- 1Laboratory of Animal Husbandry, School of Veterinary Medicine, Faculty of Health Sciences, Aristotle University of Thessaloniki, Thessaloniki, Greece
- 2Centre for Research and Technology Hellas, Institute of Applied Biosciences, Thessaloniki, Greece
- 3Department of Genetics, Development and Molecular Biology, School of Biology, Aristotle University of Thessaloniki, Thessaloniki, Greece
- 4Genomics and Epigenomics Translational Research (GENeTres), Center for Interdisciplinary Research and Innovation (CIRI-AUTH), Balkan Center, Thessaloniki, Greece
- 5Laboratory of Anatomy and Physiology of Farm Animals, Department of Animal Science, School of Animal Biosciences, Agricultural University of Athens, Athens, Greece
- 6Scotland’s Rural College, Easter Bush, Midlothian, United Kingdom
Genetic selection for higher productivity increased dairy sheep susceptibility to diseases and environmental stressors, challenging their health and welfare status and production efficiency. Improving resilience to such stressors can enhance their ability to face these challenges without compromising productivity. Our objective was to estimate genomic heritability and perform genome-wide association studies (GWAS) to detect SNPs and candidate genes associated with three proxy traits for resilience (milk somatic cell count—SCC, lactation persistency—LP, body condition score—BCS) of Chios and Frizarta dairy ewes. We used genome-wide genotypes of 317 Chios and 346 Frizarta ewes. Individual records of milk yield and BCS, and milk samples were collected monthly for two consecutive milking periods; samples were analyzed to determine SCC. The LP was calculated as the regression coefficient of daily milk yield on days from lambing. Within breed, variance components analyses and GWAS were performed using genomic relatedness matrices in single-trait animal linear mixed models. Genomic-based heritability estimates were relatively high (BCS: h2 = 0.54 and 0.55, SCC: h2 = 0.25 and 0.38, LP: h2 = 0.43 and 0.45, for Chios and Frizarta ewes, respectively), compared to previous pedigree-based studies. The GWAS revealed 7 novel SNPs associated with the studied traits; one genome-wide and two suggestive significant SNPs for SCC (Frizarta: rs403061409, rs424064526 and rs428540973, on chromosomes 9, 1 and 12, respectively), one suggestive significant SNP for BCS (Chios: rs424834097 on chromosome 4) and three suggestive significant SNPs for LP (Frizarta: rs193632931 and rs412648955 on chromosomes 1 and 6, Chios: rs428128299 on chromosome 3). Nineteen candidate genes were detected: two for BCS (Chios: POT1, TMEM229A), thirteen for SCC (Frizarta: NTAQ1, ZHX1, ZHX2, LOC101109545, HAS2, DERL1, FAM83A, ATAD2, RBP7, FSTL1, CD80, HCLS1, GSK3B) and four for LP (Frizarta: GRID2, FAIM, CEP70—Chios: GRIP1). Present results show that resilience in the studied dairy sheep breeds is heritable and advance existing knowledge on the genomic background of SCC, LP, and BCS. Future research will quantify effects of different alleles of significant SNPs on the studied traits and search for possible correlations among traits to facilitate their effective incorporation in breeding programs aiming to improve resilience.
1 Introduction
Dairy sheep production in the Mediterranean basin has been practiced over thousands of years and still constitutes an important part of the animal production sector with socioeconomic and cultural impact (Paraskevopoulou et al., 2020). A large variety of local dairy sheep breeds have evolved over the course of time driven by either natural or artificial selection processes, perfectly adapted to their local environment and traditional management practices. However, increasing demand for higher productivity led to intensification of production and strong selection pressure of sheep. In the previous decades, many local sheep breeds that failed to quickly adapt and adequately increase their milk production have been replaced by foreign improved ones (Argyriadou et al., 2020). Genetic selection focusing only on higher productivity adversely affected other important traits; improved dairy sheep breeds are likely to be less resilient to environmental stressors and more susceptible to diseases (Mucha et al., 2022). As a result, their health and welfare are easily compromised, affecting the overall efficiency of production. Resilience is defined as the animal’s ability to withstand challenging environmental stressors without compromising productivity (De Barbieri et al., 2023). The importance of animal resilience became more evident with the increasing incidence of climate change-related phenomena that further challenge the efficiency of farm animal production. Emergence of infectious diseases, aridification of grasslands, limited feedstock availability and heat/cold stress of the animals are consequences that underline the need for breeding more resilient animals (Cheng et al., 2022). In this context, the genetic potential of resilient local sheep breeds that are well-adapted to harsh environmental conditions and low-input rearing systems is gaining scientific interest (Dumont et al., 2020).
The genetic study of sheep resilience has so far involved diverse traits regarding resistance to disease, resilience to weather conditions, longevity and resource use efficiency (Sánchez-Molano et al., 2020; Tsartsianidou et al., 2021; Hine et al., 2022; Mucha et al., 2022; Machefert et al., 2023). In dairy sheep, resilience is mainly associated with good udder health combined with longevity, extended lactations and adequately high milk performance. Mastitis is one of the biggest health issues in dairy sheep farms with significant economic impact since it is associated with production loss, poor milk quality, high culling rate and compromised ewe welfare (Gelasakis et al., 2015). Immune response to intramammary infections induces inflammation characterized by neutrophil migration into the udder and increased epithelial cell apoptosis that lead to high milk somatic cell count (SCC). Increased milk SCC is typical of both clinical and subclinical mastitis, whereas low milk SCC is indicative of resilience to mastitis. Selecting for lower milk SCC, which is a heritable trait, succeeded in limiting mastitis incidence (Rupp et al., 2009). Therefore, genetic and genomic architecture of SCC is currently a major topic of scientific interest (Rupp et al., 2015; Banos et al., 2017; Sutera et al., 2021; Öner et al., 2021; Mohammadi et al., 2022). However, existing differences among populations as well as the polygenic inheritance of the trait complicate safe conclusions and breed-specific studies are necessary.
Lactation persistency (LP) is another important dairy trait, which represents the decline rate of milk yield following the peak of lactation; a lower rate is related to extended and more productive lactations (Stefanon et al., 2002). The observed decrease in milk yield as lactation progresses is associated with the rate of epithelial cell apoptosis in the mammary gland. Balancing intrinsic factors (innate regulating mechanisms, hormone excretion, reproductive stage, etc.), environmental parameters (photoperiod, milking type and frequency, etc.) and stressors (poor nutrition, mastitis, etc.) that affect cell death and proliferation processes, determines lactation length and persistency (Stefanon et al., 2002; Capuco et al., 2003; Pulina et al., 2007). Therefore, extended lactations may be indicative of dairy animals that are resilient to such environmental stressors. Lactation persistency is a heritable trait (Pulina et al., 2007), however genetic and especially genomic studies on dairy sheep are scarce (Pollott and Gootwine, 2001; Kominakis et al., 2002; Jonas et al., 2011; Carta et al., 2014).
Body condition scoring is a technique for subjectively assessing body fat reserves on live sheep. Since its conception in the 1960s it has become widely used because it is easily applicable on farm without the need for special equipment or handling, and foremost it is not biased by other tissue conformation and weight (Kenyon et al., 2014). The body condition scoring scale proposed by Russel et al. (1969) has been developed for meat sheep, however it is largely used for assessing body condition score (BCS) of dairy ewes and rams at different reproductive and lactation stages. Though BCS is a trait associated with productivity in meat sheep, it can be used as a proxy trait for resilience and efficiency in dairy sheep. Under similar nutritional management, energy balance and body fat reserves are expected to be relatively uniform among individuals of the same breed and production level; hence, deviation from an optimum BCS is an indicator of inefficient use of resources and poor resilience to environmental stressors. Although BCS is a trait of moderate heritability with improvement potential, relevant research data on dairy sheep are very limited (Machefert et al., 2023).
In the context of exploring the genetic potential for resilience in sheep breeds that are well-adapted to the local environment, the present study focuses on two highly productive local dairy sheep breeds, namely, Chios and Frizarta, reared under intensive and semi-intensive conditions in Greece. The two populations have never been under selection for resilience traits (Argyriadou et al., 2020). In the present study, we investigate the genomic basis underlying three proxy traits for resilience to disease and environmental stressors, namely, SCC, LP and BCS of Chios and Frizarta ewes. The objective of the study is twofold; i) to estimate heritability for the three resilience traits using genomic data and ii) to perform genome-wide association studies (GWAS) to detect SNP markers and candidate genes that are associated with the studied traits.
2 Materials and methods
2.1 Animals and farms
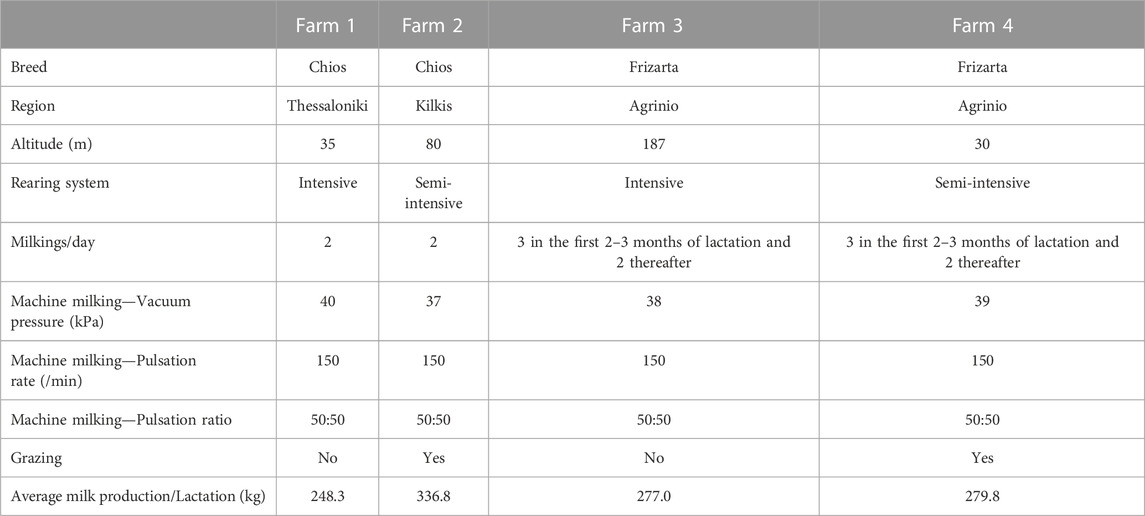
A total of 317 Chios and 346 Frizarta ewes were randomly selected from four farms (two per breed) located in northern and western Greece, respectively. Within each breed, one of the selected farms was managed intensively and the other one semi-intensively, so that the two most common sheep farming systems in Greece are represented. Selecting an equal number of representative flocks of both systems, eliminates introduction of biases related to system-specific farming practices and allows for a broader impact and practical implementation of results. Therefore, flocks were selected on the basis of being typical of the above farming systems and having the best management practices among members of the Chios Sheep Breeders’ Cooperative “Macedonia” and the Agricultural and Livestock Union of Western Greece. Ewes were fed roughages and a concentrate mix with the quantities being appropriately adjusted to meet their nutritional requirements, while they had ad libitum access to water. Furthermore, ewes in the two semi-intensively reared flocks were grazing during spring and summer months for 2–3 h daily in natural or cultivated grasslands. Details regarding the location and management practices of the farms are presented in Table 1.
2.2 Data collection and sampling
Ewes were monitored once per month for two consecutive milking periods following the weaning of lambs at 42 days postpartum. Animal handling and recording was always performed by the same qualified veterinarian. Ewe BCS was assessed by palpation of the lumbar region using the methodology described by Russel et al. (1969). The scale had scores ranging from 1 (extreme emaciation) to 5 (obesity) with 0.25 increments. The milk yield of individual ewes was recorded monthly in a single milking using designated milk meters (Waikato®, New Zealand) attached to each milking unit, which enabled also the collection of individual milk samples (an amount of approximately 50 mL) using Falcon tubes (70 mL volume capacity). Milk samples were stored in a portable freezer at 4°C and transferred to the laboratory within 24 h to assess SCC using Fossomatic™ FC (Foss, Denmark). Blood samples were collected from the jugular vein once per ewe in vacuum tubes with anticoagulant factor (EDTA, BD Vacutainer® Blood collection tubes, BD, United States) and stored at −20°C, until further processing.
2.3 Phenotypic data handling and quality control
A total of 3,856 monthly test milk yield records were collected of all ewes over both years of the study, corresponding to up to seven monthly records per ewe per milking period. Average milking lengths were 189 and 165 days for Chios and Frizarta ewes, respectively. Individual total daily milk yield was calculated from the recorded yield of one milking on each monthly recording occasion based on the following equation:
where DMY is the daily milk yield, TY is the recorded milk yield from one milking and Δt is the time period between the recorded and the previous milking (in minutes). The latter equation assumes that milk is produced evenly throughout the day and allows for calculating daily milk yield based on recording one milking per day when more than two milkings are implemented. It is used by the Chios Sheep Breeders’ Cooperative “Macedonia” for farms with high milking frequency, as an alternative to the standard methods suggested by the International Committee of Animal Recording (ICAR, 2018) that assume two milkings per day. In the present study, it was used to facilitate direct comparisons among animals since two of the farms performed three milkings per day in the first 2–3 months of the milking period.
Lactation persistency was then calculated for each year of the study, as the regression coefficient of daily milk yield (after peak of lactation) on days from lambing (Kominakis et al., 2002), according to the following linear model:
where y is the daily milk yield, DIM is the fixed effect of days from lambing, β is the regression coefficient on DIM, representing LP and α is the intercept. The overall LP for the 2 years of the study was calculated as the average of the annual values and used for further analyses.
Individual repeated measurements of SCC (n = 3,239) were used to calculate the mean for each year of the study weighted over the respective daily milk yield, according to the following formula:
where
Regarding BCS, all individual records (n = 3,784) were averaged over both years of the study. Poor body conformation was considered an indication of compromised health and welfare, which could possibly impact the expression of genetic potential (Kenyon et al., 2014); hence, ewes with average BCS less than 2 were excluded from the study (Munoz et al., 2019).
2.4 DNA extraction, animal genotyping and quality control
Genomic DNA was extracted from blood samples using the QIAamp DNA Mini and Blood Mini kit (QIAGEN, United States, RRID:SCR_008539), as previously described in detail by Tsartsianidou et al. (2021). DNA samples of Chios ewes were genotyped with the medium-density Illumina OvineSNP50 Genotyping Beadchip (Illumina Inc., San Diego, California, RRID:SCR_010233) which features 54,241 SNPs, whereas the updated Illumina OvineSNP50 Genotyping Beadchip v2 containing 53,516 SNPs was used for Frizarta samples.
Genotype quality control was performed with PLINK 1.9 software (RRID:SCR_001757; Purcell et al., 2007; Chang et al., 2015). All SNPs on non-autosomal regions were removed. Furthermore, SNPs with minor allele frequency (MAF) lower than 2%, call rate lower than 97% or deviating from Hardy-Weinberg equilibrium (HWE, p-value = 10–6) were filtered out. Sample call rate threshold was set at 90%. Chromosomal coordinates were allocated to SNPs based on the Oar_v4.0 genome assembly (The International Sheep Genomics Consortium, 2015; NCBI Assembly Archive Viewer, RRID:SCR_012917).
2.5 Principal component analysis (PCA) of genotypes
Principal Component Analysis (PCA) of animal genotypes was performed within breed using GEMMA software version 0.98.1 (Zhou and Stephens, 2012) to investigate possible population structures for subsequent analyses. The eigenvectors and eigenvalues of the decomposed matrices were then plotted in RStudio (RRID:SCR_000432; Posit team, 2022) with R version 4.1.2 (RRID:SCR_001905; R core team, 2021).
2.6 Estimation of genomic parameters
Variance components of LP, BCS and logarithmically transformed SCC were estimated within breed by residual maximum likelihood (REML) using a genomic relatedness matrix with the following single-trait animal linear mixed model:
where y is the phenotypes vector, τ is the vector of fixed effects, u is the vector of random effects, X and Z are the design matrices that associate phenotypes with fixed and random effects, respectively, whereas ε is the vector of random residual errors. Preliminary analyses were performed to identify the fixed effects for the models. The tested variables included farm (2 levels for each breed), lactation number (in the first year of the study—6 levels, representing lactation number 1–5 and ≥6), recording years (2 levels, representing ewes with records in one or both years of the study), lambing season (3 levels—representing ewes lambing in 1: Summer-Autumn i.e., first lambing group of each year; 2: Winter-Spring i.e., second lambing group of each year; or 3: either first or second lambing group depending on the year) and prolificacy (covariate, representing the average prolificacy for both years of the study). Principal Components (PCs) from PCA accounting for measurable proportions of variance were also tested. Only variables with a statistically significant effect (p < 0.05) on each studied trait were retained in the final models of statistical analyses. The Akaike Information Criterion, the Bayesian Information Criterion and the magnitude of the residual variance were also considered as goodness-of-fit indicators, to compare tested models. The random additive genetic effect of ewe and residual effect were included in all models. In the Chios analyses, the model for BCS included lactation number, lambing season and the first two PCs as fixed effects; the model for SCC included lactation number and the first PC as fixed effects; the model for LP included farm and lambing season as fixed effects. In the Frizarta analyses, the model for BCS included farm and lambing season as fixed effects; respectively, farm and lactation number were included as fixed effects in the SCC model; the model for LP included the fixed effects of lambing season and recording years.
All data analyses were performed with ASReml software version 4.2 (Gilmour et al., 2021). The centered genomic relatedness matrix for each breed was created from post quality control genomic data with GEMMA software version 0.98.1 (Zhou and Stephens, 2012). Further formatting and inversion of the matrices was completed with R version 4.1.2 (RRID:SCR_001905; R core team, 2021) and package “AGHmatrix” (Amadeu et al., 2016). Finally, heritability for each trait was estimated as the ratio of the additive genetic to the total phenotypic variance. Statistical significance (α = 0.05) of heritability estimates was assessed based on the magnitude of the respective standard errors, with the two-tailed Student’s t-distribution.
2.7 Genome-wide association study
Within breed GWAS were conducted to identify SNPs associated with the studied traits. Model [4] was used in each case with the addition of wβ, where w is the vector of SNPs and β the associated effects. Furthermore, all GWAS models on Frizarta and Chios sheep traits included the first and first three PCs, respectively, to account for population structure. Genome-wide significance threshold was set at p < 0.05. A suggestive significance threshold was also set expecting one false positive association to occur once per GWAS (Duggal et al., 2008). After Bonferroni correction for multiple comparisons, the final genome-wide threshold values were p < 1.14E-06 and p < 1.05E-06 for Chios and Frizarta sheep, corresponding to the negative common logarithms (-log10) of 5.94 and 5.98. Corrected suggestive thresholds were p < 2.27E-05 and p < 2.11E-05 (corresponding to -log10 of 4.64 and 4.68) for Chios and Frizarta sheep, respectively. Analyses were performed with GEMMA software version 0.98.1 (Zhou and Stephens, 2012). Genome-wide plots of −log10(p-values) illustrating associations of SNPs with the studied traits and the respective Quantile-Quantile (Q-Q) plots for quality assessment of results were created in RStudio (RRID:SCR_000432; Posit team, 2022) with R version 4.1.2 (RRID:SCR_001905; R core team, 2021) and statistical R package “qqman” (Turner, 2018).
2.8 Gene annotation analysis
The NCBI Genome Data Viewer (RRID:SCR_002474, Rangwala et al., 2021) was used to determine the type of genomic regions in which the genome-wide and suggestive significant SNPs were located according to Oar_v4.0 genome assembly. Furthermore, we searched for candidate genes within regions of 1 Mb upstream and downstream of all significant SNPs, according to the levels of linkage disequilibrium reported by Tsartsianidou et al. (2021) and Kominakis et al. (2017) for the same Chios and Frizarta populations as the ones described here. AnimalQTLdb (RRID:SCR_001748; Hu et al., 2022) was used to investigate previously reported QTLs associated with the studied traits or located in common regions with the ones reported herein. To explore the biological functions of all annotated genes and the associated traits we used the NCBI Gene (RRID:SCR_002473), Ensembl release version 110 (RRID:SCR_002344; Cunningham et al., 2022) and UniProt (RRID:SCR_002380; The UniProt Consortium, 2023) databases and performed a literature review across species.
3 Results
3.1 Descriptive statistics and quality control
Descriptive statistics of phenotypic data after quality control are presented in Table 2.
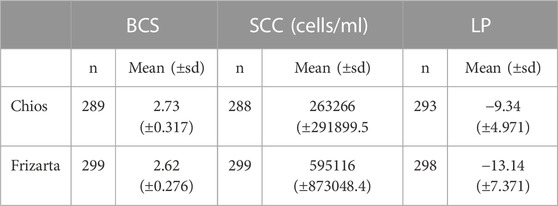
TABLE 2. Descriptive statistics (number of observations and mean values) of body condition score (BCS), milk somatic cell count (SCC) and lactation persistency (LP) after quality control. n: number of observations; sd: standard deviation.
A total of 10,243 and 6,065 SNPs were removed after quality control of Chios and Frizarta genotypes, respectively. Specifically for Chios genotypes, i) 2,157 SNPs were unmapped or located in non-autosomal regions, ii) 1,945 SNPs had call rate lower than 97%, iii) 236 deviated from HWE and iv) 5,905 had MAF lower than 2%; the respective numbers for Frizarta sheep were i) 2,144, ii) 1,999, iii) 244 and iv) 1,678 SNPs. The final datasets included 43,998 and 47,451 SNPs for Chios and Frizarta sheep, respectively. No samples were removed due to low call rate.
3.2 Population structure
Results of PCA on Chios and Frizarta genomic relatedness matrices are demonstrated in Supplementary Figures S1 and S2, respectively. Approximately 14% of the total variation observed in Chios population was explained by the first three PCs (Supplementary Figure S1A). Regarding the Frizarta population, the first PC accounted for circa 5.5% of the total variation, whereas all the other components individually accounted for less than 2.6% (Supplementary Figure S2A). Plotting the first and second or third PC revealed that population structure was associated with farm, in Chios population (Supplementary Figure S1B). Consequently, the first three PCs were considered in the following Chios sheep GWAS to account for population structure. In Frizarta, plotting the first and second PCs revealed population structure attributed to farm, as well (Supplementary Figure S2B). The first PC was fitted as covariate in the GWAS models for Frizarta sheep to adjust for population structure.
3.3 Genomic heritability
Heritability estimates for all the studied traits are presented in Table 3. High heritability estimates for BCS were observed (h2 = 0.54 and 0.55, for Chios and Frizarta sheep, respectively) and moderate to high for LP (h2 = 0.43 and 0.45) and SCC (h2 = 0.25 and 0.38). All estimates were statistically significant (α = 0.05), except for heritability of SCC in Chios sheep.
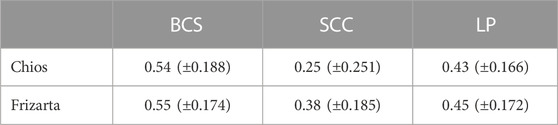
TABLE 3. Heritability estimates (± standard error in parentheses) of body condition score (BCS), milk somatic cell count (SCC) and lactation persistency (LP).
3.4 Genome-wide association study
Results from GWAS of BCS, SCC and LP are presented in Figures 1–3, respectively, and the corresponding Q-Q plots in Supplementary Figures S3–S5. Different genomic regions are involved in the inheritance of the studied traits in Chios and Frizarta populations; however, in all cases a polygenic mode of inheritance is implied. In total, one genome-wide and six suggestive significant SNPs were detected, two of the latter in Chios and four in Frizarta analyses (Table 4). Regarding LP, regions of interest were located on chromosomes 1 (rs193632931) and 6 (rs412648955) for Frizarta sheep and chromosome 3 (rs428128299) for Chios sheep. One genome-wide (rs403061409 on chromosome 9) and two suggestive significant SNPs (rs424064526 and rs428540973 on chromosomes 1 and 12, respectively) were associated with SCC in Frizarta sheep. In Chios, no significant SNP associations with SCC were detected, consistently with the non-significant heritability estimate for this trait; however, two SNPs on chromosomes 3 and 7 (rs416035680 and rs419886966, respectively) almost reached the suggestive significant threshold (p-value = 3.45E-05 and 3.54E-05, respectively). Finally, one suggestive significant SNP (rs424834097) located on chromosome 4 was associated with BCS in Chios sheep, whereas no significant associations were detected in Frizarta.
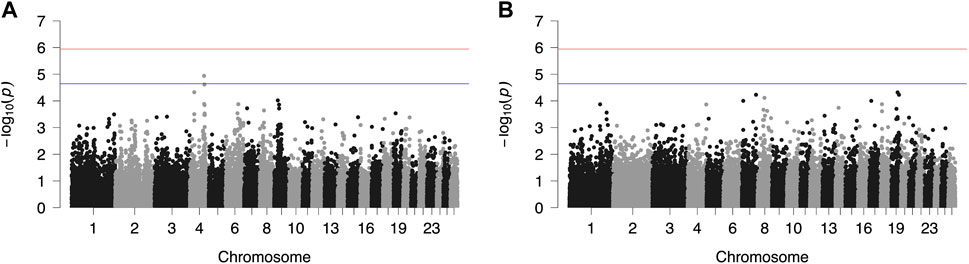
FIGURE 1. Genome-wide plots of −log10 (p-values) illustrating associations of single nucleotide polymorphisms (SNPs) with body condition score (BCS) of Chios (A) and Frizarta (B) sheep. Red and blue lines indicate the genome-wide and suggestive significance thresholds, respectively.
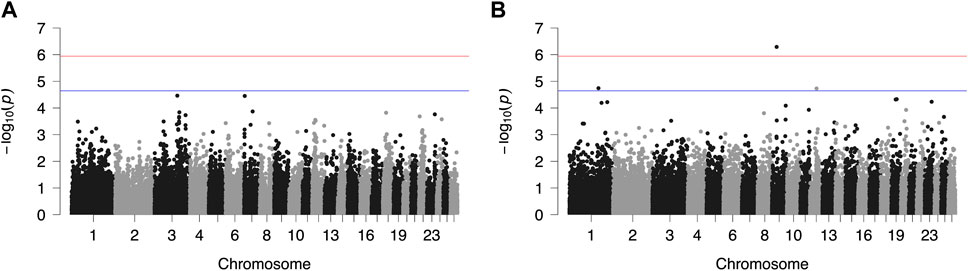
FIGURE 2. Genome-wide plots of −log10 (p-values) illustrating associations of single nucleotide polymorphisms (SNPs) with logarithmically transformed milk somatic cell count (SCC) of Chios (A) and Frizarta (B) sheep. Red and blue lines indicate the genome-wide and suggestive significance thresholds, respectively.
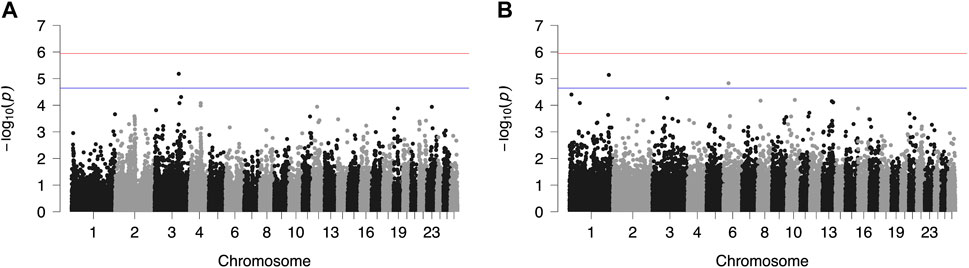
FIGURE 3. Genome-wide plots of −log10 (p-values) illustrating associations of single nucleotide polymorphisms (SNPs) with lactation persistency (LP) of Chios (A) and Frizarta (B) sheep. Red and blue lines indicate the genome-wide and suggestive significance thresholds, respectively.
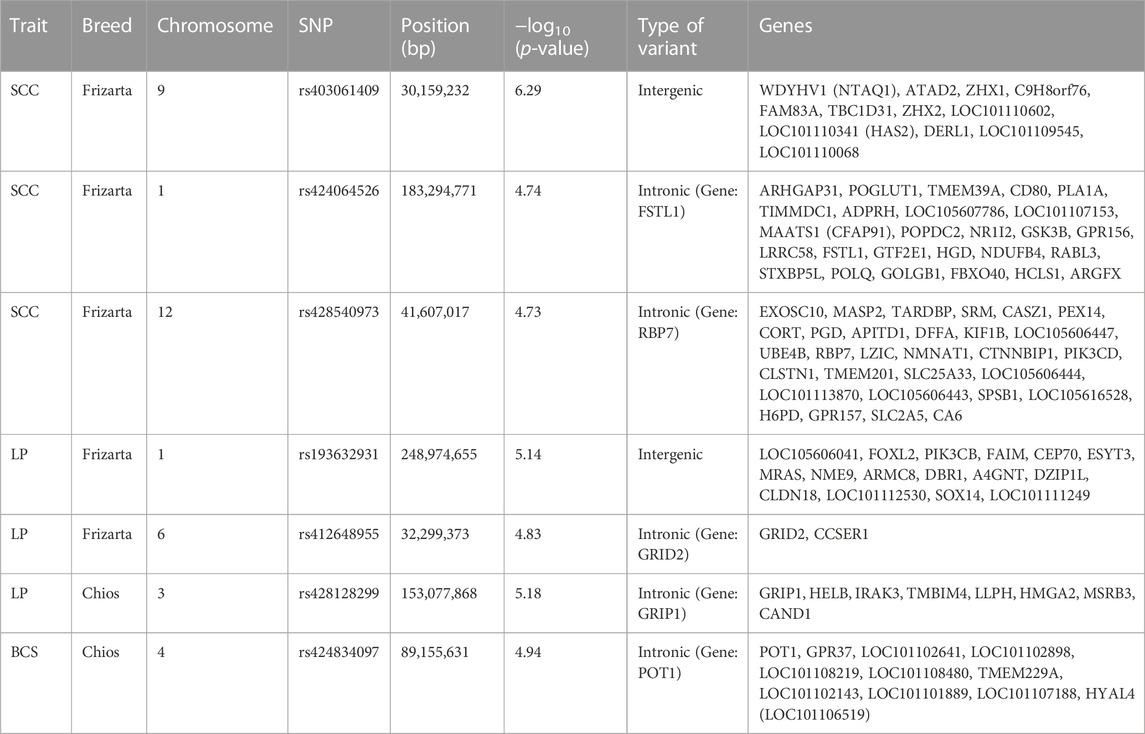
TABLE 4. Single nucleotide polymorphisms (SNPs) significantly associated with body condition score (BCS), milk somatic cell count (SCC) and lactation persistency (LP) at genome-wide (p < 1.14E-06 for Chios and p < 1.05E-06 for Frizarta sheep) and suggestive (p < 2.27E-05 for Chios and p < 2.11E-05 for Frizarta sheep) thresholds and genes located within 1 Mb upstream or downstream of the significant SNPs.
3.5 Gene annotation analysis
The genome-wide significant SNP (rs403061409) associated with SCC in Frizarta sheep was located in an intergenic region approximately 530 kb from the nearest gene (ZHX2—zinc fingers and homeoboxes 2). Almost all suggestive significant SNPs were located within introns of protein coding genes. Specifically, GRIP1 (glutamate receptor interacting protein 1) and GRID2 (glutamate ionotropic receptor delta type subunit 2) genes included two SNPs, rs428128299 and rs412648955, associated with LP in Chios and Frizarta sheep, respectively. The rs424064526 and rs428540973, that were associated with SCC in Frizarta sheep, were located within FSTL1 (follistatin like 1) and RBP7 (retinol binding protein 7) genes, respectively. Last, rs424834097, associated with BCS in Chios sheep, was located within POT1 (Protection of telomeres protein 1) gene. The only exception was rs193632931 associated with LP in Frizarta sheep, which was located in an intergenic region approximately 600 bp downstream of the nearest gene (DZIP1L—DAZ interacting zinc finger protein 1 like). According to the AnimalQTLdb, none of the significant SNPs were in regions of previously reported QTL associated with the studied traits in sheep. Within 1 Mb upstream or downstream of significant SNPs, a total of 105 genes were identified, 86 of which refer to Frizarta and 19 to Chios sheep; most of the genes were protein coding, 9 were pseudogenes and 4 were non-coding RNA genes (Supplementary Tables S1–S4).
4 Discussion
4.1 Genomic heritability
The present heritability estimates of SCC were higher than the ones reported in previous studies on the same or other dairy sheep breeds. Specifically, pedigree-based heritability estimates of SCC test-day records for Chios sheep were low to moderate and ranged from 0.09 to 0.18 depending on the stage of lactation at which records were collected (Ligda et al., 2003; Bramis et al., 2014; Banos et al., 2017). Slightly higher heritability estimates of 0.30–0.44 were reported by Psifidi et al. (2014) towards the end of lactation (between the 19th and 23rd week of lactation). To our knowledge, no studies have so far reported heritability estimates of SCC for Frizarta sheep. In other studies, heritability of somatic cell score (which is based on a binary logarithmic transformation of SCC) was also lower than the SCC heritability estimates presented here and ranged from 0.05 to 0.15 in Valle del Belice (Sutera et al., 2021), French Lacaune (Rupp et al., 2003) and blond-faced Manech sheep (Aguerre et al., 2022). However, heritability estimates of somatic cell score for Spanish Assaf and Sarda sheep (and their crosses with Lacaune) and of logarithmically transformed SCC for Churra sheep ranged from 0.21 to 0.28, similar to the present results for Chios sheep (Casu et al., 2010; Sánchez-Mayor et al., 2019; Pelayo et al., 2021). Nevertheless, it should be noted that in the present study SCC heritability estimate for Chios sheep had a high standard error and was not statistically significant (α = 0.05). Further investigation on a larger sample size may be necessary to allow safe conclusions regarding heritability of SCC in the studied Chios population.
Research data on heritability of BCS in dairy sheep are scarce. Aguerre et al. (2018) have reported BCS heritability estimates of 0.25 for blond-faced Manech sheep. The present estimates were also higher compared to the ones reported for meat and wool sheep, most of which ranged from 0.04 to 0.32 (Walkom and Brown, 2017; Macé et al., 2018; Snyman and Fisher, 2019; Marques et al., 2020; Oliveira et al., 2021; Rodrigues et al., 2021; Hickey et al., 2022; Ramos et al., 2023a). However, one study reported estimates of New Zealand Merino ewes up to 0.66 depending on the reproductive stage (Tait et al., 2018).
In the present study, heritability of LP lies close to the upper limit of previously reported estimates that ranged from 0.11 to 0.46, depending on the population and the definition of the trait which is largely inconsistent among studies (Pollott and Gootwine, 2001; Carta et al., 2014; Martinez Boggio et al., 2022). Different approaches for its estimation include calculating ratios of milk yield at different stages of lactation using either test-day or cumulative yields and estimation of specific parameters based on lactation curve modelling (Pulina et al., 2007). Kominakis et al. (2002) have used an approach similar to the present one and estimated heritability of 0.15 for Greek local Boutsko ewes, based on the regression coefficient expressing the milk yield decline across lactation.
Overall, the present heritability estimates were higher or similar to the highest estimates reported in the literature. However, in the present study, heritability of the studied traits was calculated based on genomic relatedness matrices of Chios and Frizarta populations instead of pedigree data which have been used in the above-mentioned studies. The accuracy of sheep pedigree has been questioned due to the specific characteristics and limitations of sheep rearing systems and reproduction practices that often result in false or missing parentage data. On the contrary, genomic relatedness matrices accurately capture family structure and relationships between animals (Hayes et al., 2009; Legarra et al., 2014), resulting in higher heritability estimates compared to pedigree-based approaches (Legarra et al., 2014; Cesarani et al., 2019). Differences regarding trait definitions, statistical models and analytical approaches may also contribute to the observed discrepancies among studies, as described above. Furthermore, deviating heritability estimates may reflect diverse inheritance patterns and genetic potential among sheep breeds, especially those used for different production purposes. Nevertheless, studied traits are heritable and therefore amenable to improvement through selective breeding. To facilitate inclusion in genetic improvement programs, further studies are needed to investigate possible favorable or antagonistic correlations between the studied and other important resilience or production traits.
4.2 Genome-wide association study and gene annotation
Population structure revealed by PCA was associated with the farm of origin of animals in both breeds, in accordance with the fact that targeted selection and mating practices is mainly performed on farm level. The effect was less intense in Frizarta sheep since artificial insemination is performed in some ewes (less than 30% of the flocks); in this case, the use of common sires between farms allows for more genetic admixture.
Previous GWAS for mastitis-related traits of different Mediterranean dairy sheep breeds have reported several SNPs and QTLs associated with SCC and somatic cell score, located on chromosomes 1–3, 7–8, 10, 16 and 19 (Rupp et al., 2015; Banos et al., 2017; Sutera et al., 2021; Öner et al., 2021), indicating a polygenic mode of inheritance and the involvement of different genes depending on the breed of study. For Chios sheep, Banos et al. (2017) reported associations of SCC with SNPs on chromosomes 2, 3, 16 and 19, unlike the present study that found none to be statistically significant. However, we found one SNP on chromosome 3 (rs416035680) that approached the suggestive significant threshold (p = 3.45E-05), indicating relevance to previous results of Banos et al. (2017). The main differences between the two studies are the mastitis customized 960-SNP array that was used by Banos et al. (2017), including SNPs on targeted chromosomes only, combined with the bigger sample size (n = 609) that increased statistical power. Similarly, no common regions with previously reported associations of SCC in Frizarta sheep were identified; Kominakis et al. (2019) have found significant SNPs on chromosomes 2, 18, 19 and 22. Methodological differences between studies may partly contribute to the discrepancy of results. However, a larger within breed genomic variability is suspected that may be captured in studies involving larger sample sizes. On chromosome 1, three significant SNPs for Valle del Belice sheep (Sutera et al., 2021) and one QTL for Spanish Churra sheep (Gutiérrez-Gil et al., 2018) have been associated with somatic cell score, consistently to present results for Frizarta sheep. Nevertheless, all were located far from the suggestive significant SNP (rs424064526) reported herein, the closest being circa 52 Mb apart. Furthermore, two QTLs on chromosome 9 were associated with somatic cell score (Mohammadi et al., 2022), one of which was located close (circa 500 kb) to the only genome-wide significant SNP (rs403061409 on chromosome 9) that was associated with SCC in Frizarta sheep, herein. Among genes found within 1 Mb of the latter SNP, WDYHV1 (NTAQ1), ZHX1 and ZHX2 are involved in apoptosis and immune response pathways (Wang et al., 2009; Ma et al., 2016). Furthermore, the identified LOC101109545 gene which encodes the 60S ribosomal protein L13a, is also involved in apoptosis and plays a crucial role in regulating inflammation through its expression in macrophages (Poddar et al., 2013). Moreover, LOC101110341 (HAS2) is involved in synthesis of hyaluronan, which is associated with mammary growth during gestation (Tolg et al., 2017). The latter gene along with ZHX2, DERL1, FAM83A and ATAD2 have been associated with intense symptoms, resistance to chemotherapy and bad prognosis of human breast cancer and metastatic canine mammary adenocarcinomas, implying interference in mammary cell growth and immune regulation (Kalashnikova et al., 2010; Klopfleisch et al., 2010; Lee et al., 2012; Fang et al., 2021; Choi et al., 2022). In accordance with the above, the suggestive significant SNP rs428540973 on chromosome 12, which was associated with SCC in Frizarta sheep, is located within RBP7; downregulation of its expression in human breast cancer has been associated with bad prognosis (Lin et al., 2022). Interestingly, the suggestive SNP for SCC on chromosome 1, rs424064526, is located within the intronic region of FSTL1, which is involved in pathogenetic pathways promoting tissue inflammation and subsequent fibrosis (Andreichenko et al., 2019; Li et al., 2021). Other possibly relevant genes located within 1 Mb around the above suggestive significant SNPs are CD80, HCLS1, GSK3B. The first of these genes encodes the cluster differentiation 80 protein, which plays an important role in T-cell migration to the mammary gland during infection (Bharathan and Mullarky, 2011). Similarly, leukocyte migration is also regulated by hematopoietic cell-specific lyn-substrate 1, an actin-binding protein encoded by HCLS1. Moreover, the latter protein is involved in differentiation of myeloid cells and cases of underexpression have been associated with human congenital neutropenia (Samareh-Abolhasani et al., 2013; Castro-Ochoa et al., 2018). Finally, GSK3B is indirectly involved in regulating mammary cell multiplication, since it affects the activation process of other proteins that are responsible for cell cycle regulation (Musumeci et al., 2015). In dairy sheep, high SCC is associated with mastitis (either clinical or subclinical), which is caused by the immune response of the mammary gland to intramammary pathogens leading to increased cell apoptosis, udder inflammation and fibrosis (Gelasakis et al., 2015). Considering the physiological function of the above genes and their role in mammary pathogenesis or resistance to disease, we suggest that they may participate in SCC regulation.
Research data on the genomic basis of LP in dairy sheep are scarce; to our knowledge, only one study has reported a relevant QTL on chromosome 11 for Awassi × Merino dual purpose crosses (Jonas et al., 2011). In the present study, suggestive significant SNP rs412648955 affecting LP of Frizarta ewes was located on chromosome 6, in the intronic region of GRID2. This gene is mainly expressed in the cerebellum and has been associated with gonadotropin secretion from pituitary gland and reproductive traits in sheep and cattle (Sugimoto et al., 2010; Chen et al., 2022), and secretion of GnRH in female mice (Todman et al., 2005). Interestingly, a photoperiodically regulated effect of GnRH, inducing prolactin secretion, has been observed in ewes and mares (Henderson et al., 2008). The above observations, collectively, suggest a possible association between GRID2 expression and milk production and persistency. Another study (Nazar et al., 2022) reported association of GRID2 with conformation of the udder central suspensory ligament in cows; poor udder conformation is often related to incomplete milking, which increases the risk of mastitis and reduces milk production, thereby negatively affecting LP. Closely located to the suggestive significant SNP rs193632931 associated with LP in Frizarta sheep (chromosome 1), FAIM gene is involved in processes inhibiting cell apoptosis and its expression in sheep skin has been associated with superfine wool quality (Tian et al., 2014); expression in the mammary gland may have a protective effect against apoptosis, therefore improving LP (Capuco et al., 2003). Another candidate gene in the same region is CEP70. Hao et al. (2020) reported expression of CEP70 gene in the mammary gland of two local Chinese sheep breeds. The differentially lower expression level in the breed with the highest milk production indicates a possible adverse effect of CEP70 expression on milk production and persistency. For Chios sheep, the suggestive significant SNP associated with LP, rs428128299 on chromosome 3, was located in the intronic region of GRIP1, a gene with significant expression in the uterus of sheep and cattle during oestrus (Hlaing et al., 2001). Considering the adverse effect of oestrus on milk production (King, 1977; Dobson et al., 2007), a possible indirect effect of GRIP1 on LP could be underlying the latter association.
Previously reported QTLs associated with BCS of meat and wool sheep are located on multiple chromosomes across the genome. All studies reported regions on chromosomes 1, 2 and 10 (Amorim et al., 2018; Macé et al., 2022; Ramos et al., 2023b). In addition, Amorim et al. (2018) reported a relevant QTL on chromosome 4. In the present study, the suggestive significant SNP associated with BCS in Chios sheep (rs424834097) located on chromosome 4 was not included in the QTL region in the study of Amorim et al. (2018). The observed differences compared to the present results probably reflect the distinctive body conformation and body fat mobilization processes among dairy and other types of sheep. The rs424834097 SNP is located within the intronic region of POT1, which encodes a protein that regulates telomere length and preserves the overall integrity of chromosomal DNA. Telomere length is reduced under oxidative stress conditions; therefore, it is considered an indicator of cell aging. Metabolic stress throughout lactation in dairy cows, obesity in women and poor body condition of female mink have been related to shorter telomere length (Valdes et al., 2005; Moreno-Navarrete et al., 2010; Boudreau et al., 2014; Laubenthal et al., 2016), supporting the present findings that associate BCS with POT1. TMEM229A gene, which is located in the proximity of rs424834097, has been previously associated with body conformation traits in cattle and high-performance phenotypes in athletes (Boulygina et al., 2020; Abdalla et al., 2021). While mechanisms underlying the latter observations are not fully elucidated, results point to the possibility of TMEM229A being a candidate gene for BCS.
Overall, the results of the present study indicate that resilience traits of highly productive dairy ewes are heritable and report 7 novel SNP markers and 19 candidate genes for BCS (rs424834097 and POT1, TMEM229A), SCC (rs403061409, rs424064526, rs428540973 and NTAQ1, ZHX1, ZHX2, LOC101109545, HAS2, DERL1, FAM83A, ATAD2, RBP7, FSTL1, CD80, HCLS1, GSK3B) and LP (rs193632931, rs412648955, rs428128299 and GRID2, FAIM, CEP70, GRIP1) of Chios and Frizarta ewes, on chromosomes 1, 3, 4, 6, 9 and 12. The absence of common SNPs or genes between the two breeds and discrepancies from previously reported genomic regions associated with the studied traits in other breeds, underline their polygenic inheritance and the need for breed-specific studies that will facilitate the effective incorporation of such traits in breeding programs. The present study is expected to expand existing knowledge regarding the inheritance of such traits and contribute towards breeding more resilient animals. Studies on a larger sample size would be beneficial to confirm our findings. Further investigation of the present findings will focus on the different alleles of significant SNPs and quantification of their genetic contribution and impact on the associated traits. Moreover, possible correlations among the studied and other important production traits constitute another key aspect that needs to be investigated. Such studies will facilitate identification and selection of favorable alleles, and inclusion of the studied traits as objectives in breeding programs aiming to improve resilience without compromising productivity.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://zenodo.org/deposit/8344546, DOI: 10.5281/zenodo.8344546.
Ethics statement
The animal studies were approved by 1) The Research and Ethics Committee, School of Veterinary Medicine, Faculty of Health Sciences, Aristotle University of Thessaloniki, 54124, Thessaloniki, Greece 2) The Regional Department of Veterinary Medicine of Central Macedonia, Directorate of Veterinary Medicine, General Directorate of Agricultural Economy and Veterinary Medicine, 54627, Thessaloniki, Greece. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
AA: Conceptualization, Formal Analysis, Software, Investigation, Methodology, Visualization, Writing–original draft. SM: Investigation, Methodology, Writing–review and editing. SV: Investigation, Writing–review and editing. VT: Investigation, Methodology, Writing–review and editing. AT: Methodology, Resources, Writing–review and editing. AG: Conceptualization, Methodology, Supervision, Writing–review and editing. GB: Conceptualization, Methodology, Supervision, Writing–review and editing. GA: Conceptualization, Funding acquisition, Methodology, Resources, Project administration, Supervision, Writing–review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by the European Union’s H2020 research and innovation program for the projects SMARTER—“SMAll RuminanTs breeding for Efficiency and Resilience” (Grant Number 772787; https://www.smarterproject.eu/) and iSAGE—“Innovation for Sustainable Sheep and Goat Production in Europe” (Grant Number 679302; https://www.isage.eu/).
Acknowledgments
We would like to thank the Chios Sheep Breeders’ Cooperative Macedonia, the Agricultural and Livestock Union of Western Greece and the farmers for their collaboration. Results presented in this work have been produced using the AUTH Compute Infrastructure and Resources. The authors would like to acknowledge the support provided by the Scientific Computing Office throughout the progress of this research work.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1294573/full#supplementary-material
Abbreviations
SNP, Single nucleotide polymorphism; BCS, Body condition score; SCC, Somatic cell count; LP, Lactation persistency; GWAS, Genome-wide association study; PCA, Principal component analysis; PC, Principal component; MAF, Minor allele frequency; HWE, Hardy-Weinberg equilibrium.
References
Abdalla, I. M., Lu, X., Nazar, M., Arbab, A. A., Xu, T., Yousif, M. H., et al. (2021). Genome-wide association study identifies candidate genes associated with feet and leg conformation traits in Chinese Holstein cattle. Animals 11 (8), 2259. doi:10.3390/ani11082259
Aguerre, S., Astruc, J.-M., Legarra, A., Bordes, L., Prevot, F., Grisez, C., et al. (2022). Unfavorable genetic correlations between fecal egg count and milk production traits in the French blond-faced Manech dairy sheep breed. Genet. Sel. Evol. 54 (1), 14. doi:10.1186/s12711-022-00701-1
Aguerre, S., Jacquiet, P., Brodier, H., Bournazel, J. P., Grisez, C., Prévot, F., et al. (2018). Resistance to gastrointestinal nematodes in dairy sheep: genetic variability and relevance of artificial infection of nucleus rams to select for resistant ewes on farms. Vet. Parasitol. 256, 16–23. doi:10.1016/j.vetpar.2018.04.004
Amadeu, R. R., Cellon, C., Olmstead, J. W., Garcia, A. A., Resende, M. F., and Muñoz, P. R. (2016). AGHmatrix: R package to construct relationship matrices for autotetraploid and diploid species: a blueberry example. Plant Genome 9 (3). doi:10.3835/plantgenome2016.01.0009
Amorim, S. T., Kluska, S., Berton, M. P., de Lemos, M. V., Peripolli, E., Stafuzza, N. B., et al. (2018). Genomic study for maternal related traits in Santa Inês sheep breed. Livest. Sci. 217, 76–84. doi:10.1016/j.livsci.2018.09.011
Andreichenko, I. N., Tsitrina, A. A., Fokin, A. V., Gabdulkhakova, A. I., Maltsev, D. I., Perelman, G. S., et al. (2019). 4-methylumbelliferone prevents liver fibrosis by affecting hyaluronan deposition, FSTL1 expression and cell localization. Int. J. Mol. Sci. 20 (24), 6301. doi:10.3390/ijms20246301
Argyriadou, A., Gelasakis, A. I., Banos, G., and Arsenos, G. (2020). Genetic improvement of indigenous Greek sheep and goat breeds. J. Hell. Vet. Med. Soc. 71 (1), 2063. doi:10.12681/jhvms.22967
Banos, G., Bramis, G., Bush, S. J., Clark, E. L., McCulloch, M. E. B., Smith, J., et al. (2017). The genomic architecture of mastitis resistance in dairy sheep. BMC Genomics 18 (1), 624. doi:10.1186/s12864-017-3982-1
Bharathan, M., and Mullarky, I. K. (2011). Targeting mucosal immunity in the battle to develop a mastitis vaccine. J. Mammary Gland. Biol. Neoplasia 16 (4), 409–419. doi:10.1007/s10911-011-9233-1
Boudreau, L., Benkel, B., Astatkie, T., and Rouvinen-Watt, K. (2014). Ideal body condition improves reproductive performance and influences genetic health in female mink. Anim. Reprod. Sci. 145 (1–2), 86–98. doi:10.1016/j.anireprosci.2014.01.004
Boulygina, E. A., Borisov, O. V., Valeeva, E. V., Semenova, E. A., Kostryukova, E. S., Kulemin, N. A., et al. (2020). Whole genome sequencing of elite athletes. Biol. Sport 37 (3), 295–304. doi:10.5114/biolsport.2020.96272
Bramis, G., Arsenos, G., Psifidi, A., and Banos, G. (2014). “Genetic parameters of mastitis related traits in dairy sheep,” in Proceedings of the Annual Conference of the British Society of Animal Science, Nottingham, UK, 194.
Capuco, A. V., Ellis, S. E., Hale, S. A., Long, E., Erdman, R. A., Zhao, X., et al. (2003). Lactation persistency: insights from mammary cell proliferation studies. Anim. Sci. J. 81, 18–31. doi:10.2527/2003.81suppl_318x
Carta, A., Casu, S., Usai, M. G., and Salaris, S. (2014). “Heritability of persistency traits and their genetic correlations with milk yield and udder morphology in dairy sheep,” in 39th ICAR session (Berlin, Germany.
Castro-Ochoa, K. F., Guerrero-Fonseca, I. M., and Schnoor, M. (2018). Hematopoietic cell-specific lyn substrate (HCLS1 or HS1): a versatile actin-binding protein in leukocytes. J. Leukoc. Biol. 105 (5), 881–890. doi:10.1002/jlb.mr0618-212r
Casu, S., Sechi, S., Salaris, S. L., and Carta, A. (2010). Phenotypic and genetic relationships between udder morphology and udder health in dairy ewes. Small Rumin. Res. 88 (2–3), 77–83. doi:10.1016/j.smallrumres.2009.12.013
Cesarani, A., Gaspa, G., Correddu, F., Cellesi, M., Dimauro, C., and Macciotta, N. P. P. (2019). Genomic selection of milk fatty acid composition in Sarda dairy sheep: effect of different phenotypes and relationship matrices on heritability and breeding value accuracy. J. Dairy Sci. 102 (4), 3189–3203. doi:10.3168/jds.2018-15333
Chang, C. C., Chow, C. C., Tellier, L. C., Vattikuti, S., Purcell, S. M., and Lee, J. J. (2015). Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience 4 (1), 7–0047. doi:10.1186/s13742-015-0047-8
Chen, S., Guo, X., He, X., Di, R., Zhang, X., Zhang, J., et al. (2022). Insight into pituitary lncRNA and mRNA at two estrous stages in small tail Han sheep with different FecB genotypes. Front. Endocrinol. 12, 789564. doi:10.3389/fendo.2021.789564
Cheng, M., McCarl, B., and Fei, C. (2022). Climate change and livestock production: a literature review. Atmosphere 13 (1), 140. doi:10.3390/atmos13010140
Choi, B.-H., Ryoo, I., Sim, K. H., Ahn, H., Lee, Y. J., and Kwak, M.-K. (2022). High levels of hyaluronic acid synthase-2 mediate Nrf2-driven chemoresistance in breast cancer cells. Biomol. Ther. 30 (4), 368–379. doi:10.4062/biomolther.2022.074
Cunningham, F., Allen, J. E., Allen, J., Alvarez-Jarreta, J., Amode, M. R., Armean, I. M., et al. (2022). Ensembl 2022. Nucleic Acids Res. 50 (D1), D988–D995. doi:10.1093/nar/gkab1049
De Barbieri, I., Navajas, E., Douhard, F., Conington, J., Ramos, Z., and Ciappesoni, G. (2023). PL-8 a review of sheep resilience. Animal - Sci. Proc. 14 (1), 11–12. doi:10.1016/j.anscip.2023.01.009
Dobson, H., Smith, R., Royal, M., Knight, C., and Sheldon, I. (2007). The high-producing dairy cow and its reproductive performance. Reprod. Domest. Anim. 42 (s2), 17–23. doi:10.1111/j.1439-0531.2007.00906.x
Duggal, P., Gillanders, E. M., Holmes, T. N., and Bailey-Wilson, J. E. (2008). Establishing an adjusted P-value threshold to control the family-wide type 1 error in genome wide association studies. BMC Genomics 9 (1), 516. doi:10.1186/1471-2164-9-516
Dumont, B., Puillet, L., Martin, G., Savietto, D., Aubin, J., Ingrand, S., et al. (2020). Incorporating diversity into animal production systems can increase their performance and strengthen their resilience. Front. Sustain. Food Syst. 4, 109. doi:10.3389/fsufs.2020.00109
Fang, W., Liao, C., Shi, R., Simon, J. M., Ptacek, T. S., Zurlo, G., et al. (2021). ZHX2 promotes HIF1α oncogenic signaling in triple-negative breast cancer. eLife 10, e70412. doi:10.7554/elife.70412
Gelasakis, A. I., Mavrogianni, V. S., Petridis, I. G., Vasileiou, N. G. C., and Fthenakis, G. C. (2015). Mastitis in sheep – the last 10 years and the future of research. Vet. Microbiol. 181 (1–2), 136–146. doi:10.1016/j.vetmic.2015.07.009
Gilmour, A. R., Gogel, B. J., Cullis, B. R., Welham, S. J., and Thompson, R. (2021). ASReml user guide release 4.2 functional specification. Hemel Hempstead, HP2 4TP, UK: VSN International Ltd.
Gutiérrez-Gil, B., Esteban-Blanco, C., Suarez-Vega, A., and Arranz, J. J. (2018). Detection of quantitative trait loci and putative causal variants affecting somatic cell score in dairy sheep by using a 50k SNP chip and whole-genome sequencing. J. Dairy Sci. 101 (10), 9072–9088. doi:10.3168/jds.2018-14736
Hao, Z., Zhou, H., Hickford, J. G. H., Gong, H., Wang, J., Hu, J., et al. (2020). Identification and characterization of circular RNA in lactating mammary glands from two breeds of sheep with different milk production profiles using RNA-seq. Genomics 112 (3), 2186–2193. doi:10.1016/j.ygeno.2019.12.014
Hayes, B. J., Visscher, P. M., and Goddard, M. E. (2009). Increased accuracy of artificial selection by using the realized relationship matrix. Genet. Res. 91 (1), 47–60. doi:10.1017/s0016672308009981
Henderson, H. L., Hodson, D. J., Gregory, S. J., Townsend, J., and Tortonese, D. J. (2008). Gonadotropin-releasing hormone stimulates prolactin release from lactotrophs in photoperiodic species through a gonadotropin-independent mechanism. Biol. Reprod. 78 (2), 370–377. doi:10.1095/biolreprod.107.064063
Hickey, S. M., Bain, W. E., Bilton, T. P., Greer, G. J., Elmes, S., Bryson, B., et al. (2022). Impact of breeding for reduced methane emissions in New Zealand sheep on maternal and health traits. Front. Genet. 13, 910413. doi:10.3389/fgene.2022.910413
Hine, B. C., Acton, G. A., Elks, D. J., Niemeyer, D. D. O., Bell, A. M., Colditz, I. G., et al. (2022). Targeting improved resilience in Merino sheep – correlations between immune competence and health and fitness traits. Animal 16 (7), 100544. doi:10.1016/j.animal.2022.100544
Hlaing, M., Nam, K., Lou, J., Pope, W. F., and Nephew, K. P. (2001). Evidence for expression of estrogen receptor cofactor messenger ribonucleic acid in the ovary and uterus of domesticated animals (sheep, cow and pig). Life Sci. 68 (12), 1427–1438. doi:10.1016/s0024-3205(01)00937-7
Hu, Z.-L., Park, C. A., and Reecy, J. M. (2022). Bringing the Animal QTLdb and CorrDB into the future: meeting new challenges and providing updated services. Nucleic Acids Res. 50 (D1), D956–D961. doi:10.1093/nar/gkab1116
International Committee for Animal Recording (2018). ICAR recording guidelines. International agreement of recording practices. Available at: https://www.icar.org/Guidelines/16-Dairy-Sheep-and-Goats.pdf (Accessed August 2023).
Jonas, E., Thomson, P. C., Hall, E. J., McGill, D., Lam, M. K., and Raadsma, H. W. (2011). Mapping quantitative trait loci (QTL) in sheep. IV. Analysis of lactation persistency and extended lactation traits in sheep. Genet. Sel. Evol. 43 (1), 22. doi:10.1186/1297-9686-43-22
Kalashnikova, E. V., Revenko, A. S., Gemo, A. T., Andrews, N. P., Tepper, C. G., Zou, J. X., et al. (2010). ANCCA/ATAD2 overexpression identifies breast cancer patients with poor prognosis, acting to drive proliferation and survival of triple-negative cells through control of B-Myb and EZH2. Cancer Res. 70 (22), 9402–9412. doi:10.1158/0008-5472.can-10-1199
Kenyon, P., Maloney, S., and Blache, D. (2014). Review of sheep body condition score in relation to production characteristics. N. Z. J. Agric. Res. 57 (1), 38–64. doi:10.1080/00288233.2013.857698
King, J. (1977). The effect of oestrus on milk production in cows. Vet. Rec. 101 (6), 107–108. doi:10.1136/vr.101.6.107
Klopfleisch, R., Schütze, M., Linzmann, H., Brunnberg, L., and Gruber, A. D. (2010). Increased Derlin-1 expression in metastases of canine mammary adenocarcinomas. J. Comp. Pathol. 142 (1), 79–83. doi:10.1016/j.jcpa.2009.06.006
Kominakis, A., Hager-Theodorides, A. L., Zoidis, E., Saridaki, A., Antonakos, G., and Tsiamis, G. (2017). Combined GWAS and ‘guilt by association’-based prioritization analysis identifies functional candidate genes for body size in sheep. Genet. Sel. Evol. 49 (1), 41. doi:10.1186/s12711-017-0316-3
Kominakis, A., Saridaki, A., and Antonakos, G. (2019). Novel candidate genes for somatic cell count in Frizarta dairy sheep. IJGG 7 (4), 103–109. doi:10.11648/j.ijgg.20190704.13
Kominakis, A. P., Rogdakis, E., and Koutsotolis, K. (2002). Genetic aspects of persistency of milk yield in Boutsico dairy sheep. Asian-Aust. J. Anim. Sci. 15 (3), 315–320. doi:10.5713/ajas.2002.315
Laubenthal, L., Hoelker, M., Frahm, J., Dänicke, S., Gerlach, K., Südekum, K.-H., et al. (2016). Short communication: telomere lengths in different tissues of dairy cows during early and late lactation. J. Dairy Sci. 99 (6), 4881–4885. doi:10.3168/jds.2015-10095
Lee, S.-Y., Meier, R., Furuta, S., Lenburg, M. E., Kenny, P. A., Xu, R., et al. (2012). FAM83A confers EGFR-TKI resistance in breast cancer cells and in mice. J. Clin. Investig. 122 (9), 3211–3220. doi:10.1172/jci60498
Legarra, A., Baloche, G., Barillet, F., Astruc, J. M., Soulas, C., Aguerre, X., et al. (2014). Within- and across-breed genomic predictions and genomic relationships for western Pyrenees dairy sheep breeds Latxa, Manech, and Basco-Béarnaise. J. Dairy Sci. 97 (5), 3200–3212. doi:10.3168/jds.2013-7745
Li, X., Fang, Y., Jiang, D., Dong, Y., Liu, Y., Zhang, S., et al. (2021). Targeting FSTL1 for multiple fibrotic and systemic autoimmune diseases. Mol. Ther. 29 (1), 347–364. doi:10.1016/j.ymthe.2020.09.031
Ligda, C., Papadopoulos, T., Mavrogenis, A., and Georgoudis, A. (2003). “Genetic parameters for test day milk traits and somatic cell counts in Chios dairy sheep,” in Options Méditerranéennes: Série A. Séminaires Méditerranéens; n. 55, “Breeding programmes for improving the quality and safety of products. New traits, tools, rules and organization” (Zaragoza, CIHEAM, 55–59.
Lin, H., Han, Q., Wang, J., Zhong, Z., Luo, H., Hao, Y., et al. (2022). Methylation-mediated silencing of RBP7 promotes breast cancer progression through PPAR and PI3K/AKT pathway. J. Oncol. 2022, 9039110. doi:10.1155/2022/9039110
Ma, X., Huang, M., Wang, Z., Liu, B., Zhu, Z., and Li, C. (2016). ZHX1 inhibits gastric cancer cell growth through inducing cell-cycle arrest and apoptosis. J. Cancer 7 (1), 60–68. doi:10.7150/jca.12973
Macé, T., González-García, E., Foulquié, D., Carrière, F., Pradel, J., Durand, C., et al. (2022). Genome-wide analyses reveal a strong association between LEPR gene variants and body fat reserves in ewes. BMC Genomics 23 (1), 412. doi:10.1186/s12864-022-08636-z
Macé, T., González-García, E., Pradel, J., Parisot, S., Carrière, F., Douls, S., et al. (2018). Genetic analysis of robustness in meat sheep through body weight and body condition score changes over time. Anim. Sci. J. 96 (11), 4501–4511. doi:10.1093/jas/sky318
Machefert, C., Robert-Granié, C., Lagriffoul, G., Parisot, S., Allain, C., Portes, D., et al. (2023). Opportunities and limits of commercial farm data to study the genetic determinism of feed efficiency throughout lactation in dairy sheep. Animal 17, 100951. In press. doi:10.1016/j.animal.2023.100951
Marques, C. B., Goldberg, V., and Ciappesoni, G. (2020). Genetic parameters for production traits, resistance and resilience to nematode parasites under different worm burden challenges in Corriedale sheep. Vet. Parasitol. 287, 109272. doi:10.1016/j.vetpar.2020.109272
Martinez Boggio, G., Meynadier, A., Buitenhuis, A. J., and Marie-Etancelin, C. (2022). Host genetic control on rumen microbiota and its impact on dairy traits in sheep. Genet. Sel. Evol. 54 (1), 77. doi:10.1186/s12711-022-00769-9
Mohammadi, H., Farahani, A. H., Moradi, M. H., Mastrangelo, S., Di Gerlando, R., Sardina, M. T., et al. (2022). Weighted single-step genome-wide association study uncovers known and novel candidate genomic regions for milk production traits and somatic cell score in Valle del Belice dairy sheep. Animals 12 (9), 1155. doi:10.3390/ani12091155
Moreno-Navarrete, J. M., Ortega, F., Sabater, M., Ricart, W., and Fernández-Real, J. M. (2010). Telomere length of subcutaneous adipose tissue cells is shorter in obese and formerly obese subjects. Int. J. Obes. 34 (8), 1345–1348. doi:10.1038/ijo.2010.49
Mucha, S., Tortereau, F., Doeschl-Wilson, A., Rupp, R., and Conington, J. (2022). Animal Board invited review: meta-analysis of genetic parameters for resilience and efficiency traits in goats and sheep. Animal 16 (3), 100456. doi:10.1016/j.animal.2022.100456
Munoz, C. A., Campbell, A. J. D., Hemsworth, P. H., and Doyle, R. E. (2019). Evaluating the welfare of extensively managed sheep. PLoS One 14 (6), e0218603. doi:10.1371/journal.pone.0218603
Musumeci, G., Castrogiovanni, P., Szychlinska, M. A., Aiello, F. C., Vecchio, G. M., Salvatorelli, L., et al. (2015). Mammary gland: from embryogenesis to adult life. Acta histochem. 117 (4–5), 379–385. doi:10.1016/j.acthis.2015.02.013
Nazar, M., Abdalla, I. M., Chen, Z., Ullah, N., Liang, Y., Chu, S., et al. (2022). Genome-wide association study for udder conformation traits in Chinese Holstein cattle. Animals 12 (19), 2542. doi:10.3390/ani12192542
Oliveira, E. J., Savegnago, R. P., Freitas, A., Freitas, L. A., Paz, A. C., El Faro, L., et al. (2021). Genetic parameters for body weight and morphometric traits in Santa Ines sheep using Bayesian inference. Small Rumin. Res. 201, 106446. doi:10.1016/j.smallrumres.2021.106446
Öner, Y., Serrano, M., Sarto, P., Iguácel, L. P., Piquer-Sabanza, M., Estrada, O., et al. (2021). Genome-wide association studies of somatic cell count in the Assaf breed. Animals 11 (6), 1531. doi:10.3390/ani11061531
Paraskevopoulou, C., Theodoridis, A., Johnson, M., Ragkos, A., Arguile, L., Smith, L., et al. (2020). Sustainability assessment of goat and sheep farms: a comparison between European countries. Sustainability 12 (8), 3099. doi:10.3390/su12083099
Pelayo, R., Gutiérrez-Gil, B., Garzón, A., Esteban-Blanco, C., Marina, H., and Arranz, J. J. (2021). Estimation of genetic parameters for cheese-making traits in Spanish Churra sheep. J. Dairy Sci. 104 (3), 3250–3260. doi:10.3168/jds.2020-19387
Poddar, D., Basu, A., Baldwin, W. M., Kondratov, R. V., Barik, S., and Mazumder, B. (2013). An extraribosomal function of ribosomal protein L13a in macrophages resolves inflammation. J. Immunol. 190 (7), 3600–3612. doi:10.4049/jimmunol.1201933
Pollott, G. E., and Gootwine, E. (2001). A genetic analysis of complete lactation milk production in improved Awassi sheep. Livest. Prod. Sci. 71 (1), 37–47. doi:10.1016/s0301-6226(01)00239-1
Psifidi, A., Bramis, G., Arsenos, G., and Banos, G. (2014). “Genetic parameters and genomic markers associated with mastitis resistance in dairy sheep,” in Proceedings of the 10th World Congress of Genetics Applied to Livestock Production, Vancouver, Canada.
Pulina, G., Nudda, A., Macciotta, N. P., Battacone, G., Rassu, S. P. G., and Cannas, A. (2007). Non-nutritional factors affecting lactation persistency in dairy ewes: a review. Ital. J. Anim. Sci. 6 (2), 115–141. doi:10.4081/ijas.2007.115
Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M. A. R., Bender, D., et al. (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. AJHG 81 (3), 559–575. doi:10.1086/519795
Ramos, Z., Garrick, D. J., Blair, H. T., De Barbieri, I., Ciappesoni, G., Montossi, F., et al. (2023a). Genetic and phenotypic relationships between ewe reproductive performance and wool and growth traits in Uruguayan Ultrafine Merino sheep. Anim. Sci. J. 101, skad071. doi:10.1093/jas/skad071
Ramos, Z., Garrick, D. J., Blair, H. T., Vera, B., Ciappesoni, G., and Kenyon, P. R. (2023b). Genomic regions associated with wool, growth and reproduction traits in Uruguayan Merino sheep. Genes 14 (1), 167. doi:10.3390/genes14010167
Rangwala, S. H., Kuznetsov, A., Ananiev, V., Asztalos, A., Borodin, E., Evgeniev, V., et al. (2021). Accessing NCBI data using the NCBI Sequence Viewer and Genome Data Viewer (GDV). Genome Res. 31 (1), 159–169. doi:10.1101/gr.266932.120
R Core Team (2021). R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing.
Rodrigues, F. N., Sarmento, J. L., Leal, T. M., Araújo, A. M., and Figueiredo Filho, L. A. (2021). Genetic parameters for worm resistance in Santa Inês sheep using the Bayesian animal model. Anim. Biosci. 34 (2), 185–191. doi:10.5713/ajas.19.0634
Rupp, R., Bergonier, D., Dion, S., Hygonenq, M. C., Aurel, M. R., Robert-Granié, C., et al. (2009). Response to somatic cell count-based selection for mastitis resistance in a divergent selection experiment in sheep. J. Dairy Sci. 92 (3), 1203–1219. doi:10.3168/jds.2008-1435
Rupp, R., Lagriffoul, G., Astruc, J. M., and Barillet, F. (2003). Genetic parameters for milk somatic cell scores and relationships with production traits in French Lacaune dairy sheep. J. Dairy Sci. 86 (4), 1476–1481. doi:10.3168/jds.s0022-0302(03)73732-1
Rupp, R., Senin, P., Sarry, J., Allain, C., Tasca, C., Ligat, L., et al. (2015). A point mutation in suppressor of cytokine signalling 2 (SOCS2) increases the susceptibility to inflammation of the mammary gland while associated with higher body weight and size and higher milk production in a sheep model. PLOS Genet. 11 (12), e1005629. doi:10.1371/journal.pgen.1005629
Russel, A. J., Doney, J. M., and Gunn, R. G. (1969). Subjective assessment of body fat in live sheep. J. Agric. Sci. 72 (3), 451–454. doi:10.1017/s0021859600024874
Samareh-Abolhasani, B., Welte, K., and Skokowa, J. (2013). NAMPT/NAD+/SIRT1-triggered deacetylation of hematopoietic cell-specific lyn-substrate 1 (HCLS1) in congenital neutropenia. Blood 122 (21), 441. doi:10.1182/blood.v122.21.441.441
Sánchez-Mayor, M., Pong-Wong, R., Gutiérrez-Gil, B., Garzón, A., de la Fuente, L. F., and Arranz, J. J. (2019). Phenotypic and genetic parameter estimates of cheese-making traits and their relationships with milk production, composition and functional traits in Spanish Assaf sheep. Livest. Sci. 228, 76–83. doi:10.1016/j.livsci.2019.08.004
Sánchez-Molano, E., Kapsona, V. V., Oikonomou, S., McLaren, A., Lambe, N., Conington, J., et al. (2020). Breeding strategies for animal resilience to weather variation in meat sheep. BMC Genet. 21 (1), 116. doi:10.1186/s12863-020-00924-5
Snyman, M. A., and Fisher, A. D. (2019). Genetic parameters for traits associated with resistance to Haemonchus contortus in a South African Dohne Merino sheep flock. Small Rumin. Res. 176, 76–88. doi:10.1016/j.smallrumres.2019.01.004
Stefanon, B., Colitti, M., Gabai, G., Knight, C. H., and Wilde, C. J. (2002). Mammary apoptosis and lactation persistency in dairy animals. J. Dairy Sci. 69 (1), 37–52. doi:10.1017/s0022029901005246
Sugimoto, M., Sasaki, S., Watanabe, T., Nishimura, S., Ideta, A., Yamazaki, M., et al. (2010). Ionotropic glutamate receptor AMPA 1 is associated with ovulation rate. PLoS ONE 5 (11), e13817. doi:10.1371/journal.pone.0013817
Sutera, A. M., Moscarelli, A., Mastrangelo, S., Sardina, M. T., Di Gerlando, R., Portolano, B., et al. (2021). Genome-wide association study identifies new candidate markers for somatic cells score in a local dairy sheep. Front. Genet. 12, 643531. doi:10.3389/fgene.2021.643531
Tait, I. M., Kenyon, P. R., Garrick, D. J., Pleasants, A. B., and Hickson, R. E. (2018). Genetic and phenotypic correlations between production traits and adult body condition scores in New Zealand Merino ewes. N. Z. J. Anim. Sci. Prod. 78, 71–75.
The International Sheep Genomics Consortium (2015). NCBI assembly Oar_v4.0. Available at: https://www.ncbi.nlm.nih.gov/assembly/GCF_000298735.2/?shouldredirect=false (Accessed August 29, 2023).
The UniProt Consortium (2023). Uniprot: the universal protein knowledgebase in 2023. Nucleic Acids Res. 51 (D1), D523–D531. doi:10.1093/nar/gkac1052
Tian, Y. Z., Huang, X. X., Di, J., Tian, K. C., Wu, W. W., Xu, X. M., et al. (2014). “Comparative study of 13 candidate genes applying multi-reference normalization to detect the expression of different fineness in skin tissues of wool sheep,” in Proceedings of the 10th World Congress of Genetics Applied to Livestock Production, Vancouver, Canada.
Todman, M. G., Han, S.-K., and Herbison, A. E. (2005). Profiling neurotransmitter receptor expression in mouse gonadotropin-releasing hormone neurons using green fluorescent protein-promoter transgenics and microarrays. Neuroscience 132 (3), 703–712. doi:10.1016/j.neuroscience.2005.01.035
Tolg, C., Yuan, H., Flynn, S. M., Basu, K., Ma, J., Tse, K. C., et al. (2017). Hyaluronan modulates growth factor induced mammary gland branching in a size dependent manner. Matrix Biol. 63, 117–132. doi:10.1016/j.matbio.2017.02.003
Tsartsianidou, V., Sánchez-Molano, E., Kapsona, V. V., Basdagianni, Z., Chatziplis, D., Arsenos, G., et al. (2021). A comprehensive genome-wide scan detects genomic regions related to local adaptation and climate resilience in Mediterranean domestic sheep. Genet. Sel. Evol. 53 (1), 90. doi:10.1186/s12711-021-00682-7
Turner, S. D. (2018). Qqman: an R package for visualizing GWAS results using Q-Q and Manhattan plots. J. Open Source Softw. 3 (25), 731. doi:10.21105/joss.00731
Valdes, A., Andrew, T., Gardner, J., Kimura, M., Oelsner, E., Cherkas, L., et al. (2005). Obesity, cigarette smoking, and telomere length in women. Lancet 366 (9486), 662–664. doi:10.1016/s0140-6736(05)66630-5
Walkom, S. F., and Brown, D. J. (2017). Genetic evaluation of adult ewe bodyweight and condition: relationship with lamb growth, reproduction, carcass and wool production. Anim. Prod. Sci. 57 (1), 20–32. doi:10.1071/an15091
Wang, H., Piatkov, K. I., Brower, C. S., and Varshavsky, A. (2009). Glutamine-specific N-terminal amidase, a component of the N-end rule pathway. Mol. Cell 34 (6), 686–695. doi:10.1016/j.molcel.2009.04.032
Keywords: dairy sheep, Chios breed, Frizarta breed, genome-wide association, resilience, milk somatic cell count, body condition score, lactation persistency
Citation: Argyriadou A, Michailidou S, Vouraki S, Tsartsianidou V, Triantafyllidis A, Gelasakis A, Banos G and Arsenos G (2023) A genome-wide association study reveals novel SNP markers associated with resilience traits in two Mediterranean dairy sheep breeds. Front. Genet. 14:1294573. doi: 10.3389/fgene.2023.1294573
Received: 14 September 2023; Accepted: 10 November 2023;
Published: 22 November 2023.
Edited by:
Juan José Arranz, University of León, SpainReviewed by:
Thiruvenkadan Aranganoor Kannan, Tamil Nadu Veterinary and Animal Sciences University, IndiaMohsen Gholizadeh, Sari Agricultural Sciences and Natural Resources University, Iran
Copyright © 2023 Argyriadou, Michailidou, Vouraki, Tsartsianidou, Triantafyllidis, Gelasakis, Banos and Arsenos. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Angeliki Argyriadou, YXJneXJpYW5AdmV0LmF1dGguZ3I=
 Angeliki Argyriadou
Angeliki Argyriadou Sofia Michailidou
Sofia Michailidou Sotiria Vouraki
Sotiria Vouraki Valentina Tsartsianidou
Valentina Tsartsianidou Alexandros Triantafyllidis3,4
Alexandros Triantafyllidis3,4 Athanasios Gelasakis
Athanasios Gelasakis Georgios Banos
Georgios Banos Georgios Arsenos
Georgios Arsenos