
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 07 May 2024
Sec. Genetics of Common and Rare Diseases
Volume 15 - 2024 | https://doi.org/10.3389/fgene.2024.1367399
Background: Numerous studies have demonstrated a positive association between the level of tissue inhibitor of metalloproteinase 3 (TIMP3) and chronic kidney disease (CKD). Nevertheless, whether those associations reflect causal links still to be determined. This study intended to research the causal relationship of TIMP3 with CKD and markers of kidney function, such as creatinine-based estimated glomerular filtration rate (eGFRcrea), cystatin C-based estimated glomerular filtration rate (eGFRcys), eGFRcrea in diabetics (eGFRcrea (DM)) and eGFRcrea in non diabetics (eGFRcrea (No DM)).
Methods: In this study, we investigated the causal relationships between TIMP3 and CKD and kidney function markers using a two-sample Mendelian randomization (MR) technique. We used summary level datasets for TIMP3 and CKD from genome-wide association studies that we were able to access through the study by Suhre K and Pattaro C.
Results: We found that TIMP3 had a significant positive causal effect on the risk of CKD (Inverse variance weighted (IVW):odds ratio (OR):0.962, 95% confidence interval (CI): (0.936-0.988),P:0.005). However TIMP3 levels had no significant effect on risk of eGFRcys (PIVW: 0.114),eGFRcrea (PIVW:0.333). After grouping patients based on their diabetes status, we found that genetically higher levels of TIMP3 had a significant impact on eGFRcrea in participants without diabetes (OR:1.003,95%CI (1.001-1.006),P IVW:0.007), but not in participants with diabetes (PIVW = 0.057). Heterogeneity and pleiotropy analyses were carried out to verify the accuracy of the MR findings. Their findings were all not statistically significant.
Conclusion: Our study suggests that TIMP3 may be causally associated with CKD and eGFRcrea (No DM)in people of European ancestry. Strategies aimed to increase TIMP3 levels may provide new ways to delay the deterioration of renal function.
Recently, chronic kidney disease (CKD) has drawn increasing interest in the field of human health (Shi et al., 2018). The treatment for CKD often imposes a significant finacial burden on both society and families (Xie et al., 2018). Renal function test markers serve as indicators to evaluate function of the kidney. Indicators of kidney function that are most frequently utilized include estimated glomerular filtration rate (eGFR). It comprises two components: eGFRcrea, which is based on creatinine, and eGFRcys, which is based on cystatin C (Gowda et al., 2010; Dahlén and Björkhem-Bergman, 2022). Then, depending on whether the patient had diabetes or not, the eGFRcrea was divided into two types in our study:eGFRcrea in diabetics (eGFRcrea (DM)) and eGFRcrea in non-diabetics (eGFRcrea (No DM)).
Tissue inhibitors of metalloproteinases (TIMPs) are endogenous protein regulators who modulate the activity of matrix metalloproteinases (MMPs) (Fan and Kassiri, 2020). TIMP3 suppresses all MMPs and possesses the broadest inhibitory scope against metalloproteinases (Brew and Nagase, 2010). Furthermore, TIMP3 expression in the kidney is higher when compared to other TIMPs (Catania et al., 2007). According to earlier research, TIMP3 is essential for the development of chronic kidney illnesses, including renal carcinoma, diabetic nephropathy, chronic nephritis, and other kidney diseases (Kassiri et al., 2009; Fiorentino et al., 2013; Liu and Zhang, 2021; Zhou et al., 2022). However, no randomized controlled trials (RCTs) have been carried out to investigate the connection between CKD and TIMP3.
Mendelian randomization (MR) is a method of epidemiological research that has undergone significant developments in the last few years (Bowden and Holmes, 2019). Genetic variation is used as an instrumental variable (IV) for the estimation of the causal relationship between exposure and outcome (Emdin et al., 2017). The advantage of MR is that it creates a scenario similar to randomized controlled trial, which effectively minimized the influence of residual confounders and reverse causality (Davey Smith and Hemani, 2014). In light of this, the purpose of our work was to use a two-sample MR technique to examine the relationship between TIMP3, CKD and renal function (measured by biochemical markers and the presence or absence of diabetes). Two large-sample genome-wide association studies (GWAS) of European ancestry provided summary statistics that we used.
Figure 1 provides a concise view of the two-sample MR design between TIMP3 and CKD, as well as kidney function. We performed MR analyses using summary statistics from a GWAS to investigate the impact of TIMP3 on the risk of CKD and kidney function. In order to ensure the validity of IVs, Assumptions needed to be met in three key areas (Davies et al., 2018): (i)Exposure should be significantly influenced by genetic variants; (ii)it is imperative that the genetic variants employed are free from any confounding factors; (iii)It is only the exposure indirectly that affects the outcome, and not other biological pathways.
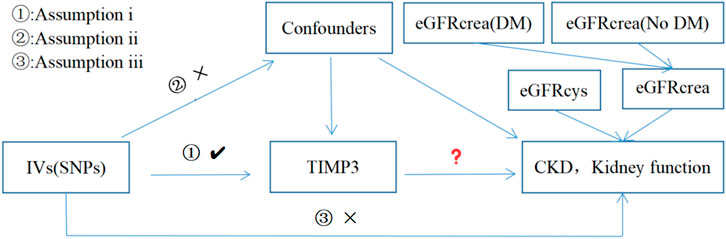
Figure 1. Three assumptions for IVs in MR analysis. Abbreviations: TIMP3, tissue inhibitor of metalloproteinase 3; SNPs, single nucleotide polymorphisms; IVs, instrumental variables; CKD, chronic kidney disease; eGFRcrea, estimated glomerular filtration rate; eGFRcys, cystatin C estimated glomerular filtration rate; DM, diabetes mellitus.
The TIMP3 summary-level data used in this investigation was obtained from a cohort study of approximately 1,000 individuals, which was made available through the Suhre K study (Suhre et al., 2017). All summary statistics pertaining to CKD and kidney function were obtained from the study of Pattaro C (Pattaro et al., 2016) which exclusively used continuous biomarkers, including CKD (N = 117165), eGFRcys (N = 32834), eGFRcrea (N = 133413),eGFRcrea (No DM) (N = 118448), and eGFRcrea (DM) (N = 11522). This study’s data are all of European descent. Moreover, the open-access GWAS dataset at https://gwas.mrcieu.ac.uk/contains the summary data for both GWAS studies. Further details regarding each dataset can be found in Table 1.
We will use the following criteria as SNP screening for TIMP3 in order to ensure the effectiveness of IVs.First, the genome-wide significance was set at p < 5e-8. Then, in order to mitigate the impact of linkage imbalance, we conducted the caking process (r2:0.1; window:10,000 KB) (Purcell et al., 2007) reference 1,000 genomes European Panel (Auton et al., 2015). We ended up with seven SNPs in Table 2. When reconciling exposure and outcome data, palindromic SNPS with moderate allele frequencies were removed. A MR Steiger filter test was then performed and SNPs suggesting reverse causality were removed (Hemani et al., 2017). Furthermore, PhenoScanner V2 was searched for phenotypes linked to these SNPs (Kamat et al., 2019). Here, we discovered that there was no significant correlation between any of these IVs and possible confounders (risk factors for renal impairment). And ensure that the F statistic is > 10, excluding the possibility of weak instrumental variable bias (Burgess and Thompson, 2011).
Examined the genetic association between IVs of TIMP3 level and the risk of CKD using a two-sample MR analysis. We used inverse variance weighting (IVW) (Pierce et al., 2011) as the main analysis method, MR-Egger, weighted median (WM), weighted model, and simple model as supplementary methods (Bowden et al., 2015; Bowden et al., 2016; Hartwig et al., 2017; Hemani et al., 2018). Then, we evaluated the second and third hypotheses indirectly by performing a sensitivity analysis. First, we used the Cochrans Q test to determine the inter-IV heterogeneity (Cohen et al., 2015). Secondly, the MR-PRESSO global test (Verbanck et al., 2018) and MR-Egger regression (Bowden et al., 2015) were used to test the horizontal pleiotropy of IVs. Finally, a leave-out analysis was conducted to determine the potential bias effect of a single SNP on MR Estimates (Burgess et al., 2017). We conducted MR Analysis using the TwoSampleMR R package (version 0.5.6) in RStudio software (version 4.3.1). We consider p < 0.05 as statistically significant.
The forest plots (Figure 2) showed the causal effect of TIMP3 on kidney function. The IVW analysis revealed that TIMP3 has a causal impact on the probability of developing CKD (IVW: OR = 0.962, 95% CI(0.936-0.988), p = 0.005; WM: OR = 0.969, 95% CI(0.94-0.999), p = 0.045). Furthermore, we found a causal relationship between TIMP3 and eGFRcream (No DM) (IVW:OR = 1.003,95%CI (1.001-1.006),p = 0.007). Results of the forest and scatter plots for each outcome were shown in Figures 3, 4.
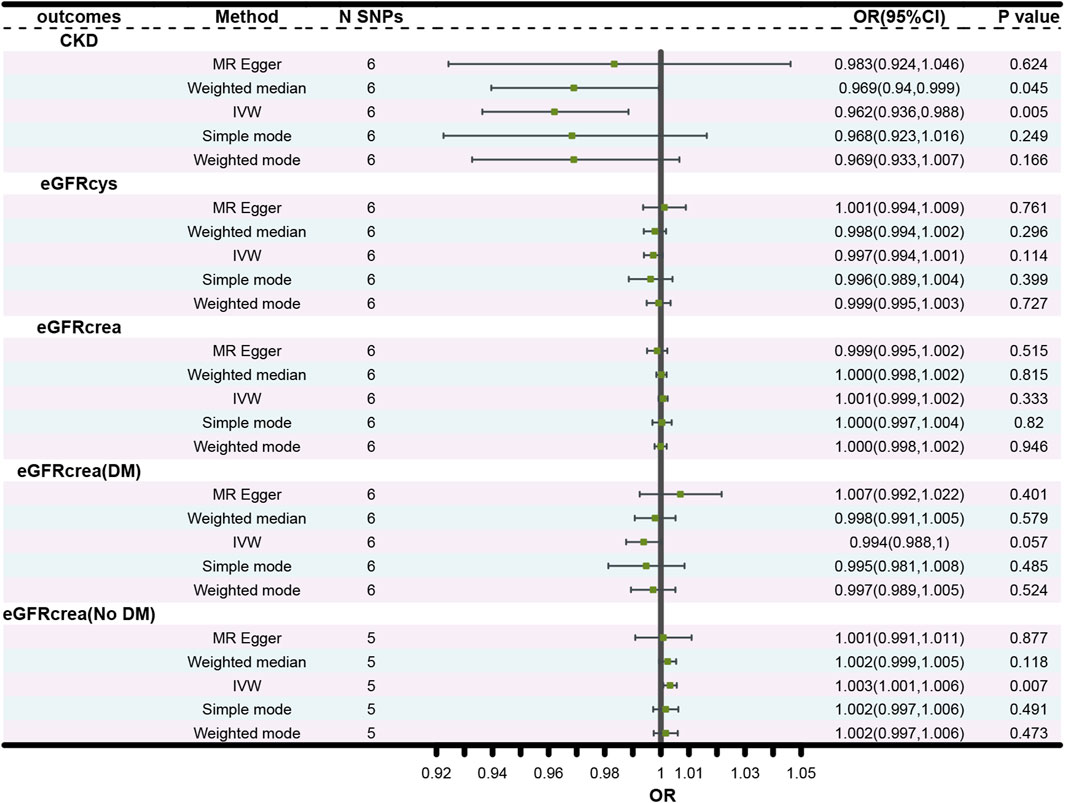
Figure 2. Forest plot of the main MR study investigating the causal effect of TIMP3 on CKD and kidney function. Abbreviations: OR, odds ratio; CI, confidence interval; IVW, Inverse variance weighted; N SNPs, number of single nucleotide polymorphisms; CKD, chronic kidney disease; eGFRcrea, estimated glomerular filtration rate; eGFRcys, cystatin C estimated glomerular filtration rate; DM, diabetes mellitus.
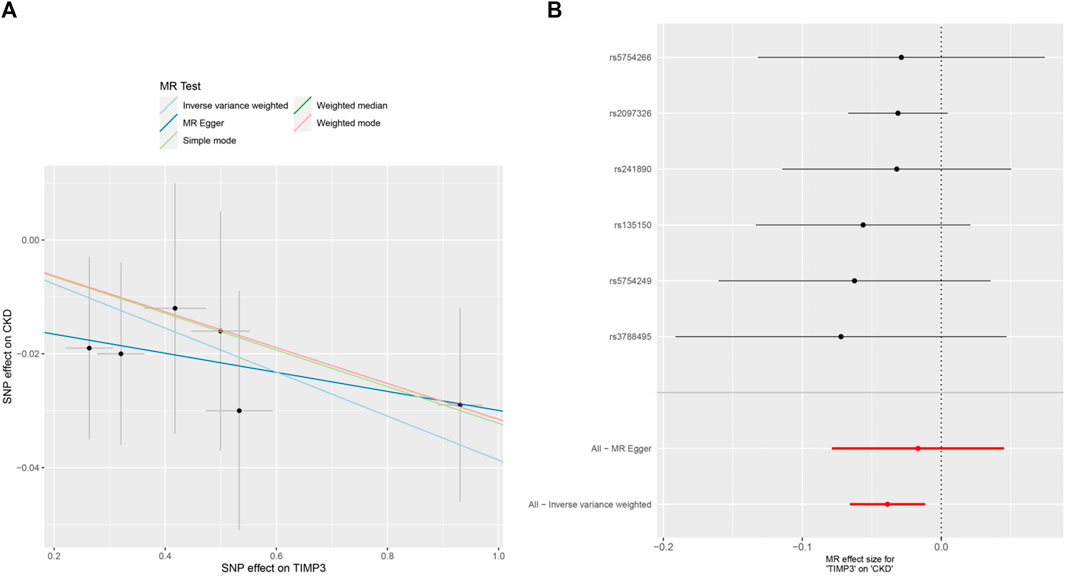
Figure 3. MR analysis visualizations of the effect of TIMP3 on CKD. (A) Scatter plot of the effect size for each SNP on TIMP3 and CKD, (B) Forest plotof the effect size for each SNP on TIMP3 and CKD. Abbreviations:CKD:chronic kidney disease, TIMP3:tissue inhibitor of metalloproteinase 3,SNP:single nucleotide polymorphism.
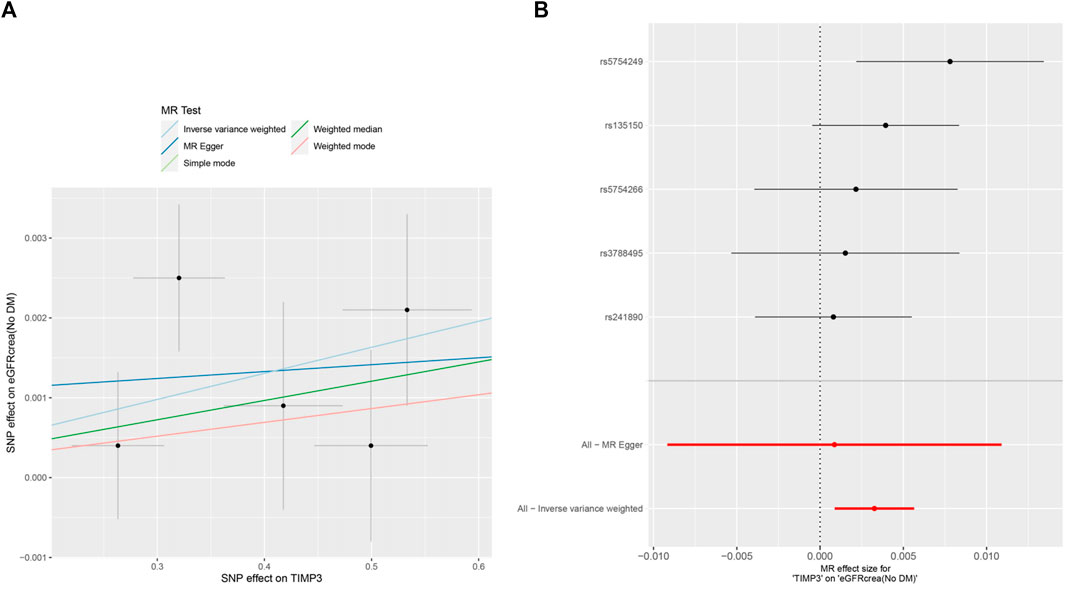
Figure 4. MR analysis visualizations of the effect of TIMP3 on eGFRcrea (No DM). (A) Scatter plot of the effect size for each SNP on TIMP3 and eGFRcrea (No DM). (B) Forest plotof the effect size for each SNP on TIMP3 and eGFRcrea (No DM). Abbreviations:eGFRcrea (No DM), creatinine-based estimated glomerular filtration rate in non-diabetics; TIMP3, tissue inhibitor of metalloproteinase 3,SNP:single nucleotide polymorphism.
Subsequently,a sensitivity analysis was then performed to evaluate how reliable our findings were. Initially, all p values above 0.05 in the Cochran’s Q test results showed no discernible heterogeneity among the independent variables (Table 3). Secondly, there was no indication of pleiotropy for either CKD or eGFR, as shown by the MR-PRESSO global test and MR-Egger regression with all p > 0.05 (Table 3). Finally, the leave-one analysis revealed that the outcomes were not influenced by any SNPs(Figure 5).
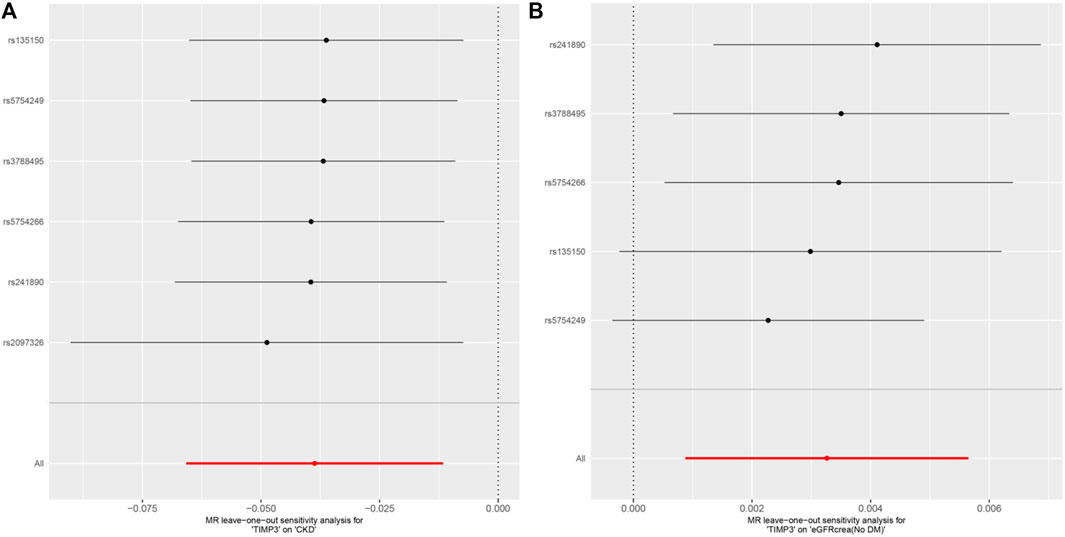
Figure 5. MR analysis visualizations of the effect of TIMP3 on CKD and eGFRcrea (No DM). (A) Scatter plot of the effect size for each SNP on TIMP3 and eGFRcrea (No DM). (B) Forest plotof the effect size for each SNP on TIMP3 and eGFRcrea (No DM). Abbreviations:CKD, chronic kidney disease; eGFRcrea (No DM), creatinine-based estimated glomerular filtration rate in non-diabetics; TIMP3, tissue inhibitor of metalloproteinase 3; SNP, single nucleotide polymorphism.
There was no causal association between TIMP3 and the other types of eGFR. The p values for all 3 MR methods were greater than 0.05 (Figure 2).
In this comprehensive two-sample MR analysis of the associations between TIMP3 and kidney function, we have provided the first suggestive evidence that higher TIMP3 levels decrease CKD, and we observed a significant negative causal relationship between TIMP3 and eGFRcrea (No DM). However, we did not find any genetic associations between TIMP3 and the other kidney function biomarkers.
Dysregulation of MMPs in CKD has been well-established. Studies using mouse models have indicated that genetic targeting of selected MMPs may have detremental effects on kidney function (Wozniak et al., 2021). Numerous studies have demonstrated that MMPs may cause rupture of the glomerular basement membrane, digestion of ECM components that are linked to renal scarring and fibrosis (Zakiyanov et al., 2019), as well as the activation of inflammatory cells and induction of autophagy in kidney tissue (Toba and Lindsey, 2019; Zheng et al., 2022). However, TIMP3, an inhibitor of MMPs (Brew and Nagase, 2010), can reduce the damaging effects of MMPs on the kidney. Renal fibrosis, a common outcome of CKD, plays a pivotal role in the development of renal insufficiency (Liu, 2011). In animal models of kidney injury, such as unilateral urethral obstruction (UUO), the absence of TIMP3 has been found to activate multiple signalling pathways, including TGFβ/Smad, TACE/TNF-α, and mitogen-activated protein kinase pathways. This leads to heightened renal fibrosis, injury, and apoptosis following UUO (Wang et al., 2014). Another study also demonstrated that high expression of TIMP3 alleviates kidney damage through the TGFβ pathway (Chatterjee et al., 2023). In human kidney disease, TIMP3 is primarily upregulated in proximal renal tubules and vascular compartments to mitigate interstitial nephritis and fibrosis, potentially serving as a protective mechanism to minimize kidney damage (Kassiri et al., 2009). Therefore, the findings of this study align with previous research and suggest that an elevated level of TIMP3 has a safeguarding impact on CKD. As with psoriatic arthritis, pharmacogenomics may be useful in selecting etanercept for treatment (Murdaca et al., 2017). Some studies have shown that pracinostat increases the expression of TIMP3, which plays an important role in inhibiting disease progression in human glioma (Chen et al., 2022). This may provide valuable insights for the treatment of CKD.
Diabetic nephropathy is a key factor in the development of CKD (Molitch et al., 2003), with its defining features being an increase in albuminuria, decline in the glomerular filtration rate (GFR), and loss of podocytes. In a retrospective study, the potential roles of TIMP3 and its related noncoding RNA in the progression of diabetic nephropathy were investigated (Casagrande et al., 2021). An example of the moderation of podocellular damage in diabetic nephrotic disease is the overexpression of non-coding RNA (lincRNA) 4930556M19Rik which downregulates miR-27a-3p and upregulates TIMP3 (Fan and Zhang, 2020). Deficiencies in TIMP3 worsen diabetic renal injury, characterized by mesangial dilation and increased microalbuminuria, and affecting the progression of diabetic nephropathy through changes to Akt, ERK1/2, and PKC signaling pathways (Basu et al., 2012). Another study has confirmed that TIMP-3 deficiency contributes to the development of diabetic nephropathy through FoxO1/STAT1 interactions (Fiorentino et al., 2013). These studies have indicated that TIMP3 has potential as a biomarker or therapeutic target for renal diabetes (Casagrande et al., 2021). In this study, high levels of TIMP3 were found to increase eGRFcrea (No DM), but no causal relationship was identified for eGRFcrea (DM). Therefore, additional genetic research is needed to clarify the correlation between blood TIMP3 levels and kidney function in diabetic patients.
The current study has a number of advantages. First of all, it constitutes the initial MR study to examine the plausible impact of escalated TIMP3 levels in reducing renal harm and promoting eGFRcrea (No DM) in a particular population subset, thereby establishing a causal link. Secondly, to reduce the possibility of confounding effects and reverse causality, this MR study makes use of two enormous sets of GWAS data from European populations. Thirdly, kidney function is less susceptible to other factors, and the MR Analysis provides insight into the long-term effects of genetically determined TIMP3 levels on that risk.
There are limitations to this study that need to be acknowledged. Firstly, every individual we have researched is of European ancestry. Hence, further research is needed to ascertain whether the generalization of findings is possible for individuals of other ethnicities. Secondly, due to a lack of identified instrumental variables, we had to adopt a more lenient screening criterion (r2:0.1) to investigate the causal connection between TIMP-3 and renal function. This may potentially affect the reliability of our findings. Additionally, due to our reliance on pooled GWAS data and the absence of complete clinical subject information, we were unable to conduct a subgroup analysis to explore possible variations further.
Our study has shown that high levels of TIMP3 are linked with a lower risk of chronic kidney disease and an increased eGFRcrea (No DM). These results indicate a potential new method for preserving renal function.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Ethical approval was not required for the studies involving humans because The human studies did not require additional ethical approval, the publicly available GWAS summary datasets were collected in studies that had already received the necessary ethical approval. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
HC: Writing–original draft, Writing–review and editing, Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization. LC: Investigation, Methodology, Writing–original draft. YC: Validation, Writing–original draft. QG: Resources, Visualization, Writing–original draft. SL: Conceptualization, Writing–review and editing.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Auton, A., Brooks, L. D., Durbin, R. M., Garrison, E. P., Kang, H. M., Korbel, J. O., et al. (2015). A global reference for human genetic variation. Nature 526, 68–74. doi:10.1038/nature15393
Basu, R., Lee, J., Wang, Z., Patel, V. B., Fan, D., Das, S. K., et al. (2012). Loss of TIMP3 selectively exacerbates diabetic nephropathy. Am. J. Physiol. Ren. Physiol. 303, F1341–F1352. doi:10.1152/ajprenal.00349.2012
Bowden, J., Davey Smith, G., and Burgess, S. (2015). Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 44, 512–525. doi:10.1093/ije/dyv080
Bowden, J., Davey Smith, G., Haycock, P. C., and Burgess, S. (2016). Consistent estimation in mendelian randomization with some invalid instruments using a weighted median estimator. Genet. Epidemiol. 40, 304–314. doi:10.1002/gepi.21965
Bowden, J., and Holmes, M. V. (2019). Meta-analysis and Mendelian randomization: a review. Res. Synth. Methods 10, 486–496. doi:10.1002/jrsm.1346
Brew, K., and Nagase, H. (2010). The tissue inhibitors of metalloproteinases (TIMPs): an ancient family with structural and functional diversity. Biochim. Biophys. Acta 1803, 55–71. doi:10.1016/j.bbamcr.2010.01.003
Burgess, S., Bowden, J., Fall, T., Ingelsson, E., and Thompson, S. G. (2017). Sensitivity analyses for robust causal inference from mendelian randomization analyses with multiple genetic variants. Epidemiology 28, 30–42. doi:10.1097/EDE.0000000000000559
Burgess, S., and Thompson, S. G. (2011). Bias in causal estimates from Mendelian randomization studies with weak instruments. Stat. Med. 30, 1312–1323. doi:10.1002/sim.4197
Casagrande, V., Federici, M., and Menghini, R. (2021). TIMP3 involvement and potentiality in the diagnosis, prognosis and treatment of diabetic nephropathy. Acta Diabetol. 58, 1587–1594. doi:10.1007/s00592-021-01766-y
Catania, J. M., Chen, G., and Parrish, A. R. (2007). Role of matrix metalloproteinases in renal pathophysiologies. Am. J. Physiol. Ren. Physiol. 292, F905–F911. doi:10.1152/ajprenal.00421.2006
Chatterjee, E., Rodosthenous, R. S., Kujala, V., Gokulnath, P., Spanos, M., Lehmann, H. I., et al. (2023). Circulating extracellular vesicles in human cardiorenal syndrome promote renal injury in a kidney-on-chip system. JCI Insight 8. doi:10.1172/jci.insight.165172
Chen, M., Zhang, L., Zhan, R., and Zheng, X. (2022). The novel histone deacetylase inhibitor pracinostat suppresses the malignant phenotype in human glioma. Mol. Biol. Rep. 49, 7507–7519. doi:10.1007/s11033-022-07559-y
Cohen, J. F., Chalumeau, M., Cohen, R., Korevaar, D. A., Khoshnood, B., and Bossuyt, P. M. (2015). Cochran's Q test was useful to assess heterogeneity in likelihood ratios in studies of diagnostic accuracy. J. Clin. Epidemiol. 68, 299–306. doi:10.1016/j.jclinepi.2014.09.005
Dahlén, E., and Björkhem-Bergman, L. (2022). Comparison of creatinine and cystatin C to estimate renal function in geriatric and frail patients. Life (Basel) 12, 846. doi:10.3390/life12060846
Davey Smith, G., and Hemani, G. (2014). Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum. Mol. Genet. 23, R89–R98. doi:10.1093/hmg/ddu328
Davies, N. M., Holmes, M. V., and Davey Smith, G. (2018). Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. Bmj 362, k601. doi:10.1136/bmj.k601
Emdin, C. A., Khera, A. V., and Kathiresan, S. (2017). Mendelian randomization. Jama 318, 1925–1926. doi:10.1001/jama.2017.17219
Fan, D., and Kassiri, Z. (2020). Biology of tissue inhibitor of metalloproteinase 3 (TIMP3), and its therapeutic implications in cardiovascular pathology. Front. Physiol. 11, 661. doi:10.3389/fphys.2020.00661
Fan, H., and Zhang, W. (2020). Overexpression of linc 4930556M19Rik suppresses high glucose-triggered podocyte apoptosis, fibrosis and inflammation via the miR-27a-3p/metalloproteinase 3 (TIMP3) Axis in diabetic nephropathy. Med. Sci. Monit. 26, e925361. doi:10.12659/MSM.925361
Fiorentino, L., Cavalera, M., Menini, S., Marchetti, V., Mavilio, M., Fabrizi, M., et al. (2013). Loss of TIMP3 underlies diabetic nephropathy via FoxO1/STAT1 interplay. EMBO Mol. Med. 5, 441–455. doi:10.1002/emmm.201201475
Gowda, S., Desai, P. B., Kulkarni, S. S., Hull, V. V., Math, A. A., and Vernekar, S. N. (2010). Markers of renal function tests. N. Am. J. Med. Sci. 2, 170–173.
Hartwig, F. P., Davey Smith, G., and Bowden, J. (2017). Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int. J. Epidemiol. 46, 1985–1998. doi:10.1093/ije/dyx102
Hemani, G., Tilling, K., and Davey Smith, G. (2017). Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS Genet. 13, e1007081. doi:10.1371/journal.pgen.1007081
Hemani, G., Zheng, J., Elsworth, B., Wade, K. H., Haberland, V., Baird, D., et al. (2018). The MR-Base platform supports systematic causal inference across the human phenome. Elife 7, e34408. doi:10.7554/eLife.34408
Kamat, M. A., Blackshaw, J. A., Young, R., Surendran, P., Burgess, S., Danesh, J., et al. (2019). PhenoScanner V2: an expanded tool for searching human genotype-phenotype associations. Bioinformatics 35, 4851–4853. doi:10.1093/bioinformatics/btz469
Kassiri, Z., Oudit, G. Y., Kandalam, V., Awad, A., Wang, X., Ziou, X., et al. (2009). Loss of TIMP3 enhances interstitial nephritis and fibrosis. J. Am. Soc. Nephrol. 20, 1223–1235. doi:10.1681/ASN.2008050492
Liu, D., and Zhang, W. (2021). Pioglitazone attenuates lupus nephritis symptoms in mice by modulating miR-21-5p/TIMP3 Axis: the key role of the activation of peroxisome proliferator-activated receptor-γ. Inflammation 44, 1416–1425. doi:10.1007/s10753-021-01426-x
Liu, Y. (2011). Cellular and molecular mechanisms of renal fibrosis. Nat. Rev. Nephrol. 7, 684–696. doi:10.1038/nrneph.2011.149
Molitch, M. E., DeFronzo, R. A., Franz, M. J., Keane, W. F., Mogensen, C. E., Parving, H. H., et al. (2003). Diabetic nephropathy. Diabetes Care 26 (Suppl. 1), S94–S98. doi:10.2337/diacare.26.2007.s94
Murdaca, G., Negrini, S., Magnani, O., Penza, E., Pellecchio, M., and Puppo, F. (2017). Impact of pharmacogenomics upon the therapeutic response to etanercept in psoriasis and psoriatic arthritis. Expert Opin. Drug Saf. 16, 1173–1179. doi:10.1080/14740338.2017.1361404
Pattaro, C., Teumer, A., Gorski, M., Chu, A. Y., Li, M., Mijatovic, V., et al. (2016). Genetic associations at 53 loci highlight cell types and biological pathways relevant for kidney function. Nat. Commun. 7, 10023. doi:10.1038/ncomms10023
Pierce, B. L., Ahsan, H., and Vanderweele, T. J. (2011). Power and instrument strength requirements for Mendelian randomization studies using multiple genetic variants. Int. J. Epidemiol. 40, 740–752. doi:10.1093/ije/dyq151
Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M. A., Bender, D., et al. (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575. doi:10.1086/519795
Shi, Z., Taylor, A. W., Riley, M., Byles, J., Liu, J., and Noakes, M. (2018). Association between dietary patterns, cadmium intake and chronic kidney disease among adults. Clin. Nutr. 37, 276–284. doi:10.1016/j.clnu.2016.12.025
Suhre, K., Arnold, M., Bhagwat, A. M., Cotton, R. J., Engelke, R., Raffler, J., et al. (2017). Connecting genetic risk to disease end points through the human blood plasma proteome. Nat. Commun. 8, 14357. doi:10.1038/ncomms14357
Toba, H., and Lindsey, M. L. (2019). Extracellular matrix roles in cardiorenal fibrosis: potential therapeutic targets for CVD and CKD in the elderly. Pharmacol. Ther. 193, 99–120. doi:10.1016/j.pharmthera.2018.08.014
Verbanck, M., Chen, C. Y., Neale, B., and Do, R. (2018). Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 50, 693–698. doi:10.1038/s41588-018-0099-7
Wang, Z., Famulski, K., Lee, J., Das, S. K., Wang, X., Halloran, P., et al. (2014). TIMP2 and TIMP3 have divergent roles in early renal tubulointerstitial injury. Kidney Int. 85, 82–93. doi:10.1038/ki.2013.225
Wozniak, J., Floege, J., Ostendorf, T., and Ludwig, A. (2021). Key metalloproteinase-mediated pathways in the kidney. Nat. Rev. Nephrol. 17, 513–527. doi:10.1038/s41581-021-00415-5
Xie, Y., Bowe, B., Mokdad, A. H., Xian, H., Yan, Y., Li, T., et al. (2018). Analysis of the Global Burden of Disease study highlights the global, regional, and national trends of chronic kidney disease epidemiology from 1990 to 2016. Kidney Int. 94, 567–581. doi:10.1016/j.kint.2018.04.011
Zakiyanov, O., Kalousová, M., Zima, T., and Tesař, V. (2019). Matrix metalloproteinases in renal diseases: a critical appraisal. Kidney Blood Press Res. 44, 298–330. doi:10.1159/000499876
Zheng, C. M., Lu, K. C., Chen, Y. J., Li, C. Y., Lee, Y. H., and Chiu, H. W. (2022). Matrix metalloproteinase-7 promotes chronic kidney disease progression via the induction of inflammasomes and the suppression of autophagy. Biomed. Pharmacother. 154, 113565. doi:10.1016/j.biopha.2022.113565
Zhou, X., Liu, G., Xu, M., Ying, X., Li, B., Cao, F., et al. (2022). Comprehensive analysis of PTEN-related ceRNA network revealing the key pathways WDFY3-AS2 - miR-21-5p/miR-221-3p/miR-222-3p - TIMP3 as potential biomarker in tumorigenesis and prognosis of kidney renal clear cell carcinoma. Mol. Carcinog. 61, 508–523. doi:10.1002/mc.23396
Keywords: mendelian randomization, GWAS, TIMP3, CKD, kidney function
Citation: Chen H, Chen L, Chen Y, Guo Q and Lin S (2024) Exploring the genetic causal association of TIMP3 on CKD and kidney function: a two-sample mendelian randomization. Front. Genet. 15:1367399. doi: 10.3389/fgene.2024.1367399
Received: 08 January 2024; Accepted: 18 April 2024;
Published: 07 May 2024.
Edited by:
Giovanni Malerba, University of Verona, ItalyReviewed by:
Swetha Ramadesikan, Nationwide Children’s Hospital Columbus OH, United StatesCopyright © 2024 Chen, Chen, Chen, Guo and Lin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shirong Lin, bGVvcGFyZDg2MkAxMjYuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.