
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet., 09 March 2023
Sec. RNA
Volume 14 - 2023 | https://doi.org/10.3389/fgene.2023.985764
This article is part of the Research TopicRNA Modifications and Epitranscriptomics - Volume IIView all 15 articles
 Xiaojuan Fei1
Xiaojuan Fei1 Meilin Jin1
Meilin Jin1 Zehu Yuan2
Zehu Yuan2 Taotao Li1
Taotao Li1 Zengkui Lu3
Zengkui Lu3 Huihua Wang1
Huihua Wang1 Jian Lu4
Jian Lu4 Kai Quan5
Kai Quan5 Junxiang Yang6
Junxiang Yang6 Maochang He6
Maochang He6 Tingpu Wang7
Tingpu Wang7 Yuqin Wang8*
Yuqin Wang8* Caihong Wei1*
Caihong Wei1*There is a genetic difference between Hu sheep (short/fat-tailed sheep) and Tibetan sheep (short/thin-tailed sheep) in tail type, because of fat metabolism. Previous studies have mainly focused directly on sheep tail fat, which is not the main organ of fat metabolism. The function of miRNAs in sheep liver fat metabolism has not been thoroughly elucidated. In this study, miRNA-Seq was used to identify miRNAs in the liver tissue of three Hu sheep (short/fat-tailed sheep) and three Tibetan sheep (short/thin-tailed sheep) to characterize the differences in fat metabolism of sheep. In our study, Hu sheep was in a control group, we identified 11 differentially expressed miRNAs (DE miRNAs), including six up-regulated miRNAs and five down-regulated miRNAs. Miranda and RNAhybrid were used to predict the target genes of DE miRNAs, obtaining 3,404 target genes. A total of 115 and 67 GO terms as well as 54 and 5 KEGG pathways were significantly (padj < 0.05) enriched for predicted 3,109 target genes of up-regulated and 295 target genes of down-regulated miRNAs, respectively. oar-miR-432 was one of the most up-regulated miRNAs between Hu sheep and Tibetan sheep. And SIRT1 is one of the potential target genes of oar-miR-432. Furthermore, functional validation using the dual-luciferase reporter assay indicated that the up-regulated miRNA; oar-miR-432 potentially targeted sirtuin 1 (SIRT1) expression. Then, the oar-miR-432 mimic transfected into preadipocytes resulted in inhibited expression of SIRT1. This is the first time reported that the expression of SIRT1 gene was regulated by oar-miR-432 in fat metabolism of sheep liver. These results could provide a meaningful theoretical basis for studying the fat metabolism of sheep.
MicroRNAs (miRNAs) are a kind of small RNA, whose length is about 22 nt (nucleotide). Previous studies revealed that miRNAs have distinctive biological characteristics in proliferation, differentiation, metabolism, and disease (Lin et al., 2020). In animals and plants, miRNAs are involved in the regulation of post-transcriptional gene expression. miRNAs usually bind to the 3'UTR region of mRNA to inhibit the post-transcriptional translation of target genes and enhance the degradation or repress the translation of mRNAs (Rouleau et al., 2017). In Chinese indigenous sheep, sheep can be divided into short/thin-tailed sheep, long/thin-tailed sheep, short/fat-tailed sheep, long/fat-tailed sheep, and fat-buttock sheep, because of the degree of fat deposition along the tail vertebra and the length of the tail vertebra (Lu et al., 2020). Hu sheep (short/fat-tailed sheep) and Tibetan sheep (short/thin-tailed sheep) are two Chinese indigenous sheep breeds with different tail types. Tail fat is the main energy source for sheep migration, drought, and food deprivation (Luo et al., 2021). However, studies mainly focus directly on tail fat to study fat metabolism, which is not the main organ of fat metabolism (Zhou et al., 2017; Li et al., 2020). The liver is a primary organ of fat metabolism, fat metabolization in the liver is equally important to its metabolism in fat tissue. Triglyceride is one of the lipids mostly formed in the liver, whose metabolism is mainly controlled through liver parenchyma cells. And the degree of fat deposition in fat tissue depends on the fat flow in the liver for fat synthesis. (Carotti et al., 2020). There are differences in the liver of sheep with different tail types that can reflect the underlying mechanism of sheep fat metabolism.
With the development of high-throughput sequencing technology, miRNA-Seq has been widely used in the omics analysis of humans (Zheng et al., 2016), mice (Peng et al., 2013), chickens (Sikorska et al., 2021) and cows (Zhang et al., 2019; Chen et al., 2020) species. And researchers showed that miRNA has an important function in fat metabolism (Deng et al., 2020). Many studies have explored the role of miRNA in liver fat metabolism disease models to clarify the process of disease occurrence. In a non-alcoholic fatty liver disease (NAFLD) mouse model, Lin et al. identified that miR-29a not only made body weight gain decrease, but also the subcutaneous, visceral, and intestinal fat accumulation and hepatocellular steatosis (Jeon and Carr., 2020). In the non-alcoholic steatohepatitis (NASH) mouse model, inhibiting the expression of miR-21 decreased liver injury, inflammation, and fibrosis (SOARES et al., 2016). In a high-fat-induced mouse model, miR-378 targeted AMPK to promote the occurrence of liver fibrosis and inflammation (Lin et al., 2019). Meanwhile, researchers have analyzed the expression patterns of miRNA in the liver of pigs (Li et al., 2021) and cows (Liang et al., 2017) across periods. These studies represented a foundation for further understanding the molecular regulatory mechanisms of liver tissue fat metabolism.
Because there is a genetic difference between Hu sheep (short/fat-tailed sheep) and Tibetan sheep (short/thin-tailed sheep) in tail type, comparing their livers’ miRNA features may find miRNAs affecting the fat metabolism of Hu sheep (short/fat-tailed sheep) and Tibetan sheep (short/thin-tailed sheep). Our results could provide a theoretical basis for further study of the fat metabolism between different sheep breeds.
All animal experiments were approved by the Science Research Department of the Institute of Animal Sciences, Chinese Academy of Agriculture Sciences (IAS-CAAS). Ethical approval complied with the Animal Ethics Committee of the IAS-CAAS (No. IAS 2019-49). Samples of liver tissues were collected from three Hu sheep (short/fat-tailed sheep, Yongdeng, Gansu, China) and three Tibetan sheep (short/thin-tailed sheep, Yushu, Qinghai, China). Samples from Hu sheep are named HG1, HG2, and HG3, respectively. Samples from Tibetan sheep are named ZG1, ZG2, and ZG3, respectively. All sheep were males and slaughtered at age 1.5. All samples were frozen in liquid nitrogen in 1.5 mL RNase-free freezing tubes and stored at −80°C for use. Trizol (Invitrogen, Carlsbad, CA, United States) was used to extract total RNA. A NanoDrop2000 spectrophotometer (Thermo Fisher Scientific, Wilmington, MA, United States) was used to quantify RNA purity at 260 and 280 nm. Six libraries were constructed with a commercial sequencing provider: BGI (Mortazavi et al., 2008; Wang et al., 2009). An Agilent 2,100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, United States) was used to examine the integrity of the library. All FASTQ sequencing files have been stored in the Sequence Read Archive (accession numbers PRJNA785102).
The cleaning of the raw data was performed based on: 1) poor quality sequencing reads, 2) reads with 5′ adaptors and without 3’ adaptors; 3) reads without insert segments; and 5) reads containing poly A; and 6) reads longer than 18 nucleotides. To ensure that each small RNA had a unique label, according to the order of possible ribosomal RNA, small conditional RNA, small nucleolar RNA, small nuclear RNA (snRNA), and transfer RNA sequences to annotate (Balaskas et al., 2020). The sheep reference genome Oar_v3.1 (https://www.ebi.ac.uk/ena/browser/view/GCA_000298735.1, accessed on 20 February 2021) and miRbase21.0 (http://www.mirbase.org, accessed on 20 February 2021) was used to map clean reads with Bowtie2 (Langmead et al., 2009).
MiRDeep2 software was used to predict novel miRNAs (Kern et al., 2020). The expression of miRNA was calculated by absolute numbers counting of molecules using unique molecular identifiers (Pflug and Haeseler., 2018). Moreover, the lengths of small RNAs (sRNAs) and the proportion of miRNAs were calculated. The “oar-miR-" and “novel_mir” terms identify known miRNAs and novel miRNAs, respectively. Hu sheep is set as a control, DESeq2 software was used to perform the differential expression analysis, in which the statistical significance was set at a fold discover rate (FDR) adjusted p-value (padj ≤0.05) by Benja-mini-Hochberg and |Log2Foldchange| > 0.5.
Miranda (John et al., 2004) and RNAhybrid (Lin et al., 2022) were used to find more accurate targets of differentially expressed miRNA (DE miRNA). g: Profiler was used for genes function enrichment analysis, in which the statistical significance was set at a fold discover rate (FDR) adjusted p-value (padj ≤0.05) by Benjamini–Hochberg (Raudvere et al., 2019). There are 3,109 target genes of upregulated and 295 target genes of downregulated DE miRNAs were annotated with Gene Ontology (GO) (http://www.geneontology.org/, accessed on 19 January 2022) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) (http://www.genome.jp, accessed on 19 January 2022), respectively.
Steam-loop real-time qPCR was used to validate miRNA sequencing data from seven randomly selected miRNAs (oar-miR-432, novel_mir70, novel_mir21, nov-el_mir64, novel_mir58, oar-miR-19b, and oar-miR-29b). The total RNA of each sample was reversed transcribed with a miRNA 1st Strand cDNA Synthesis Kit. RT-qPCR was performed on a LightCycler® 480II qPCR system using miRNA universal SYBR qPCR Master Mix (Vazyme, Nanjing, China). U6 was used as the reference gene. To detect the expression of SIRT1, HiScript III 1st Strand cDNA Synthesis Kit (+gDNA wiper) and ChamQ universal SYBR qPCR Master Mix (Vazyme, Nanjing, China) were used. And beta-actin was used as the reference gene. The reverse transcription and PCR primer sequences are listed in Supplementary Table S1. The relative expression levels of miRNA and mRNA were calculated using 2−ΔΔCT (Rao et al., 2013).
To verify the target relationship of SIRT1 and oar-miR-432, Xho I and NotI restriction enzyme cutting sites were amplified with the wild-type 3'UTR of the SIRT1. The primers are listed in Supplementary Table S1. The wild-type 3'UTR of the SIRT1 was ligated to vectors and named psiCHECK2-SIRT1-3'UTR-WT.
Using a Site-Directed Mutagenesis Kit (Thermo Fisher Scientific, MA, United States), the mutant-type 3'UTR of SIRT1 was obtained and named psiCHECK2-SIRT1-3'UTR-MT. PsiCHECK2-SIRT1-3'UTR-WT, psiCHECK2-SIRT1-3'UTR-MT, or pure vectors were co-transfected with oar-miR-432 mimics; pure vectors were co-transfected with negative control (NC) or oar-miR-432 mimics into 293T (Pan et al., 2018). After incubation for 6 h, the culture medium was changed. After 48 h of incubation, the relative luciferase activity in the cells was measured using a Dual-Luciferase Reporter Assay System (Promega, Promega, WI, United States). Each treatment was performed 4 times for each group. All plasmid, oar-miR-432 mimics, and negative control were synthesized by GenePharma (Shanghai, China).
Sheep preadipocytes were isolated from the tail fat of a 70-day-old Hu sheep fetus by collagenase digestion. Preadipocyte transfection and culture were according to our previous method (Jin et al., 2022). When the cell showed contact inhibition, we collected cells and extracted protein.
Proteins from cell were extracted with RIPA buffer and separated on SDS-PAGE gel including 4% concentrated glue and 12% separation gel. After transfer, the PVDF blot membranes were blocked and then probed with rabbit polyclonal antibody against SIRT1 (1: 1,000, Proteintech, Chicago, IL, United States) at 4°C overnight. Alpha-tubulin poly-clonal antibody (1:3,000, Abclonal, Beijing, China) was used as an internal reference. These blots were further conjugated with a goat anti-rabbit IgG secondary antibody (1:1,000, Proteintech, Chicago, IL, United States) labeled with HRP via incubation and revealed with an ECL kit (Engreen, Beijing, China), and exposed to X-ray films. Blot intensity quantification was performed using ImageJ software (1.51j8) (Rha and Gyeol Yoo, 2015).
The data were processed by SPSS 20.0 two-tailed Student’s t-test (Singh et al., 2019). All the results are presented as means ± standard deviation. Furthermore, * indicates statistically significant (p < 0.05). ** indicates statistically significant (p < 0.01).
The results of the miRNA-Seq data after quality control are displayed in Table 1. The clean tag count of each sample ranged from 27 to 28 million, and the Q20 of clean tags ranged from 98.20% to 98.50%. About 88.63%–92.75% of the clean reads were mapped to the sheep reference genome.
In this study, 134 known miRNAs and 275 novel miRNAs were identified from HG1; 132 known miRNAs and 291 novel miRNAs were identified from HG2; 137 known miRNAs and 298 novel miRNAs were identified from HG3; 132 known miRNAs and 295 novel miRNAs were identified from ZG1; 133 known miRNAs and 198 novel miRNAs were identified from ZG2; and 129 known miRNAs and 273 novel miRNAs were identified from ZG3 (Supplementary Table S2).
We found 379 novel miRNAs and 139 known miRNAs. Hu sheep is set as a control, based on the padj ≤0.05, we detected 11 DE miRNAs in ZG compared with HG (Figure 1 and Supplementary Table S3). There are six upregulated miRNAs, including novel_mir471, oar-miR-432, novel_mir21, novel_mir59, novel_mir394 and, novel_mir70. There are five downregulated miRNAs, including oar-miR-29b, novel_mir58, novel_mir54, oar-miR-19b, and novel_mir64. Three miRNAs were reported that were associated with fat metabolism.
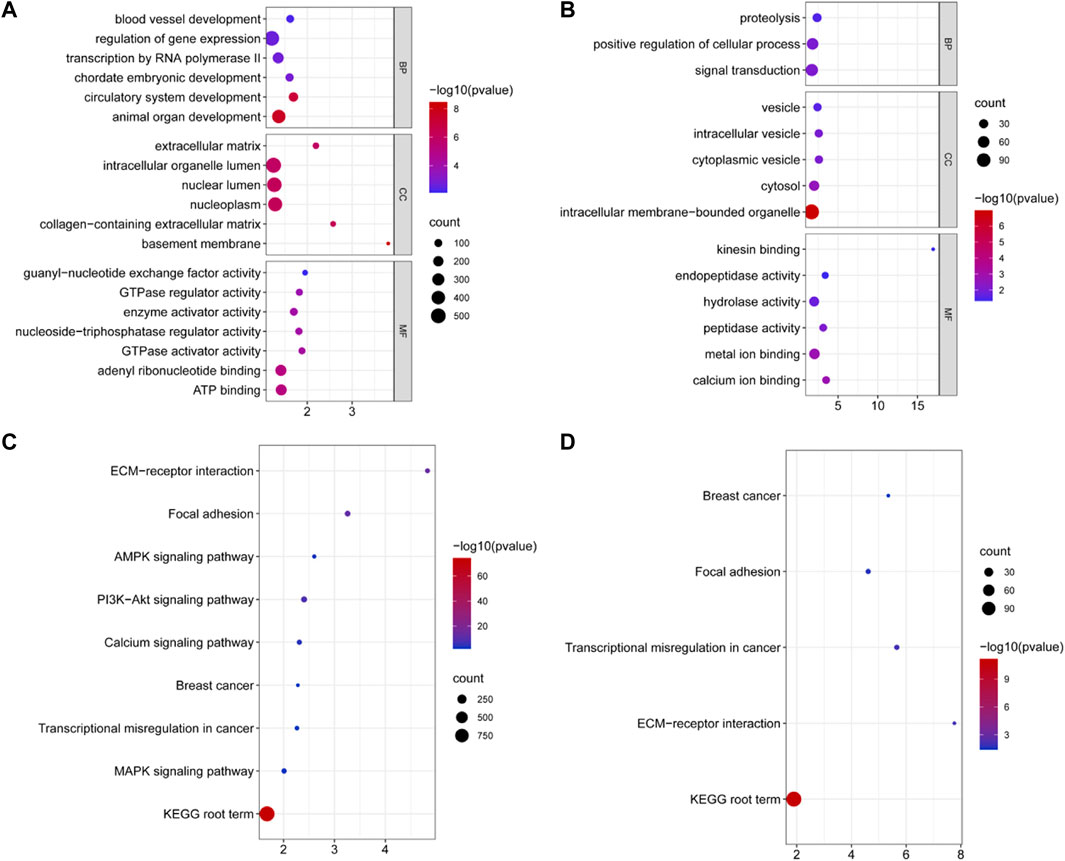
FIGURE 1. The volcano plots of all expressed miRNAs in the livers of Hu sheep (short/fat-tailed sheep) and Tibetan sheep (short/thin-tailed sheep). The x-axis denotes the values of log2 (fold-change), whereas the y-axis denotes the −log10 (padj). The colored dots represent the expressed miRNAs, with blue indicating downregulated miRNAs and red indicating upregulated miRNAs (padj ≤0.05). The black dots indicate that the miRNAs are not statistically significant (padj >0.05).
Miranda and RNAhybrid software were used to predict the target genes of DE miRNAs, resulting in 3,404 predicted target genes (Supplementary Table S4). GO annotation enrichment was used to describe the functions of the target genes of upregulated and downregulated DE miRNAs. These were involved in cellular components (CCs), molecular function (MF), and biological processes (BP), including animal organ development, intracellular organelle lumen, ATP binding, intracellular vesicles, and kinesin and calcium ion binding (Figures 2A,B and Supplementary Table S5). A total of 115 GO terms were significantly enriched by target genes of the upregulated DE miRNAs, and 54 terms were significantly enriched by target genes of the downregulated DE miRNAs. DE miRNAs were used in a KEGG pathway enrichment analysis. Based on all the target genes of upregulated and downregulated miRNAs, 67 and 5 KEGG pathways were significantly enriched, respectively (Supplementary Table S6). As shown in Figures 2C,D, the ECM–receptor interaction signaling pathway, KEGG root term signaling pathway, transcriptional regulation in the cancer signaling pathway, the focal adhesion signaling pathway, and the breast cancer signaling pathway were simultaneously enriched. Other signaling pathways related to fat metabolism were enriched, including the PI3K-Akt signaling pathway, calcium signaling pathway, AMPK signaling pathway, and MAPK signaling pathway, which are related to fat metabolism.
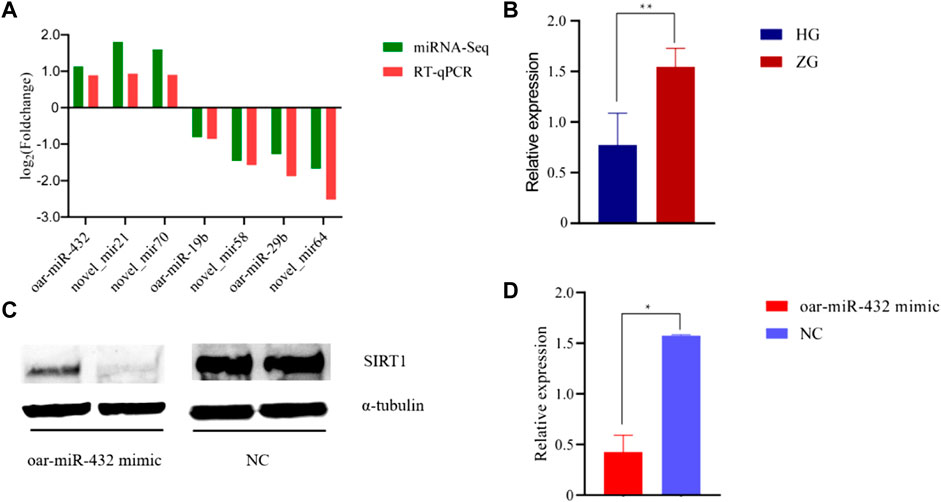
FIGURE 2. Significantly enriched Gene Ontology and KEGG for the target genes of DE miRNAs. (A) Some GO terms of target genes of upregulated DE miRNAs for BP, CC, and MF in two groups. (B) GO terms of target genes of downregulated DE miRNAs for BP, CC, and MF in two groups. The x-axis displays enrichment, and the y-axis rep-resents the GO terms. The filled colored circles display each statistically significant GO term. The size of the circles represents the gene number. (C) Signal pathway of the target genes of upregulated DE miRNAs in two groups. (D) Some signal pathways of the target genes of upregulated DE miRNAs in two groups. The x-axis displays the enrich-ment factor of the target genes, and the y-axis represents the KEGG pathway. The filled colored circles represent each statistically significant KEGG pathway. The size of the circles represents the number of genes.
The RT-qPCR technique was used to validate the sequencing results. Seven miRNAs were randomly selected for RT-qPCR verification. The validation results are displayed in Figure 3A and Supplementary Table S7.
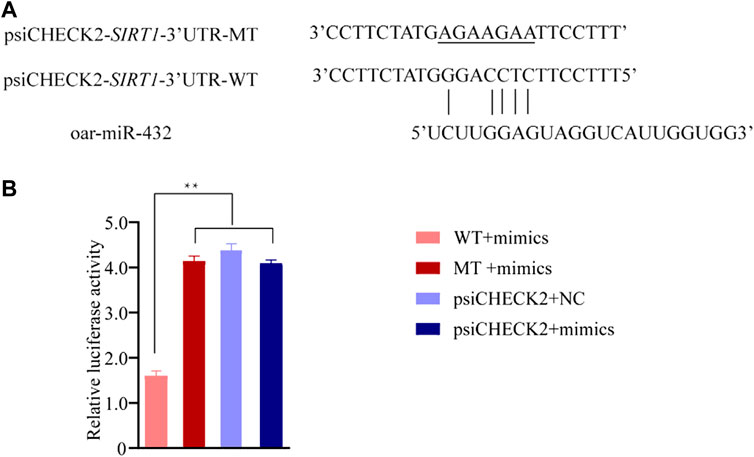
FIGURE 3. The results of RT-qPCR and Western blot. (A) RNA-Seq and RT-qPCR results of seven differentially expressed miRNAs in ZG compared with HG. (B) RT-qPCR results of SIRT1 in HG and ZG. (C) (D) Western blot results of SIRT1 in preadipocytes. NC exhibits negative control.
Eight randomly selected monoclonals and vector universal primers were used to identify the wild-type psiCHECK2 plasmid by polymerase chain reaction (PCR) (Supplementary Figure S1) and sequencing. The sequencing primers are shown in Supplementary Table S1. Site-directed mutation was used to obtain the mutant-type psiCHECK2 plasmid. The sequencing results of wild-type psiCHECK2 plasmid and mutant-type psiCHECK2 are in Supplementary Table S8 and Supplementary Table S9. Eventually, the plasmids were constructed successfully.
A dual-luciferase reporter assay indicated that oar-miR-432 significantly suppressed the luciferase activities for co-transfection with SIRT1 3'UTR wild-types, although did not affect the mutant types of SIRT1 3'UTR or blank vectors (Figure 4B and Supplementary Table S10). These results initially confirmed the direct interactions between oar-miR-432 and SIRT1.
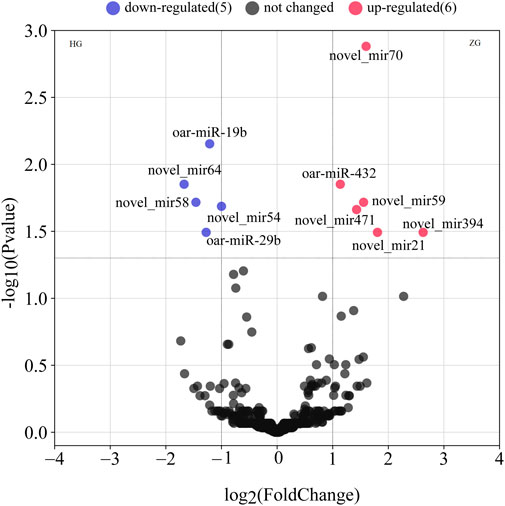
FIGURE 4. Result of the luciferase reporter assay. (A) Potential binding site between oar-miR-432 and SIRT13'UTR. The underlined sequences represent the mutant sites. (B) WT exhibits the psiCHECK2-SIRT1-3'UTR-WT. MT exhibits psiCHECK2-SIRT1-3'UTR-MT. psiCHECK2 exhibits psiCHECK2 pure vectors. Mimics exhibits oar-miR-432 mimics. NC exhibits negative control. **: indicates statistically significant (p < 0.01).
The RT-qPCR results showed that the expression trends in oar-miR-432 and SIRT1 were contrasting. oar-miR-432 was highly expressed in the liver tissue of Hu sheep, while the SIRT1 was highly expressed in the liver tissue of Tibetan sheep (Figure 3B, Supplementary Table S7).
Oar-miR-432 mimics and negative control were transfected into preadipocytes. Then we detected the expression of oar-miR-432 and SIRT1. The expression of oar-miR-432 was increased by oar-miR-432 mimics (Jin et al., 2022). The result of the Western blot showed the expression of SIRT1 was inhibited by oar-miR-432 mimics (Figures 3C,D, Supplementary Table S11, Supplementary Figure S2, Supplementary Figure S3).
Thus far, miRNA expression has been studied in the liver tissues of buffalos (Rha and Gyeol Yoo, 2015), dairy cows (Bu et al., 2017), mice (Seclaman et al., 2019), rats (Wang et al., 2017), pigeons (Wang et al., 2020), pigs (Kai et al., 2019), chickens (Xu et al., 2019), and geese (Zheng et al., 2015). RNA-Seq was used to construct 41 pairs of ceRNA networks on liver tissue from three Holstein cows, which provide new insight into resolving bovine lipid metabolism (Liang et al., 2017). In bovine hepatocytes, miR-27a-5p inhibited calcium sensing receptor (CASR) expression, triacylglycerol (TAG) accumulation was significantly suppressed, and low very density lipoprotein (VLDL) secretion was reduced (Yang et al., 2018). established miRNA-mRNA regulatory networks related to lipid deposition and metabolism in the livers of Landrace pigs with the extreme backfat thickness (Kai et al., 2019). RNA-Seq was used to construct miRNA-mRNA networks between Jinhua and Landrace pigs (Huang et al., 2019). These studies provided new insights into the molecular mechanisms to explore fat metabolism in pigs. Also, the study found there was a lncRNA-FNIP2/miR-24-3p/FNIP2 axis, which can regulate lipid metabolism in Sanghuang chicken liver (Guo et al., 2021).
In this study, we used high-throughput sequencing to identify the expression of miRNA in the livers of Hu sheep and Tibetan sheep. This study complements the current understanding of miRNA expression patterns in sheep livers and will help future research on the specific role of miRNA in regulating fat metabolism. In our study, we identified 11 differential miRNAs. miR-432, miR-19b, and miR-29b are associated with fat metabolism, and a previous study showed that miR-432 inhibits milk fat synthesis by targeting stearoyl CoA desaturase (SCD) and LPL in ovine mammary epithelial cells. Additionally, miR-432 inhibits the proliferation of ovine mammary epithelial cells (Hao et al., 2021). Transcriptome analysis revealed that miR-432 was differentially expressed in the backfat of cattle; the protein kinase AMP-activated catalytic subunit alpha 1/2 (PRKAA1/2) and peroxisome proliferator-activated receptor alpha (PPARA) were regulation targets to modulate lipid and fatty acid metabolism (Sun et al., 2014). Interestingly, miR-432 was differentially expressed in tail fat between Hu sheep and Tibetan sheep, which could have an important function in sheep fat metabolism (Fei et al., 2022). In mice SVF cells, miR-19b had an inhibitory effect on the browning process of adipose tissue (Lv et al., 2018). Researchers found that miR-29b can regulate blood sugar in adult mice, representing a target for treating metabolism disease (Hung et al., 2019). Additionally, miR-29b inhibits the differentiation of pig muscle and subcutaneous preadipocytes through targeted regulation complement component 1 (C1q) and TNF-related protein 6 (CTRP6) (Wu et al., 2021). Ma et al. found that lncRNAs, including TCONS_00372,767 and TCONS_00171,926, were related to fat metabolism among Lanzhou fat-tailed sheep, small-tailed Han sheep, and Tibetan sheep, and constructed two co-expression networks of differentially expressed mRNA and lncRNA (Ma et al., 2018). The research conducted by Cheng et al. showed that there were differences in the livers of Mongolian and Lanzhou fat-tailed sheep through RNA-Seq, which provided a reference for researching the sheep genome (Cheng et a., 2016).
Hu sheep set as a control to identify DE miRNAs. The extracellular matrix (ECM)–receptor interaction signaling pathway was significantly enriched by the target genes of upregulated DE miRNAs and downregulated DE miRNAs. The main constituents of the ECM–receptor interaction signaling pathway in adipose tissue include collagen (type I, IV, and VI), fibronectin (FN), laminin (LN1,8), hyaluronan, and proteoglycan (Lee et al., 2013). The functional analysis showed differently expressed genes in the subcutaneous and intramuscular fat of cattle were enriched in ECM–receptor interaction signaling pathway. In the study of San et al., some genes which affected intramuscular fat (IMF) deposition was significantly enriched in the ECM–receptor interaction signaling pathway (San et al., 2021). In our study, the target genes of upregulated DE miRNAs were enriched in the PI3K-Akt signaling pathway, calcium signaling pathway, the AMPK signaling pathway, and MAPK signaling pathway, which are associated with fat metabolism (Fu et al., 2022). In our study, forkhead boxO3 (FoxO3) was enriched in the PI3K/AKT signaling pathway and AMPK signal pathway. In mice fed high-glucose and high-sucrose diets, FoxO3 promoted hepatic triglyceride synthesis and hepatic triglyceride accumulation in the liver by positively regulating the sterol regulatory element binding transcription factor 1 (SREBP1c) (Wang et al., 2019). Additionally, SIRT1 was enriched in the AMPK signal pathway. SIRT1 plays an important biological role in regulating liver lipid metabolism, oxidative stress, and inflammation, and can be used as a therapeutic target for the treatment of alcoholic and non-alcoholic fatty liver diseases (Ding et al., 2017). It has been shown that vitamin D can activate the AMPK/SIRT1 pathway to inhibit the accumulation of fat in C2C12 skeletal muscle cells (Chang and Kim., 2019). miR-29 can regulate SIRT1 to inhibit fat deposits in mouse livers (Kurtz et al., 2015). Additionally, Liang et al. that dietary cholesterol can promote the occurrence of steatohepatitis through the calcium signaling pathway (Liang et al., 2018). In a diabetic mouse model, the ginsenoside metabolite compound K inhibits the activation of the NLR family pyrin domain containing 3 (NLRP3) through the NF-κB/p38 signaling pathway (Song et al., 2018). Previous studies have shown that in human liver fat cells, transforming growth factor-beta 1 (TGF-β1) regulates the platelet-derived growth factor receptor beta (PDGFD-β) subunit to maintain the activation and proliferation of fat cells (Pinzani et al., 1995). In our previous study, these pathways were enriched significantly, including ECM–receptor interaction signaling pathway, PI3K-Akt signaling pathway, calcium signaling pathway, AMPK signaling pathway, and MAPK signaling pathway (Fei et al., 2022). All of the results showed that these pathways could have a vital function in sheep fat metabolism.
In this research, our goal was to preliminarily determine how oar-miR-432 and SIRT1 regulate fat metabolism. In our current study, we use dual-luciferase reporter assays to verify the binding relationship between miR-432 and the target gene SIRT1. The expression of SIRT1 was detected in the liver tissues of Hu sheep and Tibetan sheep. RT-qPCR results showed that the expression of SIRT1 in Tibetan sheep was significantly higher than that in Hu sheep. We transfected oar-miR-432 in preadipocytes, and we found oar-miR-432 can inhibit the expression of SIRT1 at the protein level. This is the first time reported that the expression of SIRT1 gene was regulated by oar-miR-432 in fat metabolism of sheep liver. The regulation of the process leading from mRNA to protein is generally very complex. Studies have shown that gene repression could be changed due to the post-transcriptional regulation of miRNA (Pasquier and Gardès., 2016). Our study showed that oar-miR-432 downregulated the expression of SIRT1 at the transcriptional level in sheep liver tissue. Meanwhile, the result of Western blot showed that oar-miR-432 can downregulated the expression of SIRT1 protein in preadipocytes. Our study indicated that p53 is independent of the oar-miR-432 SIRT1 gene regulation.
In summary, our results provide a comprehensive expression profile of miRNA in the livers between two different sheep breeds. The DE miRNAs reported in this article may play an important role in sheep fat metabolism. We have verified that oar-miR-432 can target the regulation gene SIRT1 in sheep. This study provides a reference for further research addressing the modulation of fat metabolism in different sheep breeds.
The datasets presented in this study can be found in online repositories. The sequencing files have been stored in the Sequence Read Archive (accession numbers PRJNA785102).
The animal study was reviewed and approved by Ethical approval was in compliance with the Animal Ethics Committee of the Institute of Animal Sciences, Chinese Academy of Agriculture Sciences(IAS-CAAS).
Conceptualization, CW and YW; methodology, ZL, ZY, and HW; software, JL, KQ, MH, and ZL; validation, XF, MJ, and TL; formal analysis, JY and TW; writing—original draft preparation, XF; writing—review and editing, ZY.
This research was supported by China Agriculture Research System of MOF and MARA (CARS-38). National Natural Science Foundation of China (32272851).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.985764/full#supplementary-material
Balaskas, P., Green, J., Haqqi, T., Dyer, P., Kharaz, Y. A., Fang, Y., et al. (2020). Small non-coding RNAome of ageing chondrocytes. Int. J. Mol. Sci. 21 (16), 5675. doi:10.3390/ijms21165675
Bu, D., Bionaz, M., Wang, M., Nan, X., Ma, L., and Wang, J. (2017). Transcriptome difference and potential crosstalk be-tween liver and mammary tissue in mid-lactation primiparous dairy cows. PLoS. One. 12 (3), e0173082. doi:10.1371/journal.pone.0173082
Carotti, S., Aquilano, K., Valentini, F., Ruggiero, S., Alletto, F., Morini, S., et al. (2020). An overview of deregulated lipid metabolism in nonalcoholic fatty liver disease with special focus on lysosomal acid lipase. Am. J. Physiol. Gastrointest. Liver. Physiol. 319 (4), G469–G480. doi:10.1152/ajpgi.00049.2020
Chang, E., and Kim, Y. (2019). Vitamin D ameliorates fat accumulation with AMPK/SIRT1 activity in C2C12 skeletal muscle cells. Nutrients 11 (11), 2806. doi:10.3390/nu11112806
Chen, X., Raza, S. H. A., Cheng, G., Ma, X., Wang, J., and Zan, L. (2020). Bta-miR-376a targeting KLF15 interferes with adipogenesis signaling pathway to promote differentiation of qinchuan beef cattle preadipocytes. Anim. (Basel) 10 (12), 2362. doi:10.3390/ani10122362
Cheng, X., Zhao, S., Yue, Y., Liu, Z., Li, H., and Wu, J. (2016). Comparative analysis of the liver tissue transcriptomes of Mongolian and Lanzhou fat-tailed sheep. Genet. Mol. Res. 15 (2), gmr8572. doi:10.4238/gmr.15028572
Deng, K., Ren, C., Fan, Y., Liu, Z., Zhang, G., Zhang, Y., et al. (2020). miR-27a is an important adipogenesis reg-ulator associated with differential lipid accumulation between intramuscular and subcutaneous adipose tissues of sheep. Domest. Anim. Endocrinol. 71, 106393. doi:10.1016/j.domaniend.2019.106393
Ding, R., Bao, J., and Deng, C. (2017). Emerging roles of SIRT1 in fatty liver diseases. Int. J. Biol. Sci. 13 (7), 852–867. doi:10.7150/ijbs.19370
Fu, Y., Jia, R., Xu, L., Su, D., Li, Y., Liu, L., et al. (2022). Fatty acid desaturase 2 affects the milk-production traits in Chinese Holsteins. Anim. Genet. 53 (3), 422–426. doi:10.1111/age.13192
Fei, X., Jin, M., Wang, Y., Li, T. T., Lu, Z., Yuan, Z., et al. (2022). Tran-scriptome reveals key microRNAs involved in fat deposition between different tail sheep breeds. PLoS One 17 (3), e0264804. doi:10.1371/journal.pone.0264804
Guo, L., Chao, X., Huang, W., Li, Z., Luan, K., Ye, M., et al. (2021). Whole transcriptome analysis reveals a potential regulatory mechanism of LncRNA-FNIP2/miR-24-3p/FNIP2 Axis in chicken adipogenesis. Front. Cell. Dev. Biol. 9, 653798. doi:10.3389/fcell.2021.653798
Hao, Z., Luo, Y., Wang, J., Hickford, J. G. H., Zhou, H., Hu, J., et al. (2021). MicroRNA-432 inhibits milk fat synthesis by targeting SCD and LPL in ovine mammary epithelial cells. Food. Funct. 12 (19), 9432–9442. doi:10.1039/d1fo01260f
Hu, F., Wang, M., Xiao, T., Yin, B., He, L., Meng, W., et al. (2015). miR-30 promotes thermogenesis and the development of beige fat by targeting RIP140. Diabetes 64 (6), 2056–2068. doi:10.2337/db14-1117
Huang, M., Chen, L., Shen, Y., Chen, J., Guo, X., and Xu, N. (2019). Integrated mRNA and miRNA profile ex-pression in livers of Jinhua and Landrace pigs. Asian-Australas. J. Anim. Sci. 32 (10), 1483–1490. doi:10.5713/ajas.18.0807
Hung, Y., Kanke, M., Kurtz, C., Cubit, T., Bunaciu, R., Miao, J., et al. (2019). Acute suppression of insulin resistance-associated hepatic miR-29 in vivo improves glycemic control in adult mice. Physiol. Genomics. 51 (8), 379–389. doi:10.1152/physiolgenomics.00037.2019
Jeon, S., and Carr, R. (2020). Alcohol effects on hepatic lipid metabolism. J. Lipid. Res. 61 (4), 470–479. doi:10.1194/jlr.R119000547
Jin, M., Fei, X., Li, T., Lu, Z., Chu, M., Di, R., et al. (2022). Oar-miR-432 regulates fat differentiation and promotes the expression of BMP2 in ovine preadipocytes. Front. Genet. 13, 844747. doi:10.3389/fgene.2022.844747
John, B., Enright, A., Aravin, A., Tuschl, T., Sander, C., and Marks, D. (2004). Human MicroRNA targets. PLoS. Biol. 2 (11), e363. doi:10.1371/journal.pbio.0020363
Kai, X., Zhao, X., Ao, H., Chen, S., Yang, T., Tan, Z., et al. (2019). Transcriptome analysis of miRNA and mRNA in the livers of pigs with highly diverged backfat thickness. Sci. Rep. 9 (1), 16740. doi:10.1038/s41598-019-53377-x
Kern, F., Amand, J., Senatorov, I., Isakova, A., Backes, C., Meese, E., et al. (2020). miRSwitch: detecting microRNA arm shift and switch events. Nucleic. acids. Res. 2020, 48, (W1), W268–W274. doi:10.1093/nar/gkaa323
Kurtz, C., Fannin, E., Toth, C., Pearson, D., Vickers, K., and Sethupathy, P. (2015). Inhibition of miR-29 has a significant lipid-lowering benefit through suppression of lipogenic programs in liver. Sci. Rep. 5, 12911. doi:10.1038/srep12911
Langmead, B., Trapnell, C., Pop, M., and Salzberg, S. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome. Biol. 10 (3), R25. doi:10.1186/gb-2009-10-3-r25
Lee, H., Jang, M., Kim, H., Kwak, W., Park, W., Hwang, J., et al. (2013). Comparative transcriptome analysis of adipose tissues reveals that ECM-Receptor interaction is involved in the de-pot-specific adipogenesis in cattle. PLoS One 8 (6), e66267. doi:10.1371/journal.pone.0066267
Li, B., Yang, J., Gong, Y., Xiao, Y., Zeng, Q., Xu, K., et al. (2021). Integrated analysis of liver transcriptome, miRNA, and proteome of Chinese indigenous breed ningxiang pig in three developmental stages uncovers significant miRNA-mRNA-Protein networks in lipid metabolism. Front. Genet. 12, 709521. doi:10.3389/fgene.2021.709521
Li, Q., Lu, Z., Jin, M., Fei, X., Quan, K., Liu, Y., et al. (2020). Verification and analysis of sheep tail type-associated PDGF-D gene polymorphisms. Animals 10 (1), 89. doi:10.3390/ani10010089
Liang, J., Teoh, N., Xu, L., Pok, S., Li, X., Chu, E. S. H., et al. (2018). Dietary cholesterol promotes steatohepatitis related hepatocellular carcinoma through dysregulated metabolism and calcium signaling. Nat. Commun. 9 (1), 4490. doi:10.1038/s41467-018-06931-6
Liang, R., Han, B., Li, Q., Yuan, Y., Li, J., and Sun, D. (2017). Using RNA sequencing to identify putative competing endogenous RNAs (ceRNAs) potentially regulating fat metabolism in bovine liver. Sci. Rep. 7 (1), 6396. doi:10.1038/s41598-017-06634-w
Lin, H., Wang, F., Yang, Y., and Huang, Y. (2019). MicroRNA-29a suppresses CD36 to ameliorate high fat diet-induced steatohepatitis and liver fibrosis in mice. Cells 8 (10), 1298. doi:10.3390/cells8101298
Lin, Y., Dan, H., and Lu, J. (2020). Overexpression of microRNA-136-3p alleviates myocardial injury in coronary artery disease via the rho A/ROCK signaling pathway. Kidney. blood. Press. Res. 45 (3), 477–496. doi:10.1159/000505849
Lin, Z., Tang, Y., Li, Z., Yu, C., Yang, C., Liu, L., et al. (2022). miR-24-3p dominates the proliferation and differentiation of chicken intramuscular preadipocytes by blocking ANXA6 expression. Genes 13 (4), 635. doi:10.3390/genes13040635
Lu, Z., Liu, J., Han, J., and Yang, B. (2020). Association between BMP2 functional polymorphisms and sheep tail type. Anim. (Basel) 10 (4), 739. doi:10.3390/ani10040739
Luo, R., Zhang, X., Wang, L., Zhang, L., Li, G., and Zheng, Z. (2021). GLIS1, a potential candidate gene affect fat depo-sition in sheep tail. Mol. Biol. Rep. 48 (5), 4925–4931. doi:10.1007/s11033-021-06468-w
Lv, Y., Yu, J., Sheng, Y., Huang, M., Kong, X., Di, W., et al. (2018). Glucocorticoids suppress the browning of adipose tissue via miR-19b in male mice. Endocrinology 159 (1), 310–322. doi:10.1210/en.2017-00566
Ma, L., Zhang, M., Jin, Y., Erdenee, S., Hu, L., Chen, H., et al. (2018). Comparative transcriptome profiling of mRNA and lncRNA related to tail adipose tissues of sheep. Front. Genet. 9, 365. doi:10.3389/fgene.2018.00365
Mortazavi, A., Williams, B., McCue, K., Schaeffer, L., and Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 5 (7), 621–628. doi:10.1038/nmeth.1226
Pan, Y., Jing, J., Qiao, L., Liu, J., An, L., Li, B., et al. (2018). MiRNA-seq reveals that miR-124-3p inhibits adi-pogenic differentiation of the stromal vascular fraction in sheep via targeting C/EBPα. Domest. Anim. Endocrinol. 65, 17–23. doi:10.1016/j.domaniend
Pasquier, C., and Gardès, J. (2016). Prediction of miRNA-disease associations with a vector space model. Sci. Rep. 6, 27036. doi:10.1038/srep27036
Peng, Y., Xiang, H., Chen, C., Zheng, R., Chai, J., Peng, J., et al. (2013). MiR-224 impairs adipocyte early differentiation and regulates fatty acid metabolism. Int. J. Biochem. Cell. Biol. 45 (8), 1585–1593. doi:10.1016/j.biocel.2013.04.029
Pflug, F., and Haeseler, v. (2018). TRUmiCount: Correctly counting absolute numbers of molecules using unique molecular identifiers. Bioinformatics 34 (18), 3137–3144. doi:10.1093/bioinformatics/bty283
Pinzani, M., Gentilini, A., Caligiuri, A., Franco, R., Pellegrini, G., Milani, S., et al. (1995). Transforming growth factor-beta 1 regulates platelet-derived growth factor receptor beta subunit in human liver fat-storing cells. Hepatology 21, 232–239. doi:10.1016/0270-9139(95)90433-6
Rao, X., Huang, X., Zhou, Z., and Lin, X. (2013). An improvement of the 2ˆ(-delta delta CT) method for quantitative re-al-time polymerase chain reaction data analysis. Biostat. Bioinforma. Biomath. 3 (3), 71–85.
Raudvere, U., Kolberg, L., Kuzmin, I., Arak, T., Adler, P., Peterson, H., et al. (2019). g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic. acids. Res. 47 (W1), W191–W198. doi:10.1093/nar/gkz369
Rha, E., Gyeol Yoo, J. M. K., and Yoo, G. (2015). Volume measurement of various tissues using the image J software. J. Craniofac Surg. 26 (6), e505–e506. doi:10.1097/SCS.0000000000002022
Rouleau, S., Glouzon, J., Brumwell, A., Bisaillon, M., and Perreault, J. (2017). 3' UTR G-quadruplexes regulate miRNA binding. RNA 23 (8), 1172–1179. doi:10.1261/rna.060962.117
San, J., Du, Y., Wu, G., Xu, R., Yang, J., and Hu, J. (2021). Transcriptome analysis identifies signaling pathways related to meat quality in broiler chickens - the extracellular matrix (ECM) receptor interaction signaling pathway. Poult. Sci. 100 (6), 101135. doi:10.1016/j.psj.2021.101135
Seclaman, E., Balacescu, L., Balacescu, O., Bejinar, C., Udrescu, M., Marian, C., et al. (2019). MicroRNAs me-diate liver transcriptome changes upon soy diet intervention in mice. J. Cell. Mol. Med. 23 (3), 2263–2267. doi:10.1111/jcmm.14140
Sikorska, M., Siwek, M., Slawinska, A., and Dunislawska, A. (2021). miRNA profiling in the chicken liver under the influence of early microbiota stimulation with probiotic, prebiotic, and synbiotic. Genes (Basel). 12 (5), 685. doi:10.3390/genes12050685
Singh, S., Golla, N., Sharma, D., Singh, D., and Onteru, S. K. (2019). Buffalo liver transcriptome analysis suggests immune tolerance as its key adaptive mechanism during early postpartum negative energy balance. Funct. Integr. Genomics. 19 (5), 759–773. doi:10.1007/s10142-019-00676-1
Soares, D. O., Amaral, N., Cruz, E. M. N., DE, M. E. L. O. M. A. I. A. B., and Malagoli, R. O. C. H. A. R. (2016). Noncoding RNA profiles in tobacco- and alcohol-associated diseases. Genes 8 (1), 6. doi:10.3390/genes8010006
Song, W., Wei, L., Du, Y., Wang, Y., and Jiang, S. (2018). Protective effect of ginsenoside metabolite compound K against di-abetic nephropathy by inhibiting NLRP3 inflammasome activation and NF-κB/p38 signaling pathway in high-fat di-et/streptozotocin-induced diabetic mice. Int. Immunopharmacol. 63, 227–238. doi:10.1016/j.intimp.2018.07.027
Sun, J., Zhang, B., Lan, X., Zhang, C., Lei, C., and Chen, H. (2014). Comparative transcriptome analysis reveals significant differences in MicroRNA expression and their target genes between adipose and muscular tissues in cattle. PLoS. One. 9 (7), e102142. doi:10.1371/journal.pone.0102142
Wang, H., Shao, Y., Yuan, F., Feng, H., Li, N., Zhang, H., et al. (2017). Fish oil feeding modulates the expression of hepatic MicroRNAs in a western-style diet-induced nonalcoholic fatty liver disease rat model. Biomed. Res. Int. 2017, 2503847. doi:10.1155/2017/2503847
Wang, L., Zhu, X., Sun, X., Yang, X., Chang, X., Xia, M., et al. (2019). FoxO3 reg-ulates hepatic triglyceride metabolism via modulation of the expression of sterol regulatory-element binding protein 1c. Lipids. Health. Dis. 18 (1), 197. doi:10.1186/s12944-019-1132-2
Wang, X., Yan, P., Liu, L., Luo, Y., Zhao, L., Liu, H., et al. (2020). MicroRNA expression profiling reveals potential roles for microRNA in the liver during pigeon (Columba livia) development. Poult. Sci. 99 (12), 6378–6389. doi:10.1016/j.psj.2020.09.039
Wang, Z., Gerstein, M., and Snyder, M. (2009). RNA-seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 10 (1), 57–63. doi:10.1038/nrg2484
Wu, W., Xu, K., Li, M., Zhang, J., and Wang, Y. (2021). MicroRNA-29b/29c targeting CTRP6 influences porcine adipogenesis via the AKT/PKA/MAPK Signalling pathway. Adipocyte 10 (1), 264–274. doi:10.1080/21623945.2021.1917811
Xu, E., Zhang, L., Yang, H., Shen, L., Feng, Y., Ren, M., et al. (2019). Transcriptome profiling of the liver among the prenatal and postnatal stages in chickens. Poult. Sci. 98 (12), 7030–7040. doi:10.3382/ps/pez434
Yang, W., Tang, K., Wang, Y., and Zan, L. (2018). MiR-27a-5p increases steer fat deposition partly by targeting cal-cium-sensing receptor (CASR). Sci. Rep. 8 (1), 3012. doi:10.1038/s41598-018-20168-9
Zhang, Y., Wang, Y., Wang, H., Ma, X., and Zan, L. (2019). MicroRNA-224 impairs adipogenic differentiation of bovine preadipocytes by targeting LPL. Mol. Cell. Probes. 44, 29–36. doi:10.1016/j.mcp.2019.01.005
Zheng, F., Zhang, J., Luo, S., Yi, J., Wang, P., Zheng, Q., et al. (2016). miR-143 is associated with proliferation and apoptosis involving ERK5 in HeLa cells. Oncol. Lett. 12 (4), 3021–3027. doi:10.3892/ol.2016.5016
Zheng, Y., Jiang, S., Zhang, Y., Zhang, R., and Gong, D. (2015). Detection of miR-33 expression and the verification of its target genes in the fatty liver of geese. Int. J. Mol. Sci. 16 (6), 12737–12752. doi:10.3390/ijms160612737
Keywords: miRNA, liver, fat metabolism, Hu sheep, Tibetan sheep
Citation: Fei X, Jin M, Yuan Z, Li T, Lu Z, Wang H, Lu J, Quan K, Yang J, He M, Wang T, Wang Y and Wei C (2023) MiRNA-Seq reveals key MicroRNAs involved in fat metabolism of sheep liver. Front. Genet. 14:985764. doi: 10.3389/fgene.2023.985764
Received: 04 July 2022; Accepted: 27 February 2023;
Published: 09 March 2023.
Edited by:
Ernesto Picardi, University of Bari Aldo Moro, ItalyReviewed by:
Ian James Martins, University of Western Australia, AustraliaCopyright © 2023 Fei, Jin, Yuan, Li, Lu, Wang, Lu, Quan, Yang, He, Wang, Wang and Wei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Caihong Wei, d2VpY2FpaG9uZ0BjYWFzLmNu; Yuqin Wang, V2FuZ3lxNjgzNkAxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.