- 1Department of Anesthesiology and Perioperative Medicine, University of Pittsburgh, Pittsburgh, PA, United States
- 2Department of Obstetrics and Gynecology, Corewell Health William Beaumont University Hospital, Royal Oak, MI, United States
- 3Department of Radiology and Imaging Sciences, Indiana University School of Medicine, Indianapolis, IN, United States
- 4Department of Psychiatry, Yale School of Medicine, New Haven, CT, United States
- 5Department of Molecular and Functional Genomics, Geisinger, Danville, PA, United States
- 6Department of Botany, Bioinformatics and Climate Change Impacts Management, School of Science, Gujarat University, Ahmedabad, India
- 7Department of Life Sciences, School of Sciences, Gujarat University, Ahmedabad, India
- 8Arthritis and Clinical Immunology Program, Oklahoma Medical Research Foundation, Oklahoma City, OK, United States
Background: Neonatal opioid withdrawal syndrome (NOWS), arises due to increased opioid use during pregnancy. Cytochrome P450 (CYP) enzymes play a pivotal role in metabolizing a wide range of substances in the human body, including opioids, other drugs, toxins, and endogenous compounds. The association between CYP gene methylation and opioid effects is unexplored and it could offer promising insights.
Objective: To investigate the impact of prenatal opioid exposure on disrupted CYPs in infants and their anticipated long-term clinical implications.
Study Design: DNA methylation levels of CYP genes were analyzed in a cohort of 96 placental tissues using Illumina Infinium MethylationEPIC (850 k) BeadChips. This involved three groups of placental tissues: 32 from mothers with infants exposed to opioids prenatally requiring pharmacologic treatment for NOWS, 32 from mothers with prenatally opioid-exposed infants not needing NOWS treatment, and 32 from unexposed control mothers.
Results: The study identified 20 significantly differentially methylated CpG sites associated with 17 distinct CYP genes, with 14 CpGs showing reduced methylation across 14 genes (CYP19A1, CYP1A2, CYP4V2, CYP1B1, CYP24A1, CYP26B1, CYP26C1, CYP2C18, CYP2C9, CYP2U1, CYP39A1, CYP2R1, CYP4Z1, CYP2D7P1 and), while 8 exhibited hypermethylation (CYP51A1, CYP26B1, CYP2R1, CYP2U1, CYP4X1, CYP1A2, CYP2W1, and CYP4V2). Genes such as CYP1A2, CYP26B1, CYP2R1, CYP2U1, and CYP4V2 exhibited both increased and decreased methylation. These genes are crucial for metabolizing eicosanoids, fatty acids, drugs, and diverse substances.
Conclusion: The study identified profound methylation changes in multiple CYP genes in the placental tissues relevant to NOWS. This suggests that disruption of DNA methylation patterns in CYP transcripts might play a role in NOWS and may serve as valuable biomarkers, suggesting a future pathway for personalized treatment. Further research is needed to confirm these findings and explore their potential for diagnosis and treatment.
Introduction
In the United States, opioid use disorder (OUD) is on the rise among women of reproductive age and is a major health problem. Infants exposed to opioids during pregnancy are likely to develop a drug withdrawal syndrome known as neonatal opioid withdrawal (NOWS) also called neonatal abstinence syndrome (NAS). NOWS incidence in the United States varies between 1.5 and 8.0 for every 1,000 hospital births and has increased by 433% from 2004 to 2014 (Jilani et al., 2019). Treatment of infants with NOWS often requires prolonged hospitalization in neonatal intensive care units (NICUs) for drug withdrawal treatment, but there is no effective preventative treatment. Symptoms of withdrawal most commonly appear within the first 24–48 h after birth, depending on the type of opioid exposure; however, not all infants exposed to opioids during pregnancy experience withdrawal or require medication treatment for withdrawal symptoms. Determining the type and level of care an infant exposed to opioids will need and the risk factors for developing NOWS, remain unanswered clinical questions.
One of the main barriers to the development of an effective NOWS treatment plan is due to the lack of biomarker(s) to help predict which infants will develop NOWS, as well as our incomplete understanding of the pathogenesis of this syndrome. Currently, neonatologists face a challenge in assessing the severity of NOWS in infants, as there is a dearth of research on the molecular aspects of this condition, including algorithms, genetics, and epigenetics. This hinders understanding of the underlying issues in infants exposed to opioids before birth, further exacerbated by the absence of FDA-approved medications for NOWS treatment. The diagnosis is currently based on a history of opioid exposure in utero and symptoms in the newborn consistent with opioid withdrawal. While there are two tests approved by the FDA at present, which measure drug concentration in urine and meconium (Gray et al., 2009; Gray et al., 2010), confirmation of exposure to opioids alone cannot help determine the risk of developing NOWS and the need for treatment.
DNA methylation is an epigenetic modification where a methyl group (CH3) is added to DNA cytosine bases, typically at CpG sites. DNA methyltransferase enzymes catalyze this process, and these changes can be either inherited or acquired during one’s lifetime. This modification silences genes at promoters, influencing tissue-specific regulation, development, and genomic stability. Aberrant DNA methylation is linked to diseases, notably cancer, and DNA demethylation via TET enzymes also impacts gene regulation and development (Schuebel et al., 2016). Pharmacoepigenetics pertains to genetic variations in pharmacogenetics caused by epigenetic modifications influencing an individual’s response to medical treatments (Gomez and Ingelman-Sundberg, 2009).
Numerous factors during pregnancy impact fetal growth, organ development, and overall health (Li et al., 2019; Metpally et al., 2019). These encompass nutrition, stress, toxins, smoking, addiction, and infections. The presence of these toxins can impact the placenta and modify gene expression (Radhakrishna et al., 2021a), potentially increasing the likelihood of future health risks as altered epigenetics shape later life outcomes (Radhakrishna et al., 2023).
Cytochrome P450 (CYPs) enzymes play a critical role in the metabolism of many medications (Lynch and Price, 2007). CYP enzymes are involved in opioid metabolism, however, the rate of opioid metabolism varies widely among individuals, and some have much faster metabolisms of drugs than others (Smith, 2009). The CYP enzymes can be inhibited or induced by drugs, such that inhibition reduces metabolism and induction increases it (Bibi, 2008). Their activity is also affected by factors such as age, gender, environment, lifestyle (smoking, alcohol, and opioids) genetic and epigenetic variations (Lin and Lu, 2001; Rostami-Hodjegan et al., 2004).
It has been reported that neonates with severe NOWS, exposed to methadone prenatally displayed higher levels of homozygosity for CYP2B6 alleles at the 516 and 785 genetic loci (Mactier et al., 2017). Buprenorphine is an opioid that is approved by the FDA for treating opioid use disorder, acute pain, and chronic pain (Spreen et al., 2022). Buprenorphine is converted into its active metabolite, norbuprenorphine, by CYP3A, and SNPs in this gene may help predict the severity of buprenorphine-induced NOWS.
This study utilized a genome-wide methylation analysis to identify novel dysregulated CYPs in the placentas of mothers with opioid use disorder (OUD). These newly discovered CYPs could serve as potential markers to stratify the risk of opioid-exposed infants developing symptoms of NOWS before the symptoms become apparent. Gene ontology and pathway enrichment analyses were also conducted on the significant differentially methylated genes to gain deeper insights into their biological significance.
Materials and methods
The research study was approved by the Institutional Review Board of Beaumont Health System, Royal Oak, MI, United States (HIC#: 2019-086). Pregnant women were retrospectively identified by chart review from William Beaumont Hospital, Royal Oak, MI. Informed consent was waived for this study because it exclusively involved the collection of discarded placental tissues from the subjects, in addition to obtaining limited de-identified basic demographic data from the hospital medical records. We collected demographic and clinical-pathological data, including age, sex, ethnicity, gestational age, and detailed information on drug exposure, as described in our previously published work (Radhakrishna et al., 2021b). Patients with OUD were diagnosed using the Diagnostic and Statistical Manual of Mental Disorders, Fifth Edition, or DSM-5, assessment criteria (Hasin et al., 2013).
A total of 96 formalin-fixed, paraffin-embedded (FFPE) placental tissue biopsies were very carefully collected and processed. The placental tissue samples were divided into 3 groups. Group-1 consisted of 32 prenatally opioid-exposed newborns that required treatment for NOWS (+Opioids/+NOWS), Group-2 had 32 prenatally opioid-exposed newborns that did not require treatment for NOWS (+Opioids/-NOWS), and Group-3 was the control group of newborns with no opioid exposure in utero and no NOWS (-Opioids/-NOWS, control). Neonatologists examined newborns diagnosed with NOWS using the clinical criteria specified in the ICD-10 under code P96.1. Infants born to mothers with a history of opioid or illicit drug use were monitored in the inpatient unit for 4–5 days to observe for signs of NOWS. The infant was scored using the Finnegan Neonatal Abstinence Scoring Tool (FNAST). This scoring was done by the postpartum nurses and/or NICU nurses. If the scores met the criteria for pharmacologic treatment, the baby was transferred to the NICU for further monitoring, scoring, and treatment. Parent involvement was encouraged to optimize non-pharmacologic treatment as the mainstay before and during and treatment course whether the baby is getting pharmacologic treatment or not. The Finnegan Neonatal Abstinence Scoring Tool (FNAST) was used to determine when the pharmacologic management with morphine should be initiated.
A previous publication detailed the utilization of Illumina methylationEPIC 850 k arrays (Illumina, Inc., San Diego, CA, United States). These arrays encompass over 850,000 individual CpG sites across the entire genome, offering single-nucleotide precision. Moreover, the arrays incorporate various cytochrome P450 genes (Radhakrishna et al., 2021b). We retrieved differentially methylated CG dinucleotide data from previously unpublished DNA methylation data of CYP-encoding genes (Radhakrishna et al., 2021b). From those, CG dinucleotides met several criteria: 1) at least a 5% difference in methylation level between prenatally opioid-exposed cases and controls, 2) FDR-corrected p-value <0.05, and 3) no overlap with single nucleotide polymorphisms to avoid potential confounding factors, were selected.
Then, to confirm the possibility of altered expression of CYP-encoding genes due to the differential methylation of the promoter region. A detailed account of sample selection, diagnosis, and analysis was explained in previously published work (Radhakrishna et al., 2021b).
Statistical and bioinformatic analysis
Data (IDAT files) were normalized using Genome Studio software functional normalization and determined Cytosine methylation levels (ß-value) for each CpG site. Before analysis, we have removed all CpG-probes that have missing ß-values. Differential methylation was assessed by comparing the ß-values for the cytosine at each CpG locus in NOWS versus controls. To avoid confounding factors, we have removed probes associated with sex-chromosomes, non-specific probes, and probes targeting CpG sites within 10 bp of SNPs (each of which listed dbSNP entries within 10 bp of the CpG site) (Liu et al., 2013; Wilhelm-Benartzi et al., 2013; Zhang et al., 2013), further, SNPs with a minor allele frequency ≤ 0.05 were only considered for forwarding analysis.
Significantly differently methylated CpG sites between NOWS and controls were defined based on preset cutoff criteria FDR p < 0.05. Multiple CpG sites within a gene were resolved by the selection of the CpG with the highest AUC ROC ranking and the lowest p-value. The p-value for methylation differences between case and control groups at each locus was calculated as previously described (Vishweswaraiah et al., 2019; Radhakrishna et al., 2021b). Raw and FDR p-values corrected for multiple testing (Benjamini–Hochberg test) were calculated. Area under the receiver operating characteristic (AUC-ROC) AUC for combinations of loci was calculated using the ‘R’ program “ROCR” package (v3.5.0), based on methylation levels at the most significantly differently methylated CpG loci.
Biological functional enrichment analysis
The most used annotation database in enrichment Gene Ontology (GO), and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were conducted with CpG methylation changes associated with NOWS with a statistical significance of p-value <0.05.
Network interaction analysis using STRING
Following the identification of 16 crucial genes from the methylome results, a comprehensive investigation was conducted to examine potential interactions among these genes. The tool employed for this purpose was the STRING network analysis, a well-established bioinformatics resource for mapping possible interactions between genes. A non-functional cytochrome P450 pseudogene, CYP2D7P1, was excluded from the analysis. The remaining list of 16 genes of interest, which demonstrated significant hypermethylation or hypomethylation changes in response to prenatal opioid exposure, were inputted into the STRING database (http://string-db.org) using a multiple protein search. This network was structured with a medium interaction score threshold of 0.40. Interaction sources such as text mining, experiments, databases, co-expression, and co-occurrence were selected for their unique contributions. Text mining and co-occurrence offer indirect associations from literature and an evolutionary perspective, experiments and databases contribute direct, known interactions, while co-expression discovers functionally linked genes through shared expression patterns (Figure 1).
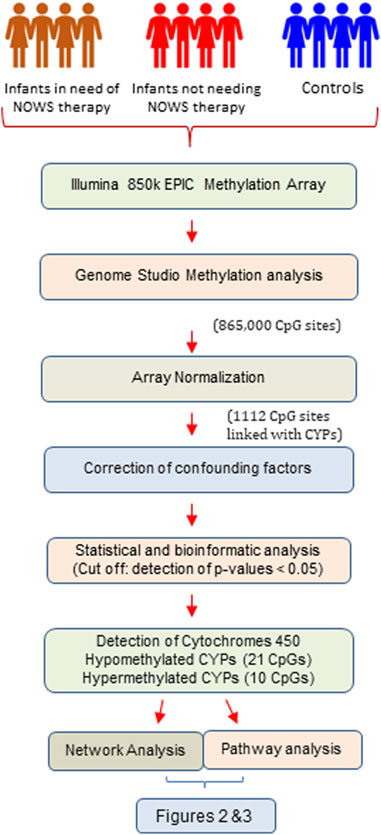
FIGURE 1. The flow chart illustrates the systematic workflow of genome-wide DNA methylation analysis.
Results
The demographic characteristics of the NOWS and control groups were compared, and no significant differences were found. We have identified a total of 1,112 CpGs exclusively associated with CYPs across all four configuration studies after array normalization but before the removal of probes associated with confounding factors (Supplementary Tables S1–S4). The analysis performed with four configurations revealed CpGs with p-values <0.05 as 316, 222, 305, and 169 in analysis-1 (+Opioids/+NOWS versus + Opioids/-NOWS); analysis-2 (+Opioids/+NOWS), + (+Opioids/-NOWS), versus (-Opioids/-NOWS, control); analysis-3 (+Opioids/+NOWS), versus (-Opioids/-NOWS, control); and analysis-4 (+Opioids/-NOWS), versus (-Opioids/-NOWS, control), respectively. Additionally, the dataset comprised CpGs with FDR p-values <0.05, signifying significant differential methylation.
There were 20 CpG sites with FDR p-values <0.05, showing methylation differences of ±0.05 associated with 17 distinct CYP genes. Among these, 14 CpGs displayed hypomethylation across genes (CYP19A1, CYP1A2, CYP4V2, CYP1B1, CYP24A1, CYP26B1, CYP26C1, CYP2C18, CYP2C9, CYP2U1, CYP39A1, CYP2R1, CYP4Z1, and CYP2D7P1), while eight exhibited hypermethylation (CYP51A1, CYP1A2, CYP26B1, CYP2R1, CYP2U1, CYP4X1, CYP2W1, and CYP4V2). Notably, genes such as CYP1A2, CYP26B1, CYP2R1, CYP2U1, and CYP4V2 exhibited both hyper and hypomethylation Tables 1–4).
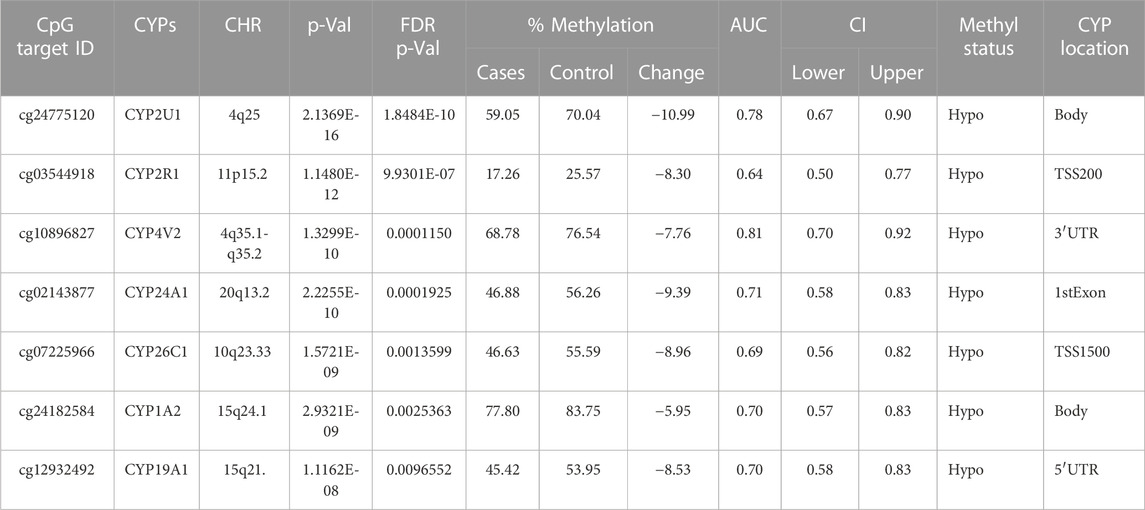
TABLE 1. Detailed overview of CYP genes methylated status and their biological functions/associated diseases. Differentially methylated CpG markers are provided, with information about genes, the corresponding methylated CpG sites chromosome location, AUC, and the percentage of difference in methylation for each CpG and its biological significance based on FDR adjusted-p values <0.05 provided with (AUC) ≥0.75 for NOWS detection. (Hypo: Hypumethylation; Hyper: Hypermethylation; TSS200 denotes -200 base pairs to transcription start site; TSS1500 denotes -1500 base pairs to transcription start site). The analysis of +Opioids/+NOWS versus + Opioids/-NOWS. Significantly differentially methylated CYPs in NOWS.
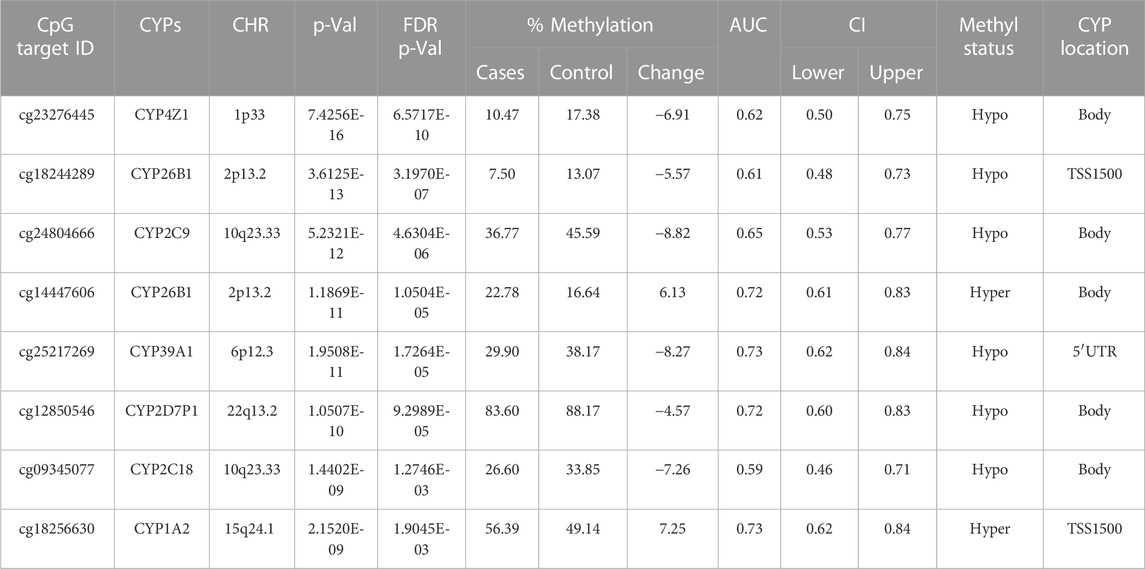
TABLE 2. Detailed overview of CYP genes methylated status and their biological functions/associated diseases. Differentially methylated CpG markers are provided, with information about genes, the corresponding methylated CpG sites chromosome location, AUC, and the percentage of difference in methylation for each CpG and its biological significance based on FDR adjusted-p values <0.05 provided with (AUC) ≥0.75 for NOWS detection. (Hypo: Hypumethylation; Hyper: Hypermethylation; TSS200 denotes -200 base pairs to transcription start site; TSS1500 denotes -1500 base pairs to transcription start site). Analysis of (+Opioids/+NOWS), + (+Opioids/-NOWS), versus (-Opioids/-NOWS, control). Details of CpG targets significantly differentially methylated CYPs NOWS.
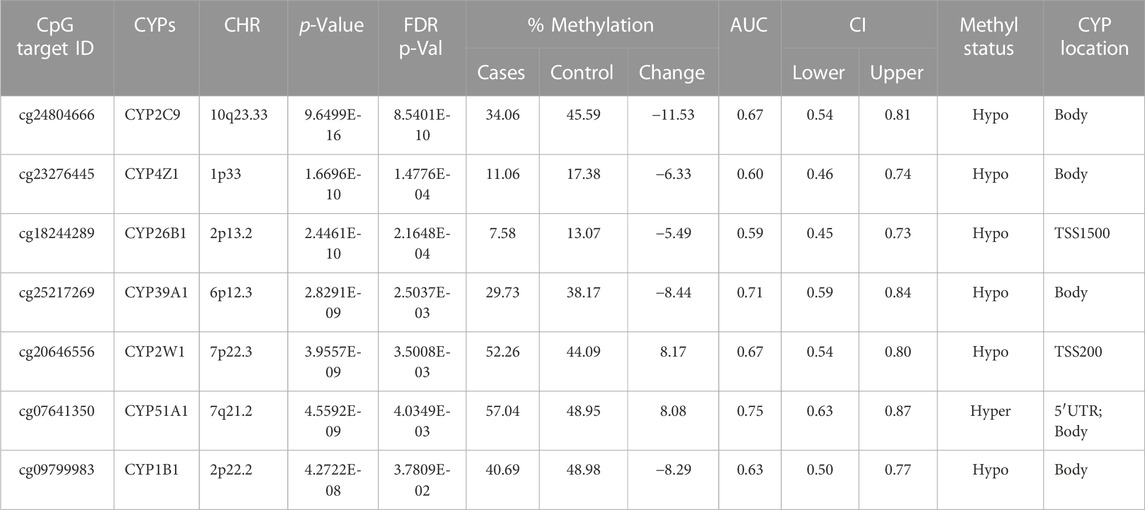
TABLE 3. Detailed overview of CYP genes methylated status and their biological functions/associated diseases. Differentially methylated CpG markers are provided, with information about genes, the corresponding methylated CpG sites chromosome location, AUC, and the percentage of difference in methylation for each CpG and its biological significance based on FDR adjusted-p values <0.05 provided with (AUC) ≥0.75 for NOWS detection (Hypo: Hypumethylation; Hyper: Hypermethylation; TSS200 denotes -200 base pairs to transcription start site; TSS1500 denotes -1500 base pairs to transcription start site). Analysis of (+Opioids/+NOWS), versus (-Opioids/-NOWS, control). Details of CpG targets significantly differentially methylated CYPs NOWS.
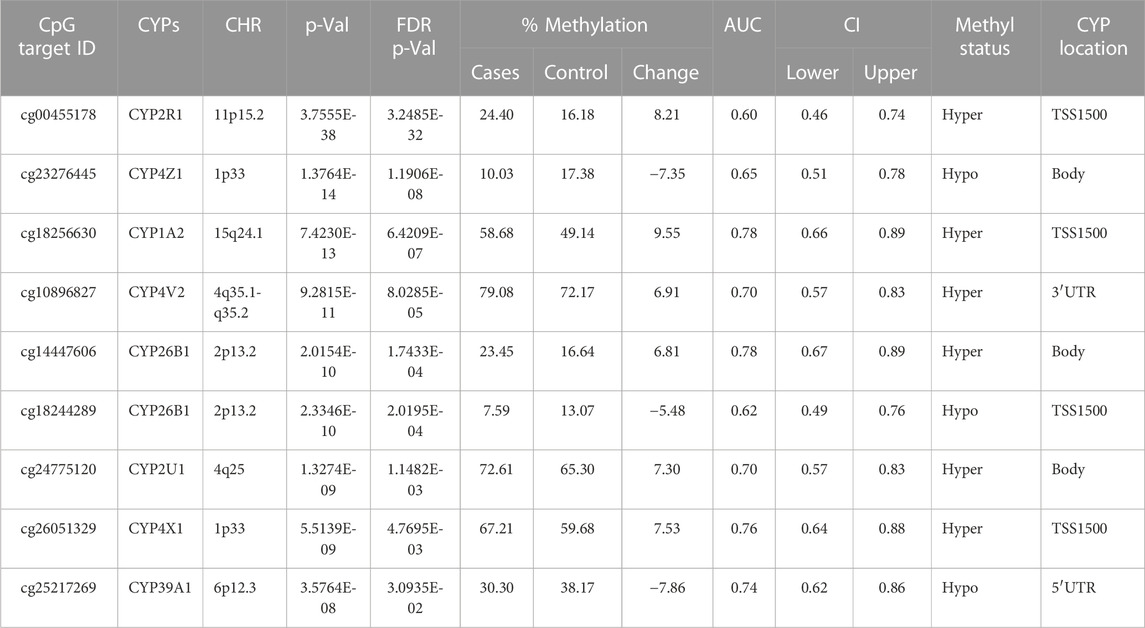
TABLE 4. Detailed overview of CYP genes methylated status and their biological functions/associated diseases. Differentially methylated CpG markers are provided, with information about genes, the corresponding methylated CpG sites chromosome location, AUC, and the percentage of difference in methylation for each CpG and its biological significance based on FDR adjusted-p values <0.05 provided with (AUC) ≥0.75 for NOWS detection (Hypo: Hypumethylation; Hyper: Hypermethylation; TSS200 denotes -200 base pairs to transcription start site; TSS1500 denotes -1500 base pairs to transcription start site). Analysis of (+Opioids/-NOWS), versus (-Opioids/-NOWS, control) with details of CpG targets significantly differentially methylated CYPs.
Gene interaction network analysis
The STRING network analysis generated an interconnected network among the 16 key genes, presenting 54 edges in total. This network, visualized in Figure 2, exhibits the complex interplay between these genes. The network statistics displayed an average node degree of 6.75 and an average local clustering coefficient of 0.459. The expected number of edges was 1, which contrasted significantly with the observed results, confirming the proteins are indeed biologically interconnected as a group. This was further validated by the PPI enrichment p-value of <1.0e-16, suggesting that the interactions observed within this network significantly exceed random expectations. Analysis of individual genes highlighted their significance as central nodes in the network, demonstrating extensive interactions with numerous other genes. This indicates a key role of these genes within the network, particularly in their response to prenatal opioid exposure.
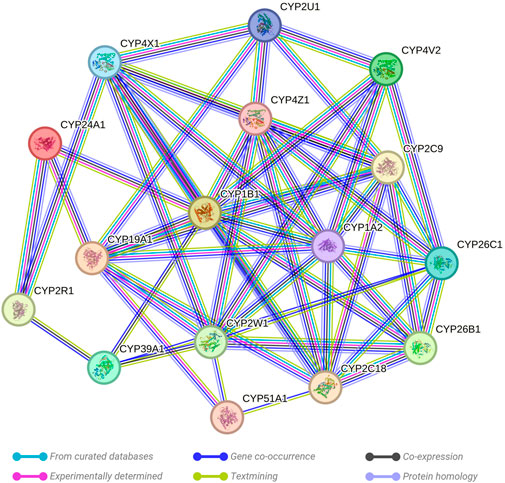
FIGURE 2. Evaluation of the links existing between the evaluated CYP genes. A critical assessment and integration of protein–protein was provided by the STRING database, including direct (physical) and indirect (functional) interactions. String predicted both physical and functional interactions between proteins. Number of nodes: 16, Number of edges: 54, Average node degree: 6.75, Avg. local clustering coefficient: 0.459, Expected number of edges: 1, and PPI enrichment p-value: <0.0001.
The findings offer insight into potential cooperative or antagonistic relationships among the selected genes. Changes in methylation status, either hypermethylation or hypomethylation, could impact the expression of these genes, thereby affecting metabolic and physiological responses to prenatal opioid exposure. These changes may further contribute to the development or severity of NOWS.
Pathway and gene ontology analysis of dysregulated CYPs
To identify the KEGG pathway-associated genes and potential GO (Gene Ontology) classifications, terms related to biological processes (BP), molecular functions (MF), and signaling pathways were analyzed. As a result of the pathway analysis, 5 major dysregulated pathways associated with NOWS were identified.
Table 5 illustrates the functional enrichment analysis of the complex network, including some of the following pathways that were over-represented: hsa01100: Metabolic pathways, hsa05204: Chemical carcinogenesis - DNA adducts, hsa00100: Steroid biosynthesis, hsa00140: Steroid hormone biosynthesis, hsa00980: Metabolism of xenobiotics by cytochrome P450, and hsa04726: Serotonergic synapse. The biological processes (BPs) of the 16 dysregulated CYPs focused predominantly on the xenobiotic metabolic process, sterol metabolic process, vitamin metabolic process, organic acid metabolic process, omega-hydroxylase P450 pathway, epoxygenase P450 pathway, retinoic acid catabolic process, estrogen metabolic process, retinol metabolic process, aflatoxin metabolic process and fatty acid omega-oxidation as shown in Table 6. Regarding the cellular components (CCs), the study revealed a significant connection between these dysregulated CYPs and the endoplasmic reticulum membrane, as well as intracellular membrane-bound organelles, as demonstrated in Table 7. Additionally, Table 8 provided enriched molecular functions (MF) associated with the 16 differentially methylated CYPs. These functions encompassed iron ion binding, heme binding, monooxygenase activity, oxidoreductase activity, steroid hydroxylase activity, aromatase activity, caffeine oxidase activity, retinoic acid 4-hydroxylase activity, arachidonic acid 14,15-epoxygenase activity, arachidonic acid epoxygenase activity, and cytochrome P450 fatty acid omega-hydroxylase activity, along with estrogen 16-alpha-hydroxylase activity.
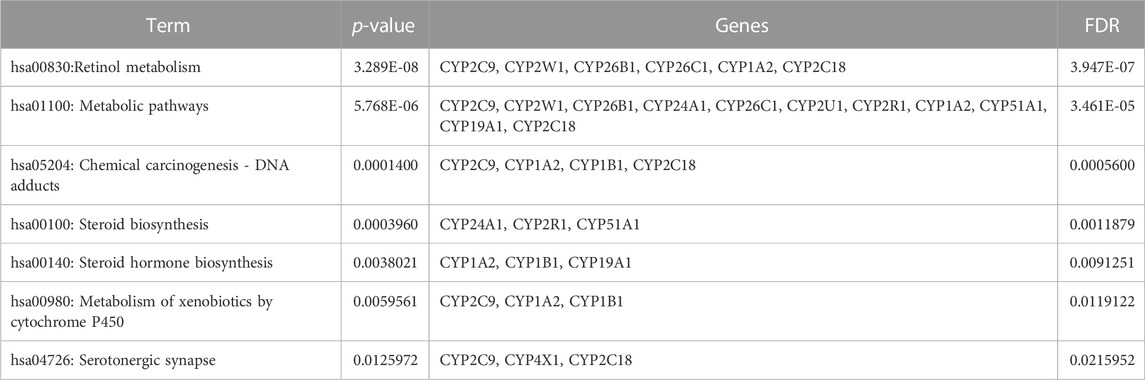
TABLE 5. Gene ontology (GO) terms associated with differentially methylated genes that are associated with NOWS such as KEGG pathway enrichment for the 12 CYP genes with altered methylation patterns linked to NOWS.
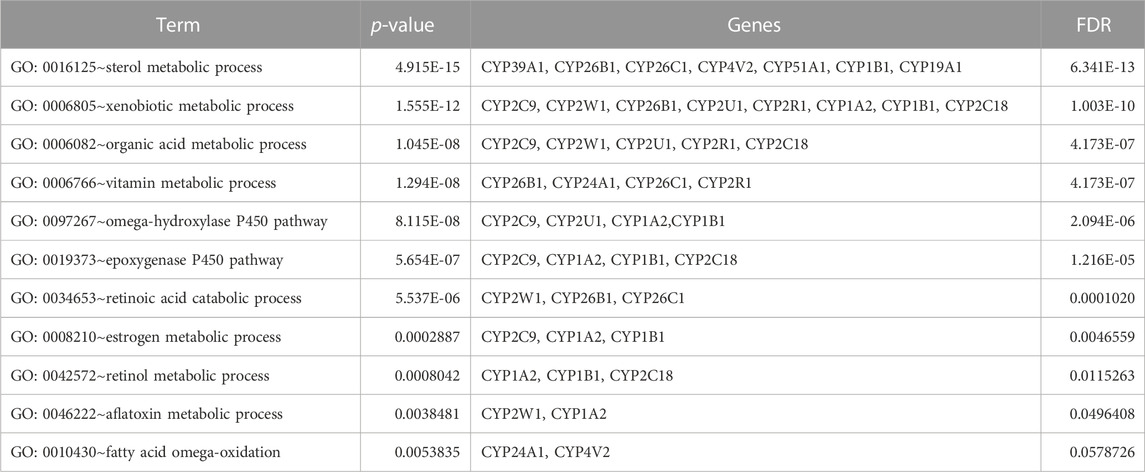
TABLE 6. Gene ontology (GO) terms associated with differentially methylated genes that are associated with NOWS such as Biological processes (BP) enrichment analysis in NOWS.

TABLE 7. Gene ontology (GO) terms associated with differentially methylated genes that are associated with NOWS such as Cellular Component (CC) enrichment analysis.
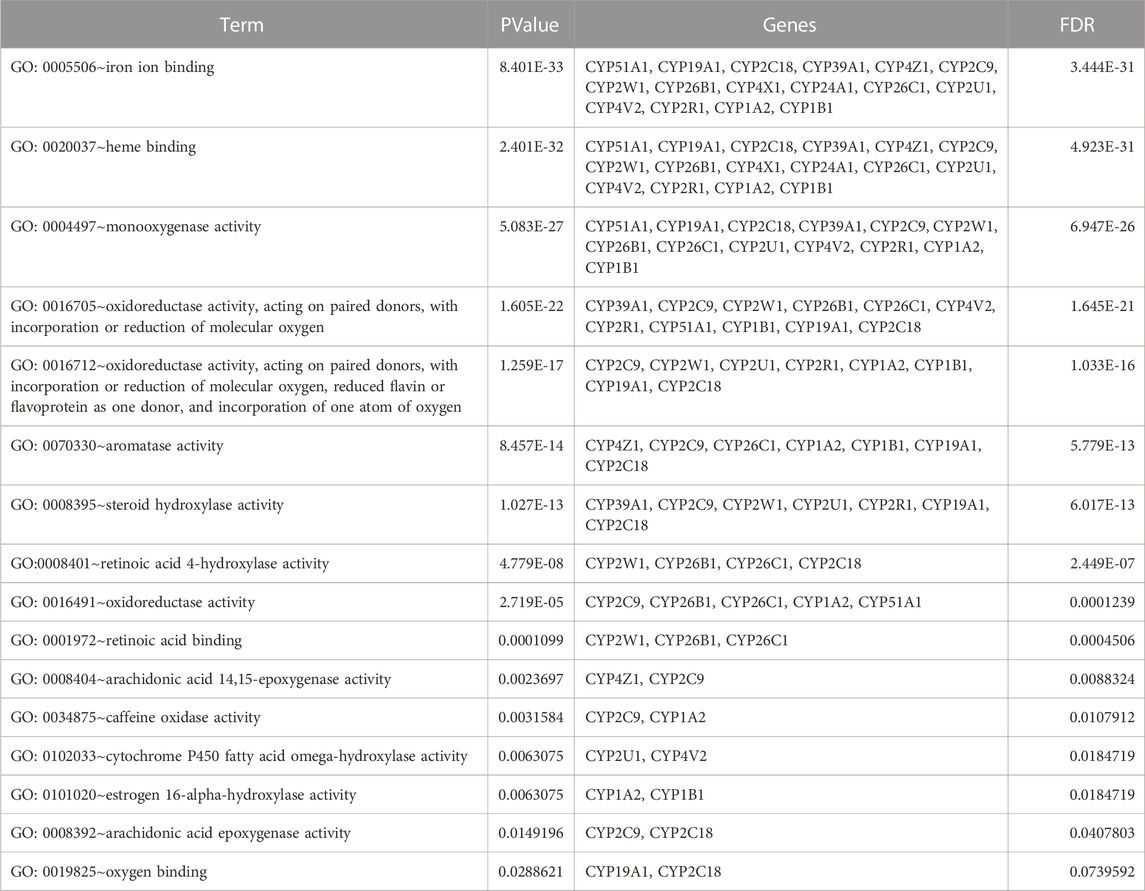
TABLE 8. Gene ontology (GO) terms associated with differentially methylated genes that are associated with NOWS such as KEGG pathway enrichment for the 12 CYP genes with altered methylation patterns linked to NOWS.
Discussion
Currently, there is no effective way to diagnose and treat NOWS before it occurs. This is partially due to asymptomatic manifestation in NOWS infants because not all infants require pharmacotherapy, as well as its complicated molecular mechanisms involved in the long-term effects. Infants with severe symptoms of NOWS have a greater risk of adverse long-term developmental outcomes including lower IQ, poor educational testing performance, lower attention, meeting disability criteria, and requiring additional classroom services when compared to children with prenatal opioid exposure who did not develop NOWS or healthy controls (Oei et al., 2017; Fill et al., 2018). A significant challenge is distinguishing the neurobiological effects of opioid agonists from various other confounding factors, including tobacco, alcohol, nonmedical drugs, environmental influences, and additional medical risks (Logan et al., 2013; Radhakrishnan et al., 2022a; Radhakrishnan et al., 2022b). The effects of opioids during pregnancy extend beyond nociceptive processes to include effects on gastrointestinal, endocrine, and autonomic functions as well as cognition, behavior, and reward circuitry for the mother and developing fetus, and subsequently the offspring (Radhakrishnan et al., 2021; Radhakrishnan et al., 2022a; Radhakrishnan et al., 2022b; Radhakrishnan et al., 2022c; Vishnubhotla et al., 2022; Wise et al., 2023).
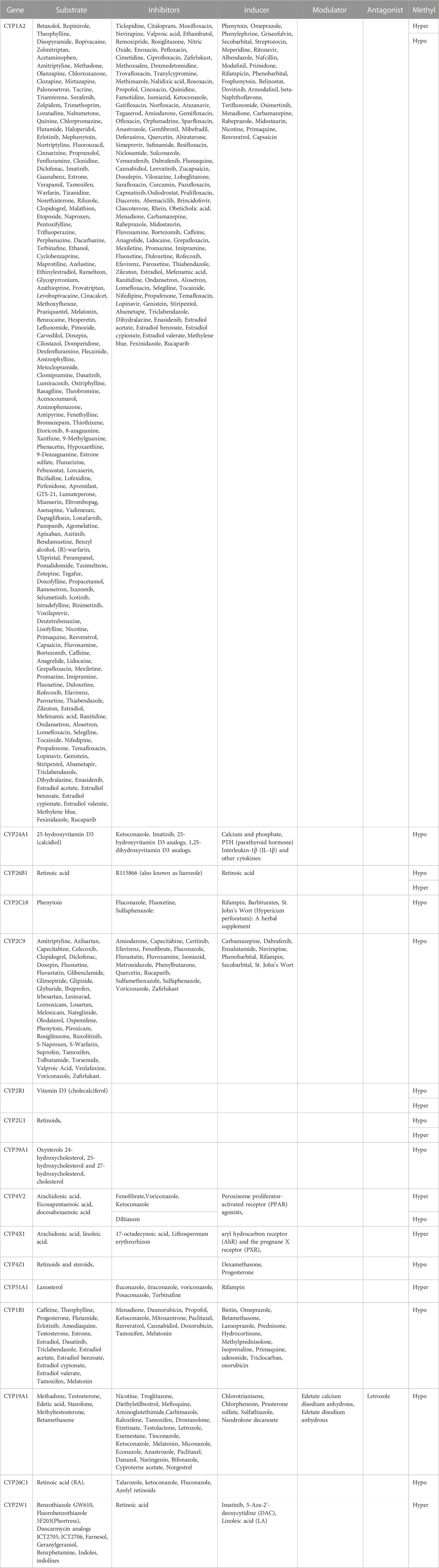
Through diverse comparative analyses, the study identified 20 CpG sites with significant differences in their methylation patterns. These studies aimed to unravel the epigenetic and molecular dysregulation associated with NOWS. The investigations employed various methods, including the following approaches. An overview of CYP enzyme substrates, inhibitors, and inducers with clinical significance in NOWS patients and controls is provided in Table 9.
Distinguishing NOWS from prenatal opioid exposure without NOWS
This comparison aims to identify the epigenetic and molecular differences between newborns with NOWS requiring treatment, and infants exposed to opioids prenatally without withdrawal symptoms and those not requiring treatment for NOWS. This analysis identifies factors leading to withdrawal in specific infants (Table 1).
Distinguishing prenatal opioid abuse versus normal controls (OUD detection)
This comparison focuses on identifying epigenetic markers associated with maternal opioid abuse during pregnancy. We analyzed to identify the differences between infants born to mothers with opioid use disorder (OUD) and infants born to mothers without substance abuse issues. Comprehending these variances aids in identifying prenatal opioid exposure and evaluating the impact of maternal opioid abuse on the newborn’s molecular and epigenetic traits (Table 2).
Distinguishing NOWS versus unexposed controls
In this comparison, we examined the epigenetic alterations in newborns with NOWS in comparison to infants born to mothers who did not use opioids during pregnancy. Through this analysis, we pinpointed NOWS-related epigenetic changes, offering crucial insights into the mechanisms driving withdrawal symptoms in newborns (Table 3).
Distinguishing opioid-induced epigenetic changes
This comparison studied the epigenetic changes in newborns who were exposed to opioids during prenatal development in comparison with infants born to mothers who refrained from opioid use during pregnancy. We discovered opioid-induced epigenetic changes, offering crucial insights into the molecular mechanisms influenced by early exposure to opioids (Table 4).
Drug metabolism by genetically polymorphic enzymes can have significant clinical implications relating to drug toxicity, therapeutic failure, drug-drug interactions, disease susceptibility, abuse liability as well and the number of drugs consumed by people who take drugs. The present study reveals that 20 CpGs align with 17 distinct CYPs, each showcasing unique methylation patterns. Figure 3 illustrates how epigenetic regulation affects drug metabolism and transport with details of dysregulated CYPs.
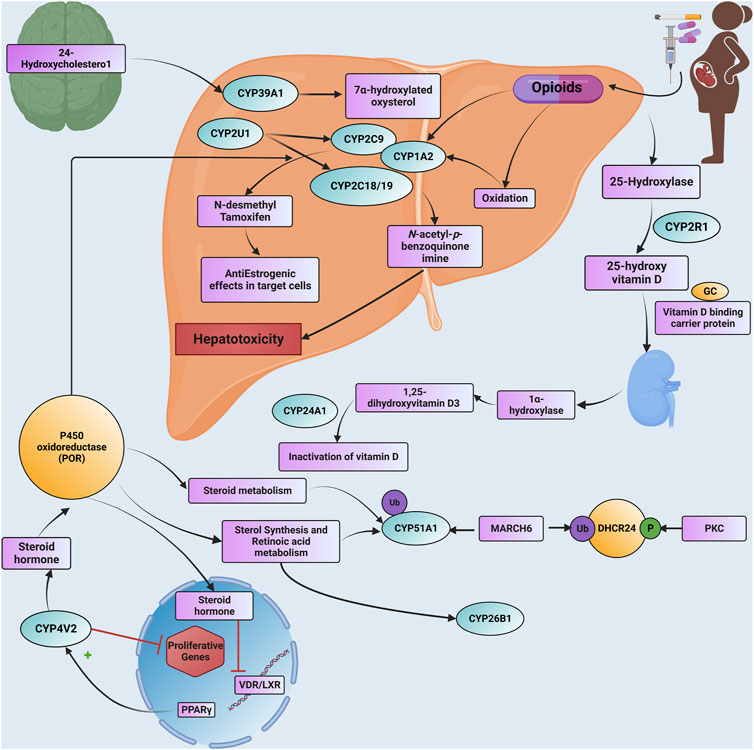
FIGURE 3. A detailed Schematic illustration is provided of how epigenetic modifications influence drug metabolism and transport through CYP genes, including specific information about dysregulated CYP enzymes. Additionally, the text outlines the pathogenetic events associated with NOWS and highlights specific CYP genes that undergo significant methylation. Abbreviations. GC: GROUP-SPECIFIC COMPONENT; Ub: ubiquitination; P: phosphorylation; DHCR24: delta (24)-sterol reductase; MARCH6: Membrane Associated Ring-CH-Type Finger 6; PKC: Protein kinase C; PPARG: Peroxisome Proliferator-Activated Receptor Gamma; VDR: Vitamin D Receptor; LXR: Liver X Receptor
Depending on circumstances, CYP-active drugs can inhibit, induce, or act as substrates for enzymes. Inhibition curbs metabolism, while induction enhances it (McDonnell and Dang, 2013). Methylation-driven disruptions in CYPs within the placenta may potentially contribute to various abnormalities observed in both NOWS and OUD cases. Insufficient data exists regarding the enduring impacts of maternal opioid usage during pregnancy on infants diagnosed with NOWS. Intermittent anomalies arising from dysregulated CYPs in infants experiencing NOWS may not manifest immediately, but could instead become noticeable later in life. These conditions encompass ailments such as Non-alcoholic Fatty Liver Disease (NAFLD), type 2 diabetes, cardiovascular diseases, and various psychiatric disorders, among others. To explore this hypothesis, we require extended follow-up studies.
CYP1A2
CYP1A2 is primarily expressed in the liver (Zanger and Schwab, 2013), but it has also been detected in the brain, suggesting a potential role in protecting the brain from xenobiotics (Yun et al., 1998). Additionally, CYP1A2 plays a critical role in converting chemical pollutants from cigarette smoke into carcinogenic compounds (Taşçıoğlu et al., 2021b). Individuals who smoke tend to be more prone to self-report opioid use disorders in comparison to non-smokers (Young-Wolff et al., 2017). Notably, a link has been established between the genetic variation rs762551 in the CYP1A2 gene and an increased susceptibility to lung cancer among smokers (Taşçıoğlu et al., 2021a), Moreover, a noteworthy correlation has emerged connecting CYP1A2 with drug addiction (Zhang et al., 2016).
CYP4V2
CYP4V2 is a key enzyme that is associated with NAFLD, a significant health concern affecting 30% of adults in the United States (Garduno and Wu, 2021). NAFLD is marked by dysbiosis, a condition impacting non-alcohol consumers, and is characterized by the accumulation of fat, liver inflammation, fibrosis, and cirrhosis. Opioid use is more common in NAFLD with cirrhosis, high BMI, and psychiatric disorders (Moon et al., 2021).
CYP51A1
The CYP51A1 in humans, also known as lanosterol 14α-demethylase is the most evolutionarily conserved member of the CYP450 (Rezen et al., 2004), and is pivotal in hepatic cholesterol synthesis (Lewinska et al., 2013). CYP51A1 removes two methyl groups from lanosterol during oxidative reactions. Cholesterol is essential for life, but elevated serum cholesterol levels raise the risk of conditions like high blood pressure, type 2 diabetes, coronary heart disease (CHD), stroke, and cardiovascular disease (CVD). It has been reported that opioid use and cholesterol levels are related since cholesterol levels affect opioid signaling in cell models (Zheng et al., 2012; Kazemi et al., 2021). Preterm birth, affecting 20%–40% of infants exposed to opioids during pregnancy, poses a substantial risk factor for NOWS (Allocco et al., 2016; Sheikh et al., 2016a). A previous study found that CYP51A1 gene variations are associated with preterm birth among women (Sheikh et al., 2016b).
CYP24A1
The CYP24A1 gene encodes a 24-hydroxylase enzyme that regulates active vitamin D levels in the body by converting the active form, 1,25-dihydroxy vitamin D3 or calcitriol, into an inactive form when it is no longer required (Prosser and Jones, 2004). Opioid addiction and ultraviolet ray addiction (Kemeny et al., 2021), occur more frequently in individuals with insufficient vitamin D levels, suggesting a potential role for this vitamin in neurodevelopment and safeguarding dopaminergic pathways in the adult brain (Eserian, 2013), Additionally, vitamin D is vital for preventing neonatal hypoxic-ischemic brain damage (Anjum et al., 2018). This vitamin is further strongly known for its role in cognitive impairment, dementia, psychosis, and autism (Weller et al., 2021). Transcription from the human CYP24A1 gene is increased by calcium ions and by excess amounts of 1,25-dihydroxy vitamin D3 (Nebert and McKinnon, 1994; Norman et al., 2001). Variations in the CYP24A1 gene lead to an accumulation of vitamin D3 and an associated hypervitaminosis D phenotype (Griffin et al., 2020), Hypervitaminosis D is linked to a range of severe health hazards, including reduced appetite, constipation, depressive symptoms, and memory impairment—similar issues experienced by individuals with chronic opioid use (Nelson and Camilleri, 2015).
CYP39A1
The endoplasmic reticulum protein, oxysterol 7-α-hydroxylase 2 encoded by CYP39A1, is pivotal in converting cholesterol to bile acids. It is primarily expressed in the liver and converts 24-hydroxycholesterol (24OHC) into a 7-alpha-hydroxylated form (Li-Hawkins et al., 2000). Aberrant CYP39A1 expression may contribute to neurodegenerative diseases like Alzheimer’s (Matsuoka et al., 2020), however, its role in NOWS is not yet known. CYP39A1 protein is involved in the pathway of cholesterol degradation, which is part of steroid metabolism (Stiles et al., 2014). Cholesterol is a structural component of membranes, as well as a precursor for bile acid and steroid hormone synthesis (Kenworthy, 2002). Individuals with OUD may experience concurrent mental health issues like depression and anxiety, which can impact their hormonal systems and stress responses.
CYP26B1
The CYP26A1, CYP26B1, and CYP26C1 enzymes are a group of enzymes that are highly conserved in vertebrates, metabolizing retinoic acid (RA), a vitamin A derivative, and potentially playing a role in xenobiotic metabolism. Vitamin A is vital for embryonic development, organ formation, immunity, and eye health (Bastos Maia et al., 2019). Currently, retinoids are used to treat a variety of skin conditions, and antiaging skincare, as well as slow the effects of photoaging (Szymanski et al., 2020), addiction, and abuse causing premature aging, wrinkles, and other skin problems (Inci et al., 2017). Furthermore, retinoic acid impacts the adult brain; animal studies have shown that isotretinoin (retinoids) inhibits neurogenesis in the hippocampus when administered (Bremner and McCaffery, 2008). Interestingly, isotretinoin induces apoptosis in different types of cells in the body and decreases the enzyme telomerase reverse transcriptase (TERT) activity (Das et al., 2022). Increased TERT expression restores telomerase activity, suggesting that transcriptional control may play a role in a wide array of diseases, including cancer and aging (Colebatch et al., 2019). Lack of Telomerase/TERT expression leads to progressive telomere erosion in dividing human cells, while critically shortened telomeres induce permanent growth arrest, increased incidence of diseases, and poor survival (Trybek et al., 2020). The impaired TERT could be another contributing factor, as opioid addicts typically have a much shorter lifespan than non-addicts (Smyth et al., 2007; Lewer et al., 2020). Yet, the life expectancy of an infant with NOWS remains unknown.
CYP2U1
CYP2U1 is expressed in the amygdala and prefrontal cortex of humans and is involved in brain and immune functions. CYP2U1 is involved in cytokine production, long-chain fatty acid synthesis, drug absorption, and the metabolism of foreign substances and eicosanoids. However, alcohol consumption and smoking can impair its function. Amygdalae, integral components of the limbic system, have a significant role in memory, decision-making, and the processing of emotional responses, including fear, anxiety, and aggression (Amunts et al., 2005). There is a link between anxiety and drug addiction due to perturbations of the anxiety and fear centers in the brain (Brady et al., 2013).
CYP2R1
CYP2R1 is an enzyme involved in vitamin D biosynthesis that is significantly hypermethylated in this study. Deficiency in vitamin D may lead to many health conditions, including chronic pain, addiction to opioids (Kemeny et al., 2021), sleep disorders, and irritability (Archontogeorgis et al., 2018). Infants with NOWS are commonly seen with symptoms such as irritability, lethargy, muscle weakness, frequent respiratory problems, and decreased sleep (Patrick et al., 2020).
CYP4X1 and CYP4Z1
The hypomethylated CYP4X1 and CYP4Z1 genes together are presumed to be involved in human breast and ovarian cancers (Blahos et al., 1986; Savas et al., 2005; Murray et al., 2010). Chronic opioid use is more common among cancer patients, possibly because of confounding anxiety and/or depression that often accompany cancer diagnosis (Marcusa et al., 2017).
CYP2C18
CYP2C18 participates in the metabolism of retinoic acid processes (Thatcher and Isoherranen, 2009). Isotretinoin, a retinoid compound, is derived from Vitamin A. Reports indicate that employing synthetic vitamin A (retinoids) like isotretinoin (accutane) during pregnancy may result in miscarriage, premature birth, and an array of developmental abnormalities in the embryo and fetus (Ross et al., 2000). Opioid use during pregnancy is linked to poor fetal growth, preterm birth, stillbirth, and birth defects, as well as NOWS (Yazdy et al., 2015; Lind et al., 2017).
CYP2C9
CYP2C9 enzyme plays a very important role in the metabolism of vitamin K antagonist anticoagulants, such as S-warfarin, S-acenocoumarol, and phenprocoumon, commonly used to treat thromboembolic diseases (Daly and King, 2006; Chen et al., 2018). Smoking is prevalent among 95% of opioid users, and tobacco contains high levels of vitamin K (Chun et al., 2009). It has been reported that opioid users are at a greater risk of ischemic stroke (Lee et al., 2013). Another key gene, VKORC1, encodes the VKORC1 (vitamin K epoxide reductase) protein, which is a key enzyme in the vitamin K cycle and was also hypomethylated in the current study (Radhakrishna et al., 2021b).
CYP1B1 and CYP19A1
The interplay of CYP1B1 and CYP19A1 in estrogen metabolism and biosynthesis is pivotal in understanding their impact on addiction development. CYP1B1, a key player in estrogen metabolism, facilitates the conversion of estrone (E1) and estradiol (E2) into diverse metabolites (Tsuchiya et al., 2004). Estrogen has an impact on critical neurotransmitter systems in the brain, specifically dopamine, serotonin, and opioids, which are intricately connected to the reward circuitry associated with the development of addiction (Krolick et al., 2018; Martinez et al., 2016). The aromatase enzyme, also known as CYP19A1, plays a crucial role in estrogen synthesis. CYP19A1-mediated alterations in estrogen levels play a significant role in influencing diverse responses to substances of abuse, potentially accentuating gender-related variations in vulnerability to addiction (Moran-Santa Maria et al., 2014).
Study Limitations
Our study has several limitations. First and foremost, we require additional investigation into the role of the dysmethylated CpGs we’ve identified to establish whether changes in methylation at these CpG sites might impact gene transcription. Additionally, we lack prior addiction history information for women who used opioids during pregnancy. Another significant limitation of this study is that the diagnosis of NOWS predominantly depends on assessing clinical signs and symptoms in newborns right after birth. Nevertheless, numerous clinical manifestations stemming from CYP dysregulations may only surface later in a child’s growth and development. Consequently, linking these initial clinical indicators in infants to potential long-term consequences can pose a formidable challenge. Furthermore, it is important to note that methylation patterns are tissue-specific. Just because certain genes are methylated in the placenta does not necessarily imply the same methylation status in the liver of newborns, a crucial site for drug metabolism.
Conclusion
In summary, our cohort study revealed dysregulation in several crucial CYP genes linked to NOWS. These genes are closely involved in the metabolism of drugs, vitamin K, vitamin D, vitamin A, and bile acids. Moreover, they are linked to various health conditions, such as Autism, Alzheimer’s disease, non-alcoholic fatty liver disease, and emotional-behavioral dysregulation in infants. The study has significant implications for understanding the long-term effects of CYP dysregulation. Nevertheless, further investigation is necessary to fully comprehend the underlying molecular mechanisms responsible for the observed CYP gene dysregulations following prenatal opioid exposure. This deeper understanding could potentially open doors for advancements in personalized medicine down the line. Further research is required to explore their feasibility, effectiveness, and integration with other relevant factors.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics statement
The studies involving humans were approved by the institutional review board, Beaumont Health. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
UR: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing–original draft, Writing–review and editing. SS: Conceptualization, Data curation, Formal Analysis, Methodology, Software, Validation, Visualization, Writing–original draft, Writing–review and editing. RR: Formal Analysis, Methodology, Software, Visualization, Writing–original draft, Writing–review and editing. AF: Data curation, Formal Analysis, Methodology, Software, Supervision, Validation, Visualization, Writing–original draft, Writing–review and editing. SM: Supervision, Validation, Visualization, Writing–original draft, Writing–review and editing, Conceptualization, Data curation, Formal Analysis, Methodology, Software. RM: Conceptualization, Formal Analysis, Methodology, Software, Supervision, Validation, Visualization, Writing–original draft, Writing–review and editing. SP: Conceptualization, Formal Analysis, Methodology, Software Development, Supervision, Validation, Visualization, Writing–original draft, Writing–review and editing. RMR: Formal Analysis, Methodology, Software, Validation, Visualization, Writing–original draft, Writing–review and editing. SV: Investigation, Methodology, Software, Validation, Visualization, Writing–review and editing. RB-S: Conceptualization, Data curation, Investigation, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing–original draft. SN: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Methodology, Software, Validation, Visualization, Writing–review and editing.
Funding
The authors declare financial support was received for the research, authorship, and/or publication of this article. Efforts of Drs. Radhakrishna, Sadhasivam, and RadhakrishnanUR, SS, and RR are supported by their collaborative NIH grants (R01HD096800 PI: S. Sadhasivam), R01DA059321 PI: R. Radhakrishnan, and R03DA056797 PI; R. Radhakrishnan. Dr. Sadhasivam’s efforts are also supported by R43DA058430, R44DA055407, R44DA056280, R41DA053877 and R43HL167661, R01DA054513 (MPI: Chelly/Sadhasivam) and U01TR003719 (PI: Sadhasivam). Dr. Sadhasivam is the Founder and Chief Medical Officer of OpalGenix, Inc., and a consultant for Neuroptics, Inc.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1292148/full#supplementary-material
SUPPLEMENTARY TABLE S1 | The analysis of +Opioids/+NOWS versus +Opioids/-NOWS.
SUPPLEMENTARY TABLE S2 | Analysis of (+Opioids/+NOWS), + (+Opioids/-NOWS), versus (-Opioids/-NOWS, control).
SUPPLEMENTARY TABLE S3 | Analysis of (+Opioids/+NOWS), versus (-Opioids/-NOWS, control).
SUPPLEMENTARY TABLE S4 | Analysis of (+Opioids/-NOWS), versus (-Opioids/-NOWS, control).
References
Allocco, E., Melker, M., Rojas-Miguez, F., Bradley, C., Hahn, K. A., and Wachman, E. M. (2016). Comparison of neonatal abstinence syndrome manifestations in preterm versus term opioid-exposed infants. Adv. Neonatal Care 16 (5), 329–336. doi:10.1097/ANC.0000000000000320
Amunts, K., Kedo, O., Kindler, M., Pieperhoff, P., Mohlberg, H., Shah, N. J., et al. (2005). Cytoarchitectonic mapping of the human amygdala, hippocampal region and entorhinal cortex: intersubject variability and probability maps. Anat. Embryol. Berl. 210 (5-6), 343–352. doi:10.1007/s00429-005-0025-5
Anjum, I., Jaffery, S. S., Fayyaz, M., Samoo, Z., and Anjum, S. (2018). The role of vitamin D in brain health: a mini literature review. Cureus 10 (7), e2960. doi:10.7759/cureus.2960
Archontogeorgis, K., Nena, E., Papanas, N., and Steiropoulos, P. (2018). The role of vitamin D in obstructive sleep apnoea syndrome. Breathe (Sheff) 14 (3), 206–215. doi:10.1183/20734735.000618
Bastos Maia, S., Rolland Souza, A. S., Costa Caminha, M. F., Lins da Silva, S., Callou Cruz, R., Carvalho Dos Santos, C., et al. (2019). Vitamin A and pregnancy: a narrative review. Nutrients 11 (3), 681. doi:10.3390/nu11030681
Bibi, Z. (2008). Role of cytochrome P450 in drug interactions. Nutr. Metabolism 5 (1), 27. doi:10.1186/1743-7075-5-27
Blahos, J., Care, A. D., and Fox, J. (1986). Effect of betamethasone and calcium and phosphorus deficient diets on the intestinal absorption of calcium and phosphate. Acta Univ. Carol. Med. (Praha) 32 (5-6), 429–433.
Brady, K. T., Haynes, L. F., Hartwell, K. J., and Killeen, T. K. (2013). Substance use disorders and anxiety: a treatment challenge for social workers. Soc. Work Public Health 28 (3-4), 407–423. doi:10.1080/19371918.2013.774675
Bremner, J. D., and McCaffery, P. (2008). The neurobiology of retinoic acid in affective disorders. Prog. Neuropsychopharmacol. Biol. Psychiatry 32 (2), 315–331. doi:10.1016/j.pnpbp.2007.07.001
Chen, X., Jin, D. Y., Stafford, D. W., and Tie, J. K. (2018). Evaluation of oral anticoagulants with vitamin K epoxide reductase in its native milieu. Blood 132 (18), 1974–1984. doi:10.1182/blood-2018-05-846592
Chun, J., Haug, N. A., Guydish, J. R., Sorensen, J. L., and Delucchi, K. (2009). Cigarette smoking among opioid-dependent clients in a therapeutic community. Am. J. Addict. 18 (4), 316–320. doi:10.1080/10550490902925490
Colebatch, A. J., Dobrovic, A., and Cooper, W. A. (2019). TERT gene: its function and dysregulation in cancer. J. Clin. Pathol. 72 (4), 281–284. doi:10.1136/jclinpath-2018-205653
Daly, A. K., and King, B. P. (2006). Contribution of CYP2C9 to variability in vitamin K antagonist metabolism. Expert Opin. Drug Metab. Toxicol. 2 (1), 3–15. doi:10.1517/17425255.2.1.3
Das, K., Daveluy, S., Kroumpouzos, G., Agarwal, K., Podder, I., Farnbach, K., et al. (2022). Efficacy and toxicity of classical immunosuppressants, retinoids and biologics in hidradenitis suppurativa. J. Clin. Med. 11 (3), 670. doi:10.3390/jcm11030670
Eserian, J. K. (2013). Vitamin D as an effective treatment approach for drug abuse and addiction. J. Med. Hypotheses Ideas 7 (2), 35–39. doi:10.1016/j.jmhi.2013.02.001
Fill, M. A., Miller, A. M., Wilkinson, R. H., Warren, M. D., Dunn, J. R., Schaffner, W., et al. (2018). Educational disabilities among children born with neonatal abstinence syndrome. Pediatrics 142 (3), e20180562. doi:10.1542/peds.2018-0562
Garduno, A., and Wu, T. (2021). Tobacco smoke and CYP1A2 activity in a US population with normal liver enzyme levels. Int. J. Environ. Res. Public Health 18 (5), 2225. doi:10.3390/ijerph18052225
Gomez, A., and Ingelman-Sundberg, M. (2009). Pharmacoepigenetics: its role in interindividual differences in drug response. Clin. Pharmacol. Ther. 85 (4), 426–430. doi:10.1038/clpt.2009.2
Gray, T. R., Choo, R. E., Concheiro, M., Williams, E., Elko, A., Jansson, L. M., et al. (2010). Prenatal methadone exposure, meconium biomarker concentrations and neonatal abstinence syndrome. Addiction 105 (12), 2151–2159. doi:10.1111/j.1360-0443.2010.03097.x
Gray, T. R., Kelly, T., LaGasse, L. L., Smith, L. M., Derauf, C., Haning, W., et al. (2009). Novel biomarkers of prenatal methamphetamine exposure in human meconium. Ther. Drug Monit. 31 (1), 70–75. doi:10.1097/FTD.0b013e318195d7cb
Griffin, T. P., Joyce, C. M., Alkanderi, S., Blake, L. M., O'Keeffe, D. T., Bogdanet, D., et al. (2020). Biallelic CYP24A1 variants presenting during pregnancy: clinical and biochemical phenotypes. Endocr. Connect. 9 (6), 530–541. doi:10.1530/EC-20-0150
Hasin, D. S., O'Brien, C. P., Auriacombe, M., Borges, G., Bucholz, K., Budney, A., et al. (2013). DSM-5 criteria for substance use disorders: recommendations and rationale. Am. J. Psychiatry 170 (8), 834–851. doi:10.1176/appi.ajp.2013.12060782
Inci, R., Kelekci, K. H., Oguz, N., Karaca, S., Karadas, B., and Bayrakci, A. (2017). Dermatological aspects of synthetic cannabinoid addiction. Cutan. Ocul. Toxicol. 36 (2), 125–131. doi:10.3109/15569527.2016.1169541
Jilani, S. M., Frey, M. T., Pepin, D., Jewell, T., Jordan, M., Miller, A. M., et al. (2019). Evaluation of state-mandated reporting of neonatal abstinence syndrome - six States, 2013-2017. MMWR Morb. Mortal. Wkly. Rep. 68 (1), 6–10. doi:10.15585/mmwr.mm6801a2
Kazemi, M., Bazyar, M., Naghizadeh, M. M., Dehghan, A., Rahimabadi, M. S., Chijan, M. R., et al. (2021). Lipid profile dysregulation in opium users based on Fasa Persian cohort study results. Sci. Rep. 11 (1), 12058. doi:10.1038/s41598-021-91533-4
Kemeny, L. V., Robinson, K. C., Hermann, A. L., Walker, D. M., Regan, S., Yew, Y. W., et al. (2021). Vitamin D deficiency exacerbates UV/endorphin and opioid addiction. Sci. Adv. 7 (24), eabe4577. doi:10.1126/sciadv.abe4577
Kenworthy, A. (2002). Peering inside lipid rafts and caveolae. Trends Biochem. Sci. 27 (9), 435–437. doi:10.1016/s0968-0004(02)02178-3
Krolick, K. N., Zhu, Q., and Shi, H. (2018). Effects of estrogens on central nervous system neurotransmission: implications for sex differences in mental disorders. Prog. Mol. Biol. Transl. Sci. 160, 105–171. doi:10.1016/bs.pmbts.2018.07.008
Lee, C. W., Muo, C. H., Liang, J. A., Sung, F. C., and Kao, C. H. (2013). Association of intensive morphine treatment and increased stroke incidence in prostate cancer patients: a population-based nested case-control study. Jpn. J. Clin. Oncol. 43 (8), 776–781. doi:10.1093/jjco/hyt080
Lewer, D., Jones, N. R., Hickman, M., Nielsen, S., and Degenhardt, L. (2020). Life expectancy of people who are dependent on opioids: a cohort study in New South Wales, Australia. J. Psychiatr. Res. 130, 435–440. doi:10.1016/j.jpsychires.2020.08.013
Lewinska, M., Zmrzljak, U. P., and Rozman, D. (2013). Low nucleotide variability of CYP51A1 in humans: meta-analysis of cholesterol and bile acid synthesis and xenobiotic metabolism pathways. Acta Chim. Slov. 60 (4), 875–883.
Li, S., Chen, M., Li, Y., and Tollefsbol, T. O. (2019). Prenatal epigenetics diets play protective roles against environmental pollution. Clin. Epigenetics 11 (1), 82. doi:10.1186/s13148-019-0659-4
Li-Hawkins, J., Lund, E. G., Bronson, A. D., and Russell, D. W. (2000). Expression cloning of an oxysterol 7alpha-hydroxylase selective for 24-hydroxycholesterol. J. Biol. Chem. 275 (22), 16543–16549. doi:10.1074/jbc.M001810200
Lin, J. H., and Lu, A. Y. (2001). Interindividual variability in inhibition and induction of cytochrome P450 enzymes. Annu. Rev. Pharmacol. Toxicol. 41, 535–567. doi:10.1146/annurev.pharmtox.41.1.535
Lind, J. N., Interrante, J. D., Ailes, E. C., Gilboa, S. M., Khan, S., Frey, M. T., et al. (2017). Maternal use of opioids during pregnancy and congenital malformations: a systematic review. Pediatrics 139 (6), e20164131. doi:10.1542/peds.2016-4131
Liu, Y., Aryee, M. J., Padyukov, L., Fallin, M. D., Hesselberg, E., Runarsson, A., et al. (2013). Epigenome-wide association data implicate DNA methylation as an intermediary of genetic risk in rheumatoid arthritis. Nat. Biotechnol. 31 (2), 142–147. doi:10.1038/nbt.2487
Logan, B. A., Brown, M. S., and Hayes, M. J. (2013). Neonatal abstinence syndrome: treatment and pediatric outcomes. Clin. Obstet. Gynecol. 56 (1), 186–192. doi:10.1097/GRF.0b013e31827feea4
Lynch, T., and Price, A. (2007). The effect of cytochrome P450 metabolism on drug response, interactions, and adverse effects. Am. Fam. Physician 76 (3), 391–396.
Mactier, H., McLaughlin, P., Gillis, C., and Osselton, M. D. (2017). Variations in infant CYP2B6 genotype associated with the need for pharmacological treatment for neonatal abstinence syndrome in infants of methadone-maintained opioid-dependent mothers. Am. J. Perinatol. 34 (9), 918–921. doi:10.1055/s-0037-1600917
Marcusa, D. P., Mann, R. A., Cron, D. C., Fillinger, B. R., Rzepecki, A. K., Kozlow, J. H., et al. (2017). Prescription opioid use among opioid-naive women undergoing immediate breast reconstruction. Plast. Reconstr. Surg. 140 (6), 1081–1090. doi:10.1097/PRS.0000000000003832
Martinez, L. A., Gross, K. S., Himmler, B. T., Emmitt, N. L., Peterson, B. M., Zlebnik, N. E., et al. (2016). Estradiol facilitation of cocaine self-administration in female rats requires activation of mGluR5. eNeuro 3 (5), ENEURO.0140–16.2016. doi:10.1523/ENEURO.0140-16.2016
Matsuoka, H., Katayama, M., Ohishi, A., Miya, K., Tokunaga, R., Kobayashi, S., et al. (2020). Orphan nuclear receptor RORα regulates enzymatic metabolism of cerebral 24S-hydroxycholesterol through CYP39A1 intronic response element activation. Int. J. Mol. Sci. 21 (9), 3309. doi:10.3390/ijms21093309
McDonnell, A. M., and Dang, C. H. (2013). Basic review of the cytochrome p450 system. J. Adv. Pract. Oncol. 4 (4), 263–268. doi:10.6004/jadpro.2013.4.4.7
Metpally, R. P., Krishnamurthy, S., Moran, K. M., Weller, A. E., Crist, R. C., Reiner, B. C., et al. (2019). The imperative of clinical and molecular research on neonatal opioid withdrawal syndrome. Mol. Psychiatry 24 (11), 1568–1571. doi:10.1038/s41380-019-0522-1
Moon, A. M., Watkins, S. E., Lok, A. S., Firpi-Morell, R. J., Trinh, H. N., Kupec, J. T., et al. (2021). Opioid use is more common in nonalcoholic fatty liver disease patients with cirrhosis, higher BMI, and psychiatric disease. Dig. Dis. 39 (3), 247–257. doi:10.1159/000511074
Moran-Santa Maria, M. M., Flanagan, J., and Brady, K. (2014). Ovarian hormones and drug abuse. Curr. Psychiatry Rep. 16 (11), 511. doi:10.1007/s11920-014-0511-7
Murray, G. I., Patimalla, S., Stewart, K. N., Miller, I. D., and Heys, S. D. (2010). Profiling the expression of cytochrome P450 in breast cancer. Histopathology 57 (2), 202–211. doi:10.1111/j.1365-2559.2010.03606.x
Nebert, D. W., and McKinnon, R. A. (1994). Cytochrome P450: evolution and functional diversity. Prog. Liver Dis. 12, 63–97.
Nelson, A. D., and Camilleri, M. (2015). Chronic opioid induced constipation in patients with nonmalignant pain: challenges and opportunities. Ther. Adv. Gastroenterol. 8 (4), 206–220. doi:10.1177/1756283X15578608
Norman, A. W., Ishizuka, S., and Okamura, W. H. (2001). Ligands for the vitamin D endocrine system: different shapes function as agonists and antagonists for genomic and rapid response receptors or as a ligand for the plasma vitamin D binding protein. J. Steroid Biochem. Mol. Biol. 76 (1-5), 49–59. doi:10.1016/s0960-0760(00)00145-x
Oei, J. L., Melhuish, E., Uebel, H., Azzam, N., Breen, C., Burns, L., et al. (2017). Neonatal abstinence syndrome and high school performance. Pediatrics 139 (2), e20162651. doi:10.1542/peds.2016-2651
Patrick, S. W., Barfield, W. D., Poindexter, B. B., Committee On, F., Newborn, C. O. S. U., and Prevention, (2020). Neonatal opioid withdrawal syndrome. Pediatrics 146 (5), e2020029074. doi:10.1542/peds.2020-029074
Prosser, D. E., and Jones, G. (2004). Enzymes involved in the activation and inactivation of vitamin D. Trends Biochem. Sci. 29 (12), 664–673. doi:10.1016/j.tibs.2004.10.005
Radhakrishna, U., Nath, S. K., Uppala, L. V., Veerappa, A., Forray, A., Muvvala, S. B., et al. (2023). Placental microRNA methylome signatures may serve as biomarkers and therapeutic targets for prenatally opioid-exposed infants with neonatal opioid withdrawal syndrome. Front. Genet. 14, 1215472. doi:10.3389/fgene.2023.1215472
Radhakrishna, U., Nath, S. K., Vishweswaraiah, S., Uppala, L. V., Forray, A., Muvvala, S. B., et al. (2021a). Maternal opioid use disorder: placental transcriptome analysis for neonatal opioid withdrawal syndrome. Genomics 113 (6), 3610–3617. doi:10.1016/j.ygeno.2021.08.001
Radhakrishna, U., Vishweswaraiah, S., Uppala, L. V., Szymanska, M., Macknis, J., Kumar, S., et al. (2021b). Placental DNA methylation profiles in opioid-exposed pregnancies and associations with the neonatal opioid withdrawal syndrome. Genomics 113 (3), 1127–1135. doi:10.1016/j.ygeno.2021.03.006
Radhakrishnan, R., Brown, B. P., Haas, D. M., Zang, Y., Sparks, C., and Sadhasivam, S. (2022a). Pilot study of fetal brain development and morphometry in prenatal opioid exposure and smoking on fetal MRI. J. Neuroradiol. 49 (1), 53–58. doi:10.1016/j.neurad.2020.12.004
Radhakrishnan, R., Elsaid, N. M. H., Sadhasivam, S., Reher, T. A., Hines, A. C., Yoder, K. K., et al. (2021). Resting state functional MRI in infants with prenatal opioid exposure-a pilot study. Neuroradiology 63 (4), 585–591. doi:10.1007/s00234-020-02552-3
Radhakrishnan, R., Vishnubhotla, R. V., Guckien, Z., Zhao, Y., Sokol, G. M., Haas, D. M., et al. (2022b). Thalamocortical functional connectivity in infants with prenatal opioid exposure correlates with severity of neonatal opioid withdrawal syndrome. Neuroradiology 64 (8), 1649–1659. doi:10.1007/s00234-022-02939-4
Radhakrishnan, R., Vishnubhotla, R. V., Zhao, Y., Yan, J., He, B., Steinhardt, N., et al. (2022c). Global brain functional network connectivity in infants with prenatal opioid exposure. Front. Pediatr. 10, 847037. doi:10.3389/fped.2022.847037
Rezen, T., Debeljak, N., Kordis, D., and Rozman, D. (2004). New aspects on lanosterol 14alpha-demethylase and cytochrome P450 evolution: lanosterol/cycloartenol diversification and lateral transfer. J. Mol. Evol. 59 (1), 51–58. doi:10.1007/s00239-004-2603-1
Ross, S. A., McCaffery, P. J., Drager, U. C., and De Luca, L. M. (2000). Retinoids in embryonal development. Physiol. Rev. 80 (3), 1021–1054. doi:10.1152/physrev.2000.80.3.1021
Rostami-Hodjegan, A., Amin, A. M., Spencer, E. P., Lennard, M. S., Tucker, G. T., and Flanagan, R. J. (2004). Influence of dose, cigarette smoking, age, sex, and metabolic activity on plasma clozapine concentrations: a predictive model and nomograms to aid clozapine dose adjustment and to assess compliance in individual patients. J. Clin. Psychopharmacol. 24 (1), 70–78. doi:10.1097/01.jcp.0000106221.36344.4d
Savas, U., Hsu, M. H., Griffin, K. J., Bell, D. R., and Johnson, E. F. (2005). Conditional regulation of the human CYP4X1 and CYP4Z1 genes. Arch. Biochem. Biophys. 436 (2), 377–385. doi:10.1016/j.abb.2005.02.022
Schuebel, K., Gitik, M., Domschke, K., and Goldman, D. (2016). Making sense of epigenetics. Int. J. Neuropsychopharmacol. 19 (11), pyw058. doi:10.1093/ijnp/pyw058
Sheikh, I. A., Ahmad, E., Jamal, M. S., Rehan, M., Assidi, M., Tayubi, I. A., et al. (2016a). Spontaneous preterm birth and single nucleotide gene polymorphisms: a recent update. BMC Genomics 17 (9), 759. doi:10.1186/s12864-016-3089-0
Sheikh, I. A., Ahmad, E., Jamal, M. S., Rehan, M., Assidi, M., Tayubi, I. A., et al. (2016b). Spontaneous preterm birth and single nucleotide gene polymorphisms: a recent update. BMC Genomics 17 (Suppl. 9), 759. doi:10.1186/s12864-016-3089-0
Smith, H. S. (2009). Opioid metabolism. Mayo Clin. Proc. 84 (7), 613–624. doi:10.1016/S0025-6196(11)60750-7
Smyth, B., Hoffman, V., Fan, J., and Hser, Y. I. (2007). Years of potential life lost among heroin addicts 33 years after treatment. Prev. Med. 44 (4), 369–374. doi:10.1016/j.ypmed.2006.10.003
Spreen, L. A., Dittmar, E. N., Quirk, K. C., and Smith, M. A. (2022). Buprenorphine initiation strategies for opioid use disorder and pain management: a systematic review. Pharmacotherapy 42, 411–427. doi:10.1002/phar.2676
Stiles, A. R., Kozlitina, J., Thompson, B. M., McDonald, J. G., King, K. S., and Russell, D. W. (2014). Genetic, anatomic, and clinical determinants of human serum sterol and vitamin D levels. Proc. Natl. Acad. Sci. U. S. A. 111 (38), E4006–E4014. doi:10.1073/pnas.1413561111
Szymanski, L., Skopek, R., Palusinska, M., Schenk, T., Stengel, S., Lewicki, S., et al. (2020). Retinoic acid and its derivatives in skin. Cells 9 (12), 2660. doi:10.3390/cells9122660
Taşçıoğlu, N., Saatçi, Ç., Emekli, R., Tuncel, G., Eşel, E., and Dundar, M. (2021a). Investigation of cytochrome p450 CYP1A2, CYP2D6, CYP2E1 and CYP3A4 gene expres-sions and polymorphisms in alcohol with-drawal. Turk. J. Clin. Psychiatry 24, 298–306. doi:10.5505/kpd.2021.60938
Taşçıoğlu, N., Saatçi, Ç., Emekli, R., Tuncel, G., Eşel, E., and Dundar, M. (2021b). Investigation of cytochrome p450 CYP1A2, CYP2D6, CYP2E1 and CYP3A4 gene expressions and polymorphisms in alcohol withdrawal (eng). J. Clin. Psy 24 (3), 298–306. doi:10.5505/kpd.2021.60938
Thatcher, J. E., and Isoherranen, N. (2009). The role of CYP26 enzymes in retinoic acid clearance. Expert Opin. Drug Metab. Toxicol. 5 (8), 875–886. doi:10.1517/17425250903032681
Trybek, T., Kowalik, A., Góźdź, S., and Kowalska, A. (2020). Telomeres and telomerase in oncogenesis. Oncol. Lett. 20 (2), 1015–1027. doi:10.3892/ol.2020.11659
Tsuchiya, Y., Nakajima, M., Kyo, S., Kanaya, T., Inoue, M., and Yokoi, T. (2004). Human CYP1B1 is regulated by estradiol via estrogen receptor. Cancer Res. 64 (9), 3119–3125. doi:10.1158/0008-5472.can-04-0166
Vishnubhotla, R. V., Zhao, Y., Wen, Q., Dietrich, J., Sokol, G. M., Sadhasivam, S., et al. (2022). Brain structural connectome in neonates with prenatal opioid exposure. Front. Neurosci. 16, 952322. doi:10.3389/fnins.2022.952322
Vishweswaraiah, S., Swierkowska, J., Ratnamala, U., Mishra, N. K., Guda, C., Chettiar, S. S., et al. (2019). Epigenetically dysregulated genes and pathways implicated in the pathogenesis of non-syndromic high myopia. Sci. Rep. 9 (1), 4145. doi:10.1038/s41598-019-40299-x
Weller, A. E., Crist, R. C., Reiner, B. C., Doyle, G. A., and Berrettini, W. H. (2021). Neonatal opioid withdrawal syndrome (NOWS): a transgenerational echo of the opioid crisis. Cold Spring Harb. Perspect. Med. 11 (3), a039669. doi:10.1101/cshperspect.a039669
Wilhelm-Benartzi, C. S., Koestler, D. C., Karagas, M. R., Flanagan, J. M., Christensen, B. C., Kelsey, K. T., et al. (2013). Review of processing and analysis methods for DNA methylation array data. Br. J. Cancer 109 (6), 1394–1402. doi:10.1038/bjc.2013.496
Wise, R. L., Brown, B. P., Haas, D. M., Sparks, C., Sadhasivam, S., Zhao, Y., et al. (2023). Placental volume in pregnant women with opioid use: prenatal MRI assessment. J. Matern. Fetal Neonatal Med. 36 (1), 2157256. doi:10.1080/14767058.2022.2157256
Yazdy, M. M., Desai, R. J., and Brogly, S. B. (2015). Prescription opioids in pregnancy and birth outcomes: a review of the literature. J. Pediatr. Genet. 4 (2), 56–70. doi:10.1055/s-0035-1556740
Young-Wolff, K. C., Klebaner, D., Weisner, C., Von Korff, M., and Campbell, C. I. (2017). Smoking status and opioid-related problems and concerns among men and women on chronic opioid therapy. Clin. J. Pain 33 (8), 730–737. doi:10.1097/AJP.0000000000000461
Yun, C. H., Park, H. J., Kim, S. J., and Kim, H. K. (1998). Identification of cytochrome P450 1A1 in human brain. Biochem. Biophys. Res. Commun. 243 (3), 808–810. doi:10.1006/bbrc.1998.8171
Zanger, U. M., and Schwab, M. (2013). Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol. Ther. 138 (1), 103–141. doi:10.1016/j.pharmthera.2012.12.007
Zhang, C., Wang, L., Chen, L., Ren, W., Mei, A., Chen, X., et al. (2013). Two novel mutations of the NCSTN gene in Chinese familial acne inverse. J. Eur. Acad. Dermatol Venereol. 27 (12), 1571–1574. doi:10.1111/j.1468-3083.2012.04627.x
Zhang, H., Yang, Q., Zheng, W., Ouyang, Y., Yang, M., Wang, F., et al. (2016). CYP gene family variants as potential protective factors in drug addiction in Han Chinese. J. Gene Med. 18 (8), 147–153. doi:10.1002/jgm.2887
Keywords: cytochromes, biomarker, opioid use, neonatal opioid withdrawal syndrome CYP19A1, CYP1A2, CYP4V2, CYP1B1, CYP24A1
Citation: Radhakrishna U, Sadhasivam S, Radhakrishnan R, Forray A, Muvvala SB, Metpally RP, Patel S, Rawal RM, Vishweswaraiah S, Bahado-Singh RO and Nath SK (2024) Placental cytochrome P450 methylomes in infants exposed to prenatal opioids: exploring the effects of neonatal opioid withdrawal syndrome on health horizons. Front. Genet. 14:1292148. doi: 10.3389/fgene.2023.1292148
Received: 11 September 2023; Accepted: 24 November 2023;
Published: 04 January 2024.
Edited by:
Anupam Basu, The University of Burdwan, IndiaReviewed by:
Bicheng Yang, Jiangxi Maternal and Child Health Hospital, ChinaValentina Pilipenko, Cincinnati Children’s Hospital Medical Center, United States
Copyright © 2024 Radhakrishna, Sadhasivam, Radhakrishnan, Forray, Muvvala, Metpally, Patel, Rawal, Vishweswaraiah, Bahado-Singh and Nath. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Uppala Radhakrishna, dXBwYWxhckBwaXR0LmVkdQ==
 Uppala Radhakrishna
Uppala Radhakrishna Senthilkumar Sadhasivam1
Senthilkumar Sadhasivam1 Rupa Radhakrishnan
Rupa Radhakrishnan Ariadna Forray
Ariadna Forray Srinivas B. Muvvala
Srinivas B. Muvvala Raghu P. Metpally
Raghu P. Metpally Rakesh M. Rawal
Rakesh M. Rawal Sangeetha Vishweswaraiah
Sangeetha Vishweswaraiah Swapan K. Nath
Swapan K. Nath