- 1College of Life Science and Technology, Tarim University, Alar, Xinjiang, China
- 2Key Laboratory of Utilization of Livestock and Forage Resources in Circum-Tarim Region, Ministry of Agriculture and Rural Affairs, Tarim University, Alar, Xinjiang, China
- 3College of Animal Science and Technology, Tarim University, Alar, Xinjiang, China
- 4Key Laboratory of Tarim Animal Husbandry Science and Technology, Xinjiang Production and Construction Corps, Alar, Xinjiang, China
Local sheep in the northeastern Tarim Basin can adapt to dry and low-rainfall regional environments. In this study, three local sheep breeds in the northeastern Tarim Basin, LOP (LOP) sheep, Bayinbuluke (BYK) sheep, and Kunlun (KUN, also known as the Qiemo sheep) sheep, and three introduced sheep breeds, Suffolk (SUF) sheep, Dorset (APD) sheep, and Texel (TEX) sheep, were analyzed for genetic diversity, population structure, and selective signature using the Illumina OvineSNP50K BeadChip. We found that LOP, BYK, and KUN had lower observed heterozygosity and expected heterozygosity than TEX, SUF, and ADP, which were differentiated based on geographic distribution. We performed fixation index (FST) analysis on three local sheep breeds in the northeastern Tarim Basin (LOP, BYK, and KUN) and introduced sheep breeds (TEX, SUF, and ADP) to measure genetic differentiation. Nucleotide diversity (PI) analysis was performed on single-nucleotide polymorphism (SNP) data of LOP, BYK, and KUN. A total of 493 candidate genes were obtained by taking the intersection at a threshold of 5%. Among them, SMAD2, ESR2, and HAS2 were related to reproductive traits. PCDH15, TLE4, and TFAP2B were related to growth traits. SOD1, TSHR, and DNAJB5 were related to desert environmental adaptation. Analyzing the genetic patterns of local sheep in the northeastern Tarim Basin can protect the germplasm resources of local sheep and promote the development and utilization of sheep genetic resources.
1 Introduction
Sheep were domesticated in the Fertile Crescent approximately 11,000 years ago to provide humans with meat, milk, skin, and wool (Zeder, 2008). Adaptation to different agricultural environments and various production goals produced genetic variation in different directions (Kijas et al., 2012). Genome-wide genetic variation markers allow selective intervention in breeding and evaluation at an early stage with great accuracy so that good production traits in sheep can be better transmitted to their offspring (Hayes et al., 2013). Through genome sequencing, Rupp et al. (2015) discovered that SOCS2 is mediated by the JAK/STAT signaling pathway, which affects inflammatory responses, growth, and milk production. Zhao et al. (2022) identified that the genes HBB, PRDX2, GPX1, GSTA1, COL14A1, and LTBP4 contributed to the adaptation of Tibetan sheep to the hypoxic environment of the plateau. Li et al. (2014) identified genes (TYRP, ASIP, and MITF) associated with sheep coat color using a combination of whole-genome association analysis and selective scanning. The study was conducted on 99 Finnsheep with different coat colors (white, black, gray, and brown). Xu et al. (2023) modeled a regulatory network regulating adipose homeostasis in the tail of sheep using a genome-wide selective signature and obtained adipocyte-associated genes, namely, BMP2, PDGFD, and VEGFA. Zhao et al. (2020) screened 73 candidate genes associated with tail fat formation, reproduction, and growth traits using three local Chinese sheep breeds based on the selective signature using the Ovine Infinium HD SNP BeadChip. The Illumina OvineSNP50K BeadChip has been widely used to study genetic diversity and the development of economically important traits in sheep. Ghoreishifar et al. (2019) studied that the genetic diversity of Iranian Zandi sheep is relatively low. Baazaoui et al. (2021) identified candidate genes, such as CDS2, PROKR1, and BMP2, associated with tail fat in sheep from semi-arid regions through the reduction of heterozygosity (ROH) and fixation index (FST) analysis.
The northeastern part of the Tarim Basin, located around the Taklamakan Desert, is characterized by significant temperature fluctuations, long sunlight exposure, low rainfall, and complex terrain (Sweet-Jones et al., 2021). Under these unique geographical and cultural conditions, excellent local breeds (LOP, BYK, and KUN) have emerged. These breeds exhibit traits such as resistance to stress and disease, growth, and reproduction (such as estrus period, ovulation rate, mating conception rate, birth weight of lambs, vitality, and survival rate) even in extreme desert environments, making them highly valuable for research purposes. In this study, genomic selection markers were used to detect genomic differences between three local sheep breeds (LOP, BYK, and KUN) in the northeastern part of the Tarim Basin and three introduced sheep breeds (SUF, APD, and TEX). The aim of this study was to explore the genetic diversity and population structure of local sheep in the Tarim Basin and identify molecular markers that are well suited for adaptation to desert environments. This research reveals the genetic imprints left in the sheep genome, providing a theoretical basis for the conservation and development of local sheep in the Tarim Basin.
2 Materials and methods
2.1 Animal care
This work was conducted following the specifications of the Ethics Committee of the Tarim University of Science and Technology (SYXK 2020-009).
2.2 Animal collection
A total of 465 samples from six sheep breeds were selected for this study. Among them, 62 samples were collected from local sheep in the northeastern Tarim Basin, including 27 KUN samples (from the conservation farm in Zhema County, Bayinbuluke Region, Xinjiang), 20 BYK samples (from the conservation farm in Bayinbuluke Region, Xinjiang), and 15 LOP samples (from the conservation farm in Bayinbuluke, Bayinbuluke Region, Xinjiang), and 403 samples were collected from the International Sheep Genomics Consortium (ISGC) (http://www.Sheephapmap.org), including 152 SUF samples from Australia and Ireland, 148 TEX samples from New Zealand, Germany, and Scotland, and 103 APD samples from Australia.
2.3 Genotyping and data quality control
Blood samples were collected using blood collection tubes containing EDTAK2, and DNA samples were extracted using a DNA kit (TIANGEN, Beijing, China). The DNA samples were genotyped using the Illumina OvineSNP50 BeadChip at Beijing Compassion Agricultural Science and Technology Co. Quality control of single-nucleotide polymorphism (SNP) genotype data was performed using PLINK v.1.9 to remove unqualified SNPs from the samples (individual detection rate > 0.95, SNP detection rate > 0.95, Hardy–Weinberg equilibrium (HWE) p-value ≥ 10−6, minor allele frequency (MAF) < 5%, and call rate < 90%).
2.4 Genetic diversity and population structure
PLINK v.1.90 (Purcell et al., 2007) was used to calculate the observed heterozygosity (HO), expected heterozygosity (He), and inbreeding coefficient (F). After quality control, principal component analysis (PCA) was conducted on the SNP data. The genetic distances of the six sheep breeds were calculated to observe the clustering of the samples and explore the effects of genetic differences and geographic differences on the populations. VCF2Dis (https://github.com/BGI-shenzhen/VCF2Dis) was used to calculate the P-distance matrix. Based on this matrix, the neighbor-joining tree (NJ tree) phylogenetic relationships were constructed using ATGC: FastME. Such phylogenetic relationships were visualized and analyzed using the iTOL tool (Letunic and Bork, 2016). Ancestral component analysis was performed using ADMIXTURE (Patters et al., 2012), where population stratification can be calculated based on genome-wide genetic variation and the proportion of the genome accounted for by the variation in each individual from each of the K ancestors.
2.5 Selective signature
In this study, FST analysis was performed on the SNP data of the three local sheep in the northeastern Tarim Basin (LOP, BYK, and KUN) and introduced sheep breeds (TEX, SUF, and ADP) (in sliding 50-kb windows with 20-kb steps). The FST analysis was performed using VCFtools 0.1.13 (Danecek et al., 2011; Shen et al., 2020). In each comparison, the top 5% genomic regions with the highest scores overlapped were considered to be potential selective signatures.
Here, MSP represents the observed mean square errors for the three local sheep in the northeastern Tarim Basin (LOP, BYK, and KUN) and introduced sheep breeds (TEX, SUF, and ADP), which is calculated as follows:
MSG represents the observed mean square errors for loci within sheep populations, which is calculated as
In the aforementioned equations, i is the subpopulations (where i = 1,···, s), ni is the sample size in subpopulation i, PAi is the frequency of the SNP allele A in the ith subpopulation, and nc is the mean sample size across samples (Nosrati et al., 2019).
Nucleotide diversity (PI) analysis is an essential measure of population diversity. PI was calculated separately for the LOP, BYK, and KUN sheep populations. Finally, the 5% high-ranking values were taken in ascending order to intersect with the FST results. The loci were annotated concerning the sheep genome Ovis Oar_v4.0 (http://www.ncbi.nlm.nih.gov/gene). Gene function annotations were prepared using the NCBI and OMIM databases (http://www.ncbi.nlm.nih.gov/omim).
where S denotes the number of segregating loci and hj denotes the heterozygosity of the j segregating locus.
2.6 Enrichment analysis for candidate genes
First, PI and FST results were selected for crossover analysis and subsequently annotated with the sheep genome Ovis Oar_v4.0 (Conway et al., 2017). Finally, Gene Ontology (GO) enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis were performed on the crossover genes using g:Profiler (https://biit.cs.ut.ee/). Among them, GO enrichment was used to predict and elucidate gene products in terms of molecular functions, biological processes, and cellular components (Ashburner et al., 2000). KEGG pathway enrichment was used to identify the major biochemical and signaling pathways in which genes are involved (Kanehisa et al., 2016).
3 Results
3.1 SNP statistics
A total of 493 individuals and 48,889 SNPs were selected for downstream analysis. Among them, there were 48,198 informative SNPs in the BYK breed, 47,530 informative SNPs in the KUN breed, 48,001 informative SNPs in the LOP breed, 49,873 informative SNPs in the SUF breed, 49,766 informative SNPs in the TEX breed, and 48,795 informative SNPs in the APD breed.
3.2 Genetic diversity
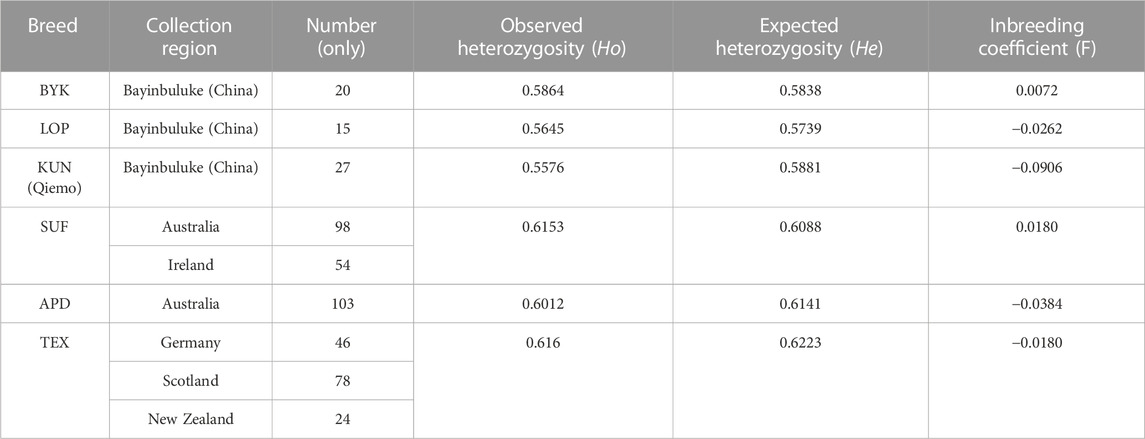
Genetic diversity analysis (Table 1) revealed that the three local sheep breeds in the northeastern Tarim Basin, KUN, BYK, and LOP, showed inbreeding coefficients (F) ranging from −0.0906 (KUN) to 0.0072 (BYK), with an average value of −0.0365. Ho ranged from 0.5576 (KUN) to 0.5864 (BYK), with a mean value of 0.5695. He ranged from 0.5739 (LOP) to 0.5881 (KUN), with a mean value of 0.5819. F of sheep breeds in other countries ranged from −0.0384 (APD) to 0.0180 (SUF), with a mean value of −0.0128. Ho ranged from 0.6012 (APD) to 0.6160 (TEX), with a mean value of 0.6108. He ranged from 0.6088 (SUF) to 0.6223 (TEX), with a mean value of 0.6151.
3.3 Population structure
The SNP data of 465 sheep were subjected to PCA (Figure 1). The results showed that six sheep breeds were categorized into four subgroups, in which Xinjiang local sheep BYK, KUN, and LOP were clustered together. BTK, LOP and KUN were partially mixed, and KUN extend outwards. Considering the evolutionary process among varieties, we further analyzed the population structure of different populations using ADMIXTURE software (Figure 2). When K = 2–6, LOP, BYK, and KUN sheep all had similar ancestral components, unlike TEX, SUF, and ADP. When K = 6, KUN had a more independent ancestral component, and the BYK and LOP ancestral components were more similar. When K = 6, the Cross Validation (CV) value is the lowest, which can best explain the mixed results. We constructed an NJ tree based on the six sheep breeds (Figure 3). The NJ tree could distinguish between local sheep in the northeastern Tarim Basin and TEX, SUF, and ADP based on the geographic distance, and the results were consistent with the PCA results.
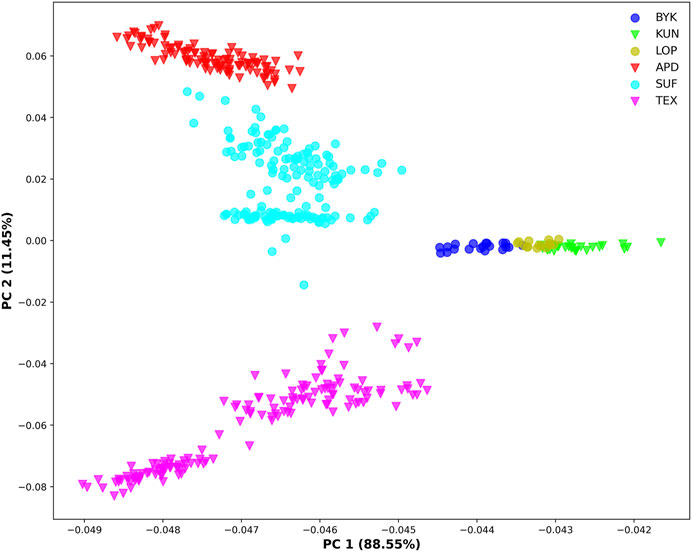
FIGURE 1. Principal component analysis of six sheep breeds (X-axis represents PC1, and Y-axis represents PC2).
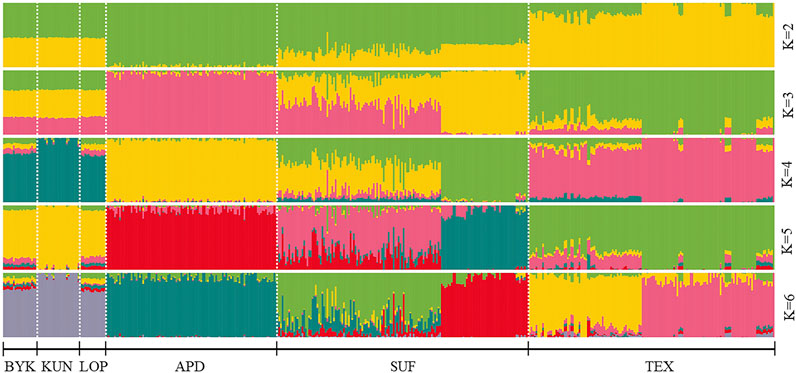
FIGURE 2. ADMIXTURE analysis of six sheep breeds. The results of the inferred numbers of clusters K = 2–6 are shown. Distinct colors represent different ancestral components.
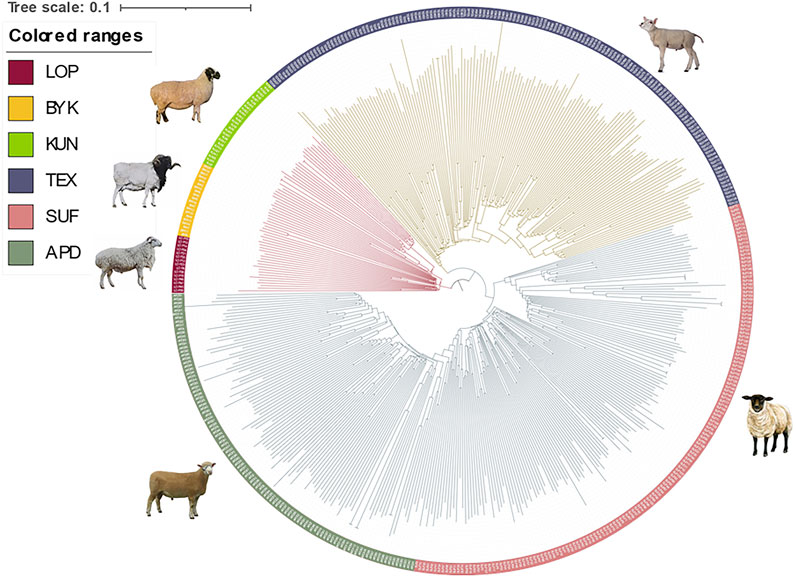
FIGURE 3. Results of the NJ tree of six sheep breeds. The indigenous sheep breeds in Xinjiang could be distinguished from those in other countries, and the relationships between BYK and LOP were the closest.
3.4 Selective signature
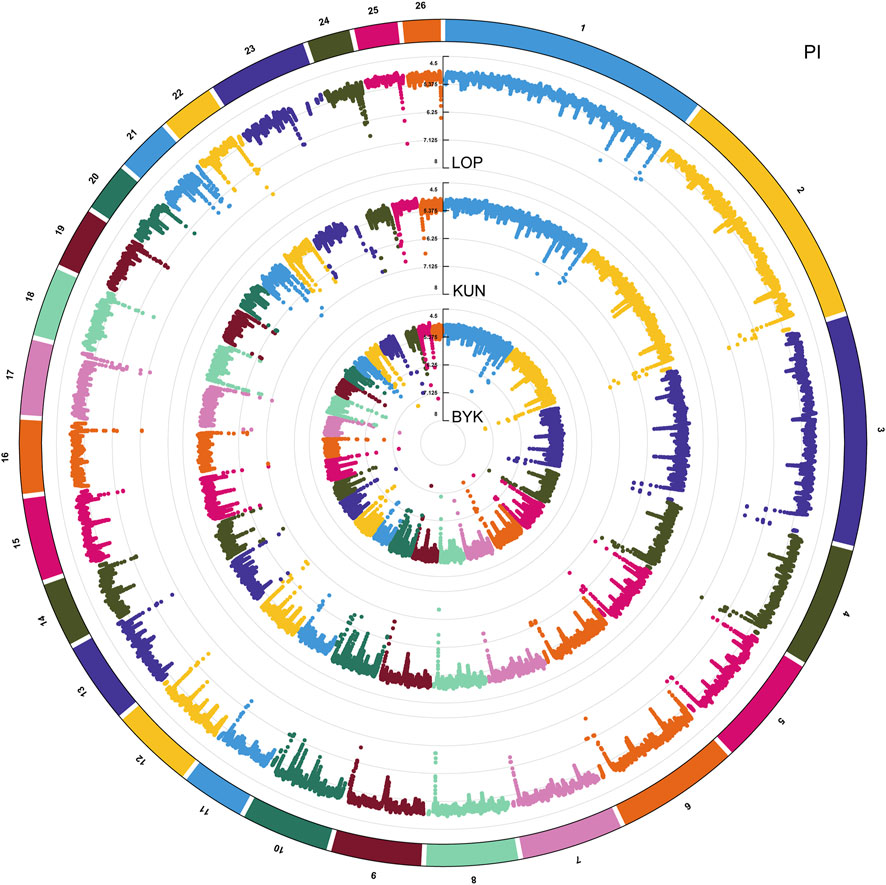
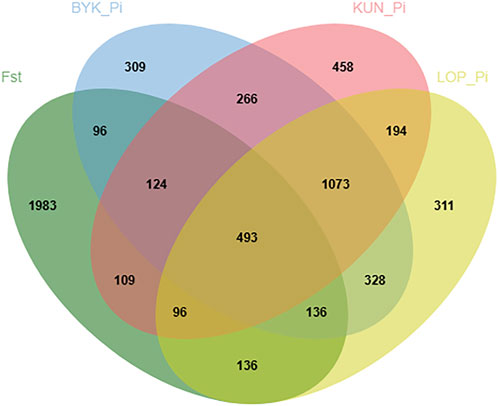
According to the descending order of FST values, the 5% high-ranking values were regarded as the selection region, and a total of 3,173 genes were obtained after FST screening (Figure 4; Supplementary Table S1). The PI values were in ascending order, and the 5% high-ranking values were regarded as the selection region. The LOP, BYK, and KUN PI values screened 2,825, 2,767, and 2,813 candidate genes, respectively (Figure 5; Supplementary Tables S2–S4). After the deletion of duplicate genes, the genes screened by PI and FST were taken for intersection, and a total of 493 genes were obtained (Figure 6). These genes include SMAD2, ESR2, and HAS2, which are related to reproductive traits. PCDH15, TLE4, and TFAP2B are related to growth traits. SOD1, TSHR, and DNAJB5 are related to desert environmental adaptation.
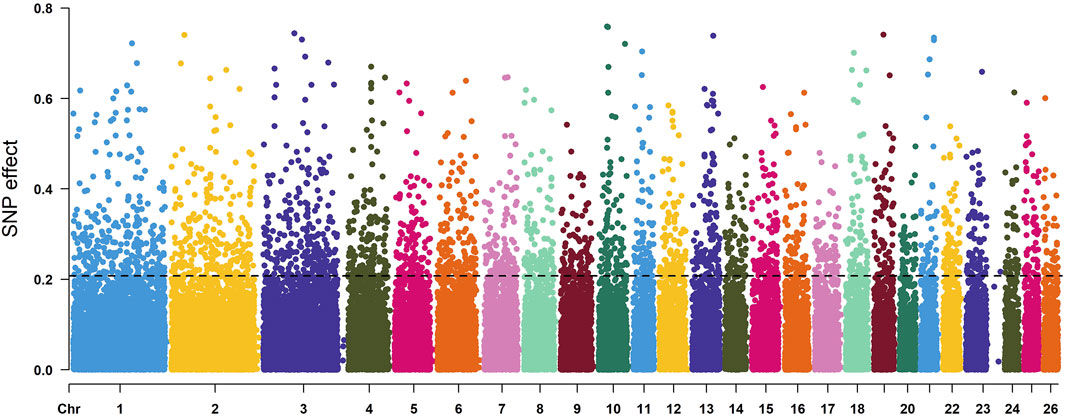
FIGURE 4. FST detection results for six sheep breeds. The black line represents the 5% threshold line.
3.5 Enrichment analysis for candidate genes
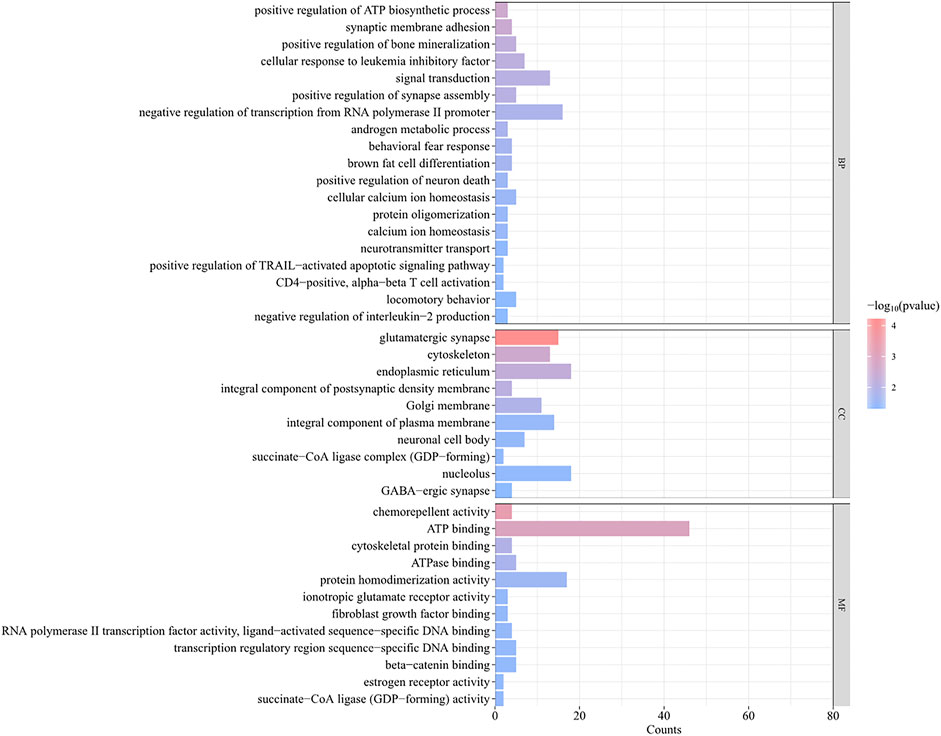
A total of 493 candidate genes were analyzed for GO enrichment and KEGG pathway analyses. GO analysis revealed that 20 genes were enriched in biological processes, 12 genes were enriched in molecular functions, and 10 genes were enriched in cellular components (Figure 7; Supplementary Table S5), including the positive regulation of ATP biosynthetic process, GABAergic synapse, negative regulation of interleukin-2 production, CD4-positive, alpha–beta T-cell activation, negative regulation of transcription from the RNA polymerase II promoter, and chemorepellent activity. The KEGG pathway analysis revealed that eight pathways (Table 2), including metabolic pathways, axon guidance, and cell adhesion molecules, were enriched.
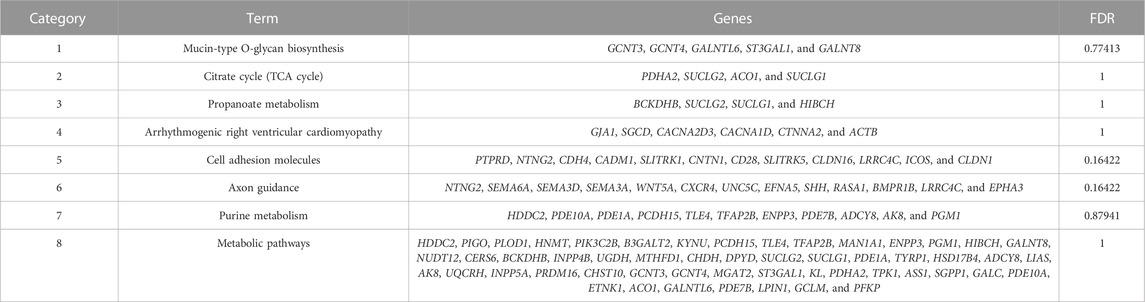
TABLE 2. Details of the enrichment pathways of 493 genes obtained by the intersection of FST and PI results.
4 Discussion
4.1 Genetic diversity and population structure
We selected the genetic patterns of LOP, BYK, and KUN for analysis to conserve and develop the genetic resources of indigenous sheep germplasms in the northeastern margin of the Tarim Basin. The results showed that the inbreeding coefficients and heterozygosity of the three indigenous sheep in the northeastern margin of the Tarim Basin were lower than those of TEX, SUF, and ADP. The reason for this was that the indigenous sheep lived in the hinterland of the desert environment, where transportation was inconvenient and there was little communication with the outside world. Introducing other breeds was more expensive, leading to less crossbreeding between other sheep breeds and the indigenous sheep of northeastern Xinjiang, resulting in a low degree of heterozygosity. Therefore, we suggest that, in the subsequent breeding of breeds, the local area needs to develop a scientific breeding plan to introduce other good genes to improve genetic diversity. Population structure analysis showed that Xinjiang native sheep were genetically distant from other sheep. The three native sheep in the northeastern margin of the Tarim Basin were close to each other and separated from TEX, SUF, and ADP. The main reason was that LOP, BYK, and KUN lived in desert environments and had long been geographically distant from the introduced sheep breeds. There was a genetic isolation phenomenon, and there were also breed protection effects. PCA and NJ trees could differentiate sheep breeds of different regions according to geographic distances. The results of ADMIXTURE showed that when K = 4–6, BYK and LOP sheep in the northeastern edge of the Tarim Basin are more closely related, mainly because they have similar ancestral components and genetic distances. This result is consistent with the findings of PCA and the evolutionary tree.
4.2 Selection sweep methods
The genome-selective sweeping method detects the origin evolution and gene exchange of species (Li et al., 2020) and explores the genetic mechanism of livestock in environmental change. Different sheep breeds have different molecular genetic markers, and molecular markers with specific significance can be obtained using genome-selective sweeping methods. Native sheep in the northeastern margin of the Tarim Basin have environmental adaptations different from those of sheep in other regions living under extreme desert conditions, and their outstanding advantage over other sheep is their resistance to hot temperatures, humidity, disease, parasites, and food shortages. In this study, SNP data of three indigenous sheep breeds in the northeastern Tarim Basin (BYK, LOP, and KUN) and introduced sheep breeds (SUF, APD, and TEX) were analyzed using the genome-selective scanning (GSS) method. PI analysis was performed on BYK, LOP, and KUN. A total of 493 genes were obtained under 5% threshold conditions, and the biology of these genes was revealed. The enrichment results showed genetic evidence and physiological mechanisms for the adaptation of sheep to desert environments in the northeastern margin of the Tarim Basin. The following section discusses only the leading candidate genes and their potential roles in desert adaptation and immune and reproductive traits.
4.3 Reproduction-related genes
The reproduction-related trait is one of the most important economic traits in sheep, including follicular development, ovulation, fertilization, embryonic development, and other physiological processes regulated by several factors and genes, and its underlying mechanism is very complex. Sheep from the northeastern margin of the Tarim Basin have undergone natural and artificial selection in the Taklamakan Desert to develop a stable reproductive capacity. Through selective scanning analysis, we identified some candidate genes related to reproduction, including SMAD2, ESR2, HAS2, DMC1, GRM1, and TSHR. SMAD2 is an important transcription factor downstream of the TGF-β/Smad pathway and is highly expressed in uterine and ovarian tissues. SMAD2 mediates various physiological processes and is a significant candidate for influencing litter size traits in Tibetan sheep. Genes can be used as molecular genetic markers to assist in selecting improved reproductive characteristics in the initial stages of sheep breeding (Li et al., 2022). Maintaining the survival of dilated oocytes through SMAD2 improves the fertilization and subsequent development of closed oocytes during oocyte–oocyte complex (COC) maturation, thereby enhancing maternal reproductive performance (Wu et al., 2021). Estrogen receptor 2 (ESR2) is an essential gene involved in the expression of estrogen receptors, which is expressed in the hypothalamus–pituitary–ovary axis of sheep. The hypothalamus–pituitary–ovarian axis and its expression in single-lamb populations were higher than those in multi-lamb populations. Based on the dose effect of ESR2, it was hypothesized that overexpression of the ESR2 gene in pituitary tissues inhibits LH release, affects follicle development and maturation, and promotes reproductive performance in sheep (Sánchez-Criado et al., 2012; Khristi et al., 2018; Xu et al., 2018). Disrupted meiotic cDNA1 (DMC1) controls ovarian meiosis and follicular development in sheep (Mandon-Pépin et al., 2003), and hyaluronan synthase 2 (HAS2) is present in dominant follicles in mammals and affects the expansion of the oocyte mound cells to promote oocyte maturation (Zhang et al., 2018). Native sheep utilize these genes, directly and indirectly, to increase reproductive rates and achieve multiple fecundities, which is essential for population continuation in harsh environments. Our study is consistent with that of Luna-Nevárez et al. (2021) who found that the thyroid-stimulating hormone receptor (TSHR) gene affects heat tolerance in pregnant Columbia–Rambouillet ewes in a desert environment (Luna-Nevárez et al., 2021). However, there are also unique genetic mechanisms for reproductive traits in native sheep in the Tarim Basin. Liu et al. (2022) identified the TSHR gene as a candidate gene for litter size in twin-bearing Pishan sheep in Xinjiang. Ming Li et al. (2023) suggested that the 29-bp nucleotide sequence variation within the TSHR gene may be a candidate for improving reproductive traits in sheep through marker-assisted selection (MAS). Zhu et al. (2022) also suggested that GRM1 was the dominant genotype for litter size in Kazakh, Chinese Merino, and Lake sheep and was significantly associated with lambing traits. Native sheep have evolved a more unique and stable reproductive system to ensure pregnancy stability in harsh desert environments.
4.4 Fat- and growth-related genes
We also identified some genes related to lipid metabolism, which has been reported to be essential for maintaining energy balance in thermoregulatory responses and is a critical biological process in the immune system under heat stress (Das et al., 2016). Sheep exposure to hyperthermia could regulate body temperature more efficiently and survive heat stress (Romero et al., 2013). Several KEGG pathways of interest related to energy homeostasis and lipid metabolism were significantly enriched in our study, such as energy metabolism and the tricarboxylic acid cycle. These pathways include CERS6, SUCLG2, MGAT2, PRDM16, and PPARGC1A genes. Ceramide synthase 6 (CERS6) alleviates insulin resistance, increases systemic energy expenditure, and maintains glucose homeostasis by improving β-oxidation capacity in liver and brown adipose tissues (Raichur et al., 2019). Succinate-CoA ligase (SUCLG2) regulates the conversion of propionate to pyruvate, promotes gluconeogenesis via PGC-1α, and maintains glucose homeostasis in hepatic gluconeogenesis (Wang et al., 2022a). Monoacylglycerol acyltransferase 2 (MGAT2) is a crucial enzyme highly expressed in the human small intestine and liver and regulates triglyceride uptake and homeostasis (Cheng et al., 2022). The peroxisome proliferator-activated receptor gamma coactivator 1 alpha (PPARGC1A) has been associated with obesity and related metabolic disorders, affecting adipocyte differentiation and lipid metabolism (Zhang et al., 2022). In practice, these adaptive genes induce energy homeostasis in response to relatively high energy demands under heat stress conditions. They regulate hypothermia caused by significant diurnal and nocturnal colds, making it easier for sheep to adapt to harsh desert environments.
Our study also identified some genes related to meat production. Zlobin et al. (2023) identified protocadherin 15 (PCDH15), a gene associated with meat productivity, between domesticated sheep (Ovis aries) and pan sheep (Ovis ammon), which is consistent with a super-dominant model of inheritance. Kizilaslan et al. (2022) studied the genetic structure of growth- and linear-type traits in Akkaraman sheep and proposed 22 genes, among which transducin-like enhancer of split-4 (TLE4) and transcription factor AP-2 β (TFAP2B) were same as the genes found in this study (Kizilaslan et al., 2022). Abousoliman showed that the activation rate of muscle stem cells is determined by the level of paired box 3 (Pax3), and alternative polyadenylation of Pax3 controls muscle stem cell fate and muscle function (de Morree et al., 2019). Genome-wide analysis for early growth-related traits of the locally adapted Egyptian Barki sheep revealed 10 genes related to early growth traits, among which SLC16A7 and TFAP2B were same as the genes found in our study (Abousoliman et al., 2021).
4.5 Genes related to desert adaptation
Recently, it has been shown that functional genes play a role in epigenetic temperature regulation mechanisms that lead to heat adaptation-mediated adaptive behaviors and protective responses in cells (Akerman et al., 2016) and that the foremost adaptation of the desert environment is the heat stress response. Four genes associated with heat stress were identified in our study. Superoxide dismutase 1 (SOD1) is a gene that regulates heat tolerance and maintains cellular oxidative homeostasis from superoxide radicals generated by stress in desert environments, such as heat and humidity (Khan et al., 2020). The DnaJ (Hsp40) homolog subfamily B member 5 (DNAJB5) gene affects heat tolerance in Dolan sheep, which is consistent with our study results. The DNAJB5 gene chaperone complex controls polarized growth by repressing Hsf1-driven heat stress-associated transcription (Vjestica et al., 2013; Wang et al., 2019). The TSHR gene regulates HSPs, which are believed to function as cellular thermometers for environmental adaptation in heat-stressed small ruminants (Luna-Nevárez et al., 2021). The phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 beta (PIK3C2B) gene is categorized in a cluster of genes compatible with the PI3K isoform. This subtype induces energetic homeostasis in response to high energy demands under heat stress conditions and is significantly related to avian mechanisms of acclimation to heat stress (Kim et al., 2017). Xie et al. (2023) concluded that the PIK3C2B gene is involved in glycerophospholipid biosynthesis, lipid modification, phospholipid biosynthesis, and phosphatidylinositol metabolism. It is more abundantly expressed in the function and pathway of lipids, and the PIK3C2B gene is one of the critical genes affecting fat deposition or lipid metabolism of swine (Xie et al., 2023). By combining the aforementioned ideas, we suggest that the PIK3C2B gene may maintain energy homeostasis by affecting fat metabolism, maintaining energy balance, and regulating body temperature, which in turn enables sheep to adapt better to the heat stress environment and maintain energy homeostasis.
Sheep are subjected to various diseases and parasites in their natural wild environment. Our study detected that genes such as CCL26, CTLA-4, PTPN13, PKHD1, ICOS, and ARPP21 are associated with immunity, and genes such as WNT5A, TIMP3, and BIN1 are associated with parasite resistance. C–C ligand 26 (CCL26) protects against localized infections, participates in the immune response, and plays a role in resistance to tick infestation and disturbance (Thutwa et al., 2021). Mature eosinophils are activated by CCL26 and migrate to the infection site, acting as an immunizing agent (E et al., 2019). In in vitro, when B7 molecules on the surface of antigen-presenting cells bind to cytotoxic T-lymphocyte-associated antigen 4 (CTLA-4) on the surface of T cells, a cost stimulus signal for T-cell activation is generated, which affects the humoral immune response in the body (Linsley et al., 1992). In a study on lambs in Spain, it was found that temperature and humidity affected the infectivity of Mycoplasma, with higher temperatures and lower humidity resulting in higher infection rates. This is quite similar to our environment, and the same protein tyrosine phosphatase non-receptor 13 (PTPN13) gene has been found in the immune function pathway (Fernández et al., 2016; Mousel et al., 2021). Polycystic kidney and liver disease 1 (PKHD1), inducible T-cell co-stimulatory factor (ICOS), and cAMP-regulated phosphoprotein 21 (ARPP21) enhance mastitis resistance. Innate and acquired immune responses trigger the production of T-assisted type 2 cytokines (Th2), which participate in the immune response. Benavides et al. described that they play multiple roles in innate and acquired immune response mechanisms and cytokine signaling, engage in hemostatic regulation and mucosal defense, and are essential for protecting sheep against parasitic invasion (Benavides et al., 2016; Banos et al., 2017; Gutiérrez-Gil et al., 2018; Öner et al., 2021).
5 Conclusion
In this study, we analyzed the genetic patterns and genomic differences between native sheep breeds in the northeastern Tarim Basin and introduced sheep breeds by genome-selective scanning. In addition, a genomic selection signaling map of the native sheep population in the Tarim Basin was constructed, revealing the genes and their network regulatory mechanisms associated with stress and disease resistance, growth, and reproductive traits. This study will help improve the production capacity of sheep and provide theoretical support for the conservation and development of sheep germplasm genetic resources in extreme desert environments.
Data availability statement
The data presented in the study are deposited in the Figshare repository, accession number http://doi.org/10.6084/m9.figshare.24461143.
Ethics statement
The animal study was approved by the Ethics Committee of the Tarim University of Science and Technology (SYXK 2020-009). The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
JW: formal analysis, methodology, writing–original draft, visualization, and writing–review and editing. JS: methodology, writing–review and editing, and writing–original draft. RY: software, visualization, and writing–original draft. C-lZ: software and writing–original draft. XL: formal analysis and writing–review and editing. ZH: writing–review and editing. WZ: writing–review and editing. SL: conceptualization, funding acquisition, visualization, writing–original draft, and writing–review and editing. QG: conceptualization, funding acquisition, project administration, supervision, writing–original draft, and writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was funded by the Bintuan Science and Technology Program (Grant No. 2022CB001-09) and GWAS-based Screening of Candidate Genes for Polyembryony Traits in Xinjiang Lop Sheep (Grant No. BSGJSYS202304).
Conflict of interest
Authors C-lZ, XL, ZH, WZ, SL, and QG were employed by Xinjiang Production and Construction Corps.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1281601/full#supplementary-material
References
Abousoliman, I., Reyer, H., Oster, M., Murani, E., Mohamed, I., and Wimmers, K. (2021). Genome-wide analysis for early growth-related traits of the locally adapted Egyptian Barki sheep. Genes 12 (8), 1243. doi:10.3390/genes12081243
Akerman, A. P., Tipton, M., Minson, C. T., and Cotter, J. D. (2016). Heat stress and dehydration in adapting for performance: good, bad, both, or neither? Temperature 3 (3), 412–436. doi:10.1080/23328940.2016.1216255
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., et al. (2000). Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25 (1), 25–29. doi:10.1038/75556
Baazaoui, I., Bedhiaf-Romdhani, S., Mastrangelo, S., and Ciani, E. (2021). Genome-wide analyses reveal population structure and identify candidate genes associated with tail fatness in local sheep from a semi-aridarea. Animal 15 (4), 100193. doi:10.1016/j.animal.2021.100193
Banos, G., Bramis, G., Bush, S. J., Clark, E. L., McCulloch, M. E., Smith, J., et al. (2017). The genomic architecture of mastitis resistance in dairy sheep. BMC Genomics 18 (1), 624. doi:10.1186/s12864-017-3982-1
Benavides, M. V., Sonstegard, T. S., and Van Tassell, C. (2016). Genomic regions associated with sheep resistance to gastrointestinal nematodes. Trends Parasitol. 32 (6), 470–480. doi:10.1016/j.pt.2016.03.007
Cheng, D., Zinker, B. A., Luo, Y., Shipkova, P., De Oliveira, C. H., Krishna, G., et al. (2022). MGAT2 inhibitor decreases liver fibrosis and inflammation in murine NASH models and reduces body weight in human adults with obesity. Cell Metab. 34 (11), 1732–1748.e5. doi:10.1016/j.cmet.2022.10.007
Conway, J. R., Lex, A., and Gehlenborg, N. (2017). UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics 33 (18), 2938–2940. doi:10.1093/bioinformatics/btx364
Danecek, P., Auton, A., Abecasis, G., Albers, C. A., Banks, E., DePristo, M. A., et al. (2011). The variant call format and VCFtools. Bioinformatics 27 (15), 2156–2158. doi:10.1093/bioinformatics/btr330
Das, R., Sailo, L., Verma, N., Bharti, P., Saikia, J., Kumar, R., et al. (2016). Impact of heat stress on health and performance of dairy animals: a review. A Rev. Vet. World. 9 (3), 260–268. doi:10.14202/vetworld.2016.260-268
de Morree, A., Klein, J. D., Gan, Q., Farup, J., Urtasun, A., Kanugovi, A., et al. (2019). Alternative polyadenylation of Pax3 controls muscle stem cell fate and muscle function. Science 366 (6466), 734–738. doi:10.1126/science.aax1694
Estrada-Reyes, Z. M., Rae, O., Postley, C., Jiménez Medrano, M. B., Leal Gutiérrez, J. D., Mateescu, R. G., et al. (2019). Association study reveals Th17, Treg, and Th2 loci related to resistance to Haemonchus contortus in Florida Native sheep1. J. Anim. Sci. 97 (11), 4428–4444. doi:10.1093/jas/skz299
Fernández, S., Galapero, J., Rey, J., Pérez, C. J., Ramos, A., Rosales, R., et al. (2016). Investigations into the seasonal presence of Mycoplasma species in fattening lambs. Vet. J. 212, 80–82. doi:10.1016/j.tvjl.2016.04.006
Ghoreishifar, S. M., Moradi-Shahrbabak, H., Parna, N., Davoudi, P., and Khansefid, M. (2019). Linkage disequilibrium and within-breed genetic diversity in Iranian Zandi sheep. Arch. Anim. Breed. 62 (1), 143–151. doi:10.5194/aab-62-143-2019
Gutiérrez-Gil, B., Esteban-Blanco, C., Suarez-Vega, A., and Arranz, J. J. (2018). Detection of quantitative trait loci and putative causal variants affecting somatic cell score in dairy sheep by using a 50K SNP chip and whole-genome sequencing. J. Dairy. Sci. 101 (10), 9072–9088. doi:10.3168/jds.2018-14736
Hayes, B. J., Lewin, H. A., and Goddard, M. E. (2013). The future of livestock breeding: genomic selection for efficiency, reduced emissions intensity, and adaptation. Trends Genet. 29 (4), 206–214. doi:10.1016/j.tig.2012.11.009
Kanehisa, M., Sato, Y., Kawashima, M., Furumichi, M., and Tanabe, M. (2016). KEGG as a reference resource for gene and protein annotation. Nucleic. acids. Res. 44 (D1), D457–D462. doi:10.1093/nar/gkv1070
Khan, Z. A., Mishra, C., and Jyotiranjan, T. (2020). In silico analysis of caprine superoxide dismutase 1 (SOD1) gene. Genomics 112 (1), 212–217. doi:10.1016/j.ygeno.2019.01.016
Khristi, V., Chakravarthi, V. P., Singh, P., Ghosh, S., Pramanik, A., Ratri, A., et al. (2018). ESR2 regulates granulosa cell genes essential for follicle maturation and ovulation. Mol. Cell. Endocrinol. 474, 214–226. doi:10.1016/j.mce.2018.03.012
Kijas, J. W., Lenstra, J. A., Hayes, B., Boitard, S., Porto Neto, L. R., San Cristobal, M., et al. (2012). Genome-wide analysis of the world's sheep breeds reveals high levels of historic mixture and strong recent selection. PLoS Biol. 10 (2), e1001258. doi:10.1371/journal.pbio.1001258
Kim, J. M., Lim, K. S., Byun, M., Lee, K. T., Yang, Y. R., Park, M., et al. (2017). Identification of the acclimation genes in transcriptomic responses to heat stress of White Pekin duck. Cell. Stress. Chaperones. 22, 787–797. doi:10.1007/s12192-017-0809-6
Kizilaslan, M., Arzik, Y., White, S. N., Piel, L. M., and Cinar, M. U. (2022). Genetic parameters and genomic regions underlying growth and linear type traits in Akkaraman sheep. Genes 13 (8), 1414. doi:10.3390/genes13081414
Letunic, I., and Bork, P. (2016). Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic. acids. Res. 44 (W1), W242–W245. doi:10.1093/nar/gkw290
Li, M. H., Tiirikka, T., and Kantanen, J. (2014). A genome-wide scan study identifies a single nucleotide substitution in ASIP associated with white versus non-white coat-colour variation in sheep (Ovis aries). Heredity 112 (2), 122–131. doi:10.1038/hdy.2013.83
Li, X., Yang, J. I., Shen, M., Xie, X. L., Liu, G. J., Xu, Y. X., et al. (2020). Whole-genome resequencing of wild and domestic sheep identifies genes associated with morphological and agronomic traits. Nat. Commun. 11 (1), 2815. doi:10.1038/s41467-020-16485-1
Li, M., He, N., Sun, R., Deng, Y., Wen, X., and Zhang, J. (2022). Expression and polymorphisms of SMAD1, SMAD2 and SMAD3 genes and their association with litter size in Tibetan sheep (Ovis aries). Genes 13 (12), 2307. doi:10.3390/genes13122307
Li, M., Zhou, Q., Pan, Y., Lan, X., Zhang, Q., Pan, C., et al. (2023). Screen of small fragment mutations within the sheep thyroid stimulating hormone receptor gene associated with litter size. Anim. Biotechnol. 34 (3), 658–663. doi:10.1080/10495398.2021.1992415
Linsley, P. S., Wallace, P. M., Johnson, J., Gibson, M. G., Greene, J. L., Ledbetter, J. A., et al. (1992). Immunosuppression in vivo by a soluble form of the CTLA-4 T cell activation molecule. Science 257 (5071), 792–795. doi:10.1126/science.1496399
Liu, L. L., Meng, J., Ma, H. Y., Cao, H., and Liu, W. J. (2022). Candidate genes for litter size in Xinjiang sheep identified by Specific Locus Amplified Fragment (SLAF) sequencing. Anim. Biotechnol. 1, 3053–3062. doi:10.1080/10495398.2022.2131561
Luna-Nevárez, G., Pendleton, A. L., Luna-Ramirez, R. I., Limesand, S. W., Reyna-Granados, J. R., and Luna-Nevárez, P. (2021). Genome-wide association study of a thermo-tolerance indicator in pregnant ewes exposed to an artificial heat-stressed environment. J. Therm. Biol. 101, 103095. doi:10.1016/j.jtherbio.2021.103095
Mandon-Pépin, B., Oustry-Vaiman, A., Vigier, B., Piumi, F., Cribiu, E., and Cotinot, C. (2003). Expression profiles and chromosomal localization of genes controlling meiosis and follicular development in the sheep ovary. Biol. Reprod. 68 (3), 985–995. doi:10.1095/biolreprod.102.008557
Mousel, M. R., White, S. N., Herndon, M. K., Herndon, D. R., Taylor, J. B., Becker, G. M., et al. (2021). Genes involved in immune, gene translation and chromatin organization pathways associated with Mycoplasma ovipneumoniae presence in nasal secretions of domestic sheep. PLoS. One. 16 (7), e0247209. doi:10.1371/journal.pone.0247209
Nosrati, M., Asadollahpour Nanaei, H., Amiri Ghanatsaman, Z., and Esmailizadeh, A. (2019). Whole genome sequence analysis to detect signatures of positive selection for high fecundity in sheep. Reprod. Domest. Anim. 54 (2), 358–364. doi:10.1111/rda.13368
Öner, Y., Serrano, M., Sarto, P., Iguácel, L. P., Piquer-Sabanza, M., Estrada, O., et al. (2021). Genome-wide association studies of somatic cell count in the assaf breed. Animals 11 (6), 1531. doi:10.3390/ani11061531
Patterson, N., Moorjani, P., Luo, Y., Mallick, S., Rohland, N., Zhan, Y., et al. (2012). Ancient admixture in human history. Genetics 192 (3), 1065–1093. doi:10.1534/genetics.112.145037
Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M. A., Bender, D., et al. (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81 (3), 559–575. doi:10.1086/519795
Raichur, S., Brunner, B., Bielohuby, M., Hansen, G., Pfenninger, A., Wang, B., et al. (2019). The role of C16: 0 ceramide in the development of obesity and type 2 diabetes: CerS6 inhibition as a novel therapeutic approach. Mol. Metab. 21, 36–50. doi:10.1016/j.molmet.2018.12.008
Romero, R. D., Montero Pardo, A., Montaldo, H. H., Rodríguez, A. D., and Hernández Cerón, J. (2013). Differences in body temperature, cell viability, and HSP-70 concentrations between Pelibuey and Suffolk sheep under heat stress. Trop. Anim. Health .Prod. 45, 1691–1696. doi:10.1007/s11250-013-0416-1
Rupp, R., Senin, P., Sarry, J., Allain, C., Tasca, C., Ligat, L., et al. (2015). A point mutation in suppressor of cytokine signalling 2 (Socs2) increases the susceptibility to inflammation of the mammary gland while associated with higher body weight and size and higher milk production in a sheep model. PLoS Genet. 11 (12), e1005629. doi:10.1371/journal.pgen.1005629
Sánchez-Criado, J. E., Trudgen, K., Millán, Y., Blanco, A., Monterde, J., Garrido-Gracia, J. C., et al. (2012). Estrogen receptor (ESR) 2 partially offsets the absence of ESR1 in gonadotropes of pituitary-specific Esr1 knockout female mice. Reproduction 143 (4), 549–558. doi:10.1530/REP-11-0214
Shen, J., Hanif, Q., Cao, Y., Yu, Y., Lei, C., Zhang, G., et al. (2020). Whole genome scan and selection signatures for climate adaption in yanbian cattle. Front. Genet. 11, 94. doi:10.3389/fgene.2020.00094
Sweet-Jones, J., Lenis, V. P., Yurchenko, A. A., Yudin, N. S., Swain, M., and Larkin, D. M. (2021). Genotyping and whole-genome resequencing of Welsh sheep breeds reveal candidate genes and variants for adaptation to local environment and socioeconomic traits. Front. Genet. 12, 612492. doi:10.3389/fgene.2021.612492
Thutwa, K., van Wyk, J. B., Dzama, K., Scholtz, A. J., and Cloete, S. W. (2021). Expression of cytokine genes at tick attachment and control sites of Namaqua Afrikaner, Dorper and South African Mutton Merino sheep. Vet. Parasitol. 291, 109384. doi:10.1016/j.vetpar.2021.109384
Vjestica, A., Zhang, D., Liu, J., and Oliferenko, S. (2013). Hsp70-Hsp40 chaperone complex functions in controlling polarized growth by repressing Hsf1-driven heat stress-associated transcription. PLoS Genet. 9 (10), e1003886. doi:10.1371/journal.pgen.1003886
Wang, W., Zhang, X., Zhou, X., Zhang, Y., La, Y., Zhang, Y., et al. (2019). Deep genome resequencing reveals artificial and natural selection for visual deterioration, plateau adaptability and high prolificacy in Chinese domestic sheep. Front. Genet. 10, 300. doi:10.3389/fgene.2019.00300
Wang, G., Zhang, J., Wu, S., Qin, S., Zheng, Y., Xia, C., et al. (2022a). The mechanistic target of rapamycin complex 1 pathway involved in hepatic gluconeogenesis through peroxisome-proliferator-activated receptor γ coactivator-1α. Anim. Nutr. 11, 121–131. doi:10.1016/j.aninu.2022.07.010
Wang, Q., Li, H., Tajima, K., Verkerke, A. R., Taxin, Z. H., Hou, Z., et al. (2022b). Post-translational control of beige fat biogenesis by PRDM16 stabilization. Nature 609 (7925), 151–158. doi:10.1038/s41586-022-05067-4
Wu, F. J., Wang, Y. W., and Luo, C. W. (2021). Human bone morphogenetic protein 8A promotes expansion and prevents apoptosis of cumulus cells in vitro. Mol. Cell. Endocrinol. 522, 111121. doi:10.1016/j.mce.2020.111121
Xie, Q., Zhang, Z., Chen, Z., Sun, J., Li, M., Wang, Q., et al. (2023). Integration of selection signatures and protein interactions reveals NR6A1, PAPPA2, and PIK3C2B as the promising candidate genes underlying the characteristics of licha black pig. Biology 12 (4), 500. doi:10.3390/biology12040500
Xu, S. S., Gao, L., Xie, X. L., Ren, Y. L., Shen, Z. Q., Wang, F., et al. (2018). Genome-wide association analyses highlight the potential for different genetic mechanisms for litter size among sheep breeds. Front. Genet. 9, 118. doi:10.3389/fgene.2018.00118
Xu, Y. X., Wang, B., Jing, J. N., Ma, R., Luo, Y. H., Li, X., et al. (2023). Whole-body adipose tissue multi-omic analyses in sheep reveal molecular mechanisms underlying local adaptation to extreme environments. Commun. Biol. 6 (1), 159. doi:10.1038/s42003-023-04523-9
Zeder, M. A. (2008). Domestication and early agriculture in the Mediterranean Basin: origins, diffusion, and impact. Proc. Natl. Acad. Sci. U. S. A. 105 (33), 11597–11604. doi:10.1073/pnas.0801317105
Zhang, R. N., Pang, B., Xu, S. R., Wan, P. C., Guo, S. C., Ji, H. Z., et al. (2018). The CXCL12-CXCR4 signaling promotes oocyte maturation by regulating cumulus expansion in sheep. Theriogenology 107, 85–94. doi:10.1016/j.theriogenology.2017.10.039
Zhang, X., Liu, C., Kong, Y., Li, F., and Yue, X. (2022). Effects of intramuscular fat on meat quality and its regulation mechanism in Tan sheep. Front. Nutr. 9, 908355. doi:10.3389/fnut.2022.908355
Zhao, F., Deng, T., Shi, L., Wang, W., Zhang, Q., Du, L., et al. (2020). Genomic scan for selection signature reveals fat deposition in Chinese indigenous sheep with extreme tail types. Animals 10 (5), 773. doi:10.3390/ani10050773
Zhao, P., Li, S., He, Z., Zhao, F., Wang, J., Liu, X., et al. (2022). Physiology and proteomic basis of lung adaptation to high-altitude hypoxia in Tibetan sheep. Animals 12 (16), 2134. doi:10.3390/ani12162134
Zhu, M., Zhang, H., Yang, H., Zhao, Z., Blair, H. T., Zhai, M., et al. (2022). Polymorphisms and association of GRM1, GNAQ and HCRTR1 genes with seasonal reproduction and litter size in three sheep breeds. Reprod. Domest. Anim. 57 (5), 532–540. doi:10.1111/rda.14091
Keywords: sheep, genetic diversity, population structure, selective signature, environmental adaptation
Citation: Wang J, Suo J, Yang R, Zhang C-l, Li X, Han Z, Zhou W, Liu S and Gao Q (2023) Genetic diversity, population structure, and selective signature of sheep in the northeastern Tarim Basin. Front. Genet. 14:1281601. doi: 10.3389/fgene.2023.1281601
Received: 22 August 2023; Accepted: 23 October 2023;
Published: 09 November 2023.
Edited by:
Tiago Almeida de Oliveira, State University of Paraíba, BrazilReviewed by:
Hua Yang, Xinjiang Academy of Agricultural and Reclamation Sciences (XAARS), ChinaNemat Hedayat, University of Mohaghegh Ardabili, Iran
Copyright © 2023 Wang, Suo, Yang, Zhang, Li, Han, Zhou, Liu and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shudong Liu, bGl1c2h1ZG9uZzYzQDEyNi5jb20=; Qinghua Gao, Z3FoZGt5QDEyNi5jb20=
 Jieru Wang
Jieru Wang Jiajia Suo
Jiajia Suo Ruizhi Yang1
Ruizhi Yang1 Cheng-long Zhang
Cheng-long Zhang Shudong Liu
Shudong Liu