
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 08 January 2024
Sec. Livestock Genomics
Volume 14 - 2023 | https://doi.org/10.3389/fgene.2023.1277647
Investigations into the correlation between growth characteristics and DNA methylation levels, along with genetic variations, can provide fundamental insights to enhance growth performance in groupers. The Myostatin (mstn) gene plays a vital role in regulating skeletal muscle development and growth. This study scrutinized the DNA methylation levels of the mstn gene across hybrid groupers (E. fuscoguttatus (♀) × E. polyphekadion (♂)) and their parental species, to evaluate its impact on growth attributes in grouper fish. The nucleotide sequence of the mstn gene was directly sequenced in the hybrid grouper, exhibiting different growth performance to identify the single nucleotide polymorphisms (SNPs) of the mstn gene and explore their correlation with growth characteristics. The findings revealed no significant differences in global DNA methylation levels within muscle tissue among the hybrid grouper and parents. However, significant differences in DNA methylation sites were discovered between the hybrid grouper and E. polyphekadion at sites 824 and 1521 (located at exon 2 and intron 2, respectively), and between E. fuscoguttatus and E. polyphekadion at site 1521. These variations could potentially influence the mRNA expression of the mstn gene. The study also identified that SNP g.1003 T > C in exon 2 of the mstn gene was significantly associated with various growth traits including body weight, total length, body length, head length, caudal peduncle height, and body height (p < 0.01). Specimens with the TT genotype at site 1003 demonstrated superior growth performance compared to those with the TC genotype. Furthermore, microstructural analyses of muscle tissue showed that the average area and diameter of muscle fibers in TT genotype individuals were significantly greater than those in TC genotype individuals. Therefore, this research provides robust evidence linking the DNA methylation level and polymorphisms of the mstn gene with growth traits, which could be beneficial for grouper breeding programs.
The grouper is a highly prized marine fish, thanks to its excellent nutritional content and limited availability in natural waters. It is a leading aquaculture species in the southern coastal regions of China. In 2021, grouper aquaculture production exceeded 200,000 tons (FAO, 2022). Hainan presently stands as China’s major grouper producer, with the production of grouper reaching 70,178 tons in 2021, according to the China Fishery Statistics Yearbook. This figure closely trails the production from Guangdong. The primary aquaculture species in Hainan is the hybrid grouper (Epinephelus fuscoguttatus (♀) × Epinephelus polyphekadion (♂)) (Cao et al., 2022). Given the direct impact of growth traits on farming benefits, understanding the molecular mechanisms driving growth in hybrid groupers is imperative.
DNA methylation, a primary mechanism that regulates gene expression (Ding et al., 2012; Si et al., 2016; Li et al., 2017), can induce phenotypic changes (Jaenisch and Bird, 2003; Angers et al., 2010). An increasing body of evidence underscores the influence of DNA methylation on growth in animals (Zhong et al., 2014; Yang and Li, 2021). Single nucleotide polymorphisms (SNPs) are another crucial resource for genetic association studies (Zhang et al., 2015; Liu et al., 2016). A significant correlation between SNP markers and traits suggests a possible association between these markers and certain characteristics (Doerge, 2002). The discovery of growth-related SNP markers in several fish species, including Danio rerio, Oncorhynchus mykiss, Hemibagrus wyckioides, and Epinephelus coioides (Salem et al., 2012; Ulloa et al., 2015; Guo et al., 2016; Zhou et al., 2021), implies that selective breeding based on these markers holds promise for enhancing growth traits.
Myostatin (mstn, also known as GDF-8), a member of the transforming growth factor-β superfamily, expresses in various fish tissues including gills, brain, ovaries, and skeletal muscle (Rodgers et al., 2001; Ko et al., 2007). The mstn gene acts as a negative regulator of muscle growth in skeletal muscle by suppressing the hypertrophy and hyperplasia of muscle cells (McPherron et al., 1997; Lee and McPherron, 1999). This mechanism has been identified in several aquatic species, such as O. mykiss (Nazari et al., 2016), Litopenaeus vannamei (Qian et al., 2013), Argopecten irradians (Meng et al., 2017), Echinoidea (Lapraz et al., 2006), and Paralichthys olivaceus (Zhong et al., 2008). Numerous studies have explored the relationships between various nucleotide polymorphisms of the mstn gene and growth traits in commercial species. These investigations provide valuable genetic markers for the genetic improvement of aquatic species (Sun et al., 2012; Peñaloza et al., 2013; Meng et al., 2017). Currently, the sequence variants and methylation status of the mstn gene in hybrid grouper remain under-explored. Therefore, this study aims to examine the correlation between growth traits and the methylation status, expression pattern, and polymorphisms of the mstn gene in grouper, in an effort to unearth potential markers for assisted selection.
Specimens of E. fuscoguttatus, E. polyphekadion, and the hybrid grouper were procured and reared at the Hainan Chenhai Aquatic Co. Ltd. In Hainan Province, China. To ensure optimal conditions, the health of the fish and environmental parameters were closely monitored on a daily basis. All experimental procedures strictly adhered to the guidelines stipulated by Administration of Affairs Concerning Animal Experimentation of China.
Muscle tissues were utilized for genomic DNA extraction in E. fuscoguttatus (five individuals), E. polyphekadion (five individuals), and the hybrid grouper (six individuals). Genomic DNA (1 μg) was processed using the ZYMO EZ DNA Methylation-Gold Kit (Zymo Research, Irvine, CA, United States), and one-tenth of the elution products were employed as templates. PCR primers were developed using the MethPrimer software package (www.urogene.org/methprimer/index.html), which was guided by the nucleotide sequence of the mstn gene (Cao et al., 2023) (refer to Supplementary Figure S1). PCR amplification was performed for 35 cycles, with an annealing duration of 30 s at 50–60°C, using the KAPA HiFi HotStart Uracil + ReadyMix PCR Kit (Kapa Biosystems, Wilmington, MA, USA). For each sample, bisulfite sequencing PCR (BSP) products (∼200 bp in length) were evenly pooled, 5′-phosphorylated, 3′-dA-tailed, and ligated to a barcoded adapter using T4 DNA ligase (NEB). Barcoded libraries from all samples were pooled uniformly and utilized for pair-end sequencing using the Illumina platform (Gao et al., 2015). The raw reads were first processed to remove adapters and filter out low-quality sequences using Trimmomatic-0.36, yielding clear reads. These clean reads were then aligned to the reference sequence (mstn gene) using BSMAP (Version 2.73). The methylation levels of individual cytosines were calculated as the ratio of the total number of methylated CpG cytosines to the number of sequenced clones. The global DNA methylation level was calculated as the average methylation levels of all CpG sites in each sample. For the methylation level of each CpG site, global DNA methylation level of mstn gene, comparisons between two groups were conducted using a two-tailed Fisher’s Exact Test with p < 0.05 indicating a statistically significant difference (Gao et al., 2014).
The hybrid grouper was reared in seawater tank (4 m × 4 m) under identical conditions and for the same duration. After 195 days of culture, their body weight and morphological indicators were measured. Individuals with slow-growth and fast-growth were selected and divided into two groups (slow-growth group and fast-growth group) according to the value of body weight. The mean value of body weight in slow-growth group and fast-growth group was 72.67 g and 196.67 g, respectively. In these two groups, individuals were excluded when their body weight was deviate significantly from the mean body weight. Finally, 51 specimens (24 individuals derived from the slow-growth group, 27 individuals derived from the fast-growth group) were collected to explore the potential influence of SNPs in the mstn gene on the growth traits of hybrid groupers. Genomic DNA was extracted from muscle tissues using the Animal Genomic DNA Extraction Kit as per the manufacturer’s instructions (Sangon Biotech, Shanghai). The DNA samples’ concentration and quality were assessed through 1% agarose gel electrophoresis. Three pairs of PCR primers, designed using Primer 5.0 and synthesized by Sangon Biotech (Guangzhou, China), were employed to clone and identify SNP sites in the mstn gene (Supplementary Table S1). Genotyping was carried out through direct sequencing (Sangon Biotech, Guangzhou) using an Applied Biosystems ABI 3730 L Genetic Analyzer (Applied Biosystems group, US), and the resulting sequences were analyzed via Chromas.
Genotype frequencies and gene frequencies at SNP sites were computed according to the following formulas (Lyu et al., 2020): PAA = NAA/N (PAA: frequency of AA genotype; NAA: number of AA genotype specimens; N: total number of hybrid groupers); PA=(2NAA + NAG)/2N, PG=(2NGG + NAG)/2N (PA, PG: frequencies of A and G alleles; NAA, NAG, and NGG: number of individuals with AA, AG, and GG genotypes; N: total number of hybrid groupers). The polymorphic information content (PIC) and expected heterozygosity (He) were computed using Curves 3.0.7 software, Linkage disequilibrium (LD) was calculated with Haploview 4.2 software. Ultimately, only two SNPs (minor allele frequency >0.01, p-value for Hardy-Weinberg equilibrium test >0.05, PIC >0.2) were selected for further analysis.
Pearson correlation coefficients were determined using SPSS 17.0 to elucidate correlations between various growth traits. The associations between the SNP site and growth traits were analyzed using the Generalized Linear Model (GLM) procedure in the R package.
The relative expression levels of mstn mRNA were analyzed using muscle cDNA sourced from E. fuscoguttatus, E. polyphekadion, and the hybrid grouper. To facilitate this, specific primers were designed using the Primer 5.0 software (Supplementary Table S1). Quantitative PCR (qPCR) was conducted on a Step One Plus™ Real-Time PCR System (Applied Biosystems) utilizing the ChamQ SYBR qPCR Master Mix (Vazyme), with protocols adhering to manufacturer guidelines. The reaction mix, with a total volume of 10 μL, was composed of 5 µL of 2 × qPCR mix, 0.25 µL of both forward and reverse primers, 2 µL of DNA template, and 2.5 µL of RNase-free water. The qPCR reaction was initiated with a pre-denaturation step at 95°C for 30 s, followed by 40 cycles of denaturation at 95°C for 10 s and annealing/extension at 60°C for 30 s. The β-actin gene was used as a reference gene for normalization. Each sample was run in triplicate using the same primers. The relative gene expression was determined using the 2−ΔΔCT method (Livak and Schmittgen, 2013).
White muscle fiber samples were obtained from the dorsal region of hybrid groupers, each having different genotypes at site 1,003 (three individuals each from TT and TC genotypes). All samples were dissected into pieces of 0.2 cm³ and subjected to sequential dehydration through a range of alcohol concentrations. Subsequently, the tissues were processed with an alcohol:xylene (1:1) solution, followed by 100% xylene, and then embedded in paraffin wax. The paraffin-embedded samples were sliced into serial sections for Hematoxylin and Eosin (HE) staining. Three fields of view were chosen for each sample to measure the muscle fiber cross-sectional area and diameter using Image-Pro Plus 6.0 software. Statistical analyses of the data were carried out using the Analysis of Variance (ANOVA) test in SPSS 17.0 software.
The entire length of the mstn gene, including a 1,468 base pair (bp) upstream fragment adjacent to the transcription start site (TSS), three exons, and two introns (Figure 1), was evaluated to assess the global level of DNA methylation across the hybrid grouper and its parental species. The data revealed that the comprehensive DNA methylation levels of the mstn gene were relatively low in all groupers, with no significant differences (p > 0.05) observed between the hybrid grouper (0.0855), E. fuscoguttatus (0.1009), and E. polyphekadion (0.1047) (Figure 2A). This suggests that methylation does not vary significantly based on species. However, there were notable differences in the mRNA expression levels of the mstn gene between E. polyphekadion, E. fuscoguttatus, and the hybrid grouper (Figure 2B).
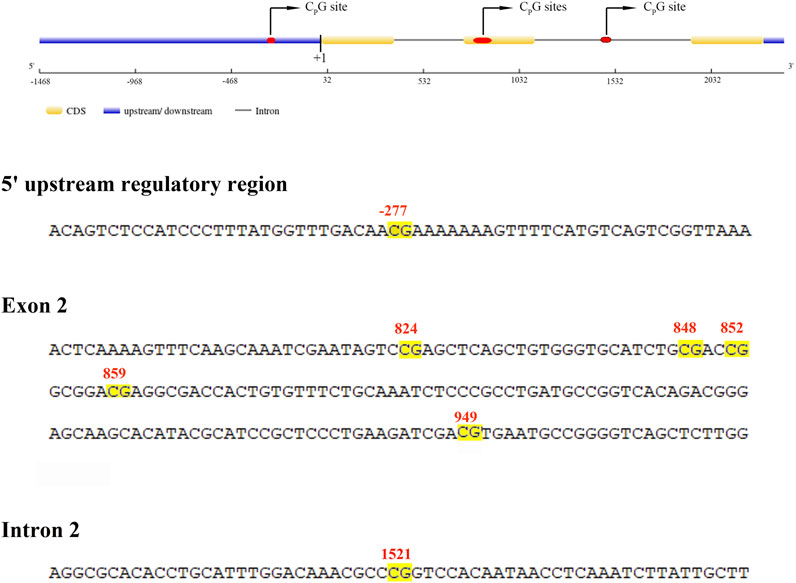
FIGURE 1. Schematic organization and partial sequence of mstn gene. The location of the CpG sites are highlighted by a yellow background.
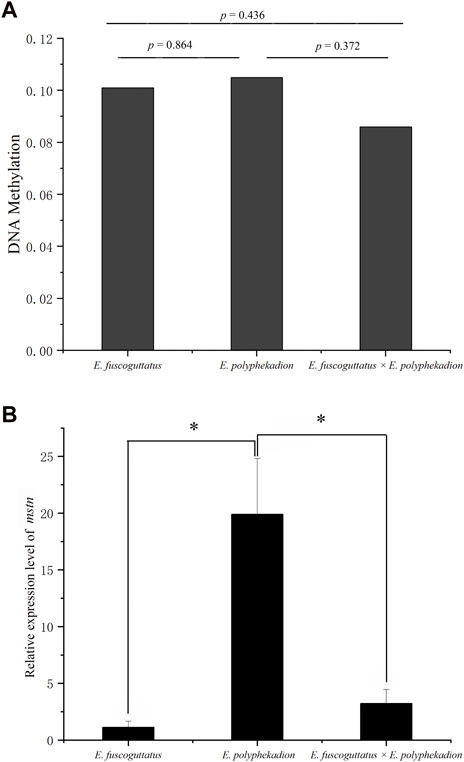
FIGURE 2. (A) The global of DNA methylation levels of mstn gene in Epinephelus fuscoguttatus, Epinephelus polyphekadion and the hybrid grouper; (B) Expression levels of mstn gene in Epinephelus fuscoguttatus, Epinephelus polyphekadion and the hybrid grouper. * indicates there is a significant difference between two groups.
Additionally, the CpG sites in the 5′upstream regulatory region and the exon and intron of the mstn gene were examined to discern differences in methylation levels between the hybrid grouper and its parent species. Notable regional differences in mstn gene DNA methylation were detected in the 5′upstream regulatory region (position −227), exon 2 (positions 824, 848, 852, 859, and 949), and intron 2 (position 1,521) (Figure 3). Significant methylation differences between E. fuscoguttatus and E. polyphekadion were found at positions −227, 949, and 1,521. At site −227, the DNA methylation level in E. polyphekadion (0.0066) was lower than that in E. fuscoguttatus (0.0080). Conversely, the DNA methylation patterns at sites 949 and 1,521 showed an inverse relationship between E. fuscoguttatus (0.0052 and 0.4730, respectively) and E. polyphekadion (0.0076 and 0.7672, respectively).
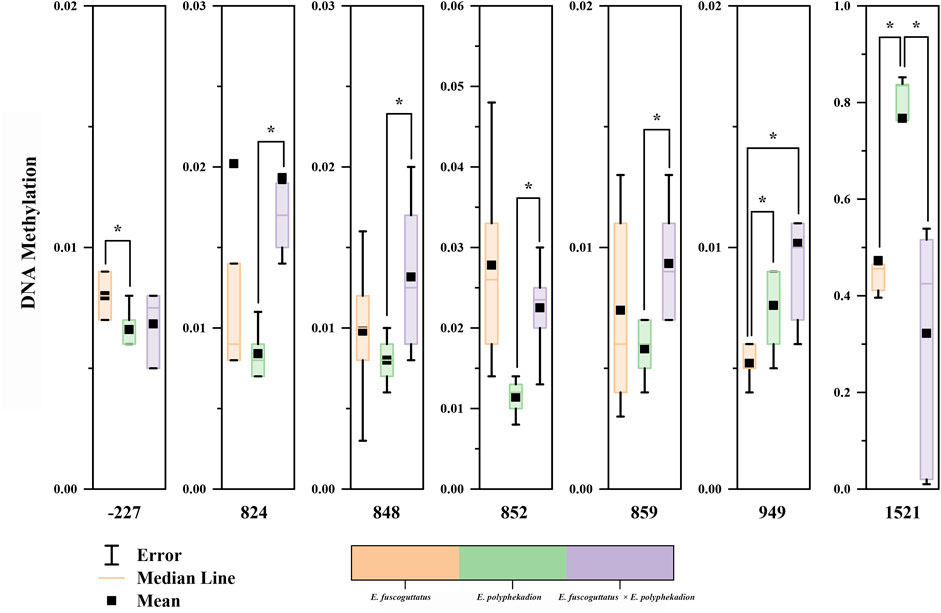
FIGURE 3. The site-specific DNA methylation levels of mstn gene in Epinephelus fuscoguttatus, Epinephelus polyphekadion and the hybrid grouper. X-axis represents the position of the cytosine in the CpG dinucleotide context relative to the transcription start site. * indicates there is a significant difference between two groups.
The DNA methylation levels exhibited significant differences between the hybrid grouper and its male parent at positions 824, 848, 852, 859, and 1,521. Generally, the hybrid grouper demonstrated significantly higher DNA methylation levels at all these positions, compared to its male parent (except at position 1,521). The hybrid grouper and the female parent only displayed a significant difference at one CpG site (position 949), where the DNA methylation level in the hybrid grouper (0.0102) exceeded that of the female parent (0.0052).
The relative expression level results revealed no significant change between the hybrid grouper and E. fuscoguttatus, suggesting that the DNA methylation level at site 949 does not influence relative expression. However, potential impacts from the methylation of other CpG sites are considered herein. The methylation levels at CpG sites 824 and 1,521 exhibited a notable correlation with the expression of the mstn gene (Figure 4). Figure 4A illustrates the association between gene expression and the methylation level at site 824. A negative correlation was observed between mstn mRNA levels and DNA methylation (R = −0.85, p < 0.05) when comparing the hybrid grouper and E. polyphekadion. Conversely, the DNA methylation level of mstn was positively correlated with gene expression at site 1,521 (Figures 4B, C). The correlation coefficients between CpG methylation and gene expression were 0.97 and 0.87 when comparing E. fuscoguttatus and E. polyphekadion, and the hybrid grouper and E. polyphekadion, respectively.
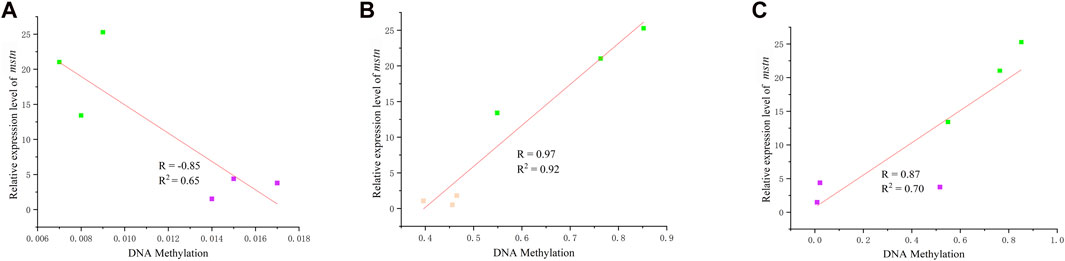
FIGURE 4. The correlation between DNA methylation and gene mRNA expression. (A) Correlation between DNA methylation at site 824 and gene mRNA expression in Epinephelus polyphekadion and the hybrid grouper. (B) Correlation between DNA methylation at site 1,521 and gene mRNA expression in Epinephelus polyphekadion and Epinephelus fuscoguttatus. (C) Correlation between DNA methylation at site 1,521 and gene mRNA expression in Epinephelus polyphekadion and the hybrid grouper. Orange represents Epinephelus fuscoguttatus, green represents Epinephelus polyphekadion and purple represents the hybrid grouper.
To identify SNP sites associated with growth traits, this study analyzed loci within or near the coding region of the mstn gene in hybrid grouper. SNPs that did not comply with the Hardy-Weinberg equilibrium were excluded from this investigation. The mean values and ranges for each trait in the hybrid grouper are presented in Table 1. Pearson correlation coefficients were computed to reveal relationships among various traits (Table 2). Significant differences were observed in the Pearson correlation coefficients between two distinct traits, indicating a strong positive correlation among all traits. Notably, the coefficients between total length and both body weight and body length were extremely close to 1, suggesting a high correlation between these attributes. Details regarding the SNPs of the mstn gene are provided in Table 3. Two SNPs (g.818 A>G, g.1003 T>C) (Supplementary Figure S2) adhered to the Hardy-Weinberg equilibrium (p > 0.05). These two SNPs exhibited polymorphism with minor allele frequencies >1% and a high level of heterozygosity (PIC >0.5).
We further conducted experiments to ascertain potential impacts of SNPs within muscle DNA. Notably, we found that SNP g.1003 showed a significant association with several growth attributes, including body weight, total length, body length, head length, body height, and caudal peduncle height (p < 0.01; refer to Table 4). Individuals possessing the TT genotype for SNP g.1003 demonstrated superior growth performance compared to those with the TC genotype (p < 0.01).
In accordance with the genotyping of the SNP g.1003 in mstn gene, we selected hybrid groupers with TT and TC genotypes, three individuals each for microscopic examination (Figure 5; Table 5). As indicated in Table 5, the average cross-sectional area and diameter of muscle fibers in the hybrid groupers with the TT genotype (1,969.03 ± 8.07 µm2, 25.02 ± 0.91 µm, respectively) were significantly greater than those of the groupers with the TC genotype (1,330.34 ± 23.87 µm2, 21.43 ± 0.49 µm) (p < 0.01). Hematoxylin and Eosin (HE) staining results showed that muscle fibers in individuals with the TT genotype were generally thicker than those in individuals with the TC genotype.
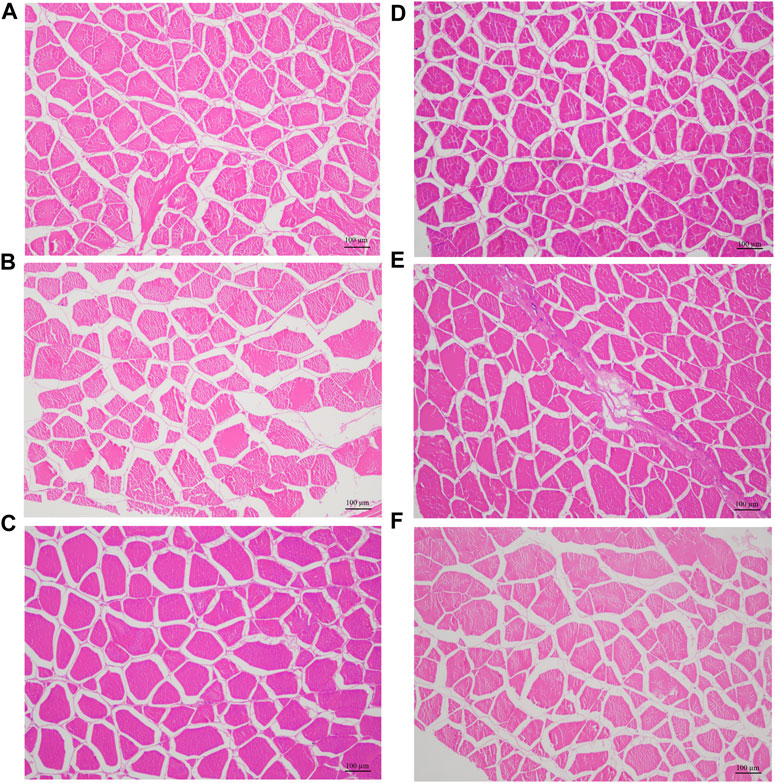
FIGURE 5. Histological characteristic of myofiber in six hybrid grouper, three with TT genotype (A–C) and three with TC genotype (D–F).
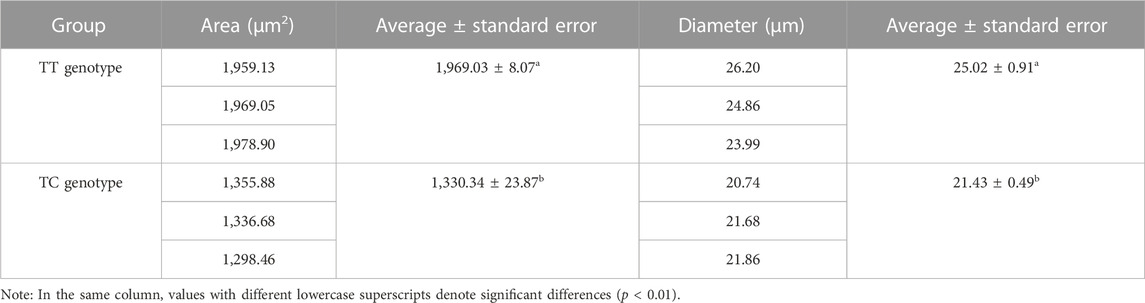
TABLE 5. The measurement of the muscle fiber cross-sectional area and diameter in the hybrid grouper with different growth rate.
Hybridization can induce alterations in genes, epigenetics, and phenotypic traits (Baack and Rieseberg, 2007). Epigenetic factors, which are chemically stable, influence gene expression, thereby modifying the phenotype during animal development (Ngô et al., 1996; Santos et al., 2005; Potok et al., 2013; Zhang et al., 2019). The mstn gene, acting as a powerful inhibitor of muscle growth, predominantly expresses in skeletal muscle (Rios et al., 2002). Several studies have documented a negative correlation between the methylation status of the mstn gene and its expression (Liu et al., 2010; Yao et al., 2017). In this study, the global methylation levels of the mstn gene were generally low, with no significant differences observed between the hybrid grouper, E. fuscoguttatus, and E. polyphekadion. However, significant variations in the mstn gene’s expression level were evident between E. fuscoguttatus and E. polyphekadion, and between the hybrid grouper and E. polyphekadion. Mark et al. (1998) revealed that methylation of around 60% of a 5-CpG island could completely inhibit gene expression, while a lower degree of methylation only reduces gene transcription and expression (Mark et al., 1998). Therefore, we deduce that the main role of mstn gene methylation in this study is to decrease gene transcription and expression.
A comparative analysis of each CpG site’s methylation status unveiled significant differences in methylation levels at three CpG sites between E. fuscoguttatus and E. polyphekadion. A correlation between methylation level and gene expression was observed only at site 1,521 (R = 0.97). This positive correlation was also obvious between the hybrid grouper and E. polyphekadion (R = 0.87). Typically, high DNA methylation patterns indicate low mRNA expression. A significant positive correlation between gene expression and DNA methylation has been observed in animals and plants, such as Mus musculus (Kobayashi et al., 2012), Sus scrofa (Xiao et al., 2014), sacred lotus (Li et al., 2021). DNA methylation on promoter or gene body can have complex regulatory roles, in some cases inhibiting gene expression and in others inducing expression (Lang et al., 2017; Huang et al., 2019). Because DNA methylation might cause alterations in chromatin structure, thereby modifying the interactions between DNA and activating or repressing transcription factors (or complexes) (Santos et al., 2005). Consequently, methylation might enhance transcription by preventing the binding of negative regulatory factors (Lai et al., 2010), or increasing the binding of positive regulatory factors (Lorincz et al., 2004; Kulis et al., 2012). In addition, methylation status at site 824 displayed a negative correlation with gene expression between the hybrid grouper and E. polyphekadion (R = −0.85). DNA methylation in exons might collaboratively regulate splicing (Schwartz et al., 2009; Schwartz and Ast, 2010). Based on these foundings in this study, it is speculated that the influence of DNA methylation is not determined by single CpG site. Further molecular genetic studies are needed to validate this hypothesis.
The mstn gene has been extensively investigated as a potential genetic marker for growth traits. Studies suggest that SNPs in the mstn gene can influence growth performance (Sun et al., 2012; Wang et al., 2014; Nazari et al., 2016; Yang et al., 2018). In this study, we identified two SNPs (g.818 A>G, g.1003 T>C) in exon 2 of the hybrid grouper’s mstn gene at Hardy-Weinberg Equilibrium (HEW). Only one SNP (g.1003 T>C) displayed significant associations with six growth traits, with the TT genotype predominating over the two genotypes. Both non-coding and coding regions’ SNPs have been linked to the regulation of growth performance (Yu et al., 2007; Liu et al., 2012; Sun et al., 2012; Nazari et al., 2016). Sequence variants in the non-coding region could affect RNA cleavage, stability, translation, export, and intracellular localization, ultimately altering gene expression (Pesole et al., 2001; Antonarakis and Cooper, 2003). Variants in the coding region could modify amino acid composition, leading to changes in protein structure (Niu et al., 2015). In our results, the g.1003 T>C was synonymous, and did not result in an amino acid change. Synonymous SNPs are considered to be functionally meaning (Sauna et al., 2007; Kerna et al., 2013). It can influence secondary structures of mRNA and thereby alter the length of pause cycles during translation, the overall rate of translation, or protein folding (Kimchi-Sarfaty et al., 2007; Bartoszewski et al., 2010). This ubiquitous long pauses can result in translational frame shifting and to protein misfolding (Wen et al., 2008). Synonymous mutation might be associated with a growth-related quantitative trait loci (QTL), thus contributing to growth performance modulation (Liu et al., 2012; Sun et al., 2012). In conclusion, polymorphisms in the mstn gene could serve as molecular markers to improve the growth of hybrid grouper, facilitating marker-assisted selective breeding.
Growth is a crucial economic trait in farmed grouper. The mstn gene is a significant growth factor that negatively regulates skeletal muscle development and growth. Investigations into the correlation between growth traits, DNA methylation levels, and genetic variations can provide fundamental insights to enhance grouper growth performance. Consequently, this study examined the DNA methylation level in a hybrid grouper (E. fuscoguttatus (♀) × E. polyphekadion (♂)) and its parent species. Significant differences in DNA methylation sites were discovered between the hybrid grouper and E. polyphekadion at sites 824 and 1,521 (located at exon 2 and intron 2, respectively), and between E. fuscoguttatus and E. polyphekadion at site 1,521. These differences can potentially influence the mRNA expression of the mstn gene. Moreover, this study screened SNPs of the mstn gene associated with growth traits in the hybrid grouper. The results revealed an SNP at position g.1003T > C in the mstn gene. The TT genotype was found to be dominant at this site and demonstrated superior growth performance compared to individuals with the TC genotype. Histological examination via HE staining further corroborated these findings, showing that the average area and diameter of muscle fibers in individuals with the TT genotype were greater than those in individuals with the TC genotype.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: GenBank database (BioProject ID: PRJNA1009792).
The animal study was approved by Administration of Affairs Concerning Animal Experimentation of China. The study was conducted in accordance with the local legislation and institutional requirements.
LC: Writing–original draft. JM: Writing–review and editing, Visualization. PC: Writing–review and editing, Validation. XH: Writing–review and editing, Validation. NY: Writing–review and editing, Validation. YL: Writing–review and editing, Validation. HH: Writing–review and editing, Supervision.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The research was supported by Hainan Provincial Natural Science Foundation of China (321QN263), The Major Science and Technology plan of Hainan Province (ZDKJ2021017), National Natural Science Foundation of China (32160861), The Innovation Platform for Academicians of Hainan Province (YSPTZX202103), State Key Laboratory of Developmental Biology of Freshwater Fish (2020KF001), Scientific Research Foundation of Hainan Tropical Ocean University (RHDRC202010), the Youth Project of Yazhou Bay Innovation Institute of Hainan Tropical Ocean University (2022CXYQNXM06) and National Natural Science Foundation of China (32002389).
We would like to sincerely thank many researchers who help to complete this manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1277647/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | Graphical map of mstn nucleotide sequence in the hybrid grouper.
SUPPLEMENTARY FIGURE S2 | Sequencing maps of SNPs of mstn gene in sites 818 (A,B) and 1003 (C,D), respectively. All the sequences were reverse complementary sequences.
Angers, B., Castonguay, E., and Massicotte, R. (2010). Environmentally induced phenotypes and DNA methylation: how to deal with unpredictable conditions until the next generation and after. Mol. Ecol. 19, 1283–1295. doi:10.1111/j.1365-294X.2010.04580.x
Antonarakis, S. E., and Cooper, D. N. (2003). “Mutations in human genetic diseases: nature and consequences,” in Nature encyclopedia of the human genome (London: Nature Publishing Group), 227–253.
Baack, E. J., and Rieseberg, L. H. (2007). A genomic view of introgression and hybrid speciation. Curr. Opin. Genet. Dev. 17, 513–518. doi:10.1016/j.gde.2007.09.001
Bartoszewski, R. A., Jablonsky, M., Bartoszewska, S., Stevenson, L., Dai, Q., Kappes, J., et al. (2010). A synonymous single nucleotide polymorphism in _F508CFTR alters the secondary structure of the mRNA and the expression of the mutant protein. J. Biol. Chem. 10, 28741–28748. doi:10.1074/jbc.M110.154575
Cao, L., Chen, P., Hou, X., Ma, J., Yang, N., Xu, Y., et al. (2022). Genetic characteristics and growth patterns of the hybrid grouper derived from the hybridization of Epinephelus fuscoguttatus (female) × Epinephelus polyphekadion (male). J. Fish Biol. 102, 328–339. doi:10.1111/jfb.15263
Cao, L., Chen, P., Zhang, Y., Fu, X., and Huang, H. (2023). Sequence and bioinformatics analysis on MSTN gene of the hybrid grouper derived from (Epinephelus fuscoguttatus ×Epinephelus polyphekadion). Agric. Biotechnol. 12, 46–49+54.
Ding, Y. X., He, F., Wen, H. S., Li, J. F., Qian, K., Chi, M. L., et al. (2012). Polymorphism in exons CpG rich regions of the cyp17-II gene affecting its mRNA expression and reproductive endocrine levels in female Japanese flounder (Paralichthys olivaceus). General & Comp. Endocrinol. 179, 107–114. doi:10.1016/j.ygcen.2012.08.003
Doerge, R. W. (2002). Mapping and analysis of quantitative trait loci in experimental populations. Nat. Revs. Genet. 3, 43–52. doi:10.1038/nrg703
Gao, F., Liang, H., Lu, H., Wang, J., Xia, M., Yuan, Z., et al. (2015). Global analysis of DNA methylation in hepatocellular carcinoma by a liquid hybridization capture-based bisulfite sequencing approach. Clin. Epigenetics 7, 86. doi:10.1186/s13148-015-0121-1
Gao, F., Zhang, J., Jiang, P., Gong, D., Wang, J.-W., Yudong, X., et al. (2014). Marked methylation changes in intestinal genes during the perinatal period of preterm neonates. Bmc Genomics 15, 716. doi:10.1186/1471-2164-15-716
Guo, L., Xia, J., Mingming, L., Sen, Y., Haoran, L., and Meng, Z. (2016). Targeted sequencing of myogenic regulatory factors and myostatins reveals an association between MSTN-1 and interorbital distance in orange-spotted grouper,Epinephelus coioides(Hamilton, 1822). J. World Aquac. Soc. 47, 741–753. doi:10.1111/jwas.12303
Huang, H., Liu, R., Niu, Q., Tang, K., Zhang, B. O., Zhang, H., et al. (2019). Global increase in DNA methylation during orange fruit development and ripening. Proc. Natl. Acad. Sci. U. S. A. 116, 1430–1436. doi:10.1073/pnas.1815441116
Jaenisch, R., and Bird, A. (2003). Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 33, 245–254. doi:10.1038/ng1089
Kerna, I., Kisand, K., Tamm, A., and Tamm, A. (2013). Synonymous SNP influences Adam12 mRNA expression level in synovial tissue. J. Mol. Genet. Med. 7, 321–322. doi:10.4172/1747-0862.1000057
Kimchi-Sarfaty, C., Oh, J. M., Kim, I. W., Sauna, Z. E., Calcagno, A. M., Ambudkar, S. V., et al. (2007). A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science 26, 525–528. doi:10.1126/science.1135308
Ko, C. F., Chiou, T. T., Chen, T. T., Wu, J. L., Chen, J. C., and Lu, J. K. (2007). Molecular cloning of myostatin gene and characterization of tissue-specific and developmental stage-specific expression of the gene in orange spotted grouper, Epinephelus coioides. Mar. Biotechnol. 9, 20–32. doi:10.1007/s10126-006-6059-8
Kobayashi, H., Sakurai, T., Imai, M., Takahashi, N., Fukuda, A., Obata, Y., et al. (2012). Contribution of intragenic DNA methylation in mouse gametic DNA methylomes to establish oocyte-specific heritable marks. PLoS Genet. 8, e1002440. doi:10.1371/journal.pgen.1002440
Kulis, M., Queiros, A. C., Beekman, R., and Martin-Subero, J. I. (2012). Intragenic DNA methylation in transcriptional regulation, normal differentiation and cancer. Biochim. Biophys. Acta 106, 248–253. doi:10.1016/j.bbagrm.2013.08.001
Lai, A. Y., Fatemi, M., Dhasarathy, A., Malone, C., Sobol, S. E., Geigerman, C., et al. (2010). DNA methylation prevents CTCF-mediated silencing of the oncogene BCL6 in B cell lymphomas. J. Exp. Med. 207, 1939–1950. doi:10.1084/jem.20100204
Lang, Z., Wang, Y., Tang, K., Tang, D., Datsenka, T., Cheng, J., et al. (2017). Critical roles of DNA demethylation in the activation of ripening-induced genes and inhibition of ripening-repressed genes in tomato fruit. Proc. Natl. Acad. Sci. U. S. A. 114, E4511–E4519. doi:10.1073/pnas.1705233114
Lapraz, F., Röttinger, E., Duboc, V. R. R., Duloquin, L., Walton, K., Wu, S., et al. (2006). RTK and TGF-beta signaling pathways genes in the sea urchin genome. Dev. Biol. 300, 132–152. doi:10.1016/j.ydbio.2006.08.048
Lee, S.-J., and McPherron, A. C. (1999). Myostatin and the control of skeletal muscle mass: commentary. Curr. Opin. Genet. Dev. 9, 604–607. doi:10.1016/s0959-437x(99)00004-0
Li, H., Yang, X., Wang, Q., Chen, J., and Shi, T. (2021). Distinct methylome patterns contribute to ecotypic differentiation in the growth of the storage organ of a flowering plant (sacred lotus). Mol. Ecol. 30, 2831–2845. doi:10.1111/mec.15933
Li, S., He, F., Wen, H., Li, J., Si, Y., Liu, M., et al. (2017). Low salinity affects cellularity, DNA methylation, and mRNA expression of igf1 in the liver of half smooth tongue sole (Cynoglossus semilaevis). Fish Physiology Biochem. 43, 1587–1602. doi:10.1007/s10695-017-0395-7
Liu, G., Zong, K., Zhang, L., and Cao, S. (2010). Dietary methionine affect meat qulity and myostatin gene exon 1 region methylation in skeletal muscle tissues of broilers. Agric. Sci. China 9, 1338–1346. doi:10.1016/s1671-2927(09)60224-8
Liu, L., Yu, X., and Tong, J. (2012). Molecular characterization of myostatin (MSTN) gene and association analysis with growth traits in the bighead carp (Aristichthys nobilis). Mol. Biol. Rep. 39, 9211–9221. doi:10.1007/s11033-012-1794-6
Liu, S., Palti, Y., Gao, G., and Rexroad, C. E. (2016). Development and validation of a SNP panel for parentage assignment in rainbow trout. Aquaculture 452, 178–182. doi:10.1016/j.aquaculture.2015.11.001
Livak, K. J., and Schmittgen, T. D. (2013). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi:10.1006/meth.2001.1262
Lorincz, M. C., Dickerson, D. R., Schmitt, M., and Groudine, M. (2004). Intragenic DNA methylation alters chromatin structure andelongation efficiency in mammalian cells. Nat. Struct. Mol. Biol. 11, 1068–1075. doi:10.1038/nsmb840
Lyu, S., Yang, P., Liu, Y., Song, T., Zhang, Z., Shi, Q., et al. (2020). Genetic effects of MOGAT1 gene SNP in growth traits of Chinese cattle. Gene 769, 145201. doi:10.1016/j.gene.2020.145201
Mark, L. G., Toshiro, H., Christina, M. B. M., Pao, M., Tsai, Y. C., Gonzales, F. A., et al. (1998). The role of DNA methylation in expression of the p19/p16 locus in human bladder cancer cell lines. Cancer Res. 58, 1245–1252.
Mcpherron, C., Lawler, M., and Lee, S. J. (1997). Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature 387, 83–90. doi:10.1038/387083a0
Meng, X., Wang, H., Qiu, X., Liu, H., and Wang, X. (2017). SNPs of myostatin (MSTN) gene and their association with growth traits in three bay scallop (Argopecten irradians) populations. Aquac. Res. 48, 531–536. doi:10.1111/are.12900
Nazari, S., Valiolah, J., Pourkazemi, M., Miandare, H. K., and Abdolhay, H. A. (2016). Association between myostatin gene (MSTN-1) polymorphism and growth traits in domesticated rainbow trout (Oncorhynchus mykiss). Agri Gene 1, 109–115. doi:10.1016/j.aggene.2016.08.003
Ngô, V., Gourdji, D., and Laverrière, J. N. (1996). Site-specific methylation of the rat prolactin and growth hormone promoters correlates with gene expression. Mol. Cell. Biol. 16, 3245–3254. doi:10.1128/mcb.16.7.3245
Niu, D., Wang, L., Bai, Z., Xie, S., Zhao, H., and Li, J. (2015). Identification and expression characterization of the myostatin (MSTN) gene and association analysis with growth traits in the razor clam Sinonovacula constricta. Gene 555, 297–304. doi:10.1016/j.gene.2014.11.020
Peñaloza, C., Hamilton, A., Guy, D. R., Bishop, S. C., and Houston, R. D. (2013). A SNP in the 5′ flanking region of the myostatin-1b gene is associated with harvest traits in Atlantic salmon (Salmo salar). BMC Genet. 14, 112. doi:10.1186/1471-2156-14-112
Pesole, G., Mignone, F., Gissi, C., Grillo, G., Licciulli, F. L., and Liuni, S. (2001). Structural and functional features of eukaryotic mRNA untranslated regions. Gene 276, 73–81. doi:10.1016/s0378-1119(01)00674-6
Potok, M. E., Nix, D. A., Parnell, T. J., and Cairns, B. R. (2013). Reprogramming the maternal zebrafish genome after fertilization to match the paternal methylation pattern. Cell 153, 759–772. doi:10.1016/j.cell.2013.04.030
Qian, Z., Mi, X., Wang, X., He, S., Liu, Y., Hou, F., et al. (2013). cDNA cloning and expression analysis of myostatin/GDF11 in shrimp, Litopenaeus vannamei. Comp. Biochem. Physiology Part A Mol. Integr. Physiology 165, 30–39. doi:10.1016/j.cbpa.2013.02.001
Rios, R., Carneiro, I., Arce, V. M., and Devesa, J. (2002). Myostatin is an inhibitor of myogenic differentiation. Am. J. Physiology 282, 993–999. doi:10.1152/ajpcell.00372.2001
Rodgers, B. D., Weber, G. M., Sullivan, C. V., and Levine, M. A. (2001). Isolation and characterization of myostatin complementary deoxyribonucleic acid clones from two commercially important fish: Oreochromis mossambicus and Morone chrysops. Endocrinology 142, 1412–1418. doi:10.1210/endo.142.4.8097
Salem, M., Vallejo, R. L., Leeds, T. D., Palti, Y., Liu, S., Sabbagh, A., et al. (2012). RNA-seq identifies SNP markers for growth traits in rainbow trout. Plos One 7, e36264. doi:10.1371/journal.pone.0036264
Santos, K. F., Mazzola, T. N., and Carvalho, H. F. (2005). The prima donna of epigenetics: the regulation of gene expression by DNA methylation. Braz. J. Med. Biol. Res. 38, 1531–1541. doi:10.1590/s0100-879x2005001000010
Sauna, Z. E., Kimchi-Sarfaty, C., Ambudkar, S. V., and Gottesman, M. M. (2007). The sounds of silence: synonymous mutations affect function. Pharmacogenomics 8, 527–532. doi:10.2217/14622416.8.6.527
Schwartz, S., and Ast, G. (2010). Chromatin density and splicing destiny: on the cross-talk between chromatin structure and splicing. EMBO J. 29, 1629–1636. doi:10.1038/emboj.2010.71
Schwartz, S., Meshorer, E., and Ast, G. (2009). Chromatin organization marks exon-intron structure. Nat. Struct. Mol. Biol. 16, 990–995. doi:10.1038/nsmb.1659
Si, Y., Ding, Y., He, F., Wen, H., Li, J., Zhao, J., et al. (2016). DNA methylation level of cyp19a1a and Foxl2 gene related to their expression patterns and reproduction traits during ovary development stages of Japanese flounder (Paralichthys olivaceus). Gene 575, 321–330. doi:10.1016/j.gene.2015.09.006
Sun, Y., Yu, X., and Tong, J. (2012). Polymorphisms in myostatin gene and associations with growth traits in the common carp (Cyprinus carpio L.). Int. J. Mol. Sci. 13, 14956–14961. doi:10.3390/ijms131114956
Ulloa, P. E., Rincón, G., Islas-Trejo, A., Araneda, C., Iturra, P., Neira, R., et al. (2015). RNA sequencing to study gene expression and SNP variations associated with growth in zebrafish fed a plant protein-based diet. Mar. Biotechnol. 17, 353–363. doi:10.1007/s10126-015-9624-1
Wang, Y., Wang, X., Meng, X., Wang, H., Jiang, Z., and Qiu, X. (2014). Identification of two SNPs in myostatin (MSTN) gene of Takifugu rubripes and their association with growth traits. Mol. Cell. Probes 28, 200–203. doi:10.1016/j.mcp.2014.03.006
Wen, J. D., Lancaster, L., Hodges, C., Zeri, A., Yoshimura, S. H., Noller, H. F., et al. (2008). Following translation by single ribosomes one codon at a time. Nature 3, 598–603. doi:10.1038/nature06716
Xiao, J., Wang, H., Ma, J., He, M., Long, K., and Jiang, A. (2014). Methylation of CpG island within second intron increased G-protein coupled receptor 120 (GPR120) Mrna transcript in Porcine (Sus scrofa) subcutaneous and visceral adipose tissue. J. Agric. Biotechnol. 22, 992–1000.
Yang, H., and Li, Q. (2021). The DNA methylation level is associated with the superior growth of the hybrid crosses in the Pacific oyster Crassostrea gigas. Aquaculture 547, 737421. doi:10.1016/j.aquaculture.2021.737421
Yang, Y., Lan, Z., Shu, H., Zhou, H., Jiang, X., Hou, L., et al. (2018). Association between expression levels and growth trait-related SNPs located in promoters of the MC4R and MSTN genes in Spinibarbus hollandi. Genes & Genomics 40, 1119–1125. doi:10.1007/s13258-018-0666-4
Yao, L., Zhu, L., Mao, X., Sulaiman, Y., and Aniwashi, J. (2017). Analysis of DNA methylation and mRNA expression level of MSTN gene in xinjiang bashiby sheep. China Animal Husb. Veterinary Med. 44, 2987–2993.
Yu, L., Tang, H., Wang, J., Wu, Y., Zou, L., Jiang, Y., et al. (2007). Polymorphisms in the 5′ regulatory region of myostatin gene are associated with early growth traits in Yorkshire pigs. Sci. CHINA Life Sci. 50, 642–647. doi:10.1007/s11427-007-0075-4
Zhang, H. W., Yin, S. W., Zhang, L. J., Hou, X. Y., and Zhang, G. S. (2015). Development and validation of single nucleotide polymorphism markers in Odontobutis potamophila from transcriptomic sequencing. Genet. Mol. Res. GMR 14, 2080–2085. doi:10.4238/2015.March.20.18
Zhang, Y., Shen, W., Cao, M., Li, J., Zheng, B., Lou, Z., et al. (2019). Dynamic alterations in methylation of global DNA and growth-related genes in large yellow croaker (Larimichthys crocea) in response to starvation stress. Comp. Biochem. Physiology Part B Biochem. Mol. Biol. 227, 98–105. doi:10.1016/j.cbpb.2018.09.006
Zhong, H., Xiao, J., Chen, W., Zhou, Y., Tang, Z., Guo, Z., et al. (2014). DNA methylation of pituitary growth hormone is involved in male growth superiority of Nile tilapia (Oreochromis niloticus). Part B, Biochem. Mol. Biol. 171, 42–48. doi:10.1016/j.cbpb.2014.03.006
Zhong, Q., Zhang, Q., Chen, Y., Sun, Y., Qi, J., Wang, Z., et al. (2008). The isolation and characterization of myostatin gene in Japanese flounder (Paralichthys olivaceus): ubiquitous tissue expression and developmental specific regulation. Aquaculture 280, 247–255. doi:10.1016/j.aquaculture.2008.04.015
Keywords: growth trait, mstn gene, SNP, DNA methylation, grouper
Citation: Cao L, Ma J, Chen P, Hou X, Yang N, Lu Y and Huang H (2024) Exploring the influence of DNA methylation and single nucleotide polymorphisms of the Myostatin gene on growth traits in the hybrid grouper (Epinephelus fuscoguttatus (female) × Epinephelus polyphekadion (male)). Front. Genet. 14:1277647. doi: 10.3389/fgene.2023.1277647
Received: 15 August 2023; Accepted: 13 December 2023;
Published: 08 January 2024.
Edited by:
Yulin Jin, Emory University, United StatesCopyright © 2024 Cao, Ma, Chen, Hou, Yang, Lu and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hai Huang, aHVhbmdoYWk3NEAxMjYuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.