- 1Yantai Key Laboratory of Characteristic Agricultural Bioresource Conservation & Germplasm Innovative Utilization, College of Life Sciences, Yantai University, Yantai, China
- 2School of Life Sciences, Henan University, Kaifeng, Henan, China
- 3Institute of Cereal and Oil Crops, Hebei Academy of Agricultural and Forestry Sciences, Hebei Key Laboratory of Crop Genetics and Breeding, Shijiazhuang, China
- 4Crop Research Institute, Shandong Academy of Agricultural Sciences, Jinan, China
- 5Institute of Grain and Oil Crops, Yantai Academy of Agricultural Science, Yantai, China
Powdery mildew is one of the most severe diseases affecting wheat yield and quality and is caused by Blumeria graminis f. sp. tritici (Bgt). Host resistance is the preferred strategy to prevent this disease. However, the narrow genetic basis of common wheat has increased the demand for diversified germplasm resources against powdery mildew. Wheat relatives, especially the secondary gene pool of common wheat, are important gene donors in the genetic improvement of common wheat because of its abundant genetic variation and close kinship with wheat. In this study, a series of 137 wheat relatives, including 53 Triticum monococcum L. (2n = 2x = 14, AA), 6 T. urartu Thumanjan ex Gandilyan (2n = 2x = 14, AA), 9 T. timopheevii Zhuk. (2n = 4x = 28, AAGG), 66 T. aestivum subsp. spelta (2n = 6x = 42, AABBDD), and 3 Aegilops speltoides (2n = 2x = 14, SS) were systematically evaluated for their powdery mildew resistance and composition of Pm genes. Out of 137 (60.58%) accessions, 83 were resistant to Bgt isolate E09 at the seedling stage, and 116 of 137 (84.67%) wheat relatives were resistant to the mixture of Bgt isolates at the adult stage. This indicates that these accessions show a high level of resistance to powdery mildew. Some 31 markers for 23 known Pm genes were used to test these 137 accessions, and, in the results, only Pm2, Pm4, Pm6, Pm58, and Pm68 were detected. Among them, three Pm4 alleles (Pm4a, Pm4b, and Pm4f) were identified in 4 T. subsp. spelta accessions. q-RT PCR further confirmed that Pm4 alleles played a role in disease resistance in these four accessions. The phylogenetic tree showed that the kinship of Pm4 was close to Pm24 and Sr62. This study not only provides reference information and valuable germplasm resources for breeding new wheat varieties with disease resistance but also lays a foundation for enriching the genetic basis of wheat resistance to powdery mildew.
Introduction
Wheat (Triticum aestivum L.) is one of the foremost staple crops in the world, contributing significantly to global food security (Igrejas and Branlard, 2020). However, its productivity is consistently threatened by myriad biotic and abiotic factors, among which wheat powdery mildew is prominent. Powdery mildew, caused by Blumeria graminis f. sp. tritici (Bgt), primarily affects wheat leaves by severely impairing photosynthesis, nutrient uptake, and overall yield, leading to diminished grain quality and quantity. In severe cases, yield losses can reach up to 40% (Savary et al., 2019). Traditionally, the management of wheat powdery mildew has relied on chemical pesticides, which present challenges such as environmental pollution and high costs. Therefore, more environmentally friendly, economical, and sustainable approaches are needed to control wheat powdery mildew. The development of wheat cultivars with inherent resistance to powdery mildew has emerged as a pivotal solution to combat this issue.
Since the first cataloged wheat powdery mildew resistance (Pm) gene Pm1 was identified in 1952, 69 formally cataloged Pm genes have been successively reported in the primary, secondary, and tertiary gene banks of wheat (McIntosh et al., 2020; He et al., 2021; Li et al., 2023). Among them, the 13 Pm genes Pm1-5, Pm8, Pm12, Pm17, Pm21, Pm24, Pm41, Pm60, and Pm69, and two multiple resistance loci Pm38/Yr18/Lr34/Sr57/Ltn1/Sb1 and Pm46/Yr46/Lr67/Sr55/Ltn3 have been cloned (Krattinger et al., 2009; Brunner et al., 2010; Hurni et al., 2013; Moore et al., 2015; Sánchez-Martín et al., 2016; Singh et al., 2018; Xing et al., 2018; Zou et al., 2018; Li et al., 2020; Lu et al., 2020; Xie et al., 2020; Hewitt et al., 2021; Sánchez-Martín et al., 2021; Li et al., 2023; Zhu et al., 2023). Although so many Pm genes have been reported, most of them cannot be directly used for production because of linkage drag or other bad agronomic traits, such as Pm55 derived from Dasypyrum villosum L. and Pm56 derived from rye (Secale cereale L.) (Zhang et al., 2016; Hao et al., 2018). Therefore, a long-term goal for fighting this disease is to mine more novel genes against powdery mildew and apply them in wheat breeding projects.
As an allohexaploid species, common wheat has three subgenomes (2n = 6x = AABBDD) derived from three wheat relatives over the long process of domestication. The wheat relatives constitute one or two subgenomes of wheat which constitute the secondary gene banks for wheat improvement. In modern wheat breeding, due to long-term artificial selection, the primary gene pool of common wheat has been fully developed and utilized, making it difficult to breed new breakthrough wheat cultivars. In comparison with primary gene banks, secondary gene banks may have larger genetic variation. Compared to tertiary gene banks, genes derived from secondary gene banks can be more fully recombined into the wheat genome, avoiding the problem of poor genetic compensation which often occurs in breeding utilization of tertiary gene sources in the form of translocations. Therefore, the unique characteristics of secondary gene sources make it easier to achieve a balance between comprehensive agronomic traits and disease resistance in production. For example, Pm4 from T. monococcum L. (2n = 2x = 14, AA), Pm6 from T. timopheevii Zhuk (2n = 4x = 28, AAGG), and Pm50 from T. dicoccum L. (2n = 4x = 28, AABB) were all introduced into common wheat, significantly improving wheat resistance to powdery mildew (Mohler et al., 2013; Wan et al., 2020; Sanchez-Martin et al., 2021). The Aegilops and Triticum genera are quite similar in genetic relationship and are easy to cross with common wheat. They are also widely used in the genetic improvement of wheat. For example, Pm13 on chromosome 3Sl of Ae. longissima has been applied in wheat disease resistance breeding (Zhang et al., 2014). Ae. speltoides (2n = 2x = 14, SS) is a donor of tetraploid and hexaploid. Pm12, Pm32, and Pm53 derived from Ae. speltoides, which have also been introduced into wheat’s genetic background (Hsam et al., 2003; Petersen et al., 2015; Zhu et al., 2023).
Accurate identification of these resistance genes in specific genotypes is a key step in rapidly and effectively applying them in wheat breeding programs. Over recent decades, traditional breeding methods have developed some resistant wheat varieties, but they demand extensive time and effort. However, recent progress in molecular biology and molecular marker technology has provided novel tools and strategies in breeding for wheat disease resistance. Molecular marker detection is a technique based on wheat genomic DNA that enables the precise and efficient identification and selection of wheat plants that carry resistance genes. Since a growing number of resistance genes have been identified and cloned, and functional markers, which have been developed based on polymorphic SNPs/Indels within their full-length sequences, could directly detect genotype variations accurately in practical application (Liu et al., 2012). For resistance genes that have not been cloned, such as Pm6, Pm58, and Pm68, closely linked markers are the preferred method of detecting them, (Ji et al., 2008; He et al., 2021; Xue et al., 2022). Previous research has identified the resistance phenotypes and gene compositions in various germplasm resources. For instance, Gebrewahid et al. (2020) identified the leaf rust resistance and Lr genes in 50 bread wheat cultivars derived from Ethiopia by molecular marker analysis.
In our lab, 137 wheat relatives, including 53 T. monococcum L. (2n = 2x = 14, AA), 6 T. urartu Thumanjan ex Gandilyan (2n = 2x = 14, AA), 9 T. timopheevii Zhuk. (2n = 4x = 28, AAGG), 66 T. aestivum subsp. spelta (2n = 6x = 42, AABBDD), and 3 Ae. speltoides (2n = 2x = 14, SS) were provided by the International Maize and Wheat Improvement Center (CIMMYT). To systematically evaluate the powdery mildew resistance and explore elite Pm genes/alleles in these germplasms, we 1) investigated their powdery mildew resistance at the seedling and adult stages, 2) identified the composition of Pm genes by molecular marker detection, 3) analyzed the haplotype of the cloned Pm genes in these accessions, and 4) explored the relationship between Pm genes and disease resistance.
Materials and methods
Plant materials and pathogens
A total of 137 wheat relatives, including 53 T. monococcum L. (2n = 2x = 14, AA), 6 T. urartu Thumanjan ex Gandilyan (2n = 2x = 14, AA), 9 T. timopheevii Zhuk (2n = 4x = 28, AAGG), 66 T. aestivum subsp. spelta (2n = 6x = 42, AABBDD), and 3 Ae. speltoides (2n = 2x = 14, SS) were provided by CIMMYT and used to test their reaction patterns to the Bgt isolate E09 (Supplementary Table S1). The wheat cultivar Tainong 18 has been identified as being susceptible to all tested Bgt isolates, so it was used as the susceptible check in phenotype identification. Ten extra single spore-derived Bgt isolates—F01, F06, F19, F21, F26, F28, E17, E18, E21, and E23-1—with different avirulent/virulent patterns were used for testing against the reaction patterns of E09-resistant accessions. These isolates were preserved on the susceptible Tainong 18 seedlings which were placed in independent glass tubes. To avoid cross infection, these tubes were covered with three layers of gauze.
Resistance identification to powdery mildew at the seedling stage
The 137 wheat relatives were first evaluated for their resistance to the Bgt isolate E09, a moderate virulent isolate mainly in the wheat production regions of North China. The resistant genotypes were further tested for their resistance to ten extra Bgt isolates. The reaction patterns of the tested accessions were determined in the greenhouse of Yantai University, Yantai, China. Five to eight seeds of each accession were sown in a 72-cell (4.0 × 4.0 × 4.2 cm) rectangular tray (54 × 28 × 4.2 cm). In the sowing design, Tainong 18, as the susceptible check, was planted randomly with three cells in each tray. These trays were placed into separate growth chambers with a daily cycle of 22°C/14 h/light and 18°C/10 h/darkness and were infected with different virulent Bgt isolates. Two weeks later, when the seedlings had grown to the one-leaf and one-heart stage, they were inoculated with fresh conidiospores propagated on Tainong 18 seedlings and cultured in conditions of 18°C/24 h/darkness with 100% humidity and then cultivated with a daily cycle of 22°C/14 h/light and 18°C/10 h/darkness. About 2 weeks later, when the spores had fully spread on the first leaf of Tainong 18, infection types (ITs) on each seedling were estimated based on a 0–4 scale. The evaluation standard was described as per Si et al. (1992). Among them, ITs 0, 1, and 2 were regarded as resistant genotypes, and ITs 3 and 4 as susceptible genotypes. All experimental procedures were repeated three times to ensure the accuracy and reliability of the data.
Resistance identification to powdery mildew at the adult stage
The 137 wheat relatives were sown in the experiment field at Yantai University (Yantai, China, 121.39′E, 37.52′N) to evaluate their resistance to powdery mildew at the adult stage in 2021–2023. In mid-October of each year, ten seeds per accession were bunch-planted, with a 0.30-m space between bunches. Tainong 18, as the susceptible check, was sown around the plots to promote the spread of spores. In early April of the next year, when the temperature had successively risen above 10°C, Tainong 18 was inoculated with a mixture of Bgt isolates (including E09, F01, F06, F19, F21, F26, F28, E17, E18, E21, and E23-1). They were then irrigated to maintain humidity to promote the onset of powdery mildew. In early June, when Tainong 18 was highly susceptible, ITs of the tested genotypes were assessed and recorded twice at an interval of 7–10 days on the 0–9 scale, in which ITs 0–2 were regarded as highly resistant, ITs 3 and 4 as moderately resistant, ITs 5 and IT 6 as moderately susceptible, and ITs 7–9 as highly susceptible (Saari and Prescott, 1975).
Molecular marker detection
Genomic DNA was isolated from the young seedlings of the tested genotypes using the protocol in Sharp et al. (1988) with minor modification. Specific primers of 23 reported Pm genes (Pm1-5, Pm6, Pm8, Pm12, Pm21, Pm24, Pm35, Pm37, Pm41, Pm42, Pm45, Pm47, Pm52, Pm58, Pm59, Pm60, Pm61, Pm68, and Pm69) were used to test all 137 wheat accessions in this study (Supplementary Table S2). PCR amplification was performed according to Jin et al. (2023). PCR products of most primers were detected in 8% non-denaturing polyacrylamide gels with 19:1, 29:1, or 39:1 ratios of acrylamide and bis-acrylamide, and were silver stained (Santos et al., 1993). The rest of the PCR products were separated by agarose gel with a concentration of 1.5% with nucleic acid dyestuff ultra GelRed (Vazyme Biotech Co., Ltd. China), and were then observed using the Gel Documentation System (Gel Doc XR+, BIO-RAD, Hercules, CA, United States) (Gebrewahid et al., 2020). See Supplementary Table S2 for the marker information and corresponding positive control of tested Pm genes.
Homologous cloning and sequence analysis
Total RNA was isolated from the young leaf tissues using TRIzol reagent (Invitrogen, United States) following product instructions. Residual DNA was then removed, and the corresponding cDNA was synthesized using the FastKing gDNA Dispelling RT SuperMix kit (Tiangen) thus: a 10-µL reaction mixture containing 50 ng–2 µg RNA, 2 µL 5× gDNA buffer and ddH2O to 10 μL, 42°C for 3 min, then adding 2 µL 10× King RT buffer, 1 µL FastKing RT Enzyme Mix, 2 µL FQ-RT Primer Mix and ddH2O to 10 μL, 42°C for 15 min, 95°C for 3 min. The Pm4 alleles were first cloned using primers GH398/GH399, GH400/GH401, GH398/GH407, and GH407/GH400 by nested PCR (Supplementary Table S2) and were then sequenced by Sanger sequencing (Sangon Biotech) and compared to the reported Pm4a-4e (GenBank, Pm4b_V1: MT783929, Pm4b_V2: MT783930) alleles with Seqman software (DNASTAR, Madison, American). For the new haplotypes, the DNA sequences were translated to amino acids with Editseq software (DNASTAR, Madison, American) for further analysis. The cDNA sequences of cloned disease-resistant genes were taken from the National Centre for Biotechnology Information (NCBI) database and were used for analysis by constructing the phylogenetic tree using the neighbor-joining method with the Poisson model in the MEGA7 software (Kumar et al., 2016).
qRT-PCR analysis
The qRT-PCR procedure was performed using ChamQ universal SYBR qPCR Master Mix (Vazyme, China) on the Roche LightCycler® 480 II real-time PCR system (Roche, Switzerland) with the specific primers Pm4-qPCR and TaActin. The expression pattern of each gene was calculated as a fold change using the comparative CT method (Livak and Schmittgen, 2001). Three biological replicates and three technical replications were performed to ensure data reliability. The TaActin gene was used as the standardized internal control.
Results
Phenotype identification against Bgt isolates at the seedling stage
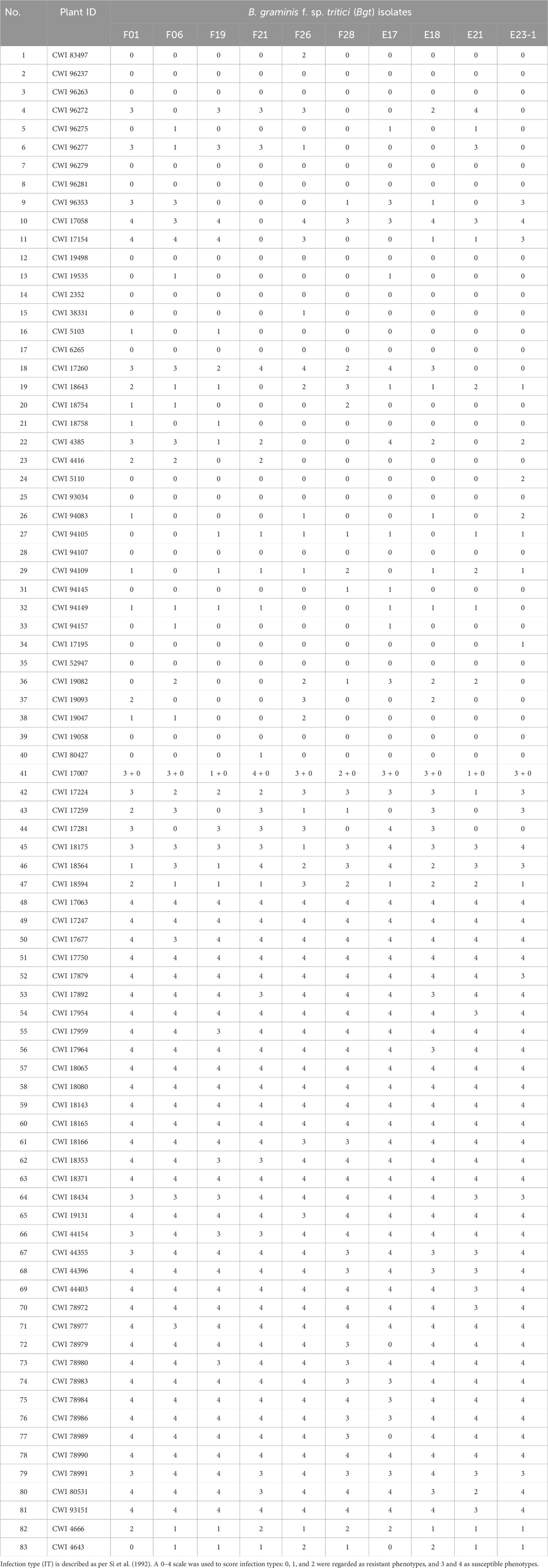
When inoculated with the Bgt isolate E09, 83 of 137 (60.6%) wheat relatives, including 34 out of 53 T. monococcum L., 5 of 6 T. urartu Thumanjan ex Gandilyan, 8 of 9 T. timopheevii Zhuk., 34 of 66 T. aestivum subsp. spelta, and 2 of 3 Ae. speltoides were resistant with ITs 0–2, whereas the remaining 54 (39.4%) were susceptible with ITs 3–4 (Figure 1A, Supplementary Table S1). The proportion of the resistant genotypes was high, and these accessions were expected to be valuable gene donors for wheat improvement against powdery mildew. Notably, among 83 resistant genotypes, 70 showed immunity with IT 0, suggesting their significant resistance against powdery mildew. Then, those 83 E09-resistant genotypes were tested on the resistance spectrum with ten extra Bgt isolates. The results showed that they have different reaction patterns to these ten Bgt isolates (Figure 2; Table 1). A total of 32 accessions showed resistance to the ten Bgt isolates tested, such as CWI 18477, CWI 96263, CWI 96237, CWI 96281, and CWI 96275; four were resistant to nine of the ten tested Bgt isolates; 70 showed resistance to tested Bgt isolates which were diversified from one to eight. However, 31 accessions were susceptible to all these Bgt isolates (Table 1). This suggests that these wheat relatives have abundant genetic diversity in responding to powdery mildew.
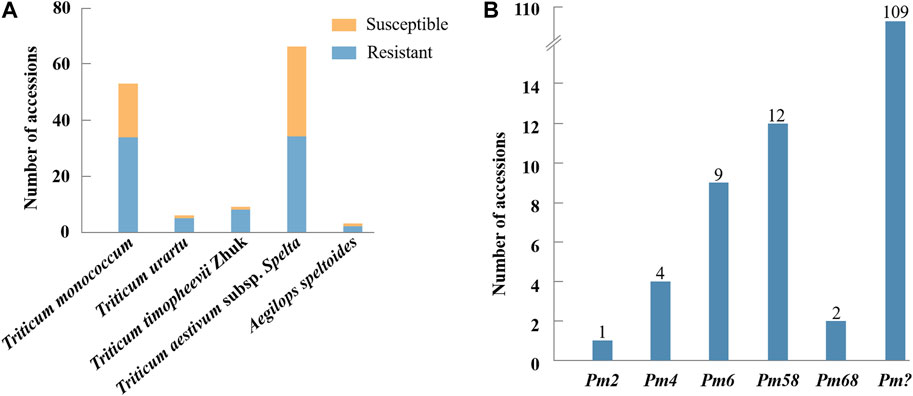
FIGURE 1. Resistance assessment of 137 wheat relatives to Blumeria graminis f. sp. tritici isolate E09 at the seedling stage (A). Molecular marker detection of 137 wheat relatives (B). “?” indicates untested Pm genes in the present study.
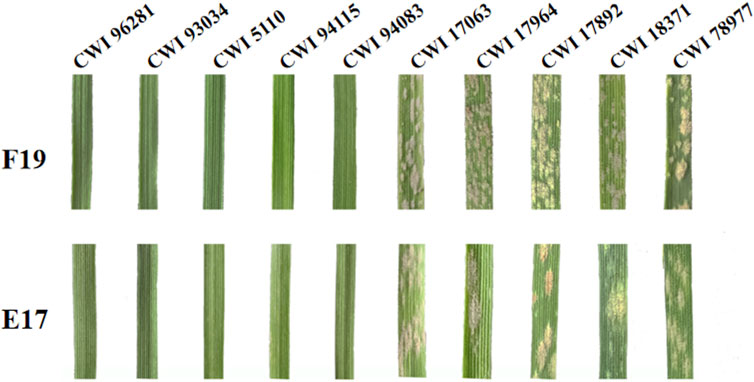
FIGURE 2. Reaction patterns of T. monococcum L. (CWI 96281, CWI 93034, CWI 5110, CWI 94115, and CWI 94083) and T. aestivum subsp. spelta (CWI 17063, CWI 17964, CWI 17892, CWI 18371, and CWI 78977) with powdery mildew B. graminis f. sp. tritici (Bgt) isolates F19 and E17.
Phenotype identification against Bgt isolates at the adult stage
The 137 wheat relatives were sown in the field at the adult stage from 2021 to 2023 to determine the resistance to powdery mildew of the mixture of Bgt isolates E09, F01, F06, F19, F21, F26, F28, E17, E18, E21, and E23-1. The susceptible control Tainong 18 was highly susceptible in the two cropping seasons. The results demonstrated that most of the tested accessions showed resistance to the powdery mildew, including 103 that were highly resistant and 13 moderately resistant. The remaining 21 wheat relatives were susceptible (Supplementary Table S1). Of these, most of the T. timopheevii Zhuk., T. aestivum subsp. spelta, and Ae. speltoides were resistant, suggesting that they were valuable in wheat disease resistance breeding.
Molecular marker identification
The functional markers of 12 cloned Pm genes Pm1-5, Pm8, Pm12, Pm21, Pm24, Pm41, Pm60, and Pm69, and closely linked or co-segregated markers of the 11 other Pm genes Pm6, Pm35, Pm37, Pm42, Pm45, Pm47, Pm52, Pm58, Pm59, Pm61, and Pm68 were used to test 137 wheat relatives to detect the absence/presence of their alleles. The results showed that Pm58 was detected in 12 T. aestivum subsp. Spelta with the highest frequency, including CWI 17370, CWI 18434, CWI 18439, CWI 18475, CWI 18476, CWI 44154, CWI 44215, CWI 44218, CWI 44409, CWI 80462, CWI 80503, and CWI 92915. Pm6, which was introgressed into common wheat from chromosome 2G of T. timopheevii, was detected in 9 T. timopheevii Zhuk. Pm4, Pm68, and Pm2 were detected in 4, 2, and 1 T. aestivum subsp. spelta accession, respectively (Figure 2B, Supplementary Table S3). The remaining markers did not produce specific fragments in any of the 137 wheat relatives, suggesting that these accessions most likely did not carry these genes (Figure 3). Notably, all these markers did not produce specific target bands in 109 out of 137 tested wheat accessions, suggesting that these accessions might carry other novel genes or gene combinations (Pm?) (Figure 2B).
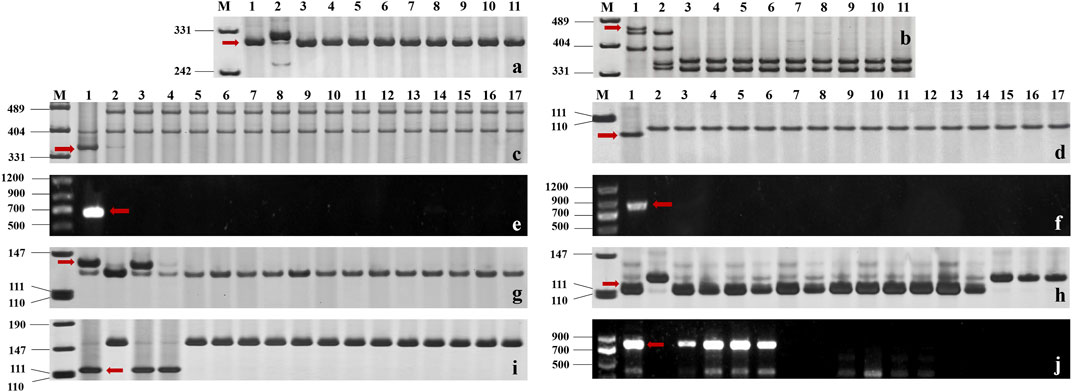
FIGURE 3. (A, B) Amplification patterns of specific markers CIT02g-18 for Pm6 (A) and Pm37-82 for Pm37 (B) in part of T. timopheevii Zhuk. M: pUC19/Msp I; Lane 1: Coker747 (A) and 66 (B); Lane 2: ChineseSpring; and Lanes 3–11: CWI 80427, CWI 80540, CWI 17007, CWI 17224, CWI 17259, CWI 17281, CWI 18175, CWI 18564, and CWI 18594. (C–F) Amplification patterns of specific markers MBH1 for Pm12 (C), STS-Pm24 for Pm24 (D), sfr43 for Pm8 (E), and Pm60-S1 for Pm60 (F) in part of T. monococcum L. M: pUC19/Msp I (C–D), DNA marker DL1200 (E–F); Lane 1: Yangmai 5 (C), Chiyacao (D), Disponent (E), and EK516 (F); Lane 2: ChineseSpring; and Lanes 3–17: CWI 96237, CWI 96263, CWI 96272, CWI 96275, CWI 17058, CWI 17154, CWI 18949, CWI 19498, CWI 19529, CWI 4393, CWI 4416, CWI 5110, CWI 93034, CWI 93289, and CWI 94083. (G–J): Amplification patterns of specific markers Pm2b-map-3 for Pm2 (G), Xsts24035 for Pm58 (H), Xdw12 for Pm68 (I), and JS717/718 for Pm4 (J) in part of T. aestivum subsp. spelta. M: pUC19/Msp I (G–J), DNA marker DL1200 (J); Lane 1: KM2939 (G), AY40 (H), TRI 1796 (I), and VPM1 (J); Lane 2: ChineseSpring; Lanes 3–17 (G): CWI 18468, CWI 14929, CWI 17063, CWI 17247, CWI 17324, CWI 17370, CWI 17677, CWI 17750, CWI 17879, CWI 17892, CWI 17942, CWI 17942, CWI 17954, CWI 17959, and CWI 17964; Lanes 3–17 (H): CWI 17370, CWI 18434, CWI 18439, CWI 18475, CWI 18476, CWI 44154, CWI 44215, CWI 44218, CWI 44409, CWI 80462, CWI 80503, CWI 92915, CWI 96275, CWI 96277, and CWI 96279; Lanes 3–17 (I): CWI 18434, CWI 18439, CWI 14929, CWI 17063, CWI 17247, CWI 17324, CWI 17370, CWI 17677, CWI 17750, CWI 17879, CWI 17892, CWI 17942, CWI 17942, CWI 17954, and CWI 17959; and Lanes 3–17 (J): CWI 18477, CWI 78977, CWI 78976, CWI 80531, CWI 14929, CWI 17063, CWI 17247, CWI 17324, CWI 17370, CWI 17677, CWI 17750, CWI 17879, CWI 17892, CWI 17942, and CWI 17942. Red arrows indicate specific bands.
Homologous cloning and sequence analysis
Only the cloned genes Pm2 and Pm4 were detected in the marker detection. Previous studies reported that Pm2 in the released rartuidy wheat cultivars were all of the Pm2a haplotype (Yu et al., 2022). Therefore, we focus on the haplotype identification of Pm4 locus in this study. Using the nest PCR, we cloned the Pm4 alleles from the four putative Pm4-carrying T. subsp. spelta accessions. Sequence alignment suggested that CWI 18477 carried Pm4a, CWI 78976 and CWI 78977 carried Pm4b, and CWI 80531 carried Pm4f (Figure 4). The alleles of Pm4c, Pm4d, Pm4e, Pm4g, and Pm4h were not detected. Additionally, comparison of the Pm4 CDS sequence with a series of 54 cloned disease-resistant genes showed that Pm4 was not related to the NLR genes but close to Pm24 and Sr62, which included tandem kinase domain (Figure 5).
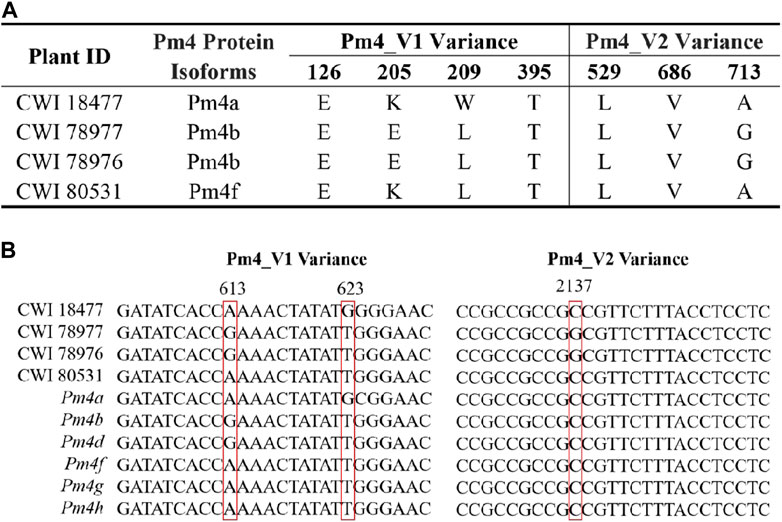
FIGURE 4. Protein sequence comparison of the Pm4 variants (A) and sequence alignments of CWI 1877, CWI 78977, CWI 78976, CWI 80531, and Pm4 alleles (B). Red box indicates nucleotide variant sites.
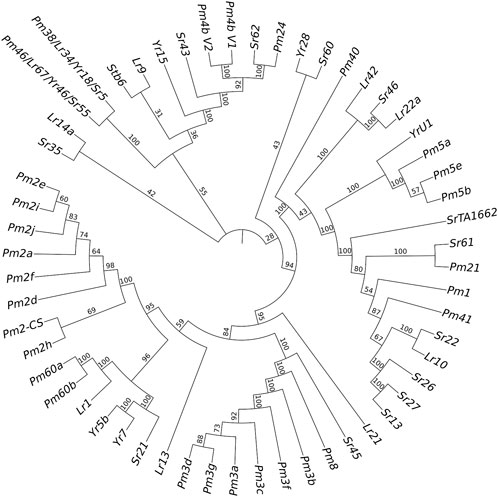
FIGURE 5. Phylogenetic tree construction results of powdery mildew (Pm), stem rust (Sr), leaf rust (Lr), and yellow rust (Yr) resistant genes.
Expression pattern analysis of Pm4 alleles. To identify the relationship between powdery mildew resistance and gene transcription levels, we used qRT-PCR to monitor the expression patterns of Pm4 alleles in those four putative Pm4-carrying T. aestivum subsp. spelta accessions after inoculating with Bgt isolate E09 at different times. As in Figure 6, the expression levels of all Pm4 alleles were significantly upregulated after inoculation. It is supposed that Pm4 alleles played a key role in disease resistance in these wheat genetic backgrounds. In addition, no significant differences between Pm4-V1 and Pm4-V2 were observed most of the time after inoculation, suggesting both two transcript splices were important.
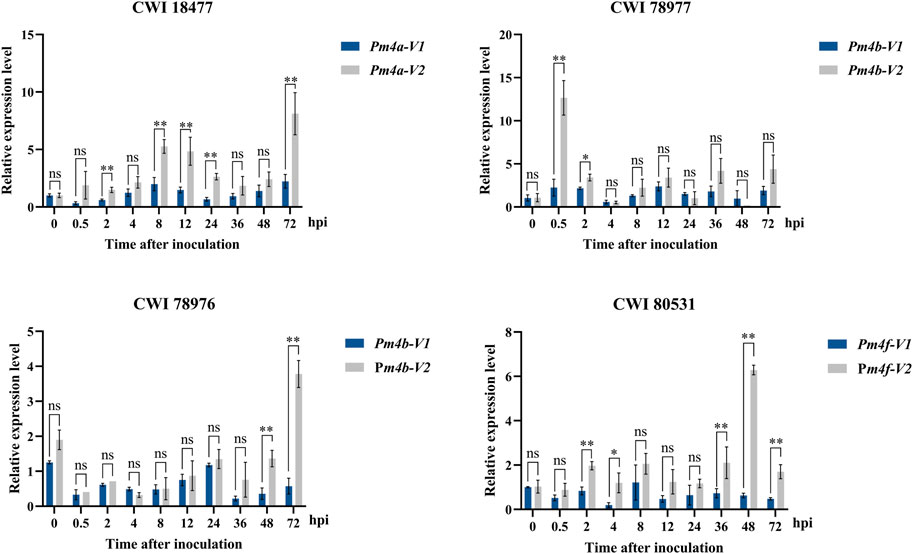
FIGURE 6. Expression patterns of the Pm4-V1 and Pm4-V2 splice variants in wheat CWI 18477, CWI 78977, CWI 78976, and CWI 80531 during a 72-h time course. Error bars represent SD based on three biological replicates. Statistical analysis used a t-test at p < 0.05 (*p < 0.05, **p < 0.01, ns: not significant). TaActin was used as the internal control.
Discussion
Wheat contributes to the diet of approximately 35% of the world’s population and is of vital importance in guaranteeing global food security. Powdery mildew threatens wheat yield and quality. Fortunately, some common wheat accessions and their relatives carry genes that allow resistance to powdery mildew during the growing period. The narrow genetic basis of common wheat has expanded the demand for diversified germplasm resources against powdery mildew. In general, the genes that were derived from wild relatives of common wheat cannot be directly applied to wheat breeding due to the undesirable linkage drags or poor agronomic traits of the donors, such as rye (S. cereale L, 2n = 2x = 14, RR) and Agropyron cristatum (2n = 2x = 14, VV) (Zhu et al., 2023). By comparison, the secondary gene bank of common wheat is an important gene donor in the genetic improvement of common wheat because of its abundant genetic variation and close kinship with wheat. They thus make it easier to balance disease resistance and comprehensive agronomic traits. In this study, a series of 137 wheat relatives, including 53 T. monococcum L., 6 T. rartu Thumanjan ex Gandilyan, 9 T. timopheevii Zhuk., 66 T. aestivum subsp. spelta, and 3 Ae. Speltoides, were systematically evaluated for their powdery mildew resistance and composition of Pm genes.
T. monococcum L. and T. rartu Thumanjan ex Gandilyan are diploid wild ancestral species of common wheat. They are also valuable germplasms for wheat improvement. Previous studies reported that T. rartu contained more abundant nucleotide-binding and leucine-rich repeat domain (NB-LRR) genes than maize, sorghum, or rice, making it a good source for Pm resistance genes (Ling et al., 2013). Zou et al. (2018) identified a new Pm gene, Pm60, from the resistant T. urartu variety PI428309 using the strategies of genetic mapping and RNA-sequencing. Another study also showed that a Pm-resistant locus in T. urartu was mapped to chromosome arm 7AL in a similar position to Pm60 (Qiu et al., 2005). Additionally, T. monococcum L. is also an important donor of Pm genes. Several Pm genes, such as Pm1b (Hu et al., 1997), Mlm80 (Yao et al., 2007), Mlm 2033 (Yao et al., 2007), pm 2026 (Xu et al., 2008), Pm4d (Schmolke et al., 2012), and Pm25 (Shi et al., 1998), were identified in T. monococcum L. In our study, 34 out of 53 T. monococcum L. (64.15%) and five of six T. urartu Thumanjan ex Gandilyan (83.33%) were resistant to Bgt isolate E09 at the seedling stage. Fortunately, 52 accessions were resistant to the mixture Bgt isolates at the adult stage. After marker detection, all 57 accessions carried none of the 23 tested Pm genes. We speculate that there may be other unknown Pm genes in these accessions, which provide potential new Pm genes to be mined.
Ae. speltoides is the closest extant relative of the wheat B subgenome which provides resistance to multiple diseases (Luo et al., 2005). To date, Pm12, Pm32, and Pm53 derived from Ae. speltoides have also been introduced into the wheat genetic background. In our study, Ae. speltoides accessions CWI 4643 and CWI 4666 from Iraq were resistant to all ten Bgt isolates but did not carry any of the tested Pm genes, indicating their potential value in resistance breeding for wheat. Additionally, Ae. speltoides has also served as a resistance source against leaf rust, including Lr28, Lr35, Lr36, Lr47, and Lr51 (Helguera et al., 2005), and stem rust, including Sr32, Sr39, Sr47, and SrAes7t (Klindworth et al., 2012). More than 200 Ae. speltoides accessions collected from five areas of Israel showed high resistance to stem rust, leaf rust, and stripe rust in both Israel and Minnesota, USA (Anikster et al., 2005). It is therefore likely that additional genes for disease resistance for use in wheat breeding could be found in Ae. speltoides.
Pm4 is an important Pm gene that has already been widely distributed across parts of China and other countries because of its resistance to Bgt isolates (Wang et al., 2005). Eight alleles (Pm4a–Pm4h) at the Pm4 locus have been successively reported (Sánchez-Martín et al., 2021). Sánchez-Martín et al. (2021) recently cloned Pm4b using the MutChromSeq strategy. The functional marker JS717/JS718 was designed and could be used to diagnose Pm4 alleles. In our study, Pm4 was detected in 4 T. aestivum subsp. spelta accessions. After homologous cloning and sequence alignment, CWI 18477 carried Pm4a, CWI 78976 and CWI 78977 carried Pm4b, and CWI 80531 carried Pm4f. The alleles of Pm4c, Pm4d, Pm4e, Pm4g, and Pm4h were not detected. Curiously, CWI 78976 and CWI 78977 carried Pm4b but exhibited contrasting levels of resistance to Bgt isolate E09. This cannot be explained solely by the differences in genetic backgrounds; there may be some other unknown Pm genes interacting with each other to provide potential resistance.
Notably, the diagnostic markers of 23 tested Pm genes did not produce specific target bands in 109 of 137 tested wheat accessions, suggesting that these accessions might carry other novel genes or gene combinations. There is therefore an urgent need to further analyze the resistant gene(s) in those accessions that did not carry tested Pm gene(s) by fine mapping and other technologies. Except for Pm genes, wheat relatives contain abundant yield, stress resistance, and resistant genes. Therefore, these germplasm resources can be better applied in wheat resistance breeding (Wang et al., 2023).
In summary, this study evaluated the powdery mildew resistance and composition of Pm genes of 137 wheat relatives. The results provided reference information and germplasms for wheat improvement against powdery mildew.
Conclusion
A total of 137 wheat relatives, including 53 T. monococcum, 6 Triticum urartu, 9 T. timopheevii Zhuk., 66 T. aestivum subsp. spelta, and 3 Ae. speltoides, were systematically evaluated their powdery mildew resistance and composition of Pm genes. Our study provided reference information and abundant gene resources for wheat resistance molecular breeding.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding authors.
Author contributions
JW, HX, YQ, RH, XS, YZ, BX, ZQ, XH, YJ, RL, and JZ: writing–original draft. CL and PM: writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was financially supported by the National Natural Science Foundation of China (32072053) and (32301923), the Key Research and Development Project of Shandong Province (2022LZG002-4), the Co-construction State Key Laboratory of Wheat and Maize Crop Science (SKL2022KF09), the Key Research and Development Project of Yantai City (2023ZDCX023), and the Wheat Industrial Technology System of Shandong Province.
Acknowledgments
We are grateful to Prof. Yilin Zhou, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, for providing the donors with the known Pm genes and Blumeria graminis f. sp. tritici isolates.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were editorial board members of Frontiers at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2024.1342239/full#supplementary-material
References
Anikster, Y., Manisterski, J., and Leonard, K. J. (2005). Resistance to leaf rust, stripe rust, and stem rust in Aegilops spp. in Israel. Plant Dis. 89, 303–308. doi:10.1094/PD-89-0303
Bie, T. D., Zhao, R. H., Zhu, S. Y., Chen, S. L., Cen, B., Zhang, B. Q., et al. (2015). Development and characterization of marker MBH1 simultaneously tagging genes Pm21 and PmV conferring resistance to powdery mildew in wheat. Mol. Breed. 35, 189–198. doi:10.1007/s11032-015-0385-3
Brunner, S., Hurni, S., Streckeisen, P., Mayr, G., Albrecht, M., Yahiaoui, N., et al. (2010). Intragenic allele pyramiding combines different specificitiesof wheat Pm3 resistance alleles. Plant J. 64, 433–445. doi:10.1111/j.1365-313X.2010.04342.x
Gebrewahid, T. W., Zhang, P. P., Yao, Z. J., Li, Z. F., and Li, Z. F. (2020). Identification of leaf rust resistance genes in bread wheat cultivars from Ethiopia. Plant Dis. 104, 2354–2361. doi:10.1094/PDIS-12-19-2606-RE
Hao, M., Liu, M., Luo, J. T., Fan, C. L., Yi, Y. J., Zhang, L. Q., et al. (2018). Introgression of powdery mildew resistance gene Pm56 on rye chromosome arm 6RS into wheat. Front. Plant Sci. 9, 1040. doi:10.3389/fpls.2018.01040
He, H. G., Liu, R. K., Ma, P. T., Du, H. N., Zhang, H. H., Wu, Q. H., et al. (2021). Characterization of Pm68, a new powdery mildew resistance gene on chromosome 2BS of Greek durum wheat TRI 1796. Theor. Appl. Genet. 134, 53–62. doi:10.1007/s00122-020-03681-2
Helguera, M., Vanzetti, L., Soria, M., Khan, I. A., Kolmer, J., and Dubcovsky, J. (2005). PCR markers for Triticum speltoides leaf rust resistance gene Lr51 and their use to develop isogenic hard red spring wheat lines. Crop Sci. 45, 728–734. doi:10.2135/cropsci2005.0728
Hewitt, T., Müller, M. C., Molnár, I., Mascher, M., Holušová, K., Šimková, H., et al. (2021). A highly differentiated region of wheat chromosome 7AL encodes a Pm1a immune receptor that recognizes its corresponding AvrPm1a effector from Blumeria graminis. New Phytol. 229, 2812–2826. doi:10.1111/nph.17075
Hsam, S. L. K., Lapochkina, I. F., and Zeller, F. J. (2003). Chromosomal location of genes for resistance to powdery mildew in common wheat (Triticum aestivum L. em Thell.) 8 Gene Pm32 in a wheat-Aegilops speltoides translocation line. Euphytica 133, 367–370. doi:10.1023/A:1025738513638
Hu, X. Y., Ohm, H. W., and Dweikat, I. (1997). Identification of RAPD markers linked to the gene Pm1 for resistance to powdery mildew in wheat. Theor. Appl. Genet. 94, 832–840. doi:10.1007/s001220050484
Hua, W., Liu, Z. J., Zhu, J., Xie, C. J., Yang, T. M., Zhou, Y. L., et al. (2009). Identification and genetic mapping of Pm42, a new recessive wheat powdery resistance gene derived from wild emmer (Triticum turgidum var. discoccoides). Theor. Appl. Genet. 119, 223–230. doi:10.1007/s00122-009-1031-4
Hurni, S., Brunner, S., Buchmann, G., Herren, G., Jordan, T., Krukowski, P., et al. (2013). Rye Pm8 and wheat Pm3 are orthologous genes and show evolutionary conservation of resistance function against powdery mildew. Plant J. 76, 957–969. doi:10.1111/tpj.12345
Igrejas, G., and Branlard, G. (2020). The importance of wheat. Berlin, Germany: Springer. doi:10.1007/978-3-030-34163-3_1
Ji, X. L., Xie, C. J., Ni, Z. F., Yang, T., Nevo, E., Fahima, T., et al. (2008). Identifcation and genetic mapping of a powdery mildew resistance gene in wild emmer (Triticum dicoccoides) accession IW72 from Israel. Euphytica 159, 385–390. doi:10.1007/s10681-007-9540-1
Jin, Y. L., Shi, F. Y., Liu, W. H., Fu, X. Y., Gu, T. T., Han, G. H., et al. (2021). Identification of resistant germplasm and detection of genes for resistance to powdery mildew and leaf rust from 2978 wheat accessions. Plant Dis. 105, 3900–3908. doi:10.1094/PDIS-03-21-0532-RE
Jin, Y. L., Xiao, L. N., Zheng, J. P., Su, F. Y., Yu, Z. Y., Mu, Y. J., et al. (2023). Genetic analysis and molecular identification of the powdery mildew resistance in 116 elite wheat cultivars/lines. Plant Dis. 107, 3801–3809. doi:10.1094/PDIS-04-23-0792-RE
Klindworth, D. L., Niu, Z., Chao, S., Friesen, T. L., Jin, Y., Faris, J. D., et al. (2012). Introgression and characterization of a Goatgrass gene for a high level of resistance to Ug99 stem rust in tetraploid wheat. G3-Genes Genomes Genet. 2, 665–673. doi:10.1534/g3.112.002386
Krattinger, S. G., Lagudah, E. S., Spielmeyer, W., Singh, R. P., Huerta-Espino, J., McFadden, H., et al. (2009). A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 323, 1360–1363. doi:10.1126/science.1166453
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi:10.1093/molbev/msw054
Li, M. M., Dong, L. L., Li, B. B., Wang, Z. Z., Xie, J. Z., Qiu, D., et al. (2020). A CNL protein in wild emmer wheat confers powdery mildew resistance. New Phytol. 228, 1027–1037. doi:10.1111/nph.16761
Li, Y. H., Wei, Z. Z., Sela, H. N., Govta, L., Klymiuk, V., Roychowdhury, R., et al. (2023). Dissection of a rapidly evolving wheat resistance gene cluster by long-read genome sequencing accelerated the cloning of Pm69. Plant Commun. 5, 100646. doi:10.1016/j.xplc.2023.100646
Ling, H. Q., Zhao, S. C., Liu, D. C., Wang, J. Y., Su, H., Zhang, C., et al. (2013). Draft genome of the wheat A-genome progenitor. Triticum urartu 496, 87–90. doi:10.1038/nature11997
Liu, Z. J., Zhu, J., Cui, Y., Liang, Y., Wu, H. B., Song, W., et al. (2012). Identification and comparative mapping of a powdery mildew resistance gene derived from wild emmer (Triticum turgidum var. dicoccoides) on chromosome 2BS. Theor. Appl. Genet. 124, 1041–1049. doi:10.1007/s00122-011-1767-5
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi:10.1006/meth.2001.1262
Lu, P., Guo, L., Wang, Z. Z., Li, B. B., Li, J., Li, Y. H., et al. (2020). A rare gain of function mutation in a wheat tandem kinase confers resistance to powdery mildew. Nat. Commun. 11, 680. doi:10.1038/s41467-020-14294-0
Luo, M. C., Deal, K., Yang, Z. L., and Dvorak, J. (2005). Comparative genetic maps reveal extreme crossover localization in the Aegilops speltoides chromosomes. Theor. Appl. Genet. 111, 1098–1106. doi:10.1007/s00122-005-0035-y
Ma, H. Q., Kong, Z. X., Fu, B. S., Li, N., Zhang, L. X., Jia, H. Y., et al. (2011). Identification and mapping of a new powdery mildew resistance gene on chromosome 6D of common wheat. Theor. Appl. Genet. 123, 1099–1106. doi:10.1007/s00122-011-1651-3
McIntosh, R. A., Dubcovsky, J., Rogers, W. J., Xia, X. C., and Raupp, W. J. (2020). Catalogue of gene symbols for wheat 2020 supplement. Available at: https://wheat.pw.usda.gov/GG3/WGC.
Miranda, L. M., Murphy, J. P., Marshall, D., Cowger, C., and Leath, S. (2007). Chromosomal location of Pm35, a novel Aegilops tauschii derived powdery mildew resistance gene introgressed into common wheat (Triticum aestivum L). Theor. Appl. Genet. 14, 1451–1456. doi:10.1007/s00122-007-0530-4
Mohler, V., Bauer, C., Schweizer, G., Kempf, H., and Hartl, L. (2013). Pm50: a new powdery mildew resistance gene in common wheat derived from cultivated emmer. J. Appl. Genet. 54, 259–263. doi:10.1007/s13353-013-0158-9
Moore, J. W., Herrera-Foessel, S., Lan, C. X., Schnippenkoetter, W., Ayliffe, M., Huerta-Espino, J., et al. (2015). A recently evolved hexose transporter variant confers resistance to multiple pathogens in wheat. Nat. Genet. 47, 1494–1498. doi:10.1038/ng.3439
Petersen, S., Lyerly, J. H., Worthington, M. L., Parks, W. R., Cowger, C., Marshall, D. S., et al. (2015). Mapping of powdery mildew resistance gene Pm53 introgressed from Aegilops speltoides into soft red winter wheat. Theor. Appl. Genet. 128, 303–312. doi:10.1007/s00122-014-2430-8
Qiu, Y. C., Zhou, R. H., Kong, X. Y., Zhang, S. S., and ang Jia, J. Z. (2005). Microsatellite mapping of a Triticum urartu Tum. Derived powdery mildew resistance gene transferred to common wheat (Triticum aestivum L). Theor. Appl. Genet. 111, 1524–1531. doi:10.1007/s00122-005-0081-5
Saari, E. E., and Prescott, J. M. (1975). A scale for appraising the foliar intensityof wheat disease. Plant Dis. Rep. 59, 377–381.
Sánchez-Martín, J., Steuernagel, B., Ghosh, S., Herren, G., Hurni, S., Adamski, N., et al. (2016). Rapid gene isolation in barley and wheat by mutant chromosome sequencing. Genome Biol. 17, 221. doi:10.1186/s13059-016-1082-1
Sánchez-Martín, J., Widrig, V., Herren, G., Wicker, T., Zbinden, H., Gronnier, J., et al. (2021). Wheat Pm4 resistance to powdery mildew is controlled by alternative splice variants encoding chimeric proteins. Nat. Plants. 7, 327–341. doi:10.1038/s41477-021-00869-2
Santos, F. R., Pena, S. D., and Epplen, J. T. (1993). Genetic and population study of a Y-linked tetranucleotide repeat DNA polymorphism with a simple non-isotopic technique. Hum. Genet. 90, 655–656. doi:10.1007/BF00202486
Savary, S., Willocquest, L., Pethybridge, S. J., Esker, P., Mxroberts, N., and Nelson, A. (2019). The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 3, 430–439. doi:10.1038/s41559-018-0793-y
Schmolke, M., Mohler, V., Hartl, L., Zeller, F., and Hsam, S. L. K. (2012). A new powdery mildew resistance allele at the Pm4 wheat locus transferred from einkorn (Triticum monococcum). Mol. Breed. 29, 449–456. doi:10.1007/s11032-011-9561-2
Sharp, P. J., Kreis, M., Shewry, P. R., and Gale, M. D. (1988). Location of β-amylase sequences in wheat and its relatives. Theor. Appl. Genet. 75, 286–290. doi:10.1007/bf00303966
Shi, A. N., Leath, S., and Murphy, J. P. (1998). A major gene for powdery mildew resistance transferred to common wheat from wild einkorn wheat. Phytopathology 88, 144–147. doi:10.1094/PHYTO.1998.88.2.144
Si, Q. M., Zhang, X. X., Duan, X. Y., Sheng, B. Q., and Zhou, Y. L. (1992). On gene analysis and classification of powdery mildew (Erysiphe graminis f. sp. Tritici) resistant wheat varieties. Acta Agron. Sin. 22, 349–355.
Singh, S. P., Hurni, S., Ruinelli, M., Brunner, S., Sanchez-Martin, J., Krukowski, P., et al. (2018). Evolutionary divergence of the rye Pm17 and Pm8 resistance genes reveals ancient diversity. Plant Mol. Biol. 98, 249–260. doi:10.1007/s11103-018-0780-3
Sun, H., Hu, J., Song, W., Qiu, D., Cui, L., Wu, P., et al. (2018). Pm61: a recessive gene for resistance to powdery mildew in wheat landrace Xuxusanyuehuang identified by comparative genomics analysis. Theor. Appl. Genet. 131, 2085–2097. doi:10.1007/s00122-018-3135-1
Tan, C. C., Li, G. Q., Cowger, C., Carver, B. F., and Xu, X. Y. (2018). Characterization of Pm59, a novel powdery mildew resistance gene in Afghanistan wheat landrace PI 181356. Theor. Appl. Genet. 131, 1145–1152. doi:10.1007/s00122-018-3067-9
Tommasini, L., Yahiaoui, N., Srichumpa, P., and Keller, B. (2006). Development of functional markers specific for seven Pm3 resistance alleles and their validation in the bread wheat gene pool. Theor. Appl. Genet. 114, 165–175. doi:10.1007/s00122-006-0420-1
Wan, W. T., Xiao, J., Li, M. L., Tang, X., Wen, M. X., Cheruiyot, A. K., et al. (2020). Fine mapping of wheat powdery mildew resistance gene Pm6 using 2B/2G homoeologous recombinants induced by the ph1b mutant. Theor. Appl. Genet. 133, 1265–1275. doi:10.1007/s00122-020-03546-8
Wang, B., Meng, T., Xiao, B., Yu, T. Y., Yue, T. Y., Jin, Y. L., et al. (2023). Fighting wheat powdery mildew: from genes to fields. Theor. Appl. Genet. 136, 196. doi:10.1007/s00122-023-04445-4
Wang, Z. L., Li, L. H., He, Z. H., Duan, X. Y., Zhou, Y. L., Chen, X. M., et al. (2005). Seedling and adult plant resistance to powdery mildew in Chinese bread wheat cultivars and lines. Plant Dis. 89, 457–463. doi:10.1094/PD-89-0457
Wu, P. P., Hu, J. H., Zou, J. W., Qiu, D., Qu, Y. F., Li, Y. H., et al. (2019). Fine mapping of the wheat powdery mildew resistance gene Pm52 using comparative genomics analysis and the Chinese Spring reference genomic sequence. Theor. Appl. Genet. 132, 1451–1461. doi:10.1007/s00122-019-03291-7
Xie, J. Z., Guo, G. H., Wang, Y., Hu, T. Z., Wang, L. L., Li, J. T., et al. (2020). A rare single nucleotide variant in Pm5e confers powdery mildew resistance in common wheat. New Phytol. 228, 1011–1026. doi:10.1111/nph.16762
Xing, L. P., Hu, P., Liu, J. Q., Witek, K., Zhou, S., Xu, J. F., et al. (2018). Pm21 from Haynaldia villosa encodes a CC-NBS-LRR protein conferring powdery mildew resistance in wheat. Mol. Plants. 11, 874–878. doi:10.1016/j.molp.2018.02.013
Xu, H. X., Yao, G. Q., Xiong, L., Yang, L. L., Jiang, Y. M., Fu, B. S., et al. (2008). Identification and mapping of Pm2026: a recessive powdery mildew resistance gene in an einkorn (Triticum monococcum L.) accession. Theor. Appl. Genet. 117, 471–477. doi:10.1007/s00122-008-0791-6
Xue, S. L., Hu, S. S., Chen, X., Ma, Y. Y., Lu, M. X., Bai, S. L., et al. (2022). Fine mapping of Pm58 from Aegilops tauschii conferring powdery mildew resistance. Theor. Appl. Genet. 135, 1657–1669. doi:10.1007/s00122-022-04061-8
Yao, G. Q., Zhang, J. L., Yang, L. L., Xu, H. X., Jiang, Y. M., Xiong, L., et al. (2007). Genetic mapping of two powdery mildew resistance genes in einkorn (Triticum monococcum L.) accessions. Theor. Appl. Genet. 114, 351–358. doi:10.1007/s00122-006-0438-4
Yu, Z. Y., Xiao, L. N., Su, F. Y., Liu, W., Luo, F. Y., Han, R., et al. (2022). Mining of wheat Pm2 alleles for goal-oriented marker-assisted breeding. Front. Plant Sci. 13, 912589. doi:10.3389/fpls.2022.912589
Zhang, J. G., Dong, N., Yan, W. L., and Ru, Z. G. (2014). Screening for SSR markers linked to wheat powdery mildew resistance gene Pm13. J. Henan Aric Sci. 43, 62–66. doi:10.15933/j.cnki.1004-3268.2014.10.050
Zhang, R. Q., Sun, B. X., Chen, J., Cao, A. Z., Xing, L. P., Feng, Y. G., et al. (2016). Pm55, a developmental-stage and tissue-specific powdery mildew resistance gene introgressed from Dasypyrum villosum into common wheat. Theor. Appl. Genet. 129, 1975–1984. doi:10.1007/s00122-016-2753-8
Zhao, F. K., Li, Y. H., Yang, B. J., Yuan, H. B., Jin, C., Zhou, L. X., et al. (2020). Powdery mildew disease resistance and marker-assisted screening at the Pm60 locus in wild diploid wheat Triticum urartu. Crop J. 8, 252–259. doi:10.1016/j.cj.2019.09.007
Zhu, S. Y., Liu, C., Gong, S. J., Chen, Z. Z., Chen, R., Liu, T. L., et al. (2023). Orthologous genes Pm12 and Pm21 from two wild relatives of wheat show evolutionary conservation but divergent powdery mildew resistance. Plant Commun. 4, 100472. doi:10.1016/j.xplc.2022.100472
Keywords: powdery mildew, markers, resistance identification, haplotype analysis, wheat relatives
Citation: Wang J, Xu H, Qie Y, Han R, Sun X, Zhao Y, Xiao B, Qian Z, Huang X, Liu R, Zhang J, Liu C, Jin Y and Ma P (2024) Evaluation and identification of powdery mildew-resistant genes in 137 wheat relatives. Front. Genet. 15:1342239. doi: 10.3389/fgene.2024.1342239
Received: 21 November 2023; Accepted: 02 January 2024;
Published: 24 January 2024.
Edited by:
Jia-Yu Xue, Nanjing Agricultural University, ChinaCopyright © 2024 Wang, Xu, Qie, Han, Sun, Zhao, Xiao, Qian, Huang, Liu, Zhang, Liu, Jin and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pengtao Ma, cHRtYUB5dHUuZWR1LmNu; Yuli Jin, eXVsaWppbkB5dHUuZWR1LmNu; Cheng Liu, bGNoNjY4ODQwN0AxNjMuY29t
†These authors have contributed equally to this work
 Jiaojiao Wang1†
Jiaojiao Wang1† Hongxing Xu
Hongxing Xu Ran Han
Ran Han Jiadong Zhang
Jiadong Zhang Cheng Liu
Cheng Liu Yuli Jin
Yuli Jin Pengtao Ma
Pengtao Ma