- Food Security and Safety Focus Area, Faculty of Natural and Agricultural Sciences, North-West University, Mmabatho, South Africa
Legumes are highly nutritious in proteins and are good food for humans and animals because of their nutritional values. Plant growth-promoting bacteria (PGPR) are microbes dwelling in the rhizosphere soil of a plant contributing to the healthy status, growth promotion of crops, and preventing the invasion of diseases. Root exudates produced from the leguminous plants’ roots can lure microbes to migrate to the rhizosphere region in other to carry out their potential activities which reveals the symbiotic association of the leguminous plant and the PGPR (rhizobia). To have a better cognition of the PGPR in the rhizosphere of leguminous plants, genomic analyses would be conducted employing various genomic sequences to observe the microbial community and their functions in the soil. Comparative genomic mechanism of plant growth-promoting rhizobacteria (PGPR) was discussed in this review which reveals the activities including plant growth promotion, phosphate solubilization, production of hormones, and plant growth-promoting genes required for plant development. Progress in genomics to improve the collection of genotyping data was revealed in this review. Furthermore, the review also revealed the significance of plant breeding and other analyses involving transcriptomics in bioeconomy promotion. This technological innovation improves abundant yield and nutritional requirements of the crops in unfavorable environmental conditions.
Introduction
The reduction in food crop production nowadays results from the inability to tame wild species, the potential for average condition of weather, and population growth. The production of crops in sustainable agriculture is highly required to feed the population as it increases tremendously in this 21st century (Albahri et al., 2023). Promoting the quantity of crop production is crucial to preserve its nutritive value. Various challenges were encountered in the production of crops due to the application of chemical derivatives, which reveal the prevalence of chemical hazards in the environment (Nguyen et al., 2023). Thus, the a demand for sustainable means of promoting a friendly approach to improving the agricultural system. Integrating lesser crops, also known as underutilized crops, into potential food systems is highly significant. The process of breeding, including molecular and conventional approaches, has been utilized to improve the production of food crops, yet to meet up with food production is far fetch (Jabran et al., 2023).
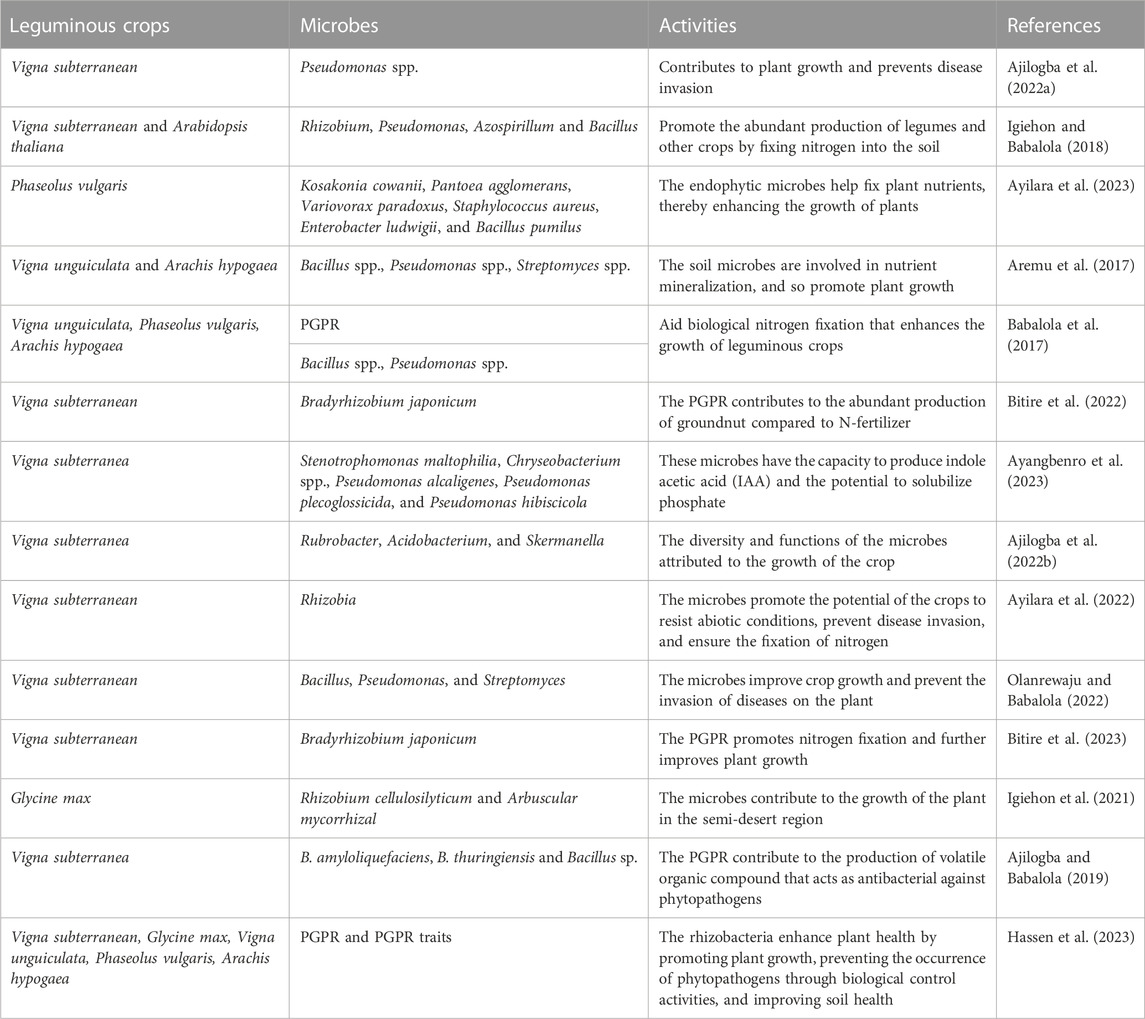
Implementing genome sequences is significant for comprehending the biochemical and physiological methods of regulating plant stimulus to biotic and abiotic stresses and plant traits (Raja and Vidya, 2023). The phylogenesis of sequence technologies has led to the production of voluminous plant genome data (Xia et al., 2023). This has produced the possibility of introducing technology in crop promotion via technological advances. In this modern technology of life science, novel observation has been found in clarifying genetic functions and their pathways in line with omic technologies and bioinformatics tools employed to analyze sequence data (Li et al., 2023a). The process has been utilized in genome-wide association studies (GWAS), genomic selection, quantitative linkage locus (QTLs), population genetics, genome editing, marker-assisted breeding, and SNP genotyping detection. The mentioned processes have been modified for the category of crops. Crops, including legumes (Adeleke and Babalola, 2022) and groundnuts (Ajilogba et al., 2022a) have been suggested to assist in the food security sector in sub-Saharan Africa. These crops contain proteins and minerals that make them essential in contributing to the healthy living of people feeding on them, thereby reducing malnutrition and food insecurity. Legumes are majorly cultivated in West African countries (Nigeria, Senegal, Cote d’Ivoire, Togo), and other eastern countries like India and Indonesia (Ajilogba et al., 2022a).
The crops are well known for their potential to fix nitrogen, improving soil fertility (Kebede, 2021). They are also known to undergo crop rotation and their ability to be cultivated without the application of chemical products (Pergner and Lippert, 2023). In rural and other isolated areas, the application of chemical fertilizers is not imminent. So, these crops were relied upon for the fixation of nitrogen into the soil. The richness in nutrients, the diversity of genetic resources, and mineral composition were some of the activities of the crop to improve the soil. Legumes nodulate with the aid of nitrogen-fixing bacteria and further promote nitrogen fixation in the soil (Cheng et al., 2023).
The diversification of crops via the involvement of underutilized crops especially legumes into significant cropping systems will promote food availability (Tan et al., 2020). However, the activities taking place in soil have revealed the immense impact on the production of crops and the effect of various environmental stresses. The rhizosphere soil of these crops contains some microbes that carry out the potential to improve the growth of plants either directly or indirectly. Among these microbes are bacteria that exist as beneficial or pathogenic. The beneficial types are plant growth-promoting microbes (PGPM) that contribute to plant growth and abundant production of crops. There is a symbiotic relationship between these soil microbes, especially rhizobacteria and leguminous plants (Fasusi et al., 2023). These rhizobacteria penetrate the root nodules of the leguminous plants and convert nitrogen from its free state into nitrate that the plant can utilize by nitrogen fixation process. This process assists legumes to flourish especially where nitrogen is deficient in the soil and likewise improves the soil by enhancing the health status of other plants while practicing crop rotation (Hyder et al., 2023). The potential of bacteria species in the rhizosphere consists of the advantages to humans like the significant function they perform in biogeochemical cycles of the major elements like carbon, nitrogen, and sulfur and minor elements including iron, Nickel, and Mercury (Martínez-Martínez et al., 2023; Noor et al., 2023). The activities improve the engagement of microbes, including plant growth-promoting rhizobacteria (PGPR) and plant growth-promoting fungi (PGPF), in changeable energy embedded in the soil. Microbes manufacture phytohormones like Auxins and other substances like vitamins, enzymes, and other materials contributing to the growth of plants and inhibition of phytopathogens (Solomon et al., 2023). The communities of microbes in the rhizosphere are attributed to the features of soil, plant type, and plant growth stages.
The association of PGPR and leguminous crops has emphasized the potential of genomics. This gives tremendous insight into microbial functions and their genetic makeup. Researchers have determined the specific genes used in the process of nitrogen fixation and likewise the growth-promoting characteristics (Chieb and Gachomo, 2023; Fahde et al., 2023; Kim et al., 2023). Through the study of genomics, scientists identified and characterized major genes that partake in this mutualistic relationship of legumes and rhizobia. This study also presents the discovery of signaling molecules used in communication and recognition between rhizobia and legumes (Khan et al., 2023). A typical example is observed in the production of chemical substances called flavonoids by leguminous plant roots that attract rhizobia to the plant and nod factors, a chemical substance produced by the rhizobia activate the production of nodules in leguminous roots. In addition, genomics also unveil some PGPR which researcher analyze and make comparison of their genetic variation that contribute to plant growth-promotion potential. This knowledge does assist to optimize and choose various microbial strains that contribute to nitrogen fixation (Yao et al., 2023).Biotechnological and novel genomic processes in plants can provide solutions that can reveal the effectiveness of the objectives (Figueiredo et al., 2023). Novel techniques comprising biotechnology and the production of bio-based material can perform the function of improving agricultural sustainability. This is because they are safer for consumer use and for the environment. Biotechnology proffers resolutions for various problems of the modern world by inserting bioenergy, biopharmaceuticals, biofuels, biomaterials, foods, and its products for selling.
This review reveals the importance of plant genomics in bioeconomy promotion and the possibilities brought about by plant breeding techniques. Due to the challenges of underdevelopment and changes in the average weather conditions, the problem of food scarcity has not been resolved. So, the advancement of crop production suggests how to combat the problems. The review further reveals the advantages of the beneficial microbes in crop growth promotion and describes the application of omic techniques to characterize plant-microbe interaction.
Legume-PGPR interaction
Plants preserve the complex environment by stimulating root exudates into the soil environment (Paponov et al., 2023). Plants produce substances that are products of photosynthesis, especially in the roots embedded in the soil. As a result, the region of soil containing carbon increases the population of microbes, unlike bulk soil which lacks carbon substances. A higher concentration of microbial communities in the soil is a result of interaction between plants and the roots that reveal various associations (Wang et al., 2023b). Some attributes measure the amount and grade of root exudates, plant growth stages (McLaughlin et al., 2023), abiotic stress (high temperature and drought) (Shaposhnikov et al., 2023), soil type (Wang et al., 2023a), phytopathogen invasion (Tortella et al., 2023), and the status of plant feeding (Heuermann et al., 2023). There will be a proposition of promoting beneficial microbes if the production of root exudates is feasible. Amino acids, sugar, phenolics, organic acids, etc., are some of the materials present in the root exudates (Agarwal et al., 2023). These carbon source substances draw the advantageous and nonadvantageous microbes into the rhizosphere soil region (Magnoli and Bever, 2023). The advantageous microbes keep plants safe against phytopathogens via direct and indirect mechanisms (Fahde et al., 2023). Yet, the carbon compounds likewise draw pathogenic microbes, which eventually lead to competition for nutrients, cause diseases in plants, and contribute to rhizobiome structuring (Adedayo and Babalola, 2023). Some plants have shown how microbial communities dwelling in their rhizosphere attract microbes that harbor protection on plants and inhibit pathogen invasions in the rhizosphere (Nwachukwu and Babalola, 2022). The physiochemical and biological features carry out significant functions in the association of plants and microorganisms. Even though phytopathogens have an immense effect on plant health, some beneficial microbes living in the rhizosphere or the tissues of plants contend with phytopathogens for nutrients and space, so end up in biocontrol activities beneficial against pathogenic organisms (Cao et al., 2023).
PGPR are found and multiply in the rhizosphere and tissues of healthy plants and improve plant growth, and abundant crop production, prevent plants against phytopathogens, and inhibit or eradicate biotic and abiotic stresses (Ojuederie et al., 2019; Olanrewaju et al., 2019). A direct form of microbial interaction with plant host is obtained in growth promotion, while the indirect form involves biological control potential against phytopathogens. Direct mechanisms involve phytohormone stimulation that reveals inhibition or promotion of the growth of plant roots, induces systemic resistance, promotes the acquisition of nutrients, and prevents crops from biotic and abiotic stress (Olanrewaju et al., 2017). Due to these properties, PGPR was accepted by researchers as an alternative to chemical fertilizers and other derivatives. Thus, PGPR, including Bacillus thuringiensis, contributes to abundant crop production and improves the health of plants, as reported by (Adeniji et al., 2021). Developing plants decrease the availability of Iron (Fe) by assimilating Fe and produce exudates that lure beneficial microbes into the soil to assimilate the Fe (Hyder et al., 2023). Certain PGPR is responsible for the production of siderophores in Fe-stressed environments and is enriched to inhibit phytopathogens via competition for Fe (Sarker et al., 2021). Plants utilize iron-bound siderophores that contribute to their growth (Swarnalatha et al., 2022). This reveals that the microorganisms producing Siderophore can avail the Fe needed by plants because Fe is a component of chlorophyll that is highly required for the photosynthesis process (Molnár et al., 2023). Another form applied to the reduction of plant diseases is the capability of microbes inhabiting the rhizosphere to inhibit soil or compost from disallowing the invasion of phytopathogen.
Symbiontic association of leguminous plant and rhizosphere microbes
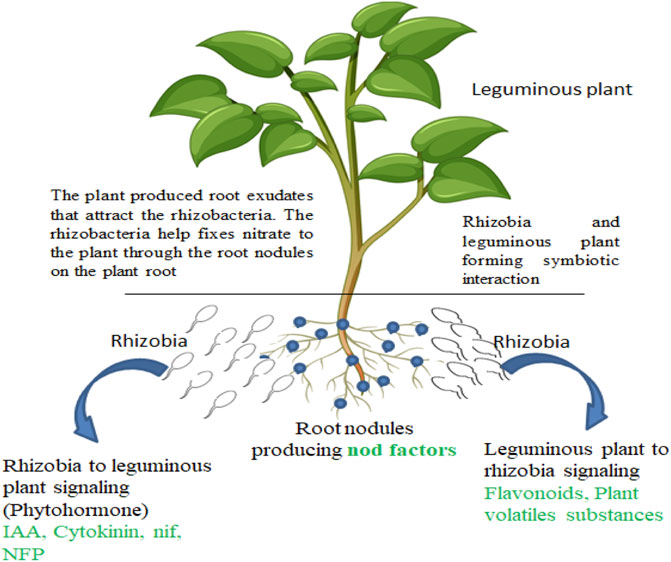
Rhizosphere microbes and leguminous plants have beneficial relationships (Figure 1). Their interaction inhibits or eradicates the invasion of the phytopathogens on the plants. Rhizobia species, including Azorhizobium, Allorhizobium, Bradyrhizobium, Mesorhizobium, and Sinorhizobium, are common examples inhabiting the rhizosphere of leguminous plants (Babalola et al., 2017). The symbiotic association leads to the acquisition of nitrogen in the soil. Leguminous plants produce nodules and fix nitrogen acquired in the soil during the symbiotic association with Bradyrhizobium strains. The production of a nodule in legume plants while interacting with rhizosphere microbes leads to the production of some compounds like flavonoids, betaines, and aldonic acid in the form of root exudates. The exudates signal the PGPR in a mutual association that otherwise promotes the production of the nod gene that stimulates nodulation by associating with the nod proteins present in the PGPR cell wall (Wang et al., 2018; Walker et al., 2020). In the root hair of leguminous plants, the physical effects were revealed when the PGPR (rhizobia) responded to the production of oligosaccharide nod factors. This results in the production of nodules that eventually promote nitrogen fixation. PGPR, especially rhizobia, contributes to plant growth and improves the development of seed by producing nod factors.
Non-symbiotic association of leguminous plants and rhizosphere microbes
Another form of association undergone by leguminous plants is the stimulation of phytohormones by the PGPR and the promotion of plant growth and abundant production of crops. Besides, leguminous plants do produce phenolics that aid the inhibition of phytopathogens, provision of nutrients to plants, and beneficial microbial growth (Egamberdieva et al., 2023). Phenolic compounds produced arbitrate the nod gene elicitors production. Nod gene elicitors control the stimulation of nod factors on the plant roots (Ahmed et al., 2020). After being collected in the rhizosphere, it results in the biological synthesis of flavonoids, which leads to the production of a higher concentration of phytoalexins for plant protection against spoilage organisms, and more reported in Table 1. Riboflavin (Vitamin B2) can likewise be produced by rhizobia and change photochemically (Lopez et al., 2019). Rhizobia has been reported to inhibit the growth of bacterial and fungal spoilage organisms affecting other crops, including soybean (Paravar et al., 2023), okra (Sharma, 2019), tomato (Adedayo et al., 2023a; Adedayo et al., 2023b), sunflower (Adeleke et al., 2021), Bambara groundnut (Ajilogba et al., 2022c), etc. Root exudates, including organic acid anions and phytosiderophores, are significant in the availability of minerals and their presence in agricultural soil (Wang et al., 2023c).
Molecular mechanisms applied in the study of PGPR
Certain biochemical and cultured-based tests have been employed for a long time to study PGPR (Utami et al., 2022). With recent progression in molecular technology, various gene technological data, including genomics, metagenomics, transcriptomics, and proteomics data, are uploaded on global websites (Lata et al., 2023). Genomic sequence analysis of PGPR is grouped into two groups, and they are whole genome sequencing (WGS) and specific genome sequencing (SGS) (Simlat et al., 2023).
WGS reveals all the chromosomes and plasmids present in the PGPR. WGS-employed next-generation sequencing (NGS) shows the appropriate genetic makeup of organisms (Kurotani et al., 2023). Whereas SGS revealed only a certain part of the genome employed for comparison and characterization (Lim et al., 2023). The particular gene in this group is 16S rRNA used to identify bacteria origin to specific levels of organization (Abramova et al., 2023). Another form of identifying the bacteria is an intra-genomic spacer (IGS) 16S-23S, repetitive sequence-based PCR (rep-PCR) that amplify and sequence highly conserved repeats. Restriction fragment length polymorphism (RLFP) analysis has been used to categorize PGPR (Li et al., 2023b).
Some genes, including atpD, dnaK, dnaJ, gap, glnll, gltA, gyrG, pnp, recA, rpoA, and thrC were employed in strain typing involved in multilocus sequence analysis (Swarnalakshmi et al., 2020). Absolute primers are employed to intensify genomic sequences randomly, known as random amplified polymorphic DNA sequencing. To amplify particular genes together with a secondary mode of sequencing using restriction profiling and denaturing gradient gel electrophoresis (DGGE) to reduce the assembly of mismatches in limited regions or variation in genome GC content (Nakatsu, 2021). The process of fixing nitrogen in the soil by the biological process is a significant method of promoting PGPR with the aid of nitrogenase enzymes producing genes like nifD, nifK, and nifH genes (Özdoğan et al., 2022).
Moreover, PGPR partakes in the solubilization of phosphorus in the production of gluconic acid, which makes use of glucose dehydrogenase. Apart from the mentioned characteristics, PGPR stimulates phytohormones like gibberellin, ethylene, indole acetic acid (IAA), abscisic acid, and cytokinin (Ojuederie et al., 2019; Koza et al., 2022). However, certain genes like ipdC and amlE are responsible for IAA production, as found in other phytohormones (Adedayo et al., 2023a). For iron utilization, siderophores were produced by PGPR which is regulated by the Siderophore gene. Pseudomonas spp. possess membrane receptor that codes for the pupA gene, which transports the Siderophore complex in the cell (Sah et al., 2021).
Plant growth-promoting genes contributing to the growth of leguminous plant
Leguminous plants have a special interaction with PGPR especially rhizobia as discussed above. This interaction yields the fixation of atmospheric nitrogen to a plant-utilizable form that contributes to the healthy status of the plant, which is important for plants that have no access to nitrate assimilation (Figure 1). The growth-promoting genes are mainly associated with the production and upkeep of mutualistic association between rhizobia and leguminous plants and the following genes discussed below are responsible for this process.
Nodulation (nod) genes: are the genes used for starting and regulating the production of nodules which is the complex body where the organism (rhizobia) dwells (Malik et al., 2020). The genes examples are nodA, nodB, nodC, and nodD.
Nitrogen fixation (nif) genes: they are involved in the production and roles of nitrogen-fixing nodules. They also carry out potential action in synthesizing nitrogenase enzyme that converts natural nitrogen gas to ammonia and their example are nifD, nifH, and nifK (Adedayo et al., 2023a)
Auxin biosynthetic genes: are genes responsible for the production of hormones that contribute to the growth of plants. The stimulation of these hormones is promoted by some rhizobia thereby contributing to the elongation of plant roots and production of the root nodules. The typical examples include indole-3-acetamide hydrolase (iaah), an enzyme responsible for the production of auxin, and indole acetic acid (IAA) which is a hormone (Adedayo et al., 2023a).
Cytokinin biosynthesis genes: these are responsible for the production of plant hormone Cytokinin. The hormone contributed to the division of cells as well as their differentiation (Šmeringai et al., 2023). Various rhizobia are producing this hormone that contributes to the production of nodules. These genes also contributed to the biological synthesis of cytokines with the aid of the isopentenyl transferase (ipt) enzyme for plant growth.
Plant receptor genes: these are genes constituting particular receptors to identify and associate with the signals produced by rhizobia. Examples include nod factor rhizobia did produces LysM Receptor Kinase (NFR) and Nod Factor Perception (NFP) genes (Geurts and Huisman, 2023). The genes explained above contribute to structural signaling pathways and their association to produce advantageous and mutualistic associations between leguminous plants and PGPR (rhizobia). They also enhance the development of the plants by supplying them with fixed nitrogen and assimilation of nutrients in the rhizosphere soil.
Plant breeding
The integration of plant and animal production systems to promote environmental quality and natural resources to improve economic viability is defined as sustainable agriculture (Fatima et al., 2023; Liu, 2023). It is also defined as the application of technologies and practices that do not harm the food quality, environment, and production (Gamage et al., 2023). The significant issue of minimizing the application of non-renewable material, e.g., phosphates, crop genotypes, and promoting sustainability of agriculture (Jha et al., 2023). The utilization of modern methods of genetic breeding, manipulation and redesign, and agricultural and ecological management incorporating crop selection is sustainable development (Hemalatha et al., 2023). Agricultural practices nowadays are based on different forms that contain input-intensive systems that make use of energy, fertilizer, oil, water, etc., unlike low-input systems, which bring about low production of crops.
So, scientists’ stance should focus on plant breeding which will cut down the rate at which energy is utilized for plant production yet ascertain the production of high-quality food products. Due to climate change and the growth of the population around the globe, plant breeding requires water and, likewise, reduces the rate of water loss in other to promote the potential of sustaining water deprivation and avail food for developing populations. To promote the effectiveness of water use and production of abundant yield, the following features are required, and they include; physiological and molecular modification, osmoregulation, improved plant root system, and improved stomatal conductivity (Woźniak-Gientka and Tyczewska, 2023). Energy in the form of oil and gas is important for modern-day high-input agricultural systems. However, the energy implored for the production of crops during the period of harvest and post-harvest incorporates the application of farm machinery for cultivation, preparation of soil, harvesting, transport of the yield, and post-harvest storage. Although, it has been noted that 15% of utilized energy is demanded by the application of chemical derivatives on farmland and transportation, according to Attfield et al. (2023). The application of chemical fertilizers is regarded as not the best means when studying sustainable agriculture. It is significant to note the breeding process to improve the resistivity and tolerance in agricultural practices to boost sustainability, where the application of chemical pesticides is reduced (Esuola et al., 2016).
Various scientific processes that aid plant feature identification include genetic engineering, hybridization, tissue culture, and plant breeding (Woźniak-Gientka and Tyczewska, 2023). The benefit of genome editing (CRISPR-Cas9) is to supply high target specificity with high precision and speed (Sahel et al., 2023). Furthermore, applying this technique does not yield transgenic plants due to its inability to transfer genes to the host plants. Applying this technique, some features, including biotic and abiotic features, were tolerated to obtain new varieties of crops to improve the abundance of agricultural products (Anguita-Maeso et al., 2023). The process involved in agrobiotechnology is employed to produce Biofertilizers, biopesticides, bioherbicides, etc., in the agricultural field (Woźniak-Gientka and Tyczewska, 2023). These compounds aid in the regulation of soil toxicity and are safe for environmental use. The growth and development of plants mainly depend on the availability of macroelements, including nitrogen (N), phosphorus (P), and potassium (K) (Bitire et al., 2023), and microelements like molybdenum (Mo), iron (Fe), zinc (Zn), manganese (Mn), copper (Cu), boron (B), etc. (Banerjee et al., 2023).
Plant breeding techniques improve the production of crops
The challenge of making available food crops to feed the developing population is the economic, social, and environmental systems of farming that oppose the adaptation of climate, degradation of the agricultural field, and loss invented on biodiversity of the microbial communities. The production of food to support the population is not sufficient as the population increases. Due to this challenge, some procedures were adopted in new breeding techniques to enhance the production of crops through genetically modified organisms (GMOs) (Rahman et al., 2023). However, the usual form of biotechnology in agricultural practices is the cultivation of new crops, also called genetically modified or engineered crops. Some of the genome-edited crops do not shelter foreign DNA (Olanrewaju et al., 2021). CRISPR-Cas9 is a genome-editing tool employed in plant study and its growth globally to improve the features of crop quality, activities against chemical derivatives like pesticides and herbicides, and disease resistance (Marone et al., 2023). Employing a CRISPR technology breeder could promote tolerance to abiotic stresses like extreme temperature and drought and improve nutrient content. Legumes have been monitored upon the application of this technology (Puppala et al., 2023). Climate change has caused extreme heat and drought that affect the production of crops to feed the global populace (Omotoso et al., 2023). CRISPR/Cas9 tools have been employed in crop plantation that requires water for their growth and cognition of significant genes to control drought stress (Banerjee et al., 2023).
Transcriptomics analysis of PGPR dwelling in the rhizosphere of leguminous plants
This involves the genetic expression study pattern of the rhizobia to obtain knowledge about their interaction and their roles in the rhizosphere soil. PGPR are known to be involved in the significant function of improving the growth of plants and likewise promoting the availability of the nutrients required via nitrogen fixation and solubilization of phosphate (Adedayo et al., 2022).
To carry out the analysis of transcriptomes on samples, scientists isolate the specific bacteria species in the soil sample of the leguminous plants. The microbes were lysed to extract the RNA from the bacterial cell and further purified (Saleh-Lakha et al., 2005). The RNA is a precursor for transcribed genes for a specific period and as a starting medium for downstream analysis. Then, the mRNA is converted to complementary strand DNA (cDNA) with the help of an enzyme called reverse transcriptase (Ying, 2003). The cDNA stands for the expressed gene of the microbial isolate.
High throughput sequencing technologies like RNA Seq can be used to collect sequence data of cDNA molecules and further subjected to analyzing the data using bioinformatics tools (Shiau et al., 2023). This constitutes aligned sequences to the reference database or genome of the recognized genes to confirm the genes expressed (Tables 2, 3). Also, the analysis of differential gene expression can be investigated to analyze various conditions of the stages of gene expression, like different rhizobia strains, rhizosphere soil they inhabit, and time range. The differentially identified genes can make available the knowledge of molecular procedures of rhizobacteria’s mutualistic association with the leguminous plants (Li et al., 2023c). These genes can contribute to the following mechanisms including; plant growth promotion, nitrogen fixation, stress tolerance, and nutrient cycling, among other functional characteristics of interest. This analysis can likewise be joined together with other high throughput omics technologies like metabolomics and proteomics to get more knowledge of PGPR community structure, functions, and potential influence on abundant production of legumes and improvement of leguminous plant health. On a short note, transcriptomics technology can assist scientists in decrypting the genetic makeup of plant-microbe interactions and make available cogent information for sustainable agriculture. The characteristics of Paenibacillus polymyxa (Table 2) and P. agglomerans (Table 3) were revealed showing various characteristics of the PGPR ranging from growth features to genomic size of various strains strain isolated. Pantoea agglomerans displayed various potential including plant growth promotion, phosphate solubilizing potential, secretion of hormones like IAA and Siderophore, and synthesis of acetoin and 2,3-butanediol according to Shariati et al. (2017).
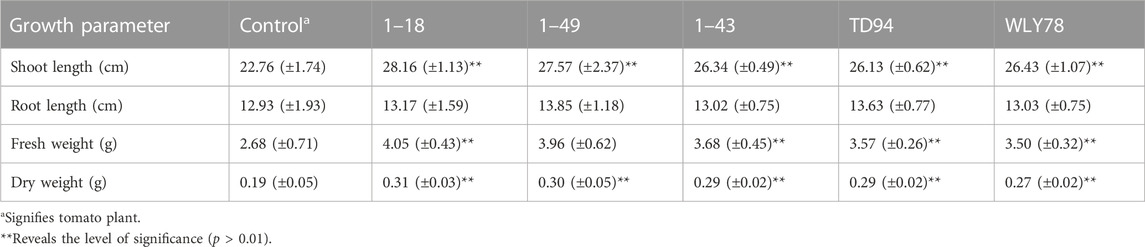
TABLE 2. Plant growth-promoting potential of Paenibacillus polymyxa contributing to the growth of crop plants like tomatoes (Xie et al., 2016).
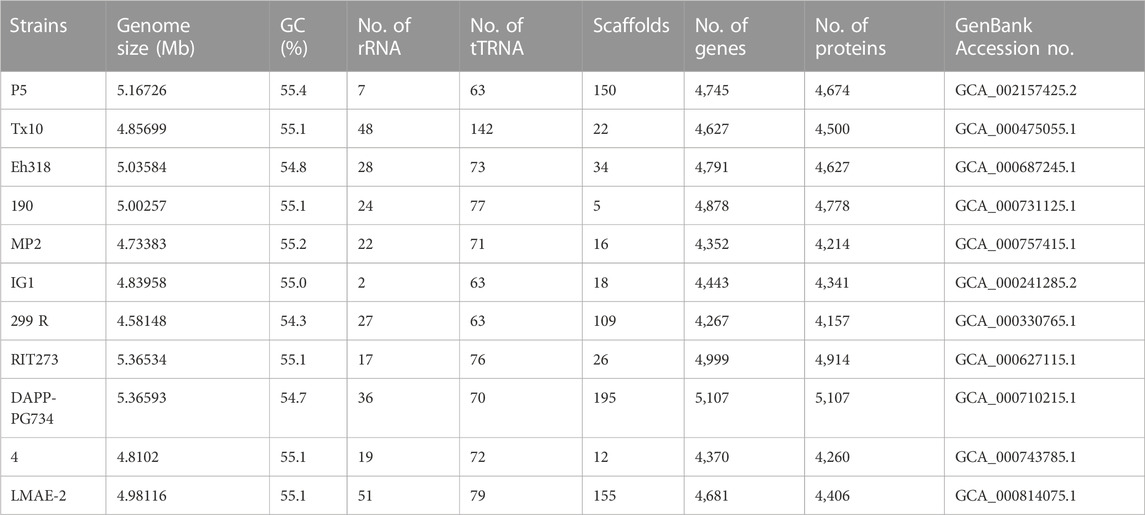
TABLE 3. Comparative genomic analysis of Pantoea agglomerans strain as plant growth-promoting rhizobacteria according to Shariati et al. (2017).
Conclusion
The sustainability of agricultural soil concerns the agricultural development and application of fertilizers to improve farming activities. Major nutrients required to boost the fertility of the soil and encourage plant growth, including nitrogen, phosphorus, and potassium, were discussed in the context. Nevertheless, PGPR contributes to the health status of plants, thereby contributing to the healthy growth of plants and improving the abundant production of crops and generally regarded as safe for humans, plants, and the environment. Due to these features, they possess, they are alternatives to chemical fertilizers. The diversity of microorganisms in soil contributes to plant growth and improves their various ecosystem functions in the soil. The plant root produces some substances called root exudate that attract soil microbes and reveal the microbial diversity inhabiting the soil. So, high throughput sequencing involving genomics is used to identify unculturable microbes. However, some factors, including soil types and plant genotypes, control the activities of PGPR in the soil. So, genetic variation of advantageous PGPR interaction with plants can be incorporated into plant breeding to boost the production of legumes for agricultural sustainability.
Author contributions
AA: Data curation, Resources, Writing–original draft, and editing. OB: Conceptualization, Funding acquisition, Supervision, Validation, Visualization, Writing–review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The National Research Foundation of South Africa grant (UID132595) for the funding support granted to OB.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abramova, A., Berendonk, T. U., and Bengtsson-Palme, J. (2023). A global baseline for qPCR-determined antimicrobial resistance gene prevalence across environments. Environ. Int. 178, 108084. doi:10.1016/j.envint.2023.108084
Adedayo, A. A., and Babalola, O. O. (2023). Fungi that promote plant growth in the rhizosphere boost crop growth. J. Fungi 9, 239. doi:10.3390/jof9020239
Adedayo, A. A., Babalola, O. O., Prigent-Combaret, C., Cruz, C., Stefan, M., Kutu, F., et al. (2022). The application of plant growth-promoting rhizobacteria in Solanum lycopersicum production in the agricultural system: a review. PeerJ 10, e13405. doi:10.7717/peerj.13405
Adedayo, A. A., Fadiji, A. E., and Babalola, O. O. (2023a). Unraveling the functional genes present in rhizosphere microbiomes of Solanum lycopersicum. PeerJ 11, e15432. doi:10.7717/peerj.15432
Adedayo, A. A., Romdhane, S. B., and Babalola, O. O. (2023b). Metagenomic study of the tomato rhizosphere soil microbiome: current state and prospect. AIMS Geosci. 9, 330–356. doi:10.3934/geosci.2023018
Adeleke, B. S., Ayangbenro, A. S., and Babalola, O. O. (2021). Bacterial community structure of the sunflower (Helianthus annuus) endosphere. Plant Signal. Behav. 16, 1974217. doi:10.1080/15592324.2021.1974217
Adeleke, B. S., and Babalola, O. O. (2022). Meta-omics of endophytic microbes in agricultural biotechnology. Biocatal. Agric. Biotechnol. 42, 102332. doi:10.1016/j.bcab.2022.102332
Adeniji, A. A., Ayangbenro, A. S., and Babalola, O. O. (2021). Genomic exploration of Bacillus thuringiensis MORWBS1.1, candidate biocontrol agent, predicts genes for biosynthesis of zwittermicin, 4,5-DOPA dioxygenase extradiol, and quercetin 2,3-dioxygenase. Mol. Plant-Microbe Interactions® 34, 602–605. doi:10.1094/mpmi-10-20-0272-sc
Agarwal, P., Vibhandik, R., Agrahari, R., Daverey, A., and Rani, R. (2023). Role of root exudates on the soil microbial diversity and biogeochemistry of heavy metals. Appl. Biochem. Biotechnol. 1–21. doi:10.1007/s12010-023-04465-2
Ahmed, A., Tariq, A., and Habib, S. (2020). “Interactive biology of auxins and phenolics in plant environment,” in Plant phenolics in sustainable agriculture: volume 1. Editors R. Lone, R. Shuab, and A. N. Kamili (Singapore: Springer), 117–133. doi:10.1007/978-981-15-4890-1_5
Ajilogba, C. F., and Babalola, O. O. (2019). GC–MS analysis of volatile organic compounds from Bambara groundnut rhizobacteria and their antibacterial properties. World J. Microbiol. Biotechnol. 35, 83. doi:10.1007/s11274-019-2660-7
Ajilogba, C. F., Olanrewaju, O. S., and Babalola, O. O. (2022a). Improving Bambara groundnut production: insight into the role of omics and beneficial bacteria. Front. Plant Sci. 13, 836133. doi:10.3389/fpls.2022.836133
Ajilogba, C. F., Olanrewaju, O. S., and Babalola, O. O. (2022b). Plant growth stage drives the temporal and spatial dynamics of the bacterial microbiome in the rhizosphere of Vigna subterranea. Front. Microbiol. 13, 825377. doi:10.3389/fmicb.2022.825377
Ajilogba, F. C., Babalola, O. O., Adebola, P., and Adeleke, R. (2022c). Bambara groundnut rhizobacteria antimicrobial and biofertilization potential. Front. Plant Sci. 13, 854937. doi:10.3389/fpls.2022.854937
Albahri, G., Alyamani, A. A., Badran, A., Hijazi, A., Nasser, M., Maresca, M., et al. (2023). Enhancing essential grains yield for sustainable food security and bio-safe agriculture through latest innovative approaches. Agronomy 13, 1709. doi:10.3390/agronomy13071709
Anguita-Maeso, M., Navas-Cortés, J. A., and Landa, B. B. (2023). Insights into the methodological, biotic and abiotic factors influencing the characterization of xylem-inhabiting microbial communities of olive trees. Plants 12, 912. doi:10.3390/plants12040912
Aremu, B. R., Alori, E. T., Kutu, R. F., and Babalola, O. O. (2017). “Potentials of microbial inoculants in soil productivity: an outlook on african legumes,” in Microorganisms for green revolution: volume 1: microbes for sustainable crop production. Editors D. G. Panpatte, Y. K. Jhala, R. V. Vyas, and H. N. Shelat (Singapore: Springer), 53–75. doi:10.1007/978-981-10-6241-4_3
Attfield, P. V., Bell, P. J. L., and Grobler, A. S. (2023). Reducing carbon intensity of food and fuel production whilst lowering land-use impacts of biofuels. Fermentation 9, 633. doi:10.3390/fermentation9070633
Ayangbenro, A. S., Adem, M. R., and Babalola, O. O. (2023). Bambara nut root-nodules bacteria from a semi-arid region of South Africa and their plant growth-promoting traits. Int. J. Microbiol. 2023, 8218721. doi:10.1155/2023/8218721
Ayilara, M. S., Abberton, M., Oyatomi, O. A., Odeyemi, O., and Babalola, O. O. (2022). Potentials of underutilized legumes in food security. Front. Soil Sci. 2, 1–12. doi:10.3389/fsoil.2022.1020193
Ayilara, M. S., Adeleke, B. S., and Babalola, O. O. (2023). Bioprospecting and challenges of plant microbiome research for sustainable agriculture, a review on soybean endophytic bacteria. Microb. Ecol. 85, 1113–1135. doi:10.1007/s00248-022-02136-z
Babalola, O. O., Olanrewaju, O. S., Dias, T., Ajilogba, C. F., Kutu, F. R., and Cruz, C. (2017). “Biological nitrogen fixation: the role of underutilized leguminous plants,” in Microorganisms for green revolution: volume 1: microbes for sustainable crop production. Editors D. G. Panpatte, Y. K. Jhala, R. V. Vyas, and H. N. Shelat (Singapore: Springer), 431–443. doi:10.1007/978-981-10-6241-4_20
Banerjee, S., Roy, P., Nandi, S., and Roy, S. (2023). Advanced biotechnological strategies towards the development of crops with enhanced micronutrient content. Plant Growth Regul. 100, 355–371. doi:10.1007/s10725-023-00968-4
Bitire, T. D., Abberton, M., Oyatomi, O., and Babalola, O. O. (2022). Effect of Bradyrhizobium japonicum strains and inorganic nitrogen fertilizer on the growth and yield of Bambara groundnut (Vigna subterranea (L.) verdc) accessions. Front. Sustain. Food Syst. 6, 1–12. doi:10.3389/fsufs.2022.913239
Bitire, T. D., Abberton, M., Oyatomi, O., and Babalola, O. O. (2023). Yield response of accessions of Bambara groundnut (Vigna subterranea (L) Verdc) inoculated with Bradyrhizobium japonicum strains. Front. Sustain. Food Syst. 7, 1–18. doi:10.3389/fsufs.2023.1142123
Cao, M., Narayanan, M., Shi, X., Chen, X., Li, Z., and Ma, Y. (2023). Optimistic contributions of plant growth-promoting bacteria for sustainable agriculture and climate stress alleviation. Environ. Res. 217, 114924. doi:10.1016/j.envres.2022.114924
Cheng, Z., Meng, L., Yin, T., Li, Y., Zhang, Y., and Li, S. (2023). Changes in soil rhizobia diversity and their effects on the symbiotic efficiency of soybean intercropped with maize. Agronomy 13, 997. doi:10.3390/agronomy13040997
Chieb, M., and Gachomo, E. W. (2023). The role of plant growth promoting rhizobacteria in plant drought stress responses. BMC Plant Biol. 23, 407. doi:10.1186/s12870-023-04403-8
Egamberdieva, D., Eshboev, F., Shukurov, O., Alaylar, B., and Arora, N. K. (2023). Bacterial bioprotectants: biocontrol traits and induced resistance to phytopathogens. Microbiol. Res. 14, 689–703. doi:10.3390/microbiolres14020049
Esuola, C. O., Babalola, O. O., Heine, T., Schwabe, R., Schlömann, M., and Tischler, D. (2016). Identification and characterization of a FAD-dependent putrescine N-hydroxylase (GorA) from Gordonia rubripertincta CWB2. J. Mol. Catal. B Enzym. 134, 378–389. doi:10.1016/j.molcatb.2016.08.003
Fahde, S., Boughribil, S., Sijilmassi, B., and Amri, A. (2023). Rhizobia: a promising source of plant growth-promoting molecules and their non-legume interactions: examining applications and mechanisms. Agriculture 13, 1279. doi:10.3390/agriculture13071279
Fasusi, O. A., Babalola, O. O., and Adejumo, T. O. (2023). Harnessing of plant growth-promoting rhizobacteria and arbuscular mycorrhizal fungi in agroecosystem sustainability. CABI Agric. Biosci. 4, 26–15. doi:10.1186/s43170-023-00168-0
Fatima, A., Singh, V. K., Babu, S., Singh, R. K., Upadhyay, P. K., Rathore, S. S., et al. (2023). Food production potential and environmental sustainability of different integrated farming system models in northwest India. Front. Sustain. Food Syst. 7, 959464. doi:10.1007/s10508-022-02464-8
Figueiredo, M., Dias, A., Neves, J., and Vicente, H. (2023). Assessment of literacy to biotechnological solutions for environmental sustainability in Portugal. Sustainability 15, 10056. doi:10.3390/su151310056
Gamage, A., Gangahagedara, R., Gamage, J., Jayasinghe, N., Kodikara, N., Suraweera, P., et al. (2023). Role of organic farming for achieving sustainability in agriculture. Farming Syst. 1, 100005. doi:10.1016/j.farsys.2023.100005
Geurts, R., and Huisman, R. (2023). Innovations in two genes kickstarted the evolution of nitrogen-fixing nodules. Curr. Opin. Plant Biol. 9, 102446. doi:10.1016/j.pbi.2023.102446
Hassen, A. I., Babalola, O. O., and Carlson, R. (2023). “Rhizobacterial-Mediated interactions for enhanced symbiotic performance of the root nodule rhizobia in legumes,” in Sustainable agrobiology: design and development of microbial consortia. Editors D. K. Maheshwari, and S. Dheeman (Singapore: Springer Nature Singapore), 41–56. doi:10.1007/978-981-19-9570-5_3
Hemalatha, P., Abda, E. M., Shah, S., Venkatesa Prabhu, S., Jayakumar, M., Karmegam, N., et al. (2023). Multi-faceted CRISPR-Cas9 strategy to reduce plant based food loss and waste for sustainable bio-economy – a review. J. Environ. Manag. 332, 117382. doi:10.1016/j.jenvman.2023.117382
Heuermann, D., Döll, S., Schweneker, D., Feuerstein, U., Gentsch, N., and Von Wirén, N. (2023). Distinct metabolite classes in root exudates are indicative for field- or hydroponically-grown cover crops. Front. Plant Sci. 14, 1122285. doi:10.3389/fpls.2023.1122285
Hyder, S., Rizvi, Z. F., Los Santos-Villalobos, S. D., Santoyo, G., Gondal, A., Khalid, N., et al. (2023). Applications of plant growth-promoting rhizobacteria for increasing crop production and resilience. J. Plant Nutr. 46, 2551–2580. doi:10.1080/01904167.2022.2160742
Igiehon, N. O., and Babalola, O. O. (2018). Rhizosphere microbiome modulators: contributions of nitrogen fixing bacteria towards sustainable agriculture. Int. J. Environ. Res. Public Health 15, 574. doi:10.3390/ijerph15040574
Igiehon, N. O., Babalola, O. O., Cheseto, X., and Torto, B. (2021). Effects of Rhizobia and Arbuscular mycorrhizal fungi on yield, size distribution and fatty acid of soybean seeds grown under drought stress. Microbiol. Res. 242, 126640. doi:10.1016/j.micres.2020.126640
Jabran, M., Ali, M. A., Zahoor, A., Muhae-Ud-Din, G., Liu, T., Chen, W., et al. (2023). Intelligent reprogramming of wheat for enhancement of fungal and nematode disease resistance using advanced molecular techniques. Front. Plant Sci. 14, 1132699. doi:10.3389/fpls.2023.1132699
Jha, U. C., Nayyar, H., Parida, S. K., Beena, R., Pang, J., and Siddique, K. H. M. (2023). Breeding and genomics approaches for improving phosphorus-use efficiency in grain legumes. Environ. Exp. Bot. 205, 105120. doi:10.1016/j.envexpbot.2022.105120
Kebede, E. (2021). Contribution, utilization, and improvement of legumes-driven biological nitrogen fixation in agricultural systems. Front. Sustain. Food Syst. 5, 1–18. doi:10.3389/fsufs.2021.767998
Khan, M., Ali, S., Al Azzawi, T. N., Saqib, S., Ullah, F., Ayaz, A., et al. (2023). The key roles of ROS and RNS as a signaling molecule in plant–microbe interactions. Antioxidants 12, 268. doi:10.3390/antiox12020268
Kim, K., Song, I.-G., Yoon, H., and Park, J.-W. (2023). Sub-micron microplastics affect nitrogen cycling by altering microbial abundance and activities in a soil-legume system. J. Hazard. Mater. 460, 132504. doi:10.1016/j.jhazmat.2023.132504
Koza, N. A., Adedayo, A. A., Babalola, O. O., and Kappo, A. P. (2022). Microorganisms in plant growth and development: roles in abiotic stress tolerance and secondary metabolites secretion. Microorganisms 10, 1528. doi:10.3390/microorganisms10081528
Kurotani, K.-I., Hirakawa, H., Shirasawa, K., Tanizawa, Y., Nakamura, Y., Isobe, S., et al. (2023). Genome sequence and analysis of Nicotiana benthamiana, the model plant for interactions between organisms. Plant Cell Physiology 64, 248–257. doi:10.1093/pcp/pcac168
Lata, S., Sharma, S., and Kaur, S. (2023). OMICS approaches in mitigating metal toxicity in comparison to conventional techniques used in cadmium bioremediation. Water, Air, & Soil Pollut. 234, 148. doi:10.1007/s11270-023-06145-7
Li, H., Tahir Ul Qamar, M., Yang, L., Liang, J., You, J., and Wang, L. (2023a). Current progress, applications and challenges of multi-omics approaches in sesame genetic improvement. Int. J. Mol. Sci. 24, 3105. doi:10.3390/ijms24043105
Li, W., Li, W.-B., Xing, L.-J., and Guo, S.-X. (2023b). Effect of arbuscular mycorrhizal fungi (AMF) and plant growth-promoting rhizobacteria (PGPR) on microorganism of phenanthrene and pyrene contaminated soils. Int. J. Phytoremediation 25, 240–251. doi:10.1080/15226514.2022.2071832
Li, X., Zhang, X., Zhao, Q., and Liao, H. (2023c). Genetic improvement of legume roots for adaption to acid soils. Crop J. 11, 1022–1033. doi:10.1016/j.cj.2023.04.002
Lim, H. J., Choi, M. J., Byun, S. A., Won, E. J., Park, J. H., Choi, Y. J., et al. (2023). Whole-genome sequence analysis of Candida glabrata isolates from a patient with persistent fungemia and determination of the molecular mechanisms of multidrug resistance. J. Fungi 9, 515. doi:10.3390/jof9050515
Liu, X. (2023). Sustainable intensification: a historical perspective on China’s farming system. Farming Syst. 1, 100001. doi:10.1016/j.farsys.2023.100001
Lopez, B. R., Palacios, O. A., Bashan, Y., Hernández-Sandoval, F. E., and De-Bashan, L. E. (2019). Riboflavin and lumichrome exuded by the bacterium Azospirillum brasilense promote growth and changes in metabolites in Chlorella sorokiniana under autotrophic conditions. Algal Res. 44, 101696. doi:10.1016/j.algal.2019.101696
Magnoli, S. M., and Bever, J. D. (2023). Plant productivity response to inter- and intra-symbiont diversity: mechanisms, manifestations and meta-analyses. Ecol. Lett. 26, 1614–1628. doi:10.1111/ele.14274
Malik, G., Singh, N., and Hooda, S. (2020). “Nitrogen stress in plants and the role of phytomicrobiome,” in Phyto-microbiome in stress regulation. Editors M. Kumar, V. Kumar, and R. Prasad (Singapore: Springer Singapore), 295–322. doi:10.1007/978-981-15-2576-6_15
Marone, D., Mastrangelo, A. M., and Borrelli, G. M. (2023). From transgenesis to genome editing in crop improvement: applications, marketing, and legal issues. Int. J. Mol. Sci. 24, 7122. doi:10.3390/ijms24087122
Martínez-Martínez, J. G., Rosales-Loredo, S., Hernández-Morales, A., Arvizu-Gómez, J. L., Carranza-Álvarez, C., Macías-Pérez, J. R., et al. (2023). Bacterial communities associated with the roots of Typha spp. and its relationship in phytoremediation processes. Microorganisms 11, 1587. doi:10.3390/microorganisms11061587
Mclaughlin, S., Zhalnina, K., Kosina, S., Northen, T. R., and Sasse, J. (2023). The core metabolome and root exudation dynamics of three phylogenetically distinct plant species. Nat. Commun. 14, 1649. doi:10.1038/s41467-023-37164-x
Molnár, Z., Solomon, W., Mutum, L., and Janda, T. (2023). Understanding the mechanisms of Fe deficiency in the rhizosphere to promote plant resilience. Plants 12, 1945. doi:10.3390/plants12101945
Nakatsu, C. H. (2021). “4 - microbial genetics,” in Principles and applications of soil Microbiology. Editors T. J Gentry, J. J Fuhrmann, and D. A Zuberer. Third Edition (Elsevier), 89–109. doi:10.1016/B978-0-12-820202-9.00004-6
Nguyen, M.-K., Lin, C., Nguyen, H.-L., Hung, N. T. Q., La, D. D., Nguyen, X. H., et al. (2023). Occurrence, fate, and potential risk of pharmaceutical pollutants in agriculture: challenges and environmentally friendly solutions. Sci. Total Environ. 899, 165323. doi:10.1016/j.scitotenv.2023.165323
Noor, W., Majeed, G., Lone, R., Tyub, S., Kamili, A. N., and Azeez, A. (2023). “Interactive role of phenolics and PGPR in alleviating heavy metal toxicity in wheat,” in Plant phenolics in abiotic stress management. Editors R. Lone, S. Khan, and A. Mohammed Al-Sadi (Singapore: Springer Nature Singapore), 287–320. doi:10.1007/978-981-19-6426-8_14
Nwachukwu, B. C., and Babalola, O. O. (2022). Metagenomics: a tool for exploring key microbiome with the potentials for improving sustainable agriculture. Front. Sustain. Food Syst. 6, 1–15. doi:10.3389/fsufs.2022.886987
Ojuederie, O. B., Olanrewaju, O. S., and Babalola, O. O. (2019). Plant growth promoting rhizobacterial mitigation of drought stress in crop plants: implications for sustainable agriculture. Agronomy 9, 712. doi:10.3390/agronomy9110712
Olanrewaju, O. S., Ayangbenro, A. S., Glick, B. R., and Babalola, O. O. (2019). Plant health: feedback effect of root exudates-rhizobiome interactions. Appl. Microbiol. Biotechnol. 103, 1155–1166. doi:10.1007/s00253-018-9556-6
Olanrewaju, O. S., Ayilara, M. S., Ayangbenro, A. S., and Babalola, O. O. (2021). Genome mining of three plant growth-promoting Bacillus species from maize rhizosphere. Appl. Biochem. Biotechnol. 193, 3949–3969. doi:10.1007/s12010-021-03660-3
Olanrewaju, O. S., and Babalola, O. O. (2022). Plant growth-promoting rhizobacteria for orphan legume production: focus on yield and disease resistance in Bambara groundnut. Front. Sustain. Food Syst. 6, 1–8. doi:10.3389/fsufs.2022.922156
Olanrewaju, O. S., Glick, B. R., and Babalola, O. O. (2017). Mechanisms of action of plant growth promoting bacteria. World J. Microbiol. Biotechnol. 33, 197. doi:10.1007/s11274-017-2364-9
Omotoso, A. B., Letsoalo, S., Olagunju, K. O., Tshwene, C. S., and Omotayo, A. O. (2023). Climate change and variability in sub-Saharan Africa: a systematic review of trends and impacts on agriculture. J. Clean. Prod. 414, 137487. doi:10.1016/j.jclepro.2023.137487
Özdoğan, D. K., Akçelik, N., and Akçelik, M. (2022). Genetic diversity and characterization of plant growth-promoting effects of bacteria isolated from rhizospheric soils. Curr. Microbiol. 79, 132. doi:10.1007/s00284-022-02827-3
Paponov, M., Flate, J., Ziegler, J., Lillo, C., and Paponov, I. A. (2023). Heterogeneous nutrient supply modulates root exudation and accumulation of medicinally valuable compounds in Artemisia annua and Hypericum perforatum. Front. Plant Sci. 14, 1174151. doi:10.3389/fpls.2023.1174151
Paravar, A., Piri, R., Balouchi, H., and Ma, Y. (2023). Microbial seed coating: an attractive tool for sustainable agriculture. Biotechnol. Rep. 37, e00781. doi:10.1016/j.btre.2023.e00781
Pergner, I., and Lippert, C. (2023). On the effects that motivate pesticide use in perspective of designing a cropping system without pesticides but with mineral fertilizer—a review. Agron. Sustain. Dev. 43, 24. doi:10.1007/s13593-023-00877-w
Puppala, N., Nayak, S. N., Sanz-Saez, A., Chen, C., Devi, M. J., Nivedita, N., et al. (2023). Sustaining yield and nutritional quality of peanuts in harsh environments: physiological and molecular basis of drought and heat stress tolerance. Front. Genet. 14, 1121462. doi:10.3389/fgene.2023.1121462
Rahman, S. U., Mccoy, E., Raza, G., Ali, Z., Mansoor, S., and Amin, I. (2023). Improvement of soybean; A way forward transition from genetic engineering to new plant breeding technologies. Mol. Biotechnol. 65, 162–180. doi:10.1007/s12033-022-00456-6
Raja, B., and Vidya, R. (2023). Application of seaweed extracts to mitigate biotic and abiotic stresses in plants. Physiology Mol. Biol. Plants 29, 641–661. doi:10.1007/s12298-023-01313-9
Sah, S., Krishnani, S., and Singh, R. (2021). Pseudomonas mediated nutritional and growth promotional activities for sustainable food security. Curr. Res. Microb. Sci. 2, 100084. doi:10.1016/j.crmicr.2021.100084
Sahel, D. K., Vora, L. K., Saraswat, A., Sharma, S., Monpara, J., D'souza, A. A., et al. (2023). CRISPR/Cas9 genome editing for tissue-specific in vivo targeting: nanomaterials and translational perspective. Adv. Sci. 10, 2207512. doi:10.1002/advs.202207512
Saleh-Lakha, S., Miller, M., Campbell, R. G., Schneider, K., Elahimanesh, P., Hart, M. M., et al. (2005). Microbial gene expression in soil: methods, applications and challenges. J. Microbiol. Methods 63, 1–19. doi:10.1016/j.mimet.2005.03.007
Sarker, A., Ansary, M. W., Hossain, M. N., and Islam, T. (2021). Prospect and challenges for sustainable management of climate change-associated stresses to soil and plant health by beneficial rhizobacteria. Stresses 1, 200–222. doi:10.3390/stresses1040015
Shaposhnikov, A. I., Belimov, A. A., Azarova, T. S., Strunnikova, O. K., Vishnevskaya, N. A., Vorobyov, N. I., et al. (2023). The relationship between the composition of root exudates and the efficiency of interaction of wheat plants with microorganisms. Appl. Biochem. Microbiol. 59, 330–343. doi:10.1134/S000368382303016X
Shariati, J. V., Malboobi, M. A., Tabrizi, Z., Tavakol, E., Owlia, P., and Safari, M. (2017). Comprehensive genomic analysis of a plant growth-promoting rhizobacterium Pantoea agglomerans strain P5. Sci. Rep. 7, 15610. doi:10.1038/s41598-017-15820-9
Sharma, D. K. (2019). Enumerations on seed-borne and post-harvest microflora associated with okra [Abelmoschus esculentus (L.) Moench] and their management. GSC Biol. Pharm. Sci. 8, 119–130. doi:10.30574/gscbps.2019.8.2.0149
Shiau, C.-K., Lu, L., Kieser, R., Fukumura, K., Pan, T., Lin, H.-Y., et al. (2023). High throughput single cell long-read sequencing analyses of same-cell genotypes and phenotypes in human tumors. Nat. Commun. 14, 4124. doi:10.1038/s41467-023-39813-7
Simlat, M., Ptak, A., Jaglarz, A., Szewczyk, A., Dziurka, M., and Gurgul, A. (2023). Seeds of Stevia rebaudiana bertoni as a source of plant growth-promoting endophytic bacteria with the potential to synthesize rebaudioside A. Int. J. Mol. Sci. 24, 2174. doi:10.3390/ijms24032174
Šmeringai, J., Schrumpfová, P. P., and Pernisová, M. (2023). Cytokinins - regulators of de novo shoot organogenesis. Front. Plant Sci. 14, 1239133. doi:10.3389/fpls.2023.1239133
Solomon, W., Mutum, L., Janda, T., and Molnár, Z. (2023). Potential benefit of microalgae and their interaction with bacteria to sustainable crop production. Plant Growth Regul. 101, 53–65. doi:10.1007/s10725-023-01019-8
Swarnalakshmi, K., Yadav, V., Tyagi, D., Dhar, D. W., Kannepalli, A., and Kumar, S. (2020). Significance of plant growth promoting rhizobacteria in grain legumes: growth promotion and crop production. Plants 9, 1596. doi:10.3390/plants9111596
Swarnalatha, G. V., Goudar, V., Reddy, E. C. R. G. S., Al Tawaha, A. R. M., and Sayyed, R. Z. (2022). “Siderophores and their applications in sustainable management of plant diseases,” in Secondary metabolites and volatiles of PGPR in plant-growth promotion. Editors R. Z. Sayyed, and V. G. Uarrota (Cham: Springer International Publishing), 289–302. doi:10.1007/978-3-031-07559-9_14
Tan, X. L., Azam-Ali, S., Goh, E. V., Mustafa, M., Chai, H. H., Ho, W. K., et al. (2020). Bambara groundnut: an underutilized leguminous crop for global food security and nutrition. Front. Nutr. 7, 601496. doi:10.3389/fnut.2020.601496
Tortella, G., Rubilar, O., Pieretti, J. C., Fincheira, P., De Melo Santana, B., Fernández-Baldo, M. A., et al. (2023). Nanoparticles as a promising strategy to mitigate biotic stress in agriculture. Antibiotics 12, 338. doi:10.3390/antibiotics12020338
Utami, D., Meale, S. J., and Young, A. J. (2022). A pan-global study of bacterial leaf spot of chilli caused by xanthomonas spp. Plants 11, 2291. doi:10.3390/plants11172291
Walker, L., Lagunas, B., and Gifford, M. L. (2020). Determinants of host range specificity in legume-rhizobia symbiosis. Front. Microbiol. 11, 585749. doi:10.3389/fmicb.2020.585749
Wang, C., Wang, J., Niu, X., Yang, Y., Malik, K., Jin, J., et al. (2023a). Phosphorus addition modifies the bacterial community structure in rhizosphere of Achnatherum inebrians by influencing the soil properties and modulates the Epichloë gansuensis-mediated root exudate profiles. Plant Soil 1–18. doi:10.1007/s11104-023-06133-0
Wang, Q., Liu, J., and Zhu, H. (2018). Genetic and molecular mechanisms underlying symbiotic specificity in legume-rhizobium interactions. Front. Plant Sci. 9, 313–318. doi:10.3389/fpls.2018.00313
Wang, Q., Wang, D., Agathokleous, E., Cheng, C., Shang, B., and Feng, Z. (2023b). Soil microbial community involved in nitrogen cycling in rice fields treated with antiozonant under ambient ozone. Appl. Environ. Microbiol. 89, e0018023. doi:10.1128/aem.00180-23
Wang, Q., Zhang, P., Zhao, W., Li, Y., Jiang, Y., Rui, Y., et al. (2023c). Interplay of metal-based nanoparticles with plant rhizosphere microenvironment: implications for nanosafety and nano-enabled sustainable agriculture. Environ. Sci. Nano 10, 372–392. doi:10.1039/d2en00803c
Woźniak-Gientka, E., and Tyczewska, A. (2023). Genome editing in plants as a key technology in sustainable bioeconomy. EFB Bioeconomy J. 3, 100057. doi:10.1016/j.bioeco.2023.100057
Xia, H., Deng, H., Li, M., Xie, Y., Lin, L., Zhang, H., et al. (2023). Chromosome-scale genome assembly of a natural diploid kiwifruit (Actinidia chinensis var. deliciosa). Sci. Data 10, 92. doi:10.1038/s41597-023-02006-4
Xie, J., Shi, H., Du, Z., Wang, T., Liu, X., and Chen, S. (2016). Comparative genomic and functional analysis reveal conservation of plant growth promoting traits in Paenibacillus polymyxa and its closely related species. Sci. Rep. 6, 21329. doi:10.1038/srep21329
Yao, K., Wang, Y., Li, X., and Ji, H. (2023). Genome-wide identification of the soybean LysM-RLK family genes and its nitrogen response. Int. J. Mol. Sci. 24, 13621. doi:10.3390/ijms241713621
Keywords: abiotic stress, genome editing, genotyping, PGPR interaction, plant breeding
Citation: Adedayo AA and Babalola OO (2023) Genomic mechanisms of plant growth-promoting bacteria in the production of leguminous crops. Front. Genet. 14:1276003. doi: 10.3389/fgene.2023.1276003
Received: 11 August 2023; Accepted: 19 October 2023;
Published: 03 November 2023.
Edited by:
Mohamed Hijri, Montreal University, CanadaReviewed by:
Bulbul Ahmed, Mohammed VI Polytechnic University, MoroccoMohini Prabha Singh, Punjab Agricultural University, India
Copyright © 2023 Adedayo and Babalola. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Olubukola Oluranti Babalola, b2x1YnVrb2xhLmJhYmFsb2xhQG53dS5hYy56YQ==
†ORCID: Afeez Adesina Adedayo, orcid.org/0000-0001-5388-2877; Olubukola Oluranti Babalola, orcid.org/0000-0003-4344-1909
 Afeez Adesina Adedayo
Afeez Adesina Adedayo Olubukola Oluranti Babalola
Olubukola Oluranti Babalola