- 1Department of Oral Pathology, Peking University School and Hospital of Stomatology and National Center of Stomatology and National Clinical Research Center for Oral Diseases & National Engineering Research Center of Oral Biomaterials and Digital Medical Devices, Beijing, China
- 2Research Unit of Precision Pathologic Diagnosis in Tumors of the Oral and Maxillofacial Regions, Chinese Academy of Medical Sciences (2019RU034), Beijing, China
- 3Department of Pulmonary and Critical Care Medicine, Peking University Third Hospital, Beijing, China
- 4Central Laboratory, Peking University School and Hospital of Stomatology, Beijing, China
- 5National Clinical Research Center for Oral Diseases and National Engineering Laboratory for Digital and Material Technology of Stomatology and Beijing Key Laboratory of Digital Stomatology, Beijing, China
Background: There is inconsistent evidence regarding the accuracy of GNAS mutations identification for the diagnosis of FD/MAS. This study was performed to estimate the prevalence and diagnostic accuracy of GNAS mutations detection and to preliminarily investigate the genotype-phenotype correlation in FD patients.
Methods: Five electronic databases were searched from 1995 to 2024 using search terms related to GNAS and fibrous dysplasia. Observational studies of FD patients undergoing GNAS mutation detection in FD were included.
Results: A total of 878 FD patients were included. The pooled prevalence of GNAS mutations in FD based on the random effects model was 74% (95% CI = 64%–83%). Regarding diagnostic accuracy, a sensitivity of 0.83 (95% CI, 0.65–0.96), specificity of 0.99 (95% CI, 0.98–1.00) and the area under the receiver operating characteristic curve of 98.38% were found. Additionally, meta-analysis and Fisher’s test showed the GNAS mutation types were significantly associated with FD types (OR = 3.51, 95% CI = 1.05 to 11.72; p < 0.05).
Conclusion: A high detection rate of GNAS mutations occurred in FD, and its detection is reliable for diagnosing FD. Additionally, GNAS mutation type was types were significantly associated with FD type.
Systematic Review Registration: Identifier CRD42024553469.
1 Introduction
Fibrous dysplasia (FD) is a rare, genetic but noninheritable bone disorder caused by a postzygotic mutation of GNAS located on chromosome 20q13.3 (Weinstein et al., 1991), which presents in three forms: monostotic FD, which occurs in one bone; polyostotic FD, which involves multiple bones; and McCune–Albright syndrome, which simultaneously contains polyostotic FD, skin hyperpigmentation and hyperfunctioning endocrinopathies (Collins et al., 2012). In this disorder, nonfunctional fibrous tissue with an abnormal quality and structure replaces normal bone tissue, which can cause pain and bone deformities and increase the risk of fracture, and these manifestations initially occur during childhood (Liens et al., 1994).
The GNAS mutations identified in FD patients result in a significantly elevated level of cyclic adenosine monophosphate (cAMP) in FD patients. As a result, this mutation is regarded as the leading cause of FD (Parvanescu et al., 2014). The specific location of the mutation is arginine 201 in exon region 8, which is usually substituted by either histidine (R201H) or cysteine (R201C). However, it is unclear whether different GNAS mutation variants could influence the regulation of GαS activity to different extents. Zhadina et al. found no significant genotype-phenotype correlation for the severity of FD in a retrospective analysis of clinical data obtained as part of a long-standing natural history study (Zhadina et al., 2021). However, the relevant evidence has not been further documented by new studies.
In addition, several studies identified GNAS mutations within FD, but other diseases that need to be distinguished from FD, for example, ossifying fibroma (OF) and low-grade central osteosarcoma (lgc-OSA), do not show GNAS mutations, which indicates the possible role of GNAS mutation analysis in the differential diagnosis of FD. However, without a comprehensive analysis with more data, it is still not clear what the exact prevalence and diagnostic accuracy of GNAS mutations detection are in FD.
In this study, we performed a meta-analysis to evaluate the prevalence and diagnostic accuracy of GNAS mutations detection in FD. We also investigated whether the different GNAS mutation types could be linked with the different FD forms. We hypothesized that GNAS mutation detection could be a very reliable test for FD diagnosis and possesses a very high diagnostic accuracy, especially specificity, and that the R201H mutation type is likely a predominant pathogenic variant in FD. To our knowledge, this study is the largest to date on the subject and the first undertaken to investigate the prevalence, diagnostic accuracy and genotype-phenotype correlation of GNAS mutations in FD to date. This study allows us to understand the prevalence and diagnostic accuracy of GNAS mutations detection in FD, which could help doctors distinguish FD from other diseases requiring differential diagnosis and prioritize variants that cause a greater burden of disease when developing targeted therapy for FD.
2 Materials and methods
2.1 Literature sources and search strategy
This study followed the Meta-analysis of Observational Studies in Epidemiology (MOOSE) guidelines from the screening protocol to the pooled analysis (Stroup et al., 2000) and was registered in PROSPERO (CRD42024553469). The literature search was performed in the PubMed, Web of Science, Cochrane Library and CNKI (China National Knowledge Infrastructure) databases to identify full-text articles up to 1 Jan 2024. The key search items were as follows: fibrous dysplasia, GNAS. There was no language restriction. Furthermore, the citations of potentially relevant studies from the retrieved articles were reviewed. The details of the PubMed and Web of Science search strategies are listed in the Supplement-Appendix as examples. Three independent investigators (Authors 1–3) screened each abstract against the inclusion criteria; conflicts resulted in inclusion at this stage. Full-text records for relevant abstracts were retrieved, and each was independently reviewed by two investigators (Author 1, 2). Two investigators then used standardized forms to extract the study endpoints.
2.2 Inclusion and exclusion criteria
The inclusion criteria for the studies to be considered in this study were as follows: (1) articles related to GNAS mutations in fibrous dysplasia; (2) studies with studies with five samples or >5 samples; and (3) studies that involved human subjects. The rationale behind establishing a threshold for sample size in included studies stems from the potential for a low sample size to introduce significant bias in prevalence estimates. In this study, we opted for “studies with five samples or >5” over a more stringent criterion of “studies with 10 samples or >10” due to our recognition of FD as a rare disease, which poses challenges in acquiring an adequate number of cases.
The exclusion criteria were as follows: (1) case reports, review articles and meeting abstracts; (2) animal studies; (3) studies with fewer than 5 FD cases; and (4) studies that did not provide essential information concerning sequencing methods and tissue sources.
2.3 Quality assessment
Quality assessments of the cross-sectional studies were performed according to the Agency for Healthcare Research and Quality (AHRQ) modified scale for observational studies, which includes 11 items (Rockville, 2008). For the 11 items, two reviewers (Authors 1 and 2) selected on of three options, including Yes, no and unclear. “Yes” was for one point, while “No” and “Unclear” were zero points (Zhou et al., 2021). The total score obtained was for the quality of each study (0–3: low quality; 4–7: moderate quality; 8–11: high quality). Additionally, the risk of bias (ROB) of the included diagnostic studies was determined by the Quality Assessment of Diagnostic Accuracy Studies-2 (QUADAS-2) tool (Whiting et al., 2011).
2.4 Data collection
According to a previously created data collection form, two reviewers (Author 1 and 2) independently extracted the data from the included studies. The following data were collected: the country of the study; authors and year of publication; the sample size of FD patients; incidence of GNAS positive mutation; FD types; FD lesion location; GNAS mutation detection methods; the tissue source of DNA extraction; and GNAS mutation types.
2.5 Statistical methods
Data analysis was performed with R version 4.2.0 and Review Manager 5.4.0. The “meta” (version 6.0, univariate model) and “metadiag4” (version 2.1.1, bivariate model) packages were used to calculate the pooled prevalence and diagnostic accuracy of GNAS mutations, respectively (Leeflang, 2014), and the Review Manager was used to analyze the genotype-phenotype correlation with odds ratios. The data on the prevalence of GNAS mutations were tested for normality. If the test showed a p-value > 0.05, the data were considered to conform to a normal distribution, and there was no need to convert the original prevalence data. However, if the test showed a p-value < 0.05, it was necessary to convert the data using methods such as logarithmic conversion, logit conversion, arcsine conversion and the Freeman-Tukey dual arcsine conversion methods; then, the normality of each conversion was verified in sequence until the converted data followed a standard normal distribution. Statistical heterogeneity was assessed by the I2 test at a = 0.1. A random-effects model was adopted. The causes of heterogeneity were explored by meta-regression. We performed a subgroup analyses according to sample size, detection methods and source of DNA acquisition. The associations between genotypes and phenotypes were evaluated by Fisher’s exact test and meta-analysis. The statistical significance of the hypothesis test was set at p < 0.05, and 95% CIs were calculated using the Clopper-Pearson method. Forest plots were used to show pooled outcomes, and a summary receiver operating characteristic curve (SROC) was generated using a Bayesian bivariate hierarchical model.
3 Results
3.1 Characteristics of the included studies
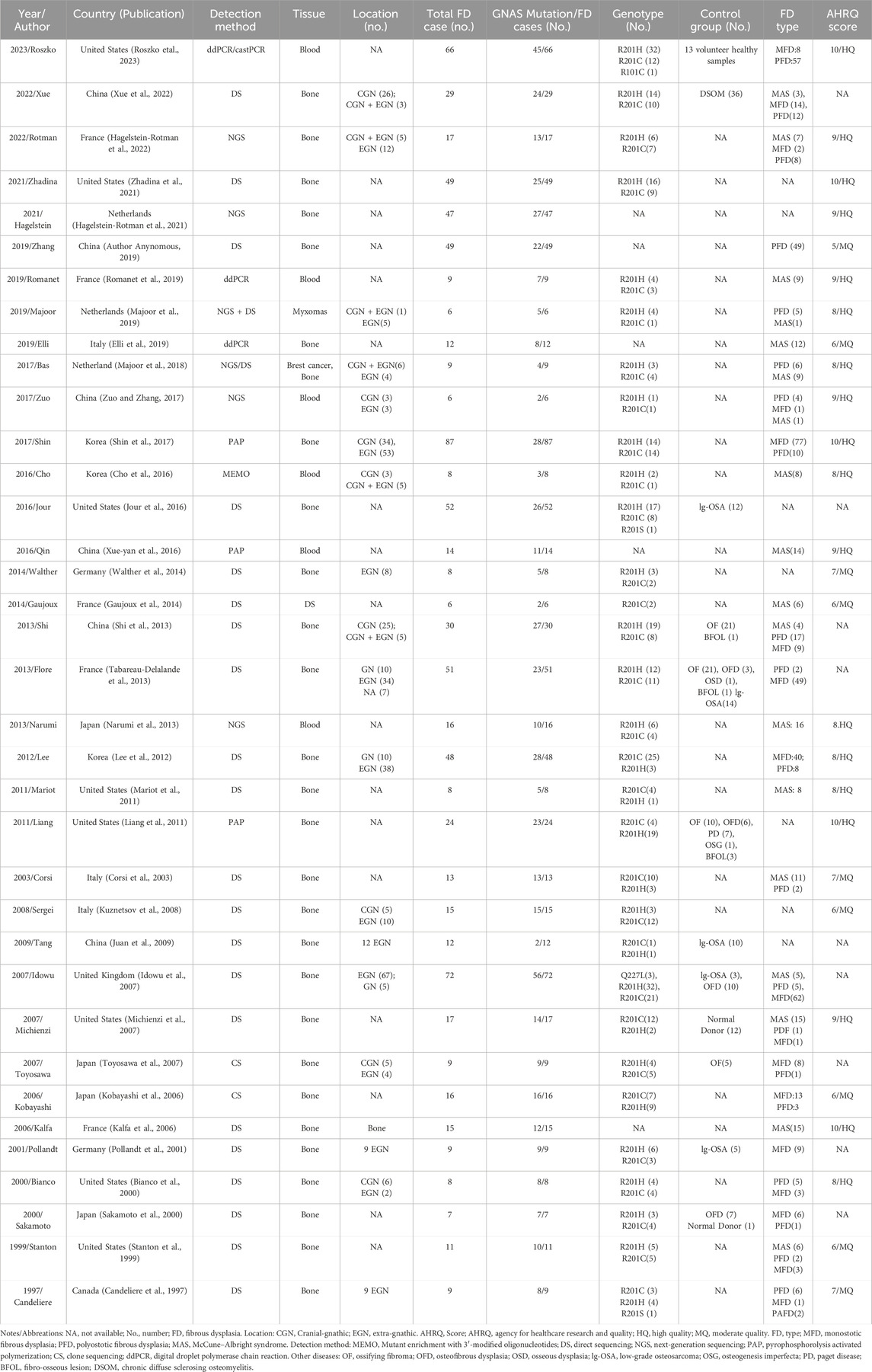
The literature search flow chart is shown in Figure 1. A total of 901 studies were obtained from five electronic databases (PubMed, 170; Cochrane, 22; Web of Science, 450; Embase, 250; CNKI, 9), of which 395 were duplicates. Then, 470 articles were excluded because they did not meet the inclusion criteria (Figure 1). Finally, 36 studies containing 878 FD patients were included in this meta-analysis after reading the full texts (Candeliere et al., 1997; Stanton et al., 1999; Bianco et al., 2000; Sakamoto et al., 2000; Pollandt et al., 2001; Corsi et al., 2003; Kalfa et al., 2006; Kobayashi et al., 2006; Idowu et al., 2007; Michienzi et al., 2007; Toyosawa et al., 2007; Kuznetsov et al., 2008; Juan et al., 2009; Liang et al., 2011; Mariot et al., 2011; Lee et al., 2012; Narumi et al., 2013; Shi et al., 2013; Tabareau-Delalande et al., 2013; Gaujoux et al., 2014; Walther et al., 2014; Cho et al., 2016; Jour et al., 2016; Xue-yan et al., 2016; Shin et al., 2017; Zuo and Zhang, 2017; Majoor et al., 2018; Author Anynomous, 2019; Elli et al., 2019; Majoor et al., 2019; Romanet et al., 2019; Hagelstein-Rotman et al., 2021; Zhadina et al., 2021; Hagelstein-Rotman et al., 2022; Xue et al., 2022; Roszko et al., 2023). Twenty-three studies employed conventional sequencing methods (CS), 21 of which of studies utilized direct sequencing and 2 of which studies employed clone sequencing. The remaining studies adopted modified sequencing approaches (MS), including next-generation sequencing (NGS, six studies), pyrophosphorolysis activated polymerization (PAP, three studies), and advanced PCR technology (four studies). In terms of DNA acquisition sources, 28 studies extracted DNA from bone tissue, six studies obtained DNA from blood, and two studies retrieved DNA from myxoma in patients with FD. All included articles were published from 1997 to 2023 and in English, except for three articles in Chinese. The characteristics of each study are presented in Table 1, and the AHRQ and QUADAS-2 quality assessments of the included studies are shown in the last column in Table 1 and Figure 2, respectively.

Figure 2. Risk of bias and applicability concerns for diagnostic studies on summary (A) and each included study (B) plots.
3.2 Quantitative synthesis
3.2.1 Prevalence of GNAS mutations in FD
The GNAS mutation data for FD patients were available for 36 studies (n = 878 patients). The overall pooled prevalence of GNAS mutations in FD patients based on the random effects model was 74% (95% CI = 64%–83%, Figure 3), with a high heterogeneity (I2 = 87%, p < 0.01). The observed heterogeneity can be attributed to two factors, namely, diverse sample types for DNA acquisition and distinct sequencing methods. To address this, a subgroup analysis was conducted based on the sample types and sequencing methods. This analysis revealed four subgroups: conventional sequencing with bone tissues (CS + Bone), modified sequencing with bone tissue (MS + Bone), modified sequencing with blood (MS + Blood), and multiple sequencing methods with tumor tissue (MT + Tumor).
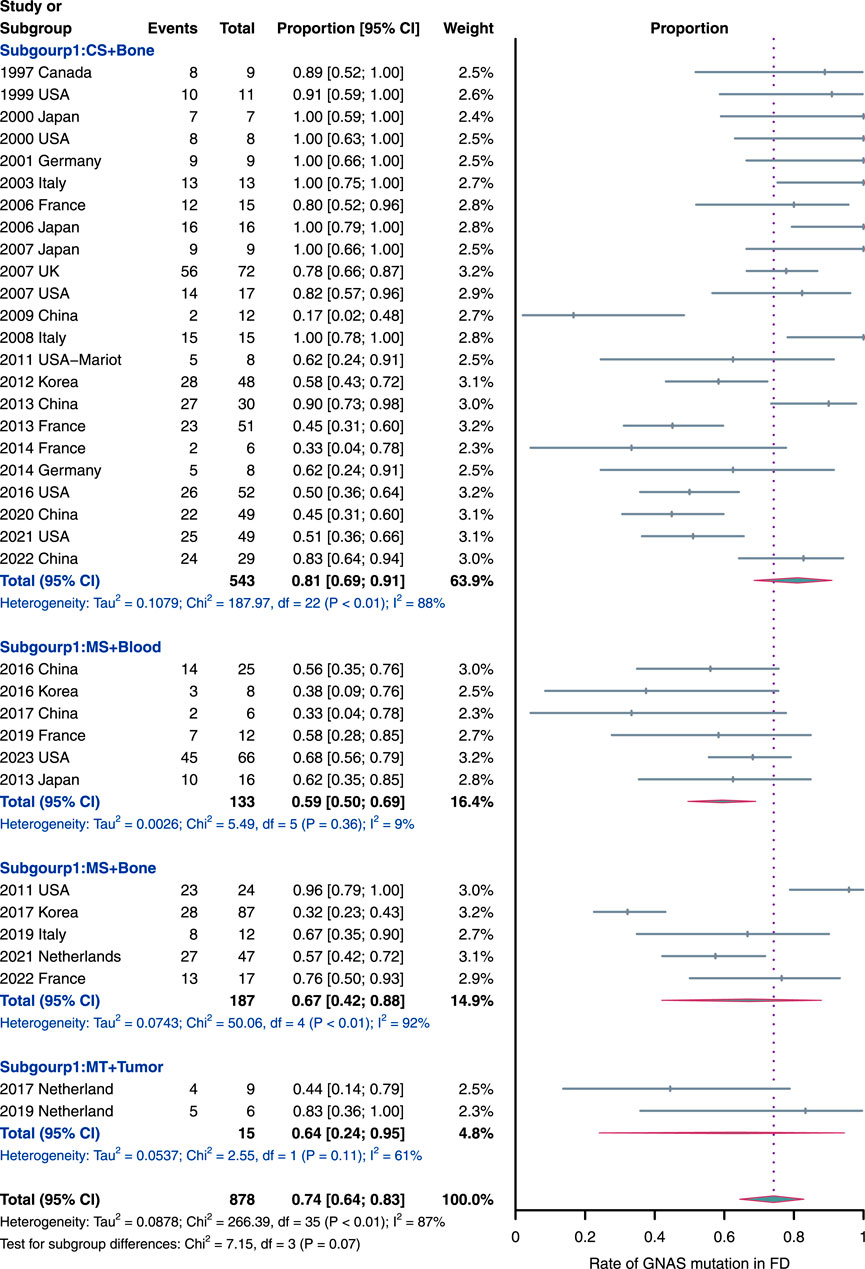
Figure 3. Rate of GNAS mutation detection in FD samples. The subgroups were as follow: MS, modified sequencing methods, including pyrophosphorolysis activated polymerization, digital droplet polymerase chain reaction, mutant enrichment with 3′-modified oligonucleotides and next-generation sequencing methods; CS, conventional sequencing methods, including direct or clone sequencing methods; and MT, multiple sequencing methods used in each study.
Notably, the “CS + Bone” subgroup emerged as the largest, comprising 543 samples from 23 studies, suggesting its current prevalence. This preference may be attributed to its straightforward process and cost-effectiveness. Simultaneously, the “CS + Bone” subgroup exhibited the highest prevalence of GNAS mutations (81%, 95% CI = 69%–91%) compared to the other subgroups. Additionally, it is worth highlighting that the “MS + Blood” subgroup (n = 133; six studies) displayed the lowest prevalence of GNAS mutations (59%, 95% CI = 50% to 69%) with a very low heterogeneity (I2 = 9%, p = 0.36), diverging significantly from the overall pooled outcomes mentioned earlier. The prevalence of GNAS mutations in ‘MS + Bone’ and ‘MT + Tumor’ subgroups was 67% (95% CI = 42%–88%; n = 187) and 64% (95% CI = 24%–95%; n = 15; two studies), respectively. In addition, we also performed an unpaired t-test to compare the detection rate between “CS + Bone” and “MS + Bone,” and there is no statistically significant difference (p-value > 0.05, Supplementary Material).
3.2.2 Diagnostic accuracy of GNAS mutations for FD
In total, 10 of 35 included studies containing differentiated cases of FD were included for this diagnostic accuracy evaluation analysis (in the ninth column of Table 1). In addition, normal donors and patients in the control group were not included in this meta-analysis, as this could have exaggerated the accuracy of the diagnoses. All included studies had a negative correlation coefficient of the threshold effect (r = 0.52, p = 0.11).
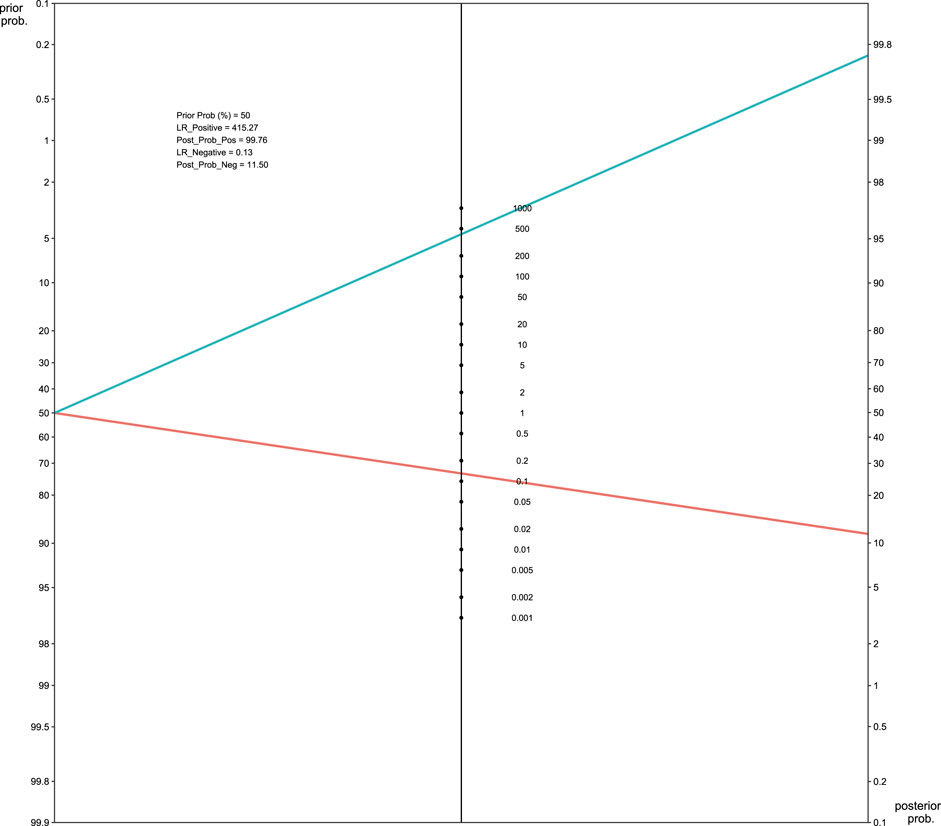
The pooled sensitivity and the 95% confidence interval for GNAS mutations for the FD were 0.83 (95% = 0.65 to 0.96, Figure 4A) while the pooled specificity was 0.99 (95% = 0.98 to 1, Figure 4B). The positive likelihood ratio (LR+) was 415.27 (95% CI, 13.82 to 1,555.30, Supplement Figure 1A), the negative likelihood ratio (LR-) was 0.13 (95% CI, 0.07 to 0.19, Supplement Figure 1B), and the diagnostic odds ratio by log was 7.23 (95% = 4.33 to 9.08, Supplement Figure 1C). The area under the SROC curve was 98.38% (Figure 4C), which indicates excellent for a diagnostic test (Mandrekar, 2010). Figure 5 displayed the Fagan’s Normogram, showing that the post-test probability of LR+ is 99.76%, whereas the post-test probability of LR-is 11.5%.
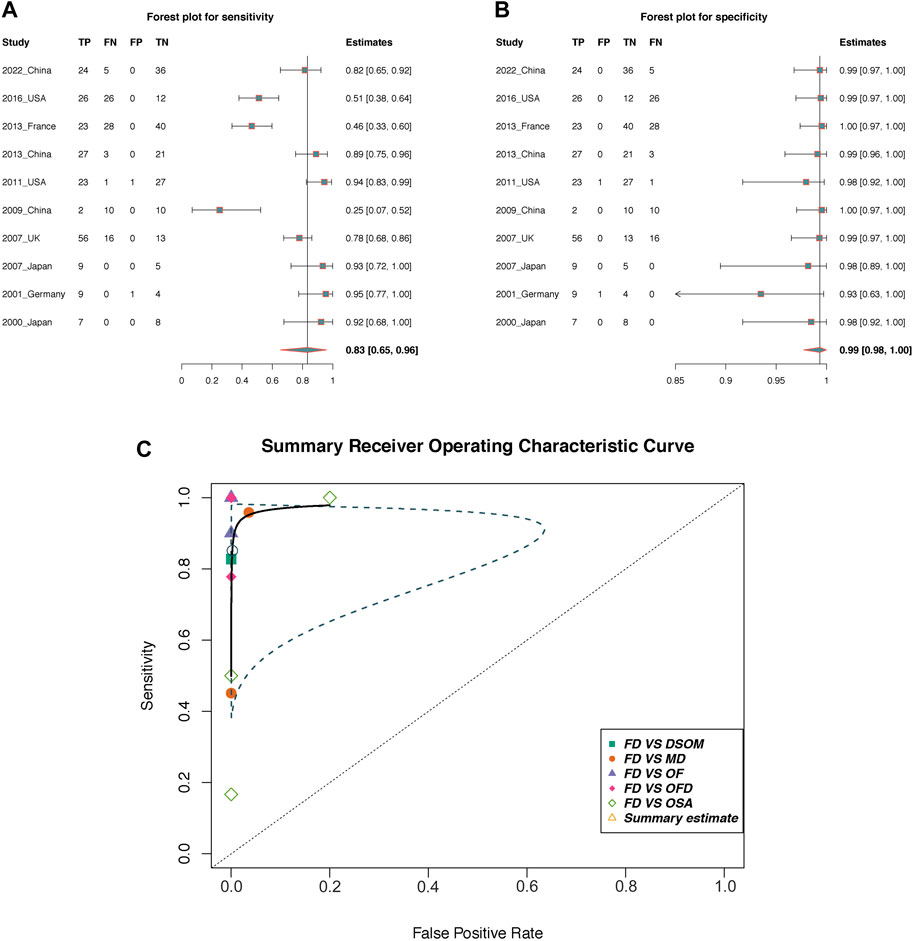
Figure 4. The diagnostic accuracy of GNAS mutation in fibrous dysplasia. (A,B) Forest plots of sensitivity and specificity; (C) Summary receiver operating curve (SROC). TP, True positive; FP, False positive; TN, True negative; FN, False negative; OF, Ossifying fibroma; OFD, osteofibrous dysplasia; OSA, low-grade osteosarcoma; DSOM, chronic diffuse sclerosing osteomyelitis; MD, multiple diseases; Blue dotted line area is the confidence interval.
3.2.3 Genotype-phenotype correlations in FD
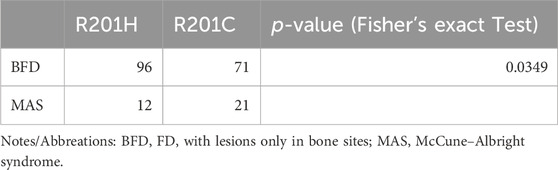
To investigate the correlation between the genotypes and phenotypes of FD/MAS, we divided FD into two categories: FD with lesions only in bone sites (BFD, including MFD and PFD) and MAS. Therefore, we investigated whether R201H mutations were more frequent on BFD than were R201C mutations. The occurrence and fraction of R201H and R201C in BFD and MAS are shown in Table 2 and Figure 6A, and the Fisher’s exact test result revealed a significant association of different mutation types with different FD types (p < 0.05, Table 2), and R201H mainly occurred in BFD while R201C mainly occurred in MAS (Figure 6A).
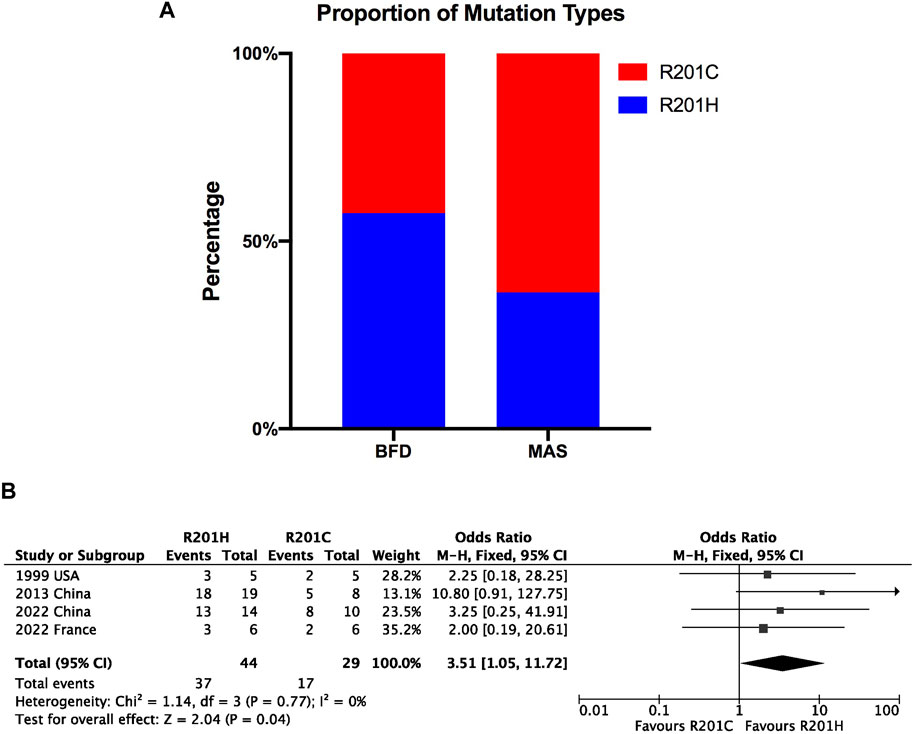
Figure 6. Bar plot for proportion of mutation types (R201H and R201C) in BFD and MAS (A) and forest plot for genotype-phenotype correlation in FD (B).
We also included four studies that simultaneously recorded the specific mutation types in BFD and MAS for meta-analysis. The pooled results also showed a significant association of different mutation types with different FD types (OR = 3.51; 95% CI = 1.05 to 11.72; p = 0.04) with no heterogeneity (Figure 6B), but after the sensitivity analysis, the data were not stable, especially after excluding the 2013 China study (Shi et al., 2013), after which the association was not significant (OR = 2.42; 95% CI = 0.58 to 10; p = 0.96); this indicates that more relevant evidence is still needed.
4 Discussion
Mutation detection methods can be a reliable assistant tool for pathologic diagnosis when confusing microscopic features are observed (Delaney et al., 2009), and currently FD has been found to uniquely contain GNAS mutations. The GNAS mutations were first identified in patients with McCune-Albright syndrome in 1991 (DiCaprio and Enneking, 2005). Since then, more studies have continued to update the incidence data of GNAS mutations in FD. In this study, we found that the pooled prevalence of GNAS mutations in FD patients was 74% (95% CI, 64%–83%), which was obtained from 34 articles published from 1997 to 2022. A mutation with such a high incidence can provide us with an understanding of FD and plays a pivotal role in two aspects: understanding the disease mechanism and enabling diagnosis. Regarding the mechanism, the current view is that GNAS mutations can cause a disruption in GTPase activity and constitutive Gs-α activation (Robinson et al., 2016), which leads to the overproduction of cAMP that causes hyperproliferation and incomplete differentiation of marrow stromal cells to abnormal osteoblasts in FD and inhibits osteoblasts-specific genes as well as stimulating cytokines that promote bone resorption by osteoclasts (Riddle and Bui, 2013).
While the overall pooled prevalence appears elevated, it is important to observe, as indicated by the forest plot, that the mutation rates in certain cohorts fall below the 50% threshold. For negative results, it cannot be concluded that the patient does not have a GNAS mutation status, as FD is a mosaic disease, and the location of cells with DNA mutations is not uniform. In certain instances, the presence of undetectable GNAS mutations in FD cases might find an explanation in the mosaic nature of FD. This mosaic pattern entails lower proportions of mutated cells when juxtaposed with their non-mutated counterparts. Alternatively, such occurrences could be attributed to the emergence of novel mutations within the context of fibrous dysplasia. This diversity in genetic landscapes could contribute to instances where GNAS mutations remain elusive in the diagnostic process. Therefore, the detection rate of GNAS is somewhat unstable because we do not know the proportion of tissue obtained that is occupied by cells rich in GNAS mutations. In addition, an alternative perspective posits that sequencing methods can impact the detection rate of GNAS, including the conventional direct sequencing approach. This method necessitates DNA of high quality and quantity, coupled with a mutant threshold of approximately 20% within the total population (Liang et al., 2011). However, controversy surrounds this viewpoint. Presently, the prevailing belief is that the variable influencing sequencing technology’s impact on detection rates lies in the diverse sources of DNA. Imanaka et al. (Imanaka et al., 2007) suggested subjecting DNA from blood to more advanced sequencing methods, such as PAP, NGS, and ddPCR. This recommendation arises from the inability of direct sequencing to detect mutations in peripheral blood leukocytes adequately (de Sanctis et al., 2017). Therefore, in comparison to bone tissue, employing a modified sequencing method in blood significantly improves the detection rate of GNAS mutations. Interestingly, in this study, those literature extracting DNA from peripheral blood all employed a modified sequencing method as opposed to DS, aligning with the aforementioned theoretical viewpoint. Furthermore, within the “DS + Bone” subgroup, serveral cohorts still exhibited a detection rate of less than 50%, highlighting mosaicism of FD as a significant factor contributing to negative results.
In the included literature, the utilization of modified sequencing technology for FD has progressively emerged since 2011. However, based on the findings of this study, the inherent mosaicism of FD and the cost-effective achievement of 80% accuracy by conventional direct sequencing, the authors contend that conventional direct sequencing remains the optimal method for molecular detection of FD when the specimen is bone. For the tissue selection, the bone affected by FD would be the most appropriate tissue source for detecting DNA mutations in FD patients (Candeliere et al., 1997). Given that it serves as the primary site of the ailment, the concentration of mutations therein will be at its peak. In addition, several studies have attempted to ascertain GNAS mutations by analyzing skin or thyroid tissue samples (Xing et al., 2022). However, given the paucity of relevant literature and data, these analyses were not incorporated into the present study. In this study, the detection rate of GNAS mutations in bone tissue was higher than that in blood tissue, which indicated that bone is a more effective tissue for GNAS mutation testing. In conclusion, the conventional direct sequencing method, when combined with bone tissue, represents the optimal choice.
Regarding diagnosis, a differential diagnosis between FD and many bone lesions, whose specific diseases have been described in the introduction, is needed. Taking DSMO and OF as examples, the overlapping clinicopathological features that appear in some of special cases could lead to difficulties in obtaining a diagnosis, and the therapies and prognoses of these two diseases are extremely different (Shi et al., 2013; Xue et al., 2022). Therefore, it is essential to accurately distinguish FD from other bone lesions. However, the specific diagnostic accuracy of the GNAS mutation detection method is still unclear. In this study, the pooled sensitivity was 83%, the specificity was almost 100%, and the specificity was more consistent than the sensitivity, which means that GNAS mutations are probably a unique characteristic in FD, as other bone lesions do not possess GNAS mutations. Furthermore, low-grade central osteosarcoma (lgcOSA) is one of the most important differential diagnoses of FD (Walther et al., 2014), this study showed that only one lgcOSA case (1/15) had a GNAS mutation, which also indicates the ideal unique specificity of GNAS mutations in FD. The SROC in this study showed a great value of 98.5%, which indicates that GNAS mutation detection is a reliable diagnostic method. However, from the results' perspective, the sensitivity is less stable than the specificity, aligning with the previously discussed the overall pooled rate. In essence, when the detection outcome is wild type, additional discrimination measures are necessary due to the mosaicism of FD. In addition, the value of LR+ > 10 or LR− < 1 indicates the more likely or less likely the disease occurred, respectively. This finding concluded that GNAS mutation has an increase and slight decrease in the likelihood of detecting FD, with the value of LR + being 415.27 and LR− of 0.13, correspondingly.
GNAS mutations in FD mainly include R201H (arginine replaced by histidine) and R201C (arginine replaced by cysteine). In this study, there was a significant association between genotypes and different phenotypes of FD/MAS. More specifically, R201H mutations mainly occur in FD confined to the bone site, and R201C mutations mainly occur in MAS. However, this finding needs to be updated in the future, as the results were unstable after sensitivity analysis. The R201H variant may confer a selective advantage by enhancing the proliferative or survival capacity of affected cells in vivo (Landis et al., 1989). Theoretically, it is difficult to facilitate a preference for developing one mutation type over the other via a biochemical mechanism because R201C and R201H variants in GNAS consistently occur in the same codons (CGT-TGT and CGT-CAT, respectively) (Riminucci et al., 2006). Therefore, more evidence is warranted to further explore the genotype-phenotype correlation in FD.
Several reasons can explain the heterogeneity observed in this study. Different sample sizes could lead to instability in the prevalence results, especially when the sample sizes are small. In this study, the prevalence of GNAS mutations in subgroups with large samples (65%) was lower than that in the subgroups with moderate and small sample sizes (85% and 86%, respectively), but none of them deviated significantly from the scope of the overall result. Additionally, different mutation detection methods and sources of DNA acquisition could also lead to heterogeneity. For the detection methods, the direct sequence technique was mostly applied in this study, but it can be matched with different PCR technologies, such as conventional PCR, nested PCR and protein nucleic acid-based PCR. However, there is still no comprehensive study to explore the specific differences in GNAS mutation detection rate between PCR with DS and other detection methods, including ddPCR, PAP and NGS, which could be an interesting factor to consider in selecting a detection method for GNAS mutations in the future.
To our knowledge, this study is the first to investigate the prevalence, diagnostic accuracy and the genotype-phenotype correlation of GNAS mutations in FD patients. However, there are a few limitations that inevitably should be concerned. First, the sample size of the included studies was inconsistent. Since FD is a rare disease, some small sample studies were included, and the proportion of GNAS mutations in several of these small sample studies was up to 100%, so larger studies are needed, preferably studies with sample sizes greater than 30, to update the GNAS mutation rate. Second, the included studies failed to evaluate the impact of different sample sources or sequencing methods on the detection rate for GNAS mutations, as barely studies evaluated the detection rate in different sequence methods or sample sources by comparing with control groups. This issue will be the focus of research on the efficacy of GNAS detection. Therefore, this article can only explore their differences vertically and cannot compare them horizontally. Third, the included literature of exploring the genotype-phenotype correlation in different FD types, especially MAS, is limited, as many included studies did not report the occurrence of specific GNAS mutations for corresponding FD/MAS, so more studies containing classification records are warranted. Finally, the included studies were all observational, which can have associated biases, such as selection bias. Finally, this study was unable to discuss the incidence of other rare mutation types, such as Q227 (Idowu et al., 2007), which were not included in this article for analysis due to insufficient records.
5 Conclusion
In conclusion, this study showed that GNAS mutations have a very high pooled prevalence of 74% in FD, which indicates that these mutations are responsible for a larger proportion of FD patients, and pooled diagnostic accuracy analysis showed that GNAS mutation detection is a reliable and efficient method for differential diagnosis from other bone lesions that can help doctors achieve early and accurate detection of FD in unclear cases. However, the detection rate of GNAS mutations exhibits instability, mirroring the mosaicism of FD. Consequently, in cases where a negative result is obtained, alternative methods must be employed to pursue further identification. Additionally, we preliminarily found a significant association between mutation variants in FD types, which is essential for the future development of targeted therapies.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author contributions
A-BZ: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Software, Validation, Visualization, Writing–original draft, Writing–review and editing. J-YZ: Supervision, Conceptualization, Methodology, Validation, Writing–original draft. JX: Conceptualization, Methodology, Visualization, Writing–original draft. Z-CW: Conceptualization, Supervision, Writing–review and editing, Formal Analysis, Investigation, Validation. Z-XX: Conceptualization, Methodology, Resources, Writing–original draft. L-SS: Project administration, Resources, Supervision, Writing–review and editing. T-JL: Funding acquisition, Project administration, Resources, Supervision, Writing–review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by the Chinese National Nature Science Foundation (81671006, 30901680) and the CAMS Innovation Fund for Medical Sciences (2019-I2M-5-038).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2024.1377716/full#supplementary-material
References
Author Anynomous (2019) Analysis of clinical data of fibrous dysplasia of bone and detection of GNAS gene in multiple fibrous dysplasia of bone. Hebei Medical University.
Bianco, P., Riminucci, M., Majolagbe, A., Kuznetsov, S. A., Collins, M. T., Mankani, M. H., et al. (2000). Mutations of the GNAS1 gene, stromal cell dysfunction, and osteomalacic changes in non-McCune-Albright fibrous dysplasia of bone. J. bone mineral Res. official J. Am. Soc. Bone Mineral Res. 15, 120–128. doi:10.1359/jbmr.2000.15.1.120
Candeliere, G. A., Roughley, P. J., and Glorieux, F. H. (1997). Polymerase chain reaction-based technique for the selective enrichment and analysis of mosaic arg201 mutations in G alpha s from patients with fibrous dysplasia of bone. Bone 21, 201–206. doi:10.1016/s8756-3282(97)00107-5
Cho, E. K., Kim, J., Yang, A., Ki, C. S., Lee, J. E., Cho, S. Y., et al. (2016). Clinical and endocrine characteristics and genetic analysis of Korean children with McCune-Albright syndrome: a retrospective cohort study. Orphanet J. rare Dis. 11, 113. doi:10.1186/s13023-016-0496-x
Collins, M. T., Singer, F. R., and Eugster, E. (2012). McCune-Albright syndrome and the extraskeletal manifestations of fibrous dysplasia. Orphanet J. rare Dis. 7 (Suppl. 1), S4. doi:10.1186/1750-1172-7-s1-s4
Corsi, A., Collins, M. T., Riminucci, M., Howell, P. G. T., Boyde, A., Robey, P. G., et al. (2003). Osteomalacic and hyperparathyroid changes in fibrous dysplasia of bone: core biopsy studies and clinical correlations. J. bone mineral Res. official J. Am. Soc. Bone Mineral Res. 18, 1235–1246. doi:10.1359/jbmr.2003.18.7.1235
Delaney, D., Diss, T. C., Presneau, N., Hing, S., Berisha, F., Idowu, B. D., et al. (2009). GNAS1 mutations occur more commonly than previously thought in intramuscular myxoma. Mod. pathology official J. U. S. Can. Acad. Pathology 22, 718–724. doi:10.1038/modpathol.2009.32
de Sanctis, L., Galliano, I., Montanari, P., Matarazzo, P., Tessaris, D., and Bergallo, M. (2017). Combining real-time COLD- and MAMA-PCR TaqMan techniques to detect and quantify R201 GNAS mutations in the McCune-albright syndrome. Hormone Res. Paediatr. 87, 342–349. doi:10.1159/000463384
DiCaprio, M. R., and Enneking, W. F. (2005). Fibrous dysplasia. Pathophysiology, evaluation, and treatment. J. bone Jt. Surg. Am. volume 87, 1848–1864. doi:10.2106/jbjs.D.02942
Elli, F. M., de Sanctis, L., Bergallo, M., Maffini, M. A., Pirelli, A., Galliano, I., et al. (2019). Improved molecular diagnosis of McCune-albright syndrome and bone fibrous dysplasia by digital PCR. Front. Genet. 10, 862. doi:10.3389/fgene.2019.00862
Gaujoux, S., Salenave, S., Ronot, M., Rangheard, A. S., Cros, J., Belghiti, J., et al. (2014). Hepatobiliary and Pancreatic neoplasms in patients with McCune-Albright syndrome. J. Clin. Endocrinol. metabolism 99, E97–E101. doi:10.1210/jc.2013-1823
Hagelstein-Rotman, M., Appelman-Dijkstra, N. M., Boyce, A. M., Chapurlat, R., Dur, N. B. J., Gensburger, D., et al. (2022). Extent of extraskeletal manifestations of fibrous dysplasia/McCune-albright syndrome in patients with mazabraud's syndrome. Calcif. tissue Int. 110, 334–340. doi:10.1007/s00223-021-00918-0
Hagelstein-Rotman, M., Meier, M. E., Majoor, B. C. J., Cleven, A. H. G., Dijkstra, P. D. S., Hamdy, N. A. T., et al. (2021). Increased prevalence of malignancies in fibrous dysplasia/McCune-albright syndrome (FD/MAS): data from a national referral center and the Dutch national pathology registry (PALGA). Calcif. tissue Int. 108, 346–353. doi:10.1007/s00223-020-00780-6
Idowu, B. D., Al-Adnani, M., O'Donnell, P., Yu, L., Odell, E., Diss, T., et al. (2007). A sensitive mutation-specific screening technique for GNAS1 mutations in cases of fibrous dysplasia: the first report of a codon 227 mutation in bone. Histopathology 50, 691–704. doi:10.1111/j.1365-2559.2007.02676.x
Imanaka, M., Iida, K., Nishizawa, H., Fukuoka, H., Takeno, R., Takahashi, K., et al. (2007). McCune-Albright syndrome with acromegaly and fibrous dysplasia associated with the GNAS gene mutation identified by sensitive PNA-clamping method. Intern. Med. Tokyo, Jpn. 46, 1577–1583. doi:10.2169/internalmedicine.46.0048
Jour, G., Oultache, A., Sadowska, J., Mitchell, T., Healey, J., Nafa, K., et al. (2016). GNAS mutations in fibrous dysplasia: a comparative study of standard sequencing and locked nucleic acid PCR sequencing on decalcified and nondecalcified formalin-fixed paraffin-embedded tissues. Appl. Immunohistochem. Mol. Morphol. AIMM 24, 660–667. doi:10.1097/pai.0000000000000242
Juan, T., Hong-ye, Z., Li, Z., Hui-zhen, Z., and Zhi-ming, J. (2009). Abnormal expression of c-myc,p53,p16 protein and GNAS1 gene mutation in fibrous dysplasia. Chin. J. Pathology 38, 292–297. doi:10.3760/cma.j.issn.0529-5807.2009.05.002
Kalfa, N., Philibert, P., Audran, F., Ecochard, A., Hannon, T., Lumbroso, S., et al. (2006). Searching for somatic mutations in McCune-Albright syndrome: a comparative study of the peptidic nucleic acid versus the nested PCR method based on 148 DNA samples. Eur. J. Endocrinol. 155, 839–843. doi:10.1530/eje.1.02301
Kobayashi, K., Imanishi, Y., Koshiyama, H., Miyauchi, A., Wakasa, K., Kawata, T., et al. (2006). Expression of FGF23 is correlated with serum phosphate level in isolated fibrous dysplasia. Life Sci. 78, 2295–2301. doi:10.1016/j.lfs.2005.09.052
Kuznetsov, S. A., Cherman, N., Riminucci, M., Collins, M. T., Robey, P. G., and Bianco, P. (2008). Age-dependent demise of GNAS-mutated skeletal stem cells and "normalization" of fibrous dysplasia of bone. J. bone mineral Res. official J. Am. Soc. Bone Mineral Res. 23, 1731–1740. doi:10.1359/jbmr.080609
Landis, C. A., Masters, S. B., Spada, A., Pace, A. M., Bourne, H. R., and Vallar, L. (1989). GTPase inhibiting mutations activate the alpha chain of Gs and stimulate adenylyl cyclase in human pituitary tumours. Nature 340, 692–696. doi:10.1038/340692a0
Lee, S. E., Lee, E. H., Park, H., Sung, J. Y., Lee, H. W., Kang, S. Y., et al. (2012). The diagnostic utility of the GNAS mutation in patients with fibrous dysplasia: meta-analysis of 168 sporadic cases. Hum. Pathol. 43, 1234–1242. doi:10.1016/j.humpath.2011.09.012
Leeflang, M. M. (2014). Systematic reviews and meta-analyses of diagnostic test accuracy. Clin. Microbiol. Infect. official Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 20, 105–113. doi:10.1111/1469-0691.12474
Liang, Q., Wei, M., Hodge, L., Fanburg-Smith, J. C., Nelson, A., Miettinen, M., et al. (2011). Quantitative analysis of activating alpha subunit of the G protein (Gsα) mutation by pyrosequencing in fibrous dysplasia and other bone lesions. J. Mol. diagnostics JMD 13, 137–142. doi:10.1016/j.jmoldx.2010.10.003
Liens, D., Delmas, P. D., and Meunier, P. J. (1994). Long-term effects of intravenous pamidronate in fibrous dysplasia of bone. Lancet London, Engl. 343, 953–954. doi:10.1016/s0140-6736(94)90069-8
Majoor, B. C., Boyce, A. M., Bovée, J. V., Smit, V. T., Collins, M. T., Cleton-Jansen, A. M., et al. (2018). Increased risk of breast cancer at a young age in women with fibrous dysplasia. J. bone mineral Res. official J. Am. Soc. Bone Mineral Res. 33, 84–90. doi:10.1002/jbmr.3286
Majoor, B. C. J., van de Sande, M. A. J., Appelman-Dijkstra, N. M., Leithner, A., Jutte, P. C., Vélez, R., et al. (2019). Prevalence and clinical features of mazabraud syndrome: a multicenter European study. J. bone Jt. Surg. Am. volume 101, 160–168. doi:10.2106/jbjs.18.00062
Mandrekar, J. N. (2010). Receiver operating characteristic curve in diagnostic test assessment. J. Thorac. Oncol. official Publ. Int. Assoc. Study Lung Cancer 5, 1315–1316. doi:10.1097/JTO.0b013e3181ec173d
Mariot, V., Wu, J. Y., Aydin, C., Mantovani, G., Mahon, M. J., Linglart, A., et al. (2011). Potent constitutive cyclic AMP-generating activity of XLαs implicates this imprinted GNAS product in the pathogenesis of McCune-Albright syndrome and fibrous dysplasia of bone. Bone 48, 312–320. doi:10.1016/j.bone.2010.09.032
Michienzi, S., Cherman, N., Holmbeck, K., Funari, A., Collins, M. T., Bianco, P., et al. (2007). GNAS transcripts in skeletal progenitors: evidence for random asymmetric allelic expression of Gs alpha. Hum. Mol. Genet. 16, 1921–1930. doi:10.1093/hmg/ddm139
Narumi, S., Matsuo, K., Ishii, T., Tanahashi, Y., and Hasegawa, T. (2013). Quantitative and sensitive detection of GNAS mutations causing mccune-albright syndrome with next generation sequencing. PloS One 8, e60525. doi:10.1371/journal.pone.0060525
Parvanescu, A., Cros, J., Ronot, M., Hentic, O., Grybek, V., Couvelard, A., et al. (2014). Lessons from McCune-Albright syndrome-associated intraductal papillary mucinous neoplasms:: GNAS-activating mutations in pancreatic carcinogenesis. JAMA Surg. 149, 858–862. doi:10.1001/jamasurg.2014.535
Pollandt, K., Engels, C., Kaiser, E., Werner, M., and Delling, G. (2001). Gsalpha gene mutations in monostotic fibrous dysplasia of bone and fibrous dysplasia-like low-grade central osteosarcoma. Virchows Archiv Int. J. pathology 439, 170–175. doi:10.1007/s004280100453
Riddle, N. D., and Bui, M. M. (2013). Fibrous dysplasia. Archives pathology laboratory Med. 137, 134–138. doi:10.5858/arpa.2012.0013-RS
Riminucci, M., Saggio, I., Robey, P. G., and Bianco, P. (2006). Fibrous dysplasia as a stem cell disease. J. bone mineral Res. official J. Am. Soc. Bone Mineral Res. 21 (Suppl. 2), P125–P131. doi:10.1359/jbmr.06s224
Robinson, C., Collins, M. T., and Boyce, A. M. (2016). Fibrous dysplasia/McCune-albright syndrome: clinical and translational perspectives. Curr. Osteoporos. Rep. 14, 178–186. doi:10.1007/s11914-016-0317-0
Rockville, M. D. (2008). Methods guide for effectiveness and comparative effectiveness reviews. Agency Healthc. Res. Qual. (US).
Romanet, P., Philibert, P., Fina, F., Cuny, T., Roche, C., Ouafik, L., et al. (2019). Using digital droplet polymerase chain reaction to detect the mosaic GNAS mutations in whole blood DNA or circulating cell-free DNA in fibrous dysplasia and McCune-albright syndrome. J. Pediatr. 205, 281–285. doi:10.1016/j.jpeds.2018.09.070
Roszko, K. L., Guthrie, L., Li, X., Collins, M. T., de Castro, L. F., and Boyce, A. M. (2023). Identification of GNAS variants in circulating cell-free DNA from patients with fibrous dysplasia/McCune albright syndrome. J. Bone Mineral Res. Official J. Am. Soc. Bone Mineral Res. 38, 443–450. doi:10.1002/jbmr.4766
Sakamoto, A., Oda, Y., Iwamoto, Y., and Tsuneyoshi, M. (2000). A comparative study of fibrous dysplasia and osteofibrous dysplasia with regard to Gsalpha mutation at the Arg201 codon: polymerase chain reaction-restriction fragment length polymorphism analysis of paraffin-embedded tissues. J. Mol. diagnostics JMD 2, 67–72. doi:10.1016/s1525-1578(10)60618-6
Shi, R. R., Li, X. F., Zhang, R., Chen, Y., and Li, T. J. (2013). GNAS mutational analysis in differentiating fibrous dysplasia and ossifying fibroma of the jaw. Mod. Pathol. 26, 1023–1031. doi:10.1038/modpathol.2013.31
Shin, S. J., Lee, S. J., and Kim, S. K. (2017). Frequency of GNAS R201H substitution mutation in polyostotic fibrous dysplasia: pyrosequencing analysis in tissue samples with or without decalcification. Sci. Rep. 7, 2836. doi:10.1038/s41598-017-03093-1
Stanton, R. P., Hobson, G. M., Montgomery, B. E., Moses, P. A., Smith-Kirwin, S. M., and Funanage, V. L. (1999). Glucocorticoids decrease interleukin-6 levels and induce mineralization of cultured osteogenic cells from children with fibrous dysplasia. J. bone mineral Res. official J. Am. Soc. Bone Mineral Res. 14, 1104–1114. doi:10.1359/jbmr.1999.14.7.1104
Stroup, D. F., Berlin, J. A., Morton, S. C., Olkin, I., Williamson, G. D., Rennie, D., et al. (2000). Meta-analysis of observational studies in epidemiology: a proposal for reporting. Meta-analysis of Observational Studies in Epidemiology (MOOSE) group. Jama 283, 2008–2012. doi:10.1001/jama.283.15.2008
Tabareau-Delalande, F., Collin, C., Gomez-Brouchet, A., Decouvelaere, A. V., Bouvier, C., Larousserie, F., et al. (2013). Diagnostic value of investigating GNAS mutations in fibro-osseous lesions: a retrospective study of 91 cases of fibrous dysplasia and 40 other fibro-osseous lesions. Mod. pathology official J. U. S. Can. Acad. Pathology, Inc 26, 911–921. doi:10.1038/modpathol.2012.223
Toyosawa, S., Yuki, M., Kishino, M., Ogawa, Y., Ueda, T., Murakami, S., et al. (2007). Ossifying fibroma vs fibrous dysplasia of the jaw: molecular and immunological characterization. Mod. pathology official J. U. S. Can. Acad. Pathology, Inc 20, 389–396. doi:10.1038/modpathol.3800753
Walther, I., Walther, B. M., Chen, Y., and Petersen, I. (2014). Analysis of GNAS1 mutations in myxoid soft tissue and bone tumors. Pathology, Res. Pract. 210, 1–4. doi:10.1016/j.prp.2013.09.003
Weinstein, L. S., Shenker, A., Gejman, P. V., Merino, M. J., Friedman, E., and Spiegel, A. M. (1991). Activating mutations of the stimulatory G protein in the McCune-Albright syndrome. N. Engl. J. Med. 325, 1688–1695. doi:10.1056/nejm199112123252403
Whiting, P. F., Rutjes, A. W. S., Westwood, M. E., Mallett, S., Deeks, J. J., Reitsma, J. B., et al. (2011). QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann. Intern. Med. 155, 529–536. doi:10.7326/0003-4819-155-8-201110180-00009
Xing, Z., Tao, G., Pan, W., Wu, D., Pan, T., Wan, L., et al. (2022). Case report: surgical treatment of McCune-Albright syndrome with hyperthyroidism and retrosternal goiter: a case report and literature review. Front. Surg. 9, 921427. doi:10.3389/fsurg.2022.921427
Xue, J., Jia, K., Li, T., Zhang, J., and An, J. (2022). GNAS mutation analysis assists in differentiating chronic diffuse sclerosing osteomyelitis from fibrous dysplasia in the jaw. Mod. pathology official J. U. S. Can. Acad. Pathology, Inc 35, 1334–1340. doi:10.1038/s41379-022-01103-w
Xue-yan, Q., et al. (2016). Analysis of clinical features and related genes variation in 41 girls with McCune-Albright syndrome. Chin. J. Endocrinol. Metab. 32, 4.
Zhadina, M., Roszko, K. L., Geels, R. E. S., de Castro, L. F., Collins, M. T., and Boyce, A. M. (2021). Genotype-phenotype correlation in fibrous dysplasia/McCune-albright syndrome. J. Clin. Endocrinol. metabolism 106, 1482–1490. doi:10.1210/clinem/dgab053
Zhou, T., Guan, H., Wang, L., Zhang, Y., Rui, M., and Ma, A. (2021). Health-related quality of life in patients with different diseases measured with the EQ-5D-5L: a systematic review. Front. public health 9, 675523. doi:10.3389/fpubh.2021.675523
Keywords: GNAS, fibrous dysplasia, diagnostic accuracy, meta-analyses, McCune Albright
Citation: Zhang A-B, Zhang J-Y, Xue J, Wu Z-C, Xu Z-X, Sun L-S and Li T-J (2024) The prevalence, diagnostic accuracy and genotype-phenotype correlation of GNAS mutations in fibrous dysplasia: a meta-analysis. Front. Genet. 15:1377716. doi: 10.3389/fgene.2024.1377716
Received: 05 February 2024; Accepted: 17 May 2024;
Published: 29 July 2024.
Edited by:
Luis A. Fernandez De Castro Diaz, National Institutes of Health (NIH), United StatesReviewed by:
Eduardo Fernandez-Rebollo, University of Southern Denmark, DenmarkDiana Ovejero Crespo, Hospital del Mar Medical Research Institute (IMIM), Spain
Copyright © 2024 Zhang, Zhang, Xue, Wu, Xu, Sun and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li-Sha Sun, bGlzYV9zdW5AYmptdS5lZHUuY24=; Tie-Jun Li, bGl0aWVqdW4yMkB2aXAuc2luYS5jb20=
 Ao-Bo Zhang
Ao-Bo Zhang Jian-Yun Zhang1,2
Jian-Yun Zhang1,2 Li-Sha Sun
Li-Sha Sun