
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 06 March 2024
Sec. Genomics of Plants and the Phytoecosystem
Volume 15 - 2024 | https://doi.org/10.3389/fgene.2024.1372644
“Heteromorphic self-incompatibility” (HetSI) in plants is a mechanism of defense to avoid self-pollination and promote outcrossing. However, the molecular mechanism underlying HetSI remains largely unknown. In this study, RNA-seq was conducted to explore the molecular mechanisms underlying self-compatible (SC, “T × P” and “P × T”) and self-incompatible (SI, “T × T” and “P × P”) pollination in the two types of flowers of Plumbago auriculata Lam. which is a representative HetSI plant. By comparing “T × P” vs. “T × T”, 3773 (1407 upregulated and 2366 downregulated) differentially expressed genes (DEGs) were identified, 1261 DEGs between “P × T” and “P × P” (502 upregulated and 759 downregulated). The processes in which these DEGs were significantly enriched were “MAPK (Mitogen-Activated Protein Kinases-plant) signaling pathway”, “plant-pathogen interaction”,“plant hormone signal transduction”, and “pentose and glucuronate interconversion” pathways. Surprisingly, we discovered that under various pollination conditions, multiple notable genes that may be involved in HetSI exhibited distinct regulation. We can infer that the HetSI strategy might be unique in P. auriculata. It was similar to “sporophytic self-incompatibility” (SSI) but the HetSI mechanisms in pin and thrum flowers are diverse. In this study, new hypotheses and inferences were proposed, which can provide a reference for crop production and breeding.
Mate selection is a critical factor for ensuring successful reproduction, with self-incompatibility (SI) being one of the predominant mate selection strategies in plants. SI plays a crucial role in promoting out-crossing to achieve genetic diversity by rejecting self-pollen (Lian et al., 2021). SI can be classified into two categories: homomorphic and heteromorphic. Homomorphic SI (HomSI) plants have only one type of flower, while heteromorphic SI (HetSI) plants have distyly or tristyly flowers (Jain et al., 2023). SI plants have developed two systems to prevent incorrect pollen deposition: sporophytic self-incompatibility (SSI) and gametophytic self-incompatibility (GSI) (Wang et al., 2023). Based on the S genotype of the sporophyte, the SSI is determined, pollen germination failure occurs on the stigma’s surface, preventing the pollen from penetrating the style tissue (Pauline et al., 2023). In GSI, haploid pollen itself determines the phenotype, and pollen tube growth can be arrested before entering the ovary (Sassa, 2016; Pauline et al., 2023). Notably, GSI systems and homomorphic systems have been studied more extensively than heteromorphic systems (McCubbin, 2008). Studies have reported that in Solanaceae, Rosaceae and Plantaginaceae, GSI is based on the SLF (SFB)/SRNase system, and in Brassicaceae SSI is based on the SP11 (SCR)/SRK system (Lv et al., 2022; Goring et al., 2023). Significant advancements have been made in comprehending the molecular foundation of HetSI, particularly in Primula and Fagopyrum (Banović et al., 2010; Li et al., 2023). Studies have demonstrated that HetSI in both types of plants shares the same HetSI controller-S locus and exhibits similar pollen‒stigma interactions (Matsui and Yasui, 2020).
A frequently encountered HetSI plant with dry stigmas is Plumbago auriculata Lam. (Plumbaginaceae) (Hu et al., 2019). The Plumbaginaceae family comprises approximately 650–1000 species, with a significant number of them being HetSI plants (Mucina and Hammer, 2019). These plants serve as excellent research systems for HetSI studies. Through artificial pollination, we found that once pollination occurs with compatible pollen, a series of signaling events are triggered, including pollen adhesion, germination, pollen tube elongation, and fertilization, all of which occur within 4 h. However, no pollen tubes were discovered in SI pollinations (Hu et al., 2019). The metabolomic study revealed that mature styles stored abundant nutrients required for the energy-consuming process in the pollen tube growth. Despite the failure of fertilization in SI pollinations, the styles still contained sufficient energy reserves to support the growth of other SC (self-compatible) pollen grains (Figure 1). Moreover, the morphology and metabolome of SI in pin and thrum flower styles were found to be distinct (Hu et al., 2019; Hu et al., 2020). This led us to hypothesize that different flower types may have varying mechanisms for SI. However, data concerning the SI reaction at the molecular level in P. auriculata are still very scarce. To gain a deeper understanding of the P. auriculata HetSI mechanism at the genomic level, RNA-seq analysis was conducted on stigmas after different pollination events. This study’s transcriptome analysis offers valuable insights into the potential molecular mechanisms influencing P. auriculata HetSI and aims to provide new evidance that support the understanding of the interaction between the pollen and stigma in HetSI.
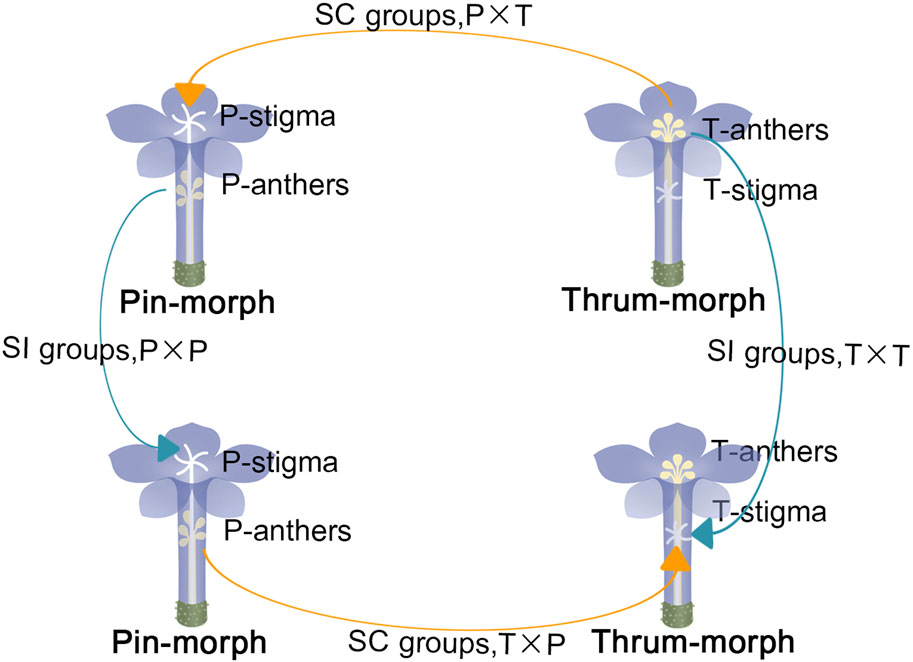
FIGURE 1. “Thrum-morph flower” (T) and “pin-morph flower” (P). the connotation of “P × T”: pin-morph stigmas that are pollinated with thrum pollens; the connotation of “P × P”: pin-morph stigmas that are pollinated with pin pollens; the connotation of “T × P”: thrum-morph stigmas pollinated with pin pollens; the connotation of “T × T”: thrum-morph stigmas pollinated with thrum pollens; “Self-incompatible (SI) pollinations” are shown by the arrows of blue color, and “self-compatible (SC) pollinations” are indicated by orange arrows.
Pin and thrum flowers of P. auriculata were cultivated separately in the greenhouse at Sichuan Normal University, located in Sichuan, China. The day before dehiscence, floral buds of pollen receptor flowers were chosen at random, emasculated, and bagged.
At 8:00 a.m., pistils were manually pollinated with SC pollination: “P × T” (pin pistils inter - pollinated with thrum pollen), “T × P” (thrum pistils inter - pollinated with pin pollen) and SI pollination: “T × T” (thrum pistils intra - pollinated with thrum pollen), “P × P” (pin pistils intra - pollinated with pin pollen). They were collected 2 h later, at 10:00 a.m. (Hu et al., 2019; Hu et al., 2020). Fifty milligrams of each sample was processed for analysis (Figure 1). For every sample, three biological replicates were employed. Upon instant freezing in liquid nitrogen, samples were kept at the temperature of −80 °C in order to extract RNA.
With a “RNAprep Pure Plant Kit” (Tiangen, China) and “TRlzol Reagent” (Life Technologies, United States), from every sample, entire RNA was extracted. Using an “Agilent 2100 Bioanalyzer” (Agilent Technologies, United States) and “Nanodrop 2000” (Thermo Fisher Scientific, United States), RNA’s quality was checked. The “Hieff NGS Ultima Dual-mode mRNA Library Prep Kit for Illumina” (Yeasen Biotechnology, China) was used to construct the cDNA library following the manufacturer’s proposals. Utilizing a “NovaSeq6000” (Illumina, United States), the cDNA libraries were sequenced. Finally, clean data were obtained through “in-house Perl scripts”. Sequence duplication, GC content, Q20, and Q30 values were calculated.
For the sake of determining the pertinent expression standards of SI and SC pollinations, the “DESeq R package” (1.10.1) was utilized. “The false discovery rate” (FDR) was controlled for the resultant “p values” by applying Benjamini and Hochberg (1995). Genes that DESeq identified as differentially expressed (DEGs) have an adjusted p-value <0.05.
Functional enrichment for the DEGs was accessed through the following databases: KOG/COG/eggNOG (Clusters of Orthologous Groups of proteins), NR (NCBI nonredundant protein sequences), Pfam (Protein family), KEGG (Kyoto Encyclopedia of Genes and Genomes), Swiss-Prot (Swiss-Prot protein sequence database), and GO (Gene Ontology) enrichment analysis.
A total of 15 DEGs were analyzed by RT‒qPCR to verify the dependability of RNA-seq. The “TUREscript 1st Stand cDNA SYNTHESIS Kit” (Aidlab, China) was used for first-stand cDNA synthesis. By utilizing a “qTOWER 2.2 Real-Time Thermocyclers” (Analytik Jena, GER.) with three biological replicates, RT‒qPCR was performed. With the GAPDH gene serving as an internal control, the 2−ΔΔCT method was used to calculate the relative quantification of gene expression (Livak and Schmittgen, 2001).
For the purpose of exploring the molecular mechanisms underlying interactions in pollen-stigma of P. auriculata, SC group (“T × P” and “P × T”) and SI group (“P × P” and “T × T”) pollinated styles were collected, and “Illumina NovaSeq technology” was employed to investigate the stigma transcriptome. Per sample from transcriptome sequencing generated 6.00 Gb of clean data, with a “Q-score” larger than the Q30 value in more than 93.85% of cases. Furthermore, every sample’s mapping ratio against the reference genome with the scope from 78.89% to 82.48%, as the Table 1 revealed. The data demonstrated robust sequencing and mapping results in our study. After assembly, we extracted 85249 unigenes, 44407 of which were annotated in the reference genome. In addition, we used the “R2 value” to evaluate the repeat correlation, which showed a high intragroup similarity.

TABLE 1. The sequencing data from RNA-seq analysis of different (“P × P”, “P × T”, “T × T” & “T × P”) styles of pollination.
Fold change (FC) > 2 as well as FDR <0.01 were employed to quantify the transcript expression and identify DEGs. The comparison of “T × P” vs. “T × T” generated 3773 (1407 upregulated and 2366 downregulated) DEGs. There were 1261 DEGs between “P × T” and “P × P” (502 upregulated and 759 downregulated) DEGs (Figures 2A, B). Among them, 392 DEGs were shared by both comparison groups (Figure 2C).
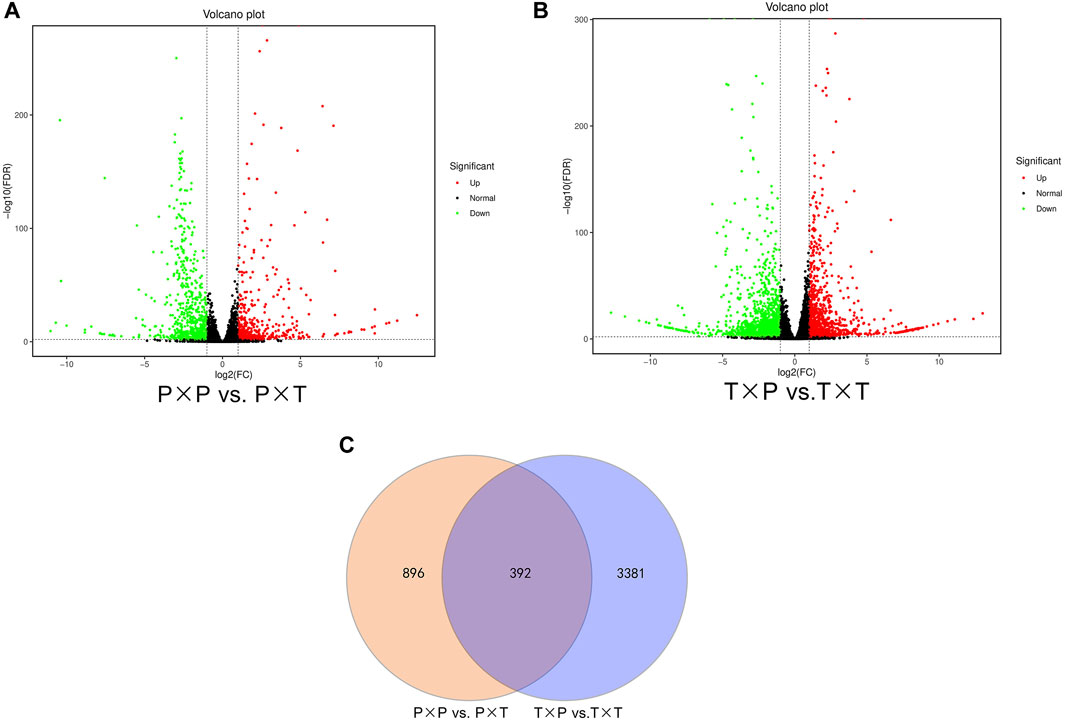
FIGURE 2. Statistics of DEGs in different pollination (A,B) Volcano plot of the identified DEGs in comparing “T × P” vs. “T × T” as well as “P × T” vs. “P × P”; (C) Venn diagrams of common and specific DEGs between “T × P” vs. “T × T” and “P × T” vs. “P × P”.
Within the COG, KOG, Nr, Swiss-Prot, Pfam and eggNOG databases, 1173, 1882, 3303, 2723, 2791, and 2924 DEGs were annotated successfully in the context of a comparison between “T × P” vs. “T × T”, and the results showed many DEGs classified in the transporting process of the carbohydrate and metabolism and signal transduction mechanisms. Additionally, protein turnover, posttranslational modification, chaperons and lipid transport revealed a great number of genes in the KOG and COG databases. For “P × T” and “P × P” (COG: 418; KOG: 618; Nr: 1121; Swiss-Prot: 916; Pfam: 983; eggNOG: 996), DEGs were enriched in the classifications above as well (Table 2; Supplementary Figure S1).
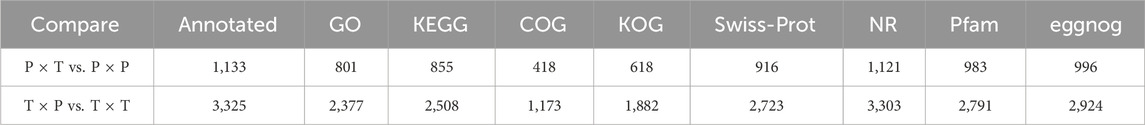
TABLE 2. Functional enrichment analysis of all DEGs by GO, COG, KOG, KEGG, NR, Pfam, eggnog, and Swiss-Prot.
To further analyze the function of the DEGs, we annotated the DEGs using “Gene Ontology” (GO) for analysis. The DEGs were categorized into the following GO primary functions, namely, “molecular function” (MF), “biological process” (BP), and “cellular component” (CC). In total, 2377 and 801 DEGs were assigned GO annotations in the “T × P” vs. “T × T” and “P × T” vs. “P × P” comparison groups, respectively (Figure 3; Supplementary Table S1). For BP, the three most abundant terms in all groups were cellular process (1312 and 445) (numbers in parentheses represent the number of unigenes in “T × P” vs. “T × T” and “P × T” vs. “P × P”, respectively), metabolic process (1218 and 393) and single-organism process (1127 and 393). For CC, these terms were cell (1477 and 471), cell part (1477 and 471) and organelle (1096 and 304). For MF, the terms were binding (1217 and 396), catalytic activity (1247 and 436), and transporter activity (200 and 91) (Table 3). In the “T × P” vs. “T × T” comparison group, the two highest secondary functions of BP were the multiorganism process and response to external stimulus. Cell periphery and protein kinase activity were abundant in CC and MF, respectively. In “P × T” vs. “P × P”, the most overrepresented GO terms in BP were protein phosphorylation and transmembrane transport, and the terms were membrane in CC and protein kinase activity in MF (Supplementary Figure S2).
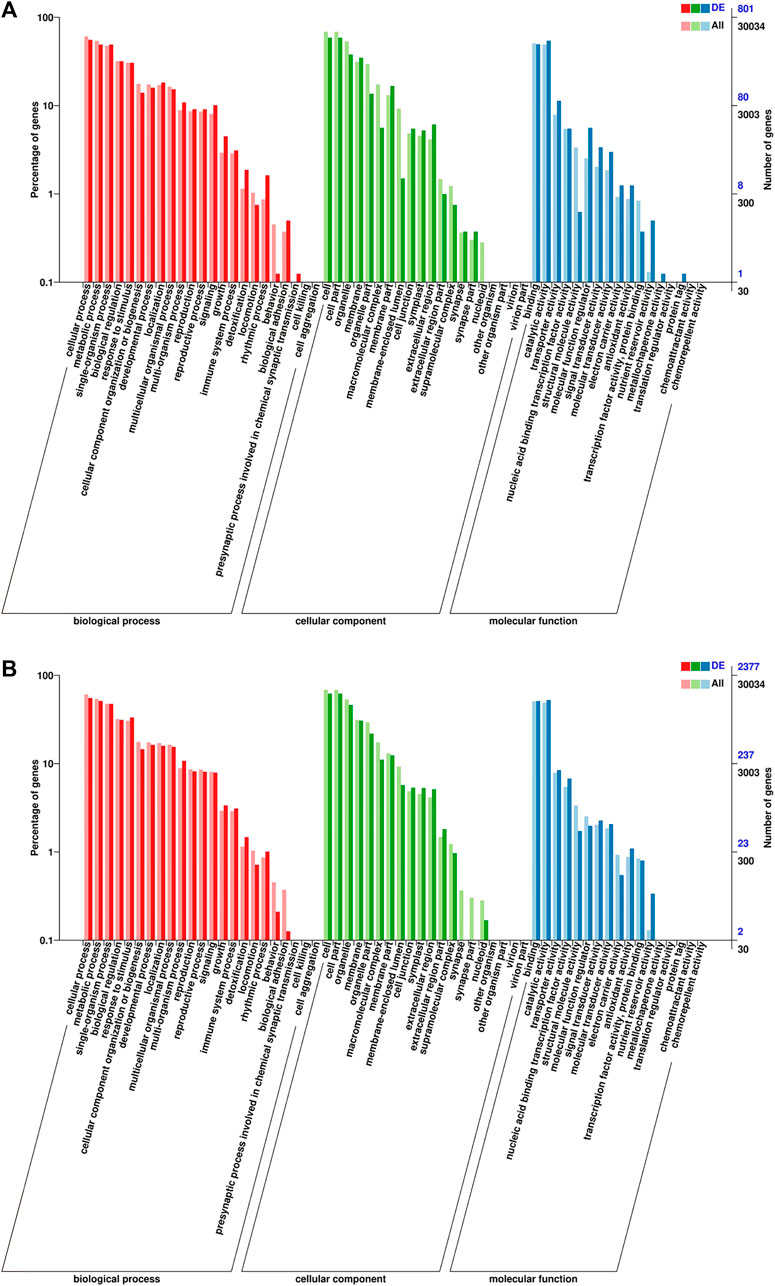
FIGURE 3. The GO terms classification of DEGs in (A) “P × T” vs. “P × P” and (B) “T × P” vs. “T × T”.

TABLE 3. The number of total genes and DEGs of GO terms in “P × T” vs. “P × P” and “T × P” vs. “T × T”.
To investigate the effect of DEGs on pathways in more detail, based on the KEGG database, we also carried out pathway enrichment analysis of DEGs. According to the results for both upregulated genes of the “T × P” vs. “T × T” comparison group, DEGs were gathered in the “transduction of plant hormone signal”, “plant-pathogen interaction”, “protein processing in endoplasmic reticulum”, “endocytosis” and “starch and sucrose metabolism”. For downregulated genes, those containing the greatest number were “plant‒pathogen interaction”, “MAPK (plant) signaling pathway”, “pentose and glucuronate interconversions”, “plant hormone signal transduction” and “starch and sucrose metabolism”. For “P × T” vs. “P × P”, among the upregulated genes were “MAPK (plant) signaling pathway”, “plant-pathogen interaction” and “plant hormone signal transduction”. The processes in which downregulated genes were enriched were “pentose and glucuronate interconversions”, “plant-pathogen interaction” and “metabolism of starch and sucrose” as showcased in the Table 4.
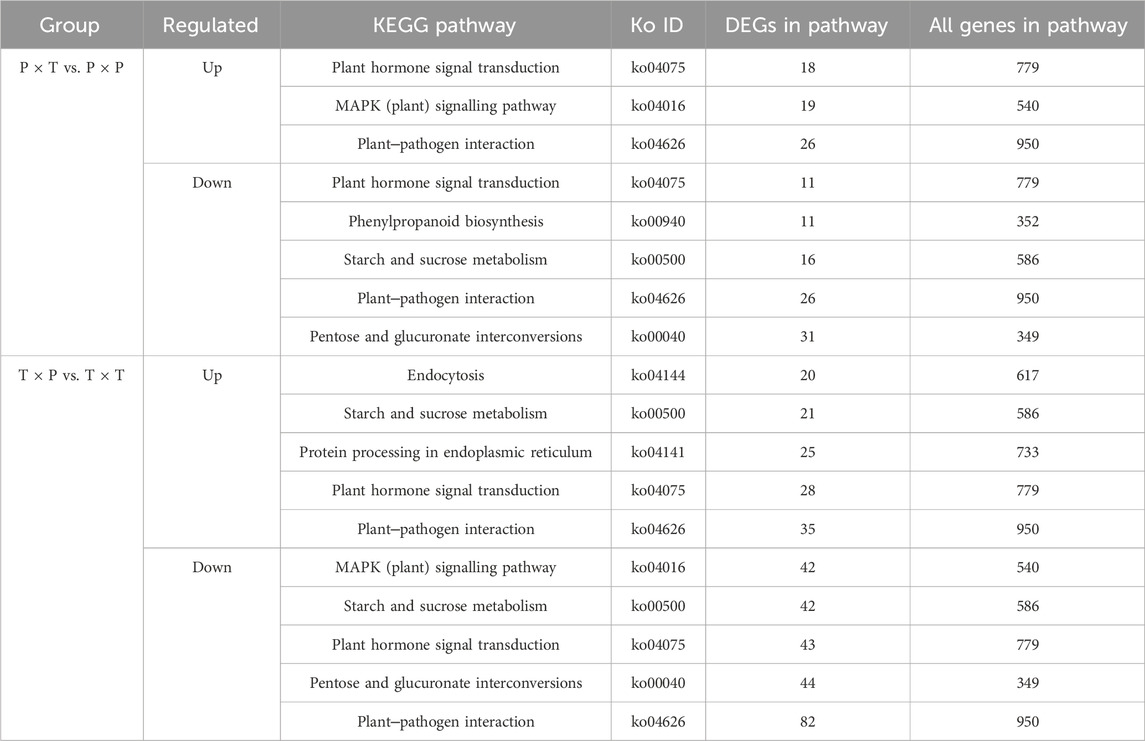
TABLE 4. Enrichment of up- and downregulated DEGs in specific KEGG pathways among stigmas of different P. auriculata pollination.
To obtain further insights, all genes were categorized based on the GO or KEGG databases. Notably, 10 genes were annotated to the plant SI S1 (self-incompatibility protein S1), while 3 genes were annotated to SRK with the full name of “S-locus receptor kinase”, PSEUDOSRKA with the full name of “putative inactive G-type lectin S-receptor-like serine/threonine-protein kinase SRK” as the major determinants of SI. ARC1 (Arm repeat containing 1), THL1/2 (thioredoxin H-like), Exo70A1 (Exocyst compounds), MLPK (M-locus protein kinase), and MOD (Aquaporin) were also found as the downstream regulators of SI. MPK3 (Mitogen-activated protein kinase 3) and MKK2/3 (Mitogen-activated protein kinase kinase 2/3) which belonged to MAPK family, were interacting with FERON (Receptor-like protein kinase FERONIA) to regulate pollen receptivity. OFT23 (O-fucosyltransferase 23) and KCS6 (3-ketoacyl-CoA synthase 6) involved in pollen-stigma interaction; BABL (Basic blue protein), FIMB5 (Fimbrin-5), PLRX4, namely “Pollen-specific leucine-rich repeat extensin-like protein 4”, TFIIB1 (Transcription initiation factor IIB) and AL11A (Pectin Methylesterases) were regulators of pollen tube growth (Table 5).
It was RT‒qPCR that was conducted to verify DEGs’ expression. These selected DEGs were pertinent to self-incompatibility (g.21343, Self-incomp-S1; g.244950, Self-incomp-S1; g.31658, Self-incomp-S1), signal transduction (g.184704, P2C76; g.127811, PUB19; g.148390, PLDD1; g.225308, GDI; g.123279, SAPK2), ABC transporters (g.153399, ABCG1; g.54574, ABCC10; g.87588, ABCG5), plant‒pathogen interaction (g.33756, CML16; g.116435, KCS12) and lipid transport and metabolism (g.163007, FAR; g.236952, HIDM) (Supplementary Table S2). The expression level and correlation analysis showed a consistent trend between RNA-seq and RT‒qPCR analysis (Figure 4).
Studies of crucifers (SSI) have revealed that the S-locus regulates SSI identification, and over a hundred S-haplotypes were present in Brassica napus. It is believed that the SRK-SCR recognition via phosphorylation and degradation facilitates the pollen‒pistil interaction (Gu et al., 1998; Goring et al., 2014) and the SI signal transducers include SRK, SLG, SCR/SP11, and MLPK (Takasaki et al., 2000; Takayama et al., 2000; Murase et al., 2004; Kakita et al., 2007b; Kitashiba and Nasrallah, 2014). Nevertheless, SRK-ARC1 interaction was not functioning in Corylus, indicating that the SSI system was distinct between Corylus and Brassica (Hou et al., 2022). Additionally, the Fagopyrum genome did not contain SRK or SLG (Banović et al., 2010). The following SSI component genes were identified in this HetSI study: SRK, SRK6, PSEUDOSRKA, ARC1, MLPK, THL1/2, and Exo70A1. Furthermore, ARC1, MLPK, THL1/2, Exo70A1, and MOD were also intimately associated with the S-locus. It was speculated that the SI signal in Brassica was transmitted via the “SRK-ARC1-Exo70A1” pathway (Wang et al., 2023). With the fall of self-pollen, SRK and SP11/SCR engaged to generate a “heterotetrameric assembly” (2:2 eSRK: SP11/SCR), and ARC1 interacted with the SRK and the domains of MLPK kinase to regulate SI through phosphorylation (Abhinandan et al., 2023). Subsequently, phosphorylated ARC1 is linked to Exo70A1 (Goring et al., 2014; Doucet et al., 2016). To prevent pollen hydration, the “ACR1-Exo70A1 complex” undergoes translocation and degradation at the “26S proteasome”, which causes the channels of MOD to open. (Maurel et al., 2008; Wang et al., 2023). Serving as a potential inhibitor to prevent spontaneous activation, THL1/2 interacted with SRK (Cabrillac et al., 2001). It is conceivable that the mechanism in HetSI might be similar to SSI. Remarkably, SI S1 was exclusively detected in GSI plants such as Citrus and Papaver (Miao et al., 2015; Goring et al., 2023). This is the first HetSI research of its kind. In the GSI study, the SI S1 protein could significantly inhibit the pollen germination frequency (Miao et al., 2015). Besides, the SI S1 protein of Citrus was upregulated after self-pollination, whereas its expression remained relatively low following cross-pollination. According to the findings, SI S1 was downregulated after SI pollination in both thrum and pin types, which was an opposite trend to that of Citrus (Miao et al., 2015). The results suggest that SI S1 may exhibit diverse role in the HetSI mechanism of P. auriculata. After recognition by the “SRK-ARC1-Exo70A1” pathway, MAPKs, in conjunction with FERON, determined pollen receptivity by regulating the ROS level on the stigma surface (Huang et al., 2023). KCS6 was involved in the accumulation of pollen coat and affected water transfer between pollen and stigma, which afterward regulated pollen hydration (Zhang et al., 2021b). In Arabidopsis, OFT23 may promote pollen tube penetration (Smith et al., 2018). BABL, FIMB5, PLRX4, TFIIB1, and AL11A were also identified as candidates for HetSI (Table 5) and previously reported as regulators of pollen tube formation (Baumberger et al., 2003; Wu et al., 2010; Zhou et al., 2013; Tang et al., 2020; Zhang et al., 2021b).
Furthermore, in addition to coding RNAs (mRNAs), non-coding RNAs (ncRNAs) have been identified as playing multifaceted roles in pollen-stigma recognition. In Brassica, a trans-acting small non-coding RNA (sRNA) Smi (SP11 methylation inducer), has been implicated in facilitating the SRK-SP11 interaction through specific promoter methylation (Finnegan et al., 2011). MicroRNAs (miRNAs), specifically 18-nt miRNAs (novel_miR_49), have been recognized as potential candidates influencing SI in blueberry (Vaccinium sp.). Additionally, miRNAs associated with phytohormone signal transduction have been proposed to play a role in pollen-stigma interactions in SI (Yang et al., 2022). Furthermore, overexpression of the long non-coding RNA (lncRNA) bra-miR5718HG has been shown to inhibit pollen tube growth in Brassica campestris (Shi et al., 2024). However, researches in this field remains largely in its infancy and requires further studies to achieve a more comprehensive understanding of SI mechanisms.
Our previous research demonstrated that HetSI in P. auriculata is characterized by a strict rejection of SI pollen by the dry-type stigma, where no pollen tubes penetrate the papillae. Notably, 2 hours post SC pollination, pollen-stigma cross-linking was evident, with distinct morphological differences observed between the crosses “P × T” and “T × P”. In “T × P”, a “foot” structure penetrated the stigma, unlike in “P × T”, where germinated pollen adhered to the stigma without forming a “foot” structure, and the surrounding papillae became less globular (Hu et al., 2019). Additionally, SC pollen exhibited a faster growth rate on the thrum morphotype compared to the pin morphotype. Metabolomic analyses revealed metabolic variations in the SI responses of pin and thrum flowers (Hu et al., 2019; 2020).
Through RNA-seq and RT‒qPCR, we explored HetSI mechanisms, finding a higher quantity of DEGs in “T × P” vs. “T × T” than in “P × T” vs. “P × P”, suggesting a more complex HetSI mechanism in the former. This indicates that the cross-direction between pin and thrum flowers may influence pollen recognition and germination, with thrum flowers potentially exhibiting a more complex and rapid germination process to offset the pollination disadvantages posed by their lower style position. Furthermore, in GSI studies, MAPK signaling was shown to induce programmed cell death (PCD) to irreversibly inhibit SI pollen (Zhou et al., 2020), whereas in sporophytic self-incompatibility (SSI) research, MAPKs were essential for maintaining stigma receptivity to compatible pollen. Specifically, MPK3/4 and MKK2/3 phosphorylate Exo70A1, localizing it at plasma membranes to regulate exocytosis and facilitate SC pollination in Arabidopsis (Jamshed et al., 2020). In our study, DEGs in the MAPK (plant) signalling pathway were downregulated in “T × P” vs. “T × T” but exhibited the opposite trend in“P × T” vs. “P × P” (Table 4). The identification of female S determinants, such as SRK in Brassica and Arabidopsis for SSI, and SI S1 in Papaver for GSI, along with critical components like PGAP1, in our results (Goring et al., 2023), suggests that P. auriculata combines both SSI and GSI characteristics, similar to another HetSI species, Primula maximowiczii (Lu et al., 2018). HetSI in pin flowers may share pathways with Arabidopsis SSI, where decreased MAPKs in “T × P” vs. “T × T” could inhibit MAPKs - Exo70A1 interaction and reduce pistil receptivity. Conversely, in thrum flowers, resembling GSI in Papaver (Goring et al., 2023), the opposite trend of MAPKs in “P × T” vs. “P × P” could trigger PCD in the stigma and facilitate HetSI.
Comparing SC with SI, most of the DEGs were detected in the “plant‒pathogen interaction”, “plant hormone signal transduction”, “MAPK (plant) signaling”, and “pentose and glucuronate interconversion” pathways. Pathogen–plant interactions are involved in the plant immune system, and entailed specific cell-cell recognition processes that selectively inhibit certain pathogens while facilitating others (Muñoz-Sanz et al., 2020). Two contrasting interaction patterns were included in pathogen-plant interactions that share a similar mechanism with pollen-pistil interactions in SI, and defence-related genes function in both pathogen and SI pollen. SRK-like S-domain RLKs have been implicated in pathogen resistance, recognizing extrinsic ligands (Sanabria et al., 2008; Catanzariti et al., 2015). Numerous stigma-specific genes encode proteins for stress and defence, according to studies in Brassica and Oryza sativa. Additionally, some common cis-regulatory elements of stigma-specific genes are shared with stress-responsive genes (Li et al., 2007). In our comparative group, 950 genes were enriched in the “pathogen–plant pathway”, second only to “carbon metabolism”. A total of 866 genes were in the “defence response (immune system process) pathway”, which was related to GO terms (Table 4; Supplementary Table S3). The results support the hypothesis that pathogen-plant and pollen-stigma interactions might involve some common signalling pathways.
A vital component of P. auriculata pollination is plant hormone signal transduction. Brassinosteroids (BRs) were previously demonstrated to be important phytohormones for reproductive processes in plants (Li and He, 2020). Research on Primula has revealed that female self-incompatibility is influenced by the brassinosteroid-inactivating enzyme, cytochrome P450 CYP734A50 (Huu et al., 2022). To enhance self-fertilization, loss of independent function mutations in CYP734A50 and exogenous BR treatment altered both female compatibility and style morphology (Huu et al., 2022). Consequently, it has been concluded that female self-incompatibility in Primula is mediated by the maintenance of low brassinosteroid (BR) levels within the style (Huu et al., 2022). In our study, CYP92A6 (Typhasterol) in the brassinosteroid biosynthesis pathway decreased sharply in both the “T × P” vs. “T × T” and “P × T” vs. “P × P” comparisons, and CYP85A1 showed the same trend in “T × P” vs. “T × T”, indicating that BR decreased in styles to modify the mate recognition of SI (Supplementary Figure S3). ARF (Auxin response factor), EIN (Ethylene-insensitive protein) and ABF 2 (Abscisic acid-insensitive 5-like protein 5) overexpression indicated that auxin, ethylene and abscisic acid signalling may participate in HetSI. Auxin is likely to participate in pollen-pistile recognition and pollen tube growth (Matsuda et al., 2015; Zhao et al., 2015) and is involved in the SI response in Olea europaea (Solfanelli et al., 2006), Petunia hybrida (Kovaleva and Zakharova, 2003), and Theobroma cacao (Hasenstein and Zavada, 2001); ethylene/abscisic acid were also found to play a vital role in the pollen-pistil interaction in the SI of Petunia (Kovaleva and Zakharova, 2003). In addition, we found that the “ATP-binding cassette (ABC)” transporter B and G family protein participated in the HetSI of P. auriculata. ABCG and ABCB transporters were previously demonstrated to implicate in auxin transport (Balzan et al., 2014; Chang et al., 2016; Liu et al., 2020) for the regulation of male gametophyte development and self-incompatibility, particularly by facilitating pollen tube growth (Gao et al., 2019; Fu et al., 2023). From all above, we can infer that plant hormone plays a pivotal role in HetSI.
Pentose and glucuronate interconversion pathways and the citric acid cycle were energy sources for pollen tube growth. DEGs involved in the interconversions of pentose and glucuronate pathway was upregulated in SC (Table 4). In the TCA cycle (citrate cycle) pathways-related pdhC (pyruvate dehydrogenase E2 component), MDH1 (Malate dehydrogenase), pdhB (pyruvate dehydrogenase E1 component subunit beta), and ACLY (ATP citrate (pro-S)-lyase) were dramatically downregulated in “T × P” vs. “T × T”; MDH1, pdhC, and ACLY exhibited consistent trends in the cross-comparison “P × T” vs. “P × P”. The aforementioned nutrients linked to energy need to be metabolized to create acetyl-CoA, which must brought into the cycle of TCA to enhance ATP generation to facilitate the growth of the pollen tube (Obermeyer et al., 2013). Moreover, pollen germination was deferred in comparison to “T × P” pollinations. The level of energy-related genes was more pronounced in “T × P” vs. “T × T”, and growing pollen tubes could also consume more supplements in “T × P” for faster pollen tube growth.
Fatty acids might play the same role in HetSI; they can provide a nutrient-rich environment for pollen hydration and germination, and the accumulation of fatty acids could abrogate the SI response (Qin et al., 2022). The degradation of fatty acids is an oxidative process channeled into gluconeogenesis via the glyoxylate cycle with concomitant energy release (Hernández et al., 2020). In the “T × P” vs. “T × T” comparison, ACOX1 (acyl-CoA oxidase), HADH (3-hydroxyacyl-CoA dehydrogenase), ECHA (enoyl-CoA hydratase), and HADHA (enoyl-CoA hydratase) in the fatty acid degradation pathway were sharply downregulated. Additionally, ACSL (long-chain acyl-CoA synthetase) showed the same trend in the “P × T” vs. “P × P” comparison, which indicated lower energy needs in SI pollination (Supplementary Figure S4). Moreover, fatty acid degradation might also involve the constituents of surface lipid polymers (cutin and suberine) on the stigma (Heizmann et al., 2000). To facilitate the growth of the pollen tube, lipids assumed at high levels in SC pollinations in our metabolomic study (Hu et al., 2019). In the process of this study, lipid transport and metabolism were also enriched. Lipids play a crucial part in the pollen-stigma capture’s hydrophobic nature (Heizmann et al., 2000) and membrane signalling during the growth of the pollen tube (Potocký et al., 2003), which have a great influence on the pollen-pistil interaction of HetSI.
Flavonoids also serve as energy-related nutrients for pollen tube growth and impact sexual reproduction. According to Lan et al.’s (2017) research, flavonoid accumulation in stigmas could break the SI in Brassica. Flavonoids may also be involved in cell wall loosening on stigmas, which can facilitate SC pollen tube penetration (Wu et al., 2023). In our preliminary metabolic study, the overall content of flavonoids in SC significantly increased (Hu et al., 2019). In this study, we found a dramatic decline in CHS (Chalcone synthase), CHI (Chalcone isomerase), and DFR (Dihydroflavonol 4-reductase) in “T × P”vs. “T × T”, FLS (Flavanol synthase), CHS and DFR in “P × T” vs. “P × P”. These genes all play a significant regulatory role in the accumulation of flavonoids (Nakatsuka et al., 2005) and displayed a similar trend with metabolomic results.
Using high-throughput RNA-seq, we carried out a transcriptomic comparative investigation of SC and SI pollination in Plumbago auriculata in this research. The S-locus in P. auriculata HetSI was further identified. The SSI-related candidates SRK, SRK6, PSEUDOSRKA, ARC1, MLPK, THL1/2 and Exo70A1 and MOD were found involved in HetSI. In addition, SI S1 was one of the key factors in GSI, however, it appeared with an opposite manner in HetSI. Comparing SC with SI, many notable genes potentially involved in HetSI were abundant in “MAPK (plant) signaling pathway”, “plant hormone signal transduction”, “plant-pathogen interaction”, and “pentose and glucuronate interconversions” KEGG pathways. Furthermore, we found that the gene expression of HetSI in different styles was distinct, and the HetSI mechanism seems more complicated in thrum morphotypes. These results offer complementary data for investigating the specific mechanism of HetSI. However, high-throughput techniques can only screen candidates involved in HetSI, molecular interactions in HetSI is still uncertain. Therefore, this research lays a basis for further exploration of molecular interactions in HetSI.
The datasets presented in this study can be found in the Sequence Read Archive (SRA) Repository (https://www.ncbi.nlm.nih.gov/sra). Accession number PRJNA1074424.
DH: Funding acquisition, Investigation, Writing–original draft, Writing–review and editing. DL: Data curation, Project administration, Resources, Supervision, Writing–review and editing. SY: Conceptualization, Methodology, Writing–review and editing. SG: Project administration, Supervision, Visualization, Writing–review and editing. TL: Conceptualization, Methodology, Writing–review and editing. WL: Funding acquisition, Methodology, Writing–original draft. TX: Software, Writing–review and editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by Sichuan Science and Technology Program (2021YJ0497), Science and Technology Research Program of Chongqing Municipal Education Commission (KJQN202303240).
The authors are grateful to Sichuan Science and Technology Department and Chongqing Municipal Education Commission for supporting this work.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2024.1372644/full#supplementary-material
Abhinandan, K., Hickerson, N. M., Lan, X., and Samuel, M. A. (2023). Disabling of ARC1 through CRISPR–Cas9 leads to a complete breakdown of self-incompatibility responses in Brassica napus. Plant Commun. 13 (2), 100504. doi:10.1016/j.xplc.2022.100504
Abhinandan, K., Sankaranarayanan, S., Macgregor, S., Goring, D. R., and Samue, M. A. (2022). Cell–cell signaling during the Brassicaceae self-incompatibility response. Trends Plant Sci. 27, 472–487. doi:10.1016/j.tplants.2021.10.011
Balzan, S., Johal, G. S., and Carraro, N. (2014). The role of auxin transporters in monocots development. Front. Plant Sci. 5, 393. doi:10.3389/fpls.2014.00393
Banović, B., Miljuš-Đukić, J., Konstantinović, M., and Maksimović, V. (2010). A search of Brassica SI-involved orthologs in buckwheat leads to novel buckwheat sequence identification: MLPK possibly involved in SI response. Arch. Biol. Sci. 62, 315–321. doi:10.2298/ABS1002315B
Baumberger, N., Doesseger, B., Guyot, R., Diet, A., Parsons, R. L., Clark, M. A., et al. (2003). Whole-genome comparison of leucine-rich repeat extensins in Arabidopsis and rice. A conserved family of cell wall proteins form a vegetative and a reproductive clade. Plant Phys. 131, 1313–1326. doi:10.1104/pp.102.014928
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B (Methodol.) 57, 289–300. doi:10.1111/j.2517-6161.1995.tb02031.x
Cabrillac, D., Cock, J. M., Dumas, C., and Gaude, T. (2001). The S-locus receptor kinase is inhibited by thioredoxins and activated by pollen coat proteins. Nature 410, 220–223. doi:10.1038/35065626
Catanzariti, A. M., Lim, G. T. T., and Jones, D. A. (2015). The tomato I-3 gene: a novel gene for resistance to Fusarium wilt disease. New Phytol. 207, 106–118. doi:10.1111/nph.13348
Chang, Z., Chen, Z., Yan, W., Xie, G., Lu, J., Wang, N., et al. (2016). An ABC transporter, OsABCG26, is required for anther cuticle and pollen exine formation and pollen-pistil interactions in rice. Plant Sci. 253, 21–30. doi:10.1016/j.plantsci.2016.09.006
Doucet, J., Lee, H. K., and Goring, D. R. (2016). Pollen acceptance or rejection: a tale of two pathways. Trends Plant Sci. 21, 1058–1067. doi:10.1016/j.tplants.2016.09.004
Finnegan, E. J., Liang, D., and Wang, M. B. (2011). Self-incompatibility: Smi silences through a novel sRNA pathway. Trends Plant Sci. 16 (5), 238–241. doi:10.1016/j.tplants.2011.01.002
Fu, Y., Zhang, H., Ma, Y., Li, C., Zhang, K., and Liu, X. (2023). A model worker: multifaceted modulation of AUXIN RESPONSE FACTOR3 orchestrates plant reproductive phases. Front. Plant Sci. 14, 1123059. doi:10.3389/fpls.2023.1123059
Fujii, S., Shimosato-Asano, H., Kakita, M., Kitanishi, T., Iwano, M., and Takayama, S. (2020). Parallel evolution of dominant pistil-side self-incompatibility suppressors in Arabidopsis. Nat. Commun. 11, 1404. doi:10.1038/s41467-020-15212-0
Gao, C., Wang, Y., and Qu, H. (2019). Study of auxin regulation of pollen tube growth through calcium channels in Pyrus pyrifolia. Plant Growth Regul. 89, 99–108. doi:10.1007/s10725-019-00522-1
Goring, D. R., Bosch, M., and Franklin-Tong, V. E. (2023). Contrasting self-recognition rejection systems for self-incompatibility in Brassica and Papaver. Curr. Biol. 33, R530–R542. doi:10.1016/j.cub.2023.03.037
Goring, D. R., Indriolo, E., and Samuel, M. A. (2014). The ARC1 E3 ligase promotes a strong and stable self-incompatibility response in Arabidopsis species: response to the Nasrallah and Nasrallah commentary. Plant Cell 26, 3842–3846. doi:10.1105/tpc.114.131243
Gu, T., Mazzurco, M., Sulaman, W., Matias, D. D., and Goring, D. R. (1998). Binding of an arm repeat protein to the kinase domain of the S-locus receptor kinase. Proc. Natl. Acad. Sci. U. S. A. 95, 382–387. doi:10.1073/pnas.95.1.382
Hasenstein, K. H., and Zavada, M. S. (2001). Auxin modification of the incompatibility response in Theobroma cacao. Physiol. Plant. 112, 113–118. doi:10.1034/j.1399-3054.2001.1120115.x
Heizmann, P., Luu, D. T., and Dumas, C. (2000). Pollen-stigma adhesion in the Brassicaceae. Ann. Bot. 85, 23–27. doi:10.1006/anbo.1999.1057
Hernández, M. L., Lima-Cabello, E., Alché, J. D. D., Martínez-Rivas, J. M., and Castro, A. J. (2020). Lipid composition and associated gene expression patterns during pollen germination and pollen tube growth in olive (Olea europaea L.). Plant Cell Physiol. 61, 1348–1364. doi:10.1093/pcp/pcaa063
Hou, S., Zhao, T., Yang, Z., Yang, D., Li, Q., Liang, L., et al. (2022). Molecular cloning and yeast two-hybrid provide new evidence for unique sporophytic self-incompatibility system of Corylus. Plant Biol. J. 24, 104–116. doi:10.1111/plb.13347
Hu, D., Gao, S., Li, W., Lei, T., Liu, Y., Li, Q., et al. (2020). Dissecting the distally response to pollination using metabolite profiling in heteromorphic incompatibility system interactions of Plumbago auriculata Lam. Acta Physiol. Plant. 42, 122. doi:10.1007/s11738-020-03111-2
Hu, D., Li, W., Gao, S., Lei, T., Hu, J., Shen, P., et al. (2019). Untargeted metabolomic profiling reveals that different responses to self and cross pollination in each flower morph of the heteromorphic plant Plumbago auriculata. Plant Physiol. biochem. 144, 413–426. doi:10.1016/j.plaphy.2019.10.010
Huang, J., Yang, L., Yang, L., Wu, X., Cui, X., Zhang, L., et al. (2023). Stigma receptors control intraspecies and interspecies barriers in Brassicaceae. Nature 614, 303–308. doi:10.1038/s41586-022-05640-x
Huu, C. N., Plaschil, S., Himmelbach, A., Kappel, C., and Lenhard, M. (2022). Female self-incompatibility type in heterostylous Primula is determined by the brassinosteroid-inactivating cytochrome P450 CYP734A50. Curr. Biol. 32, 671–676.e5. doi:10.1016/j.cub.2021.11.046
Jain, S., Maurya, P., Jain, S., Kumar, V., Amulya, S., Kiran, B., et al. (2023). Incompatibility systems in fruit crops: applications and achievements. Int. J. Environ. Clim. Chang. 13, 2653–2663. doi:10.9734/IJECC/2023/v13i92496
Jamshed, M., Sankaranarayanan, S., Abhinandan, K., and Samuel, A. M. (2020). Stigma receptivity is controlled by functionally redundant MAPK pathway components in Arabidopsis. Molec. Plant 13, 1582–1593. doi:10.1016/j.molp.2020.08.015
Kakita, M., Murase, K., Iwano, M., Matsumoto, T., Watanabe, M., Shiba, H., et al. (2007b). Two distinct forms of M-locus protein kinase localize to the plasma membrane and interact directly with S-locus receptor kinase to transduce self-incompatibility signaling in Brassica rapa. Plant Cell 19, 3961–3973. doi:10.1105/tpc.106.049999
Kakita, M., Shimosato, H., Murase, K., Isogai, A., and Takayama, S. (2007a). Direct interaction between S-locus receptor kinase and M-locus protein kinase involved in Brassica self-incompatibility signaling. Plant Biotechnol. 24, 185–190. doi:10.5511/plantbiotechnology.24.185
Kitashiba, H., and Nasrallah, J. B. (2014). Self-incompatibility in Brassicaceae crops: lessons for interspecific incompatibility. Breed. Sci. 64, 23–37. doi:10.1270/jsbbs.64.23
Kovaleva, L., and Zakharova, E. (2003). Hormonal status of the pollen-pistil system at the progamic phase of fertilization after compatible and incompatible pollination in Petunia hybrida L. Sex. Plant Reprod. 16, 191–196. doi:10.1007/s00497-003-0189-1
Lan, X., Yang, J., Kumar, A., Nie, Y., Li, X., Li, Y., et al. (2017). Flavonoids and ROS play opposing roles in mediating pollination in ornamental kale (Brassica oleracea var. acephala). Mol. Plant. 10, 1361–1364. doi:10.1016/j.molp.2017.08.002
Li, J., Li, P., Li, J., Zhang, L., and Zhang, X. (2023). Morphological characteristics and molecular markers of distally in Primula obconica. Euphytica 219, 72. doi:10.1007/s10681-023-03198-x
Li, M., Xu, W., Yang, W., Kong, Z., and Xue, Y. (2007). Genome-wide gene expression profiling reveals conserved and novel molecular functions of the stigma in rice. Plant Physiol. 144, 1797–1812. doi:10.1104/pp.107.101600
Li, Z., and He, Y. (2020). Roles of brassinosteroids in plant reproduction. Int. J. Mol. Sci. 21, 872. doi:10.3390/ijms21030872
Lian, X., Zhang, S., Huang, G., Huang, L., Zhang, J., and Hu, F. (2021). Confirmation of a gametophytic self-incompatibility in Oryza longistaminata. Oryza Longistaminata. Front. Plant. Sci. 12, 576340. doi:10.3389/fpls.2021.576340
Liu, L., Zhao, L., Chen, P., Cai, H., Hou, Z., Jin, X., et al. (2020). ATP binding cassette transporters ABCG1 and ABCG16 affect reproductive development via auxin signalling in Arabidopsis. Plant J. 102 (6), 1172–1186. doi:10.1111/tpj.14690
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi:10.1006/meth.2001.1262
Lu, W., Bian, X., Yang, W., Cheng, T., Wang, J., Zhang, Q., et al. (2018). Transcriptomics investigation into the mechanisms of self-incompatibility between pin and thrum morphs of Primula maximowiczii. Int. J. Mol. Sci. 19, 1840. doi:10.3390/ijms19071840
Lv, S., Qiao, X., Zhang, W., Li, Q., Wang, P., Zhang, S., et al. (2022). The origin and evolution of RNase T2 family and gametophytic self-incompatibility system in plants. Genome Biol. Evol. 14, evac093. doi:10.1093/gbe/evac093
Matsuda, T., Matsushima, M., Nabemoto, M., Osaka, M., Sakazono, S., Masuko-Suzuki, H., et al. (2015). Transcriptional characteristics and differences in Arabidopsis stigmatic papilla cells pre- and post-pollination. Plant Cell Physiol. 56, 663–673. doi:10.1093/pcp/pcu209
Matsui, K., and Yasui, Y. (2020). Buckwheat heteromorphic self-incompatibility: Genetics, genomics and application to breeding. Breed. Sci. 70, 32–38. doi:10.1270/jsbbs.19083
Maurel, C., Verdoucq, L., Luu, D. T., and Santoni, V. (2008). Plant aquaporins: membrane channels with multiple integrated functions. Annu. Rev. Plant Biol. 59, 595–624. doi:10.1146/annurev.arplant.59.032607.092734
McCubbin, A. (2008). “Heteromorphic self-incompatibility in primula: twenty-first century tools promise to unravel a classic nineteenth century model system,” in Self-incompatibility in flowering plants: evolution, diversity, and mechanisms. Editor V. E. Franklin-Tong (Berlin, Heidelberg: Springer), 289–308.
Miao, H., Ye, Z., Hu, G., and Qin, Y. (2015). Comparative transcript profiling of gene expression between self-incompatible and self-compatible mandarins by suppression subtractive hybridization and cDNA microarray. Mol. Breed. 35, 47. doi:10.1007/s11032-015-0204-x
Mucina, L., and Hammer, T. (2019). Limonium dagmarae (Plumbaginaceae), a new species from Namaqualand coast, South Africa. Phytotaxa 403, 71–85. doi:10.11646/phytotaxa.403.2.1
Muñoz-Sanz, J. V., Zuriaga, E., Cruz-García, F., McClure, B., and Romero, C. (2020). Self-(in)compatibility systems: target traits for crop-production, plant breeding, and biotechnology. Front. Plant Sci. 11, 195. doi:10.3389/fpls.2020.00195
Murase, K., Shiba, H., Iwano, M., Che, F. S., Watanabe, M., Isogai, A., et al. (2004). A membrane-anchored protein kinase involved in Brassica self-incompatibility signaling. Science 303, 1516–1519. doi:10.1126/science.1093586
Nakatsuka, T., Nishihara, M., Mishiba, K., and Yamamura, S. (2005). Temporal expression of flavonoid biosynthesis-related genes regulates flower pigmentation in gentian plants. Plant Sci. 168, 1309–1318. doi:10.1016/j.plantsci.2005.01.009
Obermeyer, G., Fragner, L., Lang, V., and Weckwerth, W. (2013). Dynamic adaption of metabolic pathways during germination and growth of lily pollen tubes after inhibition of the electron transport chain. Plant Physiol. 162, 1822–1833. doi:10.1104/pp.113.219857
Pauline, M., Servane, B., Corentin, D., Francisco, C., Juan-Pablo, M., Stanley, L., et al. (2023). Intra- and inter-specific reproductive barriers in the tomato clade. Front. Plant. Sci. 14, 1326689. doi:10.3389/fpls.2023.1326689
Potocký, M., Eliás, M., Profotová, B., Novotná, Z., Valentová, O., and Zárský, V. (2003). Phosphatidic acid produced by phospholipase D is required for tobacco pollen tube growth. Planta 217, 122–130. doi:10.1007/s00425-002-0965-4
Qin, H., Li, H., Abhinandan, K., Xun, B., Yao, K., Shi, J., et al. (2022). Fatty acid biosynthesis pathways are downregulated during stigma development and are critical during self-incompatible responses in ornamental kale. Int. J. Mol. Sci. 23, 13102. doi:10.3390/ijms232113102
Sanabria, N., Goring, D., Nürnberger, T., and Dubery, I. (2008). Self/nonself perception and recognition mechanisms in plants: a comparison of self-incompatibility and innate immunity. New Phytol. 178, 503–514. doi:10.1111/j.1469-8137.2008.02403.x
Sassa, H. (2016). Molecular mechanism of the S-RNase-based gametophytic self-incompatibility in fruit trees of Rosaceae. Breed. Sci. 66, 116–121. doi:10.1270/jsbbs.66.116
Shi, D., Huang, H., Zhang, Y., Qian, Z., Du, J., Huang, L., et al. (2024). The roles of non-coding RNAs in male reproductive development and abiotic stress responses during this unique process in flowering plants. Plant Sci. 341, 111995. doi:10.1016/j.plantsci.2024.111995
Smith, D. K., Jones, D. M., Lau, J. B. R., Cruz, E. R., Brown, E., Harper, J. F., et al. (2018). A putative protein o-Fucosyltransferase facilitates pollen tube penetration through the stigma-style interface. Plant Phys. 176, 2804–2818. doi:10.1104/pp.17.01577
Solfanelli, C., Bartolini, S., Vitagliano, C., and Lorenzi, R. (2006). Immunolocalization and quantification of IAA after self- and free-pollination in Olea europaea L. Sci. Hortic. 110, 345–351. doi:10.1016/j.scienta.2006.06.026
Takasaki, T., Hatakeyama, K., Suzuki, G., Watanabem, M., Isogai, A., and Hinata, K. (2000). The S receptor kinase determines self-incompatibility in Brassica stigma. Nature 403, 913–916. doi:10.1038/35002628
Takayama, S., Shiba, H., Iwano, M., Shimosato, H., Che, F. S., Kai, N., et al. (2000). The pollen determinant of self-incompatibility in Brassica campestris. Proc. Natl. Acad. Sci. U. S. A. 97, 1920–1925. doi:10.1073/pnas.040556397
Tang, C., Zhu, X., Qiao, X., Gao, H., Li, Q., Wang, P., et al. (2020). Characterization of the pectin methyl-esterase gene family and its function in controlling pollen tube growth in pear (Pyrus bretschneideri). Genomics 112, 2467–2477. doi:10.1016/j.ygeno.2020.01.021
Wang, F., Li, Y., Li, G., and Cheng, S. (2023). Genetic components of self-incompatibility in brassica vegetables. Horticulturae 9, 265. doi:10.3390/horticulturae9020265
Wu, H. M., Xie, D. J., Jia, P. F., Tang, Z. S., Shi, D. Q., Shui, G. H., et al. (2023). Homeostasis of flavonoids and triterpenoids most likely modulates starch metabolism for pollen tube penetration in rice. Plant Biotechnol. J. 21, 1757–1772. doi:10.1111/pbi.14073
Wu, Y., Yan, J., Zhang, R., Qu, X., Huang, S., Chen, N., et al. (2010). Arabidopsis FIMBRIN5, an actin bundling factor, is required for pollen germination and pollen tube growth. Plant Cell 22, 3745–3763. doi:10.1105/tpc.110.080283
Yang, Q., Fu, Y., Zhang, T., Peng, S., and Deng, J. (2022). Identification of microRNAs related to phytohormone signal transduction and self-incompatibility of rabbiteye blueberry pollen. J. Am. Soc. Hortic. 147 (6), 300–311. doi:10.21273/JASHS05143-21
Zhang, J., Yue, L., Wu, X., Liu, H., and Wang, W. (2021a). Function of small peptides during male-female crosstalk in plants. Front. Plant Sci. 12, 671196. doi:10.3389/fpls.2021.671196
Zhang, Z., Zhan, H., Lu, J., Xiong, S., Yang, N., Yuan, H., et al. (2021b). Tapetal 3-Ketoacyl-aoenzyme a synthases are involved in pollen coat lipid accumulation for pollen-stigma interaction in Arabidopsis. Front. Plant Sci. 12, 770311. doi:10.3389/fpls.2021.770311
Zhao, P., Zhang, L., and Zhao, L. (2015). Dissection of the style's response to pollination using transcriptome profiling in self-compatible (Solanum pimpinellifolium) and self-incompatible (Solanum chilense) tomato species. BMC Plant Biol. 15, 119. doi:10.1186/s12870-015-0492-7
Zhou, J., Liang, Y., Niu, Q., Chen, L., Zhang, X., and Ye, D. (2013). The Arabidopsis general transcription factor TFIIB1 (AtTFIIB1) is required for pollen tube growth and endosperm development. J. Exp. Bot. 8, 2205–2218. doi:10.1093/jxb/ert078
Keywords: heteromorphic self-incompatibility, pollination, pollen-stigma interaction, transcriptome, Plumbago auriculata Lam
Citation: Hu D, Lin D, Yi S, Gao S, Lei T, Li W and Xu T (2024) Comparative stigmatic transcriptomics reveals self and cross pollination responses to heteromorphic incompatibility in Plumbago auriculata Lam.. Front. Genet. 15:1372644. doi: 10.3389/fgene.2024.1372644
Received: 18 January 2024; Accepted: 12 February 2024;
Published: 06 March 2024.
Edited by:
Milind B. Ratnaparkhe, ICAR Indian Institute of Soybean Research, IndiaReviewed by:
Aroldo Cisneros, National Polytechnic Institute (IPN), MexicoCopyright © 2024 Hu, Lin, Yi, Gao, Lei, Li and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shouli Yi, c2hvdWxpeWlAc2ljbnUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.