- 1Núcleo Milenio MASH, Facultad de Ciencias Biológicas, Pontificia Universidad Católica de Chile, Santiago, Chile
- 2IRL 3614 Evolutionary Biology and Ecology of Algae, Centre National de la Recherche Scientifique (CNRS), Sorbonne Université, Pontificia Universidad Católica de Chile, Universidad Austral de Chile, Station Biologique, Roscoff, France
- 3GEMA Center for Genomics, Ecology and Environment, Universidad Mayor, Santiago, Chile
- 4Instituto de Ciencias Ambientales y Evolutivas, Facultad de Ciencias, Universidad Austral de Chile, Valdivia, Chile
- 5Núcleo Milenio MASH, Instituto de Ciencias Ambientales y Evolutivas, Facultad de Ciencias, Universidad Austral de Chile, Valdivia, Chile
- 6Centro FONDAP de Investigación de Ecosistemas Marinos de Altas Latitudes (IDEAL), Valdivia, Chile
Historical vicariance events, linked to the existence of stable physical barriers to gene flow, generate concordant genetic breaks in co-distributed species while stochastic processes (e.g., costal uplift) could cause species-specific genetic breaks as a result of local strong demographic bottlenecks or extinction. In Chile, previous studies show that the area of the 30°S-33°S could correspond to a stable barrier to gene flow that have affected the genetic structure of various algae and marine invertebrates. Here we sequenced two organellar genes (COI and rbcL) in four taxonomically accepted co-distributed red seaweeds species characterized by a low dispersal potential: Mazzaella laminarioides, M. membranacea, Asterfilopsis disciplinalis, and Ahnfeltiopsis vermicularis. Our results revealed the existence of ten strongly differentiated linages in the taxa studied. Strong genetic breaks, concordant in both space and time (divergence estimated to have occurred some 2.9–12.4 million years ago), were observed between taxa distributed across the 33°S. Conversely, in the Central/South part of the Chilean coast, the localization of the genetic breaks/sub-structure observed varied widely (36°S, 38°S, 39°S, and 40°S). These results suggest that a major historical vicariance event has modeled the genetic structure of several Chilean marine organisms in the north of the Chilean coast during the mid-Miocene, while more recent stochastic events and genetic drift could be the driving forces of genetic divergence/structuration in the central-southern part of the coast.
1 Introduction
Comparative phylogeography can be seen as a bridge linking the study of macro-scale biogeographic processes and local patterns of evolution at the within species level (Avise, 2000). The discipline has allowed to successfully evaluate the importance of historic and contemporary events in the evolutionary history of co-distributed species (Bermingham and Moritz, 1998; Arbogast and Kenagy, 2001; Bagley and Johnson, 2014). In the marine realm, main currents or strong upwelling systems can generate concordant genetic breaks in species of fishes, invertebrates, and algae (Kelly and Palumbi, 2010; Marko et al., 2010; Guillemin et al., 2016; Hiller and Lessios, 2020; Shelley et al., 2020). The imprint of oceanographic barriers on species genetic composition depends on both the type of barrier (i.e., hard or soft barriers; Cowman and Bellwood, 2013) and the species intrinsic characteristics. Species intrinsic characteristics, especially the ones directly related with the dispersal capacity, strongly affect the potential effect of soft barriers on genetic divergence (Kelly and Palumbi, 2010; Francisco et al., 2018; Ayres-Ostrock et al., 2019; Morales-González et al., 2019).
Other factors, as the rate of extinction and colonisation in the area under study (Fraser et al., 2018) or the population size (directly linked to the strength of genetic drift; Irwin, 2002; Kuo and Avise, 2005) can also affect a species response to a barrier to gene flow. These studies have corroborated the strong effect of drift on genetic differentiation, showing that species with small population size could be affected by very shallow and/or transitory barrier to gene flow. Stochastic events, as volcanic eruptions, floods, tsunami, or coastal uplift after earthquakes, generate demographic bottlenecks or even local extinctions (Castilla et al., 2010; Hsu et al., 2007; Pujolar et al., 2011; Schiel et al., 2019). These have lasting effects on population structure, especially in organisms with low dispersal capacity, but could be highly species-specific (Worthington Wilmer et al., 2011; Miura et al., 2017; Brante et al., 2019; Becheler et al., 2020; Parvizi et al., 2020). Indeed, the effect of drift and stochastic events could lead to lack of congruence in the spatial genetic structure of co-distributed species (Avise, 2000).
Comparative phylogeographic studies on marine organisms in the southeast Pacific mainly focused on vicariant processes at the origin of phylogenetic breaks concordant with major biogeographic barriers (Haye et al., 2014; Guillemin et al., 2016). The Chilean coast, spread over more than 4,500 km from 18°S to 56°S, is highly heterogeneous (variation in seawater temperature, salinity, upwelling regimes, and coastal topography between other) and encompasses two biogeographic breaks located at 30–33°S and 41–42°S (Camus, 2001; Thiel et al., 2007). The southern biogeographic break is related to the bifurcation of the Antarctic Circumpolar Current (ACC) and important seawater characteristics and topographic changes in the coastline occurring at 42°S has been proposed as important barriers to dispersal for marine organisms (Camus, 2001; Fernández et al., 2000). The biogeographic break at 30–33°S seems to be more permeable and taxa dependent (Santelices, 1980; Meneses and Santelices, 2000; Camus, 2001). For marine invertebrates and brown algae, the biogeographic break is reported at 30°S while for red algae the change in the community composition is observed at 32–33°S (Santelices, 1980; Meneses and Santelices, 2000; Camus, 2001). The 30–33°S area is an important transition zone in oceanographic features, with a weak but persistent upwelling in the north, and a strong but seasonal upwelling in the south (Broitman et al., 2001; Thiel et al., 2007; Tapia et al., 2014).
Numerous phylogeographic studies have corroborated the concordance between biogeographic and genetic breaks in Chilean marine invertebrates (Sanchez et al., 2011; Brante et al., 2012; Varela and Haye, 2012; Morales-González et al., 2019) and seaweeds (Tellier et al., 2009; Fraser et al., 2010; Macaya and Zucarello, 2010; Tellier et al., 2011; Montecinos et al., 2012). Along the coast located in between the two main biogeographic breaks, genetic breaks have been observed in some species. However, both the divergence time and the position of the genetic breaks seem to be more species specific (Montecinos et al., 2012; Haye et al., 2014; Guillemin et al., 2016; Morales-González et al., 2019; Quesada-Calderón et al., 2021). The coastline located between 36°S and 42°S has been regularly affected by earthquakes and coastal uplift that generated large but local mortality of intertidal organisms (Castilla, 1988; Castilla et al., 2010; Jaramillo et al., 2012; Hernández-Miranda et al., 2014; Brante et al., 2019). In addition, this area is characterized by environmental heterogeneity including variation in sea surface temperature and coastal marine currents (Atkinson et al., 2002) and large sandy beaches that can interrupt gene flow in organisms with low dispersal capacity (Fraser et al., 2010; Morales-González et al., 2019). It has been proposed that these local and transitory barriers to gene flow could have affected the genetic structure of species potentially strongly affected by genetic drift.
The present work aims to compare the genetic structure of the following four ecologically similar taxonomically accepted species of red algae: M. laminarioides, M. membranacea, Asterfilopsis disciplinalis (previously named Ahnfeltiopsis furcellata or Gymnogongrus disciplinalis, see Calderon and Boo, 2016), and Ahnfeltiopsis vermicularis (Calderon and Boo, 2017). Ahnfeltiopsis vermicularis has been historically reported in Chile under the names Ahnfeltia durvillei (Ramírez, 1991) and Ahnfeltiopsis durvillei (Hoffmann and Santelices, 1997), both names known as synonyms of Gymnogongrus durvillei (Guiry and Guiry, 2024). Molecular data associated with Chilean specimens collected under these names (Fredericq and Lopez-Bautista, 2002) appear as Ahnfeltiopsis vermicularis in the recent phylogeny of South American Phyllophoraceae published by Calderon and Boo (2017). These four species belong to the order Gigartinales, they are non-buoyant and present a limited dispersal capacity (Norton, 1992; Silva and DeCew, 1992; Hoffmann and Santelices, 1997). They are co-distributed along an extended part of the Chilean coast and are encountered in sympatry on rocky substrates located at high - mid intertidal (Santelices et al., 1989; Camus, 1992; Ramírez, 2010; Calderon and Boo, 2016). Previous studies in M. laminarioides have shown the existence of two strong genetic breaks in this species: one located at 33°S and concordant with a biogeographic break and one located at 37–39°S, in a part of the coast particularly affected by stochastic events such as earthquakes and coastal uplifts (Montecinos et al., 2012). Using sequences from two molecular markers, the mitochondrial COI and the cloroplastic rbcL, we tested for the existence of genetic breaks in three other species of red algae (i.e., M. membranacea, As. disciplinalis, and Ah. vermicularis) and searched for patterns of genetic divergence possibly congruent in time and space with the ones already observed in M. laminarioides. Our sampling cover part of the Chilean coast located between 28°55′S and 41°52′S and the algae were collected in 13 localities separated by around 100 km. The sampling includes the 30–33°S biogeographic break and the central-south part of the coastline characterized by mega-disturbance events and environmental heterogeneity. We hypothesized that all four co-distributed species of red algae will present congruent genetic breaks at 33°S, due to the effect of past vicariant events linked to the establishment of stable barrier to gene flow, while genetic divergence along the central-south part of the coastline, if detected, will be species specific. Because of the deep divergence within M. laminarioides, it has been proposed that the three genetic groups are incipient genetic species (Montecinos et al., 2012). We used species delimitation analyses to test if the genetic groups encountered could be considered as potentially new putative genetic species.
2 Materials and methods
2.1 Sampling
We sampled four species of red algae, M. laminarioides (N = 33), M. membranacea (N = 152), As. disciplinalis (N = 176), and Ah. vermicularis (N = 157), when present, in 13 sampling sites along the Chilean coast. Sites ranged from Los Burros 28°55’S to Chiloe 41°52’S (Supplementary Table S1). For M. laminarioides, 16 individuals from Loberia (38°39’S-73°29’W) and 17 individuals from Nigue (39°17’S-73°13’W) were added to the 217 individuals previously sampled by Montecinos et al. (2012) (Supplementary Table S1). Samples were assigned to the four taxonomically accepted species using the external diagnostic characters described in Hoffmann and Santelices, (1997). All samples were stored in silica gel to ensure tissue preservation before DNA extraction.
2.2 DNA extraction, PCR amplification, sequencing, and alignment
For each sample, dry tissue was grounded in a MiniBeadBeater 24 (BioSpec Products, Inc. Bortlesville, United States) and DNA was isolated using the E. Z.N.A tissue DNA extraction kit following the provider instructions (Omega Biotek, Inc. Georgia, United States). A partial sequence of the mitochondrial Cytochrome c Oxidase I gen (COI) was amplified using the primers GazF1 and GazR1. Primers and PCR conditions were as described in Saunders, (2005). Additionally, a partial sequence of the chloroplast Ribulose bisphosphate carboxylase large chain gen (rbcL) was amplified for a subsampling of M. laminarioides, M. membranacea, As. disciplinalis, and Ah. vermicularis. The primers rbcL-F and rbcL-R (Hommersand et al., 1994) and the PCR conditions described in Fredericq and Lopez-Bautista (2002) were used for the rbcL amplification. PCR products were purified using the UltraCleanTM DNA Purification kit (MO BIO Laboratories, Carlsbad, CA, United States) and sequenced with an ABI Automatic Sequencer at the AUSTRAL-OMICS core-facility (Universidad Austral de Chile, Chile). Sequences were edited using Chromas v.2.33 (McCarthy, 1997) and aligned using Mega v.5 (Tamura et al., 2011).
In the case of M. laminarioides, 217 COI and 9 rbcL sequences already available in GenBank were added to our dataset (Montecinos et al., 2012).
2.3 Phylogenetic reconstruction
For the COI, 24 sequences of Mazzaella and 48 sequences of the family Phyllophoracea were downloaded from GenBank and added to our data set (Supplementary Table S2). In the same way, 33 sequences of Mazzaella and 83 sequences of the family Phyllophoracea were added to our rbcL data set (Supplementary Table S2). For both the COI and rbcL data sets, Chondracanthus and Gigartina sequences were used as outgroup for the Mazzaella phylogenetic reconstructions while Mazzaella, Chondracanthus, and Chondrus sequences were used as outgroup for the Asterfilopsis and Ahnfeltiopsis phylogenetic reconstructions (Supplementary Table S2). Duplicated haplotypes were removed from the data sets using DnaSP v6.12.03 prior to analyses (Rozas et al., 2017).
Phylogenetic reconstructions were performed independently for Mazzaella, Asterfilopsis, and Ahnfeltiopsis with the Maximum Likelihood (ML) method and Bayesian Inference (BI), using the COI and rbcL markers. ML analyses were performed using IQTree v1.6.12 (Trifinopoulos et al., 2016). The best-fit substitution model was selected using the Bayesian Information Criterion (BIC) implemented in IQTree v1.6.12 (Kalyaanamoorthy et al., 2017). The selected models for the COI were TIM3 + F + G4 for Mazzaella and TIM + F + I + G4 for Asterfilopsis and Ahnfeltiopsis. The selected models for the rbcL were TIM + F + I + G4 for Mazzaella, Asterfilopsis, and Ahnfeltiopsis. Statistical support was estimated using 1,000 ultrafast bootstrap replicates and the nonparametric SH-aLRT single branch test. BI analyses were performed using MrBayes v3.1.2 (Huelsenbeck and Ronquist, 2001). Four independent analyses were run. Four chains and five million of generations were used for each analysis. Trees and parameters were sampled every 1,000 generations and the default parameters were used to fit temperature and swapping. The first 25% of the sampled trees were discarded as burn-in to ensure stabilization. The remaining trees were used to compute a consensus topology and posterior probability values. Split frequencies (variance among the four independent runs) were below 0.003, confirming that the posterior probability distribution was accurately sampled.
2.4 Species delimitation analyses based on genetic data sets
To test the possible existence of cryptic putative genetic species within the four species sampled, we conducted three species delimitation analyses for Mazzaella, Asterfilopsis, and Ahnfeltiopsis using the COI and the rbcL datasets independently (see Supplementary Table S2 for more information about the list of sequences used in species delimitation analyses). First, the Automatic Barcode Gap Discovery (ABGD) was run online at http://wwwabi.snv.jussieu.fr/public/abgd/abgdweb.html. ABGD compare intraspecific and interspecific pairwise genetic distances to delimit species (Puillandre et al., 2012). We computed Kimura two-parameter (K2P) genetic distances and used default ABGD settings. Second, a General Mixed Yule Coalescent (GMYC) analysis was run. GMYC identifies a threshold value for the shift in branching rate from coalescent lineage branching to interspecific diversification on an ultrametric tree and explicitly delimits “independently evolving” clusters (i.e., putative species; Monaghan et al., 2009; Pons et al., 2006). Branch lengths were estimated under a relaxed log-normal clock using the Bayesian analysis implemented in BEAST v1.8.2 (Drummond et al., 2012). A coalescent (constant size) prior was used, and Markov Chains Monte Carlo (MCMC) were run for 20 million generations. Trees were sampled each 1,000 generations with a 10% burn-in. A visual inspection of MCMC progression was performed to corroborate stabilization using Tracer v1.8 (Drummond et al., 2012). An ultrametric tree was constructed using Tree Annotator v1.8 (Drummond et al., 2012). Since the multiple-thresholds approach tends to overestimate the number of delineated species (Fujisawa and Barraclough, 2013) only the single-threshold (Pons et al., 2006) versions of GMYC was fitted on the ultrametric tree using the SPLITS package for R.
The bPTP analysis was performed using a bifurcated phylogenetic input tree as implemented in bPTP web server (https://species.h-its.org; Zhang et al., 2013). The parameters were set as following: Markov chain Monte Carlo = 500,000 generations; thinning = 100; burn-in = 0.1; and the convergence were thoroughly checked to ensure that the result was reliable. The most plausible delimitations are reported with their associated Bayesian support values; higher Bayesian support value of a node indicates that all descendants from this node are more likely to be conspecific.
2.5 Divergence time estimation
Divergence time between putative genetic species of Mazzaella, Asterfilopsis, and Ahnfeltiopsis was estimated using BEAST v2.5 (Bouckaert et al., 2019) with a single concatenated matrix with two partitions, the COI and the rbcL. One sequence of each putative genetic species was used for the analysis, only putative genetic species for which both COI and the rbcL were available were used in the analysis (see Supplementary Table S2). A relaxed uncorrelated log-normal clock and Yule model were applied as tree priors for each partition. Substitution models GTR + I + G and GTR + I were applied for the COI and the rbcL, respectively. Best fit models were selected using BIC in jModelTest (Darriba et al., 2012). Three chains were run for 500 million generations and trees were sampled every 5,000 generations. Mutation rates of 0.350% (Hu et al., 2010) and 0.127% (Kamiya et al., 2004) were used for the COI and rbcL, respectively. Since the fossil record of Florideophyceae is exceptionally scarce (Yang et al., 2016), time of divergence between Gigartinaceae and Phyllophoraceae was calibrated at 113 Ma, as proposed in TimeTree (Kumar et al., 2022). Calibration point was set as normal distribution with 113 Ma as the mean and 5% of the mean as standard deviation. Convergence of model parameters was estimated by plotting the marginal posterior probabilities versus generations in Tracer v1.8 (Rambaut et al., 2018). Effective sample size values were estimated for each parameter to ensure adequate mixing of the MCMC (ESSs >200). The final consensus tree was produced using TreeAnnotator v1.8 (Bouckaert et al., 2019).
2.6 Genetic diversity estimates
The genetic diversity within each putative genetic species (see results for more details) and within each population of Mazzaella, Asterfilopsis, and Ahnfeltiopsis was evaluated using five indices: the number of haplotypes (nH), gene diversity (H), nucleotide diversity (π), number of polymorphic sites (S), and number of private haplotypes (Hpriv). These calculated for COI dataset in DNASP 4.10.3 (Rozas et al., 2003). As only a few haplotypes, at most, were detected within putative genetic species for the rbcL marker it was not used for genetic diversity analyses.
2.7 Phylogeographic structure
For Mazzaella, Asterfilopsis, and Ahnfeltiopsis, genealogical relationship between COI sequences were reconstructed using Median-Joining algorithm implemented in Network v5.0.1.1 (Bandelt et al., 1999). Population genetic structure was examined by pairwise ΦST between populations within each putative genetic species; and significance of the ΦST values was estimated using 1,000 permutations (Excoffier et al., 1992). For three putative genetic species (i.e., M. membranacea, As. disciplinalis, and Ah. vermicularis; see results section for more details), analyses of molecular variance (AMOVA) were conducted in Arlequin (Excoffier and Lischer, 2010) to detect spatial partitioning of genetic variance within sampling sites, among sampling sites within genetic groups and among genetic groups. Levels of significance were tested using 1,000 permutations (Excoffier and Lischer, 2010). As for genetic diversity analyses, the rbcL marker was not used for phylogeographic structure analyses.
2.8 Demographic history
For each putative genetic species of Mazzaella, Asterfilopsis, and Ahnfeltiopsis, three complementary approaches were used to infer their historical demography. Tajima’s D (Tajima, 1989) and Fu’s Fs (Fu, 1997) tests were used to assess significant excess of rare alleles, performing 10,000 bootstraps replicated using Arlequin v3.5 (Excoffier and Lischer, 2010). Moreover, the observed mismatch distribution was compared with expected values under a model of sudden demographic expansion (Rogers and Harpending, 1992), and statistical significance was tested with the sum of squared deviation (SSD) and the Harpending’s Raggedness index (Rag) after 10,000 bootstraps using Arlequin v3.5 (Excoffier and Lisher, 2010). Finally, Bayesian skyline plots (BSP) were constructed using BEAST v2.5 (Bouckaert et al., 2019). The best-fit substitution models, selected using BIC in IQTree v1.6.12 (Kalyaanamoorthy et al., 2017), were: TPM3 + F for M. membranacea, HKY + F for As. disciplinalis, HKY + F + I for Ah. vermicularis and HKY + F for Ah. sp. 2. Bayesian skyline plots were constructed using 100 million iterations. Posterior distribution of parameters was approximated by Markov Chain Monte Carlo (MCMC) sampled every 10,000 iterations after a discarded burn in of five million iterations. A relaxed log-normal molecular clock was used, with a substitution rate of 0.35% per million years estimated for COI (Hu et al., 2010) to test for historical changes of effective population size. BSP reconstructions were visualized in Tracer v1.8 (Rambaut et al., 2018).
3 Results
A partial COI segment of 572 pb was successfully sequenced in 250 M. laminarioides, 152 M. membranacea, 176 As. disciplinalis, and 157 Ah. vermicularis. Partial rbcL sequences of 919 pb were sequenced for nine, four, eleven and eight individuals of M. laminarioides, M. membranacea, As. disciplinalis, and Ah. vermicularis, respectively.
3.1 Phylogeny and species delimitation
The phylogenetic analyses based on COI and rbcL sequences from Chile formed distinct well supported monophyletic lineages within all four species, except for As. disciplinalis, for which all rbcL sequences from Chile form a unique discrete monophyletic lineage while the COI sequences from Chile form a paraphyletic group (Figures 1, 2). Both the COI and rbcL separate M. laminarioides into three putative genetic species (M. laminarioides North, M. laminarioides Center, and M. laminarioides South; names follow Montecinos et al., 2012; Figure 1) and M. membranacea into two putative genetic species (M. membranacea and M. sp. 1; Figure 1). For the rbcL marker, all sequences of M. membranacea and M. sp. 1 from Chile were closely related to M. “affinis” and M. sp. 3 from Chile and Peru, respectively (Figure 1). COI and rbcL sequences of individuals identified as M. “membranacea” and collected in Tristan Island (South Atlantic) appear as sister clade to the sequences obtained from Peru and Chile (Figure 1). For the COI, haplotypes C77 and C78 of As. disciplinalis were retrieved as a basal polytomy in respect to the rest of the sequences obtained (Figure 2). Individuals collected under the name Ahnfeltiopsis vermicularis were separated in four well defined genetic lineages for the COI; Ah. vermicularis, Ah. sp. 1, Ah. sp. 2, and Ah. sp. 3 (Figure 2). Ahnfeltiopsis vermicularis (including the sequence AF388560 from South Africa), Ah. sp. 1 and Ah. sp. 2 also appear as distinct lineages in the rbcL phylogenetic tree; rbcL fragment of Ah. sp. 3 could not be amplified. All Asterfilopsis and Ahnfeltiopsis sequences obtained in our study clustered with close by Phyllophoracea species from the Southern Hemisphere (Asterfilopsis: with four newly described species of Asterfilopsis from Peru; Ahnfeltiopsis: with Acletoa tarazonae KY924618 from Peru and Gymnogongrus sp. AF388564 from Chile; Calderon and Boo, 2016; Calderon and Boo, 2017; Figure 2).
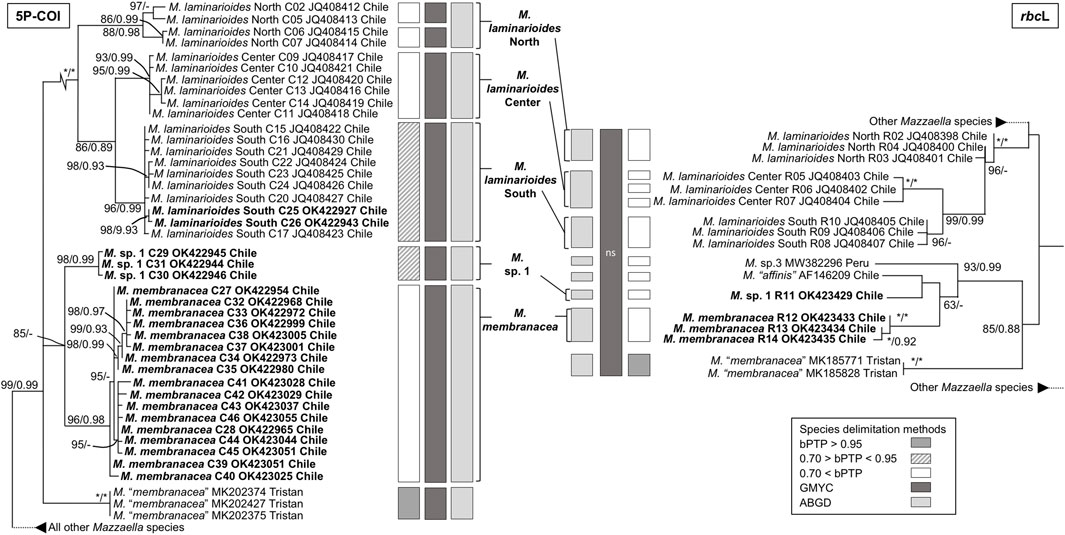
FIGURE 1. Maximun likelihooh phylogram of Mazzaella species inferred from COI and rbcL sequences. ML bootstrap values (≥60) and BI posterior probabilities (≥90) are indicated near branches; asterisk (*) indicates full support. Sequences obtained in our study are in bold. The results of three species delimitation methods (bPTP, GMYC, ABGD; see material and methods for more details) are indicated for both the COI and the rbcL markers. For the GYMC species delimitation method, analyses for which the model considering the existence of distinct species was not statistically significantly better than the null model (i.e., leading to the lumping of all sequences used) are indicated by “ns”.
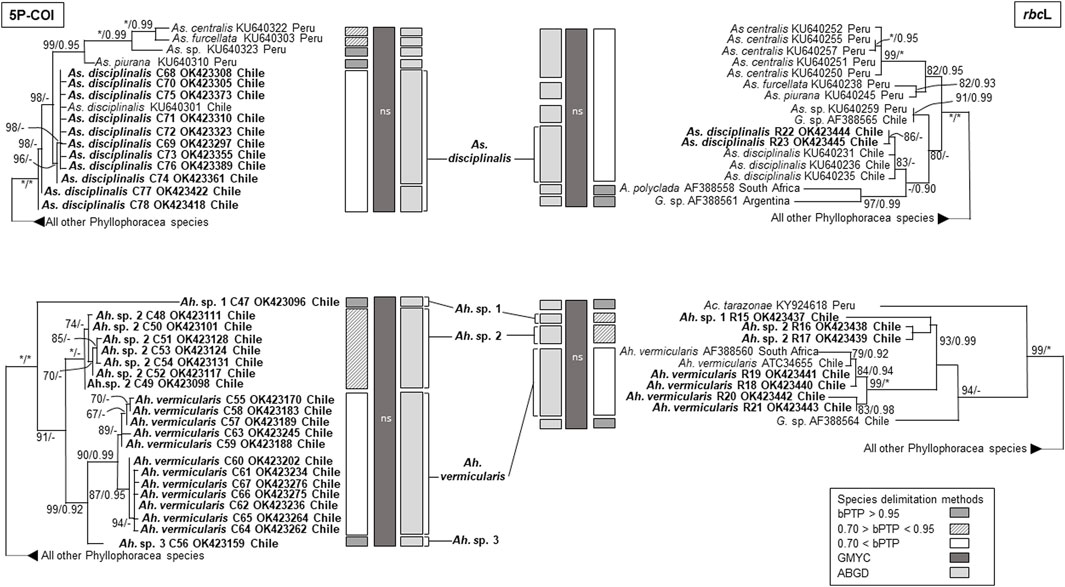
FIGURE 2. Maximun likelihooh phylogram of Asterfilopsis and Ahnfeltiopsis species inferred from COI and rbcL sequences. ML bootstrap values (≥60) and BI posterior probabilities (≥90) are indicated near branches; asterisk (*) indicates full support. Sequences obtained in our study are in bold. The results of three species delimitation methods (bPTP, GMYC, ABGD; see material and methods for more details) are indicated for both the COI and the rbcL marker. For the GYMC species delimitation method, analyses for which the model considering the existence of distinct species was not statistically significantly better than the null model (i.e., leading to the lumping of all sequences used) are indicated by “ns”.
In congruence with the well supported lineages detected in the tree reconstructions, ABGD results for both the COI and the rbcL markers suggest the existence of five putative genetic species in our Mazzaella data set: M. laminarioides North, M. laminarioides Center, M. laminarioides South, M. membranacea, and M. sp. 1 (Figure 1). For the COI, the single-threshold GMYC model (LGMY = 294.241 > L0 = 289.290, p < 0.005) was mostly congruent, but separated M. laminarioides North in two clusters (Figure 1). The GMYC analysis of the rbcL phylogeny could not reject the null hypothesis of a single species (LGMY = 32.544 > L0 = 32.083, p = 0.63; Figure 1). In Mazzaella, the separation of M. “membranacea” from Tristan Island was sustained by all three species delineation methods for the COI and by two of the three methods used for the rbcL (Figure 1). For Asterfilopsis, the ABGD analysis using sequences of COI support the separation of the haplotypes C77 and C78 from the rest sequences of As. disciplinalis obtained in Chile (including As. disciplinalis KU640301 from Chile sequenced by Calderon and Boo, 2016; Figure 2). This result was not supported by the GMYC analyses (LGMY = 94.777 > L0 = 94.025, p = 0.47) nor the ones realized using the bPTP method (Figure 2). For the rbcL, only the ABGD analysis was able to clearly separate recently diverged species from Chile and Peru and grouped all As. disciplinalis sequences from Chile (i.e., the ones obtained in our study and the ones already published in Calderon and Boo, 2016; Figure 2). For Ahnfeltiopsis, the ABGD analyses for the COI and rbcL are congruent and support the existence of four putative genetic species in our data set: Ah. vermicularis, Ah. sp. 1, Ah. sp. 2, and Ah. sp. 3 (rbcL sequence of Ah. sp. 3 not available for the analysis; Figure 2). The bPTP analyses support the existence of these putative genetic species (Figure 2). The GMYC analyses for the COI and the rbcL markers could not reject the null hypothesis of a single species (LGMY = 137.733 > L0 = 135.077, p = 0.07, and LGMY = 53.266 > L0 = 52.813, p = 0.63, for COI and rbcL, respectively; Figure 2).
3.2 Divergence times
The chronogram reconstructed with BEAST is shown in Figure 3; node age estimations exhibited relatively large confidence intervals. According to this, the separation between M. laminarioides North and M. laminarioides Center/South occurred 7.37 Ma (95% Credibility Intervals, CI: 13.22–1.82 Ma), while the separation between M. laminarioides Center and M. laminarioides South occurred 3.48 Ma (CI: 6.84–0.01 Ma). For M. membranacea and M. sp. 1, the divergence times was estimated to have occurred at 5.14 Ma (CI: 9.12–0.06 Ma). Mazzaella “membranacea” collected in Tristan Island and M. membranacea from Chile and M. sp. 1 were differentiated around 10.14 Ma (CI: 16.03–4.31 Ma). For Ahnfeltiopsis, the separation between Ah. sp. 2 and Ah. vermicularis was dated at approximately 7.44 (CI: 12.59–0.77 Ma) and the separation between Ah. sp. 1 and Ah. sp. 2/Ah. vermicularis at approximately 12.46 Ma (CI: 20.19–5.39 Ma). This last divergence time could be biased since Ah. sp. 3 was not included in the analysis (i.e., no rbcL sequence available).
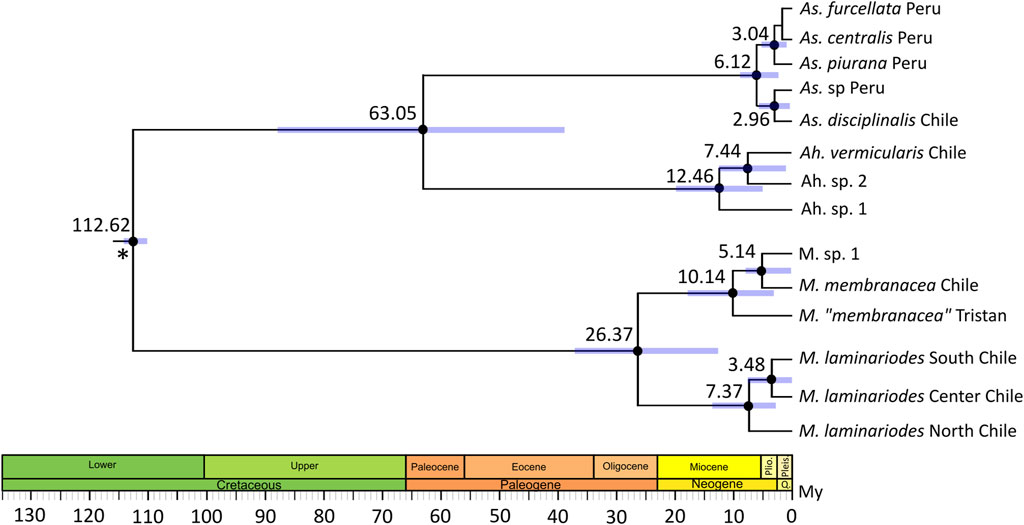
FIGURE 3. Divergence time estimates using Bayesian relaxed molecular clock (BEAST) inferred from a two-partitioned nucleotide matrix. A 572 bp COI segment and a 919bp rbcL segment were used. Black dots indicate 90% relevant nodes. Blue bars show 95% credibility intervals for node ages. Scale axis is in millions of years (My). Asterisk represent calibration point, time of divergence between Gigartinaceae and Phyllophoraceae was set to 113 My (Kumar et al., 2022). Accession numbers of the sequences used in the time-calibrated tree analysis are listed in Supplementary Table S2.
3.3 Diversity and distribution of putative genetic species
Mazzaella laminarioides North, Center, and South are well separated along the Chilean coast with M. laminarioides North distributed between the sampling sites LBR and MAI (28°55’S-32°37’S), M. laminarioides Center between the sampling sites MAT and TIR (34°05’S-37°38’S), and M. laminarioides South between the sampling sites LOB and CHI (38°39′S-41°52’S) (Figure 4; Supplementary Table S1). Mazzaella laminarioides South has been reported previously as spanning all the way down to Tierra del Fuego (∼54°S; Montecinos et al., 2012; southern sites not included in the present study). Mazzaella membranacea is a species presenting a large distribution in Chile, with individuals sampled from ∼31°S down to ∼41°S (from POS to CHI, except in MAI; Figure 4; Supplementary Table S1). The species has been mostly reported in the center south of Chile (Ramírez, 1991), where our samples have been taken; but some reports exist for Tierra del Fuego (Ramírez, 2010). Contrastingly, M. sp. 1 was encountered only in two sampling sites: MAI and TIR (32°37’S and 37°38’S, respectively; Figure 4; Supplementary Table S1). Asterfilopsis disciplinalis was encountered from MAT to CHI (34°05’S-41°52’S; Figure 4; Supplementary Table S1). Three of the four putative genetic species of Ahnfeltiopsis are fairly well separated along the Chilean coast with Ah. sp. 1 present only in LBR (28°55’S), Ah. sp. 2 in POS and MAI (32°37’S-32°37’S) and Ah. vermicularis from MAT to CHI (34°05’S-41°52’S) (Figure 4; Supplementary Table S1). Ahnfeltiopsis vermicularis extend to the coasts of South Africa (AF388560: Hondeklip Bay, Northern Cape Peninsula; Figure 2). The only sample named Ahnfeltiopsis sp. 3 was located in PMU (34°23’S; Figure 4; Supplementary Table S1).
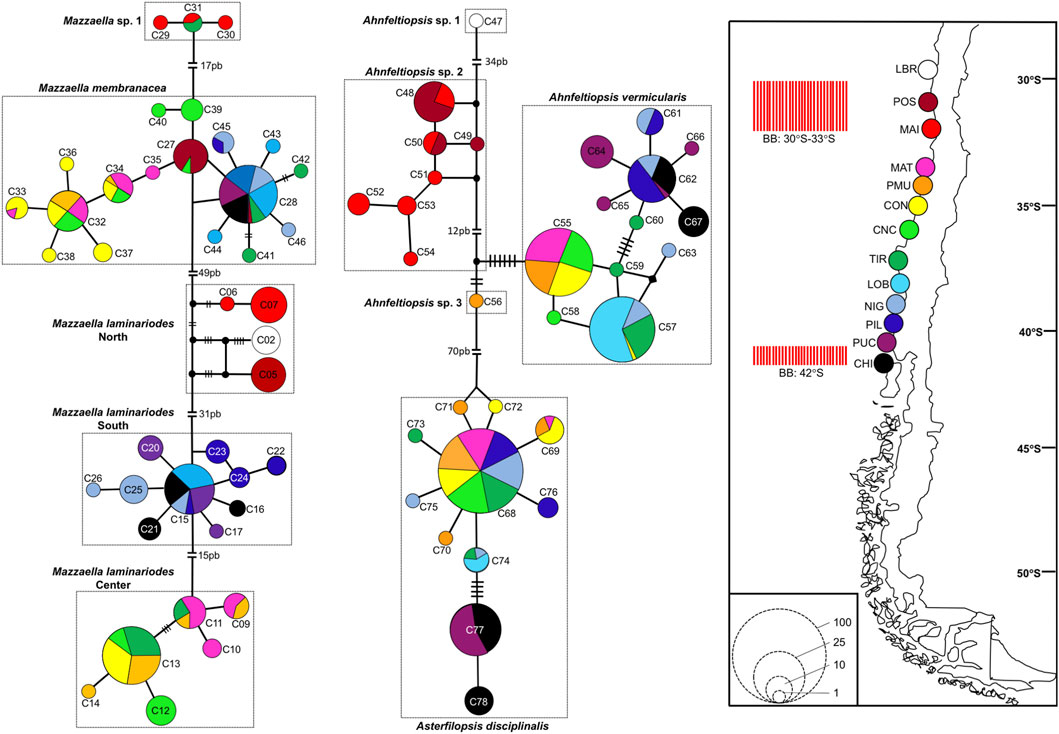
FIGURE 4. Haplotype network inferred from COI sequences in Mazzaella, Asterfilopsis, and Ahnfeltiopsis Chilean species. Map show the sampling sites and the two biogeographic breaks (BB red bars) described for the Chilean coast (Camus, 2001; Thiel et al., 2007). In the networks, each circle represents a haplotype. Circle size is proportional to the haplotype frequency while colors correspond to sampling sites where the haplotype was observed. Black circles represent hypothetical un-sampled haplotypes. For haplotypes separated by more than one mutational step, the number of steps is indicated by black bars (for values <7), or by the number of base pairs, bp (for valuer ≥7). Haplotype number as in Supplementary Table S3. Population abbreviations: Los Burros (LBR), Puerto Oscuro (POS), Maitencillo (MAI), Matanza (MAT), Pichilemu (PMU), Constitución (CON), Concepción (CNC), Tirua (TIR), Loberia (LOB), Nigue (NIG), Pilolcura (PIL), Pucatrihue (PUC), Chiloé (CHI); more details in Supplementary Table S1.
Results reported in Table 1 show that all six commonly sampled putative genetic species show a high genetic diversity. Fairly similar and high values of gene diversity were reported for these species, while nucleotide diversity was more variable. For M. laminarioides North, M. laminarioides Center, M. laminarioides South, M. membranacea, As. disciplinalis and Ah. vermicularis values obtained for H varied between 0.56 and 0.78 while values obtained for π varied between 0.0021 and 0.0103 (Table 1). In Ah. sp. 2, a putative genetic species restricted to POS and MAI, estimates of genetic diversity (36 individuals sequenced; H = 0.61; π = 0.0022; Table 1) were fairly similar to the ones obtained for the more widely distributed genetic species. For M. sp. 1, a putative genetic species for which only nine individuals were sequenced, a low level of genetic diversity was observed (H = 0.41; π = 0.0008; Table 1). In four of the six more widely distributed genetic species no clear geographic pattern of changes in genetic diversity were observed along the Chilean coast (i.e., M. laminarioides North, M. laminarioides Center, M. laminarioides South and As. disciplinalis; Supplementary Table S1). However, in M. membranacea the most southern sites of PUC and CHI were less genetically diverse (H = 0.00; π = 0.00) than the northernmost sites located from POS to PIL (0.13 < H < 0.76; 0.0002 < π < 0.0039), while in Ahfeltiopsis vermicularis the genetic diversity of the northern part of the species distribution seems to be slightly lower than the one observed in the south (MAT-LOB: 0.00 < H < 0.26; 0.00 < π < 0.0014; NIG-CHI: 0.34 < H < 0.76; 0.0006 < π < 0.0089; Supplementary Table S2).
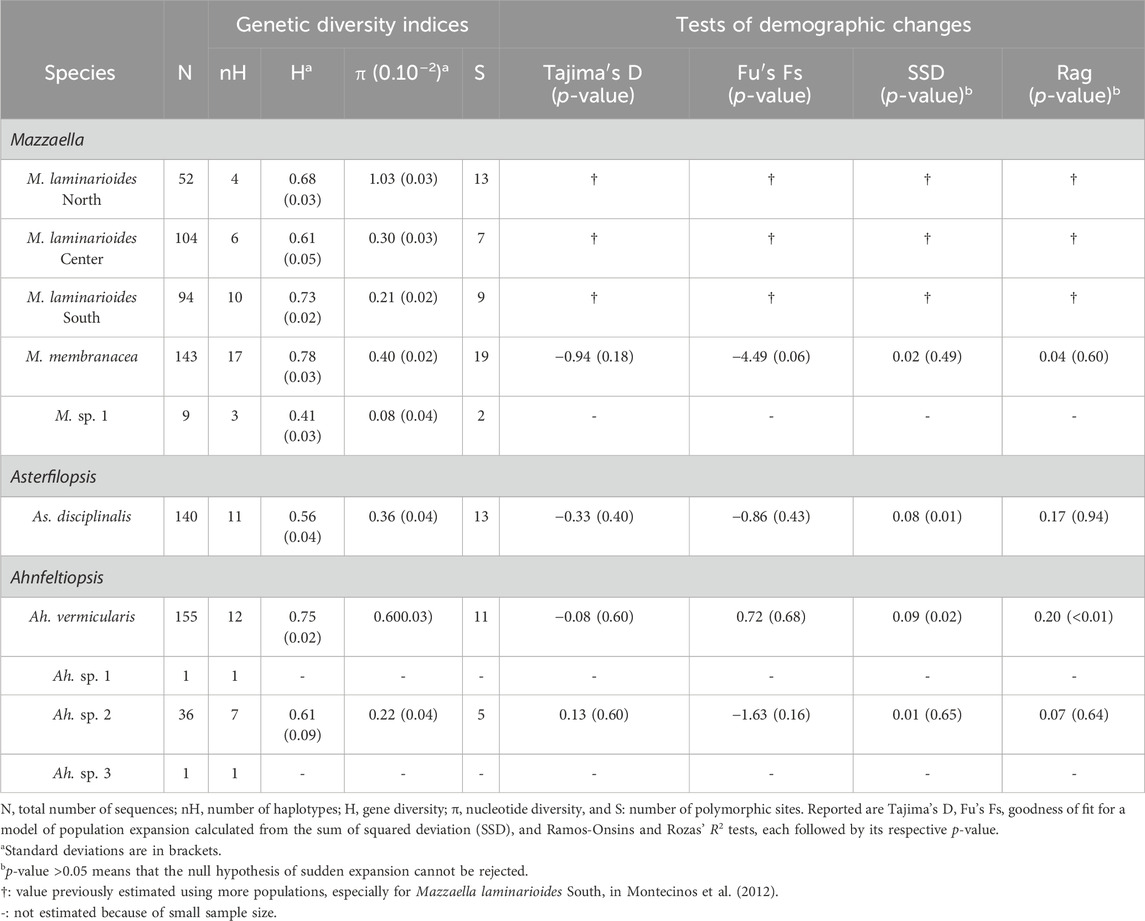
TABLE 1. Indices of genetic diversity and test of historical demographic change based on COI sequences obtained for each Mazzaella, Asterfilopsis, and Ahnfeltiopsis species sampled along the Chilean coast.
3.4 Existence of substructure in species presenting a large distribution in Chile
For M. laminarioides Center and South, no clear genetic structuring was observed, with common haplotypes shared among various sampling sites (C11 and C13 for M. laminarioides Center and C15 for M. laminarioides South; Figure 4). Contrastingly, in M. laminarioides North no haplotypes were shared among sampling sites (Figure 4; Supplementary Table S3). These haplotypes were separated by at least nine mutations (Figure 4), leading to a strong genetic substructure (0.97 < ΦST values calculated among localities <1.00; Supplementary Table S4). In M. membranacea, except for one sample from POS for which the most common haplotype sequenced in the southern part of the species distribution was observed (haplotype C28; Figure 4), haplotypes sequenced in the northern part of the species distribution (i.e., from POS to CNC; 31°24’S-36°31'S) where distinct than the ones from the south (i.e., TIR to CHI; 37°38’S-41°52'S). Only one mutation separated C28, sampled in all six southernmost populations, from C27 encountered in CNC and POS (Figure 4; Supplementary Table S3). Pairwise ΦST calculated among localities located between POS and CNC ranged from 0.03 to 0.88 (ΦST = 0.41 ± 0.31) and the one calculated among localities located between TIR and CHI ranged from 0.00 to 0.14 (ΦST = 0.06 ± 0.05) (Table 2; Supplementary Table S4). Pairwise ΦST calculated among localities from the two distinct genetic groups ranged from 0.47 to 0.91 (ΦST = 0.78 ± 0.13; Table 2; Supplementary Table S4). The results from AMOVA indicated that the majority of variation was observed between genetic groups (i.e., 62%; Table 3). Mazzaella sp. 1 did not present any sign of genetic structure, with the haplotype C31 shared between MAI and TIR (Figure 4; Supplementary Table S3).
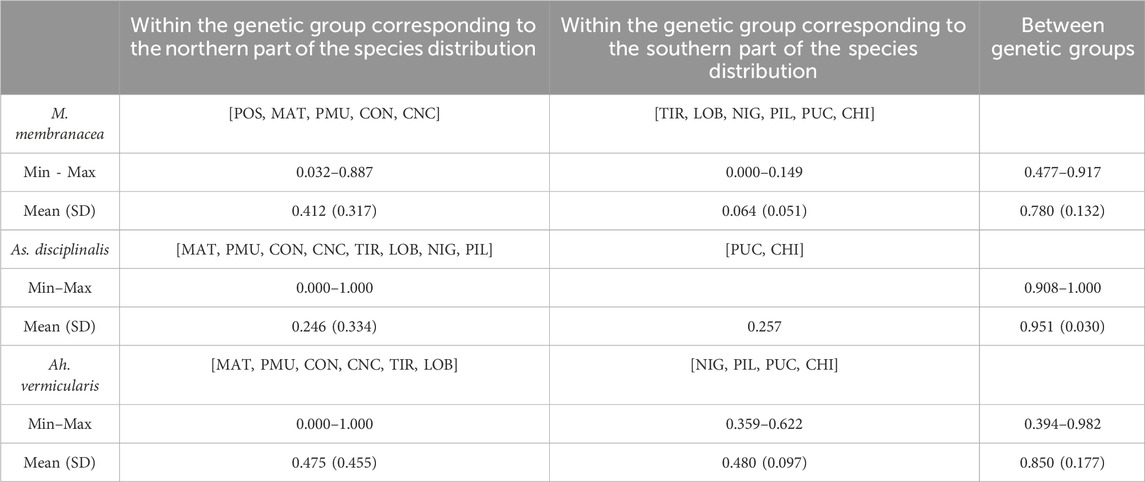
TABLE 2. Pairwise ΦST between sampling sites estimated from COI data set in Mazzaella membranacea, Asterfilopsis disciplinalis, and Ahnfeltiopsis vermicularis. Sampling sites included in each genetic group are indicated between square brackets.
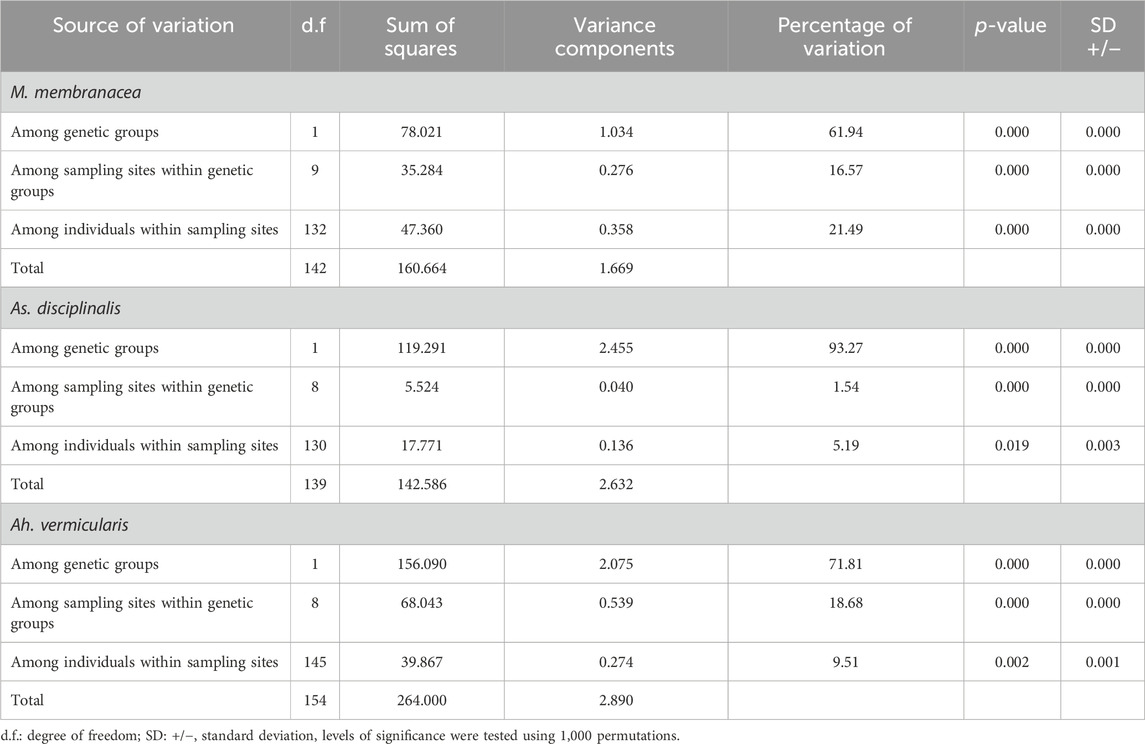
TABLE 3. Analysis of Molecular Variance (AMOVA) implemented to explore the spatial partitioning of variance within sampling sites, among sampling sites within genetic groups and among genetic groups in Mazzaella membranacea, Asterfilopsis disciplinalis, and Ahnfeltiopsis vermicularis.
Asterfilopsis disciplinalis showed a clear genetic break between localities of PIL (39°4’S) and PUC (40°32'S), with four fixed mutations separating haplotypes from the northern and southern part of the species distribution (Figure 4). Pairwise ΦST calculated among localities from the two distinct genetic groups (0.91 < ΦST < 1.00; Supplementary Table S4) were higher than the ones calculated among localities located within each group (0.00 < ΦST < 1.00; Supplementary Table S4). The AMOVA shows that the largest fraction of the variation is distributed between genetic groups (i.e., 93%; Table 3), supporting the high level of genetic differentiation between the two genetic groups (ΦST = 0.95 ± 0.03; Table 2). The COI sequences obtained in Calderon and Boo, (2016) from Pichilemu (GenBank accession number KU640301) and Valdivia (GenBank accession number KU640298), both from sites located north of the 39°S genetic break, support our results. KU640301 correspond to the haplotype C68 and KU640298 to a new rare haplotype separated only by one mutation from C68.
In Ah. vermicularis, haplotypes encountered in the northern part of the species distribution (from MAT to LOB; 34°05’S-38°39'S) were separated from the ones sampled in the south (i.e., from NIG to CHI; 39°17’S-41°52'S) by one mutation (Figure 4). The pairwise ΦST values calculated among localities located between MAT and LOB ranged from 0.00 to 1.00 (ΦST = 0.47 ± 0.45) and the one calculated among localities located between NIG and CHI ranged from 0.35 to 0.62 (ΦST = 0.48 ± 0.09) (Table 2; Supplementary Table S4). The ΦST calculated among localities from the two distinct genetic groups ranged from 0.39 to 0.98 (ΦST = 0.85 ± 0.17) (Table 2; Supplementary Table S4). The AMOVA shows that the largest fraction of the variation is distributed between genetic groups (i.e., 72%; Table 3). Ahnfeltiopsis sp. 2 showed a reticulated network with two shared haplotypes between POS and MAI (haplotypes C48 and C50; Figure 4) and the ΦST calculated between these two sites was of 0.34 (Supplementary Table S4).
3.5 Historical demography
For M. laminarioides, tests of historical demographic change have already been reported for the three putative genetic species in Montecinos et al. (2012). This study revealed that M. laminarioides North had passed through a recent bottleneck while signs of population growth were detected for M. laminarioides South, linked with rapid post-glacial expansion. Mazzaella laminarioides Center did not reveal any clear pattern of effective population size fluctuations throughout time.
For M. membranacea, As. disciplinalis, Ah. vermicularis, and Ah. sp. 2, the mismatch distributions are bimodal, with SSD and Rag index that generally did not allow to reject the hypothesis of a sudden expansion except for the SSD in the case of As. disciplinalis and both the SSD and the Rag index in the case of Ah. vermicularis (Table 1; Figure 5). Non-significant values were obtained for the Fu’s Fs and Tajima’s D tests for the four putative genetic species (Table 1). For Ah. vermicularis and Ah. sp. 2, BSP showed no evidence of clear demographic changes (Figure 5). Contrastingly, in M. membranacea and As. disciplinalis recent demographic increase are detected in the BSP analyses (more or less 25,000 years ago, Figure 5). These results are supported by fairly high negative values of Fu’s Fs and Tajima’s D in the case of these two putative genetic species (Table 1). However, only mild increase in Ne (i.e., effective population size), at most, were detected in the BSP.
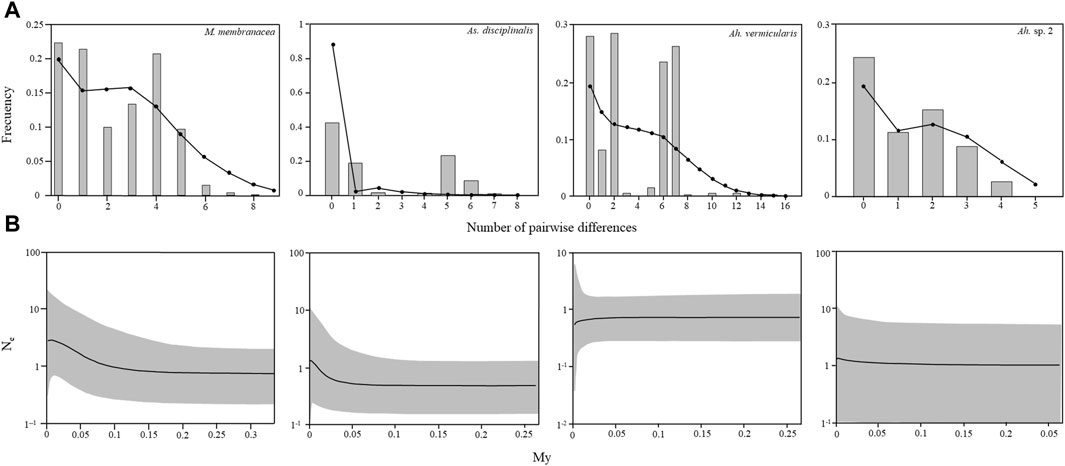
FIGURE 5. Mismatch distribution and Bayesian Skyline plots calculated with COI sequences for Mazzaella membranacea, Asterfilopsis disciplinalis, Ahnfeltiopsis vermicularis, and Ah. sp. 2. (A) Observed mismatch distribution, represented by grey bars, and expected mismatch distribution under a spatial expansion model represented by black circles. (B) Changes in effective population size estimated by Bayesian Skyline plots. Solid lines are the median posterior effective population size through time; gray area indicates the 95% confidence interval for each estimate.
4 Discussion
4.1 Widespread phylogeographical breaks in the marine flora of northern Chile: ancient vicariant origin maintained by upwelling regimes
Based on oceanographic patterns and the distributional limit of marine species, two biogeographical barriers, located at 30–33°S and 42°S, have been detected along the Chilean coast (Camus, 2001; Thiel et al., 2007). In the case of seaweeds, an important biogeographic break has been observed at 42°S for brown, green, and red seaweeds, while the northern biogeographical barrier is more taxa dependent, and was observed at 30° S for brown seaweeds, and between 32°S and 33°S for red seaweeds (Santelices, 1980; Meneses and Santelices, 2000). Our result showed the existence of deep phylogeographic breaks for Mazzaella at 33°S and Ahnfeltiopsis at 30°S and 33°S. Indeed, at 33°S, we observed the separation between the putative genetic species M. laminarioides North and M. laminarioides Center, and Ah. sp. 2 and Ah. vermicularis. The species Ah. sp. 1 and Ah. sp. 2 were located on both side of the 30°S biogeographic barrier. The divergence between M. laminarioides North and M. laminarioides Center was estimated to have occurred around 7 Ma (CI: 13–2 Ma) and the divergence between Ah. sp. 2 and Ah. vermicularis around 7 Ma (CI: 13–1 Ma) while Ah. sp. 1 spitted from and Ah. sp. 2/Ah. vermicularis some 12 Ma (CI: 20–5 Ma). These estimations support the hypothesis that an old vicariant event could have strongly structured the genetic diversity of marine organisms at 30–33°S (Tellier et al., 2009; Haye et al., 2014; Guillemin et al., 2016). Distinct sister species or highly genetically divergent lineages separated by the 30°S biogeographic barrier have been observed in intertidal kelps of the genus Lessonia (L. berteroana and L. spicata: González et al., 2012; Tellier et al., 2009), and marine invertebrates with direct development (Acanthina monodon: Sánchez et al., 2011; Crepipatella dilatata; Brante et al., 2012; Excirolana braziliensis; Varela and Haye, 2012; Orchestoidea tuberculata, Scurria scurra, and Tegula atra; Haye et al., 2014), or pelagic larvae (Stichaster striatus: Haye et al., 2014; Notochthamalus scabrosus; Zakas et al., 2009). Palaeoceanographic reconstructions have inferred important changes in the surface temperature (SST) and ocean circulation in northern Chile during the Neogene and the Quaternary (Tsuchi, 2002; Kaiser et al., 2005; Villafaña and Rivadeneira, 2018). For example, a deep impact on population demography of marine organisms (Cárdenas et al., 2008) have been associated with of the decrease of SST and the activation of the upwelling systems along the coast of northern Chile and Peru that took place during mid Miocene (∼15–12 Ma) (Tsuchi, 2002). Additionally, it has been proposed that the Quaternary glacial/interglacial cycles that begun at the end of the Plio-Pleistocene transition (i.e., some 2.5 Ma) were associated with oscillations in the position of the West Wind Drift (WWD) and main ocean currents, with the WWD reaching the Chilean coast around 33°S during glacial periods (Mohtadi and Hebbeln, 2004; Kaiser et al., 2005; Mohtadi et al., 2008). These historical changes in oceanographic features could potentially be at the origin of strong historical barriers to gene flow along the northern part of the Chilean coast. In various studies, it has been suggested that the phylogeographic breaks observed at 30–33°S have been maintained up to the present time in species with low dispersal capacity by somewhat more porous barriers to gene flow linked with changes in the currents, wind, and sea surface temperature patterns in this area (Tellier et al., 2009; Sánchez et al., 2011; Montecinos et al., 2012). Currently, the 30–33°S region is characterized by latitudinal change in the type of upwellings that module the dynamics of intertidal communities of northern Chile, with strong and seasonal upwellings south of this area and more permanent but weak upwellings in the northern part of the coast (Broitman et al., 2001; Navarrete et al., 2005; Tapia et al., 2014). Changes in coastline topography (i.e., headlands) are observed at both 30°S and 33°S and were associated with the existence of strong upwelling centers at these latitudes (Figueroa and Moffat, 2000; Aiken et al., 2008). These upwelling centers have been proposed as important dispersal barriers for marine invertebrate taxa due to offshore displacement of propagules and abrupt change in chemical structure of the water column, as the oxygen concentration, for example, (Navarrete et al., 2005; Lara et al., 2019).
4.2 Shallow and taxa dependent genetic sub-structuring patterns in central-southern Chile: potential effect of habitat discontinuities and stochastic events
Contrasting from the 30°S and the 33°S in northern Chile, the genetic breaks observed between 33°S and 42°S generally correspond to shallow intraspecific divergence and their locations vary greatly among the marine species studied in this area (locations of genetic divergence detected in red algae: 34°S, 36°S-37°S, 39°S, and 40°S; Cid-Alda et al., 2023; Lindstrom et al., 2015; Montecinos et al., 2012; present study; brown algae: 36°S-37°S Fraser et al., 2010; and marine invertebrates: 34°S, 35°S, 36°S-37°S, 39°S and 40°S; Ewers-Saucedo et al., 2016; Guinez et al., 2016; Lara et al., 2019; Peluso et al., 2023; Quesada-Calderon et al., 2021; Sáenz-Agudelo et al., 2022). The nearly linear central-southern coastline, largely dominated by rocky shores, is interrupted by extensive sandy beaches in this region, as, for example, from Punta Morguilla to Quidico (37°43’S-38°14'S; 62 km long beach) or from Loberia to Nigue (38°39’S-39°17'S; 73 km long beach) (Thiel et al., 2007). Additionally, the coastline is interrupted by the freshwater discharge of large rivers originating in the Andes Range south of 33°S such as the Maule (35°18'S), Itata (36°23'S), Bio Bio (36°49'S), Imperial (38°47'S), and Valdivia (39°5’S) rivers (Saldías et al., 2012). Both sandy beaches and river mouths, correspond to important barriers to gene flow for marine species (Lara et al., 2019), particularly for sessile organisms presenting low dispersal capacity, as seaweeds (Fraser et al., 2010; Montecinos et al., 2012; Cid-Alda et al., 2023).
We did not find any congruence in time (i.e., numbers of fixed mutations) nor space among the phylogeographic breaks or patterns of genetic sub-structure observed in Mazzaella, Asterfilopsis, and Ahnfeltiopsis. A deep phylogeographic break that occurred a few million years ago (i.e., 3 Ma, CI: 7–0 Ma) was observed between M. laminarioides Center and M. laminarioides South at 39°S. Contrastingly, only much more recent intraspecific sub-structuration patterns were detected within M. membranacea, Ah. vermicularis, and As. disciplinalis located at 36°S, 39°S, and 40°S, respectively. Depending on the species, moderate to high genetic differentiation among genetic clusters (0.78 < mean ΦST between genetic groups <0.95) were observed within M. membranacea, As. disciplinalis, and Ah. vermicularis. These sub-structuring patterns do not support the existence of a unique vicariant event driving the genetic structure of marine organisms in central-southern Chile but suggest more species-specific evolutionary history.
Since the late Pliocene, the part of the Chilean coast located between 37°S and 41°S has been affected by seismic activity causing constant and extensive coastline uplifts (Folguera et al., 2011) and sediment rerouting thought the Andean drainage systems (Rehak et al., 2008). Consequently, the central-southern Chilean coastline present a high level of habitat heterogeneity, very dynamic over time. In red algae, characterized generally by poor dispersive ability and small effective population sizes (Faugeron et al., 2001; Engel et al., 2004; Cid-Alda et al., 2023), dynamic coastal and oceanographic features as the ones described between 37°S and 41°S can lead to patchy distribution, with isolated populations rapidly diverging due to genetic drift (Irwin et al., 2001; Irwin, 2002; Kuo and Avise, 2005). Large-scale mortality of intertidal and subtidal communities as a result of tectonically uplifted shoreline has been widely recorded in central-southern Chile (Castilla, 1988; Castilla et al., 2010; Jaramillo et al., 2012). Local/regional extirpations followed by recolonizations from sources populations not necessarily located nearby can lead to genetic anomalies that present the same signature as classical phylogeographic breaks but are not linked to the existence of any barrier to gene flow (Parvizi et al., 2019; Vaux et al., 2021; Parvizi et al., 2022; Vaux et al., 2022; Vaux et al., 2023). In Chile, it has been shown that regional disturbance events can rapidly reshape the pattern of distribution of genetic diversity in coastal species (Brante et al., 2019; Becheler et al., 2020). It is possible that short-term mesoscale disturbances, as the ones occurring between 37°S and 41°S in Chile due to near-shore fault rupture events causing coastal uplift (Folguera et al., 2011), could have contributed to the generation of the specific genetic sub-structuring patterns observed in the four red algae under study.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
OH: Conceptualization, Formal Analysis, Methodology, Writing–original draft, Writing–review and editing. AM: Methodology, Writing–review and editing. FS-E: Writing–review and editing; Methodology; and Formal analysis. M-LG: Conceptualization, Methodology, Writing–original draft, Writing–review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by ANID CONICYT (Fondo Nacional de Desarrollo Científico y Tecnológico FONDECYT, Chile) under grants no. 1130797 and 1221477. OH acknowledges support from BECA DE DOCTORADO NACIONAL Grant no. 21120791 and FONDECYT POSDOCTADO Grant no. 3240090 and M-LG from the Núcleo Milenio MASH grant NCN 2021-033 (ANID, Chile).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2024.1336427/full#supplementary-material
References
Aiken, C. M., Castillo, M. I., and Navarrete, S. A. (2008). A simulation of the Chilean coastal Current and associated topographic upwelling near Valparaíso, Chile. Cont. l Shelf Res. 28 (17), 2371–2381. doi:10.1016/j.csr.2008.05.006
Arbogast, B. S., and Kenagy, G. J. (2001). Comparative phylogeography as an integrative approach to historical biogeography. J. Biogeogr. 28, 819–825. doi:10.1046/j.1365-2699.2001.00594.x
Atkinson, L. P., Valle-Levinson, A., Figueroa, D., De Pol-Holz, R., Gallardo, V. A., Schneider, W., et al. (2002). Oceanographic observations in Chilean coastal waters between Valdivia and Concepción. J. Geophys. Res. Earth Surf. 107 (18), 1–13. doi:10.1029/2001JC000991
Avise, J. C. (2000). Phylogeography: the history and formation of species. United States: Harvard University Press.
Ayres-Ostrock, L. M., Valero, M., Mauger, S., Oliveira, M. C., Plastino, E. M., Guillemin, M. L., et al. (2019). Dual influence of terrestrial and marine historical processes on the phylogeography of the Brazilian intertidal red alga Gracilaria caudata. J. Phycol. 55 (5), 1096–1114. doi:10.1111/jpy.12892
Bagley, J. C., and Johnson, J. B. (2014). Phylogeography and biogeography of the lower Central American Neotropics: diversification between two continents and between two seas. Biol. Rev. 89 (4), 767–790. doi:10.1111/brv.12076
Bandelt, H. J., Forster, P., and Röhl, A. (1999). Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48. doi:10.1093/oxfordjournals.molbev.a026036
Becheler, R., Guillemin, M. L., Stoeckel, S., Mauger, S., Saunier, A., Brante, A., et al. (2020). After a catastrophe, a little bit of sex is better than nothing: genetic consequences of a major earthquake on asexual and sexual populations. Evol. Appl. 13 (8), 2086–2100. doi:10.1111/eva.12967
Bermingham, E., and Moritz, C. (1998). Comparative phylogeography: concepts and applications. Mol. Ecol. 7, 367–369. doi:10.1046/j.1365-294x.1998.00424.x
Bouckaert, R., Vaughan, T. G., Barido-Sottani, J., Duchêne, S., Fourment, M., Gavryushkina, A., et al. (2019). BEAST 2.5: an advanced software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 15 (4), e1006650. doi:10.1371/journal.pcbi.1006650
Brante, A., Fernández, M., and Viard, F. (2012). Phylogeography and biogeography concordance in the marine gastropod Crepipatella dilatata (Calyptraeidae) along the southeastern Pacific coast. J. Hered. 103, 630–637. doi:10.1093/jhered/ess030
Brante, A., Guzmán-Rendón, G., Barría, E. M., Guillemin, M. L., Vera-Escalona, I., and Hernández, C. E. (2019). Post-disturbance genetic changes: the impact of the 2010 mega-earthquake and tsunami on Chilean Sandy Beach fauna. Sci. Rep. 9 (1), 14239. doi:10.1038/s41598-019-50525-1
Broitman, B., Navarrete, S., Smith, F., and Gaines, S. (2001). Geographic variation of southeastern Pacific intertidal communities. Mar. Eco. Prog. Ser. 224, 21–34. doi:10.3354/meps224021
Calderon, M. S., and Boo, S. M. (2016). Phylogeny of Phyllophoraceae (Rhodophyta, Gigartinales) reveals Asterfilopsis gen. Nov. From the southern Hemisphere. Phycologia 55 (5), 543–554. doi:10.2216/16-9.1
Calderon, M. S., and Boo, S. M. (2017). The Phyllophoraceae (Gigartinales, Rhodophyta) from Peru with descriptions of Acletoa tarazonae gen. and sp. nov. and Gymnogongrus caespitosus sp. nov. Phycologia 56 (6), 686–696. doi:10.2216/16-126.1
Camus, P. A. (1992). Size-specific reproductive parameters in red algae: a comparative analysis for two sympatric species from Central Chile. Oecologia 92, 450–456. doi:10.1007/BF00317472
Camus, P. A. (2001). Biogeografía marina de Chile continental. RCHN 74, 587–617. doi:10.4067/s0716-078x2001000300008
Cárdenas, L., Viard, F., and Castilla, J. C. (2008). Paleobiogeography and taxonomy of the genus Concholepas (Lamarck, 1801): a review and new evidences. RCHN 81 (3), 417–436. doi:10.4067/s0716-078x2008000300010
Castilla, J. C. (1988). Earthquake-caused coastal uplift and its effects on rocky intertidal kelp communities. Science 242, 440–443. doi:10.1126/science.242.4877.440
Castilla, J. C., Manríquez, P. H., and Camaño, A. (2010). Effects of rocky shore coseismic uplift and the 2010 Chilean mega-earthquake on intertidal biomarker species. Mar. Ecol. Prog. Ser. 418, 17–23. doi:10.3354/meps08830
Cid-Alda, F. P., Montecinos, A. E., and Guillemin, M. L. (2023). A temporal and spatial study of genetic structure in four species of bladed Bangiales (Rhodophyta) from the southeastern Pacific coast of Chile. J. Phycol. 59, 712–724. doi:10.1111/jpy.13343
Cowman, P. F., and Bellwood, D. R. (2013). Vicariance across major marine biogeographic barriers: temporal concordance and the relative intensity of hard versus soft barriers. Proc. R. Soc. B Biol. Sci. 280, 20131541. doi:10.1098/rspb.2013.1541
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9 (8), 772. doi:10.1038/nmeth.2109
Engel, C. R., Destombe, C., and Valero, M. (2004). Mating system and gene flow in the red seaweed Gracilaria gracilis: effect of haploid-diploid life history and intertidal rocky shore landscape on fine-scale genetic structure. Heredity 92, 289–298. doi:10.1038/sj.hdy.6800407
Ewers-Saucedo, C., Pringle, J. M., Sepúlveda, H. H., Byers, J. E., Navarrete, S. A., and Wares, J. P. (2016). The oceanic concordance of phylogeography and biogeography: a case study in Notochthamalus. Ecol. Evol. 6 (13), 4403–4420. doi:10.1002/ece3.2205
Excoffier, L., and Lischer, H. E. L. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Res. 10, 564–567. doi:10.1111/j.1755-0998.2010.02847.x
Excoffier, L., Smouse, P. E., and Quattro, J. (1992). Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131 (2), 479–491. doi:10.1093/genetics/131.2.479
Faugeron, S., Valero, M., Destombe, C., Martínez, E. A., and Correa, J. A. (2001). Hierarchical spatial structure and discriminant analysis of genetic diversity in the red alga Mazzaella laminarioides (Gigartinales, Rhodophyta). J. Phycol. 37, 705–716. doi:10.1046/j.1529-8817.2001.01072.x
Figueroa, D., and Moffat, C. (2000). On the influence of topography in the induction of coastal upwelling along the Chilean coast. Geophys. Res. Lett. 27 (23), 3905–3908. doi:10.1029/1999GL011302
Folguera, A., Orts, D., Spagnuolo, M., Vera, E. R., Litvak, V., Sagripanti, L., et al. (2011). A review of late cretaceous to quaternary palaeogeography of the southern Andes. Proc. R. Soc. B Biol. Sci. 103 (2), 250–268. doi:10.1111/j.1095-8312.2011.01687.x
Francisco, P. M., Mori, G. M., Alves, F. M., Tambarussi, E. V., and de Souza, A. P. (2018). Population genetic structure, introgression, and hybridization in the genus Rhizophora along the Brazilian coast. Ecol. Evol. 8 (6), 3491–3504. doi:10.1002/ece3.3900
Fraser, C. I., Davies, I. D., Bryant, D., and Waters, J. M. (2018). How disturbance and dispersal influence intraspecific structure. J. Ecol. 106 (3), 1298–1306. doi:10.1111/1365-2745.12900
Fraser, C. I., Thiel, M., Spencer, H. G., and Waters, J. M. (2010). Contemporary habitat discontinuity and historic glacial ice drive genetic divergence in Chilean kelp. BMC Evol. Biol. 10, 203–212. doi:10.1186/1471-2148-10-203
Fredericq, S., and Lopez-Bautista, J. M. (2002). Characterization and phylogenetic position of the red alga Besa papillaeformis Setchell: an example of progenetic heterochrony. Constancea 83, 1–12.
Fu, Y. X. (1997). Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147, 915–925. doi:10.1093/genetics/147.2.915
Fujisawa, T., and Barraclough, T. G. (2013). Delimiting species using single-locus data and the Generalized Mixed Yule Coalescent approach: a revised method and evaluation on simulated data sets. Syst. Biol. 62 (5), 707–724. doi:10.1093/sysbio/syt033
González, A., Beltrán, J., Hiriart-Bertrand, L., Flores, V., de Reviers, B., Correa, J. A., et al. (2012). Identification of cryptic species in the Lessonia nigrescens complex (Phaeophyceae, Laminariales). J. Phycol. 48 (5), 1153–1165. doi:10.1111/j.1529-8817.2012.01200.x
Guillemin, M.-L., Valero, M., Tellier, F., Macaya, E. C., Destombe, C., and Faugeron, S. (2016). “Phylogeography of seaweeds in the South East Pacific: complex evolutionary processes along a latitudinal gradient,” in Seaweed phylogeography. Editors Z. M. Hu, and C. Fraser (Cham: Springer), 251–277.
Guiry, M. D., and Guiry, G. M. (2024). AlgaeBase. World-wide electronic publication. Galway: National University of Ireland.
Haye, P. A., Segovia, N. I., Muñoz-Herrera, N. C., Gálvez, F. A., Martínez, A., Meynard, A., et al. (2014). Phylogeographic structure in benthic marine invertebrates of the southeast pacific coast of Chile with differing dispersal potential. PLoS ONE 9, e88613. doi:10.1371/journal.pone.0088613
Hernández-Miranda, E., Cisterna, J., Díaz-Cabrera, E., Veas, R., and Quiñones, R. A. (2014). Epibenthic macrofaunal community response after a mega-earthquake and tsunami in a shallow bay off central-south Chile. Mar. Biol. 161, 681–696. doi:10.1007/s00227-013-2370-x
Hiller, A., and Lessios, H. A. (2020). Marine species formation along the rise of Central America: the anomuran crab Megalobrachium. Mol. Ecol. 29 (2), 413–428. doi:10.1111/mec.15323
Hoffmann, A., and Santelices, B. (1997). Flora marina de Chile. Santiago, Chile: Ediciones Universidad Católica de Chile, 434.
Hommersand, M. H., Fredericq, S., and Freshwater, D. W. (1994). Phylogenetic systematics and biogeography of the Gigartinaceae (Gigartinales, Rhodophyta) based on sequence analysis of rbcL. Bot. Mar. 37, 193–203. doi:10.1515/botm.1994.37.3.193
Hu, Z., Guiry, M. D., Critchley, A. T., and Duan, D. (2010). Phylogeographic patterns indicate transatlantic migration from Europe to North America in the red seaweed Chondrus crispus (Gigartinales, Rhodophyta). J. Phycol. 46 (5), 889–900. doi:10.1111/j.1529-8817.2010.00877.x
Huelsenbeck, J. P., and Ronquist, F. (2001). MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755. doi:10.1093/bioinformatics/17.8.754
Irwin, D. E. (2002). Phylogeographic breaks without geographic barriers to gene flow. Evolution 56, 2383–2394. doi:10.1111/j.0014-3820.2002.tb00164.x
Irwin, D. E., Bensch, S., and Price, T. D. (2001). Speciation in a ring. Nature 409, 333–337. doi:10.1038/35053059
Jaramillo, E., Dugan, J. E., Hubbard, D. M., Melnick, D., Manzano, M., Duarte, C., et al. (2012). Ecological implications of extreme events: footprints of the 2010 earthquake along the Chilean coast. PLoS ONE 7, e35348. doi:10.1371/journal.pone.0035348
Kaiser, J., Lamy, F., and Hebbeln, D. (2005). A 70-kyr sea surface temperature record off southern Chile (Ocean Drilling Program Site 1233). Paleoceanography 20, 1–15. doi:10.1029/2005PA001146
Kamiya, M. (2004). Phylogeography of Caloglossa leprieurii and related species (Delesseriaceae, Rhodophyta) based on the rbcL gene sequences. Jpn J Phycol 52, 147–151.
Kelly, R. P., and Palumbi, S. R. (2010). Genetic structure among 50 species of the northeastern Pacific rocky intertidal community. PLoS ONE 5, e8594. doi:10.1371/journal.pone.0008594
Kumar, S., Suleski, M., Craig, J. M., Kasprowicz, A. E., Sanderford, M., Li, M., et al. (2022). TimeTree 5: an expanded resource for species divergence times. Mol. Biol. Evol. 39 (8), msac174. doi:10.1093/molbev/msac174
Kuo, C. H., and Avise, J. C. (2005). Phylogeographic breaks in low-dispersal species: the emergence of concordance across gene trees. Genetica 124, 179–186. doi:10.1007/s10709-005-2095-y
Lara, C., Saldías, G. S., Cazelles, B., Rivadeneira, M. M., Haye, P. A., and Broitman, B. R. (2019). Coastal biophysical processes and the biogeography of rocky intertidal species along the south-eastern Pacific. J. Biogeogr. 46 (2), 420–431. doi:10.1111/jbi.13492
Lindstrom, S. C., Gabrielson, P. W., Hughey, J. R., Macaya, E. C., and Nelson, W. A. (2015). Sequencing of historic and modern specimens reveals cryptic diversity in Nothogenia (Scinaiaceae, Rhodophyta). Phycologia 54 (2), 97–108. doi:10.2216/14-077.1
Macaya, E. C., and Zuccarello, G. C. (2010). Genetic structure of the giant kelp Macrocystis pyrifera along the southeastern pacific. Mar. Ecol. Prog. Ser. 420, 103–112. doi:10.3354/meps08893
Marko, P. B., Hoffman, J. M., Emme, S. A., McGovern, T. M., Keever, C. C., and Cox, L. N. (2010). The “expansion-contraction” model of Pleistocene biogeography: rocky shores suffer a sea change? Mol. Ecol. 19, 146–169. doi:10.1111/j.1365-294X.2009.04417.x
Meneses, I., and Santelices, B. (2000). Patterns and breaking points in the distribution of benthic algae along the temperate Pacific coast of South America. RCHN 73, 615–623. doi:10.4067/s0716-078x2000000400006
Miura, O., Kanaya, G., Nakai, S., Itoh, H., Chiba, S., Makino, W., et al. (2017). Ecological and genetic impact of the 2011 Tohoku earthquake tsunami on intertidal mud snails. Sci. Rep. 7, 44375. doi:10.1038/srep44375
Mohtadi, M., and Hebbeln, D. (2004). Mechanisms and variations of the paleoproductivity off northern Chile (24°S-33°S) during the last 40,000 years. Paleoceanography 19. doi:10.1029/2004PA001003
Mohtadi, M., Rossel, P., Lange, C. B., Pantoja, S., Böning, P., Repeta, D. J., et al. (2008). Deglacial pattern of circulation and marine productivity in the upwelling region off central-south Chile. EPSL 272 (1-2), 221–230. doi:10.1016/j.epsl.2008.04.043
Monaghan, M. T., Wild, R., Elliot, M., Fujisawa, T., Balke, M., Inward, D. J. G., et al. (2009). Accelerated species inventory on Madagascar using coalescent-based models of species delineation. Syst. Biol. 58, 298–311. doi:10.1093/sysbio/syp027
Montecinos, A., Broitman, B. R., Faugeron, S., Haye, P. A., Tellier, F., and Guillemin, M. L. (2012). Species replacement along a linear coastal habitat: phylogeography and speciation in the red alga Mazzaella laminarioides along the south east pacific. BMC Evol. Biol. 12, 97. doi:10.1186/1471-2148-12-97
Morales-González, S., Giles, E. C., Quesada-Calderón, S., and Sáenz-Agudelo, P. (2019). Fine-scale hierarchical genetic structure and kinship analysis of the ascidian Pyura chilensis in the southeastern Pacific. Ecol. Evol. 9 (17), 9855–9868. doi:10.1002/ece3.5526
Navarrete, S. A., Wieters, E. A., Broitman, B. R., and Castilla, J. C. (2005). Scales of benthic-pelagic coupling and the intensity of species interactions: from recruitment limitation to top-down control. PNAS 102 (50), 18046–18051. doi:10.1073/pnas.0509119102
Norton, T. A. (1992). Dispersal by macroalgae. Brit. Phycol. J. 27, 293–301. doi:10.1080/00071619200650271
Parvizi, E., Craw, D., and Waters, J. M. (2019). Kelp DNA records late Holocene paleoseismic uplift of coastline, southeastern New Zealand. EPSL 520, 18–25. doi:10.1016/j.epsl.2019.05.034
Parvizi, E., Fraser, C. I., Dutoit, L., Craw, D., and Waters, J. M. (2020). The genomic footprint of coastal earthquake uplift. Proc. R. Soc. B Biol. Sci. 287, 20200712. doi:10.1098/rspb.2020.0712
Parvizi, E., Fraser, C. I., and Waters, J. M. (2022). Genetic impacts of physical disturbance processes in coastal marine ecosystems. J. Biogeogr. 49 (10), 1877–1890. doi:10.1111/jbi.14464
Peluso, L., Broitman, B. R., Lardies, M. A., Nespolo, R. F., and Sáenz-Agudelo, P. (2023). Comparative population genetics of congeneric limpets across a biogeographic transition zone reveals common patterns of genetic structure and demographic history. Mol. Ecol. 32 (14), 3812–3825. doi:10.1111/mec.16978
Pons, J., Barraclough, T. G., Gomez-Zurita, J., Cardoso, A., Duran, D. P., Hazell, S., et al. (2006). Sequence-based species delimitation for the DNA taxonomy of undescribed insects. Syst. Biol. 55 (4), 595–609. doi:10.1080/10635150600852011
Puillandre, N., Lambert, A., Brouillet, S., and Achaz, G. (2012). ABGD, automatic Barcode Gap Discovery for primary species delimitation. Mol. Ecol. 21, 1864–1877. doi:10.1111/j.1365-294X.2011.05239.x
Pujolar, J. M., Vincenzi, S., Zane, L., Jesensek, D., De Leo, G. A., and Crivelli, A. J. (2011). The effect of recurrent floods on genetic composition of marble trout populations. PLoS ONE 6 (9), e23822. doi:10.1371/journal.pone.0023822
Quesada-Calderón, S., Giles, E. C., Morales-González, S., and Sáenz-Agudelo, P. (2021). Pinpointing genetic breaks in the southeastern Pacific: phylogeography and genetic structure of a commercially important tunicate. J. Biogeogr. 48 (10), 2604–2615. doi:10.1111/jbi.14227
Rambaut, A., Drummond, A. J., Xie, D., Baele, G., and Suchard, M. A. (2018). Posterior summarization in bayesian phylogenetics using tracer 1.7. Syst. Biol. 67, 901–904. doi:10.1093/sysbio/syy032
Ramírez, M. E. (1991). Catálogo de las algas marinas bentónicas de la costa temperada del Pacífico de Sudamérica. Monogr. Biológicas 5, 1–437.
Ramírez, M. E. (2010). Flora marina bentónica de la región austral de Sudamérica y la Antártica. An. del Inst. Patagon. 38 (1), 57–71. doi:10.4067/S0718-686X2010000100003
Rehak, K., Strecker, M. R., and Echtler, H. P. (2008). Morphotectonic segmentation of an active forearc, 37°–41°S, Chile. Geomorphology 94 (1-2), 98–116. doi:10.1016/j.geomorph.2007.05.002
Rogers, A. R., and Harpending, H. (1992). Population growth makes waves in the distribution of pairwise genetic differences. Mol. Biol. Evol. 9, 552–569. doi:10.1093/oxfordjournals.molbev.a040727
Rozas, J., Sánchez-Del Barrio, J. C., Messeguer, X., and Rozas, R. (2003). DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19, 2496–2497. doi:10.1093/bioinformatics/btg359
Sáenz-Agudelo, P., Peluso, L., Nespolo, R., Broitman, B. R., Haye, P. A., and Lardies, M. A. (2022). Population genomic analyses reveal hybridization and marked differences in genetic structure of Scurria limpet sister species with parapatric distributions across the South Eastern Pacific. Ecol. Evol. 12 (5), 88888–e8913. doi:10.1002/ece3.8888
Saldías, G. S., Sobarzo, M., Largier, J., Moffat, C., and Letelier, R. (2012). Seasonal variability of turbid river plumes off central Chile based on high-resolution MODIS imagery. Remote Sens. Environ. 123, 220–233. doi:10.1016/j.rse.2012.03.010
Sánchez, R., Sepúlveda, R. D., Brante, A., and Cárdenas, L. (2011). Spatial pattern of genetic and morphological diversity in the direct developer Acanthina monodon (Gastropoda: Mollusca). Mar. Ecol. Prog. Ser. 434, 121–131. doi:10.3354/meps09184
Santelices, B. (1980). Phytogeographic characterization of the temperate coast of Pacific South America. Phycologia 19, 1–12. doi:10.2216/i0031-8884-19-1-1.1
Santelices, B., Camus, P., and Hoffmann, A. (1989). Ecological studies for harvesting and culturing Gymnogongrus furcellatus (Rhodophyta, Gigartinales) in central Chile. J. Appl. Phycol. 1, 171–181. doi:10.1007/BF00003881
Saunders, G. W. (2005). Applying DNA barcoding to red macroalgae: a preliminary appraisal holds promise for future applications. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 360, 1879–1888. doi:10.1098/rstb.2005.1719
Schiel, D. R., Alestra, T., Gerrity, S., Orchard, S., Dunmore, R., Pirker, J., et al. (2019). The Kaikōura earthquake in southern New Zealand: loss of connectivity of marine communities and the necessity of a cross-ecosystem perspective. Aquat. Conserv. Mar. Freshw. 29 (9), 1520–1534. doi:10.1002/aqc.3122
Shelley, J. J., Swearer, S. E., Dempster, T., Adams, M., Le Feuvre, M. C., Hammer, M. P., et al. (2020). Plio-Pleistocene sea-level changes drive speciation of freshwater fishes in north-western Australia. J. Biogeogr. 47 (8), 1727–1738. doi:10.1111/jbi.13856
Silva, P. C., and DeCew, T. C. (1992). Ahnfeltiopsis, a new genus in the Phyllophoraceae (Gigartinales, rhodophyceae). Phycologia 31, 576–580. doi:10.2216/i0031-8884-31-6-576.1
Tajima, F. (1989). Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123, 585–595. doi:10.1093/genetics/123.3.585
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi:10.1093/molbev/msr121
Tapia, F. J., Largier, J. L., Castillo, M., Wieters, E. A., and Navarrete, S. A. (2014). Latitudinal discontinuity in thermal conditions along the nearshore of central-northern Chile. PLoS ONE 9, e110841. doi:10.1371/journal.pone.0110841
Tellier, F., Meynard, A. P., Correa, J. A., Faugeron, S., and Valero, M. (2009). Phylogeographic analyses of the 30°S south-east pacific biogeographic transition zone establish the occurrence of a sharp genetic discontinuity in the kelp Lessonia nigrescens: vicariance or parapatry? Mol. Phylogenet. Evol. 53, 679–693. doi:10.1016/j.ympev.2009.07.030
Tellier, F., Tapia, J., Faugeron, S., Destombe, C., and Valero, M. (2011). The Lessonia nigrescens species complex (Laminariales, Phaeophyceae) shows strict parapatry and complete reproductive isolation in a secondary contact zone. J. Phycol. 47 (4), 894–903. doi:10.1111/j.1529-8817.2011.01019.x
Thiel, M., Macaya, E. C., Acuña, E., Arntz, W. E., Bastias, H., Brokordt, K., et al. (2007). The Humboldt current system of northern-central Chile. Oceanogr. Mar. Biol. 45, 195–344. doi:10.1201/9781420050943.ch6
Trifinopoulos, J., Nguyen, L. T., von Haeseler, A., and Minh, B. Q. (2016). W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44 (W1), W232–W235. doi:10.1093/nar/gkw256
Tsuchi, R. (2002). Neogene evolution of surface marine climate in the Pacific and notes on related events. RMCG 19 (3), 260–270.
Varela, A. I., and Haye, P. A. (2012). The marine brooder Excirolana braziliensis (Crustacea: isopoda) is also a complex of cryptic species on the coast of Chile. RCHN 85, 495–502. doi:10.4067/s0716-078x2012000400011
Vaux, F., Craw, D., Fraser, C. I., and Waters, J. M. (2021). Northward range extension for Durvillaea poha bull kelp: response to tectonic disturbance? J. Phycol. 57 (5), 1411–1418. doi:10.1111/jpy.13179
Vaux, F., Fraser, C. I., Craw, D., Read, S., and Waters, J. M. (2023). Integrating kelp genomic analyses and geological data to reveal ancient earthquake impacts. J. R. Soc. Interface. 20, 20230105. doi:10.1098/rsif.2023.0105
Vaux, F., Parvizi, E., Craw, D., Fraser, C. I., and Waters, J. M. (2022). Parallel recolonizations generate distinct genomic sectors in kelp following high-magnitude earthquake disturbance. Mol. Ecol. 31 (18), 4818–4831. doi:10.1111/mec.16535
Villafaña, J. A., and Rivadeneira, M. M. (2018). The modulating role of traits on the biogeographic dynamics of chondrichthyans from the Neogene to the present. Paleobiology 44 (2), 251–262. doi:10.1017/pab.2018.7
Worthington Wilmer, J., Murray, L., Elkin, C., Wilcox, C., Niejalke, D., and Possingham, H. (2011). Catastrophic floods may pave the way for increased genetic diversity in endemic artesian spring snail populations. PLoS ONE 6 (12), e28645. doi:10.1371/journal.pone.0028645
Yang, E. C., Boo, S. M., Bhattacharya, D., Saunders, G. W., Knoll, A. H., Fredericq, S., et al. (2016). Divergence time estimates and the evolution of major lineages in the Florideophyte red algae. Sci. Rep. 6 (1), 21361. doi:10.1038/srep21361
Zakas, C., Binford, J., Navarrete, S. A., and Wares, J. P. (2009). Restricted gene flow in Chilean barnacles reflects an oceanographic and biogeographic transition zone. Mar. Ecol. Prog. Ser. 394, 165–177. doi:10.3354/meps08265
Keywords: comparative phylogeography, southeast pacific, rhodophyta, coastal uplift, actual and historical barriers to gene flow
Citation: Huanel OR, Montecinos AE, Sepúlveda-Espinoza F and Guillemin M-L (2024) Impact of persistent barrier to gene flow and catastrophic events on red algae evolutionary history along the Chilean coast. Front. Genet. 15:1336427. doi: 10.3389/fgene.2024.1336427
Received: 10 November 2023; Accepted: 23 February 2024;
Published: 08 March 2024.
Edited by:
Christophe William Vieira, Jeju National University, Republic of KoreaReviewed by:
Hwan Su Yoon, Sungkyunkwan University, Republic of KoreaTrevor Bringloe, The University of Melbourne, Australia
Zi-Min Hu, Yantai University, China
Copyright © 2024 Huanel, Montecinos, Sepúlveda-Espinoza and Guillemin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marie-Laure Guillemin, bWFyaWVsYXVyZS5ndWlsbGVtaW5AZ21haWwuY29t
 Oscar R. Huanel
Oscar R. Huanel Alejandro E. Montecinos4
Alejandro E. Montecinos4 Marie-Laure Guillemin
Marie-Laure Guillemin