- 1Jiangxi Provincial Institute of Traditional Chinese Medicine, Jiangxi Research Center for Protection and Development of Traditional Chinese Medicine Resources, Key Laboratory of Germplasm Selection and Breeding of Chinese Medicinal Materials, Nanchang, Jiangxi, China
- 2College of Life Sciences, Xinyang Normal University, Xinyang, China
- 3Wuhan Botanical Garden, Chinese Academy of Sciences, Wuhan, Hubei, China
The flower coloration of Brassica crops possesses significant application and economic value, making it a research hotspot in the field of genetics and breeding. In recent years, great progress has been made in the research on color variation and creation of Brassica crops. However, the underlying molecular mechanisms and evolutional processes of flower colors are poorly understood. In this paper, we present a comprehensive overview of the mechanism of flower color formation in plants, emphasizing the molecular basis and regulation mechanism of flavonoids and carotenoids. By summarizing the recent advances on the genetic mechanism of flower color formation and regulation in Brassica crops, it is clearly found that carotenoids and anthocyanins are major pigments for flower color diversity of Brassica crops. Meantime, we also explore the relationship between the emergence of white flowers and the genetic evolution of Brassica chromosomes, and analyze the innovation and multiple utilization of Brassica crops with colorful flowers. This review aims to provide theoretical support for genetic improvements in flower color, enhancing the economic value and aesthetic appeal of Brassica crops.
1 Introduction
Flowers of angiosperms serve as reproductive organs, flower coloration is a crucial trait for the survival and reproduction of plants, which can not only attract insects to pollination, but also maintains the energy balance under different lighting conditions to protect flower organs (Clegg and Durbin, 2000; Ariizumi et al., 2014). As an apparent trait, flower color has been widely used as a marker or target trait across genetics, molecular biology, ecology, evolutionary biology and crop breeding (Liu et al., 2004). Additionally, given its significant ornamental value, creating novel and diverse flower colors has remained a primary objective in flower breeding (Li et al., 2023). Flower color is mainly attributed to the pigments present in petals, carotenoids and flavonoids are two main large classes of natural pigments contributing to most of colors in plants like yellow, orange and red (Jin et al., 2014; Ohmiya, 2013). In the last two decades, the corresponding biosynthetic pathways have been well characterized and elucidated, which provide a foundation for understanding the development and molecular basis of flower color (Koes et al., 2005; Ruiz-Sola and Rodriguez-Concepcion, 2012).
Brassica species are grown worldwide as important oil crops, vegetables and feed crops. There are mainly six representative cultivated Brassica crops including three diploid species, B. rapa (2n = 20, AA), B. nigra (2n = 16, BB), and B. oleracea (2n = 18, CC), and three allotetraploid species, B. juncea (2n = 36, AABB), B. napus (2n = 38, AACC), and B. carinata (2n = 34, BBCC). These three allotetraploids originate from interspecific hybridizations among the remaining three diploid species, resulting in a more complex genetic basis (Nagaharu, 1935). Brassica crops exhibit flower color diversity that more than 63 different colored rape varieties have been obtained in China (Xiao et al., 2021). However, to date the molecular mechanisms underlying flower color diversity of Brassica crops remain elusive. Early studies reported that anthocyanins are responsible for the red and purple coloration on leaves, stems, petals and floral meristem of Brassica crops, while the total content of carotenoids showed significant differences between yellow-flowered and white- flowered varieties (Chiu and Li, 2012; Zhang et al., 2015; Li et al., 2023). In recent years, with multi-omics approach, numerous flavonoids (including anthocyanins) and carotenoids have been isolated and characterized from different-colored petals of several Brassica crops (Li et al., 2015; Yin et al., 2019; Ye et al., 2022), and a number of structural and regulatory genes involved in the flavonoids and carotenoids biosynthetic pathways have been identified as flower color controlled genes (Chen et al., 2023; Guan et al., 2023). Thus, much have been learned about the mechanism of flavonoids and carotenoids on flower color formation in Brassica crops. This paper is expected to summarize the regulation mechanisms of flower color formation in Brassica crops, as well as elucidating the relationship between different phenotypic and genotypic evolutions, which are crucial for harnessing and cultivating diverse flower colors in Brassica species and beyond.
2 The formation mechanism of flower color in plants
The coloration of petals is determined mainly by the presence and relative abundances of three pigment classes including carotenoids, flavonoids, and betalains (Zhao et al., 2022). Carotenoids and flavonoids, in particular, govern the hues exhibited by a vast array of plants, while betalains exist specifically in Caryophyllales plants (except Caryophyllaceae and Molluginaceae), and chlorophyll in cells would affect the final color (Zhao and Tao, 2015). The diversity in petal coloration among different plants arises from the unique combinations and concentrations of these pigments. The biosynthetic pathways responsible for the formation of these pigments are intricately regulated by a range of structural and regulatory genes. Variations in the types, quantities, and sequence characteristics of these encoding genes, as well as differences in their expression patterns within cells, constitute the molecular basis for the observed chromatic aberrations in petals (Ohmiya, 2013). By understanding the genetic and biochemical mechanisms underlying petal coloration, it can gain insights into the remarkable diversity of plant phenotypes and their adaptation strategies in nature.
2.1 Carotenoids biosynthetic pathways and key genes
Carotenoids are a class of fat-soluble pigments widely distributed in flowers, fruits, leaves and roots, primarily control the yellow and orange to red colorations. These pigments are a group of 40-carbon triterpenes composed of eight isoprene basic units linked end-to-end. They are synthesized through the isoprene pathway and are typically localized in plastid or chromoplast (Zhao et al., 2022). The backbone of carotenoids consists of a main chain containing nine double bonds, with each end of the chain being cycled to form a β-violone ring. Carotenoids can be categorized into two groups, non-oxygenated carotenoids (Carotenes, such as α-carotene, β-carotene, etc.) and oxygenated carotenoids (Xanthophyll, such as zeaxanthin, lutein, antheraxanthin, etc.). Oxygenated carotenoids further encompass bicyclic oxides and monocyclic oxides (Grotewold, 2006; Schweiggert et al., 2012). Xanthophylls exhibiting light yellow color, which include lutein, zeaxanthin, antheraxanthin, violaxanthin, and neoxanthin, are the main carotenoids imparting yellow colorations of petals and flesh (Jin et al., 2014; Niaz et al., 2023).
Carotenoids biosynthesis in plants occurs via the isoprene pathway. Isopentenyl pyrophosphate (IPPC5) serves as the fundamental five-carbon building block for the biosynthesis of carotenoids, which is synthesized via the methylerythritol phosphate pathway (MEP) within plastid. Geranylgeranyl diphosphate (GGPP) is the direct precursor of carotenoid biosynthesis (Walter and Strack, 2011). Under the catalysis of phytoene synthase (PSY), two GGPP molecules condense to form the phytoene which contains 40-carbon atoms (Figure 1). PSY is recognized as one of the key rate-limiting enzymes in the carotenoid synthesis pathway. Subsequently, phytoene undergoes the introduction of four conjugated double bonds through the sequential action of phytoene desaturase (PDS), ε-Carotene isomerase (Z-ISO), ε-carotene desaturase (ZDS), and carotenoid isomerase (CRTISO), ultimately leading to the formation of lycopene (Park et al., 2002; Li et al., 2007).
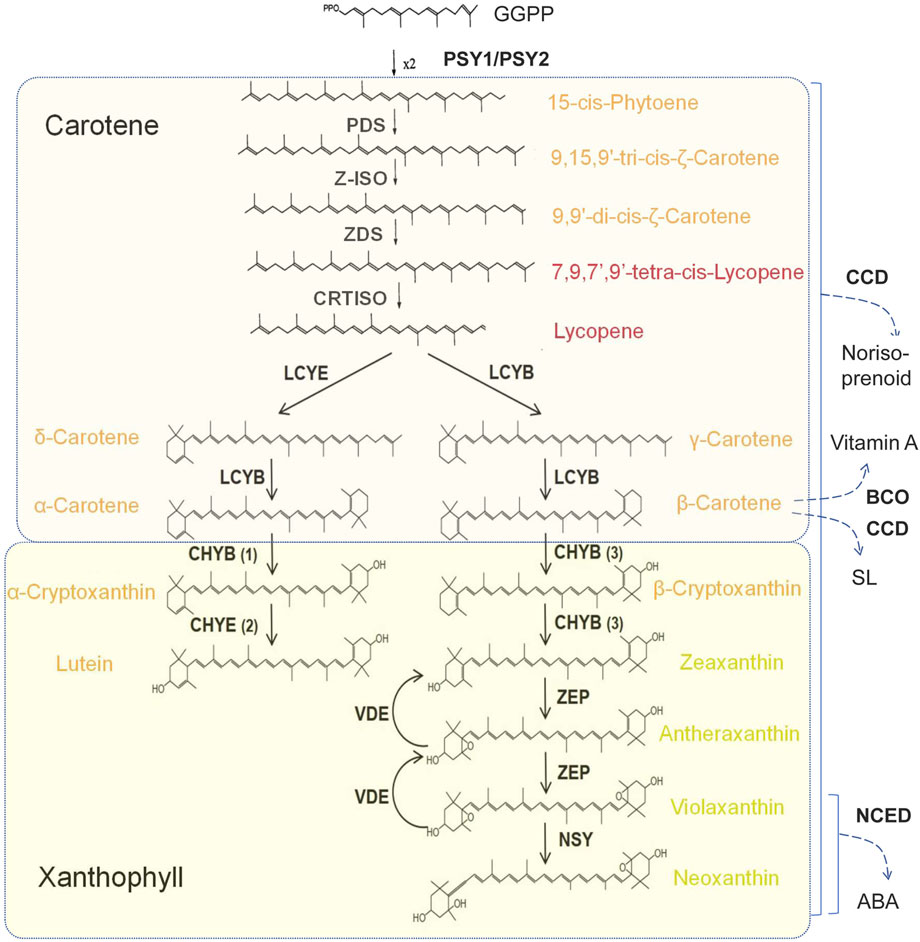
Figure 1. Carotenoids synthesis pathways in plants. PSY, Phytoene synthase; PDS, Phytoenedesaturase; Z-ISO, ε-Carotene isomerase; ZDS, ε-Carotenedesaturase; LCYE, Lycopene δ-cyclase; LCYB, Lycopene β-cyclase; CHYB, β-Carotene hydroxylase; CHYE, ε-Carotene hydroxylase; ZEP, Zeaxanthin epoxidase; VDE, Violaxanthin depoxidase; NSY, Neoxanthin synthase; CCD, Carotenoid cleavage dioxygenases; BCO, β-carotene-15,15′-oxygenase; NCED, Nine-cis epoxycarotenoid dioxygenase.
Lycopene cyclization represents a critical branch point in the carotenoid biosynthesis pathway. There are two lycopene cyclases in plants that can catalyze the formation of double bonds at distinct positions of cyclohexane, resulting in two different products (Zhao et al., 2011). Lycopene β-cyclase (LYCB) catalyzes the conversion of lycopene to γ-carotene, which contains a β-ring, while lycopene ε-cyclase (LYCE) converts lycopene to δ-carotene, containing an ε-ring (Zhao et al., 2011). Subsequently, δ-carotene is further catalyzed by LYCB to yield α-carotene and its derivatives, which possess one ε-ring and one β-ring. In contrast, γ-carotene is catalyzed by LYCB to form β-carotene and its derivatives, while β-carotene contains two β-rings (Niaz et al., 2023).
Both α-carotene and β-carotene serve as precursors for the biosynthesis of xanthophylls. Lutein is produced by the addition of a hydroxyl group to each of the two violone rings of α-carotene, catalyzed by the joint action of ε-carotene hydroxylase (CHYE) and β-carotene hydroxylase (CHYB) (Tian et al., 2004; Kebede and Rahman, 2014). Similarly, β-carotene can be catalyzed with CHYB to produce zeaxanthin by adding a hydroxyl group to each β- rings (Zhang et al., 2022). Subsequently, zeaxanthin is converted into antheraxanthin, a single-ended epoxide, and violaxanthin, a double-ended epoxide, under the action of zeaxanthin epoxidase (ZEP) (Welsch et al., 2018; Niaz et al., 2023). Conversely, violaxanthin can be converted back to antheraxanthin and then to zeaxanthin by the catalytic action of violaxanthin de-poxidase (VDE), which has opposing functionality to ZEP (Walter and Strack, 2011; Jahns and Holzwarth, 2012). Furthermore, the epoxide ring at one end of violaxanthin is cleaved and transformed into neoxanthin by the catalysis of neoxanthin synthase (NSY/NSX) (Neuman et al., 2014). In addition, carotenoids undergo degradation by dioxygenase carotenoid cleavage dioxygenase (CCD) or 9-cis-epoxycarotenoid dioxygenase (NCED), and enter into the biosynthesis pathways leading to the production of abscisic acid (ABA), vitamin A, or strigolactone (SL) (Maoka, 2020).
The expression of genes related to carotenoid synthesis and degradation metabolism plays a pivotal role in determining petal color formation in certain plants (Tian et al., 2003; Zhu et al., 2010). For instance, in the case of lily, the yellow hue of petals is tightly linked to the abundance of gene expression for enzymes such as PSY, PDS, ZDS, CRTISO and other genes (Yamagishi et al., 2010). High expression of CmCCD4a in white chrysanthemum (Chrysanthemum morifolium Ramat.) leads to rapid degradation of carotenoids, thereby resulting in the exhibition of white petals (Ohmiya et al., 2006). Conversely, the mutation of the CmCCD4a in yellow chrysanthemum causes the accumulation of carotenoids in petals, resulting in yellow flowers (Yoshioka et al., 2012). Chromoplast is the main organelle for the biosynthesis and storage of carotenoids in petals (Li and Yuan, 2013). Within the chromoplast, carotenoids accumulate within plastoglobulus (PGs), which are specialized lipoprotein particles (Ytterberg et al., 2006). Notably, lutein accumulation is primarily in the form of esters with fatty acids, rather than in its free (non-esterified) form (HorneroMéndez and Mínguez-Mosquera, 2000). Mutations in genes involved in carotenoid esterification would affect the development of PGs and the accumulation of pigments in PGs (Ariizumi et al., 2014), suggesting that carotenoid esterification is also extremely important for the formation of flower colors.
2.2 Flavonoids biosynthetic pathways and key genes
Flavonoids are diverse water-soluble pigments which control the orange, red, purple, and blue colors in petals of most plants. As important secondary metabolites in plants, flavonoids are involved in various growth and development processes. The basic structure of flavonoids is C6-C3-C6 rings (Figure 2), two benzene rings with a phenol hydroxyl group are connected to each other by a central three carbon atom (He and Giusti, 2010). The anabolic pathway for flavonoids begins with the phenylpropanoid pathway, which produces phenylalanine as a precursor. Phenylalanine is then converted into cinnamic acid by the enzyme phenylalanine ammonia-lyase (PAL). Subsequently, cinnamic acid is hydroxylated to form coumaric acid, which enters the flavonoid biosynthetic pathway (Appelhagen et al., 2014; Li, 2014; Shen et al., 2022). Flavonoids encompass a diverse array of compounds, including flavones, ehaleones, flavonols, anthocyanidins, proanthocyanidins, flavanones and isoflavones. Among them, anthocyanins are mainly related to flower color, and the corresponding flavonoid metabolic pathway is called anthocyanin pathway (Harborne, 1986; Saito et al., 2013).
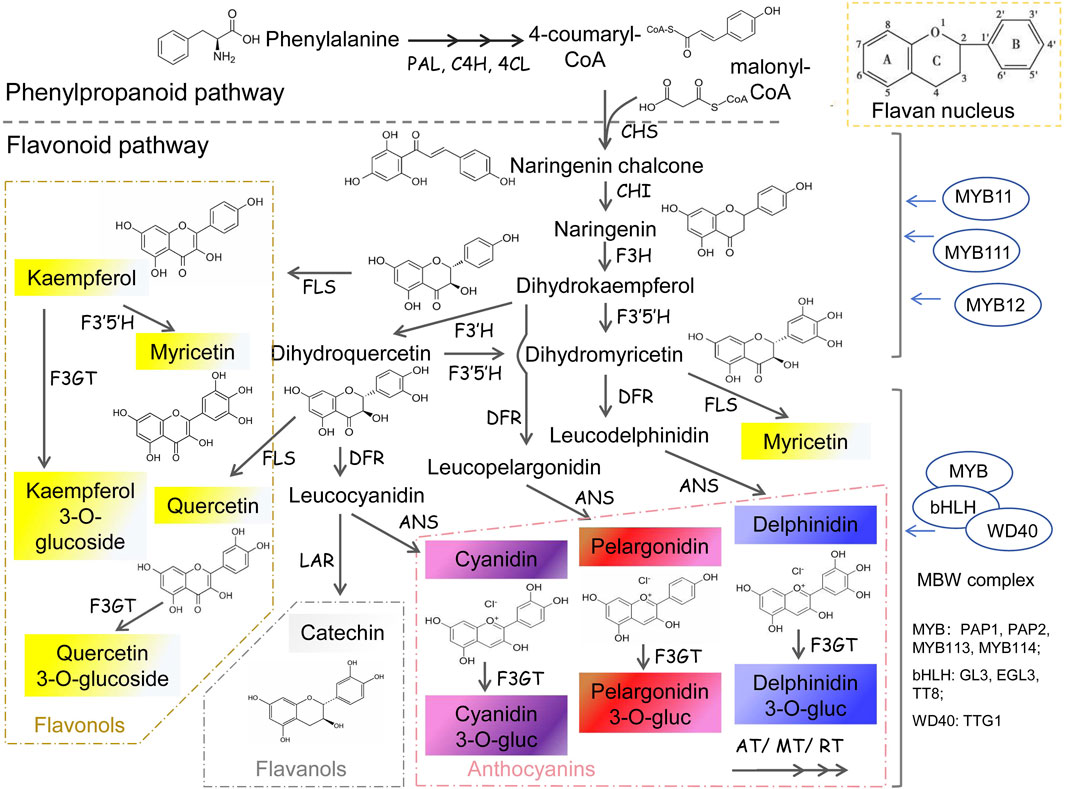
Figure 2. Flavonoids synthesis pathway in plants PAL, phenylalanine ammonialyase; C4H, cinnamate 4-hydroxylase; 4CL, 4-coumaric acid CoA ligase; CHS, chalcone synthase; CHI, chalcone isomerase; F3H, flavanone 3-hydroxylase; F3′5′H, flavonoid 3′ 5′-hydroxylase; F3′H, flavonoid3′-hydroxylase; FLS, flavonol synthase; DFR, Dihydro flavonol reductase; LAR, leucoanthocyanidin reductase; ANS, Anthocyanidin Synthase; F3GT, flavonoid 3-Glucosyl transferase; AT, Acyltransferase; MT, Methyl transferase; RT, Rhamnosyl transferase.
The anthocyanin pathway is highly specialized and involves specific genes encoding enzymes that catalyze key steps in the biosynthetic process. The resulting anthocyanins can further be categorized based on the number of hydroxyl groups present in their B ring. For instance, pelargonidins, which carry a single hydroxyl group, typically impart pink, bright red, or orange hues. Cyanidins, with two hydroxyl groups, predominantly yield purple or red colors. Delphinidins, containing three hydroxyl groups, are responsible for the predominantly blue hues observed in petals (Figure 2).
The regulation of these biosynthetic genes is crucial for controlling the production and accumulation of anthocyanins, thereby influencing the final color phenotype of flowers. Mutations or alterations in these genes can lead to significant changes in petal color, providing valuable insights into the genetic control of flavonoid biosynthesis and its impact on plant phenotype.
From naringenin, various subclasses of flavonoids can be produced through a series of enzymatic reactions catalyzed by specific genes. Multiple genes regulate flavonoid synthesis and their mutations can lead to alterations in flavonoid type and content (Appelhagen et al., 2014). Nowadays, the flavonoid synthesis pathway is well-characterized, and its regulation is not only determined by enzyme-encoding genes but also modulated by various transcription factors, either alone or as part of MBW complexes (Shen et al., 2022). For example, flavonol synthase (FLS) catalyzes the conversion of naringenin into flavonols, while dihydroflavonol 4-reductase (DFR) and anthocyanidin synthase (ANS) are involved in the biosynthesis of anthocyanins. Other key genes involved in flavonoid biosynthesis include flavonoid 3′-hydroxylase (F3H), and flavonoid 3′,5′-hydroxylase (F3′5′H), which modify the structure of flavonoids by introducing hydroxyl groups or methyl groups. For instance, roses, carnations, and chrysanthemums lack the F3′5′H gene, preventing the synthesis of delphinium pigment and resulting in the absence of natural blue petal resources in these plants (Dai and Hong, 2016).
The stability of anthocyanin is closely related to petal color duration. Transferase coding enzymes including GT (Glucosyl transferase), AT (Acyl transferase), MT (Methyl transferase), and RT (Rhamnosyl transferase) catalyze the modifications of anthocyanins in different ways, thereby enhancing their stability within cells (Bombarely et al., 2016; Zhao et al., 2021). And the different positions of acylation and methylation of anthocyanins will also affect the flower color. Subsequently, the synthesized anthocyanins will be transported to the vacuole, a process facilitated by the collaborative action of GST (Glutathione S-transferase, TT19), MATE (Multidrug andtoxic compound extrusion, TT12), and ABC (ATP-binding cassette transporter) transporters (Marinova et al., 2007; Sun et al., 2012; Behrens et al., 2019). Consequently, both the pH level of the vacuole and the presence of metal ions or other types of flavonoids within it impact the coloration of anthocyanins (Hondo et al., 1992).
3 Formation and regulation mechanisms of flower color in Brassica crops
Brassica crops exhibit flower color diversity. The predominant flower color of Brassica crops is yellow, although mutant and interspecific hybridization has led to the emergence of distinct petal colors (Chen et al., 1988; Zhang et al., 2000). To date, the reported color variations can mainly be divided into two groups, the yellow series color including white, milky white, milky yellow, yellow, golden yellow, lemon yellow, orange, and yellow-white chimera, and the red series color like orange-red, pink, apricot, red and purple, et al. Recently, significant progress has been made in elucidating the molecular mechanism of flower color formation and regulation in Brassica crops.
3.1 The inheritance of flower color in Brassica crops
The flower color inheritance patterns vary with species even individual plants (Supplementary Table S1). In Brassica rapa, yellow flower is dominant, it is reported that compared with yellow flowers, the white, milky white, light yellow and orange flowers are recessive and controlled by 1-2 pairs of genes (Kebede and Rahman, 2014; Zhang et al., 2019; Zhang et al., 2020; Guan et al., 2023). While in B. oleracea, the yellow flower is the recessive one, white flower is controlled by a dominant single gene (Han et al., 2019). As for B. juncea, the white, milky and light-yellow flowers were controlled by two pairs of recessive genes (Rawat and Anand, 1986; Zhang et al., 2018a). The white flower of B. carinata is not completely dominant compared with yellow flower, while the milky white and light-yellow flower are recessive and are controlled by a single pair of genes (Jambhulkar and Raut, 1955; Getinet et al., 1993; Tian, 2011).
Brassica napus is one of the most important oil crops and also planted as vegetable, and fodder crops worldwide. The inheritance of flower color in B. napus exhibits considerable complexity. Comparative studies have demonstrated that, compared to yellow flower, the white flowers are dominant or incompletely dominant, and governed by one or two loci (Huang et al., 2014; Zhang et al., 2015; Huang et al., 2017). Milky white is recessive relative to milky yellow or yellow, golden yellow and orange flower is recessive relative to yellow flower, light yellow is incomplete dominant relative to lemon yellow, these traits are all controlled by two pairs of genes (Zhang et al., 2000; Liu et al., 2020; Jia et al., 2021). Yellow-white chimeric materials are presumed to be controlled by locally expressed recessive albino genes and exhibit simple recessive traits independent of yellow inheritance or single gene maternal effects (Yu et al., 2004; Yin and Guan, 2013). The studies of red series color flowers primarily focus on B. napus. Compared to yellow flower, several studies suggest that the orange and red flower are controlled by two pairs of recessive genes (Dan, 2016; Yao et al., 2017), while other investigations indicate that the orange-red, apricot, and red colors are governed by a single dominant gene (Guo et al., 2021; Ye et al., 2022; Chen et al., 2023). In addition, the red flower trait is recessive when compared with the yellow flower, and was found to be gradually altered in F2 population, indicating that the red flower was a quantitative trait of B. napus (Jiang, 2021).
Collectively, these findings suggest that the molecular mechanisms underlying single flower color in Brassica crops are influenced by multiple genetic pathways. Consequently, further research is required to elucidate the genetic mechanisms that underlie the diverse color phenotypes observed in these Brassica crops. What’s more, by analyzing the inheritance pattern of yellow and white flowers of Brassica crops, it is found that in species sharing the A genome, such as B. rapa and B. juncea, the white flower trait is governed by one or two recessive loci, and the yellow flower phenotype is dominant over the white flower (Rahman, 2001; Zhang et al., 2019; Yang et al., 2021; Guan et al., 2023). Conversely, in species harboring the C genome, including B. oleracea, B. napus and B. carinata, the white flower is dominant over yellow flower, and is controlled by one or two nuclear genes (Pearson, 1929; Liu et al., 2004; Huang et al., 2014; Zhang et al., 2015; Han et al., 2019). These observations suggest a strong correlation between flower color inheritance in Brassica crops and the genetic relationships among the A, B, and C genomes. However, there are relatively few genetic studies on other flower colors, it needs more results to verify.
3.2 Genetic regulation of yellow series flowers in Brassica crops
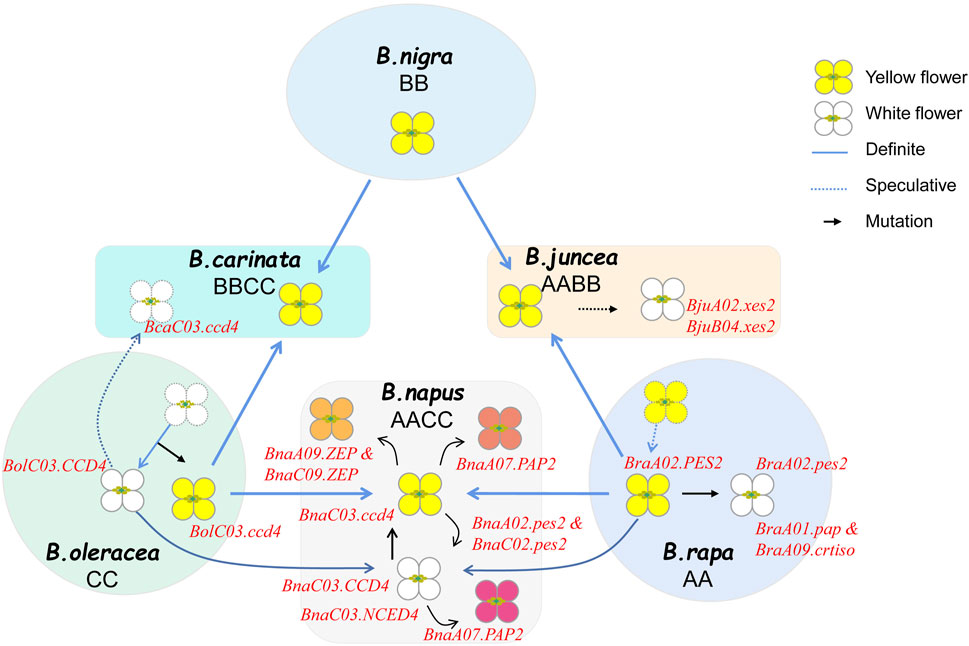
The predominant flower color in Brassica crops is yellow, while the most common color variations are from white to orange due to the decrease or accumulation of yellow pigments. To date, several genes that govern the development of yellow series flowers have been identified (Figure 3). In B. rapa, two recessive genes, BrWf1 and BrWf2 that jointly control the formation of white flower, encode a plastid-lipid associated protein (PAP) and a carotenoid isomerase (CRTISO), respectively (Zhang et al., 2019). It is reported that the mutation of BrA01. PAP (Bra013602) decrease the accumulation of carotenoid by blocking PG formation and the mutation of BrA09. CRTISO (Bra031539) decrease the flux of carotenoid biosynthesis pathway, when two genes mutant together, the flower color appeared white. The single nuclear gene BrWF3 and BraFC that controls the white flower trait were found to be the same gene, Bra032957, a homolog to Arabidopsis PES2 involved in fatty acid phytyl-ester synthesis in chloroplasts. A premature stop codon mutation within Bra032957 disrupted the gene function, leading to increase in abnormal chromoplasts with irregularly structured PG and decrease in xanthophylls in the white petals of flowers (Yang et al., 2021; Guan et al., 2023). Knockout mutants of the two orthologous genes (BnaA02. PES2-2 and BnaC02. PES2-2) of BraFC were constructed in B. napus, and showed white petals (Guan et al., 2023). In B. juncea, two independent recessive genes, Bjpc1(BjuA02. xes2) and Bjpc2 (BjuB04. xes2, BjuB027334), controlling the white flower trait, are also homologous to AtPES2 (Zhang et al., 2018a; Zhang et al., 2018b; Li et al., 2023), which mean that the carotenoid esterification contribute a lot in the formation of white flowers in Brassica crops.
Carotenoid lytic dioxygenase genes also play important roles in the formation of white flowers. Until now, In B. oleracea and B. napus, all reported white flower loci were mapped to the chromosome C03, and CCD4, a carotenoid cleavage dioxygenase 4 gene was confirmed to be the single nuclear gene controlling the white flower formation (Zhang et al., 2015; Han et al., 2019; Yang et al., 2019). Zhang et al. (2015) reported that BnaC03. CCD4 (BnaC03G0619000NO) cleaves δ-carotene and/or α-carotene into volatile α-ionone, which partitions most of the flux in the carotenoid biosynthesis pathway and results in white flowers with decreased yellowish pigments like lutein. And the insertions and deletions occurring in the coding regions of BnaC03. CCD4 disrupted its function and resulted in a significantly higher accumulation of carotenoids in petals of yellow B. oleracea, B. napus and B. carinata, which became the prevalent flower color across these species (Zhang et al., 2015). Jia et al. (2021) also confirmed that BnaC03. NCED4 (BnaC03G0710000ZS, a homologous gene of BnaC03. CCD4) can turn rape petals from yellow to white by degrading the carotenoids content. In B. oleracea, the results of gene localization and functional analysis showed that BolC.cpc-1(Bol029878) or BoCCD4 (Bol029878) is the candidate gene for white flower formation, which is highly expressed in white cabbage, but extremely low in yellow flower, and when mutant, the white flowers turn yellow (Han et al., 2019; Xu et al., 2019; Yang et al., 2019).
The reported novel flower colors of Brassica crops primarily encompass gold-yellow, and orange hues. It has been observed that as the petal color transitions from white to yellow and then to golden yellow, the total carotenoid content increases (Zeng, 2013). Notably, the content of lycopene exhibits the most significant difference between golden yellow with other colors. This differential lycopene content may be attributed to the highest expression of BnPSY and relatively low expression of BnLYCE, leading to a significantly higher synthesis of lycopene in golden yellow flowers compared to other colors (Zeng, 2013). BnaA08. PDS3 encodes phytoene desaturase 3, one of the rate-limiting enzymes in carotenoid biosynthesis pathways, and the mutation of BnaA08. PDS3 (BnaA08g17170D) results in decreased carotenoids and causes yellowish-white petals in Brassica napus (Zhao et al., 2021). The genetic loci associated with orange flower color have been primarily mapped on chromosome A09 and C09 (Yao et al., 2017; Zhang et al., 2019; Ding et al., 2019; Zhang, 2021). Specifically, BnaA09. ZEP (BnaA09g07610D) and BnaC09. ZEP (BnaC09g075550D) have been identified as the candidate genes that control the orange flower trait, as these genes were typically expressed in yellow petals but not in orange petals (Liu et al., 2020; Zhang, 2021). Functionally, these two genes exhibit redundancy in carotenoid synthesis, facilitating the restoration of orange flower color to yellow (Liu et al., 2020). RNA-sequencing studies have further revealed that β-carotene is the primary contributor to the formation of orange flowers. Decreased expression levels of CHYB and ZEP in orange petals lead to an accumulation of β-carotene (Zhang et al., 2019). This accumulation of β-carotene, in conjunction with the genetic regulation of ZEP, contributes to the distinct orange hue observed in Brassica crops. In summary, carotenoids play a pivotal role in determining flower coloration in Brassica crops, and of these pigments are intricately involved in the transition among yellow series color.
3.3 Genetic regulation of red series flower in Brassica crops
As is well-established, anthocyanins significantly influence the accumulation of red pigments in plants. In Brassica crops, BnaA07. PAP2 (BnaA07G0287000ZS) has been identified as a key regulator of the orange-red, apricot and red color in B. napus flowers (Jia et al., 2021; Ye et al., 2022; Chen et al., 2023). This gene modulates anthocyanin biosynthesis by inducing the expression of Bna. DFR and Bna. ANS, and the combined accumulation of anthocyanins and carotenoids in petals leads to a transition of flower color from orange to red. Knockout of BnaA07. PAP2 disrupts anthocyanin synthesis, resulting in a shift in petal color from orange-red to yellow (Jia et al., 2021). Besides, it has been reported that the key genes of anthocyanin biosynthesis, ANS, DFR, and UF3GT, are significantly more highly expressed in red and apricot petals compared to white and yellow petals. The RNA interference of BnaA03. ANS would alter petal colors from raspberry red to beige red and zinc yellow under different interference levels (Hao et al., 2022). The expression of BnaA10.F3′H (BnaA10g23330D) affects the synthesis of downstream peonidin and delphinidin and is a key gene regulating the purple color of petals in B. napus (Li et al., 2023). Despite these advancements, the precise molecular mechanism underlying the formation of red flowers remains incompletely understood. Future studies are needed to further elucidate the complex interactions between anthocyanins and other genetic and environmental factors that govern flower coloration in Brassica crops.
3.4 Carotenoids and anthocyanins contribute to the flower color diversity of Brassica crops
It is confirmed that carotenoids are the key pigments for yellow series flowers, whereas anthocyanins promote the formation of red series flower in Brassica crops. Manipulating the endogenous pigment levels within cells can facilitate transitions in petal color, ranging from white to yellow or red. The total carotenoids content is a culmination of biosynthesis, degradation and stable storage processes (Yang et al., 2019). While several factors involved in these processes have been verified as regulators of the petal color of Brassica crops, the intricate molecular mechanisms underlying the flower color formation through the flavonoids biosynthesis pathway in Brassica crops remain largely enigmatic.
Transcription factors (TFs) play a pivotal role in orchestrating the expression of structural genes involved in carotenoid and flavonoid biosynthesis, thus influencing flower color (Hopkins and Rausher, 2011). MYB, WD40, and bHLH are three key transcription factor families that has reported to significantly affected petal color formation (Ramsay and Glover, 2005). Notably, an R2R3-MYB transcription factor encoding gene, BnaA07. PAP2 was cloned through map-based cloning strategy, and the introduction of BnaA07. PAP2In-184-317 into yellow-flowered B. napus triggered a widespread transcriptional activation of the anthocyanin biosynthetic pathway, ultimately resulted in apricot flowers (Ye et al., 2022). Moreover, two additional genes, a MYB (BnaMYBL2) and a bHLH (BnaTT8) gene, exhibited similar expression pattern of BnaA07. PAP2 during flower developments in different-colored petals. This consistency underscores the importance of MYB and bHLH in regulating anthocyanin-based color formation in Brassica (Hao et al., 2022). The carotenoid biosynthetic pathway is regulated primarily by a diverse array of TF families, including R2R3-MYB, MADS-box, NAC, bHLH, SBP-box, AP2/ERF, HD-ZIP, NF-Y, and WRKY (Stanley and Yuan, 2019). Utilizing multi-omics approach, amounts of transcription factors involved in regulating carotenoid metabolism have been identified which showed dramatical differential expression patterns between white-flowered and yellow-flowered petals across all development stages (Jia et al., 2021; Zhang et al., 2022). Among these genes, six TFs, including BnMYB106 (BnaCnng29120D), BnMADS (BnaC02g00490D), BnHD-ZIP (BnaA09g18250D, BnaCnng02160D), BnNFYA1 (BnaA03g04040D), and BnWRKY22 (BnaCnng02000D), showed expression patterns that align with white flower gene BnNCED4 (Jia et al., 2021). Similarly, the expressive pattern of two key TF-encoding genes, Bo2g151880 (WRKY) and Bo3g024180 (SBP) were consistent with the BoCCD4 (Zhang et al., 2022). This suggests that these TFs may interact with CCD4 to jointly regulate carotenoid biosynthesis in B. napus and B. oleracea respectively, but the interaction between them remain to be explored. Collectively, these studies provides new insights into the molecular regulatory mechanism of carotenoids and flavonoids underlying petal color formation in Brassica crops.
Notably, recent studies also revealed an intriguing interaction between carotenoids and anthocyanins, resulting in the production of red flowers in B. napus (Liu et al., 2020; Hao et al., 2022). Previous studies reported that most of the epicatechin, quercetin, and isorhamnetin derivates were found in red and pink petals of B. napus, while kaempferol derivates were in yellow and pale white petals (Yin et al., 2019). And the metabolome analysis has further elucidated that apricot coloration resulted from the accumulation of yellow lutein and red anthocyanins, whereas pink color arises from the combined presence of colorless carotene and red anthocyanins (Ye et al., 2022). These findings suggest that the interplay between the carotenoids and flavonoids biosynthesis pathways significantly impacts flower color formation in Brassica crops. Thus, manipulating the abundance ratio of key carotenoids and flavonoids, including anthocyanins, through the regulation of involved genes or pH-regulating genes, may serve as a promising strategy for the creation of novel flower colors in these crops.
4 The genetic mechanisms underlying the evolution of yellow and white flower colors in Brassica crops
As is well known, the A, B and C genomes of Brassica are closely related, exhibiting high synteny within groups, particularly between A and C genomes (Prakash et al., 2009). In species sharing the A genome, the yellow flower phenotype is typically dominant over the white flower. Conversely, in species harbor the C genome, the white flower prevails. The similarity in genetic pattern of flower colors among Brassica crops may suggests a conserved evolution of key regulatory genes. For instance, the white-flower controlling gene CCD4 has been cloned on chromosome C03 of B. oleracea, B. napus and B. carinata, the fatty acid phytyl-ester function of A02. PES2 is conserved in B. rapa, B. napus and B. juncea, and their homologous genes on B or C genome are functional, jointly contributing to the white flower formation. These observations point to the varying genetic mechanisms underlying flower color evolution in Brassica crops as being closely tied to the evolutionary history of these species. Further studies into the genetic mechanisms driving these patterns could provide valuable insights into the molecular basis of flower color evolution in Brassica and potentially other plant species.
B. napus as an allopolyploid originated from the hybridization of B. rapa with B. oleracea exhibits a more complex genetic basis for flower colors. The early reported petal color mutations including natural or artificial synthesized in B. napus is white, which was supposed to be transferred from white cabbages (Heyn, 1977; Chen et al., 1988; Qi and Fu, 1992). Zhang et al. (2002) made the crosses between the yellow-petalled B. rapa with white-petalled B. oleracea and cream–yellow-petalled B. oleracea to produce resynthesized B. napus (2n = 38, AACC or CCAA) and sesquidiploids (2n = 29, AAC or CAA). The petal color parameters of different genome compositions revealed that the C-genome white petal gene was partially epistatic over the A-genome yellow petal gene. The degree of white color increases with an increased dosage of C-genome and with the presence of the B. oleracea cytoplasm. Currently, the formation of yellow and white flower of B. napus is one of the most extensively studied topics, and several key genes have been cloned from both A and C genomes. Consequently, the evolutionary relationship between yellow and white flower colors in B. rapa, B. oleracea, and B. napus has been established. For instance, Zhang et al. (2015) cloned the white flower gene BnaC3. CCD4 and identified four variations (M1-M4) in Brassica crops, which destroyed the gene function resulting in yellow-flowered lines. All of the four mutation types presented in B. oleracea with yellow flowers, but only M1 type existed in B. napus, and only M1 type and M4 type existed in B. carinata. This might suggest that the white flower gene mutated originally in B. oleracea and was subsequently preserved in interspecific hybrids, leading to yellow flowers of B. oleracea, B. napus and B. carinata, which has been confirmed in the research of Han et al. (2019). Collectively, in consideration of the inheritance patterns of yellow and white flowers of Brassica crops, it is likely that the yellow B. rapa and white B. oleracea might be the ancestral type, and in the origin center of B. napus, the yellow B. rapa crossed with the yellow-flowered B. oleracea, hence the yellow B. napus formed. In summary, the significance of considering genetic relationships and evolutionary history becomes evident when examining phenotypic traits in Brassica crops. Future research should further explore the genetic mechanisms controlling flower color and their impact on crop evolution and domestication.
5 Innovation and multiple utilization of Brassica crops with colorful flowers
The variations in flower color among Brassica crops primarily arise from natural variation, artificial mutagenesis, interspecific hybridization, and genetically modified (Li et al., 2022). Among these methods, interspecific hybridization is the most commonly employed for enhancing crop characteristics, and the target flower colors could be transferred into other Brassica crops (Li et al., 2023). For example, there are no red petals in natural variation of rapeseed but radish does. The rape accessions with orange-red flowers were obtained from the high generation backcross progenies of B. napus and the wide hybrids between B. napus and Raphanus sativus L. (Chen et al., 2023). However, it is noted that wide hybridization can also result in the unintentional transfer of undesirable traits, attributed to linkage drag. To address this, transgenics and gene editing techniques can be employed to achieve directional improvement in flower color. White flowers have been innovated by knocking out two homologous of BraA02.PES2-2 in the yellow-flowered B. napus with the CRISPR/Cas9 system (Guan et al., 2023). Furthermore, the introduction of specific metabolic enzymes, controlled by a flower-specific promoter, can create novel color phenotypes without compromising other traits, as demonstrated by Zhu et al. (2010). For instance, the ectopic overexpressing of OvPAP2 gene driven by the petal-specific promoter XY355 leads to red anthers and red petals in B. napus (Fu et al., 2018).
Novel flower colors in Brassica crops play a pivotal role in enhancing their popularization and application. Flower color serves as a key morphological marker in Brassica crops. Due to the color variations on leaves, stems and petals, Brassica crops have been used as ornamental plants in gardens and landscapes, as well as cut flowers in pots and containers. The increasing demand for visually appealing plants underscores the need to cultivate more Brassica crops with colorful flowers. Moreover, Brassica crops, particularly those with colorful petals, are rich in nutrients and bioactive compounds, including carotenoids, flavonoids, vitamins, minerals, and antioxidants, et al. These compounds make them a valuable source of natural nutrients and health-promoting compounds in functional foods, dietary supplements, and nutraceutical products (Zayed et al., 2022). Besides, these crops may also exhibit resistance to environmental stress (Verma et al., 2023). Studies have shown that certain Brassica species can accumulate heavy metals and other contaminants from soil and water (Zhang et al., 2023). Planting these crops in contaminated areas could provide a sustainable and cost-effective method for removing harmful substances from the environment. The unique aesthetic appeal, nutritional content, and environmental remediation potential of Brassica crops with colorful petals highlight their significance in modern horticulture and agriculture.
6 Conclusion and perspective
Flower color, a key morphological trait, is governed by multiple loci with complex genetic basis in Brassica crops. We outline the recent progress of different flower colors formation and regulation of six representative Brassica crops. These crops have a closely genetic relationship, which can help to elucidate the genetic mechanisms of flower color formation and evolution. It is found that carotenoids are the basic pigments which control the common colors of Brassica crops, yellow and white series color of petals, and anthocyanins promote the formation of red series flowers. Based on the cloned key genes involving in the carotenoids and anthocyanins anabolic pathways, it is possible to create more colorful resources with biotechnological methods and approaches, such as hybridization, transgenics and gene editing. Thereby, it can further increase the application value and economic value of Brassica crops. To gain deeper understanding of flower color evolution, we delve into the genetic relationship among B. rapa, B. oleracea and B. napus. By analyzing the inheritance patterns within the A and C genomes, we postulate that the yellow-flowered B. rapa and white-flowered B. oleracea might be the ancestral types. By the way, there are relatively few studies on the flower color formation and evolution of B. nigra and B. carinata that sharing the B genome.
Flower color, easily observable without specialized instrumentation or reagents, serves as a convenient morphological marker with diverse applications in plant breeding. It is frequently used to assess the natural outcross rate of varieties, verify the purity of parents and hybrids in heterosis utilization systems, and as a linkage marker for targeted breeding traits (Yin and Guan, 2013). A successful selection and breeding of an excellent restorer line with orange flowers, facilitated by the close linkage of molecular markers to the orange flower color genes Bnpc1 and Bnpc2, has demonstrated its utility. This novel flower color enables seed purity reach to 98% during seed production (Zhang, 2021). Given the limited number of flower color-controlling genes reported, it would be highly beneficial to integrate these genes with other superior agronomic trait-related genes during breeding screening in the future. These strategies hold promise for the development of Brassica crops with enhanced performance and reduced environmental risk. In conclusion, the exploration of flower color genetics and its application in Brassica crops offers promising avenues for future research, with potential to enhance breeding efficiency, economic value, and crop resilience.
Author contributions
XL: Conceptualization, Formal Analysis, Investigation, Writing–original draft, Writing–review and editing. MZ: Data curation, Investigation, Writing–review and editing. QG: Data curation, Investigation, Writing–review and editing. JL: Data curation, Investigation, Writing–review and editing. HF: Writing–review and editing, Supervision, Validation. XW: Conceptualization, Formal Analysis, Funding acquisition, Project administration, Writing–review and editing. ZG: Conceptualization, Formal Analysis, Writing–original draft, Writing–review and editing, Project administration, Supervision.
Funding
The authors declare financial support was received for the research, authorship, and/or publication of this article. This work is funded by China Agriculture Research System of MOF and MARA (CARS-21), the China Postdoctoral Science Foundation (2020M682440), The Top-notch Talent Postdoctor Introduction Program of Hubei Province and the Postdoctoral Fellowship Program of CPSF (GZB20230825).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2024.1396875/full#supplementary-material
References
Appelhagen, I., Thiedig, K., Nordholt, N., Schmidt, N., Huep, G., Sagasser, M., et al. (2014). Update on transparent testa mutants from Arabidopsis thaliana: characterisation of new alleles from an isogenic collection. Planta 240 (5), 955–970. doi:10.1007/s00425-014-2088-0
Ariizumi, T., Kishimoto, S., Kakami, R., Maoka, T., Hirakawa, H., Suzuki, Y., et al. (2014). Identification of the carotenoid modifying gene PALE YELLOW PETAL 1 as an essential factor in xanthophyll esterification and yellow flower pigmentation in tomato (Solanum lycopersicum). Plant J. 79, 453–465. doi:10.1111/tpj.12570
Behrens, C. E., Smith, K. E., Iancu, C. V., Choe, J. Y., and Dean, J. V. (2019). Transport of anthocyanins and other flavonoids by the Arabidopsis ATP-binding cassette transporter AtABCC2. Sci. Rep. 9 (1), 437. doi:10.1038/s41598-018-37504-8
Bombarely, A., Moser, M., Amrad, A., Bao, M., Bapaume, L., Barry, C. S., et al. (2016). Insight into the evolution of the Solanaceae from the parental genomes of Petunia hybrida. Nat. Plants 2 (6), 16074. doi:10.1038/NPLANTS.2016.74
Chen, B., Heneen, W., and Jönsson, R. (1988). Independent inheritance of erucic acid content and flower colour in the C-genome of Brassica napus L. Plant Breed. 100, 147–149. doi:10.1111/j.1439-0523.1988.tb00230.x
Chen, D., Jin, Q., Pan, J., Liu, Y., Tang, Y., E, Y., et al. (2023). Fine mapping of genes controlling pigment accumulation in oilseed rape (Brassica napus L.). Mol. Breed. 43 (3), 19. doi:10.1007/s11032-023-01365-5
Chiu, L., and Li, L. (2012). Characterization of the regulatory network of BoMYB2 in controlling anthocyanin biosynthesis in purple cauliflower. Planta 236 (4), 1153–1164. doi:10.1007/s00425-012-1665-3
Clegg, M. T., and Durbin, M. L. (2000). Flower color variation: a model for the experimental study of evolution. Proc. Natl. Acad. Sci. U. S. A. 97 (13), 7016–7023. doi:10.1073/pnas.97.13.7016
Dai, S. L., and Hong, Y. (2016). Molecular breeding for flower colors modification on ornamental plants based on the mechanism of anthocyanins biosynthesis and coloration. Sci. Agric. Sin. 49 (3), 529–542. doi:10.3864/j.issn.0578-1752.2016.03.011
Dan, Y. B. (2016). “Primary mapping of the orange flower gene and central leaf color gene in Brassica napus L,” (Xining: Qinghai University). Master's thesis.
Ding, G., Chen, L., Zou, X., Li, S., Xiong, J., Zou, X., et al. (2019). QTL-seq genetic analysis and InDel marker development of orange petal color gene in Brassica napus. Mol. Plant Breed. 17, 3983–3992. (in Chinese with English abstract). doi:10.13271/j.mpb.017.003983
Fu, W., Chen, D., Pan, Q., Li, F., Zhao, Z., Ge, X., et al. (2018). Production of red-flowered oilseed rape via the ectopic expression of Orychophragmus violaceus OvPAP2. Plant Biotechnol. J. 16 (2), 367–380. doi:10.1111/pbi.12777
Getinet, A., Rakow, G., and Downey, R. K. (1993). Inheritance of a cream petal mutant in Ethiopian mustard. Can. J. Plant Sci. 73, 1075–1076. doi:10.4141/cjps93-140
Grotewold, E. (2006). The genetics and biochemistry of floral pigments. Annu. Rev. Plant Biol. 57, 761–780. doi:10.1146/annurev.arplant.57.032905.105248
Guan, Z., Li, X., Yang, J., Zhao, J., Wang, K., Hu, J., et al. (2023). The mechanism of white flower formation in Brassica rapa is distinct from that in other Brassica species. Theor. Appl. Genet. 19 (6), 133. doi:10.1007/s00122-023-04344-8
Guo, Q. Q., Zhou, R., Chen, X., Chen, L., Li, T. N., and Wang, R. (2021). Location and InDel markers for candidate interval of the orange petal gene in <italic&gt;Brassica napus&lt;/italic&gt; L. by next generation sequencing. Acta Agron. Sin. 47 (11), 2163–2172. doi:10.3724/SP.J.1006.2021.04236
Han, F., Cui, H., Zhang, B., Liu, X., Yang, L., Zhuang, M., et al. (2019). Map-based cloning and characterization of BoCCD4, a gene responsible for white/yellow petal color in B. oleracea. BMC Genomics 20, 242. doi:10.1186/s12864-019-5596-2
Hao, P., Liu, H., Lin, B., Ren, Y., Huang, L., Jiang, L., et al. (2022). BnaA03ANS identified by metabolomics and RNA-seq partly played irreplaceable role in pigmentation of red rapeseed (Brassica napus) petal. Front. Plant Sci. 13, 940765. doi:10.3389/fpls.2022.940765
Harborne, J. B. (1986). Nature, distribution and function of plant flavonoids. Prog. Clin. Biol. Res. 213 (213), 15–24.
He, J., and Giusti, M. M. (2010). Anthocyanins: natural colorants with health-promoting properties. Annu. Rev. Food Sci. Technol. 1, 163–187. doi:10.1146/annurev.food.080708.100754
Hondo, T., Yoshida, K., Nakagawa, A., Kawai, T., Tamura, H., and Goto, T. (1992). Structural basis of blue-colour development in flower petals from Commelina communis. Nature 358 (6386), 515–518. doi:10.1038/358515a0
Hopkins, R., and Rausher, M. D. (2011). Identification of two genes causing rein-forcement in the Texas wildflower Phlox drummondii. Nature 469, 411–414. doi:10.1038/nature09641
Hornero-Méndez, D., and Mínguez-Mosquera, M. I. (2000). Carotenoid pigments in Rosa mosqueta hips, an alternative carotenoid source for foods. J. Agric. Food Chem. 48, 825–828. doi:10.1021/jf991136n
Huang, M., Zhang, J., Chen, P., Song, Y., and Shun, H. (2017). The inheritance of white flower characters in Brassica napus L. Jiangsu Agric. Sci. 45 (20), 83–84. doi:10.15889/j.issn.1002-1302.2017.20.018
Huang, Z., Ban, Y., Bao, R., Zhang, X., Xu, A., and Ding, J. (2014). Inheritance and gene mapping of the white flower in Brassica napus L. New Zeal J. Crop Hort. 42, 111–117. doi:10.1080/01140671.2013.863211
Jahns, P., and Holzwarth, A. R. (2012). The role of the xanthophyll cycle and of lutein in photoprotection of photosystem II. BBA-Bioenergetics. 1817, 182–193. doi:10.1016/j.bbabio.2011.04.012
Jambhulkar, S. J., and Raut, R. N. (1955). Inheritance of flower colour and leaf waxiness in Brassica carinata A. Br. Crucif. Newslett. 17, 66–67.
Jia, L. D., Wang, J. S., Wang, R., Duan, M. Z., Qiao, C. L., Chen, X., et al. (2021). Comparative transcriptomic and metabolomic analyses of carotenoid biosynthesis reveal the basis of white petal color in Brassica napus. Planta 253, 8. doi:10.1007/s00425-020-03536-6
Jiang, N. N. (2021). “Preliminary localization of red flower genes and screening of candidate genes in Brassica napus,” (Nanchang: Jiangxi Agricultural University). Master's thesis.
Jin, Q., Bi, Y., Liu, X., and Wan, F. (2014). Recent advances on research of carotenoid metabolism and functions. Chinese Journal of Animal Nutrition 26 (12), 3561–3571. doi:10.3969/j.issn.1006-267x.2014.12.003
Kebede, B., and Rahman, H. (2014). Quantitative trait loci (QTL) map** of silique length and petal colour in Brassica rapa. Plant Breeding 133 (5), 604–614. doi:10.1111/pbr.12193
Koes, R., Verweij, W., and Quattrocchio, F. (2005). Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 10 (5), 236–242. doi:10.1016/j.tplants.2005.03.002
Li, F., Murillo, C., and Wurtzel, E. T. (2007). Maize Y9 encodes a product essential for 15-cis-δ-carotene isomerization. Plant Physiol. 144, 1181–1189. doi:10.1104/pp.107.098996
Li, F. Y., Gong, Y. Y., Mason, A. S., Liu, Q., Huang, J., Ma, M., et al. (2023). Research progress and applications of colorful Brassica crops. Planta 258 (2), 45. doi:10.1007/s00425-023-04205-0
Li, L., and Yuan, H. (2013). Chromoplast biogenesis and carotenoid accumulation. Arch. Biochem. Biophys. 539, 102–109. doi:10.1016/j.abb.2013.07.002
Li, P., Zhang, S., Zhang, S., Li, F., Zhang, H., Cheng, F., et al. (2015). Carotenoid biosynthetic genes in Brassica rapa: comparative genomic analysis, phylogenetic analysis, and expression profiling. BMC Genomics 16, 492. doi:10.1186/s12864-015-1655-5
Li, R., Zeng, Q., Zhang, X., Jing, J., Ge, X., Zhao, L., et al. (2023). Xanthophyll esterases in association with fibrillins control the stable storage of carotenoids in yellow flowers of rapeseed (Brassica juncea). New Phytol. 240 (1), 285–301. doi:10.1111/nph.18970
Li, S. (2014). Transcriptional control of flavonoid biosynthesis: fine-tuning of the MYB-bHLH-WD40 (MBW) complex. Plant Signal Behav. 9 (1), e27522. doi:10.4161/psb.27522
Li, S., Chang, T., Li, X., Peng, Z., Guan, C., and Guan, M. (2022). Regulatory mechanisms of rapeseed petal color formation: current research status and future perspectives. Oil Crop Sci. 7 (4), 174–179. doi:10.1016/j.ocsci.2022.11.005
Li, S., Li, X., Wang, X., Chang, T., Peng, Z., Guan, C., et al. (2023). Flavonoid synthesis-related genes determine the color of flower petals in Brassica napus L. Int. J. Mol. Sci. 30 (7), 6472. doi:10.3390/ijms24076472
Liu, X. P., Tu, J. X., Chen, B. Y., and Fu, T. D. (2004). Identification of the linkage relationship between the flower colour and the content of erucic acid in the resynthesized Brassica napus L. Yi Chuan Xue Bao. 31, 357–362.
Liu, Y., Ye, S., Yuan, G., Ma, X., Heng, S., Yi, B., et al. (2020). Gene silencing of BnaA09.ZEP and BnaC09.ZEP confers orange color in Brassica napus flowers. Plant J. 104, 932–949. doi:10.1111/tpj.14970
Maoka, T. (2020). Carotenoids as natural functional pigments. J. Nat. Med. 74 (1), 1–16. doi:10.1007/s11418-019-01364-x
Marinova, K., Pourcel, L., Weder, B., Schwarz, M., Barron, D., Routaboul, J. M., et al. (2007). The Arabidopsis MATE transporter TT12 acts as a vacuolar flavonoid/H+ -antiporter active in proanthocyanidin-accumulating cells of the seed coat. Plant Cell 19 (6), 2023–2038. doi:10.1105/tpc.106.046029
Nagaharu, U. (1935). Genomic analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jpn. J. Bot. 7, 389–452.
Neuman, H., Galpaz, N., Cunningham, F. X., Zamir, D., and Hirschberg, J. (2014). The tomato mutation nxd1 reveals a gene necessary for neoxanthin biosynthesis and demonstrates that violaxanthin is a sufficient precursor for abscisic acid biosynthesis. Plant J. 78 (1), 80–93. doi:10.1111/tpj.12451
Niaz, M., Zhang, B., Zhang, Y., Yan, X., Yuan, M., Cheng, Y., et al. (2023). Genetic and molecular basis of carotenoid metabolism in cereals. Theor. Appl. Genet. 136 (3), 63. doi:10.1007/s00122-023-04336-8
Ohmiya, A. (2013). Qualitative and quantitative control of carotenoid accumulation in flower petals. Sci. Hortic. 163, 10–19. doi:10.1016/j.scienta.2013.06.018
Ohmiya, A., Kishimoto, S., Aida, R., Yoshioka, S., and Sumitomo, K. (2006). Carotenoid cleavage dioxygenase (CmCCD4a) contributes to white color formation in chrysanthemum petals. Plant Physiol. 142, 1193–1201. doi:10.1104/pp.106.087130
Park, H., Kreunen, S. S., Cuttriss, A. J., DellaPenna, D., and Pogson, B. J. (2002). Identification of the carotenoid isomerase provides insight into carotenoid biosynthesis, prolamellar body formation, and photomorphogenesis. Plant Cell 14, 321–332. doi:10.1105/tpc.010302
Pearson, O. H. (1929). A dominant white flower color in Brassica oleracea L. Am. Nat. 63, 561–565. doi:10.1086/280291
Prakash, S., Bhat, S. R., Quiros, C. F., and Kirti, P. (2009). “Brassica and its close allies:cytogenetics and evolution,” in Plant breeding reviews (Hoboken, NJ, USA: John Wiley & Sons, Inc.). doi:10.1002/9780470593783
Qi, C. K., and Fu, S. Z. (1992). Inheritance of white flower traits in brassica napus. Chin. J. Oil Crop Sci. (03), 60–62.
Rahman, M. H. (2001). Inheritance of petal colour and its independent segregation from seed colour in Brassica rapa. Plant Breed. 120, 197–200. doi:10.1046/j.1439-0523.2001.00607.x
Ramsay, N. A., and Glover, B. J. (2005). MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 10, 63–70. doi:10.1016/j.tplants.2004.12.011
Rawat, D. S., and Anand, I. J. (1986). Inheritance of flower colour in mustard mutant. Indian J. Agr. Sci.
Ruiz-Sola, M. Á., and Rodríguez-Concepción, M. (2012). Carotenoid biosynthesis in Arabidopsis: a colorful pathway. Arab. Book 10, e0158. e0158.749. doi:10.1199/tab.0158
Saito, K., Yonekura-Sakakibara, K., Nakabayashi, R., Higashi, Y., Yamazaki, M., Tohge, T., et al. (2013). The flavonoid biosynthetic pathway in Arabidopsis: structural and genetic diversity. Plant Physiol. Biochem. 72, 21–34. doi:10.1016/j.plaphy.2013.02.001
Schweiggert, R. M., Mezger, D., Schimpf, F., Steingass, C. B., and Carle, R. (2012). Influence of chromoplast morphology on carotenoid bioaccessibility of carrot, mango, papaya, and tomato. Food Chem. 135 (4), 2736–2742. doi:10.1016/j.foodchem.2012.07.035
Shen, N., Wang, T., Gan, Q., Liu, S., Wang, L., and Jin, B. (2022). Plant flavonoids: classification, distribution, biosynthesis, and antioxidant activity. Food Chem. 383, 132531. doi:10.1016/j.foodchem.2022.132531
Stanley, L., and Yuan, Y. W. (2019). Transcriptional regulation of carotenoid biosynthesis in plants: so many regulators, so little consensus. Front. Plant Sci. 10, 1017. doi:10.3389/fpls.2019.01017
Sun, Y., Li, H., and Huang, J. R. (2012). Arabidopsis TT19 functions as a carrier to transport anthocyanin from the cytosol to tonoplasts. Mol. Plant 5 (2), 387–400. doi:10.1093/mp/ssr110
Tian, L., Magallanes-Lundback, M., Musetti, V., and DellaPenna, D. (2003). Functional analysis of beta- and epsilon-ring carotenoid hydroxylases in Arabidopsis. Plant Cell 15 (6), 1320–1332. doi:10.1105/tpc.011403
Tian, L., Musetti, V., Kim, J., Magallanes-Lundback, M., and DellaPenna, D. (2004). The Arabidopsis LUT1 locus encodes a member of the cytochrome P450 family that is required for carotenoid ε-ring hydroxylation activity. Proc. Natl. Acad. Sci. U. S. A. 101, 402–407. doi:10.1073/pnas.2237237100
Tian, L. S. (2011). “Genetic analysis, cloning and characterization of the related genes for allogenous white flower character in Brassica napus L,” (Sichuan: Sichuan Agricultural University). Master's thesis.
Verma, T., Bhardwaj, S., Raza, A., Djalovic, I., Prasad, P., and Kapoor, D. (2023). Mitigation of salt stress in Indian mustard (Brassica juncea L.) by the application of triacontanol and hydrogen sulfide. Plant Signal Behav. 18 (1), 2189371. doi:10.1080/15592324.2023.2189371
Walter, M. H., and Strack, D. (2011). Carotenoids and their cleavage products: biosynthesis and functions. Nat. Prod. Rep. 28, 663–692. doi:10.1039/c0np00036a
Welsch, R., Zhou, X., Yuan, H., Álvarez, D., Sun, T., Schlossarek, D., et al. (2018). Clp protease and OR directly control the proteostasis of phytoene synthase, the crucial enzyme for carotenoid biosynthesis in Arabidopsis. Mol. Plant 11 (1), 149–162. doi:10.1016/j.molp.2017.11.003
Xiao, M., Wang, H., Li, X., Mason, A., and Fu, D. (2021). Rapeseed as an ornamental. Horticulture 8, 27. doi:10.3390/horticulturae8010027
Xu, X., Luo, W., Guo, J., Chen, H., Akram, W., Xie, D., et al. (2019). Fine mapping and candidate gene analysis of the yellow petal gene ckpc in Chinese kale (Brassica oleracea L. var. alboglabra Bailey) by whole-genome resequencing. Mol. Breed. 39, 96–106. doi:10.1007/s11032-019-1011-6
Yamagishi, M., Kishimoto, S., and Nakayama, M. (2010). Carotenoid composition and changes in expression of carotenoid biosynthetic genes in tepals of Asiatic hybrid lily. Plant Breed. 129 (1), 100–107. doi:10.1111/j.1439-0523.2009.01656.x
Yang, H. R., Liu, J., Li, J. N., Qin, W., and Mei, J. Q. (2019). OTL mapping and variation analysis of candidate gene for flower color in Brassica oleracea L. J. Plant Genet. Resour. 20 (5), 1271–1277. doi:10.13430/j.cnki.jpgr.20181228001
Yang, S., Tian, X., Wang, Z., Wei, X., Zhao, Y., Su, H., et al. (2021). Fine mapping and candidate gene identification of a white flower gene BrWF3 in Chinese cabbage (Brassica rapa L. ssp. pekinensis). Front. Plant Sci. 7 (12), 646222. doi:10.3389/fpls.2021.646222
Yao, Y., Li, K., Liu, H., Duncan, R. W., Guo, S. M., Xiao, L., et al. (2017). Whole-genome re-sequencing and fine mapping of an orange petal color gene (Bnpc1) in spring Brassica napus L. to a 151-kb region. Euphytica 213 (8), 165.
Ye, S., Hua, S., Ma, T., Ma, X., Chen, Y., Wu, L., et al. (2022). Genetic and multi-omics analyses reveal BnaA07.PAP2In-184–317 as the key gene conferring anthocyanin-based color in Brassica napus flowers. J. Exp. Bot. 73 (19), 6630–6645. doi:10.1093/jxb/erac312
Yin, C. F., and Guan, C. Y. (2013). A review of rapaseed flower color. Crop Res. 27 (4), 403–410. doi:10.3969/j.issn.1001-5280.2013.04.27
Yin, N., Wang, S., Jia, L., Zhu, M., Yang, J., Zhou, B., et al. (2019). Identification and characterization of major constituents in different-colored rapeseed petals by UPLC-HESI-MS/MS. J. Agric. Food Chem. 9 (40), 11053–11065. doi:10.1021/acs.jafc.9b05046
Yoshioka, S., Aida, R., Yamamizo, C., Shibata, M., and Ohmiya, A. (2012). The carotenoid cleavage dioxygenase 4 (CmCCD4a) gene family encodes a key regulator of petal color mutation in chrysanthemum. Euphytica 184, 377–387. doi:10.1007/s10681-011-0602-z
Ytterberg, A. J., Peltier, J. B., and van Wijk, K. J. (2006). Protein profiling of plastoglobules in chloroplasts and chromoplasts. A surprising site for differential accumulation of metabolic enzymes. Plant Physiol. 140, 984–997. doi:10.1104/pp.105.076083
Yu, C. Y., Hu, S. W., Zhang, C. H., and Yu, Y. J. (2004). The discovery of a novel flower color mutation in male sterile rapeseed (Brassica napus L.). Hered. 03 26, 330–332. doi:10.1300/J064v24n01_09
Zayed, A., Sheashea, M., Kassem, I. A. A., and Farag, M. (2022). Red and white cabbages: an updated comparative review of bioactives, extraction methods, processing practices, and health benefits. Crit. Rev. Food Sci. Nutr. 63, 7025–7042. doi:10.1080/10408398.2022.2040416
Zeng, D. Z. (2013). “Contents of major carotenoids and expression of the relevant genes in the petals with different color in Brassica napus L,” (Sichuan: Sichuan Agricultural University). Master's thesis.
Zhang, B., Liu, C., Wang, Y., Yao, X., Wang, F., Wu, J., et al. (2015). Disruption of a CAROTENOID CLEAVAGE DIOXYGENASE 4 gene converts flower colour from white to yellow in Brassica species. New Phytol. 206, 1513–1526. doi:10.1111/nph.13335
Zhang, B., Lu, C., Kakihara, F., and Kato, M. (2002). Effect of genome composition and cytoplasm on petal colour in resynthesized amphidiploids and sesquidiploids derived from crosses between Brassica rapa and Brassica oleracea. Plant Breed. 121 (4), 297–300. doi:10.1046/j.1439-0523.2002.722295.x
Zhang, J. F., Pu, H. C., and Qi, C. K. (2000). Inheritance of flower color character in oilseed rape (Brassica napus L.). Chin. J. Oil Crop Sci. 4, 1–4. doi:10.3321/j.issn:1007-9084.2000.03.001
Zhang, N., Chen, L., Ma, S., Wang, R. F., He, Q., Tian, M., et al. (2020). Fine mapping and candidate gene analysis of the white flower gene Brwf in Chinese cabbage (Brassica rapa L.). Sci. Rep. 10, 6080. doi:10.1038/s41598-020-63165-7
Zhang, N., Zhang, H., Ren, Y., Chen, L., Zhang, J., and Zhang, L. (2019). Genetic analysis and gene mapping of the orange flower trait in Chinese cabbage (Brassica rapa L.). Mol. Breed. 39, 76–11. doi:10.1007/s11032-019-0984-5
Zhang, Q., Wang, L., Xiao, Y., Liu, Q., Zhao, F., Li, X., et al. (2023). Migration and transformation of Cd in four crop rotation systems and their potential for remediation of Cd-contaminated farmland in southern China. Sci. Total Environ. 885, 163893. doi:10.1016/j.scitotenv.2023.163893
Zhang, X. X., Li, R., Chen, L., Niu, S. L., Chen, L., Gao, J., et al. (2018a). Fine-mapping and candidate gene analysis of the Brassica juncea white-flowered mutant Bjpc2 using the whole-genome resequencing. Mol. Genet. Genomics 293, 359–370. doi:10.1007/s00438-017-1390-5
Zhang, X. X., Li, R. H., Chen, L., Niu, S. L., Li, Q., Xu, K., et al. (2018b). Inheritance and gene mapping of the white flower trait in Brassica juncea. Mol. Breed. 38, 20–29. doi:10.1007/s11032-017-0771-0
Zhang, Y. (2021). Fine Mapping of Orange Color Gene Bnpc2 and Application of Linkage Marker of Orange Color Gene in Brassica napus L. (Qinghai: Qinghai University).
Zhang, Y., Jin, J., Zhu, S., Sun, Q., Zhang, Y., Xie, Z., et al. (2022). Citrus β-carotene hydroxylase 2 (BCH2) participates in xanthophyll synthesis by catalyzing the hydroxylation of β-carotene and compensates for BCH1 in citrus carotenoid metabolism. Hortic. Res. 10 (3), uhac290. doi:10.1093/hr/uhac290
Zhao, C., Safdar, L., Xie, M., Shi, M., Dong, Z., Yang, L., et al. (2021). Mutation of the PHYTOENE DESATURASE 3 gene causes yellowish white petals in Brassica napus. Crop J. 9, 1124–1134. doi:10.1016/j.cj.2020.10.012
Zhao, D., and Tao, J. (2015). Recent advances on the development and regulation of flower color in ornamental plants. Front. Plant Sci. 6, 261. doi:10.3389/fpls.2015.00261
Zhao, D., Zhou, C., Sheng, Y., Liang, G., and Tao, J. (2011). Molecular cloning and expression of phytoene synthase, lycopene beta-cyclase, and beta-carotene hydroxylase genes in persimmon (Diospyros kaki L.) fruits. Plant Mol. Biol. Rep. 29, 345–351. doi:10.1007/s11105-010-0238-5
Zhao, X., Zhang, Y., Long, T., Wang, S., and Yang, J. (2022). Regulation mechanism of plant pigments biosynthesis: anthocyanins, carotenoids, and betalains. Metabolites 12 (9), 871. doi:10.3390/metabo12090871
Zhao, Y. W., Wang, C. K., Huang, X. Y., and Hu, D. G. (2021). Anthocyanin stability and degradation in plants. Plant Signal Behav. 16 (12), 1987767. doi:10.1080/15592324.2021.1987767
Keywords: flower coloration, Brassica crops, carotenoids, flavonoids, formation, evolution
Citation: Li X, Zheng M, Gan Q, Long J, Fan H, Wang X and Guan Z (2024) The formation and evolution of flower coloration in Brassica crops. Front. Genet. 15:1396875. doi: 10.3389/fgene.2024.1396875
Received: 06 March 2024; Accepted: 13 May 2024;
Published: 30 May 2024.
Edited by:
Guohu Chen, Anhui Agricultural University, ChinaReviewed by:
Jianjun Zhao, Hebei Agricultural University, ChinaZhansheng Li, Chinese Academy of Agricultural Sciences, China
Copyright © 2024 Li, Zheng, Gan, Long, Fan, Wang and Guan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoqing Wang, 19589423@qq.com; Zhilin Guan, guanzhilin@wbgcas.cn
 Xuewei Li
Xuewei Li