- 1Hebei Key Laboratory of Crop Stress Biology, College of Agronomy and Biotechnology, Hebei Normal University of Science and Technology, Qinhuangdao, China
- 2Research Center of Rural Vitalization, Hebei Normal University of Science and Technology, Qinhuangdao, China
E3 ubiquitin ligases are central modifiers of plant signaling pathways that regulate protein function, localization, degradation, and other biological processes by linking ubiquitin to target proteins. E3 ubiquitin ligases include proteins with the U-box domain. However, there has been no report about the foxtail millet (Setaria italica L. Beauv) U-box gene family (SiPUB) to date. To explore the function of SiPUBs, this study performed genome-wide identification of SiPUBs and expression analysis of them in response to saline-alkali stress. A total of 70 SiPUBs were identified, which were unevenly distributed on eight chromosomes. Phylogenetic and conserved motif analysis demonstrated that SiPUBs could be clustered into six subfamilies (I–VI), and most SiPUBs were closely related to the homologues in rice. Twenty-eight types of cis-acting elements were identified in SiPUBs, most of which contained many light-responsive elements and plant hormone-responsive elements. Foxtail millet had 19, 78, 85, 18, and 89 collinear U-box gene pairs with Arabidopsis, rice, sorghum, tomato, and maize, respectively. Tissue specific expression analysis revealed great variations in SiPUB expression among different tissues, and most SiPUBs were relatively highly expressed in roots, indicating that SiPUBs may play important roles in root development or other growth and development processes of foxtail millet. Furthermore, the responses of 15 SiPUBs to saline-alkali stress were detected by qRT-PCR. The results showed that saline-alkali stress led to significantly differential expression of these 15 SiPUBs, and SiPUB20/48/70 may play important roles in the response mechanism against saline-alkali stress. Overall, this study provides important information for further exploration of the biological function of U-box genes.
1 Introduction
Ubiquitin/26S proteasome pathway is one of the most important protein degradation pathways in cells (Wang et al., 2018). This pathway can regulate all aspects of plant growth and development and the degradation of short-lived regulatory proteins (Santner and Estelle, 2010). Ubiquitin proteasome degradation pathway consists of ubiquitin (Ub), ubiquitin activing enzyme (E1), ubiquitin conjugating enzyme, E2), ubiquitin protein ligating enzyme (E3), 26S proteasome (proteasome), deubiquitinating enzymes (DUBs), and protein substrates (March and Farrona, 2018). Ubiquitin is a very important molecule in this pathway. As a marker protein, ubiquitin participates in the processes of recognition, labeling, and targeted degradation of proteins. Ubiquitin ligase E3 is required for ubiquitin activation and transfer and plays a key role in protein ubiquitination (Trujillo, 2018). Ubiquitin ligase E3 finally binds ubiquitin to the target protein to form ubiquitinated target protein. These ubiquitination targets are then snipped into small peptide chains or free amino acids by the proteasome. Among the enzymes in the ubiquitin degradation pathway, ubiquitin ligase E3 has the highest variety and quantity, including Really Interesting New Gene (RING), U-box domain protein, and homology to the E6AP C-Terminus (HECT) (Mandal et al., 2018).
U-box is a protein domain playing an important role in the ubiquitin/26S proteasome pathway. It was first discovered in yeast Ub Fusion Degradation 2 (UFD2) (Van et al., 2022). The U-box domain is a unique domain composed of about 70 amino acids and transfers ubiquitin from E2s to target proteins through salt bridges, ion chelation, and hydrogen bonding (Qi et al., 2017). It can recognize and bind target proteins and attach ubiquitin to these target proteins, thereby targeting them into ubiquitinated proteins that participate in the cell degradation process. Previous studies have found a large number of U-box genes in many species, and many studies have reported that U-box genes are responsive to abiotic stresses such as light, drought, and salt in a variety of plants, indicating that U-box gene family plays an important role in different stages of plant growth (Bergler and Hoth, 2011; Woodson et al., 2015; Adler et al., 2017; Wang et al., 2020b). Therefore, identifying the unique roles of different U-box proteins in different stress processes will help us understand the development of plant resistance (Li et al., 2017; Kim et al., 2021; Tang et al., 2022).
Foxtail millet (Setaria italica L. Beauv), which belongs to the Poaceae family, is one of the oldest cultivated crops in China. It was domesticated from wild foxtail grass and has a planting history of approximately 10,000 years, playing a crucial role in the agricultural civilization history of arid regions in northern China (Diao and Jia, 2017). As the main cultivated crop in dry green agriculture, it has the characteristics of small genome, short life cycle, self-pollination, drought resistance, and can grow under low fertility conditions, making it a model plant for C4 cereal crop research (Muthamilarasan and Prasad, 2014; Yang et al., 2020). With the completion of foxtail millet genome sequencing in 2012 and release of related sequence information, there has been increasing genome-based research on foxtail millet breeding (Bennetzen et al., 2012). Environmental stress has adverse effects on plant growth and development (Cramer et al., 2011). Excessive salinity in the soil has a great negative impact on plant growth and productivity, leading to large reduction in grain yield (Shrivastava and Kumar, 2015). Current studies have shown that U-box gene plays an important role in resistance of rice, sorghum and other plants (Hu et al., 2018; Yang et al., 2021; Fang et al., 2022). For example, plants overexpressing OsPUB15 in rice have higher salt tolerance than the wild type (Park et al., 2011). In Arabidopsis, PUB13 inactivation leads to elevated concentrations of the defense hormone salicylic acid, spontaneous cell death, and early flowering (Trujillo, 2018). In wheat, TaPUB1 induces the expression of target genes, thereby enhancing antioxidant capacity under stress conditions (Wang et al., 2020b). Overexpression of the GmPUB8 gene during seed germination and post-germination growth stages leads to hypersensitivity to salt and drought stress in soybean (Wang et al., 2016).The U-box gene family may play an important role in foxtail millet stress response. So far, there has been no report about the identification and expression analysis of U-box gene family (SiPUB) in foxtail millet. In this study, SiPUBs were identified and their expression was analyzed under saline-alkali stress, and the results may provide an important theoretical basis for the functional analysis of U-box genes.
2 Materials and methods
2.1 Plant materials and treatments
The salt-alkali tolerant foxtail millet variety, Jikegu3 (JK3), and the salt-alkali sensitive foxtail millet variety, Bao175 (B175), were selected as experimental materials based on previous studies. Seeds of uniform size were selected and sterilized with NaClO for 5 min, washed with distilled water 5 times, soaked in distilled water at room temperature for 24 h, sown, placed in artificial climate tank (Day/night duration: 12 h/12 h; Day/night temperature: 28°C/22°C; Humidity: 65%) and cultured until Sanye One stage, and 75% seawater (Bohai Sea water, water taken from the coast of Qinhuangdao Sea Port Area, China) were treated with salt and alkali stress. Both materials were sampled at 0, 6, 12, 24, and 48 h after treatment, and a mixture of leaves (second and third leaves) from the plants was collected. Each treatment was replicated three times. The samples were placed in sterile 5-mL centrifuge tubes, labeled, immediately frozen in liquid nitrogen, and stored at −80°C until RNA extraction.
2.2 Identification of SiPUB members
The genome and protein sequences of foxtail millet were obtained from the Ensemble database (https://asia.ensembl.org/index.html). The gene sequences and protein sequences of Arabidopsis U-box gene family were downloaded from the Arabidopsis database (http://www.arabidopsis.org/browse/genefamily/pub.jsp). Pfam (http://pfam.xfam.org/), BLAST, SMART (http://smart.embl.de/smart/batch.pl), and NCBI-CDD (https://www.ncbi.nlm.nih.gov/cdd) online databases were used to confirm the presence of complete conserved domains.
First, all U-box domain sequences (PF04564) from different species were downloaded from the Pfam database (http://pfam.xfam.org/). A hidden Markov model (HMM) was constructed using Hmmer2.3.2 (http://hmmer.janelia.org/) and used to search for U-box gene members in the foxtail millet protein database on Ensemble, with a condition of an E-value lower than 1 × 10−5. Then, 64 Arabidopsis U-box gene sequences and protein sequences were downloaded from the TAIR database (https://www.arabidopsis.org/), and a BLAST comparison was performed using TBtools software. The intersection of the BLAST alignment results and HMM results yielded the gene and protein sequences of the U-box gene family. Based on the above results, NCBI-CDD database (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi) and SMART website (http://smart.embl-heidelberg.de/) were used for domain prediction. The sequences that did not contain the U-box domain were removed, resulting in the identification of all U-box members. Physicochemical properties such as molecular weight and isoelectric point were analyzed by TBtools, and subcellular localization was analyzed by WOLF (https://wolfpsort.hgc.jp/).
The resulting U-box family members were named according to their Chromosomal position, such as SiPUB1: “Si” represents S. italica, “PUB” is the abbreviation for the gene family, and “1”denotes the sequence number based on their position on the Chromosomal.
2.3 Gene structure and chromosomal localization of SiPUBs
The DNA sequences of identified SiPUBs were downloaded from the ensemble database. GSDS online software (http://gsds.gao-lab.org/index.PHP) and TBtools analysis software (https://github.com/CJChen/TBtools) were used to respectively detect the structural and conserved motifs of SiPUB exons and introns. At the same time, gene location information was obtained, and the chromosomal mapping of genes was presented using MapChart software.
2.4 Systematic evolution and protein domain analysis of SiPUBs
MEGA_11.0.13 was applied to perform multiple sequence alignment of protein sequences in SiPUBs, and the phylogenetic tree was constructed by NJ (Neighbor-Joining) method. The protein domains of SiPUBs were analyzed using the online tool SMART.
2.5 Promoter analysis and gene ontology annotation of SiPUBs
The 2000bp sequence upstream of SiPUB promoter was extracted and uploaded to the SiPUB PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) web site for cis-element prediction. Gene ontology (GO) annotation for SiPUBs was performed using R 4.3.1.
2.6 Analysis of collinearity and duplication events
The U-box protein sequences of Arabidopsis, rice, sorghum, maize, tomato and foxtail millet were analyzed by interspecific and intraspecific collinearity, and the gene duplication events of foxtail millet were analyzed by MCScanX. TBtools was used to visualize the results.
2.7 Protein-protein interaction
The functional protein-protein interaction network model of U-box proteins was integrated using Web String (https://string-db.org/), with the default confidence parameter value of 0.400.
2.8 Tissue expression pattern of SiPUBs
The tissue-specific expression patterns of SiPUBs were studied by NCBI Short Read Archive database (https://www.ncbi.nlm.nih.gov/sra/) access to different groups of the transcriptome data. Heat map analysis was used in TBtools to map gene expression heat maps using log2 (TPM +1) scales.
2.9 Total RNA extraction and cDNA synthesis
Total RNA was extracted using the SteadyPure Plant RNA Extraction Kit from Accurate Biotechnology (Hunan) Co., Ltd. The quality and concentration of RNA were evaluated using a micro-volume nucleic acid and protein analyzer and RNase-free agarose gel electrophoresis. After confirming the quality and concentration of RNA, the Evo M-MLV Reverse Transcription Premix Kit from Accurate Biotechnology (Hunan) Co., Ltd. was employed to reverse transcribe it into cDNA following the instructions. The cDNA was then stored at −20°C for future use.
2.10 RT-qPCR analysis
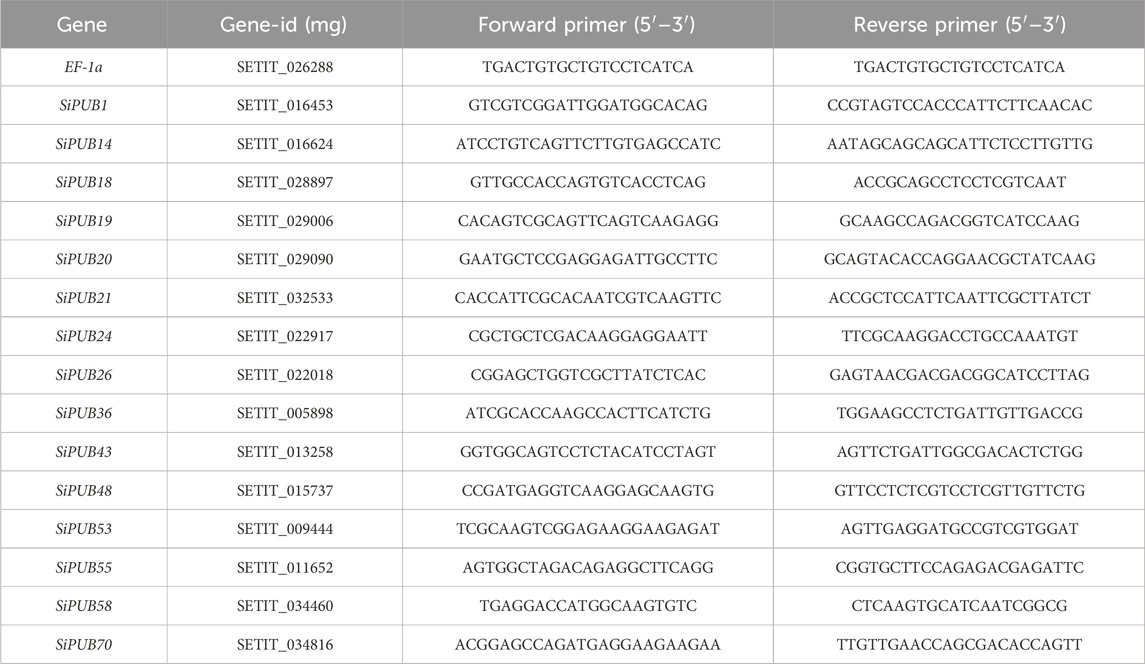
Gene primers were designed using Primer Premier 6, with foxtail millet EF-1a gene serving as the internal reference gene. The primer specificity was screened through BLAST website comparison, and the alignment data were obtained from NCBI (Table 1). The qPCR system was prepared using the SYBR Green Premix Pro Taq HS qPCR Kit from Accurate Biotechnology (Hunan) Co., Ltd. The qPCR system was run in a fluorescence quantitative PCR instrument. To ensure the accuracy of the results, three biological replicates and three technical replicates were performed. The relative expression levels of RNA transcripts were calculated using the 2−ΔΔCT method.
2.11 Protein structure analysis
Protein structure analysis was carried out for SiPUBs, and secondary structure analysis of proteins in SiPUBS was performed using SOPMA (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_sopma.html). The tertiary structure was predicted based on homology modeling method of online software SWISS-MODEL (http://swissmodel.expasy.org/interactive).
3 Results
3.1 Identification and physicochemical property analysis of SiPUBs
By using the Arabidopsis U-box gene family protein sequences as references, we conducted a BLAST comparison with the foxtail millet genome to screen candidate U-box genes, and 2016 gene family members were obtained. The Hidden Markov Model (HMM) file (PF04564) of the U-box family was obtained from the Pfam database and the conserved protein sequences of the species were summarized, and 79 gene family members were obtained. By combining the results of the two methods, 78 gene family members were obtained. Protein sequence domains were detected using SMART and CDD, and redundancies and deletions were removed. Finally, 70 U-box genes with complete U-box domains were identified (Supplementary Table S1), which were named as SiPUB1–SiPUB70 according to their position on chromosomes. The 70 SiPUBs showed great variations in protein sequences and physicochemical properties, with the amino acid (aa) length ranging from 275 aa (SiPUB45) to 1029 aa (SiPUB65) and the molecular weight ranging from 30946.21 Da to 115391.56 Da. The isoelectric point (pI) ranged from 5.17 to 8.98, but was lower than 7 for most proteins, indicating that most SiPUBs are rich in acidic amino acids and belong to acidic proteins. The instability index ranged from 32.99 to 68.16, and the aliphatic index was from 68.17 to 111.56. The most hydrophilic protein was SiPUB9 (−0.617), and the most hydrophobic protein was SiPUB59 (0.317). An online WOLF server was used to predict the localization of 70 SiPUBs in cells. The results showed different locations of SiPUBs in cells, and most of them were located in chloroplasts and cytoplasm. The subcellular localization results indicated that SiPUBs play a key role in biological processes such as plant growth and development.
3.2 Chromosomal localization of SiPUBs
Based on the information in a chromosomal distribution map of SiPUBs was obtained using the MapChart software. Figure 1 shows that SiPUBs are distributed on eight chromosomes in an uneven manner. Among different chromosomes, chromosome I had the largest number of SiPUBs (up to 17), followed by chromosome IX (13), while chromosome II had the fewest SiPUBs (only 4). Moreover, five tandem gene clusters involving 12 genes were found in foxtail millet genome, which were distributed on chromosomes I, II, III, IV, and IX, respectively, but no tandem gene cluster was found on chromosomes V, VI, and VII.
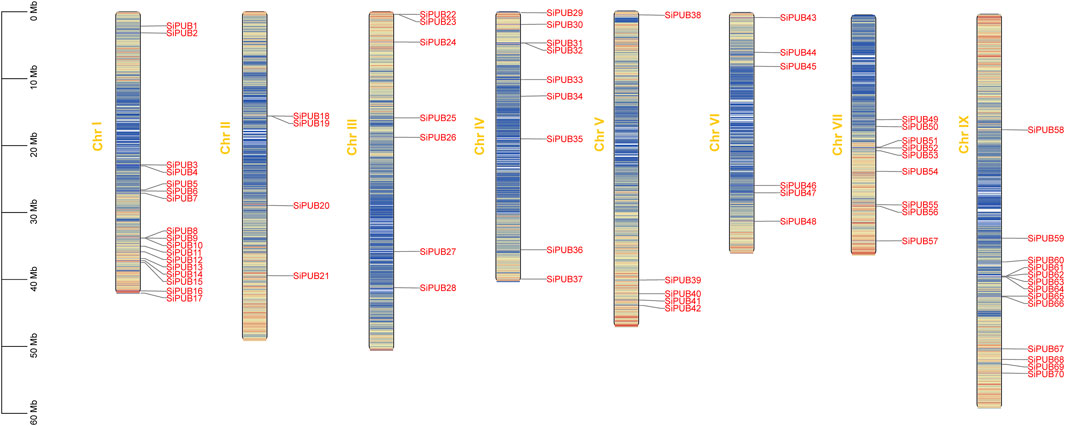
FIGURE 1. Chromosomal localization of SiPUBs. The varying colors of chromosomes indicate gene density.
3.3 Secondary structure analysis of SiPUBs
As shown in Supplementary Table S2, the secondary structure of SiPUBs includes alpha-helix, ex-tended strand, beta-sheet, and random coil. Alpha-helix (26.97%–70.86%) and random coil (21.58%–44.52%) were relatively more abundant in the protein sequences, while extended strand (2.70%–13.92%) and beta-sheet (1.36%–7.73%) were relatively less abundant.
3.4 Phylogenetic analysis of SiPUBs
To study the evolutionary relationship of SiPUB proteins, a neighbor-joining (NJ) tree was constructed with U-box proteins from foxtail millet (70 proteins), rice (77 proteins) and Arabidopsis (61 proteins). According to the phylogenetic relationships and domain composition, these proteins could be divided into six subfamilies, including U-box only (I), UFD2 specific motif + U-box (II), TPR + U-box (III), Kinase + U-box (IV), U-box + WD40 (V), and U-box + ARM (VI). Subfamilies I, II, III, IV, V, and VI contained 23, 1, 2, 16, 2, and 26 members, respectively (Figure 2). Of the six subfamilies, subfamily VI (U-box + ARM) included the most genes (26 genes). In general, the U-box genes of foxtail millet were more closely related to homologues in rice than to those in Arabidopsis, and the U-box genes with similar genetic structures were clustered together.
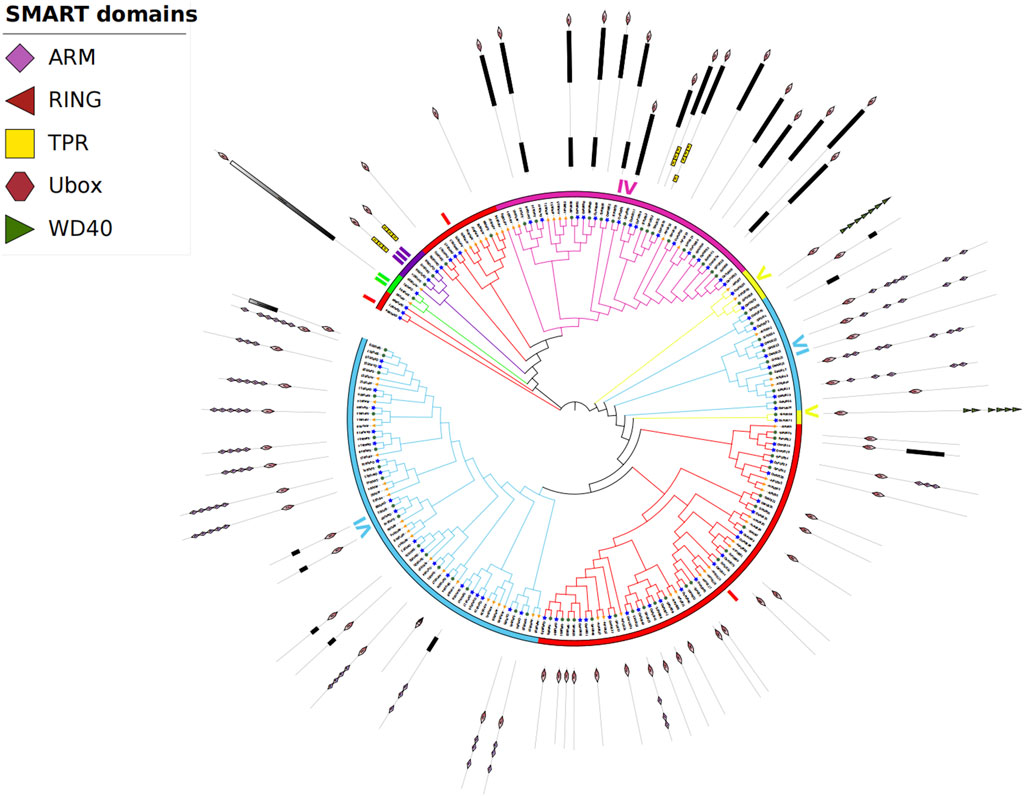
FIGURE 2. Phylogenetic tree of U-box proteins from foxtail millet, Arabidopsis, and rice. Different colors indicate different subfamilies. The three species are represented by three different shapes.
3.5 Gene structure and conserved motif analysis of SiPUBs
To further understand the possible structural evolutionary relationships of SiPUBs, we analyzed the phylogenetic tree, conserved motifs, and gene structure of SiPUBs. As shown in Figure 3, each family member had a different number of motifs, and members in the same subfamily generally had similar conserved motifs. The genes in subfamily I contained both motif 1 and motif 2 and some other motifs, while Subfamily II (SiPUB65) only contained motif 1 and motif 2. Subfamily III (SiPUB24 and SiPUB45) contained only motif 1, 2, and 19. The conserved motifs in subfamily IV included motif 1, 2, 3, 7, 10, and 11. SiPUB58 in subfamily V contained only motif 5, while SiPUB38 contained motif 1, 2, 5, 14, and 16. Most members in subfamily VI contained motif 1, 2, 6, 8, 9, 13, 14, 16, and 18. Moreover, a total of 68 genes contained both motif 1 and motif 2, and 69 genes contained motif 1.
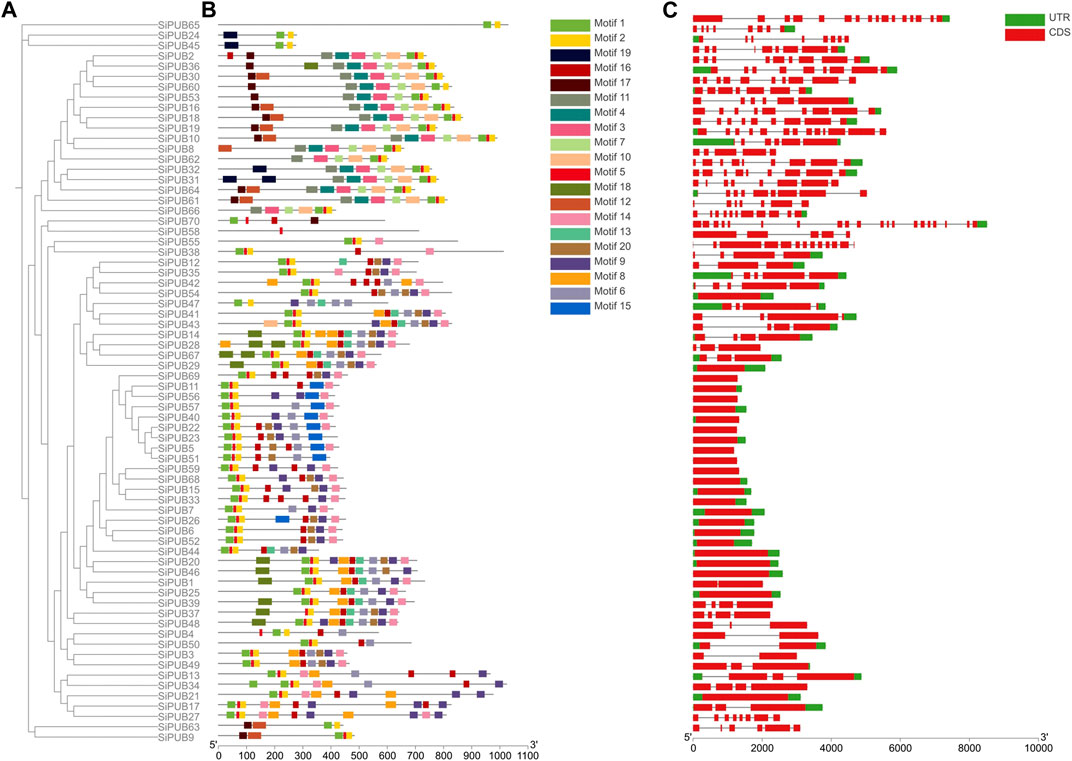
FIGURE 3. Gene structure and conserved motif analysis of SiPUBs. (A) Phylogenetic tree of 70 SiPUBs. (B) Conserved motifs of SiPUBs, with different colored squares representing different motifs. (C) Analysis of SiPUB gene structure. The red squares represent the coding sequence (CDS), the green squares represent the untranslated region (UTR), and the black lines between the two represent introns.
To further explore the diversity of SiPUB gene structure, we analyzed the intron-exon structure of SiPUBs. As shown in Figure 3, members in the same subfamily had similar intron arrangement, and most genes were broken genes, with the number of introns ranging from 0 to 19, and 24 genes having no introns. The structure of subfamily I genes was relatively simple, with most of members containing no introns, and only a few members contained 2–4 introns. Members in subfamily IV had 4–12 introns, with most genes containing eight introns. The genes in subfamily V had the largest number of introns, such as SiPUB58 with 19 introns and SiPUB38 with 12 introns. The introns in members from subfamily VI ranged from 0 to 3, with most genes containing 0 or 3 introns.
3.6 Promoter analysis and functional enrichment of SiPUBs
In order to better understand the mechanisms of gene regulation, we identified the cis-acting elements on each gene that can be used to study different environmental stress responses and tissue specificity. Finally, we identified 28 types of cis-acting elements, including light-responsive elements, plant hormone-responsive elements, abiotic stress-responsive elements, tissue-specific cell cycle elements and circadian rhythm control elements. SiPUBs showed differences in the type and number of cis-acting elements, among which SiPUB23 had the most types of cis-acting elements (16 types), followed by SiPUB18, SiPUB53, and SiPUB64 (15 types), and SiPUB61 had the largest number of cis-acting elements (51), followed by SiPUB25 (49). Sixty-nine genes contained light-responsive elements, while 68 genes had plant hormone-responsive elements, and it can be speculated that plant hormones may regulate these genes. The predicted stress-responsive elements mainly included those involved in anaerobic induction regulation, low temperature response, hypoxia induction, drought response, and trauma response. Among them, cis-acting elements involved in anaerobic induction regulation accounted for the largest proportion (53 genes), while trauma response elements were only found in SiPUB13, SiPUB18, and SiPUB44. There were 11 genes involved in endosperm specific negative expression; only 13 genes were involved in seed-specific regulation; while 37 genes were involved in molecular tissue-specific activation and expression (Figure 4). These results suggested that SiPUBs may be involved in foxtail millet’s response to biotic and abiotic stresses as well as tissue-specific responses.
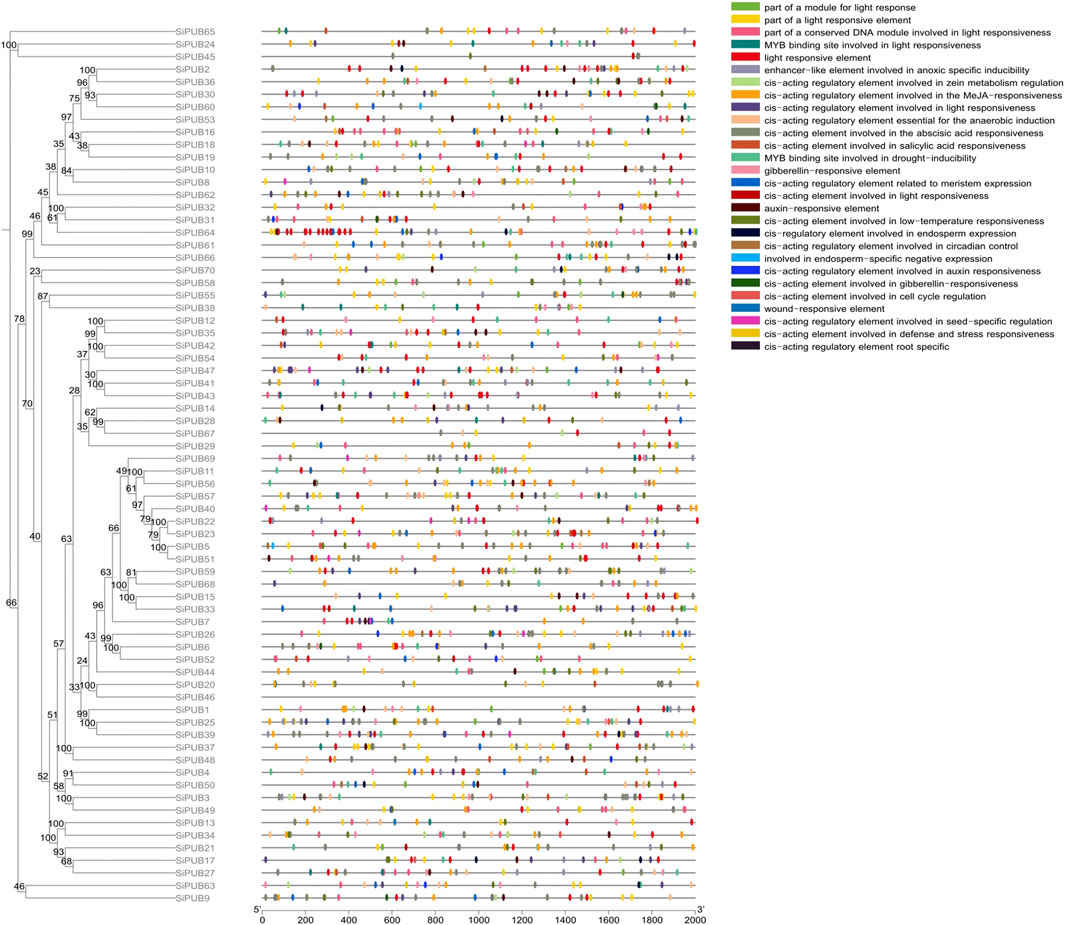
FIGURE 4. SiPUB promoter analysis. Rootless trees were generated by the neighbor-joining (NJ) method using MEGA11 software. Numbers next to branches indicate 1,000 bootstrap replicates as a percentage. Different colored squares represent different promoters. Detailed comments are included in the top right panel.
GO enrichment analysis showed that 36 of the 70 SiPUBs were classified as “biological processes” and 33 were classified as “molecular functions.” Among the “biological processes,” 34 genes were enriched in protein ubiquitination and protein modification through small protein pathways, respectively. Furthermore, 33 genes were enriched in the “molecular function” pathway of ubiquitin-protein transferase activity and ubiquitin-like protein transferase activity (Figure 5 and Supplementary Table S3).
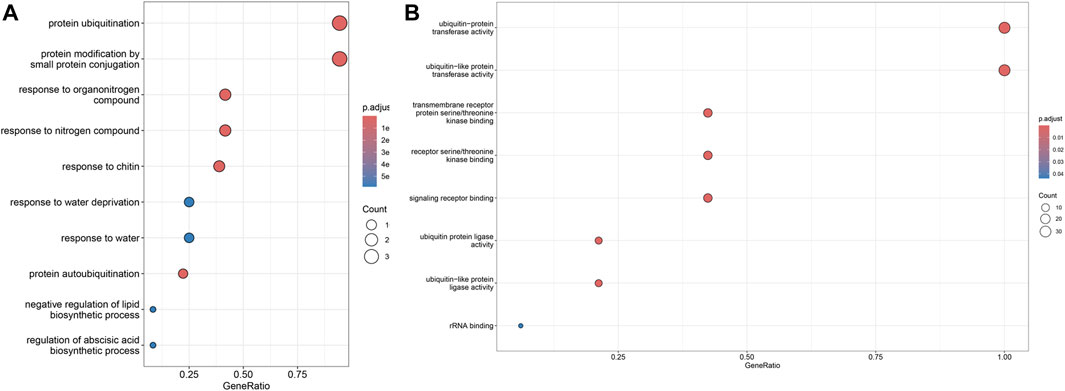
FIGURE 5. GO enrichment analysis. The figure shows top ten enrichment pathways. (A) Enrichment pathway of SiPUBs in biological processes. (B) Enrichment pathway of SiPUBs in molecular function.
3.7 Collinearity analysis of SiPUBs
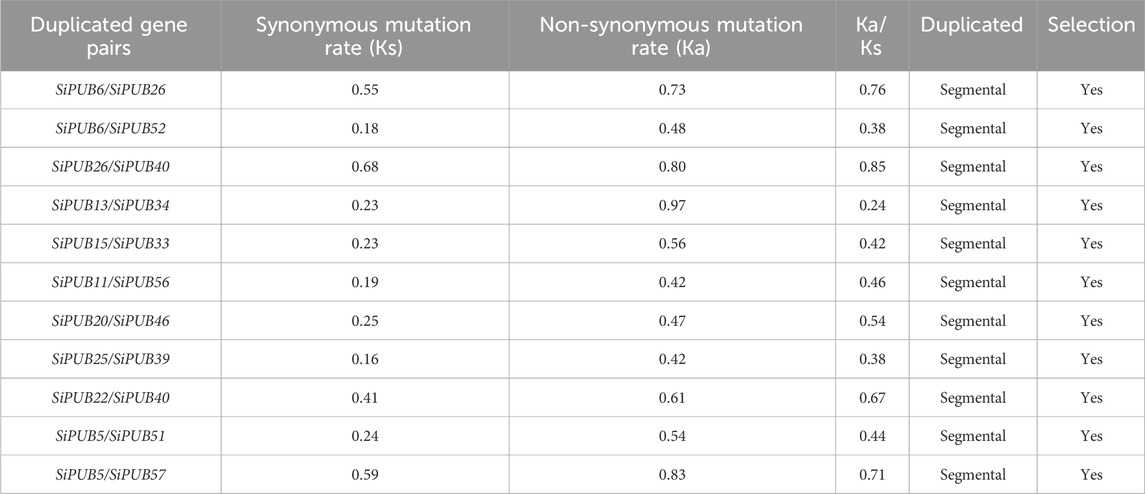
To further understand the expansion of SiPUB family, we analyzed the fragment duplication gene pairs of foxtail millet. A total of 11 duplicate gene pairs of U-box genes were identified on the foxtail millet chromosome (SiPUB6 and SiPUB26, SiPUB13 and SiPUB34, SiPUB15 and SiPUB33, SiPUB5 and SiPUB51, SiPUB6 and SiPUB52, SiPUB11 and SiPUB56, SiPUB5 and SiPUB57, SiPUB20 and SiPUB46, SiPUB25 and SiPUB39, SiPUB26 and SiPUB40, SiPUB22 and SiPUB40) (Figure 6). Among different chromosomes, chromosome I contained the most pairs of genes and the highest collinearity. Ks, Ka, and Ka/Ks of 11 U-box duplicate gene pairs in foxtail millet were calculated and analyzed (Table 2). The results showed that the Ka/Ks ratio of four duplicate gene pairs was much lower than 1, and the remaining seven duplicate gene pairs had Ka/Ks ratio close to 1. It can be speculated that gene duplication events may have contributed to the evolution of the SiPUB family.
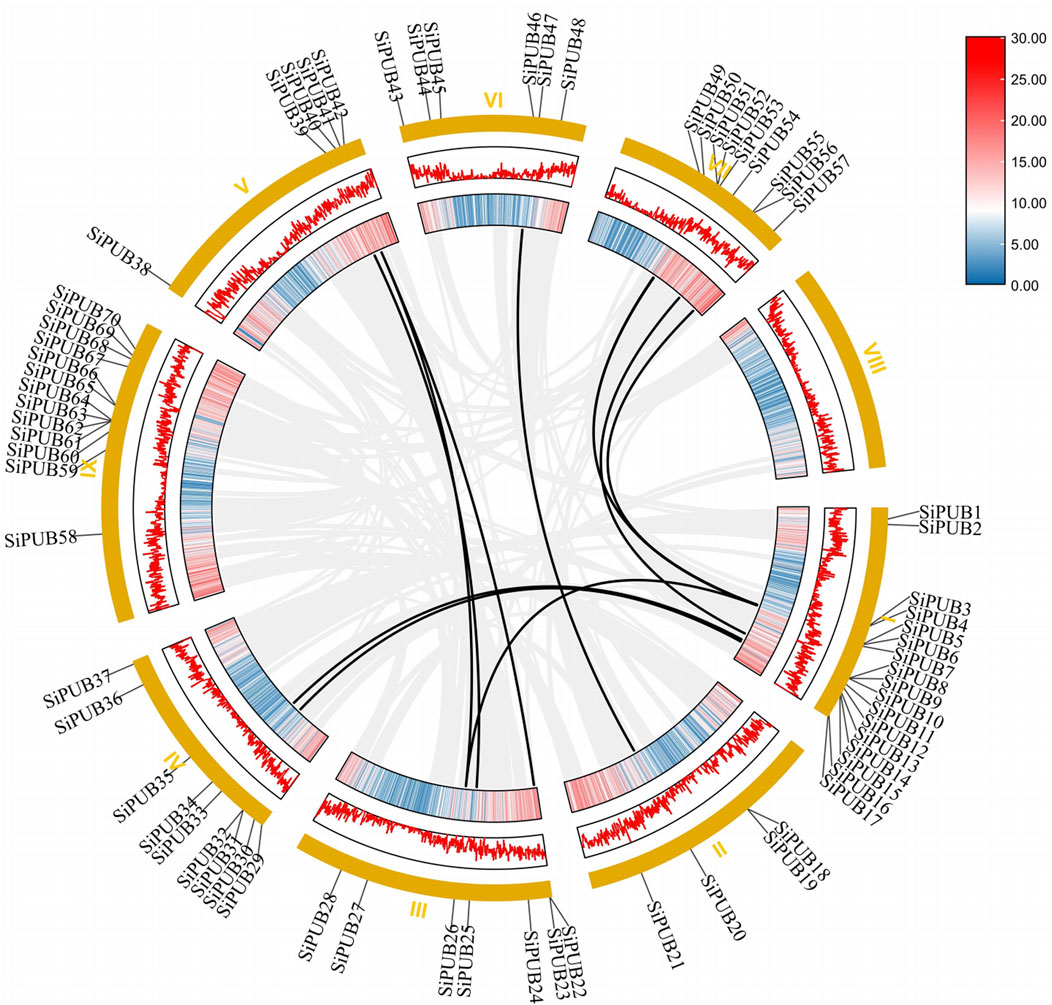
FIGURE 6. Collinearity analysis of SiPUBs. Chromosomes are shown on the outside, with yellow representing different chromosomes. The different colors of the inner circle represent the gene density of the chromosome (the gene density increases from blue to red), and the black line represents the fragment of repeating gene pairs of the foxtail millet.
In order to further elucidate the evolutionary relationship of SiPUBs, we constructed collinearity maps of U-box genes from foxtail millet and Arabidopsis, rice, and sorghum. (Figure 7). The results showed that 19 pairs of U-box genes had collinearity between foxtail millet and Arabidopsis, among which chromosome 2 in Arabidopsis had the most collinear genes. Moreover, 78 pairs of U-box genes were collinear between foxtail millet and rice, among which chromosome I of foxtail millet and chromosome 2 of rice had the most collinear genes. The U-box genes had the highest collinearity between foxtail millet and wheat (164 pairs), and chromosome I of foxtail millet and chromosome 7D of wheat had the most collinear genes. There were 85 pairs of U-box genes with collinearity between foxtail millet and sorghum, among which chromosome I of foxtail millet and chromosome 4 of sorghum had the most collinear genes. Collinearity of U-box genes was not found on chromosome 4 of Arabidopsis, chromosome 11, chromosome 7 of rice, chromosome 5 of sorghum.
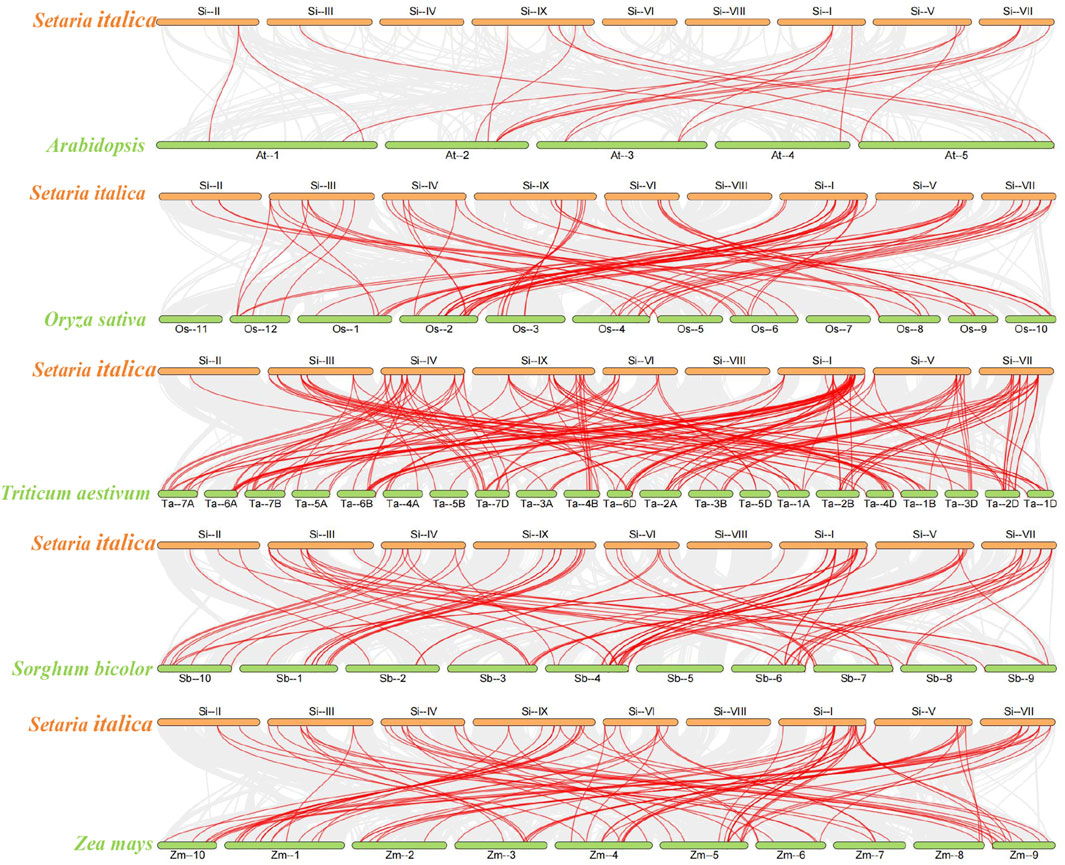
FIGURE 7. Analysis of U-box collinearity between foxtail millet and Arabidopsis, rice, sorghum, wheat, and maize. The gray line represents collinearity between all genes across species, and the red line represents collinearity between U-box genes across species.
3.8 Protein network interaction prediction in SiPUBs
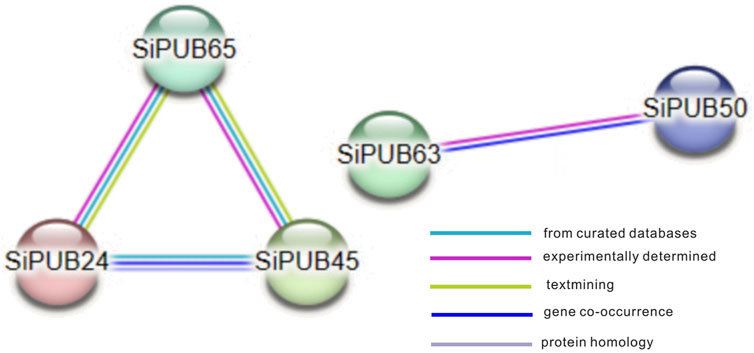
To better understand the possible interactions between SiPUBs, the interaction between SiPUBs was predicted online using the STRING website. As shown in Figure 8, there were certain interactions among SiPUBs, including paired interactions among SiPUB65, SiPUB24, and SiPUB45, and interaction between SiPUB63 and SiPUB50.
3.9 Expression patterns of SiPUBs in different tissues
Based on plant databases, the transcription levels of SiPUBs in different tissues (roots, stems, leaves, and inflorescences) were analyzed. Differences in the expression of SiPUBs were found among different tissues (Figure 9). Among them, SiPUB1/3/28/67 were highly expressed in leaves. SiPUB3/6/12/25/28/33/58 were highly expressed in roots. SiPUB3/14/25/28/58 were highly expressed in the stem and SiPUB2/58 had high expression in the inflorescence. The relative expression patterns of SiPUBs in different tissues predicted their complex role in foxtail millet growth and development.
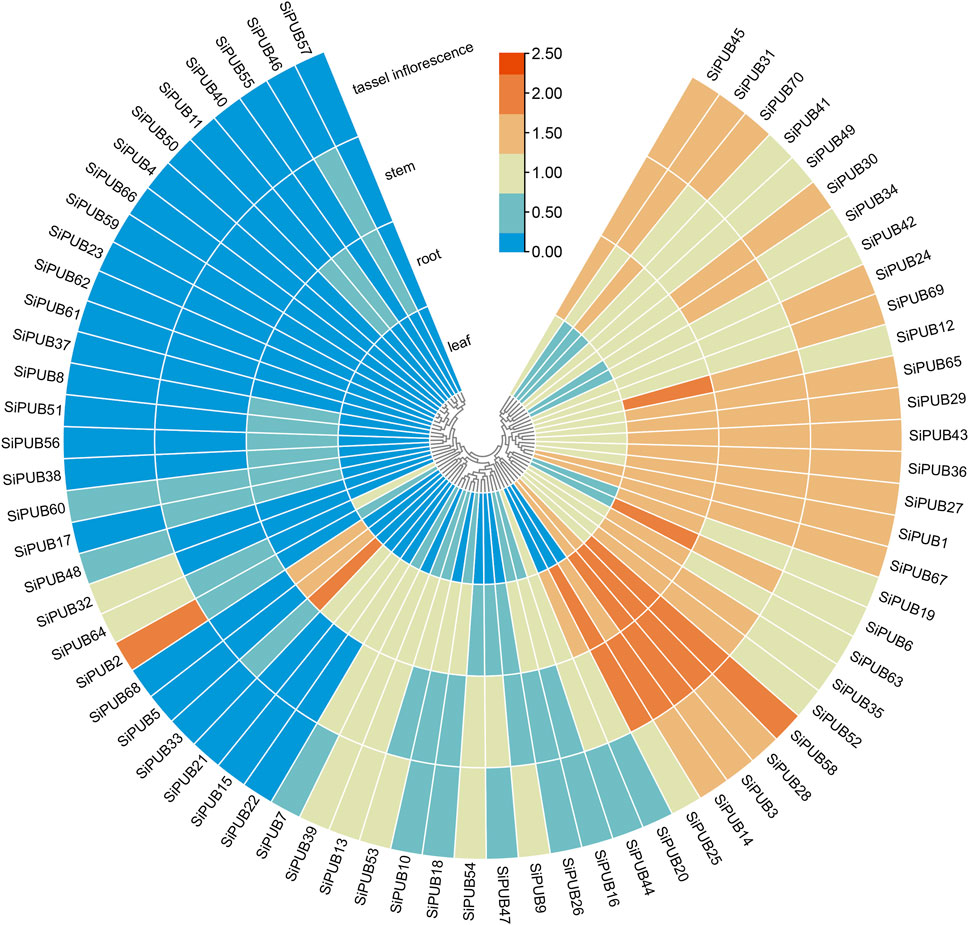
FIGURE 9. Heatmap of tissue-specific expression of SiPUBs. The color bar represents the log2 expression level of each gene (FPKM, fragments per kilobase of exon per million fragments mapped). Color bar annotation is included at the top of the image. The heatmap is colored according to expression values, with blue, yellow, and red representing at low, medium, and high transcription abundance, respectively.
3.10 Responses of SiPUBs to saline-alkali stress detected by qRT-PCR
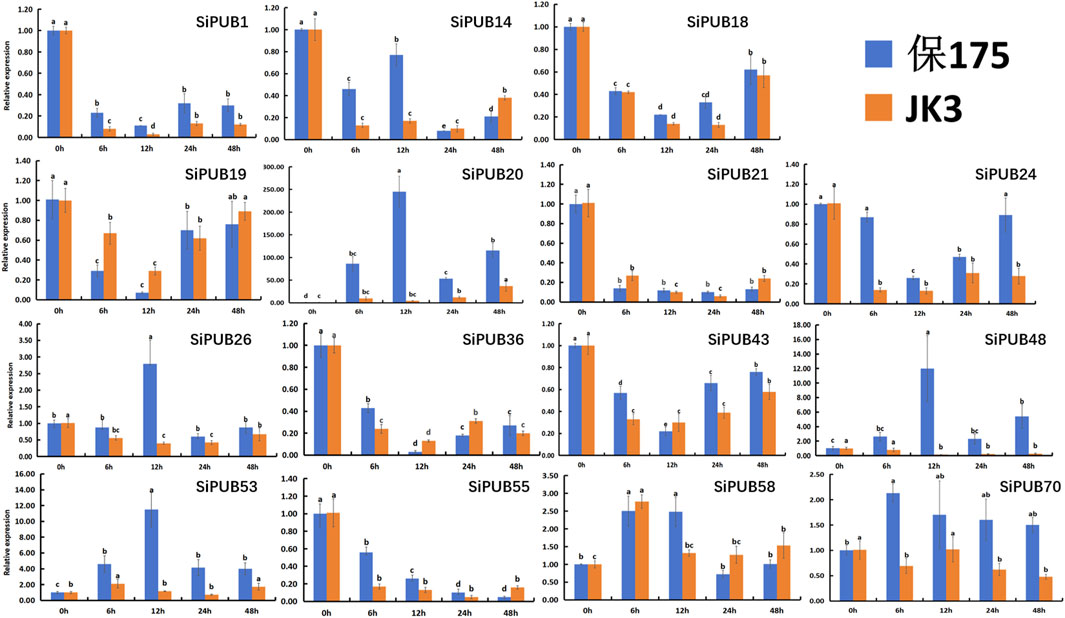
The results of tissue expression patterns showed that SiPUBs were expressed in all tissues of foxtail millet, and most of the genes were expressed or highly expressed in leaves. Leaves play a very important role in plants and are the main organs for photosynthesis, gas exchange, and transpiration. Therefore, they can most directly reflect the growth state of plants under abiotic stress. At present, the roles of U-box gene family members in foxtail millet have not been reported in detail, and the gene expression level of SiPUBs under salt-alkali stress remains clear. In order to explore the key role of SiPUBs in salt-alkali tolerance, 15 SiPUBs in leaf tissues under saline-alkali stress were randomly selected for qRT-PCR detection. The results showed that there were significant changes in the expression levels of these SiPUBs in B175 (sensitive) and JK3 (tolerant) under saline-alkali stress (Figure 10). In B175, the relative expression of SiPUB1/18/19/21/24/36/43 first decreased and then increased with the extension of treatment time. The relative expression of SiPUB14 firstly decreased, then increased, then decreased, and finally increased. The relative expression levels of SiPUB20/48/58 first increased, then decreased, and finally increased, and became significantly higher than the initial levels at 48 h. The relative expression of SiPUB26/53/70 first increased and then decreased, and the relative expression of SiPUB53/70 at 48 h was significantly higher than the initial level. The relative expression of SiPUB55 showed a consistent decreasing trend. For JK3, along with the extension of treatment time, the relative expression of SiPUB1/14/18/19/21/24/26/43/55 showed a first declining and then rising trend; that of SiPUB20 showed a consistently increasing trend, and became significantly higher than the initial level at 48 h. The relative expression of SiPUB36/70 first decreased, then increased, and finally decreased. The relative expression of SiPUB48 always showed a decreasing trend. The relative expression levels of SiPUB53/58 first increased, then decreased, and finally increased, and became higher than the initial level at 48 h. The changes in the relative expression levels of SiPUB1/18/19/21/24/43 (decreasing and then increasing) and SiPUB58 (increasing, decreasing, and then increasing) were consistent between the two varieties, but there were significant differences in the relative expression levels. In addition, the relative expression level of SiPUB20 in the two varieties was upregulated compared with that in the control, and was higher in B175 than in JK3 at different treatment time. For example, its expression level in B175 was 57.81 times that in JK3 at 12 h. Compared with that of the control, the relative expression of SiPUB48/70 was upregulated in B175, but downregulated in JK3. The relative expression of SiPUB48 in B175 was 92.23 times higher than that in JK3 at 12 h. At 6 h, the relative expression of SiPUB70 in B175 was 3.09 times that in JK3. With the extension of saline-alkali treatment, the relative expression of SiPUB26 was only upregulated in B175 at 12 h, which was 6.98 times that of JK3, but was downregulated in the two varieties at all other time points.
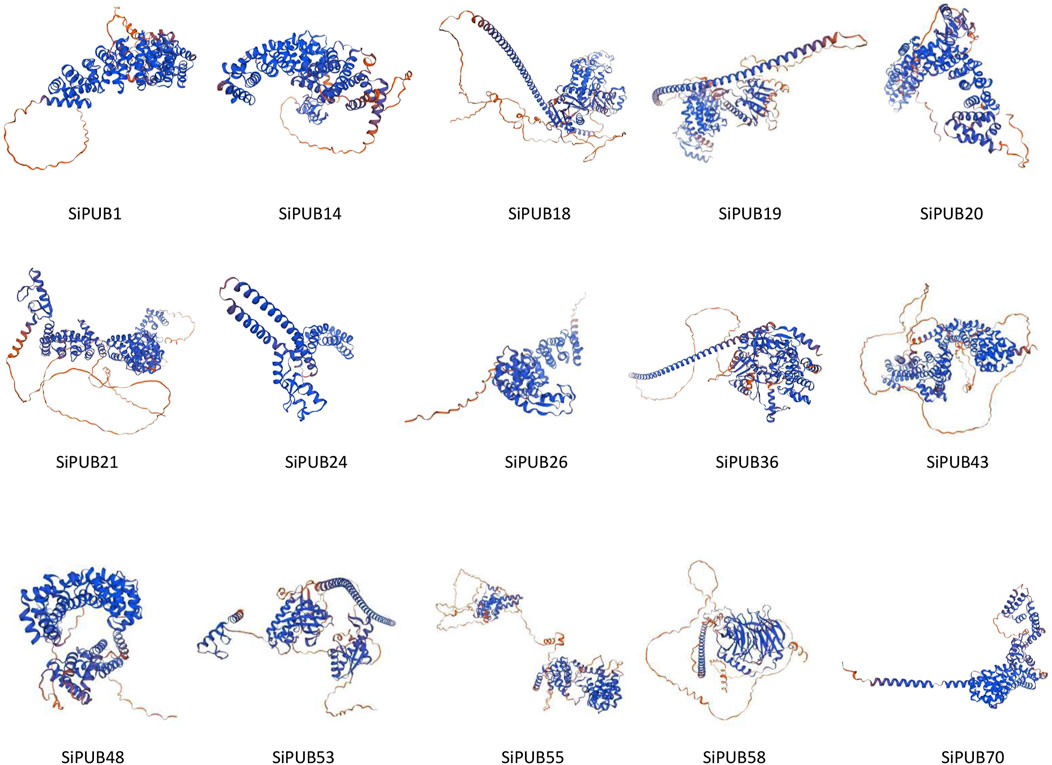
3.11 Protein structure analysis of SiPUBs
The prediction of the tertiary structure of the protein showed that the coverage of the target sequence and the model sequence was 73% or more, and the conservation of SiPUBs was not strong, and there were obvious differences among all proteins (Figure 11). The consistency of SiPUB21 and SiPUB58 genes with the model protein reached 100%, and the tertiary structure was predicted to have a generally good consistency with the model protein.
4 Discussion
4.1 SiPUBs are conserved during evolution
Plant U-box proteins constitute a small protein family with a U-box domain (Azevedo et al., 2001; Wiborg et al., 2008; Chen and Hellmann, 2013). The U-box domain consists of approximately 70 amino acids and maintains its conformation through interactions with components such as hydrogen bonds and salt bridges (Aravind and Koonin, 2000; Andersen et al., 2004). U-box E3 is involved in a variety of biological processes, such as development, self-incompatibility, and response to hormones, and are widely associated with plant stress response (Yee and Goring, 2009; Lee and Kim, 2011; Lyzenga and Stone, 2012; Chen and Hellmann, 2013; Duplan and Rivas, 2014; Stone, 2014). U-box genes play important roles in response to drought (Cho et al., 2006; Cho et al., 2008; Liu et al., 2011; Seo et al., 2012), salt (Cho et al., 2006; Ni et al., 2010; Bergler and Hoth, 2011), temperature stress (Stone et al., 2003; Cho et al., 2006), oxidative stress (Park et al., 2011), and low phosphate stress (Hur et al., 2012). At present, U-box genes have been proved to have great biological significance in a variety of plants such as Arabidopsis (Lu et al., 2008). However, there has been no report on the U-box genes in foxtail millet. In this study, SiPUBs were systematically analyzed through bioinformatics analysis, and a total of 70 members were identified. The molecular weights of SiPUBs ranged from 30946.21 Da to 115391.56 Da, and the pI ranged from 5.17 to 8.98. In addition to the U-box domain, SiPUBs also contain the ARM, WD40 and TPR secondary domains, which are mainly used to mediate the specific recognition of U-box protein and substrate protein and maintain the basic function of the family, enriching the diversity of genes (Lu et al., 2020). Gene structure analysis also revealed that most SiPUBs contain multiple introns, indicating changes in their structure during evolution, and differences in intron/exon structure can often promote the evolution of multi-gene families, which may lead to different functions of members within the same subfamily (Rogozin et al., 2003). Similar results have also been obtained for the U-box gene families of other species such as cotton (Lu et al., 2020), banana (Hu et al., 2018), and sorghum (Fang et al., 2022). The number of exons varied greatly from 1 to 20, which may be attributed to the directed evolution of function and structure of U-box genes throughout the long evolutionary history. Furthermore, gene structure and phylogenetic analysis indicated that U-box genes in different branches of the same subfamily have different numbers of exons, introns, and protein sequence lengths. However, orthologous genes in the same branch showed highly similar physicochemical properties and gene structure. These results indicated that SiPUBs are highly conserved during evolution, but at the same time, the gene functions were diversified for adaptation to environmental changes and population continuation.
4.2 Tandem duplication events are the main driving force of SiPUB evolution
Gene duplication is the primary force driving the expansion of gene families over a period of time under the regulation of environmental and biological factors. Gene duplication is a universal phenomenon in plants, including tandem duplication, fragment duplication, and whole genome duplication (Kondrashov et al., 2002; Conant and Wolfe, 2008). Chromosomal localization analysis and collinearity analysis showed that gene duplication is the main cause of SiPUB diversity. Among the 70 SiPUBs, 12 were generated by tandem duplication, which is one of the main reasons for the expansion of this gene family. A total of 11 duplication SiPUB gene pairs were identified on foxtail millet chromosomes. Similar results were obtained in other species as well. In tomato, the U-box gene family was found to experience ten duplication events (Sharma and Taganna, 2020). In cotton, 19 U-box genes were derived from tandem duplication (Lu et al., 2020). In cabbage, there are 24 pairs of tandem duplicated U-box genes (Hu et al., 2019). These results indicate that subfunctionalization or neofunctionalization of duplicate genes can lead to the expansion of gene families as well as structural organization and diversification of gene expression. Over time, the preserved sequences may undergo new functionalization to produce differentiation.
4.3 Different subfamilies of SiPUBs have differential functions
The conserved U-box domain of SiPUBs is often associated with ARM repeats, WD40 repeats, TRR structures, and other domains. SiPUBs can be divided into six subfamilies according to phylogenetic relationships and domain composition. Due to the conserved nature of genes, genes with similar or identical functions are located in the same subfamily, which lays a solid foundation for studying the functions of SiPUBs.
There is only one UFD2 gene in foxtail millet and Arabidopsis (AtUFD2). AtUFD2 protein contains a conserved domain similar to yeast UFD2 protein, which can interact with aaa ATPase CDC48 and participates in the regulation of cell cycle, death, organelle formation, and other physiological activities (Zhou et al., 2021). In Arabidopsis, AtCHIP in the TPR + U-box subfamily is involved in regulating abiotic stress response. Under low temperature and dark conditions, AtCHIP alters the activity of PP2A in stress response signal transduction, thus responding to abiotic stress (Luo et al., 2006). The U-box + ARM subfamily is the most extensively studied subfamily. AtPUB18, AtPUB19, AtPUB46, and AtPUB48, which belong to the U-box + ARM subfamily, play important roles in drought stress response (Bergler and Hoth, 2011; Adler et al., 2017). AtPUB18 and AtPUB19 are conserved proteins containing PUB-ARM, a type of E3 ubiquitin ligase. During the early development of Arabidopsis, AtPUB18 and AtPUB19 inhibit seed germination under salt stress, and the salt stress response would disappear in the absence of PUB-ARM proteins, indicating that ubiquitination is a key element stimulating salt sensitivity (Bergler and Hoth, 2011). AtPUB17 positively regulates the processes of cell apoptosis and plant defense (Sadanandom, 2007), while AtPUB9 regulates lateral root development under phosphate starvation conditions (Sankaranarayanan and Samuel, 2015). On this basis, it can be inferred that other genes in the U-box + ARM subfamily may have similar or overlapping functions. The U-box only subfamily (subfamily I in this study) has a simple gene structure and plays important roles in plant growth, development, and abiotic stress response. Previous studies have shown that apple callus overexpressing MdPUB24 and Arabidopsis seedlings with ectopic expression of MdPUB24 exhibited significant decreases in growth compared with the wild type under salt stress conditions, indicating that MdPUB24 negatively regulates salt stress response (Qi et al., 2017). Based on the homology of genes in the evolutionary relationship, it can be speculated that homologous genes of Arabidopsis and foxtail millet in the same subfamily may also participate in similar regulatory pathways.
4.4 Potential function and expression analysis of SiPUBs
Protein ubiquitination is a post-translational modification involved in various physiological processes in plants and is required for many signaling pathways in many important cellular processes such as plant growth, development and eukaryotic stress response (You et al., 2016; Ye et al., 2018). Ma et al., 2021 showed that MYB6 and VDAL are effector factors with the same regulatory effect on verticillium wilt resistance; PUB25 and PUB26 can degrade MYB6 through ubiquitination; and VDAL competes with MYB6 to bind PUB25 and PUB26 to promote resistance. Previous studies have also found that PUB25 and PUB26 have two forms of ubiquitination modification of MYB15 protein at low temperatures. At the early stages of cold stress, PUB25 and PUB26 promote the degradation of MYB15 via high-level K48 linkages and stabilize ICE1 by promoting its K63-linked ubiquitination, thereby facilitating the inhibition of MYB15 by ICE1 and leading to activation of CBF expression. Upon prolonged cold stress, PUB25 and PUB26 enhance K48-linked modification on ICE1 to target it to the 26S proteasome for degradation while simultaneously adding K63 linkages to MYB15, allowing it to bind CBF promoters and repress their expression (Wang et al., 2023). The E3 ubiquitin ligases PUB12 and PUB13 directly interact with BRI1 and ubiquitinate BRI1, and PUB12/PUB13-mediated ubiquitination regulates endocytosis and intracellular degradation of BRI1 (Zhou et al., 2018). However, there have been few reports on the function of U-box (PUB) gene in foxtail millet, but it can be inferred from previous studies that U-box gene family in foxtail millet may have similar functions.
Promoter analysis can effectively predict the gene family associations in stress-related mechanism, hormone regulation, and development (Sharma and Taganna, 2020). Similar patterns were observed in the promoter analysis of purple clover by Yang et al. (2017), indicating a common association of the U-box gene family with plant stress and hormone regulatory pathways. Similar to previous studies, this study conducted promoter function prediction analysis on SiPUBs and found that most SiPUBs contain a large number of light-responsive elements, indicating that SiPUBs may be involved in the regulation of plant photosynthesis. Moreover, a large number of plant hormone-responsive elements such as auxin, abscisic acid, and gibberellin were found in most of the genes, suggesting that SiPUBs may be involved in a series of other processes such as the growth and development of foxtail millet plants and the ripening of fruit ears.
Previous studies have demonstrated that Arabidopsis PUB4 mutant exhibited higher levels of cell proliferation and division in the root and shoot apical meristem (Kinoshita et al., 2015). In this study, expression analysis of SiPUBs in different tissues showed that SiPUBs had obvious tissue specificity, most of which had the relatively high expression in roots. Similarly, U-box genes also showed obvious tissue specificity in banana, with high expression levels in the root (Hu et al., 2018). This pattern of expression has also been found in other plants such as soybean (Wang et al., 2020a) and sorghum (Fang et al., 2022). There results suggest that U-box genes may affect root development or other growth and development processes of plants.
As a matter of fact, previous studies have demonstrated that U-box genes are involved in plant growth and development. For example, TaPUB4 has been found to participate in the regulation of pollen development by modulating the metabolism of sucrose and starch in the anther (Zhang et al., 2017). Since the expression levels of genes in different tissues and cell types can reveal their functions in organisms and roles in different physiological processes, we conducted qRT-PCR analysis on 15 SiPUBs under saline-alkali stress in this study. The results showed that all 15 genes were induced to express under saline-alkali stress, and the relative expression levels of some genes were significantly different between B175 and JK3. In B175, the expression level of SiPUB20 from subfamily VI was more significantly induced under salt and alkali stress than that of other genes, and its expression level was 53.34–245.1 folds upregulated compared with the control, and the expression level varied greatly at different treatment time. The relative expression level of SiPUB20 in JK3 was 4.20–36.83 folds higher than that in B175 at different treatment time. The expression level of SiPUB20 was the most different between JK3 and B175, and the relative expression level of B175 was 57.81 folds that of JK3 after 12 h of treatment. The relative expression levels of both SiPUB48 in subfamily VI and SiPUB70 in subfamily I were upregulated in B175 while downregulated in JK3. Previous studies have found that TaPUB1 in wheat can enhance plant salt tolerance and drought resistance. Additionally, TaPUB15 is induced by salt, abscisic acid, low temperature, and polyethylene glycol (PEG) treatment (Zhang et al., 2017). Hwang et al. (2014) demonstrated that inhibition of AtPUB30 in Arabidopsis can enhance salt tolerance during germination. Cho et al. (2006) found that CaPUB1 from pepper could increase plant salt tolerance when overexpressed in Arabidopsis. Han et al. (2019) indicated that MdPUB29 in apple may positively regulate salt tolerance. Further analysis in this study showed that the expression levels of nine out of 15 SiPUB genes in the two varieties were lower than those of the control under saline-alkali treatment, and the downregulation degree in saline-alkali tolerant variety JK3 was higher than that in saline-alkali sensitive variety B175. Previous studies have found that Arabidopsis transgenic plants overexpressing pub22 and pub23 are sensitive to drought stress, and pub22 and pub23 functionally deficient mutant plants are significantly more drought-tolerant, and pub22 and pub23 double mutant plants are even more drought-tolerant. These results indicated that PUB22 and PUB23 play a negative regulatory role in water stress response (Cho et al., 2008). Similar to these previous studies, this study suggested that U-box genes may play a critical role in the response against saline-alkali stress through negative regulation. Furthermore, subcellular localization analysis predicted that SiPUB20 is localized on membrane-bound organelles or plasma membrane, indicating that it may be involved in saline-alkali response by changing membrane composition or ion transport. Moreover, the localization of SiPUB48/70 in the nucleus suggests their potential involvement in transcriptional regulation or nucleus transport processes in response to saline-alkali stress.
5 Conclusion
In this study, a total of 70 SiPUBs were identified, all of which contained the U-box conserved domain. The 70 SiPUBs were distributed on eight chromosomes of foxtail millet, forming five tandem gene clusters, and could be divided into six subfamilies based on the phylogenetic relationships and domain composition. SiPUBs contained a total of 28 types of cis-acting elements, including light-responsive elements, plant hormone-responsive elements, abiotic stress-responsive elements, tissue-specific cell cycle, and circadian rhythm control, suggesting that SiPUBs may be involved in abiotic stress responses. Most SiPUBs showed high expression in roots, stems, leaves, and flowers, suggesting their crucial roles in plant development. Saline-alkali stress resulted in significantly differential expression of 15 randomly selected SiPUBs. Gene expression pattern analysis indicted that SiPUB20/48/70 may play important roles in the response to saline-alkali stress. The results of this study lay a foundation for further exploring the mechanism for the response of SiPUBs to saline-alkali stress, and provide reference for molecular breeding of saline-alkali tolerance in foxtail millet.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author contributions
XZ: Data curation, Formal Analysis, Investigation, Methodology, Software, Writing–original draft, Visualization. YL: Conceptualization, Supervision, Writing–review and editing. JW: Conceptualization, Supervision, Writing–review and editing. YZ: Writing–original draft, Investigation, Visualization. HW: Writing–original draft, Data curation, Formal Analysis. YH: Conceptualization, Funding acquisition, Methodology, Project administration, Supervision, Visualization, Writing–review and editing, Validation. XL: Conceptualization, Funding acquisition, Project administration, Visualization, Writing–review and editing, Validation.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by the Hebei Agriculture Research System (HBCT2023050404), the Key Research and De-velopment Program of Hebei Province (22326314D), and the Scientific Research Foundation of Hebei Normal University of Science and Technology (2021YB007).
Acknowledgments
The authors would like to thank the two reviewers for their helpful comments.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2024.1356807/full#supplementary-material
References
Adler, G., Konrad, Z., Zamir, L., Mishra, A. K., Raveh, D., and Bar-Zvi, D. (2017). The Arabidopsis paralogs, PUB46 and PUB48, encoding U-box E3 ubiquitin ligases, are essential for plant response to drought stress. BMC Plant Biol. 17, 8. doi:10.1186/s12870-016-0963-5
Andersen, P., Kragelund, B. B., Olsen, A. N., Larsen, F. H., Chua, N. H., Poulsen, F. M., et al. (2004). Structure and biochemical function of a prototypical Arabidopsis U-box domain. J. Biol. Chem. 279, 40053–40061. doi:10.1074/jbc.m405057200
Aravind, L., and Koonin, E. V. (2000). The U-box is a modified RING finger a common domain in ubiquitination. Curr. Biol. 10, 132–134. doi:10.1016/s0960-9822(00)00398-5
Azevedo, C., Santos-Rosa, M. J., and Shirasu, K. (2001). The U-box protein family in plants. Trends Plant Sci. 6, 354–358. doi:10.1016/s1360-1385(01)01960-4
Bennetzen, J. L., Schmutz, J., Wang, H., Percifield, R., Hawkins, J., Pontaroli, A. C., et al. (2012). Reference genome sequence of the model plant Setaria. Nat. Biotechnol. 30, 555–561. doi:10.1038/nbt.2196
Bergler, J., and Hoth, S. (2011). Plant U-box armadillo repeat proteins AtPUB18 and AtPUB19 are involved in salt inhibition of germination in Arabidopsis. Plant Biol. 13, 725–730. doi:10.1111/j.1438-8677.2010.00431.x
Chen, L., and Hellmann, H. (2013). Plant E3 ligases: flexible enzymes in a sessile world. Mol. Plant 6, 1388–1404. doi:10.1093/mp/sst005
Cho, S. K., Chung, H. S., Ryu, M. Y., Park, M. J., Lee, M. M., Bahk, Y. Y., et al. (2006). Heterologous expression and molecular and cellular characterization of CaPUB1 encoding a hot pepper U-box E3 ubiquitin ligase homolog. Plant Physiol. 142, 1664–1682. doi:10.1104/pp.106.087965
Cho, S. K., Ryu, M. Y., Song, C., Kwak, J. M., and Kim, W. T. (2008). Arabidopsis PUB22 and PUB23 are homologous U-Box E3 ubiquitin ligases that play combinatory roles in response to drought stress. Plant Cell 20, 1899–1914. doi:10.1105/tpc.108.060699
Conant, G. C., and Wolfe, K. H. (2008). Turning a hobby into a job: how duplicated genes find new functions. Nat. Rev. Genet. 9, 938–950. doi:10.1038/nrg2482
Cramer, G. R., Urano, K., Delrot, S., Pezzotti, M., and Shinozaki, K. (2011). Effects of abiotic stress on plants: a systems biology perspective. BMC Plant Biol. 11, 163. doi:10.1186/1471-2229-11-163
Diao, X., and Jia, G. (2017). Origin and domestication of foxtail millet. Berlin, Germany: Springer.
Duplan, V., and Rivas, S. (2014). E3 ubiquitin-ligases and their target proteins during the regulation of plant innate immunity. Front. Plant Sci. 5, 42. doi:10.3389/fpls.2014.00042
Fang, Y., Du, Q., Yang, Q., Jiang, J., Hou, X., Yang, Z., et al. (2022). Identification, characterization, and expression profiling of the putative U-box E3 ubiquitin ligase gene family in Sorghum bicolor. Front. Microbiol. 13, 942302. doi:10.3389/fmicb.2022.942302
Han, P. l., Dong, Y. h., Jiang, H., Hu, D. g., and Hao, Y. j. (2019). Molecular cloning and functional characterization of apple U-box E3 ubiquitin ligase gene MdPUB29 reveals its involvement in salt tolerance. J. Integr. Agr. 18, 1604–1612. doi:10.1016/s2095-3119(19)62594-3
Hu, D., Xie, Q., Liu, Q., Zuo, T., Zhang, H., Zhang, Y., et al. (2019). Genome-Wide distribution, expression and function analysis of the U-box gene family in Brassica oleracea L. Genes 10, 1000. doi:10.3390/genes10121000
Hu, H., Dong, C., Sun, D., Hu, Y., and Xie, J. (2018). Genome-Wide identification and analysis of U-box E3 Ubiquitin-Protein ligase gene family in banana. Int. J. Mol. Sci. 19, 3874. doi:10.3390/ijms19123874
Hur, Y. J., Yi, Y. B., Lee, J. H., Chung, Y. S., Jung, H. W., Yun, D. J., et al. (2012). Molecular cloning and characterization of OsUPS, a U-box containing E3 ligase gene that respond to phosphate starvation in rice (Oryza sativa). Mol. Biol. Rep. 39, 5883–5888. doi:10.1007/s11033-011-1399-5
Hwang, J. H., Seo, D. H., Kang, B. G., Kwak, J. M., and Kim, W. T. (2014). Suppression of Arabidopsis AtPUB30 resulted in increased tolerance to salt stress during germination. Plant Cell Rep. 34, 277–289. doi:10.1007/s00299-014-1706-4
Kim, E. J., Lee, S. H., Park, C. H., Kim, S. H., Hsu, C. C., Xu, S., et al. (2021). Corrigendum to: plant U-Box 40 mediates degradation of the brassinosteroid-responsive transcription factor BZR1 in Arabidopsis roots. Plant Cell 33, 2900. doi:10.1093/plcell/koab127
Kinoshita, A., ten Hove, C. A., Tabata, R., Yamada, M., Shimizu, N., Ishida, T., et al. (2015). A plant U-box protein, PUB4, regulates asymmetric cell division and cell proliferation in the root meristem. Development 142, 444–453. doi:10.1242/dev.113167
Kondrashov, F. A., Rogozin, I. B., Wolf, Y. I., and Koonin, E. V. (2002). Selection in the evolution of gene duplications. Genome Biol. 3, RESEARCH0008. doi:10.1186/gb-2002-3-2-research0008
Lee, J. H., and Kim, W. T. (2011). Regulation of abiotic stress signal transduction by E3 ubiquitin ligases in Arabidopsis. Mol. Cells 31, 201–208. doi:10.1007/s10059-011-0031-9
Li, Q., Li, L., Liu, Y., Lv, Q., Zhang, H., Zhu, J., et al. (2017). Influence of TaGW2-6A on seed development in wheat by negatively regulating gibberellin synthesis. Plant Sci. 263, 226–235. doi:10.1016/j.plantsci.2017.07.019
Liu, Y. C., Wu, Y. R., Huang, X. H., Sun, J., and Xie, Q. (2011). AtPUB19, a U-box E3 ubiquitin ligase, negatively regulates abscisic acid and drought responses in Arabidopsis thaliana. Mol. Plant 4, 938–946. doi:10.1093/mp/ssr030
Lu, X., Shu, N., Wang, D., Wang, J., Chen, X., Zhang, B., et al. (2020). Genome-wide identification and expression analysis of PUB genes in cotton. BMC Genomics 21, 213. doi:10.1186/s12864-020-6638-5
Lu, Y., Guan, N., and Li, C. (2008). Advance in the study on U-box proteins of Arabidopsis thaliana. Mol. Plant Breed. 6, 941–948. doi:10.1016/S1872-2067(08)60045-5
Luo, J., Shen, G., Yan, J., He, C., and Zhang, H. (2006). AtCHIP functions as an E3 ubiquitin ligase of protein phosphatase 2A subunits and alters plant response to abscisic acid treatment. Plant J. 46, 649–657. doi:10.1111/j.1365-313x.2006.02730.x
Lyzenga, W. J., and Stone, S. L. (2012). Abiotic stress tolerance mediated by protein ubiquitination. J. Exp. Bot. 63, 599–616. doi:10.1093/jxb/err310
Ma, A., Zhang, D., Wang, G., Wang, K., Li, Z., Gao, Y., et al. (2021). Verticillium dahliaeeffector VDAL protects MYB6 from degradation by interacting with PUB25 and PUB26 E3 ligases to enhance Verticillium wilt resistance. Plant Cell 33, 3675–3699. doi:10.1093/plcell/koab221
Mandal, A., Sharma, N., Muthamilarasan, M., and Prasad, M. (2018). Ubiquitination: a tool for plant adaptation to changing environments. Nucl. 61, 253–260. doi:10.1007/s13237-018-0255-6
March, E., and Farrona, S. (2018). Plant deubiquitinases and their role in the control of gene expression through modification of histones. Front. Plant Sci. 8, 2274. doi:10.3389/fpls.2017.02274
Muthamilarasan, M., and Prasad, M. (2014). Advances in Setaria genomics for genetic improvement of cereals and bioenergy grasses. Theor. Appl. Genet. 128, 1–14. doi:10.1007/s00122-014-2399-3
Ni, X., Tian, Z., Liu, J., Song, B., Li, J., Shi, X., et al. (2010). StPUB17, a novel potato UND/PUB/ARM repeat type gene, is associated with late blight resistance and NaCl stress. Plant Sci. 178, 158–169. doi:10.1016/j.plantsci.2009.12.002
Park, J., Yi, J., Yoon, J., Cho, L. H., Ping, J., Jeong, H. J., et al. (2011). OsPUB15, an E3 ubiquitin ligase, functions to reduce cellular oxidative stress during seedling establishment. Plant J. 65, 194–205. doi:10.1111/j.1365-313X.2010.04416.x
Qi, C. H., Zhao, X. Y., and Han, P. L. (2017). Functional identification of salt tolerance and ABA sensitivity of apple U-box E3 ubiquitin ligase MdPUB24. Acta Hortic. Sin. 44, 2255–2264. doi:10.16420/j.issn.0513-353x.2017-0430
Rogozin, I. B., Wolf, Y. I., Sorokin, A. V., Mirkin, B. G., and Koonin, E. V. (2003). Remarkable interkingdom conservation of intron positions and massive, lineage-specific intron loss and gain in eukaryotic evolution. Curr. Biol. 13, 1512–1517. doi:10.1016/s0960-9822(03)00558-x
Sadanandom, A. (2007). The U-box protein AtPUB17 is a functional ortholog of NtACRE276 and its E3 ubiquitin ligase activity is required for plant cell death and defence. Comp. Biochem. Physiol. 146, S203. doi:10.1016/j.cbpa.2007.01.450
Sankaranarayanan, S., and Samuel, M. A. (2015). A proposed role for selective autophagy in regulating auxin-dependent lateral root development under phosphate starvation in Arabidopsis. Plant Signal. Behav. 10, e989749. doi:10.4161/15592324.2014.989749
Santner, A., and Estelle, M. (2010). The ubiquitin-proteasome system regulates plant hormone signaling. Plant J. 61, 1029–1040. doi:10.1111/j.1365-313X.2010.04112.x
Seo, D. H., Ryu, M. Y., Jammes, F., Hwang, J. H., Turek, M., Kang, B. G., et al. (2012). Roles of four Arabidopsis U-box E3 ubiquitin ligases in negative regulation of abscisic acid-mediated drought stress responses. Plant Physiol. 160, 556–568. doi:10.1104/pp.112.202143
Sharma, B., and Taganna, J. (2020). Genome-wide analysis of the U-box E3 ubiquitin ligase enzyme gene family in tomato. Sci. Rep. 10, 9581. doi:10.1038/s41598-020-66553-1
Shrivastava, P., and Kumar, R. (2015). Soil salinity: a serious environmental issue and plant growth promoting bacteria as one of the tools for its alleviation. Saudi J. Biol. Sci. 22, 123–131. doi:10.1016/j.sjbs.2014.12.001
Stone, S. L. (2014). The role of ubiquitin and the 26S proteasome in plant abiotic stress signaling. Front. Plant Sci. 5, 135. doi:10.3389/fpls.2014.00135
Stone, S. L., Anderson, E. M., Mullen, R. T., and Goring, D. R. (2003). ARC1 is an E3 ubiquitin ligase and promotes the ubiquitination of proteins during the rejection of self-incompatible Brassica pollen. Plant Cell 15, 885–898. doi:10.1105/tpc.009845
Tang, X., Ghimire, S., Liu, W., Fu, X., Zhang, H., Sun, F., et al. (2022). Genome-wide identification of U-box genes and protein ubiquitination under PEG-induced drought stress in potato. Physiol. Plant 174, e13475. doi:10.1111/ppl.13475
Trujillo, M. (2018). News from the PUB: plant U-box type E3 ubiquitin ligases. J. Exp. Bot. 69, 371–384. doi:10.1093/jxb/erx411
Van, D., Andrew, P., and Christopher, P. H. (2022). Structural basis of ubiquitylation. Curr. Opin. Struc. Biol. 12, 822–830. doi:10.1016/s0959-440x(02)00389-5
Wang, N., Liu, Y., Cai, Y., Tang, J., Li, Y., and Gai, J. (2020a). The soybean U-box gene GmPUB6 regulates drought tolerance in Arabidopsis thaliana. Plant Physiol. bioch. 155, 284–296. doi:10.1016/j.plaphy.2020.07.016
Wang, N., Liu, Y., Cong, Y., Wang, T., Zhong, X., Yang, S., et al. (2016). Genome-wide identification of soybean U-box E3 ubiquitin ligases and roles of GmPUB8 in negative regulation of drought stress response in Arabidopsis. Plant Cell Physiol. 57 (6), 1189–1209. doi:10.1093/pcp/pcw068
Wang, W., Wang, W., Wu, Y., Li, Q., Zhang, G., Shi, R., et al. (2020b). The involvement of wheat U-box E3 ubiquitin ligase TaPUB1 in salt stress tolerance. J. Integr. Plant Biol. 62 (5), 631–651. doi:10.1111/jipb.12842
Wang, X., Zhang, X., Song, C.-P., Gong, Z., Yang, S., and Ding, Y. (2023). PUB25 and PUB26 dynamically modulate ICE1 stability via differential ubiquitination during cold stress in Arabidopsis. Plant Cell 35, 3585–3603. doi:10.1093/plcell/koad159
Wang, Z., Tian, X., Zhao, Q., Liu, Z., Li, X., Ren, Y., et al. (2018). The E3 ligase DROUGHT HYPERSENSITIVE negatively regulates cuticular wax biosynthesis by promoting the degradation of transcription factor ROC4 in rice. Plant Cell 30, 228–244. doi:10.1105/tpc.17.00823
Wiborg, J., O'Shea, C., and Skriver, K. (2008). Biochemical function of typical and variant Arabidopsis thaliana U-box E3 ubiquitin-protein ligases. Biochem. J. 413, 447–457. doi:10.1042/bj20071568
Woodson, J. D., Joens, M. S., Sinson, A. B., Gilkerson, J., Salomé, P. A., Weigel, D., et al. (2015). Ubiquitin facilitates a quality-control pathway that removes damaged chloroplasts. Sci 350, 450–454. doi:10.1126/science.aac7444
Yang, G., Ying, G., Wang, Z., Pan, W., Linghu, B., Pan, Y., et al. (2021). Genome-wide identification and expression analysis of U-box gene family in wild emmer wheat (Triticum turgidum L. ssp. dicoccoides). Gene 799, 145840. doi:10.1016/j.gene.2021.145840
Yang, Z., Zhang, H., Li, X., Shen, H., Gao, J., Hou, S., et al. (2020). A mini foxtail millet with an Arabidopsis-like life cycle as a C4 model system. Nat. Plants 6, 1167–1178. doi:10.1038/s41477-020-0747-7
Yang, Z. M., Song, J., Mo, X., Yang, H., Yue, L., and Mo, B. (2017). The U-box family genes in Medicago truncatula: key elements in response to salt, cold, and drought stresses. Plos One 12, e0182402. doi:10.1371/journal.pone.0182402
Ye, Q., Wang, H., Su, T., Wu, W.-H., and Chen, Y.-F. (2018). The ubiquitin E3 ligase PRU1 regulates WRKY6 degradation to modulate phosphate homeostasis in response to low-pi stress in Arabidopsis. Plant Cell 30, 1062–1076. doi:10.1105/tpc.17.00845
Yee, D., and Goring, D. R. (2009). The diversity of plant U-box E3 ubiquitin ligases: from upstream activators to downstream target substrates. J. Exp. Bot. 60, 1109–1121. doi:10.1093/jxb/ern369
You, Q., Zhai, K., Yang, D., Yang, W., Wu, J., Liu, J., et al. (2016). An E3 ubiquitin ligase-BAG protein module controls plant innate immunity and broad-spectrum disease resistance. Cell Host Microbe 20, 758–769. doi:10.1016/j.chom.2016.10.023
Zhang, M., Zhang, G. Q., Kang, H. H., Zhou, S. M., and Wang, W. (2017). TaPUB1, a putative E3 ligase gene from wheat, enhances salt stress tolerance in transgenic nicotiana benthamiana. Plant Cell Physiol. 58, 1673–1688. doi:10.1093/pcp/pcx101
Zhou, J., Hu, Y., Li, J., Yu, Z., and Guo, Q. (2021). Genome-Wide identification and expression analysis of the plant U-box protein gene family in Phyllostachys edulis. Front. Genet. 12, 710113. doi:10.3389/fgene.2021.710113
Keywords: foxtail millet, U-box, gene family, saline-alkali stress, qRT-PCR
Citation: Zhou X, Li Y, Wang J, Zhao Y, Wang H, Han Y and Lin X (2024) Genome-wide identification of U-box gene family and expression analysis in response to saline-alkali stress in foxtail millet (Setaria italica L. Beauv). Front. Genet. 15:1356807. doi: 10.3389/fgene.2024.1356807
Received: 16 December 2023; Accepted: 22 January 2024;
Published: 16 February 2024.
Edited by:
Yang Yang, Shanxi Agricultural University, ChinaReviewed by:
Vikas Sharma, Sant Baba Bhag Singh University, IndiaBian Jianxin, Peking University, China
Jia-Gang Wang, Shanxi Agricultural University, China
Copyright © 2024 Zhou, Li, Wang, Zhao, Wang, Han and Lin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yucui Han, eXVjdWloYW44NEAxNjMuY29t; Xiaohu Lin, eGlhb2h1bGluMjAwOEAxNjMuY29t
 Xiaoke Zhou1
Xiaoke Zhou1 Jian Wang
Jian Wang Yucui Han
Yucui Han