- 1Department of Rheumatology and Immunology, The Affiliated Hospital of Inner Mongolia Medical University, Hohhot, China
- 2Inner Mongolia Key Laboratory for Pathogenesis and Diagnosis of Rheumatic and Autoimmune Diseases, The Affiliated Hospital of Inner Mongolia Medical University, Hohhot, China
The Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR) mediated Cas9 nuclease system has been extensively used for genome editing and gene modification in eukaryotic cells. CRISPR/Cas9 technology holds great potential for various applications, including the correction of genetic defects or mutations within the human genome. The application of CRISPR/Cas9 genome editing system in human disease research is anticipated to solve a multitude of intricate molecular biology challenges encountered in life science research. Here, we review the fundamental principles underlying CRISPR/Cas9 technology and its recent application in neurodegenerative diseases, cardiovascular diseases, autoimmune related diseases, and cancer, focusing on the disease modeling and gene therapy potential of CRISPR/Cas9 in these diseases. Finally, we provide an overview of the limitations and future prospects associated with employing CRISPR/Cas9 technology for diseases study and treatment.
1 Introduction
The technology of genome editing facilitates precise manipulation of DNA sequences within the gene, enabling stable modification of genetic information through targeted knockouts, insertions, and replacements. There are primarily three genome editing technologies available: zinc finger nucleases (ZFN), transcription activator like effector nucleases (TALEN), and clustered regularly interspersed short palindromic repeats (CRISPR/Cas9) (Gaj et al., 2013), and the CRISPR/Cas9 genome editing technology being the most widely employed at present.
The CRISPR/Cas9 is an acquired immune system found in most bacteria and all archaea (Wiedenheft et al., 2012), which functions to protect against phages or foreign invading agents from swallowing plasmids (Barrangou et al., 2007). By utilizing a humanized Cas9 nuclease and guided RNA (gRNA) specifically designed for target gene, this system enables precise editing of any sequence or genome of eukaryotic cells (Jinek et al., 2012), thus becoming the third-generation genome editing technology after ZFNs and TALENs. Under the guidance of specific gRNA, the Cas9 nuclease cut, replace or inserts DNA sequences of organisms to precisely achieve the intended purpose of genome reediting (Cong et al., 2013).
The CRISPR/Cas9 system possesses several advantages, including high editing efficiency, ease of operation, cost effectiveness, and diversified recognition sites, etc. (Doudna and Charpentier, 2014; Knott and Doudna, 2018). It can be used in human genome editing (Mali et al., 2013) to treat genetic defects or gene mutation diseases (Wang et al., 2020; Weber et al., 2020; Zhang et al., 2020); editing plant genes to create more resilient plants (Seth and Harish, 2016; Soyk et al., 2017); eliminating pathogens by deleting their genes, such as the genomes of bacteria or viruses, in order to eliminate infectious diseases, offers hope for a complete cure for such diseases (Ebina et al., 2013; Yin et al., 2017; Ling et al., 2020).
The CRISPR/Cas9 system has been rapidly developed in all areas of the life science, is causing a seismic change in biomedical research, and is sweeping biology laboratories around the world with its remarkable efficiency and operational simplicity. Widely regarded as the most successful genome editing tool, we review the relevant research of CRISPR/Cas9 genome editing technology in some human diseases (focusing on neurodegenerative diseases, cardiovascular diseases, autoimmune diseases, and cancer) in recent years, and analyzed current applications and future perspective of CRISPR/Cas9 in these diseases.
2 Feature and mechanism of CRISPR/Cas9
According to the different CRISPR/Cas sites, CRISPR systems can be divided into six different types, and the commonly used CRISPR/Cas9 genome editing system is the Type II CRISPR system, with three functional regions on the CRISPR site: The trans activating CRISPR RNA (trancrRNA) region, the different Cas genes region, and the CRISPR array region, three functional regions are linearly arranged on a single DNA strand (Figure 1). The Cas gene region is used to guide the formation of nucleases that cut DNA sequences, while the CRISPR array region contains repeat sequences and spacer sequences, and the spacer sequences will be inserted for transcription when foreign genes invade (Gupta et al., 2019).
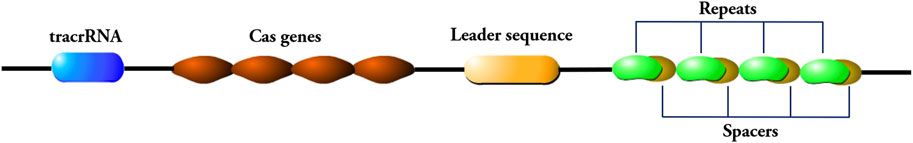
Figure 1. Components of the CRISPR/Cas9 genome editing system. The CRISPR/Cas9 system consists of a trans-activating crRNA (trancrRNA) region, a Cas gene region, and a CRISPR array region. The CRISPR array region consists of repeat sequences and spacer sequences. The repeat sequence is connected to the leader sequence adjacent to the 5′end of the locus, and the 5′end of the leader sequence is connected to the Cas gene region.
In the CRISPR/Cas9 system, matured CRISPR RNA (crRNA) combines with trans activating crRNA (tracrRNA) to form sgRNA (Deltcheva et al., 2011; Jiang and Doudna, 2017). The sgRNA forms a complex with the Cas9 protein. Cas9 protein is guided with sgRNA through base complementary pairing to bind to the specific locus of the genome. Subsequently, the Cas9 protein recognizes the protospacer adjacent motif (PAM) of the target gene and starts to cut the target DNA at the third base upstream of the PAM, producing double strand break (DSB) (Zaboikin et al., 2017; Iyer et al., 2019). The genome that produces DSB is subsequently repaired through either non-homologous end joining (NHEJ) or homologous recombination (HR) (Anzalone et al., 2020) (Figure 2). The NHEJ repair is imprecise, which can cause the insertion and deletion of bases, resulting in the change of the open reading frame. HR repair is precise and can achieve site specific repair of genes when homologous templates are introduced (Maruyama et al., 2015; Yeh et al., 2019).
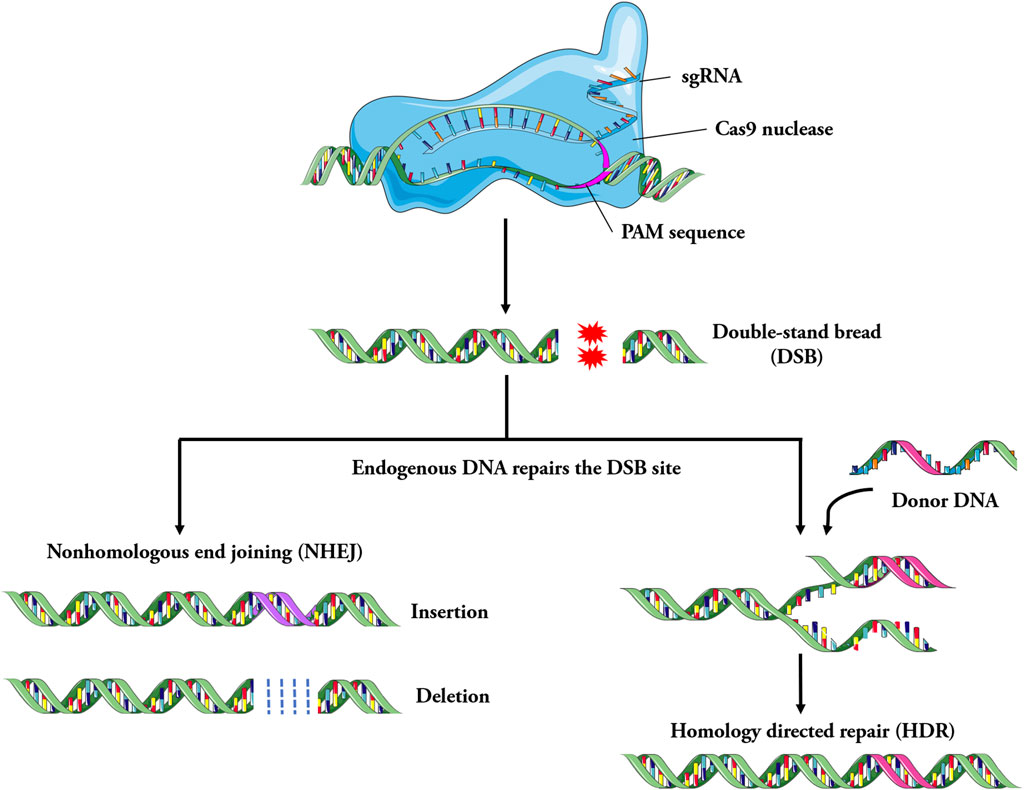
Figure 2. Schematic of the mechanism involved in CRISPR/Cas9 system. Specially designed sgRNA (guide RNA), that match the genomic DNA sequence containing the mutation, bind to the Cas9 endonuclease to form the Cas9-sgRNA complex. Under the guidance of sgRNA, Cas9 searches for target DNA and cut it, then double strand breaks (DSB) are generated, it leads to the activation of DNA repair mechanism. The genome is repaired by non-homologous end joining (NHEJ) or homologous recombination repair (HDR). In general, the imprecise NHEJs cause the insertion and deletion of bases, resulting in the change of the open reading frame, while the more precise HDR systems can lead to achieve site-specific repair of genes, such as disease-causing point mutations.
At present, Cas9 from S. yogenes (SpCas9) is the most common CRISPR/Cas9 effector protein. Natural Cas protein still has many limitations in editing efficiency and specificity, which significantly affect its practical application. Researchers have developed different Cas9 variants to achieve better editing effect and reduce off-target activity. Previous studies have shown that two catalytic domains in Cas9, RuvC and HNH, are responsible for the cleavage of one DNA strand, respectively. By inactivating one of the domains, Cas9 nickase (nCas9) that catalyzes single strand breaks can be generated (Ran et al., 2013). Compared with wild-type Cas9 protein, the use of nCas9 can reduce off-target effects (Shen et al., 2014). Yin et al. (Yin et al., 2022a) fused SpCas9 with the optimized human recombinant protein TREX2 to generate a new gene editing enzyme, Cas9TX, which can inhibit structural abnormalities such as chromosomal translocations during gene editing and significantly improve the safety of gene editing. Qi et al. mutagenized the catalytic nuclease domain of the Cas9 protein. Two mutations were introduced to obtain a catalytically inactive Cas9 protein, also known as “dead Cas9” (Qi et al., 2013). Unlike Cas9, dCas9 does not cause irreversible changes in the genome, but only affects transcription at the target site, resulting in reversible gene silencing. It has also been shown that the fusion of dCas9 to the FokI domain reduces off-target cleavage (Guilinger et al., 2014; Tsai et al., 2014; Aouida et al., 2015; Nakagawa et al., 2015; Havlicek et al., 2017; Saifaldeen et al., 2020). The tool simultaneously binds two dCas9-FokI monomers at the target site to trigger DSBs, and the probability of two adjacent dCas9-FokI fusion molecules to bind to the off-target site nonspecifically is significantly lower than that of single molecule binding (Aouida et al., 2015; Nakagawa et al., 2015). This minimizes the risk of off-target effects. Data from Aouida et al. suggest that fdCas9 endonuclease variant is a superior platform for genome editing applications in eukaryotic systems, including mammalian cells (Aouida et al., 2015) and that all engineered fdCas9 variants have demonstrated promising gene editing activity in human cells thus far (Havlicek et al., 2017; Saifaldeen et al., 2020). The development of various Cas variants has significantly enriched the CRISPR editing tools, and improved the editing efficiency and accuracy.
The research and application of the CRISPR/Cas9 system in human diseases encompass the establishment of in vivo disease models, in vitro cell models research and gene therapy, etc. The change of open reading frame caused by NHEJ repair can be used to establish gene knockout model, while gene replacement through homologous recombination holds potential for achieving gene therapy.
3 CRISPR/Cas9 genome editing for neurodegenerative diseases modeling
Neurodegenerative diseases (NDs) are a broad category of diseases caused by the degeneration of neurons (Dugger and Dickson, 2017). Aging is the primary risk factor for NDs, and patients are often accompanied by cognitive decline and motor dysfunction, which can lead to severe disability or even death (Wyss-Coray, 2016). Common examples of NDs include Alzheimer’s disease, Parkinson’s disease, and Huntington’s disease. The correlations between genetic mutations and neurodegenerative diseases, and the combination of CRISPR/Cas9 with other technologies, have enabled the creation of various animal models that mimic genetic defects and provide important insights into their pathogenesis (Yang et al., 2021; Yin et al., 2022b). In fact, the construction of animal disease models also holds significant implications for investigating other diseases. Hence, we summarize the various disease animal models constructed by CRISPR/Cas9 technology (Supplementary Table S1), highlighting the respective strengths and limitations of each model in accurately recapitulating disease pathology. The focus of this section will be on the research progress and recent applications status of the CRISPR/Cas9 system in several common neurodegenerative diseases.
3.1 Alzheimer’s disease (AD)
AD is the most common NDs, and the neurotoxic accumulation of amyloid β-protein (Aβ) and hyperphosphorylation of Tau protein to form double helix filament are the main driving factors (Lanctot et al., 2017). Animal models play a crucial role in studying AD pathogenesis and developing treatments. However, most of the previous AD mouse models did not exhibit significant neurodegeneration (De Plano et al., 2022). CRISPR/Cas9 technology was used to create new AD models and show more accurate disease phenotypes (Lu et al., 2021). Bart De Strooper et al. established an animal model of AD using CRISPR/Cas9 technology by introducing three independent point mutations (G676R, F681Y and R684H) of APP gene. They transformed endogenous mouse APP genes into human versions and established a humanized animal model. This will help to reveal novel mechanisms of disease pathogenesis (Serneels et al., 2020). Tan et al. used CRISPR/Cas9 to introduce a deficiency in the translational start codon of exon one of the Mapt gene encoding Tau protein in a new animal model that was produced in the pure C57Bl/6J background (Tan et al., 2018).
The onset and progression of the majority of AD cases are sporadic, and in fact, only in a small number of familial AD cases (<1%), mutations in related genes lead to the production of amyloid precursor protein (APP), which promotes APP processing to produce Aβ. Nevertheless, dysregulation of Aβ metabolism is involved in both sporadic and familial AD, suggesting that limiting Aβ production may offer a treatment independent of the disease (Bhardwaj et al., 2022). Studies using the CRISPR/Cas9 system have been reported to help knock out Swedish APP mutations in patient derived fibroblasts and found a 60% reduction in Aβ, the editing efficiencies of the two designed sgRNAs for APPSW in cells were 34.3% and 14%, respectively (Gyorgy et al., 2018). Sun et al. targeted the C-terminal of endogenous APP gene via CRISPR/Cas9, the robust editing of APP was observed in human iPSC derived neurons (81.8%) and mouse brains, with no detectable off-target effects. Their study shifted the processing of APP from amyloidosis related APP-β cleavage to upregulated neuroprotective APP-α cleavage, thereby reducing the production of Aβ, and showed that this method was effective in reducing the production of Aβ (Sun et al., 2019).
3.2 Parkinson’s disease (PD)
PD, the second most common NDs, is caused by degeneration, loss of dopaminergic neurons in the substantia nigra of the midbrain or the formation of Lewy bodies (Karimian et al., 2020). Like AD, most cases of PD are sporadic. Mutations in genes encoding alphasynuclein, PINK1, Parkin, LRRK2, and others have been found in 10%–15% of familial PD cases. The editing of classical PD related genes in mice has not yielded successful results (Qu et al., 2023), and the simultaneous knockout of three Parkinson’s disease related genes, including Parkin/PINK1/Dj-1, did not result in any observed neurodegenerative phenotype in mice (Kitada et al., 2009). The researchers constructed large animal models that were more closely resembled humans in size and physiology. Zhou et al. transfected the CRISPR/Cas9 expression vector targeting PARK2 and PINK1 into Porcine fetal fibroblasts (PFF) (Zhou et al., 2015), and successfully constructed a PD model pig by somatic cell nuclear transfer (SCNT). Zhu et al. used CRISPR/Cas9 combined with nuclear transplantation (SNCT) technology to breed a Bama mini pig PD model expressing SNCA gene E46K, H50Q and G51D mutations (Zhu et al., 2018). However, the PD model pigs did not exhibit significant neuronal loss, while the primate models appeared to be better at reproducing typical Parkinson’s pathological features (Yang et al., 2019; Li et al., 2021a; Yang et al., 2022).
CRISPR/Cas9 technology has also been used for the deletion of mutant gene expression or direct restoration of known PD causing mutations. The A53T mutation of the SNCA gene is one of the most studied mutations in PD, and rats carrying the A53T mutation and overexpression of alpha synuclein exhibit a phenotype resembling PD, which can be prevented through CRISPR/Cas9 mediated deletion of the mutant gene (Yoon et al., 2022). Furthermore, LRRK2 mutation represents the most common genetic cause of both sporadic and familial PD. Qing et al. successfully reduced the incidence of familial and sporadic PD by editing mutant LRRK2 using CRISPR/Cas9, thereby presenting a potential avenue for treating both forms of PD, they performed unbiased detection of the top 8 potential off-target sites predicted by the COD algorithm, and Sanger sequencing analysis showed no off-target indels (Qing et al., 2017).
3.3 Huntington’s disease (HD)
HD is a fatal ND characterized by severe psychiatric symptoms and cognitive impairment (Bates et al., 2015), which is usually inherited in an autosomal dominant manner and is primarily caused by abnormal triple amplification of CAG (cytosine adenine guanine) in the Huntington (HTT) gene on chromosome 4p16.3 (Yang et al., 2020a; Tabrizi et al., 2020). This amplification is translated into a polyglutamine (polyQ) repeat in the disease protein Huntington (HTT) (Dianov and Hubscher, 2013). The amplification of polyQ causes HTT to misfold and accumulate in the patient’s brain, resulting in preferential loss of spinous neurons in the striatum. Various transgenic mouse models of HD have been established, and it has been found that N-terminal fragments of mutant HTT with amplified polyQ repeats can accumulate in the brain, affecting motor and neuronal function (Crook and Housman, 2011; Farshim and Bates, 2018). However, in these mouse models, there was no significant selective loss of mid-spinous neurons when HD knockout (Crook and Housman, 2011). In 2018, Yan et al. used CRISPR/Cas9 technology to introduce 150 CAG repeats into endogenous porcine HTT gene and successfully generated the first porcine HD model. Notably, when the full length mutant HTT carrying 150Q was expressed endogenously in the HD porcine model, it causes significant selective neurodegeneration and movement disorders, which effectively generalizes the typical pathological and clinical features of HD patients (Yan et al., 2018).
Considering its primary genetic cause, HD is considered an ideal ND for the application of CRISPR/Cas9 technology, and studies have suggested that reducing the production of mutant HTT alleles could be an effective therapeutic approach (Ekman et al., 2019; Lopes et al., 2020). Furthermore, Jing et al. inhibited the expression of endogenous Hsp70 binding protein in the striatum of HD140QKI mice by CRISPR/Cas9, and the accumulation of HTT was significantly reduced, which indicated that CRISPR/Cas9 could play a certain role in the treatment of Huntington’s disease (Jing et al., 2021). Therefore, the removal of the mutated HTT protein has important therapeutic implications for HD, and identifying a treatment for any one neurological disease will have broad implications for the development of treatments for a large number of patients with neurological diseases.
4 CRISPR/Cas9 for cardiovascular diseases modeling and gene therapy
Cardiovascular disease is one of the chronic diseases that serious threaten human life caused by the combination of genes and environment (Morton et al., 2022), its incidence is rising year by year, and the proportion of population deaths is quite high, and it has become the primary disease that threatens human health (McKenna and Judge, 2021). CRISPR/Cas9 genome editing technology has ushered in a new era of cardiovascular disease research and offering the possibility for genetic correction of this ailment (Liu and Olson, 2022).
Inherited heart diseases, such as congenital heart disease (CHD), hypertrophic cardiomyopathy (HCM), and Duchenne muscular dystrophy (DMD), are caused by mutations in either a single gene or multiple genes (Austin et al., 2019; Yadav et al., 2019). There has been a lack of relatively reliable models of heart disease (Strong and Musunuru, 2017), and now with the aid of CRISPR/Cas9 technology, researchers have developed corresponding in vivo and in vitro models to study hereditary heart diseases. In the in vivo disease models, Alankarage et al. created CRISPR/Cas9 gene edited mouse models for functional analysis of novel missense variants identified from patients with congenital heart disease (CHD), revealing the effects of multiple congenital abnormalities on CHD (Alankarage et al., 2020). Johansen et al. used CRISPR/Cas9 and AAV9 mediated delivery of short guide RNAs targeting 3 genes (Myh6, Sav1, and Tbx20) critical for cardiac physiology to generate cardiomyocyte specific Cas9 mice, and demonstrated that Cas9 expression did not affect cardiac function or gene expression (Johansen et al., 2017). Liu et al. used CRISPR/Cas9 genome editing technology to verify the dual genes that cause hypoplastic left heart syndrome (HLHS) in mice, indicating that HLHS can be inherited through combination mode, thus providing a new paradigm for the complex genetics of congenital heart disease (Liu et al., 2017a). Other heart disease models constructed by researchers using CRISPR/Cas9 in recent years are shown in Supplementary Table S1. In the in vitro cell models, Li et al. used CRISPR/Cas9 technology to knock out MLP (striated muscle associated protein) in H9 human embryonic stem cell line to generate the corresponding defects type (Li et al., 2019), which later developed into HCM. Jaffre et al. successfully established the HCM model of Noonan syndrome on patient derived cardiomyocytes by using CRISPR/Cas9 generated isogenic control iPSC derived cardiomyocytes (Jaffre et al., 2019).
CRISPR/Cas9 technology is also commonly used to study the role of different genes or proteins in the mechanism of inherited heart diseases. Chang et al. constructed two FHL2 gene knockout ES cell lines using CRISPR/Cas9 technology (Chang et al., 2018), which could be used for mechanism research and drug screening of dilated cardiomyopathy (DCM). Xie et al. applied CRISPR/Cas9 gene editing technology to generate hESC related to TOF (tetralogy of Fallot), revealing the myocardial hypertrophy derived from human embryonic stem cells through HB-EGF signal transduction by rare mutations of TOF (Xie et al., 2019). Li et al. used CRISPR/Cas9 technology to create RAD (Ras associated with diabetes) deficient human ES cell lines, demonstrating that elevated intracellular calcium levels and abnormal calcium regulation were the core mechanisms of RAD deficiency leading to cardiac hypertrophy (Li et al., 2020). Roche et al. used CRISPR/Cas9 to introduce Brugada syndrome (BrS) mutations in human induced hiPSC-CMs, which, as a genetic background independent of patients, revealed the mechanism of sodium channel inactivation triggering BrS (de la Roche et al., 2019). By constructing PLN mutant hiPSC-CM cell line, Smith et al. confirmed that the mutation could lead to delayed response of cardiomyocytes to β-receptor agonists (Smith et al., 2018). Ang et al. combined iPSC with CRISPR/Cas9 technologies to establish a congenital heart disease model related to GATA4 gene mutation in vitro, aiming at exploring the pathogenesis caused by this gene mutation (Ang et al., 2016).
CRISPR/Cas9 shows promise as a potential treatment for cardiovascular diseases meanwhile. In the case of DMD, somatic genome editing therapy for the disease has demonstrated promising outcomes in both animal models and in vitro human models. Nelson et al. respectively loaded CRISPR-SaCas9 and two sgRNAs into AAV vectors and used the AAV-CRISPR gene editing system to target the deletion of exon 23 in DMD gene of MDX model mice (Nelson et al., 2019), resulting in the expression of dystrophin with normal functionality. The newborn mice injected intravenously with the AAV-CRISPR system still had higher gene editing efficiency and maintained high expression of restored dystrophin protein 1 year later. They found that the expression of SaCas9 in various types of muscle cell, while also detected genome editing events in other tissues at levels slightly above the detection limit (∼0.1%). Furthermore, deep sequencing analysis of the top 10 predicted off-target sites showed no significant increase in off-target cutting after 1 year. Amoasii et al. coupled Cas9 with sgRNA to target regions near the DMD gene exon 51 splicing receptor sites, and used adeno-associated viruses to deliver the CRISPR gene editing component to four dogs, where dystrophin recovered to 3%–90% of normal levels in the heart muscle. The dogs that received the highest dose had levels of dystrophin that were 92% of normal. The muscle tissue of treated dogs also improved (Amoasii et al., 2018), the analysis of predicted off-target sites revealed no specific off-target gene editing above background levels in treatment animals. Moretti et al. used a Cas9 mediated exon excision method to restore the DMD reading frame, which enabled short but functional dystrophin expression and improved bone and myocardial failure in DMD pigs, and this method prevented susceptibility of DMD cardiomyocytes to arrhythmias (Moretti et al., 2020). Although no mutations were detected at the five most likely predicted off-target sites, and no genome editing events were observed in the liver and kidneys, it is important to further evaluate possible off-target mutagenesis in longer term animal studies.
For cardiovascular diseases, CRISPR/Cas9 technology has just begun to be widely used, there is no doubt that for some cardiovascular diseases with high mortality, the application of CRISPR/Cas9 technology will be faster and more convenient to create cell and animal models, which is conducive to the research of cardiovascular development and disease pathogenesis.
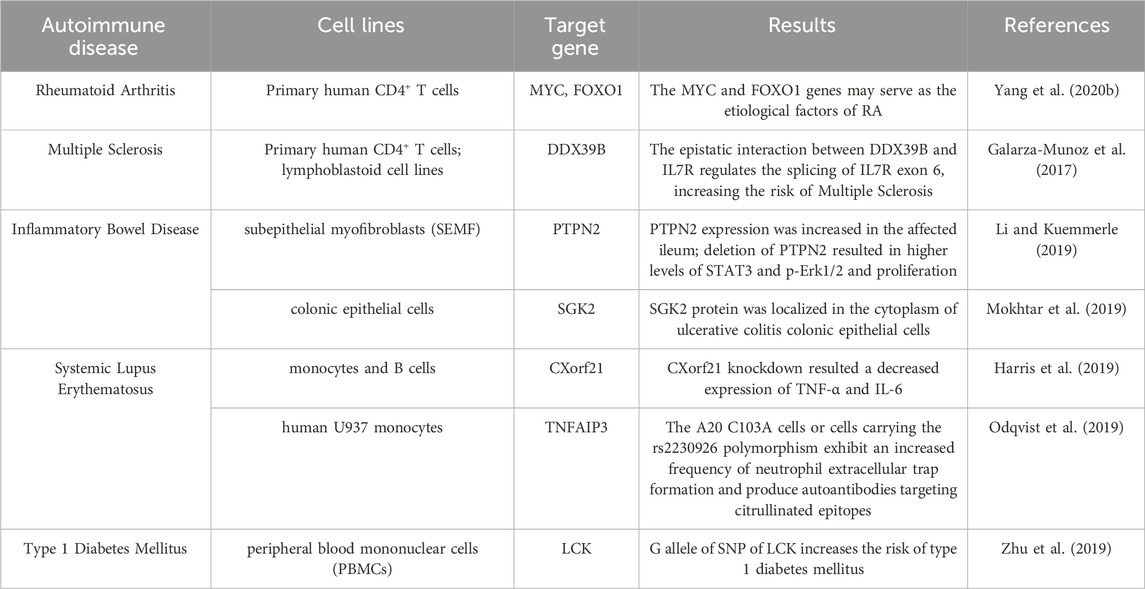
5 CRISPR/Cas9 application for autoimmune-related diseases
The primary characteristic of autoimmune diseases is that the disorders function of the immune system, the body is unable to distinguish between its own antigens and foreign antigens, and mistakes the normal components of the body for foreign substances to attack, resulting in long term inflammation (Marrack et al., 2001). The genetic background underlying autoimmune diseases is intricate, involving multiple genes and pathways in their pathogenesis. Various genetic factors can lead to the development of autoimmune diseases such as rheumatoid arthritis, systemic lupus erythematosus, type 1 diabetes, and multiple sclerosis (Cooper et al., 1999; Lee et al., 2022). CRISPR/Cas9 provides numerous novel ideas for research and treatment of autoimmune diseases including the screening of autoimmune disease regulatory genes, the construction of disease models, and the attempt to be used in the treatment of autoimmune diseases.
The CRISPR/Cas9 system has been used to identify regulatory sequences for certain important autoimmune risk loci, such as CD69 and IL2RA, the latter of which is associated with Crohn’s disease (Huang et al., 2017b; Simeonov et al., 2017). Studies have demonstrated that the destabilization of Treg cells can promote the development of autoimmunity, primarily characterized by the downregulation of transcription factor Foxp3 and the acquisition of proinflammatory properties. Cortez et al. have developed a combined CRISPR based screening platform for primary mouse Treg cell phenotype study, using this technology to perform targeted loss of function screening analysis of approximately 500 intranuclear factors, thereby identifying gene regulatory programs that can promote or interfere with Foxp3 expression (Cortez et al., 2020). Additionally, researchers used CRISPR whole genome screening technology to perform a comprehensive search across the whole genome in mouse model of type 1 diabetes, aiming to identify modifying factors that influence the survival of islet β-cells (Cai et al., 2020).
CRISPR/Cas9 has also been used in the construction of some autoimmune disease models. Researchers used CRISPR/Cas9 technology to generate Trex1 D18N/D18N point mutant mice, which display a multitude of human autoimmune related symptoms, including significantly declined survival time and damage to various organs such as the heart and kidneys. Additionally, augmented levels of autoantibodies were observed (Xiao et al., 2019). The other mouse models of autoimmune diseases are also presented in Supplementary Table S1. There are also many studies on the application of CRISPR/Cas9 to construct in vitro cell models of autoimmune diseases. Sevim et al. constructed a model for familial hemophagocytic lymphohistiocytosis 2 (FHL2) in order to assess the effectiveness of coculturing with mesenchymal stromal cells as an alternative therapy when bone marrow transplantation is unavailable (Sevim et al., 2018). The autoimmune regulatory gene (AIRE) has also been investigated using CRISPR technology, revealing that disruption of adhesion between the thymic medullary epithelial cells and thymus cells is one mechanism through which AIRE interference leads to autoimmunity (Speck-Hernandez et al., 2018). Furthermore, another study employing CRISPR/Cas9 technology on A20 demonstrated that genetic variation in the A20 deubiquitinase (DUB) domain increased the risk of systemic lupus erythematosus (SLE) and rheumatoid arthritis (Odqvist et al., 2019).
Gene therapy is a promising treatment for autoimmune diseases, while there were limited researches on the use of CRISPR/Cas9 in treating such conditions (Lee et al., 2022). Nevertheless, we cannot disregard the potential application of CRISPR/Cas9 in autoimmune diseases. Differentiated immune cells can be genetically modified using CRISPR technology. For instance, a recessive mutation in IL2RA led to reduced levels of IL2 receptor α chains in FOXP3 Tregs from a patient with a family history of autoimmune diseases. After targeted modification with CRSIPR, it found that the patient’s T cells began to express normal levels of IL2RA (Roth et al., 2018), the researchers also conducted targeted loci amplification (TLA) sequencing and observed no evidence of off-target integration above the detection limit (∼1% of alleles). In relation to rheumatoid arthritis (RA), Yang et al.'s study suggests that MYC and FOXO1 genes are implicated (Yang et al., 2020b), making these genes suitable candidates for targeted treatment using CRISPR/Cas9. Additionally, CRISPR/Cas9 has also been used to modify genes that may affect FOXP3 expression, enabling predominance of Treg expression and promoting the effect of adoptive Treg therapy in rheumatoid arthritis (Safari et al., 2018).
To date, numerous studies using CRISPR/Cas9 to treat autoimmune diseases have been conducted in cells (Table 1), and reports on clinical trials involving human subjects are very limited. However, it is undeniable that CRISPR/Cas9 plays a beneficial role in the regulation of autoimmune diseases, so it is necessary to conduct further comprehensive research, particularly more extensive in vivo investigations.
6 Application of CRISPR-Cas9 in cancer research
In recent years, CRISPR/Cas9 has become more widely used in cancer research and treatment, and there have been some encouraging advances, first reflected in the establishment of cancer animal models to study the pathogenesis of cancer. The creation of animal models for cancer is an important approach to studying the functional aspects associated with cancer genes. In the previous animal model establishment process, manipulation techniques were applied at the level of zygote stage to accomplish gene editing operations of embryonic stem cells with homologous recombination technology. The clinical application of CRISPR/Cas9 has outstanding simplification advantages for cancer animal models. Xue et al. using CRISPR/Cas9 technology successfully constructed a mouse model for liver cancer by simultaneously targeting tumor suppressor genes PTEN and P53 in mouse hepatocytes (Xue et al., 2014). Mou et al. injected Kirsten rat sarcoma viral oncogene (KRAS) gene templates along with sgRNA targeting tumor suppressor gene TP53 into wild-type B6 mice (Mou et al., 2019), resulted in tumors being observed in their livers 1 month later, thus successfully establishing a tumor model. Tyler et al. achieved successful construction of a mouse model for small cell lung cancer (SCLC) by simultaneously knocking out tumor suppressor genes Trp53 and Rb1 using CRISPR/Cas9 technology (Ng et al., 2020), their study provides an applicable mouse model of humanized SCLC that is suitable for studying the pathogenesis of SCLC. The construction of mouse models of other cancer using CRISPR/Cas9 technology is illustrated in Supplementary Table S1.
The findings from cellular and animal model studies suggest that CRISPR/Cas9 technology holds promise as a potentially effective approach for cancer treatment. The activation of oncogenes and the mutation of tumor suppressor genes make cancer cells expand and metastasize indefinitely, therefore, using CRISPR/Cas9 gene editing in cancer cells is expected to alter the biological characteristics of cancer cells, and provide a novel strategy for tumor treatment. Currently, several strategies using CRISPR/Cas9 for gene editing in cancer cells: 1) Introduction of tumor suppressor genes using CRISPR/Cas9 to inhibit tumor growth. Moses et al. used CRISPR/Cas9 along with trans-activator VP64p65-Rta (VPR) to activate the expression of PTEN in cancer cells, demonstrated that the activation of PTEN can significantly inhibit the downstream carcinogenic pathway (Moses et al., 2019). 2) Correction of deleterious mutations using CRISPR/Cas9 to suppress tumor development. Koo et al. used adenovirus (AdV) to deliver Cas9 protein and sgRNA targeting the EGFR oncogene with a single nucleotide missense mutation (CTG > CGG) (Koo et al., 2017). This method enabled to distinguish cancer causing mutations from wild-type EGFR alleles and precisely eliminate cancer causing mutant EGFR alleles. The frequency of EGFR mutant alleles in h1975 tumors was determined to be 50% (±5.3%), 40% (±2.8%), and 35% (±4.1%) at 7, 9, and 11 days post initial injection using Ad/sg EGFR and Ad/Cas9 vectors. Furthermore, no off-target effects were observed among the identified 17 potential off-target sites. Cheung et al. used the CRISPR/Cas9 system to target the point mutation L858R of EGFR (Cheung et al., 2018), the study used PAM to distinguish cancer mutations from normal mutations, providing high specificity in the degree of single nucleotide substitutions, and observed reduced expression of EGFR and inhibited cell proliferation in cells harboring the L858R mutation, providing novel insights into using CRISPR/Cas9 to selectively eliminate oncogenic mutations for cancer treatment. 3) Using CRISPR/Cas9 to target and knockout cancer related genes for tumors treatment, Zhen et al. used CRISPR/Cas9 to knockout E6 and E7 protein related genes that play a crucial role in the development and maintenance of cervical cancer, and the results showed that the knockout seriously damaged the survival ability of cancer cells (Zhen et al., 2016). Fusion genes are specific to cancer cells and are involved in many tumors, the researchers used CRISPR/Cas9 technology to insert the “suicide gene” HSV1-tk into the location of the MAN2A1-FER fusion gene break point, and found that the “suicide gene” can inhibit tumor growth, thus the method may be used as a viable strategy to treat tumors carrying the fusion gene (Chen et al., 2017).
Additionally, alterations in the gene expression of immune cells may reduce their ability to defend against and eliminate tumors. The utilization of CRISPR/Cas9 for gene editing in immune cells can enhance their cytotoxicity against tumors. The current strategies mainly involve: 1) Engineered T cells to eliminate tumors by knockout immune checkpoints. As an immune checkpoint, the high expression of PD-1 on T cells will inhibit the ability of T cells to initiate immune response, thereby rendering tumor cells susceptible to evading from the immune system. In 2020, Lu et al. injected CRISPR edited T cells targeting PD-1 into patients with advanced non-small-cell lung cancer (NSCLC). Clinical research results indicated a gene editing efficiency ranging from 8.7% to 31.2%, accompanied by significant reduction in PD-1 expression levels, thereby demonstrating that clinical application of CRISPR/Cas9 edited T cells is generally safe and feasible (Lu et al., 2020). Dong et al. found that DHX37 also serves as a crucial immune checkpoint (Dong et al., 2019), and knocking out DHX37 in CD8-positive T cells can enhance the effectiveness of adoptive immunotherapy against triple-negative breast cancer. 2) Using CRISPR/Cas9 to modify the antigen recognition capability of CAR-T cells. CAR-T cell immunotherapy employs genetic engineering to modify T lymphocytes to express chimeric antigen receptors that capable of identifying tumor cells and activating T cells simultaneously. Traditional CAR-T therapy exhibits certain defects, such as the limited number of T cells to be treated and the short in vivo survival duration. Stadtmauer et al. modified CAR-T cells with CRISPR/Cas9, demonstrated that edited CAR-T cells could persist for 9 months in patients with blood related cancers and maintain stable cytotoxicity compared to less than 7 days survival observed in original CAR-T cells (Stadtmauer et al., 2020).
The current research on utilizing CRISPR/Cas9 for gene editing of cancer cells to target tumors is primarily in the stages of basic research, and its direct application in clinical treatment remains limited. Clinical trials involving CRISPR/Cas9 are predominantly conducted in vitro, necessitating the isolation of cells from patients. Subsequently, the CRISPR/Cas9 system is employed for gene correction before being reintroduced into the patient. This process is influenced by various factors such as delivery systems and editing efficiency. However, with continuous advancements in gene editing technology, it is anticipated that CRISPR/Cas9 will possess significant potential to advance both cancer research and treatment.
7 Limitations of CRISPR/Cas9 in disease treatment and application
Although CRISPR/Cas9 is a well-established gene editing tool and has been extensively utilized in the research and treatment of various diseases, its clinical application is still in its early stages. There are several challenges that need to be addressed, including off-target effects, delivery methods, efficiency, and safety.
Off-target effects are a limitation of CRISPR/Cas9 technology and are considered a significant risk in in vivo gene therapy (Du et al., 2023). These effects occur when Cas9 proteins bind to PAM-like sequences or when guide RNAs (gRNAs) bind to unexpected nucleotide sequences at the target site. It has been reported that their efficiency exceeds 50% (Zhang et al., 2015). Off-target cleavage can result in unintended editing of normal genes, potentially disrupting their function, causing undesirable mutations, cytotoxicity, and even diseases. Selecting and designing the optimal sgRNA sequence is crucial to minimize or eliminate off-target effects. Studies have demonstrated that the structure of sgRNA influences its targeting ability, and increasing the GC content at the end of the sgRNA sequence’s PAM (Protospacer adjacent Motif) improves its targeting efficiency (Ren et al., 2014). Additionally, shortening the length of the sgRNA sequence enhances its sensitivity to residual target DNA mismatches, significantly reducing off-target effects (Hazafa et al., 2020). Furthermore, the development of high fidelity SpCas9 variants is considered the most promising strategy to address off-target effects in the context of enhanced Cas9 protein (Kulcsar et al., 2020). It is important to note that off-target effects remain unresolved in vivo and are closely associated with delivery techniques. Most clinical trials have focused on gene editing in vitro using patient derived cells, this approach reduces the risk of off-target effects and metastatic challenges but may not be suitable for all diseases.
To enhance the specificity of gene editing and achieve effective and precise therapy, it is crucial for CRISPR/Cas9 components to overcome various physical barriers and directly enter target cells (Xu et al., 2021). Additionally, the gene editing process necessitates the simultaneous delivery of functional Cas9 protein and sgRNA into the nucleus (Chen et al., 2020). Therefore, the delivery method plays a crucial role in CRISPR/Cas9 mediated editing therapy. Previously, physical and viral vectors have been explored for delivering CRISPR/Cas9 components. Physical delivery methods such as microinjection, electroporation, and hydrodynamic drug delivery exhibit high application efficiency in vitro but do not meet the requirements for in vivo applications (Liu et al., 2017b). Viral vectors also face limitations in their transformation into clinical grade therapeutic agents, including immunogenic reactions, limited packaging capacity, off-target effects, and high production costs (Tong et al., 2019; Xu et al., 2019). Non-viral vectors based on nanotechnology and materials science have been utilized in cancer therapy due to their low immunogenicity, high biocompatibility, and optimal payload capacity (Parra-Nieto et al., 2021). However, during the delivery process, CRISPR/Cas9 loaded nanoparticles encounter blood barriers, including degradation of CRISPR/Cas9 components by various enzymes in plasma (Kim et al., 2011), the clearance of nanocarriers by mononuclear phagocyte system or macrophages (de Lazaro and Mooney, 2021), and the filtration of nanocarriers by glomeruli (Han et al., 2014). Compared to the gene editing efficiency in vitro, the in vivo CRISPR/Cas9 editing efficiency is significantly lower, partially due to the low delivery efficiency of nanocarriers (Xu et al., 2021). Therefore, the lack of a safe and efficient delivery system remains the major obstacle to the clinical application of CRISPR/Cas9.
The immune system’s reaction triggered by the Cas9 protein or vector in the host represents a significant challenge in the application of CRISPR/Cas9 technology (Du et al., 2023). The Cas9 protein, originating from Streptococcus pyogenes, is considered an external antigen that can provoke an immune response upon introduction into the body (Charlesworth et al., 2019). While extreme immune responses to Cas9 are not extensively documented, the presence of Cas9 antibodies in humans indicates the potential exacerbation of hazards associated with CRISPR/Cas9 based gene therapy (Li et al., 2021b). Researchers are increasingly focusing on the immunogenicity caused by vectors, particularly viral vectors, as humans may have been previously exposed to them (Du et al., 2023). Thus, the immune response generated by the CRISPR/Cas9 gene editing framework constitutes a primary detrimental factor in the context of in vivo gene therapy using CRISPR technology.
CRISPR/Cas9 is an effective gene editing tool, but it is not yet an ideal clinical treatment. CRISPR requires a higher level of safety. Although CRISPR/Cas9 mediated gene editing has shown promising results in clinical studies, more in depth studies are needed to ensure that CRISPR/Cas9 can be safely and effectively applied to treat human diseases.
8 Conclusion and future perspectives
The discovery and research of the CRISPR/Cas9 system has changed the process of genome editing in the field of life sciences. In comparison to the ZFN and TALEN systems, it offers some incomparable advantages. In theory, every eight bases in the genome can find a site that can be edited with CRISPR/Cas9, so the technique can operate on almost any gene. Moreover, CRISPR/Cas9 system has more scalability, CRISPR/Cas9 gene editing technology can achieve a single base editing at the level of cells and organisms (Rees and Liu, 2018; Porto et al., 2020), is the most promising gene editing technology. Furthermore, one of its key strengths lies in its easy of use nature, enabling that almost any laboratory can do the work with this system.
From the various research reports and literatures on CRISPR/Cas9 system in recent years, it is not difficult to find that the application and advancement of this technology will likely help people to have a deeper understanding of the pathogenesis of diseases and provide more directions for targeted treatment of diseases. In this review, we only focus CRISPR/Cas9 research applications in neurodegenerative diseases, cardiovascular diseases, autoimmune related diseases, and cancer (summarized in Figure 3). In fact, CRISPR/Cas9 researches in variety of other human diseases have been reported. Today, advocates precision medicine and personalized medicine are being promoted, the development of the technology can also better promote the implementation of such medical treatment, so as to solve a series of problems in clinical treatment. Notably, compared with traditional genome editing technology, CRISPR/Cas9 genome editing technology also highlights great advantages in the establishment of animal models, the application of this technology can ensure the accurate and time saving establishment of disease animal models, thereby greatly facilitating the development of new drugs and the study of drug mechanism of action.
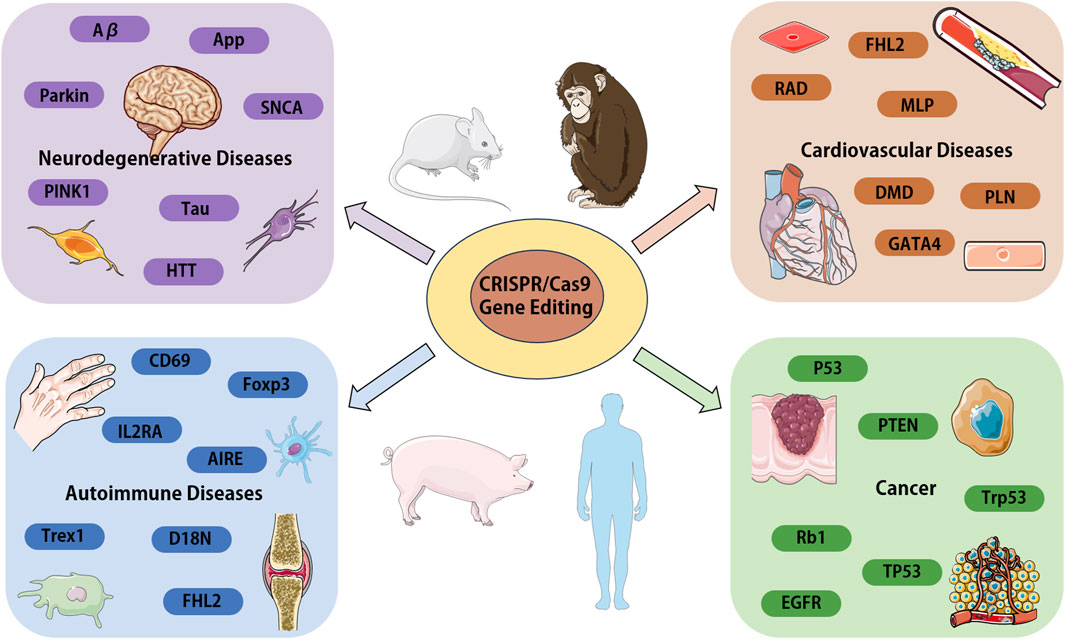
Figure 3. Overview of the CRISPR/Cas9 systems application in various human diseases. We focus on the genes and proteins associated with disease, which could be target for disease modeling or the candidates for gene therapy. Aβ, amyloid β-protein; APP, amyloid precursor protein; PARK2, parkin RBR E3 ubiquitin protein ligase; PINK1, PTEN-induced putative kinase 1; SNCA, a-synuclein; Tau, microtubule associated protein tau; HTT, huntington protein; FHL2, four and a half LIM domains 2; RAD, radish; MLP, membrane lipoprotein precursor; GATA4, GATA binding protein 4; DMD, Duchenne muscular dystrophy gene; PLN, phospholamban; IL2RA, interleukin 2 receptor subunit alpha; CD69, CD69 molecule; Foxp3, forkhead box P3; AIRE, autoimmune regulator; Trex1, three prime repair exonuclease 1; EGFR, epidermal growth factor receptor; p53, tumor suppressor gene; PTEN, phosphatase and tensin homolog; Trp53, transformation related protein 53; Rb1, RB transcriptional corepressor 1; TP53, tumor protein p53.
Although CRISPR/Cas9 technology is widely used for gene knockout or knock-in labeling, large fragment deletion, and transcriptional regulation, and has become an important tool for exploring the pathological mechanisms of diseases. However, certain challenges still need to be addressed, including the highly concerning off-target effects, delivery system optimization (van Haasteren et al., 2020), and enhancement of gene editing accuracy and efficiency. Consequently, the clinical application of CRISPR/Cas9 technology in disease still faces certain challenges (Behr et al., 2021). However, CRISPR/Cas9 technology is constantly being improved and optimized, such as by modifying the Cas9 protein, so that it does not cut the double strand DNA, but only cut the single strand, under the guidance of a specific double sgRNA, the off-target efficiency will be greatly reduced. Therefore, in the future, CRISPR/Cas9 technology will be widely used in the mechanism research and clinical treatment of diseases, and finally overcome the treatment problems of some diseases, and bring hope to more patients.
In summary, CRISPR/Cas9 is an innovative genome editing tool with significant potential for development. With the continuous advancement of basic research and clinical applications, CRISPR/Cas9 will bring new hope for disease treatment across various fields. Genome editing technology is still developing, and its integration with disease treatment is progressively close, thus expected to make substantial contributions to human health.
Author contributions
M-LZ: Writing–original draft, Writing–review and editing. H-BL: Funding acquisition, Validation, Writing–review and editing. YJ: Funding acquisition, Validation, Writing–original draft, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (82260138, 82260327); Natural Science Foundation of Inner Mongolia (2022QN08022); The Scientific Research Project of Colleges and Universities, Inner Mongolia Department of Education (NJZZ22678, NJZZ23014); The Inner Mongolia Health Commission’s Medical and Health Science and Technology Program (Class A) (202201267).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2024.1364742/full#supplementary-material
References
Alankarage, D., Jo, S., Pachter, N., Slavotinek, A., Selleri, L., Shieh, J. T., et al. (2020). Functional characterization of a novel PBX1 de novo missense variant identified in a patient with syndromic congenital heart disease. Hum. Mol. Genet. 29, 1068–1082. doi:10.1093/hmg/ddz231
Amoasii, L., Hildyard, J. C. W., Li, H., Sanchez-Ortiz, E., Mireault, A., Caballero, D., et al. (2018). Gene editing restores dystrophin expression in a canine model of Duchenne muscular dystrophy. Science 362, 86–91. doi:10.1126/science.aau1549
Ang, Y. S., Rivas, R. N., Ribeiro, A. J. S., Srivas, R., Rivera, J., Stone, N. R., et al. (2016). Disease model of GATA4 mutation reveals transcription factor cooperativity in human cardiogenesis. Cell. 167, 1734–1749. doi:10.1016/j.cell.2016.11.033
Anzalone, A. V., Koblan, L. W., and Liu, D. R. (2020). Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 38, 824–844. doi:10.1038/s41587-020-0561-9
Aouida, M., Eid, A., Ali, Z., Cradick, T., Lee, C., Deshmukh, H., et al. (2015). Efficient fdCas9 synthetic endonuclease with improved specificity for precise genome engineering. PLoS One 10, e0133373. doi:10.1371/journal.pone.0133373
Austin, K. M., Trembley, M. A., Chandler, S. F., Sanders, S. P., Saffitz, J. E., Abrams, D. J., et al. (2019). Molecular mechanisms of arrhythmogenic cardiomyopathy. Nat. Rev. Cardiol. 16, 519–537. doi:10.1038/s41569-019-0200-7
Barrangou, R., Fremaux, C., Deveau, H., Richards, M., Boyaval, P., Moineau, S., et al. (2007). CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712. doi:10.1126/science.1138140
Bates, G. P., Dorsey, R., Gusella, J. F., Hayden, M. R., Kay, C., Leavitt, B. R., et al. (2015). Huntington disease. Nat. Rev. Dis. Prim. 1, 15005. doi:10.1038/nrdp.2015.5
Baxa, M., Hruska-Plochan, M., Juhas, S., Vodicka, P., Pavlok, A., Juhasova, J., et al. (2013). A transgenic minipig model of Huntington's Disease. J. Huntingt. Dis. 2, 47–68. doi:10.3233/JHD-130001
Behr, M., Zhou, J., Xu, B., and Zhang, H. (2021). In vivo delivery of CRISPR-Cas9 therapeutics: progress and challenges. Acta Pharm. Sin. B 11, 2150–2171. doi:10.1016/j.apsb.2021.05.020
Bhardwaj, S., Kesari, K. K., Rachamalla, M., Mani, S., Ashraf, G. M., Jha, S. K., et al. (2022). CRISPR/Cas9 gene editing: new hope for Alzheimer's disease therapeutics. J. Adv. Res. 40, 207–221. doi:10.1016/j.jare.2021.07.001
Blasco, R. B., Karaca, E., Ambrogio, C., Cheong, T. C., Karayol, E., Minero, V. G., et al. (2014). Simple and rapid in vivo generation of chromosomal rearrangements using CRISPR/Cas9 technology. Cell. Rep. 9, 1219–1227. doi:10.1016/j.celrep.2014.10.051
Cai, E. P., Ishikawa, Y., Zhang, W., Leite, N. C., Li, J., Hou, S., et al. (2020). Genome-scale in vivo CRISPR screen identifies RNLS as a target for beta cell protection in type 1 diabetes. Nat. Metab. 2, 934–945. doi:10.1038/s42255-020-0254-1
Cai, J., Wu, H., Wang, C., Chen, Y., Zhang, D., Guan, S., et al. (2023). Sec1 regulates intestinal mucosal immunity in a mouse model of inflammatory bowel disease. BMC Immunol. 24, 51. doi:10.1186/s12865-023-00578-9
Chang, C. W., Chang, C. C., Hsia, K. C., and Tsai, S. Y. (2018). Generation of FHL2 homozygous knockout lines from human embryonic stem cells by CRISPR/Cas9-mediated ablation. Stem Cell. Res. 27, 21–24. doi:10.1016/j.scr.2017.12.015
Charlesworth, C. T., Deshpande, P. S., Dever, D. P., Camarena, J., Lemgart, V. T., Cromer, M. K., et al. (2019). Identification of preexisting adaptive immunity to Cas9 proteins in humans. Nat. Med. 25, 249–254. doi:10.1038/s41591-018-0326-x
Chen, F., Alphonse, M., and Liu, Q. (2020). Strategies for nonviral nanoparticle-based delivery of CRISPR/Cas9 therapeutics. Wiley Interdiscip. Rev. Nanomed Nanobiotechnol 12 (3), e1609. doi:10.1002/wnan.1609
Chen, Z. H., Yu, Y. P., Zuo, Z. H., Nelson, J. B., Michalopoulos, G. K., Monga, S., et al. (2017). Targeting genomic rearrangements in tumor cells through Cas9-mediated insertion of a suicide gene. Nat. Biotechnol. 35, 543–550. doi:10.1038/nbt.3843
Cheung, A. H., Chow, C., Zhang, J., Zhou, Y., Huang, T., Ng, K. C., et al. (2018). Specific targeting of point mutations in EGFR L858R-positive lung cancer by CRISPR/Cas9. Lab. Investig. 98, 968–976. doi:10.1038/s41374-018-0056-1
Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. doi:10.1126/science.1231143
Cooper, G. S., Miller, F. W., and Pandey, J. P. (1999). The role of genetic factors in autoimmune disease: implications for environmental research. Environ. Health Perspect. 107 (Suppl. 5), 693–700. doi:10.1289/ehp.99107s5693
Cortez, J. T., Montauti, E., Shifrut, E., Gatchalian, J., Zhang, Y., Shaked, O., et al. (2020). CRISPR screen in regulatory T cells reveals modulators of Foxp3. Nature 582, 416–420. doi:10.1038/s41586-020-2246-4
Crook, Z. R., and Housman, D. (2011). Huntington's disease: can mice lead the way to treatment? Neuron 69, 423–435. doi:10.1016/j.neuron.2010.12.035
de la Roche, J., Angsutararux, P., Kempf, H., Janan, M., Bolesani, E., Thiemann, S., et al. (2019). Comparing human iPSC-cardiomyocytes versus HEK293T cells unveils disease-causing effects of Brugada mutation A735V of Na(V)1.5 sodium channels. Sci. Rep. 9, 11173. doi:10.1038/s41598-019-47632-4
de Lazaro, I., and Mooney, D. J. (2021). Obstacles and opportunities in a forward vision for cancer nanomedicine. Nat. Mater 20, 1469–1479. doi:10.1038/s41563-021-01047-7
Deltcheva, E., Chylinski, K., Sharma, C. M., Gonzales, K., Chao, Y., Pirzada, Z. A., et al. (2011). CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471, 602–607. doi:10.1038/nature09886
De Plano, L. M., Calabrese, G., Conoci, S., Guglielmino, S. P. P., Oddo, S., and Caccamo, A. (2022). Applications of CRISPR-cas9 in Alzheimer's disease and related disorders. Int. J. Mol. Sci. 23, 8714. doi:10.3390/ijms23158714
Dianov, G. L., and Hubscher, U. (2013). Mammalian base excision repair: the forgotten archangel. Nucleic Acids Res. 41, 3483–3490. doi:10.1093/nar/gkt076
Dong, M. B., Wang, G., Chow, R. D., Ye, L., Zhu, L., Dai, X., et al. (2019). Systematic immunotherapy target discovery using genome-scale in vivo CRISPR screens in CD8 T cells. Cell. 178, 1189–1204. doi:10.1016/j.cell.2019.07.044
Doudna, J. A., and Charpentier, E. (2014). Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346, 1258096. doi:10.1126/science.1258096
Du, Y., Liu, Y., Hu, J., Peng, X., and Liu, Z. (2023). CRISPR/Cas9 systems: delivery technologies and biomedical applications. Asian J. Pharm. Sci. 18, 100854. doi:10.1016/j.ajps.2023.100854
Dugger, B. N., and Dickson, D. W. (2017). Pathology of neurodegenerative diseases. Cold Spring Harb. Perspect. Biol. 9, a028035. doi:10.1101/cshperspect.a028035
Ebina, H., Misawa, N., Kanemura, Y., and Koyanagi, Y. (2013). Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Sci. Rep. 3, 2510. doi:10.1038/srep02510
Egorova, T. V., Zotova, E. D., Reshetov, D. A., Polikarpova, A. V., Vassilieva, S. G., Vlodavets, D. V., et al. (2019). CRISPR/Cas9-generated mouse model of Duchenne muscular dystrophy recapitulating a newly identified large 430 kb deletion in the human DMD gene. Dis. Model. Mech. 12, dmm037655. doi:10.1242/dmm.037655
Ekman, F. K., Ojala, D. S., Adil, M. M., Lopez, P. A., Schaffer, D. V., and Gaj, T. (2019). CRISPR-Cas9-Mediated genome editing increases lifespan and improves motor deficits in a Huntington's disease mouse model. Mol. Ther. Nucleic Acids 17, 829–839. doi:10.1016/j.omtn.2019.07.009
Elso, C. M., Scott, N. A., Mariana, L., Masterman, E. I., Sutherland, A. P. R., Thomas, H. E., et al. (2019). Replacing murine insulin 1 with human insulin protects NOD mice from diabetes. PLoS One 14, e0225021. doi:10.1371/journal.pone.0225021
Farshim, P. P., and Bates, G. P. (2018). Mouse models of Huntington's disease. Methods Mol. Biol. 1780, 97–120. doi:10.1007/978-1-4939-7825-0_6
Gaj, T., Gersbach, C. A., and Barbas, C. F. (2013). ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 31, 397–405. doi:10.1016/j.tibtech.2013.04.004
Galarza-Munoz, G., Briggs, F. B. S., Evsyukova, I., Schott-Lerner, G., Kennedy, E. M., Nyanhete, T., et al. (2017). Human epistatic interaction controls IL7R splicing and increases multiple sclerosis risk. Cell. 169, 72–84. doi:10.1016/j.cell.2017.03.007
Guilinger, J. P., Thompson, D. B., and Liu, D. R. (2014). Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat. Biotechnol. 32, 577–582. doi:10.1038/nbt.2909
Gupta, D., Bhattacharjee, O., Mandal, D., Sen, M. K., Dey, D., Dasgupta, A., et al. (2019). CRISPR-Cas9 system: a new-fangled dawn in gene editing. Life Sci. 232, 116636. doi:10.1016/j.lfs.2019.116636
Gyorgy, B., Loov, C., Zaborowski, M. P., Takeda, S., Kleinstiver, B. P., Commins, C., et al. (2018). CRISPR/Cas9 mediated disruption of the Swedish APP allele as a therapeutic approach for early-onset Alzheimer's disease. Mol. Ther. Nucleic Acids 11, 429–440. doi:10.1016/j.omtn.2018.03.007
Han, J., Wang, Q., Zhang, Z., Gong, T., and Sun, X. (2014). Cationic bovine serum albumin based self-assembled nanoparticles as siRNA delivery vector for treating lung metastatic cancer. Small 10, 524–535. doi:10.1002/smll.201301992
Harris, V. M., Koelsch, K. A., Kurien, B. T., Harley, I. T. W., Wren, J. D., Harley, J. B., et al. (2019). Characterization of cxorf21 provides molecular insight into female-bias immune response in SLE pathogenesis. Front. Immunol. 10, 2160. doi:10.3389/fimmu.2019.02160
Havlicek, S., Shen, Y., Alpagu, Y., Bruntraeger, M. B., Zufir, N. B., Phuah, Z. Y., et al. (2017). Re-Engineered RNA-guided FokI-nucleases for improved genome editing in human cells. Mol. Ther. 25, 342–355. doi:10.1016/j.ymthe.2016.11.007
Hazafa, A., Mumtaz, M., Farooq, M. F., Bilal, S., Chaudhry, S. N., Firdous, M., et al. (2020). CRISPR/Cas9: a powerful genome editing technique for the treatment of cancer cells with present challenges and future directions. Life Sci. 263, 118525. doi:10.1016/j.lfs.2020.118525
Heckl, D., Kowalczyk, M. S., Yudovich, D., Belizaire, R., Puram, R. V., McConkey, M. E., et al. (2014). Generation of mouse models of myeloid malignancy with combinatorial genetic lesions using CRISPR-Cas9 genome editing. Nat. Biotechnol. 32, 941–946. doi:10.1038/nbt.2951
Holohan, D. R., Van Gool, F., and Bluestone, J. A. (2019). Thymically-derived Foxp3+ regulatory T cells are the primary regulators of type 1 diabetes in the non-obese diabetic mouse model. PLoS One 14, e0217728. doi:10.1371/journal.pone.0217728
Huang, H., Fang, M., Jostins, L., Umicevic Mirkov, M., Boucher, G., Anderson, C. A., et al. (2017b). Fine-mapping inflammatory bowel disease loci to single-variant resolution. Nature 547, 173–178. doi:10.1038/nature22969
Huang, J., Chen, M., Whitley, M. J., Kuo, H. C., Xu, E. S., Walens, A., et al. (2017a). Generation and comparison of CRISPR-Cas9 and Cre-mediated genetically engineered mouse models of sarcoma. Nat. Commun. 8, 15999. doi:10.1038/ncomms15999
Iyer, S., Suresh, S., Guo, D., Daman, K., Chen, J. C. J., Liu, P., et al. (2019). Precise therapeutic gene correction by a simple nuclease-induced double-stranded break. Nature 568, 561–565. doi:10.1038/s41586-019-1076-8
Jacobsen, J. C., Bawden, C. S., Rudiger, S. R., McLaughlan, C. J., Reid, S. J., Waldvogel, H. J., et al. (2010). An ovine transgenic Huntington's disease model. Hum. Mol. Genet. 19, 1873–1882. doi:10.1093/hmg/ddq063
Jaffre, F., Miller, C. L., Schanzer, A., Evans, T., Roberts, A. E., Hahn, A., et al. (2019). Inducible pluripotent stem cell-derived cardiomyocytes reveal aberrant extracellular regulated kinase 5 and mitogen-activated protein kinase kinase 1/2 signaling concomitantly promote hypertrophic cardiomyopathy in RAF1-associated noonan syndrome. Circulation 140, 207–224. doi:10.1161/CIRCULATIONAHA.118.037227
Jakobsen, J. E., Johansen, M. G., Schmidt, M., Dagnaes-Hansen, F., Dam, K., Gunnarsson, A., et al. (2013). Generation of minipigs with targeted transgene insertion by recombinase-mediated cassette exchange (RMCE) and somatic cell nuclear transfer (SCNT). Transgenic Res. 22, 709–723. doi:10.1007/s11248-012-9671-6
Jiang, F., and Doudna, J. A. (2017). CRISPR-Cas9 structures and mechanisms. Annu. Rev. Biophys. 46, 505–529. doi:10.1146/annurev-biophys-062215-010822
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., and Charpentier, E. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. doi:10.1126/science.1225829
Jing, L., Cheng, S., Pan, Y., Liu, Q., Yang, W., Li, S., et al. (2021). Accumulation of endogenous mutant huntingtin in astrocytes exacerbates neuropathology of Huntington disease in mice. Mol. Neurobiol. 58, 5112–5126. doi:10.1007/s12035-021-02451-5
Johansen, A. K., Molenaar, B., Versteeg, D., Leitoguinho, A. R., Demkes, C., Spanjaard, B., et al. (2017). Postnatal cardiac gene editing using CRISPR/Cas9 with AAV9-mediated delivery of short guide RNAs results in mosaic gene disruption. Circ. Res. 121, 1168–1181. doi:10.1161/CIRCRESAHA.116.310370
Kan, S. H., Huang, J. Y., Harb, J., Rha, A., Dalton, N. D., Christensen, C., et al. (2022). CRISPR-mediated generation and characterization of a Gaa homozygous c.1935C>A (p.D645E) Pompe disease knock-in mouse model recapitulating human infantile onset-Pompe disease. Sci. Rep. 12, 21576. doi:10.1038/s41598-022-25914-8
Karimian, A., Gorjizadeh, N., Alemi, F., Asemi, Z., Azizian, K., Soleimanpour, J., et al. (2020). CRISPR/Cas9 novel therapeutic road for the treatment of neurodegenerative diseases. Life Sci. 259, 118165. doi:10.1016/j.lfs.2020.118165
Kim, J. K., Howard, M. D., Dziubla, T. D., Rinehart, J. J., Jay, M., and Lu, X. (2011). Uniformity of drug payload and its effect on stability of solid lipid nanoparticles containing an ester prodrug. ACS Nano 5, 209–216. doi:10.1021/nn102357y
Kitada, T., Tong, Y., Gautier, C. A., and Shen, J. (2009). Absence of nigral degeneration in aged parkin/DJ-1/PINK1 triple knockout mice. J. Neurochem. 111, 696–702. doi:10.1111/j.1471-4159.2009.06350.x
Kitagawa, A., Kizub, I., Jacob, C., Michael, K., D'Alessandro, A., Reisz, J. A., et al. (2020). CRISPR-mediated single nucleotide polymorphism modeling in rats reveals insight into reduced cardiovascular risk associated with mediterranean G6PD variant. Hypertension 76, 523–532. doi:10.1161/HYPERTENSIONAHA.120.14772
Knott, G. J., and Doudna, J. A. (2018). CRISPR-Cas guides the future of genetic engineering. Science 361, 866–869. doi:10.1126/science.aat5011
Koo, T., Yoon, A. R., Cho, H. Y., Bae, S., Yun, C. O., and Kim, J. S. (2017). Selective disruption of an oncogenic mutant allele by CRISPR/Cas9 induces efficient tumor regression. Nucleic Acids Res. 45, 7897–7908. doi:10.1093/nar/gkx490
Kulcsar, P. I., Talas, A., Toth, E., Nyeste, A., Ligeti, Z., Welker, Z., et al. (2020). Blackjack mutations improve the on-target activities of increased fidelity variants of SpCas9 with 5'G-extended sgRNAs. Nat. Commun. 11, 1223. doi:10.1038/s41467-020-15021-5
La, Y., Wong, W., Peng, K., Tian, Z., Pan, J., Sun, R., et al. (2023). Decreased imiquimod-induced psoriasis-like skin inflammation in a novel mvd(F250S/+) knock-in mouse model. Inflammation 46, 1575–1586. doi:10.1007/s10753-023-01828-z
Lanctot, K. L., Amatniek, J., Ancoli-Israel, S., Arnold, S. E., Ballard, C., Cohen-Mansfield, J., et al. (2017). Neuropsychiatric signs and symptoms of Alzheimer's disease: new treatment paradigms. Alzheimers Dement. (N Y) 3, 440–449. doi:10.1016/j.trci.2017.07.001
Lee, M. H., Shin, J. I., Yang, J. W., Lee, K. H., Cha, D. H., Hong, J. B., et al. (2022). Genome editing using CRISPR-cas9 and autoimmune diseases: a comprehensive review. Int. J. Mol. Sci. 23, 1337. doi:10.3390/ijms23031337
Li, C., Brant, E., Budak, H., and Zhang, B. (2021b). CRISPR/Cas: a Nobel Prize award-winning precise genome editing technology for gene therapy and crop improvement. J. Zhejiang Univ. Sci. B 22, 253–284. doi:10.1631/jzus.B2100009
Li, C., and Kuemmerle, J. (2019). P119 the role of ptpn2 snp in the pathogenesis of fibrosis in crohn’s disease. Inflamm. Bowel Dis. 25, S57–S58. doi:10.1093/ibd/izy393.130
Li, H., Wu, S., Ma, X., Li, X., Cheng, T., Chen, Z., et al. (2021a). Co-Editing PINK1 and DJ-1 genes via adeno-associated virus-delivered CRISPR/Cas9 system in adult monkey brain elicits classical parkinsonian phenotype. Neurosci. Bull. 37, 1271–1288. doi:10.1007/s12264-021-00732-6
Li, X., Lu, W. J., Li, Y., Wu, F., Bai, R., Ma, S., et al. (2019). MLP-deficient human pluripotent stem cell derived cardiomyocytes develop hypertrophic cardiomyopathy and heart failure phenotypes due to abnormal calcium handling. Cell. Death Dis. 10, 610. doi:10.1038/s41419-019-1826-4
Li, Y., Chang, Y., Li, X., Li, X., Gao, J., Zhou, Y., et al. (2020). RAD-deficient human cardiomyocytes develop hypertrophic cardiomyopathy phenotypes due to calcium dysregulation. Front. Cell. Dev. Biol. 8, 585879. doi:10.3389/fcell.2020.585879
Lin, X., Pelletier, S., Gingras, S., Rigaud, S., Maine, C. J., Marquardt, K., et al. (2016). CRISPR-Cas9-Mediated modification of the NOD mouse genome with Ptpn22R619W mutation increases autoimmune diabetes. Diabetes. Diabetes. 65, 2134–2138. doi:10.2337/db16-0061
Ling, L., Mulaka, M., Munro, J., Dass, S., Mather, M. W., Riscoe, M. K., et al. (2020). Genetic ablation of the mitoribosome in the malaria parasite Plasmodium falciparum sensitizes it to antimalarials that target mitochondrial functions. J. Biol. Chem. 295, 7235–7248. doi:10.1074/jbc.RA120.012646
Liu, C., Zhang, L., Liu, H., and Cheng, K. (2017b). Delivery strategies of the CRISPR-Cas9 gene-editing system for therapeutic applications. J. Control Release 266, 17–26. doi:10.1016/j.jconrel.2017.09.012
Liu, N., and Olson, E. N. (2022). CRISPR modeling and correction of cardiovascular disease. Circ. Res. 130, 1827–1850. doi:10.1161/CIRCRESAHA.122.320496
Liu, X., Yagi, H., Saeed, S., Bais, A. S., Gabriel, G. C., Chen, Z., et al. (2017a). The complex genetics of hypoplastic left heart syndrome. Nat. Genet. 49, 1152–1159. doi:10.1038/ng.3870
Lopes, C., Tang, Y., Anjo, S. I., Manadas, B., Onofre, I., de Almeida, L. P., et al. (2020). Mitochondrial and redox modifications in Huntington disease induced pluripotent stem cells rescued by CRISPR/Cas9 CAGs targeting. Front. Cell. Dev. Biol. 8, 576592. doi:10.3389/fcell.2020.576592
Lu, L., Yu, X., Cai, Y., Sun, M., and Yang, H. (2021). Application of CRISPR/Cas9 in Alzheimer's disease. Front. Neurosci. 15, 803894. doi:10.3389/fnins.2021.803894
Lu, Y., Xue, J., Deng, T., Zhou, X., Yu, K., Deng, L., et al. (2020). Safety and feasibility of CRISPR-edited T cells in patients with refractory non-small-cell lung cancer. Nat. Med. 26, 732–740. doi:10.1038/s41591-020-0840-5
Lyu, Q., Dhagia, V., Han, Y., Guo, B., Wines-Samuelson, M. E., Christie, C. K., et al. (2018). CRISPR-Cas9-Mediated epitope tagging provides accurate and versatile assessment of myocardin-brief report. Arterioscler. Thromb. Vasc. Biol. 38, 2184–2190. doi:10.1161/ATVBAHA.118.311171
Mali, P., Yang, L., Esvelt, K. M., Aach, J., Guell, M., DiCarlo, J. E., et al. (2013). RNA-guided human genome engineering via Cas9. Science 339, 823–826. doi:10.1126/science.1232033
Marinaccio, C., Suraneni, P., Celik, H., Volk, A., Wen, Q. J., Ling, T., et al. (2021). LKB1/STK11 is a tumor suppressor in the progression of myeloproliferative neoplasms. Cancer Discov. 11, 1398–1410. doi:10.1158/2159-8290.CD-20-1353
Marrack, P., Kappler, J., and Kotzin, B. L. (2001). Autoimmune disease: why and where it occurs. Nat. Med. 7, 899–905. doi:10.1038/90935
Maruyama, T., Dougan, S. K., Truttmann, M. C., Bilate, A. M., Ingram, J. R., and Ploegh, H. L. (2015). Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat. Biotechnol. 33, 538–542. doi:10.1038/nbt.3190
Matsuzaka, Y., Tanihata, J., Ooshima, Y., Yamada, D., Sekiguchi, M., Miyatake, S., et al. (2020). The nSMase2/Smpd3 gene modulates the severity of muscular dystrophy and the emotional stress response in mdx mice. BMC Med. 18, 343. doi:10.1186/s12916-020-01805-5
McKenna, W. J., and Judge, D. P. (2021). Epidemiology of the inherited cardiomyopathies. Nat. Rev. Cardiol. 18, 22–36. doi:10.1038/s41569-020-0428-2
Men, H., Hankins, M. A., Bock, A. S., Beaton, B. P., Davis, D. J., Chesney, K. L., et al. (2021). Mutational analyses of novel rat models with targeted modifications in inflammatory bowel disease susceptibility genes. Mamm. Genome 32, 173–182. doi:10.1007/s00335-021-09868-2
Min, Y. L., Li, H., Rodriguez-Caycedo, C., Mireault, A. A., Huang, J., Shelton, J. M., et al. (2019). CRISPR-Cas9 corrects Duchenne muscular dystrophy exon 44 deletion mutations in mice and human cells. Sci. Adv. 5, eaav4324. doi:10.1126/sciadv.aav4324
Mokhtar, N., Den Low, E. N., Yahaya, A., and Ali, R. A. R. (2019). Characterisation of sgk2 gene using crispr/cas9 gene editing tool in ulcerative colitis. J. Gastroenterol. Hepatol. 34, 274. doi:10.1111/jgh.14879
Moretti, A., Fonteyne, L., Giesert, F., Hoppmann, P., Meier, A. B., Bozoglu, T., et al. (2020). Somatic gene editing ameliorates skeletal and cardiac muscle failure in pig and human models of Duchenne muscular dystrophy. Nat. Med. 26, 207–214. doi:10.1038/s41591-019-0738-2
Morin, F., Singh, N., Mdzomba, J. B., Dumas, A., Pernet, V., and Vallieres, L. (2021). Conditional deletions of hdc confirm roles of histamine in anaphylaxis and circadian activity but not in autoimmune encephalomyelitis. J. Immunol. 206, 2029–2037. doi:10.4049/jimmunol.2000719
Morton, S. U., Quiat, D., Seidman, J. G., and Seidman, C. E. (2022). Genomic frontiers in congenital heart disease. Nat. Rev. Cardiol. 19, 26–42. doi:10.1038/s41569-021-00587-4
Moses, C., Nugent, F., Waryah, C. B., Garcia-Bloj, B., Harvey, A. R., and Blancafort, P. (2019). Activating PTEN tumor suppressor expression with the CRISPR/dCas9 system. Mol. Ther. Nucleic Acids 14, 287–300. doi:10.1016/j.omtn.2018.12.003
Motwani, M., McGowan, J., Antonovitch, J., Gao, K. M., Jiang, Z., Sharma, S., et al. (2021). cGAS-STING pathway does not promote autoimmunity in murine models of SLE. Front. Immunol. 12, 605930. doi:10.3389/fimmu.2021.605930
Mou, H., Ozata, D. M., Smith, J. L., Sheel, A., Kwan, S. Y., Hough, S., et al. (2019). CRISPR-SONIC: targeted somatic oncogene knock-in enables rapid in vivo cancer modeling. Genome Med. 11, 21. doi:10.1186/s13073-019-0627-9
Nakagawa, Y., Sakuma, T., Sakamoto, T., Ohmuraya, M., Nakagata, N., and Yamamoto, T. (2015). Production of knockout mice by DNA microinjection of various CRISPR/Cas9 vectors into freeze-thawed fertilized oocytes. BMC Biotechnol. 15, 33. doi:10.1186/s12896-015-0144-x
Nelson, C. E., Wu, Y., Gemberling, M. P., Oliver, M. L., Waller, M. A., Bohning, J. D., et al. (2019). Long-term evaluation of AAV-CRISPR genome editing for Duchenne muscular dystrophy. Nat. Med. 25, 427–432. doi:10.1038/s41591-019-0344-3
Ng, S. R., Rideout, W. M., Akama-Garren, E. H., Bhutkar, A., Mercer, K. L., Schenkel, J. M., et al. (2020). CRISPR-mediated modeling and functional validation of candidate tumor suppressor genes in small cell lung cancer. Proc. Natl. Acad. Sci. U. S. A. 117, 513–521. doi:10.1073/pnas.1821893117
Odqvist, L., Jevnikar, Z., Riise, R., Oberg, L., Rhedin, M., Leonard, D., et al. (2019). Genetic variations in A20 DUB domain provide a genetic link to citrullination and neutrophil extracellular traps in systemic lupus erythematosus. Ann. Rheum. Dis. 78, 1363–1370. doi:10.1136/annrheumdis-2019-215434
Orozco, R. C., Marquardt, K., Mowen, K., and Sherman, L. A. (2021). Proautoimmune allele of tyrosine phosphatase, PTPN22, enhances tumor immunity. J. Immunol. 207, 1662–1671. doi:10.4049/jimmunol.2100304
Oser, M. G., Sabet, A. H., Gao, W., Chakraborty, A. A., Schinzel, A. C., Jennings, R. B., et al. (2019). The KDM5A/RBP2 histone demethylase represses NOTCH signaling to sustain neuroendocrine differentiation and promote small cell lung cancer tumorigenesis. Genes. Dev. 33, 1718–1738. doi:10.1101/gad.328336.119
Parra-Nieto, J., Del Cid, M. A. G., de Carcer, I. A., and Baeza, A. (2021). Inorganic porous nanoparticles for drug delivery in antitumoral therapy. Biotechnol. J. 16, e2000150. doi:10.1002/biot.202000150
Platt, R. J., Chen, S., Zhou, Y., Yim, M. J., Swiech, L., Kempton, H. R., et al. (2014). CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 159, 440–455. doi:10.1016/j.cell.2014.09.014
Porto, E. M., Komor, A. C., Slaymaker, I. M., and Yeo, G. W. (2020). Base editing: advances and therapeutic opportunities. Nat. Rev. Drug Discov. 19, 839–859. doi:10.1038/s41573-020-0084-6
Presa, M., Racine, J. J., Dwyer, J. R., Lamont, D. J., Ratiu, J. J., Sarsani, V. K., et al. (2018). A hypermorphic nfkbid allele contributes to impaired thymic deletion of autoreactive diabetogenic CD8(+) T cells in NOD mice. J. Immunol. 201, 1907–1917. doi:10.4049/jimmunol.1800465
Qi, L. S., Larson, M. H., Gilbert, L. A., Doudna, J. A., Weissman, J. S., Arkin, A. P., et al. (2013). Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 152, 1173–1183. doi:10.1016/j.cell.2013.02.022
Qiao, Y., Wang, J., Karagoz, E., Liang, B., Song, X., Shang, R., et al. (2019). Axis inhibition protein 1 (Axin1) deletion-induced hepatocarcinogenesis requires intact β-catenin but not notch cascade in mice. Hepatology 70, 2003–2017. doi:10.1002/hep.30556
Qing, X., Walter, J., Jarazo, J., Arias-Fuenzalida, J., Hillje, A. L., and Schwamborn, J. C. (2017). CRISPR/Cas9 and piggyBac-mediated footprint-free LRRK2-G2019S knock-in reveals neuronal complexity phenotypes and α-Synuclein modulation in dopaminergic neurons. Stem Cell. Res. 24, 44–50. doi:10.1016/j.scr.2017.08.013
Qu, J., Liu, N., Gao, L., Hu, J., Sun, M., and Yu, D. (2023). Development of CRISPR Cas9, spin-off technologies and their application in model construction and potential therapeutic methods of Parkinson's disease. Front. Neurosci. 17, 1223747. doi:10.3389/fnins.2023.1223747
Racine, J. J., Stewart, I., Ratiu, J., Christianson, G., Lowell, E., Helm, K., et al. (2018). Improved murine MHC-deficient HLA transgenic NOD mouse models for type 1 diabetes therapy development. Diabetes 67, 923–935. doi:10.2337/db17-1467
Ran, F. A., Hsu, P. D., Lin, C. Y., Gootenberg, J. S., Konermann, S., Trevino, A. E., et al. (2013). Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 154, 1380–1389. doi:10.1016/j.cell.2013.08.021
Rees, H. A., and Liu, D. R. (2018). Base editing: precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 19, 770–788. doi:10.1038/s41576-018-0059-1
Ren, X., Yang, Z., Xu, J., Sun, J., Mao, D., Hu, Y., et al. (2014). Enhanced specificity and efficiency of the CRISPR/Cas9 system with optimized sgRNA parameters in Drosophila. Cell. Rep. 9, 1151–1162. doi:10.1016/j.celrep.2014.09.044
Roper, J., Tammela, T., Cetinbas, N. M., Akkad, A., Roghanian, A., Rickelt, S., et al. (2017). In vivo genome editing and organoid transplantation models of colorectal cancer and metastasis. Nat. Biotechnol. 35, 569–576. doi:10.1038/nbt.3836
Roth, T. L., Puig-Saus, C., Yu, R., Shifrut, E., Carnevale, J., Li, P. J., et al. (2018). Reprogramming human T cell function and specificity with non-viral genome targeting. Nature 559, 405–409. doi:10.1038/s41586-018-0326-5
Sachi, N., Kamiyama, N., Saechue, B., Ozaka, S., Dewayani, A., Ariki, S., et al. (2023). CCL20/CCR6 chemokine signaling is not essential for pathogenesis in an experimental autoimmune encephalomyelitis mouse model of multiple sclerosis. Biochem. Biophys. Res. Commun. 641, 123–131. doi:10.1016/j.bbrc.2022.11.088
Safari, F., Farajnia, S., Arya, M., Zarredar, H., and Nasrolahi, A. (2018). CRISPR and personalized Treg therapy: new insights into the treatment of rheumatoid arthritis. Immunopharmacol. Immunotoxicol. 40, 201–211. doi:10.1080/08923973.2018.1437625
Saifaldeen, M., Al-Ansari, D. E., Ramotar, D., and Aouida, M. (2020). CRISPR FokI dead Cas9 system: principles and applications in genome engineering. Cells 9, 2518. doi:10.3390/cells9112518
Seita, Y., Morimura, T., Watanabe, N., Iwatani, C., Tsuchiya, H., Nakamura, S., et al. (2020). Generation of transgenic cynomolgus monkeys overexpressing the gene for amyloid-β precursor protein. J. Alzheimers Dis. 75, 45–60. doi:10.3233/JAD-191081
Serneels, L., T'Syen, D., Perez-Benito, L., Theys, T., Holt, M. G., and De Strooper, B. (2020). Modeling the β-secretase cleavage site and humanizing amyloid-beta precursor protein in rat and mouse to study Alzheimer's disease. Mol. Neurodegener. 15, 60. doi:10.1186/s13024-020-00399-z
Seth, K., and Harish, H. (2016). Current status of potential applications of repurposed Cas9 for structural and functional genomics of plants. Biochem. Biophys. Res. Commun. 480, 499–507. doi:10.1016/j.bbrc.2016.10.130
Sevim, H., Kocaefe, Y. C., Onur, M. A., Uckan-Cetinkaya, D., and Gurpinar, O. A. (2018). Bone marrow derived mesenchymal stem cells ameliorate inflammatory response in an in vitro model of familial hemophagocytic lymphohistiocytosis 2. Stem Cell. Res. Ther. 9, 198. doi:10.1186/s13287-018-0941-y
Shen, B., Zhang, W., Zhang, J., Zhou, J., Wang, J., Chen, L., et al. (2014). Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat. Methods 11, 399–402. doi:10.1038/nmeth.2857
Simeonov, D. R., Gowen, B. G., Boontanrart, M., Roth, T. L., Gagnon, J. D., Mumbach, M. R., et al. (2017). Discovery of stimulation-responsive immune enhancers with CRISPR activation. Nature 549, 111–115. doi:10.1038/nature23875
Smith, J. G. W., Owen, T., Bhagwan, J. R., Mosqueira, D., Scott, E., Mannhardt, I., et al. (2018). Isogenic pairs of hiPSC-CMs with hypertrophic cardiomyopathy/LVNC-associated ACTC1 E99K mutation unveil differential functional deficits. Stem Cell. Rep. 11, 1226–1243. doi:10.1016/j.stemcr.2018.10.006
Sondergaard, L. V., Ladewig, J., Dagnaes-Hansen, F., Herskin, M. S., and Holm, I. E. (2012). Object recognition as a measure of memory in 1-2 years old transgenic minipigs carrying the APPsw mutation for Alzheimer's disease. Transgenic Res. 21, 1341–1348. doi:10.1007/s11248-012-9620-4
Soyk, S., Muller, N. A., Park, S. J., Schmalenbach, I., Jiang, K., Hayama, R., et al. (2017). Variation in the flowering gene SELF PRUNING 5G promotes day-neutrality and early yield in tomato. Nat. Genet. 49, 162–168. doi:10.1038/ng.3733
Speck-Hernandez, C. A., Assis, A. F., Felicio, R. F., Cotrim-Sousa, L., Pezzi, N., Lopes, G. S., et al. (2018). Aire disruption influences the medullary thymic epithelial cell transcriptome and interaction with thymocytes. Front. Immunol. 9, 964. doi:10.3389/fimmu.2018.00964
Stadtmauer, E. A., Fraietta, J. A., Davis, M. M., Cohen, A. D., Weber, K. L., Lancaster, E., et al. (2020). CRISPR-engineered T cells in patients with refractory cancer. Science 367 (6481), eaba7365. doi:10.1126/science.aba7365
Strong, A., and Musunuru, K. (2017). Genome editing in cardiovascular diseases. Nat. Rev. Cardiol. 14, 11–20. doi:10.1038/nrcardio.2016.139
Sun, J., Carlson-Stevermer, J., Das, U., Shen, M., Delenclos, M., Snead, A. M., et al. (2019). CRISPR/Cas9 editing of APP C-terminus attenuates β-cleavage and promotes α-cleavage. Nat. Commun. 10, 53. doi:10.1038/s41467-018-07971-8
Tabrizi, S. J., Flower, M. D., Ross, C. A., and Wild, E. J. (2020). Huntington disease: new insights into molecular pathogenesis and therapeutic opportunities. Nat. Rev. Neurol. 16, 529–546. doi:10.1038/s41582-020-0389-4
Tan, D. C. S., Yao, S., Ittner, A., Bertz, J., Ke, Y. D., Ittner, L. M., et al. (2018). Generation of a new tau knockout (tauΔex1) line using CRISPR/Cas9 genome editing in mice. J. Alzheimers Dis. 62, 571–578. doi:10.3233/JAD-171058
Tong, S., Moyo, B., Lee, C. M., Leong, K., and Bao, G. (2019). Engineered materials for in vivo delivery of genome-editing machinery. Nat. Rev. Mater 4, 726–737. doi:10.1038/s41578-019-0145-9
Tsai, S. Q., Wyvekens, N., Khayter, C., Foden, J. A., Thapar, V., Reyon, D., et al. (2014). Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat. Biotechnol. 32, 569–576. doi:10.1038/nbt.2908
van Haasteren, J., Li, J., Scheideler, O. J., Murthy, N., and Schaffer, D. V. (2020). The delivery challenge: fulfilling the promise of therapeutic genome editing. Nat. Biotechnol. 38, 845–855. doi:10.1038/s41587-020-0565-5
Wang, L., Li, L., Ma, Y., Hu, H., Li, Q., Yang, Y., et al. (2020). Reactivation of γ-globin expression through Cas9 or base editor to treat β-hemoglobinopathies. Cell. Res. 30, 276–278. doi:10.1038/s41422-019-0267-z
Wang, X., Cao, C., Huang, J., Yao, J., Hai, T., Zheng, Q., et al. (2016). One-step generation of triple gene-targeted pigs using CRISPR/Cas9 system. Sci. Rep. 6, 20620. doi:10.1038/srep20620
Weber, L., Frati, G., Felix, T., Hardouin, G., Casini, A., Wollenschlaeger, C., et al. (2020). Editing a gamma-globin repressor binding site restores fetal hemoglobin synthesis and corrects the sickle cell disease phenotype. Sci. Adv. 6, eaay9392. doi:10.1126/sciadv.aay9392
Wiedenheft, B., Sternberg, S. H., and Doudna, J. A. (2012). RNA-guided genetic silencing systems in bacteria and archaea. Nature 482, 331–338. doi:10.1038/nature10886
Wu, T. T., Ma, Y. W., Zhang, X., Dong, W., Gao, S., Wang, J. Z., et al. (2020). Myocardial tissue-specific Dnmt1 knockout in rats protects against pathological injury induced by Adriamycin. Lab. Investig. 100, 974–985. doi:10.1038/s41374-020-0402-y
Wyss-Coray, T. (2016). Ageing, neurodegeneration and brain rejuvenation. Nature 539, 180–186. doi:10.1038/nature20411
Xia, Y., Hu, J., Li, X., Zheng, S., Wang, G., Tan, S., et al. (2022). Investigating the pathogenesis of MYH7 mutation Gly823Glu in familial hypertrophic cardiomyopathy using a mouse model. J. Vis. Exp. doi:10.3791/63949
Xiao, N., Wei, J., Xu, S., Du, H., Huang, M., Zhang, S., et al. (2019). cGAS activation causes lupus-like autoimmune disorders in a TREX1 mutant mouse model. J. Autoimmun. 100, 84–94. doi:10.1016/j.jaut.2019.03.001
Xie, Y., Ma, A., Wang, B., Peng, R., Jing, Y., Wang, D., et al. (2019). Rare mutations of ADAM17 from TOFs induce hypertrophy in human embryonic stem cell-derived cardiomyocytes via HB-EGF signaling. Clin. Sci. (Lond) 133, 225–238. doi:10.1042/CS20180842
Xu, R. C., Wang, F., Sun, J. L., Abuduwaili, W., Zhang, G. C., Liu, Z. Y., et al. (2022). A novel murine model of combined hepatocellular carcinoma and intrahepatic cholangiocarcinoma. J. Transl. Med. 20, 579. doi:10.1186/s12967-022-03791-z
Xu, X., Liu, C., Wang, Y., Koivisto, O., Zhou, J., Shu, Y., et al. (2021). Nanotechnology-based delivery of CRISPR/Cas9 for cancer treatment. Adv. Drug Deliv. Rev. 176, 113891. doi:10.1016/j.addr.2021.113891
Xu, X., Wan, T., Xin, H., Li, D., Pan, H., Wu, J., et al. (2019). Delivery of CRISPR/Cas9 for therapeutic genome editing. J. Gene Med. 21, e3107. doi:10.1002/jgm.3107
Xue, W., Chen, S., Yin, H., Tammela, T., Papagiannakopoulos, T., Joshi, N. S., et al. (2014). CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature 514, 380–384. doi:10.1038/nature13589
Yadav, S., Sitbon, Y. H., Kazmierczak, K., and Szczesna-Cordary, D. (2019). Hereditary heart disease: pathophysiology, clinical presentation, and animal models of HCM, RCM, and DCM associated with mutations in cardiac myosin light chains. Pflugers Arch. 471, 683–699. doi:10.1007/s00424-019-02257-4
Yan, S., Tu, Z., Liu, Z., Fan, N., Yang, H., Yang, S., et al. (2018). A huntingtin knockin pig model recapitulates features of selective neurodegeneration in Huntington's disease. Cell. 173, 989–1002. doi:10.1016/j.cell.2018.03.005
Yang, H., Yang, S., Jing, L., Huang, L., Chen, L., Zhao, X., et al. (2020a). Truncation of mutant huntingtin in knock-in mice demonstrates exon1 huntingtin is a key pathogenic form. Nat. Commun. 11, 2582. doi:10.1038/s41467-020-16318-1
Yang, J., McGovern, A., Martin, P., Duffus, K., Ge, X., Zarrineh, P., et al. (2020b). Analysis of chromatin organization and gene expression in T cells identifies functional genes for rheumatoid arthritis. Nat. Commun. 11, 4402. doi:10.1038/s41467-020-18180-7
Yang, W., Chen, X., Li, S., and Li, X. J. (2021). Genetically modified large animal models for investigating neurodegenerative diseases. Cell. Biosci. 11, 218. doi:10.1186/s13578-021-00729-8
Yang, W., Guo, X., Tu, Z., Chen, X., Han, R., Liu, Y., et al. (2022). PINK1 kinase dysfunction triggers neurodegeneration in the primate brain without impacting mitochondrial homeostasis. Protein Cell. 13, 26–46. doi:10.1007/s13238-021-00888-x
Yang, W., Liu, Y., Tu, Z., Xiao, C., Yan, S., Ma, X., et al. (2019). CRISPR/Cas9-mediated PINK1 deletion leads to neurodegeneration in rhesus monkeys. Cell. Res. 29, 334–336. doi:10.1038/s41422-019-0142-y
Yeh, C. D., Richardson, C. D., and Corn, J. E. (2019). Advances in genome editing through control of DNA repair pathways. Nat. Cell. Biol. 21, 1468–1478. doi:10.1038/s41556-019-0425-z
Yin, C., Zhang, T., Qu, X., Zhang, Y., Putatunda, R., Xiao, X., et al. (2017). In Vivo excision of HIV-1 provirus by saCas9 and multiplex single-guide RNAs in animal models. Mol. Ther. 25, 1168–1186. doi:10.1016/j.ymthe.2017.03.012
Yin, J., Lu, R., Xin, C., Wang, Y., Ling, X., Li, D., et al. (2022a). Cas9 exo-endonuclease eliminates chromosomal translocations during genome editing. Nat. Commun. 13, 1204. doi:10.1038/s41467-022-28900-w
Yin, P., Li, S., Li, X. J., and Yang, W. (2022b). New pathogenic insights from large animal models of neurodegenerative diseases. Protein Cell. 13, 707–720. doi:10.1007/s13238-022-00912-8
Yoon, H. H., Ye, S., Lim, S., Jo, A., Lee, H., Hong, F., et al. (2022). CRISPR-Cas9 gene editing protects from the a53t-SNCA overexpression-induced pathology of Parkinson's disease in vivo. CRISPR J. 5, 95–108. doi:10.1089/crispr.2021.0025
Zaboikin, M., Zaboikina, T., Freter, C., and Srinivasakumar, N. (2017). Non-homologous end joining and homology directed DNA repair frequency of double-stranded breaks introduced by genome editing reagents. PLoS One 12, e0169931. doi:10.1371/journal.pone.0169931
Zhang, X., Chen, L., Zhu, B., Wang, L., Chen, C., Hong, M., et al. (2020). Increasing the efficiency and targeting range of cytidine base editors through fusion of a single-stranded DNA-binding protein domain. Nat. Cell. Biol. 22, 740–750. doi:10.1038/s41556-020-0518-8
Zhang, X. H., Tee, L. Y., Wang, X. G., Huang, Q. S., and Yang, S. H. (2015). Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol. Ther. Nucleic Acids 4, e264. doi:10.1038/mtna.2015.37
Zhen, S., Lu, J. J., Wang, L. J., Sun, X. M., Zhang, J. Q., Li, X., et al. (2016). In vitro and in vivo synergistic therapeutic effect of cisplatin with human Papillomavirus16 E6/E7 CRISPR/Cas9 on cervical cancer cell line. Transl. Oncol. 9, 498–504. doi:10.1016/j.tranon.2016.10.002
Zhou, B., Yang, W., Li, W., He, L., Lu, L., Zhang, L., et al. (2020). Zdhhc2 is essential for plasmacytoid dendritic cells mediated inflammatory response in psoriasis. Front. Immunol. 11, 607442. doi:10.3389/fimmu.2020.607442
Zhou, X., Xin, J., Fan, N., Zou, Q., Huang, J., Ouyang, Z., et al. (2015). Generation of CRISPR/Cas9-mediated gene-targeted pigs via somatic cell nuclear transfer. Cell. Mol. Life Sci. 72, 1175–1184. doi:10.1007/s00018-014-1744-7
Zhu, J., Hay, A. N., Potter, A. A., Richwine, M. W., Sproule, T., LeRoith, T., et al. (2020). Abrogated AID function prolongs survival and diminishes renal pathology in the BXSB mouse model of systemic lupus erythematosus. J. Immunol. 204, 1091–1100. doi:10.4049/jimmunol.1900501
Zhu, Q., Wang, J., Zhang, L., Bian, W., Lin, M., Xu, X., et al. (2019). LCK rs10914542-G allele associates with type 1 diabetes in children via T cell hyporesponsiveness. Pediatr. Res. 86, 311–315. doi:10.1038/s41390-019-0436-2
Keywords: CRISPR/Cas9, diseases modeling, gene therapy, neurodegenerative diseases, cardiovascular diseases, autoimmune-related diseases, cancer
Citation: Zhang M-L, Li H-B and Jin Y (2024) Application and perspective of CRISPR/Cas9 genome editing technology in human diseases modeling and gene therapy. Front. Genet. 15:1364742. doi: 10.3389/fgene.2024.1364742
Received: 03 January 2024; Accepted: 11 March 2024;
Published: 11 April 2024.
Edited by:
Wenxia He, University of North Carolina at Chapel Hill, United StatesReviewed by:
Dhruva Katrekar, Arc Institute, United StatesMustapha Aouida, Hamad bin Khalifa University, Qatar
Copyright © 2024 Zhang, Li and Jin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong-Bin Li, TGhid2I3M0AxMjYuY29t; Yong Jin, MjAyMTIzMDhAaW1tdS5lZHUuY24=
 Man-Ling Zhang
Man-Ling Zhang Hong-Bin Li1,2*
Hong-Bin Li1,2*