- 1Laboratory for Plant Genetics and Crop Improvement, Division of Crop Biotechnics, Department of Biosystems, University of Leuven, Leuven, Belgium
- 2KU Leuven Plant Institute (LPI), University of Leuven, Leuven, Belgium
- 3Laboratory for Molecular Plant Hormone Physiology, Division of Crop Biotechnics, Department of Biosystems, University of Leuven, Leuven, Belgium
The S-RNase gene plays an essential role in the gametophytic self-incompatibility (GSI) system of Pyrus. It codes for the stylar-expressed S-RNase protein which inhibits the growth of incompatible pollen tubes through cytotoxicity and the induction of programmed cell death in the pollen tube. While research on the Pyrus GSI system has primarily focused on the S-RNase gene, there is still a lack of insight into its spatiotemporal expression profile and the factors that regulate it. Previous studies have suggested that S-RNase expression in the style is influenced by pollination and is dependent on the compatibility type. We here continue on this basic hypothesis by analyzing the spatiotemporal expression of the S-RNase alleles in Pyrus communis “Conference” styles in response to different types of pollination; namely, upon full- and semi-compatible pollination and upon incompatible selfing. The results revealed that temporal dynamics of S-RNase expression are influenced by the pollen’s compatibility type, indicating the presence of a signaling mechanism between pollen and style to control S-RNase production during pollen tube growth. In our experiment, S-RNase expression continuously decreased after cross-pollination and in the unpollinated control. However, after a fully incompatible pollination, S-RNase expression remained constant. Finally, semi-compatible pollination showed a initially constant S-RNase expression for both alleles followed by a strong decrease in expression. Based on these results and previous findings, we propose a regulatory mechanism to explain the effect of pollination and the associated compatibility type on S-RNase expression in the style. This proposed mechanism could be used as a starting point for future research.
1 Introduction
Upon germination of the pear (Pyrus communis) pollen on the stigma, the pollen tube passes through the specialized style transmitting tract (TT) which comprises a dense extracellular matrix (ECM) containing factors that can promote or hinder pollen tube growth. These factors are established by the continuous signal exchanges occurring between the growing pollen tubes and the style. Some examples of such signals include the chemo-attractants produced by the ovule that serve as guidance cues for the pollen tube, alterations in the metabolic status of the stylar transmitting tract in response to pollen tube germination and growth, and pollination-induced pistil aging and ovule maturation (Dresselhaus and Franklin-tong, 2013; Lora et al., 2016; Joly et al., 2019; Mandrone et al., 2019). Self-incompatibility is another important type of pollen-style interaction. In pear (Pyrus genus), inhibition of self-fertilization is regulated at the genetic level by the S-RNase-dependent gametophytic self-incompatibility (GSI) system, which is determined by a single locus; the S-locus. This locus comprises the female S-RNase gene (Sassa et al., 1992) and multiple male S-locus F-box brother (SFBB) genes (Sassa et al., 2007) that are tightly linked in one linkage block. The S-RNase protein is the female-specific GSI component and is expressed solely in the pistil. The S-RNase is a member of the RNase T2 family, but while the RNase activity of this protein is known to be required for the pistil S function in Solanaceae (Huang et al., 1994), it remains unclear whether only RNA degradation triggers pollen tube growth inhibition since the S-RNases of pear and apple were also shown to interact with other targets such as actin (Liu et al., 2007), phospholipase C (Qu et al., 2017), and pyrophosphatase (Li et al., 2018). Upon pollen germination, the stylar S-RNase proteins enter the cytosol of each growing pollen tube non-selectively (Luu et al., 2000; Goldraij et al., 2006; Hui et al., 2014), however, they only inhibit the growth of incompatible pollen tubes (De Franceschi et al., 2012). In contrast, the SFBB genes, as male-specific GSI component, are expressed in the growing pollen tube where they encode different F-box proteins that target specific S-RNases that enter the pollen tube from the transmitting tract. The molecular mechanism of GSI regulation by S-RNases and SFBBs is described in the non-self-recognition model. In this model, it is proposed that growing pollen tubes express many different SFBBs that specifically interact with invading non-self-S-RNases (Kubo et al., 2010), whereby they act as a subunit of the SCF complex to ubiquitinate and mark these S-RNases for degradation by the 26S proteasome, hence ensuring continued pollen tube growth in case of a compatible interaction (Hua et al., 2008). In case of a non-compatible pollination (e.g, selfing), the SFFBs in the pollen tube cannot interact with the invading self-type S-RNases and mark them for degradation, so that these S-RNases remain present and can exert their cytotoxic function to inhibit pollen tube growth. This non-self-recognition model of GSI was first proposed in Petunia (Kubo et al., 2010) after which it was also adopted for apple and pear based on the identification of multiple SFBB genes at the S-locus of Maleae (De Franceschi et al., 2011), the observed competitive interaction in heteroallelic pollen (Qi et al., 2011; De Franceschi et al., 2012), and the characterization of the self-compatible mutant S4sm haplotype (Okada et al., 2008).
The S-RNase gene is believed to be expressed in the cells of the stylar transmitting tract and released into the extracellular matrix (Sassa et al., 1993; McClure et al., 2011). However, there is only limited information available on the exact spatiotemporal expression profile of this gene and the factors that regulate or influence its expression. Studies on different species with S-RNase-dependent GSI have suggested that pollination may influence S-RNase expression in the style with effects depending on the compatibility type. For instance, Solanum chacoense was found to exhibit a 60% decrease in S-RNase mRNA levels in the styles 24 h after cross-pollination, compared to a more moderate 25% decrease after selfing (Liu et al., 2009). Based on these findings, the authors proposed a positive feedback mechanism in which compatible pollen tubes simultaneously degrade the S-RNase protein in their cytoplasm and send out a signal to reduce S-RNase expression, hence reinforcing the compatible reaction (Liu et al., 2009). A decreased S-RNase expression after cross-pollination and higher expression after self-pollination was also found in lemon, a member of the Rutaceae family which was recently also found to harbor a similar type of S-RNase-dependent GSI (Honsho, 2023). Expression analysis of the S3-RNase in lemon styles showed ten times higher expression after self-pollination compared to cross-pollination 20 h after pollination (Li et al., 2022). In the Pyrus bretschneideri cultivar “Dangshan,” the expression of both S-RNase alleles (PbS7 and PbS34) also decreased after cross-pollination and this occurred faster and more pronounced as compared to no pollination or self-pollination (Shi et al., 2017). These results indicate that the compatibility type of pollination affects S-RNase expression in the style and suggests the presence of a yet unknown signaling mechanism that affects the molecular control of the GSI system. However, up till now, experimental data in pear or related species, such as apple, is very limited and a possible mechanism has as of yet not been proposed. While the GSI system in Solanaceae and Rosaceae share many similarities, there are also important differences, such as the involvement of non-S-locus factors HT-B and 120K in Solanaceae (Goldraij et al., 2006), which have not been observed in Maleae. Therefore, it cannot simply be assumed that the positive feedback mechanism proposed in S. chacoense by Liu et al. (2009) also applies to pear.
In this study, we used the P. communis cultivar “Conference” as a model to monitor allele-specific S-RNase expression in unpollinated and pollinated flowers at different time points following (no) pollination. S-RNase expression was assessed in two regions of the style (upper and lower) and the impact of the three compatibility types was assessed: compatible, incompatible, and semi-compatible pollination. Based on the obtained results and information from previous studies, we propose a mechanistic model for feedback regulation of S-RNase expression depending on the pollination compatibility type in P. communis that could serve as a valuable starting point to further elucidate the molecular cues and pathways underpinning the intricate GSI system in pear.
2 Materials and methods
2.1 Plant material
The plant material used for this study included mature potted trees of P. communis cultivars “Conference,” “Légipont” and “Bartlett” (“Williams Bon Chrétien”). In order to have available pollen donors for performing the different types of in situ pollination, one tree of each pear variety was brought into the greenhouse approximately 2 weeks before the expected flowering time and was forced into flowering under controlled conditions. Light intensity in the greenhouse was maintained at >240 W/m2 between 7:00–19:00 (using supplemental Son-T lights) and the temperature was held within the range of 15°C–27°C, at an average of 21.5°C. For all three cultivars, approximately 100 flowers were harvested at the balloon stage for pollen collection. Unopened anthers were collected using forceps into a small Petri dish and air dried for 48 h until opening to release mature pollen grains. Dried pollen grains were stored until pollination at room temperature in air-tight containers containing silica gel. Pollinations were performed on flowers of the cultivar “Conference.” Five adult potted trees of cultivar “Conference” were brought into the greenhouse at the onset of flowering and were further maintained under the same conditions outlined above to minimize environmental effects during the experiment.
2.2 Pollinations and style tissue sampling
Controlled pollinations were all performed using “Conference” as pollen acceptor. On each of the five “Conference” trees, four individual branches were labeled, each to receive one of the four possible pollination types. Flowers on these branches were pruned and only flowers in the same developmental stage (balloon stage) and having the same visually assessed fitness (>6 flowers in the cluster, uniform flower size, and no visible signs of disease or insect damage) were maintained for controlled pollination. The different pollination types included no pollination (UP), self-pollination (Self), cross-pollination (Cross), and semi-compatible pollination (Semi). For the three pollination treatments (Self, Semi, and Cross), pollen was collected before the start of the experiment from forced “Conference” (Self, PcS108-PcS121), “Légipont” (Semi, PcS102-PcS108) and “Bartlett” (Cross, PcS101-PcS102) trees, respectively. Flowers of the “Conference” pollen acceptor trees were emasculated (except for the Self-treatment) and hand-pollinated at the balloon stage by manually applying the dried pollen to the stigmas using thin, soft bristle paint brushes. New, individually packaged brushes were used for each pollination type to avoid cross-contamination. Following emasculation and pollination, flowers were immediately bagged. Sampling of “Conference” styles was performed at different time points both before and after pollen application; including the white bud stage (Figure 1C, c), balloon stage (Figure 1C, e), and 1, 2, and 3 days after pollination (dpp) which was performed at balloon stage. White bud and balloon stages were sampled before pollination and were included to assess S-RNase expression in the early developmental stages before anthesis and without pollination. For each time point and pollination type a total number of 30 flowers was harvested. Flowers were sampled randomly from the five “Conference” trees with each sampling moment including flowers from at least three different trees. From each flower, the styles were dissected from the pistil and cut at two different heights using sterilized surgical scissors (Figures 1A, B). Styles were cut halfway, and the upper and lower halves of the styles were collected separately in an Eppendorf tube. In order to have sufficient material for mRNA extraction, isolated style parts from six flowers were pooled into one sample. This process was repeated five times for each pollination type and sampling time point to consistently obtain five biological replicates per treatment. Samples were immediately flash-frozen and stored at −80°C to minimize RNA degradation.
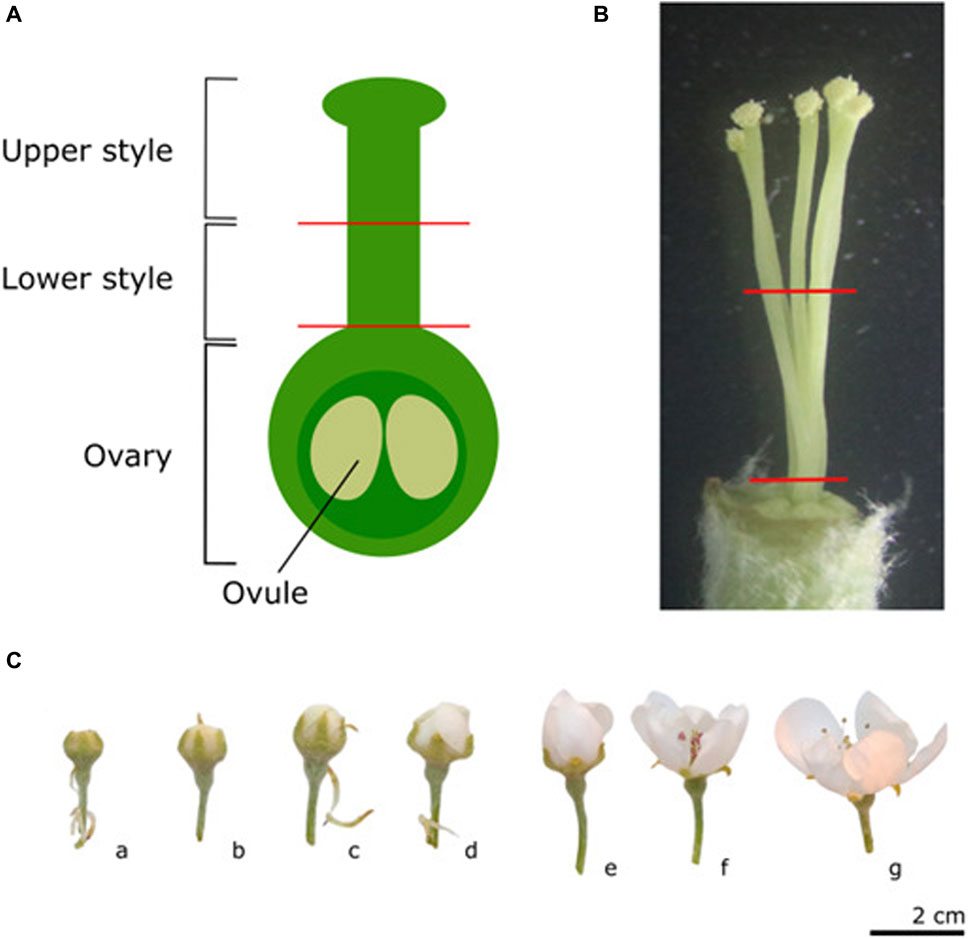
Figure 1. Schematic representation of the different parts of the pistil used for sampling (A), and a reference picture of the five pear styles of cultivar “Conference” to indicate the section sites used for tissue sampling (B). Representative images of the different phenological stages of flower development (C). a) Green bud stage: single flowers are visible but still closed. b-c) White/pink bud stage: petals start to appear and petal color is cultivar-dependent during this stage, the styles may still be very short. d) Start of the balloon stage: flowers are still closed and stamens are still tightly packed around the style. e) Balloon stage: petals are still closed, but can easily be moved aside. f) Anthesis: anthers are pink and closed but the petals have opened. In this stage, flowers can no longer be considered unpollinated since insects carrying pollen from nearby trees may have visited the flower; g) Open flower: anthers have dehisced.
2.3 Validation of the S-genotype by PCR amplicon sequencing
S-genotypes of the “Conference,” “Bartlett” and “Légipont” trees were validated before the start of the experiment to ensure the desired compatibility relations. DNA was extracted from young leaves using the Nucleospin® plant II DNA extraction kit (Macherey-Nagel) following the manufacturer’s guidelines. DNA concentration and quality were checked using a Nanodrop 2000 spectrophotometer (Thermo Scientific). Consensus and allele-specific primers (Supplementary Table S1) as described in Sanzol (2009) were used to amplify the S-RNase allele sequences based on the expected S-genotypes as derived from literature, being PcS108-PcS121 for “Conference” (Goldway et al., 2009), PcS102-PcS108 for “Légipont” (Quinet et al., 2014) and PcS101-PcS102 for “Bartlett” (Goldway et al., 2009). Multiple primers were used to obtain the full coding sequence of each S-allele. PCR-based S-genotyping using consensus primers was performed as described in Sanzol (2009). Allele-specific PCRs were performed in a T100TM Thermal Cycler (Bio-Rad) using a standard PCR program: hot-start of 2 min at 95°C; 35 cycles of 30 s at 95°C, 1 min at a primer-dependent annealing temperature (Supplementary Table S1) and 2 min at 72°C; followed by a final cycle of 10 min at 72°C. The PCR mix for each sample contained 50 ng of DNA, 5 µL of 10X DreamTaq buffer (Thermo Scientific), 0.2 mM of dNTP mixture, 0.5 µM of each primer, and 1.25U DreamTaq DNA polymerase (Thermo Scientific), supplemented with dH2O to reach a final volume of 50 µL. The resulting PCR products were separated by gel electrophoresis using 1% agarose gels containing 10 mg/mL UltraPureTM Ethidium Bromide (Invitrogen) after which they were visualized using the Gel Doc EZ documentation system (Bio-Rad). PCR amplicon bands with the correct fragment length were cut out of the gel, purified using the GeneJet PCR purification kit (Thermo Scientific), and subsequently Sanger sequenced in both directions using corresponding PCR primers (LGC Genomics, Hoddesdon, United Kingdom). Resulting amplicon sequences were trimmed and corresponding forward and reverse fragments of the different overlapping S-allele parts were aligned using Geneious® v11.1.5 software to obtain a complete consensus gDNA sequence of each S-allele. These PCR amplicon sequences were then subjected to a nucleotide BLAST analysis against the NCBI nucleotide database (http://blast.ncbi.nlm.nih.gov) to determine the corresponding S-RNase allele.
2.4 RNA extraction and cDNA synthesis
Total RNA was extracted from the collected style fragments using the Nucleospin® RNA Plant and Fungi kit (Macharey-Nagel) according to the manufacturer’s instructions. Frozen samples were prepared for extraction by adding the lysis buffer and by thoroughly crushing the sample with a decontaminated pestle. Following RNA extraction, putative DNA contamination was removed using the DNA-free™ DNA removal kit (Thermo Scientific) according to the manufacturer’s protocol. The integrity of resulting RNA samples was checked by agarose gel staining (1.5% gel stained with ethidium bromide) and RNA content was quantified using a Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific) by measuring the absorption at 260 nm. For each sample, 455 ng of purified RNA was reverse transcribed into complementary DNA (cDNA) using the SuperScript™ Reverse Transcriptase kit (Thermo Scientific) according to the manufacturer’s protocol after which the resulting cDNA was stored at −20°C.
2.5 Quantitative RT-PCR
To specifically detect and quantify the expression of the PcS108 and PcS121 alleles in “Conference” pistils, new primer pairs were designed for each allele using the NCBI primer blast tool (Supplementary Table S1). The RT-qPCR reaction mixtures contained 4 µL cDNA, 0.4 µM of each primer, and 1X SYBR Green master mix (BioRad), and were supplemented with dH2O to reach a final volume of 20 µL. The RT-qPCR program consisted of 40 cycles of 15 s at 95°C and 15 s at 60°C, followed by melting curve analysis from 65°C to 95°C with 0.5°C increments at 5 s/step. The qPCR reaction was performed in the CFX-96 Touch Real-Time PCR detection system (BioRad). At the end of each qPCR run, a high-resolution melting curve analysis was performed to check product specificity. The specificity of the primer pairs was additionally determined by Sanger sequencing (LGC Genomics) of the purified PCR product (GeneJet PCR Purification kit) for three independent samples of each primer pair. S-RNase allele expression measurements were normalized against the average expression of two reference genes, namely, Histone H3 (XM_009367550.2) and EF1α (AY338250) (Liu et al., 2018). Primer specifications for both reference genes are listed in Supplementary Table S1. A calibration curve was run in triplex in each run consisting of a two-fold serial dilution ranging up to 2–5 of a mixed cDNA sample of all samples in the experiment. The qPCR reactions were performed over different plates in an “all samples” approach where only one gene was tested per plate. To correct for any offset between plates, three inter-run calibrators (IRC) were measured per plate. Raw Cq values for all tested genes were corrected for plate-specific primer efficiency, as calculated from the calibration curves using the method described in Ganger et al. (2017), and normalized across different plates using the IRCs. The dCq value for each sample was calculated by subtracting the mean Cq value of the reference genes from the corresponding raw Cq value of the sample.
2.6 Statistics and data analysis
All statistical tests were performed in R (version R 4.2.0). Before any statistical processing, normality of the subpopulations in the data was determined using the Shapiro-Wilk test. Similarly, heteroscedasticity was evaluated through the Bartlett’s test of homogeneity of variances. Differences in the spatiotemporal expression of the two S-alleles across all time points in the unpollinated flowers were evaluated separately for each S-RNase allele using a two-way ANOVA including the fixed factors “time” and “position” and their interaction effects. The significant interaction effect between time and position for PcS108 was further investigated via a simple main effects analysis using the Phia package (De Rosario-Martínez, 2024). In addition, a three-way ANOVA was performed to analyze the effect of the pollination treatment (i.e., compatibility type), stylar position, and time on the expression of each S-RNase allele. Significant interaction effects were further investigated via a simple main effects analysis using the Phia package (De Rosario-Martínez, 2024). Multiple comparisons within the simple-effect analysis and evaluation of significance of differences were performed using Tukey’s HSD post hoc test with a significance level of 0.05. Results were plotted below as mean expression values and 95% confidence intervals.
3 Results
3.1 Validation of the S-genotype of the experimental pear cultivars
The PCR using consensus primers PyComC1F and PyComC5R yielded two bands close together at 680 bp (PcS108) and 661 bp (PcS121) for “Conference,” two bands at 1,306 bp (PcS101) and 1,723 bp (PcS102) for “Bartlett,” and two bands at the expected sizes of 1,723 bp (PcS102) and 680 bp (PcS108) for “Légipont” (Supplementary Figure S1), strongly matching the expected S-genotypes of the three cultivars (Sanzol, 2009). Sanger sequencing of the purified PCR products using allele-specific primers followed by Blast analysis of the resulting nucleotide sequences confirmed the identities of the four expected S-RNase alleles. An overview of the obtained consensus sequences and their alignment to their reference sequence is included in Supplementary Figures S2–S5.
3.2 Spatio-temporal expression dynamics of both S-RNase alleles in unpollinated pistils
As a first analysis, the basal spatio-temporal expression profile of the two “Conference” S-alleles was evaluated across all time points in unpollinated styles (UP treatment). Under these conditions, both S-RNase alleles show highest expression before anthesis (i.e., B and WB stages) after which their relative transcript level gradually decreases in the subsequent days. In the case of unpollinated styles the abbreviations 1dpp, 2dpp and 3dpp refer to days after balloon stage rather than days after pollination since no pollination was performed in this case. A two-way ANOVA statistical test hereby showed a significant main effect of time for both PcS108 [F (4,33) = (23,237), p = 3,327e-09] and PcS121 [F (4,33) = (62,650), p = 6,003e-15] and a significant interaction between time and position for PcS108 [F (4,33) = (3,875), p = 1,092e-02]. This indicates that the temporal expression pattern of the PcS108 allele depends on the position in the style, and thus differs between the higher and lower part of the style. A significant difference in PcS108 expression between the upper and lower part of the style was measured at 1 dpp (p = 8,115e-03), however, not at the other time points. In contrast, no significant differences in spatial expression were observed for PcS121 (Figure 2).
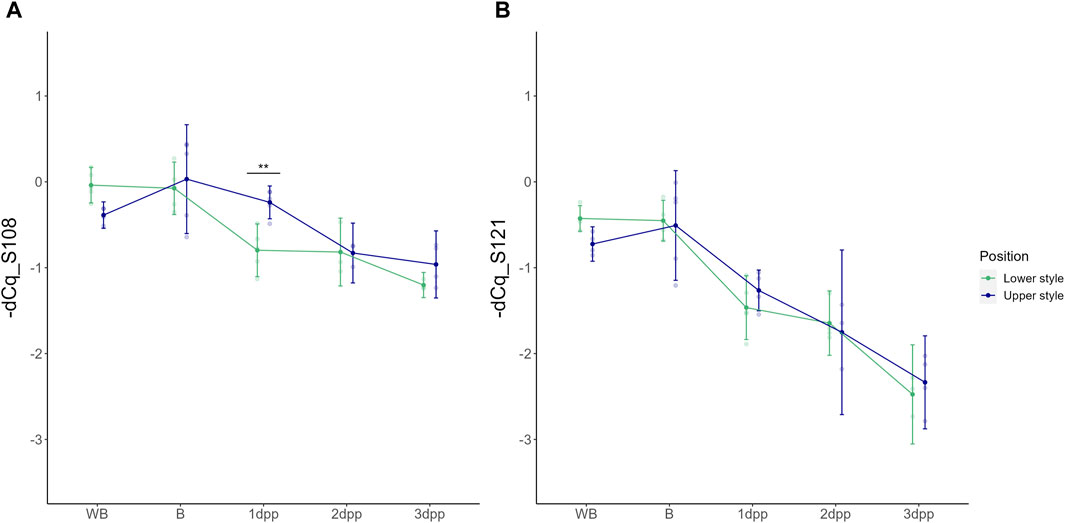
Figure 2. Temporal dynamics of the PcS108 (A) and PcS121 (B) S-RNase expression levels denoted as -dCq values in both the upper and lower half of the style across five time points throughout development of an unpollinated style of the cultivar “Conference.” Data points represent mean values, error bars represent 95% confidence intervals. Asterisks mark the time points at which there is a significant difference between S-RNase allele expression in the upper versus the lower part of the style (p < 0.05) with *: p < 0.05, **: p < 0.01 and ***: p < 0.001.
3.3 S-RNase expression profile in the pistil differs depending on the pollination type
The effect of pollination events with different compatibility types (compatible, incompatible, and semi-compatible) on the temporal expression dynamics of PcS108 and PcS121 was assessed at three successive time points following pollination for both the upper and lower region of the style.
Expression dynamics of both S-RNase alleles were highly similar for each type of pollination treatment, generally showing a gradual reduction in transcript levels upon pollination, though with marked differences in temporal dynamics between the different compatibility types (Figures 3, 4). For PcS108, the three-way ANOVA revealed that all three fixed factors, i.e., pollination treatment, position in the style, and time have a significant effect on the relative expression level, with significant interactions between treatment and time [F (6,70) = (9,911), p = 6,743e-08], and treatment and position in the style [F (3,70) = (4,005), p = 1,086e-02]. Similarly, for PcS121, significant main effects for all three independent factors were retrieved, as well as significant interactions between treatment and time [F (6,70) = (15,180), p = 4,771e-11] and treatment and position [F (3,70) = (4,549), p = 5,706e-03].
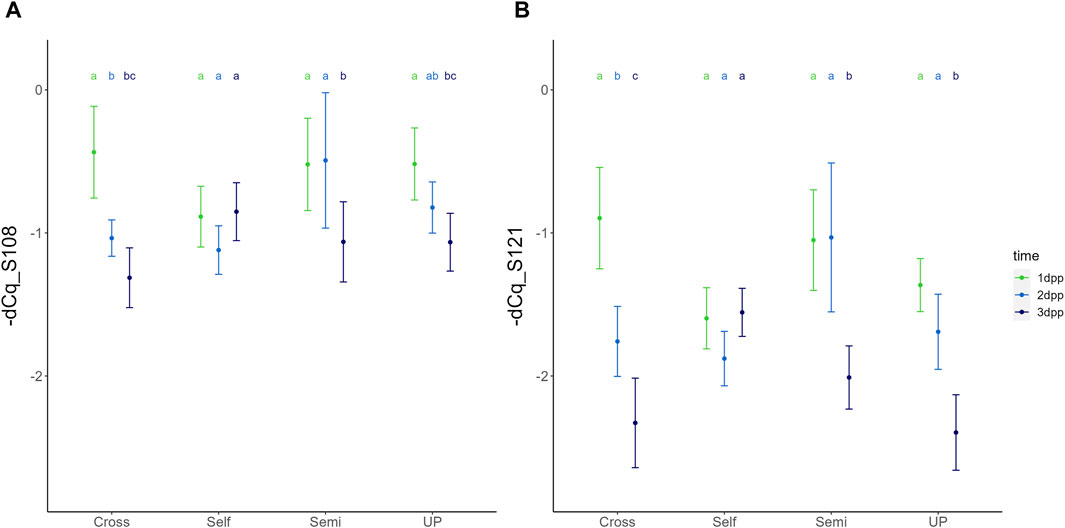
Figure 3. Temporal dynamics of the expression of PcS108 (A) and PcS121 (B) S-RNase alleles in the styles of the cultivar “Conference” upon different types of pollination treatments, including unpollinated (UP), cross-pollination (Cross), selfing (Self) and semi-compatible pollination (Semi). Data points represent mean values, error bars represent 95% confidence intervals. Letters above the data points indicate significant differences between time points for specific pollination treatments as determined by the Tukey post-hoc test with a significance level of 5% (α = 0.05).
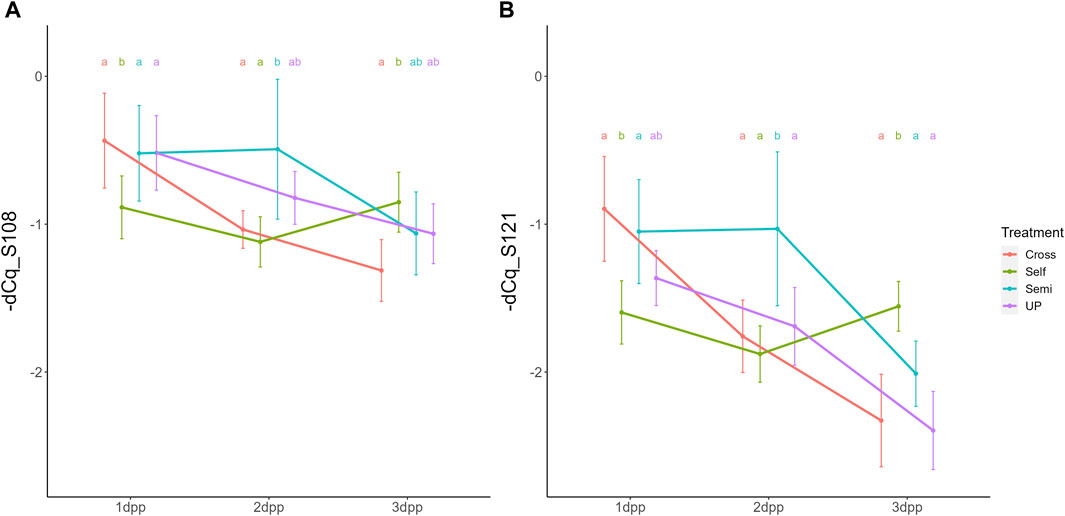
Figure 4. Temporal dynamics of the expression of PcS108 (A) and PcS121 (B) S-RNase alleles in the styles of the cultivar “Conference” upon different types of pollination treatments, including unpollinated (UP), cross-pollination (Cross), selfing (Self) and semi-compatible pollination (Semi). Data points represent mean values, error bars represent 95% confidence intervals. Letters above the data points indicate significant differences between treatments at a specific time point as determined by the Tukey post-hoc test with a significance level of 5% (α = 0.05).
The significant interaction between treatment and time indicates that the temporal expression profile of both S-RNase alleles depends on the pollination treatment, with both S-RNase alleles responding highly similar for each of the compatibility type pollinations. Figure 3 shows the comparison of the PcS108 (A) and PcS121 (B) expression levels between time points for each pollination treatment. Reversely, Figure 4 shows the comparison of the PcS108 (A) and PcS121 (B) expression levels between pollination treatments for each time point. In the complete absence of pollination (UP), expression of both S-RNase alleles progressively and significantly drops representing the baseline expression dynamics of each S-RNase allele (Figure 3). A similar progressive and significant reduction in S-RNase expression is observed for both alleles after cross-pollination (Cross). In contrast, following self-pollination, the expression of both S-RNase alleles shows a constant trend (Figure 3), after an initially lower expression at 1 dpp that is significant compared to the other treatments for both S-RNase alleles (Figure 4). This constant S-RNase expression after self-pollination is reflected by a significantly higher expression of the two S-RNase alleles at 3 days following selfing as compared to cross-pollination (for PcS108) and compared to all other treatments (for the PcS121 allele) (Figure 4). These results suggest that whereas S-RNase expression keeps decreasing after full-compatible cross-pollination, i.e., mimicking the baseline expression occurring in the absence of pollination (UP), S-RNase expression is maintained after self-pollination. Strikingly, following a semi-compatible pollination, the expression profile of both S-RNase alleles exhibits a constant level until 2 days after pollination followed by a delayed drop in expression that only occurs at 3 dpp (Figure 3). This maintained expression is reflected by a significantly higher transcript level of both S-RNase alleles at 2 dpp as compared to the other pollination treatments (Figure 4).
Parallel to the interaction between pollination treatment and time, there was also a significant interaction effect between treatment and position within the style (i.e., lower versus upper part) for both S-RNase alleles. The interaction plot between both variables is provided in Figure 5 and shows that the difference in S-RNase expression between treatments depends on the position in the style and that the effect of the pollination treatment is largest in the upper style region. Significant differences between treatments, taking together all time points, were only observed in the upper region of the style with the semi-compatible pollination being significantly different from the cross- and self-pollination treatment. This difference was not observed in the lower style. This result suggests that the delayed drop in expression after semi-compatible pollination as described above is only apparent in the upper region of the style, but not in the lower region.
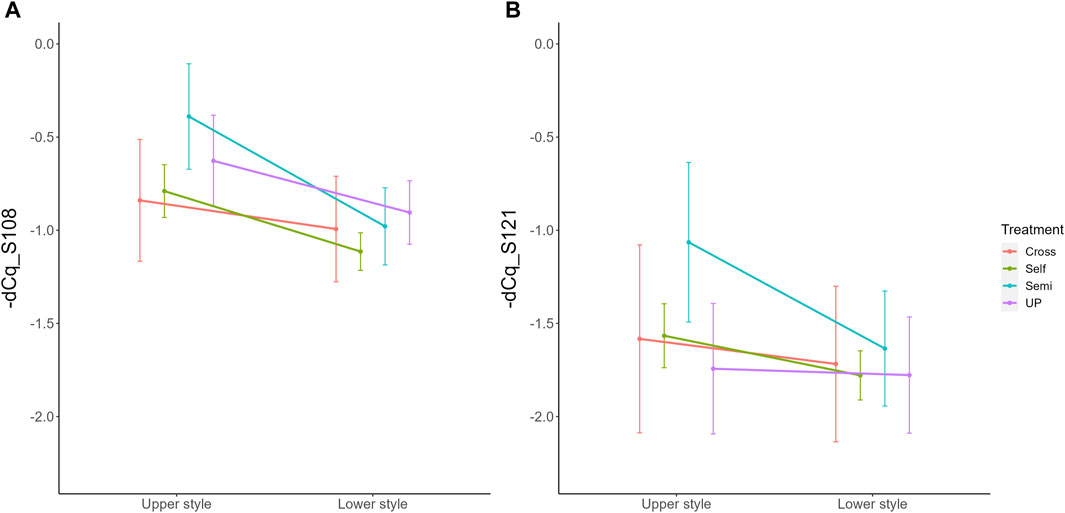
Figure 5. Interaction plots for the significant interaction effect between pollination treatment and position for both PcS108 (A) and PcS121 (B) S-RNase expression levels in styles of the cultivar “Conference”. Data points represent mean values, error bars represent 95% confidence intervals. Letters above the data points indicate significant differences between treatments over all time points per style region as determined by the Tukey post-hoc test with a significance level of 5% (α = 0.05).
4 Discussion
4.1 Towards a mechanistic model for pollination-dependent S-RNase expression in Pyrus
In this study, we examined the effect of pollination compatibility type on the allele-specific S-RNase expression in the upper and lower region of the style in the P. communis cultivar “Conference.” We found that in unpollinated styles S-RNase expression peaks at the balloon stage and gradually decreases after anthesis. A gradual decrease in “basal” S-RNase expression after anthesis was also observed in previous studies in S. chacoense (O’Brien et al., 2002) and P. bretschneideri (Shi et al., 2017), corroborating our findings. We also uncovered that this “baseline” expression profile in unpollinated styles did not significantly differ between the upper and lower region of the style. This is in line with a previous study in apple which found homogenous S-RNase expression along the entire length of the transmitting tract (Certal et al., 1999), but not with measurements in S. chacoense where elevated S-RNase expression was observed in the upper half of the style (O’Brien et al., 2002).
The results of our expression analysis also showed that the temporal S-RNase expression profile of both alleles, namely, PcS108 and PcS121, is highly similar for each individual pollination treatment. The fact that both alleles follow the same expression pattern, even upon semi-compatible pollination, suggests that any signal mediating feedback regulation of the S-RNase expression during the occurrence of a pollen-pistil (in) compatibility reaction does not operate in an allele-specific manner but instead acts on both S-RNase alleles. However, we did notice that PcS108 expression was consistently higher compared to PcS121 for all pollination/no pollination treatments.
The temporal S-RNase expression profiles of the unpollinated and cross-pollinated styles show a decreasing trend, whereas the expression profile after self-pollination shows constant expression. After semi-compatible pollination, the S-RNase expression in the upper region of the style also remained stable, though significantly higher compared to the other pollination types, followed by a delayed drop in expression at 3 dpp. Decreased S-RNase expression after cross-pollination and maintained expression after self-pollination have previously also been described in S. chacoense (O’Brien et al., 2002; Liu et al., 2009), lemon (Zhang et al., 2015; Li et al., 2022), and P. bretschneideri (Shi et al., 2017). Drawing upon these findings together with our own results in P. communis cv. “Conference,” we put forth a tentative hypothesis to explain variations in S-RNase allele expression patterns in pear, as influenced by the compatibility type of the pollination (Figure 6).
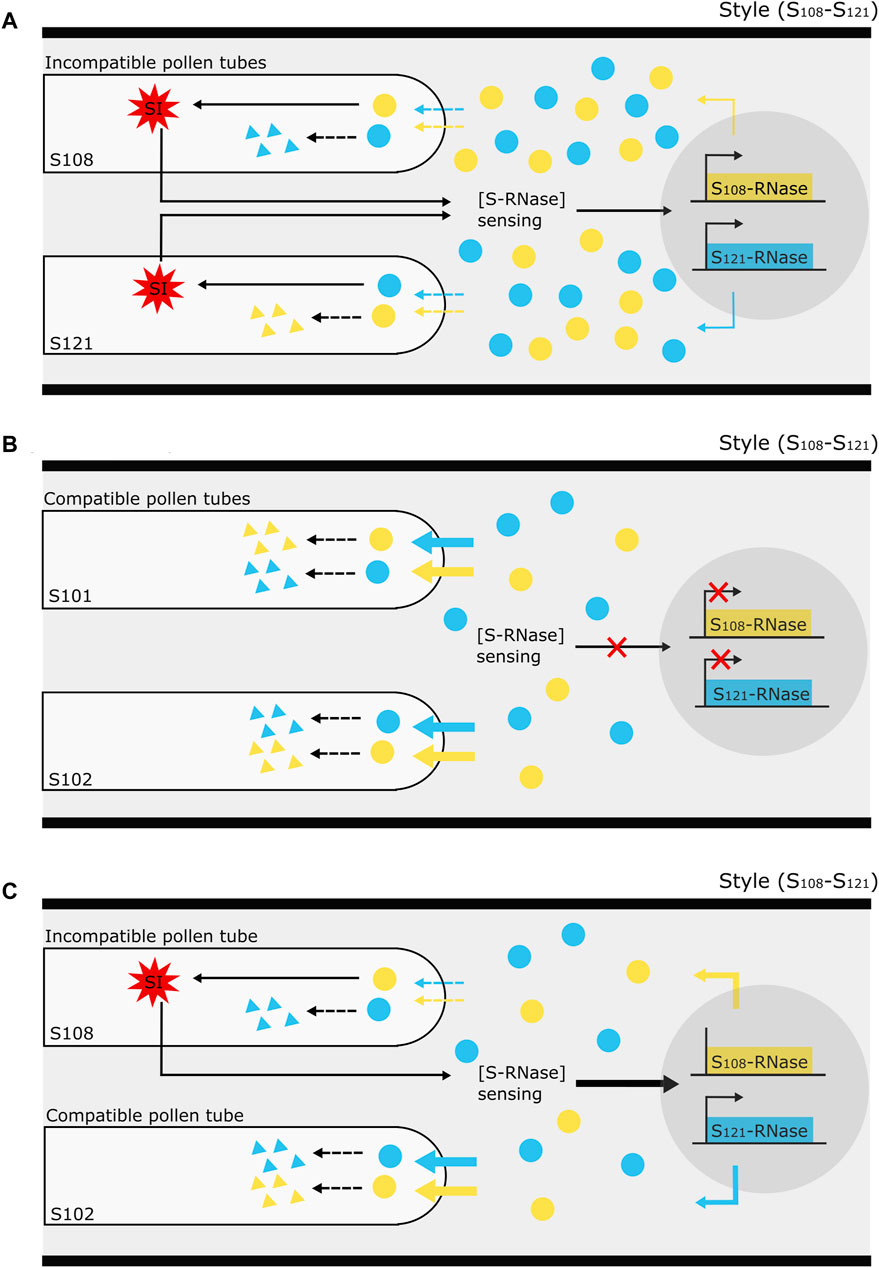
Figure 6. Illustration of the proposed mechanism underlying the effect of pollen tube (in)compatibility on S-RNase gene expression in the style of Pyrus. (A) The occurrence of an incompatibility reaction (red, multi-pointed star) in “self-type” pollen tubes sends a signal to the style to sense the stylar S-RNase protein concentration [S-RNase] and keep it at a minimum level required to maintain the self-incompatibility reaction. This minimum threshold amount is maintained by the de novo synthesis of extra S-RNase through a yet unknown system of transcriptional activation. (B) On the other hand, in the complete absence of an incompatibility reaction, such as upon cross-pollination, there is no activation of this [S-RNase] sensing mechanism to maintain S-RNase protein levels in the style, so that there is no feedback regulation on S-RNase expression and it thus follows the gradually decreasing “baseline” dynamics as occurs in the absence of pollination. Without active S-RNase replenishment from transcriptional activation, the continuous S-RNase degradation inside the compatible pollen tubes causes a general decrease in the stylar S-RNase protein level. (C) In the specific case of a semi-compatible pollination, the occurrence of an incompatibility reaction in the incompatible pollen tubes signals the style to sense the stylar S-RNase protein concentration. In the meantime, the progressive S-RNase degradation by the presence of compatible pollen tubes may strongly decrease S-RNase protein levels in the style therefore promoting S-RNase expression even further. The assumption that the depletion of non-self-S-RNase by incompatible pollen tubes is much lower than by compatible pollen tubes is schematically illustrated by dotted and full, colored arrows signifying the entrance of S-RNase protein into the pollen tubes.
First, the model presumes that the S-RNase expression in the pistil gradually decreases in the absence of any pollination event. The occurrence of an incompatibility reaction from “self-type” pollen tubes in the style serves as a signal to maintain a minimum level of S-RNase protein, i.e., by promoting the production of extra S-RNase via transcriptional activation. As such, this mechanism guarantees sustained S-RNase expression following an incompatible pollination event (Figure 6A). In the case of a fully compatible pollination (“Cross”), no incompatibility reaction occurs in the style and the stylar S-RNase protein concentration [S-RNase] sensing mechanism is not activated. As a consequence, transcriptional promotion of the S-RNase gene is not activated, and expression of both S-RNase alleles decreases (Figure 6B) similar to when no pollination event has taken place. In the case of semi-compatible pollination, the incompatibility reaction in the “self-type” pollen tubes will signal the style to keep a constant S-RNase level. However, because of the parallel presence of compatible pollen tubes in the style, there is a progressive depletion of the stylar S-RNase pool (i.e., by diffusion and degradation into the cross-type pollen tubes), triggering an enhanced activation of S-RNase gene expression (Figure 6C). As such, the model not only explains the maintained S-RNase expression level upon selfing but also accounts for the enhanced expression of both S-RNase alleles upon semi-compatible pollination events.
One major assumption in the proposed mechanistic model is that the depletion of non-self-S-RNases by incompatible pollen tubes is much lower than in the metabolically healthy compatible pollen tubes, and therefore not or only minimally affects the overall [S-RNase] pool in the pistil, hence only having low to moderate activation effects on S-RNase transcription. This would explain why, in the semi-compatible treatment, enhanced S-RNase expression is higher than upon selfing (see Figures 3, 4), and why expression dynamics of PcS121, which is degraded by both compatible and the incompatible pollen tubes, is highly similar to that of the PcS108 allele, which is solely degraded by the compatible pollen tubes.
In the semi-compatible pollination treatment, differences in the temporal S-RNase expression pattern between the upper and lower part of the style were noted. This phenomenon may be explained by the presumption that the presence of incompatible pollen tubes activates the signaling to maintain S-RNase expression in the whole style, while the depletion of the S-RNase protein pool by the compatible pollen tubes enhances this signal in a spatial manner across the length of the style to hence confer a more pronounced increase in S-RNase expression in the upper as compared to the lower region of the style. The observed drop in S-RNase expression at 3 dpp after a semi-compatible pollination can be explained by the loss of the incompatibility reaction and downstream signaling activation due to the death of incompatible pollen tubes. The fact that this drop in S-RNase expression at 3 dpp is not seen after self-pollination may be due to the fact that, in our experiment, many more incompatible pollen tubes are present after a full selfing and therefore the incompatibility reaction and related downstream signaling is much stronger and more pronounced. In addition, in a full incompatibility reaction many more pollen tubes must be inhibited before the promotive effect on S-RNase expression is seized, expectantly leading to a prolonged promotive effect on S-RNase expression compared to a semi-compatible pollination.
4.2 Validation of the putative model in the context of previous research
The mechanism described here is not the first model that has been proposed to explain differential S-RNase expression patterns and protein levels in the pistil in response to pollination compatibility type. Liu et al. (2009) suggested a positive feedback mechanism based on their observations in wild potato stating that S-RNase mRNA levels in the style decreased much faster after cross-pollination compared to self-pollination. They hypothesized that the compatible pollen tubes during their growth through the style send out a signal that reduces S-RNase expression, thereby lowering S-RNase production and reinforcing the compatible reaction. However, in this case, one would expect an increased reduction of S-RNase expression after cross-pollination compared to no pollination, which did not corroborate with our results. Interestingly, an earlier study on the same species found very similar effects of the pollination compatibility type on stylar S-RNase expression as Liu et al. (2009), however, a completely different mechanistic model was put forward to explain this “feedback” regulation (O’Brien et al., 2002). These authors suggested that the presence of dead pollen tubes or molecules liberated from the arrested pollen tubes either directly or indirectly stimulate S-RNase gene transcription, or, suppress their mRNA turnover, to ensure sufficient amounts of S-RNase protein in the style to reject putatively new incoming pollen grains from incompatible genotypes (O’Brien et al., 2002).
Our model shares the presumption that incompatible pollen tubes (or the induction of their death) form a trigger to promote transcriptional activation of both S-RNases present in the style. However, we include the additional notion that the incompatibility reaction triggers the style to maintain a minimum level of S-RNase protein, i.e., with the actual level of S-RNase enzyme in the style additionally regulating the extent of S-RNase gene expression. This presumption is supported by pollination observations in earlier research. Liu et al. (2009) found that in S. chacoense S-RNase protein levels stayed constant after self- and no pollination, but dropped significantly after semi-compatible pollination, and even more upon cross-pollination, which is in line with our proposed model. Another study in S. chacoense reported that the style requires a minimum level of S-RNase enzyme to maintain the ability to reject incompatible pollen (Qin et al., 2006), which was in agreement with the threshold hypothesis formulated earlier in Petunia (Clark et al., 1990). As part of this threshold model, it has also been suggested that style-to-style and genotype-to-genotype variations in stylar S-RNase levels could explain pseudo-compatibility due to the observed weakening of the SI response in case of reduced levels of S-RNase in the style (Qin et al., 2001; 2006). In S. chacoense this critical threshold was set at 80 ng/style or 0.06 mg mL−1 stylar extract (Qin et al., 2006). Interestingly, in Japanese pear, a minimum S-RNase protein level to ensure complete self-incompatibility was estimated at the same order of magnitude, namely, 0.1 mg mL−1 stylar extract (Hiratsuka and Zhang, 2002; Qin et al., 2006). This supports the assumption in our proposed model that there is a minimum S-RNase protein level in the style that is necessary to maintain the self-rejection capacity of the style and which is maintained in the presence of incompatible pollen tubes.
Our proposed model basically involves a minimum threshold-based feedback regulation of S-RNase gene expression by the stylar [S-RNase] pool upon the occurrence of self-type pollination and therefore presumes the existence of a signaling mechanism that promotes S-RNase expression under conditions of self-incompatibility. An important question that remains is what this signal is or how this signaling would be achieved. One possibility is that the style registers the presence of (in) compatible pollen tubes via physical or biochemical changes in the transmitting tract. For example, compatible pollen tubes may be recognized through the uptake of stylar nutrients, while the presence of ROS or other metabolites that are the result of programmed cell death (PCD) might signal the presence of incompatible pollen tubes. In Pyrus pyrifolia, S-RNase action has been associated with depolymerization of the actin cytoskeleton, mitochondrial alteration and DNA degradation in incompatible pollen tubes (“self-type”), suggesting induction of programmed cell death (PCD) (Wang et al., 2009). A transcriptome analysis in lemon comparing stylar gene expression after selfing and cross-compatible pollination revealed that genes involved in calcium-mediated signaling, and NADPH oxidase, which plays a role in ROS signaling, were upregulated following self-pollination (Zhang et al., 2015). This study also showed evidence for the involvement of phytohormone signaling transduction pathways in the SI reaction and regulation of S-RNase expression. In line with this, comparative transcriptomics between self- and cross-pollinated styles in different plant species with S-RNase-dependent GSI, including Pyrus bretschneirderi, unveil differential expression of genes involved in phytohormone synthesis and signaling, including ethylene, auxin, jasmonate, cytokinin, and gibberellins (Zhao et aal, 2015; Shi et al., 2017). Among these plant hormones, jasmonate (JA) emerges as a promising candidate for orchestrating self-incompatibility regulation. Various observations have hinted at its participation in the self-incompatibility processes as elaborated below. However, while the actual contribution of this phytohormone remains formally unproven, it’s important to note that other metabolites could also potentially play a role. For example, in P. bretschneideri the concentration of JA in the style decreases significantly upon cross-pollination, but not upon self-pollination. Moreover, a key enzyme for JA biosynthesis, allene oxide cyclase (AOC), and the JA-associated MYC transcription factor, involved in the activation of JA-mediated systemic response to wounding, were found to be significantly downregulated in cross-pollinated but not in self-pollinated styles. Further supporting the functional involvement of jasmonic acid (JA) in Pyrus gametophytic self-incompatibility (GSI), it was observed that the external application of JA-Me significantly boosted the expression of S-RNase in the style. This observation implies a potential connection between diminished S-RNase expression in compatible styles and JA signaling (Shi et al., 2017). A similar transcriptome experiment on tomato styles also revealed significant upregulation of JAZ and MYC2, as well as significantly enriched plant hormone signal transduction-related KEGG pathway, after an incompatible pollen interaction, but not after cross-pollination (Zhao et al., 2015). On the other hand, a study in apple uncovered that JA is also involved in the regulation of self-incompatibility modifiers in the pollen tubes. Gu et al. (2019) found that the uptake of S-RNase promotes the accumulation of JA in pollen tubes, which stimulates the expression of MdMYC2 and its target, a defensin gene, MdD1. This defensin gene inhibits the activity of both self- and non-self S-RNases by targeting the active site of the ribonuclease during the initial phases of pollen tube growth prior to the occurrence of self/non-self-recognition. The presumption that, in the style, S-RNase expression may be regulated by JA signaling while, in the pollen tube, S-RNase accumulation induces JA signaling seems contradictory. However, it may hint at the presence of multiple complex signaling networks that are activated independently in the style and pollen tube upon pollination and/or self-recognition. In order to get more insight into the feedback mechanism(s) that control S-RNase gene expression in Pyrus and thus regulate GSI, further research on the signaling events that occur after pollination and in response to self-incompatibility might benefit from using novel techniques that allow analysis of pollen tube and style events separately, like the analysis of male/female specific expression in the stigma upon pollination performed in Arabidopsis thaliana by Kodera et al. (2021). These insights will allow the validation of the proposed mechanistic model and will help to further unravel the intricate signaling network occurring at the style-pollen tube interface to determine the outcome of a pollination event in function of the compatibility of the parental genotypes.
5 Conclusion
Several previous studies have reported a differential effect of compatible and incompatible pollination on S-RNase expression in plant species that carry the S-RNase-dependent GSI system. In this study, allele-specific S-RNase expression analysis using RT-qPCR was performed in the pear cv. “Conference” in response to different types of pollination compatibility. Based on these results and previously reported findings, we here propose a model to explain dynamics of stylar S-RNase expression in response to pollination and compatibility type of growing pollen tubes. The model proposes that upon occurrence of an incompatibility reaction the style receives a signal to maintain a minimum critical threshold level of S-RNase protein by actively promoting its expression. In the absence of an incompatibility reaction, this signaling pathway is not activated and S-RNase expression drops gradually such as under non-pollinated conditions. More work is needed to unravel the actual mechanistic players of the proposed model.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
HC: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Writing–original draft. HP: Data curation, Formal Analysis, Investigation, Methodology, Writing–review and editing. WK: Conceptualization, Formal Analysis, Funding acquisition, Supervision, Writing–review and editing. BV: Conceptualization, Formal Analysis, Funding acquisition, Supervision, Writing–review and editing. ND: Conceptualization, Formal Analysis, Funding acquisition, Project administration, Supervision, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was financially supported by the Research Foundation of Flanders (FWO) and KU Leuven. This work was supported by the Research Fund Flanders (FWO) via a Doctoral (PhD) Strategic Basic fellowship provided to CH (Grant number: 1S02817N) and via a junior postdoc grant provided to HC (Grant number: 1230623N). In addition, support was obtained via FWO research grants to VB (FWO project G092419N and G0G0219N). Financial support was also provided by KU Leuven to DN (C14/19/073 project and STG/18/043 grant) and VB (project C14/18/056).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2024.1360332/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | Agarose gel showing the results of the S-genotyping PCR using consensus primers PyComC1F and PyComC5R (Sanzol, 2009). Electrophoresis was conducted on a 1.5% agarose gel stained with ethidium bromide. The GeneRuler Express DNA ladder was used to estimate the size of the genomic DNA fragments. Letters above the lanes denote the ID numbers of the different Pyrus cultivars used in this study: (A) “Légipont,” (B) “Conference,” (C) “Bartlett.” PCR bands of interest are marked by a red box.
SUPPLEMENTARY FIGURE S2 | Nucleotide alignment of PcS101 of cultivar “Bartlett” compared to the reference PcS101 sequence (accession no. AB236428.1).
SUPPLEMENTARY FIGURE S3 | Nucleotide alignment of PcS102 of cultivar “Bartlett” and “Légipont” compared to the reference PcS102 sequence (accession no. AB236425).
SUPPLEMENTARY FIGURE S4 | Nucleotide alignment of PcS108 of cultivar “Conference” and “Légipont” compared to the reference PcS108 sequence (accession no. AB236427).
SUPPLEMENTARY FIGURE S5 | Nucleotide alignment of PcS121 of cultivar “Conference” compared to the reference PcS121 sequence (accession no. AB236426).
References
Certal, A., Sanchez, A. M., Kokko, H., Broothaerts, W., Oliveira, M. M., and Feijó, J. A. (1999). S-RNases in apple are expressed in the pistil along the pollen tube growth path. Sex. Plant Reprod. 12, 94–98. doi:10.1007/s004970050177
Clark, K. R., Okuley, J. J., Collins, P. D., and Sims, T. L. (1990). Sequence variability and developmental expression of S-alleles in self-incompatible and pseudo-self-compatible Petunia. Plant Cell 2 (8), 815–826. doi:10.1105/tpc.2.8.815
De Franceschi, P., Dondini, L., and Sanzol, J. (2012). Molecular bases and evolutionary dynamics of self-incompatibility in the Pyrinae (Rosaceae). J. Exp. Bot. 63 (11), 4015–4032. doi:10.1093/jxb/ers108
De Franceschi, P., Pierantoni, L., Dondini, L., Grandi, M., Sanzol, J., and Sansavini, S. (2011). Cloning and mapping multiple S-locus F-box genes in European pear (Pyrus communis L.). Tree Genet. Genomes 7, 231–240. doi:10.1007/s11295-010-0327-5
De Rosario-Martínez, H. (2024). _phia: post-Hoc Interaction Analysis_, R package version 0.3-1. Available at: https://cran.r-project.org/package=phia.
Dresselhaus, T., and Franklin-tong, N. (2013). Male–female crosstalk during pollen germination, tube growth and guidance, and double fertilization. Mol. Plant 6 (4), 1018–1036. doi:10.1093/mp/sst061
Ganger, M. T., Dietz, G. D., and Ewing, S. J. (2017). A common base method for analysis of qPCR data and the application of simple blocking in qPCR experiments. BMC Bioinforma. 18 (534), 534–611. doi:10.1186/s12859-017-1949-5
Goldraij, A., Kondo, K., Lee, C. B., Hancock, C. N., Sivaguru, M., Vazquez-Santana, S., et al. (2006). Compartmentalization of S-RNase and HT-B degradation in self-incompatible Nicotiana. Nature 439 (2), 805–810. doi:10.1038/nature04491
Goldway, M., Takasaki-Yasuda, T., Sanzol, J., Mota, M., Zisovich, A., Stern, R. A., et al. (2009). Renumbering the S-RNase alleles of European pears (Pyrus communis L.) and cloning the S109 RNase allele. Sci. Hortic. 119 (4), 417–422. doi:10.1016/J.SCIENTA.2008.08.027
Gu, Z., Li, W., Doughty, J., Meng, D., Yang, Q., Yuan, H., et al. (2019). A gamma-thionin protein from apple, MdD1, is required for defence against S-RNase-induced inhibition of pollen tube prior to self/non-self recognition. Plant Biotechnol J. 17 (11), 2184–2198. doi:10.1111/pbi.13131
Hiratsuka, S., and Zhang, S.-L. (2002). Relationships between fruit set, pollen-tube growth, and S-RNase concentration in the self-incompatible Japanese pear. Sci. Hortic. 95, 309–318. doi:10.1016/S0304-4238(02)00043-2
Honsho, C. (2023). Self-incompatibility related to seedless fruit production in citrus plants. Hortic. J. 92 (1), 1–12. doi:10.2503/hortj.QH-R001
Hua, Z.-H., Fields, A., and Kao, T. (2008). Biochemical models for S-RNase-Based self-incompatibility. Mol. Plant 1 (4), 575–585. doi:10.1093/MP/SSN032
Huang, S., Lee, H. S., Karunanandaa, B., and Kao, T. H. (1994). Ribonuclease activity of Petunia inflata S proteins is essential for rejection of self-pollen. Plant Cell 6 (7), 1021–1028. doi:10.1105/TPC.6.7.1021
Hui, Y., Meng, D., Gu, Z., Li, W., Wang, A., Yang, Q., et al. (2014). A novel gene, MdSSK1, as a component of the SCF complex rather than MdSBP1 can mediate the ubiquitination of S-RNase in apple. J. Exp. Bot. 65 (12), 3121–3131. doi:10.1093/jxb/eru164
Joly, V., Tebbji, F., Nantel, A., and Matton, D. P. (2019). Pollination type recognition from a distance by the ovary is revealed through a global transcriptomic analysis. Plants 8 (185), 1–17. doi:10.3390/plants8060185
Kodera, C., Just, J., Da Rocha, M., Larrieu, A., Riglet, L., Legrand, J., et al. (2021). The molecular signatures of compatible and incompatible pollination in Arabidopsis. BMC Genomics 22 (268), 268–318. doi:10.1186/s12864-021-07503-7
Kubo, K., Entani, T., Takara, A., Wang, N., Fields, A. M., Hua, Z., et al. (2010). Collaborative non-self recognition system in S-RNase-based self-incompatibility. Science 330 (11), 796–799. doi:10.1126/science.1195243
Li, W., Meng, D., Gu, Z., Yang, Q., Yuan, H., Li, Y., et al. (2018). Apple S-RNase triggers inhibition of tRNA aminoacylation by interacting with a soluble inorganic pyrophosphatase in growing self-pollen tubes in vitro. New Phytol. 218 (2), 579–593. doi:10.1111/nph.15028
Li, Y.-Z., Zhu, J. W., Lin, W., Lan, M. Y., Luo, C., Xia, L. M., et al. (2022). Genome-wide analysis of the RNase T2 family and identification of interacting proteins of four ClS-RNase genes in “XiangShui” lemon. Int. J. Mol. Sci. 23 (10431), 10431–10519. doi:10.3390/ijms231810431
Liu, B., Morse, D., and Cappadocia, M. (2009). Compatible pollinations in Solanum chacoense decrease both S-RNase and S-RNase mRNA. PLoS ONE 4 (6), e5774. doi:10.1371/journal.pone.0005774
Liu, Z., Cheng, K., Qin, Z., Wu, T., Li, X., Tu, J., et al. (2018). Selection and validation of suitable reference genes for qRT-PCR analysis in pear leaf tissues under distinct training systems. PLoS ONE 13 (8), e0202472. doi:10.1371/journal.pone.0202472
Liu, Z., Xu, G., and Zhang, S. (2007). Pyrus pyrifolia stylar S-RNase induces alterations in the actin cytoskeleton in self-pollen and tubes in vitro. Protoplasma 232, 61–67. doi:10.1007/s00709-007-0269-4
Lora, J., Hormaza, J. I., and Herrero, M. (2016). The diversity of the pollen tube pathway in plants: toward an increasing control by the sporophyte. Front. Plant Sci. 7 (107), 107–112. doi:10.3389/fpls.2016.00107
Luu, D.-T., Qin, X., Morse, D., and Cappadocia, M. (2000). S-RNase uptake by compatible pollen tubes in gametophytic self-incompatibility. Nature 407 (10), 649–651. doi:10.1038/35036623
Mandrone, M., Antognoni, F., Aloisi, I., Potente, G., Poli, F., Cai, G., et al. (2019). Compatible and incompatible pollen-styles interaction in Pyrus communis L. show different transglutaminase features, polyamine pattern and metabolomics profiles. Front. Plant Sci. 10 (741), 741–813. doi:10.3389/fpls.2019.00741
McClure, B., Cruz-García, F., and Romero, C. (2011). Compatibility and incompatibility in S-RNase-based systems. Ann. Bot. 108 (4), 647–658. doi:10.1093/aob/mcr179
O’Brien, M., Kapfer, C., Major, G., Laurin, M., Bertrand, C., Kondo, K., et al. (2002). Molecular analysis of the stylar-expressed Solanum chacoense small asparagine-rich protein family related to the HT modifier of gametophytic self-incompatibility in Nicotiana. Plant J. 32, 985–996. doi:10.1046/j.1365-313X.2002.01486.x
Okada, K., Tonaka, N., Moriya, Y., Norioka, N., Sawamura, Y., Matsumoto, T., et al. (2008). Deletion of a 236 kb region around S4-RNase in a stylar-part mutant S4 sm-haplotype of Japanese pear. Plant Mol. Biol. 66, 389–400. doi:10.1007/s11103-007-9277-1
Qi, Y.-J., Wu, H. Q., Cao, Y. F., Wu, J., Tao, S. T., and Zhang, S. L. (2011). Heteroallelic diploid pollen led to self-compatibility in tetraploid cultivar “Sha 01” (Pyrus sinkiangensis Yü). Tree Genet. Genomes 7, 685–695. doi:10.1007/s11295-011-0366-6
Qin, X., Liu, B., Soulard, J., Morse, D., and Cappadocia, M. (2006). Style-by-style analysis of two sporadic self-compatible Solanum chacoense lines supports a primary role for S-RNases in determining pollen rejection thresholds. J. Exp. Bot. 57 (9), 2001–2013. doi:10.1093/jxb/erj147
Qin, X., Luu, D. T., Yang, Q., Maës, O., Matton, D. P., Morse, D., et al. (2001). Genotype-dependent differences in S12-RNase expression lead to sporadic self-compatibility. Plant Mol. Biol. 45, 295–305. doi:10.1023/A:1006445120648
Qu, H., Guan, Y., Wang, Y., and Zhang, S. (2017). PLC-mediated signaling pathway in pollen tubes regulates the gametophytic self-incompatibility of Pyrus species. Front. Plant Sci. 8 (1164), 1164–1217. doi:10.3389/fpls.2017.01164
Quinet, M., Kelecom, S., Raspé, O., and Jacquemart, A. L. (2014). S-genotype characterization of 13 North Western European pear (Pyrus communis) cultivars. Sci. Hortic. 165, 1–4. doi:10.1016/j.scienta.2013.10.023
Sanzol, J. (2009). Genomic characterization of self-incompatibility ribonucleases (S-RNases) in European pear cultivars and development of PCR detection for 20 alleles. Tree Genet. Genomes 5, 393–405. doi:10.1007/s11295-008-0194-5
Sassa, H., Hirano, H., and Ikehashi, H. (1992). Self-incompatibility-related RNases in styles of Japanese pear (Pyrus serotina Rehd.). Plant Cell Physiology 33 (6), 811–814. doi:10.1093/oxfordjournals.pcp.a078322
Sassa, H., Hirano, H., and Ikehashi, H. (1993). Identification and characterization of stylar glycoproteins associated with self-incompatibility genes of Japanese pear, Pyrus serotina Rehd. Mol. general Genet. 241 (1–2), 17–25. doi:10.1007/BF00280196
Sassa, H., Kakui, H., Miyamoto, M., Suzuki, Y., Hanada, T., Ushijima, K., et al. (2007). S locus F-box brothers: multiple and pollen-specific F-box genes with S haplotype-specific polymorphisms in apple and Japanese pear. Genetics 175 (4), 1869–1881. doi:10.1534/genetics.106.068858
Shi, D., Tang, C., Wang, R., Gu, C., Wu, X., Hu, S., et al. (2017). Transcriptome and phytohormone analysis reveals a comprehensive phytohormone and pathogen defence response in pear self-/cross-pollination. Plant Cell Rep. 36, 1785–1799. doi:10.1007/s00299-017-2194-0
Wang, C.-L., Xu, G. H., Jiang, X. T., Chen, G., Wu, J., Wu, H. Q., et al. (2009). S-RNase triggers mitochondrial alteration and DNA degradation in the incompatible pollen tube of Pyrus pyrifolia in vitro. Plant J. 57, 220–229. doi:10.1111/j.1365-313X.2008.03681.x
Zhang, S., Ding, F., He, X., Luo, C., Huang, G., and Hu, Y. (2015). Characterization of the “Xiangshui” lemon transcriptome by de novo assembly to discover genes associated with self-incompatibility. Mol. Genet. Genomics 290, 365–375. doi:10.1007/s00438-014-0920-7
Keywords: pear (Pyrus communis), gametophytic self-incompatibility, S-RNase, pollen-style interactions, allele-specific expression
Citation: Claessen H, Palmers H, Keulemans W, Van de Poel B and De Storme N (2024) The influence of the pollination compatibility type on the pistil S-RNase expression in European pear (Pyrus communis). Front. Genet. 15:1360332. doi: 10.3389/fgene.2024.1360332
Received: 22 December 2023; Accepted: 26 March 2024;
Published: 09 April 2024.
Edited by:
Yun Zhang, Shenyang Agricultural University, ChinaReviewed by:
Mohini Prabha Singh, Punjab Agricultural University, IndiaBojana Banović Đeri, University of Belgrade, Serbia
Copyright © 2024 Claessen, Palmers, Keulemans, Van de Poel and De Storme. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nico De Storme, bmljby5kZXN0b3JtZUBrdWxldXZlbi5iZQ==
 Hanne Claessen
Hanne Claessen Han Palmers
Han Palmers Wannes Keulemans1,2
Wannes Keulemans1,2 Bram Van de Poel
Bram Van de Poel