi:10.1105/tpc.12.11.2247
Kim, J. M., Woo, D. H., Kim, S. H., Lee, S. Y., Park, H. Y., Seok, H. Y., et al. (2012). Arabidopsis MKKK20 is involved in osmotic stress response via regulation of MPK6 activity. Plant Cell. Rep. 31 (1), 217–224. doi:10.1007/s00299-011-1157-0
Kononenko, N. V., Lazareva, E. M., and Fedoreyeva, L. I. (2023). Mechanisms of antioxidant resistance in different wheat genotypes under salt stress and hypoxia. Int. J. Mol. Sci. 24 (23), 16878. doi:10.3390/ijms242316878
Laloum, T., Martin, G., and Duque, P. (2018). Alternative splicing control of abiotic stress responses. Trends Plant Sci. 23 (2), 140–150. doi:10.1016/j.tplants.2017.09.019
Lareau, L. F., Brooks, A. N., Soergel, D. A., Meng, Q., and Brenner, S. E. (2007). The coupling of alternative splicing and nonsense-mediated mRNA decay. Adv. Exp. Med. Biol. 623, 190–211. doi:10.1007/978-0-387-77374-2_12
Li, G., Xu, D., Huang, G., Bi, Q., Yang, M., Shen, H., et al. (2022). Analysis of whole-transcriptome rna-seq data reveals the involvement of alternative splicing in the drought response of glycyrrhiza uralensis. Front. Genet. 13, 885651. doi:10.3389/fgene.2022.885651
Li, S., Yu, X., Cheng, Z., Zeng, C., Li, W., Zhang, L., et al. (2020). Large-scale analysis of the cassava transcriptome reveals the impact of cold stress on alternative splicing. J. Exp. Bot. 71 (1), 422–434. doi:10.1093/jxb/erz444
Liu, J., Shabala, S., Zhang, J., Ma, G., Chen, D., Shabala, L., et al. (2020). Melatonin improves rice salinity stress tolerance by NADPH oxidase-dependent control of the plasma membrane K+ transporters and K+ homeostasis. Plant Cell. Environ. 43 (11), 2591–2605. doi:10.1111/pce.13759
Liu, Z., Qin, J., Tian, X., Xu, S., Wang, Y., Li, H., et al. (2018). Global profiling of alternative splicing landscape responsive to drought, heat and their combination in wheat (Triticum aestivum L.). Plant Biotechnol. J. 16 (3), 714–726. doi:10.1111/pbi.12822
Marondedze, C., Thomas, L., Lilley, K. S., and Gehring, C. (2019). Drought stress causes specific changes to the spliceosome and stress granule components. Front. Mol. Biosci. 6, 163. doi:10.3389/fmolb.2019.00163
Marquez, Y., Brown, J. W., Simpson, C., Barta, A., and Kalyna, M. (2012). Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res. 22 (6), 1184–1195. doi:10.1101/gr.134106.111
Mishra, N. S., Tuteja, R., and Tuteja, N. (2006). Signaling through MAP kinase networks in plants. Arch. Biochem. Biophys. 452 (1), 55–68. doi:10.1016/j.abb.2006.05.001
Mochida, K., Sakurai, T., Seki, H., Yoshida, T., Takahagi, K., Sawai, S., et al. (2017). Draft genome assembly and annotation of Glycyrrhiza uralensis, a medicinal legume. Plant. J. 89 (2), 181–194. doi:10.1111/tpj.13385
Muthusamy, M., Yoon, E. K., Kim, J. A., Jeong, M. J., and Lee, S. I. (2020). Brassica rapa SR45a regulates drought tolerance via the alternative splicing of target genes. Genes. 11 (2), 182. doi:10.3390/genes11020182
Nykiel, M., Gietler, M., Fidler, J., Prabucka, B., and Labudda, M. (2023). Abiotic stress signaling and responses in plants. Plants 12 (19), 3405. doi:10.3390/plants12193405
Palusa, S. G., Ali, G. S., and Reddy, A. S. (2007). Alternative splicing of pre-mrnas of arabidopsis serine/arginine-rich proteins: regulation by hormones and stresses. Plant. J. 49 (6), 1091–1107. doi:10.1111/j.1365-313X.2006.03020.x
Reddy, A. S., and Shad, A. G. (2011). Plant serine/arginine-rich proteins: roles in precursor messenger rna splicing, plant development, and stress responses. Wiley Interdiscip. Rev. RNA. 2 (6), 875–889. doi:10.1002/wrna.98
Saruhashi, M., Kumar, G. T., Arai, K., Ishizaki, Y., Hagiwara, K., Komatsu, K., et al. (2015). Plant Raf-like kinase integrates abscisic acid and hyperosmotic stress signaling upstream of SNF1-related protein kinase2. Proc. Natl. Acad. Sci. U. S. A. 112 (46), E6388–E6396. doi:10.1073/pnas.1511238112
Schindler, S., Szafranski, K., Hiller, M., Ali, G. S., Palusa, S. G., Backofen, R., et al. (2008). Alternative splicing at nagnag acceptors in arabidopsis thaliana sr and sr-related protein-coding genes. BMC Genomics 9, 159. doi:10.1186/1471-2164-9-159
Shen, S., Park, J. W., Lu, Z. X., Lin, L., Henry, M. D., Wu, Y. N., et al. (2014). rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. U. S. A. 111 (51), E5593–E5601. doi:10.1073/pnas.1419161111
Soma, F., Mogami, J., Yoshida, T., Abekura, M., Takahashi, F., Kidokoro, S., et al. (2017). ABA-unresponsive SnRK2 protein kinases regulate mRNA decay under osmotic stress in plants. Nat. Plants 3, 16204. doi:10.1038/nplants.2016.204
Song, L., Pan, Z., Chen, L., Dai, Y., Wan, J., Ye, H., et al. (2020). Analysis of whole transcriptome RNA-seq data reveals many alternative splicing events in soybean roots under drought stress conditions. Genes. 11 (12), 1520. doi:10.3390/genes11121520
Syed, N. H., Kalyna, M., Marquez, Y., Barta, A., and Brown, J. W. (2012). Alternative splicing in plants--coming of age. Trends Plant Sci. 17 (10), 616–623. doi:10.1016/j.tplants.2012.06.001
Tanabe, N., Yoshimura, K., Kimura, A., Yabuta, Y., and Shigeoka, S. (2007). Differential expression of alternatively spliced mRNAs of Arabidopsis SR protein homologs, atSR30 and atSR45a, in response to environmental stress. Plant Cell. Physiol. 48 (7), 1036–1049. doi:10.1093/pcp/pcm069
Tavakkoli, E., Fatehi, F., Coventry, S., Rengasamy, P., and McDonald, G. K. (2011). Additive effects of Na+ and Cl-ions on barley growth under salinity stress. J. Exp. Bot. 62 (6), 2189–2203. doi:10.1093/jxb/erq422
Teige, M., Scheikl, E., Eulgem, T., Doczi, R., Ichimura, K., Shinozaki, K., et al. (2004). The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol. Cell. 15 (1), 141–152. doi:10.1016/j.molcel.2004.06.023
Thatcher, S. R., Danilevskaya, O. N., Meng, X., Beatty, M., Zastrow-Hayes, G., Harris, C., et al. (2016). Genome-Wide analysis of alternative splicing during development and drought stress in maize. Plant Physiol. 170 (1), 586–599. doi:10.1104/pp.15.01267
Wang, C., Chen, L., Cai, Z. C., Chen, C., Liu, Z., Liu, X., et al. (2020). Comparative proteomic analysis reveals the molecular mechanisms underlying the accumulation difference of bioactive constituents in glycyrrhiza uralensis fisch under salt stress. J. Agric. Food. Chem. 68 (5), 1480–1493. doi:10.1021/acs.jafc.9b04887
Wang, F., Jing, W., and Zhang, W. (2014). The mitogen-activated protein kinase cascade MKK1-MPK4 mediates salt signaling in rice. Plant Sci. 227, 181–189. doi:10.1016/j.plantsci.2014.08.007
Wang, L., Cao, C., Ma, Q., Zeng, Q., Wang, H., Cheng, Z., et al. (2014). RNA-seq analyses of multiple meristems of soybean: novel and alternative transcripts, evolutionary and functional implications. BMC Plant Biol. 14, 169. doi:10.1186/1471-2229-14-169
Wang, X., Wu, F., Xie, Q., Wang, H., Wang, Y., Yue, Y., et al. (2012). SKIP is a component of the spliceosome linking alternative splicing and the circadian clock in Arabidopsis. Plant. Cell. 24 (8), 3278–3295. doi:10.1105/tpc.112.100081
Wang, Y., Xu, J., Ge, M., Ning, L., Hu, M., and Zhao, H. (2020). High-resolution profile of transcriptomes reveals a role of alternative splicing for modulating response to nitrogen in maize. BMC Genomics 21 (1), 353. doi:10.1186/s12864-020-6769-8
Xiao, X., Lang, D., Yong, J., and Zhang, X. (2024). Bacillus cereus G2 alleviate salt stress in Glycyrrhiza uralensis Fisch. by balancing the downstream branches of phenylpropanoids and activating flavonoid biosynthesis. Ecotoxicol. Environ. Saf. 273, 116129. doi:10.1016/j.ecoenv.2024.116129
Xu, Y., Lu, J. H., Zhang, J. D., Liu, D. K., Wang, Y., Niu, Q. D., et al. (2021). Transcriptome revealed the molecular mechanism of glycyrrhiza inflata root to maintain growth and development, absorb and distribute ions under salt stress. BMC Plant Biol. 21 (1), 599. doi:10.1186/s12870-021-03342-6
Yang, Y., and Guo, Y. (2018). Unraveling salt stress signaling in plants. J. Integr. Plant Biol. 60 (9), 796–804. doi:10.1111/jipb.12689
Zhang, M., and Zhang, S. (2022). Mitogen-activated protein kinase cascades in plant signaling. J. Integr. Plant Biol. 64 (2), 301–341. doi:10.1111/jipb.13215
Zhang, X., He, P., Guo, R., Huang, K., and Huang, X. (2023). Effects of salt stress on root morphology, carbon and nitrogen metabolism, and yield of Tartary buckwheat. Sci. Rep. 13 (1), 12483. doi:10.1038/s41598-023-39634-0
Zhang, X. N., and Mount, S. M. (2009). Two alternatively spliced isoforms of the arabidopsis sr45 protein have distinct roles during normal plant development. Plant Physiol. 150 (3), 1450–1458. doi:10.1104/pp.109.138180
Keywords: Glycyrrhiza uralensis, salt stress response, RNA-seq analysis, splicing regulatory factor, post-transcriptional regulation
Citation: Yao H, Li G, Gao Z, Guo F, Feng J, Xiao G, Shen H and Li H (2024) Alternative splicing responses to salt stress in Glycyrrhiza uralensis revealed by global profiling of transcriptome RNA-seq datasets. Front. Genet. 15:1397502. doi: 10.3389/fgene.2024.1397502
Received: 07 March 2024; Accepted: 28 May 2024;
Published: 09 July 2024.
Edited by:
Ertugrul Filiz, Duzce University, TürkiyeReviewed by:
Satish Kumar, Directorate of Onion and Garlic Research (ICAR), IndiaGuofang Zhang, Huazhong Agricultural University, China
Copyright © 2024 Yao, Li, Gao, Guo, Feng, Xiao, Shen and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haitao Shen, c2h0c2h6LWJpb0BzaHp1LmVkdS5jbg==; Hongbin Li, bGloYkBzaHp1LmVkdS5jbg==; Jianghua Feng, eXpiQHhqbnUuZWR1LmNu
†These authors have contributed equally to this work
 Hua Yao
Hua Yao Guozhi Li
Guozhi Li Zhuanzhuan Gao1†
Zhuanzhuan Gao1† Guanghui Xiao
Guanghui Xiao Haitao Shen
Haitao Shen Hongbin Li
Hongbin Li