
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Genet., 11 June 2024
Sec. Genomics of Plants and the Phytoecosystem
Volume 15 - 2024 | https://doi.org/10.3389/fgene.2024.1296622
This article is part of the Research TopicGenomics Assisted Improvement Of Crop Plants For Adaptation To Marginal EnvironmentsView all 9 articles
 Aqsa Ijaz1‡
Aqsa Ijaz1‡ Zunaira Anwar1‡
Zunaira Anwar1‡ Ahmad Ali2†‡
Ahmad Ali2†‡ Allah Ditta1,3
Allah Ditta1,3 Muhammad Yousaf Shani1
Muhammad Yousaf Shani1 Sajjad Haidar1,3
Sajjad Haidar1,3 Boahua Wang4
Boahua Wang4 Liu Fang5
Liu Fang5 Sana Muhy-Din Khan3
Sana Muhy-Din Khan3 Muhammad Kashif Riaz Khan1,3*
Muhammad Kashif Riaz Khan1,3*Human activities and climate change have resulted in frequent and intense weather fluctuations, leading to diverse abiotic stresses on crops which hampers greatly their metabolic activities. Heat stress, a prevalent abiotic factor, significantly influences cotton plant biological activities resulting in reducing yield and production. We must deepen our understanding of how plants respond to heat stress across various dimensions, encompassing genes, RNAs, proteins, metabolites for effective cotton breeding. Multi-omics methods, primarily genomics, transcriptomics, proteomics, metabolomics, and phenomics, proves instrumental in studying cotton’s responses to abiotic stresses. Integrating genomics, transcriptomics, proteomics, and metabolomic is imperative for our better understanding regarding genetics and molecular basis of heat tolerance in cotton. The current review explores fundamental omics techniques, covering genomics, transcriptomics, proteomics, and metabolomics, to highlight the progress made in cotton omics research.
Cotton is a perennial crop that belongs to the Malvaceae family but is cultivated as an annual crop. It is commercially cultivated in the USA, China, India, Brazil, Pakistan, Uzbekistan, and Australia (Egbuta et al., 2017). It is grown in more than 100 countries, covering an extensive land area of approximately 33 million hectares. As a major cash crop, it provides around 31% of the world’s fiber. Moreover, cotton contributes to a large extent in feed and food production around the world (Nabi et al., 2015; Mollaee et al., 2019). Additionally, cotton is also used as a source of edible oil and has some applications in the biopharmaceutical industry (Sheri et al., 2023a). It is usually classified into two types based on cultivation: wild and cultivated cotton (Han et al., 2022). There are 53 known species of cotton, four commercially cultivated, while the remaining 49 grow in natural habitats such as tropical and sub-tropical regions (Peng et al., 2021). G. hirsutum, G. arboreum, G. herbaceum, and G. barbadense are commonly cultivated and primarily grown for their fiber (Peng et al., 2021). However, these four species exhibited significant variations in fiber quality like fineness, maturity, length, and strength (Egbuta et al., 2017; Anwar et al., 2023).
Even though cotton production in the world has sustained at a constant rate during the last decade, there have still been significant variations in cotton crop production among various regions (Sheri et al., 2023a). The fluctuations in environmental conditions have reduced the crop productivity and sustainability because of alterations in their development patterns and ability to cope with severe conditions. The perpetual loss of agricultural land due to increased soil salinity, desertification, population expansion resulted in urbanization has exacerbated the consequences of climate change (Raza et al., 2019).
Various environmental factors like heat, drought, rainfall, humidity, and sunlight exposure impact the productivity of crops (Mukhtar et al., 2022; Shokat et al., 2023; Victoria et al., 2023; Hafeez et al., 2024). However, among these ecological factors, high temperature stress (HTS) greatly affects the growth and development of several crops, including cotton (Yousaf et al., 2023). Singh et al. (2007) reported that the lint yield of cotton could decrease by up to 110 kg/ha if the temperature increases by 1 °C above the optimal growth range (Singh et al., 2007). The flower stage of cotton is significantly sensitive to HTS, leading to significant flower abscission, stunted plant growth, and reduction in boll weight and number of bolls (Zafar et al., 2022). The temperature ranges from 32°C to 40°C adversely affects root development and stomatal conductance. A temperature above 29°C could also reduce boll weight (Lokhande and Reddy, 2014). The temperature above 37°C significantly impacts the seed germination rate, leading to elongation in both pollen germination and pollen tube (Burke et al., 2004). Salman et al. (2019a) reported that a reduction in cotton yield could be associated with a decrease in net photosynthetic rate and sympodial branches while bolls and flowers start to shed under HTS (Salman et al., 2019a).
Alongside the reduction in cotton yield, HTS also reduces fiber fineness and uniformity and decreases lint quality by increasing short fibers (Khan et al., 2022b). HTS can detrimentally influence all stages of plant growth and their development, participating in both direct and indirect damage (Ullah et al., 2016). However, susceptibility to HTS differs amidst phenological stages and varies among species and genotypes of the same species. HTS caused various types of injuries like leaves and twigs, necrosis and chlorosis in branches, leaves, and stems, premature senescence and abscission of leaves, flower, and fruit, inhibition of root and shoot growth, alteration in leaf angle, changes in transpiration rates, seed shrinkage and alterations in phospholipid extracellular matrix components. Consequently, these injuries result in lower plant productivity (Vollenweider and Günthardt-Goerg, 2005). Even when plants are subjected to HTS, they usually close their stomata to decrease water loss by enhancing the tracheids and stomata frequency and expanding vessels of the xylem, which helps the plant cope with HTS (Abro et al., 2023). The signal transduction system is important in trigging the self-regulated and hormone-meditated mechanism under HTS in plants (QAMER et al., 2021).
Reactive oxygen species act as important signaling molecules and perform diverse functions by responding rapidly to environmental factors, but when accumulated in excess quantity, ROS becomes toxic for plants (Zahra et al., 2021). Under HTS, cell biochemistry is negatively influenced by the production of ROS, which alters the function of mitochondria and subsequently instigates oxidative damage due to lipid peroxidation (Mansoor and Naqvi, 2013; Zahra et al., 2023). Various studies reported enhanced lipid peroxidation under HTS conditions (Wu et al., 2010). HTS induces the production of ROS like OH−, H2O2, and O2−, leading to oxidative stress (Yin et al., 2008). The rubisco enzyme is responsible for the increased production of H2O2 via its oxygenase activity under HTS (Kim and Portis, 2004). ROS instigates autocatalytic peroxidation of lipids in cell membranes and pigments, causing membrane damage that influences its function and permeability (Xu et al., 2006). For example, when cotton plants experienced HTS during the reproductive stage, a higher level of ROS coupled with elevated level of lipid peroxidation was found, which is damaging for all cell organelles. Moreover, cotton plants encounter difficulty to eliminate ROS under HTS (Zhang et al., 2016; Zafar et al., 2023).
Plants have developed a two-pronged defense mechanism to protect themselves from oxidative stress. The defense system comes into action by activating various enzymes like ascorbate peroxidase and superoxide dismutase (SOD) and non-enzymatic elements like phenolic acids and flavonoids. The non-enzymatic and enzymatic systems work harmoniously to eliminate free radicals or interrupt their reactions by breaking them down (Poudel and Poudel, 2020). Plant defense mechanisms encompass various enzymatic elements, including ascorbate peroxidase (APX), ascorbate (ASC), glutathione (GSH), and superoxide dismutase (SOD). Antioxidants defend molecules from oxidation, neutralize free radicals, and transform them into less reactive forms. Thus, antioxidants play an important role in achieving a balance between ROS production and ensuring the proper functioning of cells. HTS influences antibodies in cell membranes, which makes them sensitive to denaturation under oxidative stress. Heat can affect antibodies in cell membranes, rendering them susceptible to denaturation under oxidative stress. Therefore, recognizing non-enzymatic and enzymatic systems could be associated with stress and used as an indicator of stress tolerance. Proline, a non-enzymatic antioxidant, acts as an osmoprotectant and plays a significant role in various stress signaling pathways, helping the plant endure harsh conditions. The higher level of proline accumulation under stress is crucial, as proline efficiently scavenges reactive oxygen species (ROS) from the cells (Alagoz et al., 2023). This protects the cells from damage caused by reactive oxygen species (ROS) while sustaining their regular biological functions (Alagoz et al., 2023).
Molecular signaling is triggered when newly synthesized proteins are released from the ribosome, and substrate binding domains of Heat Shock Protein (HSP70) help to identify and bind to hydrophobic amino acid residue sequence (Faust et al., 2020). The heat shock protein gene is important in plants, as it helps the plant’s cellular machinery defend against various abiotic stresses such as heat. It also performs an additional role by interacting with extended sections of protein peptides and with proteins that are folding. The interaction of HSP70 with other proteins prevents them from aggregating together, facilitates changes in their folding to reach their final shape, and controls how the proteins perform (Usman et al., 2017; Waudby et al., 2019). Many HSPs have been extracted from several organelles like plastids, the cytosol, and the endoplasmic reticulum of many different plant species under various abiotic stresses (Roy et al., 2019). These HSPs act as chaperones, proving their role in tolerance against heat and other stresses (Tang et al., 2016).
There is a dire need to understand the genetic and molecular basis of the cotton plant under HTS and how it responds to high temperatures. To sustain crop productivity in the era of climate change, private companies, breeders and publicly funded institutes must incorporate faster and more productive strategies to fast-track heat-tolerant cotton development. Traditional breeding involves crossing plants with desired traits to develop a cultivar. Breeders perpetually conduct crosses over multiple generations, involving crossing and then selection, eventually selecting plants that have the best blend of desired traits (Derbyshire et al., 2022; Ahmed et al., 2023). This could be achieved by improving traditional breeding methods through the incorporation of genomic data or the precise manipulation of genetic determinants and metabolic pathways using advanced techniques such as genome editing. Over the last decade, breeders have utilized genomic methods to improve breeding and expedite cultivar development (Ahmar et al., 2020). For instance, some crucial phenotypes are commonly seen late in the breeding cycle. It has become feasible to recognize plants much earlier by using predictive models based on marker data, potentially eliminating the need for multiple generations and several years of breeding (Fu and Yuna, 2022).
Henceforth, this manuscript aims to explore the genetic basis of cotton and the application of advanced omics techniques such as transcriptomics, genomics, proteomics, and metabolomics in studying the response of cotton species to HTS. The purpose is to comprehend how genotype, environment, and epigenetics together contribute to the development of phenotypes that are tolerant or resistant to HTS.
The ancestors of cotton (Gossypium genus) originated and grew in the semi-arid (dry-hot) habitat (Baytar et al., 2022); thus, cotton is a heat-loving plant and grows well in the tropical and subtropical regions. However, the changing climate and the increasing global temperature threaten the cotton plant’s survival and limit its sustainable productivity (Iqbal et al., 2017; Rani et al., 2022). HTS occurs at various stages of the lifecycle, potentially limiting cotton yield and quality. HTS affects cotton plants at multiple stages but is more lethal during the flowering stage. High temperature is injurious to anther and pollen, which causes male sterility and impaired pollen viability, eventually reducing boll setting and final yield (Khan et al., 2020) Exposure of cotton to high temperature for longer duration causing leaf wilting and boll shedding, and reduced photosynthesis (Yun-Ying et al., 2008; Iqbal et al., 2017). Therefore, it is important to focus on identifying genetic resources with important QTLs and developing new germplasm that carry the QTLs, which can cope with HTS (Salman et al., 2019b). High temperature or heat tolerance (HTT) of the cotton cultivars can guarantee optimum yield during stress conditions. HTT is a complex trait controlled by multiple QTLs across the genome (Rani et al., 2022). HTT is conferred by several other associated traits, i.e., stomatal conductance, canopy temperature, transpiration, and serval other biochemical traits. All of these traits are of a complex quantitative nature and regulated by several epistatic QTLs. To gain insight into the genetic and molecular mechanism of HTT, it is essential to comprehend the genetic and molecular mechanisms of the related traits. Unravelling the genetic architecture of HTT will not only be helpful for basic research, but also hold importance in breeding thermos-tolerant cotton cultivars for sustainable production under the changing climate.
The recent advances in plant biotechnology have experienced great advances, particularly in the fields of genomics, genome sequencing, and marker technology. Introducing these technologies has provided great ease to plant scientists (Ma et al., 2021a; Baytar et al., 2022). QTL mapping is widely employed in plant breeding, proving to be an effective tool for unraveling the genetic basis and architecture of complex traits and facilitating marker-assisted breeding (Holland, 2007), locating regions on the genomes affecting the phenotype of an organism and map-based cloning. With the development in the field of genomics and the advancement of molecular marker technology, the next-generation sequencing (NGS) strategies also have fast-tracked the QTL analysis (Kumar et al., 2017). QTL mapping serves as a tool to identify the genes that influence the traits, locate the gene position on the genome, step towards map-based cloning, marker-assisted selection (MAS) (Khan, 2015; Khan et al., 2022b). Linkage mapping, association mapping and bulk segregant analysis (BSA) have been used in routine to identify the QTLs and gene controlling quantitative traits in various crops. These approaches have also been utilized to identify QTLs regulation tolerance to abiotic stresses.
In cotton, QTL mapping has been extensively applied to uncover the genetic mechanism of quality and yield traits (Kushanov et al., 2021). Various studies have reported several Single Nucleotide Polymorphisms (SNPs), Quantitative Trait Loci (QTL), and QTL clusters associated with fiber quality traits (Fang et al., 2014; Islam et al., 2016; Liu et al., 2020). Similarly, the genetic basis of biotic and abiotic stresses have been focused in detail (Dabbert and Gore, 2014). Progresses have been made to identify QTL regulating resistance against wilt infection (Li et al., 2017; Zhang et al., 2019; Abdelraheem et al., 2019; Abdelraheem et al., 2020; Abdelraheem et al., 2021), Li et al. (2020) have reported QTL regulation tolerance under drought and salt stress. However, the genetics basis of HTS response and HTT in cotton remains elusive, and very few loci have been identified. The HTT associated loci have been summarized in Table 1. The majority of these loci have been identified in combined stress conditions, i.e., drought and high aerial temperature, or dry arid conditions (Ulloa et al., 2000; Saranga et al., 2004), due to fact that both stresses often happen at same time. Ulloa et al. (2000) identified two putative loci for stomatal conductance in the F3 families developed from the cross between NM24016 and TM1 (Ulloa et al., 2000). One of these loci, G3800 showed over-dominance effect on the expression of stomatal conductance in heterozygous state. Interspecific hybridization is used as an effective tool for gene/alleles introgression from the wild relative into cultivated crop species. Saranga et al. (2004) developed an F2 population (n = 406) and subsequent F3 families via interspecific cross between G. hirsutum (cv. Sivon) X G. barbadense (cv. F-177) (Saranga et al., 2004). Interestingly, 33 loci were identified in the F2 population for physiological traits, including canopy temperature and chlorophyll contents under stress conditions. Similarly, Debbert et al. (2014) identified several loci regulating fiber quality and agronomic characteristics under HTS conditions in two RIL populations (Dabbert, 2014). In a RIL population obtained from the cross between TM-1 and M24016, Pauli et al. (2016) pinpointed numerous loci associated with canopy temperature and other related traits under hot arid conditions (Pauli et al., 2016). Canopy temperature related loci were identified on Chromosome A01, A08, A09, A13, D10, and D12 Interestingly, some of these loci overlapped with other loci regulating agronomic traits. GWAS analysis coupled with transcriptome wide association analysis (TWAS), Ma et al. (2021a) identified some loci that negatively regulated affected the pollen viability and caused male sterility (Ma et al., 2021a). Further, the TWAS analysis confirmed that GhHRK1 (Ghir_A01G006180) negatively regulates anther development under HTS. Rani et al. (2022) developed an F2 population from MNH-886 (heat tolerant) and heat sanative (MNH-814) parents. This F2 population carried 17 QTLs for agronomic and physiologic trait under HTS (Rani et al., 2022). Overall, it can be summarized, that the genetics of HTT is not well studied compared to other stresses in cotton, as well as other crops. Due to the increasing areal temperature and prevalent arid dry conditions during cotton growth, it is of utmost importance to analyze the genetics of HTT and identify the important genes that confer HTT.
Transcriptome analysis reveals gene expression differences, giving insights into the functions of genes. The advancements in RNA profiling technologies, such as microarrays and RNA-seq, have made transcriptome analysis easier (Duque et al., 2013). High-throughput sequencing has enabled larger-scale transcriptome analysis (Pandit et al., 2018). Larger transcriptome datasets obtained from various individuals or conditions provide valuable data about the regulatory responses of plants to their environment. Current studies have used a transcriptomic approach to understand abiotic stress pathways in multiple crops. The transcriptomics tool helps to identify stress-responsive genes to compare stress tolerance and control gene expression (Agarwal et al., 2014). Until now, this technique has unveiled various stress responsive genes and their expression pattern under environmental stresses in numerous crops like wheat, maize, barley, sorghum, rice, cotton and soybean (Baillo et al., 2019; Javaid et al., 2022; Kopecká et al., 2023).
Several studies have deciphered the molecular responses to heat stress (HTS), which lead to indehiscence and pollen sterility, resulting in lower yield in cotton plants. Genes associated with pollen and anthers play important roles in male reproduction and the response to HTS. Peng et al. (2016) also found 4,698 genes differentially expressed after 4–8h under HTS when comparing heat-tolerant (NH) and heat-sensitive (E7) cotton genotypes. Many heat-induced differentially expressed genes (DEGs) showed higher expression in NH or were unique to NH, encoding protein kinases, transcription factors, and heat shock proteins important for thermo-tolerance. Key possible thermo-tolerance regulators included two heat shock transcription factors (AtHsfA3, AtHsfC1 homologs) and four AP2/EREBP genes (AtERF20, AtERF026, AtERF053, AtERF113 homologs) (Peng et al., 2016). Zhang et al. (2022) performed comparative transcriptomics on anther and pollen from high-temperature sensitive lines under both controlled and high temperature in cotton and identified 1,066 genes specific to anthers and 1,111 genes specific to pollen. Under heat stress, 833 genes were differentially expressed in pollen when analyzed and compared. Ten pollen-specific genes responsive to heat stress were found (Zhang et al., 2022). Ma et al. (2021b) discovered that the heat-susceptible gene GhHRK1 exhibits a negative association with HTS in various crops like cotton (Ma et al., 2021a). Interestingly, its Arabidopsis mutant imparts heat tolerance, suggesting a significant role in heat resistance. Genes involved in cytokinin, ABA, and brassinosteroid signaling like CRE1, ABF, and CYCD may regulate cotton’s heat response and sustain growth (Prerostova et al., 2020; Zhao et al., 2020). Similarly, Liang et al. (2021b) identified long non-coding RNAs (lncRNAs) targeting essential candidate genes associated with high-temperature tolerance in cotton. These candidate genes encompass chlorophyll a-b binding proteins, ribosomal proteins, and heat shock proteins. Examination of the expression profiles of the anticipated lncRNAs revealed higher expression levels in the heat-tolerant cultivar compared to the heat-sensitive cultivar under conditions of HTS. The roles of circular RNAs (circRNAs) in the growth, development, and stress response of cotton were investigated (Liang et al., 2021a; Wang et al., 2024). They performed bioinformatics analysis on RNA sequencing data from pollen grains of two near-isogenic cotton lines, NH and SH which differ in fertility stability under high temperature (HT) stress. In total, 967 circRNAs were identified in the cotton pollen grains, of which 250 were differentially expressed under high temperature stress. A detailed list of genes identified by using transcriptome in cotton under heat has been given in Table 2.
Proteomics plays an imperative role in investigating of biochemical processes and plant responses to various abiotic stresses, such as HTS (Timperio et al., 2008; Katam et al., 2020). Plant stress proteomics can be utilized to elucidate potential key regulatory genes for genetically improving cotton’s resistance to extreme heat conditions (Ahmad et al., 2016). Plants respond differently against multiple harsh environmental extremes and assist in regulating cell homeostasis. Various proteins (HSPs and HSFs) have been recognized as essential for developing stress resilience in cotton (Manna et al., 2021).
Crop plants like cotton actuate several mechanisms in response to HTS, including physio-biochemical and molecular responses (Yadav et al., 2020). According to their molecular weight, these HSPs are categorized into various categories, including small HSPs, HSP60, HSP70, HSP90, and HSP100 (Khan and Shahwar, 2020). In cotton, HSPs primarily act as molecular chaperones and are in charge of the folding of proteins, transport, accumulation, and destruction (Usman et al., 2014). For instance, Hsp90 is involved in the translocation of signaling proteins, including actin, calmodulin, and kinases. Despite this, the interaction between HSPs and HTS is strongly correlated, and the development of a transformed cotton plant with a desired gene of interest encodes the synthesis of HSP101 protein perceived in Arabidopsis thaliana. So, the transgenic cotton plant enhances pollen germination rate and pollen tube elongation under HTS conditions (Nadeem et al., 2018). For example, Hsp70 assists in preventing protein refolding from accumulating and protecting cells from damaging effects caused by endoplasmic reticulum (ER) stress, while Hsp90 plays a crucial role in the translocation of signaling proteins (kinases, calmodulin, actin, etc.) (Nadeem et al., 2018). Likewise, by breaking down misfolded proteins, tiny HSPs decrease the build-up of irreversibly unfolded proteins and support plants for adaptation against HTS (Hoter et al., 2018). A transgenic cotton plant with a gene encoding the Arabidopsis thaliana HSP101 protein was formed, providing solid evidence for the link between HT and HSP. Under high temperatures, transformed cotton plants generate pollen that has a higher germination rate and longer elongation pollen tubes. Additionally, binding proteins (BiP) directly associated with the classical HSP-70 family, bind with newly prepared proteins and keep them in folded form. BiPs are involved in protein translocation and activation of unfolded proteins response directly linked with the basic leucine zipper 28 (Bzip28) stress sensor of the endoplasmic reticulum (ER). The transfer of heat shock protein related gene (AsHSP70) in G. hirsutum was conducted via Agrobacterium, and the expression of transgenic cotton species under high-temperature constraints at the molecular, physiological, and biochemical levels only 1.9% transformation efficiency was found using genetic transformation (Batcho et al., 2021). Transgenic plant genomic DNA contained an 1800 bp fragment of AsHSP70, which was amplified by PCR. In transgenic cotton plants, polyamine oxidase increased the relative expression of AsHSP70 from 1.02 to 9.58 as the length of heat stress increased. In the transgenic plant, AsHSP70 expression was comparatively higher in the leaves than in the root and stem during the combined heat and drought stress (Anaraki et al., 2018).
HTS and severe drought occurrence pose constrains in the lint quality and quantity improvement. A complete characterization and harnessing Hsf gene family in G. hirsutum is essential for understanding the roles of several Hsfs across the genomic level (Fan et al., 2021), particularly in HTT. In cotton (G. hirsutum), heat shock transcription factor (Hsf) genes were cloned and identified using genome-wide analysis and EST assembly (GhHsf). Cloning, identification, and classification of forty (40) GhHsf genes in three primary classes (A, B, and C) based on their domains properties will assist in identifying and better understanding of the key heat shock transcription factors involved in developing heat resilience in crop plants (Wang et al., 2014; Chang et al., 2017; Liang et al., 2021b). The expression pattern of GhHsf related transcripts in the majority of tissues in cotton crop, including morphological characters such as leaves, stems, roots, and fibers synthesis way. Results of quantitative real-time PCR (qRT-PCR) aid in determine the elevated expressiveness level in developing ovules of cotton under heat stress scenario. A comparative investigation of the expression patterns between fiber less and wild-type mutants which showed that Hsfs are involved in the formation of cotton fiber. The upland cotton genome has around 80 Hsf genes owing to duplication of genome, and the D-sub genome alone has 40 Hsf-related genes. The heat shock-induced expressions in several tissues demonstrated the importance of GhHsfs for both heat stress and fiber formation (Wang et al., 2014; Huang et al., 2022).
Due to major abiotic extremes such as drought and heat stressors, it was observed that the new transcription factors, i.e., Cys-2/His-2-type zinc finger (C2H2-ZF), regulate stomatal aperture (Han et al., 2020). WRKY TFs play a crucial role in wheat response to abiotic stress. In cotton, non-coding RNAs, phytohormones, and smaller-sized peptide formation are thought to be important elements that carry out gene functions in response to abiotic stress conditions (Patra et al., 2023). Several TFs, such as dehydration-responsive element/C-repeat (DRE/CRT) and DRE/binding protein 2 (DREB2), regulate a variety of phytohormone-independent toward harsh environmental stresses in tetraploid cotton (Li et al., 2020; Sadau et al., 2024). According to transcriptome analyses conducted on cotton under heat stress, certain genes are differentially expressed (DE) to combat stress scenarios (Ma et al., 2021b). Distinct patterns of OsMADS gene expression were discovered in cotton crops that were still developing in response to drought stress (Premkumar, 2023). These transcriptome sequencing investigations may be helpful for functional evaluations as their expression pattern is differential during crop development and growth stress response. Thus, these studies highlight the significance of transcriptomics for proper crop development and response against various environmental constrains. The comprehensive analysis of the proteome under heat stress is presented in Table 3.
Plants, being sessile organisms, are unable to avoid environmental conditions. Consequently, they are constantly subjected to abiotic stresses, which can have adverse effects on their growth and development, ultimately impacting productivity and crop yields-an issue that has been previously emphasized. Thus, there is need of day to completely understand the biochemical nature of plant responses to abiotic stresses. Hence, we would comprehend and predict the metabolic activity of plants under harsh conditions (Hasanuzzaman et al., 2013; Zhang et al., 2023). In the light of recent advancements in ‘omics’ technologies, metabolomics is emerging as a promising tool of plant biology. It involves the synthesis of information from gene expression, protein interactions, and pathway regulations (Weckwerth, 2003). Metabolomics primarily focuses on widespread quantitative and qualitative investigation of all small molecules of biological organisms. It involves the thorough analytical examination of low molecular-weight compounds known as metabolites found within biological specimens, all within specified conditions (Oh et al., 2023; Shokat et al., 2023). Metabolomic analysis can target either a predefined set of compounds or encompass the entire metabolome. NMR (Nuclear Magnetic Resonance) and MS (Mass Spectrometry) are the predominant techniques utilized for generating comprehensive metabolomic profiles (Misra and van der Hooft, 2016).
Plants contain a vast array of metabolites, which can be categorized into primary and secondary types (Arif et al., 2022). Primary metabolites are crucial for synthesizing key macromolecules such as carbohydrates, lipids, and proteins, and they play essential roles in fundamental biological processes like the tricarboxylic acid cycle (TCA), glycolysis, and fatty acid biosynthesis (Razzaq et al., 2019). These metabolites serve as the final products of cellular activities, and changes in their concentrations can serve as indicators of the overall condition of the entire plant. The primary objective of investigating metabolic alterations in plants is to uncover metabolic characteristics, often referred to as metabolic biomarkers, which help restore cellular balance and normal metabolic functioning in stressed plants. These biomarkers play a crucial role in facilitating stress response and tolerance (Katam et al., 2022).
The plant metabolome, comprising all low molecular weight metabolites synthesized by plant cells under specific conditions at a given time, influences processes at the macro-molecular level. These processes can be studied using various analytical techniques (Allwood et al., 2011). Metabolomics analysis has transformed our understanding of phytochemicals by enabling comprehensive profiling and characterization of both primary and specialized secondary metabolites in plants across different environmental conditions. This is especially notable in examining responses to both abiotic and biotic stresses, as well as resistance and tolerance mechanisms (Salam et al., 2023). Metabolomics has proven valuable in various crops such as soybean, rice, corn, barley, tomato, maize, and wheat, particularly for studying stress conditions (Razzaq et al., 2019). Nonetheless, its targeted use in assessing heat stress in cotton remains untapped.
In Arabidopsis, metabolic profiling has unveiled the temporal dynamics linked with heat stress (Kaplan et al., 2004). Likewise, metabolomic investigations in grapes have explored drought and salinity stresses, revealing changes in metabolic pathways concerning gluconeogenesis, energy metabolism, and nitrogen assimilation (Cramer et al., 2007; Rabara et al., 2017 found that gradual dehydration led to changes in the root metabolome, resulting in the accumulation of 4-hydroxy-2-oxoglutaric acid and coumestrol in tobacco and soybean roots (Rabara et al., 2017). In 2017, Aayudh Das noted that essential metabolites, including carbohydrates, amino acids, lipids, cofactors, nucleotides, peptides, and secondary metabolites, exhibited varying accumulation patterns under heat and drought stress conditions in soybean leaves. Drought and heat stress were observed to impact a range of metabolites involved in essential cellular processes, including glycolysis, the tricarboxylic acid (TCA) cycle, the pentose phosphate pathway, and starch biosynthesis. These processes regulate carbohydrate metabolism, amino acid metabolism, peptide metabolism, as well as purine and pyrimidine biosynthesis. During heat stress, wheat demonstrated elevated levels of sucrose and G1p, while soybean exhibited increased concentrations of 1,3-dihydroxyacetone, ribose, glycolate, and other metabolites (Das et al., 2017). These discussions imply that metabolomics serves as a valuable tool for comprehending plant responses to stress conditions. Metabolomics offers valuable insights into how plants respond to particular stress conditions by altering their internal biochemistry, thereby mitigating harmful effects (Villate et al., 2021).
Currently, there is limited knowledge available regarding the cotton metabolome during stress conditions, particularly in the context of heat stress. Metabolomics combined with HTP-phenomics will significantly enhance our understanding of how cotton responds to heat stress. Integrating metabolomics with genomics and transcriptomics will assist in identifying potential genes and pathways responsible for generating responses.
The rise in global temperatures has elevated extreme high-temperature events, leading to an increase in abiotic stress on terrestrial plants (Zhao et al., 2020; Verma et al., 2022). Plants, being sessile organisms and anchored to soil, are unable to evade the environmental extremes (HTS in particular). Therefore, plants activate complex physiological and molecular or gene networks in response to these stresses. However, the complete understanding of these responses still needs to be understood. Beside the physiological and molecular network, epigenetic modifications and regulations are also considered important in stress condition. Recently, it has been observed that epigenetic regulations play efficient role in the plant’s survival under various abiotic stresses (Verma et al., 2022). Epigenetic modifications encompass changes in the chromatin structure of stressed genes through various chemical modifications during both transcriptional and post-transcriptional events (Kim et al., 2015; Lämke and Bäurle, 2017). Molecular mechanisms of epigenetic regulation mainly consist of DNA methylation, chromatin/histone modifications and small non-coding RNAs, etc (Grewal and Rice, 2004; Huang et al., 2016). These changes aim to alter the expression of responsive genes under stress conditions. These alterations also create stress memory that can be passed on to the succeeding generation (Friedrich et al., 2019). Hence, the understanding of epigenetic responses and its consequences become more important in crop species particularly in cotton under the increasing Arial temperature and HTS.
Several plants species have been reported to exhibit epigenetic modification in response to abiotic stresses. Compared to other stresses (drought and salinity), less information is yet available about the epigenetic regulations and modification under HTS. The Arabidopsis HDA9 interacts with the PWR protein, leading to enhanced H3K9 deacetylation of PIF4 and YUC8 (Tasset et al., 2018; Van Der Woude et al., 2019). Similarly, under normal temperatures, HDA15 suppresses warm-temperature-related genes such as ATHB2, XTR7, YUCCA8, IAA19 and IAA29, while activating these genes under HTS (Shen et al., 2019). An increased DNA methylation have been reported in B. napus under HTS (Gao et al., 2014).
In cotton, the floral development stage and high-temperature occurrence coincide, threatening the development of floral organs, i.e., anther and pollen. The epigenetic modifications in response to HTS and the affected genes have been summarized in Table 4 and illustrated in Figure 1. Min et al. (2014) analyzed another development in two cotton genotypes (84,021 and H05) under high and normal temperature (Min et al., 2014). Interestingly, several genes were reported to be associated with DNA methylation and histone modification (acetylation and deacetylation) identified under HTS. Epigenetic modification were found to be involved in anther development, and influenced by high temperature. Ma et al. (2018) observed disruption in DNA methylation (CHH methylation) under HTS conditions in heat sensitive cultivar, which lead to male infertility. Suppression of DNA methylation in sensitive lines under normal temperature also led to male sterility. Disturbing the DNA methylation alters the ROS sugar metabolism and hormonal pathways, which cause abnormalities in pollen development under high HTS. The cotton anther genome displays higher methylation CG, CHG, and CHH sequence. Global methylome shows that promotor-unmethylated genes show higher expression compared to promotor-methylated genes (Ma et al., 2018). Zhang et al. (2020) reported that high temperature induced promoter methylation changes which led to upregulation of the mitochondrial respiratory chain enzyme-associated genes GhNDUS7, GhCOX6A, GhCX5B2, and GhATPBM. The increased expression of these genes initiated a sequence of redox processes aimed at generating ATP to support normal anther development under (HTS) (Zhang et al., 2020). Li et al. (2023a) observed widespread disruption in H3K4me3 and H3K27me3 modifications in cotton anthers under High-Temperature Stress (HTS), resulting in male sterility. Eliminating H3K27me3 at the promoters of jasmonate-related genes enhanced their expression, resulting in the restoration of male fertility under HTS in heat-tolerant lines (Li et al., 2023a).
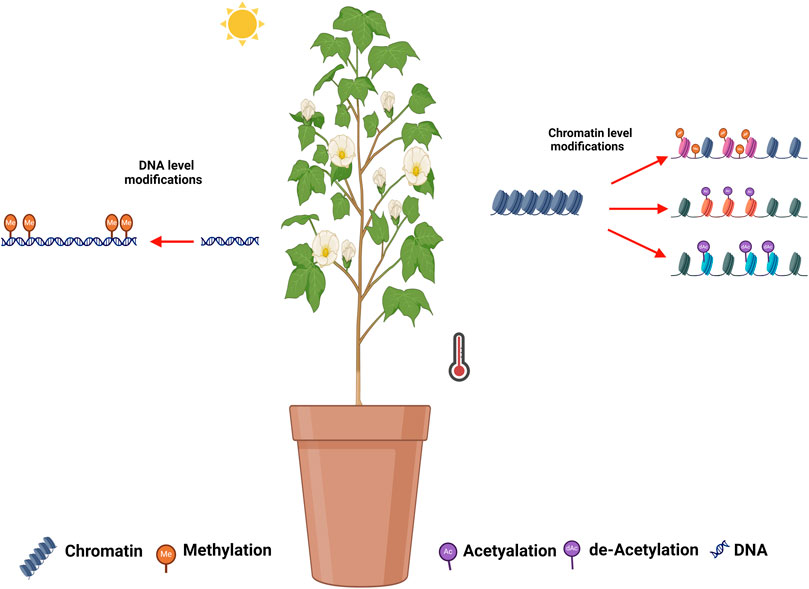
Figure 1. Illustration of epigenetic modifications during HTS in cotton. DNA methylation, histone methylation, histone acetylation, and histone de-acetylation have been reported in cotton in response to HTS in cotton.
Studies have revealed that epigenetic modifications, including acetylation and methylation, play a substantial role in regulating responses to abiotic stresses and adapting to environmental conditions. The influence of these epigenetic modifications extends significantly to agronomic traits and plant productivity. Unraveling the mechanisms of epigenetic responses in plants has the potential to bring about a significant revolution in crop breeding, especially in the context of climate change (Abdulraheem et al., 2024).
Rising global temperatures is one of the primary environmental factors that significantly impacts agricultural productivity across the globe. Enhancing our knowledge of the underlying mechanisms that govern how plants respond to heat stress will enable the development of new technologies and breeding approaches to improve plant tolerance to high temperatures (Chen et al., 2022). Previous research has identified several key traits that serve as important indicators of a plant’s tolerance to heat and drought stress (Zhao et al., 2020; Oguz et al., 2022). Evaluating and measuring these tolerance-related traits often requires specialized equipment and advanced technologies. Establishing reliable phenotypic markers of heat stress tolerance that can be evaluated at both the vegetative and reproductive stages would expedite the selection of plant germplasm with enhanced thermotolerance (Gao et al., 2020; Sarkar et al., 2021).
Continuously tracking plant growth and development after heat stress exposure using non-destructive methods like RGB imaging, thermal imaging and chlorophyll fluorescence analysis can offer important insights into physiological responses associated with photosynthetic performance and plant cooling capabilities-insights that would be difficult to gather through visual observation alone. The automated, environmentally-controlled system allows for rapid, efficient screening of large plant populations within a single experiment (Gao et al., 2020). Phenotypic traits, such as plant size, temperature, and photosynthetic efficiency have been successfully applied to evaluate plant performance under drought, salinity and chilling stress (Gao et al., 2020). In a recent study Chen et al. (2019) used high-throughput phenotyping to describe the physiological consequences of heat and drought stress at the early flowering stage of Brassica rapa, which identified corbooxylation, phosphate use and flower volume as stress tolerance associated traits (Chen et al., 2019). Hyperspectral imaging of Korean ginseng proved to significantly differentiate the heat tolerant and susceptible and genotypes with maximum precision (Abebe et al., 2023). Arabidopsis hsp101 mutants were exposed to heat stress and phenotype with high-throughput phenotyping system to monitor daily changes in plant morphology and photosynthetic activity in response. Heat stress reduced the quantum yield of PSII and enhanced leaf angle. Longer exposure influenced growth and morphology of plants (Gao et al., 2020).
Therefore, employing image-based phenotyping enables the measurement of diverse traits over time, rendering high-throughput phenotyping appropriate and precise for screening heat stress. Although high-throughput phenotyping has not yet been used to uncover the mechanism by which heat stress affects cotton plants. Cotton breeders have opted to use phenomics coupled with machine learning, which allows automated phenotyping to screen larger cotton populations at early stages and save significant resources compared to conventional phenotyping methods.
Furthermore, conventional phenotyping of cotton has been used only at the reproductive phase to identify heat-tolerant varieties. However, the conventional phenotyping is laborious, time consuming and prone to errors. Thus, the application of HTP and phenomics at earlier stage identification of cotton plants and the ability to monitor daily changes in cotton plants under stress could help identify more tolerant crop varieties in a shorter period of time.
Omics, accompanied by several modern approaches such as including metabolomics, proteomics, ionomics, and phenomics, are extensively explored in the agricultural sector to gain a deeper understanding of the molecular pathways and cell physiology that govern cell homeostasis and normal functioning (Singh et al., 2021; Yang et al., 2021). Multi-omics techniques, when combined with high-throughput methods, have proven essential for comprehending development, productivity, growth behavior against biotic and abiotic extremes in various crop (Mahmood et al., 2022; Khan et al., 2023). All the techniques mentioned above have been widely used several important crops such as cotton (Gossypium hirsutum L.), wheat (Triticum aestivum L.), soybeans (Glycine max), tomatoes (Solanum lycopersicum), barley (Hordeum vulgare L.), maize (Zea mays L.), and rice (Oryza sativa L) (Roychowdhury et al., 2023). The linkage between functional genomics and other omics tools reveals the connections between genomics and phenotypic responses under certain climatic and physiological attributes (Figure 2) (Yang et al., 2021). To assess the lines tolerant to heat stress and having resilience against male sterility induction, the dire need is to evaluate the functionality of anthers and transcriptomes.
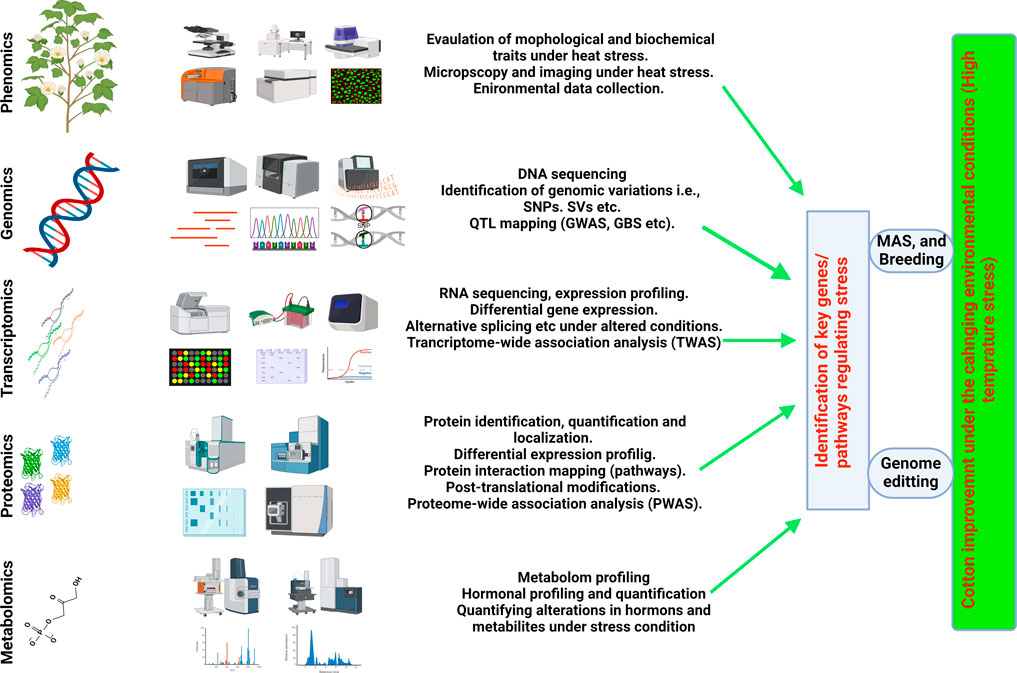
Figure 2. Integrated omics (Multiomic) approaches for understanding HTS response and understanding HTT in cotton and its possible utility in breeding HT-Cotton cultivars.
The integration of a Genome-Wide Association Study (GWAS) and transcriptome divergence analysis facilitated the identification of three loci associated with heat tolerance. These loci included expression quantitative trait loci (eQTLs) for 13,132 transcripts, 75 protein-coding genes, and 27 long noncoding RNAs. The most effective recognition of 4,820 genes linked to 13 fiber-related variables in cotton by a thorough GWAS has aided in the identification of novel genetic resources for improving fiber quality (Yasir et al., 2019; Li et al., 2021a; Ma et al., 2021b). Concurrently, enriched variants in the gene coding regions and comprehensive expression profiles for genes are provided by parallel transcriptome analysis. These enriched resources of crucial genes prove invaluable in identifying expressions of quantitative trait loci (eQTLs) that may regulate gene expression. The agronomic parameters that play a key role in increasing the growth, biomass, and productivity of cotton were identified by the combined use of GWAS and HRPF approaches (Meena et al., 2022). This reliable method superseded conventional phenomics, offering the sciences of crop genetics and breeding a potent new instrument (Yang et al., 2021) the effectiveness of agronomic features in conjunction with QTL mapping in cotton crop. Concurrently, genetic data and possible phenotyping techniques can offer insights into intricate features to enhance agricultural productivity (Kushanov et al., 2021). At present, limited knowledge is available in cotton on the integration of multiomoc approaches. Limited information of high throughput phenotyping (HTP) and HTP platforms is available in cotton. However, in future the integration of phenomics (high throughput phenotyping) with genomic, transcriptomic, proteomic and other omics will greatly resolve the genetic and molecular and metabolic mechanism of agronomic traits particularly in HTS in cotton. The Figure 2 illustrates how integrated omics (multi-omics) approaches contribute to understanding the heat stress response (HTS) and heat tolerance (HTT) in cotton, as well as their potential utility in breeding HT-Cotton cultivars.
In the review, we presented the progress in understanding the HTS and its response in cotton. To date, attempts have been made to decipher the genetic, molecular mechanism, and omics, which play a crucial role in understanding the response generated by the cotton plants to HTS. Researchers have utilized genetic (linkage and associating mapping), omics (genomics, transcriptomics, proteomics, and metabolomics), and epigenomic tools for this purpose. Integrative multi-omics has also been employed for this purpose. Together, these provide workable toolkits to develop HTS tolerant cotton cultivars that may cope with the changing environmental conditions and increasing areal temperatures. However, considering the current pace of climate change, growing global population and scarcity of resources, the current progress is lagging behind. Apparently, the pace of research needs to be improved, and progress must be revisited. In light of this review, the following suggestions can be proposed for future studies.
1. High throughput phenotyping and phenomics are gaining attention in crop physiology and breeding. However, the development of HTP platforms and its utility in in cotton is underutilized particularly in stress conditions. There is a dire need to develop HTP platform for growth modeling, non-destructive physiological and morphological assessment of cotton under various growth conditions.
2. Based on the available published information, very limited information is available about the genetics and genetic mechanism of HTS and HTT in cotton. To address this issue, much focus is needed on collecting the germplasm from across the geographical regions and systematically analyzing it under HTS. This will identify tolerant accessions and provide a gene pool for breeding and advance studies. Utilizing this material in genetic and genomics studies will help identify new genes and linked markers related to HTS responses and HTT, which can be used in marker-assisted breeding and gene pyramiding.
3. Transcriptomics and other omics have identified several genes, portions, and metabolites with pivotal roles in generating stress responses, conferring tolerance, and adjusting internal homeostasis under high stress. Several HSP and HSF encoding genes and other DEGs have been identified in cotton in response to HTS, and their pathways have been described. Very few genes have been functionally characterized and utilized in cotton improvement programs.
4. Epigenetic modifications are important in plant development and stress response. In cotton, epigenetic modification has only been studied in floral tissues, particularly in anthers, that cause male sterility. However, the HTS occurrence in cotton at the flowering time has far-reaching impact on yield. Understanding the global epigenomic alteration rather than tissue or stage-specific epigenomic alteration in response to HTS is important. Understanding global epigenomic alteration will enhance our understanding of the HTS and its response in cotton.
5. Integrated omics (or multi-omics) may prove to be more powerful than relying on a single omics tool. Integrated omics studies have been reported to have more power and precision in identifying genes in other crops (maize and rice). Utilizing integrated omics in cotton during HTS may provide a clearer understanding got HTS response, which can be translated into breeding HTT cultivars.
6. Utilization of genome editing technologies for trait engineering may be helpful. CRISPR/Cas-directed evolution (CDE) has been utilized in some crops (rice), which proved effective in generating quantitative tolerance to abiotic stresses. Therefore, identifying genes via omics and further editing with the CDE platform will be an effective and workable strategy for developing climate-resilient cotton cultivars.
AI: Writing–original draft, Writing–review and editing. ZA: Writing–original draft, Writing–review and editing. AL: Conceptualization, Visualization, Writing–review and editing. AD: Supervision, Writing–review and editing. MS: Conceptualization, Writing–original draft, Writing–review and editing. SHr: Writing–review and editing. BW: Conceptualization, Writing–review and editing. SM-U-D: Data curation, Resources, Writing–original draft. LF: Writing–review and editing. MK: Conceptualization, Supervision, Writing–review and editing.
The author(s) declare that inancial support was received for the research, authorship, and/or publication of this article. Authors are grateful to The National Key R&D Program of China (2021YFE0101200), Pakistan Science Foundation, PSF/CRP/18th Protocol (07), National Natural Science Foundation of China (32171994, 32072023, 32272090), Public Sector Development Program under the Ministry of Planning, Development & Special Initiatives (PSDP Project No. 829) and International Foundation for Science, Sweden, and COMSTECH, Islamabad (IFS-C-6500-1) for provision of funds.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abdelraheem, A., Elassbli, H., Zhu, Y., Kuraparthy, V., Hinze, L., Stelly, D., et al. (2020). A genome-wide association study uncovers consistent quantitative trait loci for resistance to Verticillium wilt and Fusarium wilt race 4 in the US Upland cotton. Theor. Appl. Genet. 133, 563–577. doi:10.1007/s00122-019-03487-x
Abdelraheem, A., Esmaeili, N., O’Connell, M., and Zhang, J. (2019). Progress and perspective on drought and salt stress tolerance in cotton. Ind. Crop. Prod. 130, 118–129.
Abdelraheem, A., Thyssen, G. N., Fang, D. D., Jenkins, J. N., Mccarty, J. C., Wedegaertner, T., et al. (2021). GWAS reveals consistent QTL for drought and salt tolerance in a MAGIC population of 550 lines derived from intermating of 11 Upland cotton (Gossypium hirsutum) parents. Mol Genet. Genomic. 296, 119–129.
Abdullah, M., Ahmad, F., Zang, Y., Jin, S., Ahmed, S., Li, J., et al. (2023). HEAT-RESPONSIVE PROTEIN regulates heat stress via fine-tuning ethylene/auxin signaling pathways in cotton. Plant Physiol. 191, 772–788. doi:10.1093/plphys/kiac511
Abdulraheem, M. I., Xiong, Y., Moshood, A. Y., Cadenas-Pliego, G., Zhang, H., and Hu, J. (2024). Mechanisms of plant epigenetic regulation in response to plant stress: recent discoveries and implications. Plants 13, 163. doi:10.3390/plants13020163
Abebe, A. M., Kim, Y., Kim, J., Kim, S. L., and Baek, J. (2023). Image-based high-throughput phenotyping in horticultural crops. Plants 12, 2061. doi:10.3390/plants12102061
Abro, A. A., Anwar, M., Javwad, M. U., Zhang, M., Liu, F., Jiménez-Ballesta, R., et al. (2023). Morphological and physio-biochemical responses under heat stress in cotton: overview. Biotechnol. Rep. 40, e00813. doi:10.1016/j.btre.2023.e00813
Agarwal, P., Parida, S. K., Mahto, A., Das, S., Mathew, I. E., Malik, N., et al. (2014). Expanding frontiers in plant transcriptomics in aid of functional genomics and molecular breeding. Biotechnol. J. 9, 1480–1492. doi:10.1002/biot.201400063
Ahmad, P., Abdel Latef, A. A., Rasool, S., Akram, N. A., Ashraf, M., and Gucel, S. (2016). Role of proteomics in crop stress tolerance. Front. Plant Sci. 7, 1336. doi:10.3389/fpls.2016.01336
Ahmar, S., Gill, R. A., Jung, K.-H., Faheem, A., Qasim, M. U., Mubeen, M., et al. (2020). Conventional and molecular techniques from simple breeding to speed breeding in crop plants: recent advances and future outlook. Int. J. Mol. Sci. 21, 2590. doi:10.3390/ijms21072590
Ahmed, S. R., Anwar, Z., Shahbaz, U., Skalicky, M., Ijaz, A., Tariq, M. S., et al. (2023). Potential role of silicon in plants against biotic and abiotic stresses. Silicon 15, 3283–3303. doi:10.1007/s12633-022-02254-w
Alagoz, S. M., Lajayer, B. A., and Ghorbanpour, M. (2023) Proline and soluble carbohydrates biosynthesis and their roles in plants under abiotic stresses. Plant stress mitigators. Germany: Elsevier.
Ali, M. M., Ali, Z., Ahmad, F., Nawaz, F., Qamar Shakil, S. A., Khan, A. A., et al. (2022). Transcript abundance of heat shock protein genes confer heat tolerance in cotton (Gossypium hirsutum L.). Pak. J. Bot. 54, 65–71. doi:10.30848/pjb2022-1(21)
Allwood, J. W., de Vos, R. C., Moing, A., Deborde, C., Erban, A., Kopka, J., et al. (2011). Plant metabolomics and its potential for systems biology research: background concepts, technology, and methodology. Methods Enzym. 500, 299–336. doi:10.1016/B978-0-12-385118-5.00016-5
Anaraki, Z. E., Tafreshi, S. A. H., and Shariati, M. (2018). Transient silencing of heat shock proteins showed remarkable roles for HSP70 during adaptation to stress in plants. Environ. Exp. Bot. 155, 142–157. doi:10.1016/j.envexpbot.2018.06.031
Anwar, Z., Ijaz, A., Ditta, A., Wang, B., Liu, F., Khan, S. M.-U.-D., et al. (2023). Genomic dynamics and functional insights under salt stress in Gossypium hirsutum L. Genes. 14, 1103. doi:10.3390/genes14051103
Arif, M. A. R., Waheed, M. Q., Lohwasser, U., Shokat, S., Alqudah, A. M., Volkmar, C., et al. (2022). Genetic insight into the insect resistance in bread wheat exploiting the untapped natural diversity. Front. Genet. 13, 828905. doi:10.3389/fgene.2022.828905
Baillo, E. H., Kimotho, R. N., Zhang, Z., and Xu, P. (2019). Transcription factors associated with abiotic and biotic stress tolerance and their potential for crops improvement. Genes. 10, 771. doi:10.3390/genes10100771
Batcho, A. A., Sarwar, M. B., Rashid, B., Hassan, S., and Husnain, T. (2021). Heat shock protein gene identified from Agave sisalana (As HSP70) confers heat stress tolerance in transgenic cotton (Gossypium hirsutum). Theor. Exp. Plant Physiology 33, 141–156. doi:10.1007/s40626-021-00200-6
Baytar, A. A., Peynircioğlu, C., Sezener, V., Frary, A., and Doğanlar, S. (2022). Association analysis of germination level cold stress tolerance and candidate gene identification in Upland cotton (Gossypium hirsutum L.). Physiology Mol. Biol. Plants 28, 1049–1060. doi:10.1007/s12298-022-01184-6
Burke, J. J., Velten, J., and Oliver, M. J. (2004). In vitro analysis of cotton pollen germination. Agron. J. 96, 359–368. doi:10.2134/agronj2004.3590
Chang, Y.-W., Chen, J.-Y., Lu, M.-X., Gao, Y., Tian, Z.-H., Gong, W.-R., et al. (2017). Cloning and expression of genes encoding heat shock proteins in Liriomyza trifolii and comparison with two congener leafminer species. PLoS one 12, e0181355. doi:10.1371/journal.pone.0181355
Chen, S., Guo, Y., Sirault, X., Stefanova, K., Saradadevi, R., Turner, N. C., et al. (2019). Nondestructive phenomic tools for the prediction of heat and drought tolerance at anthesis in Brassica species. Plant Phenomics 2019, 3264872. doi:10.34133/2019/3264872
Chen, Z., Galli, M., and Gallavotti, A. (2022). Mechanisms of temperature-regulated growth and thermotolerance in crop species. Curr. Opin. Plant Biol. 65, 102134. doi:10.1016/j.pbi.2021.102134
Cramer, G. R., Ergül, A., Grimplet, J., Tillett, R. L., Tattersall, E. A., Bohlman, M. C., et al. (2007). Water and salinity stress in grapevines: early and late changes in transcript and metabolite profiles. Funct. Integr. Genomics 7, 111–134. doi:10.1007/s10142-006-0039-y
Dabbert, T. A. (2014). Genetic analysis of cotton evaluated under high temperature and water deficit (PhD thesis: The University of Arizona). http://hdl.handle.net/10150/338975.
Dabbert, T., and Gore, M. A. (2014). Challenges and perspectives on improving heat and drought stress resilience in cotton. J. Cotton Sci. 18, 393–409.
Das, A., Rushton, P. J., and Rohila, J. S. (2017). Metabolomic profiling of soybeans (Glycine max L.) reveals the importance of sugar and nitrogen metabolism under drought and heat stress. Plants 6, 21. doi:10.3390/plants6020021
Derbyshire, M. C., Batley, J., and Edwards, D. (2022). Use of multiple omics techniques to accelerate the breeding of abiotic stress tolerant crops. Curr. Plant Biol. 32, 100262. doi:10.1016/j.cpb.2022.100262
Ding, Y., Ma, Y., Liu, N., Xu, J., Hu, Q., Li, Y., et al. (2017). Micro RNA s involved in auxin signalling modulate male sterility under high-temperature stress in cotton (Gossypium hirsutum). Plant J. 91, 977–994. doi:10.1111/tpj.13620
Duque, A. S., de Almeida, A. M., Da Silva, A. B., Da Silva, J. M., Farinha, A. P., Santos, D., et al. (2013). Abiotic stress responses in plants: unraveling the complexity of genes and networks to survive. Abiotic stress-plant responses Appl. Agric., 49–101. doi:10.5772/52779
Egbuta, M. A., Mcintosh, S., Waters, D. L., Vancov, T., and Liu, L. (2017). Biological importance of cotton by-products relative to chemical constituents of the cotton plant. Molecules 22, 93. doi:10.3390/molecules22010093
Fang, D. D., Jenkins, J. N., Deng, D. D., Mccarty, J. C., Li, P., and Wu, J. (2014). Quantitative trait loci analysis of fiber quality traits using a random-mated recombinant inbred population in Upland cotton (Gossypium hirsutum L.). BMC genomics 15, 397–415. doi:10.1186/1471-2164-15-397
Fan, K., Mao, Z., Ye, F., Pan, X., Li, Z., Lin, W., et al. (2021). Genome-wide identification and molecular evolution analysis of the heat shock transcription factor (HSF) gene family in four diploid and two allopolyploid Gossypium species. Genomics 113, 3112–3127. doi:10.1016/j.ygeno.2021.07.008
Faust, O., Abayev-Avraham, M., Wentink, A. S., Maurer, M., Nillegoda, N. B., London, N., et al. (2020). HSP40 proteins use class-specific regulation to drive HSP70 functional diversity. Nature 587, 489–494. doi:10.1038/s41586-020-2906-4
Friedrich, T., Faivre, L., Bäurle, I., and Schubert, D. (2019). Chromatin-based mechanisms of temperature memory in plants. Plant, Cell. and Environ. 42, 762–770. doi:10.1111/pce.13373
Fu, G., and Yuna, Y. (2022). Phenotyping and phenomics in aquaculture breeding. Aquac. Fish. 7, 140–146. doi:10.1016/j.aaf.2021.07.001
Gao, G., Li, J., Li, H., Li, F., Xu, K., Yan, G., et al. (2014). Comparison of the heat stress induced variations in DNA methylation between heat-tolerant and heat-sensitive rapeseed seedlings. Breed. Sci. 64, 125–133. doi:10.1270/jsbbs.64.125
Gao, G., Tester, M. A., and Julkowska, M. M. (2020). The use of high-throughput phenotyping for assessment of heat stress-induced changes in Arabidopsis. Plant Phenomics 2020, 3723916. doi:10.34133/2020/3723916
Grewal, S. I., and Rice, J. C. (2004). Regulation of heterochromatin by histone methylation and small RNAs. Curr. Opin. Cell. Biol. 16, 230–238. doi:10.1016/j.ceb.2004.04.002
Hafeez, M. B., Ghaffar, A., Zahra, N., Ahmad, N., Shair, H., Farooq, M., et al. (2024). Exogenous application of plant growth regulators improves economic returns, grain yield and quality attributes of late-sown wheat under saline conditions. Int. J. Plant Prod. doi:10.1007/s42106-024-00285-4
Han, G., Lu, C., Guo, J., Qiao, Z., Sui, N., Qiu, N., et al. (2020). C2H2 zinc finger proteins: master regulators of abiotic stress responses in plants. Front. Plant Sci. 11, 115. doi:10.3389/fpls.2020.00115
Han, P., Tian, X., Wang, Y., Huang, C., Ma, Y., Zhou, X., et al. (2022). Construction of a core germplasm bank of upland cotton (Gossypium hirsutum L.) based on phenotype, genotype and favorable alleles. Genet. Resour. Crop Evol. 69, 2399–2411. doi:10.1007/s10722-022-01379-6
Hasanuzzaman, M., Nahar, K., Alam, M. M., Roychowdhury, R., and Fujita, M. (2013). Physiological, biochemical, and molecular mechanisms of heat stress tolerance in plants. Int. J. Mol. Sci. 14, 9643–9684. doi:10.3390/ijms14059643
He, S., Zhang, Y., Wang, J., Wang, Y., Ji, F., Sun, L., et al. (2022). H3K4me2, H4K5ac and DNA methylation function in short-and long-term heat stress responses through affecting the expression of the stress-related genes in G. hirsutum. Environ. Exp. Bot. 194, 104699. doi:10.1016/j.envexpbot.2021.104699
Holland, J. B. (2007). Genetic architecture of complex traits in plants. Curr. Opin. Plant Biol. 10, 156–161. doi:10.1016/j.pbi.2007.01.003
Hoter, A., el-Sabban, M. E., and Naim, H. Y. (2018). The HSP90 family: structure, regulation, function, and implications in health and disease. Int. J. Mol. Sci. 19, 2560. doi:10.3390/ijms19092560
Huang, X., Liu, H., and Ma, B. (2022). The current progresses in the genes and networks regulating cotton plant architecture. Front. Plant Sci. 13, 882583. doi:10.3389/fpls.2022.882583
Huang, Y., Mo, Y., Chen, P., Yuan, X., Meng, F., Zhu, S., et al. (2016). Identification of SET domain-containing proteins in Gossypium raimondii and their response to high temperature stress. Sci. Rep. 6, 32729. doi:10.1038/srep32729
Iqbal, M., Ul-Allah, S., Naeem, M., Ijaz, M., Sattar, A., and Sher, A. (2017). Response of cotton genotypes to water and heat stress: from field to genes. Euphytica 213, 131–211. doi:10.1007/s10681-017-1916-2
Islam, M. S., Thyssen, G. N., Jenkins, J. N., Zeng, L., Delhom, C. D., Mccarty, J. C., et al. (2016). A MAGIC population-based genome-wide association study reveals functional association of GhRBB1_A07 gene with superior fiber quality in cotton. BMC genomics 17, 903–917. doi:10.1186/s12864-016-3249-2
Javaid, M. H., Khan, A. R., Salam, A., Neelam, A., Azhar, W., Ulhassan, Z., et al. (2022). Exploring the adaptive responses of plants to abiotic stresses using transcriptome data. Agriculture 12, 211. doi:10.3390/agriculture12020211
Kaplan, F., Kopka, J., Haskell, D. W., Zhao, W., Schiller, K. C., Gatzke, N., et al. (2004). Exploring the temperature-stress metabolome of Arabidopsis. Plant Physiol. 136, 4159–4168. doi:10.1104/pp.104.052142
Katam, R., Lin, C., Grant, K., Katam, C. S., and Chen, S. (2022). Advances in plant metabolomics and its applications in stress and single-cell biology. Int. J. Mol. Sci. 23, 6985. doi:10.3390/ijms23136985
Katam, R., Shokri, S., Murthy, N., Singh, S. K., Suravajhala, P., Khan, M. N., et al. (2020). Proteomics, physiological, and biochemical analysis of cross tolerance mechanisms in response to heat and water stresses in soybean. Plos one 15, e0233905. doi:10.1371/journal.pone.0233905
Khan, A. H., Min, L., Ma, Y., Wu, Y., Ding, Y., Li, Y., et al. (2020). High day and night temperatures distinctively disrupt fatty acid and jasmonic acid metabolism, inducing male sterility in cotton. J. Exp. Bot. 71, 6128–6141. doi:10.1093/jxb/eraa319
Khan, A. H., Wu, Y., Luo, L., Ma, Y., Li, Y., Ma, H., et al. (2022a). Proteomic analysis reveals that the heat shock proteins 70-17 and BiP5 enhance cotton male fertility under high-temperature stress by reducing the accumulation of ROS in anthers. Industrial Crops Prod. 188, 115693. doi:10.1016/j.indcrop.2022.115693
Khan, M. K. R., Ditta, A., Wang, B., Fang, L., Anwar, Z., Ijaz, A., et al. (2023) The intervention of multi-omics approaches for developing abiotic stress resistance in cotton crop under climate change. Sustainable Agriculture in the Era of the OMICs Revolution. Germany: Springer.
Khan, M. K. R., Liu, F., Wang, B., Hussain, M., Ditta, A., Anwar, Z., et al. (2022b) Breeding cotton for heat tolerance. Cotton breeding and biotechnology. USA: CRC Press.
Khan, S. (2015). QTL mapping: a tool for improvement in crop plants. Res. J. Recent Sci. 2277, 2502.
Khan, Z., and Shahwar, D. (2020). Role of heat shock proteins (HSPs) and heat stress tolerance in crop plants. Sustain. Agric. era Clim. change, 211–234. doi:10.1007/978-3-030-45669-6_9
Kim, J.-M., Sasaki, T., Ueda, M., Sako, K., and Seki, M. (2015). Chromatin changes in response to drought, salinity, heat, and cold stresses in plants. Front. Plant Sci. 6, 114. doi:10.3389/fpls.2015.00114
Kim, K., and Portis, A. R. (2004). Oxygen-dependent H2O2 production by Rubisco. Febs Lett. 571, 124–128. doi:10.1016/j.febslet.2004.06.064
Kopecká, R., Kameniarová, M., Černý, M., Brzobohatý, B., and Novák, J. (2023). Abiotic stress in crop production. Int. J. Mol. Sci. 24, 6603. doi:10.3390/ijms24076603
Kumar, J., Gupta, D. S., Gupta, S., Dubey, S., Gupta, P., and Kumar, S. (2017). Quantitative trait loci from identification to exploitation for crop improvement. Plant Cell. Rep. 36, 1187–1213. doi:10.1007/s00299-017-2127-y
Kushanov, F. N., Turaev, O. S., Ernazarova, D. K., Gapparov, B. M., Oripova, B. B., Kudratova, M. K., et al. (2021). Genetic diversity, QTL mapping, and marker-assisted selection technology in cotton (Gossypium spp.). Front. Plant Sci. 12, 779386. doi:10.3389/fpls.2021.779386
Lämke, J., and Bäurle, I. (2017). Epigenetic and chromatin-based mechanisms in environmental stress adaptation and stress memory in plants. Genome Biol. 18, 124–211. doi:10.1186/s13059-017-1263-6
Liang, Y., Gong, Z., Wang, J., Zheng, J., Ma, Y., Min, L., et al. (2021a). Nanopore-based comparative transcriptome analysis reveals the potential mechanism of high-temperature tolerance in cotton (Gossypium hirsutum L.). Plants 10, 2517. doi:10.3390/plants10112517
Liang, Y., Wang, J., Zheng, J., Gong, Z., Li, Z., Ai, X., et al. (2021b). Genome-wide comparative analysis of heat shock transcription factors provides novel insights for evolutionary history and expression characterization in cotton diploid and tetraploid genomes. Front. Genet. 12, 658847. doi:10.3389/fgene.2021.658847
Li, K., Wang, J., Kuang, L., Tian, Z., Wang, X., Dun, X., et al. (2021a). Genome-wide association study and transcriptome analysis reveal key genes affecting root growth dynamics in rapeseed. Biotechnol. Biofuels 14, 178–220. doi:10.1186/s13068-021-02032-7
Li, T., Ma, X., Li, N., Zhou, L., Liu, Z., Han, H., et al. (2017). Genome-wide association study discovered candidate genes of Verticillium wilt resistance in upland cotton (Gossypium hirsutum L.). Plant Biotechnol. J. 15, 1520–1532. doi:10.1111/pbi.12734
Liu, W., Song, C., Ren, Z., Zhang, Z., Pei, X., Liu, Y., et al. (2020). Genome-wide association study reveals the genetic basis of fiber quality traits in upland cotton (Gossypium hirsutum L.). BMC plant Biol. 20, 395–413. doi:10.1186/s12870-020-02611-0
Li, W., Chen, Y., Ye, M., Lu, H., Wang, D., and Chen, Q. (2020). Evolutionary history of the C-repeat binding factor/dehydration-responsive element-binding 1 (CBF/DREB1) protein family in 43 plant species and characterization of CBF/DREB1 proteins in Solanum tuberosum. BMC Evol. Biol. 20, 142–214. doi:10.1186/s12862-020-01710-8
Li, Y., Chen, M., Khan, A. H., Ma, Y., He, X., Yang, J., et al. (2023a). Histone H3 lysine 27 trimethylation suppresses jasmonate biosynthesis and signaling to affect male fertility under high temperature in cotton. Plant Commun. 4, 100660. doi:10.1016/j.xplc.2023.100660
Li, Y., Li, Y., Chen, Y., Wang, M., Yang, J., Zhang, X., et al. (2021b). Genome-wide identification, evolutionary estimation and functional characterization of two cotton CKI gene types. BMC Plant Biol. 21, 229–315. doi:10.1186/s12870-021-02990-y
Li, Y., Li, Y., Su, Q., Wu, Y., Zhang, R., Li, Y., et al. (2022). High temperature induces male sterility via MYB66–MYB4–Casein kinase I signaling in cotton. Plant Physiol. 189, 2091–2109. doi:10.1093/plphys/kiac213
Li, Y., Ma, H., Wu, Y., Ma, Y., Yang, J., Li, Y., et al. (2023b). Single-cell transcriptome atlas and regulatory dynamics in developing cotton anthers. Adv. Sci. 11, 2304017. doi:10.1002/advs.202304017
Lokhande, S., and Reddy, K. R. (2014). Quantifying temperature effects on cotton reproductive efficiency and fiber quality. Agron. J. 106, 1275–1282. doi:10.2134/agronj13.0531
Mahmood, U., Li, X., Fan, Y., Chang, W., Niu, Y., Li, J., et al. (2022). Multi-omics revolution to promote plant breeding efficiency. Front. Plant Sci. 13, 1062952. doi:10.3389/fpls.2022.1062952
Manna, M., Thakur, T., Chirom, O., Mandlik, R., Deshmukh, R., and Salvi, P. (2021). Transcription factors as key molecular target to strengthen the drought stress tolerance in plants. Physiol. Plant. 172, 847–868. doi:10.1111/ppl.13268
Mansoor, S., and Naqvi, F. N. (2013). Effect of heat stress on lipid peroxidation and antioxidant enzymes in mung bean (Vigna radiata L) seedlings. Afr. J. Biotechnol. 12.
Masoomi-Aladizgeh, F., Kamath, K. S., Haynes, P. A., and Atwell, B. J. (2022). Genome survey sequencing of wild cotton (Gossypium robinsonii) reveals insights into proteomic responses of pollen to extreme heat. Plant, Cell. and Environ. 45, 1242–1256. doi:10.1111/pce.14268
Masoomi-Aladizgeh, F., Najeeb, U., Hamzelou, S., Pascovici, D., Amirkhani, A., Tan, D. K., et al. (2021). Pollen development in cotton (Gossypium hirsutum) is highly sensitive to heat exposure during the tetrad stage. Plant, Cell. and Environ. 44, 2150–2166. doi:10.1111/pce.13908
Ma, Y., Min, L., Wang, J., Li, Y., Wu, Y., Hu, Q., et al. (2021a). A combination of genome-wide and transcriptome-wide association studies reveals genetic elements leading to male sterility during high temperature stress in cotton. New Phytol. 231, 165–181. doi:10.1111/nph.17325
Ma, Y., Min, L., Wang, J., Li, Y., Wu, Y., Hu, Q., et al. (2021b). A combination of genome-wide and transcriptome-wide association studies reveals genetic elements leading to male sterility during high temperature stress in cotton. New Phytol. 231, 165–181. doi:10.1111/nph.17325
Ma, Y., Min, L., Wang, M., Wang, C., Zhao, Y., Li, Y., et al. (2018). Disrupted genome methylation in response to high temperature has distinct affects on microspore abortion and anther indehiscence. Plant Cell. 30, 1387–1403. doi:10.1105/tpc.18.00074
Meena, M. R., Appunu, C., Arun Kumar, R., Manimekalai, R., Vasantha, S., Krishnappa, G., et al. (2022). Recent advances in sugarcane genomics, physiology, and phenomics for superior agronomic traits. Front. Genet. 13, 854936. doi:10.3389/fgene.2022.854936
Min, L., Li, Y., Hu, Q., Zhu, L., Gao, W., Wu, Y., et al. (2014). Sugar and auxin signaling pathways respond to high-temperature stress during anther development as revealed by transcript profiling analysis in cotton. Plant physiol. 164, 1293–1308. doi:10.1104/pp.113.232314
Misra, B. B., and van der Hooft, J. J. (2016). Updates in metabolomics tools and resources: 2014–2015. Electrophoresis 37, 86–110. doi:10.1002/elps.201500417
Mollaee, M., Mobli, A., Mutti, N. K., Manalil, S., and Chauhan, B. S. (2019). Challenges and opportunities in cotton production. Cotton Prod., 371–390. doi:10.1002/9781119385523.ch18
Mukhtar, T., Sultan, T., Hussain Munis, F., and Chaudhary, H. J. (2022). Induction of heat tolerance in tomato cultivar with heat tolerant bacteria under field condition. Asian J. Agric. Biol. 2022, 202103112.
Nabi, G., Shokat, S., Azhar, M., and Azhar, F. (2015). Genetic basis of ion uptake and proline accumulation in Gossypium hirsutum L. Bulg. J. Agric. Sci. 21, 835–842.
Nadeem, M., Li, J., Wang, M., Shah, L., Lu, S., Wang, X., et al. (2018). Unraveling field crops sensitivity to heat stress: mechanisms, approaches, and future prospects. Agronomy 8, 128. doi:10.3390/agronomy8070128
Oguz, M. C., Aycan, M., Oguz, E., Poyraz, I., and Yildiz, M. (2022). Drought stress tolerance in plants: interplay of molecular, biochemical and physiological responses in important development stages. Physiologia 2, 180–197. doi:10.3390/physiologia2040015
Oh, S.-W., Imran, M., Kim, E.-H., Park, S.-Y., Lee, S.-G., Park, H.-M., et al. (2023). Approach strategies and application of metabolomics to biotechnology in plants. Front. Plant Sci. 14, 1192235. doi:10.3389/fpls.2023.1192235
Pandit, A. A., Shah, R. A., and Husaini, A. M. (2018). Transcriptomics: a time-efficient tool with wide applications in crop and animal biotechnology. J. Pharmacogn. Phytochemistry 7, 1701–1704.
Patra, G. K., Gupta, D., Rout, G. R., and Panda, S. K. (2023). Role of long non coding RNA in plants under abiotic and biotic stresses. Plant Physiology Biochem. 194, 96–110. doi:10.1016/j.plaphy.2022.10.030
Pauli, D., Andrade-Sanchez, P., Carmo-Silva, A. E., Gazave, E., French, A. N., Heun, J., et al. (2016). Field-based high-throughput plant phenotyping reveals the temporal patterns of quantitative trait loci associated with stress-responsive traits in cotton. G3 Genes. Genomes, Genet. 6, 865–879. doi:10.1534/g3.115.023515
Peng, R., Jones, D. C., Liu, F., and Zhang, B. (2021). From sequencing to genome editing for cotton improvement. Trends Biotechnol. 39, 221–224. doi:10.1016/j.tibtech.2020.09.001
Peng, Z., He, S., Gong, W., Sun, J., Pan, Z., Sun, G., et al. (2016). Identification of candidate thermotolerance genes during early seedling stage in upland cotton (Gossypium hirsutum L.) revealed by comparative transcriptome analysis. Acta Physiol. Plant. 38, 221–316. doi:10.1007/s11738-016-2238-z
Poudel, P. B., and Poudel, M. R. (2020). Heat stress effects and tolerance in wheat: a review. J. Biol. Today’s World 9, 1–6.
Premkumar, A. (2023). Chapter-5 review on plant response to abiotic stress. Chief editor dr. Walunjkar Babasaheb Chang. 73.
Prerostova, S., Dobrev, P. I., Kramna, B., Gaudinova, A., Knirsch, V., Spichal, L., et al. (2020). Heat acclimation and inhibition of cytokinin degradation positively affect heat stress tolerance of Arabidopsis. Front. Plant Sci. 11, 87. doi:10.3389/fpls.2020.00087
Qamer, Z., Chaudhary, M. T., du, X., Hinze, L., and Azhar, M. T. (2021). Review of oxidative stress and antioxidative defense mechanisms in Gossypium hirsutum L. in response to extreme abiotic conditions. J. Cotton Res. 4, 9. doi:10.1186/s42397-021-00086-4
Rabara, R. C., Tripathi, P., and Rushton, P. J. (2017). Comparative metabolome profile between tobacco and soybean grown under water-stressed conditions. BioMed Res. Int. 2017, 3065251. doi:10.1155/2017/3065251
Rani, S., Baber, M., Naqqash, T., and Malik, S. A. (2022). Identification and genetic mapping of potential QTLs conferring heat tolerance in cotton (Gossypium hirsutum L.) by using micro satellite marker’s approach. Agronomy 12, 1381. doi:10.3390/agronomy12061381
Raza, A., Razzaq, A., Mehmood, S. S., Zou, X., Zhang, X., Lv, Y., et al. (2019). Impact of climate change on crops adaptation and strategies to tackle its outcome: a review. Plants 8, 34. doi:10.3390/plants8020034
Razzaq, A., Sadia, B., Raza, A., Khalid Hameed, M., and Saleem, F. (2019). Metabolomics: a way forward for crop improvement. Metabolites 9, 303. doi:10.3390/metabo9120303
Roychowdhury, R., Das, S. P., Gupta, A., Parihar, P., Chandrasekhar, K., Sarker, U., et al. (2023). Multi-omics pipeline and omics-integration approach to decipher plant’s abiotic stress tolerance responses. Genes. 14, 1281. doi:10.3390/genes14061281
Roy, S., Mishra, M., Dhankher, O. P., Singla-Pareek, S. L., and Pareek, A. (2019). Molecular chaperones: key players of abiotic stress response in plants. Genet. Enhanc. Crops Toler. Abiotic Stress Mech. Approaches I, 125–165. doi:10.1007/978-3-319-91956-0_6
Sadau, S. B., Liu, Z., Ninkuu, V., Guan, L., and Sun, X. (2024). DREB transcription factors are crucial regulators of abiotic stress responses in Gossypium spp. Plant Stress 11, 100350. doi:10.1016/j.stress.2024.100350
Salam, U., Ullah, S., Tang, Z.-H., Elateeq, A. A., Khan, Y., Khan, J., et al. (2023). Plant metabolomics: an overview of the role of primary and secondary metabolites against different environmental stress factors. Life 13, 706. doi:10.3390/life13030706
Saleem, M. A., Malik, W., Qayyum, A., Ul-Allah, S., Ahmad, M. Q., Afzal, H., et al. (2021). Impact of heat stress responsive factors on growth and physiology of cotton (Gossypium hirsutum L.). Mol. Biol. Rep. 48, 1069–1079. doi:10.1007/s11033-021-06217-z
Salman, M., Majeed, S., Rana, I. A., Atif, R. M., and Azhar, M. T. (2019a). Novel breeding and biotechnological approaches to mitigate the effects of heat stress on cotton. Recent approaches omics plant Resil. Clim. change, 251–277. doi:10.1007/978-3-030-21687-0_11
Salman, M., Zia, Z. U., Rana, I. A., Maqsood, R. H., Ahmad, S., Bakhsh, A., et al. (2019b). Genetic effects conferring heat tolerance in upland cotton (Gossypium hirsutum L.). J. Cotton Res. 2, 9–8. doi:10.1186/s42397-019-0025-2
Saranga, Y. E., Jiang, C. X., Wright, R., Yakir, D., and Paterson, A. (2004). Genetic dissection of cotton physiological responses to arid conditions and their inter-relationships with productivity. Plant, Cell. and Environ. 27, 263–277. doi:10.1111/j.1365-3040.2003.01134.x
Sarkar, S., Islam, A. A., Barma, N., and Ahmed, J. (2021). Tolerance mechanisms for breeding wheat against heat stress: a review. South Afr. J. Bot. 138, 262–277. doi:10.1016/j.sajb.2021.01.003
Shen, Y., Lei, T., Cui, X., Liu, X., Zhou, S., Zheng, Y., et al. (2019). Arabidopsis histone deacetylase HDA 15 directly represses plant response to elevated ambient temperature. Plant J. 100, 991–1006. doi:10.1111/tpj.14492
Sheri, V., Kumar, M., Jaconis, S., and Zhang, B. (2023a). Antioxidant defense in cotton under environmental stresses: unraveling the crucial role of a universal defense regulator for enhanced cotton sustainability. Plant Physiology Biochem. 204, 108141. doi:10.1016/j.plaphy.2023.108141
Shokat, S., GroßKINSKY, D. K., Singh, S., and Liu, F. (2023). The role of genetic diversity and pre-breeding traits to improve drought and heat tolerance of bread wheat at the reproductive stage. Food Energy Secur. 12, e478. doi:10.1002/fes3.478
Singh, R. P., Prasad, P. V., Sunita, K., Giri, S., and Reddy, K. R. (2007). Influence of high temperature and breeding for heat tolerance in cotton: a review. Adv. Agron. 93, 313–385. doi:10.1016/s0065-2113(06)93006-5
Singh, D., Chaudhary, P., Taunk, J., Singh, C. K., Singh, D., Tomar, R. S. S., et al. (2021). Fab advances in fabaceae for abiotic stress resilience: from ‘omics’ to artificial intelligence. Int. J. Mol. Sci. 22, 10535. doi:10.3390/ijms221910535
Smith, J., Wijewardene, I., Cai, Y., Esmaeili, N., Shen, G., Hequet, E., et al. (2023). Co-overexpression of RCA and AVP1 in cotton substantially improves fiber yield for cotton under drought, moderate heat, and salt stress conditions. Curr. Res. Biotechnol. 5, 100123. doi:10.1016/j.crbiot.2023.100123
Tang, T., Yu, A., Li, P., Yang, H., Liu, G., and Liu, L. (2016). Sequence analysis of the Hsp70 family in moss and evaluation of their functions in abiotic stress responses. Sci. Rep. 6, 33650. doi:10.1038/srep33650
Tasset, C., Singh Yadav, A., Sureshkumar, S., Singh, R., van der Woude, L., Nekrasov, M., et al. (2018). POWERDRESS-mediated histone deacetylation is essential for thermomorphogenesis in Arabidopsis thaliana. PLoS Genet. 14, e1007280. doi:10.1371/journal.pgen.1007280
Timperio, A. M., Egidi, M. G., and Zolla, L. (2008). Proteomics applied on plant abiotic stresses: role of heat shock proteins (HSP). J. Proteomics 71, 391–411. doi:10.1016/j.jprot.2008.07.005
Ullah, K., Khan, N., Usman, Z., Ullah, R., Saleem, F. Y., Shah, S. A. I., et al. (2016). Impact of temperature on yield and related traits in cotton genotypes. J. Integr. Agric. 15, 678–683. doi:10.1016/s2095-3119(15)61088-7
Ulloa, M., Cantrell, R., Percy, R., Zeiger, E., and Lu, Z. (2000). QTL analysis of stomatal conductance and relationship to lint yield in an interspecific cotton. J. Cotton Sci. 4, 10–18.
Usman, M. G., Rafii, M. Y., Martini, M. Y., Yusuff, O. A., Ismail, M. R., and Miah, G. (2017). Molecular analysis of Hsp70 mechanisms in plants and their function in response to stress. Biotechnol. Genet. Eng. Rev. 33, 26–39. doi:10.1080/02648725.2017.1340546
Usman, M. G., Rafii, M., Ismail, M., Malek, M., Latif, M. A., and Oladosu, Y. (2014). Heat shock proteins: functions and response against heat stress in plants. Int. J. Sci. Technol. Res. 3, 204–218.
van der Woude, L. C., Perrella, G., Snoek, B. L., van Hoogdalem, M., Novák, O., van Verk, M. C., et al. (2019). HISTONE DEACETYLASE 9 stimulates auxin-dependent thermomorphogenesis in Arabidopsis thaliana by mediating H2A. Z Deplet. Proc. Natl. Acad. Sci. 116, 25343–25354. doi:10.1073/pnas.1911694116
Verma, N., Giri, S. K., Singh, G., Gill, R., and Kumar, A. (2022). Epigenetic regulation of heat and cold stress responses in crop plants. Plant gene. 29, 100351. doi:10.1016/j.plgene.2022.100351
Victoria, O., Idorenyin, U., Asana, M., Jia, L., Shuoshuo, L., Yang, S., et al. (2023). Seed treatment with 24-epibrassinolide improves wheat germination under salinity stress. Asian J. Agric. Biol. 2023. doi:10.35495/ajab.2022.076
Villate, A., San Nicolas, M., Gallastegi, M., Aulas, P.-A., Olivares, M., Usobiaga, A., et al. (2021). Review: metabolomics as a prediction tool for plants performance under environmental stress. Plant Sci. 303, 110789. doi:10.1016/j.plantsci.2020.110789
Vollenweider, P., and Günthardt-Goerg, M. S. (2005). Diagnosis of abiotic and biotic stress factors using the visible symptoms in foliage. Environ. Pollut. 137, 455–465. doi:10.1016/j.envpol.2005.01.032
Wang, J., Sun, N., Deng, T., Zhang, L., and Zuo, K. (2014). Genome-wide cloning, identification, classification and functional analysis of cotton heat shock transcription factors in cotton (Gossypium hirsutum). BMC genomics 15, 961–1019. doi:10.1186/1471-2164-15-961
Wang, R., Zhang, M., Wang, H., Chen, L., Zhang, X., Guo, L., et al. (2024). Identification and characterization of circular RNAs involved in the fertility stability of cotton CMS-D2 restorer line under heat stress. BMC Plant Biol. 24, 32. doi:10.1186/s12870-023-04706-w
Waudby, C. A., Dobson, C. M., and Christodoulou, J. (2019). Nature and regulation of protein folding on the ribosome. Trends Biochem. Sci. 44, 914–926. doi:10.1016/j.tibs.2019.06.008
Weckwerth, W. (2003). Metabolomics in systems biology. Annu. Rev. Plant Biol. 54, 669–689. doi:10.1146/annurev.arplant.54.031902.135014
Wu, X., Ruan, Z., Gao, Y., Yin, Y., Zhou, X., Wang, L., et al. (2010). Dietary supplementation with L-arginine or N-carbamylglutamate enhances intestinal growth and heat shock protein-70 expression in weanling pigs fed a corn-and soybean meal-based diet. Amino Acids 39, 831–839. doi:10.1007/s00726-010-0538-y
Xu, S., Li, J., Zhang, X., Wei, H., and Cui, L. (2006). Effects of heat acclimation pretreatment on changes of membrane lipid peroxidation, antioxidant metabolites, and ultrastructure of chloroplasts in two cool-season turfgrass species under heat stress. Environ. Exp. Bot. 56, 274–285. doi:10.1016/j.envexpbot.2005.03.002
Yadav, S., Modi, P., Dave, A., Vijapura, A., Patel, D., and Patel, M. (2020). Effect of abiotic stress on crops. Sustain. Crop Prod. 3. doi:10.5772/intechopen.88434
Yang, Y., Saand, M. A., Huang, L., Abdelaal, W. B., Zhang, J., Wu, Y., et al. (2021). Applications of multi-omics technologies for crop improvement. Front. Plant Sci. 12, 563953. doi:10.3389/fpls.2021.563953
Yasir, M., He, S., Sun, G., Geng, X., Pan, Z., Gong, W., et al. (2019). A genome-wide association study revealed key SNPs/genes associated with salinity stress tolerance in upland cotton. Genes. 10, 829. doi:10.3390/genes10100829
Yin, H., Chen, Q., and Yi, M. (2008). Effects of short-term heat stress on oxidative damage and responses of antioxidant system in Lilium longiflorum. Plant Growth Regul. 54, 45–54. doi:10.1007/s10725-007-9227-6
Yousaf, M. I., Hussain, Q., Alwahibi, M. S., Aslam, M. Z., Khalid, M. Z., Hussain, S., et al. (2023). Impact of heat stress on agro-morphological, physio-chemical and fiber related paramters in upland cotton (Gossypium hirsutum L.) genotypes. J. King Saud University-Science 35, 102379. doi:10.1016/j.jksus.2022.102379
Yun-Ying, C., Hua, D., Li-Nian, Y., Zhi-Qing, W., Shao-Chuan, Z., and Jian-Chang, Y. (2008). Effect of heat stress during meiosis on grain yield of rice cultivars differing in heat tolerance and its physiological mechanism. Acta Agron. Sin. 34, 2134–2142. doi:10.1016/s1875-2780(09)60022-5
Zafar, M. M., Chattha, W. S., Khan, A. I., Zafar, S., Subhan, M., Saleem, H., et al. (2023). Drought and heat stress on cotton genotypes suggested agro-physiological and biochemical features for climate resilience. Front. Plant Sci. 14, 1265700. doi:10.3389/fpls.2023.1265700
Zafar, M. M., Jia, X., Shakeel, A., Sarfraz, Z., Manan, A., Imran, A., et al. (2022). Unraveling heat tolerance in upland cotton (Gossypium hirsutum L.) using univariate and multivariate analysis. Front. Plant Sci. 12, 727835. doi:10.3389/fpls.2021.727835
Zahra, N., Hafeez, M. B., Ghaffar, A., Kausar, A., al Zeidi, M., Siddique, K. H., et al. (2023). Plant photosynthesis under heat stress: effects and management. Environ. Exp. Bot. 206, 105178. doi:10.1016/j.envexpbot.2022.105178
Zahra, N., Wahid, A., Shaukat, K., Hafeez, M. B., Batool, A., and Hasanuzzaman, M. (2021). Oxidative stress tolerance potential of milk thistle ecotypes after supplementation of different plant growth-promoting agents under salinity. Plant Physiology Biochem. 166, 53–65. doi:10.1016/j.plaphy.2021.05.042
Zhang, F., Jin, X., Wang, L., Li, S., Wu, S., Cheng, C., et al. (2016). A cotton annexin affects fiber elongation and secondary cell wall biosynthesis associated with Ca2+ influx, ROS homeostasis, and actin filament reorganization. Plant Physiol. 171, 1750–1770. doi:10.1104/pp.16.00597
Zhang, J., Zhu, Y., Abdelraheem, A., Teng, Z., Thyssen, G. N., Fang, D. D., et al. (2019). “A genome-wide association study of Fusarium wilt resistance in a MAGIC population of Upland cotton,” in 2019 Beltwide Cotton Conferences, New Orleans, LA, January 16, 2019 (IEEE).
Zhang, M., Zhang, X., Guo, L., Qi, T., Liu, G., Feng, J., et al. (2020). Single-base resolution methylome of cotton cytoplasmic male sterility system reveals epigenomic changes in response to high-temperature stress during anther development. J. Exp. Bot. 71, 951–969. doi:10.1093/jxb/erz470
Zhang, R., Zhou, L., Li, Y., Ma, H., Li, Y., Ma, Y., et al. (2022). Rapid identification of pollen-and anther-specific genes in response to high-temperature stress based on transcriptome profiling analysis in cotton. Int. J. Mol. Sci. 23, 3378. doi:10.3390/ijms23063378
Zhang, Y., Xu, J., Li, R., Ge, Y., Li, Y., and Li, R. (2023). Plants’ response to abiotic stress: mechanisms and strategies. Int. J. Mol. Sci. 24, 10915. doi:10.3390/ijms241310915
Keywords: high-temperature stress, cotton, epigenetic modification, transcriptomics, multiomics
Citation: Ijaz A, Anwar Z, Ali A, Ditta A, Shani MY, Haidar S, Wang B, Fang L, Khan SM-D and Khan MKR (2024) Unraveling the genetic and molecular basis of heat stress in cotton. Front. Genet. 15:1296622. doi: 10.3389/fgene.2024.1296622
Received: 18 September 2023; Accepted: 29 April 2024;
Published: 11 June 2024.
Edited by:
Sajid Shokat, International Atomic Energy Agency, AustriaReviewed by:
Muhammad Shahid Iqbal, Ayub Agricultural Research Institute, PakistanCopyright © 2024 Ijaz, Anwar, Ali, Ditta, Shani, Haidar, Wang, Fang, Khan and Khan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Muhammad Kashif Riaz Khan, mkrkhan@gmail.com
†ORCID: Ahmad Ali, orcid.org/0000-0001-8777-5519
‡These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.